Abstract
MicroRNA (miRNA) expression is reportedly associated with clinical outcomes in childhood acute lymphoblastic leukemia (ALL). Here, we aimed at investigating whether miRNA expression is associated with clinical outcomes in pediatric ALL patients treated with the Taiwan Pediatric Oncology Group (TPOG) protocols. The expression of 397 miRNAs was measured using stem-loop quantitative real-time polymerase chain reaction miRNA arrays in 60 pediatric ALL patients treated with TPOG-ALL-93 or TPOG-ALL-97 VHR (very high-risk) protocols. In order to identify prognosis-related miRNAs, original cohort was randomly split into the training and testing cohort in a 2:1 ratio, and univariate Cox proportional hazards regression was applied to identify associations between event-free survival (EFS) and expressions of miRNAs. Four prognosis-related miRNAs were selected and validated in another independent cohort composed of 103 patients treated with the TPOG-ALL-2002 protocol. Risk score, including the impact of four prognosis-related miRNAs, was calculated for each patients, followed by grouping patients into the high or low risk-score groups. Irrespective of the training, testing, or validation cohort, risk-score group was significantly associated with EFS and overall survival (OS). Risk-score group combining with clinical characteristics including the age onset (≥10 years), white blood cell counts (≥100 × 109/L), cell type (T- or B-cell), sex, and risk groups of the treatment protocols were used as predictors of EFS using the multivariate Cox proportional hazards regression. Results showed that the risk-score group was the strongest predictor. In the validation cohort, hazard ratios (HRs) of the risk-score group were 7.06 (95% CI=1.93-25.84, p-value =0.003) and 14.03 (95% CI=3.34-59.04, p-value =0.003) for EFS and OS, respectively. High risk-score group had higher risk of having poor prognosis and risk of death than that in the low risk group. Accuracy of the prediction model for 5-year EFS could reach 0.76. For the prediction of 5-year OS, accuracy was 0.75. In conclusion, a miRNA signature was associated with clinical outcomes in childhood ALL patients treated with TPOG protocols and might be a suitable prognostic biomarker.
Keywords: Childhood ALL, microRNA signature, TPOG
Introduction
MicroRNAs (miRNAs) execute diverse functions by simultaneously targeting the mRNAs of multiple genes. Alterations in miRNAs contribute to many human malignancies [1]. In addition, miRNAs are important regulators of hematopoiesis [2-4]. miRNA-mediated control of gene dosage is critical for lineage fate determination of hematopoietic cells, and disruption of this regulation may lead to malignant transformation. Moreover, dysregulation of miRNA expression is frequently associated with cytogenetic abnormalities, and certain abnormalities directly affect the aberrant expression of miRNAs in hematologic malignancies [5-7]. Calin et al. first demonstrated that a loss of miR-15a and miR-16-1 was the target of 13q14 deletion, a region frequently lost in patients with chronic lymphoblastic leukemia (CLL) [8]. In patients expressing the aberrant fusion protein, AML1/ETO, the most common acute myeloid leukemia-associated fusion resulting from t(8;21), The fusion oncoprotein was first-ever reported to directly repress the expression of miR-223 by triggering chromatin remodeling and epigenetic silencing, which in turn blocks myeloid precursor cell differentiation [9]. miR-155 is essential for B-cell development and is aberrantly upregulated in B-cell malignancies, including diffuse large B-cell lymphoma, follicular lymphoma, and CLL [10-14]. miRNA expression profiles are also used to discriminate different subtypes of acute lymphoblastic leukemia (ALL) [3,15-18]. The expression of some miRNAs is associated with the clinical outcomes of acute myeloid leukemia [19-21].
Lu et al. used miRNA expression to classify cancers, including childhood ALL. Based on miRNA expression, ALL can be differentiated into major cytogenetic subtypes, including B-ALL with ETV6-RUNX1, BCR-ABL1, hyperdiploidy, and T-ALL [3]. Bueno et al. showed that the genetic and epigenetic silencing of miRNA-203 enhanced, the expression of ABL1 and BCR-ABL1 oncogenes and it might function as a tumor suppressor [22]. The expression of miR-196b was enriched in patients with KMT2A fusion leukemia, and its function is necessary for KMT2A fusion-mediated immortalization [16]. miR196b was also linked with the activation of HOXA in pediatric ALL-not restricted to KMT2A-rearranged leukemia-and directly targeted the HOXA/MEIS1 and FAS tumor suppressor genes in KMT2A-rearranged leukemia [23,24]. In a previous work, we showed that miRNA 181-a was regulated by ETV6-RUNX1 through a loop feedback [18]. Recently, Malouf et al. showed that miR-130b and miR-128a are downstream targets of MLL-AF4 and can individually drive the transition from a preleukemic stage to an acute leukemia in an murine MLL-AF4 model [25]. miRNA-497/195 is tumor-suppressive and cooperates with CDKN2A/B in pediatric ALL [26]. miRNAs are also involved in the pathogenesis of T-ALL via the NOTCH1 and MYB pathways in T-ALL [27,28].
As, miRNAs are involved in ALL pathways, it is possible that miRNA expression be a prognostic marker for childhood ALL. Several studies have shown that miRNAs are prognostic markers in childhood ALL. Zhang et al. described a miRNA signature that could be used to predict prednisone response in childhood ALL patients, which was validated using a smaller cohort [29]. Schotte et al. demonstrated a correlation between the probability of disease-free survival and the expression levels of 31 distinct miRNAs. Upregulation of miRNA-21 is a poor prognostic marker in patients with childhood B-ALL [30]. Upregulated miR-155 is associated with poor prognosis in childhood ALL and promotes cell proliferation by targeting ZNF238 [31]. The expression of miR-143/miR-182 is associated with the prognosis and risk stratification specificity of BFM-treated childhood ALL [32]. In this study, we used miRNA arrays to identify a signature in pediatric ALL patients treated with Taiwan Pediatric Oncology Group (TPOG) protocols and validated its prognostic impact.
