Abstract
During embryonic development, the olfactory sensory neurons (OSNs) and the gonadotropic-releasing hormone neurons (GNRHNs) migrate from the early nasal cavity, known as the olfactory placode, to the brain. Defects in the development of OSNs and GNRHNs result in neurodevelopmental disorders such as anosmia and congenital hypogonadotropic hypogonadism, respectively. Treatments do not restore the defective neurons in these disorders, and as a result, patients have a diminished sense of smell or a gonadotropin hormone deficiency. Human pluripotent stem cells (hPSCs) can produce any cell type in the body; therefore, they are an invaluable tool for cell replacement therapies. Transplantation of olfactory placode progenitors, derived from hPSCs, is a promising therapeutic to replace OSNs and GNRHNs and restore tissue function. Protocols to generate olfactory placode progenitors are limited, and thus, we describe, in this study, a novel in vitro model for olfactory placode differentiation in hPSCs, which is capable of producing both OSNs and GNRHNs. Our study investigates the major developmental signaling factors that recapitulate the embryonic development of the olfactory tissue. We demonstrate that induction of olfactory placode in hPSCs requires bone morphogenetic protein inhibition, wingless/integrated protein inhibition, retinoic acid inhibition, transforming growth factor alpha activation, and fibroblast growth factor 8 activation. We further show that the protocol transitions hPSCs through the anterior pan-placode ectoderm and neural ectoderm regions in early development while preventing neural crest and non-neural ectoderm regions. Finally, we demonstrate production of OSNs and GNRHNs by day 30 of differentiation. Our study is the first to report on OSN differentiation in hPSCs.
Keywords: human pluripotent stem cell, differentiation, olfactory placode, olfactory sensory neurons, gonadotropin-releasing hormone neurons
Introduction
Human pluripotent stem cells (hPSCs) are an invaluable tool to study human embryonic development [1], and their ability to produce large numbers of specialized cells support their potential as a therapy to replace damaged cells [2]. A number of preclinical studies showed that transplantation of hPSC-derived neural progenitors into animal models of disease substantially restored tissue function through replacement of neurons and glia [3,4]. Highly efficient differentiation protocols were developed for a plethora of cell types and are essential for their function in transplantations [3,4].
The olfactory tissue is a unique model to study development and neurogenesis because of its diverse population of cells and its lifetime capacity to regenerate [5,6]. During early embryonic development, olfactory sensory neurons (OSNs) and gonadotropin-releasing hormone neurons (GNRHNs) reside in the olfactory placode and travel to their designated locations in the brain [7,8]. Defects in the embryonic development of OSNs lead to congenital anosmia, the inability to detect odors [9]. Additionally, age-related olfactory dysfunction may occur in adults 40 years or older due to progressive degeneration of OSNs and is often the first symptom detected in those with neurodegeneration [9]. Olfactory dysfunction is estimated to occur in 3%–20% of the population in the United States [9]. Defects in GNRHNs cause a hormone deficiency called congenital hypogonadotropic hypogonadism (CHH), which affects 3.75 in 100,000 people and results in failure to sexually mature and reproduce [7]. Even rarer, Kallmann syndrome is the combination of anosmia and CHH [7].
Transplantation of olfactory progenitors, derived from hPSCs, have the potential to treat anosmia and CHH through replacement of OSNs and GNRHNs, respectively. The first step toward their use in the clinic is to create a model to differentiate hPSCs into olfactory placode through a sequence of molecular cues that recapitulate characteristics of their natural development. Placodes are thickenings of ectoderm, derived from a common region called the pan-placode ectoderm (PPE), which develop into sensory organs such as the ocular lens, olfactory tissue, or inner ear [10]. Previous studies demonstrated the in vitro production of lens epithelial cells, hormone-releasing pituitary cells, trigeminal neurons, inner ear hair cells, and auditory neurons in hPSCs [11–22]. However, there are no studies on OSN differentiation in hPSCs and only one study on GNRHN differentiation [23].
During gastrulation, antagonists of bone morphogenetic proteins (BMPs), nodal growth differentiation factors (NODALs), wingless/integrated proteins (WNTs), and fibroblast growth factors (FGFs) in the epiblast cells promote ectoderm differentiation [24,25]. In the early gastrula, different BMP signals distinguish between the various ectodermal regions, including the early PPE [10,26]. Subsequently, during the late gastrula, BMP inhibition is required for the completion of PPE development [10,26]. Additionally, the combination of BMP inhibition, WNT inhibition, and FGF activation promotes optimal PPE development [27]. The olfactory placode is derived from the anterior PPE and specified during late-stage gastrulation [10]. BMP inhibition plays a major role in promoting an olfactory fate versus a lens fate [28,29]. Furthermore, antagonists of WNT and retinoic acid (RA) promote anterior and rostral cell fates while inhibiting posterior placodes [30–34].
Based on evidence from these published studies, we established a novel protocol for olfactory placode differentiation and demonstrated molecular analysis supporting a role for neural olfactory placode expression and the production of OSNs and GNRHNs in hPSCs.
Materials and Methods
Stem cell cultures
The hPSC line used was iPS-IMR90-1 (passage 40–60), purchased from WiCell [35] under a Materials Transfer Agreement (WiCell agreement 20-W0304). The use of this de-identified cell line does not constitute human subjects research. Stem cells were maintained on Matrigel-coated plates (Corning, Cat. No. 354277), cultured in mTeSR1 media (Stem Cell Technologies, Cat. No. 85850), and passaged following the WiCell Feeder Independent mTeSR1 protocol. Stem cell cultures were passaged every 4–6 days using Dispase I (Life Technologies, Cat. No. 17105-041) at densities between 60% and 80% confluency and a 1:3–1:5 split ratio.
Directed differentiation of hPSCs into olfactory placode cells and neurons
To start differentiation, hPSCs were dissociated into single cells using Accutase (Stem Cell Technologies, Cat. No. 07920) and seeded at a density of 50,000 cells/cm2 in 24-well plates coated with Matrigel (Corning, Cat. No. 354277). Cells were grown in an insulin–transferrin–selenite (ITS) base differentiation media, consisting of DMEM/F12 medium (Thermo Fisher, Cat. No. 11320-033) supplemented with 10 mL/L penicillin–streptomycin (Thermo Fisher, Cat. No. 15140-122), 1.35 g/L glucose (Thermo Fisher, Cat. No. 15023021), 5 mg/L insulin solution human (Sigma-Aldrich, Cat. No. I9278), 50 mg/L apo-transferrin human (Sigma-Aldrich, Cat. No. T2036-500), and 0.03 mM sodium selenite (Sigma-Aldrich, Cat. No. S5261). Cells were treated with 10 μM Y-27632 dihydrochloride (Sigma-Aldrich, Cat. No. Y0503) for the first 24 h after seeding and differentiation media were changed every 24 h until the end of the experiment.
For olfactory placode experiments, the small molecules and growth factors added to the ITS media for directed differentiation included: 300 nM LDN-193189 (AXON Medchem, Cat. No. 1509), 1 μM SB-431542 (StemMACS, Cat. No. 130-105-336), 1 μM IWP2 (StemMACS, Cat. No. 130-105-335), 100 nM AGN193109 (Tocris, Cat. No. 5758), 10 ng/mL transforming growth factor alpha (TGFα; Stem Cell Technologies, Cat. No. 78123), and 10 ng/mL FGF8 (PeproTech, Cat. No. 100-25). Timed addition of these small molecules and growth factors are detailed in the Results section.
