Abstract
Investigation on how nature produces natural compounds with chemical and biological diversity at the genetic level offers inspiration for the discovery of new natural products and even their biological targets. The polyketide rumbrin (1) is a lipid peroxide production and calcium accumulation inhibitor, which contains a chlorinated pyrrole moiety that is a rare chemical feature in fungal natural products. Here, we identify the biosynthetic gene cluster (BGC) rum of 1 and its isomer 12E-rumbrin (2) from Auxarthron umbrinum DSM3193, and elucidate their biosynthetic pathway based on heterologous expression, chemical complementation, and isotopic labeling. We show that rumbrins are assembled by a highly reducing polyketide synthase (HRPKS) that uniquely incorporates a proline-derived pyrrolyl-CoA starer unit, and followed by methylation and chlorination. Sequent precursor-directed biosynthesis was able to yield a group of rumbrin analogues. Remarkably, inspired by the presence of a human immunodeficiency virus (HIV)-Nef-associated gene in the rum cluster, we predicted and pharmacologically demonstrated rumbrins as potent inhibitors of HIV at the nanomolar level. This work enriches the recognition of unconventional starter units of fungal PKSs and provides a new strategy for genome mining-guided drug discovery.
KEY WORDS: Rumbrins, Polyketide, Precursor-directed biosynthesis, Resistance gene, Anti-HIV activity
Graphical abstract
We reconstituted the biosynthesis of rumbrins, identified a new starter unit of PKS, and obtained nine rumbrin analogues. Guided by rumB, we predicted and pharmacologically demonstrated rumbrins as anti-HIV agents.
1. Introduction
Natural products hold vast chemical and biological diversity that cannot be matched by libraries of synthetic compounds, and contribute continually in the fields of biology, chemistry, and medicine1. Through biosynthetic evolutionary optimization, natural products tend to be the perfect sources of drug discovery2. Understanding the biosynthetic mechanisms of natural products is a key to guiding rational manipulation of nature's biosynthetic machinery to produce natural compounds and their engineered variants1. Besides, a deep comprehension of the biosynthetic logic and enzymatic mechanism has created a new field for genome mining-guided discovery of new and bioactive natural products3, 4, 5, 6, 7, 8. For example, bioinformatic analysis or functional verification of a putative self-resistance gene in a BGC was able to connect the encoded natural products to a potential target9.
As an important class of natural products in pharmacological science, polyketides have yielded a large number of new drugs for clinical application4,10, such as lovastatin, rapamycin, and daunomycin. Such success is partly due to the significant variety of polyketide backbones and extensive mechanisms were reported for polyketide scaffolds diversification. For example, a set of catalytic domains including β-ketoreductases, enoyl reductases, and dehydratases can selectively modify the polyketide backbone and a variety of tailoring enzymes are able to modify the polyketide scaffold after it has been assembled by a PKS. While another important diversification step, the incorporation of unconventional starter units, often occurs earlier. Although most of the polyketides are primed with acetyl-CoA, a few PKSs utilize multifarious starter units, such as propionyl acyl carrier protein (CP)11, 4-coumaroyl-CoA12, hexanoyl-CoA13, 2-methylbutyryl-CoA14, benzoyl-CoA15, 4-guanidinobutyramide16, pyrrolyl-CP17 and 4,5-dichloropyrrolyl-CP18 (Supporting Information Fig. S1), which in many cases lead to the diverse structural and biological features to the natural products.
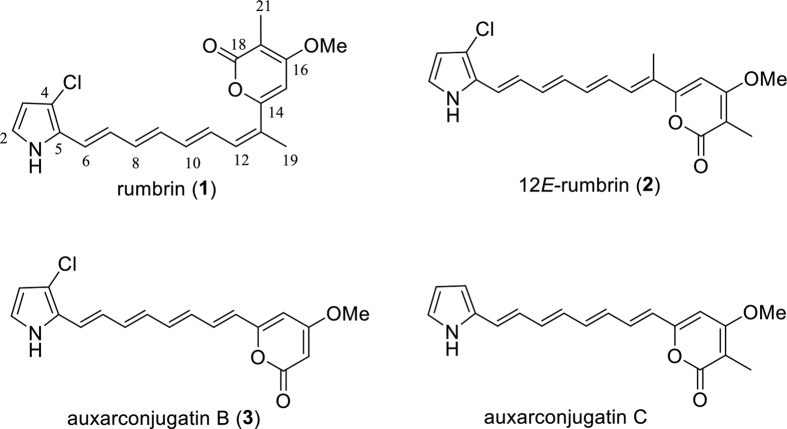
Rumbrin (1), an unusual chlor-containing polyenylpyrrole, was produced by several fungi from the terrestrial fungal family Onygenales19 (Fig. 1). Structurally, 1 and its analogs, such as 12E-rumbrin (2) and auxarconjugatin B (3), are featured in an α-pyrone and a pyrrole ring connected by a polyene linker with all E or part of Z geometrical configuration (Fig. 1). The structural diversity of rumbrins was also constructed by the presence or absence of chlorination on pyrrole moiety and the methyl substitution at different positions on the polyene chain as well as α-pyrone (Fig. 1). Members of this rare class of natural products are usually associated with potent biological activities. For example, 1, while relatively unstable, was reported to exhibit remarkable cytoprotective and anti-lipid peroxidation activities19,20. 2 displays selective cytotoxicity against ovarian cancer cell lines at micromolar level20, and 3 also show significant cytotoxicity against non-structural protein (NS-1) cell line21.
Figure 1.
Chemical structures of rumbrin (1) and its analogues.
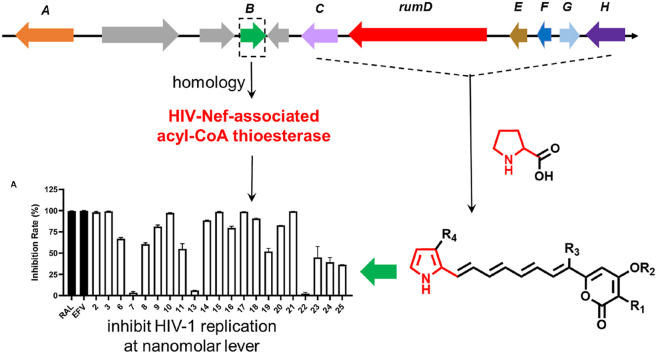
Because of the potent biological activity of 1, its biosynthesis has attracted broad interesting since its isolation. Although the biosynthetic origin of 1 was proposed and its biosynthetic precursors were subsequently suggested by labeling experiments (Supporting Information Fig. S2)22,23. The biosynthesis of 1 and 2 had never been revealed genetically. Herein, we identified the BGC rum and characterized the biosynthetic pathway of 1 and 2 through heterologous expression, chemical complementation, and isotopic labeling. Precursors-direct biosynthesis led to the production of a group of rumbrin analogs. Moreover, a putative resistance-like gene rumB showing homologous to the gene encoding an HIV-Nef-associated acyl-CoA thioesterase (hACTE-III) in the rum cluster was discovered, which led to our inspiration to evaluate rumbrins as anti-HIV agents.
2. Results and discussion
2.1. Bioinformatic analysis of the genome of Auxarthron umbrinum
Recently, we isolated 2 and 3 from the wild type A. umbrinum DSM3193, which is the native producer of 119,25. The structures of 2 and 3 were fully characterized via 13C and 1H NMR (see detail in Supporting Information Tables S4 and S6). Regrettably, 1 was not isolated but detectable in our case since it was photosensitive and readily converted to its isomer 2 (Supporting Information Fig. S3).
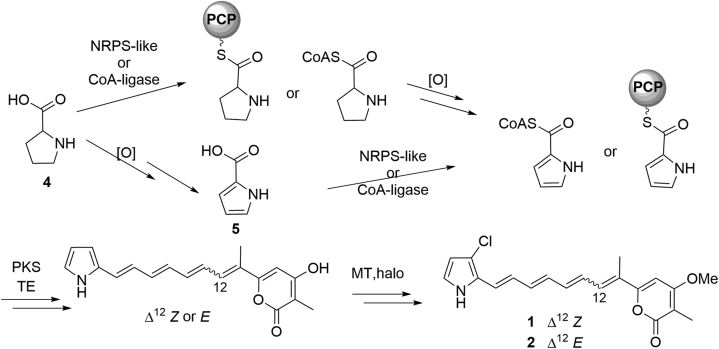
The entire carbon backbone of 1 and 2 is hypothesized to be assembled from a single HRPKS according to the isotope-labeling studies22 as well as pathways of structurally related natural products such as citreoviridin26. The pyrrole moiety in the structure of 1 and 2 was believed to be derived from proline (4) or pyrrole-2-carboxylic acid (5)23. Considering the high incorporation of 5 into 1 in previous labeling experiments22, it could be a direct precursor of 1 and 2. In addition, methylation and halogenation are also required for their biosynthesis (Fig. 2).
Figure 2.
Proposed biosynthetic pathway of 1 and 2.
For most HRPKSs, there is frequent lack of a domain for product release, instead a acyltransferase-like enzyme or discrete thioesterase (TE) is required to release the product27. For instance, a trans-acting TE, BerkB, was reported to catalyze the macrolactonization during the biosynthesis of fungal macrolide, A26771B28. We deduced that a trans-acting TE would exist in the gene cluster of 1 and 2 (Fig. 2). Furthermore, the enzymes responsible for amino acid activation and incorporation are required for constructing the aminoacylated polyketides, such as CoA-ligase and NRPS-like enzyme (Fig. 2)29, according to the known fungal HRPKS programming rules. Overall, a BGC encoding a HRPKS, a trans-acting TE, a CoA-ligase or NRPS-like enzyme, a methyltransferase, a halogenase and additional redox enzymes are necessary during the biosynthesis of 1 and 2.
