Abstract
A bovine plasminogen activator was purified from the culture supernatant of the bovine pathogen Streptococcus uberis NCTC 3858. After the final reverse-phase high-performance liquid chromatography step a single protein with a molecular mass of 32 kDa was detected in the active fraction. A partial peptide map was established, and degenerate primers were designed and used for amplification of fragments of the gene encoding the activator. Inverse PCR was subsequently used for obtaining the full-length gene. The S. uberis plasminogen activator gene (skc) encodes a protein consisting of 286 amino acids including a signal peptide of 25 amino acids. In an amino acid sequence comparison the cloned activator showed an identity of approximately 26% to the streptokinases isolated from Streptococcus equisimilis and Streptococcus pyogenes. Interestingly, the activator from S. uberis was found to lack the C-terminal domain possessed by the streptokinase from S. equisimilis. This is apparently a general feature of the streptokinases of this species; biochemical and genetic analysis of 10 additional strains of S. uberis revealed that 9 of these were highly similar to strain NCTC 3858. Sequencing of the skc gene from three of these strains indicated that the amino acid sequence of the protein is highly conserved within the species.
Plasmin is a potent serine proteinase that has an important function in physiological processes in mammals, such as degradation of extracellular matrix proteins, blood clot dissolution (fibrinolysis), cellular migration, and cancer metastasis. Plasminogen, the blood-borne zymogen of plasmin, has two physiological activators, tissue-type plasminogen activator and urokinase-type plasminogen activator. These activators are themselves serine proteinases and activate plasminogen by cleavage of a single peptide bond. However, in addition to these two physiological plasminogen activators, several pathogenic microorganisms have developed plasminogen activators which enable them to exploit host plasmin activity. The generation of plasmin activity assists the microorganism in proteolytic breakdown of fibrin and other extracellular matrix proteins, which, in turn, facilitates the bacterial penetration of normal tissue barriers and ultimately enables bacterial colonization of deep-tissue sites (reviewed in references 2 and 19). Some bacteria that produce plasminogen activators also produce plasmin(ogen) surface receptors. The binding of plasmin(ogen) to these receptors equips the bacteria with host-derived plasmin activity, and at the same time, the receptors shield the bound plasmin from physiological inhibitors (11, 12). Bacterial plasminogen activators include the streptokinase produced by a variety of pathogenic Streptococcus species and the staphylokinase produced by Staphylococcus aureus. Due to its fibrinolytic potential, streptokinase is currently used as a thrombolytic therapy drug. Streptokinase and staphylokinase have unique, but slightly different, mechanisms of plasminogen activation. Streptokinase and staphylokinase form 1:1 stoichiometric plasminogen activator complexes with plasminogen and plasmin, respectively. Streptokinase induces a conformation of the serine proteinase domain of plasminogen, which exposes the active site of the proteinase without prior proteolytic cleavage, thereby providing the streptokinase-plasminogen complex with what has been called “virgin” enzyme activity (23). In contrast, the staphylokinase-plasminogen complex is proteolytically inactive but can be transformed into the active staphylokinase-plasmin complex by activation with plasmin (4). Notably, streptokinases isolated from different strains of streptococci possess an intrinsic species specificity for their target plasminogen molecules that parallels the host range of the microorganisms (21).
Two novel plasminogen activators have recently been described. They were derived from the bovine mastitis-inducing pathogens Streptococcus uberis (14) and Streptococcus dysgalactiae (17) and showed specificity to bovine plasminogen. Mastitis is an inflammatory disease of the mammary gland. In the United Kingdom, S. uberis is responsible for around 20% of all clinical cases of bovine mastitis (3), and in Denmark, 23% of the mastitis cases in organic dairy herds could be connected with infection by S. uberis (26). Leigh (13) showed that the activity associated with the plasminogen activator secreted from S. uberis was different from that of Streptococcus pyogenes (Lancefield group A) and Streptococcus equisimilis (Lancefield group C) strains, as it activated bovine but not human plasminogen. It also differed from Lancefield group E activity by not activating porcine plasminogen (13). By activation of plasminogen to plasmin through the action of its plasminogen activator, S. uberis was also shown to be able to acquire surface-localized plasmin activity (16), and plasmin binding to the bacterial surface was susceptible to increasing concentrations of NaCl and lysine (18). For a mastitis-inducing pathogen, the production of a plasminogen activator could be of importance in two ways. In addition to generation of plasmin activity needed for degradation of extracellular matrix proteins and subsequent colonization, the activation of endogenous plasminogen present in milk would lead to hydrolysis of milk proteins and, thereby, liberation of peptides from which S. uberis could obtain essential amino acids (10).
In this study we have performed purification and partial amino acid sequencing of the plasminogen activator from S. uberis and have cloned and sequenced its gene. By sequence comparison, the plasminogen activator was shown to be related to the already-known streptokinases.
MATERIALS AND METHODS
Chemicals and reagents.
