Abstract
In 2020, the novel coronavirus, SARS-CoV-2, caused a pandemic, which is still raging at the time of writing this. Here, we present results from SpikeSeq, the first published Sanger sequencing-based method for the detection of Variants of Concern (VOC) and key mutations, using a 1 kb amplicon from the recognized ARTIC Network primers. The proposed setup relies entirely on materials and methods already in use in diagnostic RT-qPCR labs and on existing commercial infrastructure offering sequencing services. For data analysis, we provide an automated, open source, and browser-based mutation calling software (https://github.com/kblin/covid-spike-classification, https://ssi.biolib.com/covid-spike-classification). We validated the setup on 195 SARS-CoV-2 positive samples, and we were able to profile 85% of RT-qPCR positive samples, where the last 15% largely stemmed from samples with low viral count. We compared the SpikeSeq results to WGS results. SpikeSeq has been used as the primary variant identification tool on > 10.000 SARS-CoV-2 positive clinical samples during 2021. At approximately 4€ per sample in material cost, minimal hands-on time, little data handling, and a short turnaround time, the setup is simple enough to be implemented in any SARS-CoV-2 RT-qPCR diagnostic lab. Our protocol provides results that can be used to choose antibodies in a clinical setting and for the tracking and surveillance of all positive samples for new variants and known ones such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1) Delta (B.1.617.2), Omicron BA.1(B.1.1.529), BA.2, BA.4/5, BA.2.75.x, and many more, as of October 2022.
Keywords: SARS-CoV-2, COVID-19, CoV, Spike protein, Variants of concern, Surveillance, Mutations, Profiling, Coronavirus, Sequencing, Contact tracing, Omicron, BA.1, BA.2, BA.4/5, BA.2.75.x
1. Introduction
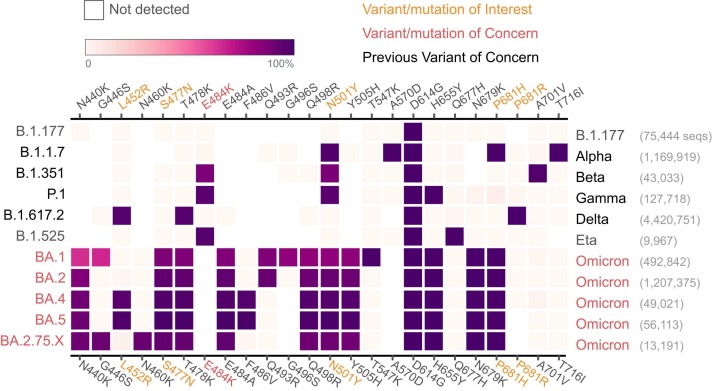
The pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a crisis faced by nearly every country in the world at the time of writing this article. Throughout 2020 and 2021, the spread of SARS-CoV-2 caused millions of deaths, and more than 600 million people have been diagnosed with COVID-19, primarily with Real-Time quantitative Polymerase Chain Reaction (RT-qPCR) diagnostic tests (Wang et al., 2021). Variants of the virus are identified by whole genome sequencing (WGS) based mutation calls and compared to the originally published genome (Wu et al., 2020) or by variant specific qPCR tests (Vogels, 2021, Vogels et al., 2020, Spiess et al., 2021). WGS of SARS-CoV-2 has led to the identification of several VOCs of SARS-CoV-2, such as the Pango (Rambaut et al., 2020) lineages B.1.1.7 (Preliminary, 2020) (Alpha), B.1.351 (Tegally et al., 2020) (Beta), P.1 (Genomic characterisation, 2021)(Gamma), B.1.617.2 (Delta), B.1.1.529 (Omicron BA.1), B.1.1.529.2 (Omicron BA.2), B.1.1.529.4/ B.1.1.529.5 (Omicron BA.4/5), and B.1.1.529.2.75.x (Omicron BA.2.75.x) (O’Toole et al., 2021a). Fig. 1 shows the mutations in the SpikeSeq amplicon and that all the shown variants can be distinguished from each other using only the SpikeSeq amplicon. Monitoring VOCs makes it possible to tailor the societal response for maximal containment of SARS-CoV-2 at the lowest cost. While WGS is crucial for surveillance work, it is not optimal for near real-time contact tracing of VOCs, as it is it is expensive, slow and requires a centralized setup and a high degree of technical and