Materials and methods
Patients and protocols
Diagnostic and/or relapsed bone marrow (BM) or peripheral blood samples were obtained from 60 children with newly diagnosed ALL from July 1996 to December 2001 at the National Taiwan University Hospital as the primary cohort. There were 50 patients with B-cell ALL and 10 patients with T-cell ALL. Forty-five patients were treated with the TPOG-ALL-93 protocol, and fifteen patients were treated with the TPOG-97-VHR protocol [33]. These 60 patients were assigned to the training (40 patients) and testing (20 patients) cohorts (Table 1). To validate the results, another 103 patients were included as the validation cohort; these included 78 B-cell and 25 T-cell ALL patients who were treated with TPOG-ALL-2002 protocol. The diagnosis of ALL was based on morphologic findings of BM aspirates and immuno-phenotype analyses of leukemic cells by flow cytometry. Conventional cytogenetic analysis were done as part of the routine work-flow [34].
Table 1.
Clinical characteristics of the 60 ALL patients in the original cohort. Patients were randomly assigned to the training (n=40) or testing cohort (n=20)
| Total | Training | Testing | p-value | |
|---|---|---|---|---|
| n (%) | n (%) | n (%) | ||
| Onset age (years) | 0.326 | |||
| <10 | 47 (78.3) | 33 (82.5) | 14 (70.0) | |
| ≥10 | 13 (21.7) | 7 (17.5) | 6 (30.0) | |
| Sex | 1.000 | |||
| Female | 32 (53.3) | 21 (52.5) | 11 (55.0) | |
| Male | 28 (46.7) | 19 (47.5) | 9 (45.0) | |
| Initial WBC (109/L) | 0.189 | |||
| <100 | 48 (80.0) | 34 (85.0) | 14 (70.0) | |
| ≥100 | 12 (20.0) | 6 (15.0) | 6 (30.0) | |
| Risk group | 0.579 | |||
| SR+HR | 39 (65.0) | 27 (67.5) | 12 (60.0) | |
| VHR | 21 (35.0) | 13 (32.5) | 8 (40.0) | |
| Genetic subtypes* | 0.245 | |||
| BCR-ABL1 | 1 (3.5) | 0 (0.0) | 1 (11.1) | |
| ETV6-RUNX1 | 8 (27.6) | 6 (30.0) | 2 (22.2) | |
| Hyperdiploidy | 8 (27.6) | 7 (35.0) | 1 (11.1) | |
| DUX4-rearranged | 1 (3.5) | 0 (0.0) | 1 (11.1) | |
| ZNF384-rearranged | 1 (3.5) | 1 (5.0) | 0 (0.0) | |
| T-ALL | 10 (34.5) | 6 (30.0) | 4 (44.4) |
A total of 31 samples had unknown subtype.
WBC, white blood cells; SR, standard risk group; HR, high-risk group; VHR, very high-risk group.
The treatment protocols and risk classifications were described previously [33-35]. The patients were prospectively assigned to one of the three risk groups (standard [SR], high [HR], and very high [VHR]) based on clinical and biological features of Intrathecal chemotherapy replaced cranial irradiation for CNS prophylaxis in HR and VHR patients in the TPOG-ALL-2002 protocol. The Institutional Review Board of the National Taiwan University Hospital approved the study, and all participants provided written informed consent in accordance with the Declaration of Helsinki. Details of the protocols and risk group assignments have been published elsewhere [33,35].
RNA extraction and miRNA detection
Total RNA was extracted from BM or peripheral blood using TRIzol (Invitrogen) at the time of diagnosis. miRNA expression profiling was performed using the ABI PRISM 7900 and stem-loop RT-qPCR miRNA arrays containing 397 mature human miRNAs (Applied Biosystems), as described previously [18]. Individual miRNAs were quantified using TaqMan miRNA assays (Applied Biosystems). All miRNA arrays were run concurrently with a calibration control (U6 snRNA).
Statistical methods
The follow-up care for these patients included physical examination and complete blood analysis after completing the chemotherapy. In case of any abnormality during physical examination or blood tests, the disease relapse possibility will be excluded after BM examination. To identify prognosis-related miRNAs and evaluate the effect of these miRNAs, 60 patients were randomly assigned to the training and testing sets at a ratio of 2:1. Univariate Cox proportional hazards regression was applied to evaluate the association between the event-free survival (EFS) and expression of each miRNA in the training set. Events were defined based on any relapse, death, or secondary malignancy. If the p-value of coefficient in the regression model was less than 0.05, then the miRNA was considered as the prognosis-related miRNA. For concluding the effect of prognosis-related miRNAs, the risk score of each patient was calculated using a linear combination of expressions of prognosis-related miRNAs, weighted by the coefficients of the regression model (Supplementary Table 1). In our study, four miRNAs were identified as prognosis-related miRNA. Therefore, the formula of risk score was defined as:
Where β is the regression coefficient. The cut-off of the risk score is the median of risk score distribution. Patients were grouped into high risk-score group if their risk scores were higher than the cut-off, whereas patients with a risk score lower than the cut-off were grouped into low risk-score group. Finally, the linear equation of risk score calculation and cut-off were also applied in the testing cohort same as the training cohort (Figure 1).
Figure 1.
