Abstract
Inducing tRNA +1 frameshifting to read a quadruplet codon has the potential to incorporate a non-canonical amino acid (ncAA) into the polypeptide chain. While this strategy is attractive for genome expansion in biotechnology and bioengineering endeavors, improving the yield is hampered by a lack of understanding of where the shift can occur in an elongation cycle of protein synthesis. Lacking a clear answer to this question, current efforts have focused on designing +1-frameshifting tRNAs with an extra nucleotide inserted to the anticodon loop for pairing with a quadruplet codon in the aminoacyl-tRNA binding (A) site of the ribosome. However, the designed and evolved +1-frameshifting tRNAs vary broadly in achieving successful genome expansion. Here we summarize recent work on +1-frameshifting tRNAs. We suggest that, rather than engineering the quadruplet anticodon-codon pairing scheme at the ribosome A site, efforts should be made to engineer the pairing scheme at steps after the A site, including the step of the subsequent translocation and the step that stabilizes the pairing scheme in the +1-frame in the peptidyl-tRNA binding (P) site.
Keywords: SufB2 tRNA, ProM tRNA, ProL tRNA, m1G37-tRNA, mcmo5U34-tRNA
Graphical Abstract
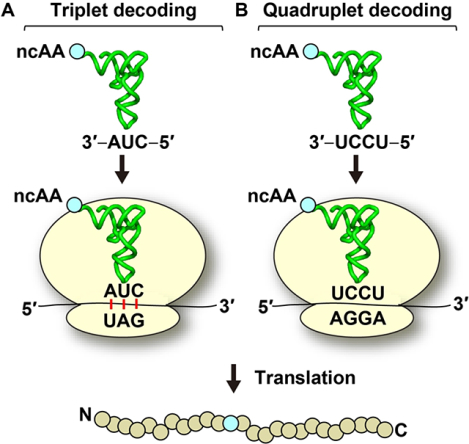
Introduction
A designer tRNA that can shift into the +1-frame at a specific quadruplet codon has the potential to expand the proteome that currently consists of the 20 canonical amino acids [1], as well as selenocysteine and pyrrolysine that are known as the 21st and 22nd amino acids, respectively [2, 3]. This designer tRNA can carry a non-canonical amino acid (ncAA) to the quadruplet codon and deliver it to protein synthesis, allowing the ribosome to use the ncAA for site-specific incorporation into the nascent polypeptide chain. The potential impact on protein bioengineering is high, permitting studies of structure and function with new chemical properties. While genome expansion is most commonly achieved with triplet decoding by suppressor tRNAs that deliver a ncAA to a premature stop codon within an open reading frame (Graphical abstract, panel A) [4], the capacity is limited. First, only two ncAAs can be inserted into one protein at a time due to the cellular need to reserve the third stop codon for termination of protein synthesis. Second, in eukaryotes, the introduction of an internal stop codon channels the mRNA for rapid degradation by the non-sense mediated mRNA decay (NMD) pathway, which is a quality control mechanism to remove the mRNA from synthesis of potentially toxic and truncated proteins [5, 6]. In contrast, genome expansion with quadruplet reading by +1-frameshifting tRNAs is not subject to these limitations [7] (Graphical abstract, panel B). It has a much higher capacity for simultaneous insertion of multiple ncAAs to multiple quadruplet codons; it is not regulated by the NMD pathway; and it can be used in combination with other strategies. Indeed, recent work has demonstrated the incorporation of multiple distinct ncAAs into one protein using a combination of triplet decoding at a premature stop codon and quadruplet decoding by read-through of a quadruplet codon [8–10]. Additionally, new strategies have been developed to use modified bacterial ribosomes to incorporate ncAAs via decoding with a dipeptidyl-tRNA as a single ribosomal event [11], thus potentially broadening the field of genome expansion.
Despite the high potential of genome expansion with +1-frameshifting tRNAs, however, the yield of the shift at a given quadruplet codon is not readily predictable, and the efficiency of the shift is low relative to triplet decoding (usually less than 3%) [10, 12]. The major constraint is imposed by the tRNA tertiary structure, which has been optimized in evolution for aminoacylation by a cognate aminoacyl-tRNA synthetase (aaRS) [13] to deliver the aminoacyl group to the ribosome using a triple anticodon-codon pairing scheme [14]. Another constraint is imposed by the ribosome itself, which has been optimized to accurately translate triplet codons of the genetic code [15, 16].
In bacterial ribosomes, the translational reading frame is defined by base pairing between the 16S rRNA of the 30S small subunit with the Shine-Dalgarno (SD) sequence of the mRNA to identify the start codon AUG. Protein synthesis is initiated by placing the initiator fMet-tRNAfMet (fMet: formyl-methionine) at the start codon AUG to establish the ribosomal peptidyl-tRNA-binding site (P site) (Figure 1). This is followed by recruiting the 50S large subunit to assemble the 70S initiation complex (70SIC), which is now poised to enter the elongation cycle. Each elongation cycle is defined by three key steps: (i) delivery of an aminoacyl-tRNA (aa-tRNA) to the mRNA codon at the ribosomal aminoacyl-tRNA-binding site (A site) via a ternary complex (TC) with the GTPase EF-Tu and GTP; (ii) synthesis of the next peptide bond in the peptidyl transferase center on the ribosome by transferring the nascent chain from the P-site tRNA to the aa-tRNA at the A site to form a pre-translocation complex (PRE); and (iii) translocation of the mRNA and the P- and A-site tRNAs to the exit (E) site and P site, respectively, to form a post-translocation complex (POST) in a reaction catalyzed by the GTPase EF-G and GTP (Figure 1). This elongation cycle continues with a new triplet codon entering the A site and the synthesis of a new peptide bond between the peptidyl moiety in the P site and the aminoacyl moiety of the A site. Protein synthesis is terminated when a stop codon appears in the A site, which is recognized by release factor 1 or 2 (RF1 or RF2) to trigger release of the polypeptide chain from the P site. Within this process, inducing tRNA +1 frameshifting at a quadruplet codon is an interruption, yielding protein products from both the 0-frame and the +1-frame translation. While both protein products have the same N-terminus or N-terminal sequence, they differ in the C-terminal sequence starting at the site of +1 frameshifting and read-through of the quadruplet codon. The relative proportions of the protein product from each are dynamically controlled.
Figure 1.
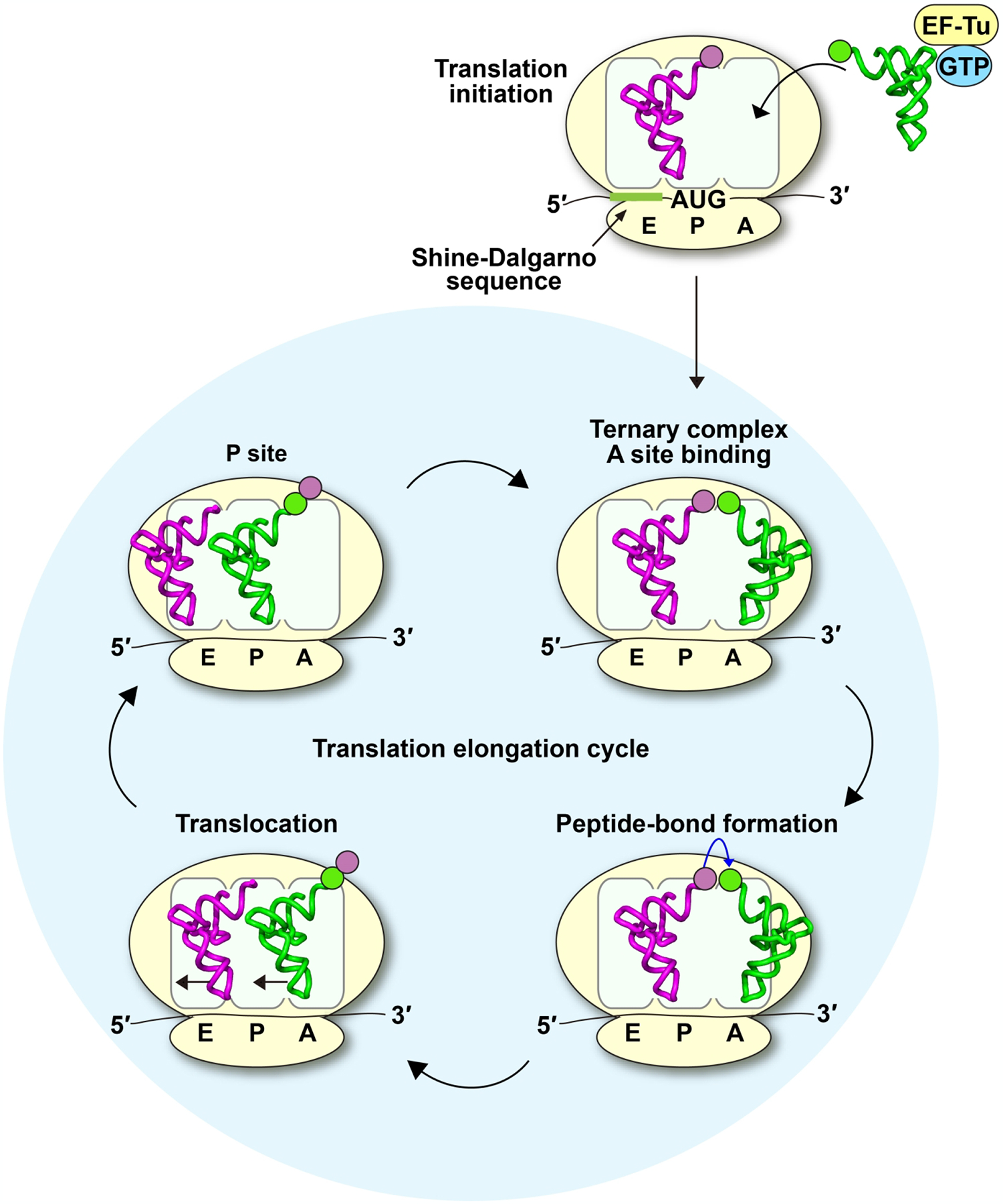
The elongation cycle of protein synthesis in bacteria. A 70SIC is formed with fMet-tRNAfMet sitting on the first AUG codon downstream from the SD sequence of the mRNA. The elongation cycle begins upon delivery of an aa-tRNA to the mRNA codon in the A site via the formation of a TC with EF-Tu and GTP. After the 30S subunit inspects the anticodon-codon pairing in the A site, GTP is hydrolyzed and the aa-tRNA is accommodated. The aminoacyl moiety of the aa-tRNA in the A site accepts the fMet moiety from the P site in a reaction catalyzed by the 50S subunit, forming the dipeptidyl-PRE complex. Subsequently, EF-G-GTP catalyzes translocation of the dipeptidyl-PRE complex into the dipeptidyl-POST complex, positioning the dipeptidyl-tRNA in the P site. A new mRNA codon enters the A site, allowing the elongation cycle to continue until it encounters a stop codon in the A site. Here, tRNAfMet is shown in purple and tRNAPro shown in green, each with the corresponding color of the aminoacyl-group, and the codon.
This review will summarize recent progress on elucidating the mechanism of +1 frameshifting on the ribosome. It is not meant to stand in for comprehensive reviews published by others in the field [16–20]. Also, we acknowledge that there has been development of exciting new technologies of genome expansion by adding new synthetic letters to the genetic code language [21, 22], or by synthesizing a new bacterial genome with a minimal set of codons for all amino acids to allow the unused codons for recoding purposes [23, 24]. Instead, this review discusses how the recent new information on +1 frameshifting can be harnessed for genome expansion at quadruplet codons.
A historical view
The key question of how a quadruplet codon is read in an elongation cycle of protein synthesis has remained elusive historically. Answering this question is necessary to provide a framework to improve reading of the quadruplet codon at the steps of the elongation cycle where it occurs. However, due to the lack of a clear answer to this question, most past and present efforts have been directed to the ribosomal A site by designing an extra nucleotide inserted to the anticodon loop of a designer tRNA to create an expanded anticodon-stem-loop (ASL) structure that can pair with a quadruplet codon in the A site [1]. This design concept came from early genetic isolation of several high-efficiency +1-frameshifting tRNAs, many of which possess an expanded ASL [16].
