Abstract
Background: DNA methylation controls the transcription of genes and is involved in the development of lung cancer. Our preliminary bioinformatics prediction revealed that sperm associated antigen 6 (SPAG6) was considerably hypermethylated in lung squamous cell carcinoma (LUSC). Thus, this study aimed to probe the mechanism underlying its hypermethylation. Methods: The effect of DNA methylation of SPAG6 on its expression in LUSC was analyzed. The contributors to SPAG6 DNA hypermethylation were sought. CCK-8, EdU, and Transwell assays were carried out to assess the malignant phenotype of LUSC cells. KEGG pathway enrichment analysis was used to screen for pathways affected by SPAG6, which were confirmed by dual-luciferase assays. Bioinformatics analysis was conducted to dissect the impact of SPAG6 on the immune response and cancer cell stemness in LUSC. Results: DNA methyltransferase 3b (DNMT3b)-mediated hypermethylation of the SPAG6 promoter in LUSC led to SPAG6 downregulation. SPAG6 reverted the malignant phenotype of LUSC cells. SPAG6 regulated the JAK/STAT pathway by inhibiting the transcription of STAT1 and STAT3. The expression of SPAG6 was positively related to immune infiltration in LUSC and inversely related to the expressions of the immunosuppressive genes CTLA4 and PDCD1. SPAG6 expression was negatively correlated with cancer cell stemness in LUSC, and its expression inhibited the expressions of Nanog, ALDH1, and Sox2, markers of cancer cell stemness. Conclusions: DNMT3b-mediated SPAG6 promoter hypermethylation activates the JAK/STAT pathway to promote LUSC progression.
Keywords: Lung squamous carcinoma, DNMT3b, SPAG6, JAK/STAT pathway, DNA methylation
Introduction
Lung cancer is a major contributor to cancer-related mortality. The 5-year overall survival of non-small cell lung cancer, the most common subtype, remains at 15% in spite of continuous research and development of novel therapeutic regimens [1]. Non-small cell lung cancer is histologically categorized into lung squamous cell carcinoma (LUSC), adenocarcinoma, and large-cell carcinoma [2]. LUSC, the most prevalent form of non-small cell lung cancer (around 30%), is also characterized by tumor heterogeneity, genetic mutations, cancer stem cells (CSCs), immune-resistance and chemoresistance [3,4]. Novel therapeutic biomarkers and strategies therefore, must be discovered to cure LUSC.
Epigenetic modification, predominantly DNA methylation, has been implicated in genomic instability in LUSC [5]. Models based on differences in DNA methylation might be beneficial to classifying the molecular subtypes of LUSC and to offering customized treatments and prognostic markers [6]. Moreover, 11 DNA methylation signatures have shown substantial sensitivity and specificity in predicting the overall survival of patients with LUSC [7]. Similarly, we obtained two cytosine and guanine (CpG) islands, cg14055896 and cg10648197 which might serve as biomarkers in LUSC using two bioinformatic algorithms. Only sperm-associated antigen 6 (SPAG6), the host gene of cg10648197, was predicted to be significantly downregulated in LUSC. SPAG6 was first identified in human testicular tissues and recognized as a cancer-testis antigen [8]. Promoter-associated CpG island methylation of the SPAG6 gene was frequently observed in bladder cancer [9]. More importantly, depletion of SPAG6 was observed in primary tumor tissues relative to matched non-malignant lung tissues of NSCLC patients, and SPAG6 is tumor-specifically methylated in NSCLC [10]. However, the modifiers that contribute to downregulation and hypermethylation of SPAG6 in NSCLC remains unclear. The methylated cytosines are almost completely produced at the CpG (5’) dinucleotide sequence, and CpG methylation is catalyzed by DNA methyltransferase proteins (DNMTs) [11]. Further prediction results in the above study showed that DNMT3b shared a binding relation with SPAG6. DNMT3b is a main DNA methyltransferase, and DNMT3b serves as a major enzyme fueling methylation of intragenic regions of genes [12]. Taken together, we hypothesized that DNMT3b modulates SPAG6 promoter methylation to downregulate its expression, thereby resulting in LUSC progression. This study sought to substantiate the mechanism for the hypermethylation of SPAG6 promoter and its involvement in the LUSC cell behavior in vitro.
Materials and methods
Publicly available databases
The hypermethylated CpG islands on the genomic chromosomes of LUSC patients, the methylation status of SPAG6 CpG islands, and the correlation between the methylation levels and the expression of SPAG6 were evaluated using SMART (http://www.bioinfo-zs.com/smartapp/). MethSurv (https://biit.cs.ut.ee/methsurv/) was applied to predict the CpG islands that can serve as LUSC biomarkers and to analyze the correlation between CpG island methylation profiles and clinical characteristics of patients. The gene expression in LUSC and the correlation between expressions of SPAG6 and the DNMTs were predicted using GEPIA (http://gepia.cancer-pku.cn/index.html). The relation between SPAG6 expression and patients’ survival was assessed using PROGgeneV2 (http://www.progtools.net/gene/index.php). The chromatin immunoprecipitation (ChIP)-sequencing (seq) database Cistrome Data Browser (http://cistrome.org/db/#/) was used to study the binding of DNMT3b to the SPAG6 promoter. Analysis of factors that were significantly correlated with SPAG6 expression in LUSC was carried out via UALCAN (http://ualcan.path.uab.edu/index.html). KEGG pathway enrichment analysis (https://www.kegg.jp/brite/query=04024&htext=br08901.keg&option=-a&node_proc=br08901_org&proc_enabled=ko&panel=collapse) was used to predict pathways enriched by co-expressed factors with SPAG6. The association between SPAG6 expression and immune cell infiltration and the expression of immune regulatory factors was analyzed via CIBERSORT (https://cibersort.stanford.edu/). Sangerbox 3.0 (http://vip.sangerbox.com/) was queried to predict the correlation between SPAG6 expression and cancer cell stemness.
Patients and sample preparation
Tissues were collected after obtaining permission from the institutional review board of the First Affiliated Hospital of Guangxi Medical University and informed consent from each recruited patient. Tumor tissues and adjacent tissues from 45 LUSC patients who underwent surgery at the First Affiliated Hospital of Guangxi Medical University from March 2019 to September 2020 were obtained. None of the recruited participants had received radiotherapy, chemotherapy, targeted therapy, or immunotherapy.
