Abstract
It has been shown that several ribonuclease (RNase) A superfamily proteins serve as ligands of receptor tyrosine kinases (RTKs), representing a new concept for ligand/receptor interaction. Moreover, recent studies indicate high clinical values for this type of ligand/RTK interactions. However, there is no structural report for this new family of ligand/receptor. In an attempt to understand how RNase and RTK may interact, we focused on the RNase1/ephrin type-A receptor 4 (EphA4) complex and predicted their structure by using the state-of-the-art machine learning method, AlphaFold and its derivative method, AF2Complex. In this model, electrostatic force plays an essential role for the specific ligand/receptor interaction. We found the R39 of RNase1 is the key residue for EphA4-binding and activation. Mutation on this residue causes disruption of an essential basic patch, resulting in weaker ligand-receptor association and leading to the loss of activation. By comparing the surface charge distribution of the RNase A superfamily, we found the positively charged residues on the RNase1 surface is more accessible for EphA4 forming salt bridges than other RNases. Furthermore, RNase1 binds to the ligand-binding domain (LBD) of EphA4, which is responsible for the traditional ligand ephrin-binding. Our model reveals the location of RNase1 on EphA4 partially overlaps with that of ephrin-A5, a traditional ligand of EphA4, suggesting steric hindrance as the basis by which the ephrin-A5 precludes interactions of RNase1 with EphA4. Together, our discovery of RNase1/EphA4 interface provides a potential treatment strategy by blocking the RNase1-EphA4 axis.
Keywords: RNase1, EphA4, AlphaFold, breast cancer
Introduction
Ribonuclease (RNase) is a large group of enzymes in both prokaryotic and eukaryotic cells that catalyze the hydrolysis of RNA into smaller pieces [1,2]. RNase can be divided into two main classes, endoribonucleases and exoribonucleases [3-5]. Endoribonuclease RNase1 belongs to the pancreatic RNase A superfamily and can be detected in human serum/plasma [6,7]. The studies of RNase in the last 50-60 years discovered numerous RNase family [8,9], however, the biological function of RNase1 have not yet been completely defined. Human RNase1 is not only essential for RNA clearance, but also involved in hemostasis, inflammation and innate immunity [7,10-13]. Recent studies indicated that RNase1 could serve as a soluble ligand for the receptor tyrosine kinase (RTK) ephrin type-A receptor 4 (EphA4) in breast cancer [14], RNase5 is an epidermal growth factor receptor (EGFR) ligand in pancreatic ductal adenocarcinoma (PDAC) [15], and RNase7 acts as a ligand of c-ros oncogene 1 (ROS1) in hepatocellular carcinoma (HCC) [16]. These findings uncover a non-canonical ligand-receptor relationship and the novel role of RNase A superfamily in tumor progression [17-19].
Erythropoietin-producing hepatocellular carcinoma (Eph) receptors are a large family of RTKs and are important in embryonic development and adult tissue homeostasis [20]. Eph receptors bind to their membrane-bound ephrin ligands on the neighboring cells to stimulate juxtacrine signals [21]. Notably, soluble ephrin-A ligands generated by metalloprotease have been detected in several cancer cell lines such as glioblastoma and breast cancer, where they activate EphA receptors and downstream signaling [22-25]. In addition, EphA4 is upregulated by the epithelial-mesenchymal transition (EMT) program in breast cancer stem cells (CSCs) and the juxtacrine signaling maintains the stem cell state through the physical interaction of tumour-associated monocytes and macrophages [26]. CSCs have been reported to promote tumorigenicity, invasiveness and immune evasion, and are ideal targets for cancer treatment [27-30]. Understanding the mechanism of autocrine/paracrine signaling involved in EphA4 activation through RNase1-binding may provide a broad vision of Eph receptor regulation and reveal therapeutic strategies against CSCs by blocking EphA4 signaling [14].
Although three RNase A superfamily proteins RNase1, RNase5 and RNase7 were reported as the respective ligand of EphA4, EGFR and ROS1, respectively [14-16], the structural information of these ligands/receptors complex is still awaiting to answer many important questions, such as the RNase-binding region on the receptor and the competition between the RNase and the canonical ligand. Moreover, the structural information could help us to design or find inhibitors to block the RNases/RTKs interactions in various of cancer cells, such as breast CSCs, PDAC and HCC in the future.
With the development of deep-learning neural networks, DeepMind made public an artificial intelligence (AI) tool called AlphaFold [31,32]. AlphaFold has been trained on hundreds of thousands of protein structures and sequences in structure database (i.e. Protein Data Bank) and genetic database. As we input a new sequence, it first searches homolog sequences in databases, suggesting the new sequence and homolog sequences are spatially close. This algorithm provides a distinctive way to estimate the distance of amino-acid pairs in the new sequence. Next, it repeatedly adjusts the 3D model based on the homolog sequence clues, continually updating its prediction. To evaluate the result, local-distance difference test (pLDDT) is used for per-residue accuracy, and usually pLDDT > 70 is confident and > 90 is highly confident [31-33].
A few months after AlphaFold publications [31,32], AF-Multimer was released for multimeric proteins complex prediction [34]. Besides, several teams aim to extend the usage of AlphaFold. ColabFold offered accelerated prediction of protein and complex structures by combining fast homology search [35]. AF2Complex optimized the algorithm by using multiple test sets and without using paired sequence alignments, significantly improved the AF-Multimer [36].
In this study, we took advantage of the structural prediction breakthrough of AlphaFold to analyze the possible RNase1/EphA4 interface and compared the ligands/receptor interfaces of RNase1 and ephrin to EphA4. We found the electrostatic force contributes to the RNase1/EphA4 association and the RNase1-binding region is close to the ephrin-binding pocket. Lastly, this structure may provide a platform for designing EphA4 inhibitors specific for RNase1 in the future.
Materials and methods
AlphaFold analysis
Prediction of human, mouse and horse RNase1/EphA4 LBD complexes as well as KYL peptide (KYLPYWPVLSSL)/EphA4 LBD complex were accomplished by AlphaFold version 2.1.2 [31,34], ColabFold [35] and AF2Complex version 1.2.2 [36] with default settings. The top pLDDT model from all predictions was analyzed in this study. The buried interface area and interface details of the complex were analyzed by PISA [37]. All protein structure graphic figures were made by PyMOL version 2.3 (Schrödinger, LLC).
Cloning, expression and purification of recombinant RNase1
We produced recombinant human RNase1 protein from E. coli by using a similar protein expression protocol referred to published studies [14]. The mutations R39A and R85A were introduced into pET15b-RNase1 wild type (WT) plasmid through site-directed mutagenesis by polymerase chain reaction (PCR). All constructs were verified by DNA sequencing. Briefly, the proteins were expressed in E. coli BL21 (DE3) strain by growing the cells in 1 L LB medium at 37°C until reaching an OD600 of 0.6-0.8 followed by induction with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The cells were then allowed to grow overnight at 16°C before harvest. The cells were lysed by sonication in buffer (20 mM Tris, 300 mM NaCl, pH 8.0) and the lysate spun down at 30,000 g on an Avanti J-26XP centrifuge (Beckman-Coulter, USA). The supernatant was loaded onto Ni-NTA resin (Qiagen, USA) pre-equilibrated with the loading buffer and eluted with the same buffer containing 250 mM imidazole. The purity of samples was checked with use of Coomassie-blue-stained sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and was > 90%.
