Abstract
Background:
Medullary Thyroid Cancer (MTC) is a very aggressive type of thyroid carcinoma. Mutation in RET proto-oncogene is demonstrated in MTC development. We aimed to knock-out of RET-oncogene using CRISPR/Cas9 genome editing method in MTC cell-lines.
Methods:
This research was conducted in Shahid Beheshti University of Medical Sciences, Tehran, Iran during 2019–2020. Four different sgRNAs were designed to target exons one, two, and four of RET-oncogene in TT and MZ-CRC-1 cell-lines using bioinformatics tools, then the CRISPR/Cas9 constructs was made. About 72-hours after cell transfection, T7EI method and DNA sequencing were used to confirm the knock-out of RET-oncogene. Expression of RET, Calcitonin genes and RET protein were evaluated by Real-time PCR and ELISA, respectively.
Results:
The results of T7E1, and DNA sequencing of transfected cells confirmed RET gene knock-out by CRISPR/Cas9. There was a significant decrease in RET gene expression and RET protein in transfected TT and MZ cells compared to controls. The rate of cell apoptosis in transfected cells was significantly increased. Calcitonin gene expression was also significantly reduced in transfected cells. p-RET, p-PI3K, p-AKT, p-MEK, p-ERK protein levels were significantly reduced in TT and MZ transfected cells.
Conclusion:
For the first time, knock-out of RET gene was performed and confirmed using CRISPR/Cas9. Inhibition of this gene leads to inhibition of the tyrosine kinase RET signal transduction pathway. Therefore, it can be one of the most effective and specific therapeutic goals in the field of Personalized Medicine in the treatment of diseases caused by over activity of RET molecular pathway.
Keywords: CRISPR/Cas9, Gene editing, Medullary thyroid cancer, RET gene, TT cell-line, MZ-CRC-1 cell-line, Iran
Introduction
Cancer is one of the most important causes of death worldwide and has a great social and economic impact on communities. According to the Global Cancer Observatory (GCO) 2020, the number of new cancer cases was 19,292,789 (9,227,487 females, 10,065,305 males) in the world. Also, the worldwide cancer mortality rate in 2020 was 9,958,133 cases (4,429,323females, 5,525,810males). “The most common types of cancer in men were lung, prostate, colorectal, stomach and liver, respectively” (1). The most common types of cancer in women were breast, colorectal, lung, cervix, and thyroid, respectively in 2020 (2). According to the GCO 2020 (3), the total number of new cases of various types of cancer in Iran was 131,191 (60,487females, 70,704males). In addition, 79,136 cancer deaths have been reported in Iran during 2020 (32,700 females, 46,436 males). The most common types of cancer in men were lung, prostate, colorectal, stomach, and liver. Moreover, breast, colorectal, stomach, lung, and thyroid were the most cancer in women (3).
Thyroid cancer is the most common malignancy of the endocrine system (4). It is divided into two types based on the origin of cells (follicular and C-cells/parafollicular cells). Follicular cell-derived thyroid cancers include papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), heart cell carcinoma, and anaplastic thyroid cancer (5), which are the most common thyroid malignancies (1). Parafollicular cell-derived thyroid cancer leads to medullary thyroid cancer (MTC) accounting for about five to ten percent of all types of thyroid cancer (6). MTC is a very invasive type of thyroid cancer. The gain of function mutation in RET proto-oncogene in the development of this disease is well known (7). RET proto-oncogene is a cadherin-like superfamily and encodes a membrane tyrosine kinase whose role is growth and differentiation signal transduction into the cells and plays a key role in neural crest, kidney, urinary tract and intestinal nerve development (8).
RET mutations have been observed in Multiple endocrine neoplasia, type 2A (MEN2A) (98%), Multiple endocrine neoplasia type 2B (MEN2B) (95%), Familial MTC (FMTC) (88%) cases (9). In sporadic MTC, up to 40%–60% of patients have somatic mutations in RET (9, 10). Moreover, 100% of Iranian hereditary and 80% of sporadic MTCs had RET mutations (11–15).
New targeted molecular therapies are now used for cancer therapy, such as tyrosine kinase inhibitors. However, these drugs still have their side effects and toxicities. They are not specific completely, and lead to the development of drug resistance mechanisms. In addition, they are costly and have a temporary effect. Vandetanib, which inhibits RET, VEGFR3, and EGFR receptors (16, 17), is effective only in MTC patients with the M918T mutation, however other types of MTC that have other RET mutations (V804L and V804M), are resistant to vandetanib. On the other hand, due to the side effects of kinase inhibitors, these drugs are used only in advanced disease cases (18, 19).
Gene therapy over the past few decades has focused on restoring defective gene function. However, even after successful gene transfer, disease-causing mutations persisted that could cause a dominant negative effect. Therefore, the best solution was to modify the mutation, or knockout the disease-causing gene (1). “Recently, clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein-9 (CRISPR/Cas9) system has emerged as a powerful technology for cancer treatment due to its high accuracy and efficiency. The new CRISPR/Cas9 gene editing method, which also won the Nobel Prize in Chemistry in 2020 (20) has become a powerful strategy for modifying organisms’ genomes and a powerful tool in tumor treatment” (21). It has made “gene knockout” processes more efficient, cost-effective and easier than before (21).