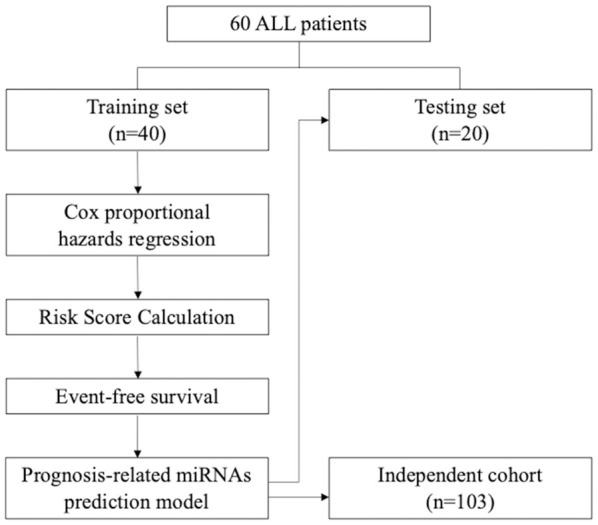
Flowchart for identifying the miRNA signature of childhood ALL for clinical outcome prediction.
To validate the identified miRNAs and prediction model, prognosis-related miRNAs were quantified by RT-qPCR experiments for an independent cohort of 103 patients. Additionally, as the experiment platform was different from the training and testing cohorts, weighting values of risk scores were recalculated using the univariate Cox proportional hazards regression. Risk score was calculated by summation of the expression values of the four prognosis-related miRNAs, weighted by the coefficients of the regression model. Patients were classified into the two groups based on the threshold value, estimated using the maxstat method.
The association between categorical variables was evaluated using the Fisher’s exact test. Survival curves were estimated using the Kaplan-Meier method, and p-values were determined using the log-rank test. In order to elucidate the impact of risk-score group and develop the prognostic prediction model, a multivariate Cox proportional hazards regression model was used and risk-score group with clinical variables, including onset age (≥10 years), white blood cell (WBC) counts (≥100 × 109/L), cell type (T- or B-cell), and sex, and risk groups of the treatment protocols as predictors. In this model development, complete case analysis was applied. Based on the risk model of the multivariate Cox proportional hazards regression model, Harrell’s concordance statistics, and time-dependent ROC (receiver operating characteristics) curves at 5-year EFS and 5-year OS were applied to assess the prediction efficiency. Harrell’s C-index near 1 indicates that the prediction model performs well in deciding which patient would have the event first. By contrast, index near 0 indicates that prediction model is worse than the coin flip. All statistical analyses were two-tailed and performed using the SAS software (version 9.4; SAS Institute, Inc., Cary, NC, USA). Statistical significance was set at P<0.05.
Results
Clinical features of the original cohort
Sixty patients were included in the original cohort. Among these, 28 were male and 32 were female (a male-to-female ratio of 0.88:1). The median age of the patients at diagnosis was 12.1 years (range, 1.4-17.4 years). The median WBC count was 69.8 × 109/L (range, 0.6-1096 × 109/L). The clinical features of the original cohort are presented in Table 1.
miRNA signature associated with clinical outcomes in patients treated with TPOG-ALL-93 or -97 VHR protocols in the original cohort
Of the 60 samples from the original cohort, 40 were used as the training set and 20 as the validation set. There were no differences in clinical parameters, such as age, risk group, and immunophenotypes, in the original cohort (Table 1). A flow chart for identifying the miRNA signature is illustrated in Figure 1. We identified that miRNAs 133a, 193a, 151, and 129 were associated with EFS as the prognosis-related miRNAs (Supplementary Table 1). After risk score calculation for each patient, summarizing the impact of the four prognosis-related miRNAs, patients were grouped into the high risk-score and low risk-score group based on the cut-off value. Patients with low risk scores had a better EFS and overall OS than those with high risk scores (95.00%, 69.47-99.28 versus 65.00%, 40.30-81.53 for 5-year EFS and 100%, 100-100, versus 70.00%, 45.05-85.25 for 5-year OS) in the training set (Figure 2A). These results were confirmed in the testing set (Figure 2B).
Figure 2.
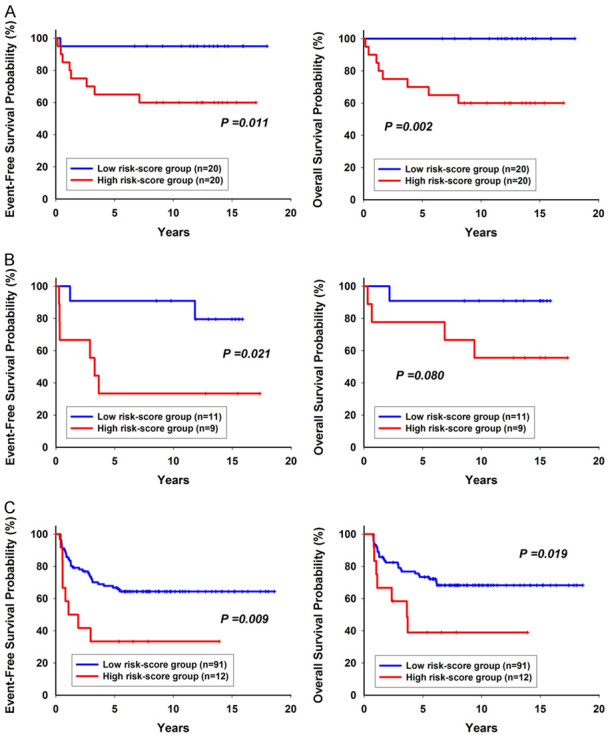
Risk-score group was significantly associated with event-free survival and overall survival (A) in the training cohort (n=40) (B) in the testing cohort (n=20) (C) in the validation cohort (n=103).