Western blotting
BT549 breast cancer cells were serum-starved overnight and stimulated with/without 1 µg/ml of RNase1 proteins for 15 min. The cells were then harvested and 30 µg of total protein was used for immunoblotting. In brief, the protein lysates were separated by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. After blocking with 5% of skim milk for 30 min, the membranes were probed with the primary antibodies against EphA4 (sc-365503, Santa Cruz Biotechnology), EphA4-pY799 (EP2751, ECM biosciences) and α-Tubulin (#2144, Cell Signaling Technology). Following overnight incubation with the primary antibodies at 4°C, the membranes were incubated with the horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 hour at the room temperature. The signals were visualized with the Trident Western HRP Substrate (GeneTex) and detected by using the iBright FL1500 Imaging System (ThermoFisher Scientific).
Results
Electrostatic force contribution to RNase1-EphA4 complex formation
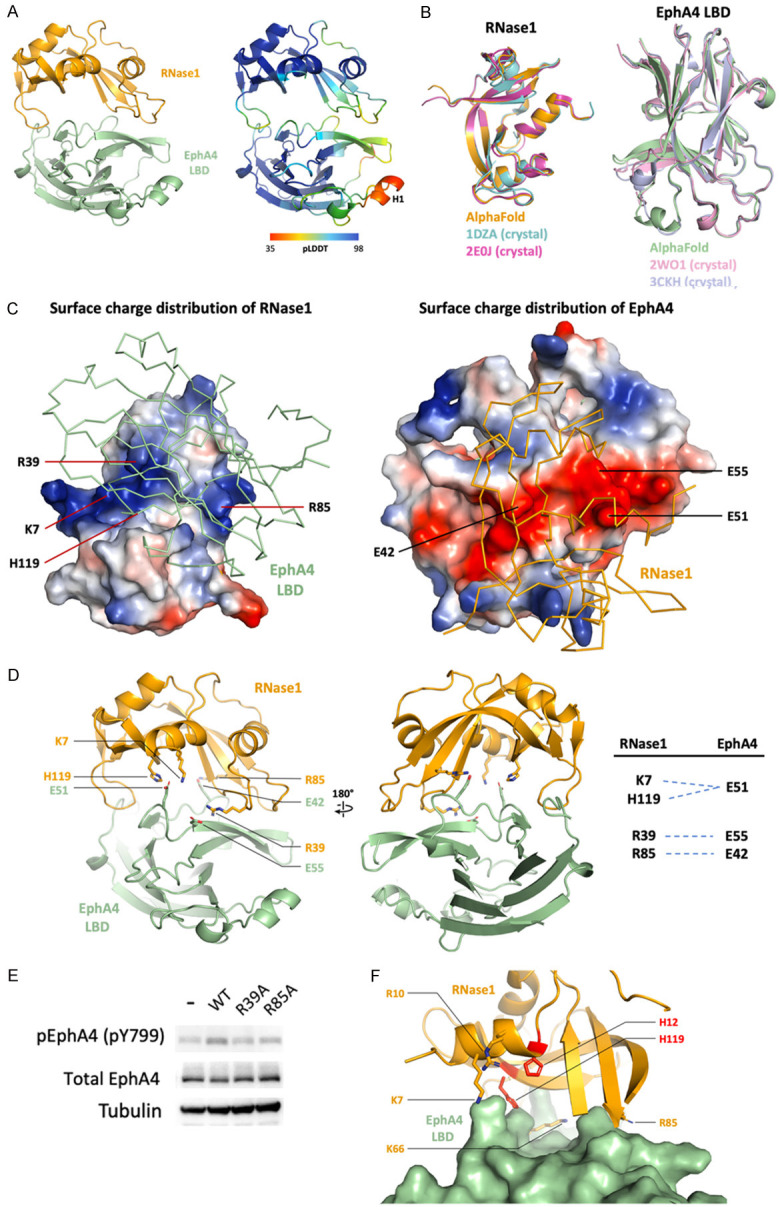
In order to predict the unbiased RNase1-EphA4 complex structure, we used AlphaFold [31], ColabFold [35] and AF2Complex [36] and compared the results parallelly. All of the predictions showed that RNase1 binds to the ligand-binding domain (LBD) of EphA4, so we input EphA4-LBD sequence together with RNase1 and predict the complex again. The output from AF2Complex gives the highest average pLDDT score, 83.64 (Figure 1A, right). In this model, 81.7% of residues are predicted to be confident (pLDDT > 70), except the H1 of EphA4 (residues 156 to 161), which was reported as a loop in many EphA4 structures (Protein Data Bank (PDB) ID: 2WO1, 3CKH and 4BK4) [38-40]. The overall structures of individual RNase1 and EphA4 are highly conserved with the previously resolved atomic-resolution structures (root-mean-square deviation (RMSD) values are closed to 0.5 Å) in the Protein Data Bank (Figure 1B). And the buried interface area between RNase1 and EphA4 is 933 Å2 as calculated with PISA [37], which is close to 12% of the total solvent-accessible area.
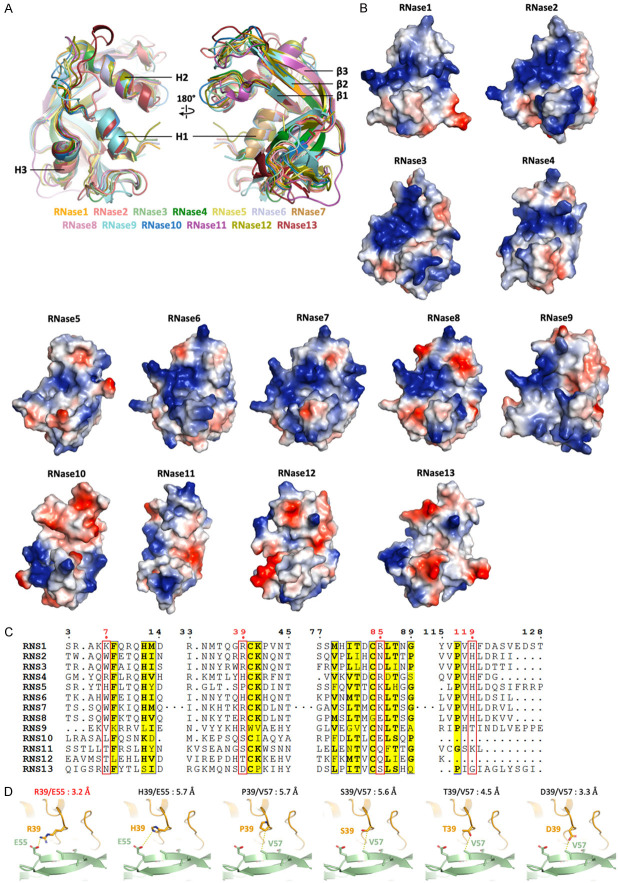
Figure 1.