Super Taq polymerase was from HT Biotechnology (Cambridge, United Kingdom), Ready to Go PCR beads were from Pharmacia (Uppsala, Sweden), oligonucleotides were from DNA Technology (Aarhus, Denmark), and all other enzymes were from New England Biolabs (Hitchin, United Kingdom). PCR was performed in a Hybaid (Middlesex, United Kingdom) ABACUS thermal cycler. Sequencing was performed with a dye terminator cycle sequencing kit from PE Applied Biosystems (Foster City, Calif.), and ProBlott membranes were from the same supplier. The Wizard DNA purification kit was from Promega (Madison, Wis.). Sequencing, ligation, transformation of Escherichia coli, DNA preparation, PCR, and other DNA-modifying processes were performed according to the manufacturers’ recommendations or standard laboratory procedures, unless otherwise indicated. For cloning of PCR products, a TOPO cloning kit from Invitrogen (Carlsbad, Calif.) was used. Prestained molecular mass marker proteins (Seeblue) and 10 to 20% Tris-glycine-polyacrylamide gels were from NOVEX (San Diego, Calif.). All protein purification columns were from Pharmacia. Modified trypsin was from Promega, S-2251 [(H-d-Val)-Leu-Lys-pNA] was from Chromogenix (Mölndal, Sweden), bovine [Asp1]plasminogen was from American Diagnostica (Greenwich, Conn.), plasminogen-depleted bovine fibrinogen was from Enzyme Research Laboratories (South Bend, Ind.), and human thrombin was from Sigma (St. Louis, Mo.). Alkaline phosphatase-conjugated swine anti-rabbit immunoglobulin G was from Dako (Glostrup, Denmark), and Nitro Blue Tetrazolium and 5-bromo-4-chloro-3-indolylphosphate p-toluidine salt were from Sigma.
Identification, growth, and fingerprinting of S. uberis strains.
Strain NCTC 3858 was obtained from the National Collection of Type Cultures (Colindale, UK). Strains 120-295-1 (=SK880), 137-391-1 (=SK881), 149-451-2 (=SK882), 156-162-1 (=SK883), and 159-684-1 (=SK884), isolated from different herds in Denmark, and strains 5793-LR (9057-7) (=SK885), 9758-34-RR (=SK886), 27-RR (=SK887), 9057-14-LR (=SK888), and 9756-296-LF (=SK889), isolated from different herds in the United States in 1994, were kindly provided by F. Aarestrup, Danish Veterinary Laboratory, Copenhagen, Denmark. All strains were grown overnight in Todd-Hewitt broth (Difco Laboratories, Detroit, Mich.). The identities of the strains were verified by standard techniques, including demonstration of β-glucuronidase and alkaline phosphatase activities. All S. uberis strains used in this study were shown to represent distinct clones as shown by digestion of genomic DNA by MspI and fragment analysis by agarose gel electrophoresis (data not shown).
Plasminogen activator activity assay.
Plasminogen activator activity was identified by the ability of the activator to activate bovine plasminogen to plasmin. The formation of plasmin was measured by its hydrolysis of the chromogenic peptidyl anilide substrate S-2251. The reaction was performed in a total of 0.2 ml containing 0.1 M Tris-HCl (pH 7.4), 0.02% Tween 80, 0.05 μM [Asp1]plasminogen, and 0.5 mM S-2251; the reaction was initiated by addition of the sample dissolved in the various elution buffers used during purification. The reaction was monitored at 405 nm over a period of 1 h in a Bio-Tek EL 311 BioKinetics Reader (Bio-Tek Instruments, Winooski, Vt.).
Purification of the plasminogen activator.
Twenty liters of bacterial culture was centrifuged at 3,000 × g at 4°C until the supernatant could be collected. The supernatant was then adjusted to 38% saturation with (NH4)2SO4 and to 0.05 (wt/vol) saturation with NaN3. The solution was stirred overnight at 4°C and centrifuged at 3,000 × g for 30 min. The precipitate was dissolved in 150 ml of H2O, dialyzed against 50 mM NH4HCO3 (pH 8.9), and frozen at −20°C. The sample was then applied to a 50-ml DEAE-Sepharose column and eluted with a gradient (0.05 to 1.0 M) of NH4HCO3 (pH 8.9). Active fractions were pooled, dialyzed against 20 mM NH4HCO3 (pH 8.0), and freeze-dried. The sample was then dissolved in 50 mM CHOOH–6 M urea (pH 4.0), applied to a Mono-S HR 10/10 HPLC column, and eluted with a 0 to 1 M NaCl gradient in the same buffer. Active fractions were dialyzed against 20 mM NH4HCO3 (pH 8.0) and lyophilized. The sample was then redissolved in 0.1% trifluoroacetic acid, applied to a reverse-phase Sephasil C8 SC 2.1/10 column, and eluted with a gradient of 0 to 80% isopropanol in 0.1% trifluoroacetic acid.
Generation of peptides and amino acid sequence analysis.
Material eluted from the Mono-S column was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted onto ProBlott membranes. The band corresponding to the plasminogen activator was excised from several lanes and processed essentially as described previously (5). The generated tryptic peptides were applied to a reverse-phase μRPC C2/C18 SC 2.1/10 SMART column and eluted with a gradient of 0 to 60% CH3CN in 0.1% trifluoroacetic acid on a SMART high-performance liquid chromatography (HPLC) system. The resolved peptide peaks were subjected to automated Edman degradation on an ABI 477A/120A protein sequencer (Applied Biosystems) by using standard programs.
Cloning of the streptokinase gene by PCR with degenerate primers and by inverse PCR.