For the time course experiment and experiments in which we differentiated OSNs and GNRHNs, neural media were used from day 12 to 30 of differentiation, and 1 μM DAPT (StemMACS, Cat. No. 130-103-924) was added from day 24 to 30 to accelerate neuronal differentiation. Neural media contained DMEM/F12 supplemented with 10 mL/L penicillin–streptomycin (Thermo Fisher, Cat. No. 15140-122), 1.35 g/L glucose (Thermo Fisher, Cat. No. 15023021), and N2 supplement-B (Stem Cell Technologies, Cat. No. 07156).
Quantitative real-time polymerase chain reaction
Total RNA was extracted using the RNeasy Micro Kit (Qiagen, Cat. No. 74004) and RNase-Free DNase Kit (Qiagen, Cat. No. 79254). RNA was reversed transcribed into cDNA using the High-Capacity RNA to cDNA Kit (Applied Biosystems, Cat. No. 4387406). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using the SybrGreen PCR Master Mix (Applied Biosystems, Cat. No. 4309155) and reactions were run on the real-time PCR thermocycler (Applied Biosystems). Primers were designed using the PrimerQuest Tool and purchased from Integrated DNA Technologies. A complete list of the primers is provided in Supplementary Table S1. Data were normalized to the gene 18S (internal control) and fold change was calculated relative to Ct values from vehicle-treated groups or day 0 stem cells in the time course experiment using the ΔΔCt method. There were five experimental replicates (n = 5) for all experiments, except the time course experiment which had four experimental replicates (n = 4).
RNA sequencing
Total RNA extracted at day 30 of differentiation (either vehicle or Final+DAPT treated, n = 3), were subjected to RNA sequencing (RNA-Seq). Preparation of RNA library and transcriptome sequencing was conducted by Novogene Co., Ltd. (UC Davis, CA). Briefly, cDNA was generated from purified, fragmented mRNA using random hexamers and used for library preparation. Paired-end reads were generated using the Illumina platform. High-quality reads obtained after quality control were mapped to the hg38 reference genome using Hisat2 (v. 2.0.5). FeatureCounts (v1.5.0-p3) was used to generate read counts and FPKM of each gene was calculated. The R package, DESeq2 (1.20.0) was used to assess differential expression between vehicle-treated and Final+DAPT-treated groups. P values obtained were adjusted using the Benjamini–Hochberg approach.
Genes were filtered to exclude low read counts (<10), and only those with an adjusted P value <0.05 and |log2FC| > 1 were considered as differentially expressed. We examined the presence of differentially expressed genes in human olfactory placode formation (GO: 0071698, 0071699, 0071700, 0030910, 0060166) using gene ontology data [36–38] and the Park and Saint-Jeannet, 2010 article [39].
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde (Alfa Aesar, Cat. No. 43368) for 10 min, permeabilized with 0.3% Triton (Sigma, Cat. No. T9284-500 ml) for 10 min, and blocked with a solution of 2% fetal bovine serum (Thermo Fisher, Cat. No. 10438-026) and 0.01% Tween in 1 × phosphate-buffered saline (PBS; EmbryoMax, Cat. No. BSS-1006-B) for 30 min. Primary and secondary antibodies were diluted in the same blocking solution. A complete list of the primary antibodies used in this study, as well as their dilution factors, is provided in Supplementary Table S2. The EMX2 antibody (Thermo Fisher, Cat. No. MA5-18821) was not included in our analyses, because staining was unsuccessful and alternative antibodies were not available. The olfactory marker protein (OMP) antibody used in this study was previously used in eight other studies [40–47] and the gonadotropin-releasing hormone 1 (GNRH1) antibody was previously used in one other study [48]. Cross-absorbed Alexa Fluor anti-mouse IgG, anti-rabbit IgG, or anti-goat IgG (Invitrogen) were used as secondary antibodies, as outlined in Supplementary Table S2.
Primary antibodies were incubated for 1 h at 37°C and secondary antibodies were incubated for 30 min at 37°C. Cells were treated with 10 μg/mL DAPI (Invitrogen) for 1 min and then washed three times with 0.01% Tween in 1 × PBS. Samples were stored in 1 × PBS and imaged on the 24-well plates. For experiments with neurons, stem cells were seeded onto coverslips and differentiated. At the end of the experiment, neurons were fixed and stained, and then coverslips were transferred to glass slides for imaging at magnification 20 × or 63 × with oil. Microscopy was performed on a LSM 800 confocal microscope (Zeiss). We performed a Z-stack and took five images per experimental group. We quantified the number of positive cells for each image and constructed figures using the HALO software. The HALO software calculated the number of positive cells for each primary antibody and DAPI-positive cells. The number of positive cells for each protein was normalized to the total number of cells in the sample (DAPI-positive cells).
Statistical analyses
Sample size was determined using G*Power software. The parameters were set to a priori, an alpha error probability of 0.05, and a statistical power of 0.80. Analyses were performed using statistical software (GraphPad Prism 5.04; GraphPad Software). All statistical tests were two-sided and data were presented as sample mean ± standard error of the mean (SEM). Differences between groups were analyzed by one-way ANOVA with Dunnett's multiple comparisons to calculate P-values (*P < 0.05, **P < 0.01, ***P < 0.001).
Results
Timed BMP inhibition induces olfactory placode expression with neural potential
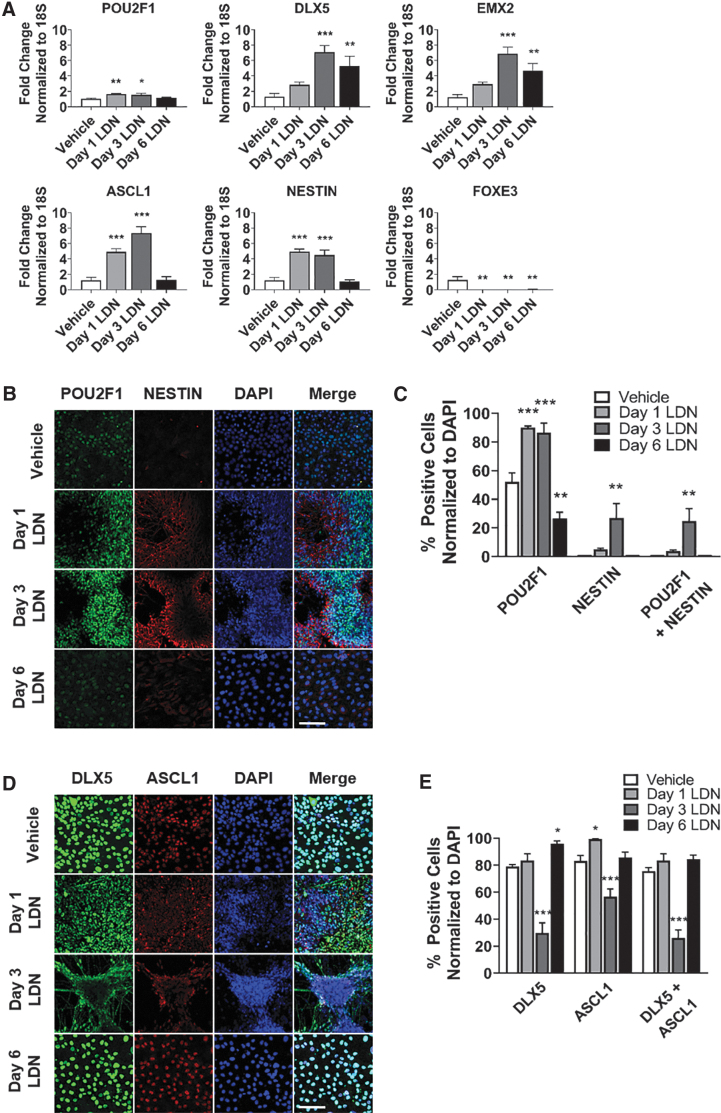
BMP signaling determines cell fate decisions between lens and olfactory placodes during late-stage gastrulation [28,29]. BMP activation promotes a lens fate, whereas BMP inhibition induces an olfactory fate [28,29]. Therefore, we tested the hypothesis that BMP inhibition induces olfactory placode in hPSCs. We treated hPSCs with LDN-193189 (LDN), a small-molecule BMP inhibitor, starting at day 1, 3, or 6 of differentiation to test the optimal timing of BMP inhibition treatment for the induction of olfactory placode. We analyzed expression of key olfactory placode and neural progenitor markers at day 24 of differentiation. Gene expression of olfactory placode (POU2F1, DLX5, EMX2) and neural progenitor (ASCL1, NESTIN) was highest in the day 3 LDN group, whereas lens placode (FOXE3) was inhibited at all LDN treatment days (Fig. 1A). In the day 3 LDN group, cells aggregated to form colonies by day 24 of differentiation.