AF2Complex prediction of RNase1/EphA4 LBD complex. A. Overall structures are colored by chains (orange: RNase1; green: EphA4 LBD) and pLDDT score (spectrum, ranged from 35 to 98). B. Superimposition of published RNase1 and EphA4 structures, respectively. Crystal structures of RNase1, PDB ID: 1DZA (cyan) and 2E0J (magenta) are superimposed to AlphaFold predicted RNase1 (orange). Crystal structures of EphA4 LBD, PDB ID: 2WO1 (pink) and 3CKH (light blue) are superimposed to AlphaFold predicted EphA4 LBD (green). C. Surface charge distribution of RNase1 and EphA4 shows the basic patch (blue) and acidic patch (red), respectively. Residues forming salt bridges are shown in sticks. D. Detailed map of RNase1 (orange)-EphA4 LBD (green) interaction. Salt bridges are listed in the panel. E. Phosphorylation of EphA4 Y799 stimulated by RNase1 WT, R39A and R85A in BT549 breast cancer cells. F. Surface representation of EphA4 showing RNA-binding residues of RNase1 are closed to the surface. Residues involving in RNA-binding (orange) and active site (red) are shown in sticks.
Electrostatic force contributes mainly to the RNase1/EphA4 complex formation. RNase1 surface possess a continuous basic patch (Figure 1C, left), which is suitable for the prolonged acidic patch on the EphA4 surface (Figure 1C, right). Four positively charged residues K7, R39, R85 and H119 of RNase1 form salt bridges with EphA4 negatively charged residues E42, E51 and E55 (Figure 1D). Most of these salt bridges observed in our model are highly conserved in mammals (K7, R85 and H119 in RNase1; E42, E51 and E55 in EphA4) (Figure 2A, 2B). Notably, R39 of the RNase1 is not conserved (S39 in mouse/rat and W39 in horse), indicating the salt bridge R39/E55 may be replaced by other types of non-covalent interaction in other species. To rule out this possibility, we predicted the mouse RNase1/EphA4 as well as horse RNase1/EphA4 by using AF2Complex. The alignment of the mouse and horse complexes shows that their interfaces are similar to human RNase1/EphA4 (Figure 2C, left). The S39 of mouse RNase1 forms a hydrogen bond with Q71 of EphA4, and W39 of horse RNase1 binds to hydrophobic residue V57 of EphA4 (Figure 2C, right), suggesting that the salt bridge R39/E55 is replaced by a hydrogen bond or hydrophobic interaction leading to variants of the interface. Nevertheless, the rest of the salt bridges are still present in the mouse and horse RNase1/EphA4 interface to stabilize the complex. In conclusion, RNase1 has potentiality as the ligand of EphA4 in other mammals with similar binding interface.
Figure 2.
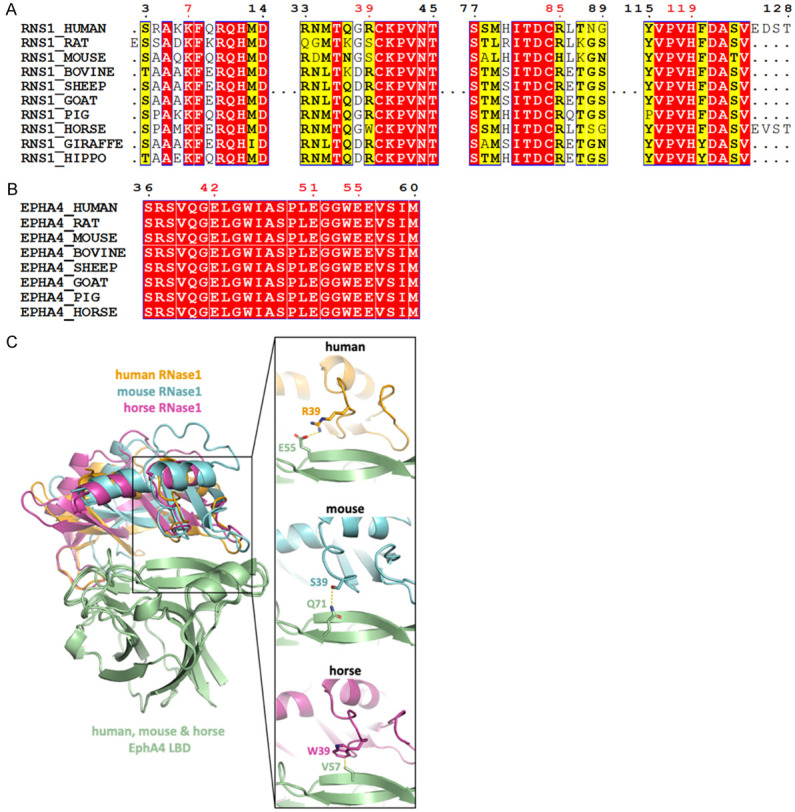
Residues forming salt bridges in the RNase1/EphA4 LBD complex are highly conserved. (A) Multiple sequence alignment of RNase1 among mammals. Sequences from UniProt [51] are RNS1_HUMAN (P07998), RNS1_RAT (P00684), RNS1_MOUSE (P00683), RNS1_BOVINE (P61823), RNS1_SHEEP (P67927), RNS1_GOAT (P67926), RNS1_PIG (P00671); RNS1_HORSE (P00674); RNS1_GIRAFFE (P00662) and RNS1_HIPPO (P00672). Sequence alignment produced by Clustal W [52] and drawn with ESPript [53]. Identities are boxed in red. Similar residues are written with black bold characters and boxed in yellow. Residues number 7, 39, 85 and 119 that are involved in EphA4-binding are labeled in red. (B) Multiple sequence alignment of EphA4 among mammals. Sequences from UniProt are EPHA4_HUMAN (P54764), EPHA4_RAT (D3ZZK3), EPHA4_MOUSE (Q03137), EPHA4_BOVINE (E1BER5), EPHA4_SHEEP (W5QGB9), EPHA4_GOAT (A0A452G0A9), EPHA4_PIG (F1SR66) and EPHA4_HORSE (F7AJ26). Sequence alignment is generated and labelled as in (A). Residues number 42, 51 and 55 that are involved in RNase1-binding are labeled in red. (C) AF2Complex prediction of mouse (cyan) and horse (magentas) RNase1/EphA4 LBD complexes. Superimposition of three complexes showing the salt bridge (R39/E55) in human (orange) is replaced by the hydrogen bond (S39/Q71) and hydrophobic interaction (W39/V57) in mouse and horse, respectively.
