Summary
During meiosis, programmed DNA double-strand breaks (DSBs) are repaired by homologous recombination. DMC1, a conserved recombinase, plays a central role in this process. DMC1 promotes DNA strand exchange between homologous chromosomes, thus creating the physical linkage between them. Its function is regulated not only by several accessory proteins but also by bivalent ions. Here, we show that whereas calcium ions in the presence of ATP cause a conformational change within DMC1, stimulating its DNA binding and D-loop formation, they inhibit the extension of the invading strand within the D-loop. Based on structural studies, we have generated mutants of two highly conserved amino acids – E162 and D317 – in human DMC1, which are deficient in calcium regulation. In vivo studies of their yeast homologues further showed that they exhibit severe defects in meiosis, thus emphasizing the importance of calcium ions in the regulation of DMC1 function and meiotic recombination.
Subject areas: Cell biology, Structural biology
Graphical abstract
Highlights
-
•
Human DMC1 undergoes a conformational change upon binding calcium ions
-
•
Calcium ions inhibit the extension of DMC1-mediated D-loop
-
•
Recombination intermediates accumulate in calcium binding-deficient dmc1 mutants
-
•
Calcium binding-deficient dmc1 mutants show meiotic defects
Cell biology; Structural biology
Introduction
Meiosis is an essential cell division program in the life cycle of sexually reproducing eukaryotic organisms. During meiosis, the number of chromosomes is reduced by one round of DNA replication followed by two rounds of chromosome segregation – meiosis I and meiosis II. In meiosis I, homologous chromosomes segregate, whereas in meiosis II, sister chromatids are separated.
The central event of meiosis is meiotic recombination. It establishes a physical connection between homologous chromosomes necessary for their proper and efficient segregation. Defects in meiotic recombination thus result in aneuploidy or apoptosis. Meiosis is initiated by the programmed induction of double-strand breaks (DSBs) throughout the genome by a topoisomerase II-like protein, SPO11 (Hunter, 2015). Following DSB formation, the dsDNA ends are further processed by resection machinery to create 3′ single-stranded DNA (ssDNA) overhangs. The extruded ssDNA tails are bound by two widely conserved RecA recombinases – DMC1 and RAD51. They form nucleoprotein filaments capable of searching and invading the genome within homologous sequences. The processing of resulting intermediates via several mechanistically different pathways can then lead to either crossover or non-crossovers outcomes (Brown and Bishop, 2014; Hunter, 2015; Kagawa and Kurumizaka, 2010).
DMC1 is a meiosis-specific protein crucial for the proper execution of meiotic recombination in many organisms (Bishop et al., 1992; Pittman et al., 1998; Schwacha and Kleckner, 1997; Yoshida et al., 1998). In mice, DMC1 is required for homolog synapsis and loss of DMC1 results in sterility (Pittman et al., 1998; Yoshida et al., 1998). Similarly, a severe phenotype has also been observed in budding yeast S. cerevisiae. Cells lacking the DMC1 gene fail to efficiently repair DSBs, leading to a delay or arrest in meiotic progression and impaired sporulation (Bishop et al., 1992). The other recombinase, RAD51, has essential functions in both mitotic and meiotic cells. However, its strand exchange activity is dispensable in meiosis where it acts as an accessory factor to DMC1 (Cloud et al., 2012).
Whereas RAD51 and its regulation are studied well, the regulation of DMC1 is less clear. Monovalent and bivalent ions have been shown to be important regulators of DMC1 as well as RAD51 function. The ion requirement is conserved within many organisms (Bugreev et al., 2005; Bugreev and Mazin, 2004; Chan et al., 2014; Kant et al., 2005; Kelso et al., 2015; Lee et al., 2005; Nimonkar et al., 2012; Rajanikant et al., 2006; Sakane et al., 2008). Structural studies from a RecA family member, Methanococcus voltae RadA, revealed that potassium as well as calcium ions stabilize the active form of RadA by changing the conformation of its largely disordered L2 region, thus increasing the affinity for DNA and assembly of nucleoprotein filaments (Qian et al., 2006a, 2006b, 2006b; Wu et al., 2005). Several studies have provided evidence that calcium ions affect the biochemical activity of both recombinases such as D-loop formation, strand exchange activity, or ATP hydrolysis (Bugreev et al., 2005; Bugreev and Mazin, 2004; Chan et al., 2014; Lee et al., 2005; Nimonkar et al., 2012). However, the mechanism of this regulation is different. In the case of RAD51, calcium ions inhibit its ATPase activity, thus keeping it in its active ATP-bound form and stabilizing the presynaptic filament (Bugreev and Mazin, 2004). On the other hand, the activation of DMC1 by calcium ions is presumably mediated by conformational changes leading to an increased ability to form a nucleoprotein filament (Bugreev et al., 2005). Nevertheless, the exact mechanism is still poorly understood.
In this study, we set out to characterize the regulation of DMC1 by bivalent ions. We show that the conformation of DMC1 significantly differs in the presence of calcium versus magnesium ions supplemented by the ATP cofactor. Surprisingly, although calcium ions stimulate DMC1-mediated D-loop formation, they inhibit the following extension of the invading strand within the D-loop structure. We have further selected two amino acids - E162 and D317 - as potential sites for calcium binding and biochemically characterized their mutant variants, which in contrast to wild type were capable of efficiently binding single-stranded DNA even without the presence of calcium ions. As both amino acids are highly conserved, we analyzed the phenotype of yeast dmc1 mutant variants in vivo. Similarly to dmc1Δ cells, both mutants exhibited defects in chromosome synapsis, processing of joint molecules, and meiotic progression. Our observations thus provide evidence that calcium ions play an important role in the regulation of DMC1.
Results
Calcium ions cause a conformational change in DMC1
Calcium ions have been shown to have a direct effect on the enzymatic activities of proteins as well as their structural and interaction properties (Bugreev et al., 2005; Bugreev and Mazin, 2004; Strynadka and James, 1991). Interestingly, the levels of calcium ions were reported to oscillate in meiotically dividing cells, suggesting its physiological role in the regulation of meiotic recombination (Carroll et al., 1994; Kaneuchi et al., 2015; McDougall and Sardet, 1995). As the biochemical activity of the meiotic recombinase DMC1 has been shown to be stimulated by calcium ions, we set out to characterize the effect of calcium and magnesium ions on its function.
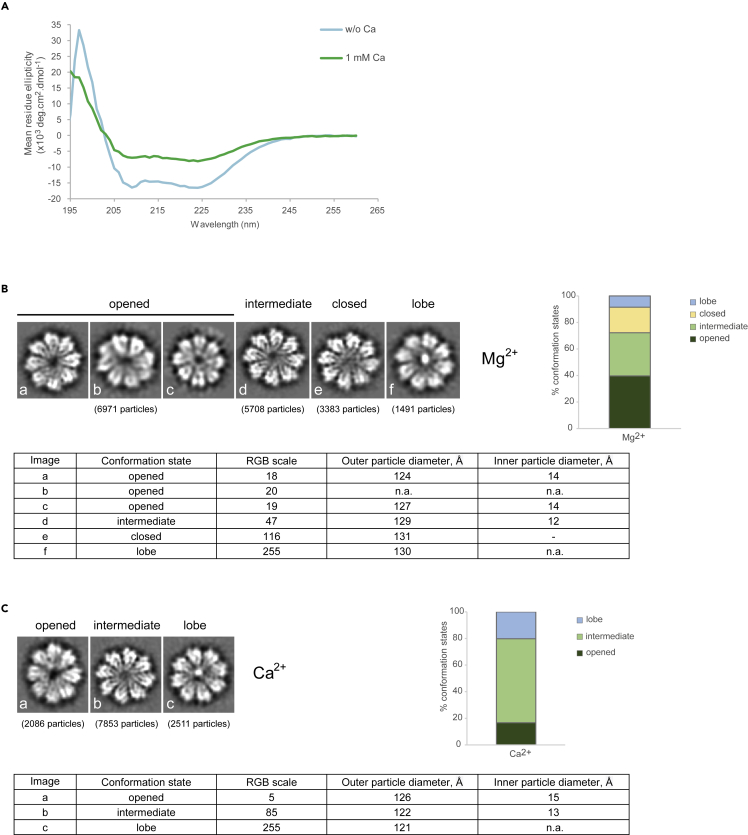
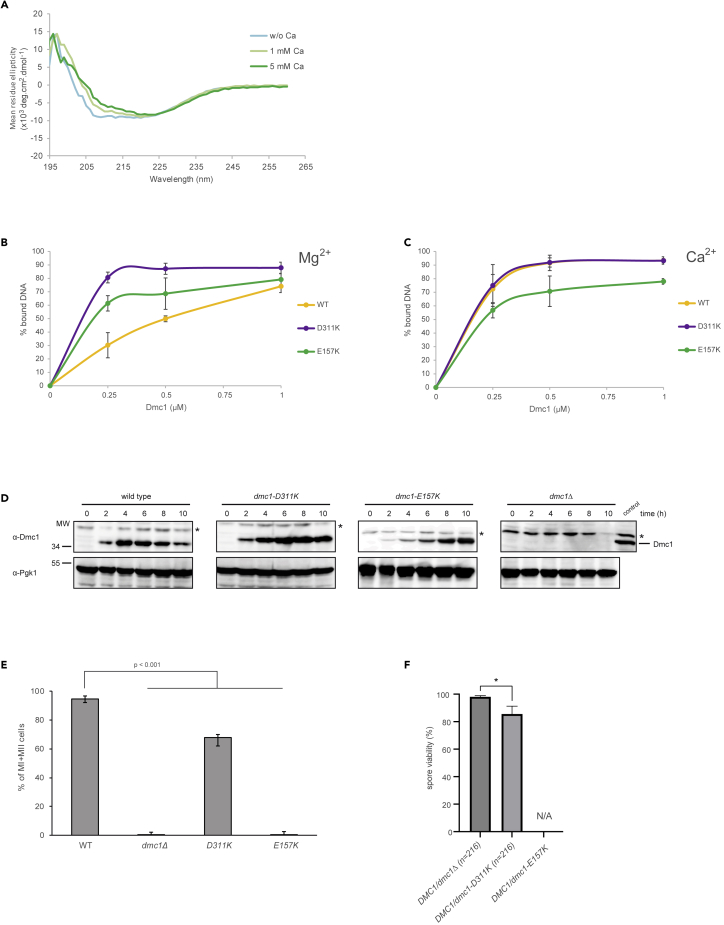
First, we analyzed the secondary structure of DMC1 in the presence or absence of calcium ions using circular dichroism. Indeed, calcium ions (1 mM) induced a significant change in the secondary structure of DMC1 (Figure 1A). We observed a similar structural effect in the presence of Mg2+, but it was less pronounced compared with Ca2+ (Figure S1). To confirm the effect of bivalent ions on the structure of DMC1 we used cryo-EM and generated 2D class averages of DMC1 and ATP in the presence of magnesium or calcium ions, respectively. The obtained 2D class averages show multiple conformation states of the ring center, which are characterized as open, intermediate, closed, or lobe state. In the presence of magnesium ions, the equilibrium is shifted toward the open and intermediate states (Figure 1B). Meanwhile, in the presence of calcium ions, the majority of particles are represented in the intermediate state. Moreover, a significant increase of particles in the lobe state but no particles in the closed state are observed (Figure 1C). In summary, these data demonstrate structural changes in DMC1 in the presence of bivalent ions and nucleotide cofactor with calcium having a more dominant effect on the structural state.
Figure 1.
Calcium ions affect the conformation of human DMC1
(A) CD spectra of the human DMC1 protein (5 μM) in the presence of ATP (0.3 mM) and absence of calcium ions (light blue line), or in the presence of 1-mM CaCl2 (green line).
(B) 2D class averages of cryo-EM particles of human DMC1 protein in the presence of MgCl2 and ATP representing “opened” (a, b, and c), “intermediate” (d), “closed” (e), and “lobe” (f) states. (C) 2D class averages of cryo-EM particles of human DMC1 protein in the presence of CaCl2 and ATP illustrating “opened” (a), “intermediate” (b), and “lobe” (c) states. Classification of individual states was based on RGB scale in range from 0 (black color) to 255 (white color), respectively. The intensity in RGB scale 1–50 corresponds to opened, 51–100 to intermediate, 101–200 to closed, and 201–300 to lobe forms, respectively. See also Figure S1.