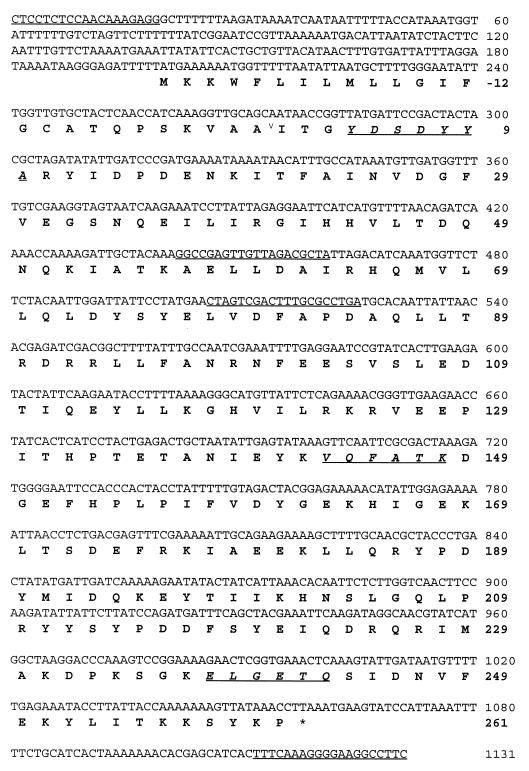
Genomic S. uberis DNA was isolated as described previously (9). Several degenerate oligonucleotides corresponding to the obtained partial amino acid sequences were synthesized. The oligonucleotides were each labeled with [γ-32P]ATP and used as probes in Southern blotting of genomic S. uberis DNA digested with AflIII (data not shown). Each oligonucleotide hybridized with a single and the same DNA fragment. The three strongest-hybridizing oligonucleotides were selected as primers (Table 1) in PCRs with 100 ng of genomic DNA, 200 μM deoxynucleoside triphosphate, 4 μM primer, 1× PCR buffer, and 2.5 U of Super Taq in a total volume of 50 μl and the following cycling parameters (35 cycles): 94°C for 60 s, 50°C for 60 s, and 72°C for 60 s, with an initial denaturation step of 300 s in the first cycle. For use in inverse PCR, genomic DNA was digested with BglII and ligated overnight at 14°C at a concentration of 10 ng/μl in PCR buffer supplemented with 67 μM ATP. The ligated DNA mixture (50 ng) containing circularized BglII fragments was then used directly as a template in the inverse PCR with the primers spanning nucleotides 506 to 525 (forward) and 461 to 442 (reverse) (see Fig. 3) and the following cycling parameters (35 cycles): 94°C for 60 s, 55°C for 60 s, and 72°C for 300 s, with an initial denaturation step of 300 s in the first cycle. Amplification of the total gene was performed by PCR with primers spanning nucleotides 1 to 19 (forward) and 1131 to 1112 (reverse) (see Fig. 3) and with 50 ng of genomic DNA, 200 μM deoxynucleoside triphosphate, 0.4 μM primer, 1× PCR buffer, and 2.5 U of Super Taq and the following cycling parameters (20 cycles): 94°C for 60 s, 55°C for 60 s, and 72°C for 60 s, with an initial denaturation step at 94°C for 300 s. All PCR products were cloned into the pCR2.1-TOPO cloning vector and sequenced with vector-specific and custom-designed primers.
TABLE 1.
Amino acid sequences of tryptic peptides and derived degenerate oligonucleotide sequences
| Peak | Amino acida sequence | Degenerate oligonucleotideb | Degen-eracy |
|---|---|---|---|
| 1c | Y11IDPDENK18 | GAYCCNGAYGARAAYAA | 64 |
| V143QFATK148 | TTNGTNGCRAAYTGNAC (reverse) | 256 | |
| 2 | G51IHHVLTDQNQK62 | ||
| 3 | I1TGYDSDYYAR11 | ||
| 4 | A57ELLDAIR64 | ||
| 5 | E238LGETQSI245 | TGNGTYTCNCCIARYTC (reverse) | 128 |
| 6 | D149GEFHPLPIF158 | ||
| N terminus | I1TGYDSDYYARYIDPD16 | TAYGAYWSIGAYTAYTAYGC (forward) | 128 |
Underlining indicates amino acids used for design of degenerate oligonucleotides.
Ambiguous bases are abbreviated as follows: N stands for A, C, T, or G; R stands for A or G; Y stands for C or T; I stands for deoxyinosine; W stands for A or T; and S stands for C or G.
The double sequence present in peak 1 could be interpreted since the sequence YIPDENK was recognized as derived from the N-terminal sequence.
FIG. 3.
Nucleotide sequence and deduced amino acid sequence (in boldface) of the streptokinase gene from S. uberis. Primers used in cloning of the gene are underlined, and peptide sequences used for design of degenerate primers are shown in italics and underlined. ∗, stop codon; ∨, signal peptide cleavage site.
Western blotting and zymography analysis of streptokinases produced by heterologous S. uberis strains.
Bacterial culture supernatants were mixed 1:1 with sample buffer (20 mM Tris [pH 6.8], 2% SDS, 20% glycerol) and heated at 95°C, and samples were subjected to SDS-PAGE on 10 to 20% Tris-glycine NOVEX gels with a standard running buffer (25 mM Tris [pH 8.3], 0.2 M glycine, 0.1% SDS). For Western blotting, the gel was blotted onto a ProBlott membrane in a transfer buffer consisting of 10 mM 3-(cyclohexylamino)-1-propanosulfonic acid (CAPS) (pH 11.0), 10% (vol/vol) methanol, and 0.05% SDS. The membrane was blocked with 2% Tween 20–0.5 M NaCl–0.05 M Tris (pH 7.4), and all subsequent washing and incubation steps were performed in 0.1% Tween 20–0.5 M NaCl–0.05 M Tris (pH 7.4). Polyclonal antibodies against the S. uberis plasminogen activator were raised in rabbits by immunization with S. uberis plasminogen activator, produced in recombinant E. coli (unpublished results), and the resulting serum was used without purification. Alkaline phosphatase-conjugated swine anti-rabbit immunoglobulin G antibodies were used as secondary antibodies. The zymography was essentially performed as described previously (1), using SDS-PAGE conditions as for Western blotting and agarose gels containing 1.7 mg of bovine fibrinogen per ml and 8 μg of bovine plasminogen per ml, and fibrin polymerization was initiated by addition of 0.03 U of human thrombin per ml.