Confirmation of the prognostic significance of the miRNA score using another cohort treated with the TPOG-ALL-2002 protocol
Among the 103 additional patients in the validation cohort, 59 were male and 44 were female (male-to-female ratio: 1.34:1). The median age of the patients at diagnosis was 12.1 years (range, 1.4-17.4 years). The median WBC count was 69.8 × 109/L (range 0.6-1096 × 109/L). The clinical characteristics of the original and validation cohort were not significantly different except the genetic subtypes (Supplementary Table 2). However, half of the patients had unknown genetic subtypes in the original cohort (Supplementary Table 2). Based on the expression profiles of the four selected miRNAs, 91 patients were classified into the low risk-score group and 12 were classified into the high risk-score group. The low risk-score group had better EFS and OS than the high risk-score group (Figure 2C). The 5-year EFS and OS rates were 66.72% (55.96-75.42) and 73.35% (62.91-81.27) for patients with a low risk score and 38.89 (12.63-64.98) and 33.33 (10.27-58.87) for patients with a high risk score, respectively.
Development of miRNA signature prediction model
In order to develop the prognostic prediction model, risk-score group in association with clinical variables, onset age (≥10 years), WBC counts (≥100 × 109/L), cell type (T- or B-cell), sex, and risk groups of the treatment protocols were used as predictors. It was used to predict EFS using the multivariate Cox proportional hazards regression model (Table 3). Irrespective of the training, testing, or validation cohort, the high risk-score group was the strongest predictive factor for unfavorable EFS (Table 2 and Supplementary Table 3). In the validation cohort, the high risk group had 7.06-fold risk to have the prognosis events than the low risk-score group (95% CI=1.93-25.84, p-value =0.003). Furthermore, for the OS prediction, results also show that the high risk-score group had 14.03-fold risk of death than the low risk-score group (95% CI=3.34-59.04, p-value =0.003, Table 2). In order to assess whether the prediction model has prediction efficacy for clinical outcome, Harrell’s concordance statistics and ROC curves at 5-year EFS and 5-year OS were applied. Harrell’s C-index for EFS were 0.80, 0.89, and 0.70 in the training, testing, and validation cohort, respectively; the prediction power of overall survival, were 0.89, 0.89, and 0.72 in the training, testing, and validation cohort, respectively (Supplementary Table 4). In terms of EFS or OS, although a lower Harrell’s C-index was determined in the validation cohort than in the other cohorts it could reach at least 0.7. Results of ROC curve at 5-year EFS also showed that accuracy could reach above 0.8 in the training and testing cohort, and accuracy in the validation cohort was 0.76 (Figure 3). Accuracy of 5-year OS prediction could reach even higher. The risk model efficiently predicted the clinical outcome, especially for 5-year OS prediction (Figure 3).
Table 3.
Multivariable analysis of 5-year event-free and overall survival
| Variable | Hazard ratio (HR) | 95% Confidence interval (CI) | p-value | |
|
| ||||
| High risk-score group | 2.73 | 1.25 | 5.95 | 0.012 |
|
| ||||
| Variable | HR | 95% CI | p-value | |
|
| ||||
| High risk-score group | 7.06 | 1.93 | 25.84 | 0.003 |
| Initial WBC ≥100 | 1.74 | 0.71 | 4.25 | 0.224 |
| T cell | 0.31 | 0.09 | 1.08 | 0.066 |
| Male | 1.81 | 0.91 | 3.63 | 0.093 |
| Age ≥10 | 1.91 | 0.95 | 3.80 | 0.068 |
| Very high risk-score group | 1.67 | 0.60 | 4.63 | 0.323 |
|
| ||||
| Variable | HR | 95% CI | p-value | |
|
| ||||
| High risk-score group | 2.61 | 1.13 | 6.01 | 0.024 |
|
| ||||
| Variable | HR | 95% CI | p-value | |
|
| ||||
| High risk-score group | 14.03 | 3.34 | 59.04 | 0.003 |
| Initial WBC ≥100 | 1.88 | 0.75 | 4.67 | 0.177 |
| T cell | 0.12 | 0.03 | 0.55 | 0.006 |
| Male | 1.80 | 0.83 | 3.89 | 0.134 |
| Age ≥10 | 2.00 | 0.96 | 4.17 | 0.064 |
| Very high risk-score group | 1.88 | 0.65 | 5.38 | 0.242 |
WBC, white blood cells.
Table 2.
Adjusted hazard ratios of predictors in the prognostic model estimated using the multivariate Cox proportional hazards regression model for the validation cohort
| Variables | Event-Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| HR | 95% CI | p-value | HR | 95% CI | p-value | |||
| High risk-score group | 7.06 | 1.93 | 25.84 | 0.003 | 14.03 | 3.34 | 59.04 | 0.003 |
| Initial WBC ≥100 | 1.74 | 0.71 | 4.25 | 0.224 | 1.88 | 0.75 | 4.67 | 0.177 |
| T cell | 0.31 | 0.09 | 1.08 | 0.066 | 0.12 | 0.03 | 0.55 | 0.006 |
| Male | 1.81 | 0.91 | 3.63 | 0.093 | 1.80 | 0.83 | 3.89 | 0.134 |
| Age ≥10 | 1.91 | 0.95 | 3.80 | 0.068 | 2.00 | 0.96 | 4.17 | 0.064 |
| VHR | 1.67 | 0.60 | 4.63 | 0.323 | 1.88 | 0.65 | 5.38 | 0.242 |
WBC, white blood cells.
Figure 3.