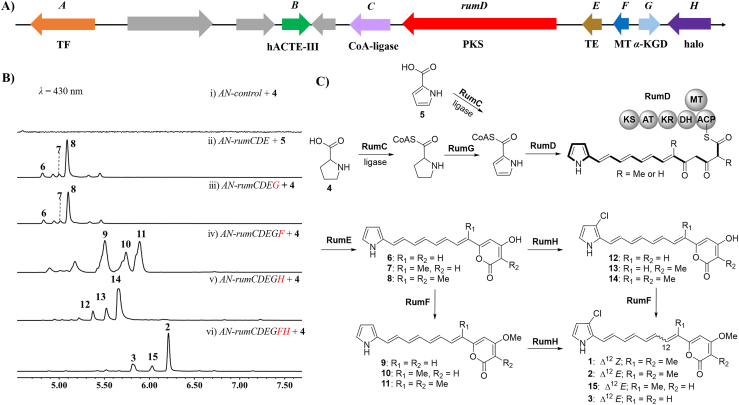
To investigate the biosynthetic pathway of 1 and 2, we sequenced the producer fungus A. umbrinum DSM3193. Bioinformatic analysis revealed that the BGC located in scaffold 127 is the only candidate qualifying the 1 and 2 biosynthesis and is named rum cluster (Fig. 3A and Table S3). With the aid of RT-PCR, we defined the rum cluster to contain 10 genes (Fig. S3), which encode a transcriptional regulatory (TF, RumA), an hACTE-III (RumB), a CoA-ligase (RumC), a HRPKS (RumD) with six domains (KS-AT-KR-DH-MT-ACP), a discrete TE (RumE), a methyltransferase (RumF), a 2-oxoglutarate-dependent dioxygenase (α-KGD, RumG), a flavin-dependent halogenase (RumH) and three uncharacterized proteins, respectively.
Figure 3.
Identification of the BGC and pathway of 1 and 2. (A) The rum gene cluster in A. umbrinum. (B) LC–MS analysis of products profiles of heterologous expression of different combinations of rum genes in A. nidulans. (C) Putative biosynthetic pathway of 1 and 2. AT, acyl transferase; ACP, acyl-carrier protein; DH, dehydratase; KS, ketoacyl synthase; KR, ketoreductase; MT, methyltransferase.
2.2. Characterization of rum cluster for rumbrin biosynthesis
According to the known mechanism for pyrrole biosynthesis, pyrrolyl-CoA or pyrrolyl-CP is usually derived from proline through oxidation. The pyrrole moiety in rumbrin was also suggested to be derived from proline based on 15N-labeled precursor22. To further confirm the biosynthetic origin of pyrrole ring in 1 and 2, we used the full labeled l-proline-[13C5,15N] to perform the feeding experiment. As a result, an increase of 6 mu in molecular weight of 2 and 3 were detected by mass spectrometry (Supporting Information Fig. S5). This revealed that five 13C and one 15N from l-proline-[13C5,15N] were all incorporated into 2 and 3, which further confirmed that the pyrrole moiety in 2 was initially originated from proline (4).
Subsequently, to confirm the rum gene cluster and elucidate the biosynthetic pathway of 1 and 2, gene knockout experiments in A. umbrinum were carried out firstly. Nevertheless, all attempts to knockout genes in the rum cluster failed. We then turned to heterologously express the rum genes in the host Aspergillus nidulans A1145. Expression of rumD, rumDE, rumCD and even rumCDE separately in A. nidulans, supplying proline (4, 2 mmol/L), did not yield any new products compared to the AN-control (data not shown). While the transformant AN-rumCDE feeding with 5 produced additional peaks 6–8 detected by LC–MS analysis (Fig. 3B, trace ii), which were later identified as 16-demethyl-auxarconjugatin D, 13-methyl-16-demethyl-auxarconjugatin D, and 16-demethyl-dechloroisorumbrin respectively, via 1D and 2D NMR (Supporting Information Tables S4 and S6). These results not only confirm the rum cluster was responsible for the biosynthesis of 1 and 2, but also supported our proposal that oxidation of proline occurred before the polyketide chain elongation (Fig. 2). The results that the strains AN-rumD, AN-rumDE and AN-rumCD, which did not produce any new products, while the strain AN-rumCDE did (Supporting Information Fig. S6, traces ii-v), also suggests that RumC activates substrate, HRPKS RumD assembles a C17 polyketide chain through six iterations of malonyl-CoA extension, and RumE catalyzes the lactonization of the C17 polyketide chain (Fig. 3C).
2.3. Oxidation catalyzed by a 2-oxoglutarate-dependent dioxygenase, RumG
It was reported that a dehydrogenase was sufficient to convert proline to pyrrole ring in bacteria30,31. While the homologous protein of RedW, which is a dehydrogenase that catalyzes the conversion from prolyl-CP to pyrrolyl-CP in bacteria (Supporting Information Fig. S7)32, was not found among the proteins encoded by the rum cluster. Instead, an α-KGD (RumG) with sequence homology to EcdG (28%), which was suggested to catalyze the hydroxylation of proline in the biosynthesis of echinocandin B, was shown in rum cluster. As the only redox enzyme in rum cluster, RumG was postulated to be involved in the oxidization of proline to form pyrrole ring. To investigate its role, the gene rumG was introduced into the strain AN-rumCDE feeding with 4. After 4 days of culture, a major product 8 together with minor products 6 and 7 were detected by LC–MS (Fig. 3B, trace iii). This supported our hypothesis that RumG catalyzed the formation of tetrahydropyrrole ring to pyrrole ring (Fig. 3C). To test whether 4 was the direct substrate of RumG, we heterologously expressed rumG in A. nidulans and cultured it in a proline-containing CD-ST media. Unfortunately, the desired product 5 was not detected (Supporting Information Fig. S8), which lead to our postulation that CoA thioester instead of free proline may be the natural substrate for RumG (Fig. 3C).
2.4. Post-PKS modification
Next, we focused on the function of the methyltransferase RumF and the halogenase RumH. Co-expression of rumCDEGF feeding with 4 led to the production of three new metabolites 9–11 (Fig. 3B, trace iv), which were purified and characterized as auxarconjugatin D (9), 4-dechloro-17-demethyl-isorumbrin (10), and 12E-dechloroisorumbrin (11), respectively. The identification of 9–11 with 16-O-methylated structures highly suggested that RumH was responsible for the O-methylation at C-16. While the strain AN-rumCDEGH produced minor products 12–13 and major product 14 (Fig. 3B, trace v), in which 13 and 14 were fully identified as 12E-demethylisorumbrin and 16-demethyl-isorumbrin by NMR, respectively. Although 12 was not purified, its molecular weight and UV indicated this is the chlorinated 6. This suggested RumH was responsible for the chlorination at C-4.
To determine whether chlorination happened after the formation of polyketide skeleton, 4 and 5 were separately fed as substrate to the culture of strain AN-rumH. As a result, no chlorinated products could be detected in both cases (Supporting Information Fig. S9). Actually, RumH was phylogenetically close to a halogenase, MalA, which also catalyzed the chlorination of an off-line substrate in the biosynthetic pathway of malbrancheamide33 (Supporting Information Figs. S10 and S11). Combined with the low incorporation of 3-chloropyrrole-2-carboxylate into 1 in the previous report22, it is reasonable that the chlorination was a post-PKS modification step.
To further determine the function order of RumH and RumF, we performed feeding experiments. Upon supplementing the mixture of 6–8 or 9–11 to A. nidulans expressing RumH, the corresponding chlorinated products were detected (Supporting Information Fig. S12, traces i and ii). Furthermore, the mixture of 6–8 or 12–14 was converted to the corresponding O-methylated products in the presence of RumF (Fig. S12, traces iii and iv). These results suggested that the methylation and halogenation could occur simultaneously during the biosynthesis of 1 and 2. As a trial, both RumF and RumH were added to the strain AN-rumCDEG feeding with 4, and the production of major 2 along with minor 3 and 15 were observed (Fig. 3B, trace vi), which also supported our proposal.
Hence, the overall biosynthetic pathway of 1 and 2 in A. umbrinum could be deduced (Fig. 3C). Beginning with the conversion of proline to prolyl-CoA by a CoA-ligase, an α-KGD oxidizes prolyl-CoA to form pyrrolyl-CoA, and the PKS and discrete TE could utilize pyrrolyl-CoA and malonyl-CoA as building blocks to construct polyketide backbone, which can be O-methylated and chlorinated to give 1 and 2.