bioinformatics expertise. In a clinical setting, clear and rapid differentiation between variants can be used for selecting therapeutic antibodies, e.g. Casirivimab/Imdevimab for Delta infected patients and retaining Sotrovimab for Omicron infected patients (Li and Gandhi, 2022). The clinical laboratories around the world can therefore benefit greatly by using our low cost SpikeSeq for routine screening of COVID-19 positive patients. SpikeSeq is a cost efficient, rapid, and simple method for SARS-CoV-2 variant mutation typing using no additional equipment or reagents to that already in use for diagnostic RT-qPCR SARS-CoV-2 testing ( Fig. 2). At a cost of approximately 4€ per reaction in materials, and approximately one and a half hours of hands-on time per 96 samples (Fig. 2), this system can readily be implemented for large scale typing of all positive samples, without the use of specialized equipment or personnel. The material cost is much cheaper than the cost of WGS and does not increase in price per sample if only a few samples are run at a time (Quick, 2020). SpikeSeq consists of an RT-PCR with a single ARTIC Network primer set, using the same enzyme mix used in diagnostic COVID-19 RT-qPCR tests, and using Sanger sequencing to sequence a 1001 bp amplicon located in the spike gene. Sanger sequencing as a technology is more than 40 years old (Sanger et al., 1977), and a large existing commercial infrastructure offers Sanger sequencing services. We provide a bioinformatics tool that enables automated basecalling, mapping and calling of mutations from Sanger electropherogram files (.ab1), fasta or fastq files. The results are reported in a user-friendly table as well as in fastq format, available as a command line interface from https://github.com/kblin/covid-spike-classification or in a browser-based app running locally from https://ssi.biolib.com/app/covid-spike-classification/run without data leaving the computer. SpikeSeq has been the primary variant profiling tool for all positive samples from the test facility at the Technical University of Denmark (DTU) and used by several other Danish hospitals, totaling > 10.000 samples through 2021.
Fig. 1.
Profiling of selected SARS-CoV-2 variants based on key mutations from N440K to T716I in a single amplicon in the spike protein. Even without the near universal D614G mutation, all current and previous VOC variants have a unique mutation profile in this amplicon window, except for BA.4/5, which are commonly reported together. While not shown, the only mutation in the primer sequences in any VOC is on the likely inconsequential K417.
Figure adapted from Outbreak.info (Tsueng et al., 2022), downloaded 2022–10–26.
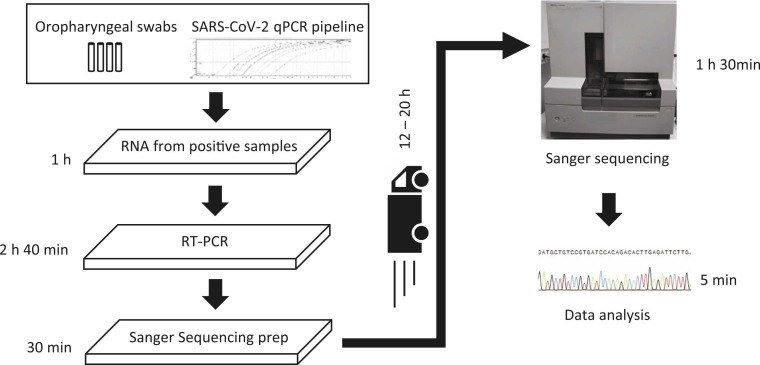
Fig. 2.
Workflow of SpikeSeq typing of SARS-CoV-2 variant mutations. Time estimates are based on a single 96 well PCR plate and no automation. The total hands-on time is one hour and 30 min, and only requires the set up of an RT-PCR from already extracted RNA from a diagnostics lab followed by transfer of 1.5 μl of the product to a new 96 well PCR plate. The plate is subsequently sent for Sanger sequencing at a commercial provider. The total cost per sample including plastware, enzymes, transport, and sequencing is approx. 4 €. After the raw data is received, data analysis takes less than five minutes for 96 samples. If Sanger sequencing is available in-house, the time to result could be less than six hours.
