Abstract.
Purpose
We developed a model integrating multimodal quantitative imaging features from tumor and nontumor regions, qualitative features, and clinical data to improve the risk stratification of patients with resectable non-small cell lung cancer (NSCLC).
Approach
We retrospectively analyzed 135 patients [mean age, 69 years (43 to 87, range); 100 male patients and 35 female patients] with NSCLC who underwent upfront surgical resection between 2008 and 2012. The tumor and peritumoral regions on both preoperative CT and FDG PET-CT and the vertebral bodies L3 to L5 on FDG PET were segmented to assess the tumor and bone marrow uptake, respectively. Radiomic features were extracted and combined with clinical and CT qualitative features. A random survival forest model was developed using the top-performing features to predict the time to recurrence/progression in the training cohort (), validated in the testing cohort () using the concordance, and compared with a stage-only model. Patients were stratified into high- and low-risks of recurrence/progression using Kaplan–Meier analysis.
Results
The model, consisting of stage, three wavelet texture features, and three wavelet first-order features, achieved a concordance of 0.78 and 0.76 in the training and testing cohorts, respectively, significantly outperforming the baseline stage-only model results of 0.67 () and 0.60 (), respectively. Patients at high- and low-risks of recurrence/progression were significantly stratified in both the training () and the testing () cohorts.
Conclusions
Our radiomic model, consisting of stage and tumor, peritumoral, and bone marrow features from CT and FDG PET-CT significantly stratified patients into low- and high-risk of recurrence/progression.
Keywords: radiomics, machine learning, lung cancer, computed tomography, positron emission tomography
1. Introduction
Imaging modalities such as CT and -fluoro-2-deoxy-D-glucose (FDG) PET-CT play an important role in the diagnosis and staging of lung cancer. Traditional tumor, node, and metastasis (TNM) staging is currently the gold standard for defining the extent of disease and for predicting prognosis. However, even when lung cancers are diagnosed early, studies have shown that 20% to 50% of patients will develop either a local or distant recurrence 5 years posttreatment.1–3 Therefore, there remains an unmet need to discover biomarkers to identify patients at a high risk of recurrence that may benefit from more aggressive or personalized treatment.
Radiomics uses medical images to obtain quantitative imaging features for applications in diagnosis, treatment selection, and response assessment in cancer patients.4–6 These radiomic features allow for a noninvasive and comprehensive characterization of tumors. Studies using radiomics for outcome prediction in non-small cell lung cancer (NSCLC) have mainly focused on single modality models, with studies demonstrating the utility of both quantitative CT and FDG PET imaging for predicting recurrence risk, disease-free survival, and overall survival in NSCLC.7–10 Previous work from our group detailed the prognostic power of tumor and bone marrow features from FDG PET in predicting recurrence in lung cancer patients. However, this study only focused on features from FDG PET images and did not include CT features.11 With promising results established using single modalities, more recent studies have aimed to combine information from different modalities and areas outside the tumor in an attempt to improve overall model performance. Peritumoral regions on CT and FDG PET images have been shown to have significant prognostic power when it comes to predicting outcomes.11,12 Additionally, qualitative features that describe the location, geometry, and appearance of the tumor and lung tissue have also been used to predict outcomes in lung cancer and have been shown to be complementary to radiomic features.9
A study incorporating both tumor and nontumor features from CT and FDG PET, along with clinical and qualitative features, has not yet been explored. Our study builds on prior work analyzing the utility of FDG PET tumor, peritumoral, and bone marrow radiomics features for recurrence prediction. We hypothesize that adding CT radiomic and qualitative features to FDG PET and clinical features could stratify high-risk NSCLC patients beyond traditional TNM staging alone.
2. Materials and Methods
2.1. Patient Selection
The dataset used in this study was de-identified and publicly available on The Cancer Imaging Archive (TCIA).13–15 Local institutional review board approval (IRB) was obtained to conduct this study.
The study included 136 NSCLC patients from the retrospective NSCLC-radiogenomics cohort obtained from TCIA previously analyzed by Mattonen et al.11 Each patient was referred for upfront surgical resection between 2008 and 2012. Patients had preoperative diagnostic CT and FDG PET-CT scans acquired. Patients were excluded from the study if their diagnostic CT scan was unavailable (), which resulted in 135 patients being used in the analysis. A total of six clinical features were collected and included in the dataset: age, sex, ethnicity, smoking history, pathological staging [American Joint Committee on Cancer (AJCC) seventh edition], and tumor histology (adenocarcinoma, squamous cell carcinoma, or non-small cell cancer not otherwise specified). Recurrence or progression was used as our outcome of interest. Recurrence was defined as local, regional, or distant for curable lung cancer (stages I to IIIA). Progression was the outcome of interest for patients with incurable lung cancer (stage IIIB or IV). Additionally, patients received adjuvant treatment based on the standard clinical guidelines. Time to recurrence/progression was defined as the time between the preoperative diagnostic CT scan and date of the event or last known follow-up. We used a random number generator to randomly split the dataset into training (75%, ) and testing (25%, ) cohorts. A chi-square () test was used to examine differences in the categorical clinical features (i.e., sex, stage, ethnicity, smoking status, histology) between the training and the testing cohorts, and an independent two-sample -test was used to assess differences for continuous variables with a normal distribution (i.e., age).
2.2. Imaging
Preoperative diagnostic CT scans were acquired at one of three medical centers as previously described.14,15 Images were primarily acquired using one of three GE scanner types: GE Discovery CT750HD, GE LightSpeed16, or GE LightSpeedVCT (GE Healthcare, Waukesha, Wisconsin). Preoperative FDG PET-CT scans were acquired using one of four scanners: Discovery LS PET/CT, Discovery VCT (GE Healthcare, Waukesha, Wisconsin); Biograph mCT (Siemens Healthcare, Erlangen, Germany); or Allegro/Gemini TF PET/CT (Phillips Healthcare, Cleveland, Ohio). CT and FDG PET images were generated using similar protocols at one of three Stanford medical centers. The median time between CT and FDG PET-CT image acquisition for the patients in the dataset was 18 days. Complete details on the imaging protocols can be found in the Appendix.