2.5. Precursor-directed biosynthesis
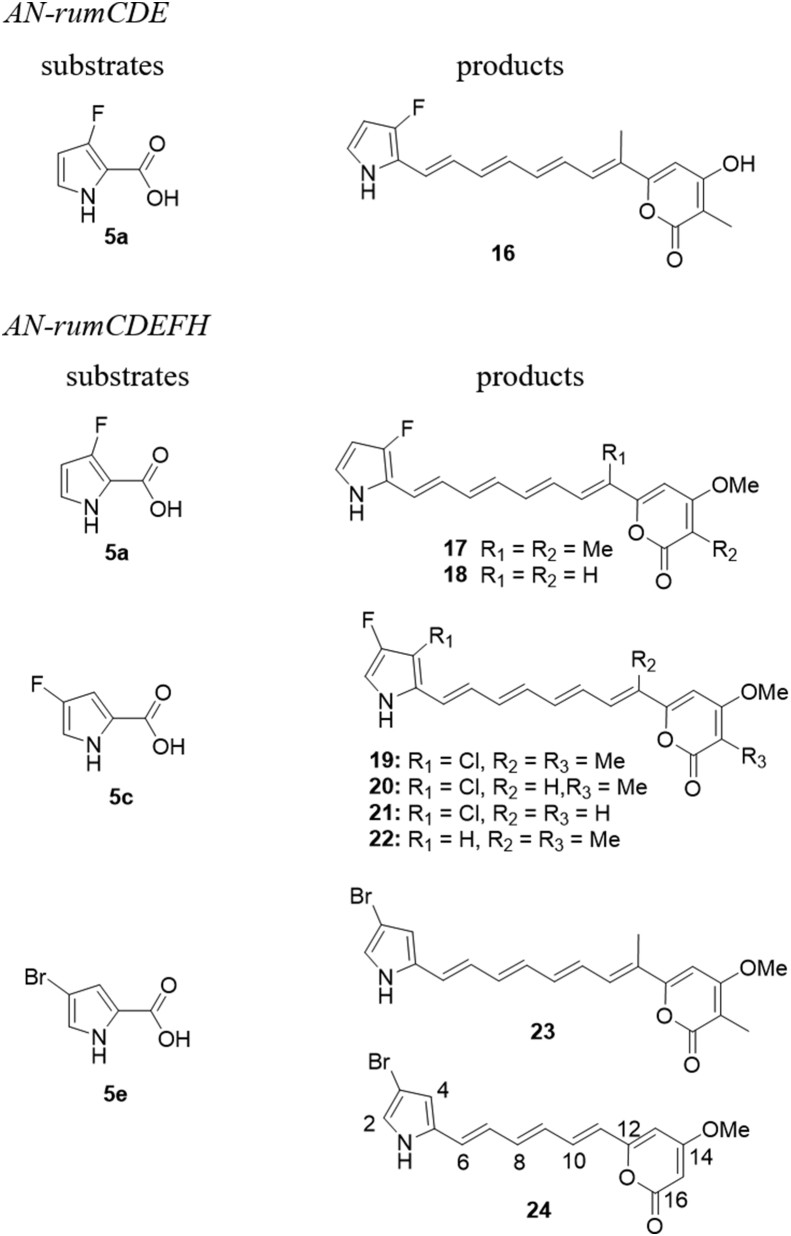
As a feasible strategy, precursor-directed biosynthesis is frequently used to construct new functional groups to improve structural diversity, which usually impacts the bioactivity of the compound34 or provide a functionalizable handle for structural modification35. Encouraged by fact that CoA-ligase RumC was able to recognize different substrates as mentioned earlier23, a panel of commercially available analogues of 5 (Supporting Information Fig. S13) were separately fed into the culture of the strain AN-rumCDE and AN-rumCDEFH, to produce rumbrin derivatives. As a result, among all the substrates fed, 4-bromo-, 4-fluoro-, and 3-fluoro-pyrrole-2-carboxylic acids can be converted into new products, while switching the pyrrole ring to thiophen, furan, or pyridine ring did not yield any new products, as detected by LC–MS (Supporting Information Fig. S14). Finally, a total of nine products (16–24, Fig. 4) were isolated and structurally characterized by MS and NMR.
Figure 4.
Precursor-directed biosynthesis of rumbrin analogs.
2.6. Resistance-like gene-directed anti-HIV evaluation
Self-resistance genes are continually reported from microbial natural product BGC7,36,37. This phenomenon has been frequently used to identify BGCs of natural products with known bioactivities, as well as to predict bioactivity and molecular target of natural products8,38, 39, 40, 41, 42. Remarkably, phylogenetic analyses revealed that RumB, differs from classical TE, belongs to TesB-like subfamily (Supporting Information Figs. S15 and S16) and shows homolog to Escherichia coli thioesterase II (32%), and human thioesterase II (hTE, 33%)43. It seems that RumB does not involve in the biosynthetic steps toward 1 and 2 based on bioinformatic analysis. Indeed, when rumB was introduced into the strain AN-rumCDEFGH, neither any additional products were produced nor yield of rumbrins increased (data not shown). The results from bioinformatic analysis and heterologous expression inspired our suspicion that rumB may function as a resistance-like gene.
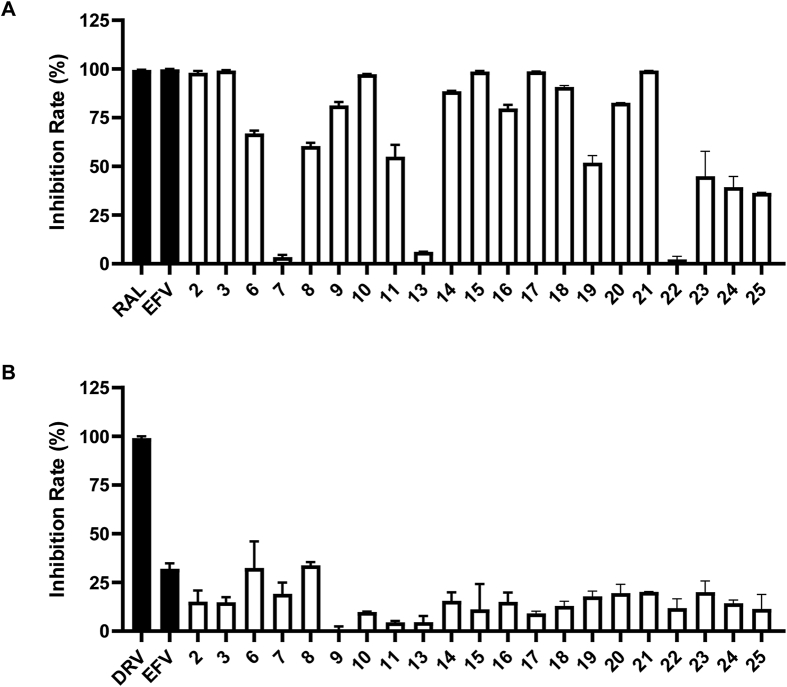
Of note, hTE has been reported to be an important cellular partner of HIV-1 Nef and suggested to play several roles in HIV-1 infectivity44,45. Therefore, the possible interplay between hTE and the compounds from rum cluster suggests their potential anti-HIV-1 bioactivity. Subsequently, all the rumbrin analogs obtained from this study were evaluated for their inhibition activity against HIV-1. As a result, most of the test compounds significantly inhibited the HIV-1 infection in the early stage of a one-cycle HIV-1 infection assay (Fig. 5A). In detail, compounds 2–3, 9–10, 14–15, 17–18, and 20–21 possess over 80% inhibition rate at 10 μmol/L concentration and IC50 among 20.9–376.6 nmol/L (Supporting Information Table S11). It seems that the presence of 16-O-methyl and/or 4-Cl group contribute significantly to their anti-HIV activity. To assess the effect of the compounds upon the late stage of HIV-1 replication, which includes the steps from gene expression to virus production, HEK293T cells were transfected with pNL4-3luc.R-E- and VSVg in the presence of test compounds or DMSO. Then, supernatant virus particles were collected and infected SupT1 cells. Meanwhile, rumbrins (Fig. 5B) had little effect on HIV-1 infection in the late stage. Taken together, these data suggest that rumbrins potently interrupt the early step of viral replication.
Figure 5.
Rumbrins potentially inhibit the early stage of HIV-1 replication. (A) HIV-1 NL4-3luc.R-E- pseudoviruses were used to infect SupT1 cells in the presence of test compounds. The infected cells were lysed followed by the measurement of luciferase activity. EFV and RAL were used as controls; (B) HEK293T cells were transfected with pNL4-3luc.R-E- in the presence of test compounds or DMSO. The resulting virus particles were collected and infected SupT1 cells, followed by measuring luciferase activity to determine viral infectivity. As controls, protease inhibitors (PIs) darunavir (DRV), as well as inactive control EFV were also tested; Data represent the mean ± SD of three independent experiments.
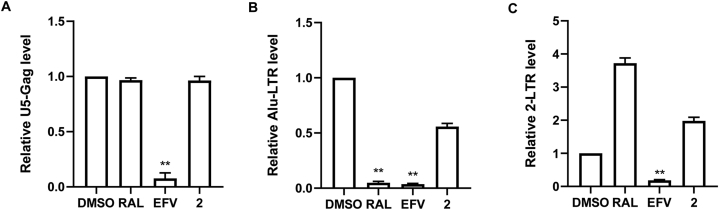
To explore the mechanism how these compounds inhibit the early stage of viral replication, which includes viral entry, reverse transcription and integration, we next assess their effect upon viral reverse transcription and integration using relative quantitative PCR (RQ-PCR). Primers were designed to detect U5-Gag, Alu-LTR and 2-LTR sequences, which were indicative of reverse transcription products, integrated viral DNA and 2-LTR-containing DNA circles, respectively. SupT1 cells were infected with pseudotyped HIV-1 in the treatment of DMSO, EFV, RAL, or the most active compound 2. As shown in Fig. 6A, 2 did not affect the level of U5-Gag products, suggesting that 2 had no inhibitory effect upon reverse transcription. In contrast, compound 2 markedly reduced the level of integrated viral DNA, supporting an inhibition on viral integration (Fig. 6B).
