Abstract
Topologically associating domains (TADs) are thought to play an important role in preventing gene misexpression by spatially constraining enhancer–promoter contacts. The deleterious nature of gene misexpression implies that TADs should, therefore, be conserved among related species. Several early studies comparing chromosome conformation between species reported high levels of TAD conservation; however, more recent studies have questioned these results. Furthermore, recent work suggests that TAD reorganization is not associated with extensive changes in gene expression. Here, we investigate the evolutionary conservation of TADs among 11 species of Drosophila. We use Hi-C data to identify TADs in each species and employ a comparative phylogenetic approach to derive empirical estimates of the rate of TAD evolution. Surprisingly, we find that TADs evolve rapidly. However, we also find that the rate of evolution depends on the chromatin state of the TAD, with TADs enriched for developmentally regulated chromatin evolving significantly slower than TADs enriched for broadly expressed, active chromatin. We also find that, after controlling for differences in chromatin state, highly conserved TADs do not exhibit higher levels of gene expression constraint. These results suggest that, in general, most TADs evolve rapidly and their divergence is not associated with widespread changes in gene expression. However, higher levels of evolutionary conservation and gene expression constraints in TADs enriched for developmentally regulated chromatin suggest that these TAD subtypes may be more important for regulating gene expression, likely due to the larger number of long-distance enhancer–promoter contacts associated with developmental genes.
Keywords: Drosophila, genome organization, topologically associating domains
Introduction
Within the nucleus, chromosomes are arranged in a nested hierarchy of 3D organization, from chromosome territories down to nucleosomes. Individual chromosomes are arranged into topologically associating domains (TADs) of interacting chromatin with many 3D contacts occurring within domains and few contacts between domains. TADs have been detected in a variety of metazoan species, and TAD-like structures have also been observed in plants and bacteria (Szabo et al. 2019). Architectural proteins with insulating properties bind to specific motifs at TAD boundaries, which specify the location of these domains. In vertebrates, TADs form via extrusion of a chromatin loop through a cohesin ring. Convergently oriented CTCF sites specify TAD boundaries by blocking this loop extrusion (Sanborn et al. 2015; Fudenberg et al. 2016; Rao et al. 2017). Although there is a Drosophila ortholog of CTCF, recent work suggests that this protein does not play a major role in TAD formation in Drosophila (Kaushal et al. 2021). Instead, the insulator proteins M1BP and BEAF-32, in conjunction with CP190 or Chromator, seem to play a more important role in TAD boundary formation in flies (Szabo et al. 2019; Bag et al. 2021).
The prevailing view regarding the role of TADs in genome function is that they act to partition the genome into regulatory units. Such partitioning constrains chromatin looping so that intra-TAD enhancer/promoter contacts are more common than inter-TADs contacts, thereby reducing aberrant contacts that could lead to gene misexpression (Robson et al. 2019). A role for TADs in preventing gene misexpression implies that they should be evolutionarily conserved. Indeed, studies in both vertebrates and flies have reported strong conservation of TAD organization across millions of years of evolution (Dixon et al. 2012; Phillips-Cremins et al. 2013; Krefting et al. 2018; Lazar et al. 2018; Renschler et al. 2019). Recent work, however, has questioned these results as well as the role of TADs in controlling gene expression. In particular, between humans and chimps, TADs and TAD boundaries have been shown to be less conserved than any other regulatory phenotype (Eres et al. 2019). Furthermore, less than 10% of differentially expressed genes between these two species were associated with differences in Hi-C contacts (Eres et al. 2019).
In this study, we address whether TADs are evolutionarily conserved using a comparative genomics and phylogenetic approach to estimate the rate of TAD evolution across 11 species of Drosophila. Surprisingly, we find that TAD substitutions occur relatively rapidly, suggesting that, overall, there is little selection to preserve TAD organization. However, we also find that TADs enriched for developmentally regulated chromatin evolve significantly slower than TADs enriched for active chromatin, consistent with increased evolutionary constraint on these TAD subtypes. We find no evidence of increased constraint on gene expression levels for the TADs that exhibit higher conservation among species, consistent with recent studies questioning the importance of TADs with respect to gene regulation. Overall, these results suggest that TADs may be more evolutionarily labile than previously suggested and raise the possibility that most TADs may only play a minor role in the regulation of gene expression.
Results
Genome Scaffolding
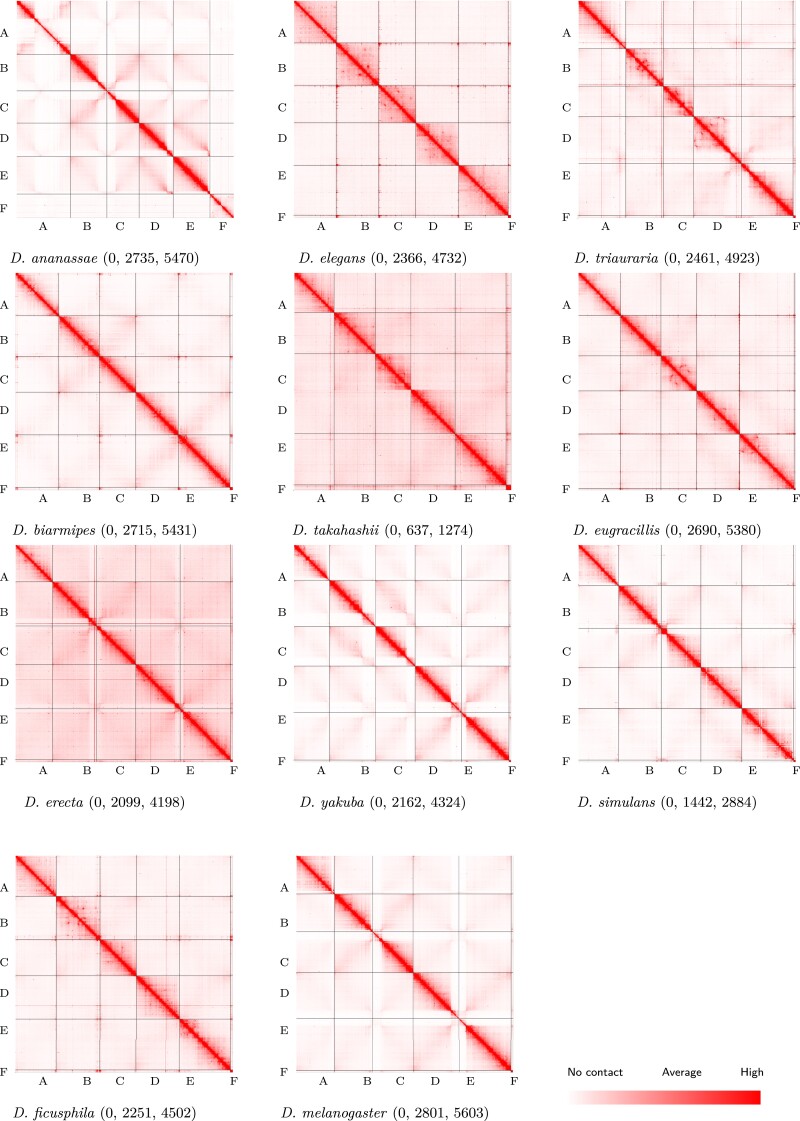
We generated Hi-C datasets from the embryos of 11 species from the melanogaster species group (Bock 1980) with divergence times between 4 and 26 Ma (Suvorov et al. 2021), whose genomes range from nearly colinear (e.g., Drosophila melanogaster vs. D. simulans) to highly rearranged (e.g., D. melanogaster vs. D. ananassae) (Bhutkar et al. 2008). The reference genome assemblies for these species vary in their level of fragmentation. We therefore used our Hi-C data to arrange the genome contigs of each species (except for D. melanogaster) into scaffolds that represent each of the six chromosome arms conserved across Drosophila (Muller elements A–F) (see Methods) (fig. 1). The resulting assemblies ranged in size from 120 to 179 Mb (mean of 136 Mb) (supplementary table S1, Supplementary Material online).
Fig. 1.
Hi-C contact maps. Chromosome-length assemblies were generated via Hi-C scaffolding for 9 of 11 Drosophila species shown here. The version 6 D. melanogaster assembly already contains chromosome-length scaffolds and the D. triauraria assembly was scaffolded in a prior study (Torosin et al. 2020). Scaffolding was performed using 3D-DNA (Dudchenko et al. 2017) and contact frequencies were visualized using Juicebox (Durand, Robinson, et al. 2016). Contact maps are colored according to contact frequencies between bins with darker red representing more contacts. For each contact map, the numerical values following the species name indicate the lowest, average, and highest number of contacts, respectively.