PCR and sequence analysis of skc genes in heterologous S. uberis strains.
The 3′ region of the skc gene was amplified with primers (0.4 μM) spanning nucleotides 506 to 525 (forward) and 1131 to 1112 (reverse), 1 ng of genomic DNA, Ready to Go PCR beads, and the following cycling parameters (30 cycles): 94°C for 60 s, 52°C for 60 s, and 72°C for 90 s, with an initial denaturation step at 94°C for 300 s (nucleotide numbering refers to Fig. 3). The full-length skc gene was amplified with primers spanning nucleotides 1 to 19 (forward) and 1131 to 1112 (reverse) and otherwise the same conditions as described above. PCR fragments were analyzed by agarose gel electrophoresis. The PCR fragments containing the full-length skc gene were purified of PCR components by using a Wizard DNA purification column and sequenced on both strands with appropriate primers.
Nucleotide sequence accession numbers.
The sequences for the skc genes from strains SK882, SK884, and SK889 have been deposited in the EMBL nucleotide sequence database under accession no. AJ131604, AJ131605, and AJ131631, respectively.
RESULTS AND DISCUSSION
Purification, generation of peptides, and amino acid sequence analysis.
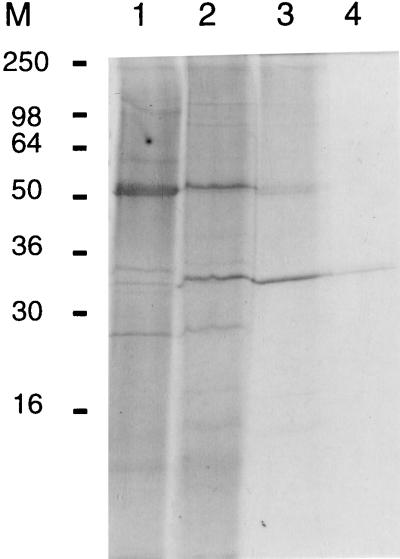
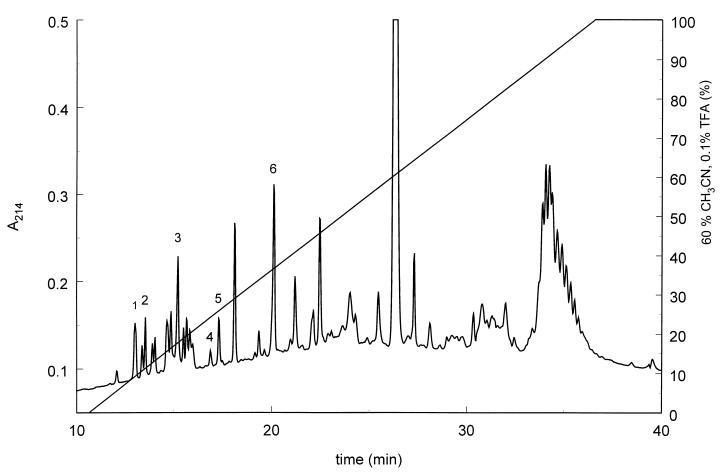
The bovine plasminogen activator from S. uberis NCTC 3858 was purified from the culture supernatant by a combination of ammonium sulfate precipitation, DEAE–ion-exchange chromatography, denaturing Mono-S HPLC, and reverse-phase HPLC. Interestingly, the plasminogen activator appeared to be a very stable protein, as demonstrated by the fact that activity survived treatment with strong denaturing agents, such as 6 M urea, 8 M guanidinium hydrochloride, and 60% formic acid (data not shown), or passage over reverse-phase columns. The activator was identified as a 32-kDa protein, since only this band was present in the active fraction eluted from the C8 column (Fig. 1). The N-terminal sequence derived from this band (Table 1) did not show any similarities to known sequences as revealed by BLAST homology searching. The recovery of protein from the reverse-phase C8 column was insufficient for generation of suitable amounts of peptides for use in amino acid sequencing, and therefore, tryptic degradation on the blot was carried out on material from the Mono-S column. These tryptic peptides were separated on a C2/C18 reverse-phase HPLC column (Fig. 2), and N-terminal sequence analysis was performed (Table 1).
FIG. 1.
Reducing SDS-PAGE of material from different steps during purification of the plasminogen activator from S. uberis. Lanes: 1, ammonium sulfate precipitate; 2, pooled active fractions from DEAE–ion-exchange chromatography; 3, pooled active fractions from Mono-S HPLC; 4, the most active fraction from C8 Sephasil reverse-phase HPLC. M, molecular mass markers; molecular masses are in kilodaltons.
FIG. 2.
Tryptic peptides derived from degradation on the blot were separated by HPLC with a narrow-bore C2/C18 reverse-phase column. The sequences of labeled peaks are shown in Table 1. TFA, trifluoroacetic acid.
Cloning of the gene for the plasminogen activator.