Figure 6.
12E-Rumbrin (2) inhibits HIV-1 replication by blocking viral integration. (A–C) VSV-G pseudotyped NL4-3Luc R-E- viruses were used to infect SupT1 cells with the treatment of DMSO, 4 nmol/L EFV, 25 nmol/L RAL, or 2. HIV-1 RT DNA (A), integrated DNA (B), and 2-LTR circles (C) were quantified by RQ-PCR using the corresponding primers (Table S1). ∗P < 0.05.
2-LTR circles form at a low level in the nucleus through the action of cellular nonhomologous DNA end joining, blockage to HIV-1 integration would result in an increasing level of 2-LTR circles after nuclear entry of HIV-1 cDNA46. Therefore, the level of 2-LTR circles is considered as an indicative for the nuclear import of pre-integration complex. Indeed, the number of 2-LTR circles was 3.7-fold higher in RAL-treated cells compared with DMSO-treated control cells (Fig. 6C). The number of 2-LTR circles in 2 group was 2 times higher than that of the DMSO group (Fig. 6C). Together, these data suggest that compound 2 inhibit HIV-1 replication by blocking viral integration, while other possible anti-HIV mechanism awaits further investigation.
2.7. Discussion
Proline-derived pyrrolyl-CP, an unconventional starter unit of PKS, was reported to be originated from an NRPS-like pathway in bacteria. The selective adenylation of proline by a prolyl-AMP ligase, loading onto the discrete CP, yielding prolyl-S-CP, which was then converted into pyrrolyl-CP by prolyl-S-CP dehydrogenase, in an FAD-dependent manner (Fig. S7)17. Nonetheless, pyrrolyl act as a starter unit has never been verified genetically in fungal PKS systems. In our case, while we did not completely clarify the biosynthesis mechanism of pyrrolyl-CoA, we suggested a new pathway for pyrrolyl-CoA formation which is completely different from the pyrrolyl-CP biosynthesis well-known in bacteria. In detail, a CoA-ligase RumC activates substrate, then the α-KGD RumG catalyze the oxidation of both sides of the prolyl-CoA or, alternatively, hydroxylate C4 and followed by spontaneous dehydration to form Δ4-pyrrolinyl-2-carboxyl-CoA intermediate, which was oxidized to the pyrrolyl-CoA spontaneously. Comparing to the biosynthesis of pyrrole-CP in bacteria, fungi take a more convenient pathway. Our study shed light on the biosynthesis of pyrrole-containing natural compounds. Admittedly, in vitro assay was expected to completely clarify the biosynthesis mechanism of pyrrolyl-CoA and further study on the RumG is ongoing in our lab.
The presence of a self-resistance gene in a BGC opens a new window for the prediction of the bioactivity of natural products synthesized by the biosynthetic pathway. Here, we predicted that rumbrins have anti-HIV activity according to the existence of a potential self-resistance gene rumB in the rum cluster. Even though it would be more convincing if hTE is proved to be the target of rumbrins, the potent inhibition against HIV-1 by rumbrins provides evidence supporting the efficiency of self-resistance gene-guided bioactive natural product discovery. Interestingly, a gene cluster mas homologous to rum was found in Malbranchea sulfurea. The absence of homolog to rumB in mas cluster suggested the products from this cluster may not possess anti-HIV activity. Indeed, heterologous expression of the mas cluster in A. nidulans led to the production of a rumbrin analog, 25, with a shorter polyene linker (Supporting Information Fig. S17), and it showed very weak anti-HIV activity in the bioassay, which highly suggested the presence of rumB is connective to the anti-HIV activity of rumbrin. Interestingly, RumB, a protein from a soil fungus, shows homologous to hTE, which is widely distributed from prokaryotes to eukaryotes47,48. Its function in the biosynthesis of rumbrin is unknown and deserves further study.
Notably, early studies reveal that the interaction of Nef and hTE plays an important role in the down-regulation of CD4 and MHC-I47,49, while the function of hTE in HIV-1 replication is still largely unexplored and inconclusive, because of its diverse biological functions including the recycling and sorting of membrane proteins, the lipid metabolism and the β-oxidation. This work raises a possibility that hTE may involve in HIV-1 integration and act as a potential target for the development of anti-HIV therapeutics.
3. Conclusions
In summary, we successfully characterized the genetic basis for rumbrins biosynthesis, and proposed that a new starter unit containing pyrrole ring was incorporated by fungal PKS. This led to the generation of a series of rumbrin analogs by precursor-directed biosynthesis. Based on the existence of a resistance-like gene rumB homologous to the gene encoding an HIV-Nef-associated acyl-CoA thioesterase, we predicted and pharmacologically demonstrated rumbrins as potent anti-HIV agents. Our study opens a new window for gene mining-guided drug discovery.
4. Experimental
4.1. General materials
TRIZOL® Reagent (Invitrogen, Carlsbad, CA, USA) and the plant genomic DNA extraction kit (Tiangen, Beijing, China) were used to prepare the RNA and genomic DNA of A. umbrinum, respectively. DNA de novo sequencing was performed at SHBIO (Shanghai, China) using the IIIumina Hiseq Xten platform. Primer synthesis was performed by Genscript Biotech Corporation (Beijing, China). Zymoprep™ Yeast Plasmid Miniprep I Kit (Zymo Research, Orange County, CA, USA) was used to extract plasmids from yeast. Q5 high-fidelity DNA polymerase (New England Biolabs, NEB) or 2 × Hieff® PCR Master Mix (YEASEN, Shanghai, China) was used to carry out polymerase chain reactions (PCR). An Mx3000P real-time PCR system (Agilent Technologies Inc., Palo Alto, CA, USA) was used to perform real-time PCR. DNA restriction enzymes were purchased from NEB. DNA sequencing was performed by BGI Tech Solutions (Beijing Liuhe) Co., Ltd. (Beijing, China). E. coli trans1-Blue and trans2-Blue were produced by Transgen Biotech (Beijing, China). Potato dextrose broth medium (PDB) and potato dextrose agar medium (PDA) were produced by BD Difco (Sparks, NV, USA). Cells were collected by centrifuging (Sigma 3-18K, Gettingen, Germany). DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) was used to extract the total DNA of SupT1 cells. The HEK293T and SupT1 cells were purchased from ATCC.
Thermo Nicolet 5700 FT-IR spectrophotometer (Waltham, MA, USA) was used for IR spectra measurement. Bruker Avance III 800, Model Avance DMX 700 or Ascend 600 (Bruker Corp., Karlsruhe, Germany) NMR instruments was used for NMR spectra measurement with TMS as internal standard and DMSO-d6 as solvent. UV spectra were recorded using the JASCO V-650 instrument (JASCO, Corp., Tokyo, Japan). Agilent 1100 Accurate-Mass Q-TOP LC–MS spectrometer (Agilent Technologies, Ltd., Santa Clara, CA, USA) and an LTQ-FT Ultra ESI-FTICR-MS spectrometer (ThermoFisher Scientific, CA, USA) were used for recording HRESIMS data. LC–MS analyses were carried out on an Agilent Infinity II Prime LC with a DAD detector and a G6125B LC/MSD mass detector, or a Waters ACQUITY H-Class UPLC–MS (Milford, MA, USA) with a PDA detector and a QDA mass detector (ACQUITY UPLC® BEH, C18 column, 1.7 μm, 50 mm × 2.1 mm), using positive and negative mode electrospray ionization with a linear gradient of 5%–99% (v/v) acetonitrile (MeCN)/H2O in 8 min followed by 99% MeCN/H2O (both with 0.02% formic acid, v/v) for 4 min. The flow rate was 0.4 mL/min. Semi-preparative HPLC was carried out using a Silgreen ODS column (5 μm, 250 mm × 10 mm) in a LabAlliance SSI series III HPLC (Scientific Systems Inc., Philadelphia, PA, USA) with UV detector. Analytical grade solvents were used for extraction, HPLC grade MeCN and CH3OH were used for semi-preparation HPLC separation, and LC–MS grade MeCN was used for LC–MS analysis.
4.2. Strains and culture conditions
The fungus A. umbrinum DSM3193 was cultivated on PDA (BD) at 25 °C and stocked in −80 °C with 20% glycerol. For the production of secondary metabolites and RNA extraction, it was grown in PDB (BD) at 25 °C for 7 and 3 days, respectively. A. nidulans A1145 was cultured on CD plate (1.52 g/L KH2PO4, 6 g/L NaNO3, 0.52 g/L MgSO4·7H2O, 0.52 g/L KCl, 10 g/L glucose, 20 g/L agar, and 1 mL/L trace elements solution) with supplementation of 10 mmol/L uridine, 0.5 mg/L pyridoxine, 5 mmol/L uracil and 2.5 mg/L riboflavin for the collection of spores and the spores were stored at −80 °C in 20% glycerol. A. nidulans was grown on CD-ST medium (6 g/L NaNO3, 20 g/L tryptone, 20 g/L soluble starch, 1.52 g/L KH2PO4, 0.52 g/L KCl, 0.52 g/L MgSO4·7H2O, 1 mL/L trace elements, 20 g/L agar) to heterologously express the gene cluster and produce compounds. To construct the A. nidulans overexpression plasmids, Saccharomyces cerevisiae BJ5464-NpgA was used for yeast DNA recombination cloning. Yeast extract peptone dextrose (YPD) medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L dextrose) and the uracil-dropout semisynthetic medium were used for routine growth and selection of plasmids transformed into S. cerevisiae, respectively. E. coli trans1-Blue and trans2-Blue (Transgen Biotech) were used for routine cloning. E. coli strains were cultured at 37 °C in LB media (5 g/L yeast extract, 10 g/L NaCl, 10 g/L tryptone, and 20 g/L agar for solid cultivation). The concentration of ampicillin in the media was 100 μg/mL. Plasmids and primers are described in Supporting Information Tables S1 and S2.