FIG. 1.
Timed BMP inhibition induces olfactory placode expression with neural potential. (A) qRT-PCR results at day 24 for olfactory placode (POU2F1, DLX5, EMX2), neural progenitor (ASCL1, NESTIN), and lens placode (FOXE3). Values are normalized to 18S and compared with vehicle-treated group (n = 5, mean ± SEM). (B) Immunocytochemistry analysis comparing protein expression on day 24 of differentiation for POU2F1 (green) and NESTIN (red). Scale bars, 100 μM. (C) Quantification of POU2F1- and NESTIN-positive cells. Values are normalized to DAPI and compared with vehicle-treated group (n = 5, mean ± SEM). (D) Immunocytochemistry analysis comparing protein expression on day 24 of differentiation for DLX5 (green) and ASCL1 (red). Scale bars, 100 μM. (E) Quantification of DLX5- and ASCL1-positive cells. Values are normalized to DAPI and compared with vehicle-treated group (n = 5, mean ± SEM). P-values (*P < 0.05, **P < 0.01, ***P < 0.001). BMP, bone morphogenetic protein; qRT-PCR, quantitative real-time polymerase chain reaction; SEM, standard error of the mean.
POU2F1-positive cells were found in the center of the colonies and NESTIN-positive cells were expressed at the periphery of the colonies (Fig. 1B). There was a 34% increase of POU2F1-positive cells, 26% increase in NESTIN-positive cells, and a 24% increase in POU2F1 and NESTIN coexpressing positive cells in the day 3 group (Fig. 1C). DLX5 and ASCL1 were expressed at the periphery of the colonies and in the interconnecting cells of the colonies in the day 3 LDN group (Fig. 1D). There was a significant decrease in the number of DLX5 and ASCL1-positive cells in the 3-day LDN-treated group versus control; however, there was also a substantial difference in morphology of the colonies and DLX5- and ASCL1-expressing cells were localized to the periphery (Fig. 1E). Overall, the substantial increase in gene expression along with localized expression of the markers to the periphery of the colonies indicates formation of olfactory placode. EMX2 was not assessed due to inadequate detection of the antibody in the immunocytochemistry experiments.
WNT inhibition elevates olfactory placode expression and increases the number of neural progenitors
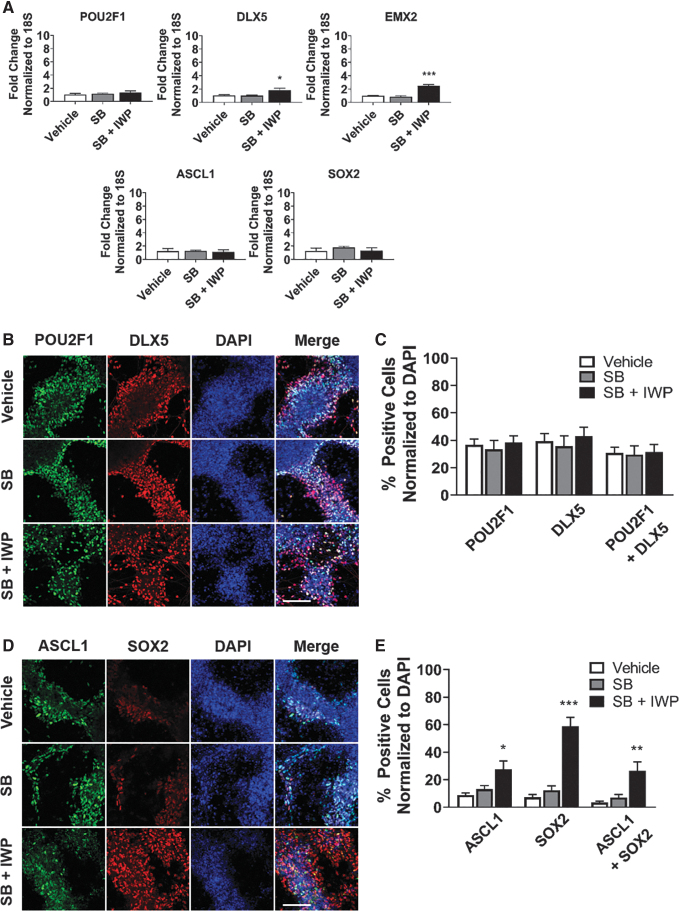
We next explored the synergy of manipulating the NODAL and WNT pathways in combination with BMP inhibition. Inhibition of the NODAL pathway promotes ectoderm by blocking endoderm and mesoderm [49–51]. The WNT pathway patterns the anterior–posterior axis, in which WNT inhibition promotes anterior cell fates [30–32]. We treated hPSCs with LDN (days 3–24) in combination with vehicle, SB-431542 (SB) to block the NODAL pathway (days 3–18), or both SB (days 3–18) and IWP2 (IWP) to block the WNT pathway (days 3–18). Expression of olfactory placode (POU2F1, DLX5, EMX2) and neural progenitor (ASCL1, SOX2) markers were analyzed at day 24 of differentiation. DLX5 and EMX2 gene expression were significantly elevated in the SB- and IWP-treated group; however, there was no difference with SB alone. SB and IWP treatment did not affect gene expression of POU2F1, ASCL1, or SOX2 (Fig. 2A).
FIG. 2.
WNT inhibition elevates olfactory placode expression and increases the number of neural progenitors. (A) qRT-PCR results at day 24 for olfactory placode (POU2F1, DLX5, EMX2) and neural progenitor (ASCL1, SOX2). Values are normalized to 18S and compared with vehicle-treated group (n = 5, mean ± SEM). (B) Immunocytochemistry analysis comparing protein expression on day 24 of differentiation for POU2F1 (green) and DLX5 (red). Scale bars, 100 μM. (C) Quantification of POU2F1- and DLX5-positive cells. Values are normalized to DAPI and compared with vehicle-treated group (n = 5, mean ± SEM). (D) Immunocytochemistry analysis comparing protein expression on day 24 of differentiation for ASCL1 (green) and SOX2 (red). Scale bars, 100 μM. (E) Quantification of ASCL1- and SOX2-positive cells. Values are normalized to DAPI and compared with vehicle-treated group (n = 5, mean ± SEM). P-values (*P < 0.05, **P < 0.01, ***P < 0.001). WNT, wingless/integrated proteins.
For the immunocytochemistry analysis, there was no significant difference in the number of POU2F1- or DLX5-positive cells in either of the treatment groups (Fig. 2B, C). There was a 42% increase in ASCL1-positive cells, a 58% increase in SOX2-positive cells, and a 51% increase in ASCL1 and SOX2 coexpressing positive cells in the SB- and IWP-treated group (Fig. 2D, E). These results indicate that WNT inhibition elevates olfactory placode gene expression and increases the number of neural progenitors. Since NODAL inhibition, without WNT inhibition, did not promote more neural progenitors, we concluded that SB treatment was not necessary and was excluded from all subsequent experiments.