2. Methods
The detailed SpikeSeq protocol was the first Sanger sequencing-based SARS-CoV-2 variant protocol published, and has been available from https://www.protocols.io/view/sanger-sequencing-of-a-part-of-the-sars-cov-2-spik-bsbdnai6 since February 10th, 2021 (Jørgensen, 2021).
2.1. Input RNA
The RNA from the diagnostic SARS-CoV-2 RT-qPCR facility at the Centre for Diagnostics at DTU was obtained from oropharyngeal swabs with specimens transferred immediately into a tube containing 3 M Guanidine Thiocyanate for instant lysis of viral particles. This ensures stability of RNA during transportation and during the process of transferring patient material from sampling tube to plate format. RNAdvance Viral Reagent Kit - Built on SPRI (Solid Phase Reversible Immobilization) bead-based technology (#C63510 Bechman Coulter, IN, USA), was used for the purification of input RNA. RNA was frozen for 1–2 days at −20 C° prior to being used for SpikeSeq.
2.2. RT-PCR and sequencing reaction
The SpikeSeq Reverse Transcription Polymerase Chain Reaction (RT-PCR) was set up in 20 μl RT-PCR reactions using 10 μl One Step PrimeScript III (Takara, Shimogyō-ku, Kyoto), 0.4 μM forward primer ( Table 1), 0.4 μM reverse primer, and 5 μl purified template RNA per reaction (Table 1). RT-PCR was performed with the following program: reverse transcription for 5 min at 52 °C, then hot start polymerase activation at 95 °C for 10 s, followed by 45 cycles of 95 °C for 5 s, 58 °C for 30 s, and 72 °C for 1 min, followed by 5 min at 72 °C. For each Sanger reaction, 1.5 μl unpurified RT-PCR product was diluted to 18.5 μl containing 1.2 µM of the nCoV-2019_76_LEFT_alt3 primer. The Sanger sequencing was performed by Eurofins Genomics (Eurofins, Luxembourg, Luxembourg).
Table 1.
Primers suggested for typing SARS-CoV-2 VOCs. The Primer melting temperature is based on the use of Takara Primescript III master mix, which contains 2 mM MgCl2. Primer melting temperature was calculated using the Oligo Analyzer from IDT (https://eu.idtdna.com/calc/analyzer, Integrated DNA Technologies, Coralville, Iowa, USA).
| Primer name | Primer sequence | Primer Tm | Primer end position in NC_045512 | SpikeSeq mutation analysis range |
|---|---|---|---|---|
| nCoV-2019_76_LEFT_alt3 | GGGCAAACTGGAAAGATTGCTGA | 65.4 ºC | 22,822 | N439 to T716 |
| nCoV-2019_78_RIGHT | TGTGTACAAAAACTGCCATATTGCA | 63.9 ºC | 238,23 |
2.3. Data analysis
For analyzing the Sanger sequencing data, we set up a workflow consisting of: 1) basecalling the raw Sanger sequencing electropherogram files using Tracy (Rausch et al., 2020), followed by 2) mapping of fastq data to the NC_045512.2 SARS-CoV-2 sequence using Bowtie2 (Langmead and Salzberg, 2012), and finally 3) mutation calling with Samtools (Li et al., 2009). The codons with known mutations were subsequently extracted and translated, and the amino acid calls compared to a list of known mutations, which is continuously updated as new variants are reported. To date, version 0.6.4 of the software includes the following mutations in the spike protein: K417N/T, N439K, N440K, G446S, Y449H/N, L452M/R, Y453F, S477N, T478K/R, E484A/K/Q, F486V, F490R, Q493K/R, G496S, Q498R, N501Y, Y505H, T547K, A570D, Q613H, D614G, A626S, H655Y, Q677E/H, N679K, P681H/R, I692V, A701V, S704L, T716I, T732A. Mutation information is primarily derived from covariant.org (Hodcroft, 2021) and outbreak.info (Julia Mullen, 2022), and reported as a csv table file, along with the basecalls in fastq format. The program is available from https://github.com/kblin/covid-spike-classification. Alternatively, Statens Serum Institut (SSI), the Danish government body for SARS-CoV-2 surveillance, has set up a self-contained web-app running the program without data upload at https://ssi.biolib.com/app/covid-spike-classification/run.