4.3. Sequencing and bioinformatic analysis
The genomic DNA of A. umbrinum was prepared using the Plant Genomic DNA Kit (Tiangen). Gene clusters were analyzed by 2ndFind (http://biosyn.nih.go.jp/2ndfind) analysis and antiSMASH (https://fungismash.secondarymetabolites.org). The open reading frames of gene clusters were predicted and manually checked by comparison with known proteins using NCBI Blast tools. Functional domains of PKS were analyzed in PKS/NRPS Analysis Website (http://nrps.igs.umaryland.edu).
4.4. General molecular biology experiments
Restriction enzyme digestion using DNA restriction enzymes recommended by the manufacturer (NEB). DNA sequencing were used to confirm the PCR products and plasmid DNAs.
4.5. Reverse transcription-PCR (RT-PCR)
Mycelia of A. umbrinum were frozen with liquid nitrogen, then broken by grinding and dissolved in 1 mL Trizol (Invitrogen). The mixture was added with 200 μL chloroform, vortexed and centrifuged for 15 min at 12,000 rpm. An equal volume of isopropanol was added to the supernatant, then RNA was precipitated and was washed with 75% ethanol and resuspended in 40 μL RNase-free water. The genomic DNA was digested with RNase-free DNase (Promega). Then the cDNA was obtained by RT-PCR using PrimeScirpt™ RT reagent kit (Takara). The cDNA template was used to detect gene expression level by PCR.
4.6. Heterologous expression of the rum gene cluster in A. nidulans
For heterologous expression in A. nidulan, plasmids pANP, pANR, and pANU, which contained auxotrophic markers for pyridoxine (pyroA), riboflavin (riboB) and uracil (pyrG), respectively, were used as vectors to insert genes. glaA, gpdA, and amyB were inserted as promoters. Genes from the rum cluster with their terminators (∼300 bp) were amplified from genomic DNA extract from A. umbrinum with overhang primers (Table S1). pANR and pANP were digested with BamHI and PacI. pANU was double-digested with BamHI and NotI. The digested vector, corresponding promoters and overlapping DNA fragments were assembled in S. cerevisiae BJ5464-NpgA by yeast homologous recombination. The correct colonies were checked by colony-PCR. The plasmids extracted from yeast were transformed into E. coli trans1-Blue or trans2-Blue for propagation. The correct E. coli were transformants checked by colony-PCR. The plasmids extracted from E. coli were checked by enzymatic digestion and sequencing.
To obtain the protoplasts of A. nidulans, the spores of A. nidulans were inoculated into 30 mL liquid GMM medium (1 mL/L trace elements, 0.52 g/L KCl, 10 g/L glucose, 0.52 g/L MgSO4·7H2O, 10 g/L yeast extract, 6 g/L NaNO3, 1.52 g/L KH2PO4), which supplemented with 5 mmol/L uracil, 10 mmol/L uridine, 2.5 mg/L riboflavin, and 0.5 mg/L pyridoxine, and germinated at 37 °C, 220 rpm for 5–7 h. The germlings were harvested by centrifugation at 4000 rpm, 4 °C for 6 min, and washed with 25 mL osmotic buffer (0.6 mol/L MgSO4, 10 mmol/L sodium phosphate buffer, pH 7.0). Then, the germlings were resuspended in 10 mL osmotic buffer which containing 20 mg yatalase (Takara) and 30 mg lysing enzymes (Sigma–Aldrich) and incubated at 80 rpm for 7–8 h at 30 °C for enzymatic digestion. The mixture was transferred to a 50 mL tube and slowly added of 10 mL trapping buffer (0.1 mol/L Tris-HCl, 0.6 mol/L sorbitol, pH 7.0). And then centrifugation at 3750 rpm, 4 °C for 15 min, the protoplasts at the middle layer were transferred to a new tube and washed with doubled volumes of STC buffer (1.2 mol/L sorbitol, 10 mmol/L Tris-HCl, 10 mmol/L CaCl2, pH 7.5). After centrifugation at 3750 rpm, 4 °C for 5 min, the protoplasts were then resuspended in a suitable volume of STC buffer for transformation. For single transformation, firstly, plasmids were added into 100 μL of protoplasts and incubated on ice for 50 min. Then the mixture was added to 600 μL PEG solution (10 mmol/L Tris-HCl, 60% PEG4000 and 50 mmol/L CaCl2, pH 7.5) and lightly mixed, incubated 20 min at 25 °C. Finally, the mixture was transferred to CD-sorbitol medium (6 g/L NaNO3, 0.52 g/L MgSO4·7H2O, 0.52 g/L KCl, 1.52 g/L KH2PO4, 10 g/L glucose, 1 mL/L trace elements solution, 20 g/L agar and 1.2 mol/L sorbitol) and cultured at 37 °C for 2 days.
The picked transformants were inoculated into solid CD medium and cultured at 37 °C for 3–4 days to harvest spores. Three empty plasmids were transferred into the strain AN-control, which was used as the control in this study. The spores were inoculated into CD-ST medium for 4 days at 25 °C. Then, the medium was extracted with ethyl acetate (EtOAc). The EtOAc extracts were evaporated to dryness, and then redissolved in methanol (CH3OH) and subjected to LC–MS analysis.
4.7. Feeding experiments
4.7.1. Feeding experiments in A. nidulans
The transformed A. nidulans strains were inoculated in CD-ST medium, with l-proline (4, 2 mmol/L) or pyrrole-2-carboxylic acid (5, 2 mmol/L), for 4 days at 25 °C. To determine the function order of RumF and RumH, a mixture of 6–8, or 9–11, or 12–14 was fed to the corresponding strains for 4 days at 25 °C. After 4 days, the medium was extracted with EtOAc and analyzed by LC–MS.
4.7.2. l-Proline-[13C5,15N] feeding studies
A. umbrinum was cultured in 30 mL YPD media and cultivated at 28 °C, 220 rpm. 2 mmol/L l-proline-[13C5,15N] or proline used as substrates were added in 3rd day. The mycelia were collected after 7 days, followed by extraction with EtOAc and analysis by LC–MS.
4.8. Precursor-directed feeding studies
Precursors (dissolved in DMSO) were introduced to the CD-ST medium of corresponding heterologously overexpressed A. nidulans strains with a final concentration of 2 mmol/L. The cultures were exhaustively extracted with EtOAc on the 4th day, and the extracts were analyzed by LC–MS.
4.9. Isolation and structural characterization of metabolites
To isolate 12E-isorumbrin (2) and auxarconjugatin A (3), A. umbrinum was cultured in 500 mL of seed medium (uracil dropout) at 180 rpm, for 2 days at 28 °C. Then the seed culture was inoculated into 10 L production medium at 220 rpm, for 10 days at 28 °C. The culture was filtered and the mycelia were extracted with EtOAc. The resulted crude residue (17 g) was subjected to MCI gel column chromatography eluted with a gradient of CH3OH/H2O (20:80, 50:50, 70:30 and 100:0) to give five subfractions (Frs. A–E). Fr. D was subjected to flash chromatography on an ODS column (solvent gradient: 0–20 min, 70% CH3OH/H2O; 20–110 min, 70%–100% CH3OH/H2O; 110–130 min, 100% CH3OH/H2O) to afford Frs. D1–D15. Fraction Fr. D9 (446.6 mg) was separated by a Sephadex LH-20 column (CH3OH) to obtain Frs. D9A–D9E. Fr. D9D (190 mg) was purified by semi-preparative HPLC (CH3OH/H2O, 80:20) to obtain 2 (6.0 mg, tR = 37.5 min) and 3 (1.0 mg, tR = 20.5 min).
12E-Isorumbrin (2): red powder; ESI-MS m/z 356 [M–H]–. 13C and 1H NMR data are shown in Tables S4 and S6.
Auxarconjugatin B (3): red powder; ESI-MS m/z 324 [M–H]–. 13C and 1H NMR data are shown in Tables S4 and S6.
To isolate compounds 6–8, the spores of strain AN-rumCDE were inoculated into 5 L CD-ST agar media containing 5 (2 mmol/L) as substrate and grown for 7 days at 25 °C. The culture was exhaustively extracted with EtOAc to give the crude extract, which was partitioned between CH3OH and petroleum ether (PE) (1:1, v/v). The CH3OH layer extract was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to afford four fractions (Frs. 1–4). Fr. 3 was separated by semi-preparative HPLC (CH3OH/H2O, 68:32) to obtain 6 (1.2 mg, tR = 19.0 min), 7 (0.8 mg, tR = 23.5 min), and 8 (7.0 mg, tR = 32.5 min).
Compound 6: red powder; UV (MeCN) λmax (log ε) 267 (4.22), 318 (4.13), 334 (4.28), 434 (4.75) nm. IR νmax 3429, 2960, 2919, 2851, 1722, 1621, 1566, 1543, 1468, 1432, 1415, 1261, 1097, 1035, 863, 802, 720 cm−1. 13C and 1H NMR data are shown in Tables S4 and S6. ESI-MS m/z 282 [M+H]+ and 280 [M–H]–. HRESIMS m/z 282.1119 [M+H]+ (Calcd. for C17H16NO3, 282.1125).