In the present study, we aimed to target and to make knockout RET gene by CRISPR/Cas9 technique in HEK-293 cell-line and MTC cell-lines TT, MZ-CRC-1.
Materials and Methods
This research was conducted in Shahid Beheshti University of Medical Sciences, Tehran, Iran during 2019–2020.
Thyroid Carcinoma Cell-lines and Cell Culture
The human medullary thyroid carcinoma cell-lines MZ-CRC1 and TT were used for cell-based assays. These two cell-lines were given as gift by Professor Robert Hofstra (the Head of Genetic Department, Erasmus University). TT-cell (MEN2A) line contains a pathogenic RET C634W mutation, MZCRC-1 (MEN2B) contains RET M918T pathogenic mutation. HEK-293 (human embryonic kidney cell-line) was purchased from National Cell Bank of Iran (Pasteur Institute of Iran, Tehran, Iran).
TT and MZ-CRC cells were cultured in complete medium DMEM/F-12 (Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12, Gibco) supplemented with 20%fetal bovine serum (FBS;Gibco). HEK-293 cell-line was cultured in RPMI medium with 10% FBS, 1% penicillin/streptomycin at 37 °C and 5% CO2 in fully humidified incubator.
Plasmid construction
The plasmids used in this study, pSpCas9(22)-2AGFP (PX458), expression vector Cas9 from S. pyogenes with 2A-EGFP, and cloning backbone for sgRNA was a gift from Tokameh Mahmoudi, Department of Biochemistry, Erasmus MC (AddgenePlasmid #48138).
Designing and cloning of sgRNAs
Single-guide RNAs for RET knockout were designed using the online CRISPR design tools web-based software such as CRISPERA, CHOP-CHOP, ECRISP, CRISPOR, CASOFFINDER.
Four SpCas9- sgRNA [5’-20nt-NGG PAM] for RET target region (exons one, two, and four) were designed and chosen based on specificity and efficiency score. The SpCas9-sgRNAs were begun with an extra G at the 5′end, the preferred start nucleotide for the U6 promoter. The synthetic complementary oligonucleotides encoding the gene-specific sgRNAs were annealed to produce short oligonucleotide duplexes fragments with 4bp overhangs. Double-strand Oligonucleotides encoding the sgRNAs were annealed and cloned into the BbsI-digested pSpCas9 (22)-2A-GFP (PX458). All sgRNAs preceded by the canonical trinucleotide 5′-NGG, the protospacer adjacent motif (1). To minimize the off-target mutations, the DNA target site should entirely match the PAM motif and the 12bp seed sequence closest to the PAM. After retrieving the sequence data of RET (NG_007489.1RefSeqGene) and designing four sgRNAs, the related constructs to express sgRNAs of the CRISPR/Cas9 nuclease are related, were generated. In this study, sequences that contained the PAM motif inside exon 1, 2, 4 of RET were identified. Then, to determine whether or not candidate sequences were unique in the genome, they were analyzed by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and Bioinformatics tools such as Cas-Offinder were used to find likely off-target sequences.
The ligation products were transformed into TOP10 bacterial cells using CaCl2 method. Colony PCR identified positive clones and confirmed by Sanger sequencing using U6 and Tracer primers.
Cell Transfection
CRISPR/Cas9 RET-KO-Plasmids, and Control CRISPR/Cas9 Plasmid (Scramble) were transfected into HEK-293, TT, and MZ-CRC-1 cell-lines. Briefly, HEK-293, TT, and MZ-CRC-1 cells were seeded into a 6-well plate. After that, the cells reached 70%–90% confluency. The culture media were removed. The cells were washed with PBS, and transfection was carried out using Lipofectamine® 3000 transfection reagent (Invitrogen, USA) to manufacturers’ protocol. Non-transfected cells and transfected cells with an equal amount of the pSpCas9 (22)-2A-GFP (PX458) without sgRNA were considered as negative controls. Seventy-two hours after transfection, successful transfection of the CRISPR plasmids was visually verified under florescent microscope. Because the vector used in this study was GFP+, the vector cells were seen as green fluorescent.
Transformation optimization was performed until its efficiency reached over 50%. Transfection efficiency was also assessed using a flow cytometer (BD FACS Calibur, BD biosciences, San-Jose, CA, USA). Besides, BD FACS Aria II Cell Sorter was used to sort GFP+ cells. After cell sorting, other experiments, including T7 Endonuclease Assay, preparation of a single clone with serial dilution of cells, and re-culture of the sorted cells were performed on the GFP+ cells. Preparation of single clone after sorting GFP+ cells was performed in two ways: by BD FACS Aria II, which sorted GFP+ cells as single cells in all 96-well plate wells. The second was serial dilution, which was done manually.
Surveyor Mutation Detection Assay, T7 Endonuclease I assay
CRISPR-Cas9 editing efficiency with the different sgRNAs was assessed by T7EI (New England BioLabs) on HEK-293, TT, and MZ-CRC-1 cells. Non-transfected cells (WT) and non-targeting sgRNA (control sgRNA) were used as negative controls.
