Abstract
The lipid nanoparticle (LNP)-encapsulated, nucleoside-modified mRNA platform has been used to generate safe and effective vaccines in record time against COVID-19. Here, we review the current understanding of the manner whereby mRNA vaccines induce innate immune activation and how this contributes to protective immunity. We discuss innate immune sensing of mRNA vaccines at the cellular and intracellular levels and consider the contribution of both the mRNA and the LNP components to their immunogenicity. A key message that is emerging from recent observations is that the LNP carrier acts as a powerful adjuvant for this novel vaccine platform. In this context, we highlight important gaps in understanding and discuss how new insight into the mechanisms underlying the effectiveness of mRNA-LNP vaccines may enable tailoring mRNA and carrier molecules to develop vaccines with greater effectiveness and milder adverse events in the future.
Keywords: mRNA vaccine, nucleoside modification, lipid nanoparticle, ionizable lipid, adjuvant, innate immunity, T cell, germinal center, dendritic cell, neutralizing antibody, SARSCoV- 2 vaccine
Lipid nanoparticle-encapsulated, nucleoside-modified mRNA vaccines produce adaptive immune responses protective against COVID-19. Here, Pardi and colleagues review the innate immune-sensing pathways of this vaccine modality and discuss research directions that may yield a next generation of mRNA vaccines with improved immunogenicity and reduced reactogenicity.
Introduction
The nucleoside-modified mRNA vaccines against coronavirus disease 2019 (COVID-19), i.e., BNT162b2 (Comirnaty) from Pfizer/BioNTech and mRNA-1273 (Spikevax) from Moderna, are the first mRNA products to receive approval from the Food and Drug Administration (FDA) or the European Medicines Agency (EMA). The two key components of these vaccines are (1) nucleoside-modified mRNA (Karikó et al., 2005), which encodes the antigenic protein of interest (in this case, the spike protein of severe acute respiratory syndrome coronavirus 2 [SARS-CoV-2]), and (2) lipid nanoparticles containing ionizable lipids (iLNPs), which enable efficient delivery of intact mRNA to the cytoplasm of cells that can then translate the encoded protein (Cullis and Hope, 2017). These nucleoside-modified mRNA-iLNP vaccines are highly effective in inducing spike-specific adaptive immune responses in humans, most notably neutralizing antibodies for protective immunity against SARS-CoV-2 infection (Baden et al., 2021; Gilbert et al., 2022; Polack et al., 2020; Wei et al., 2022) as well as T cell responses that promote humoral immunity and may contribute to preventing severe COVID-19 (Bertoletti et al., 2022; Kedzierska and Thomas, 2022; Sette and Crotty, 2021; Wherry and Barouch, 2022). However, little is known about which aspects of innate immunity are critical to drive protective immune responses and which are dispensable. Also lacking is a detailed mechanistic understanding of how innate immune pathways modulate adaptive immunity in mRNA vaccine responses. In this review, we focus on the innate immune stimulation by mRNA-iLNP vaccines, discuss recent insights gained from descriptive and hypothesis-driven studies on the dynamics and activation of innate immune cells, and explore the possible molecular mechanisms that contribute to the immunogenicity and reactogenicity of this novel vaccine platform.
Overview of the nucleoside-modified mRNA and iLNP technologies
Safe and effective vaccines must stimulate the innate immune system at an appropriate level such that they achieve a balance between immunogenicity and reactogenicity; that is, they provide the signals needed to prime and maintain adaptive immune responses while avoiding excessive local or systemic inflammation. Historically, one of the key obstacles to using synthetic mRNA as a vaccine (or therapeutic) modality has been its highly inflammatory nature, owing to the recognition of mRNA molecules by numerous innate sensors in multiple subcellular compartments (Alameh and Weissman, 2022). While RNA sensing serves a vital role in bridging innate and adaptive immune responses to viral infections, it can also impede the therapeutic potential of mRNA vaccines by suppressing the synthesis of the encoded antigen and by causing dose-limiting adverse reactions (Verbeke et al., 2019).
Decades of basic research were needed to understand how to circumvent the problems posed by innate immune recognition of mRNA. A major breakthrough came via the studies of Karikó, Weissman, and colleagues in the 2000s, who discovered that the presence of certain modified vs. unmodified ribonucleosides works as a molecular mechanism by which cellular RNA sensors discriminate self-RNA from foreign RNA (Karikó et al., 2005). In particular, replacement of uridine with naturally occurring derivatives of uridine (e.g., pseudouridine [Ψ] and its derivatives) allows mRNA to escape or mitigate detection by most innate immune sensors, resulting in reduced inflammation and greatly improved translation (Anderson et al., 2010, 2011; Karikó et al., 2008). This discovery was fundamental to the success of the two approved COVID-19 mRNA vaccines in which all uridines are replaced with N1-methylpseudouridine (m1Ψ).
Another major hurdle in mRNA vaccine development was the inefficient cytosolic delivery of mRNA in vivo. This was overcome in the 2010s using the iLNP technology for mRNA vaccine delivery (Bahl et al., 2017; Geall et al., 2012; Lutz et al., 2017; Pardi et al., 2017; Richner et al., 2017). These iLNPs generally comprise four components: a cationic “ionizable” aminolipid with a pKa of approximately 6.0 to 6.8, a polyethylene glycol (PEG)-conjugated lipid to improve colloidal stability, cholesterol, and 1,2-distearoyl-sn-glycero-3-phosphocholine (Ansell and Du, 2017; Cullis and Hope, 2017; Hassett et al., 2019). The ionizable lipid is critical to achieve the structure of the iLNP, which is not liposomal but rather has an electron-dense core containing complexes of the negatively charged mRNA associated with ionizable lipid. The ionizable lipid is also essential to achieve a nanoparticle that has a neutral surface charge at physiological pH and after cellular uptake becomes positively charged in the acidifying endosome, facilitating disruption of the endosomal membrane and escape of the mRNA into the cytosol. Both approved COVID-19 mRNA-iLNP vaccines use ionizable lipids (SM-102 in the Moderna vaccine and ALC-0315 in the Pfizer/BioNTech vaccine) that feature ester linkages in the lipid tails and were selected based on mRNA delivery efficiency, biodegradability, and tolerability (Ansell and Du, 2017; Hassett et al., 2019; Maier et al., 2013).
While nucleoside-modified mRNA was designed to be non-inflammatory, nucleoside-modified mRNA-iLNP vaccines are clearly not immunosilent, as evidenced by strong innate immune activation (Alameh et al., 2021; Li et al., 2022a; Tahtinen et al., 2022) and documentation of local and systemic adverse events following COVID-19 mRNA vaccination in humans (Rosenblum et al., 2022). Indeed, a considerable advantage of the mRNA-iLNP platform is that no further addition of adjuvant is required to elicit robust protective immune responses against various pathogens. Another strength is the potent immunogenicity of mRNA-iLNP vaccines in the elderly, because it is well documented for other vaccines that this population may need formulations with stronger adjuvants and/or higher vaccine doses to reach sufficient protection (Gartner et al., 2022). As will be further discussed, the iLNP carrier is thought to be the primary driver of the adjuvanticity and reactogenicity of mRNA-iLNP vaccines. A variety of signals are induced specifically by the iLNP component, including chemokines and pro-inflammatory cytokines (e.g., C-C motif chemokine ligand 2 [CCL2], CCL3, CCL4, granulocyte-macrophage colony-stimulating factor [GM-CSF], C-X-C motif chemokine ligand 2 [CXCL2], CXCL10 , interleukin [IL]-1β, IL-6, tumor necrosis factor [TNF], and interferon [IFN]-γ) (Alameh et al., 2021; Li et al., 2022a; Ndeupen et al., 2021; Tahtinen et al., 2022). Thus, the iLNP technology not only enables the efficient delivery of mRNA into innate immune cells following vaccine administration, but also seems to play a second key role in providing a strong adjuvant activity to this vaccine platform. This represents a paradigm shift in understanding how the current generation of mRNA vaccines activates the innate immune system and also highlights an opportunity to refine their design for safer and more effective mRNA vaccines and therapeutics in the future. It is important to note, however, that the precise contributions of the nucleoside-modified mRNA vs. iLNP components to the overall immune response to the current generation of COVID-19 mRNA vaccines have not yet been clearly ascertained (Table 1 ).
Table 1.
Features of mRNA-iLNP vaccines that may impact innate immune activation
| Component | Sensing mechanisms | Comments | References | |
|---|---|---|---|---|
| IVT mRNA |
5’ cap structure | Cap0 or ppp at the 5’ end of the mRNA are recognized by IFIT1 in ssRNA and by RIG-I in dsRNA. | Cap1 is added enzymatically or co-transcriptionally to reduce immune sensing and increase translation. | (Abbas et al., 2017; Devarkar et al., 2016; Schuberth-Wagner et al., 2015) |
| Uridine | Uridine-containing RNA is recognized by TLR7/8. | Modified uridines (for example Ψ, m1Ψ, m5U) or uridine depletion reduce sensing and improve translation. | (Andries et al., 2015; Karikó et al., 2005, 2008; Lutz et al., 2017; Tanji et al., 2015; Zhang et al., 2016) | |
| dsRNA byproducts |
dsRNA is recognized by various intracellular receptors, e.g., TLR3, RIG-I, MDA5, PKR, OAS. |
dsRNA can be removed by stringent purification or RNase III digestion. |
(Baiersdorfer et al., 2019; Foster et al., 2019; Karikó et al., 2011; Mu et al., 2018; Nelson et al., 2020) |
|
|
Component |
Proposed sensing mechanisms |
Comments |
References |
|
| iLNP | Ionizable aminolipid | Specific PRRs, inflammasome, or other IL-1β activating pathways or indirect sensing mechanisms? | The ionizable lipid is critical for the adjuvant activity of iLNPs to induce strong antibody responses, but the mechanism is unknown. | (Alameh et al., 2021; Hassett et al., 2019; Li et al., 2022a; Tahtinen et al., 2022) |
| Cholesterol | Cholesterol accumulation in myeloid cells can play a role in the activation of various inflammatory responses. | Not known whether cholesterol in iLNPs can cause or contribute to innate immune effects. | (Anand, 2020) | |
| PEG-conjugated lipid | Anti-PEG antibodies could contribute to allergic reactions. | Hypothesized to lead to adverse reactions such as anaphylaxis. | (Ju et al., 2022; Szebeni et al., 2022) |
iLNP, ionizable lipid nanoparticle; RIG-I, retinoic acid-inducible gene I; IFIT1, interferon-induced protein with tetratricopeptide repeats; dsRNA, double-stranded RNA; ssRNA, single-stranded RNA; PRR, pathogen recognition receptor; TLR, Toll-like receptor; MDA5, melanoma differentiation-associated protein 5; PKR, protein kinase R; OAS, 2’-5’-oligoadenylate synthetase; Ψ, pseudouridine; m1Ψ, N1-methylpseudouridine; m5U, 5-methyluridine; IL, interleukin; PEG, polyethylene glycol. The helper lipid component of the iLNP is not listed in the table due to the lack of sufficient information about its role in innate immune activation.