Compound 7: red powder; UV (CHCl3) λmax (log ε) 267 (4.35), 318 (4.17), 334 (4.20), 434 (4.92) nm. IR νmax 3417, 2962, 2918, 2851, 1724, 1600, 1563, 1468, 1445, 1407, 1261, 1092, 1038, 863, 802, 705 cm−1. 13C and 1H NMR data are shown in Tables S4 and S6. ESI-MS m/z 296 [M+H]+ and 294 [M–H]–. HRESIMS m/z 296.1276 [M+H]+ (Calcd. for C18H18NO3, 296.1281).
Compound 8: red powder; UV (CHCl3) λmax (log ε) 268 (3.75), 319 (3.60), 334 (3.84), 434 (4.28) nm. IR νmax 3371, 3207, 2961, 2923, 2853, 1634, 1584, 1554, 1474, 1405, 1374, 1262, 1155, 1096, 1029, 981, 800, 724 cm−1. 13C and 1H NMR data are shown in Tables S4 and S6. ESI-MS m/z 310 [M+H]+ and 308 [M–H]–. HRESIMS m/z 310.1425 [M+H]+ (Calcd. for C19H20NO3, 310.1438).
To isolate auxarconjugatin D (9), compound 10, and 12E-dechloroisorumbrin (11), the spores of strain AN-rumCDEF were cultured in 6 L CD-ST agar media containing 5 (2 mmol/L) as substrate and cultured for 7 days at 28 °C. The culture was extracted with EtOAc to give the crude extract (3.8 g), which was partitioned between CH3OH and PE (1:1, v/v). The CH3OH layer extract (3.4 g) was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to afford six fractions (Frs. 1–6). Fr. 4 (330 mg) was separated by semi-preparative HPLC (CH3OH/H2O, 72:28, 0.02% formic acid) to obtain 9 (2.2 mg, tR = 24.5 min), 10 (1.0 mg, tR = 27.0 min) and 11 (70.0 mg, tR = 37.5 min).
Auxarconjugatin D (9): red powder; ESI-MS m/z 296 [M+H]+. 13C and 1H NMR data are shown in Tables S4 and S6.
Compound 10: red powder; UV (MeCN) λmax (log ε) 269 (3.99), 338 (3.84), 454 (4.42) nm. IR νmax 3340, 2962, 2920, 2852, 1712, 1661, 1628, 1538, 1412, 1261, 1095, 1037, 863, 801, 705 cm−1. 13C and 1H NMR data are shown in Tables S4 and S7. ESI-MS m/z 310 [M+H]+ and 308 [M–H]–. HRESIMS m/z 310.1433 [M+H]+ (Calcd. for C19H20NO3, 310.1438).
12E-dechloroisorumbrin (11): red powder; ESI-MS m/z 323 [M+H]+. 13C and 1H NMR data are shown in Tables S4 and S7.
To isolate 12E-demethylisorumbrin (13) and compound 14, the spores of A. nidulans strain AN-rumCDEH were cultured in 8 L CD-ST agar media with 5 (2 mmol/L) as substrate and grown for 6 days at 28 °C. The culture was exhaustively extracted with EtOAc to obtain the crude extract (6.8 g), which was partitioned between CH3OH and n-hexane. The CH3OH layer extract was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to afford seven fractions (Frs. 1–7). Fr. 4 (320 mg) was separated by semi-preparative HPLC (MeCN/H2O, 53:47, 0.02% formic acid) to obtain 13 (1.2 mg, tR = 20.7 min) and 14 (5.83 mg, tR = 24.8 min).
12E-demethylisorumbrin (13): red powder; 13C and 1H NMR data are shown in Tables S4 and S7. ESI-MS m/z 330 [M+H]+ and 328 [M–H]−. HRESIMS m/z 328.0749 [M–H]− (Calcd. for C18H15ClNO3, 328.0746).
Compound 14: red powder; UV (MeCN) λmax: 269, 322, 339, 430 nm. 13C and 1H NMR data are shown in Tables S4 and S7. ESI-MS m/z 344 [M+H]+ and 342 [M–H]−. HRESIMS m/z 342.0904 [M–H]− (Calcd. for C19H17ClNO3, 342.0902).
To isolate auxarconjugatin E (15), the spores of A. nidulans strain AN-rumCDEFH were inoculated into 5 L CD-ST agar media with 5 (2 mmol/L) as precursor and grown for 7 days at 28 °C. The culture was exhaustively extracted with EtOAc to obtain crude extract (4.3 g), which was partitioned between CH3OH and PE (1:1, v/v). The CH3OH layer extract was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to afford seven fractions (Frs. 1–7). Fr. 4 (380 mg) was separated by semi-preparative HPLC (CH3OH/H2O, 72:28, 0.02% formic acid) to yield 15 (0.9 mg, tR = 27.0 min).
Auxarconjugatin E (15): red powder; 13C and 1H NMR data are shown in Supporting Information Tables S5 and S7. ESI-MS m/z 344 [M+H]+ and 342 [M–H]−. HRESIMS m/z 344.1042 [M+H]+ (Calcd. For C19H19O3NCl, 344.1048).
To isolate compound 16, the spores of the A. nidulans strain expressing rumCDE were inoculated into 10 L CD-ST agar media with 5a (2 mmol/L) as precursor and grown for 7 days at 28 °C. The culture was exhaustively extracted with EtOAc to obtain crude extract (11 g), which was partitioned between CH3OH and PE (1:1, v/v). The CH3OH layer extract (10 g) was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to obtain 22 fractions (Frs. 1–22). Frs. 13–19 was combined (232 mg) and separated by semi-preparative HPLC (MeCN/H2O, 53:47, 0.02% formic acid) to yield 16 (5.9 mg, tR = 19.2 min).
Compound 16: red powder; UV (MeCN) λmax: 266, 320, 332, 430 nm. 13C and 1H NMR data are shown in Tables S5 and S7. ESI-MS m/z 328 [M+H]+ and 326 [M–H]−. HRESIMS m/z 328.1339 [M+H]+ (Calcd. for C19H19FNO3, 328.1343).
To isolate compounds 17 and 18, the spores of the A. nidulans strain expressing rumCDEFH were inoculated into 5 L CD-ST solid media containing 5a (2 mmol/L), and grown for 6 days at 28 °C. The culture was exhaustively extracted with EtOAc to obtain the crude extract (3.3 g), which was redissolved with n-hexane. The residue (3 g) was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to afford eight fractions (Frs. 1–8). Fr. 6 (7 mg) was separated by semi-preparative HPLC (MeCN/H2O, 60:40, 0.02% formic acid) to yield 17 (1.28 mg, tR = 23.2 min). Fr. 4 (5 mg) was separated by semi-preparative HPLC (MeCN/H2O, 65:35, 0.02% formic acid) to yield 18 (1.15 mg, tR = 16.3 min).
Compound 17: red powder; UV (MeCN) λmax: 270, 322, 338, 437 nm. 13C and 1H NMR data are shown in Supporting Information Tables S5 and S8. ESI-MS m/z 342 [M+H]+ and 340 [M–H]−. HRESIMS m/z 342.1495 [M+H]+ (Calcd. for C20H21FNO3, 342.1500).
Compound 18: red powder; UV (MeCN) λmax: 261, 312, 329, 435 nm. 13C and 1H NMR data are shown in Tables S5 and S8. ESI-MS m/z 314 [M+H]+ and 312 [M–H]−. HRESIMS: m/z 314.1185 [M+H]+ (Calcd. for C18H17FNO3, 314.1187).
To isolate compounds 19–21 and 3-floroisorumbrin (22), the spores of the A. nidulans strain expressing rumCDEFH were inoculated into 5 L CD-ST agar media containing 5c (2 mmol/L), and grown for 6 days at 28 °C. The culture was exhaustively extracted with EtOAc to obtain the crude extract (10.2 g). The crude extract was redissolved with n-hexane and the residue (9 g) was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to afford 10 fractions (Frs. 1–10). Fr. 6 (40 mg) was separated by semi-preparative HPLC (MeCN/H2O, 53:47, 0.02% formic acid) to yield 20 (1.54 mg, tR = 28.7 min), 21 (11.4 mg, tR = 24.3 min) and 22 (3.21 mg, tR = 23.1 min). Fr. 8 (60 mg) was separated by semi-preparative HPLC (MeCN/H2O, 65:35, 0.02% formic acid) to yield 19 (23.5 mg, tR = 26.5 min).
Compound 19: red powder; UV (MeCN) λmax: 261, 325, 339, 430 nm. 13C and 1H NMR data are shown in Tables S5 and S8. ESI-MS m/z 376 [M+H]+ and 374 [M–H]−. HRESIMS m/z 376.1108 [M+H]+ (calcd for C20H20ClFNO3, 376.1110).
Compound 20: red powder; UV (MeCN) λmax: 267, 324, 339, 432 nm. 13C and 1H NMR data are shown in Tables S5 and S8. ESI-MS m/z 362 [M+H]+ and 360 [M–H]−. HRESIMS m/z 362.0955 [M+H]+ (Calcd. For C19H18ClFNO3, 362.0954).