Seventy-two hours after transfection, genomic DNA (gDNA) was extracted using salting/out Proteinase-K- method, then gDNA was amplified by target PCR primer pairs flanking the sgRNA targeted region. The PCR products were then electrophoresed on 2% agarose gel and PCR products were extracted from the gel. To perform the T7EI test, the Heteroduplex DNA formation reaction must first be performed between the edited and normal DNA. For this purpose, 400 ng of PCR product extracted from the gel was poured into each tube. Then 2μl of enzyme buffer was added and made-up of 19μl with sterile distilled water. Then, according to the Nature Protocols for T7E1 (23) DNA Heteroduplex formation was performed. After heteroduplex construction, 10units of T7EI enzyme (Bio Labs, Cat:M0302L) were added to each tube and incubated at 37 °C for 15 minutes. If a heteroduplex structure is formed between normal and mutant DNA, T7E1 cuts the double-stranded DNA. The samples were run on 4%–20% gradient polyacrylamide gel and was visualized by the silver nitrate staining method. After confirming enzymatic cleavage, a single clone was prepared using TA cloning (Bio Basic, Cat:BS435-20) from T7EI+ PCR products to sequence the edited regions.
RNA Extraction and RT-qPCR Assay for Evaluation of RET expression
Seventy-two hours after transfection, total RNA was extracted from the studied cell-lines using the RNX-Plus kit according to its instruction (Cat. No. RN7713C, Sina Clone-Company, Iran). Thermo Scientific kit (Pub. No. MAN0012757, USA) was used for cDNA synthesis.
Quantitative Real-Time PCR (RT-qPCR) assay was carried out (Ampliqon, Denmark) using Rotor-Gene 6000 instrument (Corbett Research, Sydney, Australia).
The sequences of the primer sets were used, including RET-F:5′TGAAGATGCTGAAAGAGAACG3′; RET-R:5′CCATACAATTTGATGACATGTGG3′; CALCITONINF:5′ACAAGTTTCACACGTTCCCC3′;CALCITONINR:5′CATTCTGGGGCATGCTAACA3′,GAPDHF:5′AGGCAGGGATGATGTTCTGG3′;GAPDHR:5′ TCGTGGAAGGACTCATGACC3′.The cycle conditions used for all genes consist of an initial step of 95 °C for 15 min, followed by 40 cycles of 95 °C (5sec); 54°C (30sec), 72 °C (30sec). Relative quantification (24) of gene expression was calculated using the comparative Ct method and the equation 2−ΔΔCt after normalization of the mRNA levels of the target gene with GAPDH as an endogenous housekeeping gene.
Enzyme Linked Immunosorbent Assay (ELISA)
Expression of RET at the protein level was also measured by ELISA kit (Zell Bio, Germany) as instructed by the manufacturers. Moreover, to evaluate the number of phosphorylated proteins RET, PI3K, AKT, MEK, and ERK in cell-lines treated with sgRNAs, ELISA was used. Proteins were extracted from the studied cells, using the MPERTM Mammalian Protein extraction reagent kit (Thermo Scientific, cat.78501) according to the manufacturer’s instructions with adding anti-protease cocktail (ProBlock-50, Cat#GB-326-1, Zell Bio, Germany). The protein concentrations were calculated according to the standard curves created using standard samples provided by the kit.
Apoptosis and Necrosis Assay
To examine the effect of RET knockout using CRISPR/Cas9 on apoptosis induction, apoptosis was analyzed using PE Annexin V Apoptosis Detection Kit with 7-AAD in GFP+ cells (treated with sgRNA) and FITC Annexin V Apoptosis Detection Kit with PI was also used to evaluate cell apoptosis in untreated cells (without GFP) according to the manufacturer’s instructions. Briefly, 72 hours after transfection, cells were harvested and resuspended in 500μl 1× Binding buffer. Next, 100 μL of the cell suspension (105 cells) was transferred to a 5-mL culture tube and incubated with 5μL of FITC Annexin V or PE-V Annexin and 5μL of propidium iodide (PI) for 15 min at 4 °C in the dark. The samples were subjected to fluorescence-activated cell sorting (FACS) using a BD FACS Calibur (BD-biocsencies, SanJose, CA, USA). FACS data were analyzed using Flow Jo software.
Statistical analysis
The normality of the quantitative data was evaluated using Shapiro wilk test. To compare the mean, for two groups of normal data, Student’s t-test and for more than two groups, one-way ANOVA test were used. Comparison before and after treatment paired t-test was used. Paired t-test was used to compare before and after cell treatment. Data analysis was performed using SPSS (ver. 25 (IBM Corp., Armonk, NY, USA), Graph pad Prism (ver. 8), and Med Calc (ver. 1–8) software. In all tests, statistical significance was defined as P≤0.05.
Results
Optimizing Cas9/SgRNAs Delivery of MTC Cell-lines
We aimed to optimize conditions for the co-transfection of Cas9/sgRNA expression plasmids MTC Cell-lines via lipofectamine reagent. For this purpose, we used reporter plasmid with GFP as a reporter gene in Cas9 and scramble vectors. As shown in Fig.1, fluorescence micrographs of GFP-transfected cells indicated that lipofectamine 3000 is an appropriate reagent for delivering Cas9 or scramble in MTC Cell-lines.