TAD Boundary and Domain Annotation
For each species, we generated two replicate Hi-C datasets with combined sequencing depths ranging from 184 to 346 million read pairs (supplementary table S2, Supplementary Material online). We used HiCExplorer (Ramírez et al. 2018) to identify TADs and TAD boundaries at 5 kb resolution for each replicate dataset and found strong correlations (Spearman’s rho >0.93 in all cases) (supplementary table S3, Supplementary Material online) in the TAD separation scores between replicates. To further assess reproducibility among replicate datasets, we used HiCRep (Yang et al. 2017) to calculate the stratum-adjusted correlation coefficient (SCC) between replicate datasets for each chromosome of each species. The median SCC values for each species ranged from 0.86 to 0.997 (supplementary table S4, Supplementary Material online).
We next compared TAD and TAD boundary locations between the two replicates from each species and defined high-confidence TADs and TAD boundaries as those that were found independently in each replicate. Similarly, we defined low-confidence TADs and TAD boundaries as those that were found in only one of the two replicates. Across the 11 species, we identified between 307 and 830 high-confidence TADs () and 553 and 1,016 high-confidence TAD boundaries () (supplementary table S5, Supplementary Material online).
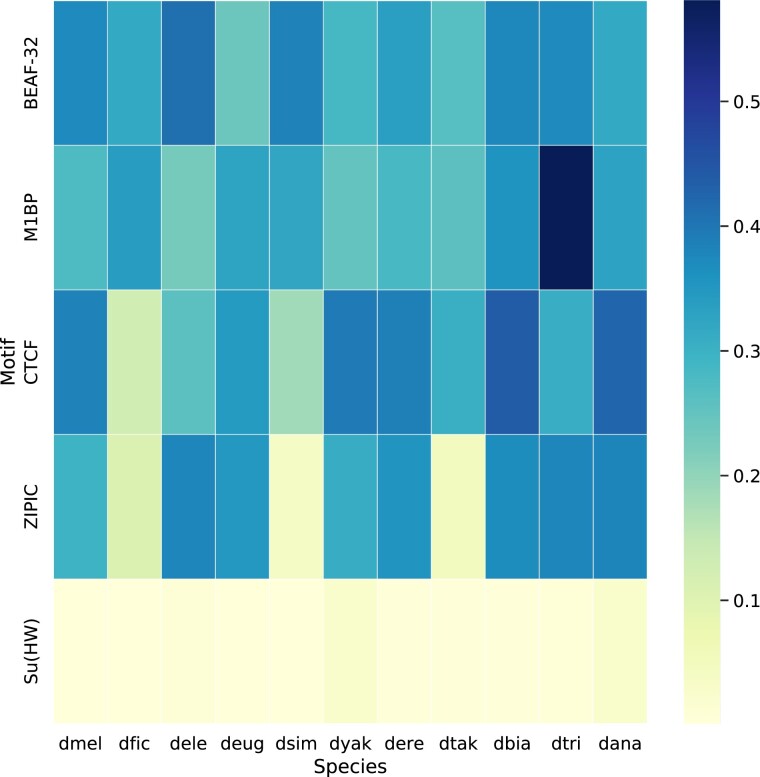
Drosophila melanogaster TAD boundaries are highly enriched for binding motifs for the insulator proteins BEAF-32 and M1BP (Ramírez et al. 2018). Binding motifs of other insulator proteins such as ZIPIC, Su(Hw), and CTCF have also been identified at D. melanogaster TAD boundaries; however, their importance in boundary specification is less clear. In particular, although CTCF plays a crucial role in TAD boundary formation in vertebrates, it seems to be less important in Drosophila (Szabo et al. 2019; Kaushal et al. 2021). To determine whether the TAD boundaries in other Drosophila species show enrichment of insulator protein binding motifs similar to those described in D. melanogaster, we examined enrichment for the following motifs across the 11 species studied here: BEAF-32, M1BP, CTCF, ZIPIC, and Su(Hw). All 11 species show consistent enrichment of BEAF-32 and M1BP motifs at TAD boundaries. Su(Hw) showed the least enrichment across all species, whereas CTCF and ZIPIC showed variable levels of enrichment (fig. 2 and supplementary table S6, Supplementary Material online). Overall, these results suggest that M1BP and BEAF-32 play a role in TAD boundary specification across the melanogaster species group while also validating our boundary predictions in these species; however, we note that other processes, such as transcription, may also contribute to boundary formation.
Fig. 2.
Boundary motif enrichment. We used AME (v. 5.3.3) from MEME suite (Bailey et al. 2009) to measure the enrichment of sequence motifs for five insulator proteins whose binding sites have previously been shown to be associated with TAD boundaries in D. melanogaster. Shown here are the percentage of boundaries containing at least one sequence motif (a value reported as %TP by AME). The heatmap shows %TP for the BEAF-32, M1BP, CTCF, ZIPIC, and Su(HW) insulator protein binding motifs at the boundaries of all 11 species used in this study. We found consistent enrichment of M1BP and BEAF-32 at TAD boundaries across all species, both of which are known to be important for TAD boundary formation in D. melanogaster (Ramírez et al. 2018). CTCF, ZIPIC, and Su(HW) motifs are found at a relatively small number of TAD boundaries in D. melanogaster (Ramírez et al. 2018). CTCF and ZIPIC show variable enrichment across species, whereas Su(HW) shows consistently weak enrichment.
TAD Orthology
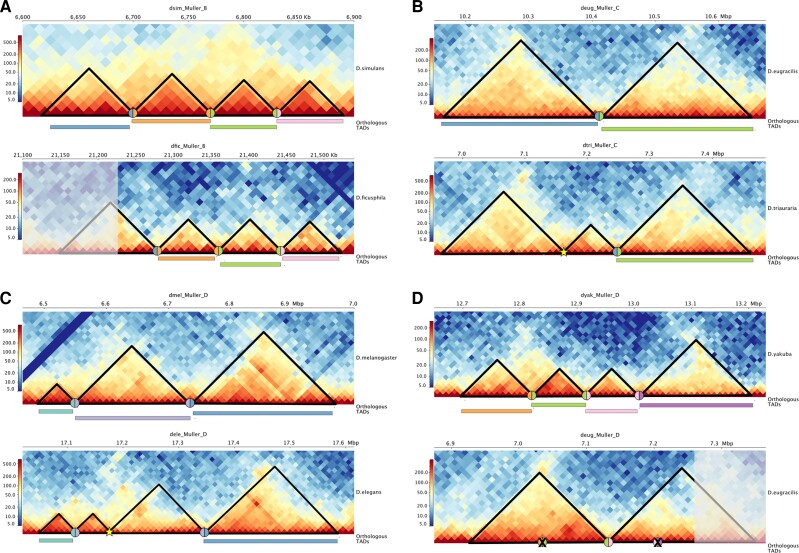
We used a whole-genome alignment liftover approach to assess TAD orthology between all pairwise interspecies comparisons (see Methods; supplementary table S7, Supplementary Material online). We then used Louvain clustering to create TAD orthogroups based on the pairwise orthology assignments (see Methods) (supplementary tables S8 and S9, Supplementary Material online). After assigning TADs to orthogroups, we examined Hi-C contact matrices to visualize specific examples of TAD gain/loss between species to confirm the accuracy of our approach (fig. 3).
Fig. 3.
TAD reorganization in Drosophila. Each panel shows Hi-C contact maps for orthologous genomic regions between two species pairs that differ in their TAD organization. For each panel, the top matrix shows the ancestral TAD configuration, whereas the bottom matrix shows the derived state. The black triangles highlight the TAD locations and the colored rectangles show TADs that are orthologous between the two species pairs. The colored circles show the locations of orthologous boundaries, whereas yellow stars show the location of novel TAD boundaries and X’s show the loss of a boundary. (A) A chromosomal rearrangement in D. ficusphila has created a novel TAD fusion compared with the ancestral state (represented here by D. simulans). The shaded portion of the leftmost TAD shows the location of the fusion. (B) The formation of a novel TAD boundary in D. triauraria has subdivided a single ancestral TAD (represented here by D. eugracilis) into two novel TADs. (C) The formation of a novel TAD boundary in D. elegans has subdivided a single ancestral TAD (represented here by D. melanogaster) into two novel TADs. (D) Loss of two TAD boundaries in D. eugracilis has produced two neighboring TAD fusions while a chromosomal rearrangement has led to an additional fusion event. The ancestral state is represented here by D. yakuba.