2.4. WGS
To compare the SpikeSeq mutation calls to WGS mutation calls, 29 samples with SpikeSeq data were subjected to WGS. The illumina based WGS method used at Rigshospitalet for the samples of this study has been described previously (Andersen et al., 2021). Sequence reads were trimmed using BBDuk (Bushnell et al., 2017) v.36.49 and aligned to Wuhan-Hu-1 reference genome (GenBank MN908947.3) using Minimap2 (Li, 2018) v.2.17. Aligned reads were sorted using Samtools (Li et al., 2009) v.1.10 and primers were trimmed, and consensus sequence called, using iVar (Grubaugh et al., 2019) v.1.3 (options “-m 10 -t 0.9 -n N” chosen for consensus calling). SARS-CoV-2 lineages were identified with pangolin (v.2.3.2) / pangoLEARN (v.2021–02–21) (O’Toole et al., 2021b) (https://github.com/cov-lineages/pangolin). From the assembled WGS consensus sequence, the amplicon sequence used for SpikeSeq was extracted and subjected to the same analysis as the Sanger data, using the --fasta switch in https://github.com/kblin/covid-spike-classification.
3. Results & discussion
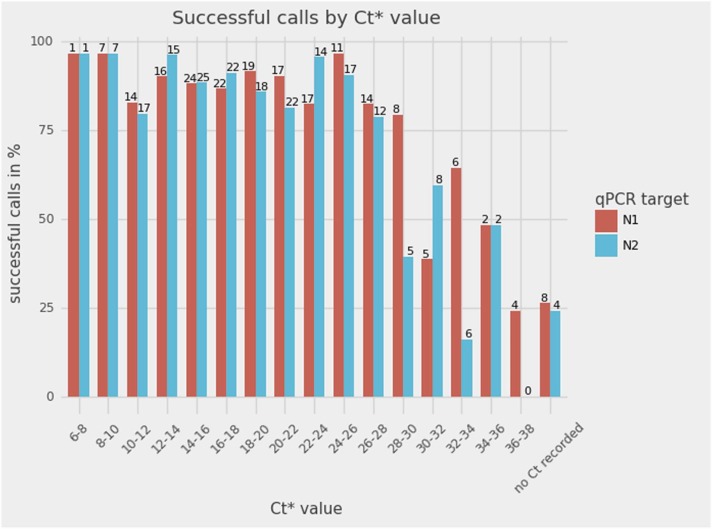
In RT-qPCR SARS-CoV-2 tests, the relative number of viral copies in a sample is estimated by the threshold cycle (Ct) value, where a higher value indicates fewer viral copies. A batch of 195 SARS-CoV-2 positive samples was used to investigate the relationship between viral load and the ability of SpikeSeq to generate mutation calls. In Fig. 3, the SpikeSeq results of the 195 samples have been binned according to the diagnostic RT-qPCR Ct value for both of the targets (N1 and N2). Overall, the success rate of SpikeSeq was 85% (166/195 Sanger reads mapped to the correct position of the SARS-CoV-2 spike gene). The difference in sensitivity observed between N1 and N2 reflects that the Ct for the two RT-qPCR targets of SARS-CoV-2 can be different, and sometimes one target is not recorded (n = 8 for N1 and n = 4 for N2). Of note is that unpurified RT-PCR product was sequenced, significantly reducing the workload involved in mutation profiling. The fraction of samples with Ct < 30 with a mappable result from Sanger sequencing is approximately 90% (Fig. 3). For samples where the Sanger read successfully mapped, 99.1% of positions had corresponding sequence information available (4770 out of 4814 positions, CSC version 0.6.4, 166 samples mapped, 29 positions, counting the positions with multiple mutations once only, see Table S1). The positions without sequence information were mainly from the ends of the sequence, as is common in Sanger sequencing data. As expected, the “No Template Control” did not yield a result. Note that the initial seven cycles in the diagnostic RT-qPCR are not fluorescence recorded and therefore do not count towards the final Ct value, which means that the sensitivity could be even greater than reported. Five of the 166 sequences had the combination N501Y, A570D, P681H, and T716I mutations, which strongly suggests infection with the B.1.1.7 lineage (Fig. 1, Table S1) based on the known circulating variants at the time. This key result shows that at least 85% of SARS-CoV-2 positive sample scan be profiled using SpikeSeq, which is comparable to WGS success rates (Baker et al., 2021).