Pharmacokinetics of nucleoside-modified mRNA-iLNP vaccines and early innate immune cell dynamics
mRNA-iLNP vaccines disseminate in the body depending on the iLNP formulation and on the route of administration (Bahl et al., 2017; Carrasco et al., 2021; Hassett et al., 2019; Liang et al., 2017a; Pardi et al., 2015). Intramuscular administration of the COVID-19 mRNA vaccines and other similarly designed vaccines results in vaccine uptake and production of the encoded antigen at the site of injection and draining lymph nodes (LNs), but limited spread of mRNA and/or lipid can also be detected in other tissues such as the liver, lungs, spleen, and non-draining LNs (European Medicines Agency, 2020, 2021; Hassett et al., 2019; Li et al., 2022a) (Figure 1 ). Administration of COVID-19 mRNA vaccines in the deltoid muscle in humans therefore likely results in most antigen production at the injection site and draining axillary, apical, and supraclavicular LNs (Roltgen et al., 2022). Similar biodistribution is seen for conventional vaccine types, such as adjuvanted protein subunits (Liang et al., 2017b; Ols et al., 2020; Segal et al., 2015). Intriguingly, a low amount of SARS-CoV-2 spike protein is found in the plasma of mice and humans receiving the BNT162b2 vaccine (Li et al., 2022a; Roltgen et al., 2022). This indicates that either the spike protein or its mRNA template (as mRNA-iLNP or cell-associated mRNA) can spread systemically after intramuscular injection.
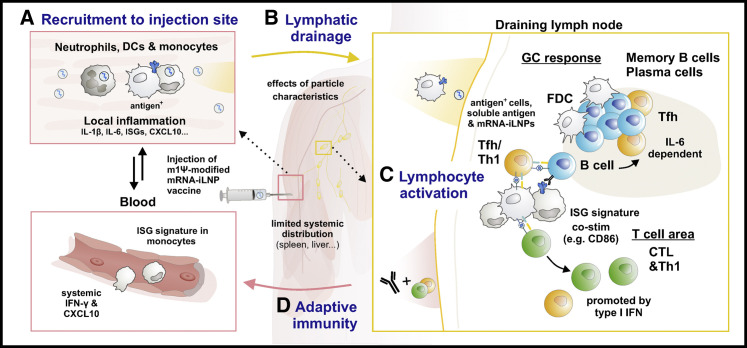
Figure 1.
Biodistribution and innate immune cell dynamics upon administration of mRNA-iLNP vaccines
(A) Intramuscular administration of nucleoside-modified mRNA-iLNP vaccines results in local inflammation, which recruits neutrophils, monocytes, and various dendritic cell (DC) subsets from the blood to the injection site by production of chemokines and other inflammatory mediators contributing to the extravasation of immune cells.
(B) mRNA-iLNPs and/or antigen-expressing cells are transported to the draining lymph node. The size and surface properties of the particles can impact biodistribution, protein absorption (opsonization), and cellular uptake.
(C) DCs and monocytes/macrophages contribute to antigen presentation and priming of T cells.
(D) T follicular helper (Tfh) cells provide help to B cells in germinal center (GC) reactions in the presence of follicular DCs, leading to affinity maturation. In mice, an important role for iLNP-induced IL-6 was found in the induction of Tfh and GC B cell responses, while type I IFNs promoted CTL responses. Abbreviations: m1Ψ: N1-methylpseudouridine, iLNP: ionizable lipid nanoparticle, IL: interleukin, IFN: interferon, ISG: interferon-stimulated gene, CXCL10: C-X-C motif chemokine ligand 10, CD86: cluster of differentiation 86, FDC: follicular dendritic cell, CTL: cytotoxic T lymphocyte, Th1: T helper 1.
In preclinical studies, protein production after mRNA-iLNP injection starts early, quickly peaks after 4–24 hours, and then gradually wanes; this can last for several days or more than a week, depending on the dose of mRNA, the encoded protein, the type of iLNP, and the route of mRNA-iLNP administration (Carrasco et al., 2021; Elia et al., 2021; Hassett et al., 2019; Pardi et al., 2015). Both spike protein and spike-encoding mRNA are detectable in germinal centers (GCs) in the draining axillary LNs for up to 60 days after the second dose of BNT162b2 or mRNA-1273 in vaccinated humans, although the duration of antigen expression needs to be further confirmed (Roltgen et al., 2022). While high and sustained antigen availability is associated with more robust antibody responses (Tam et al., 2016), it remains to be demonstrated whether sustained protein production from mRNA vaccination contributes to enhanced immune responses compared with other vaccine platforms.
Nucleoside-modified mRNA-iLNP vaccines induce robust T follicular helper (Tfh) cell responses and the formation of GCs, which are needed for persistent and high-affinity antibody production (Figure 1) (Lindgren et al., 2017; Pardi et al., 2018). In the draining LNs of BNT162b2-vaccinated humans, GC B cell and Tfh cell responses persist for as long as 6 months post-second dose with 30 μg mRNA (Mudd et al., 2022; Roltgen et al., 2022; Turner et al., 2021) and culminate in the generation of affinity-matured memory B cells and long-lived bone marrow-resident plasma cells (Kim et al., 2022). In humans, COVID-19 mRNA vaccines induce antigen-specific circulating Tfh cells, as well as CD4+ T cells with T helper 1 (Th1) polarization and IFN-γ-producing CD8+ T cells, which remain detectable for up to 6 months post-boost vaccination (Goel et al., 2021; Guerrera et al., 2021; Oberhardt et al., 2021; Painter et al., 2021; Zhang et al., 2022).
The surface modifications of iLNPs used for mRNA vaccine delivery may affect the biodistribution, lymphatic transport, and cellular uptake by innate immune cells. The surface decoration of nanoparticles with PEG is widely used as a strategy to prevent aggregation of nanomaterials and support distribution into the lymphatic system, while slowing opsonization processes and reducing clearance by phagocytic cells (Suk et al., 2016). However, iLNPs in the COVID-19 mRNA vaccines are engineered with PEG-containing lipids with a short hydrophobic anchor. Therefore, these PEG lipids are expected to rapidly desorb from the iLNPs, subsequently allowing endogenous proteins and lipids present in the extracellular space to efficiently bind and form a “biomolecular corona” around the iLNP (Dilliard et al., 2021; Mok et al., 1999). Complement fragments, immunoglobulins, and lipoproteins are among the most abundant proteins binding to mRNA-iLNPs upon exposure with blood-derived fluids (Francia et al., 2020). Although there is a dearth of information on the role of protein/iLNP interactions in lymphatic and cellular transport, it is possible that opsonization of mRNA-iLNPs drives receptor-mediated recognition and uptake by innate immune cells. For example, after intravenous delivery of mRNA-iLNPs, apolipoprotein E (ApoE) enables receptor-mediated binding and uptake of the particles by the low-density lipoprotein receptor (LDL-R) into hepatocytes (Akinc et al., 2010; Dilliard et al., 2021). Consistent with this, incubation of iLNPs with ApoE promotes mRNA delivery into dendritic cells (DCs) in cell culture experiments (Pardi et al., 2015). Particle size also dictates the cellular uptake and transport of nanoparticles into LNs (Maisel et al., 2017; Manolova et al., 2008; Oussoren and Storm, 2001; Reddy et al., 2006). Generally, PEGylated nanoparticles up to 100–200 nm can passively diffuse into the lymph to reach LNs minutes after injection, while larger particles may instead be internalized by DCs or other endocytic immune cells that migrate to LNs over the course of hours (Manolova et al., 2008; Reddy et al., 2006).
There is some insight on the role of innate immune cells in mRNA-iLNP uptake, translation, and some aspect of immunogenicity. In a rhesus macaque model, local inflammation occurs at the site of injection of either nucleoside-modified or unmodified mRNA-iLNP vaccines, with infiltration of several immune cell types such as neutrophils, DCs, and monocytes (Lenart et al., 2022; Liang et al., 2017a). Similarly, mRNA-iLNP administered intradermally in macaques or mice results in local inflammation (Liang et al., 2017a; Ndeupen et al., 2021). Neutrophils make up a large part of the immune cells infiltrating the sites of injection. Interestingly, neutrophils can efficiently internalize fluorescently labeled iLNPs but show poor production of the encoded reporter protein (Liang et al., 2017a). It is therefore possible that neutrophils could compete with other immune cells for the uptake of mRNA-iLNP vaccines. In contrast, monocytes and DC subsets efficiently take up and translate mRNA-iLNPs (Lenart et al., 2022; Liang et al., 2017a). In line with this, single-cell RNA-sequencing analysis of draining LNs from mice vaccinated with BNT162b2 shows that spike mRNA is found primarily in myeloid DC, monocyte, and macrophage cell clusters (Li et al., 2022a). In these studies, the frequencies of monocytes and/or macrophages and myeloid DC subsets are increased in draining LNs and exhibit increased expression of co-stimulatory markers such as CD80 and CD86 compared with cells in the contralateral non-draining LNs. Neutrophils are dispensable for Tfh and GC B cell responses in mice intradermally vaccinated with an mRNA-iLNP vaccine encoding influenza virus hemagglutinin (HA), with no significant difference in anti-HA serum immunoglobulin [Ig] G or protection from morbidity following influenza virus challenge (Ndeupen et al., 2022). In contrast, conventional type 1 DCs and Langerhans cells contribute (redundantly) to optimal Tfh and GC B cell responses, although mice lacking both of these cell types exhibit only moderately reduced antibody responses.
Taken together, activated monocytes, macrophages, and DCs are among the most critical cell types responsible for successful mRNA-iLNP uptake, synthesis of the encoded protein, and antigen presentation in lymphoid tissue to drive the adaptive immune response. Nonetheless, there are still many unknown aspects of mRNA-iLNP uptake by these immune cells and how physicochemical parameters such as size, surface composition, and morphology of iLNPs can affect the pharmacokinetics of these vaccines.
Immune recognition of unmodified and nucleoside-modified mRNA
The three major categories of innate immune sensing of synthetic mRNA are (1) uridine-dependent recognition of various RNA species (Tanji et al., 2015; Zhang et al., 2016), (2) recognition of double-stranded RNA (dsRNA) (Alexopoulou et al., 2001; Balachandran et al., 2000; Mitoma et al., 2013; Yoneyama et al., 2004, 2005), and (3) recognition of the 5’ end of mRNA if not properly capped (Abbas et al., 2017; Devarkar et al., 2016; Schuberth-Wagner et al., 2015). Currently, there is a notable dearth of studies that directly compares the immunogenicity and efficacy of various mRNA vaccine platforms (e.g., nucleoside-modified vs. unmodified mRNA). However, it is clear that nucleoside-modified mRNA vaccines have experienced more clinical success and widespread deployment at present (Hogan and Pardi, 2022).