Generation and characterization of DMC1 mutants with impaired ion regulation
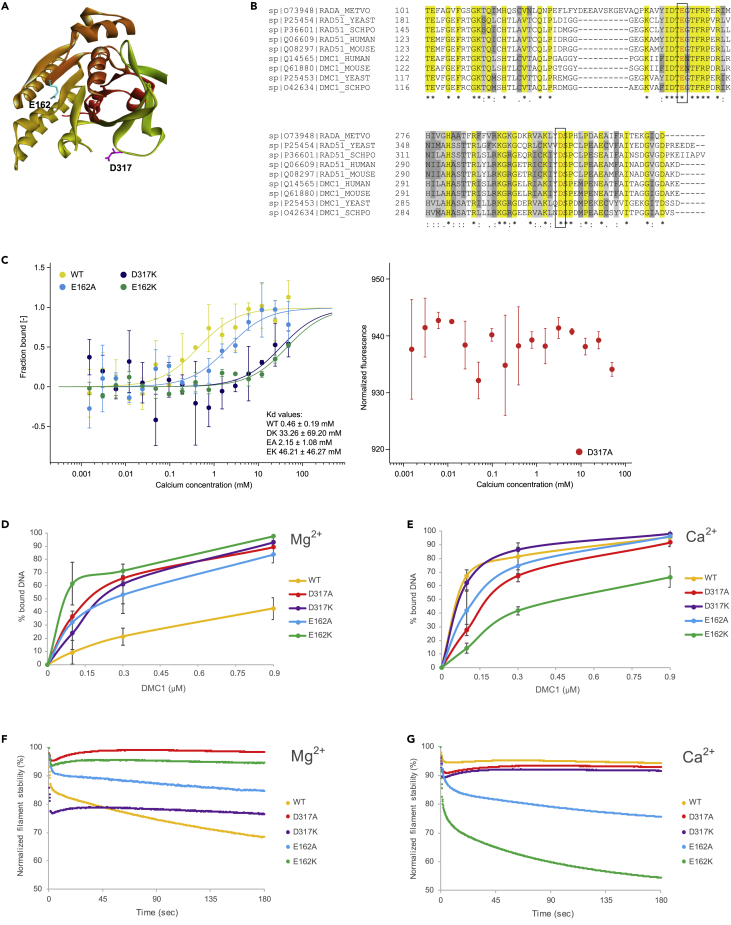
To understand the regulatory role of bivalent ions, we aimed to identify amino acids required for this DMC1 regulation. The crystal structure of archaeal recombinase RadA from M. voltae has revealed the indirect contact of highly conserved amino acids E151 and D302 (corresponding to E162 and D317 in human DMC1) with bound calcium ion (Figures 2A and 2B) (Qian et al., 2006a). Therefore, we have selected these two amino acids and constructed mutant variants either by replacing them with alanine (to keep the neutral charge) or lysine (to reverse the charge), respectively. All DMC1 variants were expressed in E. coli and purified according to the same protocol to near homogeneity.
Figure 2.
Biochemical activities of human DMC1 mutants
(A) 3D model of the human DMC1 structure with the highlighted conserved amino acids E162 (light blue) and D317 (purple). The modeling was performed based on the structure available in the protein data bank (PDB entry 4HYY).
(B) Sequence alignment of RecA family proteins. The highlighted conserved amino acids were substituted with either alanine (A) or lysine (K).
(C) Microscale thermophoresis binding curve of various DMC1 protein fractions vs calcium ion concentration (mean ± SD; n = 3).
(D) Graphical representation of DNA binding activity of DMC1 mutants in the presence of magnesium ions. Increasing concentrations of DMC1 mutants (0.1, 0.3, and 0.9 μM) were incubated with 5′-end fluorescently labelled 90-mer single-stranded DNA (pR231, 0.9-μM nucleotides), 1-mM ATP and 1-mM MgCl2 (mean ± SD; n = 3).
(E) Graphical representation of DNA binding activity of DMC1 mutants in the presence of CaCl2. Increasing concentrations of DMC1 mutants (0.1, 0.3, and 0.9 μM) were incubated with 5′-end fluorescently labelled 90-mer single-stranded DNA (pR231, 0.9-μM nucleotides), 1-mM ATP and 1-mM CaCl2 (mean ± SD; n = 3).
(F) Stability of DMC1 filaments in the presence of MgCl2 measured by bio-layer interferometry. DMC1 filaments were formed by binding DMC1 (5 μM) to 5′-biotinylated 90-mer ssDNA (1.8-μM nucleotides) pre-bound to streptavidin biosensor in the presence of 1-mM ATP and 1-mM MgCl2. The protein dissociation was measured after incubation of biosensor in a buffer containing 200-mM KCl. The data are shown as the average of three independent experiments.
(G) Stability of DMC1 filaments in the presence of calcium ion measured by BLI. DMC1 filaments were formed by binding DMC1 (5 μM) to 5′-biotinylated 90-mer ssDNA (1.8-μM nucleotides) pre-bound to streptavidin biosensor in the presence of 1-mM ATP and 1-mM CaCl2. The protein dissociation was measured after incubation of biosensor in a buffer containing 200-mM KCl. The data are shown as the average of three independent experiments. See also Figures S2 and S3.
To analyze Ca2+ binding by DMC1, we have utilized microscale thermophoresis, which measures the changes in the intrinsic fluorescence of DMC1 upon the titration of calcium ions. We were able to detect a classical sigmoid binding curve for wild-type DMC1 with the estimated Kd around 450 μM (Figure 2C). Meanwhile, all DMC1 mutants showed decreased or even abolished (D317A mutant) binding of calcium ions (Figure 2C), thus confirming the role of residues D317 and E162 in the coordination of this bivalent ion. This is also supported by the recent DMC1 filament structure where E162 residue coordinates Ca2+ (Figure S2A) (Luo et al., 2021). Furthermore, CD analysis of D317A and D317K mutants in the presence of either Ca2+ or Mg2+ revealed a similar spectrum to one observed for the wild-type protein in the presence of Ca2+ (Figures S2B and S2C), indicating that these mutants might to some extent mimic the calcium-bound state of DMC1. Accordingly, the cryo-EM analysis of the DMC1-D317K mutant in the presence of magnesium ions shows that the majority particles observed for D317K mutant represented lobe state, similar to the ones observed for the wild-type protein in the presence of Ca2+ (Figure S2D).
We next sought to characterize the effect of each mutation on the biochemical activities of DMC1 in the presence of either magnesium or calcium ions. First, we assessed the DNA binding activity of DMC1 mutants on single-stranded DNA by EMSA. As expected, we observed a significant stimulation of wild-type DMC1 binding to ssDNA in the presence of calcium ions. Surprisingly, compared with wild-type DMC1 all mutants exhibited ∼ 2-fold increased ssDNA binding in the presence of magnesium ions and, except for the E162K mutant, comparable binding efficiency as wild-type protein with calcium ions (Figures 2D, 2E, S3A, and S3B). In line with this, analysis of DNA binding in the absence of bivalent ions revealed that only D317 mutants showed significant binding to ssDNA (Figures S3C and S3D), indicating that these mutants might to some extent mimic the calcium-bound state.
Next, we tested the stability of DMC1 filaments in the presence of either magnesium or calcium ions using bio-layer interferometry (BLI) as previously used for the RAD51 filament (Xue et al., 2021). The DMC1 filaments were initially assembled on biotinylated 90-mer ssDNA bound to the streptavidin biosensor. Dissociation of the DMC1 filaments was monitored in time upon the addition of buffer containing 200-mM KCl (Figures 2F and 2G). In the presence of magnesium ion, dramatic destabilization was observed for wild-type and D317K mutants, in comparison with D317A and E162K (Figure 2F). Calcium ions have stabilized the filaments of both wild-type and D317K mutants; however, they strongly decreased the stability of the filament formed by the E162K mutant (Figure 2G), in agreement with its lower DNA binding (Figure 2E). The D317A mutant shows overall good filament stability irrespective of the ion used, indicating a loss of the regulatory effect by bivalent ions on the stability of the DMC1-DNA complex.
DMC1 mutants deficient in ion regulation are defective in D-loop extension
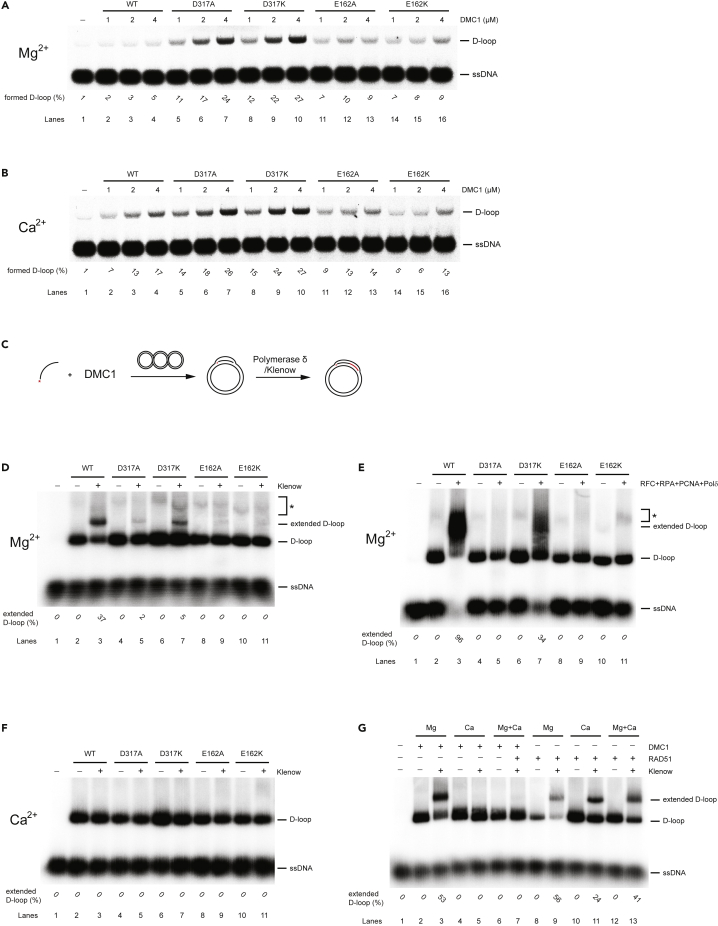
DMC1 is capable of searching for a homologous sequence to promote the formation of joint molecules (D-loops). Therefore, we further tested the ability of all DMC1 mutants to form D-loops in the presence of Mg2+ or Ca2+. In agreement with previously published data (Bugreev et al., 2005), calcium ions stimulated the D-loop formation activity of the wild-type DMC1 (Figures 3A and 3B), corresponding to 3–4 fold higher yield. However, very weak or no stimulation of calcium ions on D-loop formation was observed for E162A and E162K mutants, respectively. Interestingly, only the D317 mutants were able to efficiently form D-loops even in the presence of magnesium ions, supporting that these mutants might mimic the calcium-bound state.
Figure 3.
Calcium ions inhibit extension of D-loop promoted by human DMC1
(A) Increasing concentrations of DMC1 mutants (1, 2, and 4 μM) were incubated with fluorescently labeled 90-mer ssDNA (4.5-μM nucleotides) followed by the addition of pBluescript plasmid DNA in the presence of 1-mM MgCl2.
(B) Increasing concentrations of DMC1 mutants (1, 2, and 4 μM) were incubated with fluorescently labeled 90-mer ssDNA (4.5-μM nucleotides) followed by the addition of pBluescript plasmid DNA in the presence of 1-mM CaCl2.
(C) Schematic representation of D-loop extension assay.
(D) D-loop extension of DMC1 mutants in the presence of 1-mM MgCl2 mediated by Klenow fragment. The quantification of the stable extension product corresponds to the ration of extended D-loop to D-loop formed by wild type (lane 2).
(E) D-loop extension of DMC1 mutants (2 μM) in the presence of 1-mM MgCl2 mediated by RFC, PCNA, and polymerase δ. The quantification of the stable extension product corresponds to the ration of extended D-loop to D-loop formed by wild type (lane 2).
(F) D-loop extension of DMC1 mutants in the presence of CaCl2 mediated by Klenow fragment.
(G) D-loop extension of DMC1 or RAD51 in the presence of 1-mM MgCl2 or CaCl2 mediated by Klenow fragment. Asterisk indicates a nonspecific band. See also Figure S4.
Several genetic and biochemical studies provided evidence that the D-loop activity of DMC1 is stimulated by the HOP2-MND1 complex (Petukhova et al., 2005; Pezza et al., 2006, 2007; Zhao and Sung, 2015). To test if HOP2-MND1 can regulate D-loop formation in the context of DMC1 mutants, we performed D-loop assays in the presence of this complex. All DMC1 mutants were stimulated for D-loop formation to a certain extent in the presence of HOP2-MND1 and either magnesium or calcium ions, respectively (Figures S4A and S4B). Similarly to the D-loop assay done in the absence of HOP2-MND1, the D317K mutant exhibited the highest yield of D-loop compared with the wild-type protein.