Fig. 3.
The sensitivity of the SpikeSeq method versus the Ct values of the diagnostic RT-qPCR test (n = 195). The number above each bar refers to the number of samples in that bin. Overall, Sanger sequencing data from 85% of samples were of sufficient quality for mapping. The non-template control is not depicted, as it did not produce a Sanger read that mapped to the reference genome. * : Ct values stem from a 2-step PCR where the first 7 cycles are not fluorescence registered, possibly meaning that the Sanger sequencing assay is even more sensitive than suggested by this figure.
To verify SpikeSeq mutation calls, we focused on 29 samples where both SpikeSeq and whole genome consensus sequences were available. The sample set included 7 B.1.1.7 (Alpha), 13 B.1.177, 2 B.1.177.12, 2 B.1.177.21, 1 B.1.258.16, 2 B.1.525 and the first detected P.1 (Gamma) sample in Denmark. Of the 29 samples, 28 had identical mutation profiles in the SpikeSeq amplicon window (Supplementary table 2). One sample had mutations identical to B.1.1.7 by SpikeSeq but was classified as B.1.177.21 by WGS and had no B.1.1.7 defining mutations in the amplicon window in the WGS consensus sequence. We have not been able to identify the cause of the discrepancy, but a sample mix up can explain the observed difference.
SpikeSeq has been used as the primary variant identification tool at DTU Diagnostics, where it detected the first case of Gamma in Denmark in early March 21. In early April 2021, before Delta was officially named, SpikeSeq data was used to identify a Delta infection and alert authorities of the unusual variant, allowing contact tracing to stop further transmission. Both of these variants were later confirmed by WGS (data not shown), and highlights the ability of SpikeSeq to find known and emerging variants.
For samples where the SpikeSeq mutation profile is not a known VOC or nonVOC, we suggest that the provided fastq file is aligned to the reference and manually viewed to identify additional mutations. Then, to gain more information from the mutation profile, we suggest using the fantastic resources available at nextstrain.org/ncov (Hadfield et al., 2018) to filter the GISAID deposited genome sequences to the samples with a similar mutation profile, which will give information on the geographical spread and novelty of the mutation profile.
An Achilles heel of any PCR based assay is mutations in the primer binding regions. This is true for all major SARS-CoV-2 WGS approaches, all variant specific RT-qPCR setups, and SpikeSeq, however the three approaches are differently affected. For WGS, where a combination of 30–100 primer sets is routinely used, dropout of a single amplicon will rarely influence the analysis. For RT-qPCRs on the other hand, mutations both in the primer binding regions and in the probe region can lead to misinterpretation of the results, sometimes even in an undecipherable way. This may potentially lead to wrong mutation calls and hence variant profiling. This was observed during the transition from Alpha to Delta in 2021 with the spike position 452 not only being Leucine (L) or Arginine (R) as expected, but also Glutamine (Q) and Methionine (M) providing false positives/negatives for a common RT-qPCR variant assay in Denmark. For the SpikeSeq method, mutations in the primer binding site could lead to lowered success rate of the assay, but not to wrong mutation calling, as there is either a good quality sequence or not. The primer sites chosen for SpikeSeq turned out to encompass and allow for the detection and distinction of all current and previous VOCs, many of which came to dominate after the assay design. Position K417, which is located in the middle of the forward primer and consequently outside of the SpikeSeq analysis window, is mutated in a number of lineages, such as Beta, Gamma, and all Omicron variants. It does not strongly reduce the primer binding affinity however, since it is not close to the 3′ end of the primer (Stadhouders et al., 2010). The mutation of position K417 in some VOCs is the only mutation in the primer sequences in the current and previous VOCs, highlighting the robustness of SpikeSeq. Nevertheless, if an important SARS-CoV-2 variant should emerge with significant mutations in the primer binding sites, the relentless work from the ARTIC network generates, tests, and publishes primer sets, which can be used instead of the original SpikeSeq primer set. In this way, SpikeSeq can be adapted to be used on any conceivable variant, with validated primer sets covering the entire genome of SARS-CoV-2 provided by the ARTIC network. This would be relevant for differentiation of e.g. Omicron BA.4 and Omicron BA.5, two variants where the SpikeSeq amplicon is identical.
