Abstract
Pharmaceutical analysis is a discipline based on chemical, physical, biological, and information technologies. At present, biotechnological analysis is a short branch in pharmaceutical analysis; however, bioanalysis is the basis and an important part of medicine. Biotechnological approaches can provide information on biological activity and even clinical efficacy and safety, which are important characteristics of drug quality. Because of their advantages in reflecting the overall biological effects or functions of drugs and providing visual and intuitive results, some biotechnological analysis methods have been gradually applied to pharmaceutical analysis from raw material to manufacturing and final product analysis, including DNA super-barcoding, DNA-based rapid detection, multiplex ligation-dependent probe amplification, hyperspectral imaging combined with artificial intelligence, 3D biologically printed organoids, omics-based artificial intelligence, microfluidic chips, organ-on-a-chip, signal transduction pathway-related reporter gene assays, and the zebrafish thrombosis model. The applications of these emerging biotechniques in pharmaceutical analysis have been discussed in this review.
Key words: Biotechnology, Pharmaceutical analysis, Raw materials, Manufacturing control, Quality analysis
Graphical abstract
Pharmaceutical analysis (PA) is a discipline dominant by chemical and physical technologies, with week role of biological and information technologies. Here, the applications of emerging biotechnologies in PA are reviewed.
1. Introduction
Pharmaceutical analysis is a discipline based on chemical, physical, biological, and information technologies. Chemical substances are the basis for drugs to exert their therapeutic effects, and chemical-based analytical methods were the first to receive attention. The physical properties of a drug (e.g., uniformity and crystal form) can also affect its quality and have thus received attention. Nevertheless, biotechnology and latest information technology have not received enough attention from pharmacists.
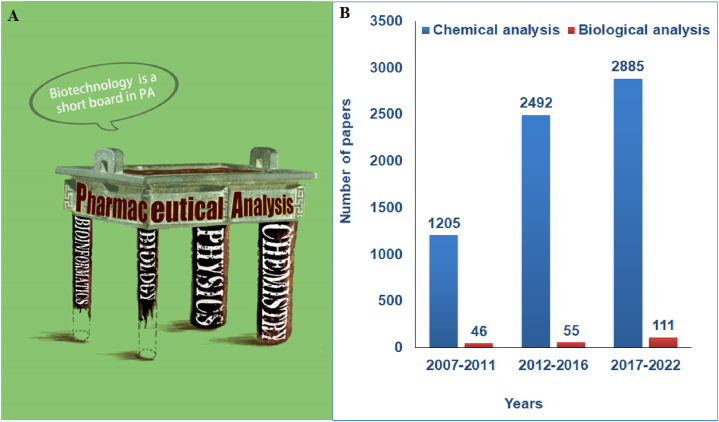
The role of biotechnology in pharmaceutical analysis is weak (Fig. 1A). The number of papers published in the last 15 years (2007–2022) included in the Web of Science was searched with the keywords “pharmaceutical analysis” combined with “chemical analysis” or “biological analysis”. The number of papers published every five years are shown in Fig. 1B, indicating that chemical analysis still plays a dominant role in pharmaceutical analysis, whereas the role of biological analysis is limited. Although chemical and physical methods show high accuracy, sensitivity, throughput, and robustness, some of the detected index components do not represent the efficacy or effectiveness of natural products, which is related to the complex action mechanism of natural products with complex composition.
Figure 1.
Overview of current pharmaceutical analysis methods.
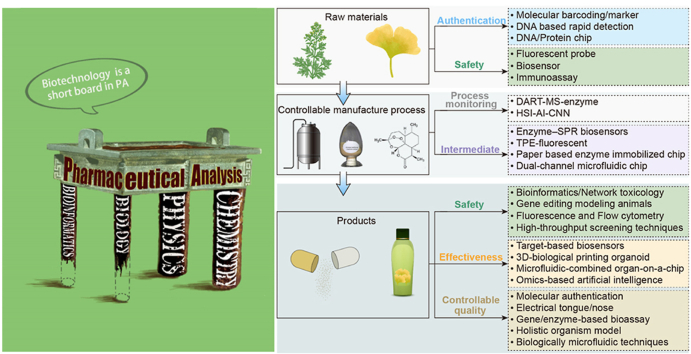
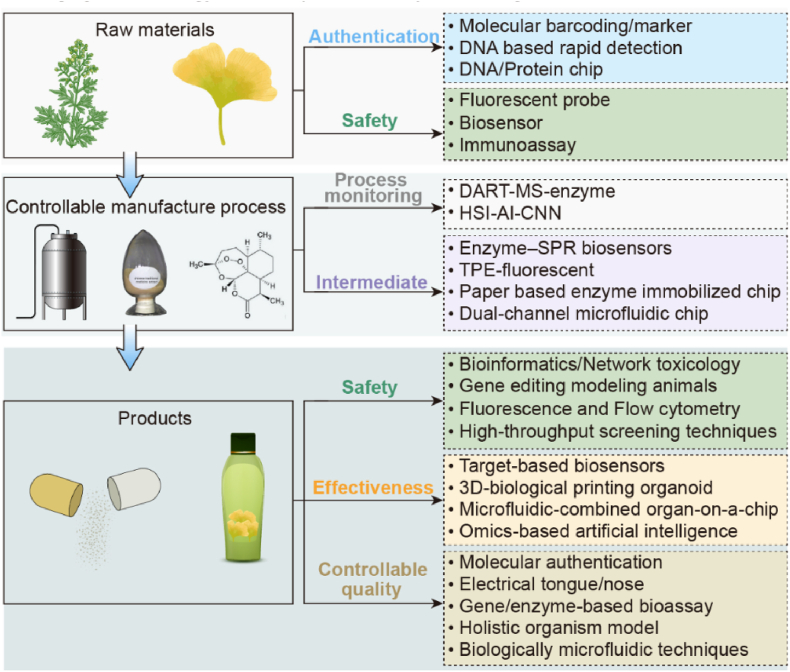
Biological analysis is the basis and an important part of life science, particularly medicine, and the development of biotechnology has advanced the field of medicine1. Because of their advantages in reflecting the overall biological effects, function, or action mechanism of drugs and providing visual and intuitive results, some biotechnologies have been gradually applied to pharmaceutical analysis (Fig. 2) from raw material to manufacturing and final products. Biological detection methods are the core evaluation techniques used to study the effectiveness, safety, and quality of drugs2. In this review, the applications of these emerging biotechnologies in pharmaceutical analysis are summarized and discussed.
Figure 2.
Emerging biotechnologies and bioinformatics for pharmaceutical analysis.
2. Application of biotechnology for raw material analysis
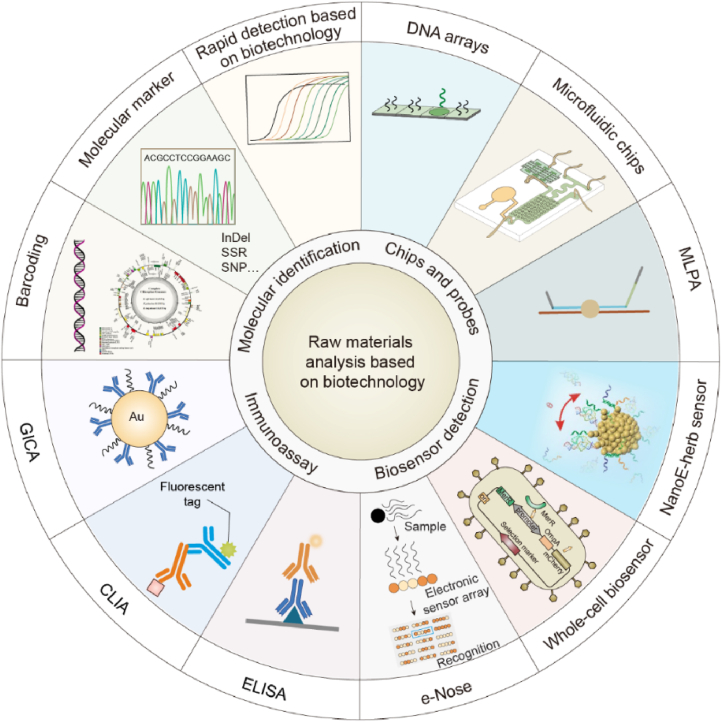
The current biotechnologies for the raw material analysis of animal and herbal medicine in the Chinese Pharmacopoeia (2020) are polymerase chain reaction (PCR) and polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP). Other novel and convenient biotechnological technologies3 are being adopted in actual application and are discussed in Table 1 and Fig. 3.
Table 1.
Application of detection methods based on biotechnology in raw materials.
| Technique | Sort | Characteristic | Application |
|---|---|---|---|
| Barcoding | DNA barcoding | Not affected by individual development | Paeonia lactiflora4, Crataegus species5 |
| DNA super-barcoding | |||
| Molecular marker | InDel, SNP, SSR, ISSR, ESTs, SRAP, RAPD, AFLP, PCR-RFLP | Wide distribution, large quantity, high conservation and high polymorphism | Zanthoxylum schinifolium6, Angelica dahurica7 |
| Rapid detection technology based on biotechnology | HAD, LAMP, RPA, HRM, RT-qPCR | Simple, rapid, efficient, sensitive and specific | Pheretima aspergillum8 |
| Chips | DNA chip, DArT, Protein chip, Microfluidic chip | Fast, highly efficient, automated, parallel, and economical | Chinemys reevesii9, heavy metal10 |
| Probes | MLPA Fluorescent probe |
High throughput, specificity, speed and simplicity | Fritillaria cirrhosa11, Hg2+12 |
| Biosensor | Electrochemical sensor | Fast, sensitive, stable, reliable, selective and reproducible | Crocus sativus13, Pb2+, Hg2+14, organophosphorus pesticides15, imidacloprid16, thiamethoxam17, glyphosate18 |
| Enzyme biosensor | |||
| Whole-cell biosensor | |||
| Nanochannel sensor | |||
| Molecularly imprinted sensors | |||
| Immunosensors | |||
| Fluorescent sensor | |||
| Electronic nose | |||
| Immunoassay | Enzyme linked immunosorbent assay | Fast, sensitive, cheap | Imidacloprid19 |
| Fluorescent immunoassay | Sensitive, easy to operate | Acetochlor20 | |
| Chemiluminescent immunoassay | Specific, fast, automatic | Parathion21, Cu2+22 | |
| Gold immunochromatographic assay | Result visualization | Carbendazim23 |
Figure 3.
Biotechnological methods to detect raw materials.
2.1. Molecular technologies for identifying the authenticity of raw materials
With the continuous upgradation of sequencing technology and the continuous decline in sequencing cost, the genetic information of many species has been obtained, and different databases have been established24,25, which lays the foundation for the development of molecular identification technology and the breeding of new varieties26. Barcoding, molecular markers, and rapid detection are technologies based on DNA and can accurately identify raw materials.
2.1.1. Barcoding
Barcoding is widely used to identify plant and animal materials. It uses a standard and relatively short sequence for species identification and is not affected by ontological development stages. Single barcodes, such as internal transcribed spacer (ITS)/ITS2, trnH-psbA, MatK, RbcL, and COI, were first used to identify the authenticity of raw materials. ITS/ITS2 was used to distinguish different species of Euphorbiaceae27 and Apocynaceae28 and identify cultivated varieties of Paeonia lactiflora4. COI can accurately identify snake medicine29. ITS2 has high identification ability for ultrafine powder or cell wall-broken powder30. Two- and three-loci combinations were also used to identify the authenticity of raw materials. matK and rbcL were used to identify Arisaematis Rhizoma, Pinelliae Tuber, and common adulterants31. matK and rbcL combined with trnH-psbA are the ideal barcodes to discriminate Kaempferia and their adulterants32. However, these barcodes have limited mutation sites and low identification ability for related species. As a super-barcode with sufficient genetic variation, the chloroplast genome has been used to identify Crataegus and its related species5. In addition, the large single-copy region of chloroplast genome is used to identify Dendrobium species33.