The substitution of certain modified nucleosides, such as pseudouridine or its m1Ψ derivative, in place of uridine in synthetic mRNA has at least two important advantages: it increases protein production from the mRNA and decreases immune sensing by a host of intracellular RNA sensors (Karikó et al., 2005, 2008). The best-described sensors of unmodified single-stranded RNA (ssRNA) and its degradative products are Toll-like receptor (TLR) 7, which binds simultaneously to guanosine and the tri-ribonucleotide UUU (Zhang et al., 2016), and TLR8 (active in humans but not mice), which binds uridine and the di-nucleotide UG (Tanji et al., 2015). Modified nucleosides can also dampen activation of various dsRNA sensors, including TLR3, retinoic acid-inducible gene I (RIG-I), melanoma differentiation-associated protein 5 (MDA5), protein kinase R (PKR), and 2’-5’-oligoadenylate synthetase (OAS) (Anderson et al., 2010, 2011; Karikó et al., 2005; Mu et al., 2018). Recognition of uridine-containing RNA is associated with increased expression of pro-inflammatory cytokines, particularly type I IFN (Karikó et al., 2005, 2011), which promotes the expression of RNA sensors even further and can lead to inhibition of antigen expression from the mRNA via the activities of PKR and OAS.
Nucleoside modification is required for high vaccine efficacy in at least some circumstances; for example, an mRNA-iLNP vaccine encoding influenza virus HA elicits strong Tfh cell, GC B cell, and antibody responses in mice only when the mRNA is m1Ψ modified and not when unmodified (Pardi et al., 2018). It is still not clear exactly whether and how nucleoside modification per se promotes GC reactions compared with unmodified mRNA, but here we present several theories that are not mutually exclusive: (1) protein from modified mRNA is generally expressed both at a higher level and for a longer duration than from unmodified mRNA. For example, firefly luciferase expression from unmodified mRNA-iLNP can be detected for 8 days after intradermal injection in mice, while luciferase from m1Ψ-modified mRNA-iLNP is detectable for 13 days and at ∼10-fold higher total levels (Pardi et al., 2018). The latter provides more favorable kinetics to drive GC reactions, which are promoted by prolonged antigen availability (Tam et al., 2016). (2) It is possible that the crucial difference between modified and unmodified mRNA is not (only) the overall protein expression, but also differential expression in key antigen-presenting cell types, such as monocytes, macrophages, and DCs (Lenart et al., 2022; Li et al., 2022a; Liang et al., 2017a). These cell types may be especially sensitive to the translation-inhibiting effects of unmodified mRNA due to higher expression of RNA sensors (e.g., TLR7/8), potentially hindering a key pathway of T cell priming: direct presentation of translated antigens to CD4+ and CD8+ T cells (Miller et al., 2015; Ochsenbein et al., 2001). (3) The cytokine milieu generated by modified mRNA-iLNP (discussed in more detail below) may be more favorable for GC reactions than that induced by unmodified mRNA-iLNP.
Despite the advantages of modified nucleosides, unmodified mRNA can also achieve robust protein expression (Thess et al., 2015) and neutralizing antibody responses (Lutz et al., 2017). CureVac developed a sequence-engineering strategy to mitigate detection by pattern recognition receptors by altering the codon usage with synonymous changes that minimize uridine and increase the GC content, thus promoting strong antigen expression and immunogenicity. This approach can certainly decrease uridine-induced inflammation after mRNA delivery but is unlikely to yield an immunosilent mRNA as all uridines cannot be removed from the mRNA sequence. Perhaps this (at least partially) explains the differences in reactogenicity and immunogenicity of CureVac’s COVID-19 mRNA vaccine vs. those of Moderna and Pfizer/BioNTech (Baden et al., 2021; Kremsner et al., 2022; Polack et al., 2020).
Another mRNA vaccine component that can significantly contribute to deleterious innate immune recognition is dsRNA. In vitro transcription (IVT) by T7 RNA polymerase tends to yield not only the desired mRNA, but also a variety of unwanted RNA species, including various short abortive transcripts as well as antisense RNAs transcribed from the promoter-less end of the DNA template (Karikó et al., 2011; Mu et al., 2018). This can lead to the formation of dsRNA, which induces a potent inflammatory response and translational blockade via the recognition of various intracellular receptors (OAS, PKR, RIG-I, MDA5, TLR3, DHX33, etc.) (Anderson et al., 2010, 2011; Durbin et al., 2016; Karikó et al., 2011; Mitoma et al., 2013). Double-stranded secondary and tertiary structures could also form based on the sequence of the mRNA. The most important dsRNA sensors with regard to translational blockade are the IFN-inducible cytosolic sensors OAS and PKR. When OAS binds dsRNA, it activates RNase L, which in turn cleaves all ssRNA within the cell, thus destroying any therapeutic mRNA. Moreover, when PKR is activated by dsRNA or other RNA ligands (Anderson et al., 2010), it phosphorylates and inactivates the translation initiation factor eIF2α, thus shutting down protein synthesis. These deleterious innate immune responses can be reduced by at least two methods. First, as mentioned above, the inclusion of modified nucleosides can reduce the activation of PKR and OAS (Anderson et al., 2010, 2011). Second, dsRNA can be removed from IVT mRNA by certain purification methods. While dsRNA is not removed by common alcohol precipitation methods, it can be significantly reduced by high performance liquid chromatography (HPLC) (Karikó et al., 2011) or simpler methods such as adsorption on cellulose (Baiersdorfer et al., 2019) or RNase III digestion (Foster et al., 2019). It appears critical to reduce dsRNA from unmodified IVT mRNA to achieve a high level of protein production in vitro in DCs or in vivo in mice, but it may be less critical for m1Ψ-modified IVT mRNA (Baiersdorfer et al., 2019; Karikó et al., 2011; Nelson et al., 2020). Incorporation of m1Ψ and elimination of dsRNA can both dampen type I IFN responses upon intravenous administration of mRNA-iLNP products into mice (Nelson et al., 2020). Interestingly, the incorporation of certain modified nucleosides such as Ψ, m1Ψ, and 5-methylcytidine can also reduce the formation of dsRNA byproducts in T7-transcribed mRNA preparations (Mu et al., 2018). Lowering the Mg2+ concentration (from 30 mM to 5 mM) in the IVT reaction mixture and performing the IVT at higher temperature (∼50–55°C vs. 37°C) with a thermostable T7 RNA polymerase also reduces the dsRNA contaminants in the IVT reaction (Mu et al., 2018; Wu et al., 2020), and it is theoretically possible that the T7 enzyme could be mutated or replaced with an RNA polymerase that produces less dsRNA, or none at all, thus facilitating the economical manufacture of high-quality, good manufacturing practice (GMP)-grade mRNA.
Finally, the 5’-capping strategy also has an impact on the performance (translatability and immune activation) of IVT mRNA. A 5’ cap that can be conveniently added by vaccinia virus capping enzyme is the cap0 structure (m7GpppN), but this is recognized by RIG-I (Devarkar et al., 2016; Schuberth-Wagner et al., 2015) and by the human IFN-induced protein with tetratricopeptide repeats 1 (IFIT1) protein, which inhibits translation by competing with the cap-binding initiation factor complex eIF4F (Abbas et al., 2017). Therefore, it is common for mRNA therapeutics to use the cap1 structure (m7GpppNm), which features methylation on the first cap-proximal nucleotide and is a signature of “self” in higher eukaryotes. Cap1 can be obtained either by 2’-O-methyltransferase treatment of cap0-mRNA (Pardi et al., 2013) or by co-transcriptional capping with the CleanCap method (Henderson et al., 2021).
To summarize, the use of modified uridines is considered a key aspect of the effectiveness of COVID-19 mRNA vaccines, although both the translatability and inflammatory capacity of the mRNA component can be influenced by multiple other factors. Unfortunately, little public information is available regarding the exact methods of RNA preparation (transcription, capping, and purification) used by Moderna, Pfizer/BioNTech, and other mRNA companies, complicating basic research in this area.
Ionizable lipid nanoparticles provide potent adjuvant activity
iLNPs function as more than delivery agents. In fact, both empty iLNPs and mRNA-iLNP complexes serve as extremely effective adjuvants. Empty iLNPs were successfully used to adjuvant vaccines containing hepatitis B virus or dengue virus protein antigens (Swaminathan et al., 2016a, 2016b), and modified mRNA-iLNPs encoding luciferase were used to strongly adjuvant an influenza HA protein vaccine (Pardi et al., 2018). More recently, empty iLNPs mixed with influenza HA or SARS-CoV-2 spike receptor binding domain (RBD) proteins were shown to induce much stronger immune responses in mice than AddaVax-adjuvanted protein vaccines (Alameh et al., 2021). Of note, iLNP-adjuvanted protein and mRNA-iLNP vaccines induce similar cellular and humoral immune responses, including strong Tfh and GC B cell responses. The ionizable lipid component is critical for this adjuvant effect, as empty LNPs without the ionizable lipid do not induce strong antibody responses (Alameh et al., 2021). A mechanistic understanding is just beginning to emerge for the potent adjuvant activity of iLNPs and the inflammatory milieu that they stimulate. In the draining LNs of immunized mice, both empty iLNPs and nucleoside-modified mRNA-iLNPs elicit the production of several chemokines (including CXCL1, CXCL2, CXCL5, CXCL10, CCL3, CCL4) and cytokines IL-1β, IL-6, leukemia inhibitory factor (LIF), and GM-CSF at levels much greater than HA protein plus AddaVax (Alameh et al., 2021). Empty iLNPs or iLNPs containing a non-coding RNA stimulate robust production of the same set of immune mediators in the skin of intradermally vaccinated mice, as well as a vigorous immune cell infiltrate in injected tissues (Ndeupen et al., 2021). The rapid and robust production of IL-6, a key regulator of Tfh cell biology in mice (Choi et al., 2013), may explain the efficient induction of Tfh cells and GC B cells by empty iLNP-adjuvanted protein and modified mRNA-iLNP vaccines. Mechanistic studies in IL-6-deficient (II6 −/−) mice and experiments utilizing anti-IL-6 blocking antibodies demonstrate a requirement for iLNP-induced IL-6 in eliciting optimal Tfh cell and GC B cell responses in mice (Alameh et al., 2021).