2.3. Region of Interest Delineation
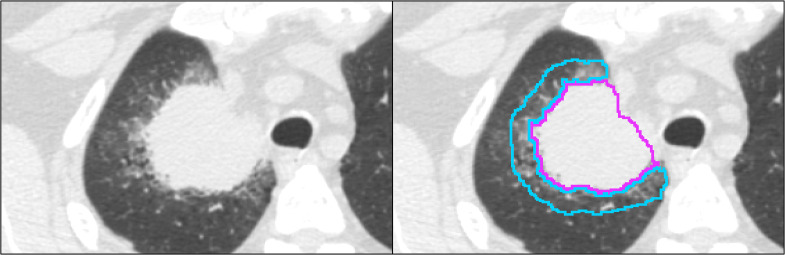
The segmentation was completed for the preoperative CT scans using our threshold-based semiautomated segmentation algorithm developed in MATLAB R2021B (The MathWorks, Natick, Massachusetts), which is publicly available on GitHub.16,17 The segmentation algorithm utilizes Otsu’s thresholding and postprocessing to remove disconnected components, connected structures (e.g., chest wall or mediastinum), and surrounding vessels (Appendix). A 1 cm three-dimensional (3D) peritumoral region extending outward from the tumor segmentation but within the lung was also defined (Fig. 1). All segmentations were performed by a PhD student (J.R.C.) with 2 years of image analysis experience after training, and visual verification was performed by a resident in radiology (O.D.) with 5 years of experience.
Fig. 1.
(a) One 1.25 mm slice of a preoperative diagnostic CT image of a patient with NSCLC. (b) The tumor segmentation is shown in pink, and the peritumoral segmentation is shown in cyan.
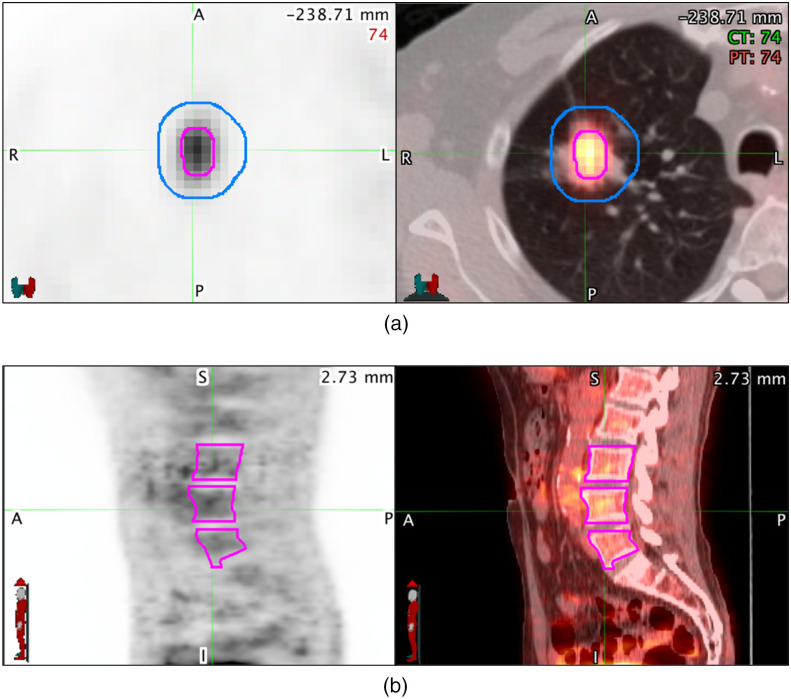
The tumor volume and bone marrow segmentations on the preoperative FDG PET-CT images were completed as part of a previous study.11 Vertebral bodies (L3 to L5) were segmented using a manual threshold tool on the CT portion of the fused FDG PET-CT images to assess bone marrow uptake on the FDG PET. Metabolic tumor volume (MTV) segmentations were completed using a semiautomatic gradient-based method (PET-edge) that is part of commercially available software (MIM Software; version 6.6, Cleveland, Ohio). In addition to the MTV, a 1 cm 3D peritumoral region extending outward from the surface of the MTV was generated to sample the surrounding uptake (Fig. 2). In total, the regions of interest (ROIs) used in the study were the tumor and peritumoral volumes from the diagnostic CT and the FDG PET-CT, along with the MTV plus peritumoral region, and bone marrow uptake volumes from the FDG PET-CT.
Fig. 2.
(a) Transaxial and (b) sagittal preoperative diagnostic PET (left) and PET/CT fusion (right) images of a patient with NSCLC. (a) The metabolic tumor volume (MTV) is shown in pink, and the peritumoral segmentation is shown in blue. (b) The vertebral bodies L3 to L5 are shown segmented in pink to sample bone marrow uptake. These regions of interest are used to extract the local, regional, and distant PET imaging features.
A randomly chosen subset () of images was used to assess the inter- and intra-observer variability of the CT segmentations. This number was determined using the power analysis calculation shown by Bujang.18 The intraobserver variability consisted of segmentations performed by a PhD student (J.R.C.) who resegmented the subset of images 6 months after the initial segmentations. Interobserver variability was assessed with segmentations performed by S.A.M, an imaging scientist with 10 years of experience. The segmentation variability was assessed using reproducibility metrics used to assess the spatial overlap and boundary similarities between segmentations, including the Dice similarity coefficient, mean absolute boundary deviation, and absolute volume difference between the contours.19 The FDG PET-CT segmentation variability was previously assessed.11,20
2.4. Feature Extraction
An open-source package, Pyradiomics (version 3.0.1), was used to extract 5050 features from the ROIs on both the CT and FDG PET images.21 Radiomic features were calculated on the tumor and peritumoral regions on CT and the tumor, peritumoral, tumor plus peritumoral, and bone marrow regions on FDG PET (Table 1). Features included first-order intensity statistics (), shape-based (), gray level co-occurrence matrix (GLCM) (), gray level run length matrix (GLRLM) (), gray level size zone matrix (GLSZM) (), neighboring gray-tone difference matrix (NGTDM) (), and gray level dependence matrix (GLDM) () on both the original and wavelet filtered images. The wavelet transformation created eight images labeled as LLL, LLH, LHL, LHH, HLL, HLH, HHL, and HHH, where H and L are the high- and low-frequency filters respectively that are applied on the , , and axes of the volume. Radiomic feature robustness was assessed for the CT segmentations using the intraclass correlation coefficient (ICC) for the randomly chosen subset of images () between the three segmentations. Robust features were defined as those with . Complete feature extraction details can be found in the Appendix. Additionally, 28 qualitative features describing characteristics from the tumor and lung tissue on the CT image were included (Table 5). These semantic assessments were assigned by two academic radiologists with expertise in lung cancer imaging as previously described.15
Table 1.