After the formation of a stable D-loop, HR further proceeds by extending the invading strand to promote DSB repair. To analyze this step, we first formed D-loops by DMC1 mutants in the presence of HOP2-MND1, followed by an extension with a bacterial polymerase (Klenow fragment) (Figure 3C). Unexpectedly, whereas the wild-type protein was able to extend 37% of D-loops, only 2% or 5% was extended for the D317A or D317K mutants, corresponding to a 18- or 7-fold reduction, respectively (Figure 3D). D-loops formed by the other two DMC1 mutants were not extended at all (Figure 3D). Next, we tested the same extension using the components of the yeast replication machinery including replication factor C (RFC), proliferating cell nuclear antigen (PCNA), and polymerase δ (Figure 3E). Whereas the extension in the context of wild-type DMC1 was very efficient, reaching almost completion (96%), DMC1 mutants showed reduced (34%) or no extension (Figure 3E). These data suggest that D-loops formed by DMC1 mutants are not accessible for polymerase. As calcium ions show structural effects on the DMC1 protein, we also performed the same assay in the presence of Ca2+. Unexpectedly, not only DMC1 mutants, but also wild-type protein was completely unable to extend the D-loop under these conditions (Figure 3F). To exclude the possibility that Ca2+ negatively affects the polymerase, we analyze D-loop extension promoted by the RAD51 recombinase. In contrast to DMC1, where D-loop extension was inhibited even in the presence of both bivalent ions, RAD51-mediated D-loop was extended in all tested conditions (Figure 3G). This suggests that Ca2+ specifically inhibits D-loop extension promoted by human DMC1.
DMC1 mutants defective in ion-regulation are impaired for ATP hydrolysis
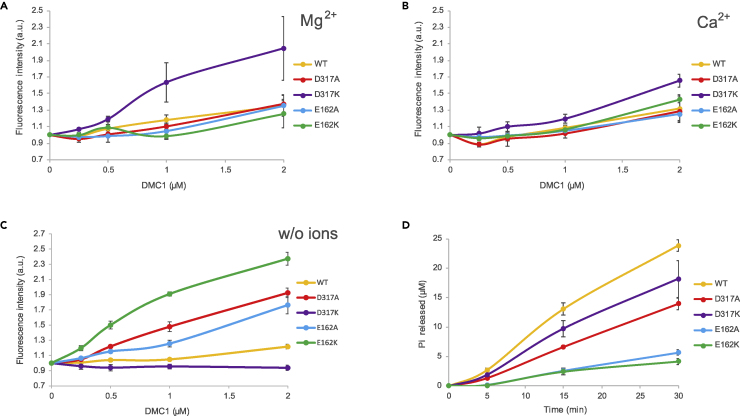
The DMC1 recombinase contains Walker motifs and can efficiently bind as well as hydrolyze ATP. We therefore tested the ATP binding of DMC1 mutants in the presence of ssDNA and either magnesium or calcium ions using a fluorescently-modified ATP analog (TNP-ATP). Efficient binding of DMC1 to TNP-ATP leads to an increase in fluorescent intensity which was measured in a concentration-dependent manner. The D317K mutation caused an increased ATP binding to DMC1 compared with the wild type in the presence of either magnesium or calcium ions, meanwhile, the other mutants bound ATP only in the absence of bivalent ions (Figures 4A–4C).
Figure 4.
ATP binding and hydrolysis of human DMC1 mutants
(A) ATP binding of human DMC1 variants measured as a change of the fluorescence emission intensity of TNP-ATP upon binding to DMC1 in the presence of ATP (1 mM) and 1-mM MgCl2 (mean ± SD; n = 3).
(B) ATP binding of human DMC1 variants measured as a change of the fluorescence emission intensity of TNP-ATP upon binding to DMC1 in the presence of ATP (1 mM) and 1-mM CaCl2 (mean ± SD; n = 3).
(C) ATP binding of human DMC1 variants measured as a change of the fluorescence emission intensity of TNP-ATP upon binding to DMC1 in the presence of ATP (1 mM) and absence of bivalent ion (mean ± SD; n = 3).
(D) ATP hydrolysis of DMC1 variants (2 μM) analyzed using the colorimetric phosphate detection assay in the presence of 90-mer ssDNA (6.3-μM nucleotides) and MgCl2 (1 mM) (mean ± SD; n = 3).
Interestingly, we observed a different behavior in the case of ATP hydrolysis measured by a colorimetric ATPase assay. D317 mutant variants could still hydrolyze ATP, however with a significantly lower efficiency compared with the wild-type protein (Figure 4D). Meanwhile, the E162 mutation caused severe defects in ATP hydrolysis, indicating that this amino acid is required for proper ATP hydrolysis.
Calcium regulation-impaired dmc1 mutants are defective in meiotic progression
To examine whether the defects observed in the in vitro system have any effect on meiosis in vivo, we used yeast S. cerevisiae as a model organism. In accordance with the human DMC1 data, we observed a change in the secondary structure of yeast Dmc1 in the presence of Ca2+ (Figure 5A). Next, we introduced selected mutations into conserved residues of yeast Dmc1 (D311K and E157K) and purified the corresponding proteins to determine whether they have the same effects on yeast Dmc1 activity. The DNA binding activity of the yeast Dmc1 mutants was also increased in the presence of Mg2+, with the highest binding affinity observed for D311K, which is in agreement with the data described above for the human variants (Figures 5B and 5C and S5A and S5B).
Figure 5.
Calcium regulation-impaired dmc1 mutants exhibit meiotic defects
(A) CD spectra of the yeast Dmc1 protein (5 μM) with ATP (0.3 mM) and in the absence (light blue line) and presence of 1-mM CaCl2 (light green line) or 5-mM CaCl2 (green line).
(B) Graphical representation of DNA binding of Dmc1 mutants (0.25, 0.5, and 1 μM) to 5′-fluorescently labeled ssDNA (0.9-μM nucleotides) in the presence of 1-mM ATP and 1-mM MgCl2 analyzed by gel-based assay. The error bars represent the standard deviation from three independent experiments.
(C) Graphical representation of DNA binding of Dmc1 mutants (0.25, 0.5, and 1 μM) to 5′-fluorescently labeled ssDNA (0.9-μM nucleotides) in the presence of 1-mM ATP and 1-mM CaCl2 analyzed by gel-based assay. The error bars represent the standard deviation from three independent experiments.
(D) Western blot analysis of Dmc1 expression from synchronized meiotic culture time points in wild-type and mutant strains. Pgk1 protein was used as a loading control. Asterisk indicates an unspecific band.
(E) DAPI analysis of meiotic divisions in dmc1 mutants. Cells were harvested after 10 h after synchronous induction of meiosis and the percentage of MI and MII cells was determined using fluorescent microscopy of fixed DAPI-stained nuclei. At least 140 cells were counted for each strain. Average values from minimum three independent cultures for each strain were plotted with error bars representing standard deviation (SD). Statistical significance was calculated using unpaired t-test, n.s. not significant.
(F) Spore viability was measured after microdissection of tetrads from diploid strains carrying one copy of DMC1 and one copy of DMC1, dmc1-D311K, dmc1-E157K, or dmc1Δ. Plotted values indicate the mean with standard deviation (two-tailed unpaired t-test; ∗p < 0.05) from three independent experiments. In sum, 216 spores were analyzed per genotype in three independent experiments. N/A, non-available as DMC1/dmc1-E157K cells failed to sporulate. See also Figures S5–S7.
This observation prompted us to study the phenotype of D311K and E157K mutants in vivo. We introduced the corresponding mutations into the S. cerevisiae genomic DNA and tested Dmc1 expression. Both mutants expressed Dmc1 4 h after transfer to sporulation media. However, in contrast to the wild-type protein, which peaked in expression at 4–6 h, both mutants accumulated Dmc1 protein in cells even after 10 h in the sporulation medium (Figures 5D and S7A). Moreover, they also exhibited severe defects in meiotic progression, especially the E157K mutant was as defective as the dmc1Δ strain (Figure 5E), indicating that both mutations negatively affect the proper progression of meiotic recombination. Interestingly, when we analyzed the effect of the mutants in a heterozygous state, the dmc1-E157K/DMC1 showed strongly impaired sporulation, indicating a dominant effect of this allele. In the case of dmc1-D311K/DMC1, we observed a more subtle effect, with reduced spore viability (Figure 5F).
Several studies have shown that deletion or overexpression of several genes involved in the regulation of meiotic recombination can, in some cases only partially, overcome the recombination defects in dmc1Δ mutant (Bishop et al., 1999; Grushcow et al., 1999; Lindgren et al., 2000; Lydall et al., 1996; Niu et al., 2009; Pak and Segall, 2002; Tsubouchi and Roeder, 2003; Xu et al., 1997). First, we analyzed the phenotype of Dmc1 mutants in a red1Δ deletion strain background. Earlier studies indicate a Dmc1-independent repair of meiotic DSBs in this genetic background (Xu et al., 1997) and failure to arrest cells in meiotic prophase in response to unrepaired DSBs (Bishop et al., 1999). Indeed, the RED1 deletion was able to completely alleviate the prophase delay conferred by dmc1-D311K. Moreover, it strongly improved the meiotic progression of dmc1-E157K mutants, even though the resultant spores were inviable (Figures S7B and S7C). Next, we deleted one of the checkpoint genes, RAD17, which is required for the meiotic arrest caused by defective repair of DSBs (Grushcow et al., 1999; Lydall et al., 1996). The prophase arrest in dmc1Δ can be bypassed by RAD17 deletion, leading to meiotic progression despite the persistence of unrepaired DSBs in cells (Lydall et al., 1996). The double mutant rad17Δ dmc1-E157K exhibited only very minor alleviation in prophase arrest. Interestingly, the rad17Δ dmc1-D311K double mutant showed even more severe defects in meiotic progression, compared with the single mutant (Figure S7D), suggesting that removal of this checkpoint is not sufficient to allow meiotic progression in the dmc1 mutants analyzed. Furthermore, the defects in dmc1Δ cells can be overcome by activation of Rad51-dependent interhomolog recombination that can be achieved by overexpression of Rad51 and phosphorylation defective Rad54-T132A (Niu et al., 2009; Tsubouchi and Roeder, 2003). In contrast to dmc1Δ cells, neither overexpression of Rad51, nor Rad54-T132A, was able to significantly improve the meiotic progression of both Dmc1 mutants (Figure S7E), indicating that the Rad51 pathway cannot substitute for these Dmc1 mutants. To further confirm this observation, we also analyzed meiotic divisions in hed1Δ cells which improves the meiotic progression in dmc1Δ cells (Figure S7F). Both hed1Δ dmc1-D311K and hed1Δ dmc1-E157K mutants exhibited similar defects in meiotic divisions as in a HED1 wild-type strain background, confirming that Rad51-mediated recombination cannot rescue the meiotic defect of these mutants (Figure S7F).
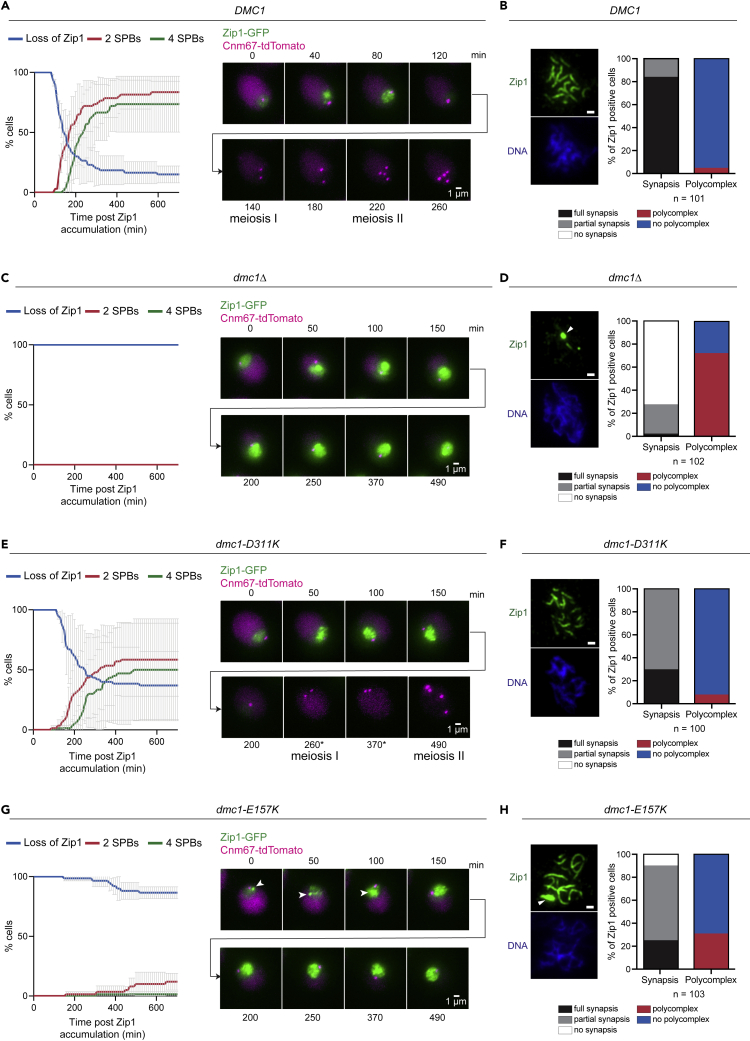
Cells lacking Dmc1 exhibit defective assembly of the synaptonemal complex (SC), resulting in the formation of Zip1-containing polycomplexes (PCs) (Lao et al., 2013). To follow SC dynamics in the dmc1 mutants, we performed live-cell imaging of meiotic cells expressing Zip1-GFP. Cnm67-tdTomato was used to visualize spindle pole bodies and monitor meiotic progression. In parallel, we also prepared chromosome spreads 5h after meiotic induction, to monitor synapsis more closely. In the control strain (DMC1), Zip1-GFP disappeared between 80-120 min after its initial accumulation (Figure 6A). The immunostaining of chromosome spreads further showed that, 5 h after the induction of meiosis, the vast majority of cells had fully synapsed chromosomes, with a small proportion containing Zip1 polycomplexes (Figure 6B). As expected (Lao et al., 2013), dmc1Δ cells exhibited persistent nuclear Zip1-GFP signal, a high frequency of Zip1 PCs, and failure in chromosome synapsis (Figures 6C and 6D). Interestingly, both dmc1 point mutants showed SC assembly defects and a delayed entry into meiosis I (Figures 6E–6H). The phenotype was significantly more severe in dmc1-E157K mutant cells, in which we observed persistent Zip1-GFP signal and an increased frequency of PCs. Taken together, both mutants showed impaired SC formation and delayed entry into meiosis I, indicating possible defects in the formation/processing of joint molecules and in the production of mature recombinants.