In the 1970s, various +1-frameshifting mutants of the Salmonella histidine (his) operon that disrupt the operon function were identified in genetic studies, each containing a single-nucleotide insertion [16]. Suppressors of these +1-frameshifting mutations were subsequently isolated, which led to the discovery of high-efficiency +1-frameshifting tRNAs that were extragenic to the operon. Sequence analysis showed that, while some suppressor tRNAs contain base substitutions, the majority carry an extra nucleotide in the anticodon loop resulting in an expanded ASL [16]. For example, the hisD gene encodes the natural sequence ACC-CCU-GAA (coding for Thr-Pro-Glu) in a +1-frameshift-prone window [25]. Insertion of a single C to the CCU codon in the window results in the hisD3018 +1-frameshift mutation [26], altering the sequence in the window to ACC-CCC-UGA (coding for Thr-Pro-stop) and producing a premature termination codon of protein synthesis (Figure 2A). A high-efficiency +1-frameshifting suppressor tRNA SufB2 was isolated with the ability to suppress the hisD3018 mutation by insertion of Pro in response to the quadruplet codon CCC-U, thus restoring the translational reading frame [27]. The high-efficiency +1 frameshifting of SufB2 is notable, at 1–5% above the background [27, 28] relative to the frameshifting efficiencies of 0.01% (or one in 104 codons) at the overall basal level [29]. Additionally, this high efficiency occurred in a cellular context where SufB2 co-exists with two natural iso-acceptor tRNAs that compete for reading of the triplet CCC within the quadruplet codon CCC-U. One of the iso-acceptors is tRNAPro(GGG) (encoded by the proL gene and hereafter referred to as the ProL tRNA) and the other is tRNAPro(U*GG) (encoded by the proM gene and hereafter referred to as the ProM tRNA) (Figure 2B). In the ProM tRNA, the wobble nucleotide U* in the U*GG anticodon stands for 5-methyl-carboxy-methoxy uridine 34 (mcmo5U34) [30], where the cmo5 core moiety enables the wobble nucleotide to pair with all four natural nucleotides [31]. Sequence analysis of SufB2 showed that it is derived from the ProL tRNA by harboring an extra nucleotide G37a in the anticodon loop [32] (Figure 2B). The expanded ASL in SufB2, as well as in many other similarly isolated +1-frameshifting suppressors [16], suggested the possibility of a quadruplet-pairing model in which the anticodon-codon interaction adopts a quadruplet-pairing scheme in the A site, which is thought to translocate to the ribosomal P site [33, 34].
Figure 2.
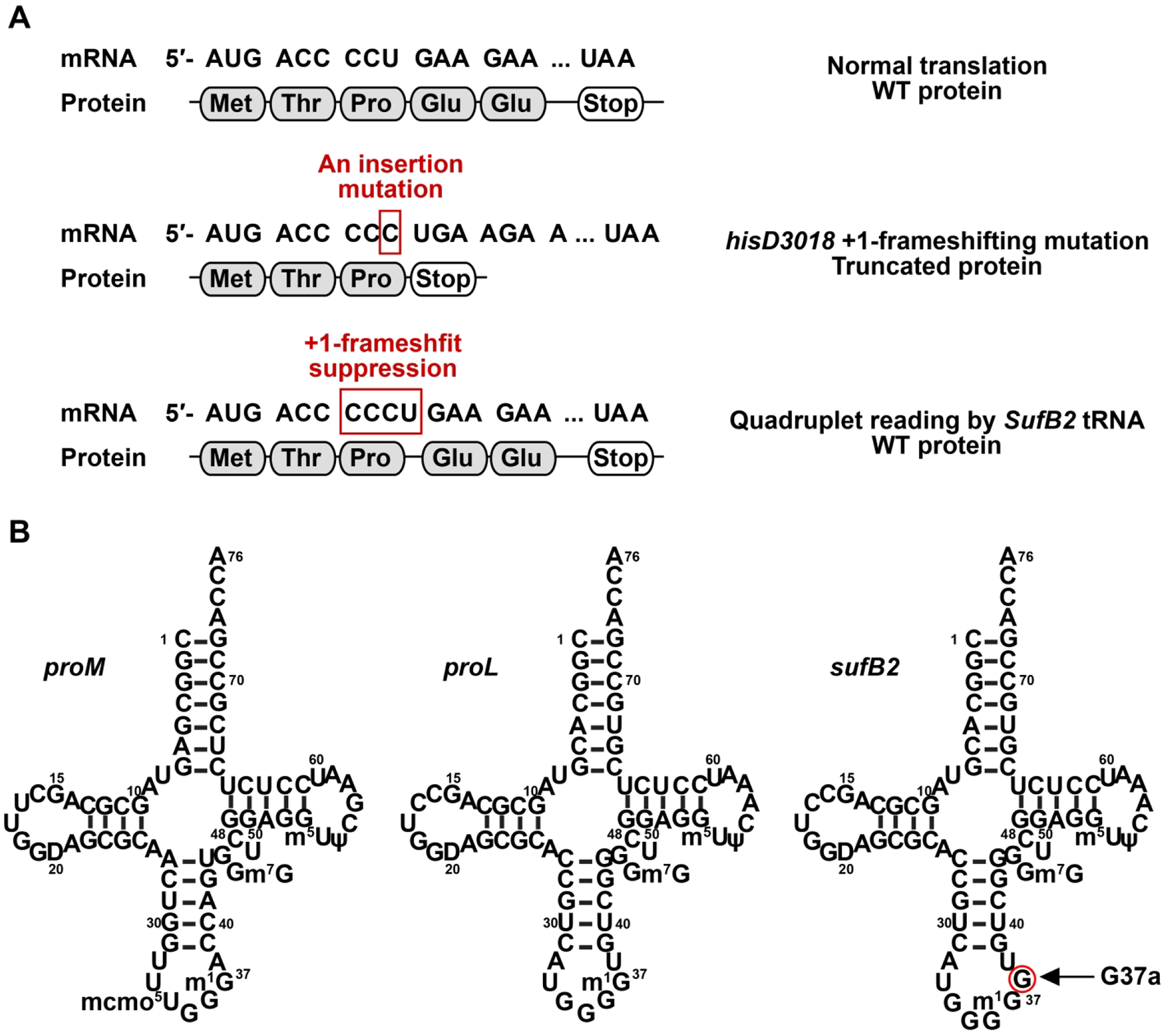
Quadruplet reading by a +1-frameshifting tRNA. (A) The hisD nucleotide sequence in Salmonella contains the +1-frameshifting window ACC-CCU-GAA (coding for Thr-Pro-Glu). The hisD3018 mutant of Salmonella harbors a single C insertion to the CCU codon, resulting in the sequence of the +1-frameshifting window ACC-CCC-UGA (coding for Thr-Pro-stop). This hisD3018 mutant is suppressed by the +1-frameshifting tRNA SufB2, which reads the quadruplet codon CCCU, thus restoring the original hisD reading frame of protein synthesis (for Thr-Pro-Glu). (B) Sequence and cloverleaf structure of Salmonella ProM, ProL, and SufB2, each in the native-state with the full complement of all natural post-transcriptional modifications. Note that SufB2 is derived from ProL by the insertion of an extra G37a nucleotide in the anticodon loop, resulting in an expanded ASL.
This quadruplet-pairing model then laid the foundation for subsequent development of +1-frameshifting tRNAs that would contain an expanded ASL. Using strategies of in vitro evolution, many labs designed random libraries of selected tRNA nucleotides in the context of an expanded ASL and screened for variants that decoded a quadruplet codon [7, 8, 10, 35–43]. This screen was typically performed in E. coli cells expressing an orthogonal pair of tRNA and aaRS that is mutually specific to each other for aminoacylation and does not cross-react with the corresponding host tRNA or host aaRS. The most frequently used orthogonal pairs include the pair of tRNAPyl-PylRS from Methanosarcina mazei (for aminoacylation with pyrrolysine in the native cellular environment) [44, 45], and the pair of tRNATyr-TyrRS from Methanococcus jannaschii [39]. For example, a +1-frameshifting tRNA with the anticodon UCUA for reading the quadruplet codon UAG-A was isolated using the orthogonal pair tRNAPyl-PylRS as a start [8]. Separately, the same anticodon-codon pair was isolated from evolving the orthogonal pair tRNATyr-TyrRS [36] (Figure 3A). Likewise, a +1-frameshifting tRNA with the anticodon UCCU for reading the quadruplet codon AGG-A was isolated by alteration of the orthogonal pair tRNATyr-TyrRS [35] (Figure 3A). Notably, all three tRNAs referenced here, tRNAPyl(UCUA), tRNATyr(UCUA), and tRNATyr(UCCU), were selected for their high +1-frameshifting efficiency relative to other similarly designed ASL-expanded tRNAs. If the only requirement is a quadruplet anticodon-codon pairing scheme in the A site, it is not clear why some tRNAs display high +1-frameshifting efficiency whereas others do not.
Figure 3.
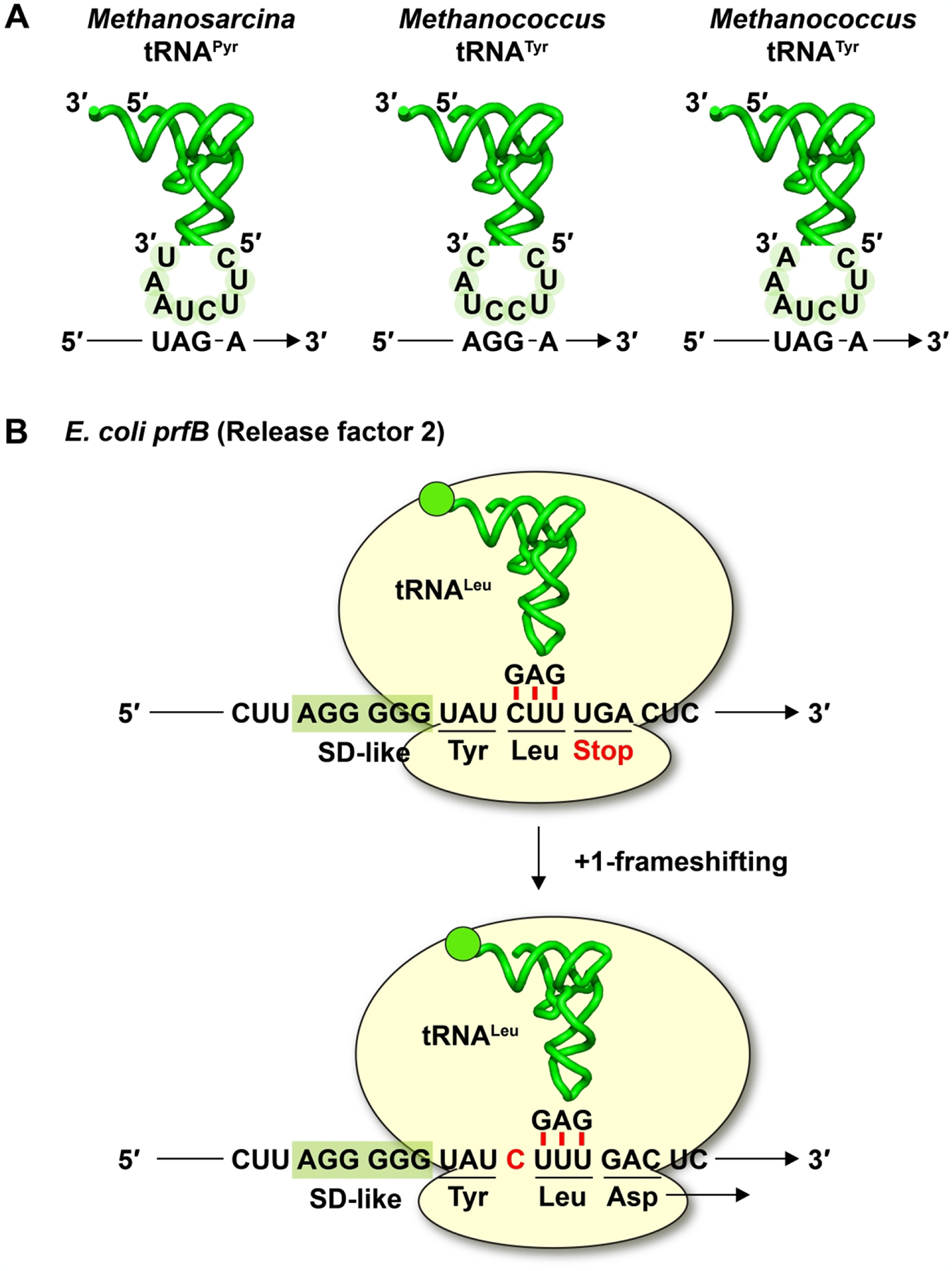
Quadruplet reading by +1 frameshifting ASL-expanded tRNAs and canonical tRNAs. (A) Examples of genetically evolved +1-frameshifting tRNAs that contain an expanded ASL: reading of the quadruplet codon UAGA by Methanosarcina tRNAPyl with the anticodon UCUA, reading of the quadruplet codon AGGA by Methanococcus tRNATyr with the anticodon UCCU, and reading of the quadruplet codon UAGA by Methanococcus tRNATyr with the anticodon UCUA. Note that the polarity of the mRNA codons is anti-parallel to that of the tRNA examples. (B) Translation of the full-length E. coli prfB gene (coding for RF2) by reading of the quadruplet codon CUUU using the canonical E. coli tRNALeu with the anticodon GAG, involving a +1-frameshifting event.