We first used our TAD orthogroups to assess the effect of increasing the stringency of TAD calling on both the number of orthogroups identified and the conservation of orthologous TADs across species. We called TADs in D. melanogaster using increasing levels of stringency and intersected each set of TAD calls with our set of TAD orthogroups (see Methods). We found that both the total number of TADs identified and the number of TADs matching those previously assigned to an orthogroup decreased with increasing levels of stringency (supplementary fig. S1A, Supplementary Material online). This result is likely due to the fact that real TADs are lost with higher stringency since the two highest stringency sets of TAD calls each contained fewer than 200 total TADs (supplementary fig. S1A, Supplementary Material online). However, we also found that the TADs that are identified using higher levels of stringency show higher levels of evolutionary conservation (supplementary fig. S1B, Supplementary Material online). This result is consistent with highly conserved TADs having boundaries that show stronger insulation.
We next assessed variation in size among TADs from the same orthogroup. Overall, orthologous TADs tend to be similar in size: on average, the largest TAD within an orthogroup is 25% larger than the smallest TAD. To examine size variation in more detail, we selected a TAD from Muller A that is among the top 10% of TADs that are both conserved among the majority of species and also show the largest variation in size (supplementary fig. S2, Supplementary Material online). This TAD is conserved across all species except for D. ficusphila and ranges in size from 135 Kb in D. melanogaster to 217.5 Kb in D. ananassae.
To examine gene content among orthologous TADs, we identified 4,560 single-copy gene orthologs conserved across all 11 species (supplementary table S10, Supplementary Material online). The same three single-copy gene orthologs (SCOs) are found within this TAD in all species with the exception of D. triauraria, where one of the SCOs is now present on Muller B. It is likely that the size variation among species for this TAD, and TADs in general, is due to differences in the amount of noncoding sequence present within the TAD, particularly repetitive elements. To test this prediction, we compared the abundance of simple repeats and transposable elements within the TAD versus the overall size of the TAD, across the ten species. We found a strong positive correlation between simple repeat abundance and TAD size (Spearman’s , ), whereas TE abundance was not correlated with TAD size (Spearman’s , ), suggesting that simple repeat expansion/contraction may be responsible for the variation in size of this TAD among species.
We next examined the relationship between repeat abundance and TAD size across all TAD orthogroups and found that both simple repeat and TE abundances are significantly positively correlated with TAD size (Spearman’s and 0.31, respectively, in both cases) (supplementary fig. S3, Supplementary Material online). These results suggest that repetitive, noncoding sequences are important contributors to variation in size among orthologous TADs.
Rate of TAD Evolution
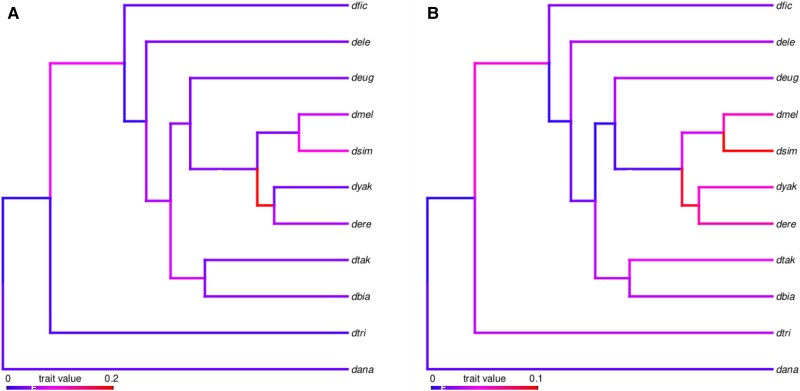
We next sought to use the pattern of TAD gain and loss among these 11 species to infer the rate of TAD evolution in Drosophila. We created a binary character matrix to track the presence/absence of orthologous TADs within each species and used a maximum-likelihood approach to estimate substitution rates for both TADs and TAD boundaries. Summing branches across the 11-species tree, we infer 0.0257 substitutions per TAD per My (95% C.I. 0.0256–0.0258) (fig. 4A) and 0.0210 substitutions per boundary per My (95% C.I. 0.0208–0.0211) (fig. 4B and supplementary table S11, Supplementary Material online).
Fig. 4.
Rate of TAD evolution. We used a maximum-likelihood approach to estimate per-branch substitution rates across the 11-species phylogeny shown here for TAD domains (A) and TAD boundaries (B). There is rate variation among branches, especially within the D. melanogaster subgroup and branches are colored according to evolutionary rate. Across the entire tree, we estimate the rate of evolution for TADs (A) as 0.0257 substitutions per TAD per My (95% C.I. 0.0256–0.0258) with per-branch rates varying from to 0.15. The rate of evolution for TAD boundaries (B) is 0.0210 substitutions per TAD per My (95% C.I. 0.0208–0.0211) with per-branch rates varying from to 0.07.
We next compared the rate of TAD evolution to the rate of boundary evolution on a branch-by-branch basis and found that TADs evolve significantly faster than boundaries (paired Wilcoxon rank sum test ). This result is further supported by the fact that 68% of branches (13 of 19) are longer in the TAD tree, whereas only 32% (6 of 19) are longer in the boundary tree. The evolutionary rates of TAD and boundary substitution are relatively rapid which suggests that overall, there may not be strong selection to preserve TAD organization over evolutionary time.
Given the relatively rapid rate of TAD evolution, we sought to compare the number of TAD gains versus losses across the phylogeny. We used the GLOOME software package (Cohen et al. 2010) to estimate the total number of gains and losses across the tree given our TAD presence/absence matrix. GLOOME infers 4,325 gains and 4,256 losses across the tree for the entire set of TADs (supplementary table S12, Supplementary Material online). The similar number of gains and losses suggests an equilibrium where TADs are turning over relatively frequently but gains are offset by losses.
TAD Evolution and Chromatin States
Genes within TADs tend to have similar expression patterns and epigenetic states (Dixon et al. 2012; Sexton et al. 2012; Schauer et al. 2017), and previous work has suggested that TADs enriched for different chromatin states evolve at different rates (Bredesen and Rehmsmeier 2019; Torosin et al. 2020). To test this prediction, we assigned each D. melanogaster high-confidence TAD to one of the five chromatin states defined by Filion et al. (2010). BLUE and BLACK chromatin states are associated with repressive, developmentally regulated chromatin, with BLUE corresponding to polycomb-repressed genes. The YELLOW chromatin state is associated with broadly expressed, transcriptionally active genes, whereas the RED chromatin state is associated with genes that are less broadly expressed compared with the YELLOW state but lack the repressive chromatin found in the BLUE and BLACK states. The GREEN chromatin state marks classic constitutive heterochromatin. Note that this analysis assumes that chromatin states are conserved between D. melanogaster and the other species studied here, however, this assumption is not unrealistic given that strong conservation of chromatin states among related species has been previously described both in Drosophila (Brown and Bachtrog 2014) and among primates (Cain et al. 2011).
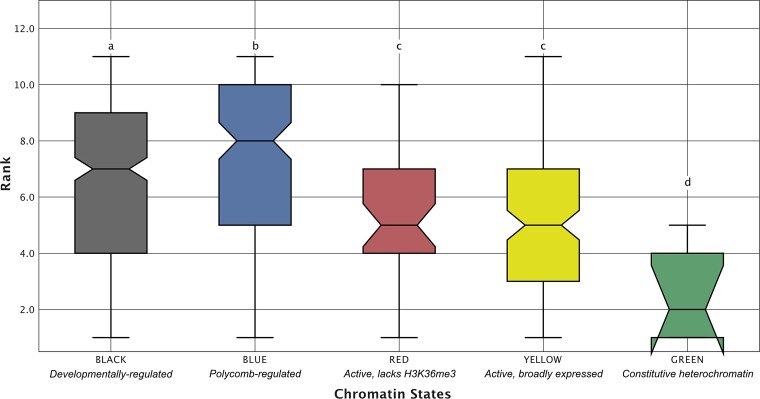
We used our orthologous TAD presence/absence matrix to rank each D. melanogaster TAD based on its level of conservation among the 11 species in our matrix, where a ranking of 1 represents a TAD that is only present in D. melanogaster and a ranking of 11 represents a TAD that has been conserved since the common ancestor of the melanogaster group (supplementary fig. S4, Supplementary Material online). We find that TADs enriched for the BLUE and BLACK chromatin states have significantly higher levels of conservation compared with TADs enriched for the YELLOW and RED chromatin states (the Wilcoxon rank sum test in all comparisons, fig. 5), whereas TADs enriched for the GREEN chromatin state are the least conserved (the Wilcoxon ranked sum test in all comparisons, fig. 5).