Compound 21: red powder; UV (MeCN) λmax: 262, 315, 330, 426 nm. 13C and 1H NMR data are shown in Tables S5 and S8. ESI-MS m/z 348 [M+H]+ and 346 [M–H]−. HRESIMS m/z 348.0795 [M+H]+ (Calcd. for C18H16ClFNO3, 348.0797).
3-Floroisorumbrin (22): red powder; 13C and 1H NMR data are shown in Tables S5 and S8. ESI-MS m/z 342 [M+H]+ and 340 [M–H]−. HRESIMS m/z 342.1502 [M+H]+ (Calcd. for C20H20FNO3, 342.1500).
To isolate 3-bromoisorumbrin (23) and compound (24), the spores of the A. nidulans strain expressing rumCDEFH were inoculated into 5 L CD-ST agar media containing 5e (2 mmol/L), and grown for 6 days at 28 °C. The culture was exhaustively extracted with EtOAc to obtain the crude extract (5.2 g), which was redissolved with n-hexane. The residue (5 g) was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to afford six fractions (Frs. 1–6). Fr. 6 (32 mg) was separated by semi-preparative HPLC (MeCN/H2O, 70:30, 0.02% formic acid) to yield 23 (1.7 mg, tR = 17.5 min). Fr. 4 (24 mg) was separated by semi-preparative HPLC (MeCN/H2O, 75:25, 0.02% formic acid) to yield 24 (1.0 mg, tR = 25.5 min).
3-Bromoisorumbrin (23): red powder; 13C and 1H NMR data are shown in Tables S5 and S8. ESI-MS m/z 403 [M+H]+ and 401 [M–H]−.
Compound 24: red powder; (MeCN) λmax: 247, 301, 414 nm. 13C and 1H NMR data are shown in Supporting Information Tables S5 and S9. ESI-MS m/z 349 [M+H]+ and 347 [M–H]−. HRESIMS m/z 348.0234 [M+H]+ (Calcd. for C16H15BrNO3, 348.0230).
To isolation of compound 25, the spores of the A. nidulans strain expressing malACDEF were inoculated into 9.5 L CD-ST agar media containing 5 (2 mmol/L), and grown for 7 days at 28 °C. The culture was exhaustively extracted with EtOAc to give the crude extract (3.5 g), which was redissolved with petroleum ether (PE). The residue (3 g) was purified by BUCHI REV Prep with a gradient subjected CH3OH/H2O (30:70 to 100:0) to afford 15 fractions (Frs. 1–15). Fr. 12 (46 mg) was separated by semi-preparative HPLC (MeCN/H2O, 38:62, 0.02% formic acid) to yield 25 (6.0 mg, tR = 18.1 min).
Compound 25: dark colored amorphous powder; UV (MeCN) λmax: 230, 295, 410 nm. 13C and 1H NMR data are shown in Supporting Information Table S6. HRESIMS m/z 292.0736 [M+H]+ (Calcd. for C15H15ClNO3, 292.0735).
4.10. Anti-HIV assay
4.10.1. Cell culture
HEK293T and SupT1 cells were maintained in DMEM and RPMI1640 supplemented with 10% fetal bovine serum (FBS), respectively.
4.10.2. Plasmids and reagents
The HIV pNL4-3Luc.R-E- vector encodes for a full-length HIV-1 proviral DNA, in which luciferase gene is inserted as a reporter and env is defective. A plasmid pHIT/G expressing the vesicular stomatitis virus glycoprotein (VSV-G) was provided by Johnny He24.
4.10.3. Viral preparation and assay for the inhibitory activity measurement of compounds
In short, 2 × 105 HEK293T cells were co-transfected with 0.4 μg of pHIT/G and 0.6 μg of pNL4-3Luc.R-E-. The pseudotyped HIV-1 viruses were harvested by filtration followed by the quantification of viral capsid protein by p24 antigen capture ELISA. The resulting viruses were used to infect a total of 5 × 105 SupT1 cells at an MOI of 1, in the presence of compounds at serial dilutions. After further incubation at 37 °C, 5% CO2 for 48 h, a firefly luciferase assay system (Promega, Madison, WI, USA) was performed to determine the inhibition rate.
4.10.4. Semi-quantitative real-time PCR
VSV-G pseudotyped NL4-3Luc.R-E- viruses were used to infect a total of 5 × 106 SupT1 cells treated with 4 nmol/L efavirenz (EFV), 25 nmol/L raltegravir (RAL) or test compounds (10 times IC50). Cells were harvested at 24 h and subjected to total DNA extraction by DNeasy Blood&Tissue Kit (Qiagen). An aliquot of each sample was analyzed by quantitative PCR. The primers used in the PCR were listed in Table S1. Copy numbers of total viral DNA, integrated DNA, and 2-LTR were normalized by that of the GAPDH gene.
Acknowledgments
This work was supported financially by the National Key Research and Development Program of China (2018YFA0901900), and the CAMS Innovation Fund for Medical Sciences (CIFMS, No. 2021-I2M-1-029, China).
Author contributions
Beifen Zhong conducted experiments, bioinformatics analysis and wrote the manuscript; Jun Wang constituted the skeleton and post-PKS modification pathway of rumbrin; Changhui Shang performed the isolations and structural elucidation of compounds from heterologous expression and wrote the manuscript; Jiajia Wen and Yujia Wang performed anti-HIV assays; Jian Bai involved in NMR data analysis; Shan Cen designed the anti-HIV assays, discussed the project and edited the manuscript; Youcai Hu designed and guided the entire project and edited the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Footnotes
Peer review under responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2022.02.005.
Contributor Information
Shan Cen, Email: shancen@imb.pumc.edu.cn.
Youcai Hu, Email: huyoucai@imm.ac.cn.
Appendix A. Supporting information
The following is the Supplementary data to this article:
References
- 1.Shen B. A new golden age of natural products drug discovery. Cell. 2015;163:1297–1300. doi: 10.1016/j.cell.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 3.Bai J., Mu R., Dou M., Yan D., Liu B., Wei Q., et al. Epigenetic modification in histone deacetylase deletion strain of Calcarisporium arbuscula leads to diverse diterpenoids. Acta Pharm Sin B. 2018;8:687–689. doi: 10.1016/j.apsb.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei Q., Bai J., Yan D., Bao X., Li W., Liu B., et al. Genome mining combined metabolic shunting and OSMAC strategy of an endophytic fungus leads to the production of diverse natural products. Acta Pharm Sin B. 2021;11:572–587. doi: 10.1016/j.apsb.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alanjary M., Kronmiller B., Adamek M., Blin K., Weber T., Huson D., et al. The antibiotic resistant target seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery. Nucleic Acids Res. 2017;45:W42–W48. doi: 10.1093/nar/gkx360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lautru S., Deeth R.J., Bailey L.M., Challis G.L. Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol. 2005;1:265–269. doi: 10.1038/nchembio731. [DOI] [PubMed] [Google Scholar]
- 7.Yan Y., Liu Q., Zang X., Yuan S., Bat-Erdene U., Nguyen C., et al. Resistance-gene-directed discovery of a natural-product herbicide with a new mode of action. Nature. 2018;559:415–418. doi: 10.1038/s41586-018-0319-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Panter F., Krug D., Baumann S., Müller R. Self-resistance guided genome mining uncovers new topoisomerase inhibitors from myxobacteria. Chem Sci. 2018;9:4898–4908. doi: 10.1039/c8sc01325j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan Y., Liu N., Tang Y. Recent developments in self-resistance gene directed natural product discovery. Nat Prod Rep. 2020;37:879–892. doi: 10.1039/c9np00050j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Zhang R., Chen X., Sun X., Yan Y., Shen X., et al. Biosynthesis of aromatic polyketides in microorganisms using type II polyketide synthases. Microb Cell Factories. 2020;19:110. doi: 10.1186/s12934-020-01367-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldman A.J., Balskus E.P. Lomaiviticin biosynthesis employs a new strategy for starter unit generation. Org Lett. 2014;16:640–643. doi: 10.1021/ol403714g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe I., Utsumi Y., Oguro S., Morita H., Sano Y., Noguchi H. A plant type III polyketide synthase that produces pentaketide chromone. J Am Chem Soc. 2005;127:1362–1363. doi: 10.1021/ja0431206. [DOI] [PubMed] [Google Scholar]
- 13.Crawford J.M., Vagstad A.L., Ehrlich K.C., Townsend C.A. Starter unit specificity directs genome mining of polyketide synthase pathways in fungi. Bioorg Chem. 2008;36:16–22. doi: 10.1016/j.bioorg.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M., Hou X.F., Qi L.H., Yin Y., Li Q., Pan H.X., et al. Biosynthesis of trioxacarcin revealing a different starter unit and complex tailoring steps for type II polyketide synthase. Chem Sci. 2015;6:3440–3447. doi: 10.1039/c5sc00116a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalaitzis J.A., Cheng Q., Thomas P.M., Kelleher N.L., Moore B.S. In vitro biosynthesis of unnatural enterocin and wailupemycin polyketides. J Nat Prod. 2009;72:469–472. doi: 10.1021/np800598t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong H., Fill T., Leadlay P.F. A common origin for guanidinobutanoate starter units in antifungal natural products. Angew Chem Int Ed Engl. 2013;52:13096–13099. doi: 10.1002/anie.201308136. [DOI] [PubMed] [Google Scholar]