Number of extracted features within each feature category.
| Regions of interest | Radiomic features () | Other () | ||||||
|---|---|---|---|---|---|---|---|---|
| Shape (Orig) | Intensity (Orig and Wav) | GLCM (Orig and Wav) | GLDM (Orig and Wav) | GLRLM (Orig and Wav) | GLSZM (Orig and Wav) | NGTDM (Orig and Wav) | ||
| CT | ||||||||
| Tumor | 14 | 162 | 216 | 126 | 144 | 144 | 45 | |
| Peritumoral | — | 162 | 216 | 126 | 144 | 144 | 45 | |
| Qualitative | — | — | — | — | — | — | — | 28 |
| PET | ||||||||
| MTV | 14 | 162 | 216 | 126 | 144 | 144 | 45 | |
| Peritumoral | — | 162 | 216 | 126 | 144 | 144 | 45 | |
| MTV with peritumoral | — | 162 | 216 | 126 | 144 | 144 | 45 | |
| Bone marrow | — | 162 | 216 | 126 | 144 | 144 | 45 | |
| Clinical | — | — | — | — | — | — | — | 6 |
Orig, Original; Wav, Wavelet; GLCM, gray level co-occurrence matrix; GLDM, gray level dependence matrix; GLRLM, gray level run length matrix; GLSZM, gray level size zone matrix; NGTDM, neighboring gray tone difference matrix
Table 5.
A total of 28 qualitative features were used in this study.15
| Category | Feature | Value |
|---|---|---|
| Nodule features | Anatomic location | Right upper lobe, right middle lobe, right lower lobe, left upper lobe, lingula, left lower lobe |
| Axial location | Central, peripheral | |
| Lung nodule | Lung mass, not specified | |
| Internal features | Internal air alveolograms/bronchograms, necrosis, cavitation | |
| Nodule margins – primary pattern | Smooth, irregular, lobulated, spiculation, poorly defined | |
| Nodule margins – secondary pattern | Smooth, irregular, lobulated, spiculation, poorly defined | |
| Nodule attenuation | Solid, pure ground glass, semiconsolidation, part-solid (≤5 mm diameter), part-solid (>5 mm diameter) | |
| Nodule shape | Round, oval, complex, polygonal | |
| Nodule calcification | No calcification, central calcification, peripheral | |
| Nodule associated findings | Attachment to pleura, attachment to vessel, attachment to bronchus, pleural retraction, entering airway, thickened adjacent bronchovascular bundle, vascular convergence, septal thickening | |
| Nodule periphery | Emphysema, fibrosis, normal, scarring | |
| Nodules in Contralateral Lung (>4 mm noncalcified) | Absent, solid, nonsolid, semiconsolidation, part-solid | |
| Nodules in nonlesion lobe same lung (>4 mm noncalcified) | Absent, solid, nonsolid, semiconsolidation, part-solid | |
| Satellite nodules in primary lesion lobe (>4 mm noncalcified) | Absent, solid, nonsolid, semiconsolidation, part-solid | |
| Centrilobular nodules – diffuse (RB type nodules) | Absent, present | |
| Lung parenchyma features | Fibrosis | Absent, present |
| Anatomic fibrosis distribution | Apicalm upper predominant, middle predominant, lower predominant, diffuse, patchy, unable to determine | |
| Axial fibrosis distribution | Subpleural, bronchovascular, both 1 and 2, random | |
| Fibrosis type | Usual interstitial pneumonia, nonspecific interstitial pneumonia, hypersensitivity pneumonitis, sarcoidosis, smoking-related, postinfectious (include oesophago-gastro-duodenoscopy), other (specify), indeterminate | |
| Lung parenchyma features | Airway abnormalities, bronchial wall thickening, airway ectasia, bronchiectasis, luminal narrowing, bronchiolar prominence, tree-in-bud, mosiac oligemia | |
| Emphysema | Absent, present | |
| Primary emphysema pattern | Centrilobular, pan-acinar, paraseptal, paracicatricial, NA | |
| Primary distribution | Upper predominant, middle predominant | |
| Lower predominant, diffuse, no predominance, patchy, no predominance | ||
| NA or unable to determine | ||
| Primary emphysema laterality | Right, left, both | |
| Secondary emphysema pattern | Centrilobular, pan-acinar, paraseptal, paracicatricial, NA | |
| Secondary emphysema distribution | Upper predominant, middle predominant | |
| Lower predominant, diffuse, no predominance, patchy, no predominance | ||
| NA or unable to determine | ||
| Secondary emphysema laterality | Right, left, both | |
| Overall emphysema severity | None, low (1% to 25%), moderate (26% to 50%), moderately high (51% to 75%), high (>75%) |
2.5. Feature Selection and Model Development
The extracted radiomics features were standardized using -score transformation of the training data prior to model building, and this transformation was applied to the testing dataset. This transformation was performed to ensure that all features were on a similar scale prior to feature selection. The extracted features from each region were combined into one feature set. After standardization, the optimal features for predicting time to recurrence/progression in the training dataset were determined from the full feature set using the least absolute shrinkage and selection operator (LASSO) with Python 3.7.4. The regularization parameter (), which controls the strength of the LASSO regression, was selected using 10-fold cross-validation on the training cohort. The that produced the highest concordance index in the training cohort was chosen. We built and trained a random survival forest (RSF) model (scikit-survival 0.15.0; RandomSurvivalForest) using these selected features in the training dataset.22 The parameters of the RSF model were as follows: number of estimators – 1000, maximum depth – 25, minimum samples per leaf – 25, minimum samples per split – 10. To summarize, feature selection and model training was performed using the training cohort, and this model was then locked. The locked model was then evaluated on the unseen testing cohort.
Statistical analysis was conducted using R, version 4.1.0.23 We used the concordance index as our evaluation metric (survcomp 1.42.0; concordance.index). Kaplan–Meier survival analysis was performed to separate high- and low-risk groups using the median risk score in the training cohort. This model was also compared with a baseline clinical model with cancer stage only to determine its incremental value in predicting time to recurrence/progression in NSCLC patients (survcomp 1.42.0; cindex.comp). Statistical significance was determined by a -value of .