Fig. 5.
Evolutionary conservation of TADs by chromatin state. To assess whether TADs enriched for different chromatin states also vary in terms of evolutionary conservation, we compared conservation ranks between TADs enriched for different chromatin states. A rank of 11 was assigned to TADs with orthologs present in all 11 species, whereas a rank of 1 represents a D. melanogaster-specific TAD. TADs enriched for BLUE and BLACK chromatin have significantly higher levels of conservation than TADs enriched for the YELLOW and RED chromatin states (the Wilcoxon ranked sum test ). Compact letter display: the Wilcoxon rank sum tests were performed for all pairwise comparisons between chromatin states. Chromatin states whose distributions of conservation rank were not significantly different from each other are those that share the same letter above the top whisker of the box.
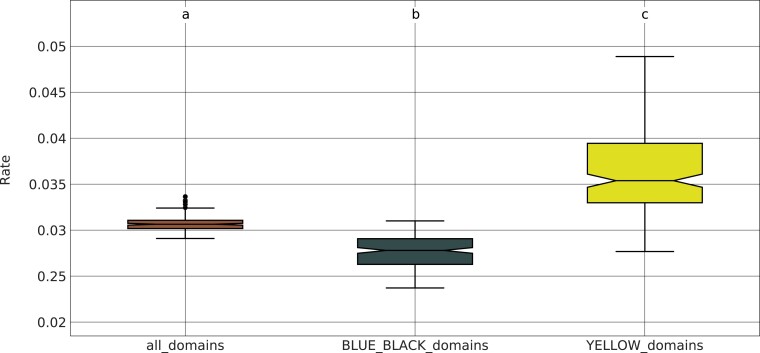
To further confirm the difference in evolutionary conservation between BLUE/BLACK and YELLOW chromatin states, we subdivided our TAD presence/absence matrix into two different submatrices: one containing only TADs that are enriched for the BLUE or BLACK state in D. melanogaster, and the other containing only TADs enriched for the YELLOW state. We re-estimated the rate of TAD evolution for each submatrix using 100 bootstrap replicates. Consistent with our previous analysis, we find that the BLUE and BLACK TADs have a significantly reduced rate of evolution compared with the YELLOW TADs (the Wilcoxon rank sum test ), whereas the rates inferred from the full matrix lie in between those from BLACK/BLUE and YELLOW (fig. 6).
Fig. 6.
Evolutionary rate comparison for active versus developmentally regulated TADs. We divided our TAD presence/absence matrix into two submatrices: one containing developmentally regulated TADs (i.e., those enriched for BLUE or BLACK chromatin states) and another containing active TADs (i.e., those enriched for the YELLOW chromatin state). We used a maximum-likelihood approach to estimate the rate of TAD substitutions separately for these two categories using 100 bootstrap replicates. Developmentally regulated TADs have a significantly reduced rate of evolution compared with active TADs (the Wilcoxon rank sum test ). We also included the complete matrix of all TAD orthogroups for comparison. The substitution rates for this matrix are significantly higher than those from the BLUE/BLACK submatrix and significantly lower than those from the YELLOW submatrix (the Wilcoxon rank sum test and , respectively).
We next investigated whether genes from highly conserved TADs are enriched for specific functional categories. We found that D. melanogaster genes associated with Gene Ontology (GO) categories related to a wide variety of developmental processes were more likely to be found in TADs with higher evolutionary conservation, for example regionalization, cellular developmental process, and anatomical structure morphogenesis were among the top most enriched terms in the Biological Process ontology (FDR , , and , respectively; supplementary tables S13 and S14, Supplementary Material online). The enrichment of these gene categories in highly conserved TADs is likely due to their association with developmentally regulated chromatin. Indeed, we find that TADs conserved across at least 9 species contain significantly more polycomb-repressed genes (Bredesen and Rehmsmeier 2019) than TADs present in three or fewer species (one-sided Fisher’s exact test ).
TAD Evolution and Gene Expression
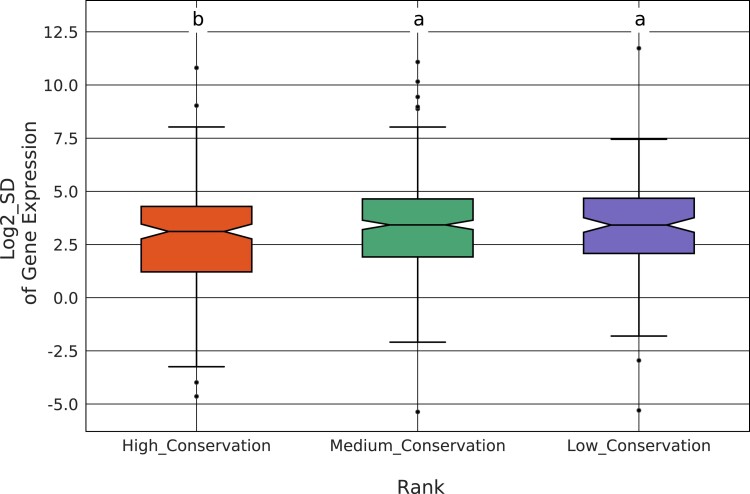
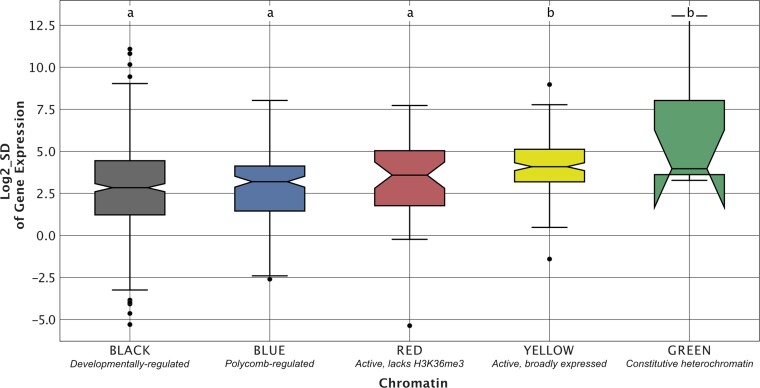
We next sought to determine whether TADs that show higher levels of evolutionary conservation also show less interspecies variation in gene expression levels. We performed mRNA-seq from embryos collected in an identical fashion to those used for Hi-C data generation (supplementary table S15, Supplementary Material online). For each single-copy gene orthogroup, we calculated the standard deviation (SD) of the expression level across the 11 species. We then compared gene expression SD between highly conserved (i.e., ranks 9–11) and lowly conserved (i.e., ranks 1–3) TADs (see supplementary table S16, Supplementary Material online for gene ortholog to TAD assignments). We found that genes from highly conserved TADs show significantly less interspecies variation in expression compared with genes from less conserved TADs (the Wilcoxon rank sum test [high vs. low: , high vs. medium ]), suggesting that TAD conservation is associated with gene expression constraint (fig. 7). Next, we compared expression variation among TADs enriched for different chromatin states. We found that BLUE and BLACK TADs show higher levels of gene expression constraint compared with YELLOW TADs (the Wilcoxon rank sum test ) (fig. 8).
Fig. 7.
Evolutionary conservation of TADs and gene expression constraint. To assess whether TAD conservation is associated with interspecies variation in gene expression, we compared the standard deviation (SD) of gene expression between TADs showing high (ranks 9–11), intermediate (ranks 4–8) and low (ranks 1–3) levels of conservation. Genes from highly conserved TADs show significantly less interspecies variation in expression compared with genes from less conserved TADs (the Wilcoxon rank sum test [high vs. low: , high vs. medium ]).
Fig. 8.
Gene expression constraint varies with TAD chromatin state. To investigate whether TADs enriched for different chromatin states show differences in interspecies variation in gene expression, we compared the standard deviation (SD) of gene expression between TADs enriched for each of five different chromatin states. We found that TADs enriched for BLUE and BLACK developmentally regulated chromatin show higher levels of gene expression constraint compared with YELLOW enriched TADs, which contain active, broadly expressed genes (the Wilcoxon rank sum test ). Compact letter display: the Wilcoxon rank sum tests were performed for all pairwise comparisons between chromatin states. Chromatin states whose distributions of conservation rank were not significantly different from each other are those that share the same letter above the top whisker of the box.