Combined RA inhibition and WNT inhibition treatment amplifies neural olfactory placode expression
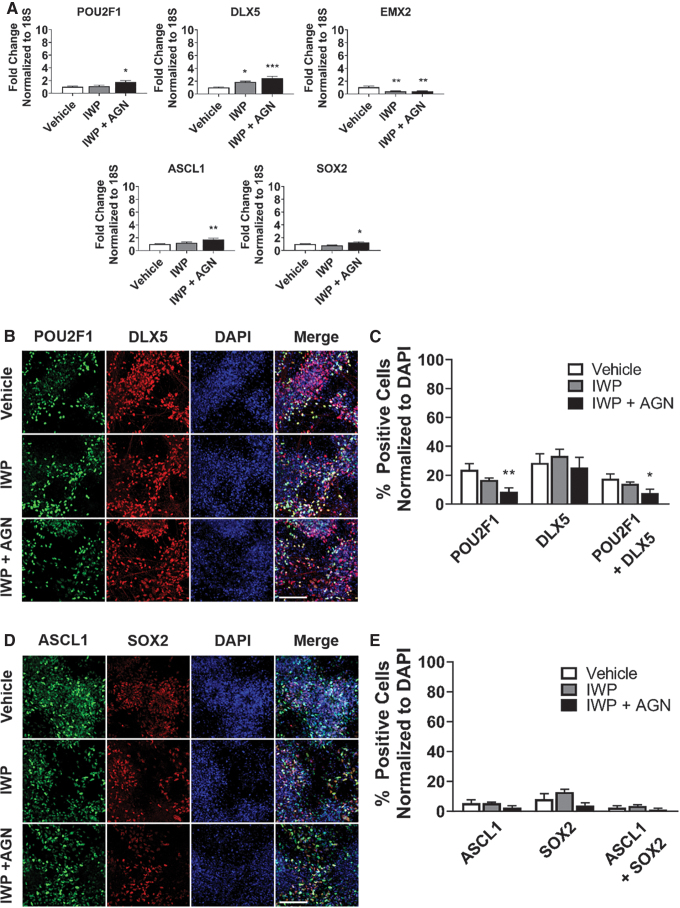
RA patterns the rostral–caudal axis of the head and is also a posterior factor, thus, its inhibition promotes the development of anterior and rostral cell fates [32–34]. We assessed the effects of RA inhibition on olfactory placode expression by using the small-molecule inhibitor, AGN193109 (AGN), to block the RA pathway. We treated hPSCs with LDN (days 3–24) in combination with vehicle, IWP (days 3–18), or both IWP (days 3–18) and AGN (days 9–18), and evaluated expression of markers at day 24 of differentiation. The combination of IWP and AGN treatment significantly increased gene expression of olfactory placode (POU2F1, DLX5) and neural progenitor (ASCL1, SOX2) markers compared with vehicle treated; however, EMX2 showed decreased gene expression following IWP or combined IWP and AGN treatment (Fig. 3A).
FIG. 3.
Combined RA inhibition and WNT inhibition treatment amplifies neural olfactory placode expression. (A) qRT-PCR results at day 24 for olfactory placode (POU2F1, DLX5, EMX2) and neural progenitor (ASCL1, SOX2). Values are normalized to 18S and compared with vehicle-treated group (n = 5, mean ± SEM). (B) Immunocytochemistry analysis comparing protein expression on day 24 of differentiation for POU2F1 (green) and DLX5 (red). Scale bars, 100 μM. (C) Quantification of POU2F1- and DLX5-positive cells. Values are normalized to DAPI and compared with vehicle-treated group (n = 5, mean ± SEM). (D) Immunocytochemistry analysis comparing protein expression on day 24 of differentiation for ASCL1 (green) and SOX2 (red). Scale bars, 100 μM. (E) Quantification of ASCL1- and SOX2-positive cells. Values are normalized to DAPI and compared with vehicle-treated group (n = 5, mean ± SEM). P-values (*P < 0.05, **P < 0.01, ***P < 0.001). RA, retinoic acid.
The number of POU2F1-positive cells and POU2F1/DLX5-coexpressing cells was reduced in the combined IWP- and AGN-treated group, but there was no difference in the number of DLX5-positive cells across groups (Fig. 3B, C). Additionally, there were no differences in the number of ASCL1- and SOX2-positive cells between groups (Fig. 3D, E). The qRT-PCR data suggest that the combined IWP and AGN treatment further amplifies neural olfactory placode gene expression. POU2F1 is a transcription factor in cell cycle regulation [52], and the reduction of POU2F1-positive cells, after RA inhibition treatment, may be an indicator of the cells exiting the cell cycle.
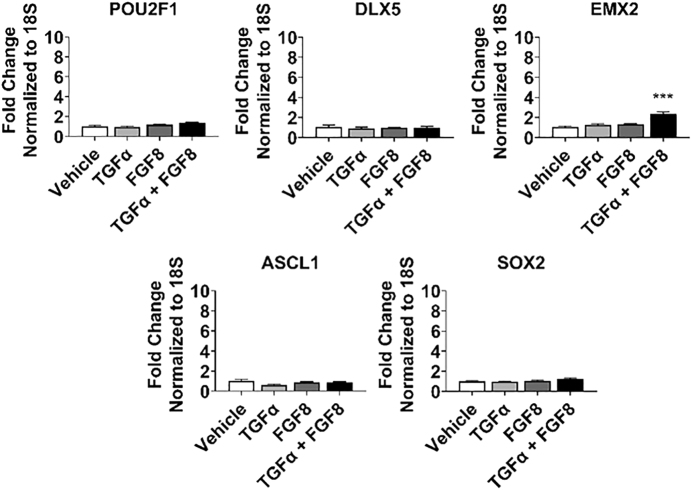
Combined TGFα and FGF8 treatment boosts EMX2 gene expression
TGFα stimulates cell proliferation in olfactory explant cultures and promotes neuronal survival [53–56]. FGF8 is expressed in the invaginating epithelium of the olfactory pit and plays a crucial role in olfactory development and neurogenesis [56–60]. We hypothesize that TGFα and FGF8 treatment in combination with BMP inhibition promotes olfactory placode differentiation and increases neural proliferation. We treated hPSCs with LDN (days 3–24) combined with vehicle, TGFα, FGF8, or both (all at days 9–24) and analyzed expression of markers at day 24 of differentiation. The combination of TGFα and FGF8 produced a significant increase in EMX2 expression; however, individually they had no effect. Additionally, TGFα and FGF8 did not alter gene expression for other olfactory placode (POU2F1, DLX5) or neural progenitor (ASCL1, SOX2) markers (Fig. 4). We conclude that the combined treatment of TGFα and FGF8 specifically boosts EMX2 expression and does not promote other olfactory genes.
FIG. 4.
Combined TGFα and FGF8 treatment boosts EMX2 gene expression. qRT-PCR results at day 24 for olfactory placode (POU2F1, DLX5, EMX2) and neural progenitor (ASCL1, SOX2). Values are normalized to 18S and compared with vehicle-treated group (n = 5, mean ± SEM). P-values (***P < 0.001). FGF, fibroblast growth factors; TGFα, transforming growth factor alpha.