3. Results
3.1. Patient Demographics
Table 2 summarizes the demographics of the 135 patients used in this study. The training cohort consisted of 101 patients (mean age ± standard deviation, years; 75% male), and the testing cohort consisted of 34 patients (mean age ± standard deviation, years; 71% male). The patients in the training and testing cohorts were well-matched for all clinical variables as shown in Table 2. The event rate in the training and testing cohorts was 28% and 29% respectively. Overall, the number of patients who experienced an event based on stage was as follows: 33% (2/6) of the stage 0 patients, 16% (13/78) of the stage I patients, 27% (7/26) of the stage II patients, 57% (12/21) of the stage III patients, and 100% (4/4) of the stage IV patients. The median time to recurrence/progression was 37 months in the training cohort and 40 months in the testing cohort. The median follow-up time for censored patients without an event was 46 months in the training cohort and 45 months in the testing cohort.
Table 2.
Patient demographics and lesion characteristics.
| Training cohort (n = 101) | Testing cohort () | Value | |
|---|---|---|---|
| Mean age [range] | 69 [43–87] | 69 [56–85] | 0.77 |
| Sex | 0.59 | ||
| Men | 76 (75%) | 24 (71%) | |
| Women | 25 (25%) | 10 (29%) | |
| Histology | 0.74 | ||
| Adenocarcinoma | 80 (79%) | 26 (76%) | |
| Squamous cell | 19 (19%) | 8 (24%) | |
| Not otherwise specified | 2 (2%) | 0 (0%) | |
| Ethnicity | 0.69 | ||
| Caucasian | 75 (74%) | 26 (76%) | |
| African American | 6 (6%) | 0 (0%) | |
| Asian | 15 (15%) | 6 (18%) | |
| Native Hawaiian/Pacific Islander | 2 (2%) | 1 (3%) | |
| Hispanic/Latino | 3 (3%) | 1 (3%) | |
| Smoking status | 0.55 | ||
| Former | 66 (65%) | 19 (56%) | |
| Current | 21 (21%) | 8 (23%) | |
| Nonsmoker | 14 (14%) | 7 (21%) | |
| Pathological stage | 0.35 | ||
| 0a | 5 (5%) | 1 (3%) | |
| I | 59 (58%) | 19 (56%) | |
| II | 16 (16%) | 10 (29%) | |
| III | 17 (17%) | 4 (12%) | |
| IV | 4 (4%) | 0 (0%) | |
| Recurrence | 0.85 | ||
| Yes | 28 (28%) | 10 (29%) | |
| No | 73 (72%) | 24 (71%) |
Pathologic stage 0 disease is defined as a carcinoma in situ as per the American Joint Committee on Cancer (AJCC) seventh edition (7th) staging system.
3.2. Segmentation and Feature Robustness
The Dice similarity coefficient, mean absolute boundary deviation, and absolute volume difference between the three semiautomatic CT segmentations in the subset of 33 patients are shown in Table 3. The CT segmentations achieved an average Dice of 0.92, boundary deviation of 0.50 mm, and volume difference of 0.54 mL. Table 4 shows the percentage of robust features from each feature type. All feature types were found to be robust with of the features in each type, demonstrating an .
Table 3.
CT semiautomatic segmentation inter- (1 versus 2) and intraobserver (1A versus 1B) variability.
| Observer | Dice similarity coefficient (DSC) | Mean absolute boundary deviation (MAD, mm) | Absolute volume difference (mL) |
|---|---|---|---|
| 1A versus 1B | 0.92 (0.09) | 0.46 (0.68) | 0.61 (2.43) |
| 1A versus 2 | 0.93 (0.11) | 0.38 (0.37) | 0.20 (1.42) |
| 1B versus 2 | 0.90 (0.14) | 0.53 (0.83) | 0.80 (3.16) |
All values represent the mean (standard deviation)
Table 4.
Number of robust radiomic features for CT semiautomatic segmentation.
| Feature type | Total number of features () | Number (%) of robust features ()a |
|---|---|---|
| Shape | 14 | 13 (93%) |
| Intensity | 162 | 155 (96%) |
| GLCM | 216 | 207 (96%) |
| GLDM | 126 | 124 (98%) |
| GLRLM | 144 | 143 (99%) |
| GLSZM | 144 | 144 (100%) |
| NGTDM | 45 | 45 (100%) |
GLCM, gray level co-occurrence matrix; GLDM, gray level dependence matrix; GLRLM, gray level run length matrix; GLSZM, gray level size zone matrix; NGTDM, neighboring gray tone difference matrix
Robust features represented with ICC > 0.8
3.3. Feature Selection
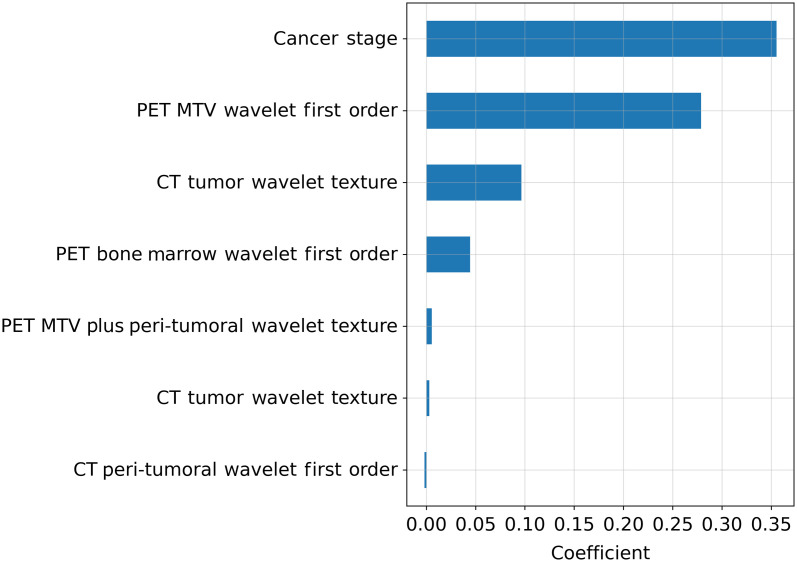
The regularization strength chosen by 10-fold cross-validation was 0.12. Using this regularization strength in the LASSO regression resulted in a total of seven features identified as the top-performing features in predicting time to recurrence/progression. Six of the top features were wavelet features with three being texture features [MTV plus peritumoral HHH GLCM maximal correlation coefficient (MCC), CT tumor LHL GLCM maximum probability, and CT tumor LHL GLDM large dependence high gray level emphasis) and three being first-order features (CT peritumoral HLH mean, MTV LHH kurtosis, and bone marrow HHL mean). These selected features were deemed to be robust in the ICC analysis. The remaining feature was the clinical feature of the cancer stage.