Cells and culture conditions
LUSC cell line NCI-H1703 (CL-0390), SK-MES-1 (CL-0213) and normal lung epithelial cells BEAS-2B (CL-0496) were obtained from Procell (Wuhan, Hubei, China). All cells were cultivated in RPMI-1640 (Gibco, Carlsbad, CA, USA) medium plus 10% FBS and 1% penicillin-streptomycin.
Decitabine (catalog number: S1200, Selleck, Houston, TX, USA) at 0.5 μM was used for a 24-h treatment. The dimethylsulfoxide (DMSO)-treated cells were set as a control. VectorBuilder (Guangzhou, Guangdong, China) provided the overexpressed DNA plasmid of SPAG6 (oe-SPAG6, accession number: NM_012443.4), short hairpin RNA (shRNA) of DNMT3b (sh-DNMT3b, sequence: 5’-ACACGCAACCAGTGGTTAATACTCGAGTATTAACCACTGGTTGCGTGT-3’) and their respective control (sh-NC sequence: 5’-GCAACAAGATGAAGAGCACCAA’). Lipofectamine 2000 was applied for all transfections according to the manufacturer’s protocol.
RT-qPCR
Total RNA from cells and tissues were isolated using TRIzol™ reagent (Thermo Fisher) and reverse transcribed into cDNA using StarScript IIIReverse Transcription Kit (GenStar BioSolutions, Beijing, China), then qPCR reactions were performed using RealStar Fast SYBR qPCR Mix (GenStar BioSolutions) on a StepOnePlus™ Real-Time Fluorescent qPCR System (Applied Biosystems, Inc., Foster City, CA, USA). All primers are listed in Table 1. The relative expression of mRNAs was calculated by 2ΔΔCT and normalized to β-actin mRNA.
Table 1.
RT-qPCR primers used in this study
| Gene | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| SPAG6 | GACAGTAGTGGATGCAGGAGCT | CCATTTCTGCCAGATCCACGGA |
| DNMT3b | TAACAACGGCAAAGACCGAGGG | TCCTGCCACAAGACAAACAGCC |
| Nanog | CTCCAACATCCTGAACCTCAGC | CGTCACACCATTGCTATTCTTCG |
| ALDH1 | CGGGAAAAGCAATCTGAAGAGGG | GATGCGGCTATACAACACTGGC |
| Sox2 | GCTACAGCATGATGCAGGACCA | TCTGCGAGCTGGTCATGGAGTT |
| β-actin | CACCATTGGCAATGAGCGGTTC | AGGTCTTTGCGGATGTCCACGT |
Note: SPAG6, Sperm Associated Antigen 6; DNMT3b, DNA-Methyltransferase 3b; ALDH1, Aldehyde Dehydrogenase 1.
Methylated DNA immunoprecipitation (MeDIP)
The DNA methylation level of SPAG6 promoter region in LUSC was measured using MeDIP kit (BersinBio, Guangzhou, Guangdong, China). For the assay in tissues, the tissues were cut into 0.3-0.5 cm3 size, homogenized with 0.5 mL Tris-ethylenediaminetetraacetic acid buffer, and centrifuged at 10,000 g for 5 min at room temperature. The cells (1 × 107 cells) were resuspended with DNA extraction buffer and lysed with 10% SDS and Protease K. DNA was then extracted and ultrasonically fragmented to 300-700 bp. The samples were divided into two parts: 0.1 mL (Input) and 0.8 mL (IP), and the Input was stored at -20°C. The DNA in the IP group was denatured at 95°C for 3 min, rapidly cooled on ice, and incubated overnight at 4°C with 3-5 μg of antibodies to 5-methylcytidine or negative antibody IgG. Protein A/G-beads were supplemented to collect the protein-DNA complexes and eluted. Then purified DNA was collected. The enrichment of the SPAG6 promoter sequence was analyzed by qPCR.
CCK-8
The proliferative capacity of LUSC cells was evaluated using CCK-8 (GK10001, Glpbio, Montclair, CA, USA). The transfected LUSC cells were cultured in 96-well plates at 3000 cells each well. Assays were conducted at appropriate time points (0, 24, 48, 72 h). After that, 10 μL of CCK8 reagent was supplemented to each well of the plate for a 2 h incubation. A microplate reader was applied to read optical density (OD) value at 450 nm.
EdU assay
The DNA synthesis of LUSC cells was assessed using Cell-Light EdU Apollo567 in Vitro Kit (C10310-1, RiboBio, Guangzhou, Guangdong, China). The transfected LUSC cells were plated in 96-well plates at 1 × 105 cells/well, and 50 μM EdU medium (100 μL) was supplemented for a 2 h incubation. The cells were fixed with 4% paraformaldehyde for 0.5 h at room temperature, permeabilized with 0.5% TritonX-100 for 10 min, and stained with 100 μL of Apollo® Staining Solution for 0.5 h at room temperature in the dark. After that, 100 μL of Hoechst 33342 Reaction Solution was supplemented into each well for a 30 min nuclear counter-staining at room temperature in darkness. Fluorescent staining was observed by confocal fluorescence microscopy (Carl Zeiss, Oberkochen, Germany).
Transwell assays
The migration and invasion of cells were measured using the Transwell® Cell Culture Insert (Corning Glass Works, Corning, N.Y., USA). When performing the invasion assay, the bottom of the apical chamber was pre-coated with Ceturegel™ Matrix LDEV-Free Matrigel (Yeasen, Shanghai, China). The apical chamber was loaded with 1 × 105 cells resuspended in serum-free medium, and the basolateral chamber was supplemented with complete medium plus 10% FBS. The cells were incubated at 37°C in 5% CO2 for 24 h. The cells on the upper surface were subsequently wiped off with a cotton swab, and the cells that migrated or invaded to the lower surface were fixed with 4% paraformaldehyde. After being stained with crystalline violet, the cells were viewed and counted under a microscope (Olympus Optical Co., Ltd., Tokyo, Japan). Matrigel pre-coating was not performed in the cell migration assay.
Dual-luciferase assay
The effect of SPAG6 on JAK/STAT pathway activity was assessed using dual-luciferase assays. STAT1 luciferase reporter plasmid (11504ES03) and STAT3 luciferase reporter plasmid (11503ES03) were purchased from Yeasen. The above luciferase reporter plasmids were co-transfected into LUSC cells with oe-NC or oe-SPAG6, respectively. After 48 h, a Dual Luciferase Reporter Gene Assay Kit (11402ES60, Yeasen) was applied to evaluate the luciferase activity of STAT1 and STAT3 reporter plasmids.