Figure 6.
Impaired meiotic recombination and meiotic progression in dmc1-E157K and dmc1-D311K mutants
(A) S. cerevisiae dmc1Δ/DMC1 cells expressing Zip1-GFP and Cnm67-tdTomato were synchronously released to undergo meiosis from G0/G1 by transfer into sporulation medium (SPM). Live cell imaging was initiated ∼3.5 h after the induction of meiosis. The time elapsed from Zip1-GFP accumulation (set as t = 0) until Zip1-GFP removal from chromosomes was quantified for 30 cells and plotted in Blue. Plotted in Red and Green are the times elapsed from Zip1 accumulation until entry into meiosis I (two Cnm67-tdTomato signals) or meiosis II (four Cnm67-tdTomato signals), respectively. Plotted is the mean of two independent experiments (Figure S8). Error bars represent the range. Representative images of the hallmark events are shown on the right.
(B) Chromosome surface spreads of cells in (A) were prepared 5 h after induction of meiosis. Chromosomes were stained for DNA (DAPI) and Zip1 (anti-Zip1 antibodies) and prophase I cells identified by positive Zip1 staining. The extent of chromosome synapsis and the frequency of polycomplexes were quantified, as indicated. A representative image is shown. More than 100 spreads were analyzed in one experiment. Scale bar, 1 μm.
(C) As in (A), for dmc1Δ/dmc1Δ homozygous mutants. (D) As in (B), for dmc1Δ/dmc1Δ homozygous mutants. White arrowhead indicates a polycomplex.
(E) As in (A), for dmc1Δ/dmc1-D311K mutants. (∗) Brightness of Cnm67-tdTomato was adjusted to facilitate visibility of the signal.
(F) As in (B), for dmc1Δ/dmc1-D311K mutants.
(G) As in (A), for dmc1Δ/dmc1-E157K mutants. White arrowhead indicates a polycomplex.
(H) As in (B), for dmc1Δ/dmc1-E157K mutants. White arrowhead indicates a polycomplex.
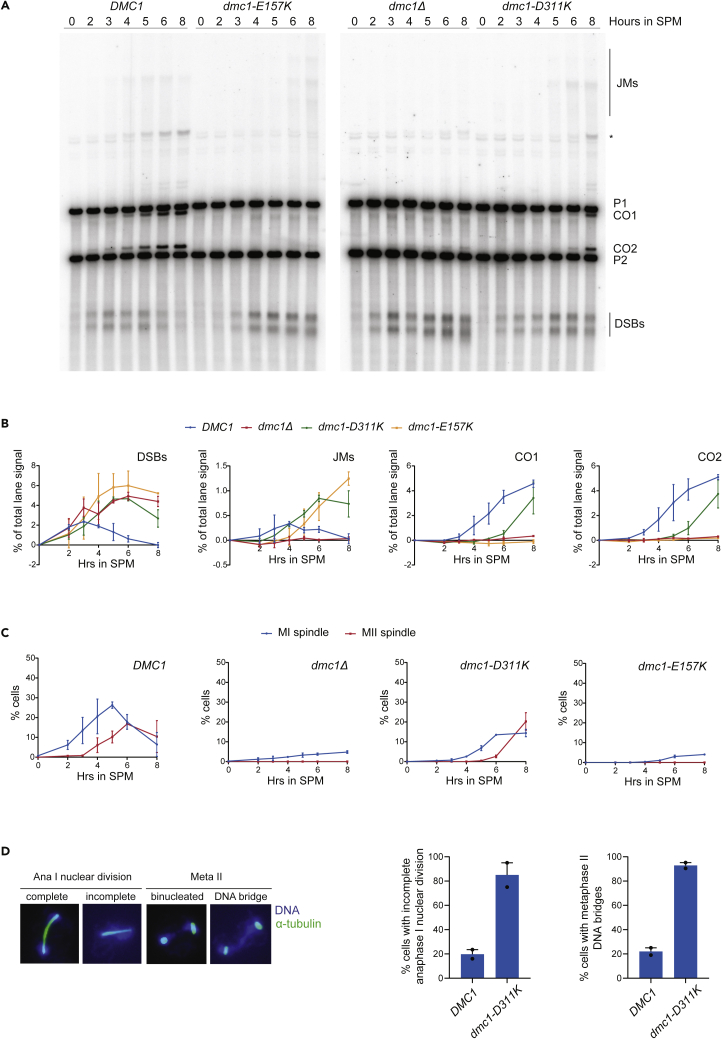
To directly determine the impact of the DMC1 mutations on DSB and crossover formation, we performed a physical analysis of recombination at the well-characterized HIS4-LEU2 hotspot (Hunter and Kleckner, 2001). In contrast to dmc1Δ, which accumulated DSBs but failed to form DNA joint molecules (JMs), as previously described (Bishop et al., 1992; Schwacha and Kleckner, 1997), both the dmc1-D311K and dmc1-E157K mutants accumulated JM intermediates, in spite of showing a noticeable delay in DSB repair (Figures 7A and S9). In both DMC1 point mutants, crossover formation was strongly impaired, with dmc1-E157K mutants lacking crossover products during the time course analysis performed and dmc1-D311K mutants showing a 3–4 h delay relative to the wild type (Figures 7A and S9). We also noted the accumulation of various forms of JMs in the dmc1-E157K mutant, and to a lower extent also in dmc1-D311K cells (Figure 7A). Whereas the fast-migrating products likely correspond to single-ended invasion intermediates (Hunter and Kleckner, 2001), the slower-migrating ones may correspond to multi-chromatid JMs (Oh et al., 2007). These results suggest that both mutants are capable of supporting homology search and generate stable D-loops but they both fail to further process/disengage them. Finally, to control for defects in cell cycle progression, we have also monitored meiotic progression by FACS analysis of DNA content and spindle morphology. Whereas all strains showed comparable kinetics of DNA replication (Figure S9), the D311K mutant entered meiotic divisions very slowly and the E157K mutant, similarly to dmc1Δ, completely fail to do so (Figure 7B). Interestingly, dmc1-D311K cells that entered meiosis I failed to complete chromosome segregation (Figures 7C and 7D), most likely due to the persistence of unresolved joint molecules. Taken together, these data clearly point to severe meiotic recombination defects in the dmc1-E157K and dmc1-D311K mutants, further highlighting the possible role of calcium ion binding in regulating this process.
Figure 7.
Defective DSB repair in dmc1-E157K and dmc1-D311K mutants during meiosis
(A) Cells from (Figure S9) were treated with psoralen to crosslink DNA for the physical analysis of recombination at the HIS4-LEU2 locus. JMs, joint molecules; P1, parental DNA 1; P2, parental DNA 2; CO1 and CO2, reciprocal recombinants from P1 and P2; DSBs, double-strand breaks; asterisk indicates ectopic recombination. The image shown is representative of two independent experiments.
(B) Dynamics of double strand break (DSB), joint molecule (JM), and crossover (CO) accumulation were quantified as fractions of the total lane signal from (A) and a biological replicate. Plotted values show the mean of two independent experiments; error bars represent the range.
(C) Kinetics of meiotic progression from strains in (A and Figure S9) determined by spindle morphology. Spindles were visualized by anti-tubulin immunofluorescent staining; meiosis I cells had a single bipolar spindle (MI spindle, blue), whereas meiosis II cells had two bipolar spindles (MII spindle, red). Plotted values show the mean of two independent experiments with error bars representing range; 200 cells were analyzed per time point in both experiments.
(D) Immunofluorescence analysis of anaphase I and metaphase II from WT and dmc1-D311K mutants. Cells were stained with anti-α-tubulin and DAPI. Representative images of anaphase I (Ana I) and metaphase II (Meta II) nuclear division are shown (left panel) and the frequency of incomplete nuclear division at these stages in WT and dmc1-D311K strains (right panel). Anaphase I nuclear division is considered complete when two fully separated DNA masses can be distinguished. Cells with two short bipolar spindles, in metaphase II, that contain DNA bridges between the two nuclei are inferred to have failed to fully undergo anaphase I. Mean values from two independent experiments are plotted; error bars represent the range. The number of analyzed anaphase I cells was n1 = 25 and n2 = 17 for WT and n1 = 20 and n2 = 12 for dmc1-D311K. For metaphase II analysis, 42–44 WT cells and 21 dmc1-D311K cells were analyzed in each experiment.
Discussion
Calcium ions effectively and selectively regulate the function of a wide spectrum of proteins in many cellular processes, including meiosis. It has been well established that calcium ion is implicated in spermatogenesis and oogenesis. During oogenesis in many animal species, Ca2+ currents regulate cell cycle control checkpoints, which direct the transition from one meiotic phase to the next phase (Whitaker and Patel, 1990). Further, Ca2+ deficiency has been shown to lead to impaired spermatogenesis and male infertility (Harchegani et al., 2019).
Several studies from various organisms demonstrated that calcium ion also functions as a universal cofactor of the meiosis-specific recombinase DMC1 and promotes its homology-search and strand exchange activity during meiotic recombination. Interestingly, human DMC1 is further regulated by potassium and magnesium ions together with ATP cofactor thus emphasizing the importance of ion regulation for proper progress of meiosis (Sehorn et al., 2004). The mechanistic understanding of these regulations remained poorly described.
In this work, we observed changes in the secondary structure of human as well as yeast Dmc1 proteins associated with ion binding. Our cryo-EM data further confirmed the structural changes within DMC1 in the presence of different bivalent ions. We have also observed very stable DMC1-ssDNA filaments in the presence of Ca2+, in contrast to Mg2+, which is in agreement with previously reported data (Luo et al., 2021; Passy et al., 1999).
In agreement, the presence of calcium ions initiates the transition of the disordered L2 region to largely ordered conformation, which promotes the assembly of active presynaptic filament in bacterial recombinase, MvRadA (Qian et al., 2006a). This protein shares a significant sequence homology with human DMC1 and RAD51 recombinases, and possesses a strand exchange activity, which can be similar to human proteins stimulated by calcium ions (Qian et al., 2006a). The crystallographic model from MvRadA showed that Ca2+ binding is stabilized by indirect contact with highly conserved amino acids E151 and D302 (part of the ATP cap) (Qian et al., 2006a) and can thus be applied to DMC1. We generated the mutants by substituting the corresponding negatively charged amino acids E162 and D317 by either neutrally or positively charged amino acids, respectively. Interestingly, all generated mutants exhibited significantly increased ssDNA binding activity in the presence of Mg2+ compared with the wild type. The efficiency of DNA binding was at a similar level as DNA binding of the wild type in the presence of Ca2+. Furthermore, this is to a certain degree also supported by increased stability of DMC1/DNA complexes, even though in E162 mutants they are unstable in the presence of calcium ions, indicating that dissociation from DNA might be more complex. These findings suggest that the selected amino acids E162 and D317 are involved in the binding or coordination of bivalent ions and that some of these mutants might represent Ca2+-bound state of DMC1 and could serve as novel separation of function mutants.