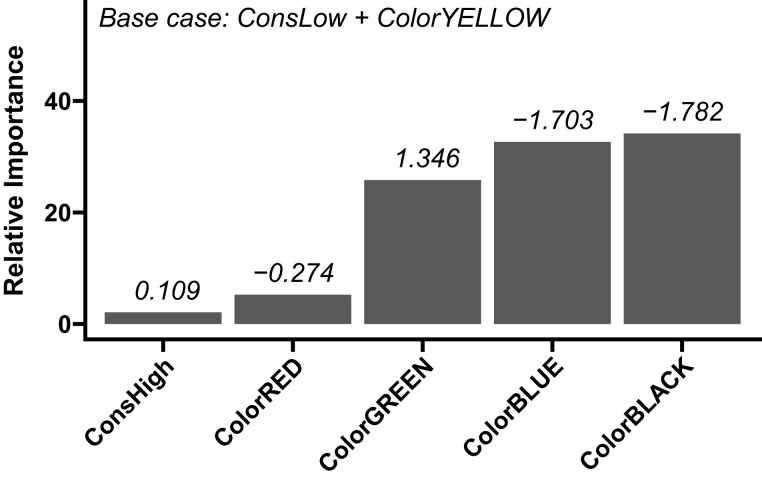
It is possible that our observed relationship between TAD conservation and gene expression variation is driven, at least in part, by the tendency of BLUE and BLACK TADs (which show higher gene expression constraint) to be more evolutionarily conserved. To address this possibility, we used partial least squares regression (PLS). PLS can be used to identify the relative importance (RI) of multiple factors in a linear model in the presence of multicollinearity or nonindependent categorical predictor variables (Chong and Jun 2005). Because of the nonindependence of chromatin state and conservation rank, we applied PLS to identify the RI of chromatin states and conservation rank as predictors of expression constraint (see Methods) (supplemental table S17, Supplementary Material online). We find that annotation as a BLUE or BLACK chromatin domain is the most robust predictor of increased expression constraint relative to annotation as a YELLOW domain, which we used as the base case in this analysis (fig. 9). In contrast, annotation as a low-conservation domain or a high conservation domain was the least robust predictor of expression constraint (fig. 9). These results suggest that TAD conservation is a relatively weak predictor of gene expression constraint compared with chromatin state. Within chromatin states, we find no difference in gene expression constraint between highly and lowly conserved TADs (fig. 10).
Fig. 9.
Importance analysis of chromatin states and TAD conservation as predictors of gene expression constraint. We estimated the RI of chromatin state or TAD conservation rank ( axis) for predicting expression constraint. Gray bars show RI (see Methods) calculated from partial least squares regression coefficients (shown above each bar). Coefficients represent expected difference in log2 of the standard deviation in gene expression associated with each variable on the axis relative to the reference case (YELLOW state and low-conservation). These results suggest that TAD conservation is a relatively weak predictor of gene expression constraint compared with chromatin state.
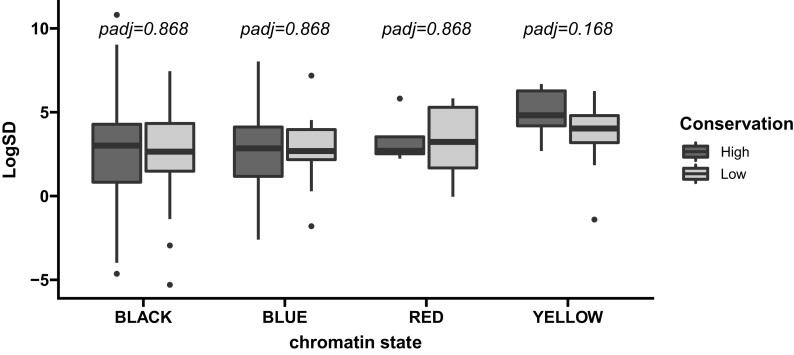
Fig. 10.
Comparison of gene expression constraint for highly and lowly conserved TADs separated by chromatin state. We compared log2 of the standard deviation in gene expression (LogSD) for low and high conservation TADs within each chromatin state. Two-sided Wilcoxon rank sum tests were used to compare LogSD between conservation classes and resulting values were corrected using the Benjamini–Hochberg procedure. Green TADs were exclusively low-conservation and were excluded.
Our analysis of gene expression constraint relied on single-copy orthologs (SCOs), which, in theory, could be enriched for housekeeping genes with high levels of expression constraint. To assess the likelihood of this possibility, we performed a GO enrichment test using the SCOs as the target set and all D. melanogaster genes as the background set. Surprisingly, rather than housekeeping genes, we found that the SCOs are enriched for developmental-related functions, for example, cellular developmental process and regulation of multicellular organismal development (FDR and , respectively; see supplementary table S18, Supplementary Material online for full table of results).
We also intersected our set of single-copy orthologs with a set of genes identified by Nourmohammad et al. (2017) whose pattern of expression across seven Drosophila species is consistent with adaptive changes in gene expression. Almost half of these genes (47%, 1,633/3,450) are present within our set of single-copy orthologs, which represents a highly significant enrichment (Fisher’s exact test ). We therefore conclude that our set of single-copy orthologs is not enriched for genes whose expression is highly constrained.
Discussion
In this study, we have combined comparative genomics and phylogenetics to empirically estimate the rate of TAD evolution. Surprisingly, we find that TADs evolve rapidly, consistent with a lack of strong selection for conservation of TAD structures. To put the numbers in perspective, our estimates of the TAD and TAD boundary substitution rates (0.0257 and 0.0210 substitutions per feature per My, respectively) are nearly an order of magnitude larger than the rate of gene duplication, which in Drosophila has been estimated to range from 0.0010 (Hahn et al. 2007) to 0.0023 (Lynch and Conery 2000) per gene per My.
We also find that the architectural proteins M1BP and BEAF-32, which are associated with TAD boundary formation in D. melanogaster, have binding motifs that are strongly enriched in the TAD boundaries of all 11 species examined here (fig. 2), suggesting that they contribute to TAD boundary formation across all species in the melanogaster species group. Interestingly, we find that TAD boundaries evolve more slowly than TADs themselves (fig. 4AandB), which could be because they represent a smaller mutational target. The slower rate of TAD boundary evolution suggests that one major route by which new TADs form is via chromosomal rearrangements that reorganize TAD structures by shuffling the locations of preexisting TAD boundaries.
Our results are consistent with our previous pairwise comparison of TAD conservation between D. melanogaster and D. triauraria where we found that the majority of TADs have been reorganized since the common ancestor of these two species 15 Ma. More broadly, our findings support other recent work that questions the long-held assumption that TADs are highly conserved. For example, a rigorous assessment of TAD conservation between humans and chimpanzees found that only 43% of TADs are conserved between the two species (Eres et al. 2019). Eres et al. (2019) and Eres and Gilad (2021) provide several explanations for why other interspecies comparisons of TAD organization may have erroneously concluded that TADs are highly conserved. For example, some studies do not directly call TADs but rather base their conclusion of strong conservation on a significant correlation of Hi-C contacts between species. Other studies use unidirectional comparisons of TAD conservation between species that differ markedly in the depth of Hi-C sequencing. In these studies, bidirectional comparisons using the same data produce much lower estimates of TAD conservation.
We have sequenced the Hi-C datasets used here at similar depths across all species and use bidirectional comparisons of TAD conservation. Furthermore, to the extent that our approach is biased, it is likely biased towards identifying higher levels of TAD conservation: we focus on TADs and TAD boundaries that are identified independently in replicate Hi-C datasets which are likely to be the most highly insulated TADs in the genome, yet we still find evidence of rapid TAD evolution. Finally, computational algorithms for TAD prediction are still being actively developed and improved (Belokopytova and Fishman 2020). There is some evidence that current approaches suffer from low accuracy (Dali and Blanchette 2017; Zufferey et al. 2018). For this reason, we used replicate Hi-C datasets and focused on TADs and TAD boundaries that were independently identified in both replicates.
Another issue that may contribute to conflicting results regarding TAD conservation is related to the approach used to measure conservation. Several previous studies have compared the percentage of TADs that were observed to be conserved between species to the percentage expected if TADs were randomly distributed across the genome. Directly estimating the rate of TAD evolution, as we have done here, or comparing TAD conservation levels to those of other genomic features, as done by Eres et al. (2019), are both approaches that allow for a more accurate determination of whether TAD organization is evolutionarily conserved.