Statistical analyses
GraphPad Prism 8.0 (GraphPad, San Diego, CA, USA) was applied to conduct these analyses. The results were reported as mean ± SD. Statistical significance of differences between two groups was calculated using paired or unpaired t-test. To compare the differences among multiple groups, one-way or two-way ANOVA test was performed, followed by Tukey’s post-hoc test. Fisher’s exact test was used to compare the association of two categories in contingency tables, and Pearson’s correlation coefficient was utilized to analyze the correlation between two variables. Significance was set at P < 0.05.
Results
Reduced expression of SPAG6 in LUSC is associated with its DNA hypermethylation
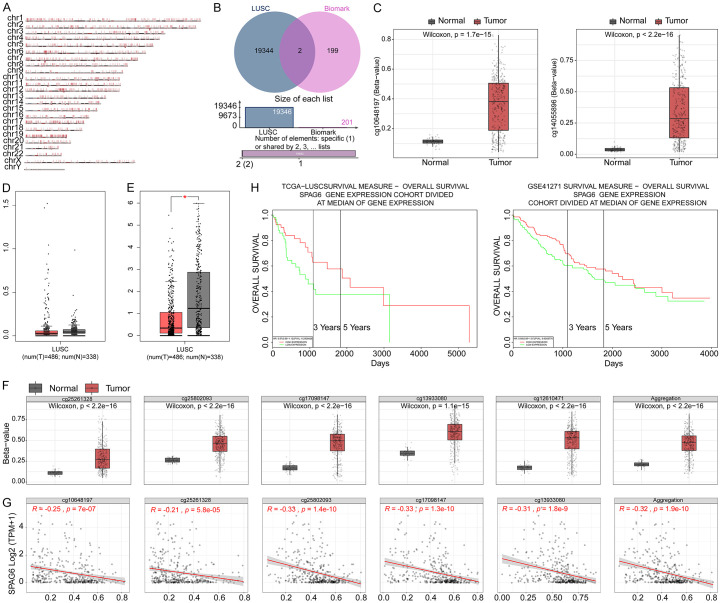
Hypermethylation data of CpG islands on the genomic chromosomes of LUSC patients were downloaded from SMART (Figure 1A). Subsequently, CpG islands that can be used as biomarkers for LUSC were downloaded from MethSurv. There are 2 intersecting CpG islands in both databases: cg14055896 and cg10648197 (Figure 1B). Both CpG islands had significantly elevated methylation levels in LUSC (Figure 1C). The expression of the host genes CACNG8 (Figure 1D) and SPAG6 (Figure 1E) in LUSC was analyzed in GEPIA. Only SPAG6 expression was downregulated in LUSC. The methylation level of the CpG island of SPAG6 was generally elevated in LUSC (Figure 1F), and the SPAG6 expression was inversely related to the methylation level of the CPG island (Figure 1G). In PROGgeneV2, patients with high SPAG6 expression had better survival than those with low SPAG6 expression (Figure 1H).
Figure 1.
Reduced expression of SPAG6 in LUSC is associated with its DNA hypermethylation. A: Hypermethylation of the genome of LUSC patients. B: Key hypermethylated CpG islands in LUSC. C: Methylation of cg14055896 and cg10648197 in LUSC. D: Querying the expression of CACNG8 in LUSC in GEPIA (T refers to tumor and N refers to normal). E: Querying the expression of SPAG6 in LUSC in GEPIA (T refers to tumor and N refers to normal). F: Methylation levels of CpG islands of SPAG6 in LUSC. G: Correlation between the expression of SPAG6 and the methylation level of CpG islands in LUSC. H: The prognostic significance of SPAG6 expression to the patients with LUSC. SPAG6, Sperm Associated Antigen 6; LUSC, Lung Squamous Cell Carcinoma; CpG, Cytosine and Guanine.
DNA methylation-mediated low expression of SPAG6 is associated with poor prognosis in patients with LUSC
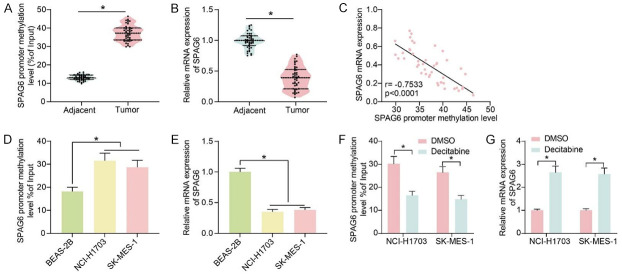
The DNA methylation levels of SPAG6 promoter region were measured by MeDIP in the collected LUSC tissues and adjacent tissues (Figure 2A). The DNA methylation of SPAG6 promoter was significantly elevated in LUSC tissues. A decrease in SPAG6 expression was noted in LUSC tissues (Figure 2B), and a substantial negative correlation between SPAG6 expression in LUSC tissues and their promoter DNA methylation levels was observed as well (Figure 2C). According to the mean value of SPAG6 expression in LUSC tissues (0.386), we divided the patients into low SPAG6 expression (n = 22) and high SPAG6 expression (n = 23) groups. Patients with low SPAG6 expression had higher tumor stage, poorer differentiation and lymph node metastasis (Table 2).
Figure 2.
SPAG6 promoter hypermethylation is associated with poor prognosis of LUSC patients. (A) DNA methylation levels in the SPAG6 promoter region in LUSC tissues and adjacent normal tissues (n = 45) by MeDIP assays. (B) Detection of SPAG6 expression in LUSC tissues and adjacent normal tissues by RT-qPCR (n = 45). (C) The correlation between SPAG6 expression in LUSC tissues and its promoter DNA methylation level (n = 45) by Pearson’s correlation analysis. (D) DNA methylation levels in the promoter region of SPAG6 in the LUSC cell lines NCI-H1703 and SK-MES-1 and human normal lung epithelial cells BEAS-2B by MeDIP. (E) Detection of SPAG6 expression in LUSC cell lines NCI-H1703 and SK-MES-1 and human normal lung epithelial cells BEAS-2B by RT-qPCR. (F) The effect of decitabine treatment on the DNA methylation level of SPAG6 in LUSC cells by MeDIP assay. (G) Effect of Decitabine treatment on the expression of SPAG6 in LUSC cells by RT-qPCR. Data represent mean ± SD from three independent experiments were analyzed using paired t-test (A, B) or one-way (D, E)/two-way ANOVA (F, G) (*P < 0.05). SPAG6, Sperm Associated Antigen 6; LUSC, Lung Squamous Cell Carcinoma.
Table 2.