A similar inflammatory signature including CCL4/, CXCL10, IL-6, IFN-α, and IFN-γ is seen in the serum of mice after vaccination with BNT162b2 (Li et al., 2022a). In mice, vaccine-stimulated type I IFN and IFN-stimulated gene (ISG) signatures in various cell types (macrophages, monocytes, DCs, natural killer [NK] cells analyzed by single-cell transcriptional profiling in draining LNs) peak at day 1 and return to baseline by day 7, whereas NK cells and T cells exhibit a continuous increase in expression of cell cycle- and transcription-associated genes (Li et al., 2022a). Of note, serum IFN-γ is higher (8.6-fold) 6 hours after administration of a second vaccine dose, coming largely from CD4+ and CD8+ T cells, compared with 6 hours after the first dose when most of the IFN-γ is derived from NK cells. In this setting, IFN-γ signaling activates antigen-presenting cells but surprisingly is not required for CD4+ and CD8+ T cell or antibody responses. Type I IFN signaling is required for strong CD8+ T cell responses but is mostly dispensable for strong binding antibody responses, although Tfh and GC B cell responses and neutralizing antibody levels were not measured (Li et al., 2022a). It is unclear which component of the mRNA-iLNP vaccine is responsible for inducing type I IFNs, but plasmacytoid DCs produce IFN-α only when exposed to mRNA-iLNPs and not empty iLNPs, suggesting that the mRNA component may be important for type I IFN induction (Liang et al., 2017a).
More limited data is available on innate immune activation in humans following mRNA-iLNP vaccination, but the studies to date have in large part agreed with the animal studies above. CXCL10 and IFN-γ are significantly elevated in the blood of human volunteers 1 day after receiving the BNT162b2 vaccine, particularly after the second immunization (Arunachalam et al., 2021). It is interesting that this study did not detect significant induction of IFN-α (measured ≥1 day post-injection) in humans, which might be explained by the fact that circulating IFN-α is highly transient (<1 day) in mice injected with the same vaccine and potentially also by differences in IFN-α responses between species (Li et al., 2022a). Consistent with findings in mice (Li et al., 2022a), the concentration of IFN-γ is higher after administration of the second vaccine dose, and this coincides with enhanced activation of myeloid cells (Arunachalam et al., 2021). The higher level of overall immune activation after the boost is in agreement with the observation that adverse effects are more frequent after administration of the second dose of both approved COVID-19 mRNA vaccines, particularly for systemic reactions (Chapin-Bardales et al., 2021; Rosenblum et al., 2022). Arunachalam et al. and Li et al. postulate that the enhanced activation of myeloid cells and T cells after administration of the second vaccine dose may reflect a crosstalk between lymphocytes and myeloid cells that involves epigenetic changes in the myeloid cells that increase their responsiveness to subsequent vaccine encounters (Arunachalam et al., 2021; Li et al., 2022a). This also raises the question of whether mRNA-iLNP vaccines are capable of inducing long-lasting boosting effects in myeloid cells (trained immunity) that may influence the protection against COVID-19 or other infectious diseases (Kobiyama and Ishii, 2022; Li et al., 2022b; Mantovani and Netea, 2020).
Human peripheral blood mononuclear cells (PBMCs) treated in vitro with m1Ψ-modified mRNA-iLNPs formulated with SM-102 lipid release IL-1β, IL-6, TNF, CCL5, granulocyte colony-stimulating factor (G-CSF), vascular endothelial growth factor-A (VEGF-A), and other molecules (Tahtinen et al., 2022). IL-1β is of particular interest given that it is viewed as a central mediator of innate immunity (Chan and Schroder, 2020), including reactogenicity and fever, and stimulates the production of many of the other inflammatory cytokines stimulated by mRNA-iLNP, such as IL-6, IFN-γ, TNF, and various chemokines (Mantovani et al., 2019). Interestingly, human PBMCs secrete lower baseline levels of IL-1 receptor antagonist (IL-1ra), a negative regulator of IL-1α/β signaling, compared with mice and non-human primates. This may explain why human cells make such potent IL-1β responses in reaction to modified mRNA-iLNP vaccines and suggests that there may be important species-dependent differences in IL-1β responses that must be considered when testing vaccines in animal models, but clearly more research is needed on this topic.
Innate immune detection of ionizable lipid nanoparticles
How are iLNPs sensed by the innate immune system? The component(s) of the vaccine that cause an innate immune response could include (1) the entire nanoparticle or other multimolecular structures, (2) individual lipids (e.g., ionizable lipid) (Alameh et al., 2021; Ndeupen et al., 2021), cholesterol (Anand, 2020), or (3) modified and/or metabolized lipid products (e.g., oxidative impurities of ionizable lipid [Packer et al., 2021] or oxidized phospholipids [Weismann and Binder, 2012]). It is also possible that endogenous molecules (e.g., ApoE or complement proteins) that bind to the iLNPs after mRNA vaccine delivery not only play a role in receptor-mediated uptake by innate immune cells, but also contribute to sensing mechanisms. Another possibility is that there is a cellular receptor, such as a TLR, that specifically recognizes iLNPs, because certain cationic liposomes and LNPs are sensed by TLR4 (Tanaka et al., 2008), TLR2 (Lonez et al., 2014), nucleotide binding domain and leucine-rich repeat pyrin domain-containing protein 3 (NLRP3) (Lonez et al., 2014), and the stimulator of interferon genes (STING) (Miao et al., 2019).
There is emerging insight on the signaling pathways that convey immune sensing of nucleoside-modified mRNA-iLNP vaccines. The adjuvant activity of iLNPs in protein subunit vaccination does not depend on MyD88 or MAVS, as shown using Myd88 −/− and Mavs −/− mouse strains. However, Myd88 −/− mice exhibit a partially blunted Tfh and GC B cell immune response to a purified, modified mRNA-iLNP vaccine (Alameh et al., 2021). Thus, it is possible that the mRNA (modified with m1Ψ and depleted of dsRNA) or its degradative products are still somehow recognized by sensors that signal via MyD88, such as TLR7. The immunogenicity of the BNT162b2 vaccine (antibody and CD8+ T cell response) is not reduced in the absence of TLR2, TLR4, TLR5, TLR7, apoptosis-associated speck-like protein containing a caspase recruitment domain [CARD] (ASC), NLRP3, cyclic GMP-AMP synthase (cGAS), or STING, suggesting that these important sensors of nucleic acids and microbial lipids, including inflammasome mediators, are not required for a strong immune response to this vaccine (Li et al., 2022a). TLR3 deficiency led to moderate (2.4-fold) decreases in neutralizing antibody titers, but not spike-binding IgG, indicating that this sensor may play a small role in adjuvanticity. It is important to note that some of these sensors could play redundant functions, in which case individual gene deletions may not reveal an effect. These data also suggest that these iLNPs and the TLR2/4-sensed cationic liposomes referenced above are sensed by different mechanisms. Of note, Mda5 −/− mice and Ifnar1 −/− mice exhibit a massive decrease in the frequency of antigen-specific CD8+ T cells, but only a mild effect on antibody responses, upon BNT162b2 vaccination. This suggests that the MDA5-IFN-α signaling pathway may be relevant to certain aspects of the immune response to nucleoside-modified mRNA-iLNP vaccines, such as the strong CD8+ T cell response noted for the BNT162b2 vaccine, but it may not be a key contributor to antibody-mediated protection. MDA5 is typically associated with sensing long dsRNA molecules or ssRNA with secondary structures (Pichlmair et al., 2009). This raises the possibility that secondary and tertiary mRNA structures or certain aspect(s) of mRNA production (for example, incomplete dsRNA removal) may play a role in the innate immune sensing of this mRNA vaccine.
Besides a sensor that can directly sense iLNPs (or their degradative products), there are a variety of ways that iLNPs could be indirectly sensed or otherwise contribute to innate immune activation. For example, a polycytidine (non-coding) RNA-iLNP triggers expression of a necroptosis-associated gene set, suggesting it could cause the release of inflammatory damage-associated molecular patterns (DAMPs) from dying cells (Ndeupen et al., 2021). Another intriguing idea is a mechanism that can sense membrane disturbances (curved membranes, cationic membrane patches, etc.) caused by fusion of iLNPs with the endosomal or plasma membrane. Such a pathway was proposed by Holm et al. after observing that various types of virus-like particles and cationic liposomes were able to induce DCs to produce IFN-β, CXCL10, and other immune mediators in a STING-dependent and membrane fusion-dependent manner (Holm et al., 2012). Additional contributions to adjuvanticity may come from complement activation (Szebeni et al., 2022), antigen-depot effects, or the possibility that iLNPs could transport mRNA to specific cell types or subcellular compartments that favor immune sensing and antigen presentation.
As mentioned earlier, an innate immune signaling pathway that is strongly implicated in the iLNP adjuvant effect is that of IL-1β, a cytokine consistently detected in humans, animals, and PBMCs exposed to mRNA-iLNPs or empty iLNPs. Conventionally, active IL-1β is secreted after the sensing cell integrates two signals: signal 1, typically provided via TLRs, leads to the transcription of pro-IL-1β, while signal 2, typically provided via the cytoplasmic sensing of pathogen-associated molecular patterns (PAMPs) and DAMPs, leads to the activation of caspase-1, which then cleaves pro-IL-1β into active IL-1β. Signal 2 is commonly achieved by activation of the inflammasome, an oligomeric structure that self-assembles in the cytoplasm in response to ligand recognition, culminating in the proteolytic activation of caspase-1. Inflammasomes can include a variety of pattern recognition receptors and adapter proteins and thus can respond to a wide variety of danger signals. To activate IL-1β, iLNPs would need to be sensed in at least two separate ways (directly and/or indirectly) to provide signal 1 and signal 2. But some of the best-described inflammasome components do not appear to be required for mRNA-iLNP adjuvanticity, namely NLRP3 and the widely used adapter molecule ASC (Li et al., 2022a). Therefore, IL-1β may be stimulated by iLNPs by an ASC-independent inflammasome (Ball et al., 2020; Hanamsagar et al., 2011) and/or a pathway that bypasses inflammasomes altogether, several of which have been described (Bossaller et al., 2012; Philip et al., 2014; Shenderov et al., 2014; Weng et al., 2014). Murine macrophages deficient in both caspase-1 and caspase-11 were no longer capable of producing IL-1β when exposed to empty iLNPs, presenting the best evidence to date that at least one of these pathways is important (Ndeupen et al., 2022). There is also precedent for IL-1β-activating pathways that are completely independent of caspase-1 and other inflammasome components. For example, cytosolic pro-IL-1β can be directly cleaved into active IL-1β by the neutrophil serine protease, proteinase 3 (Joosten et al., 2009). A similar scenario might be achieved under conditions of lysosomal rupture, which can lead to the release of cathepsin proteases into the cytosol, where they exert pro-inflammatory signaling effects (Hornung et al., 2008). Indeed, as iLNPs are designed to become positively charged and fusogenic in the acidifying endosome, it is quite plausible that they could perforate the endolysosomal membrane. Thus, the ability of iLNPs to deliver mRNA across the endolysosomal membrane may be intrinsically linked to their ability to transmit cathepsins or other endolysosomal molecules into the cytosol. This could potentially serve as a DAMP and a proxy for endolysosomal fusion, a common feature of viral infections; however, this model is currently completely speculative.