The inability to reliably predict the efficiency of quadruplet reading posed the question of whether it is necessary to form a quadruplet anticodon-codon pair in the A site. Several observations also challenged the requirement of a quadruplet anticodon-codon pair in the A site. For example, we have shown that the native-state E. coli ProM tRNA, isolated from cells with a canonical ASL, is highly prone to +1 frameshifting [46] (Figure 2B). This native ProM tRNA contains the full complement of all natural post-transcriptional modifications, including N1-methylation of G37 (m1G37) on the 3’-side of the anticodon, which would block Watson-Crick (W-C) base pairing and prevent a quadruplet anticodon-codon pairing scheme in the A site [47]. Thus, even a native tRNA, fully modified in a canonical structure and lacking an expanded ASL, can display high-efficiency +1 frameshifting relative to other canonical tRNAs.
Notably, one of the highest efficiencies of +1 frameshifting in nature is associated with expression of the full-length bacterial prfB gene (coding for RF2), which usually occurs at an efficiency of 30% but can approach nearly 100% in modified conditions [48]. Specifically, expression of the full-length prfB requires a +1-frameshifting event at the sequence window UAU-CUU-UGA-C [49] (Figure 3B), coding for Tyr-Leu-stop, where the UGA is an internal stop codon that senses the intracellular concentration of RF2. When RF2 level is high, it ensures that most of the translating ribosomes would terminate at the UGA and release a short peptide that is rapidly degraded. In contrast, when RF2 level is low, the peptidyl-tRNALeu at the CUU codon would shift to the +1-frame, allowing the translating ribosome to incorporate Asp at the GAC codon and to synthesize Tyr-Leu-Asp instead [49] (Figure 3B). In this high-efficiency mechanism, all tRNA components are in the native-state and have the canonical ASL [50], supporting the notion that it is not necessary to use a quadruplet anticodon-codon pairing in the A site to induce +1 frameshifting.
No quadruplet pairing or +1 frameshifting in the A site
An important step toward elucidating the mechanism of quadruplet decoding came from recent crystal structures of the bacterial ribosome in complex with an expanded ASL that is derived from genetically isolated +1-frameshifting tRNAs. Most of these structures are in complex with the ASL of the SufA6 +1-frameshifting tRNA [51–55], which is derived from Salmonella tRNAPro(CGG) (encoded by the proK gene and hereafter referred to as ProK) by insertion of G37a to the 3’-side of the anticodon [32]. Additionally, one structure is in complex with the ASL of the SufJ +1-frameshifting tRNA [56], which is derived from Salmonella tRNAThr(GGU) by insertion of C31.5 on the 5’-side of the anticodon [57]. In all cases, regardless of where the extra nucleotide is inserted relative to the anticodon, only the classical triplet anticodon-codon pairing scheme is observed in the A site [51, 53, 54, 56].
The lack of quadruplet pairing in the A site does not exclude the possibility of a triplet slippage event, in which the anticodon-codon pairing starts in a triplet in-frame scheme (the 0-frame) and then shifts to the +1-frame in the A site. While this triplet slippage model was discussed early [47, 58], whether it occurred in the A site, or during translocation of the +1-frameshifting tRNA from the A to the P site [48], or during tRNA occupancy within the P site [59], was not known. This question was resolved by recent kinetic assays of SufB2, which is an experimentally trackable model. While SufB2 was isolated from Salmonella [27], it has been expressed and validated for high efficiency of +1 frameshifting in E. coli [60, 61].
A series of kinetic assays, including ensemble and single-molecule measurements, have been developed to address how SufB2 undergoes +1 frameshifting at a quadruplet codon [60] (Table 1, Table 2). These assays are performed with reconstituted E. coli ribosomes that are programmed with an mRNA and supplemented with requisite tRNAs. SufB2 was prepared in one of the three forms of the modification state. The transcript-state (also known as the G37-state) is made by in vitro transcription, lacking any post-transcriptional modification. The m1G37-state is made by enzymatic synthesis of m1G37 on the transcript-state, generating a single post-transcriptional methylation site on the otherwise unmodified G37-state. The native-state SufB2 is isolated from cells, harboring the full complement of all natural post-transcriptional modifications, including m1G37.
Table 1.
Summary of E. coli tRNAs that contribute to +1 frameshifting at quadruplet codons
| tRNA | Anticodon 5’-3’ | ASL | Nucleotide 37 in the native-state | Occurrence in an elongation cycle and the kobs and yield (%) of +1 frameshifting |
|---|---|---|---|---|
| ProM | U*GG | Canonical | m1G37 | Primarily in the stalled P site [64] |
| Slow kobs at 4.8 × 10−3 s−1 [64] | ||||
| 38% in the G37-state [64] | ||||
| N.D. in the m1G37-state and in the native-state [64] | ||||
| ProL | GGG | Canonical | m1G37 | Primarily in the stalled P site [64] |
| Slow kobs at 2 × 10−3 s−1 [64] | ||||
| 26% in the G37-state [64] | ||||
| 8.1% in the native-state ([64] | ||||
| N.D. in the native-state + EF-P [64] | ||||
| SufB2 | GGG-G | Expanded | m1G37 | Primarily during translocation [60] |
| Fast kobs at 0.089 s−1 [60] | ||||
| 90% in the G37-state [60] | ||||
| 30% in the native-state [60] | ||||
Table 2.
Yield and rate constant kobs of tRNA +1 frameshifting by E. coli tRNAs
| During translocation in the G37-state | |||
|---|---|---|---|
| Pro(UGG) (ProM) | Pro(GGG) (ProL) | SufB2 | |
| Yield (%) | 5.9 | 1.2 | 90 |
| kobs (s−1) | 0.06 | 0.14 | 0.09 |
| During translocation in the m1G37-state or native-state | |||
| Yield (%) | 3.0 | 0.6 | 30 |
| kobs (s−1) | 0.03 | 0.5 | Not determined |
| During P-site occupancy in the G37-state | |||
| Yield (%) | 38 | 26 | Not determined |
| kobs (s−1) | 0.005 | 0.002 | Not determined |
| During P-site occupancy in the m1G37-state or native-state | |||
| Yield (%) | 5 | 8 | Not determined |
| kobs (s−1) | 0.00002 | 0.013 | Not determined |
All of the values are obtained from previous work [60, 61, 64] and are presented without the statistical error of each number. The mRNA coding sequence based on which these values were derived from was 5’-AUG-CCC-CGU-U. In cases where values of both the m1G37-state or the native-state are both available, those of the native-state are presented.
To maximize the ability to detect quadruplet pairing and +1 frameshifting in the A site, SufB2 was prepared in the transcript-state. These kinetic assays show that, in the A site, not only does SufB2 maintain the triplet-pairing frame, but that it also does not undergo triplet slippage or +1 frameshifting [60]. Specifically, SufB2 exhibits essentially the same kinetic parameters as ProL in reactions at the A site, starting from binding and accommodation to the decoding center to accepting peptidyl transfer from the P site to synthesize a peptide bond [60]. This 0-frame pairing at the A site, up to the reaction of peptide-bond formation, is also observed in a physical mapping study that measures the reading frame of the translating ribosome [61]. Additional experiments demonstrate that SufB2 and ProL are mutually competitive with each other in peptide-bond formation in the A site, indicating that the two tRNAs are kinetically indistinguishable [60]. To exclude the possibility of triplet slippage or +1 frameshifting in the A site, further experiments show that the ability of SufB2 to form a peptide bond is not sensitive to mutations on the 4th position of a quadruplet codon motif, but that it is sensitive to mutations on the 3rd position of the motif. This latter result provides strong evidence that SufB2 does not undergo +1 frameshifting or triplet slippage in the A site [60].
The absence of +1 frameshifting in the A site is consistent with the mechanism of the ribosome decoding center [62]. In this mechanism, three conserved nucleotides of the 16S rRNA of the 30S subunit (G530, A1492, and A1493 in the numbering of the E. coli ribosome) mutually undergo an induced-fit conformational rearrangement to select for the cognate anticodon-codon pairs and to discriminate against non-cognate pairs [62, 63]. The induced-fit selection of only the cognate pair in the correct triplet reading frame indicates that the decoding center has evolved to emphasize the fidelity of translational decoding with respect to the anticodon-codon pairing in the A site. Even with a sub-optimal anticodon-codon pairing, as presented by an expanded ASL, the decoding center has managed to maintain accuracy. This accuracy in the A site indicates that quadruplet reading of a quadruplet codon is mediated by a +1-frameshifting event downstream from the A site. Below we refer quadruplet reading as involving tRNA +1 frameshifting, which in principle can occur by triplet slippage or by a quadruplet anticodon-codon pairing scheme.
Shifting tRNA to the +1-frame can occur during translocation from the A site to the P site
Gamper et al. developed a series of ensemble kinetic assays that provide the first biochemical insight into +1 frameshifting during translocation [60, 64]. In an ensemble assay to monitor +1 frameshifting during translocation, an E. coli 70SIC, harboring the initiator fMet-tRNAfMet in the P site, was rapidly mixed with an equimolar mixture of SufB2-, Val-, and Arg-TC (where SufB2-TC indicates prolyl (Pro)-SufB2-tRNAPro-TC, Val-TC indicates valyl (Val)-tRNAVal-TC and Arg-TC indicates arginyl (Arg)-tRNAArg-TC) (Figure 4A). The mRNA coding sequence was AUG-CCC-CGU-U. As soon as the SufB2-PRE complex associated with the CCC-C codon motif was translocated to the POST in the presence of the EF-G-GTP complex, the 3rd codon that appeared in the A site was either a GUU codon encoding Val in the +1-frame or a CGU codon encoding Arg in the 0-frame (Figure 4B). Thus, the formation of an fMPV tripeptide would report on the sub-population of SufB2 that had shifted to the +1-frame during translocation, whereas the formation of an fMPR tripeptide would report on the sub-population that had remained in the 0-frame. In this design, SufB2 was in the unmodified transcript-state to maximize its potential for +1 frameshifting, whereas Val-TC and Arg-TC were in the fully modified native-state to stabilize their reading of codons [60]. The rapid co-delivery of SufB2-, Val-, and Arg-TC to the 70SIC leaves no time for +1 frameshifting of SufB2 in the P site. The results show rapid synthesis of the fMPV tripeptide to high yields (90%), indicating that a major sub-population of SufB2 has shifted to the +1-frame during translocation [60]. The notion of SufB2 undergoing +1 frameshifting during translocation has been confirmed by mapping the ribosome position on the reading frame of the mRNA [61].
Figure 4.