To validate the salt bridges observed in the prediction, we generate single mutation R39A or R85A of RNase1 to test if these mutations lost its EphA4-stimulating ability in breast cancer cells (Figure 1E). The result showed that WT RNase1 do stimulate EphA4 Y799 phosphorylation in BT549 cells as previously reported [14]. Mutation R39A obviously decreased the EphA4 phosphorylation level comparing to the WT, indicating the important role of salt bridge R39/E55. In addition, mutation R85A decreased the EphA4 phosphorylation level slightly. We speculate this is because of the basic patch selection. RNase1 surface possesses two basic patches (Figure 1C, left). K7, H119 and R39 are located in the larger basic patch and R85 in the smaller one. R39A disrupts the larger basic patch, which contributes to stronger electrostatic force, resulting in weaker ligand-receptor association and leading to the loss of activation.
It’s worth noting that RNase1 uses its RNA-binding interface to associate with EphA4. As the model shows, several RNA-binding residues are close to the EphA4 surface, such as K7, K66, R85 and H119 (Figure 1F), but the active site H12 is far from the EphA4 surface (about 6 Å), unlikely to participate in the EphA4-binding. In our previous study, the mutation H12A causes a catalytically inactive RNase1, but still binds to EphA4 and has the tumor-initiating capability [14]. This observation further supports our model, which shows H12 does not directly interact with EphA4.
Surface charge distribution of the RNases plays the key role in RTK receptor recognition
The overall architectures of RNase A superfamily are similar. We superimposed the structures/predictions from RNase1 to RNase13 and the RMSDs between these thirteen RNases are less than 1.0 Å (Figure 3A), showing high structural conservation of the RNase A superfamily. Although the overall structures within the superfamily are conserved, the surface charge distributions are variable (Figure 3B). The surfaces of the canonical RNases 1-8 contain continuous basic patches, except of RNase5, which exhibits extremely weak RNase enzymatic activity [41]. On the other hand, non-canonical members of the human RNases 9-13 harbor neither long basic patch nor RNase activities [6], indicating that both of the surface basic patch and the active site residues are important for enzymatic activity. Because RNase1, RNase5 and RNase7 can specifically bind to target RTKs (i.e. EphA4, EGFR and ROS1, respectively), the surface charge distribution among RNase A superfamily would play the key role for receptor-selection in different type of cells.
Figure 3.
Surface charge distribution of the RNases is related to its enzymatic activity and plays the key role in RTK receptor recognition. A. Superimposition of published crystal structures of RNase1 (PDB ID: 1DZA), RNase2 (PDB ID: 1HI2), RNase3 (PDB ID: 1DYT), RNase4 (PDB ID: 1RNF), RNase5 (PDB ID: 1ANG), RNase6 (PDB ID: 4X09), RNase7 (PDB ID: 2HKY), RNase8 (AlphaFold model), RNase9 (AlphaFold model), RNase10 (Phyre2 [54] model), RNase11 (Phyre2 model), RNase12 (AlphaFold model) and RNase13 (AlphaFold model). The highly conserved secondary structures are labeled. B. Surface charge distribution from RNase1 to RNase13 shows the positively- (blue) and negatively-charged (red) residues. Models are aligned to the same orientation. C. Multiple sequence alignment of human RNases 1-13. Sequences from UniProt are RNS1 (P07998), RNS2 (P10153), RNS3 (P12724), RNS4 (P34096), RNS5 (P03950), RNS6 (Q93091), RNS7 (Q9H1E1), RNS8 (Q8TDE3), RNS9 (P60153), RNS10 (Q5GAN6), RNS11 (Q8TAA1), RNS12 (Q5GAN4) and RNS13 (Q5GAN3). Sequence alignment is generated and labelled as in Figure 2A. Residues number 7, 39, 85 and 119 that involved in EphA4-binding are labeled in red. D. Comparison of the R39 of RNase1 (orange) and variants H39, P39, S39, T39 and D39 interact to E55 or V57 of EphA4 (green), respectively. The distance between two interactive residues is labelled.
Finally, the sequence alignment within human RNase A superfamily (RNases 1-13) shows that residues involving in EphA4-binding (except H119, the active site proton donor) are not conserved with other RNase A superfamily proteins (Figure 3C). R39 of RNase1, for example, can be histidine (H), proline (P), serine (S), threonine (T) or aspartic acid (D) in other RNases. We mutate R39 to five variant amino acids in the model to see if variants can still bind to EphA4 (Figure 3D). H39 may provide weaker electrostatic interaction because the distance to E55 of EphA4 is 5.7 Å. P39, S39, T39 and D39 may have van der Waals interaction with V57 and the distances ranged from 3.3 Å to 5.7 Å. These R39 variants may contribute weak to none interaction, denoting this key residue is important for corresponding EphA4 recognition.
RNase1- and ephrin-A5-binding site on EphA4 are partially overlapping
Several EphA4 and ephrin complex structures have been solved by using X-ray crystallography. Alignment of ephrin-A5/EphA4 LBD complex (PDB ID: 4BKA) and ephrin-B3/EphA4 LBD (PDB ID: 4BKF) complex shows that ephrin-A5 and B3 binding to the conserved pocket of EphA4 (Figure 4A) [40]. Next, to compare the binding sites of RNase1 and ephrin on EphA4, we superimposed the RNase1/EphA4 LBD model to the ephrin-A5/EphA4 LBD structure (Figure 4B). There is a steric hindrance region between RNase1 (residues 27-42 and 85-97) and ephrin-A5 (residues 36-39, 61-65 and 116-121) (Figure 4C) as they bind to EphA4, indicating that RNase1 and ephrin-A5 are exclusively complexed with EphA4. This structural information explains the ephrin-A5 precludes interactions of RNase1 with EphA4 [14].
Figure 4.