2.1.2. Molecular markers
Molecular markers reveal the arrangement rules of genes and the expression rules of phenotypic traits by analyzing DNA fragments with genetic information differences among organisms. They are not affected by the external environment, developmental stages, and differences among tissues and organs, and can detect various organs, tissues, and even cells of organisms in different developmental stages. These markers have the advantages of wide distribution, abundance, stable traits, convenient selection, simple operation, and rapid detection and have been widely used in the analysis and identification of herbal medicines. For example, insertion/deletion (InDel) markers distinguish different species by analyzing differences in gene insertion or deletion nucleotide fragments in the same position between related species or different individuals of the same species. Kim et al.6 accurately distinguished Zanthoxylum schinifolium and Zanthoxylum piperitum by the InDel analysis of psbZ-trnG. Simple sequence repeat (SSR) can be used to mark 2–6 bases of tandem repeats in genetic information, and this method could accurately distinguish multiple cultivars of plum varieties34. Inter-simple sequence repeat (ISSR) markers developed based on SSR can anchor 2–4 random bases at the 5′ or 3′ end of SSR as primers and anneal at a specific site. This method was used for the systematic classification and identification of Hedyotis diffusa35. Expressed sequence tags are molecular markers developed on expressed sequences and cDNA and can be used to distinguish species populations such as Ligusticum chuanxiong36. Single nucleotide polymorphism (SNP) is the DNA sequence polymorphism caused by single nucleotide variation and can be used to screen excellent varieties and identify species such as Panax ginseng37. In addition, many molecular markers are combined with PCR for species identification, such as sequence-related amplified polymorphism for identifying wild resources and cultivated varieties of Platycodon grandiflorus38, sequence-characterized amplified regions for distinguishing Angelica dahurica7 and mistletoe species39 from their related varieties, PCR-RFLP for identifying Ophiocordyceps sinensis and its powder samples, and random amplified polymorphic DNA for distinguishing Viscum coloratum40.
2.1.3. Rapid detection based on biotechnology
Most biological rapid detection techniques were developed from PCR amplification. High-resolution melting (HRM), multiplex PCR, and real-time PCR are commonly used to identify raw materials. HRM is a genotyping technique based on the formation of different morphological dissolution curves of double-stranded DNA at different dissolution temperatures. As a tool for species identification, HRM was used for plant origin identification41. HRM was also combined with DNA barcoding and SSR to distinguish Akebia quinata, Artemisia, and Glycyrrhiza uralensis42,43. Multiplex PCR with two or more pairs of primers in the same reaction system can amplify different templates and obtain different target fragments. It was used to detect Longan Arillus, Litchi Semen, and Nephelium lappaceum44. Fluorescent groups or probes were added to the real-time quantitative PCR system for real-time monitoring of the whole PCR process by the accumulation of fluorescent signals. Zhang45 identified donkey, horse, pig, and cattle and their hide gelatin by selecting the COI gene using TaqMan RT-PCR. Meanwhile, the dye quantitative RT-PCR method can be used to identify P. ginseng and Fritillaria cirrhosa.
Isothermal amplification can identify raw materials, does not depend on the PCR instrument, and can be used on site. In helicase-dependent amplification, the target fragment is melted to form a single chain under the action of a helicase. The single-chain binding protein is combined with DNA single chain, the target fragment is kept in a single-chain state, a primer is combined with the single-chain DNA to amplify the target fragment, and the amplified double-chain DNA is used as a substrate for the next amplification cycle. This method has been used for identifying the authenticity of P. ginseng and its preparations46. Loop-mediated isothermal amplification (LAMP) hybridizes six to eight fragments of the target sequence with BstDNA polymerase and four to six pairs of specific primers under constant temperature. LAMP experimental results were judged by eye by changing the color of the experimental solution or using a turbidimeter. This method has been used for the authenticity identification of Pheretima aspergillum and Cordyceps sinensis8. Recombinase polymerase amplification (RPA) features recombinant protease and forward and reverse primers that form a complex under the action of auxiliary factors. The complex searches for and binds to a target site on a target sequence. Under the action of recombinase, primer, and single-chain binding protein, the target fragment is kept in a single-chain state. The amplification of the target sequence is then promoted by DNA polymerase. Finally, the experimental results were observed by real-time monitoring or electrophoresis with fluorescent probes. This method has been used to identify Gelsemium elegans47 and is often combined with lateral flow test paper to quickly identify raw materials.
Molecular technology has been used for rapid and accurate identification of medicinal materials due to its rapid, trace, and strong specificity. However, these methods cannot identify the adulteration of non-pharmaceutical parts of medicinal materials and classify their quality grade.
2.2. Chips and probes to identify and detect raw materials
2.2.1. Chips
Chip technology has been used to detect the authenticity of raw materials, heavy metals, and pesticide residues. Gene chips integrate oligonucleotide or cDNA in high density according to the preset nucleic acid molecule hybridization derivative array and then fix it on the surface of the support carrier. After the carrier surface hybridizes with the probe, the biological signal is captured by a special device comprising semiconductor sensors, and the content is recorded synchronously and sent back to the computer for analysis. Finally, the gene function and gene phenotype characteristics of the carrier sample can be obtained. Gene chips can distinguish Bupleurum chinense from different sources to ensure its authenticity48. Diversity arrays technology was used to distinguish Eucalyptus grandis from related species. The protein chip sequentially immobilizes some known proteins on a carrier, specifically interacts with known molecules according to their molecular characteristics, and performs purification and subsequent processing to quickly and accurately screen the protein component. This method can be used for identifying Chinemys reevesii and Pheretima aspergillum9. Microfluidic chips can manipulate fluids in micro- and nanoscale spaces and miniaturize the basic functions of sample preparation, reaction, separation, and detection onto a portable microchip for the flexible combination and scale integration of multiple cell technologies on a controllable microplatform. Detection techniques using this tool, such as electrochemical microfluidics49, can detect heavy metals and pesticides to control the safety of raw materials10. Chip technology has the advantages of short detection cycle and high efficiency. However, chip technology still has shortcomings such as high false positives, requiring simplified preparation, labeling of test samples, and high cost of gene chip application.
2.2.2. Probes
Multiplex ligation-dependent probe amplification (MLPA) detects differences in gene copy number in DNA methylation, and single base variation by four steps: target DNA denaturation, probe hybridization, ligase linking of left and right probes, and PCR amplification. MLPA designed for ITS1 could realize the identification and quantitative analysis of Fritillariae cirrhosae and its common counterfeits11. The fluorescent probe is connected to the recognition group to form small molecules, which can stably emit fluorescence after being electrified. In addition, the fluorescent probe can be observed and quantified by a fluorescence index meter. The method can be used for sensitively detecting Hg2+ in samples12. The probe technology has the characteristics of high sensitivity and strong specificity. However, the difficulty in probe design and the high cost of labeling have become limiting factors in their application. Therefore, developing a simple and convenient probe detection method is necessary.
2.3. Biosensors for raw material detection
Biosensors use active substances, such as enzymes and microorganisms, as sensitive materials for recognition and convert biological information to electrical signals with high sensitivity, accuracy, and stability in real time. This technology has been used to detect the authenticity of raw materials, heavy metals, and pesticide residues. Shi designed a sequencing-free electrochemical herb sensor based on the ITS2 of Crocus sativus to identify the plant and its counterfeits13. Lei explored an efficient and simple electrochemical sensor to prepare carbon-supported X-manganate for the detection of Pb2+ and Hg2+14. Enzyme biosensors can calculate the pesticide residue level in a sample by measuring the degree to which the enzyme activity (fixed on the electrode surface) is inhibited by pesticides. Hana Fourou fixed β-galactosidase from Aspergillus oryzae on the electrochemical sensor to detect Cr4+ and Cd2+50. Enzyme biosensors based on cholinesterase enzymes, organophosphorus hydrolase, and organophosphorus acid anhydrolase were used to detect organophosphorus pesticides15. Whole-cell biosensors are based on microorganisms. Kim modified Escherichia coli strains to contain copA, zntA, and mer promoters and inserted the luciferase (lux) gene as the reporter gene into the plasmid to respond to the induction of copper, cadmium, and mercury, respectively, to detect heavy metals51. Tu designed a nanochannel sensor made of Pb2+-specific peptide modified porous anodized aluminum membrane to detect lead ions in complex traditional Chinese medicine (TCM) samples52. Pesticide residues of imidacloprid16 and thiamethoxam17 are often detected by molecularly imprinted sensors. Immunosensors are a biological sensing device based on affinity that combine immunochemical reactions with an appropriate sensor. Glyphosate, atrazine, and parathion can be detected by immunosensors18. The electronic nose can analyze trace odor substances, compare odor similarities in samples, and establish a corresponding database by pre-collecting standard samples for the prediction and evaluation of unknown samples. It is used for the identification of volatile constituents and pesticide residues53.
2.4. Immunoassay to detect harmful substances in raw materials
Immunoassay is based on specific antigen–antibody reaction and uses a detection method for antigen and labeled antigen competitive binding antibody. Immunoassay techniques used to detect harmful substances include enzyme-linked immunosorbent assay (ELISA), fluorescent immunoassay (FIA), chemiluminescent immunoassay (CLIA), and gold immunochromatographic assay (GICA). ELISA uses enzymes as tracers to mark antigens or antibodies, and the enzyme catalyzes the substrate to develop color or emit light to establish the relationship between the developed color degree of the system and the content of the substance to be tested. Ethylene thiourea54, neonicotinoid55, and imidacloprid19 were tested using ELISA. FIA uses fluorescein as a tracer to label antibodies or antigens and combines the specificity of immunological reactions with the sensitivity of fluorescence techniques. Acetochlor, metolachlor, and propisochlor were detected using this technology20. CLIA directly labels a luminescent substance of an excited state intermediate generated under excitation of a reactant on an antigen or an antibody and combines high-sensitivity chemiluminescence determination with high-specificity immune reaction. Parathion, methyl parathion, fenitrothion21, and Cu2+ 22 were detected by this method. GICA uses colloidal gold as the tracer marker. Gold particles have high electron density, and black-brown particles at the binding site of gold-labeled protein can be observed under the microscope. When these markers accumulate in large amounts at the corresponding ligands, red or pink spots can be visualized with the naked eye. This technology is often used in combination with lateral flow strips (LFS) to detect pesticide residues such as carbendazim23. In addition, colloidal silver-based lateral flow immunoassay is used to detect profenofos56.
Immunoassay is the most widely used method to detect pesticide residues. However, preparing pesticide antibodies is difficult and the development cost is high. It is only suitable to detect pesticide residues in a single product and not for multi-residue analysis. Therefore, establishing a high-throughput immunoassay technique is particularly important.
3. Application of biotechnology in pharmaceutical manufacturing control
Conventional pharmaceutical manufacturing is usually accomplished using batch processing with laboratory testing conducted on collected samples to evaluate quality. This conventional approach has been successful in providing quality medicine to some extent. Although the quality of newly introduced products has increased, concerns over pharmaceutical product quality persist due to unacceptably high product recalls. Natural products are well-known for their complex multi-compound, multi-ingredient formulation and preparations57, and large variation in the quality of products from different manufacturers or different batches by the same manufacturer. Thus, process monitoring in natural product manufacturing has been a challenging topic.