Fig. 1:
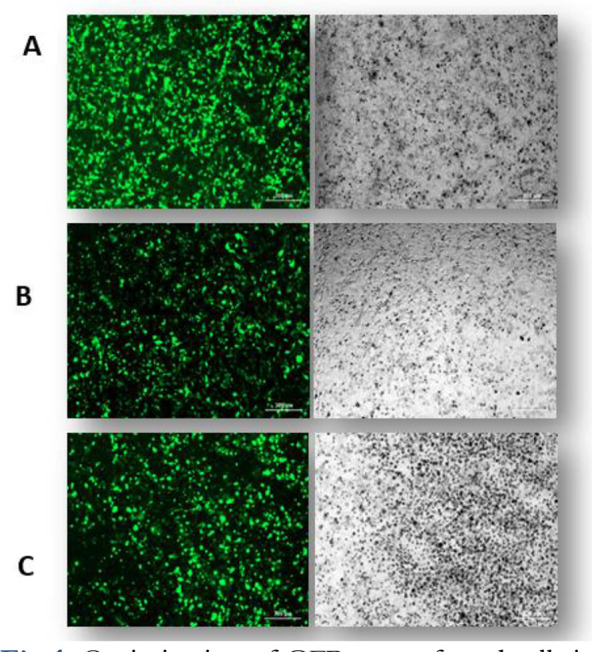
Optimization of GFP+ transfected cells in HEK-293 (1-A), TT (1-B), MZ-CRC1 (1-C) cell lines (fluorescent microscopy).
Verification of Cleavage activity of sgRNAs Targeting RET Gene in Cultured Cells
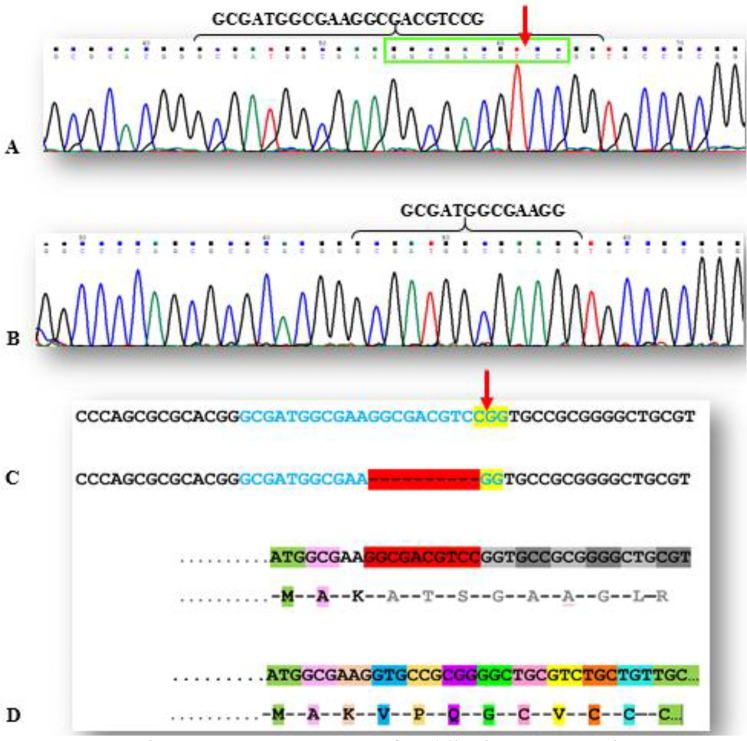
To characterize the functional effect of Cas9 nuclease and cleavage activity of sgRNAs targeting RET, we used the T7E1 assay and direct Sanger sequencing of PCR amplicons. Genomic DNA was extracted from transfected cells and screened for the presence of site-specific gene modification by PCR and T7E1 assay. As shown in Fig.2, both of the duplex DNA strands were catalyzed in the PAM sequence (for Exon one). Moreover, our data show that PCR amplicon of 580bp in RET (for Exon four) was cut into two smaller fragments (400bp, 180bp). We found that all sgRNAs successfully triggered site-specific cleavage in RET (T7E1 Data of Exons one, and two were not shown). Sanger sequencing data of indel after NHEJ in RET exon one, was shown in Figs.2,3.
Fig. 2:
A. Exon one sequence of RET in the sgRNA region
B. Knocked out sequence of RET in exon one with 10 nucleotides deletion after NHEJ.
C. Top: Exon one sgRNA sequence (blue) and PAM region (yellow), the red arrow shows Cas9 cut point in this sample. Bottom: Exon one sgRNA sequence (blue) with 10 nucleotides deletion (red dashed line) and PAM region (yellow).
D. Top: The Exon one sgRNA, is in the region that encodes the first 7 amino acids of RET protein signal peptide. Bottom: In this sample, after the removal of 10 nucleotides and the subsequent removal of 3 amino acids, RET protein reading frame is completely disrupted and also leads to premature stop codon
Fig. 3:
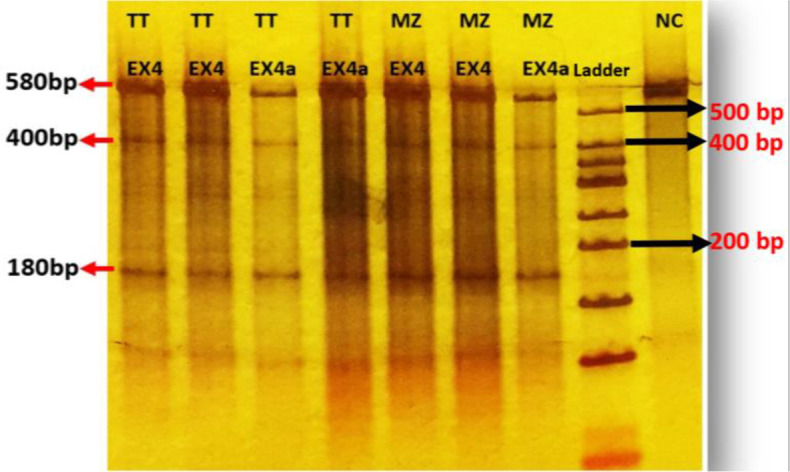
T7 positive test of transfected TT and MZ-CRC1 cells (sgRNAs exon4), electrophoresed on a 4–20% gradient polyacrylamide gel. The PCR product size is 580bp. After cleavage by the T7EI, 400 and 180bp fragments were produced. Normal control was not cleaved by the enzyme.