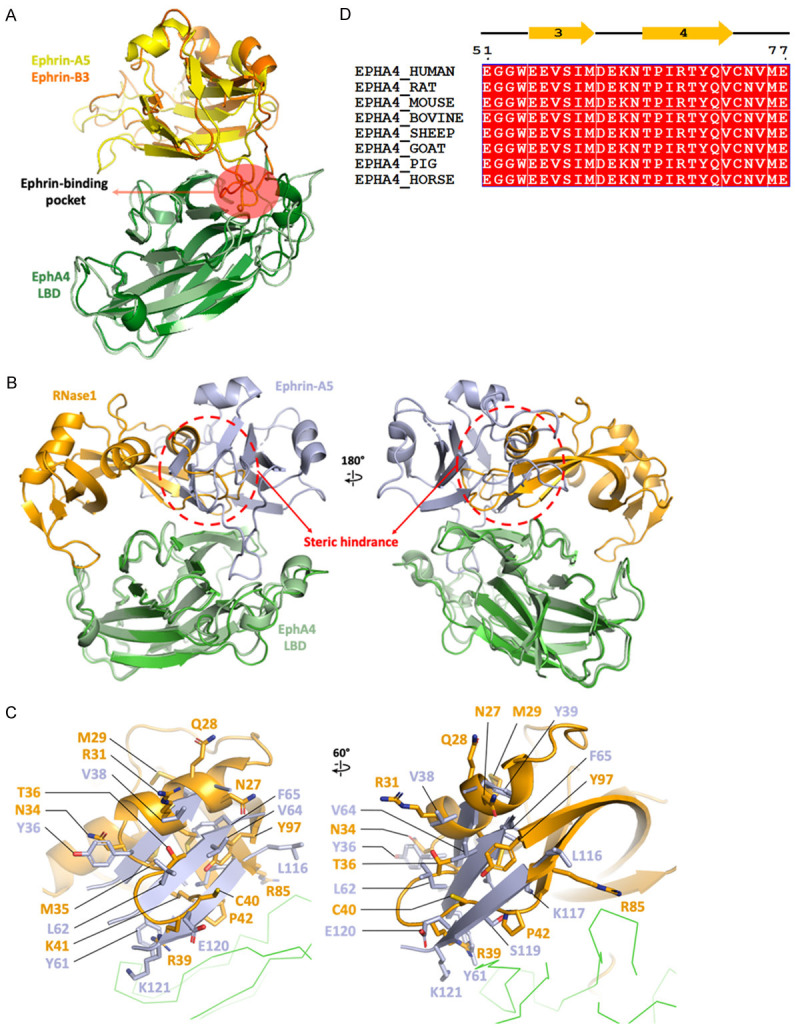
RNase1- and ephrin-A5-binding site on EphA4 are partially overlapping. A. Superimposition of published structures of ephrin-A5/EphA4 (PDB ID: 4BKA) and ephrin-B3/EphA4 (PDB ID: 4BKF) shows ephrin binding to the conserved ligand-binding pocket. Ephrin-A5, ephrin-B3 and EphA4 LBD are colored in yellow, orange and green, respectively. B. Superimposition of ephrin-A5/EphA4 (PDB ID: 4BKA) and RNase1/EphA4. The steric hindrance between ligands RNase1 and ephrin-A5 is indicated by red circle dashed. Ephrin-A5, RNase1 and EphA4 LBD are colored in light blue, orange and green, respectively. C. Details of the steric hindrance of RNase1 (orange) and ephrin-A5 (light blue). Ephrin-A5 sequence numbers are labelled as in PDB ID: 4BKA. D. Multiple sequence alignment of EphA4 from residue 51 to 77, involving β3-β4, are entirely conserved (red box). Sequences from UniProt are the same as Figure 2B.
From the previous studies, the binding affinity of RNase1/EphA4 is 92.4 nM [14] and ephrin-A5/EphA4 is 5.6 nM [14,42], showing 16.5-fold higher binding constant of ephrin-A5 to EphA4. To demonstrate the binding affinity disparity, we compared the interface area of RNase1/EphA4 with that of ephrin-A5/EphA4 by using PISA [37]. The interface area of RNase1/EphA4 LBD is 933 Å2 as described previously while that of ephrin-A5/EphA4 LBD is 1,204 Å2. Moreover, the number of residues in the interface are 25 in RNase1/EphA4 LBD and 38 in ephrin-A5/EphA4 LBD, respectively. The interface analysis shows the higher binding affinity of ephrin-A5/EphA4 is consistent to the structural feature.
The ligands overlapping region on the EphA4 surface is located in β3 and β4 (residues 55 to 72). And the sequence of this region is entirely conserved among mammals (Figure 4D), indicating β3-β4 is essential for ligand-binding, including both ephrin and RNase1. The mutation or blocking of β3-β4 has a potential to inhibit EphA4 in multiple ligand-dependent pathways.
KYL peptide blocks both RNase1 and ephrin-A5
The linear KYL peptide (KYLPYWPVLSSL) is a well-established EphA4 inhibitor, which specifically occupies the ligand-binding pocket of EphA4 and blocks the binding of ligands such as ephrin-A5 [43,44]. In our previous study, KYL also prevents the RNase1 binding to EphA4 [14]. To date, limiting structural information is available for studying KYL inhibition. More recently, the structure of a cyclic peptide antagonist, APY (APYCVYRGSWSC) complexed with EphA4 was solved by using X-ray crystallography [45] and another cyclic peptide TYY (CTYYWPLPC) docking to EphA4 was also analyzed by using Autodock4 program [46]. However, the sequence variation of these cyclic peptides may not well-explain the binding mode of KYL.
Here, we used AlphaFold to predict KYL/EphA4 LBD complex and demonstrated how the KYL inhibits EphA4 from ligands association. In this model, the KYL peptide occupies the same ligand-binding pocket of EphA4 (Figure 5A); nevertheless, the KYL peptide occupies larger surface (893 Å2) than APY (662 Å2) and creates additional steric hindrance on β5 (residues 101 to 105) of ephrin-A5 (Figure 5B, top).
Figure 5.
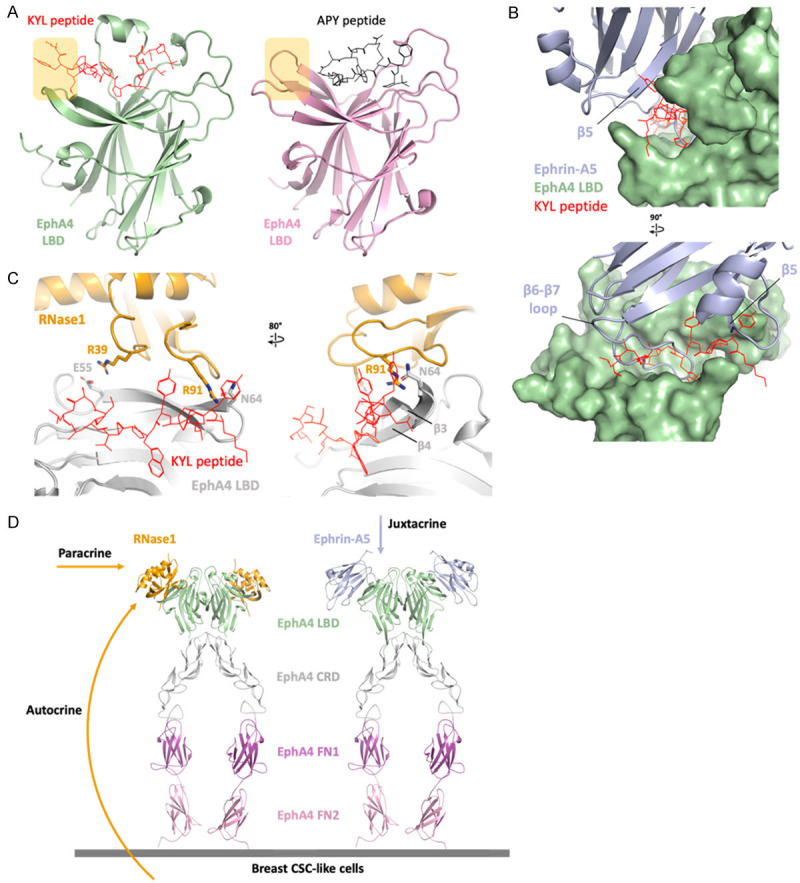
EphA4 inhibitors blocks ephrin and RNase1. A. Linear peptide KYL (red) and circular peptide APY (black) occupied the ephrin-binding site of EphA4 (AlphaFold model in green and crystal structure, PDB ID: 4W50, in pink). The extended region of the KYL peptide is indicated by yellow square. B. Superimposition of ephrin-A5/EphA4 LBD and KYL/EphA4 LBD. Surface representation of EphA4 (green) showing the KYL peptide (red) occupied the ephrin-A5 (light blue)-binding pocket. The KYL peptide blocks the β6-β7 loop as well as β5 of ephrin-A5. C. Superimposition of RNase1/EphA4 LBD and KYL/EphA4 LBD. The KYL peptide (red) blocks the hydrogen bond between R91 of RNase1 (orange stick) and N64 of EphA4 LBD (gray stick). This steric hindrance changed the conformation of β3-β4 of EphA4 and may also disrupt the salt bridge R39/E55. D. Docking the RNase1 (orange)/EphA4 LBD complex to the ephrin-A5 (light blue)/EphA4 extracellular region (ECR) complex (PDB ID: 4M4R) on the basis of the EphA4 LBD to demonstrate the role of RNase1 in paracrine/autocrine signaling. EphA4 ligand binding domain (LBD), cysteine-rich domain (CRD), fibronectin type III repeat 1 (FN1) and fibronectin type III repeat 2 (FN2) are colored in green, gray, magenta and pink, respectively.