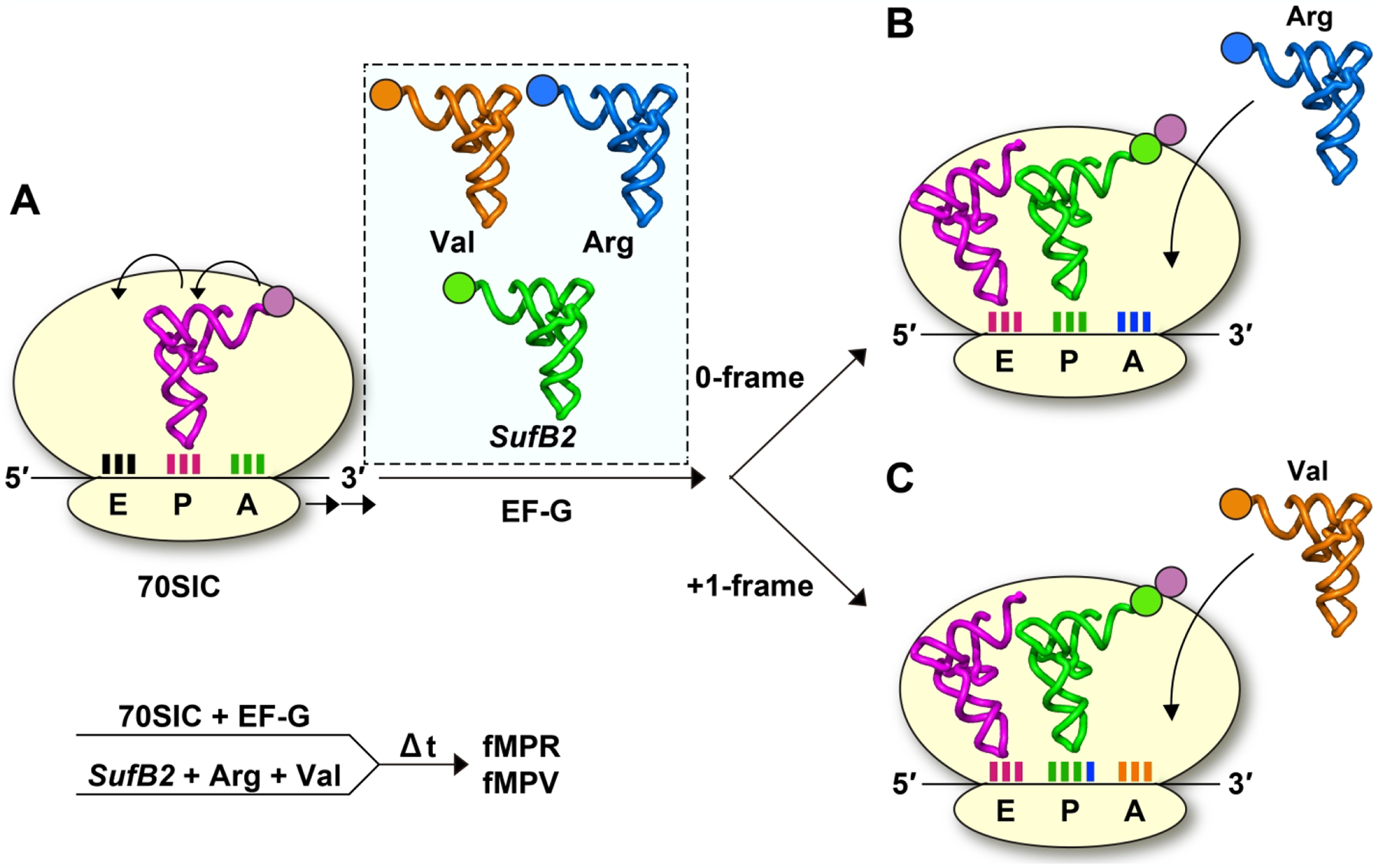
An ensemble kinetic assay to monitor +1 frameshifting during translocation. (A) A 70SIC harboring fMet-tRNAfMet on the AUG codon in the P site, programmed with the mRNA sequence AUG-CCC-CGU-U, is rapidly mixed with an equimolar mixture of the SufB2-TC, Val-TC, and Arg-TC and formation of the tripeptide fMPV or fMPR is monitored over time. The SufB2-TC is in the transcript-state, while the Val-TC and Arg-TC are in the native-state. In the presence of EF-G-GTP, the fMP-PRE complex is translocated to the P site. (B) If the fMP-PRE complex remains in the 0-frame during translocation, the CGU codon for Arg would enter the A site, allowing synthesis of fMPR. (C) If the fMP-PRE complex undergoes +1 frameshifting during translocation, the GUU codon for Val would enter the A site, allowing synthesis of fMPV. The results show 10% synthesis of fMPR but 90% synthesis of fMPV, indicating that SufB2 distributes between a small sub-population that remains in the 0-frame and a large sub-population that has shifted to the +1-frame during translocation.
Additional ensemble assays show that E. coli ProM and ProL tRNAs, each prepared in the unmodified transcript-state, also display a fractional shift to the +1-frame during translocation [64] (Table 1, Table 2). This indicates that translocation is prone to +1 frameshifting and that this propensity is not specific to SufB2 but is general to canonical tRNAs when the latter lack post-transcriptional modifications. Notably, SufB2 has a substantially higher yield of +1 frameshifting during translocation than the two canonical tRNAs [60, 64], indicating a specific ribosome response to the expanded ASL. In all three tRNAs, SufB2, ProM, and ProL, the presence of m1G37 reduces the yield of +1 frameshifting by 2–3-fold [60, 64], demonstrating the importance of the methylation to minimize changes of the reading frame. Additionally, the observed rate constant (kobs) of +1 frameshifting for all three tRNAs is comparable to the rate constants of peptide-bond formation as measured with the same in vitro translation system [60, 64]. This suggests that +1 frameshifting during translocation can pose a challenge to the quality of protein synthesis.
A single-molecule assay of SufB2 provides insight into ribosomal dynamics that regulates +1 frameshifting during translocation [60]. This assay monitors changes of the spatial interaction between the L1 and L9 proteins of the large ribosomal 50S subunit, which reports on the successive opening and closing of the L1 stalk that takes place in an elongation cycle [65–67]. The results show that the kinetic property of SufB2 is identical to that of ProL up to and including formation of the PRE complex, but that it differs substantially from ProL during translocation [60]. Specifically, the conformational dynamics of SufB2 is indistinguishable from that of ProL in early steps of translocation, which begin after peptide-bond formation and involve EF-G-independent movement of the acceptor stems of the A-site and P-site tRNAs into the P and E sites of the 50S subunit, respectively, forming the hybrid state [60]. This kinetic similarity of SufB2 to ProL supports the notion that SufB2 undergoes no triplet slippage or +1 frameshifting in the A site as established by ensemble experiments [60]. However, upon addition of the EF-G-GTP complex, the dynamics of the SufB2-bound ribosome is markedly reduced relative to the ProL-bound ribosome [60]. This reduction, by 2–3 orders of magnitude, suggests that the SufB2-bound ribosome adopts an unusual conformation that impedes rearrangements specifically occurring at late steps in translocation. The implication is that the reduced dynamics of the SufB2-bound ribosome allows time to shift the anticodon-codon pairing scheme from the 0-frame to the +1-frame. Notably, the late steps of translocation are to move the tRNA ASLs and the associated codons from the P and A sites to the E and P sites of the 30S subunit. The key events of the late steps include the severing of the decoding center from the anticodon-codon duplex in the A site [68–71], forward and reverse swiveling of the head domain [72, 73] associated with opening and closing of the E-site gate of the 30S subunit [74], reverse relative rotation of the ribosomal subunits [75, 76], and opening of the L1 stalk [65, 66, 77]. Of these, the event that is most likely concomitant with +1 frameshifting is the head domain swiveling of the 30S subunit [60], which is particularly important for movement of the ASLs and the associated codons within the 30S subunit [72–74, 78].
Cryo-EM structures of +1 frameshifting during translocation
While kinetic assays strongly suggest that tRNA +1 frameshifting occurs during translocation, these assays do not directly visualize the change of the reading frame. Direct visualization of +1 frameshifting during translocation is now successfully achieved by analysis of cryo-EM structures [79]. These cryo-EM structures examine the native-state of E. coli ProM, which is particularly prone to +1-frameshifting on the frameshifting mRNA codon motif CCC-A [46], but is stable on the non-frameshifting mRNA codon motif CCA-A. The native-state ProM tRNA contains the modified mcmo5U34 at the wobble position, which expands the capacity of base pairing to all four natural nucleotides [31]. A cryo-EM structure that captures the tRNA in the A site demonstrates the triplet 0-frame anticodon-codon pairing scheme. This is observed both with the non-frameshifting codon motif, where the mcmo5U34 is paired to A3 of the CCA codon, and with the frameshifting codon motif, where mcmo5U34 is paired to C3 of the CCC codon (Figure 5A, B). This latter structure, obtained on a frameshift-prone codon motif, confirms the lack of +1 frameshifting in the A site, although it does exhibit a notable difference from the non-frameshifting structure.
Figure 5.
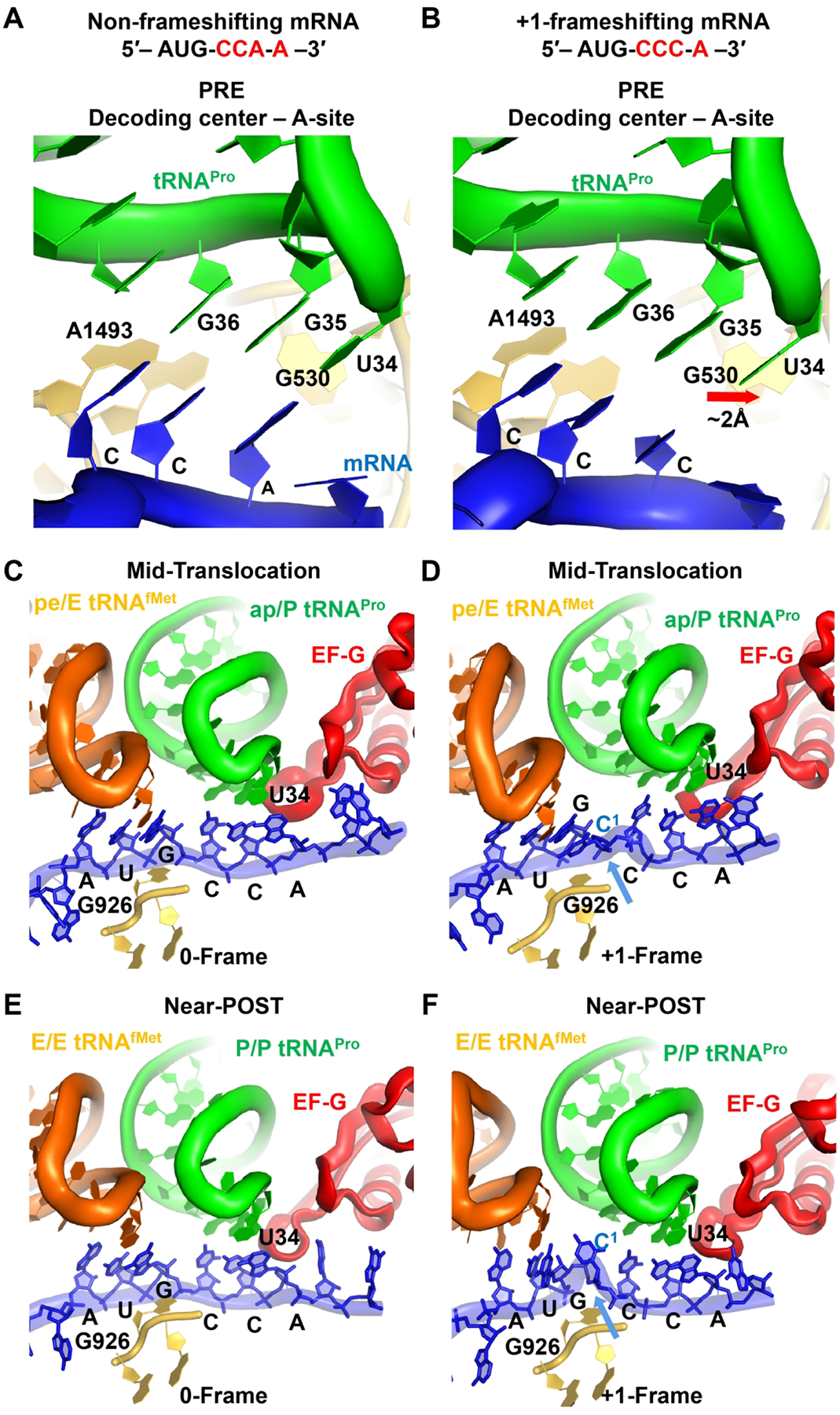
Translation by E. coli ProM of a non-frameshifting mRNA sequence AUG-CCA-A or a frameshifting mRNA sequence AUG-CCC-A as captured by cryo-EM structures. Reading of (A) the non-frameshifting sequence and (B) the frameshifting sequence in the A site; reading of (C) the non-frameshifting sequence and (D) the frameshifting sequence during mid-translocation; and reading of (E) the non-frameshifting sequence and (F) the frameshifting sequence near the end of translocation. For complexes C-F, the translocation factor EF-G and the nonhydrolyzable analog GDPcP of GTP are present.