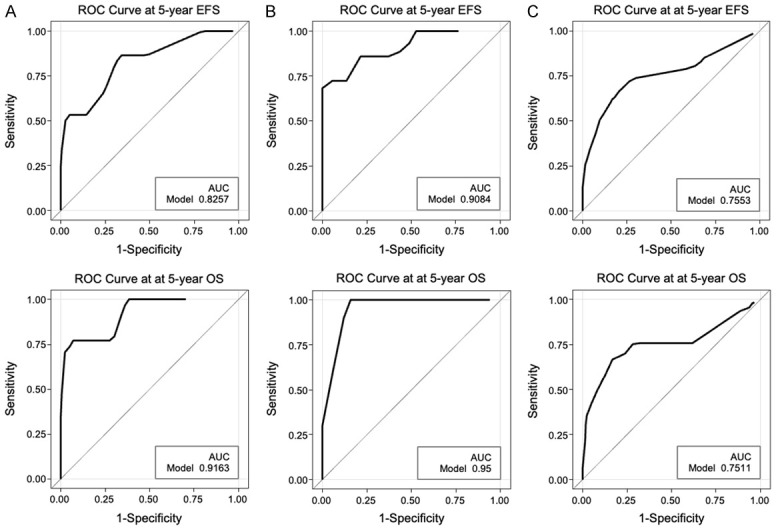
ROC curve at 5-year EFS and 5-year OS. (A) in the training cohort (n=40) (B) in the testing cohort (n=20) (C) in the validation cohort (n=103).
Discussion
In this study, we analyzed the expression levels of 397 miRNAs in pediatric ALL using specific, stem-loop RT-qPCR miRNA assays. We correlated miRNA expression profiles of 60 patients samples in the original cohort and proposed a miRNA signature associated with clinical outcomes. This signature was validated in samples from another 103 patients. The miRNA signature score included the expression of miR133a, 193a, 151, and 129. The prognostic value was determined to be an independent prognostic marker after the multivariate analysis.
There are several reports on the prognostic value of such signatures in childhood ALL. Schotte et al. used a similar assay to correlate miRNA expression and drug resistance [23]. They identified resistance to vincristine, which was characterized by an approximately 20-fold upregulation of miR-125b, miR-99a, and miR-100 (PFDR≤0.002). A combined expression profile based on 14 miRNAs that were individually associated with prognosis was highly predictive of clinical outcomes in pediatric ALL [23]. One of these 14 miRNAs, miR-193a, was also identified to be associated with clinical outcomes in this study. However, this miRNA did not retain its prognostic significance after the application of correction factors. Avigad et al. identified five miRNAs in a cohort of 48 samples [36]. The authors then used real-time quantitative PCR on a cohort of precursor B-cell ALL patients (n=138). Low expression of miR-151-5p, miR-451, and high expression of miR-1290 or a combination of all three predicted an inferior relapse-free survival. The prognostic relevance of the three miRNAs was evaluated in another B-cell ALL cohort (n=33) treated with other protocols. A significant correlation between aberrant expression of at least one of the three miRNAs and poor outcome was maintained (P<0.0001). Piatopoulou et al. examined the expression profile of miR-143 and miR-182 in 125 childhood ALL patients who received the Berlin-Frankfurt-Münster (BFM) protocol. BM levels of miR-143/miR-182 were significantly decreased in childhood ALL patients at diagnosis, and overexpression of miR-143/miR-182 at the end of induction presented a significantly higher risk for short-term relapse and death. Zamani et al. reported that the expression levels of miR-324-3p and miR-508-5p were different between samples with positive and negative MRD and could serve as potential diagnostic and multidrug-resistant biomarkers in childhood ALL [37].
Our approach was similar to that of Schotte et al. and Avigad et al. and was better than that of Piatopoulou et al. and Zamani et al., as the latter just used two miRNA expressions [23,36-38]. We used an original cohort to obtain the most significant miRNA signature and have validated this finding with a different cohort, who were treated with another protocol. Moreover, we followed the guidelines of the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis initiative [39]. Thus, the miRNA signature identified herein has a better chance to be successfully validated in a future prospective trial in Taiwan. As the predictive markers might be related to the treatment protocols and genetic background, the miRNA markers identified by different groups may be distinct [40].
Sengupta et al. identified conserved miR-193a target sites within the 3’-untranslated region of the MLL1 gene transcript [17]. MiR-193a directly targeted the 3’-untranslated region of the MLL1 mRNA. Ectopic expression of miR-193a modulated the global H3K4 mono-, di-, and tri-methylation levels. Prolonged ectopic expression of miR-193a inhibits growth and cell migration and induces apoptosis. Another similar miR-193 family, miR-193b-3p, was downregulated in several cytogenetically defined subgroups of pediatric and adult AML, and low expression served as an independent indicator of poor prognosis in pediatric AML [20]. This trend is similar to that observed in our study. In knockout mice, loss of miR-193b cooperated with Hoxa9/Meis1 during leukemogenesis, whereas restoring miR-193b expression impaired leukemic engraftment. Similarly, expression of miR-193b in AML blasts from patients diminished leukemic growth in vitro and in mouse xenografts. Mechanistically, miR-193b induces apoptosis and G1/S-phase arrest in various human AML subgroups by targeting multiple factors in the KIT-RAS-RAF-MEK-ERK (MAPK) signaling cascade and the downstream cell cycle regulator, CCND1. MiR-133a was downregulated in AMLs bearing the AML1/ETO rearrangements [41]. Fulci et al. identified miR-148, miR-151, and miR-424 as discriminative of T-lineage versus B-lineage ALL [42]. However, how these miRNAs affect drug resistance and the 5-year EFS and OS might require further investigation in the future.