As the KYL peptide binding to EphA4, the conformational change of β3-β4 disrupts the hydrogen bond between R91 of RNase1 and N64 of EphA4 and may also disrupt the salt bridge R39/E55, where E55 locates on the β3 (Figure 5C). However, comparing to ephrin-A5/EphA4 LBD complex, the KYL peptide blocks the majority of the ephrin-A5-binding region, which is a long loop between β6 and β7 of ephrin-A5 (Figure 5B, bottom). This structural information is consistent with our previous study that KYL could only partially compete with RNase1, but strongly abolish ephrin-A5 binding [43,44].
Discussion
In this study, we present the prediction of the RNase1/EphA4 LBD complex, showing the interface between the ligand-receptor is contributed mainly by four salt bridge pairs and hydrogen bonds (Table 1). We proved the R39 of RNase1 is the key residue for EphA4-binding and activation. R39A mutation disrupt a basic patch, which contributes to an essential electrostatic force, resulting in weaker ligand-receptor association and leading to the loss of activation. From the sequence alignment result, residues involved in electrostatic interaction for both the ligand and receptor are highly conserved among mammals, suggesting this novel ligand-receptor interface between RNases and RTKs could extend to other species. Besides, we found that the EphA4-binding site on RNase1 is highly overlapped with its RNA-binding site, indicating this basic patch is bifunctional in both RNA clearance and tumorigenesis. In our model, the active site proton acceptor H12 does not interact with EphA4; therefore, the catalytically inactive mutation H12A does not affect RNase1 to promote in vitro oncogenic transformation [14]. This finding is consistent to the fact that the oncogenic activities of all of the reported RNases (i.e. RNase1, RNase5 and RNase7) are ribonucleolytic activity-independent [14-16].
Table 1.
Interactional residues of RNase1 with EphA4
| Type | RNase1 residue (atom) | EphA4 residue (atom) |
|---|---|---|
| Salt bridge | K7 (NZ) | E51 (OE2) |
| Salt bridge | R39 (NH2) | E55 (OE2) |
| Salt bridge | R85 (NH2) | E42 (OE2) |
| Salt bridge | H119 (ND1) | E51 (OE1) |
| Hydrogen bond | P42 (O) | R68 (NH2) |
| Hydrogen bond | N67 (O) | S48 (OG) |
| Hydrogen bond | L86 (O) | R68 (NH2) |
| Hydrogen bond | R91 (NH2) | N64 (O) |
We also found that RNase1 and ephrin-A5 are exclusively complexed with EphA4 due to the steric hindrance, and the interface area buried in ephrin-A5/EphA4 LBD complex is 30% larger than that in RNase1/EphA4 LBD complex, leading to a 16.5-fold higher binding affinity [14]. However, the low binding affinity ligand like ephrin-B2 still plays an important role in neural development through EphA4 [38,47,48]. Moreover, weak affinity EGFR ligands such as epiregulin and epigen induce weaker EGFR dimerization, leading to cell differentiation rather than proliferation [49]. This evidence supports that ligands with low binding affinity can still have significant function.
To better understand the role of RNase1-mediated paracrine/autocrine signaling of EphA4, we dock the RNase1/EphA4 LBD complex to the ephrin-A5/EphA4 extracellular region (ECR) complex [50] on the basis of the EphA4 LBD (Figure 5D). Elevated serum RNase1 binds to EphA4 and triggers EphA4 activation in paracrine/autocrine manner in breast tumor cells, and then promotes the activity of breast tumor initiation [14]. On the other hand, classical ephrin ligands interact with EphA4 in a cell-cell contact and juxtacrine manner [21]. Thus, RNase1 and ephrins function in different settings, and RNase1 may play a critical role in breast cancer development and normal mammary tissue homeostasis due to its secretable nature.
Moreover, the structural basis of RNases and RTKs complexes may provide platforms for designing/finding inhibitors to block RNases/RTKs axes. Therefore, the study of structure of RNases/RTKs would lead novel therapeutic strategies against human malignancies. In addition, RNase1, 5 and 7 are natively secretory ligands, which can be developed to non-invasive diagnosis serum biomarkers for precision medicine as they are enriched in serum.
Acknowledgements
We thank the Ministry of Science and Technology (MOST), Taiwan, for grants MOST 110-2639-B-039-001-ASP (to M.-C.H.), MOST 110-2314-B-039-060 (to H.Y.), MOST 111-2813-C-039-074-B (to Y.-C.L.), MOST 110-2321-B-039-003 (to K.-C.H.), MOST 110-2314-B-039-010-MY2 (to K.-C.H.) and China Medical University for Yingtsai Scholar Award (CMU109-YT-06 to H.Y.).
Disclosure of conflict of interest
None.