Specifically, the non-frameshifting structure features a canonical “closed” 30S subunit, in which G530, A1492, and A1493 are in the closed state and interact with the backbone of the cognate anticodon-codon helix [79] (Figure 5A). G530 contacts A1492, resulting in a latched decoding center nearly identical to that in cognate 70S complexes formed with other tRNAs [80, 81]. By contrast, the frameshifting structure with the mcmo5U34-C3 wobble pair features an “open” 30S conformation, in which the shoulder domain of the small subunit is shifted away from the body domain (Figure 5B). This open conformation resembles transient intermediates of decoding captured by cryo-EM and is preferred when mismatches are present in the anticodon-codon duplex [82, 83]. Here G530 (at the shoulder) is retracted by ~2 Å from the closed state, shifting away from A1492 at the body domain and from the backbone of G35 of the tRNA, and resulting in a disrupted triad G530-A1492-A1493 relative to the non-frameshifting structure (Figure 5B). Thus, while the anticodon-codon helix is in the normal 0-frame of the frameshifting complex, the mcmo5U34-C3 wobble pairing shifts the 30S dynamics equilibrium toward the open conformation. It appears that the mcmo5U34 wobble nucleotide is pre-disposed to shift the pairing with the near-cognate CCC codon to pairing with the cognate CCA codon in the +1-frame.
After formation of the PRE complex, the 30S subunit spontaneously rotates as the ProM tRNAPro and the initiator tRNAfMet adopt the A/P and P/E hybrid states, respectively, in which their acceptor stems are translocated to the large 50S subunit. A cryo-EM structure that captures the PRE complex with a bound EF-G and the non-hydrolyzable GDPcP analogue of GTP during translocation is available for both the non-frameshifting mRNA and the frameshifting mRNA [79] (Figure 5C, D). In the non-frameshifting complex, EF-G binds to the rotated conformation of the complex, where the head of the 30S is swiveled by ~16°, which is coupled with translocation of the tRNA ASLs and the associated mRNA codons, placing the dipeptidyl fMP-tRNAPro between the A and P sites of the 30S subunit but maintaining the 0-frame mcmo5U34-A3 pairing scheme (Figure 5C). This 0-frame pairing is seen in other translocation intermediates [84]. In the frameshifting complex, in contrast, the dipeptidyl-tRNAPro pairs with the mRNA in the +1-frame between the A and P sites of the 30S subunit (Figure 5D). Here, mcmo5U34 of tRNAPro is base-paired with A4 of the mRNA, even though the mcmo5 modification of U34 is not well resolved, likely due to its conformational dynamics. The neighboring deacylated tRNAfMet is bound to the AUG codon near the E site, resulting in a bulged mRNA nucleotide C1 between the E and P sites. This bulged C1 is stabilized by G926 of 16S rRNA (Figure 5D), allowing the mRNA to compact and accommodate four mRNA nucleotides in the E site. The frameshifting also shifts tRNAfMet away from tRNAPro, which is compensated by a shift of EF-G loop II without affecting the rest of EF-G domain IV. The specific rearrangement of loop II is likely to interfere with the speed of translocation [70, 85], consistent with the slow translocation of SufB2 that is observed in the single-molecule study [60]. A previous crystal structure implicates the ability of the 16S rRNA nucleotides C1397 and A1503 to prevent mRNA slippage by interacting with the bases of the translocating mRNA [86, 87]. C1397 flanks the A site, while A1503 flanks the E site, both as a part of the central region of the 30S head [86, 87]. However, both nucleotides maintain the same conformation between the non-frameshifting and the frameshifting complex [79], suggesting that the compact and frameshifted mRNA can be accommodated in the regular mRNA tunnel without perturbing the head domain conformation.
A cryo-EM structure that depicts a nearly completed translocation is also captured for the non-frameshifting and frameshifting complexes [79]. The non-frameshifting complex features a small head swivel (~1°) and the dipeptidyl-tRNAPro in the P site (Figure 5E), resembling the features of the mid-translocation complex and those of other non-rotated POST complex [88]. Both the dipeptidyl-tRNAPro and the deacylated tRNAfMet have their anticodon-codon pairs in the 0-frame of the P and E sites. Notably, the frameshifting complex also exhibits the same features of the mid-translocation complex, showing that the dipeptidyl-tRNAPro is in the +1-frame of the P site and the deacylated tRNAfMet is in the E site, which also accommodates C1 and the AUG codon as a result of the +1-frameshifting event that has occurred with the dipeptidyl-tRNAPro (Figure 5F). Notably, C1 is now detached from G926, producing the structure of the canonical P site at the end of translocation.
Collectively, these cryo-EM structures support the model of ensemble and single-molecule studies in that no triplet slippage, or +1 frameshifting, occurs in the A site, but that tRNA shifts to the +1-frame during EF-G-catalyzed translocation. These cryo-EM structures are important for understanding the mechanism of +1 frameshifting for several reasons. First, they represent the first ribosomal complexes that carry a full-length +1-frameshifting tRNA, rather than just the ASL. Second, they are ribosome-tRNA complexes that carry a canonical ASL structure, rather than an expanded ASL. Third, these cryo-EM structures are formed via enzymatic reactions that successively move the +1-frameshifting tRNA from the A site to the P site via translocation, thus providing direct insight into the shift from the 0-frame to the +1-frame during translocation.
Shifting tRNA to the +1-frame can also occur in the ribosomal P site
Earlier genetic studies of +1-frameshifting tRNAs suggest that P-site re-alignment is a major cause of changes of the reading frame [16]. The hypothesis on the P site was proposed because the efficiency of +1 frameshifting measured in previous genetic studies was sensitive to the efficiency of decoding at the A site [59, 89–92], implicating a role of the A site on the reading frame of the P site. The notion of the P site being permissive of tRNA +1 frameshifting is confirmed and elaborated by recent kinetic assays of E. coli ProM, ProL, and SufB2 tRNAs and cell-based assays of SufB2 tRNA [60, 61, 64].
A series of ensemble kinetic assays identifies two mechanisms of tRNA +1 frameshifting in the P site. One mechanism involves a P-site-stalled ribosome [64]. A study of E. coli ProM and ProL tRNAs in the unmodified transcript-state demonstrates that, when stalled in the P site at a CCC-C codon motif next to an empty A site, each undergoes +1 frameshifting in the P site [64] (Figure 6A). Although both tRNAs also undergo +1 frameshifting during translocation, the yield of the shift is significantly higher in the P site upon stalling (Table 2). This provides a clear contrast to SufB2, which exhibits a high efficiency of +1 frameshifting during translocation but no evidence of the shift when stalled in the P site next to an empty A site [60]. Thus, the choice of where to explore +1 frameshifting is different, with the two canonical tRNAs primarily exploring the shift within the P site, whereas the ASL-expanded SufB2 exploring the shift during translocation (Table 1).
Figure 6.
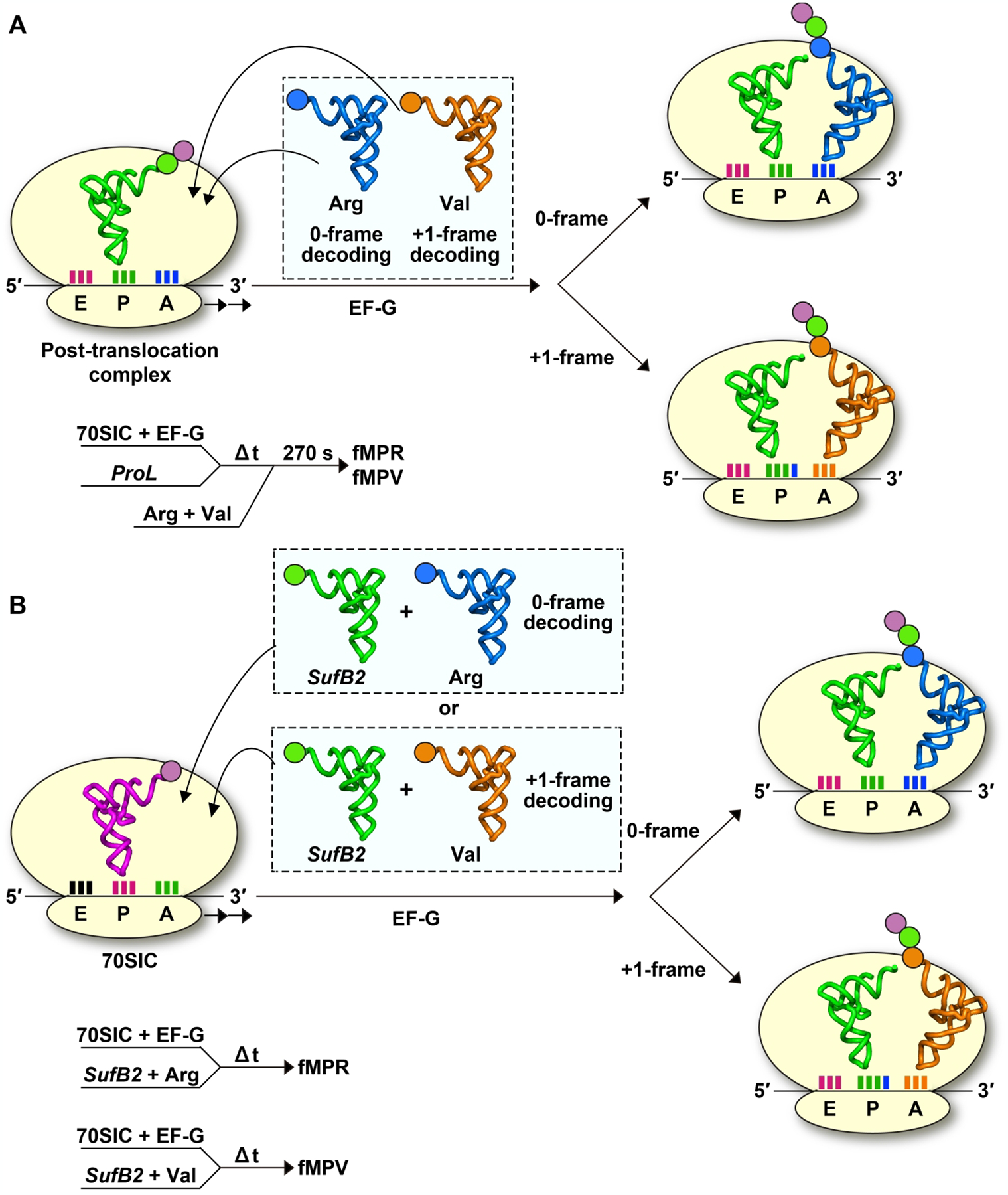
Ensemble kinetic assays to monitor +1 frameshifting of SufB2 in the P site. (A) A POST complex harboring the dipeptidyl-ProL in the P site is formed on the mRNA sequence AUG-CCC-CGU-U. The POST complex is stalled over time and any change of the reading frame in the P site is monitored by addition of an equimolar mixture of Val-TC and Arg-TC. The result identifies synthesis of fMPV, indicating evidence of +1 frameshifting of the POST complex. (B) A 70SIC harboring fMet-tRNAfMet in the P site, programmed with the same mRNA as above, is rapidly delivered with an equimolar mixture of SufB2-TC and Val-TC or with an equal mixture of SufB2-TC and Arg-TC during active protein synthesis. The result reveals complete synthesis of fMPV when the 70SIC is rapidly mixed with SufB2-TC and Val-TC, but complete synthesis of fMPR when the 70SIC is rapidly mixed with SufB2-TC and Arg-TC, providing evidence of frameshifting in the P site depending on the reading-frame occupancy of the A-site tRNA.