The identified amino acid sequences were used for design of degenerate oligonucleotides (Table 1), and these were subsequently used as primers for PCR on genomic DNA of S. uberis NCTC 3858, in order to isolate the gene encoding the plasminogen activator. The degenerate primer pair corresponding to the amino acid sequences YDSDYYA (forward) and VQFATK (reverse) yielded a single band of ∼400 bp, and the primer pair corresponding to YDSDYYA (forward) and ELGETQ (reverse) yielded a single band of ∼700 bp. By Southern blotting experiments, the degenerate oligonucleotide corresponding to the sequence DYYARY was shown to hybridize with these amplicons, indicating that the PCR products were amplified from the plasminogen activator gene. The two PCR products were then cloned into the pCR2.1-TOPO vector and sequenced. The resulting sequences of the two fragments overlapped and comprised the codons for all the sequenced peptides, and they were subsequently used for design of primers for use in inverse PCR. By using primers spanning nucleotides 461 to 442 (reverse) and 506 to 525 (forward) (Fig. 3) and the BglII-cut and religated genomic DNA as a template in a PCR, an appropriate DNA fragment of approximately 5 kbp was amplified. This fragment was subsequently cloned into plasmid pCR2.1-TOPO and partially sequenced from both ends. An open reading frame (ORF) encoding a streptokinase-like protein could be deduced from the combined sequence information for the PCR fragments amplified with degenerate primers and the fragment obtained by inverse PCR. Finally, the primers spanning nucleotides 1 to 19 (reverse) and 1131 to 1112 (reverse) (Fig. 3) were used in a PCR amplifying the full-length streptokinase gene on one DNA fragment. This PCR product was then cloned into the pCR2.1-TOPO cloning vector, and three independent clones were sequenced on both strands to control for PCR-introduced mutations (Fig. 3). The cloned streptokinase gene contains an ORF with the potential of encoding a protein of 286 amino acids. The ORF is preceded by the sequence GGAGA, which may function as a ribosome binding site (24, 25). The N-terminal amino acid sequence obtained for the secreted streptokinase was identified 25 amino acids downstream from the sequence corresponding to the ATG start codon in the ORF, indicating that Ala-Ile in positions 25 and 26 is the cleavage site for the signal peptidase. In support of this, the 25 N-terminal amino acids encoded by the ORF display features typical of a signal peptide (27). The deduced mature protein thus comprises 261 amino acids with a calculated molecular mass of 30.7 kDa, in agreement with the 32 kDa estimated by SDS-PAGE.
Diversity of the streptokinases among S. uberis strains.
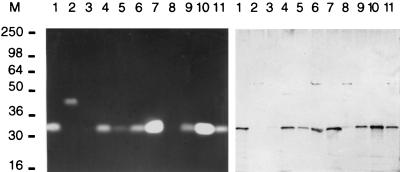
To investigate whether the purified and cloned streptokinase from S. uberis NCTC 3858 was representative of streptokinases produced by other strains of the same species, the properties of streptokinases from 10 additional S. uberis strains were investigated by Western blotting, zymography, and PCR and those for 3 of the strains were investigated by gene sequencing. Nine of the 10 strains were shown by Western blotting and zymography to produce streptokinases of similar molecular masses, although for 2 of the strains, SK881 and SK886, the amounts of streptokinase produced were insufficient to allow reproduction of lysis zones in the zymography (Fig. 4). In agreement with this result, these strains gave rise to only very faint bands in Western blotting analysis (Fig. 4). The relative intensities of the Western bands corresponded roughly to the sizes of lysis zones in the zymography and were also in accordance with activities measured by the plasminogen activation assay (data not shown). These data probably reflect different expression levels of the streptokinases among the investigated strains under the conditions used rather than different reactivities to the polyclonal antibodies or differences in catalytic strength in the plasminogen activator complexes. Further work with standardized growth of the bacteria will be needed to clarify this point.
FIG. 4.
Zymography (left) and Western blotting analysis (right). Supernatants (4 μl) from each strain were mixed with sample buffer and loaded in lanes as follows: 1, NCTC 3858; 2, SK880; 3, SK881; 4, SK882; 5, SK883; 6, SK884; 7, SK885; 8, SK886; 9, SK887; 10, SK888; and 11, SK889. M, molecular mass markers; molecular masses are in kilodaltons.
Interestingly, one strain produced a streptokinase of a higher molecular mass (∼45 kDa) (Fig. 4). The higher molecular mass of this streptokinase corresponds to the molecular masses of streptokinases from several other species and could indicate the presence of an extra domain (see below). Presumably, this streptokinase has a markedly different primary structure since no antibody cross-reactivity could be detected by Western blotting.
To evaluate the gene structures and identities of the different streptokinases, amplification of the 3′ region nucleotides (506 to 1131 [Fig. 3]) of the skc gene was attempted. All 10 strains except strain SK880 yielded a band of ∼600 bp, indicating the presence of an skc gene with a 3′ region similar to that of the skc gene of strain NCTC 3858. Attempts to amplify the full-length skc gene (nucleotides 1 to 1131) resulted in appropriate PCR fragments from all strains except strain SK880 and SK883. The genes from strains SK882, SK884, and SK889 were subsequently sequenced. These three skc genes all have the potential of encoding proteins of 286 amino acids with a very high degree of identity to that of strain NCTC 3858. The amino acid substitutions found in these strains are summarized as follows (numbering refers to strain NCTC 3858): strains SK882 and SK884 (identical) have the substitutions Val79, Gln115, Gln124, Leu211, Gln240, and His242; and strain SK889 has the substitutions Val79 and Ser211. The identities of the skc genes of strains SK882 and SK884 and their divergence from that of strain SK889 are in agreement with their geographical origin, as strains SK882 and SK884 were isolated in Denmark and SK889 was isolated in the United States.