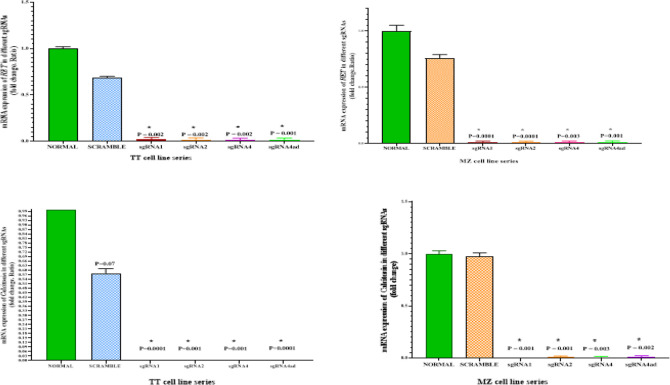
Evaluation of RET Gene Expression at mRNA Level
To confirm that RET was knockout in the studied cell-lines can influence on RET expression at mRNA level, we examined the expression by RT-qPCR. RET is highly expressed in MTC cell-lines, and knocking out of RET by CRISPR/Cas9 leads to a significant reduction of this gene in the cells. However, the scramble negative control cells did not show any significant difference in the expression of RET in both cell-lines (Fig.4).
Fig. 4:
RET (top) and Calcitonin (bottom) gene expression in TT and MZ-CRC1 cells line treated with different sgRNAs, control and scramble groups. *P<0.05
Evaluation of Calcitonin Gene Expression at mRNA Level
As calcitonin is a diagnostic marker in medullary thyroid cancer and its gene expression is significant in MTC cells, real-time PCR was used to investigate the knockout effect of RET on calcitonin gene expression in these cells. Interestingly, calcitonin gene expression in sgRNA-treated cells was very significantly lower than in the control group. However, calcitonin gene expression was not significantly different in the control groups (Fig. 4).
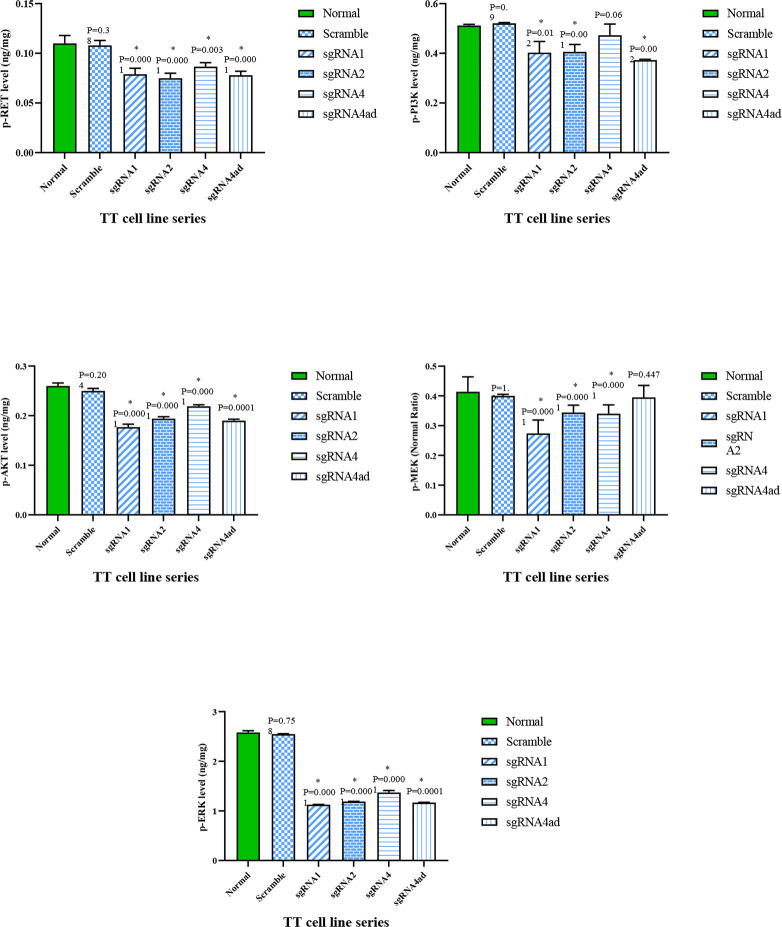
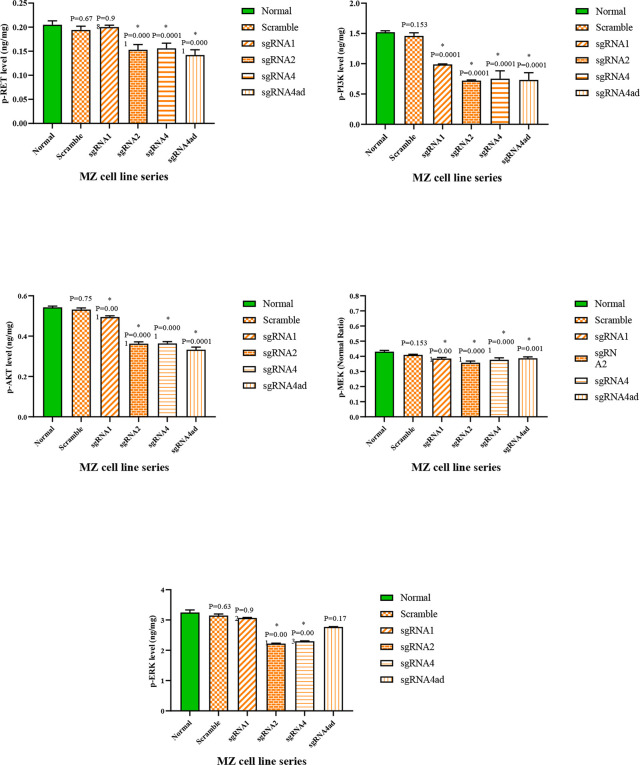
Evaluation of proteins RET, p-RET, p-PI3K, p-AKT, p-MEK, p-ERK in cell-lines by ELISA
To evaluate RET protein production level, the activity of RET protein kinase as well as two downstream signal transduction pathways of this molecule, phosphorylated RET, PI3K/AKT as well as ERK/MEK proteins were measured by ELISA method in cells treated with different sgRNAs, scramble, and negative control groups. In the group of MTC cell-lines treated with different sgRNAs compared to the scramble, and negative control groups, a significant reduction of RET, p-RET, p-PI3K, p-AKT, p-MEK, p-ERK molecules was observed (P<0.05) (Fig. 5, 6).
Fig. 5:
Phosphorylated proteins RET, PI3K, AKT, MEK, ERK in TT-cell-line treated with different sgRNAs compared to the control group
Fig. 6:
Phosphorylated proteins RET, PI3K, AKT, MEK, ERK in MZ-CRC1 cell-line treated with different sgRNAs compared to the control group
The Effect of CRISPR/Cas9-Mediated RET Knockout on Cellular Apoptosis
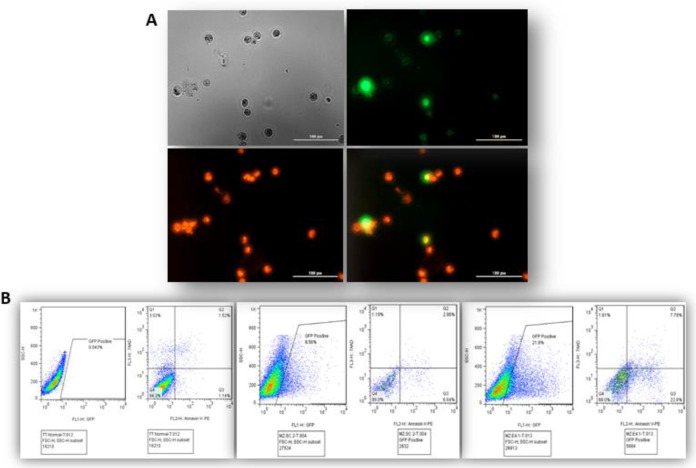
Since the inhibition of RET protein kinase can lead to apoptosis induction in the cell, knockout of RET may also lead to induction of apoptosis in the studied cells. Therefore, cell apoptosis was evaluated in treated cells with sgRNAs, scramble, and negative control cells, by flow cytometry. As demonstrated in Fig.7, CRISPR/Cas9-mediated RET knockout shows a remarkable induction of apoptosis in all cell-lines. However, scramble plasmid, did not induce apoptosis in the MTC cell-lines.
Fig. 7:
A) Fluorescence images of AnnexinV/PI cell apoptosis detection. B) Flow cytometry assay to detect apoptosis in sgRNAs treated, control, and scrambled cells
Discussion
In this study, RET gene knockout was performed by CRISPR/Cas9 gene editing method for the first time in two medullary thyroid cancer cell-lines, TT and MZ-CRC-1 cell-lines, and was confirmed at the DNA, RNA and protein levels.
RET Gene Knockout and Calcitonin Gene Expression
According to the few available studies, there are unknown molecular pathways between RET expression and calcitonin gene expression. Studies have also shown that inhibiting the tyrosine kinase RET protein molecule can reduce calcitonin secretion (22). In the present study, after treating TT and MZ-CRC-1 cell-lines of medullary thyroid cancer with sgRNAs leading to knockout of RET, the expression of calcitonin gene at RNA level decreased very significantly. Identifying the relationship between intracellular pathways and the effect of RET expression on calcitonin gene expression requires more extensive molecular studies.