Another line of evidence used to support the evolutionary conservation of TADs is an enrichment of rearrangement breakpoints at TAD boundaries, which has been observed in both vertebrates and flies (Lazar et al. 2018; Fishman et al. 2019; Liao et al. 2021). Our results may seem to be in contradiction to this observation; however, in Drosophila, only 24% of observed breakpoints overlap TAD boundaries (Liao et al. 2021). The majority of breakpoints (76%) do not overlap a TAD boundary and thus likely result in TAD reorganization (Liao et al. 2021). Polycomb-regulated chromatin has previously been shown to exhibit a strong conservation of gene order and a lack of rearrangement breakpoints (Harmston et al. 2017). It is possible that the enrichment of rearrangement breakpoints at TAD boundaries is due to selection specifically against rearrangement of polycomb-regulated TADs, which we previously observed in a pairwise comparison of 3D genome organization between D. melanogaster and D. triauraria (Torosin et al. 2020) and which evolve significantly slower than TADs enriched for YELLOW chromatin, as reported here.
Our observation that, in general, TADs tend to evolve rapidly calls into question the role of TADs in regulating gene expression. The prevailing view regarding the function of TADs is that they act to constrain enhancer/promoter contacts and prevent gene misexpression due to aberrant enhancer/promoter interactions (Robson et al. 2019). If TAD reorganization leads to gene misexpression, one would expect TADs to show high levels of evolutionary stability. Ours and other results supporting rapid evolution of TADs only make sense if TAD reorganization rarely causes gene misexpression. Recent work does indeed suggest that the relationship between changes in TAD structures and changes in gene expression levels are not as strong as previously thought (Ibrahim and Mundlos 2020). For example, TAD reorganization caused by the highly rearranged balancer chromosome in Drosophila is not associated with widespread changes in gene expression levels (Ghavi-Helm et al. 2019).
On the other hand, experimental manipulation of specific TADs, as well as naturally occurring mutations in TAD boundaries, have both been shown to lead to deleterious changes in gene expression levels (Arzate-Mejía et al. 2020). We previously proposed that there may be different functional subtypes of TADs that evolve under different selective constraints with TADs enriched for developmentally regulated genes evolving more slowly than TADs enriched for broadly expressed genes (Torosin et al. 2020). These results make sense given that many developmental genes are known to be regulated by long-distance enhancer/promoter contacts (Ghavi-Helm et al. 2014). In this study, we find additional support for slower evolution of TADs enriched for developmentally regulated genes (fig. 5). However, within these TADs, gene expression constraint is similar between those that show high conservation and those that show low conservation across all 11 species (fig. 10), suggesting that TAD reorganization is not associated with widespread changes in gene expression.
Our results are consistent with a model where TADs are an emergent property resulting from selection for the establishment and maintenance of insulating boundaries between active and inactive chromatin states. It is possible that the specific organization of TADs is less important as long as these boundaries between chromatin states are maintained. In this sense, it is notable that our results suggest that TAD boundaries evolve significantly slower than TADs themselves. Under this model, fusion and fission of TADs over evolutionary time may be tolerated as long as they occur between similar chromatin states. However, at the same time, it is likely that a distinct subset of TADs containing developmental genes regulated by long-distance enhancer/promoter contacts are evolving under purifying selection because their disruption could lead to aberrant gene expression (Ibrahim and Mundlos 2020). Such TADs would be expected to show higher levels of evolutionary conservation among species. Future work involving comparative epigenomics of chromatin states between the species studied here would help to test this model.
Methods
Genome Scaffolding and Annotation
DNase Hi-C chromosome conformation capture was performed for two biological replicates of each species using 8–16 h embryos as described in Torosin et al. (2020). The D. triauraria genome was sequenced, assembled, and annotated as described in Torosin et al. (2020). For the remaining species, reads from both biological replicates were concatenated and the Juicer software package (Durand, Shamim, et al. 2016) was used to build the Hi-C contact matrix followed by scaffolding of the reference genome contigs by 3D-DNA (Dudchenko et al. 2017) (see GitHub repository for reference assembly versions). The Juicebox software package (Durand, Robinson, et al. 2016) was used to visualize the contact matrices, assign chromosome boundaries, and export the scaffolded assembly. Strain information for each species is listed in supplementary table S19, Supplementary Material online.
To assign chromosome-length scaffolds to their corresponding Muller elements (i.e., Muller A–F) for each species, we performed a translated BLAST search of the scaffolds using FlyBase r6.21 D. melanogaster peptides as queries. We then used a custom python script to extract the chromosome-length scaffolds and rename them to the corresponding Muller Element (available in the GitHub repository). The final fasta file contained six scaffolds representing each Muller element. These fasta files were then softmasked using Repeatmasker software (Smit et al. 2013).
To annotate the scaffolded genomes, we used the UCSC liftover pipeline http://genomewiki.ucsc.edu/index.php/LiftOver˙Howto to transfer NCBI RefSeq annotations to the new assembly coordinates for the following species: D. ananassae, D. erecta, D. elegans, D. biarmipes, D. ficusphila, and D. takahashii. We annotated D. triauraria as described in Torosin et al. (2020). For D. melanogaster, D. simulans, D. eugracilis, and D. yakuba RefSeq annotations were transferred using GenomeThreader (v. 1.7.1) (Gremme et al. 2005).
Identifying TAD Boundaries and Domains
We removed adapter sequences from the Hi-C reads for each species using Trimmomatic software (Bolger et al. 2014) and split reads that contain a ligation junction. We used the BWA software package (Li and Durbin 2009) to align all Hi-C reads to each species’ repeatmasked and Hi-C scaffolded assembly. We then used HiCExplorer (v. 2.2) (Ramírez et al. 2018) to create a normalized contact frequency matrix. To examine reproducibility among replicates, we used HiCExplorer to convert the contact matrices from h5 format to cool format. We then used HiCRep (v. 1.12.2) (Yang et al. 2017) to calculate the stratum-adjusted correlation coefficient between replicate matrices, with the following parameters: resol = 5,000, h = 30, lbr = 0, ubr = 1,000,000.
To correct the matrices and find TAD boundaries and domains, we used the HiCExplorer (v. 3.6) (Ramírez et al. 2018) hicFindTads utility for each species and biological replicate. High-confidence boundaries were identified using Bedtools (Quinlan 2014) by requiring that the boundary overlap by at least 1 bp in both replicates. The midpoint of the boundary between the two replicates was used in downstream analyses. High-confidence domains were identified using Bedtools (Quinlan 2014) by requiring that the start and end coordinates lie within 5,000 bp of each other in both replicates. Boundaries and domains that overlapped in both replicates according to these criteria were considered high-confidence, whereas those identified in only one replicate were considered low confidence.
Orthology Pipeline Overview
We used the Cactus software package (Armstrong et al. 2019) to create a whole-genome alignment that includes the genome assemblies of all 11 species studied here. The Cactus package includes the halLiftover utility which allows genome coordinates from one species to be lifted-over to the coordinates of any of the other species, based on the genome alignment. We lifted-over the coordinates of each of the high-confidence TADs from each species to find their corresponding coordinates in each of the other species.
Large insertions/deletions or chromosomal rearrangements between species will cause a single set of coordinates to liftover into multiple fragments. We merged such fragments as long as their lifted-over locations were within 20 kb of each other. The purpose of the merging is so that a pair of TADs will still be considered orthologous even if there are intra-TAD chromosomal rearrangements and/or large insertions such as those from transposable elements. We then compared the coordinates of the merged fragments with the coordinates of the TADs that were identified in the target species. TADs were considered to be orthologous if the lifted-over coordinates overlapped either a high- or low-confidence TAD in the target species, and they overlapped reciprocally by at least 90%. We performed the liftovers in a pairwise fashion between all species pairs and in both directions (i.e., liftover from species A to B and from species B to A), which resulted in a large set of ortholog pairs. We then used Louvain clustering (see below) to create orthogroups based on the pairwise orthology assignments.
The clustering approach to identify orthogroups has two important properties: (1) a TAD can be assigned to an orthogroup as long as it was found to be orthologous to at least one other member of the orthogroup—it does not need to have been found to be orthologous to all other members of the group in the pairwise comparisons. (2) Because the liftovers were done in both directions, a low confidence TAD (i.e., one that was identified in only one replicate) can be assigned to an orthogroup if it was found to be orthologous to a high-confidence TAD in another species. The inclusion of low-confidence TADs in orthogroups means that occasionally, two different sets of TAD coordinates from the same species will be present in the same TAD orthogroup. This will happen when a low-confidence TAD from each replicate is found to be orthologous to a high-confidence TAD in another species but the two low-confidence TADs failed to overlap in a way that would meet the high-confidence criterion. In these cases, the coordinates of the two low-confidence TADs were merged for downstream analysis.