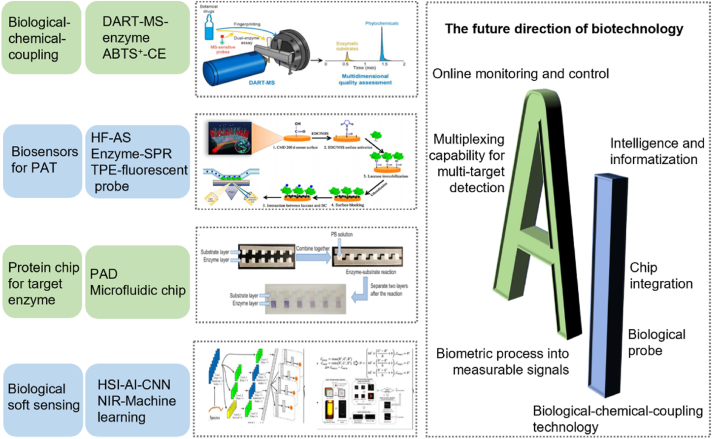
Process analysis technology (PAT) was proposed as a system for designing, analyzing, and controlling manufacturing by timely measurement (i.e., during processing) of critical quality and performance attributes of raw and in-process materials and processes to ensure the final product quality. The term “analysis” in PAT is broadly viewed to include chemical, physical, and biopharmaceutical characteristics in an integrated manner. However, most quality control techniques in pharmaceutical production focus on the analysis of physicochemical properties. Given that natural products are complex mixtures, this chemical complexity brings great difficulty for current chromatographic and spectroscopic techniques based on the chemical approach to detect all their components58. In addition, chemical information lacks enough evidence to ensure the clinical safety and efficacy of drugs because the correlation between chemical information provided by the chemical approach and overall in vivo activity has not been justified59. Many substances, particularly certain biological active ingredients and bio-pollutants, cannot be detected by chromatographic methods because of the lack of absorption in spectra. Biotechnology has been used to control the quality of natural products and synthetic pharmaceuticals. Although biotechnological approaches cannot provide chemical information (chemical components), they have the advantage of offering information on the direct bioactivity and even clinical safety and efficacy of biological products60. The application of biotechnology in pharmaceutical manufacturing control is shown in Fig. 4 and Table 2.
Figure 4.
Biotechnologies in pharmaceutical manufacturing process control and intermediate analysis.
Table 2.
Application of biotechnology in pharmaceutical manufacturing control.
| Category | Technique | Response | Application | Ref. |
|---|---|---|---|---|
| Biological-chemical-coupling technology | DART-MS-enzyme | Chemical components and biological activities | Danshen injection | 61 |
| ABTS ± CE | Major antioxidants in injection | Shuxuening injection | 62 | |
| Metabolomics-chemometrics | Bioactive-chemical quality marker combination | Pollen of T. orientalis | 63 | |
| Biosensors for PAT | Enzyme based SPR | Acceptable therapeutic monitoring | Bromocriptine | 64 |
| HF-AS | Lipase-binding ligands | Lotus leaf extracts | 65 | |
| TPE-fluorescent probe | Monitor ACE activity | Tongmaiyangxin pill | 66 | |
| FRET fluorescent acceptor | Patulin and zearalenone | Extracts of TCM | 67 | |
| Aptamer biosensor | Rapid detection of pathogenic E. coli | Licorice extracts | 68 | |
| Protein chip for target enzyme | PAD | Hypoglycemic and xanthine oxidase inhibitory activity | Mulberry, lotus and Salvia miltiorrhiza extracts | 69, 70, 71 |
| Dual-channel microfluidic chip | Multiple biomarker assay | Qishen Yiqi Pill | 72 | |
| Biological soft sensing technology | HSI-AI-CNN | Physical, chemical and biological properties | Shuxuening injection | 73 |
| NIR-machine learning | Chemical compounds and anti-inflammatory activity | Chrysanthemum extracts | 74 |
3.1. Process monitoring
3.1.1. Biological and chemical coupling for pharmaceutical processes
Currently, only a few bioassays have met the assay requirements of multiplexing, sensitivity, and speed. Detection approaches with multiplexing capabilities are highly valued for natural products because of their multi-component and multi-target characteristics. Thus, biological assays that can be integrated with chemical analyses are being explored. Such integration capability can enable parallel chemical and biological analyses on the same analytical platform, thereby reducing instrument cost and variations between instruments.
Recent advances in ambient ionization technology have opened up the opportunity of using mass spectrometry (MS) to be an innovative PAT tool in natural product manufacturing75. Li et al.61 proposed an on-demand strategy based on direct analysis in real time-mass spectrometry (DARTMS), which uses an MS probe as the substrate of the enzyme to determine the biological activity of selected thrombin and angiotensin-converting enzyme (ACE). Each probe consists of a specific peptide sequence and 1-(2-pyrimidyl) piperazine as a label with high ionization efficiency. The strategy achieved the simultaneous determination of the chemical components and biological activities of Danshen (Salvia miltiorrhiza) injection. The detection results showed that the platform can achieve multi-dimensional drug quality evaluation under real-time monitoring.
3.1.2. Biological soft sensing for quality control in pharmaceutical processes
The heterogeneous nature of natural products causes variability in raw materials and drug products, leading to uncertainty in their therapeutic consistency. Therefore, multi-dimensional quality control is essential to ensure the safety and efficacy of these drug products. Using different tools for the quality analysis of different dimensions results not only in high costs but also in variations between instruments. Soft sensing is a new and popular method in the field of monitoring76. This technology realizes the real-time estimation of the quality or activity that is difficult to directly measure by easily detectable indicators and the corresponding mathematical model. Therefore, soft sensing shows good performance on detecting difficult-to-measure quality indicators in pharmaceutical and other industrial processes.
Taking Shuxuening injection as an example, Zhong et al.73 used hyperspectral imaging (HSI) combined with artificial intelligence (AI) as a biological soft sensing technology to simultaneously monitor physical properties, chemical components, and biological activities of the injection in a non-destructive manner. The detection of chemical components in drugs often requires analytical equipment such as high-performance liquid chromatography (HPLC). Antioxidant and anticoagulant activities require the use of DPPH-free radical scavenging assay and Z-Gly-Gly-Arg-AMC acetate probe substrate. The chromaticity and visible particles of the injection are detected using image recognition algorithms. The HSI-AI method predicted the five quality indicators of total flavonol, total ginkgolides, antioxidant activity, anticoagulant activity, and visible particles. Thus, the platform can be used for the process monitoring of multiple quality characteristics in pharmaceutical production.
Biological soft-sensing techniques combined with HSI can simultaneously acquire multiple quality attributes of a sample under non-destructive conditions. It is worth noting that soft measurement is an indirect measurement technique. Therefore, standard methods must be used to detect the quality indicators of the samples in advance, and then the correlation model between the sample quality and the HSI data must be established. The upper limit of the accuracy of the soft measurement is the measurement accuracy of the standard method.
3.2. Intermediate product analysis
In the pharmaceutical process, differences in natural products and the fluctuation of operating parameters in each process unit may lead to the quality difference in intermediates between batches and affect the quality of the final product. Therefore, the intermediates produced in the process must be subjected to quality control.
3.2.1. Biological and chemical coupling for intermediate product analysis
The diverse sources and production environments of natural products lead to uncertainty about pharmaceutical products. All biological and chemical active ingredients that may affect the quality and effectiveness of medicines are comprehensively and accurately detected.
The main material basis of Shuxuening injection (SI) is its antioxidant effect. Therefore, the qualitative and quantitative analyses of antioxidants in its pharmaceutical process is of importance to ensure its quality. Ma et al.62 established an ABTS+ (2,2-azinobis-(3-ethyl benzothiazoline-6-sulfonic acid))-CE online analytical method for the monitoring of various antioxidants in the preparation of injections. ABTS+ was first integrated into the capillary and used for the simultaneous determination and separation of major antioxidants in the injection. The experimental results were verified by SI and showed that the strategy is a simple, reliable, and effective tool for the quantitative evaluation of drugs.
Some markers cannot comprehensively evaluate TCM preparations containing multiple components, thus neglecting the inherent chemical complexity and multiple mechanisms of their pharmacological activity. Wang et al.63 proposed a strategy for screening Typha orientalis quality using bioactive chemical comprehensive markers based on thrombin and a combination of metabolomics and chemometrics. This research provides ideas for creating methods to simultaneously evaluate the biological and chemical properties of drugs.
3.2.2. Biosensors for intermediate product analysis
Biosensors are small integrated analytical devices consisting of biological components in close contact with physical sensors that convert the biometric process to measurable signals. They provide specific quantitative or semi-quantitative analytical information using biometric elements that convert information from the biological domain into chemical or physical output signals77, 78, 79.
A study used a label-free biosensor based on surface plasmon resonance (SPR) technology to detect bromocriptine64. First, candidate enzymes for the bromocriptine biosensor were screened by protein ligand docking simulation and competitive inhibition experiment to verify the accuracy and reliability of the results. This enzyme SPR method can realize the rapid, real-time, and label-free monitoring of drugs. The results showed that the combination of SPR technology and different enzymes could monitor various drugs. In another study, hollow fiber-based affinity selection was proposed to detect inhibitors from medicinal plant extracts65. First, lipase from porcine pancreas was adsorbed onto the surface of polypropylene hollow fibers to form a stable ligand-harvesting matrix. Various factors related to binding capacity, including enzyme concentration, incubation time, and temperature, were then optimized using the known lipase. The proposed method can be used to detect lipase-binding ligands in lotus leaf extracts. As a typical aggregation-induced emission-based fluorophore, tetraphenylethylene (TPE) has a range of applications in biosensors and cell imaging. Wang et al.66 synthesized TPE-based fluorescent probes by two processes. One is TPE with the carboxylate group synthesized by the McMurray reaction, and the other is TPE-SDKP prepared by standard solid-phase synthesis. This probe can be used to monitor ACE activity and determine the efficiency of ACE inhibitors in extracts.
Aptamers are single-stranded oligonucleotide molecules screened by ligand evolution using an exponential enrichment system with the advantages of good stability, strong specificity, in vitro synthesis, low cost, and easy modification. They serve as an alternative target recognition element for antibodies in biosensors and can overcome the high cost of antibody-based ELISA.
Different aptamer-based sensing platforms have been developed to detect cytotoxins and contaminants in food and pharmaceuticals80, 81, 82, 83, 84, 85, 86, 87, 88, 89. Because of their great damage to human health, cytotoxin and other pollutants must be strictly detected in the pharmaceutical process. A recent study used fluorescent acceptors and achieved the simultaneous detection of patulin (PAT) and zearalenone (ZEN)67. In this study, the aptamers of PAT and ZEN were labeled with FAM and CY3, respectively, as fluorescent probes. Both aptamers were adsorbed on the surface of graphene oxide (GO) by π–π stacking, resulting in fluorescence resonance energy transfer between the fluorophore and GO. Therefore, this detection platform has good selectivity and reliability in the detection of TCM samples.
The damage caused by pathogenic E. coli to human health has attracted increasing attention68. A recent study used an aptamer-based electrochemical biosensor in licorice extracts to rapidly detect pathogenic E. coli90. To enhance the interaction between aptamer and E. coli, the sulfuration signal was first captured, followed by biotinylation of E. coli. A portion of the biotinylated aptamer was detached from the capture probe in the presence of E. coli. Residual biotinylated aptamer probes quantitatively bound to streptavidin-alkaline phosphatase. The results showed that the designed biosensor can be used to rapidly detect microbes in TCM and related fields.