Correlations between SPAG6 and clinical characteristics in 45 LUSC patients
| Clinicopathological parameters | Total patients | SPAG6 expression | p value | ||
|---|---|---|---|---|---|
|
|
|
||||
| (n = 45) | Low (n = 22) | High (n = 23) | |||
| Age | ≥ 60 | 34 | 18 | 16 | 0.491 |
| < 60 | 11 | 4 | 7 | ||
| Sex | Male | 36 | 19 | 17 | 0.459 |
| Female | 9 | 3 | 6 | ||
| Smoking history | Smokers | 33 | 15 | 18 | 0.514 |
| Nonsmokers | 12 | 7 | 5 | ||
| Tumor stage | I~II | 21 | 5 | 16 | 0.003 |
| III~IV | 24 | 17 | 7 | ||
| Differentiation | Well and moderate | 17 | 3 | 14 | 0.002 |
| Poor | 28 | 19 | 9 | ||
| Lymph node metastasis | Positive | 26 | 18 | 8 | 0.002 |
| Negative | 19 | 4 | 15 | ||
Note: SPAG6, sperm associated antigen 6; LUSC, lung squamous cell carcinoma. Fisher’s exact test was used to analyze the association of individual clinicopathological parameters.
The DNA methylation levels of the SPAG6 promoter region were examined using MeDIP in the LUSC cell lines NCI-H1703 and SK-MES-1 and human normal lung epithelial cells BEAS-2B. The DNA methylation levels of SPAG6 were also elevated in LUSC cells (Figure 2D). The mRNA expression of SPAG6 was reduced in LUSC cells (Figure 2E). Decitabine treatment significantly reduced the DNA methylation level of SPAG6 (Figure 2F) while it enhanced the expression of SPAG6 in LUSC cells (Figure 2G).
DNMT3b mediates SPAG6 promoter hypermethylation
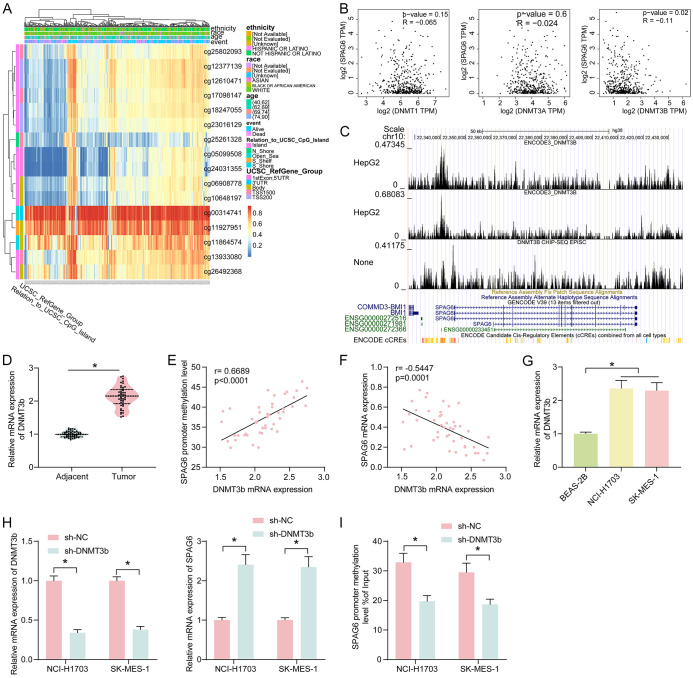
In MethSurv, we analyzed the correlation of CpG island methylation of SPAG6 with the physiological parameters of the patients (Figure 3A). The methylation level of SPAG6 in LUSC patients was not related to ethnicity, race and age. Therefore, we speculated that the cause of its DNA methylation is at the genetic level. Subsequently, the relation between the expression of DNMTs and SPAG6 expression was predicted in GEPIA (Figure 3B), and we only found a significantly negative correlation between DNMT3b and SPAG6 expression, so we speculated that DNMT3b mediated the DNA methylation in SPAG6. In the ChIP-seq database Cistrome Data Browser, we also confirmed the binding relationship between DNMT3b and SPAG6 promoter (Figure 3C).
Figure 3.
SPAG6 promoter methylation is regulated by DNMT3b. (A) Correlation between CpG island methylation status of SPAG6 and physiological data of patients. (B) Correlation between the expression of DNMTs and SPAG6 expression in LUSC was queried in GEPIA. (C) Binding relationship between DNMT3b and SPAG6 promoter region. (D) Detection of DNMT3b expression in LUSC tissues and adjacent normal tissues (n = 45) by RT-qPCR. (E) Correlation between DNMT3b expression and SPAG6 promoter DNA methylation levels in LUSC tissues (n = 45) by Person’s correlation analysis. (F) Correlation between DNMT3b expression and SPAG6 expression in LUSC tissues (n = 45) by Pearson’s correlation analysis. (G) Expression of DNMT3b in LUSC cell lines NCI-H1703 and SK-MES-1 and human normal lung epithelial cells BEAS-2B by RT-qPCR. (H) Effect of DNMT3b inhibition on the DNMT3b and SPAG6 expression in LUSC cell lines by RT-qPCR. (I) Effect of DNMT3b inhibition on the DNA methylation level of SPAG6 by MeDIP assay. Data represent mean ± SD from three independent experiments and were analyzed using paired t-test (D) or one-way (G)/two-way (H, I) ANOVA (*P < 0.05). SPAG6, Sperm Associated Antigen 6; LUSC, Lung Squamous Cell Carcinoma; DNMT, DNA Methyltransferase.
DNMT3b expression was significantly elevated in LUSC cells, shown by RT-qPCR (Figure 3D). The expression of DNMT3b was positively correlated with the DNA methylation level of SPAG6 (Figure 3E) and negatively linked to the SPAG6 expression pattern (Figure 3F). DNMT3b expression was significantly elevated in LUSC cells (Figure 3G). Inhibition of DNMT3b by transfection with a shRNA appreciably upregulated SPAG6 (Figure 3H) and decreased the SPAG6 promoter DNA methylation (Figure 3I).
Overexpression of SPAG6 reverts the malignant phenotype of LUSC cells
The oe-SPAG6 was transfected in LUSC cells (with oe-NC as control), and the overexpression efficiency was assessed using RT-qPCR (Figure 4A). The overexpression of SPAG6 considerably inhibited the proliferation of LUSC cells (Figure 4B). It was also revealed by EdU staining that SPAG6 upregulation significantly inhibited DNA synthesis in LUSC cells (Figure 4C). The expression of SPAG6 also inhibited the migration and invasion of LUSC cells (Figure 4D, 4E).
Figure 4.