Similarly to DNA binding, wild-type DMC1 is also stimulated by Ca2+ or other accessory factors such as HOP2-MND1 to efficiently promote D-loop formation. Interestingly, mutant variants of D317 were capable to execute D-loop formation in the presence of Mg2+ even without any accessory factor. This result is inconsistent with previously published data, where authors observed a decreased ability of D317K mutant to promote D-loop compared with wild type in the presence of Mg2+ (Chang et al., 2015), which might reflect different reaction conditions used. In contrast to D317 mutants, only low amount of D-loop in the presence of magnesium ion was observed for E162 mutants which could be explained by the severe defects in the ATP hydrolysis of these mutants. On the other hand, calcium ions were able to significantly stimulate DMC1-promoted D-loop formation for wild-type DMC1. A weak stimulation was also observed in the case of all studied DMC1 variants except of E162K, corresponding to its robust protein/DNA complex instability and also decreased ssDNA binding affinity in the presence of Ca2+. Interestingly, the D-loop formation of all DMC1 mutants could still be promoted by HOP2-MND1 complex suggesting that bivalent ions regulate DMC1 function by different mechanisms than HOP2-MND1.
The extension of the D-loop, which follows after its formation, was compared for all mutants in the presence of HOP2-MND1, which ensures sufficient formation of the D-loop. To our surprise, in the presence of Mg2+ we only observed the extension with wild type and to very little extent also for D317K mutant. Unexpectedly, although calcium ions stimulate the DMC1-promoted D-loop formation, they completely inhibit the following D-loop extension even for wild-type DMC1. The inhibitory effect can be observed also if both magnesium and calcium ions are present in the reaction, indicating the dominant effect of Ca2+. Contrarily, calcium ions did not affect D-loop extension promoted by human RAD51, excluding the general inhibitory effect of Ca2+ on polymerase activity and indicating different calcium-mediated mechanisms of regulation for human DMC1 and RAD51. Potentially different mechanism of Ca2+ regulation for human DMC1 and RAD51 is in agreement with data showing that in the case of RAD51, calcium ions act as an inhibitor of its ATPase activity, thus protecting RAD51 from forming stable but inactive ADP-RAD51-ssDNA complex (Bugreev and Mazin, 2004). Meanwhile, in the case of DMC1, inhibition of ATP hydrolysis by Ca2+ does not lead to the formation of a stable DMC1-ssDNA complex with ADP (Bugreev et al., 2005).
As DMC1 is evolutionarily conserved from viruses to humans and a similar effect of magnesium or calcium ions on its DNA binding activity was observed for the corresponding yeast variants, we envision that the mechanism of ion regulation might be evolutionary conserved. In agreement with this notion, S. cerevisiae dmc1-D311K and dmc1-E157K mutants exhibited significant delays in meiotic cell cycle progression past prophase I. Moreover, both mutants also displayed defective chromosome synapsis, delayed DSB repair, as well as impaired joint molecule formation and processing. The phenotype of the dmc1-D311K mutant was particularly strong, in some assays comparable with that of the dmc1Δ mutant. We note, however, that the dmc1-D311K allele is dominant when in a heterozygous setting, in contrast to dmc1Δ. This demonstrates that the Dmc1-D311K mutant protein does retain some of its properties, in such a way that Dmc1-D311K interferes with the function of wild-type Dmc1.
Some aspects of the phenotypes of the dmc1 point mutants are reminiscent of what has been previously described for csm2Δ mutants. Csm2 is a member of the Shu complex that was shown to be critical for meiotic recombination (Sasanuma et al., 2013). However, the mechanism seems different from Dmc1 mutants, as Csm2 was shown to be required for nucleotide-independent stabilization and formation of Rad51 complexes with only a mild effect on Dmc1 foci formation (Sasanuma et al., 2013). Interestingly, the E157 mutation to aspartic acid bypasses the requirement for accessory complex Mei5/Sae3 for Dmc1 recombination function in yeast (Reitz et al., 2019). However, the recombination promoted by this mutant is abnormal, it forms foci in the absence of DNA breaks, display high levels of multi-chromatid and intersister joint molecules, and ectopic recombination. In contrast to our mutant, it produces regular COs and enters meiotic division (Reitz et al., 2019), suggesting that the charge change in our mutant responsible for the loss of calcium binding has a more severe phenotype.
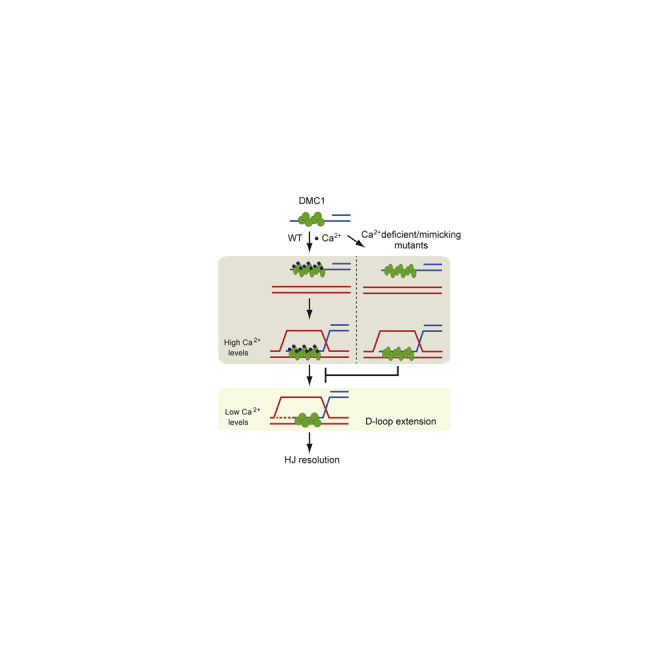
Taken together we suggest that calcium ions not only stabilize the DMC1 filament and make it more proficient for homology search and strand exchange, but they also help to regulate the accessibility of the 3′ invaded end of a D-loop to allow its extension. DMC1-mediated D-loop extension seems to control the formation of double Holliday junctions and regulate when and how the recombination intermediates are processed, representing a novel regulatory step within meiotic HR. The physiological role of calcium ions in this process is supported by its concentration-dependent effect on sporulation with an observed 12-fold increase before sporulation (Suizu et al., 1995). Natural fluctuation of calcium ions during meiosis could regulate DMC1 activity by introducing conformational changes that first enable stable binding of DNA and D-loop formation. As the Ca2+ wave passes, DMC1 conformation changes might now allow for D-loop extension. The mutants seem to have a faulty response to the ion environment leading to their issues in forming and/or processing the recombination intermediates. It would be interesting to find whether such regulation of D-loop extension can be also mediated by DMC1-interacting partner(s).
Limitations of the study
We did not demonstrate in vivo direct regulatory role of calcium ions and their oscillations or protein factor that would mimic this effect during early steps of meiotic recombination.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| α-Dmc1 | Akira Shinohara | N/A |
| anti-Pgk1 (22C5D8) | Thermo Fisher | 459250, RRID:AB_2532235 |
| Rat monoclonal anti-tubulin | Serotec | MCA78G; RRID: AB_325005 |
| Donkey anti-Rat IgG Alexa Fluor 488 | Invitrogen | A-21208; RRID: AB_141709 |
| Donkey anti-Rabbit IgG Alexa Fluor 488 | Invitrogen | A-21206; RRID:AB_2535792 |
| Rabbit polyclonal anti-Zip1 | J. Matos (MPL) | N/A |
| Bacterial and virus strains | ||
| BLR(DE3) | Novagen | 69053, Sigma-Aldrich |
| Rosetta(DE3)pLysS | Novagen | 70956, Sigma-Aldrich |
| DH5 alpha | Thermofisher Scientific | 18258012 |
| Chemicals, peptides, and recombinant proteins | ||
| IPTG | AppliChem | A1008 |
| 2-mercaptoethanol | AppliChem | A4338 |
| NP40 | AppliChem | A1694 |
| Aprotinin | AppliChem | A2132 |
| Chymostatin | AppliChem | A2144 |
| Leupeptin Hemisulfate | AppliChem | A2183 |
| Pepstatin A | AppliChem | A2205 |
| Imidazole | AppliChem | A1073 |
| ATP | AppliChem | A1348 |
| Glutaraldehyde (50% solution) | AppliChem | A4393 |
| Sodium creatine phosphate dibasic | Sigma-Aldrich | 27920 |
| Creatine kinase | Sigma-Aldrich | 10127566001 |
| Klenow fragment (3′-5′exo) | New England Biolabs | M0212 |
| BSA | New England Biolabs | B9000 |
| dATP | Thermo Fisher | R0141 |
| dGTP | Thermo Fisher | R0161 |
| dTTP | Thermo Fisher | R0171 |
| dCTP | Thermo Fisher | R0151 |
| PMSF | AppliChem | A0999 |
| VECTASHIELD Antifade Mounting Medium with DAPI | Vector Laboratories | H-1200 |
| ATP, [γ-32P], 3000 Ci/mmol | Hartmann Analytic | FP-501 |
| HIS-Select Nickel Affinity Gel | Sigma-Aldrich | P6611 |
| CHT Ceramic Hydroxyapatite | Bio-Rad | 1584000 |
| MonoQ | Cytiva | 17516701 |
| Superdex 200 Increase | Cytiva | 28990944 |
| Talon Superflow resin | Cytiva | 28957502 |
| Heparin Sepharose 6 Fast Flow resin | Cytiva | 17099801 |
| TNP-ATP | Jena Bioscience | NU-221 |
| Trioxsalen | Sigma Aldrich | T6137 |
| ProLongTM Diamond Antifade Mountant with DAPI | Thermo Fisher | P36962 |
| Concanavalin A | Sigma Aldrich | Cat#C0412 |
| Nunc Lab-Tek II Chambered Coverglass | Thermo Scientific | Cat#155409 |
| Critical commercial assays | ||
| PiColorLock phosphate detection system | Expedeon | 303–0030 |
| Qubit dsDNA broad range kit | Thermo Fisher | Q32850 |
| Experimental models: Organisms/strains | ||
| Saccharomyces cerevisiae, see Table S2 | ||
| Oligonucleotides | ||
| see Table S1 | This study | N/A |
| Recombinant DNA | ||
| pET11c/Dmc1 | Michael Sehorn | N/A |
| pET15b/DMC1 | Shigeyuki Yokoyama | N/A |
| pBluescript | Patrick Sung | N/A |
| Software and algorithms | ||
| MultiGauge V3.2 software | Fujifilm | N/A |
| OriginPro7 | OriginLab | https://www.originlab.com/ |
| softWoRx software | Applied Precision, Inc. | www.appliedprecision.com |
| MO.Affinity Analysis software | NanoTemper Technologies | https://nanotempertech.com/monolith-mo-control-software/ |
| CellQuest | BD Biosciences | https://www.bd.com/en-uk/products/molecular-diagnostics/cytometric-analysis-products |
| Visiview 2.1.4. | Visitron systems | https://www.visitron.de/products/visiviewr-software.html |
| Motion Core2 | (Zheng et al., 2017) | https://sbgrid.org/software/titles/motioncor2 |
| Gctf | (Zhang, 2016) | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| Gautomatch_v0.53 | (Zhang, 2016) | https://www2.mrc-lmb.cam.ac.uk/download/gautomatch-053/ |
| Relion-3.0 | (Zivanov et al., 2018) | https://github.com/3dem/relion |
| Other | ||
| High Precision Streptavidin biosensors (SAX) | ForteBio | 18–0037 |
| NT.LabelFree capillaries | NanoTemper Technologies | MO-Z022 |
| Vivaspin-2 (30,000MWCO) | Sigma-Aldrich | Z614254 |
| Whatman cellulose chromatography paper | Sigma-Aldrich | WHA3030917 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Lumir Krejci (lkrejci@chemi.muni.cz).
Materials availability
All materials generated in this study are available from the lead contact.
Experimental model and subject details
Yeast strains
All strains used in this study were derived from Saccharomyces cerevisiae SK1 strains NHY1210 and NHY1215 (a kind gift from Neil Hunter) and their genotypes can be found in Table S2. Strains containing a mutation in DMC1 gene were generated according to (Liu et al., 2014). The presence of desired mutation was confirmed by PCR. The deletion of specific genes was performed by polymerase chain reactions (PCR)-based methods using the kanMX6, and natMX4 markers (Goldstein and McCusker, 1999; Longtine et al., 1998).