Biosensors have the advantages of strong specificity, fast analysis speed, and high accuracy. Biosensors can be integrated with computers to automatically collect and process data, provide more scientific and accurate results, and form an automated detection system. At the same time, chip technology is increasingly combined with sensors to realize the potential of integrated detection systems.
3.2.3. Protein chip for intermediate product analysis
Paper-based microarrays are analytical devices made of paper that have been rapidly developed in recent years91, 92, 93, 94. They are low cost, easy to use, and can perform multiple chemical analyses. In addition, these paper devices have a high specific surface area that allows molecules to easily bind and adsorb proteins. Used paper devices can be incinerated to reduce the pollution caused by experimental consumables. The white paper provides an ideal background signal for observing colorimetric reactions.
Paper-based microarrays are an analytical device made of paper and have been developed rapidly in recent years91, 92, 93, 94. They are low cost, easy to use, and can perform multiple chemical analyses. In addition, these paper devices have a high specific surface area that allows molecules to easily bind and adsorb proteins. Used paper device can be incinerated to reduce the pollution caused by experimental consumables. The white paper provides an ideal background signal for observing colorimetric reactions. Gong et al.69 proposed an effective omics technology to prepare paper-based microarrays and used them to detect active components in TCM extracts. Polycaprolactone/chitosan-modified paper was prepared by 3D printing (3DP), and α-glucosidase was immobilized on the modified paper. The strategy was used to test the hypoglycemic biological activity of mulberry extracts70 and lotus extracts71 using a paper-based analytical device (PAD). Another researcher immobilized xanthine oxidase on a PAD69 and measured the xanthine oxidase inhibitory activity of S. miltiorrhiza extracts. Therefore, paper-based microarrays can be used for the rapid quality detection of natural products, intermediates, and preparations.
Biomarkers have been used as drug quality indicators to evaluate the biological parameters of formulations. A high-quality biomarker should reflect the mechanism of action of the drug and be clinically relevant. In one study, a chip-based method was proposed to evaluate the bioconsistency of natural products using target enzymes thrombin and ACEs as quality biomarkers. A dual-channel microfluidic chip was designed to perform bioassays72, where enzyme complex formation occurs in one channel and the subsequent enzymatic reaction occurs in the other channel. Magnetic beads serve as both an efficient surface for immobilizing enzymes and a controllable solid support for enhanced on-chip mixing and transport. Enzyme activity is used to indicate drug quality (or potency). In Qishen Yiqi Pills, different batches of intermediates and preparations were tested, and the results showed that the bioassay had better discriminatory power for abnormal samples than chromatographic fingerprinting.
3.2.4. Biological soft sensing technology for intermediate product analysis
Most researchers have focused on the relationship between chromatography/mass spectrometry and biological activity of natural products, whereas the relationship between spectrum and activity has often been ignored.
Near-infrared reflectance spectroscopy (NIRS) was used to detect the anti-inflammatory activity of chrysanthemum extracts74. This work used NIRS combined with anti-inflammatory activity data. A complex relationship between Q-markers and overall anti-inflammatory activity was established by introducing backpr-opagation-artificial neural network. Thus, the comprehensive approach based on NIRS can quantify multiple compounds and predict overall biological activity for drug quality control. However, network toxicology or even bioinformatics alone can make the prediction, and the results should be verified by further experiments.
4. Application of biotechnology in pharmaceutical products analysis
4.1. Safety analysis
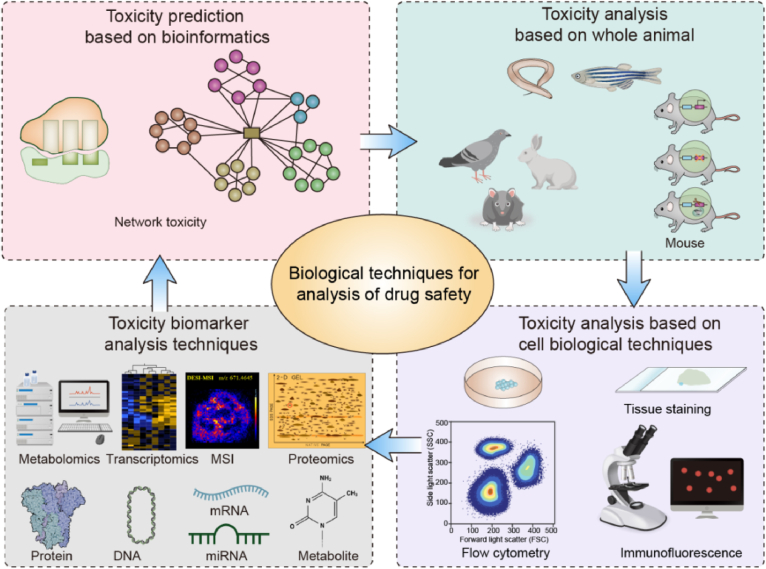
Efficacy and safety are two aspects of the properties of a drug when considering its use in disease treatment. It is necessary to consider the adverse reaction (including toxicity and side effects) of drugs when they are used. Toxicity evaluation usually comprises analyses of drug metabolic distributions, physicochemical properties, and pharmacokinetic characteristics such as genotoxicity target and organ toxicity. New drugs (including natural products and synthetic pharmaceuticals) have emerged, with rising safety problems. Traditional drug safety analysis techniques and methods can no longer meet growing needs, which requires an establishment of fast and accurate drug safety analysis techniques. Here, the progress of biological techniques used in the analysis of drug safety is summarized (Fig. 5).
Figure 5.
Overview of biological techniques used in drug safety evaluation.
4.1.1. Drug toxicity prediction based on bioinformatics
The concept of network toxicology is an important method for biopharmaceutical research developed by network pharm-acology95. Network toxicology of TCM refers to the study of the toxicological characteristics of a network model and analyzes the interaction and regulation of toxic materials in biological systems by the established network model, which plays an important role in predicting TCM toxic components95,96. Network toxicology refers to the description of the toxicological characteristics of drugs by constructing a specific network model to analyze and predict drug toxicity, thereby elucidating the toxic side effects of the drug on the human body and predicting the toxic components of natural products. Network toxicity is usually combined with other methods to detect drug toxicity. Network toxicity based on metabolomics has been used in toxicity mechanism analysis of TCMs97. For instance, the integration of toxicology networks and metabolomics was used to detect the metabolites in rat liver induced by esculentoside A, a triterpenoid saponin from Phytolacca acinosa98. However, network toxicology or even bioinformatiocs only can make the prediction, and the results should be verified by further experiments.
4.1.2. Drug toxicity analysis based on whole animal
Animal models are used to assess and avoid risk to humans from exposure to potential hazards. The pharmacopoeia suggests that drugs be tested by comparing with digitalis minimum lethality in pigeon99. Traditional animals, such as rats and mice, are commonly used animal models for drug toxicity analysis. However, the experimental animal models used in preclinical studies poorly predict drug metabolism and potential toxicity.
4.1.2.1. Transgenic animal analysis techniques
Transgenic or gene editing mice expressing labeled proteins and optical biosensors show potential for the analysis of disease etiology, pathogenesis, and the rapid large-scale screening of drug toxicity in preclinical studies100. The generation of available transgenic mouse models for long-term tracking of organelles, cells, or tissues is essential in drug toxicity analysis. Transgenic mice, such as human leukocyte antigen (HLA) mice lines and human liver chimeric mouse models, have rapidly developed over the past decade101,102. Abacavir, a human immunodeficiency virus reverse transcriptase inhibitor, and its drug reactions are highly associated with HLA alleles. Abacavir-induced liver injury was usually observed when HLA mice were treated with CpG-oligodeo-xynucleotides103.
Transgenic or gene-edited mice promote drug toxicity analysis, but this analysis requires huge workload and incurs high costs. Mouse models occasionally cannot meet the requirements of drug phenotypic screening. For example, thalidomide, which can induce birth defects in unborn children, does not cause any defects in mice104. For more comprehensive and cost-effective evaluation of drug safety, some new models have been established and more are emerging.
4.1.2.2. Disease animal model analysis techniques
Traditional animal models such as rats and mice involve high cost and long life. Therefore, alternatives to mammals for toxicology studies are warranted. With its ease of husbandry, high fecundity, small size, rapid production, development, and transparency, the young zebrafish (Danio rerio) shows potential as a model organism for detecting drug toxicity and toxicity mechanisms in vivo. The ability of in vivo screening coupled with the relevance of phenotypes to human diseases has contributed to the emergence of zebrafish as a leading model organism for whole-animal drug toxicity105,106. Fructus Psoraleae, the seed of Psoralea corylifolia, shows hepatotoxicity, and its potential hepatotoxic mechanisms were investigated in a zebrafish model107. The cardiotoxicity of four typical macrolides—azithromycin, clarithromycin, tilmicosin, and tylosin—was studied in zebrafish embryos108. The brain mitochondrial toxicity of Bacopa monnieri (Brahmi), traditionally used as a nootropic agent, was also analyzed in zebrafish109.
Caenorhabditis elegans is a small, non-pathogenic nematode with a short lifespan. Low-cost maintenance, easy cultivation, and efficient reproduction make this nematode suitable for rapid developmental toxicity testing. C. elegans assays were first used as a biological model and in environmental and chemical toxicity to obtain data from a whole animal with active digestive, reproductive, sensory, and neuromuscular systems110,111. Nowadays, this model is used in drug toxicity analysis, such as cisplatin112 and aesculin, a coumarin compound extracted from TCM Qinpi (Cortex Fraxini)113.
4.1.3. Drug toxicity analysis based on cell biological techniques
Cell morphology is altered or damaged by drug toxicity, which are phenotypically categorized based on morphological characteristics. Tissue staining has been used for pathological screening in traditional toxicological analyses, but it cannot be used to observe organelle (i.e., endoplasmic reticulum, cytoskeleton) damage or reveal the mechanisms of endogenous molecular regulation caused by toxic drugs114. Although the tissue staining experiment is still required to detect drug toxicity, more cell biological techniques have been used.
Immunofluorescence is an important technique used in the diagnosis of many infectious diseases by detecting and localizing the antigens in different tissues and cells115. A near-infrared fluorescent probe was reported to evaluate hydrazide-induced liver injury by detecting the N2H4 level116. Flow cytometry is used for counting, analyzing, and sorting cells in a cell suspension by their light-absorbing or fluorescing properties. It permits the assessment of physical (cell size, shape, and internal complexity), chemical (expression of proteins and other molecules), and biological attributes of single cells in the suspension117. Flow cytometry techniques can be used for measuring the morphological effects of both natural products118 and synthetic pharmaceuticals. It is usually used to analyze cytotoxicity induced or repressed by drugs. For instance, ginsenoside Re inhibited rot-induced cytotoxicity in SH-SY5Y cells in Drosophila could be measured by flow cytometry119.
Cell biological techniques are the fastest and most convenient analysis techniques of drug safety, but they are also limited by cell lines and states. Sometimes, cells are easily contaminated; thus, a sterile environment is required.
4.1.4. Drug toxicity biomarker analysis based on high-throughput techniques
The formal qualification of safety biomarkers is a milestone in the application of biomarkers to drug development. Biomarkers significantly contribute to new drug development because they facilitate drug toxicity assessment. With the development of high-throughput sequencing technologies or omics, drug toxicity analysis is promoted, especially to identify biomarkers.
Toxicogenomics is a branch of toxicology that applies several genomic analysis techniques to determine how chemicals, both environmental and pharmaceutical agents, react on human and ecological health120. It includes the identification of organisms associated with disease and toxic biomarkers. Equipped with innovative technologies from genomics and high-throughput methodologies, toxicogenomics provides an unprecedented opportunity for improved risk assessment and regulatory decision making.