3.4. Radiomic Model Assessment for Recurrence Risk Stratification
The LASSO coefficients for the most predictive seven features are shown in Fig. 3. Stage was the best predictor of recurrence/progression among the selected features. Five of the remaining six features had positive coefficients, whereas CT peritumoral HLH first-order mean was the only selected feature with a negative coefficient. Qualitatively, the high-risk patients had more heterogenous textures in both the CT and FDG PET tumor and peritumoral regions as well as the bone marrow compared with the low-risk patients.
Fig. 3.
Coefficients for the selected features using LASSO in the training cohort showing the directionality of the association of each feature in descending order of feature contribution. MTV, metabolic tumor volume.
The multivariate model was a significant predictor of time-to-event in the training cohort (Concordance = 0.78 [95% CI: 0.70 to 0.86], ). The locked multivariate model was evaluated on the testing cohort and was shown to be a significant predictor in that cohort (Concordance = 0.76 [95% CI: 0.59 to 0.87], ).
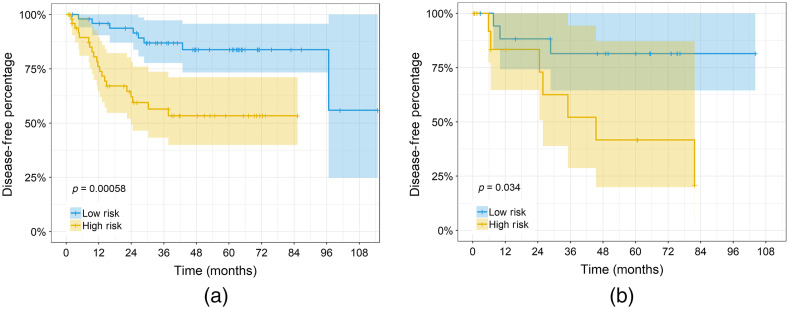
We separated patients into low- and high-risk groups using the median risk score from the RSF model in the training cohort (risk score of 4.84). Figure 4 shows the Kaplan–Meier time-to-event curves for the multivariate model. The high- and low- risk groups in both the training and testing cohorts were found to be significantly different ( and , respectively). The percentage of patients that had a recurrence at 5 years in the low-risk groups for the training and testing cohorts were 16% and 19%, respectively, versus the high-risk groups with rates of 46% and 58%, respectively.
Fig. 4.
Kaplan–Meier curves for the multivariate model risk scores in the (a) training cohort (, ) and (b) testing cohort (, ). Patients were stratified using the median risk score in the training cohort. The shaded regions show the 95% confidence intervals (CI), and + indicates censored data.
3.5. Clinical Model Assessment for Recurrence Risk Stratification and Model Comparison
The clinical stage-only model achieved a concordance of 0.67 [95% CI: 0.58 to 0.76, ] in the training cohort and, when evaluated on the testing cohort, achieved a concordance of 0.60 [95% CI: 0.48 to 0.74, ]. The multivariate radiomic model significantly outperformed the clinical stage-only model in the training cohort (0.67 [95% CI: 0.58 to 0.76] versus 0.78 [95% CI: 0.70 to 0.86], ) and testing cohort (0.60 [95% CI: 0.48 to 0.74] versus 0.76 [95% CI: 0.59 to 0.87], ).
4. Discussion
Our study investigated using the combination of local (tumor), regional (peritumoral), and distant (bone marrow) preoperative CT and FDG PET features with clinical and qualitative CT features to stratify NSCLC patients into low- and high-risks of recurrence/progression. A study investigating this combination of features to predict time to recurrence/progression and stratify patients based on their risk has not yet been presented in the literature. The results of this study suggest that the cancer stage and radiomic features from tumor and nontumor areas on CT and FDG PET can significantly stratify NSCLC patients into low- and high-risks of recurrence/progression. In both the training and testing cohorts, the multimodality radiomic model significantly outperformed stage alone. This implies that the radiomic features can augment the cancer stage, the current clinical gold standard when predicting time to recurrence/progression. We did not investigate a radiomics-only model as we wanted to determine if radiomics can significantly improve on the current clinical standard of the cancer stage. Additionally, the ability of the model to significantly separate patients into low- and high-risks of recurrence/progression demonstrates the model’s ability to identify patients who may need more aggressive treatment plans.
Most previous radiomics studies have not used more than one imaging modality for outcome prediction, and those that have only focused on tumor information.24 The features in our multimodality radiomic model that were discovered to be important in determining a patient’s risk of recurrence/progression were found in many regions of interest: tumor and peritumoral regions on FDG PET and CT, bone marrow on FDG PET, and the clinical feature stage. Our model outperformed a model presented in the literature using only PET radiomic features on the same dataset, suggesting that CT provides added value in predicting a patient’s risk of recurrence.11 Stage was the only clinical feature selected as prognostic in our model, despite other clinical features such as age and smoking status demonstrating prognostic value in the literature.11,25 Qualitative CT features were not chosen as prognostic in our model. This suggests that radiomic features may better capture these qualitative characterizations of the tumor. Additionally, qualitative features are subjective and only consist of limited categories; therefore, they may not capture the full range of appearances. Most of the selected radiomic features had a coefficient above zero, indicating that an increase in that feature is associated with an increased risk of recurrence/progression. With most of these features representing heterogeneity, this suggests that patients with more heterogeneous textures in their tumor and peritumoral regions are more likely to recur. For example, the feature MTV plus peritumoral HHH GLCM MCC measures the complexity of the texture in the MTV region. An increase in this feature, and therefore a more complex texture found in this region, was found to be associated with a higher risk of recurrence/progression. These results are in line with previous literature that has demonstrated tumor heterogeneity in CT and PET to be associated with progression and treatment failure.11,26–29
The most common models used in radiomic studies for time-to-event analysis are the Cox proportional hazards and RSF models. RSF models have been shown to provide comparable results to Cox models and have been used to predict distant metastases and risk stratify lung cancer patients.30–32 The RSF hyperparameters used in this study were chosen based on default parameters used in a previous study using a random forest classification method on the same dataset.33 Due to the high number of regions of interest and features used in this study as well as the small dataset, hyperparameter tuning for the RSF was not conducted to avoid overfitting.