Between the two canonical tRNAs, ProM is completely suppressed from +1 frameshifting in the P site by the single m1G37 methylation [64] (Table 1, Table 2), indicating that the methylation is the major determinant for this tRNA to control the reading-frame accuracy, whereas other post-transcriptional modifications have little effect. In contrast, ProL is suppressed from +1 frameshifting in the P site by the combined action of m1G37 and the elongation factor EF-P [64] (Table 1, Table 2). The reason for why EF-P is required for stabilizing ProL in the P site is not yet clear, but the factor is known to bind to the P site [93] and to have high affinity for tRNA species that have a large D-loop (e.g., 9 nucleotides) with a stable D stem such as ProL [94]. Additionally, the ability of EF-P to maintain reading-frame accuracy of ProL in the P site on a single CCC-C motif is unexpected, because the factor is best known for its ability to relieve ribosome stalling at multiple codons for Pro [95, 96]. Notably, the kobs of +1 frameshifting in the P site of the two canonical tRNAs is slow, by 1–2 orders of magnitude, relative to the rate of peptide-bond formation as measured with the same reconstituted ribosome [64] (Table 1, Table 2). Therefore, the slow shift in the P site is not comparable to the rate of active protein synthesis and it occurs only when the A site is empty.
The second mechanism of +1 frameshifting in the P site involves an actively elongating ribosome [61], where the A site is continuously occupied (Figure 6B). An ensemble kinetic study of SufB2 demonstrates that, during active protein synthesis, SufB2 is induced to change its reading frame in the P site, depending on whether the A site is being read by a +1-frame- or a 0-frame-occupying tRNA (Figure 6B). As discussed above [60], the unmodified transcript-state of SufB2 distributes between two sub-populations after translocation into the P site, with the major sub-population (90%) in the +1-frame and a minor sub-population in the 0-frame (10%). It is found that the addition of Val-TC, which would occupy the A site in the +1-frame, shifts the 0-frame sub-population into the +1-frame, thus converting the entire population into the +1-frame [61]. Conversely, the addition of Arg-TC, which would occupy the A site in the 0-frame, shifts the +1-frame sub-population into the 0-frame, thus converting the entire population into the 0-frame [61]. These separate assays demonstrate the ability of SufB2 to shift in the P site.
The +1-frameshifting mechanism during active protein synthesis is important to the field on multiple fronts. Most notably, it demonstrates that SufB2 can twice explore +1 frameshifting in one elongation cycle of protein synthesis. In the first exploration, the major sub-population of SufB2 attempts to shift during translocation (90%), while in the second exploration, the remaining sub-population attempts to shift during occupancy in the P site (10%) upon the entry of an incoming tRNA occupying the +1-frame of the A site [61]. Thus, while +1 frameshifting during translocation and during P-site occupancy was shown previously, but in separate kinetic models [60, 64], it can occur consecutively and sequentially, leading to a complete shift of the tRNA into the +1-frame at the end of the elongation cycle. Additionally, the mechanism reveals that the SufB2 shift within the P site can be bi-directional [61], shifting to the +1-frame upon occupancy of the A site by a +1-frame tRNA, while shifting to the 0-frame upon occupancy of the A site by a 0-frame tRNA. This flexibility of shift in the P site has also been demonstrated for SufB2 in a cell-based reporter assay during active protein synthesis in vivo [61]. It is also consistent with earlier cell-based studies, documenting that the change of the reading frame in the P site is modulated by the reading frame of the A site [59, 89–92]. Notably, while the SufB2 shift within the P site occurs with a only minor sub-population, the rate of the shift kobs is fast, both to the +1-frame and back to the 0-frame, as compared to the slow kobs for ProL in the P site [61] (Table 2). This difference in the rate constant probably reflects weak base pairing of SufB2 to the mRNA codon in the P site, due to the expanded ASL. It again indicates the distinct response of the ribosome dynamics to the unusual ASL structure of SufB2 relative to the canonical ASL in ProL.
Principles of +1 frameshifting at a quadruplet codon
New principles have emerged from the recent studies cited above with broad implications for translational accuracy and unconventional decoding. Most importantly, we emphasize that decoding of a quadruplet codon starts with a triplet anticodon-codon pairing scheme in the A site, followed by a +1-frameshifting event after the A site. This +1-frameshifting event primarily takes place during translocation for an ASL-expanded tRNA, such as SufB2 [60, 61], but it primarily takes place in the P site for a canonical tRNA that is frameshift-prone, such as the ProM tRNA [79]. No quadruplet anticodon-codon pairing is observed at the A site for both. However, during the dynamic movements of the ribosome in the translocation reaction, it is possible that an ASL-expanded tRNA might explore quadruplet pairing, provided that the nucleotide at position 37 is able to pair with the quadruplet codon. Thermodynamically, a quadruplet pairing during translocation might be favored over triplet slippage, due to the more stability of the anticodon-codon pairing scheme. This possibility could explain the particularly high yields of +1 frameshifting obtained with the transcript-state SufB2 relative to the fully modified native-state [60]. Based on this fundamental principle, several critical considerations are suggested below for genome expansion, which is ideally to be performed in a cell model.
First, virtually all natural tRNAs with a canonical ASL contain a purine at position 37, which is invariably modified in such a way as to prevent a quadruplet pairing scheme. Similarly, most of the genetically evolved and isolated +1-frameshifting tRNAs with an expanded ASL also contain a purine at position 37 (e.g., [8]), which is likely post-transcriptionally modified as well, as shown in the case of SufB2 [60]. For example, m1G37 is conserved in all Pro-specific tRNAs [97] and its presence substantially reduces the efficiency of +1 frameshifting for ProL, ProM, and SufB2 in all experimental conditions tested [60, 61, 64]. A rationale for the role of m1G37 in maintenance of the reading frame is provided by X-ray crystal structures of the ASL of the ProK tRNA in complex with the bacterial ribosome. These structures demonstrate that m1G37 stabilizes the ASL in an active conformation, allowing formation of the intramolecular 32–38 base pair that is commonly present in natural tRNAs [51]. This 32–38 base pair appears to position the anticodon-codon base pairing in the canonical triplet 0-frame scheme [51, 62, 80]. In contrast, loss of m1G37 eliminates the 32–38 base pair, resulting in an altered ALS conformation that is more flexible and more prone to frameshifting [51]. Thus, the removal of m1G37 reduces the energetic constraints imposed on the ASL and promotes +1 frameshifting. While the structural impact on the ASL by other post-transcriptional modifications at position 37 is not yet clear, it is likely that the same principle would hold true.
Second, all evidence points to the notion that the ribosome itself is a major determinant of +1 frameshifting. The lack of +1 frameshifting in the A site is attributed to the limited space of the decoding center and to the induced-fit selection for only the triplet anticodon-codon pairing in the 0-frame using a conserved mechanism that involves three nucleotides (G530, A1492, and A1493) of the 16S rRNA [62]. In contrast, translocation consists of a series of large conformational rearrangements of the ribosome [65], particularly the EF-G-catalyzed swiveling of the 30S head domain that controls the movement of the tRNA ASLs and the associated mRNA codons from the A and P sites into the P and E sites of the 30S subunit, respectively [72–74, 78]. This swiveling of the 30S head domain is a key conformational rearrangement in the late steps of translocation and it is implicated as the motion that permits +1 frameshifting [60]. It may also be involved in regulating programmed −1 frameshifting during translocation [98]. Similarly, the dynamics of the 30S subunit determines the efficiency of +1 frameshifting. While the 30S subunit has a different priority in the P site relative to the A site, using only one nucleotide (C1400) to inspect the anticodon-codon paring scheme and three nucleotides (A790, A1338, and A1339) to stabilize the anticodon stem [80], this subunit nonetheless performs dynamic rearrangement similar to that during translocation. Specifically, when a frameshift-prone tRNA moves into the +1-frame of the P site, the 30S head domain undergoes a large swiveling-like rotation relative to the body domain to stabilize the anticodon-codon pair in a motion similar to that during late steps of translocation [55]. This large swiveling-like rotation of the 30S head domain is observed for both a canonical ASL, lacking post-transcriptional modifications, and for an expanded ASL, to position each in the +1-frame [55]. Thus, the two steps of the elongation cycle that allow +1 frameshifting–translocation and occupancy of the P site–are supported and permitted by the ribosome conformational dynamics of each.
Third, +1 frameshifting does not necessarily require an ASL-expanded tRNA (e.g., SufB2), but can occur with some canonical tRNAs (e.g., the native-state ProM). However, the yield and mechanism differ between the two types (Table 1). For example, SufB2 preferentially makes the shift during translocation and produces high yields [60], whereas ProM and ProL preferentially make the shift in the P site and produce relatively lower yields [64]. In both cases, however, the shift occurs while each tRNA is stalled [60, 64], indicating that the ribosome dynamics must be slowed to allow time to rearrange the anticodon-codon pairing scheme from the 0-frame to the +1-frame. A similar stalling of the ribosome dynamics is also observed as a requirement for programmed −1 frameshifting [99], indicating that changes of the reading frame occur when the ribosome is dynamically constrained. During change of the reading frame, while the 30S head swiveling may drive the anticodon-codon pairing to the new frame, additional structural elements of the 30S subunit are likely required. For example, during translocation, two hinges in the 16S rRNA that comprise the 30S neck domain may be involved [100]. Hinge 1 consists of two G-U base pairs separated by a bulged G within helix 28 (h28), while hinge 2 consists of a GUCU linker between h34 and h35/36 within a three-helical junction in h38. Separately, during stabilization of the P-site tRNA, the C-terminus of the S9 protein in the 30S subunit may be involved. This C-terminus stabilizes the backbones of nucleotides 32–34 of the anticodon loop of the P-site tRNA, and its truncation leads to +1 frameshifting [59].
Perspectives of genome expansion by tRNA +1 frameshifting at quadruplet codons
The new principles that have emerged from recent work will provide an improved framework for genome expansion by tRNA +1 frameshifting at quadruplet codons. Key considerations of this improved framework are summarized (Figure 7).
Figure 7.
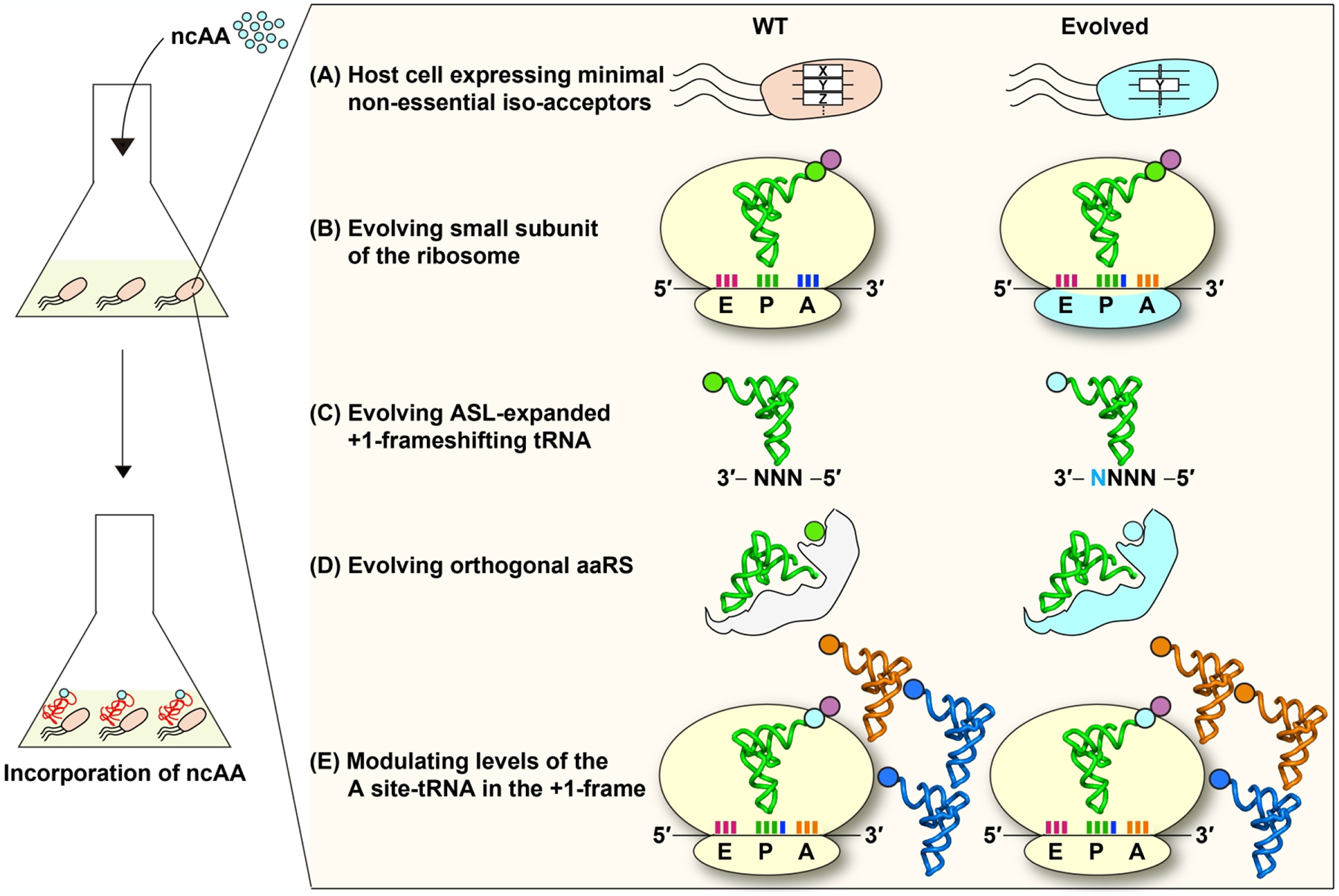
A framework for genome expansion by tRNA +1 frameshifting at quadruplet codons. For improved incorporation of the ncAA to protein synthesis, components should include (A) an evolved host cell expressing a minimal set of non-essential iso-acceptors (e.g., the X or Z gene), leaving only the essential iso-acceptor of the tRNA for the amino acid of interest; (B) an evolved ribosomal small subunit; (C) an evolved ASL-expanded +1 frameshifting tRNA; (D) an evolved orthogonal aaRS that charges the ncAA to the tRNA; and (E) increased levels of the tRNA occupying the +1-frame of the A site downstream from the quadruplet codon. Evolved components are shown in cyan.