The ability to screen numerous molecules (e.g., metabolites, proteins, or DNA) allows the identification of toxicity biomarkers, which may be used to enhance drug safety assessments and disease diagnostics. Different from measuring a single molecule at one time, high-throughput profiling simultaneously screens thousands of molecular changes. Transcriptomics or gene expression profiling measures the changes in transcript levels and reflects gene expression regulation at transcriptional and post-transcriptional levels. Vine tea is made from the tender leaves of Ampelopsis grossedentata, and its chemical composition is mainly flavonoids and polyphenolic substances with dose-related hepatotoxicity. Transcriptomic profiling revealed 34 potential toxic components and 57 potential hepatotoxic targets in vine tea121. Meanwhile, proteomic profiling measures the changes in protein levels and reflects the regulation of gene expression, protein translation, and post-translational protein stability122. The toxicities of Herba Lysimachiae (Jinqiancao), which is usually used to treat rheumatic arthralgia, were analyzed by proteomic profiling123. With DARTs-based proteomics, the potential toxic target of psoralen, a major hepatotoxic and blood entry component in Fructus Psoraleae, was identified124. Metabolomic profiles determine molecules acting as intermediates and metabolic products and identify hormones and other signaling molecules in many biological processes125. Heshouwu (Polygonum multiflorum), a famous traditional Chinese herb, is associated with significant liver injury126. Toxic components related to the drug-induced liver injury of Heshouwu and specific biomarkers were investigated using the UHPLC–MS-based metabolic approach127. Anthraquinone, which belongs to polycyclic aromatic hydrocarbons, is the main active component in Cassiae semen (Juemingzi) and shows toxic reactions. The established urinary metabolomics approach allowed the determination of aurantio-obtusin biomarkers for the early diagnosis of its toxicity128. Thus, the biomolecular coverage provided by omics profiling can be used to improve the performance of traditional toxicity prediction methods.
4.2. Effectiveness analysis
Early and effective pharmaceutical analysis methods are of great significance in the biotechnology and pharmaceutical industries. The purpose of drug quality evaluation and control is to ensure clinical efficacy and safety. Although the quality standardization of natural medicines and synthetic drugs has made significant progress in biochemical research129, its guiding or supporting role in the rationalization of clinical medication and improvement of clinical efficacy needs to be studied further.
Bioassays have become one of the most important development directions of drug quality standardization because of their technical advantages of efficacy-related properties and overall control130 To evaluate the efficacy-related biological effects expressed by test drugs acting on the biological model (whole animals, isolated organs and tissues, cells, biology-related factors, and enzymes) under specific conditions, bioassays have been used for the qualitative or quantitative evaluation of drugs131, 132, 133 (Fig. 6).
Figure 6.
Bioassays in effectiveness analysis.
Several related studies have been performed in the effectiveness analysis of both natural medicines and synthetic drugs. For instance, the rat bile duct ligation model was established to evaluate the anti-hepatic fibrosis effect134, the NRLP3 inflammasome activation model was constructed to determine the anti-inflammatory function135, and thrombin activity was used to assess the anticoagulant response136 of drugs. Many promising cutting-edge techniques, such as biosensors, organ-on-a-chip (OOAC), and 3DP, have been developed for pharmaceutical analysis (Fig. 6). By using these biotechniques, bioassays can be more efficiently and accurately developed for pharmaceutical analysis.
4.2.1. Target-based biosensors for effectiveness analysis
Biosensors are bioanalytical devices developed by integrating electronic techniques and biological molecules or systems. They hold promise in the analysis of various drugs because of their high sensitivity and low cost137. The discovery of taste type II receptor (TAS2R) agonists is valuable for treating asthma, but the development of a fast screening method to identify TAS2R agonists is challenging. Inspired by the advantages of the high-electron-mobility transistor (HEMT) sensor, Wang et al.144 developed a virtual and affinity screening strategy based on the combination of a HEMT biosensor with UPLC–MS analysis to screen TAS2R14 agonists from Platycodon grandiflorum. They obtained six potential TAS2R14 agonists, with platycodin L as a special TAS2R14 agonist. This study provided a strong reference for the direct screening of agonists or inhibitor classes of drugs from complex natural medicines138. In addition, using the ligand-induced conformational changes in G protein-coupled receptors (GPCRs) as the basis, Dong et al.139 designed a 5-HT2AR-based fluorescent biosensor termed psychLight to detect endogenous serotonin release. This sensor identified previously unknown hallucinogenic drugs and a non-hallucinogenic psychedelic analog with neural plasticity-promoting and antidepressant properties. This study provided insights into the early identification of designer drugs of abuse and the development of 5-HT2AR-dependent non-hallucinogenic therapeutics. Given the critical selectivity of GPCRs on ligands, such biosensors can be used to monitor the active components and their holistic potency of natural products in a target-based way, namely, target-based drug potency. Therefore, biosensors can be used to analyze and assess the quality of natural products with multiple components140. Because of the direct association of dopamine and uric acid in human blood serum with human emotional and physical health, the simultaneous detection of both chemicals is important for the early diagnosis and treatment of related diseases. Gong et al.141 developed a low-cost high-entropy porous CrO/CrN/C biosensor for simultaneously determining dopamine and uric acid in human serum samples. The high-density distribution of catalytic CrO/CrN nanoparticles in porous Cr-JFC/DAC2 composite provided the high surface, abundant crystal interface, and excellent conductive properties of the Cr-JFC/DAC2/Nafion/GCE bio-sensor. Under optimum conditions, the biosensor could simultane-ously detect dopamine and uric acid in the range 0.05–4 μmol/L with limits of detection of 10.65 and 17.54 nmol/L, respectively. Chen et al.142 designed a cotton thread-based multi-channel photothermal biosensor for the simultaneous detection of three breast cancer-related miRNAs, namely, miRNA-10b, miRNA-27a, and miRNA-let-7a. The biosensor has high specificity and sensitivity with the detection limits of 37, 38, and 38 pmol/L for the three miRNAs. In addition, its application in cell lysates indicated the excellent capacity of the developed multi-channel photothermal biosensor for the practical analysis of multiple targets.
4.2.2. 3D biological printing of organoids for personalized pharmaceutical analysis
As a 3D cell culture, the organoid is a physiologically relevant model for basic and clinical applications and presents a self-organizing, self-renewing, and more physiologically relevant model than conventional 2D cell cultures, thus opening new avenues for drug testing and the development of therapeutic approaches in a pre-clinical setting143. Patient-derived organoids exhibit several advantages as personalized tumor models and can be established in a short period with low cost for the identification and testing of new anticancer drugs. Schuster et al.144 developed an automated microfluidic organoid culture platform for dynamic and combinatorial drug screening, thus enabling the highly reproducible, dynamic, and robust analyses of organoids with potential to facilitate treatment decisions for personalized therapy.
As one of the most progressive innovations in pharmaceutical science, 3DP has become a revolutionary and powerful tool in the precise manufacturing of individually developed dosage forms, tissue engineering, pharmaceutical analysis, and disease modeling145. It allows a fast, efficient, and economical production of customized drug carriers with desirable shape, size, and structure from 3D computer data to provide personalized medication, various drug combinations, and complex drug release profiles. Diverse 3DP technologies include binder jetting, fused deposition modeling, pressure-assisted microsyringe, inkjet printing, vat photopolymerization, and selective laser sintering, which have been developed in drug and medicine research146. Using acrylic acid (AA) as a monomer, poly (ethylene glycol) dimethacrylate as a difunctional crosslinker, and acrylated hyperbranched polyester (AHBPE) as a multifunctional crosslinker, Chen et al.147 produced 5-fluorouracil-loaded tablets using digital light processing 3DP. They found that the printing time and drug release increased with AA content but decreased with increasing AHBPE content. In addition, they suggested the use of AHBPE as a crosslinker in vat photopolymerization 3DP of personalized drug delivery.
The effective recapitulation of the structure and function of tissues as organoids can help predict patient response for precise drug screening and further personalized drug validation and therapy in a timely manner148. For example, because patient-derived tumor organoids allow a highly cellular repository to recapitulate the essential characteristics of the original native tumors, they can be used as superior models for identifying and testing anticancer drugs or natural medicines149. To determine the effective drugs to treat glioblastoma, the most common and deadly primary brain malignancy, Chadwick et al.150 generated 4D cell culture arrays by 3DP with thermo-responsive shape memory polymer for rapid assessment of drug responses in glioblastoma patient-derived organoid-like models. They evaluated drug sensitivity, on-target activity, and synergy in drug combinations. The platform is beneficial for rapid functional drug assessments for future selection of more effective personalized therapies. Curcumin, a polyphenol compound derived from the stems of Curcuma longa, is effective in suppressing various phases of colorectal cancer (CRC) development. Chen et al.151 designed patient-derived organoids of colorectal cancer and developed a non-targeted metabonomic technique to evaluate the inhibitory effect and mechanism of curcumin on CRC organoids. This study provided a strong reference for curcumin as a potential natural drug in treating human-derived CRC-like solid tumors.
4.2.3. Microfluidic-combined OOAC for high simulation pharmaceutical analysis
OOAC mimics the physiology and functionality in human organs on a chip and has been claimed to foster a paradigm shift in drug testing and development152. It is constructed with the silicon-based organic polymer polydimethylsiloxane and has a compact size and various microchannels for early drug discovery and preclinical screening and testing. Ren et al.153 reported a “heart-on-a-chip” platform that combines microgrooves and electrical pulse stimulation to recapitulate the well-aligned structure and synchronous beating of cardiomyocytes, which could provide a high-throughput drug screening platform for preclinical drug development. A bioartificial liver consisting of a surface-engineered microfluidic silicon chip with microtrenches mimicking hepatic sinusoids has also been reported154. This artificial liver model can extend 3D primary hepatocyte culture. It allows reliable prediction of in vitro responses, including drug responses and environmental stimulus responses. It has potential applications in various fields such as drug discovery and toxicity testing, and basic pathology and physiology research155,156. Currently, a more advanced “body-on-a-chip” or “human-on-a-chip” platform simulates the physiology of the entire human body, which can serve as an alternative model system to replace animal models in drug development and analysis. However, because of the complexity of the human system, some technical challenges should be addressed in the future157.
Microfluidic chip, also known as “laboratory-on-a-chip”, is a rapidly growing versatile technology that precisely manipulates nanoliter volumes (10−9 to 10−18 L) of fluids in microchannels. With distinctive features of low consumption of reagents, fast mixing, large specific surface area, high mass, short diffusion distance, and rapid actuation and response, this technology is a powerful tool for pharmaceutical analysis from drug synthesis to drug delivery to drug evaluation158. For example, Huang et al.159 constructed an arrayed geometrically enhanced mixing chip with individual function zones to quickly and accurately evaluate the potency of five drugs. In addition, with the rapid development of digital manufacturing techniques, 3DP-assisted microfluidic chip devices have proved advantageous in improving the efficiency, speed, and control of pharmaceutical analysis160.
Microfluidic OOAC systems provide unparalleled independent control over multiple key biological, chemical, molecular, cellular, and mechanical parameters within the intestinal microenvironment, thereby enabling researchers to apply a synthetic biology approach at the cell, tissue, and organ levels that can lead to new insights into intestinal physiology and disease mechanisms. For example, “intestine-on-a-chip” can be used to analyze the molecular processes underlying various enteropathies and to advance the development of new therapies161.