Our semiautomatic segmentation algorithms use commercial (MIM) and freely available source code (The MathWorks, Natick, Massachusetts) for tumor delineation on both CT and FDG PET imaging. This minimizes any interobserver variability that may arise from segmentations performed by different users and proved to be highly reproducible and stable.16 Additionally, the extracted features were shown to be highly reproducible.11,16,20 Our MATLAB CT segmentation algorithm requires minimal user interaction and only one 2D bounding box, and it is time efficient, a key goal of segmentation.16,34
We chose to use radiomic features and traditional machine learning-based models because of their interpretability and ease of implementation on smaller datasets; however, deep learning techniques for outcome prediction in lung cancer have increased in popularity in recent years and have produced promising results when analyzing preoperative images.35–37 They have also shown that they can augment the performance of hand-crafted radiomic models.38 Therefore, future work should include exploring deep learning techniques.
The main limitation of this study is its small sample size. We used one round of sampling, determined by a random number generator, to train and evaluate our model. This may result in an over- or under-estimation of results compared with additional rounds of sampling. Therefore, the results of this study need to be validated with a larger external patient cohort to assess the reliability of the model. A second limitation is the retrospective nature of the study. Our study investigated diagnostic CT and FDG PET-CT images from multiple scanners, acquisition and reconstruction protocols, which can potentially introduce variability in the imaging data and resulting radiomic features. Radiomic features are known to be impacted by the acquisition parameters used such as the reconstruction kernel and slice thickness.39–41 However, the results achieved using multiple scanners may demonstrate the generalizability of our model. Future studies assessing the impact of different scanners and imaging acquisition parameters on the radiomic features used in this study are required. Another limitation lies in the lack of information on specific pathologic tumor markers in this dataset. In this era, histological subtypes and immunomarkers are increasingly important in the management of patients, so these are routinely reported by pathologists as prognosis can be impacted. Future studies could perform a subanalysis based on histopathology. Furthermore, the patients in this dataset were treated prior to 2012 and were, therefore, staged with the AJCC seventh edition. However, the AJCC eighth edition staging is now available, and future work should aim to investigate the impact of this change in staging.
The novelty of our study arises from the use of local, regional, and distant areas of interest for analysis. To the best of our knowledge, no study has combined both qualitative and quantitative features from the tumor, surrounding peritumoral region, and bone marrow to assess patient outcomes. We hypothesize that there may be other areas within the lungs or throughout the body that may add prognostic value to machine learning models. Future studies should aim to expand the number of regions that may provide prognostic information. Whole-body imaging for machine learning applications is not currently incorporated into clinical decision-making; our study represents a possible first step toward that goal.
5. Conclusion
We found that a multimodality radiomics model augmenting stage with quantitative features from the tumor, peritumoral region, and bone marrow on CT and PET was able to significantly stratify patients into low- and high-risk of recurrence/progression. This model can be useful in the identification of high-risk NSCLC patients to help clinicians provide more aggressive or personalized treatment options.
6. Appendices
6.1. Imaging
The FDG dose for the FDG PET-CT scans was 138.9-572.3 MBq (mean: 309.3 MBq), and the uptake time was 23.1 to 128.9 min (mean: 66.6 min). Each bed position consisted of 1 to 5 min acquisition times depending on the weight of the subject. Routine coverage of base-of-skull to midthigh was included in the image acquisition, with additional spot views when necessary. Ordered subset expectation maximization (OSEM) reconstruction was used for CT-based attenuation correction. Images were converted to SUV units normalized by the patient body weight.
The preoperative diagnostic CT scans were acquired with the following scanning parameters: 80 to 140 kVp (mean: 120 kVp), 124 to 699 mA (mean: 220 mA), and slice thickness 0.625 to 3 mm (median: 1.5 mm). Scans were acquired with subjects in the supine position with arms at their sides, from the apex of the lung to the adrenal gland within a single breath-hold.
6.2. Region of Interest Delineation
The tumor volume on CT was segmented using intensity threshold levels automatically derived by the algorithm for each patient, as previously described.16 Algorithm initialization requires the user to identify the axial slice with the longest axial tumor diameter and define a bounding box around it. The bounding box size is used to determine the number of slices required to capture above and below the selected axial slice to create a bounding cube. The resulting cube is separated into four intensity levels, in which all intensities greater than the third level are defined as tumor. The algorithm subsequently eliminates surrounding connected and disconnected structures (e.g., vessels). This is accomplished through two main processes: analysis of structure extent and analysis of structure size progression from one slice to the next. Extent is defined as the area of the structure divided by the area of the bounding box needed to encapsulate that structure. This metric captures the elongation of the structure. Structures above a particular extent were considered to be vessels. This analysis was accompanied by the structure size progression, which only allowed structures to be removed if they were less than a certain percentage in size of the segmented tumor area in the axial slices adjacent to the slice being analyzed. The algorithm also segments the lungs prior to segmenting the tumor to eliminate any mediastinum or chest wall structure in the final result. These semiautomatic segmentations were able to be edited at the user’s discretion. All segmentations used in this study were visually verified by a resident in radiology. If discrepancies were determined, manual adjustment of the segmentation was performed together. Edits were required in of cases, and these typically were a result of erroneously including vessels adjacent to the tumor.
6.3. Feature Extraction
The texture features were calculated in 3D with symmetrical matrices and a fixed bin size of 25 Hounsfield units (HU) on the CT and 0.2 SUV on the PET/CT. The CT images were resampled to a size (mm) of [1,1,1.5], and the PET/CT images were resampled to a size (mm) of [4,4,4]. Additionally, CT images had a voxel array shift applied of 1000 to prevent negative values. Shape features were not calculated within the bone marrow and peritumoral ROIs on either the CT or PET-CT.
6.4. Qualitative Features
The qualitative CT features used in this study are presented in Table 5.
Acknowledgments
The authors would like to acknowledge funding from the Gerald C. Baines Foundation, donor support through the London Health Sciences Foundation, the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grants program (RGPIN-2020-06498), the Lawson Health Research Institute’s Internal Research Fund (IRF), and the National Cancer Institute (NCI) (T32CA009515).
Biographies
Jaryd R. Christie is a CAMPEP PhD candidate in the Department of Medical Biophysics at Western University. He completed his BMSc degree at Western University in 2019 with an Honours Specialization in medical biophysics. His research focuses on outcome prediction in lung cancer patients using radiomics and machine learning on medical images.
Omar Daher is a post graduate year 5 diagnostic radiology resident at the Schulich School of Medicine and Dentistry at Western University. He completed his medical school at the Northern Ontario School of Medicine. His research interests include thoracic imaging and artificial intelligence.