A time course analysis demonstrates the dynamic expression of genes for the combined treatment plan to derive neural olfactory placode
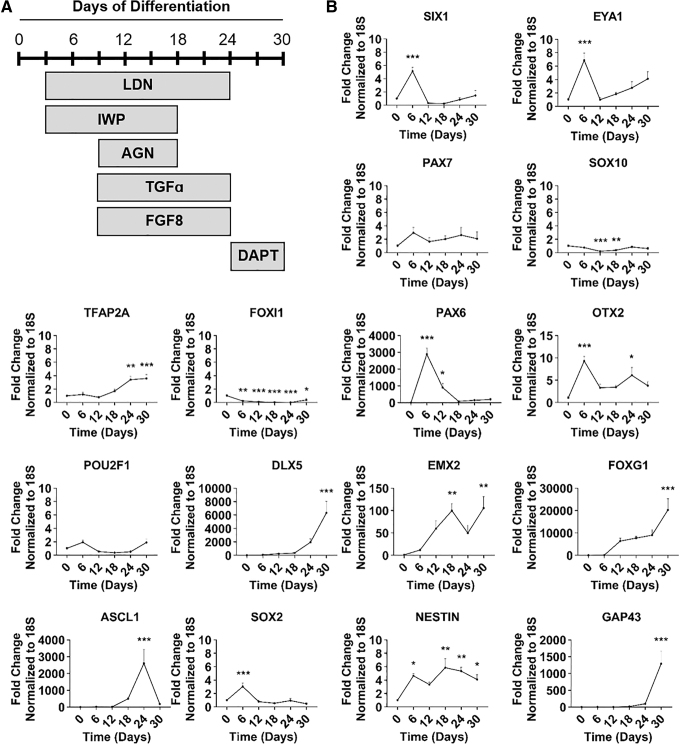
We next evaluated the dynamic expression of genes over the course of 30 days and analyzed expression every 6 days using the total combined treatment. DAPT is a γ-secretase inhibitor and was added to cells on days 24–30 to accelerate neuronal differentiation [61] (Fig. 5A). We first evaluated transcriptional markers of the four different ectodermal regions in early development: PPE, neural crest (NC), non-neural ectoderm (NNE), and neural ectoderm (NE). SIX1 and EYA1 (PPE) gene expression peaked at day 6 of differentiation and turned off by day 12, suggesting transition through the PPE. There was no increase in gene expression for PAX7 and SOX10 (NC) or TFAP2A and FOXI1 (NNE) in the day 6–12 window, showing that NC and NNE were inhibited. PAX6 (NE) had a substantial increase between days 6 and 12, indicating early induction of NE (Fig. 5B).
FIG. 5.
A time course analysis demonstrates the dynamic expression of genes for the combined treatment plan to derive neural olfactory placode. (A) Schematic representation of final combined experimental treatment plan. (B) qRT-PCR results at day 0, 6, 12, 18, 24, and 30: pan-placode ectoderm (SIX1, EYA1), neural crest (PAX7, SOX10), non-neural ectoderm (TFAP2A, FOXI1), neural ectoderm (PAX6), olfactory placode (POU2F1, DLX5, EMX2, OTX2, FOXG1), neural progenitor (ASCL1, SOX2), and immature neuron (NESTIN, GAP43). Values are normalized to 18S and compared with day 0 group (IMR90-1, n = 4, mean ± SEM). P-values (*P < 0.05, **P < 0.01, ***P < 0.001).
We next looked at transcriptional markers for olfactory placode and neural progenitors, which are expressed much later in differentiation. DLX5 (olfactory placode) peaked at day 30 and EMX2 (olfactory placode) peaked between days 18 and 30. POU2F1 (olfactory placode) stayed at low basal levels and was not induced, possibly indicating an exit from the cell cycle. OTX2 (olfactory placode) peaked early at day 6 and had a small increase at day 24, and FOXG1 (olfactory placode) had a considerable induction at day 30 of differentiation. ASCL1 (neural progenitor) peaked by day 24 of differentiation and decreased by day 30, the time at which immature neurons appeared in the culture. Surprisingly, SOX2 (neural progenitor) expression peaked at day 6 of differentiation and then remained low. NESTIN (immature neuron) was elevated between days 18 and 30, and GAP43 (immature neuron) peaked at day 30 (Fig. 5B).
Finally, we confirmed negative expression of different placodes, pluripotency, mesoderm germ layer, and endoderm germ layer. FOXE3 (lens placode), LHX3 (adenohypophyseal placode), PAX3 (trigeminal placode), and PAX8 (otic placode) all stayed at basal levels and had slight increases by day 30. POU5F1 (pluripotency) and TBXT (mesoderm) significantly decreased across all times indicating loss of pluripotency and mesodermal cell fates, respectively. SOX17 (endoderm) had a small peak at day 12 but was decreased at all other time points, indicating endoderm inhibition (Supplementary Fig. S1A). These results suggest induction of neural olfactory placode that is being driven by early expression of anterior PPE and NE.
OSNs and GNRHNs arise from hPSC-derived olfactory placode
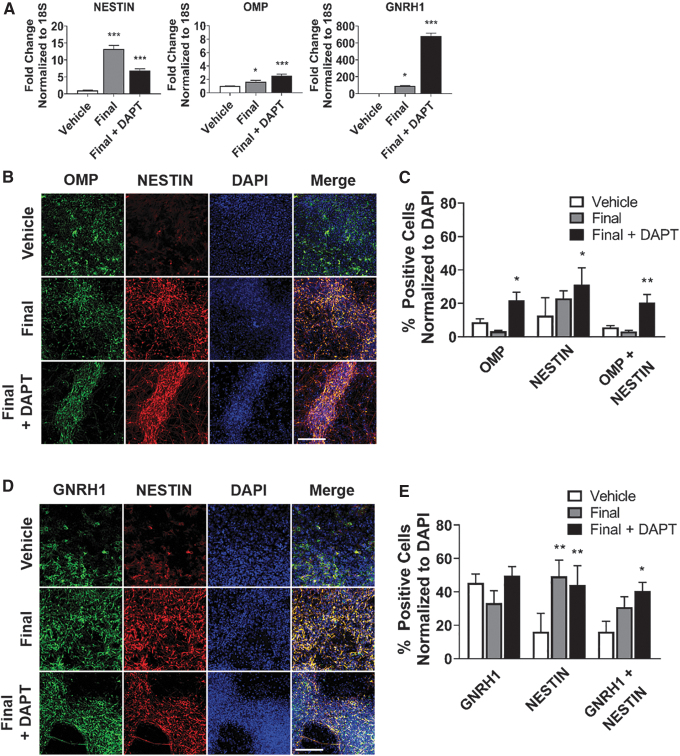
Finally, we differentiated hPSCs out to day 30 and tested for production of OSNs, which express OMP, and GNRHNs, which express GNRH1. There were three groups: vehicle treated (Vehicle), final treatment plan (Final), and final treatment plan with DAPT (Final+DAPT). The final treatment plan included: LDN (days 3–24), IWP (days 3–18), AGN (days 9–18), TGFα (days 9–24), and FGF8 (days 9–24). Gene expression for NESTIN, OMP, and GNRH1 were significantly increased in the final treated group and Final+DAPT-treated group, as assessed by qRT-PCR (Fig. 6A).
FIG. 6.
OSNs and GNRHNs arise from hPSC-derived olfactory placode. (A) qRT-PCR results at day 30 for immature neuron (NESTIN), OSN (OMP), and GNRHN (GNRH1). Values are normalized to 18S and compared with vehicle-treated group (n = 5, mean ± SEM). (B) Immunocytochemistry analysis comparing protein expression on day 30 of differentiation for OMP (green) and NESTIN (red). Scale bars, 100 μM. (C) Quantification of OMP, NESTIN, and OMP+NESTIN-positive cells. Values are normalized to DAPI and compared with vehicle-treated group (n = 5, mean ± SEM). (D) Immunocytochemistry analysis comparing protein expression on day 30 of differentiation for GNRH1 (green) and NESTIN (red). Scale bars, 100 μM. (E) Quantification of GNRH1, NESTIN, and GNRH1+NESTIN-positive cells. Values are normalized to DAPI and compared with vehicle-treated group (n = 5, mean ± SEM). P-values (*P < 0.05, **P < 0.01, ***P < 0.001). GNRH1, gonadotropin-releasing hormone 1; GNRHNs. gonadotropin-releasing hormone neuron; hPSC, human pluripotent stem cell; OMP, olfactory marker protein; OSNs, olfactory sensory neurons.