SpikeSeq is easy to automate and we estimate that the total price of these simple steps is 4€ per sample in material cost including plastics, enzymes, buffers, shipping and sequencing. This assumes that excess RNA from a diagnostic COVID-19 qPCR facility is available as input for SpikeSeq. This makes the workflow cost efficient, even when compared to multi-target RT-qPCR typing or low cost optimized WGS methods such as CoronaHiT and ARTIC LoCost (€7.5–12 and £6.22–9.75, respectively) (Baker et al., 2021). Furthermore, the SpikeSeq price scales linearly with sample number with no minimum number of samples, as opposed to WGS approaches, where large batches are required to keep the cost down. If Sanger sequencing can be performed in-house the time to result can be less than six hours from positive COVID-19 sample to variant call, which is faster than any high throughput WGS based analysis, with less computational workload and data storage. The advantages of WGS approaches compared to SpikeSeq is the much greater resolution, where it is always possible to assign lineage to a sample. Having the complete genome sequence mean that all mutations can be identified, also any consequential mutations outside of the SpikeSeq analysis window. The cheapest WGS methods offer a relatively low cost of only double the price of SpikeSeq, if the sample volume is sufficiently great and the laboratory infrastructure and analysis infrastructure is readily available.
Globally, a number of scientists have utilized the ubiquitous Sanger Sequencing technology, either to generate complete genomes (Shaibu et al., 2021, Paden et al., 2020, Moniruzzaman et al., 2020), or, in similar efforts to this, to type SARS-CoV-2 variants. These protocols, published at a later date, are targeting the S gene, using a similar approach to SpikeSeq (Daniels et al., 2021, Bezerra et al., 2021). In one (Bezerra et al., 2021), the assay is –similiarly to SpikeSeq— based on ARTIC primers, and the amplicon is 320 nt shorter than the SpikeSeq assay amplicon and sequenced from both the forward and reverse primer. This will hypothetically mean a slightly higher assay success rate, but also mean that a handful of important positions are not included in the analysis window, including H655Y, N659K, P681R/H, A701V, T716I. Surprisingly, the K417 mutations are reported in this study, however the forward primer is placed over the position, which means that K417 cannot be reliably analyzed by the assay. The data analysis in (Bezerra et al., 2021) is based on commercial software, but could easily be adapted to be used with an open source software similarly to SpikeSeq, by simply loading the Sanger electropherogram files.
In (Daniels et al., 2021), it is suggested to use several amplicons to target the spike gene instead of a single amplicon, and while this would slightly increase the resolution of Sanger sequencing based variant identification, it would come with the price of increased complexity in both primer maintenance, laboratory work, data analysis, and cost of the analysis. The analysis software in SpikeSeq is automated, open source, secure, and easy to use for non-bioinformaticians, however neither study similar to SpikeSeq (Daniels et al., 2021, Bezerra et al., 2021) includes software for users of the methods.SpikeSeq uses identical enzymes to those used in COVID-19 diagnostic testing, simplifying the supply chain. Presumably, any RT-PCR enzyme mix will perform well with SpikeSeq. Identifying emerging VOCs requires large-scale tracking of the complete SARS-CoV-2 genome, and we do not suggest substituting the WGS efforts with SpikeSeq. SpikeSeq of all SARS-CoV-2 positive samples can however enable rapid, decentralized mutation typing and association to current variants and mutations of concern and has already proven useful for identifying emerged variants like Alpha, Beta, Gamma, Delta, and Omicron (BA.1,BA.2). The proposed amplicon will potentially continue to cover all mutations in important variants arising in the future. As RT-qPCR requires modification and testing of the assay for each new emerging variant, SpikeSeq has proven to be a more robust method, since no modifications have been needed in the assay to be able to identify all VOCs to date. However, with an established assay, variant specific qPCR will often be faster and potentially cheaper than SpikeSeq if analyzing only a few mutations per sample.