In summary, a single strain, SK880, produced a plasminogen activator which could not be recognized by heterologous antibodies, and its gene was apparently too diverse to allow amplification by the primers used. Nine of the 10 investigated strains contained skc genes with similar structures and produced a streptokinase with properties similar to those of strain NCTC 3858. Based on these data, and the sequence analysis of three of the strains, the skc gene seems to be highly conserved in its species. The skc gene of strain NCTC 3858 was used for comparison with other species.
Comparison of the streptokinasesS. uberis sequence to the sequences and domain boundaries of other streptokinases.
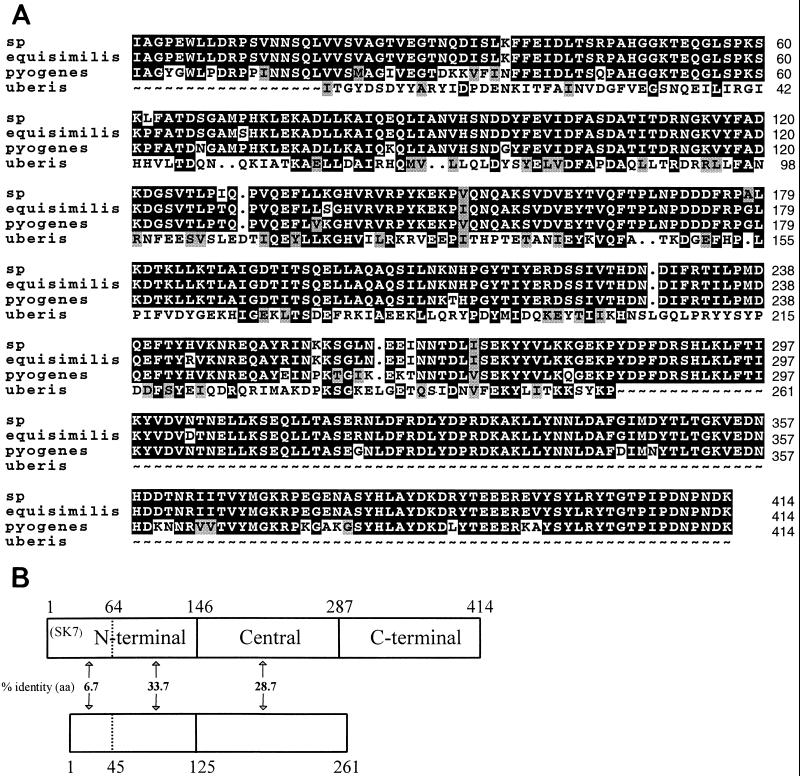
Alignment of the deduced amino acid sequence of the streptokinase from S. uberis NCTC 3858 (streptokinaseS. uberis) to the sequences of three other streptokinases, streptokinaseStreptococcus (isolated from a group G streptococcus [28]), streptokinaseS. equisimilis (20), and streptokinaseS. pyogenes (29), showed identities of 26.4, 26.0, and 25.7%, respectively; the plasminogen activator from S. uberis is thus related to the other known streptokinases (Fig. 5A). However, among the currently known streptokinases, streptokinaseS. uberis seems to be the least conserved, since the degrees of identity for six other streptokinases from serological groups A, C, and G range between 80 and 98% (8). In line with this observation, no homology to streptokinaseS. uberis could be found at the nucleotide level by BLAST searching. The fact that streptococci with different host specificities produce streptokinases that show considerable sequence diversity but conserved plasminogen activation potential indicates that generation of plasmin activity is important for the pathogenesis of these bacteria. On the other hand, the high degree of amino acid sequence diversity also indicates that only a low degree of sequence constraint is needed for the ability of the streptokinase to activate plasminogen.
FIG. 5.
(A) Multiple alignment of deduced amino acid sequences of secreted streptokinases from a group G streptococcus (sp; accession no. P10519), S. equisimilis (equisimilis; accession no. P00779), S. pyogenes (pyogenes; accession no. P96471), and S. uberis NCTC 3858 (uberis; accession no. AJ006413). Identical amino acids are indicated by black boxes, and similar amino acids are indicated by shaded boxes. (B) Schematic presentation of alignment of streptokinaseS. uberis to streptokinaseS. equisimilis. The domain assignment of streptokinaseS. equisimilis is according to reference 22. The N-terminal region (residues 1 to 64) of streptokinaseS. equisimilis is quickly removed by proteolysis (22). The degree of identity between corresponding regions is indicated, as are amino acid positions at domain boundaries. aa, amino acids.
StreptokinaseS. equisimilis was previously suggested to comprise three domains (22), and this has recently been confirmed by the elucidation of the crystal structure of the streptokinaseS. equisimilis-microplasmin plasminogen activator complex (30). Investigation of the multiple-sequence alignment (Fig. 5A) in relation to the proposed domain boundaries of streptokinaseS. equisimilis (summarized in Fig. 5B) reveals several interesting features. StreptokinaseS. equisimilis consists of 414 amino acids (∼47 kDa) and thus comprises 153 amino acids more than streptokinaseS. uberis, which is mainly due to the fact that streptokinaseS. uberis was found to lack the C-terminal domain. Besides, the first half (residues 1 to 64) of the N-terminal domain in streptokinaseS. equisimilis has no similarity (6.7% identity) to the corresponding region (residues 1 to 45) of streptokinaseS. uberis, which, in addition, is 18 amino acids shorter. In contrast, the second half of the N-terminal domain of streptokinaseS. equisimilis has a relatively high degree of identity (33.7%) to the corresponding region in streptokinaseS. uberis. The central domain of streptokinaseS. equisimilis has an identity of 28.7% to the corresponding region of streptokinaseS. uberis. In summary, the major differences between streptokinaseS. uberis and streptokinaseS. equisimilis are apparently the deletion of the C-terminal domain and the lack of identity in the N-terminal part (residues 1 to 64). This could indicate that the intrinsic species specificity determinants of the streptokinases mainly are located in the same regions. Future experiments employing domain swapping between streptokinaseS. equisimilis or streptokinaseS. pyogenes and streptokinaseS. uberis might evaluate the species specificity of separate domains of the streptokinases.