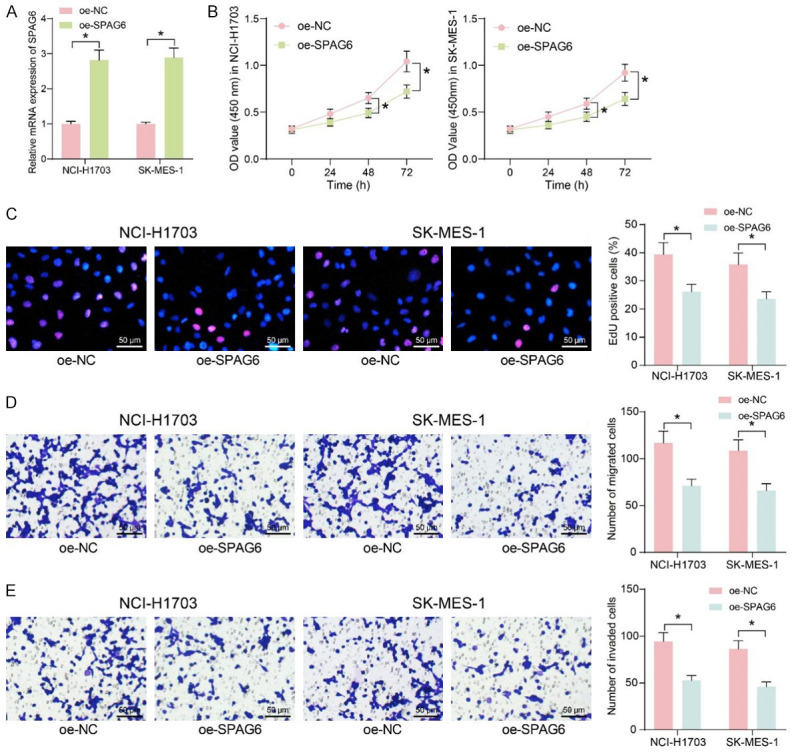
Restoration of DNMT3b inhibits migration and invasion of LUSC cells. A: The expression of SPAG6 following the transfection of overexpression vectors containing SPAG6 in LUSC cells by RT-qPCR. B: Effect of overexpression of SPAG6 on cell proliferation by CCK8 assays. C: Effect of overexpression of SPAG6 on cellular DNA synthesis by EdU staining. D: Effect of overexpression of SPAG6 on the ability of cells to migrate by Transwell assay. E: Effect of overexpression of SPAG6 on the ability of cells to invade by Transwell assay. Data represent mean ± SD from three independent experiments and were analyzed using two-way ANOVA (A-E) (*P < 0.05). SPAG6, Sperm Associated Antigen 6; LUSC, Lung Squamous Cell Carcinoma.
SPAG6 regulates JAK/STAT pathway in LUSC cells
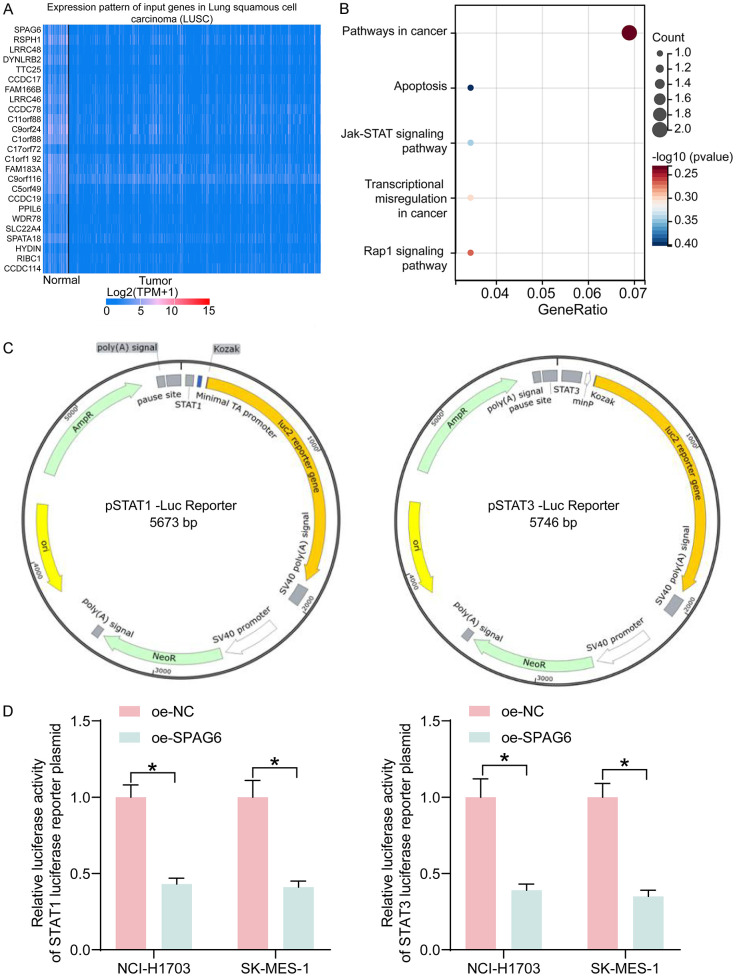
Factors with significant correlation with SPAG6 expression in LUSC were downloaded from UALCAN (Figure 5A), and the pathways enriched by these factors were analyzed by KEGG pathway enrichment, which found that JAK/STAT pathway was the key pathway (Figure 5B). The effect of SPAG6 on the JAK/STAT pathway activity was assessed using STAT1 and STAT3 luciferase reporter gene plasmids (Figure 5C). We revealed that SPAG6 overexpression significantly blocked the JAK/STAT pathway in LUSC cells by a dual-luciferase assay (Figure 5D).
Figure 5.
SPAG6 mediates the JAK/STAT pathway in LUSC. (A) Factors with significant correlation with SPAG6 expression in LUSC (only the top 25 factors were shown in the heatmap). (B) The pathway affected by SPAG6 analyzed using KEGG pathway enrichment analysis. (C) The mapping of STAT1 and STAT3 luciferase reporter gene plasmids. (D) Effect of SPAG6 on the luciferase activity of STAT1 and STAT3 luciferase reporter gene plasmids by dual-luciferase assays. Data represent mean ± SD from three independent experiments and were analyzed using two-way ANOVA (D) (*P < 0.05). SPAG6, Sperm Associated Antigen 6; LUSC, Lung Squamous Cell Carcinoma.