4. Conclusion
We show that typing of SARS-CoV-2 VOC specific mutations can be performed by SpikeSeq in a cheap and fast way, and we provide an automatic, open source, and upload-free mutation analysis software. Our SpikeSeq protocol is robust as it has been able to detect several VOCs arising after the assay was designed, namely Beta, Gamma, Delta, and Omicron (BA.1, BA.2, BA.4/5, BA.2.75.x), and many non-VOC variants as well. SpikeSeq has the potential to change how SARS-CoV-2 VOC mutations are typed globally, as it is the cheapest and simplest way proposed yet to variant type SARS-CoV-2 positive samples, without the need to constantly modify the assay. We achieved mutation calls from 85% of all samples, which is comparable to the reported WGS success rate. We do not suggest abandoning the use of WGS, as it is important for tracking emerging variants, but SpikeSeq have advantages over both WGS (speed, price, ease, data storage, small batch size) and qPCR (robustness, result resolution). SpikeSeq has been used routinely during the pandemic to type all positive samples at the High Throughput SARS-CoV-2 RT-qPCR testing facility at DTU and at the Copenhagen University Hospital, Rigshospitalet.
CRediT authorship contribution statement
Tue Sparholt Jørgensen: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Martin Schou Pedersen: Data curation, Formal analysis, Investigation, Resources, Validation, Writing – review & editing. Kai Blin: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – review & editing. Franziska Kuntke: Data curation, Investigation, Validation, Visualization, Writing – review & editing. Henrik K. Salling: Conceptualization, Investigation, Methodology, Resources, Writing – review & editing. Rasmus L. Marvig: Data curation, Formal analysis, Investigation, Resources, Validation, Writing – review & editing. Thomas Y Michaelsen: Investigation, Resources, Writing – review & editing. Mads Albertsen: Resources, Writing – review & editing. Helene Larsen: Conceptualization, Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Jeppe Hallgren and Jørn Emborg from Biolib for setting up the self-contained web-app and Kim Ng and Thor Bech Johannesen for coordination assistance from Statens Serum Institut. We thank Benni Winding Hansen for equipment access. We thank the NGS lab at The Novo Nordisk Center for Biosustainability for allowing parts of the work to be performed in their facilities. We thank Søren Michael Karst and Nikolai Kirkby for valuable discussions, and the many people maintaining public COVID19 resources, like covariants.org, nextstrain.org and outbreak.info.
This work was supported by the Poul Due Jensen Fonden (Grundfos Fonden, Corona-Danica). T.S.J. and K.B. received support by The Novo Nordisk Foundation through the grants NNF-IIMENA (NNF16OC0021746) and NNF CFB (NNF20CC0035580). We are grateful to researchers, clinicians, and public health authorities for making SARS-CoV-2 sequence data available through GISAID.
Statement of data use
All samples are pseudo anonymized before shipment to the DTU facility, and once more before shipment of samples to Eurofins genomics. For this study, Sanger reads were anonymized.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jviromet.2022.114648.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
Data will be made available on request.