Recent work has indicated that the plasminogen molecule part of the plasminogen activator complex streptokinaseS. equisimilis-plasminogen binds to the C-terminal domain of streptokinaseS. equisimilis, while the substrate plasminogen molecule binds to the N-terminal domain (32). Other studies have shown that the minimal streptokinase sequence requirement for generation of amidolytic activity is fragments containing sequence from both the central and the C-terminal domains (31). The present study, using a streptokinase lacking the C-terminal domain while retaining catalytic activity, thus demonstrates that the function of the C-terminal domain can be compensated for by mutations in the N-terminal and/or C-terminal domain. In theory, the deletion of the C-terminal domain of streptokinaseS. uberis could also suggest a novel mechanism of plasminogen activation mediated by streptokinaseS. uberis, e.g., through dimerization (14).
Potential of streptokinaseS. uberis as a vaccine agent.
Vaccination of cows against infection of S. uberis has been investigated by injection of live S. uberis (strain 0140J), but immunization proved to be efficient only against the homologous strain (6, 7). Data indicated that the key virulence determinants were not present on the surfaces of the bacteria, and it was suggested that factors produced and secreted by the bacteria in vivo could fulfill this role. In agreement with these data, promising results have been obtained, with a small number of cows, by immunization with a partially purified preparation of the S. uberis plasminogen activator and subsequent experimental challenge with a heterologous strain (15). The isolation and cloning of streptokinaseS. uberis will allow specific evaluation of its potential as a vaccine agent. The highly conserved amino acid sequences found among the vast majority of the investigated strains, as well as the observed antibody cross-reactivity, suggest that a possible vaccine would be efficacious also against most of the heterologous strains. An exciting task for the future will be to analyze at the molecular level additional plasminogen activators important for bacterial pathogenesis, e.g., from strain SK880, and to test their efficacies as vaccine agents. Also, the potential of streptokinaseS. uberis production in Lactococcus lactis (unpublished results) makes it attractive to investigate whether lactic acid bacteria could be used as vaccine carriers by mediating antigen presentation to the mucosal immune system.
ACKNOWLEDGMENTS
This work is part of the FØTEK program supported by the Danish Dairy Research Foundation (Danish Dairy Board), the Danish government, and the Danish Medical Research Council.
The kind gift of S. uberis strains from Frank Aarestrup is highly appreciated.
REFERENCES
- 1.Andreasen P A, Nielsen L S, Kristensen P, Grondahl-Hansen J, Skriver L, Danø K. Plasminogen activator inhibitor from human fibrosarcoma cells binds urokinase-type plasminogen activator, but not its proenzyme. J Biol Chem. 1986;261:7644–7651. [PubMed] [Google Scholar]
- 2.Boyle M D, Lottenberg R. Plasminogen activation by invasive human pathogens. Thromb Haemostasis. 1997;77:1–10. [PubMed] [Google Scholar]
- 3.Bramley A J, Dodd F H. Reviews of the progress of dairy science: mastitis control—progress and prospects. J Dairy Res. 1984;51:481–512. doi: 10.1017/s0022029900023797. [DOI] [PubMed] [Google Scholar]
- 4.Collen D, Schlott B, Engelborghs Y, Van Hoef B, Hartmann M, Lijnen H R, Behnke D. On the mechanism of the activation of human plasminogen by recombinant staphylokinase. J Biol Chem. 1993;268:8284–8289. [PubMed] [Google Scholar]
- 5.Fernandez J, Andrews L, Mische S M. An improved procedure for enzymatic digestion of polyvinylidene difluoride-bound proteins for internal sequence analysis. Anal Biochem. 1994;218:112–117. doi: 10.1006/abio.1994.1148. [DOI] [PubMed] [Google Scholar]
- 6.Finch J M, Hill A W, Field T R, Leigh J A. Local vaccination with killed Streptococcus uberis protects the bovine mammary gland against experimental intramammary challenge with the homologous strain. Infect Immun. 1994;62:3599–3603. doi: 10.1128/iai.62.9.3599-3603.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finch J M, Winter A, Walton A W, Leigh J A. Further studies on the efficacy of a live vaccine against mastitis caused by Streptococcus uberis. Vaccine. 1997;15:1138–1143. doi: 10.1016/s0264-410x(96)00307-6. [DOI] [PubMed] [Google Scholar]
- 8.Frank C, Steiner K, Malke H. Conservation of the organization of the streptokinase gene region among pathogenic streptococci. Med Microbiol Immunol. 1995;184:139–146. doi: 10.1007/BF00224351. [DOI] [PubMed] [Google Scholar]
- 9.Hohwy J, Kilian M. Clonal diversity of the Streptococcus mitis biovar 1 population in the human oral cavity and pharynx. Oral Microbiol Immunol. 1995;10:19–25. doi: 10.1111/j.1399-302x.1995.tb00113.x. [DOI] [PubMed] [Google Scholar]