Identifying the innate immune-sensing mechanisms of synthetic mRNA and its lipid carrier remains a major task in the mRNA vaccine field (Figure 2 ). A detailed understanding of the molecular mechanisms by which iLNPs or their components are sensed will enable improved vaccine design in the future.
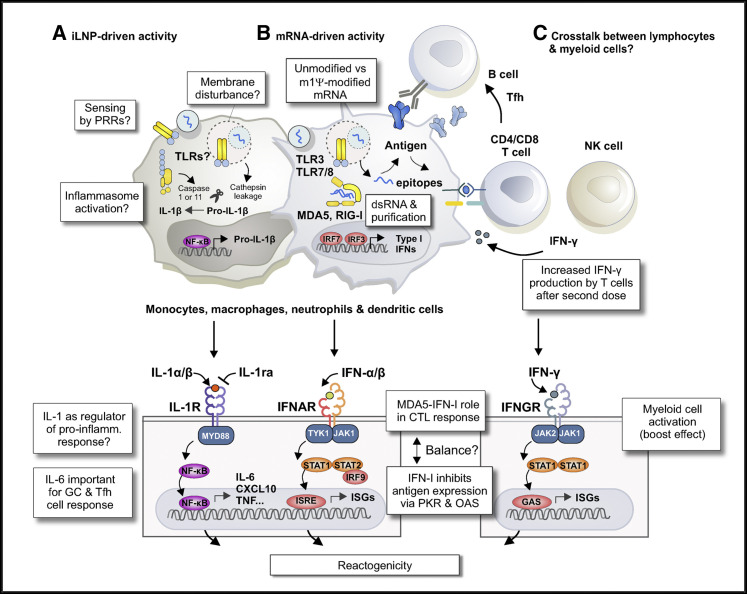
Figure 2.
Working model of the innate immune mechanisms that contribute to the immunogenicity and reactogenicity of mRNA-iLNP vaccines
(A) Uptake of empty iLNPs by innate immune cells and other cell types is sufficient to induce local and systemic inflammation, characterized by the release of pro-inflammatory cytokines such as IL-1β and IL-6.
(B) The incorporation of modified uridines and stringent IVT mRNA purification drastically lowers the recognition of IVT mRNA by TLR3/7/8 and other RNA sensors. These modifications are important to minimize the negative effects of type I IFN-stimulated RNA sensors on protein expression from the antigen-encoding mRNA and to avoid dose-limiting toxicities. The role of the MDA5-IFN-α signaling pathway in inducing CTLs to BNT162b2 in mice suggests residual type I IFN activity of the current generation of mRNA vaccines.
(C) After administration of a second vaccine dose, there is a strong boost in the T cell responses, which is associated with higher IFN-γ production. The enhanced activation of myeloid cells and T cells following boosting may reflect a crosstalk between lymphocytes and myeloid cells. Abbreviations: IVT, in vitro transcribed; iLNP, ionizable lipid nanoparticle; m1Ψ, N1-methylpseudouridine; NF-κB, nuclear factor-κB; IL-1R, interleukin-1 receptor; IL-1ra, interleukin-1 receptor antagonist; MYD88, myeloid differentiation primary response protein 88; dsRNA, double-stranded RNA; MDA5, melanoma differentiation-associated protein 5; RIG-I, retinoic acid-inducible gene I; PRR, pattern recognition receptor; IRF, interferon regulatory factor; IFN-I, type I interferon; IFNAR, interferon-α/β receptor; TYK1, leukocyte receptor tyrosine kinase; JAK, janus kinase; STAT, signal transducer and activator of transcription; ISRE, interferon-sensitive response element; PKR, protein kinase R; OAS, 2’-5’-oligoadenylate synthetase; IFNGR, interferon gamma receptor; GAS, gamma interferon activation site; ISG, interferon-stimulated gene; NK, natural killer; CTL, cytotoxic T lymphocyte; Tfh, T follicular helper; GC, germinal center; TLR, toll-like receptor; CXCL10, C-X-C motif chemokine ligand 10; TNF, tumor necrosis factor.
The path to next-generation mRNA-iLNP vaccines
Most of the innate immune activation seems to rely on the iLNP component of nucleoside-modified mRNA-iLNP vaccines, as empty iLNPs can elicit a variety of cytokines including IL-1β, IL-6, GM-CSF, and many chemokines (Alameh et al., 2021; Ndeupen et al., 2021; Tahtinen et al., 2022). With respect to the mRNA component, the m1Ψ nucleoside modification appears to improve the tolerability of mRNA-iLNP vaccines in humans by lowering innate immune recognition, with likely benefits to translation efficiency and adaptive immunity. Intriguingly, observations from several studies indicate that nucleoside-modified mRNA-iLNP vaccines (including the licensed SARS-CoV-2 mRNA vaccines) are still capable of inducing a transient type I IFN response, suggesting the possibility of complementary, or maybe synergistic, innate immune activation by the mRNA component (Alameh et al., 2021; Arunachalam et al., 2021; Li et al., 2022a; Liang et al., 2017a). Of particular interest is the finding that the MDA5-IFN-α signaling pathway is important for the CD8+ T cell response to BNT162b2 in mice (Li et al., 2022a). While it still needs to be determined what activates the MDA5 pathway, it is tempting to speculate that IVT or other manufacturing process-related impurities may play a role in this (European Medicines Agency, 2020; Mu et al., 2018), thus contributing to the capacity of COVID-19 mRNA vaccines to elicit CD8+ T cell responses. This observation may have implications for mRNA vaccine development for other infectious diseases and cancer, where CD8+ T cells are critical for effective immunity.
At present, it is unclear which innate immune pathways contribute to mRNA-iLNP-induced protective immunity vs. which contribute to their reactogenicity while being dispensable for efficacy, or whether these two aspects of the immune response can be disentangled at all. Most adverse events reported following BNT162b2 and mRNA-1273 vaccination were mild to moderate in severity and duration, commonly including injection-site pain, fatigue, headache, and chills (Baden et al., 2021; Polack et al., 2020; Rosenblum et al., 2022). So far, clinical studies suggest that there is only a weak association between the severity of these adverse events and the magnitude of antibody responses, while there is little or no correlation between the T cell response and reactogenicity (Bauernfeind et al., 2021; Held et al., 2021). These data need to be confirmed by larger and more rigorous studies. Still, these results raise the hope that the common adverse reactions to nucleoside-modified mRNA-iLNP vaccination are mechanistically dissociable from strong protective immunity. A deeper understanding of the source of reactogenicity may allow for the design of next generation mRNA-iLNP vaccines that stimulate stronger and more durable adaptive immune responses while also mitigating the moderate and severe adverse reactions experienced by vaccine recipients, including rare manifestations of anaphylaxis and acute myocarditis/pericarditis (Rosenblum et al., 2022; Szebeni et al., 2022).
Regarding the efficacy of mRNA-iLNP vaccines, one key consideration is the design of the antigen-encoding mRNA. It is likely that the protein-coding and non-coding sequence of the mRNA, as well as secondary structure formation, can be further optimized to maximize antigen expression and adaptive immune responses without increasing the vaccine dose. For instance, CureVac and GSK have started clinical development of a second-generation unmodified mRNA vaccine (CV2CoV) that differs in the 5’- and 3’-untranslated regions and poly(A) tail from the older CVnCoV to improve the spike protein expression (Gebre et al., 2022). In comparative studies, CV2CoV elicits 10- to 100-fold higher neutralizing antibodies and greater protection from SARS-CoV-2 replication than CvnCoV in non-human primates. CV2CoV also induced higher levels of IFNα2a, CXCL10, and CCL3 than CvnCoV, so it will be important to evaluate the tolerability and efficacy of this vaccine in humans, ideally compared head-to-head with the approved nucleoside-modified mRNA vaccines for COVID-19. In the future, additional types of RNA constructs such as self-amplifying (sa)RNA may also be clinically useful (Fuller and Berglund, 2020), but disappointing clinical results for two saRNA-based COVID-19 vaccines, i.e. ARCT-021 and LNP-nCoVsaRNA, seem to indicate that more efforts are needed to optimize this platform (e.g., minimizing the innate immune response) (Low et al., 2021; Pollock et al., 2022).
In parallel with the mRNA design, there are ample opportunities for improving the iLNP formulation. Most studies so far have focused on optimization of the ionizable lipid, as illustrated by how the SM-102 lipid was selected by empirical screening for improved antigen expression and tolerability in preclinical models (Hassett et al., 2019). Importantly, antigen expression alone may not accurately predict the immunogenicity of mRNA-iLNP vaccines (Hassett et al., 2019). Understanding the impact of the chemistry of the ionizable lipid on how the mRNA-iLNP formulation interacts with innate immune cells and initiates their activation could provide further insight into the optimization of this platform. Modification of other helper lipid components or the structure of mRNA-iLNPs (e.g., lipid molar ratios and lipid-to-mRNA ratio) can also be considered. iLNPs formulated with lipids conjugated with polysarcosine in place of PEG elicit reduced pro-inflammatory cytokine activity in an in vitro whole human blood assay and induce lower levels of hepatotoxicity markers after multiple intravenous administrations in mice (Nogueira et al., 2020). Both the choice of ionizable lipid and the lipid-to-mRNA ratio in the iLNP can be optimized to avoid systemic distribution of mRNA-iLNPs following intramuscular administration, which may be useful as a strategy to minimize systemic adverse events (Carrasco et al., 2021). Moreover, changing the route of administration of BNT162b2 from intramuscular to subcutaneous in a mouse model reduces the pro-inflammatory response without compromising vaccine efficacy (Syenina et al., 2022). In a phase I study for a nucleoside-modified mRNA-iLNP vaccine against influenza virus, the intradermal route was associated with higher rates of certain adverse events (Feldman et al., 2019).
Because there are many variables that can affect the reactogenicity and immunogenicity of mRNA vaccines, there are multiple paths to explore toward the development of less reactogenic and more effective vaccine candidates. This process will be greatly aided by expanding our knowledge of the interactions of mRNA-iLNP components with innate immune mechanisms.
Concluding remarks
The nucleoside-modified mRNA-iLNP platform represents a dramatic leap forward in vaccine development that has proven its value in a short time during the COVID-19 pandemic. Recent studies have helped to gain some insights into the mechanisms of action of this novel vaccine modality, and it is nearly certain that future research into the innate immune-sensing and immune-signaling pathways activated by this vaccine will yield improvements in this technology. We expect that in the coming years, these improvements will lead to a second generation of mRNA-iLNPs with enhanced efficacy, safety, and tolerability compared with BNT162b2 and mRNA-1273. A wide variety of future mRNA therapeutics may eventually become possible as we learn how to tailor mRNA and carrier molecules to preferentially activate the B cell response (most relevant to acute infections) vs. the T cell response (e.g., for chronic infections and cancer) (Sahin et al., 2020), to become completely immune silent (e.g., for gene therapy), and even to induce immunological tolerance to treat autoimmune diseases (Krienke et al., 2021).