Mohamed Abdelrazek received his MSc and PhD degrees from Cairo University. He completed his undergraduate studies and his radiology residency at Cairo University. He completed clinical fellowships in thoracic and cardiac imaging from the University of Ottawa. He is a cardiothoracic radiologist in the Department of Medical Imaging at Victoria Hospital in London, Canada and an assistant professor at the University of Western Ontario. His research interests include thoracic and cardiovascular imaging.
Perrin E. Romine is a medical oncology and senior research fellow at the University of Washington and Fred Hutchinson Cancer Research Center. Her interest lies in optimizing care delivery for people with lung and head and neck cancer, with a focus on rarer subtypes of head and neck cancer.
Richard A. Malthaner is the chair/chief of the Division of Thoracic Surgery and professor in the Departments of Surgery, Oncology, and Epidemiology and Biostatistics at the Schulich School of Medicine and Dentistry and The University of Western Ontario. He is a scientist at the Lawson Health Research Institute and one of the founding members of Canadian Surgical Technologies and Advanced Robotics (CSTAR).
Mehdi Qiabi is an assistant professor of Surgery and Oncology at Western University. He is the current program director of Western’s Thoracic Surgery residency program. He also holds roles as the Thoracic Surgery Division’s simulation director and the undergraduate program director. He is an associate scientist with the Lawson Health Research Institute. His research interests include patient-oriented outcomes, simulation, three-dimensional image-guided education, and surgical education.
Rahul Nayak is a consultant thoracic surgeon at London Health Sciences Centre. He has multiple published book chapters and peer-reviewed articles. Currently, he serves as a reviewer for the Canadian Medical Association Journal, Canadian Journal of Surgery, and Journal of Thoracic Disease. He is a guest editor for the Shanghai CHEST journal. His research interests lie in the introduction of technological augmentation in thoracic surgery as well as population-based outcomes research in thoracic diseases.
Sandy Napel received his BSES degree from SUNY Stony Brook in 1974 and his MSEE and PhD degrees in EE from Stanford University in 1976 and 1981, respectively. He was formerly VP of engineering at Imatron Inc. Currently, he is a professor of radiology and of electrical engineering and medicine at Stanford University. He coleads the Stanford Radiology 3D and Quantitative Imaging Lab and leads the Radiology Department’s Division of Integrative Biomedical Imaging Informatics.
Viswam S. Nair translates fundamental scientific discoveries into the clinic to improve the care of people with lung cancer. He studies how to best integrate molecular and imaging-based biomarkers into clinical practice to improve lung cancer detection and treatment. He is also a physician with board certifications in internal medicine, pulmonary disease, and critical care. His research focuses on biomarkers, or telltale biological markers of health or disease.
Sarah A. Mattonen is an assistant professor in the Department of Medical Biophysics at Western University in London, Canada. She received her PhD in medical biophysics from Western University and completed her postdoctoral training at Stanford University in the Department of Radiology. Her research interests include developing, evaluating, and translating quantitative image analysis tools to support diagnosis, treatment, and response assessment in oncology.
Disclosures
The authors have no relevant financial interests and no other potential conflicts of interest to disclose.
Contributor Information
Jaryd R. Christie, Email: jchris63@uwo.ca.
Omar Daher, Email: omar.daher@lhsc.on.ca.
Mohamed Abdelrazek, Email: mohamed.abdelrazek@lhsc.on.ca.
Perrin E. Romine, Email: perrinr@uw.edu.
Richard A. Malthaner, Email: richard.malthaner@lhsc.on.ca.
Mehdi Qiabi, Email: mehdi.qiabi@lhsc.on.ca.
Rahul Nayak, Email: rahul.nayak@lhsc.on.ca.
Sandy Napel, Email: snapel@stanford.edu.
Viswam S. Nair, Email: vnair@fredhutch.org.
Sarah A. Mattonen, Email: sarah.mattonen@uwo.ca.
Code, Data, and Materials Availability
The code used to segment the tumors on CT images can be found on GitHub at https://github.com/baines-imaging-mattonen-lab/CT-Lung-Tumour-Segmentation.
References
- 1.Uramoto H., Tanaka F., “Recurrence after surgery in patients with NSCLC,” Transl. Lung Cancer Res. 3(4), 242–249 (2014). 10.3978/j.issn.2218-6751.2013.12.05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ellison L. F., “Progress in net cancer survival in Canada over 20 years,” Health Rep. 29(9), 10–18 (2018). [PubMed] [Google Scholar]
- 3.Wu C.-F., et al. , “Recurrence risk factors analysis for stage I non-small cell lung cancer,” Medicine 94(32), e1337 (2015). 10.1097/MD.0000000000001337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambin P., et al. , “Radiomics: the bridge between medical imaging and personalized medicine,” Nat. Rev. Clin. Oncol. 14(12), 749–762 (2017). 10.1038/nrclinonc.2017.141 [DOI] [PubMed] [Google Scholar]
- 5.Gillies R. J., Kinahan P. E., Hricak H., “Radiomics: images are more than pictures, they are data,” Radiology 278(2), 563–577 (2016). 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li R., et al., eds., Radiomics and Radiogenomics: Technical Basis and Clinical Applications, 1st ed., Chapman and Hall/CRC; (2019). [Google Scholar]
- 7.Huang Y., et al. , “Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (I or II) non-small cell lung cancer,” Radiology 281(3), 947–957 (2016). 10.1148/radiol.2016152234 [DOI] [PubMed] [Google Scholar]
- 8.Coroller T. P., et al. , “CT-based radiomic signature predicts distant metastasis in lung adenocarcinoma,” Radiother. Oncol.: J. Eur. Soc. Therapeutic Radiol. Oncol. 114(3), 345–350 (2015). 10.1016/j.radonc.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q., et al. , “CT imaging features associated with recurrence in non-small cell lung cancer patients after stereotactic body radiotherapy,” Radiat. Oncol. 12(1), 158 (2017). 10.1186/s13014-017-0892-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oikonomou A., et al. , “Radiomics analysis at PET/CT contributes to prognosis of recurrence and survival in lung cancer treated with stereotactic body radiotherapy,” Sci. Rep. 8, 4003 (2018). 10.1038/s41598-018-22357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattonen S. A., et al. , “Bone marrow and tumor radiomics at 18F-FDG PET/CT: impact on outcome prediction in non-small cell lung cancer,” Radiology 293(2), 451–459 (2019). 10.1148/radiol.2019190357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akinci D’Antonoli T., et al. , “CT radiomics signature of tumor and peritumoral lung parenchyma to predict nonsmall cell lung cancer postsurgical recurrence risk,” Acad. Radiol. 27(4), 497–507 (2020). 10.1016/j.acra.2019.05.019 [DOI] [PubMed] [Google Scholar]
- 13.Clark K., et al. , “The cancer imaging archive (TCIA): maintaining and operating a public information repository,” J. Digit. Imaging 26(6), 1045–1057 (2013). 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bakr S., et al. , Data for NSCLC Radiogenomics Collection (Version 4) [data set], The Cancer Imaging Archive; (2017). [Google Scholar]
- 15.Bakr S., et al. , “A radiogenomic dataset of non-small cell lung cancer,” Sci. Data 5, 180202 (2018). 10.1038/sdata.2018.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie J. R., et al. , “A semi-automatic threshold-based segmentation algorithm for lung cancer delineation,” Proc. SPIE 12036, 120361T (2022). 10.1117/12.2611501 [DOI] [Google Scholar]
- 17.Christie J. R., “CT-Lung-Tumour-Segmentation.baines-imaging-mattonen-lab,” https://github.com/baines-imaging-mattonen-lab/CT-Lung-Tumour-Segmentation (2022).