Method details
Human DMC1 and its mutants
The E. coli strain BLR(DE3) was transformed with plasmid expressing DMC1 protein variant containing (His)6-affinity tag and protein expression was induced by 1 mM IPTG at 37°C for 4 h. Extract from 4.7 g of cell paste was prepared by sonication in 20 mL of cell breakage buffer containing 50 mM Tris-HCl pH 7.5, 600 mM KCl, 10% sucrose, 2 mM EDTA, 0.01% NP40, 1 mM β-mercaptoethanol, and protease inhibitor cocktail. The lysate was clarified by ultracentrifugation and the resulting supernatant was incubated with Ni-NTA agarose (Sigma-Aldrich) for 1 h at 4°C. The beads were washed with buffer T (25 mM Tris-HCl, 10% glycerol, 0.5 mM EDTA, pH 7.5, 0.01% NP40, 1 mM β-mercaptoethanol) containing 200 mM KCl. The bound proteins were eluted with buffer T containing 50 mM KCl and imidazole (from 50 to 1000 mM). Fractions containing DMC1 (150 to 1000 mM imidazole) were loaded on 2-mL hydroxyapatite column (Bio-Rad) equilibrated in T buffer containing 100 mM KCl and eluted using a 12-mL gradient of 100–900 mM KH2PO4 in T buffer. The peak fractions were applied onto MonoQ column (GE Healthcare) and DMC1 protein was eluted using a 15-mL gradient of 200–900 mM KCl in T buffer. Fractions containing DMC1 were concentrated and applied on Superdex S200 column (24 mL) equilibrated in T buffer containing 150 mM KCl. The peak DMC1 fractions were concentrated in Vivaspin-2 (30,000 MWCO) concentrator, aliquoted and stored at −80°C in small aliquots.
Yeast Dmc1 and its mutants
Yeast Dmc1 variants were purified as described elsewhere (Busygina et al., 2013) with minor modifications. Briefly, the plasmid expressing Dmc1 protein with N-terminus (His)6-affinity tag was introduced into E. coli strain Rosetta(DE3)pLysS. Protein expression was induced by 1 mM IPTG at 37°C for 3 h in 2xTY media supplemented with ampicillin (100 μg/μL). Extract from 4.7 g of cell paste was prepared by sonication in 20 mL of buffer containing 25 mM Tris-HCl pH 7.5, 500 mM KCl, 10% glycerol, 0.5 mM EDTA, 0.01% NP40, 1 mM DTT, 1 mM MgCl2, 1 mM ATP, and protease inhibitor cocktail. The lysate was clarified by ultracentrifugation and the resulting supernatant was incubated with 300 μL of Talon Resin (GE Healthcare) for 2 h at 4°C. The beads with bound proteins were washed with 10 mL of buffer T (25 mM Tris-HCl pH 7.5, 10% glycerol, 0.5 mM EDTA, 0.01% NP40, 1 mM DTT) containing 150 mM KCl followed by additional washing step with 10 mL of buffer T containing 500 mM KCl. The protein was eluted in steps with 200 and 500 mM imidazole in buffer T containing 140 mM KCl, 1 mM MgCl2, and 1 mM ATP. Fractions containing Dmc1 protein were applied onto a 1-mL Heparin column (GE Healthcare) equlibriated with buffer T containing 150 mM KCl, and eluted using a 15-mL gradient of 150–1000 mM KCl in buffer T containing 1 mM MgCl2 and 1 mM ATP. The peak fractions were pooled and loaded on a 1-mL MonoQ column (GE Healthcare) equlibriated with buffer T containing 150 mM KCl. Dmc1 protein was eluted using a 12-mL gradient of 150–550 mM KCl in buffer T containing 1 mM MgCl2 and 1 mM ATP. The fractions containing Dmc1 protein were concentrated to 3 μg/μL in a Vivaspin-2 (30,000 MWCO) concentrator and stored at −80°C in small aliquots.
Other proteins
Human RAD51, HOP2-MND1, RFC complex, PCNA and Pol δ were purified as described previously (Finkelstein et al., 2003; Pezza et al., 2006; Sebesta et al., 2011; Spirek et al., 2018).
DNA binding assay
Indicated amounts of yeast or human DMC1 variants were mixed with 5‘-end fluorescently labelled 90-mer single-stranded DNA (pR231, 0.9 μM nucleotides) in buffer DM (25 mM Tris-HCl pH 7.5, 50 mM KCl) containing 1 mM ATP and either 1 mM MgCl2 or CaCl2 as indicated. After the incubation at 37°C for 5 min, glutaraldehyde (0.05%) was added to each reaction followed by additional incubation at 37°C for 5 min. After the addition of gel loading buffer (60% glycerol, 10 mM Tris–HCl, pH 7.4 and 60 mM EDTA), the reaction mixtures were resolved in 0.9% agarose gel in 1x TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA, pH 7.5). Gels were scanned using FLA-9000 Starion (Fujifilm) and quantified by MultiGauge V3.2 software (Fujifilm). DNA binding assay with yeast Dmc1 variants and dsDNA was performed using the same conditions. dsDNA substrate was prepared using 5’-end fluorescently labelled 49-mer (pR27) and complementary oligonucleotide (pR28) as described previously (Matulova et al., 2009).
Bio-layer interferometry of DNA binding experiments
High Precision Streptavidin biosensors (SAX, ForteBio) were incubated in 200 μL of reaction buffer A (25 mM Tris-HCl pH 7.5, 50 mM KCl, 0.05% Tween-20) for at least 10 min before the experiment. Binding kinetics were measured on a BLItz instrument (ForteBio) using the BLItz Pro Software (ForteBio) according to the manufacturer’s manual. Briefly, streptavidin biosensor was incubated in buffer A for 30 s to measure the baseline value. Four μL of 5′-biotinylated 90-mer ssDNA (pR231, 1.8 μM nucleotides) was then loaded on the biosensor for 2 min followed by a brief wash (15 s) in 200 μL of buffer B (25 mM Tris-HCl pH 7.5, 50 mM KCl, 0.05% Tween-20, 1 mM ATP, and either 1 mM MgCl2 or CaCl2). Four μL of indicated DMC1 protein variant (5 μM) diluted in buffer B was loaded on the biosensor for 3 min to form a presynaptic filament. The protein dissociation (stability of the filament) was measured by further incubation of biosensor in 200 μL of buffer C (25 mM Tris-HCl pH 7.5, 200 mM KCl, 0.05% Tween-20, 1 mM ATP, and either 1 mM MgCl2 or CaCl2).
D-loop
The D-loop reaction was conducted similarly as described in (Sebesta et al., 2011). Briefly, indicated concentration of DMC1 protein was incubated for 5 min at 37°C with 5‘-end fluorescently labeled 90-mer ssDNA (pR231, 4.5 μM nucleotides) in buffer DM containing 1 mM ATP and either 1 mM MgCl2 or CaCl2. In indicated experiments, HOP2-MND1 complex was added to the reactions after 3 min of incubation. Next, pBluescript replicative form I (60 μM base pairs) was added to the reactions and incubated for 5 min at 37°C. The reactions were stopped by addition of 0.5% SDS (final) and 0.5 mg/mL proteinase K followed by incubation for 15 min at 37°C. The deproteinized samples were loaded onto a 0.8% agarose gel and analyzed as described above.
D-loop extension
The D-loop extension assay was performed essentially as described previously (Sebesta et al., 2011). Briefly, radioactively labeled 90-mer ssDNA (pR231) was incubated for 3 min at 37°C with 2 μM DMC1 or RAD51 in 10 μL of buffer B (30 mM Tris-HCl pH 7.5, 1 mM ATP, 1 mM MgCl2 or CaCl2, 50 mM KCl, 1 mM DTT, and an ATP-regenerating system consisting of 20 mM creatine phosphate and 20 μg/mL creatine kinase). Following addition of 1 μL of 5 μM HOP2-MND1, the mixtures were incubated for an additional 2 min at 37°C. The D-loop synthesis was initiated by addition of pBluescript replicative form I (35 μM base pairs) in 2 μL, and the reaction was incubated for 5 min at 37°C. Next, either 1 U of Klenow fragment (3’-5′ exo-, New England BioLabs) (0.2 μL) or 830 nM RPA (1 μL), 4.3 nM PCNA (1 μL), 12.5 nM RFC (1 μL), and 19 nM Pol δ (1 μL) in buffer O (20 mM Tris–HCl at pH 7.5, 5 mM DTT, 0.1 mM EDTA, 150 mM KCl, 40 μg/mL BSA, 8 mM MgCl2, 5% glycerol, 0.5 mM ATP, 75 μM each dGTP, dTTP, dATP, and dCTP) were added to the reactions (in final reaction volume 24.5 μL), and the mixtures were incubated for 5 min at 30°C. The reactions were stopped with 0.5% SDS (final) and 0.5 mg/mL proteinase K for 15 min at 37°C and loaded onto a 0.8% agarose gel. After electrophoresis, the gel was dried on grade 3 chromosome paper (Whatman), exposed to a phosphorimager screen, and analyzed as described above. We note that final concentration of MgCl2 in the reaction after addition of all components is either 3.3 mM or 3.7 mM (when MgCl2 is added to the initial filament formation step).
ATP binding
Increasing concentrations of DMC1 were mixed with 2 μM TNP-ATP (Jena Bioscience) in 50 μL of buffer DM containing unlabeled 90-mer ssDNA (pR231, 6.3 μM nucleotides) and 1 mM MgCl2 or CaCl2, respectively. Fluorescence emission intensity of TNP-ATP at 535 nm with an excitation wavelength of 403 nm was measured in 96-well plate at room temperature using Infinity F500 microplate reader (Tecan Group Ltd.)
ATPase activity
ATP hydrolysis was measured by a colorimetric phosphate detection assay (Innova Biosciences). DMC1 protein (2 μM) was incubated at 37°C with 100 μM ATP in buffer DM containing 1 mM MgCl2 and unlabeled 90-mer ssDNA (pR231, 6.3 μM nucleotides). After indicated time, released phosphate was detected according to a manufacturer’s protocol using Infinity F500 microplate reader (Tecan Group Ltd.).
Microscale thermophoresis
Binding affinity quantifications by microscale thermophoresis were performed using the Monolith NT.Label Free (Nanotemper Technologies). Measurements were performed by using DM buffer containing 1 mM ATP and CaCl2 and supplemented with 0.05% Tween-20. Samples were loaded into NT.LabelFree capillaries (NanoTemper Technologies). Measurements were performed at 25°C, 50% LED, medium IR laser power and constant concentration of 2 μM DMC1. Data were analysed by the MO.Affinity Analysis software (NanoTemper Technologies).
Molecular modelling
DMC1 structure is present in the Protein Data Bank (PDB) with entry 7C9C, it is homo trimer with two ATPase active sites formed by the interfaces between chain A and Chain B for the first site and Chain B and Chain C for the second site. In the PDB structure, ATP is replaced by an analogous compound ANP which differs from ATP by the replacement of the Oxygen in the phosphodiester bond between β and γ phosphates of ATP with a Nitrogen. In order to study physiological conditions, the PDB file was edited manually to replace the Nitrogen with an Oxygen. Three structures were prepared, one corresponding to the wild type (Wt), one corresponding to E162K on chain A and one corresponding to D317A on chain B. To reduce the size of the system, chain C and the nucleic acid chain were discarded. The mutations were performed by editing the PDB files with PyMol using the mutagenesis tool. To study the effect of the mutations, three short Molecular Dynamics simulations were performed using GROMACS 2020.1, the forcefield used was CHARMM36 as it was then possible to parametrize the protein, the ATP and the Calcium ion. Simulations were performed in explicit water with SPCE water model. The preparation consisted in creating a cubic periodic boundary condition, filled with SPCE waters, the system was neutralized by adding Na+ and Cl- ions. The next step was to perform a 50,000 steps steepest descent energy minimisation with a step size of 0.01. The minimisation was followed successively by NVT and NPT equilibrations for 50,000 steps each with a dt of 2 femtoseconds (NVT 100 picoseconds then NPT 100 picoseconds). Finally, a 1 nanosecond (500,000 steps at 2 femtoseconds dt) Molecular Dynamics simulation was performed on each of the three prepared structures.
Circular dichroism
All CD experiments were performed with DMC1 protein (5 μM) diluted in DM buffer containing 0.3 mM ATP and indicated concentrations of MgCl2 or CaCl2, respectively. CD spectra were measured over a range of wavelengths (185–260 nm) at room temperature on Chirascan CD spectrometer (Applied Photophysics). The raw CD spectra were evaluated using the OriginPro7 and Microsoft Office Excel softwares.
Cryo-EM grid preparation and data collection
DMC1 protein was diluted in buffer EM (50 mM Tris-HCl pH 7.5, 50 mM KCl, 1 mM ATP) containing 1 mM MgCl2 or 1 mM CaCl2 to the final concentration of 200 ng/μL or 260 ng/μL, respectively. Each sample (4 μL) was applied to R2/1 200 mesh QuantiFoil grid that had been previously glow-discharged for 30 s. The grids were blotted for 3 s at 100% humidity and 4°C and plunged into liquid ethane cooled by liquid nitrogen using FEI Vitrobot MK IV. All datasets were collected on Titan Krios TEM operated at 300 kV and equipped with Falcon II direct electron detector. The micrographs for each sample were collected with a calibrated pixel size of 1.061 or 1.07 Å per pixel and 40 frames were collected within 1 s exposure giving a total dose of 60 e−/Å2. Micrographs were recorded with a defocus −1.2 μm to −3.3 μm and 3400 to 5827 micrographs were collected for each sample.