Engineering of the host cell.
The reading of a quadruplet codon in a cell model, whether by triplet slippage or by quadruplet pairing, inevitably encounters competition between the +1-frameshifting tRNA of interest and the natural iso-acceptors that would read a triplet codon within the quadruplet codon motif. This competition is one reason why the +1-frameshifting efficiency of SufB2 in a cell model, where competition exists, is consistently reduced relative to a cell-free model, where competition does not exist [61]. Thus, we should consider engineering of a host cell model that lacks some or all of the non-essential iso-acceptors (Figure 7A). This host cell model would provide an environment that is dedicated to perform genome expansion at quadruplet codons. While the elimination of some or all of the non-essential iso-acceptors might compromise cell growth, it would improve the efficiency of decoding quadruplet codons. Notably, genome engineering to minimize competition is feasible. For example, an E. coli strain lacking RF1 has been generated [12], which eliminates the competition between RF1 and a designer tRNA for reading of the UAG stop codon, thus enabling the designer tRNA to read UAG solely as a sense codon. Similarly, recent work has also constructed E. coli strains lacking a non-essential iso-acceptor tRNA to examine the structure-activity relationship of the essential iso-acceptor [101, 102].
Engineering of the host ribosome.
While initial success in genome recoding has been achieved by engineering the anticodon-codon interactions of a +1-frameshifting tRNA at the A site [12, 38], efforts to engineer the structural elements of the host ribosome should be as, or even more, effective (Figure 7B). Such ribosome engineering for quadruplet decoding has begun with promises [39, 103]. Here we suggest that engineering should focus on the 30S head swiveling and include the structural elements that regulate translocation or those that regulate tRNA stability in the P site. This can be achieved by multiplex screening for 30S variants that exhibit high efficiencies of +1-frameshifting with +1-frameshifting tRNAs at quadruplet codons while preserving 0-frame translation by canonical tRNAs at triplet codons.
Engineering of the designer tRNA.
Between the choice of an ASL-expanded tRNA and a canonical tRNA, a higher priority should be placed for the ASL-expanded tRNA (Figure 7C). In general, the yield of +1 frameshifting is much higher for an ASL-expanded tRNA relative to its canonical counterpart, such as SufB2 relative to ProL [60]. A recent study of multiplex screening of ASL-expanded tRNAs has generated promising results [10]. Another recent study with an ASL-expanded tRNA has expanded the genetic code in the animal model of the nematode worm (Caenorhabditis elegans) [104]. Thus, continued structural work to understand how the ribosome differentiates an ASL-expanded-tRNA from a canonical tRNA would be necessary to provide a structure-based roadmap for engineering the ASL-expanded tRNA. Notably, an obvious rule of the designer tRNA is to permit base pairing between the wobble nucleotide at position 34 of the anticodon and the 4th nucleotide of the quadruplet codon motif. This is best observed in the pairing between the mcmo5U34 wobble nucleotide of the ProM tRNA and the A4 nucleotide of the CCC-A codon motif in cryo-EM structures (Figure 5D). Additionally, to minimize the effect of post-transcriptional modifications at position 37, efforts should be made to generate designer ASL-expanded tRNAs with an unmodified pyrimidine. This will require in parallel the engineering of an orthologous aaRS to pair with the designer tRNA (Figure 7D). Previous work has demonstrated the feasibility of engineering of an aaRS to pair with a mutant tRNA [105].
Targeting the elongation step that permits +1 frameshifting.
Finally, between the two steps in an elongation cycle of protein synthesis that permit +1 frameshifting, the manipulation of the P-site shift would be an easier step (Figure 7E). This can be readily achieved by increasing cellular levels of the tRNAs that would occupy the +1-frame of the A site, as has been validated by previous genetic studies for various tRNAs [59, 89–92] and by our cell-based study of SufB2 [61]. To increase the cellular level of a specific tRNA, we have developed a plasmid-based over-expression system that drives the transcription of the tRNA gene in E. coli. Transcription is driven by a strong promoter that is inducible to avoid toxicity of over-expression. Biochemical assays have confirmed the presence of selected post-transcriptional modifications in tRNAs that have been over-expressed in this cell model, including m1G37 [60, 102], mcmo5U34 [101], and dihydrouridine D17 [106]. The confirmation of the expected post-transcriptional modifications at positions 34 and 37 is important, both of which are determinants of the reading frame. A similar over-expression system can be designed for other bacterial species and for eukaryotes.
Summary.
We have used the Pro-specific ProM, ProL, and SufB2 tRNAs as examples to develop new perspectives that should improve the yield of +1 frameshifting at quadruplet codons (Table 1). These new perspectives are likely applicable to other tRNAs, although specific constraints may exist. Combining these perspectives in an engineered cell model should lead to increased success in genome expansion.
Research Highlights.
Quadruplet decoding requires a +1-frameshifting event of the quadruplet-reading tRNA.
No +1 frameshifting occurs in the ribosomal A site.
The ribosome permits +1 frameshifting during translocation and P-site occupancy.
Acknowledgements
This work was supported by Research Grant R35GM134931 to Y.M.H. from the National Institutes of General Medical Sciences (NIGMS). We thank Dr. Yuko Nakano and Dr. Jinwei Zhang for figures and discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CrediT Author Statement
Howard Gamper: Conceptualization, Writing – Review and Editing; Isao Masuda: Conceptualization, Writing – Review and Editing; Ya-Ming Hou: Conceptualization, Writing – Original Draft, Writing – Review and Editing, Supervision
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Wang K, Schmied WH, Chin JW. Reprogramming the genetic code: from triplet to quadruplet codes. Angew Chem Int Ed Engl. 2012;51:2288–97. [DOI] [PubMed] [Google Scholar]
- [2].Turanov AA, Xu XM, Carlson BA, Yoo MH, Gladyshev VN, Hatfield DL. Biosynthesis of selenocysteine, the 21st amino acid in the genetic code, and a novel pathway for cysteine biosynthesis. Adv Nutr. 2011;2:122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Brugere JF, Atkins JF, O’Toole PW, Borrel G. Pyrrolysine in archaea: a 22nd amino acid encoded through a genetic code expansion. Emerg Top Life Sci. 2018;2:607–18. [DOI] [PubMed] [Google Scholar]
- [4].Kato Y Translational Control using an Expanded Genetic Code. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Celik A, He F, Jacobson A. NMD monitors translational fidelity 24/7. Curr Genet. 2017;63:1007–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nasif S, Contu L, Muhlemann O. Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin Cell Dev Biol. 2018;75:78–87. [DOI] [PubMed] [Google Scholar]
- [7].Moore B, Persson BC, Nelson CC, Gesteland RF, Atkins JF. Quadruplet codons: implications for code expansion and the specification of translation step size. J Mol Biol. 2000;298:195–209. [DOI] [PubMed] [Google Scholar]
- [8].Wang K, Sachdeva A, Cox DJ, Wilf NM, Lang K, Wallace S, et al. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat Chem. 2014;6:393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hankore ED, Zhang L, Chen Y, Liu K, Niu W, Guo J. Genetic Incorporation of Noncanonical Amino Acids Using Two Mutually Orthogonal Quadruplet Codons. ACS Synth Biol. 2019;8:1168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].DeBenedictis EA, Carver GD, Chung CZ, Soll D, Badran AH. Multiplex suppression of four quadruplet codons via tRNA directed evolution. Nat Commun. 2021;12:5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang C, Talukder P, Dedkova LM, Hecht SM. Facilitated synthesis of proteins containing modified dipeptides. Bioorg Med Chem. 2021;41:116210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lajoie MJ, Rovner AJ, Goodman DB, Aerni HR, Haimovich AD, Kuznetsov G, et al. Genomically recoded organisms expand biological functions. Science. 2013;342:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Thyer R, Ellington AD. The Role of tRNA in Establishing New Genetic Codes. Biochemistry. 2019;58:1460–3. [DOI] [PubMed] [Google Scholar]
- [15].Stahl G, McCarty GP, Farabaugh PJ. Ribosome structure: revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem Sci. 2002;27:178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Atkins JF, Bjork GR. A gripping tale of ribosomal frameshifting: extragenic suppressors of frameshift mutations spotlight P-site realignment. Microbiol Mol Biol Rev. 2009;73:178–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dinman JD. Translational recoding signals: Expanding the synthetic biology toolbox. J Biol Chem. 2019;294:7537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 2016;44:7007–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Atkins JF. Culmination of a half-century quest reveals insight into mutant tRNA-mediated frameshifting after tRNA departure from the decoding site. Proc Natl Acad Sci U S A. 2018;115:11121–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Guo J, Niu W. Genetic Code Expansion Through Quadruplet Codon Decoding. J Mol Biol. 2021:167346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Malyshev DA, Dhami K, Lavergne T, Chen T, Dai N, Foster JM, et al. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014;509:385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hoshika S, Leal NA, Kim MJ, Kim MS, Karalkar NB, Kim HJ, et al. Hachimoji DNA and RNA: A genetic system with eight building blocks. Science. 2019;363:884–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fredens J, Wang K, de la Torre D, Funke LFH, Robertson WE, Christova Y, et al. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019;569:514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wang K, de la Torre D, Robertson WE, Chin JW. Programmed chromosome fission and fusion enable precise large-scale genome rearrangement and assembly. Science. 2019;365:922–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yourno J, Heath S. Nature of the hisD3018 frameshift mutation in Salmonella typhimurium. J Bacteriol. 1969;100:460–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yourno J, Kohno T. Externally suppressible proline quadruplet ccc U. Science. 1972;175:650–2. [DOI] [PubMed] [Google Scholar]
- [27].Riddle DL, Roth JR. Suppressors of frameshift mutations in Salmonella typhimurium. J Mol Biol. 1970;54:131–44. [DOI] [PubMed] [Google Scholar]
- [28].Riddle DL, Roth JR. Frameshift suppressors. 3. Effects of suppressor mutations on transfer RNA. J Mol Biol. 1972;66:495–506. [DOI] [PubMed] [Google Scholar]
- [29].Jorgensen F, Kurland CG. Processivity errors of gene expression in Escherichia coli. J Mol Biol. 1990;215:511–21. [DOI] [PubMed] [Google Scholar]
- [30].Sakai Y, Miyauchi K, Kimura S, Suzuki T. Biogenesis and growth phase-dependent alteration of 5-methoxycarbonylmethoxyuridine in tRNA anticodons. Nucleic Acids Res. 2016;44:509–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nasvall SJ, Chen P, Bjork GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sroga GE, Nemoto F, Kuchino Y, Bjork GR. Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNA(Pro)2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res. 1992;20:3463–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Roth JR. Frameshift suppression. Cell. 1981;24:601–2. [DOI] [PubMed] [Google Scholar]
- [34].O’Connor M Insertions in the anticodon loop of tRNA1Gln(sufG) and tRNA(Lys) promote quadruplet decoding of CAAA. Nucleic Acids Res. 2002;30:1985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee BS, Kim S, Ko BJ, Yoo TH. An efficient system for incorporation of unnatural amino acids in response to the four-base codon AGGA in Escherichia coli. Biochim Biophys Acta. 2017;1861:3016–23. [DOI] [PubMed] [Google Scholar]