Acknowledgments
N.P. is supported by the National Institute of Allergy and Infectious Diseases (R01AI146101, R01AI153064, P01AI158571) and the Katalin Karikó Fellowship Fund in Vaccine Development (University of Pennsylvania). R.V. is a post-doctoral fellow funded by the Special Research Fund (BOF) from Ghent University, Flanders, Belgium, and acknowledges support from Ghent University Concerted Research Action grant BOF21/GOA/033. K.L. is supported by the Swedish Research Council (2019-01036, 2020-05829) and the Knut and Alice Wallenberg Foundation (2021-0018). We thank Sunny Shin (University of Pennsylvania), Katalin Karikó (University of Pennsylvania and University of Szeged), and Caius G. Radu (University of California, Los Angeles) for scientific feedback and critical reading of the manuscript. The authors apologize to all colleagues whose great studies could not be cited here owing to space limitations.
Author contributions
R.V., M.J.H., K.L., and N.P. wrote the manuscript.
Declaration of interests
N.P. is named on a patent describing the use of nucleoside-modified mRNA in lipid nanoparticles as a vaccine platform (WO2016176330A1). N.P. and M.J.H. are named on a patent describing a nucleoside-modified mRNA vaccine against Zika virus (WO2018132537A1). We have disclosed those interests fully to the University of Pennsylvania and Children’s Hospital of Philadelphia, and we have in place an approved plan for managing any potential conflicts arising from licensing of our patents. R.V. is a named inventor on a patent related to mRNA vaccine design (WO2020058239A1).
References
- Abbas Y.M., Laudenbach B.T., Martinez-Montero S., Cencic R., Habjan M., Pichlmair A., Damha M.J., Pelletier J., Nagar B. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2'-O methylations. Proc. Natl. Acad. Sci. USA. 2017;114:E2106–E2115. doi: 10.1073/pnas.1612444114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alameh M.-G., Weissman D. In: In RNA Therapeutics. Giangrande P.H., de Franciscis V., Rossi J.J., editors. Academic Press; 2022. Chapter 7 - Nucleoside modifications of in vitro transcribed mRNA to reduce immunogenicity and improve translation of prophylactic and therapeutic antigens; pp. 141–169. [Google Scholar]
- Alameh M.G., Tombácz I., Bettini E., Lederer K., Sittplangkoon C., Wilmore J.R., Gaudette B.T., Soliman O.Y., Pine M., Hicks P., et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54:2877–2892.e7. doi: 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A.C., Medzhitov R., Flavell R.A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Anand P.K. Lipids, inflammasomes, metabolism, and disease. Immunol. Rev. 2020;297:108–122. doi: 10.1111/imr.12891. [DOI] [PubMed] [Google Scholar]
- Anderson B.R., Muramatsu H., Jha B.K., Silverman R.H., Weissman D., Karikó K. Nucleoside modifications in RNA limit activation of 2'-5'-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011;39:9329–9338. doi: 10.1093/nar/gkr586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson B.R., Muramatsu H., Nallagatla S.R., Bevilacqua P.C., Sansing L.H., Weissman D., Karikó K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Ansell S.M., Du X. 2017. Lipids and Lipid Nanoparticle Formulations for Delivery of Nucleic Acids. Patent Application 9738593B2. [Google Scholar]
- Arunachalam P.S., Scott M.K.D., Hagan T., Li C., Feng Y., Wimmers F., Grigoryan L., Trisal M., Edara V.V., Lai L., et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature. 2021;596:410–416. doi: 10.1038/s41586-021-03791-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J., et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25:1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baiersdörfer M., Boros G., Muramatsu H., Mahiny A., Vlatkovic I., Sahin U., Karikó K. A facile method for the removal of dsRNA contaminant from in vitro-transcribed mRNA. Mol. Ther. Nucleic Acids. 2019;15:26–35. doi: 10.1016/j.omtn.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S., Roberts P.C., Brown L.E., Truong H., Pattnaik A.K., Archer D.R., Barber G.N. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity. 2000;13:129–141. doi: 10.1016/s1074-7613(00)00014-5. [DOI] [PubMed] [Google Scholar]
- Ball D.P., Taabazuing C.Y., Griswold A.R., Orth E.L., Rao S.D., Kotliar I.B., Vostal L.E., Johnson D.C., Bachovchin D.A. Caspase-1 interdomain linker cleavage is required for pyroptosis. Life Sci. Alliance. 2020;3 doi: 10.26508/lsa.202000664. e202000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind S., Salzberger B., Hitzenbichler F., Scigala K., Einhauser S., Wagner R., Gessner A., Koestler J., Peterhoff D. Association between reactogenicity and immunogenicity after vaccination with BNT162b2. Vaccines. 2021;9:1089. doi: 10.3390/vaccines9101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoletti A., Le Bert N., Tan A.T. SARS-CoV-2-specific T cells in the changing landscape of the COVID-19 pandemic. Immunity. 2022;55:1764–1778. doi: 10.1016/j.immuni.2022.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossaller L., Chiang P.I., Schmidt-Lauber C., Ganesan S., Kaiser W.J., Rathinam V.A.K., Mocarski E.S., Subramanian D., Green D.R., Silverman N., et al. Cutting edge: FAS (CD95) mediates noncanonical IL-1beta and IL-18 maturation via caspase-8 in an RIP3-independent manner. J. Immunol. 2012;189:5508–5512. doi: 10.4049/jimmunol.1202121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco M.J., Alishetty S., Alameh M.G., Said H., Wright L., Paige M., Soliman O., Weissman D., Cleveland T.E., 4th, Grishaev A., Buschmann M.D. Ionization and structural properties of mRNA lipid nanoparticles influence expression in intramuscular and intravascular administration. Commun. Biol. 2021;4:956. doi: 10.1038/s42003-021-02441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.H., Schroder K. Inflammasome signaling and regulation of interleukin-1 family cytokines. J. Exp. Med. 2020;217 doi: 10.1084/jem.20190314. e20190314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin-Bardales J., Gee J., Myers T. Reactogenicity following receipt of mRNA-based COVID-19 vaccines. JAMA. 2021;325:2201–2202. doi: 10.1001/jama.2021.5374. [DOI] [PubMed] [Google Scholar]
- Choi Y.S., Eto D., Yang J.A., Lao C., Crotty S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J. Immunol. 2013;190:3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullis P.R., Hope M.J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 2017;25:1467–1475. doi: 10.1016/j.ymthe.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarkar S.C., Wang C., Miller M.T., Ramanathan A., Jiang F., Khan A.G., Patel S.S., Marcotrigiano J. Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc. Natl. Acad. Sci. USA. 2016;113:596–601. doi: 10.1073/pnas.1515152113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilliard S.A., Cheng Q., Siegwart D.J. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2109256118. e2109256118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin A.F., Wang C., Marcotrigiano J., Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio. 2016;7 doi: 10.1128/mBio.00833-16. 008333-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia U., Ramishetti S., Rosenfeld R., Dammes N., Bar-Haim E., Naidu G.S., Makdasi E., Yahalom-Ronen Y., Tamir H., Paran N., et al. Design of SARS-CoV-2 hFc-conjugated receptor-binding domain mRNA vaccine delivered via lipid nanoparticles. ACS Nano. 2021;15:9627–9637. doi: 10.1021/acsnano.0c10180. [DOI] [PubMed] [Google Scholar]
- European Medicines Agency . 2020. Comirnaty: EPAR - Public Assessment Report.https://www.ema.europa.eu/en/documents/assessment-report/comirnaty-epar-public-assessment-report_en.pdf [Google Scholar]
- European Medicines Agency . 2021. Spikevax: EPAR - Public Assessment Report.https://www.ema.europa.eu/en/documents/assessment-report/spikevax-previously-covid-19-vaccine-moderna-epar-public-assessment-report_en.pdf [Google Scholar]
- Feldman R.A., Fuhr R., Smolenov I., Mick Ribeiro A., Panther L., Watson M., Senn J.J., Smith M., Almarsson Ӧ., Pujar H.S., et al. mRNA vaccines against H10N8 and H7N9 influenza viruses of pandemic potential are immunogenic and well tolerated in healthy adults in phase 1 randomized clinical trials. Vaccine. 2019;37:3326–3334. doi: 10.1016/j.vaccine.2019.04.074. [DOI] [PubMed] [Google Scholar]
- Foster J.B., Choudhari N., Perazzelli J., Storm J., Hofmann T.J., Jain P., Storm P.B., Pardi N., Weissman D., Waanders A.J., et al. Purification of mRNA encoding chimeric antigen receptor is critical for generation of a robust T-cell response. Hum. Gene Ther. 2019;30:168–178. doi: 10.1089/hum.2018.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia V., Schiffelers R.M., Cullis P.R., Witzigmann D. The biomolecular corona of lipid nanoparticles for gene therapy. Bioconjug. Chem. 2020;31:2046–2059. doi: 10.1021/acs.bioconjchem.0c00366. [DOI] [PubMed] [Google Scholar]
- Fuller D.H., Berglund P. Amplifying RNA vaccine development. N. Engl. J. Med. 2020;382:2469–2471. doi: 10.1056/NEJMcibr2009737. [DOI] [PubMed] [Google Scholar]
- Gärtner B.C., Weinke T., Wahle K., Kwetkat A., Beier D., Schmidt K.J., Schwarz T.F. Importance and value of adjuvanted influenza vaccine in the care of older adults from a European perspective - a systematic review of recently published literature on real-world data. Vaccine. 2022;40:2999–3008. doi: 10.1016/j.vaccine.2022.04.019. [DOI] [PubMed] [Google Scholar]
- Geall A.J., Verma A., Otten G.R., Shaw C.A., Hekele A., Banerjee K., Cu Y., Beard C.W., Brito L.A., Krucker T., et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109:14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebre M.S., Rauch S., Roth N., Yu J., Chandrashekar A., Mercado N.B., He X., Liu J., McMahan K., Martinot A., et al. Optimization of non-coding regions for a non-modified mRNA COVID-19 vaccine. Nature. 2022;601:410–414. doi: 10.1038/s41586-021-04231-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science (New York, N.Y.) 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrera G., Picozza M., D'Orso S., Placido R., Pirronello M., Verdiani A., Termine A., Fabrizio C., Giannessi F., Sambucci M., et al. BNT162b2 vaccination induces durable SARS-CoV-2-specific T cells with a stem cell memory phenotype. Sci. Immunol. 2021;6:eabl5344. doi: 10.1126/sciimmunol.abl5344. [DOI] [PubMed] [Google Scholar]