RET Gene Knockout and Apoptosis Induction
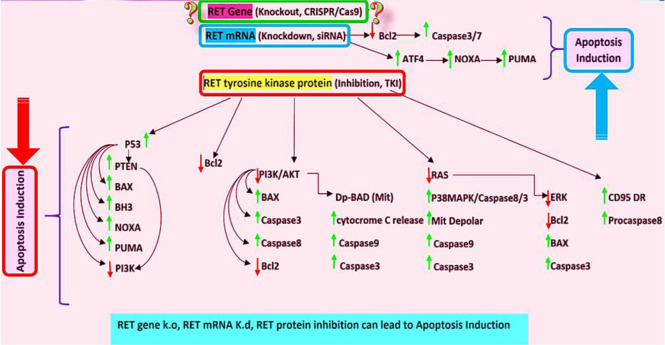
The expression of RET onco-protein in MZCRC-1 cell-line can prevent cell apoptosis. Increasing the expression of RET in these cells leads to increased expression of Fap-1 protein, which also inhibits the CD95 death receptor and procasapase8 molecules, which prevent cell apoptosis (25). In another study, RET tyrosine kinase inhibitors in TT-cells led to increased apoptosis in these cells (26). In another study, knockdown of RET using siRNA resulted in increased expression and phosphorylation of the ATF4 molecule, which is very important in promoting the process of apoptosis in the cell through the proapoptotic molecules NOXA and PUMA (26). Noxa and PUMA are proteins of the Bcl-2 family that belong to a subset of BH3-only proteins. Noxa induces apoptosis through p53-dependent and/or p53-independent mechanisms. These proteins can bind and inhibit the anti-apoptotic molecules Bcl-2, Bcl-XL, Bcl-W (27, 28). Activation of RET signal transduction pathway by GDNF/GFRalpha activates the PI3K/AKT pathway, resulting in increased BCL2, and decreased BAX and Caspase3 gene expression. The ERK pathway is also phosphorylated and activated, increasing BCL2 expression, decreasing BAX and Caspase3 expression. As a result of the above processes, cellular apoptosis is inhibited (29). Therefore, in general, inhibition of RET signal transduction pathway at the protein and RNA levels can lead to the induction of apoptosis (Fig. 8).
Fig. 8:
According to the literatures, reduced RET tyrosine kinase activity by TKIs (in protein level and downstream pathways) or by siRNA (mRNA level) lead to induction of apoptosis via activation of apoptotic molecules and inhibition of antiapoptotic molecules. It seems that RET KO, induces apoptosis pathways (Original)
In the present study, after treatment of TT and MZ-CRC-1 cells with different sgRNAs of RET, the cellular apoptosis was significantly increased, which may be related to the knockout of RET and its effect on the downstream molecular pathway in these cells.
Conclusion
Knocking out RET in MTC cells was confirmed by CRISPR/Cas9 method. As a result of knocking out RET, a decrease in the expression of this gene at the mRNA level and a decline in the activity of RET protein were confirmed. The knockout of RET, followed by a decrease in its mRNA and protein, can reduce the activity of RET protein kinase signaling pathway, and induce cellular apoptosis, which was also observed in this study. Given that modern medicine today is moving towards “Personalized Medicine” and “Precision Medicine”, the use of new therapies should be considered to achieve this critical goal. The new CRISPR/Cas9 gene editing method, is a method that could revolutionize genome editing technology.
Today, there is great hope to treat a wide range of diseases, especially genetic diseases, using the CRISPR/Cas gene editing system. Scientists hope to use this technology to provide valuable services to humanity.
Journalism Ethics considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
This work was financially supported by the Dr. Farhud Foundation.
Footnotes
Conflict of interest
The authors declare that there is no conflict of interests.
References
- 1.Babashpour SP, Saify-Nabiabad Khosro, Sabouni Haidar, Farzaneh. (2018). Identification, Cloning and Structural Analysis of Major Genes from Portulaca oleracea L. Hairy Roots that Involved in the Biosynthesis of Dopamine. Journal of Medicinal Plants and Byproducts, 2: 153–162. [Google Scholar]
- 2.WHO (2020). https://gco.iarc.fr/today/data/factsheets/populations/900-world-fact-sheets.pdf. In: WHOG.
- 3.WHO (2020). https://gco.iarc.fr/today/data/factsheets/populations/364-iran-islamic-republic-of-fact-sheets.pdf
- 4.Pellegriti G, Frasca F, Regalbuto C, et al. (2013). Worldwide increasing incidence of thyroid cancer: update on epidemiology and risk factors. J Cancer Epidemiol, 2013:965212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raza U, Saatci O, Uhlmann S, et al. (2016). The miR-644a/CTBP1/p53 axis suppresses drug resistance by simultaneous inhibition of cell survival and epithelial-mesenchymal transition in breast cancer. Oncotarget, 7(31):49859–49877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowland KJ, Moley JF. (2015). Hereditary thyroid cancer syndromes and genetic testing. J Surg Oncol, 111(1):51–60. [DOI] [PubMed] [Google Scholar]
- 7.Donis-Keller H, Dou S, Chi D, et al. (1993). Mutations in the RET proto-oncogene are associated with MEN 2A and FMTC. Hum Mol Genet, 2(7):851–6. [DOI] [PubMed] [Google Scholar]
- 8.Arighi E, Borrello MG, Sariola H. (2005). RET tyrosine kinase signaling in development and cancer. Cytokine & Growth Factor Reviews, 16(4–5):441–67. [DOI] [PubMed] [Google Scholar]