Cryo-EM data processing
Frames were corrected using MotionCor2 software to minimize the effect of beam-induced motion, and CTF parameters were estimated using Gctf software (Zhang, 2016; Zheng et al., 2017). Particle picking was done using Gautomatch_v0.53 software. Micrographs and particle coordinates were imported into Relion-3.0 for sub sequential particle extraction (box size 256 × 256 pixels) and 2D classification (Zivanov et al., 2018). In total 1 083 464 particles were extracted for DMC1 in the presence of 1 mM MgCl2, and 803 305 particles for DMC1 in the presence of 1 mM CaCl2. Each dataset went through five rounds of reference-free 2D classification and class averages showing distinguishable features were selected for next round of 2D classification. Final 2D classification with 25 iteration and tau fudge 2 provided reduced particle number and multiple 2D classes with noticeable differences for each sample.
Analysis of meiotic progression
Synchronous meiotic time course was performed as previously described (Oh et al., 2009). Cell images after DAPI staining to determine the meiotic progression of analyzed yeast strains were acquired using DeltaVision Elite microscope (Applied Precision, Inc.) equipped with a 100x objective lens. Images were analyzed by softWoRx software (Applied Precision, Inc.).
Western blot analysis
Synchronous meiotic time course was performed as previously described (Oh et al., 2009). Whole-cell extracts were prepared from samples (2 mL) taken from meiotic cultures at the indicated times. Pellets were lysed in 1 mL of fresh TCA lysis buffer (300 mM NaOH, 7.4% β-mercaptoethanol, and 2 mM PMSF) followed by precipitation of proteins by adding 160 μL of 50% trichloroacetic acid. Samples were spun down, the supernatants removed, and the pellets were washed with 1 mL of cold acetone. Pellets were then resuspended in 100 μL of SDS Laemmli buffer and samples were analyzed by SDS-PAGE. After semi-dry western blotting, proteins were incubated with primary antibodies anti-Dmc1 (1:2000, a kind gift from Akira Shinohara) and anti-Pgk1 (1:4000, Novex, clone 22C5D8). Following the incubation with the secondary HRP-conjugated anti-rabbit antibodies (1:7500, Sigma-Aldrich, A6154) or HRP-conjugated anti-mouse antibodies (1:50000, Sigma-Aldrich A0168), respectively, the proteins were visualized by chemiluminescence detection (Merck Millipore).
Meiotic time courses
Meiotic time courses (used for FACS analysis, fluorescence microscopy of nuclear division, and for physical analysis of recombination) were performed as previously described (Matos et al., 2008; Petronczki et al., 2006). In brief, cells were grown on YP-glycerol plates (2% peptone (m/V), 1% yeast extract (m/V), 2% glycerol (V/V), 2% agar (m/V)) for two days at 30°C. Colonies were selected and transferred to YPD plates (24 h, 30°C), followed by a transfer to another YPD plate to form a lawn (24 h, 30°C). Cells were further expanded on YPD plates (24 h, 30°C) and then used to inoculate pre-sporulation medium (2% peptone, 1% yeast extract, 2% KAc) to OD600 ∼ 0.3 using a 10 L fermenter system as described (Grigaitis et al., 2018, 2020). Cells were grown for 15 h at 25°C, washed with sporulation medium (SPM – 2% KAc) and inoculated into SPM to OD600 ∼ 3.5–4.0. This time is defined as t = 0 in meiotic time course experiments.
FACS analysis of DNA content
1 mL of the meiotic culture was collected and fixed in 70% ethanol, and stored at 4°C. The cells were washed and resuspended in 50 mM Tris-HCl (pH 7.5 at 25°C). The cells were treated with 400 μg/mL RNase A for 2–4 h at 37°C, followed by washing and resuspension in FACS buffer (200 mM Tris-HCl (pH 7.5 at 25°C), 211 mM NaCl, 78 mM MgCl2) containing 50 μg/mL propidium iodide. The samples were sonicated and diluted 10–20 times in 1 mL 50 mM Tris-HCl (pH 7.5 at 25°C). DNA content was measured using a FACSCalibur cytometer (Becton Dickinson) running CellQuest software.
Fluorescence microscopy of anaphase I and metaphase II
Yeast cells were processed for immunostaining as described (Matos et al., 2011). In brief, cells were fixed in 3.7% formaldehyde overnight (stored at 4°C) and treated with 100T Zymolyase. Spheroplasts were put on slides coated with poly-(L)-lysine and stained with rat anti-α-tubulin (1:300). Secondary antibody conjugated to Alexa488 was used for detection (1:300). DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI). Images were acquired with Zeiss Axio Imager M2 multiplexed with 40x 1,3NA DIC Oil EC Plan-NEOFLUAR objective and CoolSnap HQ2 camera (Visitron systems) under the control of Visiview 2.1.4. (Visitron systems) software. Images were processed using Fiji.
Immunodetection of Zip1 on chromosome surface spreads was performed as described in (Loidl et al., 1998), using rabbit anti-Zip1 (1:500, (Grigaitis et al., 2020)). Secondary antibody conjugated to Alexa Fluor 488 was used for detection (1:500, Invitrogen). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Spreads were observed on a DeltaVision Ultra epifluorescence microscope (GE Healthcare) with a 100x 1.4NA Oil UPlanSApo objective. Images were taken with a sCMOS camera controlled by AcquireUltra software (version 1.2.3) and processed with Fiji.
Live cell imaging
Zip1-GFP700 (Scherthan et al., 2007) and Cnm67-tdTomato (Matos et al., 2008) were imaged in live cells using an environmental chamber heated to 30°C and 8-well Lab-Tek II chambered coverglasses (Nunc, 155409) coated with Concanavalin A (Sigma, 2 mg/mL in dissolving solution). All live imaging experiments used conditioned sporulation media (filter-sterilized SPM from respective cultures). Approximately 3–4 h after transfer to SPM, cells were diluted to OD600 = 1.8 with conditioned SPM and 0.1 mL aliquots were loaded into concanavalin A-coated Lab-Tek chamber wells. Images were taken on a DeltaVision Ultra epifluorescence microscope (GE Healthcare) with a 60x 1.42NA Oil UPlanXApo objective and using a sCMOS camera controlled by AcquireUltra software (version 1.2.3). Z-stack images (8 sections, 1 μm apart) were acquired every 10 min for 15 h using the following exposure times and neutral density filter settings: 200 ms and 2% transmission for GFP (Excitation 475/28), 200 (experiment 1) – 300 (experiment 2) ms and 5% transmission for tdTomato (Excitation 542/27). Images were maximum intensity z-projected over the range of acquisition in Fiji. For quantifications, 30 cells of each strain were individually followed through meiosis. Time of appearance and disappearance of structured Zip1-GFP signal, as well as spindle pole body (SPB) separation, were tracked for each time point and percentages of cellular events were calculated. First appearance of structured Zip1-GFP signal was chosen as a reference and set to t = 0 in each cell, and meiotic events of all cells quantified were aligned accordingly. The time of SPB separation in meiosis I and II is defined as the first frame when two or four, respectively, distinct foci were detected.
Physical analysis of recombination at the HIS4-LEU2 locus
Southern blot analyses were performed as described previously (Arter et al., 2018; Kim et al., 2010). Briefly, 50–100 mL of cells from the cultures was treated with psoralen. DNA was crosslinked using an UVP Crosslinker CL-3000L (Analytik Jena). The energy used was 3600 mJ/cm2, with cells being kept on ice and mixed at regular intervals. Genomic DNA was extracted and the concentrations were measured using Qubit dsDNA broad range kit. DNA (∼1.5 μg) was digested with XhoI and separated by electrophoresis on 0.6% agarose gels, followed by a transfer to a Zeta probe GT membrane (Bio-Rad) by alkaline transfer. The membranes were hybridized with a radioactively labeled probe, exposed to a phosphor screen and imaged with a Typhoon scanner. Signal intensities of different recombination intermediates were quantified relative to the total lane signal using ImageQuant software. Signal at 0 h in SPM was considered background and manually removed from all measurements.
Quantification and statistical analysis
Data are provided as means ± standard deviations (SD) from the number of independent experiments performed. Quantification and statistical analysis of in vivo experiments are described in each figure legend.
Acknowledgments
We would like to thank Marek Sebesta and Martin Pacesa for purified proteins HOP2-MND1, RFC, polymerase δ, and PCNA, Jan Zalesak for initial cryo-EM study, and Alexandra Sisakova for technical support. We are also indebted to Nancy Hollingsworth for YEpFAT4-RAD51, YEpFAT4-RAD54-T132A, and pNH301 plasmids, and Neil Hunter for NHY1210 and NHY1215 strains. CIISB, Instruct-CZ Centre of Instruct-ERIC EU consortium, funded by MEYS CR infrastructure project LM2018127, is acknowledged for the financial support of the measurements at the CF Cryo-electron Microscopy and Tomography and Biophysical Techniques. Computational resources were supplied by the project “e-Infrastruktura CZ” (e-INFRA CZ LM2018140) supported by the Ministry of Education, Youth and Sports of the Czech Republic. We are also thankful to Biomolecular Interaction core facility of CEITEC and Loschmidt Laboratories for access to Prometheus NT.48 instrument and Chirascan CD spectrometer, respectively. We thank the BioOptics facility at MPL Vienna for their valuable support for the live-cell imaging experiment. We thank Michael Lichten for helpful comments on the manuscript. This work was supported by the Czech Science Foundation (GACR 21-22593X); the European Structural and Investment Funds, Operational Programme Research, Development and Education “Preclinical Progression of New Organic Compounds with Targeted Biological Activity” (Preclinprogress, CZ.02.1.01/0.0/0.0/16_025/0007381); Wellcome Trust Collaborative Grant (206292/E/17/Z); Masaryk University (MUNI/G/1594/2019); and the European Union's Horizon 2020 research and innovation programme under grant agreement No 812829. The Matos lab is supported by the Swiss National Science Foundation (155823 and 176108), European Research Council (101002629) and the University of Vienna.
Author contributions
V.A., M.S., A.J., and T.C. performed the biochemical and biophysical experiments. J.M. and A.J. performed the EM analysis. V.A., L.K., and J.M. designed the yeast experiments. V.A. and M.S. performed the yeast genetics experiments. L.O. performed the physical analyses of recombination. L.O. and A.H performed and analyzed the live cell imaging experiments. A.H performed and analyzed the chromosome spreads. R.G. performed the molecular modelling. V.A. and L.K. wrote the paper with inputs and edits from all authors.
Declaration of interests
The authors declare no competing interest.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105439.
Contributor Information
Veronika Altmannova, Email: valtmann@med.muni.cz.
Lumir Krejci, Email: lkrejci@chemi.muni.cz.
Supplemental information
Data and code availability
This study did not generate any unique datasets or code.