- Hanamsagar R., Torres V., Kielian T. Inflammasome activation and IL-1beta/IL-18 processing are influenced by distinct pathways in microglia. J. Neurochem. 2011;119:736–748. doi: 10.1111/j.1471-4159.2011.07481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held J., Esse J., Tascilar K., Steininger P., Schober K., Irrgang P., Alsalameh R., Tenbusch M., Seggewies C., Bogdan C. Reactogenicity correlates only weakly with humoral immunogenicity after COVID-19 vaccination with BNT162b2 mRNA (Comirnaty) Vaccines. 2021;9:1063. doi: 10.3390/vaccines9101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson J.M., Ujita A., Hill E., Yousif-Rosales S., Smith C., Ko N., McReynolds T., Cabral C.R., Escamilla-Powers J.R., Houston M.E. Cap 1 messenger RNA synthesis with co-transcriptional cleancap analog by in vitro transcription. Curr. Protoc. 2021;1:e39. doi: 10.1002/cpz1.39. [DOI] [PubMed] [Google Scholar]
- Hogan M.J., Pardi N. mRNA vaccines in the COVID-19 pandemic and neyond. Annu. Rev. Med. 2022;73:17–39. doi: 10.1146/annurev-med-042420-112725. [DOI] [PubMed] [Google Scholar]
- Holm C.K., Jensen S.B., Jakobsen M.R., Cheshenko N., Horan K.A., Moeller H.B., Gonzalez-Dosal R., Rasmussen S.B., Christensen M.H., Yarovinsky T.O., et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Bauernfeind F., Halle A., Samstad E.O., Kono H., Rock K.L., Fitzgerald K.A., Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten L.A.B., Netea M.G., Fantuzzi G., Koenders M.I., Helsen M.M.A., Sparrer H., Pham C.T., van der Meer J.W.M., Dinarello C.A., van den Berg W.B. Inflammatory arthritis in caspase 1 gene-deficient mice: contribution of proteinase 3 to caspase 1-independent production of bioactive interleukin-1beta. Arthritis Rheum. 2009;60:3651–3662. doi: 10.1002/art.25006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju Y., Lee W.S., Pilkington E.H., Kelly H.G., Li S., Selva K.J., Wragg K.M., Subbarao K., Nguyen T.H.O., Rowntree L.C., et al. Anti-PEG antibodies boosted in humans by SARS-CoV-2 lipid nanoparticle mRNA vaccine. ACS Nano. 2022;16:11769–11780. doi: 10.1021/acsnano.2c04543. [DOI] [PubMed] [Google Scholar]
- Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Karikó K., Muramatsu H., Ludwig J., Weissman D. Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res. 2011;39:e142. doi: 10.1093/nar/gkr695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierska K., Thomas P.G. Count on us: T cells in SARS-CoV-2 infection and vaccination. Cell reports. Cell Rep. Med. 2022;3:100562. doi: 10.1016/j.xcrm.2022.100562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Zhou J.Q., Horvath S.C., Schmitz A.J., Sturtz A.J., Lei T., Liu Z., Kalaidina E., Thapa M., Alsoussi W.B., et al. Germinal centre-driven maturation of B cell response to mRNA vaccination. Nature. 2022;604:141–145. doi: 10.1038/s41586-022-04527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobiyama K., Ishii K.J. Making innate sense of mRNA vaccine adjuvanticity. Nat. Immunol. 2022;23:474–476. doi: 10.1038/s41590-022-01168-4. [DOI] [PubMed] [Google Scholar]
- Kremsner P.G., Ahuad Guerrero R.A., Arana-Arri E., Aroca Martinez G.J., Bonten M., Chandler R., Corral G., De Block E.J.L., Ecker L., Gabor J.J., et al. Efficacy and safety of the CVnCoV SARS-CoV-2 mRNA vaccine candidate in ten countries in Europe and Latin America (HERALD): a randomised, observer-blinded, placebo-controlled, phase 2b/3 trial. Lancet Infect. Dis. 2022;22:329–340. doi: 10.1016/S1473-3099(21)00677-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krienke C., Kolb L., Diken E., Streuber M., Kirchhoff S., Bukur T., Akilli-Öztürk Ö., Kranz L.M., Berger H., Petschenka J., et al. A noninflammatory mRNA vaccine for treatment of experimental autoimmune encephalomyelitis. Science (New York, N.Y.) 2021;371:145–153. doi: 10.1126/science.aay3638. [DOI] [PubMed] [Google Scholar]
- Lenart K., Hellgren F., Ols S., Yan X., Cagigi A., Cerveira R.A., Winge I., Hanczak J., Mueller S.O., Jasny E., et al. A third dose of the unmodified COVID-19 mRNA vaccine CVnCoV enhances quality and quantity of immune responses. Mol Ther Methods Clin Dev. 2022;27:309–323. doi: 10.1016/j.omtm.2022.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Lee A., Grigoryan L., Arunachalam P.S., Scott M.K.D., Trisal M., Wimmers F., Sanyal M., Weidenbacher P.A., Feng Y., et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat. Immunol. 2022;23:543–555. doi: 10.1038/s41590-022-01163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wang Y., Sun Q., Knopf J., Herrmann M., Lin L., Jiang J., Shao C., Li P., He X., et al. Immune response in COVID-19: what is next? Cell Death Differ. 2022;29:1107–1122. doi: 10.1038/s41418-022-01015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Lindgren G., Lin A., Thompson E.A., Ols S., Röhss J., John S., Hassett K., Yuzhakov O., Bahl K., et al. Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther. 2017;25:2635–2647. doi: 10.1016/j.ymthe.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F., Lindgren G., Sandgren K.J., Thompson E.A., Francica J.R., Seubert A., De Gregorio E., Barnett S., O'Hagan D.T., Sullivan N.J., et al. Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci. Transl. Med. 2017;9:eaal2094. doi: 10.1126/scitranslmed.aal2094. [DOI] [PubMed] [Google Scholar]
- Lindgren G., Ols S., Liang F., Thompson E.A., Lin A., Hellgren F., Bahl K., John S., Yuzhakov O., Hassett K.J., et al. Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol. 2017;8:1539. doi: 10.3389/fimmu.2017.01539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonez C., Bessodes M., Scherman D., Vandenbranden M., Escriou V., Ruysschaert J.M. Cationic lipid nanocarriers activate Toll-like receptor 2 and NLRP3 inflammasome pathways. Nanomedicine. 2014;10:775–782. doi: 10.1016/j.nano.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Low J.G., de Alwis R., Chen S., Kalimuddin S., Leong Y.S., Mah T.K.L., Yuen N., Tan H.C., Zhang S.L., Sim J.X.Y., et al. A phase 1/2 randomized, double-blinded, placebo controlled ascending dose trial to assess the safety, tolerability and immunogenicity of ARCT-021 in healthy adults. medRxiv. 2021 doi: 10.1101/2021.07.01.21259831. Preprint at. [DOI] [Google Scholar]
- Lutz J., Lazzaro S., Habbeddine M., Schmidt K.E., Baumhof P., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Heidenreich R., Fotin-Mleczek M. Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ vaccines. 2017;2:29. doi: 10.1038/s41541-017-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisel K., Sasso M.S., Potin L., Swartz M.A. Exploiting lymphatic vessels for immunomodulation: rationale, opportunities, and challenges. Adv. Drug Deliv. Rev. 2017;114:43–59. doi: 10.1016/j.addr.2017.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolova V., Flace A., Bauer M., Schwarz K., Saudan P., Bachmann M.F. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- Mantovani A., Netea M.G. Trained innate immunity, epigenetics, and covid-19. N. Engl. J. Med. 2020;383:1078–1080. doi: 10.1056/NEJMcibr2011679. [DOI] [PubMed] [Google Scholar]
- Mantovani A., Dinarello C.A., Molgora M., Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity. 2019;50:778–795. doi: 10.1016/j.immuni.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L., Li L., Huang Y., Delcassian D., Chahal J., Han J., Shi Y., Sadtler K., Gao W., Lin J., et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019;37:1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- Miller M.A., Ganesan A.P.V., Luckashenak N., Mendonca M., Eisenlohr L.C. Endogenous antigen processing drives the primary CD4+ T cell response to influenza. Nat. Med. 2015;21:1216–1222. doi: 10.1038/nm.3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitoma H., Hanabuchi S., Kim T., Bao M., Zhang Z., Sugimoto N., Liu Y.J. The DHX33 RNA helicase senses cytosolic RNA and activates the NLRP3 inflammasome. Immunity. 2013;39:123–135. doi: 10.1016/j.immuni.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok K.W., Lam A.M., Cullis P.R. Stabilized plasmid-lipid particles: factors influencing plasmid entrapment and transfection properties. Biochim. Biophys. Acta. 1999;1419:137–150. doi: 10.1016/s0005-2736(99)00059-0. [DOI] [PubMed] [Google Scholar]
- Mu X., Greenwald E., Ahmad S., Hur S. An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res. 2018;46:5239–5249. doi: 10.1093/nar/gky177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudd P.A., Minervina A.A., Pogorelyy M.V., Turner J.S., Kim W., Kalaidina E., Petersen J., Schmitz A.J., Lei T., Haile A., et al. SARS-CoV-2 mRNA vaccination elicits a robust and persistent T follicular helper cell response in humans. Cell. 2022;185:603–613.e15. doi: 10.1016/j.cell.2021.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndeupen S., Bouteau A., Herbst C., Qin Z., Jacobsen S., Powers N.E., Hutchins Z., Kurup D., Diba L.Z., Watson M., et al. Langerhans cells and cDC1s play redundant roles in mRNA-LNP induced protective anti-influenza and anti-SARS-CoV-2 immune responses. PLoS Pathog. 2022;18:e1010255. doi: 10.1371/journal.ppat.1010255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndeupen S., Qin Z., Jacobsen S., Bouteau A., Estanbouli H., Igyártó B.Z. The mRNA-LNP platform's lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience. 2021;24:103479. doi: 10.1016/j.isci.2021.103479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J., Sorensen E.W., Mintri S., Rabideau A.E., Zheng W., Besin G., Khatwani N., Su S.V., Miracco E.J., Issa W.J., et al. Impact of mRNA chemistry and manufacturing process on innate immune activation. Sci. Adv. 2020;6:eaaz6893. doi: 10.1126/sciadv.aaz6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira S.S., Schlegel A., Maxeiner K., Weber B., Barz M., Schroer M.A., Blanchet C.E., Svergun D.I., Ramishetti S., Peer D., et al. Polysarcosine-functionalized lipid nanoparticles for therapeutic mRNA delivery. ACS Appl. Nano Mater. 2020;3:10634–10645. doi: 10.1021/acsanm.0c01834. [DOI] [Google Scholar]
- Oberhardt V., Luxenburger H., Kemming J., Schulien I., Ciminski K., Giese S., Csernalabics B., Lang-Meli J., Janowska I., Staniek J., et al. Rapid and stable mobilization of CD8(+) T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsenbein A.F., Sierro S., Odermatt B., Pericin M., Karrer U., Hermans J., Hemmi S., Hengartner H., Zinkernagel R.M. Roles of tumour localization, second signals and cross priming in cytotoxic T-cell induction. Nature. 2001;411:1058–1064. doi: 10.1038/35082583. [DOI] [PubMed] [Google Scholar]