- 17.Hu D.X., Withall D.M., Challis G.L., Thomson R.J. Structure, chemical synthesis, and biosynthesis of prodiginine natural products. Chem Rev. 2016;116:7818–7853. doi: 10.1021/acs.chemrev.6b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorrestein P.C., Yeh E., Garneau-Tsodikova S., Kelleher N.L., Walsh C.T. Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. Proc Natl Acad Sci U S A. 2005;102:13843–13848. doi: 10.1073/pnas.0506964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamagishi Y., Matsuoka M., Odagawa A., Kato S., Shindo K., Mochizuki J. Rumbrin, a new cytoprotective substance produced by Auxarthron umbrinum. I. Taxonomy, production, isolation and biological activities. J Antibiot (Tokyo) 1993;46:884–887. doi: 10.7164/antibiotics.46.884. [DOI] [PubMed] [Google Scholar]
- 20.Clark B.R., Capon R.J., Lacey E., Tennant S., Gill J.H. Polyenylpyrroles and polyenylfurans from an Australian isolate of the soil ascomycete Gymnoascus reessii. Org Lett. 2006;8:701–704. doi: 10.1021/ol052880y. [DOI] [PubMed] [Google Scholar]
- 21.Fang Z., Liao P.C., Yang Y.L., Yang F.L., Chen Y.L., Lam Y., et al. Synthesis and biological evaluation of polyenylpyrrole derivatives as anticancer agents acting through caspases-dependent apoptosis. J Med Chem. 2010;53:7967–7978. doi: 10.1021/jm100619x. [DOI] [PubMed] [Google Scholar]
- 22.Clark B.R., Murphy C.D. Biosynthesis of pyrrolylpolyenes in Auxarthron umbrinum. Org Biomol Chem. 2009;7:111–116. doi: 10.1039/b813236d. [DOI] [PubMed] [Google Scholar]
- 23.Clark B.R., O'Connor S., Fox D., Leroy J., Murphy C.D. Production of anticancer polyenes through precursor-directed biosynthesis. Org Biomol Chem. 2011;9:6306–6311. doi: 10.1039/c1ob05667k. [DOI] [PubMed] [Google Scholar]
- 24.Levin A., Armon-Omer A., Rosenbluh J., Melamed-Book N., Graessmann A., Waigmann E., et al. Inhibition of HIV-1 integrase nuclear import and replication by a peptide bearing integrase putative nuclear localization signal. Retrovirology. 2009;6:112. doi: 10.1186/1742-4690-6-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamagishi Y., Shindo K., Kawai H. Rumbrin, a new cytoprotective substance produced by Auxarthron umbrinum. II. Physico-chemical properties and structure determination. J Antibiot (Tokyo) 1993;46:888–891. doi: 10.7164/antibiotics.46.888. [DOI] [PubMed] [Google Scholar]
- 26.Lin T.S., Chiang Y.M., Wang C.C. Biosynthetic pathway of the reduced polyketide product citreoviridin in Aspergillus terreus var. aureus revealed by heterologous expression in Aspergillus nidulans. Org Lett. 2016;18:1366–1369. doi: 10.1021/acs.orglett.6b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skellam E. Biosynthesis of fungal polyketides by collaborating and trans-acting enzymes. Nat Prod Rep. 2022;39:754–783. doi: 10.1039/d1np00056j. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y., Bai J., Zhang L., Zhang C., Liu B., Hu Y. Self-resistance in the biosynthesis of fungal macrolides involving cycles of extracellular oxidative activation and intracellular reductive inactivation. Angew Chem Int Ed Engl. 2021;60:6639–6645. doi: 10.1002/anie.202015442. [DOI] [PubMed] [Google Scholar]
- 29.Moore B.S., Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat Prod Rep. 2002;19:70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 30.Garneau S., Dorrestein P.C., Kelleher N.L., Walsh C.T. Characterization of the formation of the pyrrole moiety during clorobiocin and coumermycin A1 biosynthesis. Biochemistry. 2005;44:2770–2780. doi: 10.1021/bi0476329. [DOI] [PubMed] [Google Scholar]
- 31.Garneau-Tsodikova S., Dorrestein P.C., Kelleher N.L., Walsh C.T. Protein assembly line components in prodigiosin biosynthesis: characterization of PigA,G,H,I,J. J Am Chem Soc. 2006;128:12600–12601. doi: 10.1021/ja063611l. [DOI] [PubMed] [Google Scholar]
- 32.Thomas M.G., Burkart M.D., Walsh C.T. Conversion of L-proline to pyrrolyl-2-carboxyl-S-PCP during undecylprodigiosin and pyoluteorin biosynthesis. Chem Biol. 2002;9:171–184. doi: 10.1016/s1074-5521(02)00100-x. [DOI] [PubMed] [Google Scholar]
- 33.Fraley A.E., Garcia-Borràs M., Tripathi A., Khare D., Mercado-Marin E.V., Tran H., et al. Function and structure of MalA/MalA', iterative halogenases for late-stage C-H functionalization of indole alkaloids. J Am Chem Soc. 2017;139:12060–12068. doi: 10.1021/jacs.7b06773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy C.D., Clark B.R., Amadio J. Metabolism of fluoroorganic compounds in microorganisms: impacts for the environment and the production of fine chemicals. Appl Microbiol Biotechnol. 2009;84:617–629. doi: 10.1007/s00253-009-2127-0. [DOI] [PubMed] [Google Scholar]
- 35.Deb Roy A., Grüschow S., Cairns N., Goss R.J. Gene expression enabling synthetic diversification of natural products: chemogenetic generation of pacidamycin analogs. J Am Chem Soc. 2010;132:12243–12245. doi: 10.1021/ja1060406. [DOI] [PubMed] [Google Scholar]
- 36.Kale A.J., McGlinchey R.P., Lechner A., Moore B.S. Bacterial self-resistance to the natural proteasome inhibitor salinosporamide A. ACS Chem Biol. 2011;6:1257–1264. doi: 10.1021/cb2002544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeh H.H., Ahuja M., Chiang Y.M., Oakley C.E., Moore S., Yoon O., et al. Resistance gene-guided genome mining: serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic pathway for fellutamide B, a proteasome inhibitor. ACS Chem Biol. 2016;11:2275–2284. doi: 10.1021/acschembio.6b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thaker M.N., Wang W., Spanogiannopoulos P., Waglechner N., King A.M., Medina R., et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol. 2013;31:922–927. doi: 10.1038/nbt.2685. [DOI] [PubMed] [Google Scholar]
- 39.Tang X., Li J., Millán-Aguiñaga N., Zhang J.J., O'Neill E.C., Ugalde J.A., et al. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem Biol. 2015;10:2841–2849. doi: 10.1021/acschembio.5b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yarlagadda V., Medina R., Johnson T.A., Koteva K.P., Cox G., Thaker M.N., et al. Resistance-guided discovery of elfamycin antibiotic producers with antigonococcal activity. ACS Infect Dis. 2020;6:3163–3167. doi: 10.1021/acsinfecdis.0c00467. [DOI] [PubMed] [Google Scholar]
- 41.O'Neill E.C., Schorn M., Larson C.B., Millán-Aguiñaga N. Targeted antibiotic discovery through biosynthesis-associated resistance determinants: target directed genome mining. Crit Rev Microbiol. 2019;45:255–277. doi: 10.1080/1040841X.2019.1590307. [DOI] [PubMed] [Google Scholar]
- 42.Almabruk K.H., Dinh L.K., Philmus B. Self-resistance of natural product producers: past, present, and future focusing on self-resistant protein variants. ACS Chem Biol. 2018;13:1426–1437. doi: 10.1021/acschembio.8b00173. [DOI] [PubMed] [Google Scholar]
- 43.Li J., Derewenda U., Dauter Z., Smith S., Derewenda Z.S. Crystal structure of the Escherichia coli thioesterase II, a homolog of the human Nef binding enzyme. Nat Struct Biol. 2000;7:555–559. doi: 10.1038/76776. [DOI] [PubMed] [Google Scholar]
- 44.Liu L.X., Margottin F., Le Gall S., Schwartz O., Selig L., Benarous R., et al. Binding of HIV-1 Nef to a novel thioesterase enzyme correlates with Nef-mediated CD4 down-regulation. J Biol Chem. 1997;272:13779–13785. doi: 10.1074/jbc.272.21.13779. [DOI] [PubMed] [Google Scholar]
- 45.Dillon S.C., Bateman A. The hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinf. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding J., Zhao J., Yang Z., Ma L., Mi Z., Wu Y., et al. Microbial natural product alternariol 5-O-methyl ether inhibits HIV-1 integration by blocking nuclear import of the pre-integration complex. Viruses. 2017;9:105. doi: 10.3390/v9050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmeira J.D.F., Argañaraz G.A., de Oliveira G., Argañaraz E.R. Physiological relevance of ACOT8-Nef interaction in HIV infection. Rev Med Virol. 2019;29:e2057. doi: 10.1002/rmv.2057. [DOI] [PubMed] [Google Scholar]
- 48.Hunt M.C., Solaas K., Kase B.F., Alexson S.E. Characterization of an acyl-coA thioesterase that functions as a major regulator of peroxisomal lipid metabolism. J Biol Chem. 2002;277:1128–1138. doi: 10.1074/jbc.M106458200. [DOI] [PubMed] [Google Scholar]
- 49.Staudt R.P., Alvarado J.J., Emert-Sedlak L.A., Shi H., Shu S.T., Wales T.E., et al. Structure, function, and inhibitor targeting of HIV-1 Nef-effector kinase complexes. J Biol Chem. 2020;295:15158–15171. doi: 10.1074/jbc.REV120.012317. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.