SPAG6 modulates the immune response in LUSC
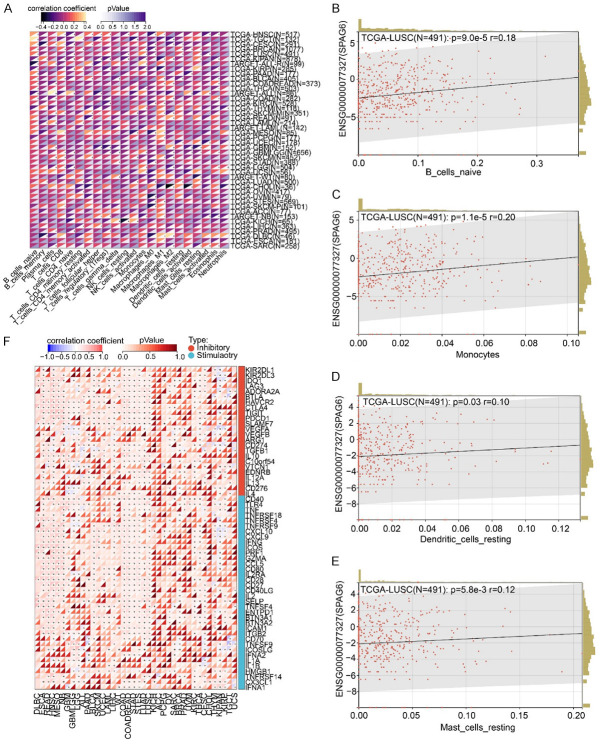
In Figure 5, we had evidenced that SPAG6 impaired the JAK/STAT pathway in LUSC. The JAK-STAT pathway is also associated with immune evasion in cancers [13]. We therefore analyzed the expression of SPAG6 in pan-cancer and its relationship with immunity in CIBERSORT. A significant correlation was found between SPAG6 expression and immune infiltration in cancers (Figure 6A). In LUSC, the SPAG6 expression was positively correlated with the infiltration of immune cells, including naive B cells (Figure 6B), monocytes (Figure 6C), dendritic cells (Figure 6D), and mast cells (Figure 6E). Further analysis showed that SPAG6 expression also correlated with the expression of immunoregulatory genes in cancers (Figure 6F). The expression of SPAG6 in LUSC was inversely related to the expression of the typical immunosuppressive gene CTLA4 and PDCD1.
Figure 6.
SPAG6 correlates with immune response in LUSC. A: Expression of SPAG6 in cancers and the correlation with immune cell infiltration. B: Correlation of SPAG6 expression with infiltration of naive B cells in LUSC. C: Correlation of SPAG6 expression with infiltration of monocytes in LUSC. D: Correlation of SPAG6 expression with infiltration of dendritic cells in LUSC. E: Correlation of SPAG6 expression with infiltration of mast cells in LUSC. F: Correlation between SPAG6 expression and expression of immunoregulatory genes in cancers. SPAG6, Sperm Associated Antigen 6; LUSC, Lung Squamous Cell Carcinoma.
SPAG6 suppresses cancer cell stemness in LUSC
Nanog, a marker of CSCs, has been suggested to induce stemness, self-renewal, metastasis, invasiveness, and chemoresistance of cancer cells through the JAK/STAT pathway [14]. The correlation between SPAG6 expression and cancer cell stemness was analyzed using Sangerbox 3.0. It was shown that SPAG6 was significantly and negatively correlated with cancer cell stemness in LUSC (Figure 7A). Furthermore, the results of RT-qPCR demonstrated that the cancer cell stemness markers Nanog, ALDH1, and Sox2 were significantly inhibited in cells overexpressing SPAG6 (Figure 7B-D).
Figure 7.
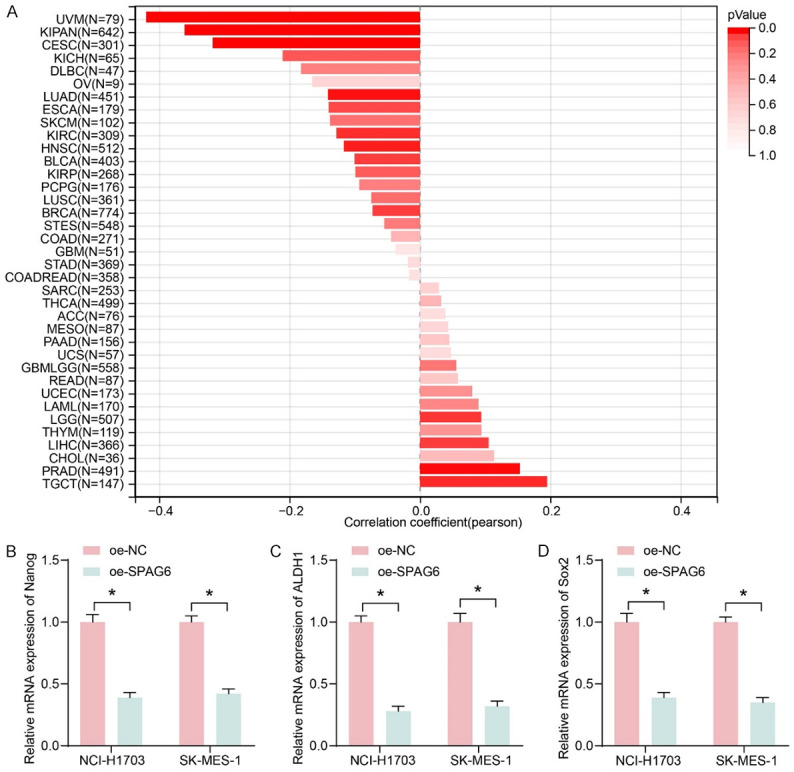
SPAG6 correlates with cancer cell stemness in LUSC. A: Correlation of SPAG6 expression with cancer stemness. B: Effect of overexpression of SPAG6 on the cancer cell stemness marker Nanog by RT-qPCR. C: Effect of overexpression of SPAG6 on the cancer cell stemness marker ALDH1 by RT-qPCR. D: Effect of overexpression of SPAG6 on the cancer cell stemness marker Sox2 by RT-qPCR. Data represent mean ± SD from three independent experiments and were analyzed using two-way ANOVA (B-D) (*P < 0.05). SPAG6, Sperm Associated Antigen 6; LUSC, Lung Squamous Cell Carcinoma; ALDH1, Aldehyde Dehydrogenase 1.