- Ols S., Yang L., Thompson E.A., Pushparaj P., Tran K., Liang F., Lin A., Eriksson B., Karlsson Hedestam G.B., Wyatt R.T., Loré K. Route of vaccine administration alters antigen trafficking but not innate or adaptive Immunity. Cell Rep. 2020;30:3964–3971.e7. doi: 10.1016/j.celrep.2020.02.111. e3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oussoren C., Storm G. Liposomes to target the lymphatics by subcutaneous administration. Adv. Drug Deliv. Rev. 2001;50:143–156. doi: 10.1016/s0169-409x(01)00154-5. [DOI] [PubMed] [Google Scholar]
- Packer M., Gyawali D., Yerabolu R., Schariter J., White P. A novel mechanism for the loss of mRNA activity in lipid nanoparticle delivery systems. Nat. Commun. 2021;12:6777. doi: 10.1038/s41467-021-26926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. e2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Pelc R.S., Muramatsu H., Andersen H., DeMaso C.R., Dowd K.A., Sutherland L.L., Scearce R.M., Parks R., et al. Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature. 2017;543:248–251. doi: 10.1038/nature21428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Muramatsu H., Weissman D., Karikó K. In vitro transcription of long RNA containing modified nucleosides. Methods Mol. Biol. 2013;969:29–42. doi: 10.1007/978-1-62703-260-5_2. [DOI] [PubMed] [Google Scholar]
- Pardi N., Tuyishime S., Muramatsu H., Kariko K., Mui B.L., Tam Y.K., Madden T.D., Hope M.J., Weissman D. Expression kinetics of nucleoside-modified mRNA delivered in lipid nanoparticles to mice by various routes. J. Control. Release. 2015;217:345–351. doi: 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip N.H., Dillon C.P., Snyder A.G., Fitzgerald P., Wynosky-Dolfi M.A., Zwack E.E., Hu B., Fitzgerald L., Mauldin E.A., Copenhaver A.M., et al. Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-kappaB and MAPK signaling. Proc. Natl. Acad. Sci. USA. 2014;111:7385–7390. doi: 10.1073/pnas.1403252111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Rehwinkel J., Kato H., Takeuchi O., Akira S., Way M., Schiavo G., Reis e Sousa C. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock K.M., Cheeseman H.M., Szubert A.J., Libri V., Boffito M., Owen D., Bern H., O'Hara J., McFarlane L.R., Lemm N.M., et al. Safety and immunogenicity of a self-amplifying RNA vaccine against COVID-19: COVAC1, a phase I, dose-ranging trial. EClinicalMedicine. 2022;44:101262. doi: 10.1016/j.eclinm.2021.101262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy S.T., Rehor A., Schmoekel H.G., Hubbell J.A., Swartz M.A. In vivo targeting of dendritic cells in lymph nodes with poly(propylene sulfide) nanoparticles. J. Control. Release. 2006;112:26–34. doi: 10.1016/j.jconrel.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Richner J.M., Himansu S., Dowd K.A., Butler S.L., Salazar V., Fox J.M., Julander J.G., Tang W.W., Shresta S., Pierson T.C., et al. Modified mRNA vaccines protect against zika virus infection. Cell. 2017;168:1114–1125.e10. doi: 10.1016/j.cell.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A., et al. Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022;185:1025–1040.e14. doi: 10.1016/j.cell.2022.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblum H.G., Gee J., Liu R., Marquez P.L., Zhang B., Strid P., Abara W.E., McNeil M.M., Myers T.R., Hause A.M., et al. Safety of mRNA vaccines administered during the initial 6 months of the US COVID-19 vaccination programme: an observational study of reports to the Vaccine Adverse Event Reporting System and v-safe. Lancet Infect. Dis. 2022;22:802–812. doi: 10.1016/S1473-3099(22)00054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U., Oehm P., Derhovanessian E., Jabulowsky R.A., Vormehr M., Gold M., Maurus D., Schwarck-Kokarakis D., Kuhn A.N., Omokoko T., et al. An RNA vaccine drives immunity in checkpoint-inhibitor-treated melanoma. Nature. 2020;585:107–112. doi: 10.1038/s41586-020-2537-9. [DOI] [PubMed] [Google Scholar]
- Schuberth-Wagner C., Ludwig J., Bruder A.K., Herzner A.M., Zillinger T., Goldeck M., Schmidt T., Schmid-Burgk J.L., Kerber R., Wolter S., et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N1-2'O-Methylated self RNA. Immunity. 2015;43:41–51. doi: 10.1016/j.immuni.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal L., Morelle D., Kaaber K., Destexhe E., Garçon N. Non-clinical safety assessment of single and repeated intramuscular administration of a human papillomavirus-16/18 vaccine in rabbits and rats. J. Appl. Toxicol. 2015;35:1577–1585. doi: 10.1002/jat.3131. [DOI] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenderov K., Riteau N., Yip R., Mayer-Barber K.D., Oland S., Hieny S., Fitzgerald P., Oberst A., Dillon C.P., Green D.R., et al. Cutting edge: Endoplasmic reticulum stress licenses macrophages to produce mature IL-1beta in response to TLR4 stimulation through a caspase-8- and TRIF-dependent pathway. J. Immunol. 2014;192:2029–2033. doi: 10.4049/jimmunol.1302549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan G., Thoryk E.A., Cox K.S., Meschino S., Dubey S.A., Vora K.A., Celano R., Gindy M., Casimiro D.R., Bett A.J. A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccine. 2016;34:110–119. doi: 10.1016/j.vaccine.2015.10.132. [DOI] [PubMed] [Google Scholar]
- Swaminathan G., Thoryk E.A., Cox K.S., Smith J.S., Wolf J.J., Gindy M.E., Casimiro D.R., Bett A.J. A tetravalent sub-unit dengue vaccine formulated with ionizable cationic lipid nanoparticle induces significant immune responses in rodents and non-human primates. Sci. Rep. 2016;6:34215. doi: 10.1038/srep34215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syenina A., Gan E.S., Toh J.Z.N., de Alwis R., Lin L.Z., Tham C.Y.L., Yee J.X., Leong Y.S., Sam H., Cheong C., et al. Adverse effects following anti-COVID-19 vaccination with mRNA-based BNT162b2 are alleviated by altering the route of administration and correlate with baseline enrichment of T and NK cell genes. PLoS Biol. 2022;20:e3001643. doi: 10.1371/journal.pbio.3001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szebeni J., Storm G., Ljubimova J.Y., Castells M., Phillips E.J., Turjeman K., Barenholz Y., Crommelin D.J.A., Dobrovolskaia M.A. Applying lessons learned from nanomedicines to understand rare hypersensitivity reactions to mRNA-based SARS-CoV-2 vaccines. Nat. Nanotechnol. 2022;17:337–346. doi: 10.1038/s41565-022-01071-x. [DOI] [PubMed] [Google Scholar]
- Tahtinen S., Tong A.J., Himmels P., Oh J., Paler-Martinez A., Kim L., Wichner S., Oei Y., McCarron M.J., Freund E.C., et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat. Immunol. 2022;23:532–542. doi: 10.1038/s41590-022-01160-y. [DOI] [PubMed] [Google Scholar]
- Tam H.H., Melo M.B., Kang M., Pelet J.M., Ruda V.M., Foley M.H., Hu J.K., Kumari S., Crampton J., Baldeon A.D., et al. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci. USA. 2016;113:E6639–E6648. doi: 10.1073/pnas.1606050113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Legat A., Adam E., Steuve J., Gatot J.S., Vandenbranden M., Ulianov L., Lonez C., Ruysschaert J.M., Muraille E., et al. DiC14-amidine cationic liposomes stimulate myeloid dendritic cells through Toll-like receptor 4. Eur. J. Immunol. 2008;38:1351–1357. doi: 10.1002/eji.200737998. [DOI] [PubMed] [Google Scholar]
- Tanji H., Ohto U., Shibata T., Taoka M., Yamauchi Y., Isobe T., Miyake K., Shimizu T. Toll-like receptor 8 senses degradation products of single-stranded RNA. Nat. Struct. Mol. Biol. 2015;22:109–115. doi: 10.1038/nsmb.2943. [DOI] [PubMed] [Google Scholar]
- Thess A., Grund S., Mui B.L., Hope M.J., Baumhof P., Fotin-Mleczek M., Schlake T. Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol. Ther. 2015;23:1456–1464. doi: 10.1038/mt.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J.S., O'Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeke R., Lentacker I., De Smedt S.C., Dewitte H. Three decades of messenger RNA vaccine development. Nano Today. 2019;28:100766. doi: 10.1016/j.nantod.2019.100766. [DOI] [Google Scholar]
- Wei J., Pouwels K.B., Stoesser N., Matthews P.C., Diamond I., Studley R., Rourke E., Cook D., Bell J.I., Newton J.N., et al. Antibody responses and correlates of protection in the general population after two doses of the ChAdOx1 or BNT162b2 vaccines. Nat. Med. 2022;28:1072–1082. doi: 10.1038/s41591-022-01721-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann D., Binder C.J. The innate immune response to products of phospholipid peroxidation. Biochim. Biophys. Acta. 2012;1818:2465–2475. doi: 10.1016/j.bbamem.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng D., Marty-Roix R., Ganesan S., Proulx M.K., Vladimer G.I., Kaiser W.J., Mocarski E.S., Pouliot K., Chan F.K.M., Kelliher M.A., et al. Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proc. Natl. Acad. Sci. USA. 2014;111:7391–7396. doi: 10.1073/pnas.1403477111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry E.J., Barouch D.H. T cell immunity to COVID-19 vaccines. Science. 2022;377:821–822. doi: 10.1126/science.add2897. [DOI] [PubMed] [Google Scholar]
- Wu M.Z., Asahara H., Tzertzinis G., Roy B. Synthesis of low immunogenicity RNA with high-temperature in vitro transcription. RNA. 2020;26:345–360. doi: 10.1261/rna.073858.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y.M., Gale M., Jr., Akira S., et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- Yoneyama M., Kikuchi M., Natsukawa T., Shinobu N., Imaizumi T., Miyagishi M., Taira K., Akira S., Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Mateus J., Coelho C.H., Dan J.M., Moderbacher C.R., Gálvez R.I., Cortes F.H., Grifoni A., Tarke A., Chang J., et al. Humoral and cellular immune memory to four COVID-19 vaccines. Cell. 2022;185:2434–2451.e17. doi: 10.1016/j.cell.2022.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Ohto U., Shibata T., Krayukhina E., Taoka M., Yamauchi Y., Tanji H., Isobe T., Uchiyama S., Miyake K., Shimizu T. Structural analysis reveals that toll-like receptor 7 is a dual receptor for guanosine and single-stranded RNA. Immunity. 2016;45:737–748. doi: 10.1016/j.immuni.2016.09.011. [DOI] [PubMed] [Google Scholar]