4.2.4. Omics-based AI for drug effectiveness analysis
AI technology provides opportunities for the design, discovery, and development of innovative drugs in different areas, from peptide synthesis to molecule design, virtual screening to molecular docking, and quantitative structure–activity relationship to drug repositioning162,163. It mainly includes deep learning and machine learning algorithms for the identification and validation of chemical compounds, target identification, drug monitoring, and drug efficacy and effectiveness assessment164,165. By exploring the capacity of AI to predict the activity of various synthesized molecules, Polykovskiy et al.166 demonstrated that AI not only improved procedural accuracy and efficiency but also enabled drug discovery. Although tremendous amount of work is required to incorporate AI tools in the pharmaceutical analysis cycle, it is thought that AI technology will bring revolutionary changes in innovative drug discovery and analysis processes. For example, by combining chemical fingerprint and bioactivity evaluation of S. miltiorrhiza using AI technology by introducing intelligent chemometric methods, the possible antibacterial components of this natural medicinal plant on Pseudomonas aeruginosa was rapidly and effectively explored167. The AI-assisted fingerprint–activity relationship can act as a powerful bioassay model to identify the bioactive components in natural medicines for omics-based pharmaceutical analysis and quality control.
AI technology can improve the efficiency and accuracy of drug discovery and development, which not only enhances process efficiency but also reduces or eliminates the requirement for clinical trials by conducting simulations, reducing costs and ethical concerns, and heralding a new age for pharmaceutical analysis.
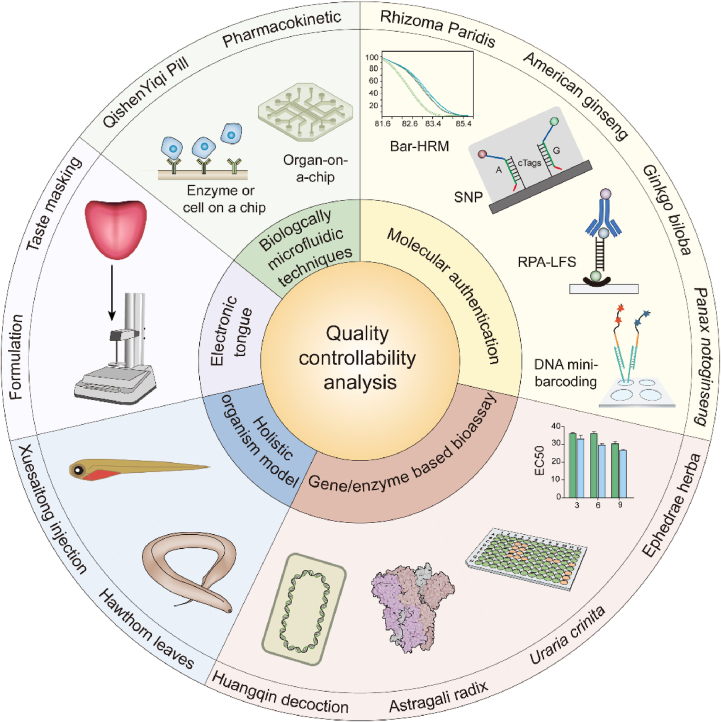
4.3. Quality controllability analysis
Some biotechnologies have been explored in drug quality evaluation due to their advantages in reflecting the overall biological effects of drugs, reflecting the drug quality related to its function, and providing visual and intuitive results. Biological detection methods are the core evaluation methods to study the effectiveness and safety of drugs as well as quality evaluation. For drug quality analysis, there are higher requirements for the sensitivity, accuracy, and throughput of techniques compared with techniques for pharmacological or toxicological research methods. Based on the biological effects of drugs, biotechnologies such as gene/enzyme/cell model organisms, microfluidic chips, and electronic tongues have been used to evaluate the quality of drugs from the perspective of activities to achieve the goal of controlling or evaluating drug quality related to its function. Some representative biological evaluation methods are described in Fig. 7.
Figure 7.
Biotechniques for pharmaceutical quality control analysis.
4.3.1. Molecular authentication for formulation analysis
DNA barcoding technology is a technique that uses standard DNA sequences (200–600 bp) as species identification markers, and usually requires DNA extraction, PCR, sequencing, and sequence analysis steps. Compared with macromolecular techniques such as protein or RNA, the DAN barcoding technique is more stable and accurate. This method has been widely used for the species identification of raw materials of natural products. However, it is difficult to obtain complete DNA barcodes from preparations of natural products because of DNA degradation, at which point DNA mini-barcoding comes into the picture. Compared with DNA barcoding, DNA mini-barcoding uses shorter DNA lengths (<200 bp), which can achieve more efficient extraction and amplification168,169. DNA mini-barcoding broadens the application of DNA barcodes and is suitable for evaluating the quality of natural product preparations. An adaptor ligation-mediated PCR protocol was derived to amplify sets of target DNA fragments isolated from Angelica sinensis and Panax notoginseng170. DNA extracted from A. sinensis and P. notoginseng were ligated with adaptors and amplified by an adaptor primer and a single universal barcode primer to avoid amplification of non-target DNA sequences and obtain partial ITS2 sequences. The results showed that various lengths of DNA fragments within the ITS2 region were amplified and could be used to identify the concerned species. Liu et al.171 developed a rapid identification method using the RPA assay to detect two species, Ginkgo biloba and Sophora japonica (as adulteration), in Ginkgo folium products. The short region in the ITS2 sequence was amplified to identify S. japonica and the short region in the rbcL sequence was amplified to identify G. biloba. During the authentication process, the RPA-LFS assay showed a higher specificity, sensitivity, and efficiency than PCR-based methods, demonstrating that this assay can be developed into an efficient tool for the rapid on-site authentication of plant species in G. biloba herbal products.
DNA metabarcoding is a technique that uses universal PCR primers to simultaneously amplify multiple DNA barcodes and identify multiple species in a single environmental sample172. Metabarcoding and single-molecule, real-time (SMRT) sequencing was used to detect the multiple ingredients in Jiuwei Qianghuo Wan173, and the results showed that with the combination of metabarcoding and SMRT sequencing, it is repeatable, reliable, and sensitive enough to detect species in TCM products. Han et al.174 developed a nucleotide signature based on an SNP site unique to American ginseng and 4F/4R unique to ginseng were designed. Twenty-four batches of ginseng products in Chinese patent medicines marketed in Beijing were tested, and five batches were counterfeit products and two batches were adulterated products. Thus, nucleotide signatures broadened the application of DNA barcoding technology in the identification of ginseng products. The method can rapidly identify ginseng products. Ririe et al.175 introduced melting curves into DNA analysis in the 1990s. The technique uses melting curves of several species and avoids sequencing. The principle of barcoding high-resolution DNA melting (Bar-HRM)176 is as follows. The double-stranded DNA used in PCR is bound to a dye. Although the fluorescence is intense when bound to the dye, the fluorescence level is low when released. After PCR, 50–500-bp-long amplicons are gradually denatured by a small temperature increase of 0.01–0.2 °C, and the fluorescent dye is slowly released from the denatured amplicons. The diminishing fluorescence with increasing temperature can be plotted as a melting curve. Bar-HRM has been used to analyze herbal material products such as Rhizoma Paridis177.
4.3.2. Electronic tongues for pharmaceutical formulation analysis
The term “electronic tongue” simulating the human gustatory system was first reported in 1996178. Electronic tongues are devices consisting of arrays of low-selective sensors and using advanced mathematical procedures for signal processing based on multivariate data analysis, principal component analysis (PCA), pattern recognition, and artificial neural networks (ANNs)179. A large number of chemical sensors were used in the design of electronic tongue sensor arrays180. Specifically, the electronic tongue system has been used for pharmaceutical analysis, such as the investigation of taste-masking efficiency, dissolution measurement, and quality control of drugs, herbal medicine, and medicinal plants for batch-to-batch product reproducibility and determination of active pharmaceutical ingredients (APIs)181,182.
New drug formulations and dosage forms are constantly being introduced with novel pharmaceutical technologies. Artificial sensing systems were thus developed as supplementary analysis methods for pharmaceutical formulations180. Weitschies et al.183 used a commercial taste-sensing device (TS-5000Z) to evaluate the different taste-masking strategies. The taste-masking efficiency of cyclosporin A (CyA) loaded self-emulsifying drug delivery system and orally disintegrating tablets was evaluated by the α-Astree electronic tongue184. Winnicka et al.185 reported a prototype of an electronic tongue for the comparison of microparticles, lyophilisates, and orodispersible tablets loaded with dihydrochloride cetirizine. The taste profiles of three diclofenac drug formulations could be differentiated using electronic tongues186. The Euclidean distances on the PCA plot were highly correlated (R2 = 0.986) with the bitterness intensities of the chemical signals of formulations of different brands187.
The electronic tongue systems were also used to differentiate quality variation in natural product preparations. Shakaff et al.188 developed a multichannel sensor incorporated with an array of the artificial lipid polymer membrane as a fingerprinting device for quality analysis of extracts from different parts, age, batch, and extraction mode of Eurycoma longifolia, with the obtained potentiometric fingerprint profiles. Various taste-masking methodologies and electronic tongue techniques were used in pharmaceutical analysis189, 190, 191. However, the benefits and drawbacks of electronic tongues for pharmaceutical applications are hardly evaluated because of the variance in the principle of chemical sensors and data processing methodologies. Therefore, the reliability of two commercially available systems (from Insent and AlphaMOS) and four laboratory prototype systems were used for the analysis of the same set of pharmaceutical samples under blind conditions192.
As an emerging and promising analysis tool in the area of pharmaceutical products, the electronic tongue has proved to be an important technology to assess and predict the taste of APIs in the pre-clinical development of pharmaceutical candidates as well as to promote the rational design of oral taste-masked dosage forms186.
4.3.3. Reporter gene transfected cell-based and enzyme-based quality analysis
Reporter genes encode proteins or enzymes that can be easily detected in the context of endogenous proteins193. Specifically, a reporter gene is connected with a target gene or regulatory sequence. The activity of the coding product of the reporter gene directly reflects the transcriptional activity or expression level of the gene in the cell194. The firefly luciferase reporter gene is a commonly used reporter gene in mammalian cells. Luciferase has high sensitivity and a wide linear detection range of 7–8 orders of magnitude. The enzyme activity can be detected by fluorescence colorimetry; thus, it is suitable for high-throughput screening with high accuracy. It has been gradually recognized and applied in the quality control of herbal medicines.
Professor Cheng's laboratory has established a series of cell lines transfected with luciferase reporter genes related to hormone, endocrine, immune, and inflammation signal pathways, and applied them to the quality evaluation of single herbal medicines195 and formulation yiv-906196, a standardized four-herb formula from a traditional formula “Huang Qin Tang” for some gastrointestinal symptoms, including diarrhea, nausea, and vomiting. This method realizes the purpose of distinguishing yiv-906 and commercial Huangqin Tang preparations, although yiv-906 and commercial Huangqin Tang preparations could not be differentiated with the chemical profile based on LC–MS fingerprints with 77 peak intensities. Therefore, this combined biological evaluation method has more distinguishing ability than the LC–MS fingerprinting method, although LC–MS has high resolution and sensitivity. At the same time, this evaluation method is highly related to the results of pharmacological evaluation in vivo and can reflect the action mechanism of yiv-906. Therefore, this method is also called mechanism-based quality control (MBQC). In addition, enzyme activity assay related to hormone, endocrine, immune, and inflammation signal pathways were used in this MBQC platform195,196. Chao et al.197 used ER-α promoter and nuclear factor erythroid-2-related factor (Nrf2) promoter transferred cell lines and COX-2 enzyme inhibitor screening assay to evaluate the quality of Uraria crinita processed by three drying methods and different temperatures. The bioassay showed that the biological activity of U. crinita oven-dried at 40 °C was the best, which was confirmed by chemical profile analysis that the main active component content in U. crinita oven-dried at 40 °C was the highest. These bioassays could be used to assess the effect of drying temperature on U. crinita quality. Cell lines stably transfected with luciferase reporter genes related to AP-1, TLR2, ER-α, Nrf2, and AR signal pathways were used to compare different processed Ephedrae Herba198 and Astragali Radix199, and this method can significantly distinguish different processed products. These studies show that bioassays can be used to evaluate differences in quality caused by different processing methods.