- 18.Bujang M. A., “A simplified guide to determination of sample size requirements for estimating the value of intraclass correlation coefficient: a review,” Arch. Orofacial Sci. 12, 1–11 (2017). 10.21315/mjms2021.28.2.2 [DOI] [Google Scholar]
- 19.Dice L. R., “Measures of the amount of ecologic association between species,” Ecology 26(3), 297–302 (1945). 10.2307/1932409 [DOI] [Google Scholar]
- 20.Mattonen S. A., et al. , “[18F] FDG positron emission tomography (PET) tumor and penumbra imaging features predict recurrence in non–small cell lung cancer,” Tomography 5(1), 145–153 (2019). 10.18383/j.tom.2018.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griethuysen J. J. M., et al. , “Computational radiomics system to decode the radiographic phenotype,” Cancer Res. 77(21), e104–e107 (2017). 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pölsterl S., “scikit-survival: a library for time-to-event analysis built on top of scikit-learn,” J. Mach. Learn. Res. 21(212), 8747–8752 (2020). [Google Scholar]
- 23.R Core Team, R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria: (2018). [Google Scholar]
- 24.Zhou Z., et al. , “Multi-objective radiomics model for predicting distant failure in lung SBRT,” Phys. Med. Biol. 62(11), 4460–4478 (2017). 10.1088/1361-6560/aa6ae5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Q., et al. , “Predictive risk factors for early recurrence of stage pIIIA-N2 non-small cell lung cancer,” Cancer Manage. Res. 13, 8651–8661 (2021). 10.2147/CMAR.S337830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Win T., et al. , “Tumor heterogeneity and permeability as measured on the CT component of PET/CT predict survival in patients with non-small cell lung cancer,” Clin. Cancer Res.: Off. J. Am. Assoc. Cancer Res. 19(13), 3591–3599 (2013). 10.1158/1078-0432.CCR-12-1307 [DOI] [PubMed] [Google Scholar]
- 27.Ganeshan B., et al. , “Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival,” Eur. Radiol. 22(4), 796–802 (2012). 10.1007/s00330-011-2319-8 [DOI] [PubMed] [Google Scholar]
- 28.Grove O., et al. , “Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma,” PLoS ONE 10(3), e0118261 (2015). 10.1371/journal.pone.0118261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de la Pinta C., Barrios-Campo N., Sevillano D., “Radiomics in lung cancer for oncologists,” J. Clin. Transl. Res. 6(4), 127–134 (2020). 10.18053/jctres.06.2020S4.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu W., et al. , “Development and validation of a predictive radiomics model for clinical outcomes in stage I non-small cell lung cancer,” Int. J. Radiat. Oncol. Biol. Phys. 102(4), 1090–1097 (2018). 10.1016/j.ijrobp.2017.10.046 [DOI] [PubMed] [Google Scholar]
- 31.Kakino R., et al. , “Application and limitation of radiomics approach to prognostic prediction for lung stereotactic body radiotherapy using breath-hold CT images with random survival forest: a multi-institutional study,” Med. Phys. 47(9), 4634–4643 (2020). 10.1002/mp.14380 [DOI] [PubMed] [Google Scholar]
- 32.Wang H., et al. , “Prognostic value of cancer antigen -125 for lung adenocarcinoma patients with brain metastasis: a random survival forest prognostic model,” Sci. Rep. 8, 5670 (2018). 10.1038/s41598-018-23946-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He B., et al. , “A biomarker basing on radiomics for the prediction of overall survival in non–small cell lung cancer patients,” Respir. Res. 19(1), 199 (2018). 10.1186/s12931-018-0887-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizzo S., et al. , “Radiomics: the facts and the challenges of image analysis,” Eur. Radiol. Exp. 2(1), 36 (2018). 10.1186/s41747-018-0068-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baek S., et al. , “Deep segmentation networks predict survival of non-small cell lung cancer,” Sci. Rep. 9, 17286 (2019). 10.1038/s41598-019-53461-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afshar P., et al. , “DRTOP: deep learning-based radiomics for the time-to-event outcome prediction in lung cancer,” Sci. Rep. 10, 12366 (2020). 10.1038/s41598-020-69106-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim H., et al. , “Preoperative CT-based deep learning model for predicting disease-free survival in patients with lung adenocarcinomas,” Radiology 296(1), 216–224 (2020). 10.1148/radiol.2020192764 [DOI] [PubMed] [Google Scholar]
- 38.Paul R., et al. , “Deep feature transfer learning in combination with traditional features predicts survival among patients with lung adenocarcinoma,” Tomography 2(4), 388–395 (2016). 10.18383/j.tom.2016.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Timmeren J. E., et al. , “Test–retest data for radiomics feature stability analysis: generalizable or study-specific?” Tomography 2(4), 361–365 (2016). 10.18383/j.tom.2016.00208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackin D., et al. , “Measuring computed tomography scanner variability of radiomics features,” Invest. Radiol. 50(11), 757–765 (2015). 10.1097/RLI.0000000000000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larue R. T. H. M., et al. , “Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study,” Acta Oncol. 56(11), 1544–1553 (2017). 10.1080/0284186X.2017.1351624 [DOI] [PubMed] [Google Scholar]