- 10.Kitt A J, Leigh J A. The auxotrophic nature of Streptococcus uberis. The acquisition of essential acids from plasmin derived casein peptides. Adv Exp Med Biol. 1997;418:647–650. [PubMed] [Google Scholar]
- 11.Kuusela P, Ullberg M, Saksela O, Kronvall G. Tissue-type plasminogen activator-mediated activation of plasminogen on the surface of group A, C, and G streptococci. Infect Immun. 1992;60:196–201. doi: 10.1128/iai.60.1.196-201.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuusela P, Saksela O. Binding and activation of plasminogen at the surface of Staphylococcus aureus. Increase in affinity after conversion to the Lys form of the ligand. Eur J Biochem. 1990;193:759–765. doi: 10.1111/j.1432-1033.1990.tb19397.x. [DOI] [PubMed] [Google Scholar]
- 13.Leigh J A. Activation of bovine plasminogen by Streptococcus uberis. FEMS Microbiol Lett. 1993;114:67–72. doi: 10.1111/j.1574-6968.1993.tb06552.x. [DOI] [PubMed] [Google Scholar]
- 14.Leigh J A. Purification of a plasminogen activator from Streptococcus uberis. FEMS Microbiol Lett. 1994;118:153–158. doi: 10.1111/j.1574-6968.1994.tb06818.x. [DOI] [PubMed] [Google Scholar]
- 15.Leigh J A. Udder health management for environmental streptococci. Madison, Wis: National Mastitis Council; 1997. Progress in the development of vaccines against environmental streptococcal mastitis; pp. 59–74. [Google Scholar]
- 16.Leigh J A, Lincoln R A. Streptococcus uberis acquires plasmin activity following growth in the presence of bovine plasminogen through the action of its specific plasminogen activator. FEMS Microbiol Lett. 1997;154:123–129. doi: 10.1111/j.1574-6968.1997.tb12633.x. [DOI] [PubMed] [Google Scholar]
- 17.Leigh J A, Hodgkinson S M, Lincoln R A. The interaction of Streptococcus dysgalactiae with plasmin and plasminogen. Vet Microbiol. 1998;61:121–135. doi: 10.1016/s0378-1135(98)00179-5. [DOI] [PubMed] [Google Scholar]
- 18.Lincoln R A, Leigh J A. Characterization of the interaction of bovine plasmin with Streptococcus uberis. J Appl Microbiol. 1998;84:1104–1110. doi: 10.1046/j.1365-2672.1998.00452.x. [DOI] [PubMed] [Google Scholar]
- 19.Lottenberg R, Minning-Wenz D, Boyle M D P. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 1994;2:20–24. doi: 10.1016/0966-842x(94)90340-9. [DOI] [PubMed] [Google Scholar]
- 20.Malke H, Roe B, Ferretti J J. Nucleotide sequence of the streptokinase gene from Streptococcus equisimilis H46A. Gene. 1985;34:357–362. doi: 10.1016/0378-1119(85)90145-3. [DOI] [PubMed] [Google Scholar]
- 21.McCoy H E, Broder C C, Lottenberg R. Streptokinases produced by pathogenic group C streptococci demonstrate species-specific plasminogen activation. J Infect Dis. 1991;164:515–521. doi: 10.1093/infdis/164.3.515. [DOI] [PubMed] [Google Scholar]
- 22.Parrado J, Conejero-Lara F, Smith R A G, Marshall J M, Ponting C P, Dobson C M. The domain organization of streptokinase: nuclear magnetic resonance, circular dichroism, and functional characterization of proteolytic fragments. Protein Sci. 1996;5:693–704. doi: 10.1002/pro.5560050414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy K N N, Markus G. Mechanism of activation of human plasminogen by streptokinase. Presence of active center in streptokinase-plasminogen complex. J Biol Chem. 1972;247:1683–1691. [PubMed] [Google Scholar]
- 24.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stormo G D, Schneider T D, Gold L M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982;10:2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaarst M, Enevoldsen C. Patterns of clinical mastitis manifestations in Danish organic dairy herds. J Dairy Res. 1997;64:23–37. doi: 10.1017/s002202999600194x. [DOI] [PubMed] [Google Scholar]
- 27.Von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walter F, Siegel M, Malke H. Nucleotide sequence of the streptokinase gene from a group-G streptococcus. Nucleic Acids Res. 1989;17:1262. doi: 10.1093/nar/17.3.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter F, Siegel M, Malke H. Nucleotide sequence of the streptokinase gene from a Streptococcus pyogenes type 1 strain. Nucleic Acids Res. 1989;17:1261. doi: 10.1093/nar/17.3.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Lin X, Loy J A, Tang J, Zhang X C. Crystal structure of the catalytic domain of human plasmin complexed with streptokinase. Science. 1998;281:1662–1665. doi: 10.1126/science.281.5383.1662. [DOI] [PubMed] [Google Scholar]
- 31.Young K C, Shi G Y, Chang Y F, Chang B I, Chang L C, Lai M D, Chuang W J, Wu H L. Interaction of streptokinase and plasminogen. Studied with truncated streptokinase peptides. J Biol Chem. 1995;270:29601–29606. doi: 10.1074/jbc.270.49.29601. [DOI] [PubMed] [Google Scholar]
- 32.Young K-C, Shi G-Y, Wu D-H, Chang L-C, Chang B-I, Ou C-P, Wu H-L. Plasminogen activation by streptokinase via a unique mechanism. J Biol Chem. 1998;273:3110–3116. doi: 10.1074/jbc.273.5.3110. [DOI] [PubMed] [Google Scholar]