There are several limitations to this study. One of the limitations of this study is its relatively small sample size. Although we performed the validation with another cohort treated with the selected ALL protocol, we did not have other samples with different genetic backgrounds treated with other regimens to validate its significance. There are several limitations of correlation of miRNA expression with clinical outcomes in childhood ALL. In childhood ALL, the RNA-based signature or genetic predictive markers are difficult to validate across protocols or study populations [43-47]. There is also a problem with miRNA expression in ALL. There are several reports discussing its prognosis and drug resistance [23,32,36,37]. However, these miRNAs were not the same, and the miRNA-based signature could be a predictive marker, but this approach might not be easily validated in larger clinical trials. Another limitation is the lack of an MRD parameter in the analysis. Lastly, the subtypes defined by RNA-seq had several novel subtypes, and in this study, there were many patients lacking detailed subtyping. In the current treatment protocol used in Taiwan, these two parameters were used, and a new prospective clinical trial to investigate the clinical significance of this score with MRD levels and novel subtypes might be needed in the future.
In conclusion, the miRNA 133a, 193a, 151, and 129 signature score was associated with clinical outcomes in the selected cohort treated with TPOG-ALL protocols and was validated. In addition to the major cytogenetic alterations, we identified a miRNA signature associated with clinical outcomes. Future larger clinical trials including complete genotyping, MRD, and these miRNAs might be worthy of further investigations.
Acknowledgements
The authors express their gratitude to all patients who participated in this study and their parents. The authors also acknowledge the efforts of the TPOG and Childhood Cancer Foundation in Taiwan and the technical support from the Pharmacogenomics Laboratory of National Core Facility for Biopharmaceuticals (NCFB), the NGS and Microarray Core Facility of NTU Centers of Genomic and Precision Medicine. This work was supported by grants from the Ministry of Science and Technology, Taiwan (MOST-106-2314-B-002-199- and MOST-107-2314-B-002-173-MY2 to YLY and MOST-110-2740-B-002-005-). This work was also supported by the “Excellent Research Projects of National Taiwan University” grant number BM01-05.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 4.O’Connell RM, Chaudhuri AA, Rao DS, Gibson WS, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc Natl Acad Sci U S A. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc Natl Acad Sci U S A. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapiro E, Russell LJ, Struski S, Cave H, Radford-Weiss I, Valle VD, Lachenaud J, Brousset P, Bernard OA, Harrison CJ, Nguyen-Khac F. A new recurrent translocation t(11;14)(q24;q32) involving IGH@ and miR-125b-1 in B-cell progenitor acute lymphoblastic leukemia. Leukemia. 2010;24:1362–1364. doi: 10.1038/leu.2010.93. [DOI] [PubMed] [Google Scholar]
- 7.Sonoki T, Iwanaga E, Mitsuya H, Asou N. Insertion of microRNA-125b-1, a human homologue of lin-4, into a rearranged immunoglobulin heavy chain gene locus in a patient with precursor B-cell acute lymphoblastic leukemia. Leukemia. 2005;19:2009–2010. doi: 10.1038/sj.leu.2403938. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazi F, Racanicchi S, Zardo G, Starnes LM, Mancini M, Travaglini L, Diverio D, Ammatuna E, Cimino G, Lo-Coco F, Grignani F, Nervi C. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 10.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: how a complex biology gets more complex. Leukemia. 2015;29:1004–1017. doi: 10.1038/leu.2014.351. [DOI] [PubMed] [Google Scholar]
- 11.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 13.Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood J, Wainscoat JS, Hatton CS. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 14.Vargova K, Curik N, Burda P, Basova P, Kulvait V, Pospisil V, Savvulidi F, Kokavec J, Necas E, Berkova A, Obrtlikova P, Karban J, Mraz M, Pospisilova S, Mayer J, Trneny M, Zavadil J, Stopka T. MYB transcriptionally regulates the miR-155 host gene in chronic lymphocytic leukemia. Blood. 2011;117:3816–3825. doi: 10.1182/blood-2010-05-285064. [DOI] [PubMed] [Google Scholar]
- 15.Almeida RS, Costa E Silva M, Coutinho LL, Garcia Gomes R, Pedrosa F, Massaro JD, Donadi EA, Lucena-Silva N. MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematol Oncol. 2019;37:103–112. doi: 10.1002/hon.2567. [DOI] [PubMed] [Google Scholar]
- 16.Popovic R, Riesbeck LE, Velu CS, Chaubey A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, Chen J, Rowley JD, Zeleznik-Le NJ. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sengupta D, Deb M, Kar S, Parbin S, Pradhan N, Patra SK. miR-193a targets MLL1 mRNA and drastically decreases MLL1 protein production: ectopic expression of the miRNA aberrantly lowers H3K4me3 content of the chromatin and hampers cell proliferation and viability. Gene. 2019;705:22–35. doi: 10.1016/j.gene.2019.04.046. [DOI] [PubMed] [Google Scholar]