Identifying and Defining Orthologous Boundaries
Repeatmasked genomes were aligned using Cactus (Armstrong et al. 2019) to generate a hal file. We used halLiftover (Hickey et al. 2013) to liftover the coordinates of the high-confidence boundaries from the query to the target species. To filter the halLiftover (Hickey et al. 2013) output, we merged “lifted-over” boundary locations in the target species that were within 5,000 bp of each other into a single feature and removed any remaining segments less than 500 bp in size. Lifted-over high-confidence boundaries from the query species that were located within 5 kb of either a high- or low-confidence boundary in the target species were considered to be orthologous. The boundary orthology pipeline was completed in both directions for all 11 species resulting in 110 orthology analyses. To identify boundary orthogroups among all 11 species, all pairwise boundary orthologs were concatenated and input to the software Cytoscape (Shannon et al. 2003) for network analysis using the Louvain clustering method and default settings. The output represents boundary orthogroups containing sets of orthologous boundaries that were found in two or more species.
Identifying and Defining Orthologous Domains
To evaluate TAD conservation, we used halLiftover to liftover the genomic coordinates of high-confidence TADs from the query to the target species. To filter the halLiftover output, we merged “lifted-over” features separated by less than 20 kb and removed all “lifted-over” segments less than 5 kb in size. Lifted-over high-confidence domains from the query species that reciprocally overlapped either a high- or low-confidence domain in the target species by at least 90% were considered to be orthologous. Those that did not meet this criteria were considered nonorthologous. Domain orthogroups were identified using Louvain clustering, as described above for the TAD boundaries.
TAD Stringency
We assessed the effect of TAD calling stringency using a single replicate of our D. melanogaster Hi-C data. We called TADs with these data using the HiCExplorer (v. 3.6) hicFindTads utility and the following values for the delta parameter: 0.01 (default value), 0.03, 0.05, 0.07, 0.09, 0.11, 0.13, and 0.15. We intersected each set of TAD calls with the set of D. melanogaster TADs we had previously assigned to TAD orthogroups. We retained TADs whose start and end coordinates were within 5 kb of each other and found the conservation rank for each of these. We then visualized the distribution of conservation ranks for each set of TAD calls.
TAD Size Variation
We calculated simple repeat and transposon abundance using the RepBase library for the Drosophila genus. For each TAD orthogroup we determined the mean and standard deviation for the following values: size, simple repeat abundance, and transposon abundance. We calculated Z-scores for each species by subtracting the mean value from the observed value and dividing by the standard deviation.
Rate of Evolution
To estimate boundary and domain evolutionary rates, we used the orthogroup assignments to create a binary character state matrix based on the presence/absence of each orthologous boundary or domain across all 11 species. For both the boundary and domain binary matrices, we inferred maximum-likelihood estimates of branch lengths using IQ-TREE (Minh et al. 2020) with topology constraints based on the species tree in Suvorov et al. (2021) and allowing for automatic model selection. We implemented a custom R script (see GitHub repository) to calculate the substitution rate of boundaries and domains per My, respectively, using branch lengths from the boundary and domain trees and divergence time estimates from Suvorov et al. (2021). We generated confidence intervals for our rate estimates by recalculating boundary and domain evolutionary rates for each of 100 bootstrap replicates.
To compare evolutionary rates among TADs enriched for different chromatin states, we followed the same methodology as described above using 100 bootstrap replicates. Drosophila melanogaster TADs were assigned chromatin states using annotations from Filion et al. (2010) based on the state that covered the largest proportion of the TAD.
To compare the number of TAD gains versus losses across the phylogeny, we used the GLOOME web server (Cohen et al. 2010) with default parameters except for the following modifications: we set Gain and Loss rates to variable and instructed GLOOME to estimate parameters with multiple random starting points.
Boundary Motif Enrichment
For each of the 11 species, we assessed high-confidence TAD boundaries for enrichment of motifs for the following insulator proteins: BEAF-32, M1BP, CTCF, ZIPIC, and Su(Hw). We used motifs identified from D. melanogaster, which we obtained from JASPAR (Fornes et al. 2020) (accession numbers in supplementary table S5, Supplementary Material online). We quantified motif enrichment at high-confidence boundaries using AME (v. 5.3.3) from the MEME suite (Bailey et al. 2009). The background dataset consisted of the genome assembly split into 5 kb sequences.
Gene Expression
Stranded mRNA-seq libraries were prepared from 8 to 16 h embryos as described in Torosin et al. (2020). RNA-seq data were aligned to their respective genomes using HISAT2 (v. 2.2.1) (Kim et al. 2019). Per gene expression values were calculated using TPMcalculator (v. 0.0.3) (Vera Alvarez et al. 2019).
Gene Expression and Chromatin Analyses
We used Orthofinder (v. 2.5.2) (Emms and Kelly 2019) to identify single-copy gene orthologs across all 11 species (see GitHub repository for peptide FASTA files). TADs were assigned to chromatin states as described previously. When comparing gene expression variation among TADs enriched for different chromatin states, genes were assigned the chromatin state of their parent TAD.
To compare gene expression variation among TADs that differ in their level of evolutionary conservation, TAD orthogroups were assigned a rank based on the number of species that shared the orthologous TAD (i.e., conserved across five species rank 5). Lineage specific TADs were assigned rank 1. Gene orthologs were assigned the rank of their parent TAD. In theory, it is possible for genes to move between TADs via translocations or retrotransposition events. In order to make our pipeline robust to such occurrences, we included a filter so that gene orthologs whose parent TADs belonged to different orthogroups were omitted from further analysis. To evaluate expression differences based on evolutionary conservation, we divided genes into three categories: low-conservation—ranks 1–3, medium-conservation—ranks 4–8, and high-conservation—ranks 9–11.
GO Enrichment
We evaluated whether genes from highly conserved TADs are enriched for specific functional categories using the GOrilla tool (Eden et al. 2009). We input a single ranked list of genes (supplementary table S14, Supplementary Material online) using the gene symbol ordered from highest to lowest by the conservation ranks assigned as described in the previous section. We used all D. melanogaster genes found in each D. melanogaster TAD orthogroup, rather than only single-copy orthologs because we found that the set of single-copy orthologs was itself enriched for development-related functions supplementary table S18, Supplementary Material online. We ran GOrilla for all three GO Ontologies: Biological Process, Molecular Function, and Cellular Component. The results for all three are reported in supplementary tables S13 and S18, Supplementary Material online.
Partial Least Squares Regression
We performed partial least squares regression (PLS) using the plsr function from the plsR package (v. 2.8-1) (Mevik and Wehrens 2007) under R 4.2.1 on Ubuntu 20.04.4. We regressed the previously described expression constraint against chromatin state and conservation group (Low or High). Cross-validation runs on one through five components revealed that three components minimized the root mean squared error of prediction. Three components were used for subsequent analysis. To rank RI of individual chromatin states or conservation rank, we calculated a relative importance metric by summing the absolute values of the PLS coefficients and recalculated each coefficient as a proportion of that total.
Supplementary Material
Acknowledgements
The authors acknowledge the Office of Advanced Research Computing (OARC) at Rutgers and The State University of New Jersey for providing access to the Amarel cluster and associated research computing resources that have contributed to the results reported here. The authors thank Wilson Leung for advice regarding approaches to transfer gene model predictions between assembly versions. Stocks obtained from the University of California San Diego, Washington University, the National Drosophila Species Stock Center, the Drosophila Genetic Reference Panel, Dr. Andrew Kern, and the Ehime University Fly Stock Center were used in this study.
Contributor Information
Nicole S Torosin, Department of Genetics, Rutgers University, Piscataway, NJ 08854, USA.
Tirupathi Rao Golla, LifeCell, Kelambakkam Main Road, Keelakottaiyur, Chennai 600127, Tamil Nadu, India.
Matthew A Lawlor, Department of Genetics, Rutgers University, Piscataway, NJ 08854, USA.
Weihuan Cao, Department of Genetics, Rutgers University, Piscataway, NJ 08854, USA.