- [36].Chatterjee A, Lajoie MJ, Xiao H, Church GM, Schultz PG. A bacterial strain with a unique quadruplet codon specifying non-native amino acids. Chembiochem. 2014;15:1782–6. [DOI] [PubMed] [Google Scholar]
- [37].Niu W, Schultz PG, Guo J. An expanded genetic code in mammalian cells with a functional quadruplet codon. ACS Chem Biol. 2013;8:1640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang N, Shang X, Cerny R, Niu W, Guo J. Systematic Evolution and Study of UAGN Decoding tRNAs in a Genomically Recoded Bacteria. Sci Rep. 2016;6:21898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Neumann H, Wang K, Davis L, Garcia-Alai M, Chin JW. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–4. [DOI] [PubMed] [Google Scholar]
- [40].Magliery TJ, Anderson JC, Schultz PG. Expanding the genetic code: selection of efficient suppressors of four-base codons and identification of “shifty” four-base codons with a library approach in Escherichia coli. J Mol Biol. 2001;307:755–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Anderson JC, Wu N, Santoro SW, Lakshman V, King DS, Schultz PG. An expanded genetic code with a functional quadruplet codon. Proc Natl Acad Sci U S A. 2004;101:7566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hohsaka T, Ashizuka Y, Taira H, Murakami H, Sisido M. Incorporation of nonnatural amino acids into proteins by using various four-base codons in an Escherichia coli in vitro translation system. Biochemistry. 2001;40:11060–4. [DOI] [PubMed] [Google Scholar]
- [43].Anderson JC, Magliery TJ, Schultz PG. Exploring the limits of codon and anticodon size. Chem Biol. 2002;9:237–44. [DOI] [PubMed] [Google Scholar]
- [44].Fekner T, Chan MK. The pyrrolysine translational machinery as a genetic-code expansion tool. Curr Opin Chem Biol. 2011;15:387–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wan W, Tharp JM, Liu WR. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta. 2014;1844:1059–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou YM. The UGG Isoacceptor of tRNAPro Is Naturally Prone to Frameshifts. Int J Mol Sci. 2015;16:14866–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Qian Q, Li JN, Zhao H, Hagervall TG, Farabaugh PJ, Bjork GR. A new model for phenotypic suppression of frameshift mutations by mutant tRNAs. Mol Cell. 1998;1:471–82. [DOI] [PubMed] [Google Scholar]
- [48].Weiss RB, Dunn DM, Shuh M, Atkins JF, Gesteland RF. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989;1:159–69. [PubMed] [Google Scholar]
- [49].Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb Symp Quant Biol. 1987;52:687–93. [DOI] [PubMed] [Google Scholar]
- [50].Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell. 2004;118:45–55. [DOI] [PubMed] [Google Scholar]
- [51].Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops. Proc Natl Acad Sci U S A. 2014;111:12740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hong S, Sunita S, Maehigashi T, Hoffer ED, Dunkle JA, Dunham CM. Mechanism of tRNA-mediated +1 ribosomal frameshifting. Proc Natl Acad Sci U S A. 2018;115:11226–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nguyen HA, Hoffer ED, Dunham CM. Importance of a tRNA anticodon loop modification and a conserved, noncanonical anticodon stem pairing in tRNACGGProfor decoding. J Biol Chem. 2019;294:5281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dunham CM, Selmer M, Phelps SS, Kelley AC, Suzuki T, Joseph S, et al. Structures of tRNAs with an expanded anticodon loop in the decoding center of the 30S ribosomal subunit. RNA. 2007;13:817–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hoffer ED, Hong S, Sunita S, Maehigashi T, Gonzalez RLJ, Whitford PC, et al. Structural insights into mRNA reading frame regulation by tRNA modification and slippery codon-anticodon pairing. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fagan CE, Maehigashi T, Dunkle JA, Miles SJ, Dunham CM. Structural insights into translational recoding by frameshift suppressor tRNASufJ. RNA. 2014;20:1944–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Bossi L, Smith DM. Suppressor sufJ: a novel type of tRNA mutant that induces translational frameshifting. Proc Natl Acad Sci U S A. 1984;81:6105–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Bossi L, Roth JR. Four-base codons ACCA, ACCU and ACCC are recognized by frameshift suppressor sufJ. Cell. 1981;25:489–96. [DOI] [PubMed] [Google Scholar]
- [59].Jager G, Nilsson K, Bjork GR. The phenotype of many independently isolated +1 frameshift suppressor mutants supports a pivotal role of the P-site in reading frame maintenance. PLoS One. 2013;8:e60246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gamper H, Li H, Masuda I, Miklos Robkis D, Christian T, Conn AB, et al. Insights into genome recoding from the mechanism of a classic +1-frameshifting tRNA. Nat Commun. 2021;12:328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gamper H, Mao Y, Masuda I, McGuigan H, Blaha G, Wang Y, et al. Twice exploration of tRNA +1 frameshifting in an elongation cycle of protein synthesis. Nucleic Acids Res. 2021;49:10046–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ogle JM, Brodersen DE, Clemons WM Jr., Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. [DOI] [PubMed] [Google Scholar]
- [63].Ogle JM, Ramakrishnan V. Structural insights into translational fidelity. Annu Rev Biochem. 2005;74:129–77. [DOI] [PubMed] [Google Scholar]
- [64].Gamper HB, Masuda I, Frenkel-Morgenstern M, Hou YM. Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat Commun. 2015;6:7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fei J, Bronson JE, Hofman JM, Srinivas RL, Wiggins CH, Gonzalez RL Jr. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A. 2009;106:15702–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fei J, Kosuri P, MacDougall DD, Gonzalez RL Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–59. [DOI] [PubMed] [Google Scholar]
- [67].Ning W, Fei J, Gonzalez RL, Jr. The ribosome uses cooperative conformational changes to maximize and regulate the efficiency of translation. Proc Natl Acad Sci U S A. 2014;111:12073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Khade PK, Joseph S. Messenger RNA interactions in the decoding center control the rate of translocation. Nat Struct Mol Biol. 2011;18:1300–2. [DOI] [PubMed] [Google Scholar]
- [70].Liu G, Song G, Zhang D, Zhang D, Li Z, Lyu Z, et al. EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon-anticodon duplex. Nat Struct Mol Biol. 2014;21:817–24. [DOI] [PubMed] [Google Scholar]
- [71].Abeyrathne PD, Koh CS, Grant T, Grigorieff N, Korostelev AA. Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome. Elife. 2016;5:doi: 10.7554/eLife.14874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Pulk A, Cate JH. Control of ribosomal subunit rotation by elongation factor G. Science. 2013;340:1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Ratje AH, Loerke J, Mikolajka A, Brunner M, Hildebrand PW, Starosta AL, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–34. [DOI] [PubMed] [Google Scholar]
- [75].Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–40. [DOI] [PubMed] [Google Scholar]
- [76].Ermolenko DN, Noller HF. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol. 2011;18:457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A. 2009;106:2571–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Guo Z, Noller HF. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci U S A. 2012;109:20391–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Demo G, Gamper HB, Loveland AB, Masuda I, Carbone CE, Svidritskiy E, et al. Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation. Nat Commun. 2021;12:4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. [DOI] [PubMed] [Google Scholar]
- [81].Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–9. [DOI] [PubMed] [Google Scholar]
- [82].Loveland AB, Demo G, Grigorieff N, Korostelev AA. Ensemble cryo-EM elucidates the mechanism of translation fidelity. Nature. 2017;546:113–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Loveland AB, Demo G, Korostelev AA. Cryo-EM of elongating ribosome with EF-Tu*GTP elucidates tRNA proofreading. Nature. 2020;584:640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ramrath DJ, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, et al. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci U S A. 2013;110:20964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. Role of domains 4 and 5 in elongation factor G functions on the ribosome. J Mol Biol. 2000;300:951–61. [DOI] [PubMed] [Google Scholar]
- [86].Zhou J, Lancaster L, Donohue JP, Noller HF. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Zhou J, Lancaster L, Donohue JP, Noller HF. Spontaneous ribosomal translocation of mRNA and tRNAs into a chimeric hybrid state. Proc Natl Acad Sci U S A. 2019;116:7813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science. 2009;326:694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Pande S, Vimaladithan A, Zhao H, Farabaugh PJ. Pulling the ribosome out of frame by +1 at a programmed frameshift site by cognate binding of aminoacyl-tRNA. Mol Cell Biol. 1995;15:298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Caulfield T, Coban M, Tek A, Flores SC. Molecular Dynamics Simulations Suggest a Non-Doublet Decoding Model of −1 Frameshifting by tRNA(Ser3). Biomolecules. 2019;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Atkins JF, Gesteland RF, Reid BR, Anderson CW. Normal tRNAs promote ribosomal frameshifting. Cell. 1979;18:1119–31. [DOI] [PubMed] [Google Scholar]
- [92].Curran JF, Yarus M. Reading frame selection and transfer RNA anticodon loop stacking. Science. 1987;238:1545–50. [DOI] [PubMed] [Google Scholar]
- [93].Blaha G, Stanley RE, Steitz TA. Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome. Science. 2009;325:966–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Katoh T, Wohlgemuth I, Nagano M, Rodnina MV, Suga H. Essential structural elements in tRNA(Pro) for EF-P-mediated alleviation of translation stalling. Nat Commun. 2016;7:11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Doerfel LK, Wohlgemuth I, Kothe C, Peske F, Urlaub H, Rodnina MV. EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science. 2013;339:85–8. [DOI] [PubMed] [Google Scholar]
- [96].Ude S, Lassak J, Starosta AL, Kraxenberger T, Wilson DN, Jung K. Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science. 2013;339:82–5. [DOI] [PubMed] [Google Scholar]
- [97].Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J. 2001;20:231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Caliskan N, Katunin VI, Belardinelli R, Peske F, Rodnina MV. Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell. 2014;157:1619–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Chen J, Petrov A, Johansson M, Tsai A, O’Leary SE, Puglisi JD. Dynamic pathways of −1 translational frameshifting. Nature. 2014;512:328–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Mohan S, Donohue JP, Noller HF. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci U S A. 2014;111:13325–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Masuda I, Takase R, Matsubara R, Paulines MJ, Gamper H, Limbach PA, et al. Selective terminal methylation of a tRNA wobble base. Nucleic Acids Res. 2018;46:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Masuda I, Matsubara R, Christian T, Rojas ER, Yadavalli SS, Zhang L, et al. tRNA Methylation Is a Global Determinant of Bacterial Multi-drug Resistance. Cell Syst. 2019;8:302–14 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wang K, Neumann H, Peak-Chew SY, Chin JW. Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat Biotechnol. 2007;25:770–7. [DOI] [PubMed] [Google Scholar]
- [104].Xi Z, Davis L, Baxter K, Tynan A, Goutou A, Greiss S. Using a quadruplet codon to expand the genetic code of an animal. Nucleic Acids Res. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Salazar JC, Ahel I, Orellana O, Tumbula-Hansen D, Krieger R, Daniels L, et al. Coevolution of an aminoacyl-tRNA synthetase with its tRNA substrates. Proc Natl Acad Sci U S A. 2003;100:13863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Betteridge T, Liu H, Gamper H, Kirillov S, Cooperman BS, Hou YM. Fluorescent labeling of tRNAs for dynamics experiments. RNA. 2007;13:1594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]