References
- 1.Irie M. Structures and functions of ribonucleases. Yakugaku Zasshi. 1997;117:561–582. doi: 10.1248/yakushi1947.117.9_561. [DOI] [PubMed] [Google Scholar]
- 2.Barnard EA. Biological function of pancreatic ribonuclease. Nature. 1969;221:340–344. doi: 10.1038/221340a0. [DOI] [PubMed] [Google Scholar]
- 3.Stevens A, Niyogi SK. Hydrolysis of oligoribonucleotides by an enzyme fraction from Escherichia coli. Biochem Biophys Res Commun. 1967;29:550–555. doi: 10.1016/0006-291x(67)90520-7. [DOI] [PubMed] [Google Scholar]
- 4.Muller WE, Schroder HC, Arendes J, Steffen R, Zahn RK, Dose K. Alterations of activities of ribonucleases and polyadenylate polymerase in synchronized mouse L cells. Eur J Biochem. 1977;76:531–540. doi: 10.1111/j.1432-1033.1977.tb11623.x. [DOI] [PubMed] [Google Scholar]
- 5.Cuchillo CM, Nogues MV, Raines RT. Bovine pancreatic ribonuclease: fifty years of the first enzymatic reaction mechanism. Biochemistry. 2011;50:7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorrentino S. The eight human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett. 2010;584:2194–2200. doi: 10.1016/j.febslet.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Lu L, Li J, Moussaoui M, Boix E. Immune modulation by human secreted RNases at the extracellular space. Front Immunol. 2018;9:1012. doi: 10.3389/fimmu.2018.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koczera P, Martin L, Marx G, Schuerholz T. The ribonuclease A superfamily in humans: canonical RNases as the buttress of innate immunity. Int J Mol Sci. 2016;17:1278. doi: 10.3390/ijms17081278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta SK, Haigh BJ, Griffin FJ, Wheeler TT. The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun. 2013;19:86–97. doi: 10.1177/1753425912446955. [DOI] [PubMed] [Google Scholar]
- 10.Eller CH, Lomax JE, Raines RT. Bovine brain ribonuclease is the functional homolog of human ribonuclease 1. J Biol Chem. 2014;289:25996–26006. doi: 10.1074/jbc.M114.566166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zernecke A, Preissner KT. Extracellular Ribonucleic Acids (RNA) enter the stage in cardiovascular disease. Circ Res. 2016;118:469–479. doi: 10.1161/CIRCRESAHA.115.307961. [DOI] [PubMed] [Google Scholar]
- 12.Simsekyilmaz S, Cabrera-Fuentes HA, Meiler S, Kostin S, Baumer Y, Liehn EA, Weber C, Boisvert WA, Preissner KT, Zernecke A. Role of extracellular RNA in atherosclerotic plaque formation in mice. Circulation. 2014;129:598–606. doi: 10.1161/CIRCULATIONAHA.113.002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabrera-Fuentes HA, Ruiz-Meana M, Simsekyilmaz S, Kostin S, Inserte J, Saffarzadeh M, Galuska SP, Vijayan V, Barba I, Barreto G, Fischer S, Lochnit G, Ilinskaya ON, Baumgart-Vogt E, Boning A, Lecour S, Hausenloy DJ, Liehn EA, Garcia-Dorado D, Schluter KD, Preissner KT. RNase1 prevents the damaging interplay between extracellular RNA and tumour necrosis factor-alpha in cardiac ischaemia/reperfusion injury. Thromb Haemost. 2014;112:1110–1119. doi: 10.1160/TH14-08-0703. [DOI] [PubMed] [Google Scholar]
- 14.Lee HH, Wang YN, Yang WH, Xia W, Wei Y, Chan LC, Wang YH, Jiang Z, Xu S, Yao J, Qiu Y, Hsu YH, Hwang WL, Yan M, Cha JH, Hsu JL, Shen J, Ye Y, Wu X, Hou MF, Tseng LM, Wang SC, Pan MR, Yang CH, Wang YL, Yamaguchi H, Pang D, Hortobagyi GN, Yu D, Hung MC. Human ribonuclease 1 serves as a secretory ligand of ephrin A4 receptor and induces breast tumor initiation. Nat Commun. 2021;12:2788. doi: 10.1038/s41467-021-23075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang YN, Lee HH, Chou CK, Yang WH, Wei Y, Chen CT, Yao J, Hsu JL, Zhu C, Ying H, Ye Y, Wang WJ, Lim SO, Xia W, Ko HW, Liu X, Liu CG, Wu X, Wang H, Li D, Prakash LR, Katz MH, Kang Y, Kim M, Fleming JB, Fogelman D, Javle M, Maitra A, Hung MC. Angiogenin/Ribonuclease 5 is an EGFR ligand and a serum biomarker for erlotinib sensitivity in pancreatic cancer. Cancer Cell. 2018;33:752–769. e758. doi: 10.1016/j.ccell.2018.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu C, Zha Z, Zhou C, Chen Y, Xia W, Wang YN, Lee HH, Yin Y, Yan M, Chang CW, Chan LC, Qiu Y, Li H, Li CW, Hsu JM, Hsu JL, Wang SC, Ren N, Hung MC. Ribonuclease 7-driven activation of ROS1 is a potential therapeutic target in hepatocellular carcinoma. J Hepatol. 2021;74:907–918. doi: 10.1016/j.jhep.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Hu GF. Emerging role of angiogenin in stress response and cell survival under adverse conditions. J Cell Physiol. 2012;227:2822–2826. doi: 10.1002/jcp.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang YN, Lee HH, Hung MC. A novel ligand-receptor relationship between families of ribonucleases and receptor tyrosine kinases. J Biomed Sci. 2018;25:83. doi: 10.1186/s12929-018-0484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HH, Wang YN, Hung MC. Functional roles of the human ribonuclease A superfamily in RNA metabolism and membrane receptor biology. Mol Aspects Med. 2019;70:106–116. doi: 10.1016/j.mam.2019.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arvanitis D, Davy A. Eph/ephrin signaling: networks. Genes Dev. 2008;22:416–429. doi: 10.1101/gad.1630408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lisabeth EM, Falivelli G, Pasquale EB. Eph receptor signaling and ephrins. Cold Spring Harb Perspect Biol. 2013;5:a009159. doi: 10.1101/cshperspect.a009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ieguchi K, Omori T, Komatsu A, Tomita T, Deguchi A, Maru Y. Ephrin-A1 expression induced by S100A8 is mediated by the toll-like receptor 4. Biochem Biophys Res Commun. 2013;440:623–629. doi: 10.1016/j.bbrc.2013.09.119. [DOI] [PubMed] [Google Scholar]
- 23.Ieguchi K, Tomita T, Omori T, Komatsu A, Deguchi A, Masuda J, Duffy SL, Coulthard MG, Boyd A, Maru Y. ADAM12-cleaved ephrin-A1 contributes to lung metastasis. Oncogene. 2014;33:2179–2190. doi: 10.1038/onc.2013.180. [DOI] [PubMed] [Google Scholar]