Christopher E Ellison, Department of Genetics, Rutgers University, Piscataway, NJ 08854, USA.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability Statement
Illumina data generated for this project is available at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) under BioProject PRJNA813540. Assemblies generated are available through Zenodo DOI:10.5281/zenodo.6306490. Complete analysis pipelines and all custom scripts described in this project can be found on GitHub at https://github.com/Ellison-Lab/TADs.
References
- Armstrong J, Hickey G, Diekhans M, Fiddes I, Novak A, Deran A, Fang Q, Xie D, Feng S, Stiller J, et al. 2020. Progressive Cactus is a multiple-genome aligner for the thousand-genome era. Nature. 587(7833):246–251. 10.1038/s41586-020-2871-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzate-Mejía RG, Josué Cerecedo-Castillo A, Guerrero G, Furlan-Magaril M, Recillas-Targa F. 2020. In situ dissection of domain boundaries affect genome topology and gene transcription in Drosophila. Nat Commun. 11(1):894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bag I, Chen S, Rosin LF, Chen Y, Liu C-Y, Yu G-Y, Lei EP. 2021. M1BP cooperates with CP190 to activate transcription at TAD borders and promote chromatin insulator activity. Nat Commun. 12(1):4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37(Web Server):W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belokopytova P, Fishman V. 2020. Predicting genome architecture: challenges and solutions. Front Genet. 11:617202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutkar A, Schaeffer SW, Russo SM, Xu M, Smith TF, Gelbart WM. 2008. Chromosomal rearrangement inferred from comparisons of 12 Drosophila genomes. Genetics 179(3):1657–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock IR. 1980. Current status of the Drosophila melanogaster species-group (Diptera). Syst Entomol. 5(4):341–356. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredesen BA, Rehmsmeier M. 2019. DNA sequence models of genome-wide Drosophila melanogaster polycomb binding sites improve generalization to independent polycomb response elements. Nucleic Acids Res. 47(15):7781–7797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EJ, Bachtrog D. 2014. The chromatin landscape of Drosophila: comparisons between species, sexes, and chromosomes. Genome Res. 24(7):1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain CE, Blekhman R, Marioni JC, Gilad Y. 2011. Gene expression differences among primates are associated with changes in a histone epigenetic modification. Genetics 187(4):1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong I-G, Jun C-H. 2005. Performance of some variable selection methods when multicollinearity is present. Chemom Intell Lab Syst. 78(1–2):103–112. [Google Scholar]
- Cohen O, Ashkenazy H, Belinky F, Huchon D, Pupko T. 2010. GLOOME: gain loss mapping engine. Bioinformatics 26(22):2914–2915. [DOI] [PubMed] [Google Scholar]
- Dali R, Blanchette M. 2017. A critical assessment of topologically associating domain prediction tools. Nucleic Acids Res. 45(6):2994–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485(7398):376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. 2017. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356(6333):92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, Aiden EL. 2016. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3(1):99–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Lieberman Aiden E. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eres IE, Gilad Y. 2021. A TAD skeptic: is 3D genome topology conserved? Trends Genet. 37(3):216–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eres IE, Luo K, Hsiao CJ, Blake LE, Gilad Y. 2019. Reorganization of 3D genome structure may contribute to gene regulatory evolution in primates. PLoS Genet. 15(7):e1008278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143(2):212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman V, Battulin N, Nuriddinov M, Maslova A, Zlotina A, Strunov A, Chervyakova D, Korablev A, Serov O, Krasikova A. 2019. 3D organization of chicken genome demonstrates evolutionary conservation of topologically associated domains and highlights unique architecture of erythrocytes’ chromatin. Nucleic Acids Res. 47(2):648–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornes O, Castro-Mondragon JA, Khan A, van der Lee R, Zhang X, Richmond PA, Modi BP, Correard S, Gheorghe M, Baranašić D, et al. 2020. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res.. 48(D1):D87–D92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep. 15(9):2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. 2019. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet. 51(8):1272–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EEM. 2014. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512(7512):96–100. [DOI] [PubMed] [Google Scholar]
- Gremme G, Brendel V, Sparks ME, Kurtz S. 2005. Engineering a software tool for gene structure prediction in higher organisms. Inf Softw Technol. 47(15):965–978. [Google Scholar]
- Hahn MW, Han MV, Han S-G. 2007. Gene family evolution across 12 Drosophila genomes. PLoS Genet. 3(11):e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmston N, Ing-Simmons E, Tan G, Perry M, Merkenschlager M, Lenhard B. 2017. Topologically associating domains are ancient features that coincide with metazoan clusters of extreme noncoding conservation. Nat Commun. 8(1):441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey G, Paten B, Earl D, Zerbino D, Haussler D. 2013. HAL: a hierarchical format for storing and analyzing multiple genome alignments. Bioinformatics 29(10):1341–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim DM, Mundlos S. 2020. The role of 3D chromatin domains in gene regulation: a multi-facetted view on genome organization. Curr Opin Genet Dev. 61:1–8. [DOI] [PubMed] [Google Scholar]
- Kaushal A, Mohana G, Dorier J, Özdemir I, Omer A, Cousin P, Semenova A, Taschner M, Dergai O, Marzetta F, et al. 2021. CTCF loss has limited effects on global genome architecture in Drosophila despite critical regulatory functions. Nat Commun. 12(1):1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 37(8):907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krefting J, Andrade-Navarro MA, Ibn-Salem J. 2018. Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 16(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar NH, Nevonen KA, O’Connell B, McCann C, O’Neill RJ, Green RE, Meyer TJ, Okhovat M, Carbone L. 2018. Epigenetic maintenance of topological domains in the highly rearranged gibbon genome. Genome Res. 28(7):983–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Zhang X, Chakraborty M, Emerson JJ. 2021. Topologically associating domains and their role in the evolution of genome structure and function in Drosophila. Genome Res. 31(3):397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155. [DOI] [PubMed] [Google Scholar]
- Mevik B-H, Wehrens R. 2007. The PLS package: principal component and partial least squares regression in R. J Stat Softw. 18:1–23. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourmohammad A, Rambeau J, Held T, Kovacova V, Berg J, Lässig M. 2017. Adaptive evolution of gene expression in Drosophila. Cell Rep. 20(6):1385–1395. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. 2013. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell 153(6):1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47:11.12.1–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 9(1):189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang S -C, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon K -R, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171(2):305–320.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renschler G, Richard G, Valsecchi CIK, Toscano S, Arrigoni L, Ramirez F, Akhtar A. 2019. Hi-C guided assemblies reveal conserved regulatory topologies on X and autosomes despite extensive genome shuffling. BioRxiv. [DOI] [PMC free article] [PubMed]
- Robson MI, Ringel AR, Mundlos S. 2019. Regulatory landscaping: how enhancer-promoter communication is sculpted in 3D. Mol Cell. 74(6):1110–1122. [DOI] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S -C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 112(47):E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer T, Ghavi-Helm Y, Sexton T, Albig C, Regnard C, Cavalli G, Furlong EE, Becker PB. 2017. Chromosome topology guides the Drosophila dosage compensation complex for target gene activation. EMBO Rep.18(10):1854–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148(3):458–472. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013. RepeatMasker Open-4.0.2013-2015.
- Suvorov A, Kim BY, Wang J, Armstrong EE, Peede D, D’Agostino ERR, Price DK, Wadell P, Lang M, Courtier-Orgogozo V, et al. 2021. Widespread introgression across a phylogeny of 155 Drosophila genomes. Curr Biol. 32(1):111–123.e5. doi: 10.1016/j.cub.2021.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Bantignies F, Cavalli G. 2019. Principles of genome folding into topologically associating domains. Sci Adv. 5(4):eaaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torosin NS, Anand A, Golla TR, Cao W, Ellison CE. 2020. 3D genome evolution and reorganization in the Drosophila melanogaster species group. PLoS Genet. 16(12):e1009229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera Alvarez R, Pongor LS, Mariño-Ramírez L, Landsman D. 2019. TPMCalculator: one-step software to quantify mRNA abundance of genomic features. Bioinformatics 35(11):1960–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Zhang F, Yardımcı GG, Song F, Hardison RC, Noble WS, Yue F, Li Q. 2017. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res. 27(11):1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey M, Tavernari D, Oricchio E, Ciriello G. 2018. Comparison of computational methods for the identification of topologically associating domains. Genome Biol. 19(1):217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Illumina data generated for this project is available at the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) under BioProject PRJNA813540. Assemblies generated are available through Zenodo DOI:10.5281/zenodo.6306490. Complete analysis pipelines and all custom scripts described in this project can be found on GitHub at https://github.com/Ellison-Lab/TADs.