- 9.Figlioli G, Landi S, Romei C, et al. (2013). Medullary thyroid carcinoma (MTC) and RET proto-oncogene: mutation spectrum in the familial cases and a meta-analysis of studies on the sporadic form. Mutat Res, 752(1):36–44. [DOI] [PubMed] [Google Scholar]
- 10.Romei C, Cosci B, Renzini G, et al. (2011). RET genetic screening of sporadic medullary thyroid cancer (MTC) allows the preclinical diagnosis of unsuspected gene carriers and the identification of a relevant percentage of hidden familial MTC (FMTC). Clin Endocrinol (Oxf), 74(2):241–7. [DOI] [PubMed] [Google Scholar]
- 11.Hedayati M, Yeganeh MZ, Sheikholeslami S, et al. (2015). Skewed mutational spectrum of RET proto-oncogene Exon10 in Iranian patients with medullary thyroid carcinoma. Tumour Biol, 36:5225–31. [DOI] [PubMed] [Google Scholar]
- 12.Hedayati M, Zarif Yeganeh M, Sheikhol Eslami S, et al. (2011). Predominant RET Germline Mutations in Exons 10, 11, and 16 in Iranian Patients with Hereditary Medullary Thyroid Carcinoma. J Thyroid Res, 2011:264248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedayati M, Zarif Yeganeh M, Sheikholeslami S, et al. (2016). Diversity of mutations in the RET proto-oncogene and its oncogenic mechanism in medullary thyroid cancer. Crit Rev Clin Lab Sci, 53(4):217–27. [DOI] [PubMed] [Google Scholar]
- 14.Sheikholeslami S, Zarif Yeganeh M, Hoghooghi Rad L, et al. (2014). Haplotype Frequency of G691S/S904S in the RET Proto-Oncogene in Patients with Medullary Thyroid Carcinoma. Iran J Public Health, 43(2):235–240. [PMC free article] [PubMed] [Google Scholar]
- 15.Yeganeh MZ, Sheikholeslami S, Hedayati M. (2015). RET proto oncogene mutation detection and medullary thyroid carcinoma prevention. Asian Pac J Cancer Prev, 16(6):2107–17. [DOI] [PubMed] [Google Scholar]
- 16.Wedge SR, Ogilvie DJ, Dukes M, et al. (2002). ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res, 62(16):4645–55. [PubMed] [Google Scholar]
- 17.Carlomagno F, Vitagliano D, Guida T, et al. (2002). ZD6474, an orally available inhibitor of KDR tyrosine kinase activity, efficiently blocks oncogenic RET kinases. Cancer Res, 62(24):7284–90. [PubMed] [Google Scholar]
- 18.Carlomagno F, Guida T, Anaganti S, et al. (2004). Disease associated mutations at valine 804 in the RET receptor tyrosine kinase confer resistance to selective kinase inhibitors. Oncogene, 23:6056–63. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira CV, Siqueira DR, Ceolin L, et al. (2013). Advanced medullary thyroid cancer: pathophysiology and management. Cancer Manag Res, 5:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farhud DD, Zarif-Yeganeh M. (2020). CRISPR Pioneers Win 2020 Nobel Prize for Chemistry. Iran J Public Health, 49:2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akram F, Ul Haq I, Ahmed Z, et al. (2020). CRISPR-Cas9, a promising therapeutic tool for cancer therapy: A review. Protein Pept Lett, 27(10):931–944. [DOI] [PubMed] [Google Scholar]
- 22.Akeno-Stuart N, Croyle M, Knauf JA, et al. (2007). The RET kinase inhibitor NVP-AST487 blocks growth and calcitonin gene expression through distinct mechanisms in medullary thyroid cancer cells. Cancer Res, 67(14):6956–64. [DOI] [PubMed] [Google Scholar]
- 23.Ran F, Hsu PD, Wright J, et al. (2013). Genome engineering using the CRISPR-Cas9 system. Nature protocols, 8(11):2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulstrode H, Johnstone E, Marques-Torrejon MA, et al. (2017). Elevated FOXG1 and SOX2 in glioblastoma enforces neural stem cell identity through transcriptional control of cell cycle and epigenetic regulators. Genes Dev, 31(8):757–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicolini V, Cassinelli G, Cuccuru G, et al. (2011). Interplay between Ret and Fap-1 regulates CD95-mediated apoptosis in medullary thyroid cancer cells. Biochem Pharmacol, 82:778–788. [DOI] [PubMed] [Google Scholar]
- 26.Koga K, Hattori Y, Komori M, et al. (2010). Combination of RET siRNA and irinotecan inhibited the growth of medullary thyroid carcinoma TT cells and xenografts via apoptosis. Cancer Sci, 101(4):941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang LN, Li JY, Xu W. (2013). A review of the role of Puma, Noxa and Bim in the tumorigenesis, therapy and drug resistance of chronic lymphocytic leukemia. Cancer Gene Ther, 20(1):1–7. [DOI] [PubMed] [Google Scholar]
- 28.Morsi RZ, Hage-Sleiman R, Kobeissy H, et al. (2018). Noxa: Role in Cancer Pathogenesis and Treatment. Curr Cancer Drug Targets, 18(10):914–928. [DOI] [PubMed] [Google Scholar]
- 29.Li W, Wei D, Lin J, et al. (2019). Dl-3-n-Butylphthalide Reduces Cognitive Impairment Induced by Chronic Cerebral Hypoperfusion Through GDNF/GFRα1/Ret Signaling Preventing Hippocampal Neuron Apoptosis. Front Cell Neurosci, 13:351. [DOI] [PMC free article] [PubMed] [Google Scholar]