- 18.Yang YL, Yen CT, Pai CH, Chen HY, Yu SL, Lin CY, Hu CY, Jou ST, Lin DT, Lin SR, Lin SW. A double negative loop comprising ETV6/RUNX1 and MIR181A1 contributes to differentiation block in t(12;21)-positive acute lymphoblastic leukemia. PLoS One. 2015;10:e0142863. doi: 10.1371/journal.pone.0142863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chuang MK, Chiu YC, Chou WC, Hou HA, Chuang EY, Tien HF. A 3-microRNA scoring system for prognostication in de novo acute myeloid leukemia patients. Leukemia. 2015;29:1051–1059. doi: 10.1038/leu.2014.333. [DOI] [PubMed] [Google Scholar]
- 20.Bhayadia R, Krowiorz K, Haetscher N, Jammal R, Emmrich S, Obulkasim A, Fiedler J, Schwarzer A, Rouhi A, Heuser M, Wingert S, Bothur S, Döhner K, Mätzig T, Ng M, Reinhardt D, Döhner H, Zwaan CM, van den Heuvel Eibrink M, Heckl D, Fornerod M, Thum T, Humphries RK, Rieger MA, Kuchenbauer F, Klusmann JH. Endogenous tumor suppressor microRNA-193b: therapeutic and prognostic value in acute myeloid leukemia. J. Clin. Oncol. 2018;36:1007–1016. doi: 10.1200/JCO.2017.75.2204. [DOI] [PubMed] [Google Scholar]
- 21.Zhu R, Lin W, Zhao W, Fan F, Tang L, Hu Y. A 4-microRNA signature for survival prognosis in pediatric and adolescent acute myeloid leukemia. J Cell Biochem. 2019;120:3958–3968. doi: 10.1002/jcb.27679. [DOI] [PubMed] [Google Scholar]
- 22.Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 23.Schotte D, De Menezes RX, Akbari Moqadam F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R, Den Boer ML. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica. 2011;96:703–711. doi: 10.3324/haematol.2010.026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, Zhang Z, Elkahloun A, Cao D, Shen C, Wunderlich M, Wang Y, Neilly MB, Jin J, Wei M, Lu J, Valk PJM, Delwel R, Lowenberg B, Le Beau MM, Vardiman J, Mulloy JC, Zeleznik-Le NJ, Liu PP, Zhang J, Chen J. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malouf C, Antunes ETB, O’Dwyer M, Jakobczyk H, Sahm F, Landua SL, Anderson RA, Soufi A, Halsey C, Ottersbach K. MiR-130b and miR-128a are essential lineage-specific co-drivers of t(4;11) MLL-AF4 acute leukemia. Blood. 2021;138:2066–2092. doi: 10.1182/blood.2020006610. [DOI] [PubMed] [Google Scholar]
- 26.Boldrin E, Gaffo E, Niedermayer A, Boer JM, Zimmermann M, Weichenhan D, Claus R, Münch V, Sun Q, Enzenmüller S, Seyfried F, Demir S, Zinngrebe J, Cario G, Schrappe M, Den Boer ML, Plass C, Debatin KM, Te Kronnie G, Bortoluzzi S, Meyer LH. MicroRNA-497/195 is tumor-suppressive and cooperates with CDKN2A/B in pediatric acute lymphoblastic leukemia. Blood. 2021;138:1953–1965. doi: 10.1182/blood.2020007591. [DOI] [PubMed] [Google Scholar]
- 27.Shu Y, Wang Y, Lv WQ, Peng DY, Li J, Zhang H, Jiang GJ, Yang BJ, Liu S, Zhang J, Chen YH, Tang S, Wan KX, Yuan JT, Guo W, Fu G, Qi XK, Liu ZD, Liu HY, Yang C, Zhang LH, Liu FJ, Yu J, Zhang PH, Qu B, Zhao H, He TC, Zou L. ARRB1-promoted NOTCH1 degradation is suppressed by oncomiR miR-223 in T-cell acute lymphoblastic leukemia. Cancer Res. 2020;80:988–998. doi: 10.1158/0008-5472.CAN-19-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mets E, Van der Meulen J, Van Peer G, Boice M, Mestdagh P, Van de Walle I, Lammens T, Goossens S, De Moerloose B, Benoit Y, Van Roy N, Clappier E, Poppe B, Vandesompele J, Wendel HG, Taghon T, Rondou P, Soulier J, Van Vlierberghe P, Speleman F. MicroRNA-193b-3p acts as a tumor suppressor by targeting the MYB oncogene in T-cell acute lymphoblastic leukemia. Leukemia. 2015;29:798–806. doi: 10.1038/leu.2014.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labib HA, Elantouny NG, Ibrahim NF, Alnagar AA. Upregulation of microRNA-21 is a poor prognostic marker in patients with childhood B cell acute lymphoblastic leukemia. Hematology. 2017;22:392–397. doi: 10.1080/10245332.2017.1292204. [DOI] [PubMed] [Google Scholar]
- 31.Liang C, Li Y, Wang LN, Zhang XL, Luo JS, Peng CJ, Tang WY, Huang LB, Tang YL, Luo XQ. Up-regulated miR-155 is associated with poor prognosis in childhood acute lymphoblastic leukemia and promotes cell proliferation targeting ZNF238. Hematology. 2021;26:16–25. doi: 10.1080/16078454.2020.1860187. [DOI] [PubMed] [Google Scholar]
- 32.Piatopoulou D, Avgeris M, Drakaki I, Marmarinos A, Xagorari M, Baka M, Pourtsidis A, Kossiva L, Gourgiotis D, Scorilas A. Clinical utility of miR-143/miR-182 levels in prognosis and risk stratification specificity of BFM-treated childhood acute lymphoblastic leukemia. Ann Hematol. 2018;97:1169–1182. doi: 10.1007/s00277-018-3292-y. [DOI] [PubMed] [Google Scholar]