Reporter gene assays are suitable for high-throughput analysis with high accuracy and sensitivity, but their repeatability may be affected by the state of transfected cell lines and it is not easy to establish the reference material as a positive control. Furthermore, the bioassay based on the luciferase reporter gene-transfected cell line or enzyme is related to the action mechanism of the drug on signal transduction pathways. The selection of luciferase reporter genes or enzymes can reflect the actual drug efficacy in vivo, which may be a combination of different biological activities for TCM with multiple functions.
4.3.4. Holistic organism-based quality analysis
Compared with cell lines or enzymes, holistic organisms are intact living organisms that can reflect the synergistic effects of a drug on different parts of an organism to determine its actual quality. Compared with mammals used for conventional pharmacological evaluation, these model organisms are relatively convenient. Therefore, model organisms are routinely used in pharmacological studies and have gradually been introduced to exploratory studies of drug quality evaluation.
The most widely used model organisms in health studies include zebrafish, C. elegans, Drosophila, E. coli, and Saccharomyces200, 201, 202, 203. In the last decade, large-scale genome sequencing has revealed that human genes are 61% similar to those of Drosophila, 43% similar to those of nematodes, and 46% similar to those of Saccharomyces. As an emerging model organism, zebrafish has a high genomic sequence similarity of 87% to humans204, and it has several biological advantages such as in vitro fertilization, embryonic transparency, and easy observation of tissue and organ development under the microscope without dissection205. Experiments can be performed in cell plates, thus requiring a small amount of samples. Therefore, this model has been recommended as a new alternative animal by the European Centre for the Validation of Alternative Methods206,207. As a whole animal model, the zebrafish has unique advantages in the screening of pharmacodynamic substances and evaluating the quality of natural drugs with complex composition and unclear pharmacological mechanisms. It may realize the convenient, rapid, and high-throughput screening of pharmacodynamic substances of TCM and establish their biological quality evaluation standard.
Zebrafish larvae with arachidonic acid (AA)-induced thrombus was used to evaluate the antithrombotic effects of Xuesaitong injection (XST) from different batches208. The results indicated that XST could significantly and dose-dependently restore the intensity of heart red blood cells in thrombotic zebrafish but decreased red blood cell accumulation in the caudal vein. Using the zebrafish thrombosis model, five abnormal batches of XST were effectively distinguished from 24 normal batches of XST. This finding was confirmed by conventional chemical methods. Therefore, this zebrafish thrombosis model can be used for the batch-to-batch consistency evaluation of XST. Quality-marker (Q-marker) is an emerging concept to evaluate TCM quality that aims to screen marker compounds that fully reflect the “five principles”104. Li et al.209 identified the potential Q-markers of Danhong injection (DHI) using an in vivo zebrafish thrombosis model for batch-to-batch quality consistency evaluation. The results showed that rosmarinic acid and p-coumaric acid, the main chemical components of DHI, showed moderate antithrombotic effects in a dose-dependent manner and could be used as potential Q-markers to evaluate the quality consistency of DHI. Gao et al.210 also examined the antithrombotic activity of Hawthorn leaf fractions by platelet aggregation assay and a zebrafish model for antithrombotic drugs. Combined HPLC–QTOF-MS and molecular modeling identified shadyside D and norhawthornoid B as key components in inhibiting platelet aggregation mainly by acting on the targets of key regulators of antiplatelet aggregation activity (P2Y1 and P2Y12).
C. elegans has also been used in drug quality evaluation. Ping et al.211 evaluated the effect of Ganoderma lucidum water extracts on oxidative stress resistance, including the five origins of G. lucidum from wild-type and forkhead box O transcription factor mutant, using C. elegans. The results showed significant differences in the effect of G. lucidum on improving resistance to oxidative stress and in the mechanisms by which different origins of G. lucidum improve the ability of nematodes to oxidative stress. This finding suggested the presence of different potent active substances in different origins of G. lucidum and proved that the use of the model organism C. elegans could effectively distinguish the biological activities of G. lucidum from different origins.
4.3.5. Biologically microfluidic technologies based-quality control methods
The microfluidic chip or micro total analysis system (uTAS) is a new technology that integrates sampling, pretreatment, reagent addition, reaction, separation, and detection on a microchip. This technology has the advantages of small size, simple operation, low sample usage, fast analysis, and easy automation of operation212, and it can be used to perform chemical separation and enzyme and cell analyses213, 214, 215. The integration of microfluidic chip and enzymes or cells may improve the repeatability of bioassays. Microfluidic chips have been rapidly developed and used in drug quality evaluation. Ho et al.216 developed a device that can quantify the API content in antimalarial drugs to evaluate their quality. The system consists of two parts, a detection reagent (probe) and a microfluidic platform, and is based on the luminol reaction between the probe and API. Pure and unqualified samples of artesunate (Arsuamoon tablets) were detected using this system, and the accuracy of the quantitative method was confirmed in comparison with the conventional 96-well plate spectrophotometric method. Li et al.72 reported a multiplex biomarker assay for assessing drug quality using the combination of microfluidics and enzymes. In this study, thrombin and ACE were used to evaluate the quality of Qishen Yiqi Pill. Potency variations were detected in different batches of intermediates and preparations of Qishen Yiqi Pill. The results showed the feasibility of using microfluidic technology for the quality evaluation of Chinese pharmaceutical preparations.
OOAC is more accurate than traditional cell-based models and is expected to replace the animal phase of drug testing to significantly reduce the time and cost of drug development. Researchers have demonstrated a fluid-coupled two-channel microfluidic human OOAC model of the intestine/liver/kidney205. The model provides the pharmacokinetic parameters of drug absorption, metabolism, and excretion in humans and predicts pharmacokinetic parameters for oral nicotine and intravenous cisplatin. With further development, this technology can be used to create chips with the combination of different organs to achieve rapid and high-throughput analysis of drugs for quality evaluation (Table 3).
Table 3.
Application of biotechnology in pharmaceutical product analysis.
| Techniques | Characteristic | Application |
|---|---|---|
| Toxicity analysis | ||
| Network toxicology | Multiple gene and target | Hepatotoxicity analysis of esculentoside A98 |
| Transcriptomics | Most widely used; high sensitivity; high resolution | The potential toxic components and hepatotoxic targets analysis of vine tea121 |
| Proteomics | High sensitivity; high resolution | The toxicities analysis of Herba Lysimachiae123 |
| Metabolomics | High sensitivity; high resolution | Toxic components related to drug-induced liver injury of Heshouwu127 |
| Mass spectrometry imaging technology | High sensitivity; Visual | The damage to liver cell of acetaminophen217 |
| Transgenic animal | Humanized; rapid large-scale screening | Drug reaction analysis of abacavir in HLA mice103 |
| Disease model animal | Short life, low cost, and efficient reproduction | The cardiotoxicity analysis of four typical MALs108 |
| Immunofluorescence and its derivative | High specificity, sensitivity, and speed | Evaluation the side effects of hydrazide drugs116 |
| Flow cytometry | Speed-up, high precision, good accuracy | The cytotoxicity of Drosophila ginsenoside119 |
| Effectiveness analysis | ||
| Target-based biosensors | High sensitivity, low cost, excellent practicability | Early identification of designer drugs of abuse, the development of 5-HT2AR-dependent non-hallucinogenic therapeutics139,141,142 |
| 3D-biological printing organoid | Self-organizing, self-renewing, physiologically relevant, automatic, economic | Dynamic and combinatorial drug screening, personalized drug delivery, assessment in tumor patient-derived organoid-like models144,147,150,151 |
| Microfluidic-combined organ-on-a-chip | Low consumption of reagents, rapid actuation and response | Drug potency evaluation, application at the cell, tissue, and organ levels, analysis of the molecular process in various enteropathies153,159,218 |
| Omics-based artificial intelligence | Improved the efficiency and accuracy, no requirement of clinical trials with low costs | Discovery of innovative drugs, identification of bioactive components in natural medicines166,167 |
| Quality controllability analysis | ||
| DNA mini-barcoding | Accurate for natural products preparation | Angelica sinensis, Panax notoginseng170,174 |
| DNA meta-barcoding | Use universal PCR primers to identify multiple species | Jiuwei Qianghuo Wan |
| Single nucleotide polymorphism | Short DNA sequence unique to a species, highly conserved, rapidly | Ginseng products in Chinese patent medicines |
| DNA melting analysis | Sensitivity, avoids sequencing | Rhizoma Paridis Herrmann170 |
| Electronic tongue | Fast and simple | Taste masking |
| Luciferase reporter transfected cell lines | Highly related to pharmacological effect | Differentiate Huangqin Tang preparations, Uraria crinita, Ephedrae Herba and Astragali Radix |
| COX-2 inhibitor; iNOS activity | Fast and simple | |
| Thrombosis model | Easy for observation, rapid and high-throughput, holistic effect | Distinguish XST; Q-marker identification200 |
| Oxidative stress | Low number of somatic cells; evolution of each cell | Distinguish Ganoderma lucidum quality |
| Chip with enzyme/cells | Automation of operation, more repeatable than cell-based models | Evaluate the quality of Qishen Yiqi Pill from different batches |
| Intestine/liver/kidney | Reducing time and cost of drug development, high-throughput | Pharmacokinetic parameters153,154,218 |
5. Summary and prospects
Biotechnology approaches are as important as physical- and chemical-based detection techniques because they can provide information on biological activity and even clinical efficacy and safety of a drug. Chemical technology and biotechnology have their pros and cons: botanicals with identical chemical spectrum may display different biological activities when bioactive constituents cannot be detected under analytical conditions, and botanicals with different chemical profiles may have the same bioactivity when the phytochemicals responsible for the difference are biologically inert. In the future, biotechnology together with chemical and physical analyses should participate in the whole life cycle of drug quality control.
Bioanalysis is important for pharmaceutical analysis and has been widely used in different areas of pharmaceutical analysis, including qualitative analysis of drugs such as species identification based on herbgenomics219, drug safety analysis such as pesticide residue detection, and effectiveness evaluation such as biological evaluation. Biotechnology has the advantages of high sensitivity and specificity, but its popularization is often limited by the complex operations and high costs. It also has the problems of poor repeatability and false-positivity, requiring much research for optimization and improvement.
At present, bioanalytical technology is applied more to biological drugs and less to natural product and synthetic drugs. This might be because most researchers in the field of pharmaceutical analysis have a background in chemistry. The importance of bioassays and their applications are ignored in current pharmaceutical analysis. With the continuous development of basic biology and the subsequent improvement of bioanalytical methods, biotechnology can play a vital role in the overall quality control of natural products and synthetic pharmaceuticals.
Acknowledgments
We would like to thank TopEdit (www.topeditsci.com) for linguistic assistance during the preparation of this manuscript. This study was supported by the National Key R&D Program of China (No. 2019YFC1711100, China) and the National Natural Science Foundation of China (No. U1812403-1, China). The author would like to thank Dr. Jiabo Wang, Weijun Kong (School of Traditional Chinese Medicine, Capital Medical University, Beijing, China).
Author contributions
Shilin Chen conceived, supervised and reviewed the manuscript; Shilin Chen, Zheng Li, Sanyin Zhang, Yuxin Zhou, Xiaohe Xiao, Pengdi Cui, Binjie Xu, Yuntao Dai, Qinghe Zhao and Shasha Kong wrote the manuscript. Yuntao Dai conceived and reviewed the manuscript. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Contributor Information
Shilin Chen, Email: slchen@icmm.ac.cn.