References
- Arter M., Hurtado-Nieves V., Oke A., Zhuge T., Wettstein R., Fung J.C., Blanco M.G., Matos J. Regulated crossing-over requires inactivation of yen1/GEN1 resolvase during meiotic prophase I. Dev. Cell. 2018;45:785–800. doi: 10.1016/j.devcel.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop D.K., Nikolski Y., Oshiro J., Chon J., Shinohara M., Chen X. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Gene Cell. 1999;4:425–444. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- Bishop D.K., Park D., Xu L., Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Brown M.S., Bishop D.K. DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb. Perspect. Biol. 2014;7:a016659. doi: 10.1101/cshperspect.a016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V., Mazin A.V. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. USA. 2004;101:9988–9993. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev D.V., Golub E.I., Stasiak A.Z., Stasiak A., Mazin A.V. Activation of human meiosis-specific recombinase Dmc1 by Ca2+ J. Biol. Chem. 2005;280:26886–26895. doi: 10.1074/jbc.M502248200. [DOI] [PubMed] [Google Scholar]
- Busygina V., Gaines W.A., Xu Y., Kwon Y., Williams G.J., Lin S.W., Chang H.Y., Chi P., Wang H.W., Sung P. Functional attributes of the Saccharomyces cerevisiae meiotic recombinase Dmc1. DNA Repair. 2013;12:707–712. doi: 10.1016/j.dnarep.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J., Swann K., Whittingham D., Whitaker M. Spatiotemporal dynamics of intracellular [Ca2+]i oscillations during the growth and meiotic maturation of mouse oocytes. Development. 1994;120:3507–3517. doi: 10.1242/dev.120.12.3507. [DOI] [PubMed] [Google Scholar]
- Chan Y.L., Brown M.S., Qin D., Handa N., Bishop D.K. The third exon of the budding yeast meiotic recombination gene HOP2 is required for calcium-dependent and recombinase Dmc1-specific stimulation of homologous strand assimilation. J. Biol. Chem. 2014;289:18076–18086. doi: 10.1074/jbc.M114.558601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.Y., Liao C.Y., Su G.C., Lin S.W., Wang H.W., Chi P. Functional relationship of ATP hydrolysis, presynaptic filament stability, and homologous DNA pairing activity of the human meiotic recombinase DMC1. J. Biol. Chem. 2015;290:19863–19873. doi: 10.1074/jbc.M115.666289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloud V., Chan Y.L., Grubb J., Budke B., Bishop D.K. Rad51 is an accessory factor for Dmc1-mediated joint molecule formation during meiosis. Science. 2012;337:1222–1225. doi: 10.1126/science.1219379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein J., Antony E., Hingorani M.M., O'Donnell M. Overproduction and analysis of eukaryotic multiprotein complexes in Escherichia coli using a dual-vector strategy. Anal. Biochem. 2003;319:78–87. doi: 10.1016/s0003-2697(03)00273-2. [DOI] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061. [DOI] [PubMed] [Google Scholar]
- Grigaitis R., Ranjha L., Wild P., Kasaciunaite K., Ceppi I., Kissling V., Henggeler A., Susperregui A., Peter M., Seidel R., et al. Phosphorylation of the RecQ helicase Sgs1/BLM controls its DNA unwinding activity during meiosis and mitosis. Dev. Cell. 2020;53:706–723.e5. doi: 10.1016/j.devcel.2020.05.016. [DOI] [PubMed] [Google Scholar]
- Grigaitis R., Susperregui A., Wild P., Matos J. Characterization of DNA helicases and nucleases from meiotic extracts of S. cerevisiae. Methods Cell Biol. 2018;144:371–388. doi: 10.1016/bs.mcb.2018.03.029. [DOI] [PubMed] [Google Scholar]
- Grushcow J.M., Holzen T.M., Park K.J., Weinert T., Lichten M., Bishop D.K. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigi Harchegani A., Irandoost A., Mirnamniha M., Rahmani H., Tahmasbpour E., Shahriary A. Possible mechanisms for the effects of calcium deficiency on male infertility. Int. J. Fertil. Steril. 2019;12:267–272. doi: 10.22074/ijfs.2019.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 2015;7:a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N., Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Kagawa W., Kurumizaka H. From meiosis to postmeiotic events: uncovering the molecular roles of the meiosis-specific recombinase Dmc1. FEBS J. 2010;277:590–598. doi: 10.1111/j.1742-4658.2009.07503.x. [DOI] [PubMed] [Google Scholar]
- Kaneuchi T., Sartain C.V., Takeo S., Horner V.L., Buehner N.A., Aigaki T., Wolfner M.F. Calcium waves occur as Drosophila oocytes activate. Proc. Natl. Acad. Sci. USA. 2015;112:791–796. doi: 10.1073/pnas.1420589112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant C.R., Rao B.J., Sainis J.K. DNA binding and pairing activity of OsDmc1, a recombinase from rice. Plant Mol. Biol. 2005;57:1–11. doi: 10.1007/s11103-004-5828-x. [DOI] [PubMed] [Google Scholar]
- Kelso A.A., Say A.F., Sharma D., Ledford L.L., Turchick A., Saski C.A., King A.V., Attaway C.C., Temesvari L.A., Sehorn M.G. Entamoeba histolytica Dmc1 catalyzes homologous DNA pairing and strand exchange that is stimulated by calcium and hop2-mnd1. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.P., Weiner B.M., Zhang L., Jordan A., Dekker J., Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao J.P., Cloud V., Huang C.C., Grubb J., Thacker D., Lee C.Y., Dresser M.E., Hunter N., Bishop D.K. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., Chang Y.C., Hong E.L., Grubb J., Chang C.S., Bishop D.K., Wang T.F. Calcium ion promotes yeast Dmc1 activity via formation of long and fine helical filaments with single-stranded DNA. J. Biol. Chem. 2005;280:40980–40984. doi: 10.1074/jbc.M505896200. [DOI] [PubMed] [Google Scholar]
- Lindgren A., Bungard D., Pierce M., Xie J., Vershon A., Winter E. The pachytene checkpoint in Saccharomyces cerevisiae requires the Sum1 transcriptional repressor. EMBO J. 2000;19:6489–6497. doi: 10.1093/emboj/19.23.6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gaines W.A., Callender T., Busygina V., Oke A., Sung P., Fung J.C., Hollingsworth N.M. Down-regulation of Rad51 activity during meiosis in yeast prevents competition with Dmc1 for repair of double-strand breaks. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J., Klein F., Engebrecht J. Genetic and morphological approaches for the analysis of meiotic chromosomes in yeast. Methods Cell Biol. 1998;53:257–285. doi: 10.1016/s0091-679x(08)60882-1. [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., 3rd, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061. [DOI] [PubMed] [Google Scholar]
- Luo S.C., Yeh H.Y., Lan W.H., Wu Y.M., Yang C.H., Chang H.Y., Su G.C., Lee C.Y., Wu W.J., Li H.W., et al. Identification of fidelity-governing factors in human recombinases DMC1 and RAD51 from cryo-EM structures. Nat. Commun. 2021;12:115. doi: 10.1038/s41467-020-20258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D., Nikolsky Y., Bishop D.K., Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Matos J., Blanco M.G., Maslen S., Skehel J.M., West S.C. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., Lipp J.J., Bogdanova A., Guillot S., Okaz E., Junqueira M., Shevchenko A., Zachariae W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135:662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Matulova P., Marini V., Burgess R.C., Sisakova A., Kwon Y., Rothstein R., Sung P., Krejci L. Cooperativity of Mus81.Mms4 with Rad54 in the resolution of recombination and replication intermediates. J. Biol. Chem. 2009;284:7733–7745. doi: 10.1074/jbc.M806192200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- McDougall A., Sardet C. Function and characteristics of repetitive calcium waves associated with meiosis. Curr. Biol. 1995;5:318–328. doi: 10.1016/s0960-9822(95)00062-5. [DOI] [PubMed] [Google Scholar]
- Nimonkar A.V., Dombrowski C.C., Siino J.S., Stasiak A.Z., Stasiak A., Kowalczykowski S.C. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J. Biol. Chem. 2012;287:28727–28737. doi: 10.1074/jbc.M112.373290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Wan L., Busygina V., Kwon Y., Allen J.A., Li X., Kunz R.C., Kubota K., Wang B., Sung P., et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell. 2009;36:393–404. doi: 10.1016/j.molcel.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.D., Jessop L., Lao J.P., Allers T., Lichten M., Hunter N. Stabilization and electrophoretic analysis of meiotic recombination intermediates in Saccharomyces cerevisiae. Methods Mol. Biol. 2009;557:209–234. doi: 10.1007/978-1-59745-527-5_14. [DOI] [PubMed] [Google Scholar]
- Oh S.D., Lao J.P., Hwang P.Y.H., Taylor A.F., Smith G.R., Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak J., Segall J. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell Biol. 2002;22:6430–6440. doi: 10.1128/mcb.22.18.6430-6440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy S.I., Yu X., Li Z., Radding C.M., Masson J.Y., West S.C., Egelman E.H. Human Dmc1 protein binds DNA as an octameric ring. Proc. Natl. Acad. Sci. USA. 1999;96:10684–10688. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Matos J., Mori S., Gregan J., Bogdanova A., Schwickart M., Mechtler K., Shirahige K., Zachariae W., Nasmyth K. Monopolar attachment of sister kinetochores at meiosis I requires casein kinase 1. Cell. 2006;126:1049–1064. doi: 10.1016/j.cell.2006.07.029. [DOI] [PubMed] [Google Scholar]
- Petukhova G.V., Pezza R.J., Vanevski F., Ploquin M., Masson J.Y., Camerini-Otero R.D. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Pezza R.J., Petukhova G.V., Ghirlando R., Camerini-Otero R.D. Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J. Biol. Chem. 2006;281:18426–18434. doi: 10.1074/jbc.M601073200. [DOI] [PubMed] [Google Scholar]
- Pezza R.J., Voloshin O.N., Vanevski F., Camerini-Otero R.D. Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 2007;21:1758–1766. doi: 10.1101/gad.1562907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman D.L., Cobb J., Schimenti K.J., Wilson L.A., Cooper D.M., Brignull E., Handel M.A., Schimenti J.C. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol. Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- Qian X., He Y., Ma X., Fodje M.N., Grochulski P., Luo Y. Calcium stiffens archaeal Rad51 recombinase from Methanococcus voltae for homologous recombination. J. Biol. Chem. 2006;281:39380–39387. doi: 10.1074/jbc.M607785200. [DOI] [PubMed] [Google Scholar]
- Qian X., He Y., Wu Y., Luo Y. Asp302 determines potassium dependence of a RadA recombinase from Methanococcus voltae. J. Mol. Biol. 2006;360:537–547. doi: 10.1016/j.jmb.2006.05.058. [DOI] [PubMed] [Google Scholar]
- Rajanikant C., Kumbhakar M., Pal H., Rao B.J., Sainis J.K. DNA strand exchange activity of rice recombinase OsDmc1 monitored by fluorescence resonance energy transfer and the role of ATP hydrolysis. FEBS J. 2006;273:1497–1506. doi: 10.1111/j.1742-4658.2006.05170.x. [DOI] [PubMed] [Google Scholar]
- Reitz D., Grubb J., Bishop D.K. A mutant form of Dmc1 that bypasses the requirement for accessory protein Mei5-Sae3 reveals independent activities of Mei5-Sae3 and Rad51 in Dmc1 filament stability. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane I., Kamataki C., Takizawa Y., Nakashima M., Toki S., Ichikawa H., Ikawa S., Shibata T., Kurumizaka H. Filament formation and robust strand exchange activities of the rice DMC1A and DMC1B proteins. Nucleic Acids Res. 2008;36:4266–4276. doi: 10.1093/nar/gkn405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H., Tawaramoto M.S., Lao J.P., Hosaka H., Sanda E., Suzuki M., Yamashita E., Hunter N., Shinohara M., Nakagawa A., Shinohara A. A new protein complex promoting the assembly of Rad51 filaments. Nat. Commun. 2013;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Wang H., Adelfalk C., White E.J., Cowan C., Cande W.Z., Kaback D.B. Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2007;104:16934–16939. doi: 10.1073/pnas.0704860104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Sebesta M., Burkovics P., Haracska L., Krejci L. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair. 2011;10:567–576. doi: 10.1016/j.dnarep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehorn M.G., Sigurdsson S., Bussen W., Unger V.M., Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429:433–437. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- Špírek M., Mlcoušková J., Belán O., Gyimesi M., Harami G.M., Molnár E., Novacek J., Kovács M., Krejci L. Human RAD51 rapidly forms intrinsically dynamic nucleoprotein filaments modulated by nucleotide binding state. Nucleic Acids Res. 2018;46:3967–3980. doi: 10.1093/nar/gky111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strynadka N.C., James M.N. Towards an understanding of the effects of calcium on protein structure and function. Curr. Opin. Struct. Biol. 1991;1:905–914. doi: 10.1016/0959-440x(91)90085-8. [DOI] [Google Scholar]
- Suizu T., Tsutsumi H., Kawado A., Suginami K., Imayasu S., Murata K. Calcium ion influx during sporulation in the yeast Saccharomyces cerevisiae. Can. J. Microbiol. 1995;41:1035–1037. doi: 10.1139/m95-143. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H., Roeder G.S. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev. Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Whitaker M., Patel R. Calcium and cell cycle control. Development. 1990;108:525–542. doi: 10.1242/dev.108.4.525. [DOI] [PubMed] [Google Scholar]
- Wu Y., Qian X., He Y., Moya I.A., Luo Y. Crystal structure of an ATPase-active form of Rad51 homolog from Methanococcus voltae. Insights into potassium dependence. J. Biol. Chem. 2005;280:722–728. doi: 10.1074/jbc.M411093200. [DOI] [PubMed] [Google Scholar]
- Xu L., Weiner B.M., Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- Xue C., Molnarova L., Steinfeld J.B., Zhao W., Ma C., Spirek M., Kaniecki K., Kwon Y., Belan O., Krejci K., et al. Single-molecule visualization of human RECQ5 interactions with single-stranded DNA recombination intermediates. Nucleic Acids Research. 2021;49:285–305. doi: 10.1093/nar/gkaa1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K., Kondoh G., Matsuda Y., Habu T., Nishimune Y., Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol. Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Sung P. Significance of ligand interactions involving Hop2-Mnd1 and the RAD51 and DMC1 recombinases in homologous DNA repair and XX ovarian dysgenesis. Nucleic Acids Res. 2015;43:4055–4066. doi: 10.1093/nar/gkv259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.