Immunocytochemistry analysis indicated that the number of OMP-positive cells was increased in the Final+DAPT-treated cells, whereas the number of GNRH1 cells did not change. However, there was an increase in NESTIN-positive cells following treatment, which resulted in colocalization, as represented by an increase in OMP+NESTIN- and GNRH1+NESTIN-positive cells (Fig. 6B, D). There were around 20% of OMP+NESTIN-coexpressed positive neurons at magnification 20 × (Fig. 6C) and around 40% of GNRH1+NESTIN-coexpressed positive neurons at magnification 20 × (Fig. 6E).
Immunocytochemistry analysis of OMP+NESTIN and GNRH1+NESTIN-coexpressed positive neurons are displayed at magnification 63 × in Supplementary Fig. S2A and B. This shows that the morphology of the GNRH1 neurons resembles that of GNRH1 neurons identified during mouse postnatal development [62]. Supplementary Fig. S3 includes a preliminary study in which we had OMP-negative and GNRH1-negative staining with NESTIN-positive cells to show no background from primary antibodies. In summary, our protocol produced a heterogeneous population of OMP and GNRHN immature neurons at day 30 of differentiation.
We performed bulk RNA-Seq on the vehicle-treated and Final+DAPT-treated groups. We evaluated 23 olfactory placode genes identified in the article by Park and Saint-Jeannet [39]. Of these, eight were found to be significantly upregulated (ASCL1, EYA1, EYA4, HES5, HEY1, LHX2, SIX4, and SIX6), and three were significantly downregulated (DLX5, EMX2, and FOXG1) (Supplementary Table S3). In addition, four olfactory placode-associated genes identified through the GO database, but not reported by Park and Saint-Jeannet, were also upregulated (HOMER2, LRRN1, SIX5, and SOX2) (Supplementary Table S3).
Discussion
In this study, we developed a methodology to generate olfactory placode, OSNs, and GNRHNs from hPSCs by investigating the major developmental signaling factors involved in olfactory placode differentiation. We showed that BMP inhibition alone elicited a strong induction of neural olfactory placode and prevented induction of lens placode, producing colonies that were similar to olfactory tissue explants harvested from rodents, in which cell proliferation is found on the periphery of the clusters [55]. The timing of BMP inhibition is critical for neural olfactory placode cells as early LDN treatment does not induce olfactory placode and late LDN treatment does not promote neurogenesis.
The dual-SMAD inhibition protocol, a well-established protocol for neuronal differentiation, includes the combination of NODAL and BMP inhibition, which blocks SMADs 1/5 and SMADs 2/3, respectively [63]. Therefore, we expected an increase in neural progenitors with the addition of SB treatment to block the NODAL pathway. However, in our study, there were no differences in gene expression, morphology, or staining pattern in the SB-treated group compared with vehicle, which suggests that BMP inhibition alone is a dominant pathway for neural olfactory placode differentiation.
The addition of IWP, to inhibit WNT signaling and promote anterior cell fates, elevated two key olfactory placode genes. Additionally, combined WNT and BMP inhibition increased the proportion of neural progenitors within the olfactory placode, which is consistent with previous reports in the animal literature that this dual inhibition promotes NE in the early gastrula [10]. The RA pathway does not play a role in the broad specification of the body plan [24], and this is reflected in our differentiation protocol where we did not observe any major changes in the morphology of olfactory placode following RA inhibition. However, gene expression was increased for two olfactory placode and two neural progenitor markers. The number of POU2F1-positive cells was reduced despite the overall increased gene expression of POU2F1, which may reflect the role of this transcription factor in cell cycle regulation, indicating the potential exiting of cells from the cell cycle and differentiating into neurons.
Our study showed increased EMX2 gene expression following combined BMP inhibition with TGFα and FGF8 treatment. There are no previous reports on the regulation of EMX2 expression by FGF8 in the animal olfactory placode development literature. However, GNRH1-expressing neurons derived from hPSCs using dual-SMAD inhibition and high concentration of FGF8 show elevated EMX2 expression at day 21 of differentiation [23]. Altogether, these studies suggest that FGF8 signaling may have a direct or indirect effect on EMX2 gene expression and may increase the developmental potential for olfactory placode.
We observed a peak in PPE and NE gene expression at day 6 of differentiation. PAX6 was at a 3,000-fold increase compared with the 6–10-fold increase of the PPE genes, suggesting there was a much higher NE induction. Our differentiation plan blocked NC and NNE expression. Four olfactory placode genes (DLX5, EMX2, OTX2, FOXG1) and three neural progenitor genes (ASCL1, NESTIN, GAP43) had significant increases by day 24 and 30, indicating induction of neural olfactory placode progenitors. SOX2 was expected to peak at day 24 as ASCL1 and SOX2 coexpression is found in neural progenitors; however, this was not seen.
Early expression of PAX6, OTX2, and SOX2 indicates the start of optic vesicle differentiation or early retina cells, which is developed from the NE [64]. However, the absence of expression at later time points indicates specificity of this protocol for olfactory placode. Interestingly, the olfactory placode was previously shown in animal models to develop in the same time window as the formation of the optic cup of the eye, as well as, arise from a continuous region of anterior PPE and anterior NE [65,66]. This may explain why we see early gene expression of a mix population for PPE, optic cup, and NE. As expected, gene expression representing the other placodes and germ layers were inhibited in this protocol.
Coexpression of OMP with NESTIN and GNRH1 with NESTIN indicates production of immature OSNs and GNRHNs. While coexpression of NESTIN and OMP is not usually immunodetectable [67,68], other studies have reported mature neural markers coexpressed with NESTIN or other proliferating neural markers [69–71]. In particular, Hendrickson et al. [70] demonstrated coexpression of NESTIN (neural progenitor) with NEUN (mature neuron) in adult rat and human brain cells. It is possible that we are capturing immature neuronal cells that are beginning to express mature neuronal markers. Future studies should include a longer window of differentiation, until ∼50–60 days to ensure mature neurons are generated, which may provide a clearer picture of the transition from immature to mature neurons.
Within our RNA-Seq data, upregulation of eight olfactory placode-associated genes from published studies, and an additional four identified from the GO database, supports the claim that our method drives differentiation toward olfactory placode. However, downregulation of three olfactory placode-associated genes (DLX5, EMX2, and FOXG1) may be representative of the time frame examined. Our time course experiment indicated upregulation of these genes, but the experimental conditions were different (control was day 0 cells in qRT-PCR experiments and vehicle-treated day 30 cells in RNA-Seq experiments). Future experiments assessing transcriptome-wide profiles over a longer time course (eg, up to 60 days), along with additional qRT-PCR validation of these same samples, may provide more clues about the regulation of olfactory placode-associated genes in our differentiation model.
Furthermore, several olfactory placode-associated genes were not found to be significantly upregulated, suggesting that increased sample numbers may provide more convincing evidence of olfactory placode derivation. Overall, we conclude that the combined treatment plan generates cells consistent with a neural olfactory placode phenotype, including immature OSNs and GNRHNs. Future studies, however, are needed to provide more evidence of olfactory placode transcriptional processes and generation of mature OSNs and GNRHNs, including increased sample numbers, additional time course experiments, and functional assays such as electrophysiology and transplantation experiments.
Conclusion
We report, in this study, the molecular analysis of neural olfactory placode induction in hPSCs and evidence for the production of OSNs and GNRHNs. This study is the first to report on the differentiation of OSNs in hPSCs. Establishing differentiation protocols with high efficiency is critical for the use of olfactory progenitors in preclinical experiments. These studies provide a baseline for the development of stem cell therapies that can treat anosmia and CHH through transplantation of hPSC-derived olfactory placode progenitors.