References
- Andersen, C.Ø. et al. A major outbreak of COVID-19 at a residential care home. Dan. Med. J. 68(10):A03210227, 10 (2021). [PubMed]
- Baker D.J., et al. CoronaHiT: high-throughput sequencing of SARS-CoV-2 genomes. Genome Med. 2021;13:21. doi: 10.1186/s13073-021-00839-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezerra M.F., et al. A Sanger-based approach for scaling up screening of SARS-CoV-2 variants of interest and concern. Infect. Genet. Evol. 2021;92 doi: 10.1016/j.meegid.2021.104910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell B., Rood J., Singer E. BBMerge – accurate paired shotgun read merging via overlap. PLOS ONE. 2017;12 doi: 10.1371/journal.pone.0185056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R.S., et al. A Sanger sequencing protocol for SARS-CoV-2 S-gene. Influenza Other Respir. Virus. 2021;15:707–710. doi: 10.1111/irv.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings - SARS-CoV-2 coronavirus / nCoV-2019 Genomic Epidemiology. Virological https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586 (2021).
- Grubaugh N.D., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20:8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., et al. Nextstrain: real-time tracking of pathogen evolution. Bioinforma. Oxf. Engl. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft, E.B. CoVariants: SARS-CoV-2 Mutations and Variants of Interest. https://covariants.org/ (2021).
- Jørgensen, T.S. Sanger sequencing of a part of the SARS-CoV-2 spike protein. (2021) doi:10.17504/protocols.io.bsbdnai6.
- Julia Mullen, Ginger Tsueng, et al. outbreak.info. outbreak.info https://outbreak.info/.
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., et al. The sequence Alignment/Map format and SAMtools. Bioinforma. Oxf. Engl. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.Z., Gandhi R.T. Realizing the potential of Anti–SARS-CoV-2 monoclonal antibodies for COVID-19 management. JAMA. 2022;327:427–429. doi: 10.1001/jama.2021.19994. [DOI] [PubMed] [Google Scholar]
- Moniruzzaman M., et al. Coding-complete genome sequence of SARS-CoV-2 isolate from bangladesh by sanger sequencing. Microbiol. Resour. Announc. 2020;9 doi: 10.1128/MRA.00626-20. e00626-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole, Á. et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Preprint at https://doi.org/10.12688/wellcomeopenres.16661.1 (2021a).
- O’Toole Á., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7:veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paden C.R., et al. Rapid, sensitive, full-genome sequencing of severe acute respiratory syndrome Coronavirus 2. Emerg. Infect. Dis. 2020;26:2401–2405. doi: 10.3201/eid2610.201800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations - SARS-CoV-2 coronavirus / nCoV-2019 Genomic Epidemiology. Virological https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563 (2020).
- Quick, J. nCoV-2019 sequencing protocol v3 (LoCost). (2020).
- Rambaut A., et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch, T., Fritz, M.H.-Y., Untergasser, A. & Benes, V. Tracy: basecalling, alignment, assembly and deconvolution of sanger chromatogram trace files. BMC Genomics 21, 230 (2020). [DOI] [PMC free article] [PubMed]
- Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaibu J.O., et al. Full length genomic sanger sequencing and phylogenetic analysis of severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) in Nigeria. PLOS ONE. 2021;16 doi: 10.1371/journal.pone.0243271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess, K. et al. Rapid surveillance platforms for key SARS-CoV-2 mutations in Denmark. 2021.10.25.21265484 Preprint at https://doi.org/10.1101/2021.10.25.21265484 (2021).
- Stadhouders R., et al. The effect of primer-template mismatches on the detection and quantification of nucleic acids using the 5′ nuclease assay. J. Mol. Diagn. 2010;12:109–117. doi: 10.2353/jmoldx.2010.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally, H. et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020.12.21.20248640 (2020) doi:10.1101/2020.12.21.20248640.
- Tsueng, G. et al. Outbreak.info Research Library: A standardized, searchable platform to discover and explore COVID-19 resources. 2022.01.20.477133 Preprint at https://doi.org/10.1101/2022.01.20.477133 (2022). [DOI] [PMC free article] [PubMed]
- Vogels C., et al. Multiplexed RT-qPCR to screen for SARS-COV-2 B.1.1.7, B.1.351, and P.1 variants of concern. PLoS Biol. 2021 doi: 10.17504/protocols.io.brrhm536. [DOI] [Google Scholar]
- Vogels C.B.F., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., et al. COVID-19 in early 2021: current status and looking forward. Signal Transduct. Target. Ther. 2021;6:1–14. doi: 10.1038/s41392-021-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.