Discussion
SPAG6 has been shown to play a crucial role in immuno-regulation, tumor cell proliferation, apoptosis, invasion and metastasis, thus participating in the occurrence and progression of a variety of human cancers [15]. A significant hypermethylation and downregulation of SPAG6 in LUSC tissues versus adjacent normal tissues was observed in this study, signifying that the depletion of SPAG6 was associated with DNMTs-related methylation modification. Moreover, SPAG6 mRNA levels were downregulated and promoter methylation was upregulated in LUSC cell lines, and ectopic expression of SPAG6 successfully reverted the malignant phenotype of LUSC cells. Furthermore, SPAG6 was verified to be regulated by DNMT3b and predicted to mediate immune evasion and cancer cell stemness in LUSC by the JAK/STAT pathway.
Decitabine is prodrug cytosine analog that is incorporated into nucleic acids as azacytosine-guanine pairs following their uptake and phosphorylation [16]. Our observation here revealed that suppression of the DNMT activity using Decitabine remarkably reduced the DNA methylation of SPAG6, while it restored the expression of SPAG6 in LUSC cells. Our findings rudimentarily proved that the depletion of SPAG6 was related to the hypermethylation of its promoter. Further online prediction showed that the hypermethylation status of SPAG6 in LUSC was modulated by DNMT3b. The high level of DNMT3b expression was drastically related to poor prognosis in young patients with non-small cell lung cancer [17]. After determining the overexpression of DNMT3b in LUSC cell lines, we transfected shRNA targeting DNMT3b into LUSC cells and found that loss of DNMT3b restored the mRNA expression of SPAG6 as Decitabine treatment. Similarly, TIMP3 downregulation by DNA methylation involves the recruitment of Sp1 into the TIMP3 promoter by DNMT3b in oral cancer [18]. In the context of LUSC, DNMT3b overexpression was correlated with hypermethylation in the TSG promoters, particularly among smoking patients [19]. Furthermore, we transfected LUSC cell lines with overexpression vectors containing SPAG6. Reduced LUSC cell malignant aggressiveness was observed in cells overexpressing SPAG6, indicating the tumor inhibitory role of SPAG6 in vitro.
Combining the KEGG enrichment analysis results, we further found that the JAK/STAT pathway was the downstream effector of SPAG6. STAT3 activation was determined in non-small cell lung cancer patients and cells, and STAT3 stimulated the inflammation and evasion of anti-tumor immunity [20]. The biomarker potential of immune cells, including naïve B cells [21], lung tumor-infiltrated monocytes [22], dendritic cells [23] and mast cells [24] has been identified in lung cancer. The emergence of immunotherapies with check point inhibitors and check point inhibitor-chemotherapy combinations have fundamentally transformed medical care for patients with advanced non-small-cell lung cancer, leading to revised treatment algorithms [25,26]. The CTLA4 immune checkpoint represents a negative regulator of T-cell immune function, and inhibition of CTLA4 contributes to the activation of the immune system, allowing development of new immunotherapies for non-small cell lung cancer [27]. PDCD1 is another important player in the mediation of the immune system responses and production of self-tolerance by suppressing T cells [28]. Here, we not only observed the direct positive correlation between SPAG6 expression and naïve B cells, monocytes, dendritic cells and mast cells, but also revealed a negative regulation of SPAG6 on CTLA4 and PDCD1 expressions through a series of bioinformatic prediction. Through online prediction website, the negative correlation between SPAG6 expression and CSCs was also noted. Pluripotency genes, including Nanog and Sox2, and drug resistance markers including ALDH1 can both serve as CSC markers in the lung [29,30]. We also identified the anti-tumor effects of SPAG6 by repressing these markers.
Altogether, this study expounded that overexpressed DNMT3b in LUSC expedites SPAG6 promoter methylation to downregulate its expression. Overexpression of SPAG6 reduces LUSC cells growth, migration, and invasion. We postulated that SPAG6 can regulate immune invasion and cancer cell stemness in LUSC by the JAK/STAT pathway. Our study does carry a limitation: we cannot exclude other upstream regulators responsible for SPAG6 loss and other potential pathways of SPAG6 in LUSC progression. Further studies are required on these issues. Still, the findings obtained from this study may be helpful in developing new therapeutic options for LUSC.
Disclosure of conflict of interest
None.
References
- 1.Schiffmann I, Greve G, Jung M, Lubbert M. Epigenetic therapy approaches in non-small cell lung cancer: update and perspectives. Epigenetics. 2016;11:858–870. doi: 10.1080/15592294.2016.1237345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang S, Huang Y, Zhao Q. Epigenetic alterations and inflammation as emerging use for the advancement of treatment in non-small cell lung cancer. Front Immunol. 2022;13:878740. doi: 10.3389/fimmu.2022.878740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh S, Javia A, Shetty S, Bardoliwala D, Maiti K, Banerjee S, Khopade A, Misra A, Sawant K, Bhowmick S. Triple negative breast cancer and non-small cell lung cancer: clinical challenges and nano-formulation approaches. J Control Release. 2021;337:27–58. doi: 10.1016/j.jconrel.2021.07.014. [DOI] [PubMed] [Google Scholar]
- 4.Sasa GBK, Xuan C, Chen M, Jiang Z, Ding X. Clinicopathological implications of lncRNAs, immunotherapy and DNA methylation in lung squamous cell carcinoma: a narrative review. Transl Cancer Res. 2021;10:5406–5429. doi: 10.21037/tcr-21-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai L, Bai H, Duan J, Wang Z, Gao S, Wang D, Wang S, Jiang J, Han J, Tian Y, Zhang X, Ye H, Li M, Huang B, He J, Wang J. Epigenetic alterations are associated with tumor mutation burden in non-small cell lung cancer. J Immunother Cancer. 2019;7:198. doi: 10.1186/s40425-019-0660-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li XS, Nie KC, Zheng ZH, Zhou RS, Huang YS, Ye ZJ, He F, Tang Y. Molecular subtypes based on DNA methylation predict prognosis in lung squamous cell carcinoma. BMC Cancer. 2021;21:96. doi: 10.1186/s12885-021-07807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Luo L, Dong J, Liu M, Zhai D, Huang D, Ling L, Jia X, Luo K, Zheng G. A prognostic 11-DNA methylation signature for lung squamous cell carcinoma. J Thorac Dis. 2020;12:2569–2582. doi: 10.21037/jtd.2020.03.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Silina K, Zayakin P, Kalnina Z, Ivanova L, Meistere I, Endzelins E, Abols A, Stengrevics A, Leja M, Ducena K, Kozirovskis V, Line A. Sperm-associated antigens as targets for cancer immunotherapy: expression pattern and humoral immune response in cancer patients. J Immunother. 2011;34:28–44. doi: 10.1097/CJI.0b013e3181fb64fa. [DOI] [PubMed] [Google Scholar]