Supplementary Material
Acknowledgment
The authors thank Dr. Wouter Koek for his consultation on the experimental design and statistical analysis to this study.
Author Disclosure Statement
All authors declare that they have no conflicts of interest.
Funding Information
This research was supported by the Douglas Foundation and the National Center for Advancing Translational Sciences of the NIH under Award Number TL1TR001119.
Supplementary Material
References
- 1. Zhu Z and Huangfu D. (2013). Human pluripotent stem cells: an emerging model in developmental biology. Development 140:705–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wu SM and Hochedlinger K. (2011). Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol 13:497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alessandrini M, Preynat-Seauve O, De Bruin K and Pepper MS. (2019). Stem cell therapy for neurological disorders. S Afr Med J 109:70–77. [DOI] [PubMed] [Google Scholar]
- 4. Song CG, Zhang YZ, Wu HN, Cao XL, Guo CJ, Li YQ, Zheng MH and Han H. (2018). Stem cells: a promising candidate to treat neurological disorders. Neural Regen Res 13:1294–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Im S and Moon C. (2015). Transcriptional regulatory network during development in the olfactory epithelium. BMB Rep 48:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brann JH and Firestein SJ. (2014). A lifetime of neurogenesis in the olfactory system. Front Neurosci 8:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim SH. (2015). Congenital hypogonadotropic hypogonadism and kallmann syndrome: past, present, and future. Endocrinol Metab (Seoul) 30:456–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Forni PE, Taylor-Burds C, Melvin VS, Williams T and Wray S. (2011). Neural crest and ectodermal cells intermix in the nasal placode to give rise to GnRH-1 neurons, sensory neurons, and olfactory ensheathing cells. J Neurosci 31:6915–6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boesveldt S, Postma EM, Boak D, Welge-Luessen A, Schopf V, Mainland JD, Martens J, Ngai J and Duffy VB. (2017). Anosmia-a clinical review. Chem Senses 42:513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Singh S and Groves AK. (2016). The molecular basis of craniofacial placode development. Wiley Interdiscip Rev Dev Biol 5:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang C, Yang Y, Brennan L, Bouhassira EE, Kantorow M and Cvekl A. (2010). Efficient generation of lens progenitor cells and lentoid bodies from human embryonic stem cells in chemically defined conditions. FASEB J 24:3274–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mengarelli I and Barberi T. (2013). Derivation of multiple cranial tissues and isolation of lens epithelium-like cells from human embryonic stem cells. Stem Cells Transl Med 2:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zimmer B, Piao J, Ramnarine K, Tomishima MJ, Tabar V and Studer L. (2016). Derivation of diverse hormone-releasing pituitary cells from human pluripotent stem cells. Stem Cell Reports 6:858–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen W, Jongkamonwiwat N, Abbas L, Eshtan SJ, Johnson SL, Kuhn S, Milo M, Thurlow JK, Andrews PW, et al. (2012). Restoration of auditory evoked responses by human ES-cell-derived otic progenitors. Nature 490:278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koehler KR, Mikosz AM, Molosh AI, Patel D and Hashino E. (2013). Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature 500:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dincer Z, Piao J, Niu L, Ganat Y, Kriks S, Zimmer B, Shi SH, Tabar V and Studer L. (2013). Specification of functional cranial placode derivatives from human pluripotent stem cells. Cell Rep 5:1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lahlou H, Lopez-Juarez A, Fontbonne A, Nivet E and Zine A. (2018). Modeling human early otic sensory cell development with induced pluripotent stem cells. PLoS One 13:e0198954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koehler KR, Nie J, Longworth-Mills E, Liu XP, Lee J, Holt JR and Hashino E. (2017). Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat Biotechnol 35:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gunewardene N, Bergen NV, Crombie D, Needham K, Dottori M and Nayagam BA. (2014). Directing human induced pluripotent stem cells into a neurosensory lineage for auditory neuron replacement. Biores Open Access 3:162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ozone C, Suga H, Eiraku M, Kadoshima T, Yonemura S, Takata N, Oiso Y, Tsuji T and Sasai Y. (2016). Functional anterior pituitary generated in self-organizing culture of human embryonic stem cells. Nat Commun 7:10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dewi CU, Mason M, Cohen-Hyams T, Killingsworth MC, Harman DG, Gnanasambandapillai V, Liyanage L and O'Connor MD. (2020). A simplified method for producing human lens epithelial cells and light-focusing micro-lenses from pluripotent stem cells. Exp Eye Res 2022:108317. [DOI] [PubMed] [Google Scholar]
- 22. Fu Q, Qin Z, Jin X, Zhang L, Chen Z, He J, Ji J and Yao K. (2017). Generation of functional lentoid bodies from human induced pluripotent stem cells derived from urinary cells. Invest Ophthalmol Vis Sci 58:517–527. [DOI] [PubMed] [Google Scholar]
- 23. Lund C, Pulli K, Yellapragada V, Giacobini P, Lundin K, Vuoristo S, Tuuri T, Noisa P and Raivio T. (2016). Development of gonadotropin-releasing hormone-secreting neurons from human pluripotent stem cells. Stem Cell Reports 7:149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tuazon FB and Mullins MC. (2015). Temporally coordinated signals progressively pattern the anteroposterior and dorsoventral body axes. Semin Cell Dev Biol 42:118–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fathi A, Eisa-Beygi S and Baharvand H. (2017). Signaling molecules governing pluripotency and early lineage commitments in human pluripotent stem cells. Cell J 19:194–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kwon HJ, Bhat N, Sweet EM, Cornell RA and Riley BB. (2010). Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet 6:e1001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Litsiou A, Hanson S and Streit A. (2005). A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development 132:4051–4062. [DOI] [PubMed] [Google Scholar]
- 28. Sjodal M, Edlund T and Gunhaga L. (2007). Time of exposure to BMP signals plays a key role in the specification of the olfactory and lens placodes ex vivo. Dev Cell 13:141–149. [DOI] [PubMed] [Google Scholar]
- 29. Bhattacharyya S, Bailey AP, Bronner-Fraser M and Streit A. (2004). Segregation of lens and olfactory precursors from a common territory: cell sorting and reciprocity of Dlx5 and Pax6 expression. Dev Biol 271:403–414. [DOI] [PubMed] [Google Scholar]
- 30. Park BY and Saint-Jeannet JP. (2008). Hindbrain-derived Wnt and Fgf signals cooperate to specify the otic placode in Xenopus. Dev Biol 324:108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Heisenberg CP, Brand M, Jiang YJ, Warga RM, Beuchle D, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, et al. (1996). Genes involved in forebrain development in the zebrafish, Danio rerio. Development 123:191–203. [DOI] [PubMed] [Google Scholar]
- 32. Saint-Jeannet JP and Moody SA. (2014). Establishing the pre-placodal region and breaking it into placodes with distinct identities. Dev Biol 389:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Janesick A, Shiotsugu J, Taketani M and Blumberg B. (2012). RIPPLY3 is a retinoic acid-inducible repressor required for setting the borders of the pre-placodal ectoderm. Development 139:1213–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nikaido M, Navajas Acedo J, Hatta K and Piotrowski T. (2017). Retinoic acid is required and Fgf, Wnt, and Bmp signaling inhibit posterior lateral line placode induction in zebrafish. Dev Biol 431:215–225. [DOI] [PubMed] [Google Scholar]
- 35. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318:1917–1920. [DOI] [PubMed] [Google Scholar]