- 24.Yang WH, Cha JH, Xia W, Lee HH, Chan LC, Wang YN, Hsu JL, Ren G, Hung MC. Juxtacrine signaling inhibits antitumor immunity by upregulating PD-L1 expression. Cancer Res. 2018;78:3761–3768. doi: 10.1158/0008-5472.CAN-18-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cha JH, Chan LC, Wang YN, Chu YY, Wang CH, Lee HH, Xia W, Shyu WC, Liu SP, Yao J, Chang CW, Cheng FR, Liu J, Lim SO, Hsu JL, Yang WH, Hortobagyi GN, Lin C, Yang L, Yu D, Jeng LB, Hung MC. Ephrin receptor A10 monoclonal antibodies and the derived chimeric antigen receptor T cells exert an antitumor response in mouse models of triple-negative breast cancer. J Biol Chem. 2022;298:101817. doi: 10.1016/j.jbc.2022.101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu H, Clauser KR, Tam WL, Frose J, Ye X, Eaton EN, Reinhardt F, Donnenberg VS, Bhargava R, Carr SA, Weinberg RA. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat Cell Biol. 2014;16:1105–1117. doi: 10.1038/ncb3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y, Shin JH, Longmire M, Wang H, Kohrt HE, Chang HY, Sunwoo JB. CD44+ cells in head and neck squamous cell carcinoma suppress T-cell-mediated immunity by selective constitutive and inducible expression of PD-L1. Clin Cancer Res. 2016;22:3571–3581. doi: 10.1158/1078-0432.CCR-15-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu JM, Xia W, Hsu YH, Chan LC, Yu WH, Cha JH, Chen CT, Liao HW, Kuo CW, Khoo KH, Hsu JL, Li CW, Lim SO, Chang SS, Chen YC, Ren GX, Hung MC. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 2018;9:1908. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badve S, Nakshatri H. Breast-cancer stem cells-beyond semantics. Lancet Oncol. 2012;13:e43–48. doi: 10.1016/S1470-2045(11)70191-7. [DOI] [PubMed] [Google Scholar]
- 30.Chang CJ, Yang JY, Xia W, Chen CT, Xie X, Chao CH, Woodward WA, Hsu JM, Hortobagyi GN, Hung MC. EZH2 promotes expansion of breast tumor initiating cells through activation of RAF1-beta-catenin signaling. Cancer Cell. 2011;19:86–100. doi: 10.1016/j.ccr.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Zidek A, Bridgland A, Cowie A, Meyer C, Laydon A, Velankar S, Kleywegt GJ, Bateman A, Evans R, Pritzel A, Figurnov M, Ronneberger O, Bates R, Kohl SAA, Potapenko A, Ballard AJ, Romera-Paredes B, Nikolov S, Jain R, Clancy E, Reiman D, Petersen S, Senior AW, Kavukcuoglu K, Birney E, Kohli P, Jumper J, Hassabis D. Highly accurate protein structure prediction for the human proteome. Nature. 2021;596:590–596. doi: 10.1038/s41586-021-03828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, Zidek A, Green T, Tunyasuvunakool K, Petersen S, Jumper J, Clancy E, Green R, Vora A, Lutfi M, Figurnov M, Cowie A, Hobbs N, Kohli P, Kleywegt G, Birney E, Hassabis D, Velankar S. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, Ronneberger O, Bodenstein S, Zielinski M, Bridgland A, Potapenko A, Cowie A, Tunyasuvunakool K, Jain R, Clancy E, Kohli P, Jumper J, Hassabis D. Protein complex prediction with AlphaFold-multimer. bioRxiv. 2021 [Google Scholar]
- 35.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;19:679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao M, Nakajima An D, Parks JM, Skolnick J. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat Commun. 2022;13:1744. doi: 10.1038/s41467-022-29394-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 38.Bowden TA, Aricescu AR, Nettleship JE, Siebold C, Rahman-Huq N, Owens RJ, Stuart DI, Jones EY. Structural plasticity of eph receptor A4 facilitates cross-class ephrin signaling. Structure. 2009;17:1386–1397. doi: 10.1016/j.str.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin H, Shi J, Noberini R, Pasquale EB, Song J. Crystal structure and NMR binding reveal that two small molecule antagonists target the high affinity ephrin-binding channel of the EphA4 receptor. J Biol Chem. 2008;283:29473–29484. doi: 10.1074/jbc.M804114200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seiradake E, Schaupp A, del Toro Ruiz D, Kaufmann R, Mitakidis N, Harlos K, Aricescu AR, Klein R, Jones EY. Structurally encoded intraclass differences in EphA clusters drive distinct cell responses. Nat Struct Mol Biol. 2013;20:958–964. doi: 10.1038/nsmb.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shapiro R, Fett JW, Strydom DJ, Vallee BL. Isolation and characterization of a human colon carcinoma-secreted enzyme with pancreatic ribonuclease-like activity. Biochemistry. 1986;25:7255–7264. doi: 10.1021/bi00371a002. [DOI] [PubMed] [Google Scholar]
- 42.Flanagan JG, Vanderhaeghen P. The ephrins and Eph receptors in neural development. Annu Rev Neurosci. 1998;21:309–345. doi: 10.1146/annurev.neuro.21.1.309. [DOI] [PubMed] [Google Scholar]
- 43.Lamberto I, Qin H, Noberini R, Premkumar L, Bourgin C, Riedl SJ, Song J, Pasquale EB. Distinctive binding of three antagonistic peptides to the ephrin-binding pocket of the EphA4 receptor. Biochem J. 2012;445:47–56. doi: 10.1042/BJ20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murai KK, Nguyen LN, Koolpe M, McLennan R, Krull CE, Pasquale EB. Targeting the EphA4 receptor in the nervous system with biologically active peptides. Mol Cell Neurosci. 2003;24:1000–1011. doi: 10.1016/j.mcn.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Lamberto I, Lechtenberg BC, Olson EJ, Mace PD, Dawson PE, Riedl SJ, Pasquale EB. Development and structural analysis of a nanomolar cyclic peptide antagonist for the EphA4 receptor. ACS Chem Biol. 2014;9:2787–2795. doi: 10.1021/cb500677x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han X, Xu Y, Yang Y, Xi J, Tian W, Duggineni S, Huang Z, An J. Discovery and characterization of a novel cyclic peptide that effectively inhibits ephrin binding to the EphA4 receptor and displays anti-angiogenesis activity. PLoS One. 2013;8:e80183. doi: 10.1371/journal.pone.0080183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brors D, Bodmer D, Pak K, Aletsee C, Schafers M, Dazert S, Ryan AF. EphA4 provides repulsive signals to developing cochlear ganglion neurites mediated through ephrin-B2 and -B3. J Comp Neurol. 2003;462:90–100. doi: 10.1002/cne.10707. [DOI] [PubMed] [Google Scholar]
- 48.Lackmann M, Boyd AW. Eph, a protein family coming of age: more confusion, insight, or complexity? Sci Signal. 2008;1:re2. doi: 10.1126/stke.115re2. [DOI] [PubMed] [Google Scholar]
- 49.Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, Valley CC, Ferguson KM, Leahy DJ, Lidke DS, Lemmon MA. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 2017;171:683–695. e618. doi: 10.1016/j.cell.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu K, Tzvetkova-Robev D, Xu Y, Goldgur Y, Chan YP, Himanen JP, Nikolov DB. Insights into Eph receptor tyrosine kinase activation from crystal structures of the EphA4 ectodomain and its complex with ephrin-A5. Proc Natl Acad Sci U S A. 2013;110:14634–14639. doi: 10.1073/pnas.1311000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]