- 9.Kitchen MO, Bryan RT, Haworth KE, Emes RD, Luscombe C, Gommersall L, Cheng KK, Zeegers MP, James ND, Devall AJ, Fryer AA, Farrell WE. Methylation of HOXA9 and ISL1 predicts patient outcome in high-grade non-invasive bladder cancer. PLoS One. 2015;10:e0137003. doi: 10.1371/journal.pone.0137003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altenberger C, Heller G, Ziegler B, Tomasich E, Marhold M, Topakian T, Mullauer L, Heffeter P, Lang G, End-Pfutzenreuter A, Dome B, Arns BM, Klepetko W, Zielinski CC, Zochbauer-Muller S. SPAG6 and L1TD1 are transcriptionally regulated by DNA methylation in non-small cell lung cancers. Mol Cancer. 2017;16:1. doi: 10.1186/s12943-016-0568-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeifer GP, Rauch TA. DNA methylation patterns in lung carcinomas. Semin Cancer Biol. 2009;19:181–187. doi: 10.1016/j.semcancer.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gagliardi M, Strazzullo M, Matarazzo MR. DNMT3B functions: novel insights from human disease. Front Cell Dev Biol. 2018;6:140. doi: 10.3389/fcell.2018.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mohan CD, Rangappa S, Preetham HD, Chandra Nayaka S, Gupta VK, Basappa S, Sethi G, Rangappa KS. Targeting STAT3 signaling pathway in cancer by agents derived from mother nature. Semin Cancer Biol. 2022;80:157–182. doi: 10.1016/j.semcancer.2020.03.016. [DOI] [PubMed] [Google Scholar]
- 14.Vasefifar P, Motafakkerazad R, Maleki LA, Najafi S, Ghrobaninezhad F, Najafzadeh B, Alemohammad H, Amini M, Baghbanzadeh A, Baradaran B. Nanog, as a key cancer stem cell marker in tumor progression. Gene. 2022;827:146448. doi: 10.1016/j.gene.2022.146448. [DOI] [PubMed] [Google Scholar]
- 15.Zheng DF, Wang Q, Wang JP, Bao ZQ, Wu SW, Ma L, Chai DM, Wang ZP, Tao YS. The emerging role of sperm-associated antigen 6 gene in the microtubule function of cells and cancer. Mol Ther Oncolytics. 2019;15:101–107. doi: 10.1016/j.omto.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saleh MH, Wang L, Goldberg MS. Improving cancer immunotherapy with DNA methyltransferase inhibitors. Cancer Immunol Immunother. 2016;65:787–796. doi: 10.1007/s00262-015-1776-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing J, Stewart DJ, Gu J, Lu C, Spitz MR, Wu X. Expression of methylation-related genes is associated with overall survival in patients with non-small cell lung cancer. Br J Cancer. 2008;98:1716–1722. doi: 10.1038/sj.bjc.6604343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su CW, Chang YC, Chien MH, Hsieh YH, Chen MK, Lin CW, Yang SF. Loss of TIMP3 by promoter methylation of Sp1 binding site promotes oral cancer metastasis. Cell Death Dis. 2019;10:793. doi: 10.1038/s41419-019-2016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA methyltransferases contributes to 5’CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 20.Mohrherr J, Uras IZ, Moll HP, Casanova E. STAT3: versatile functions in non-small cell lung cancer. Cancers (Basel) 2020;12:1107. doi: 10.3390/cancers12051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stankovic B, Bjorhovde HAK, Skarshaug R, Aamodt H, Frafjord A, Muller E, Hammarstrom C, Beraki K, Baekkevold ES, Woldbaek PR, Helland A, Brustugun OT, Oynebraten I, Corthay A. Immune cell composition in human non-small cell lung cancer. Front Immunol. 2018;9:3101. doi: 10.3389/fimmu.2018.03101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Ridder K, Locy H, Piccioni E, Zuazo MI, Awad RM, Verhulst S, Van Bulck M, De Vlaeminck Y, Lecocq Q, Reijmen E, De Mey W, De Beck L, Ertveldt T, Pintelon I, Timmermans JP, Escors D, Keyaerts M, Breckpot K, Goyvaerts C. TNF-alpha-secreting lung tumor-infiltrated monocytes play a pivotal role during Anti-PD-L1 immunotherapy. Front Immunol. 2022;13:811867. doi: 10.3389/fimmu.2022.811867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Plumas J. Harnessing dendritic cells for innovative therapeutic cancer vaccines. Curr Opin Oncol. 2022;34:161–168. doi: 10.1097/CCO.0000000000000815. [DOI] [PubMed] [Google Scholar]
- 24.Sangaletti S, Ferrara R, Tripodo C, Garassino MC, Colombo MP. Myeloid cell heterogeneity in lung cancer: implication for immunotherapy. Cancer Immunol Immunother. 2021;70:2429–2438. doi: 10.1007/s00262-021-02916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ackermann CJ, Adderley H, Ortega-Franco A, Khan A, Reck M, Califano R. First-line immune checkpoint inhibition for advanced non-small-cell lung cancer: state of the art and future directions. Drugs. 2020;80:1783–1797. doi: 10.1007/s40265-020-01409-6. [DOI] [PubMed] [Google Scholar]
- 26.Addeo A, Passaro A, Malapelle U, Luigi Banna G, Subbiah V, Friedlaender A. Immunotherapy in non-small cell lung cancer harbouring driver mutations. Cancer Treat Rev. 2021;96:102179. doi: 10.1016/j.ctrv.2021.102179. [DOI] [PubMed] [Google Scholar]
- 27.Buchbinder EI, Desai A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am J Clin Oncol. 2016;39:98–106. doi: 10.1097/COC.0000000000000239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghafouri-Fard S, Hussen BM, Mohaqiq M, Shoorei H, Baniahmad A, Taheri M, Jamali E. Interplay between non-coding RNAs and programmed cell death proteins. Front Oncol. 2022;12:808475. doi: 10.3389/fonc.2022.808475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiuthed A, Chantarawong W, Chanvorachote P. Lung cancer stem cells and cancer stem cell-targeting natural compounds. Anti-cancer Res. 2018;38:3797–3809. doi: 10.21873/anticanres.12663. [DOI] [PubMed] [Google Scholar]
- 30.Shukla S, Khan S, Sinha S, Meeran SM. Lung cancer stem cells: an epigenetic perspective. Curr Cancer Drug Targets. 2018;18:16–31. doi: 10.2174/1568009617666170206104623. [DOI] [PubMed] [Google Scholar]