- 33.Liang DC, Yang CP, Lin DT, Hung IJ, Lin KH, Chen JS, Hsiao CC, Chang TT, Peng CT, Lin MT, Chang TK, Jaing TH, Liu HC, Wang LY, Yeh TC, Jou ST, Lu MY, Cheng CN, Sheen JM, Chiou SS, Wu KH, Hung GY, Chen RL, Chen SH, Cheng SN, Chang YH, Chen BW, Ho WL, Wang JL, Lin ST, Hsieh YL, Wang SC, Chang HH, Yang YL, Huang FL, Chang CY, Chang WH, Lin KS. Long-term results of Taiwan Pediatric Oncology Group studies 1997 and 2002 for childhood acute lymphoblastic leukemia. Leukemia. 2010;24:397–405. doi: 10.1038/leu.2009.248. [DOI] [PubMed] [Google Scholar]
- 34.Yang YL, Hung CC, Chen JS, Lin KH, Jou ST, Hsiao CC, Sheen JM, Cheng CN, Wu KH, Lin SR, Yu SL, Chen HY, Lu MY, Wang SC, Chang HH, Lin SW, Su YN, Lin DT. IKZF1 deletions predict a poor prognosis in children with B-cell progenitor acute lymphoblastic leukemia: a multicenter analysis in Taiwan. Cancer Sci. 2011;102:1874–1881. doi: 10.1111/j.1349-7006.2011.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li MJ, Liu HC, Yen HJ, Jaing TH, Lin DT, Yang CP, Lin KH, Hung IJ, Jou ST, Lu MY, Hsiao CC, Peng CT, Chang TT, Wang SC, Lin MT, Chen JS, Chang TK, Hung GY, Wu KH, Yang YL, Chang HH, Chen SH, Yeh TC, Cheng CN, Lin PC, Chiou SS, Sheen JM, Cheng SN, Chen SH, Chang YH, Ho WL, Chao YH, Chen RL, Chen BW, Wang JL, Hsieh YL, Liao YM, Yang SH, Chang WH, Chao YY, Liang DC. Treatment for childhood acute lymphoblastic leukemia in Taiwan: Taiwan Pediatric Oncology Group ALL-2002 study emphasizing optimal reinduction therapy and central nervous system preventive therapy without cranial radiation. Pediatr Blood Cancer. 2017;64:234–241. doi: 10.1002/pbc.26142. [DOI] [PubMed] [Google Scholar]
- 36.Avigad S, Verly IR, Lebel A, Kordi O, Shichrur K, Ohali A, Hameiri-Grossman M, Kaspers GJ, Cloos J, Fronkova E, Trka J, Luria D, Kodman Y, Mirsky H, Gaash D, Jeison M, Avrahami G, Elitzur S, Gilad G, Stark B, Yaniv I. miR expression profiling at diagnosis predicts relapse in pediatric precursor B-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2016;55:328–339. doi: 10.1002/gcc.22334. [DOI] [PubMed] [Google Scholar]
- 37.Zamani A, Fattahi Dolatabadi N, Houshmand M, Nabavizadeh N. miR-324-3p and miR-508-5p expression levels could serve as potential diagnostic and multidrug-resistant biomarkers in childhood acute lymphoblastic leukemia. Leuk Res. 2021;109:106643. doi: 10.1016/j.leukres.2021.106643. [DOI] [PubMed] [Google Scholar]
- 38.Piatopoulou D, Avgeris M, Drakaki I, Marmarinos A, Xagorari M, Baka M, Pourtsidis A, Kossiva L, Gourgiotis D, Scorilas A. Clinical utility of miR-143/miR-182 levels in prognosis and risk stratification specificity of BFM-treated childhood acute lymphoblastic leukemia. Ann Hematol. 2018;97:1169–1182. doi: 10.1007/s00277-018-3292-y. [DOI] [PubMed] [Google Scholar]
- 39.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Br J Cancer. 2015;112:251–259. doi: 10.1038/bjc.2014.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lv M, Zhu S, Peng H, Cheng Z, Zhang G, Wang Z. B-cell acute lymphoblastic leukemia-related microRNAs: uncovering their diverse and special roles. Am J Cancer Res. 2021;11:1104–1120. [PMC free article] [PubMed] [Google Scholar]
- 41.Cammarata G, Augugliaro L, Salemi D, Agueli C, Rosa ML, Dagnino L, Civiletto G, Messana F, Marfia A, Bica MG, Cascio L, Floridia PM, Mineo AM, Russo M, Fabbiano F, Santoro A. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am J Hematol. 2010;85:331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 42.Fulci V, Colombo T, Chiaretti S, Messina M, Citarella F, Tavolaro S, Guarini A, Foà R, Macino G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009;48:1069–1082. doi: 10.1002/gcc.20709. [DOI] [PubMed] [Google Scholar]
- 43.Cario G, Stanulla M, Fine BM, Teuffel O, Neuhoff NV, Schrauder A, Flohr T, Schäfer BW, Bartram CR, Welte K, Schlegelberger B, Schrappe M. Distinct gene expression profiles determine molecular treatment response in childhood acute lymphoblastic leukemia. Blood. 2005;105:821–826. doi: 10.1182/blood-2004-04-1552. [DOI] [PubMed] [Google Scholar]
- 44.Yang YL, Lin SR, Chen JS, Lin SW, Yu SL, Chen HY, Yen CT, Lin CY, Lin JF, Lin KH, Jou ST, Hu CY, Chang SK, Lu MY, Chang HH, Chang WH, Lin KS, Lin DT. Expression and prognostic significance of the apoptotic genes BCL2L13, Livin, and CASP8AP2 in childhood acute lymphoblastic leukemia. Leuk Res. 2010;34:18–23. doi: 10.1016/j.leukres.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 45.Holleman A, den Boer ML, Kazemier KM, Janka-Schaub GE, Pieters R. Resistance to different classes of drugs is associated with impaired apoptosis in childhood acute lymphoblastic leukemia. Blood. 2003;102:4541–4546. doi: 10.1182/blood-2002-11-3612. [DOI] [PubMed] [Google Scholar]
- 46.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou X, Li J, Liu H, Pui CH, Evans WE, Naeve C, Wong L, Downing JR. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 47.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJP, Kazemier KM, Pei D, Cheng C, Pui CH, Relling MV, Janka-Schaub GE, Pieters R, Evans WE. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. New Engl J Med. 2004;351:533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