- 36. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seth Carbon ED, Benjamin Good M, Deepak Unni R, Nomi Harris L, Christopher Mungall J, Siddartha Basu, Rex Chisholm L, Robert Dodson J, et al. (2021). The gene ontology resource: enriching a GOld mine. Nucleic Acids Res 49:D325–d334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B and Lewis S. (2009). AmiGO: online access to ontology and annotation data. Bioinformatics 25:288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Park BY and Saint-Jeannet JP. (2010). Induction and Segregation of the Vertebrate Cranial Placodes. Morgan & Claypool Life Sciences, San Rafael, CA. ISBN: 9781615041039. [PubMed] [Google Scholar]
- 40. Choi R, Kurtenbach S and Goldstein BJ. (2019). Loss of BMI1 in mature olfactory sensory neurons leads to increased olfactory basal cell proliferation. Int Forum Allergy Rhinol 9:993–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brozzetti L, Sacchetto L, Cecchini MP, Avesani A, Perra D, Bongianni M, Portioli C, Scupoli M, Ghetti B, et al. (2020). Neurodegeneration-associated proteins in human olfactory neurons collected by nasal brushing. Front Neurosci 14:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Casadei E, Tacchi L, Lickwar CR, Espenschied ST, Davison JM, Muñoz P, Rawls JF and Salinas I. (2019). Commensal bacteria regulate gene expression and differentiation in vertebrate olfactory systems through transcription factor REST. Chem Senses 44:615–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiong S, Zhang S, Guan L, Chen J, Tu X, Li Q and Jiang H. (2017). Differentiation of induced pluripotent stem cells for future olfactory repair using an indirect co-culture technique. Int J Clin Exp Pathol 10:8072–8081. [PMC free article] [PubMed] [Google Scholar]
- 44. Goncalves S and Goldstein BJ. (2020). Acute N-Acetylcysteine administration ameliorates loss of olfactory neurons following experimental injury in vivo. Anat Rec (Hoboken) 303:626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bathini P, Mottas A, Jaquet M, Brai E and Alberi L. (2019). Progressive signaling changes in the olfactory nerve of patients with Alzheimer's disease. Neurobiol Aging 76:80–95. [DOI] [PubMed] [Google Scholar]
- 46. Chen M, Chen Y, Huo Q, Wang L, Tan S, Misrani A, Jiang J, Chen J, Chen S, et al. (2021). Enhancing GABAergic signaling ameliorates aberrant gamma oscillations of olfactory bulb in AD mouse models. Mol Neurodegener 16:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. de Melo GD, Lazarini F, Levallois S, Hautefort C, Michel V, Larrous F, Verillaud B, Aparicio C, Wagner S, et al. (2021). COVID-19-related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci Transl Med 13:eabf8396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cao N, Cao L, Gao M, Wang H, Zhang L and Yang L. (2021). Changes in mRNA and protein levels of gonadotropin releasing hormone and receptor in ovine thymus, lymph node, spleen, and liver during early pregnancy. Domest Anim Endocrinol 76:106607. [DOI] [PubMed] [Google Scholar]
- 49. Tremblay KD, Hoodless PA, Bikoff EK and Robertson EJ. (2000). Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development 127:3079–3090. [DOI] [PubMed] [Google Scholar]
- 50. Kubo A, Shinozaki K, Shannon JM, Kouskoff V, Kennedy M, Woo S, Fehling HJ and Keller G. (2004). Development of definitive endoderm from embryonic stem cells in culture. Development 131:1651–1662. [DOI] [PubMed] [Google Scholar]
- 51. D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E and Baetge EE. (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23:1534–1541. [DOI] [PubMed] [Google Scholar]
- 52. Segil N, Roberts SB and Heintz N. (1991). Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science 254:1814–1816. [DOI] [PubMed] [Google Scholar]
- 53. Farbman AI and Buchholz JA. (1996). Transforming growth factor-alpha and other growth factors stimulate cell division in olfactory epithelium in vitro. J Neurobiol 30:267–280. [DOI] [PubMed] [Google Scholar]
- 54. Farbman AI and Ezeh PI. (2000). TGF-alpha and olfactory marker protein enhance mitosis in rat olfactory epithelium in vivo. Neuroreport 11:3655–3658. [DOI] [PubMed] [Google Scholar]
- 55. Peterson J, Lin B, Barrios-Camacho CM, Herrick DB, Holbrook EH, Jang W, Coleman JH and Schwob JE. (2019). Activating a reserve neural stem cell population in vitro enables engraftment and multipotency after transplantation. Stem Cell Reports 12:680–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. LaMantia AS, Bhasin N, Rhodes K and Heemskerk J. (2000). Mesenchymal/epithelial induction mediates olfactory pathway formation. Neuron 28:411–425. [DOI] [PubMed] [Google Scholar]
- 57. Kawauchi S, Shou J, Santos R, Hebert JM, McConnell SK, Mason I and Calof AL. (2005). Fgf8 expression defines a morphogenetic center required for olfactory neurogenesis and nasal cavity development in the mouse. Development 132:5211–5223. [DOI] [PubMed] [Google Scholar]
- 58. Kawauchi S, Beites CL, Crocker CE, Wu HH, Bonnin A, Murray R and Calof AL. (2004). Molecular signals regulating proliferation of stem and progenitor cells in mouse olfactory epithelium. Dev Neurosci 26:166–180. [DOI] [PubMed] [Google Scholar]
- 59. Forni PE, Bharti K, Flannery EM, Shimogori T and Wray S. (2013). The indirect role of fibroblast growth factor-8 in defining neurogenic niches of the olfactory/GnRH systems. J Neurosci 33:19620–19634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maier E, von Hofsten J, Nord H, Fernandes M, Paek H, Hebert JM and Gunhaga L. (2010). Opposing Fgf and Bmp activities regulate the specification of olfactory sensory and respiratory epithelial cell fates. Development 137:1601–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Crawford TQ and Roelink H. (2007). The notch response inhibitor DAPT enhances neuronal differentiation in embryonic stem cell-derived embryoid bodies independently of sonic hedgehog signaling. Dev Dyn 236:886–892. [DOI] [PubMed] [Google Scholar]
- 62. Messina A, Langlet F, Chachlaki K, Roa J, Rasika S, Jouy N, Gallet S, Gaytan F, Parkash J, et al. (2016). A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat Neurosci 19:835–844. [DOI] [PubMed] [Google Scholar]
- 63. Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M and Studer L. (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27:275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hever AM, Williamson KA and van Heyningen V. (2006). Developmental malformations of the eye: the role of PAX6, SOX2 and OTX2. Clin Genet 69:459–470. [DOI] [PubMed] [Google Scholar]
- 65. Torres-Paz J and Whitlock KE. (2014). Olfactory sensory system develops from coordinated movements within the neural plate. Dev Dyn 243:1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Whitlock KE. (2015). The loss of scents: do defects in olfactory sensory neuron development underlie human disease? Birth Defects Res C Embryo Today 105:114–125. [DOI] [PubMed] [Google Scholar]
- 67. Doyle KL, Khan M and Cunningham AM. (2001). Expression of the intermediate filament protein nestin by sustentacular cells in mature olfactory neuroepithelium. J Comp Neurol 437:186–195. [DOI] [PubMed] [Google Scholar]
- 68. Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, et al. (2017). Deconstructing olfactory stem cell trajectories at single-cell resolution. Cell Stem Cell 20:817–830.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Brann JH, Ellis DP, Ku BS, Spinazzi EF and Firestein S. (2015). Injury in aged animals robustly activates quiescent olfactory neural stem cells. Front Neurosci 9:367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hendrickson ML, Rao AJ, Demerdash ON and Kalil RE. (2011). Expression of nestin by neural cells in the adult rat and human brain. PLoS One 6:e18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gonsalvez DG, Cane KN, Landman KA, Enomoto H, Young HM and Anderson CR. (2013). Proliferation and cell cycle dynamics in the developing stellate ganglion. J Neurosci 33:5969–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.