Yuntao Dai, Email: ytdai@icmm.ac.cn.
References
- 1.Zhang Y., Lv X., Qu J., Zhang X., Zhang M., Gao H., et al. A systematic strategy for screening therapeutic constituents of Schisandra chinensis (Turcz.) Baill infiltrated blood–brain barrier oriented in lesions using ethanol and water extracts: a novel perspective for exploring chemical material basis of herb medicines. Acta Pharm Sin B. 2020;10:557–568. doi: 10.1016/j.apsb.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramakrishnan P., Loh W.M., Gopinath S.C.B., Bonam S.R., Fareez I.M., Guad R.M., et al. Selective phytochemicals targeting pancreatic stellate cells as new anti-fibrotic agents for chronic pancreatitis and pancreatic cancer. Acta Pharm Sin B. 2020;10:399–413. doi: 10.1016/j.apsb.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., Tang Q., Cheng P., Zhu M., Zhang H., Liu J., et al. Whole-genome sequencing and analysis of the Chinese herbal plant Gelsemium elegans. Acta Pharm Sin B. 2020;10:374–382. doi: 10.1016/j.apsb.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lia Q., Xu J., Hana L., Gao C., Lu J., Dua G., et al. Evaluation of ITS2 for intraspecific identification of Paeonia lactiflora cultivars. Biotechnol Rep (Amst) 2017;8 doi: 10.1016/j.btre.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu L., Cui Y., Wang Q., Xu Z., Wang Y., Lin Y., et al. Identification and phylogenetic analysis of five Crataegus species (Rosaceae) based on complete chloroplast genomes. Planta. 2021;254:14. doi: 10.1007/s00425-021-03667-4. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y., Shin J., Cho S.S., Hwang Y.P., Choi C. Development and application of InDel markers for authentication of the Korean herbs Zanthoxylum schinifolium and Zanthoxylum piperitum. Foods. 2019;8:658. doi: 10.3390/foods8120658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noh P., Kim W.J., Yang S., ID I.P., Moon B.C. Authentication of the herbal medicine Angelicae Dahuricae Radix using an ITS sequence-based multiplex SCAR assay. Molecules. 2018;23:2134. doi: 10.3390/molecules23092134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J.J., Xiong C., Liu Y., Liang J.S., Zhou X.W. Loop-mediated isothermal amplification (LAMP): emergence as an alternative technology for herbal medicine identification. Front Plant Sci. 2016;7:1956. doi: 10.3389/fpls.2016.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han K., Wang M., Zhang L., Wang C. Application of molecular methods in the identification of ingredients in Chinese herbal medicines. Molecules. 2018;23:2728. doi: 10.3390/molecules23102728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Y., Gritsenko D., Feng S., Teh Y.C., Lu X., Xu J. Detection of heavy metal by paper-based microfluidics. Biosens Bioelectron. 2016;83:256–266. doi: 10.1016/j.bios.2016.04.061. [DOI] [PubMed] [Google Scholar]
- 11.Wang B., Zhou Y., Qin G., Nie J. Identification of adulteration in Fritillariae cirrhosae Bulbus using multiplex ligation-dependent probe amplification technology. J Pharm Anal. 2018;38:2104–2109. [Google Scholar]
- 12.Wang S., Lin B., Chen L., Li N., Xu J., Wang J., et al. Branch-migration based fluorescent probe for highly sensitive detection of mercury. Anal Chem. 2018;90:11764–11769. doi: 10.1021/acs.analchem.8b03547. [DOI] [PubMed] [Google Scholar]
- 13.Shi R., Hu Z., Lu H., Liu L., Xu L., Liu Y., et al. Hierarchical nanostructuring array enhances mid-hybridization for accurate herbal identification via ITS2 DNA barcode. Anal Chem. 2020;92:2136–2144. doi: 10.1021/acs.analchem.9b04687. [DOI] [PubMed] [Google Scholar]
- 14.Lei P., Zhou Y., Zhao S., Dong C., Shuang S. Carbon-supported X-manganate (XNi, Zn, and Cu) nanocomposites for sensitive electrochemical detection of trace heavy metal ions. J Hazard Mater. 2022;435 doi: 10.1016/j.jhazmat.2022.129036. [DOI] [PubMed] [Google Scholar]
- 15.Songa E.A., Okonkwo J.O. Recent approaches to improving selectivity and sensitivity of enzyme-based biosensors for organophosphorus pesticides: a review. Talanta. 2016;155:289–304. doi: 10.1016/j.talanta.2016.04.046. [DOI] [PubMed] [Google Scholar]
- 16.Peng S., Yang S., Zhang X., Jia J., Chen Q., Lian Y., et al. Analysis of imidacloprid residues in mango, cowpea and water samples based on portable molecular imprinting sensors. PLoS One. 2021;16 doi: 10.1371/journal.pone.0257042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng S., Wang A., Lian Y., Zhang X., Zeng B., Chen Q., et al. Smartphone-based molecularly imprinted sensors for rapid detection of thiamethoxam residues and applications. PLoS One. 2021;16 doi: 10.1371/journal.pone.0258508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang L., Liao X., Jia B., Shi L., Kang L., Zhou L., et al. Recent progress in immunosensors for pesticides. Biosens Bioelectron. 2020;164 doi: 10.1016/j.bios.2020.112255. [DOI] [PubMed] [Google Scholar]
- 19.Du M., Yang Q., Liu W., Ding Y., Chen H., Hua X., et al. Development of immunoassays with high sensitivity for detecting imidacloprid in environment and agro products using phage-borne peptides. Sci Total Environ. 2020;723 doi: 10.1016/j.scitotenv.2020.137909. [DOI] [PubMed] [Google Scholar]
- 20.Zha Y., Li Y., Hu P., Lu S., Ren H., Liu Z., et al. Duplex-specific nuclease-triggered fluorescence immunoassay based on dual-functionalized AuNP for acetochlor, metolachlor, and propisochlor. Anal Chem. 2021;93:13886–13892. doi: 10.1021/acs.analchem.1c02736. [DOI] [PubMed] [Google Scholar]
- 21.Mu X., Yao Y., Peng T., Wang D., Meng L., Wang Y., et al. Current status of organophosphorus pesticide residues in root and rhizome medicinal materials and research progress of rapid detection methods. Zhongguo Zhongyao Zazhi. 2021;46:5736–5743. doi: 10.19540/j.cnki.cjcmm.20210820.101. [DOI] [PubMed] [Google Scholar]
- 22.Ouyang H., Shu Q., Wang W., Wang Z., Yang S., Wang L., et al. An ultra-facile and label-free immunoassay strategy for detection of copper (II) utilizing chemiluminescence self-enhancement of Cu (II)-ethylenediaminetetraacetate chelate. Biosens Bioelectron. 2016;85:157–163. doi: 10.1016/j.bios.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Qin Ja, Lu Q., Wang C., Luo J., Yang M. Colloidal gold-based lateral flow immunoassay with inline cleanup for rapid on-site screening of carbendazim in functional foods. Anal Bioanal Chem. 2021;413:3725–3735. doi: 10.1007/s00216-021-03321-8. [DOI] [PubMed] [Google Scholar]
- 24.Liao B., Hu H., Xiao S., Zhou G., Sun W., Chu Y., et al. Global Pharmacopoeia Genome Database is an integrated and mineable genomic database for traditional medicines derived from eight international pharmacopoeias. Sci China Life Sci. 2022;65:809–817. doi: 10.1007/s11427-021-1968-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou G.R., Liao B.S., Li Q.S., Xu J., Chen S.L. Establishing a genomic database for the medicinal plants in the Brazilian Pharmacopoeia. Chin Med. 2021;16:71. doi: 10.1186/s13020-021-00484-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liao B., Shen X., Xiang L., Guo S., Chen S., Meng Y., et al. Allele-aware chromosome-level genome assembly of Artemisia annua reveals the correlation between ADS expansion and artemisinin yield. Mol Plant. 2022;15:1310–1328. doi: 10.1016/j.molp.2022.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Pang X., Song J., Zhu Y., Xie C., Chen S. Using DNA barcoding to identify species within Euphorbiaceae. Planta Med. 2010;76:6. doi: 10.1055/s-0030-1249806. [DOI] [PubMed] [Google Scholar]
- 28.Lv Y.N., Yang C.Y., Shi L.C., Zhang Z.L., Xu A.S., Zhang L.X., et al. Identification of medicinal plants within the Apocynaceae family using ITS2 and psbA-trnH barcodes. Chin J Nat Med. 2020;18:594–605. doi: 10.1016/S1875-5364(20)30071-6. [DOI] [PubMed] [Google Scholar]
- 29.Cao S., Guo L., Luo H., Yuan H., Chen S., Zheng J., et al. Application of COI barcode sequence for the identification of snake medicine (Zaocys) Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27:483–489. doi: 10.3109/19401736.2014.905828. [DOI] [PubMed] [Google Scholar]
- 30.Xiang L., Tang H., Cheng J., Chen Y., Deng W., Zheng X., et al. The species traceability of the ultrafine powder and the cell wall-broken powder of herbal medicine based on DNA barcoding. Acta Pharm Sin. 2015;50:1660–1667. [PubMed] [Google Scholar]
- 31.Moon B.C., Kim W.J., Ji Y., Lee Y.M., Kang Y.M., Choi G. Molecular identification of the traditional herbal medicines, Arisaematis Rhizoma and Pinelliae Tuber, and common adulterants via universal DNA barcode sequences. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15017064. [DOI] [PubMed] [Google Scholar]
- 32.Basak S., Moolam R.A., Parida A., Mitra S., Rangan L. Evaluation of rapid molecular diagnostics for differentiating medicinal Kaempferia species from its adulterants. Plant Diver. 2019;41:206–211. doi: 10.1016/j.pld.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L., Jiang Y., Liu Y., Niu Z., Xue Q., Liu W., et al. The large single-copy (LSC) region functions as a highly effective and efficient molecular marker for accurate authentication of medicinal Dendrobium species. Acta Pharm Sin B. 2020;10:1989–2002. doi: 10.1016/j.apsb.2020.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang R., Ou J., Li X., Li X., Zhang W., Fang C. Construction of identification cards of medicine Plum varieties based on simple sequence repeats markers. J Anhui Unuv Chinese Med. 2020;39:62–67. [Google Scholar]
- 35.Zhou M., Gao H., Feng Y., Zeng Z., Tian H. Analysis on genetic diversity of Hedyotis diffusa Willd. by inter simple sequence repeat (ISSR) J China Pharm Univ. 2019;50:200–205. [Google Scholar]
- 36.Yuan C., Peng F., Yang Z., Zhong W., Mou F., Gong Y., et al. EST-SSR identification, markers development of Ligusticum chuanxiong based on Ligusticum chuanxiong transcriptome sequences. Zhongguo Zhongyao Zazhi. 2017;42:3332–3340. doi: 10.19540/j.cnki.cjcmm.20170814.004. [DOI] [PubMed] [Google Scholar]
- 37.Wang H., Sun H., Kwon W.S., Jin H., Yang D.C. A PCR-based SNP marker for specific authentication of Korean ginseng (Panax ginseng) cultivar “Chunpoong”. Mol Biol Rep. 2009;37:1053–1057. doi: 10.1007/s11033-009-9827-5. [DOI] [PubMed] [Google Scholar]
- 38.Wu S., Yu Y., Yan Y., Jin D., Wu J. SRAP examination of genetic diversity of the wild Platycodon grandiflorus and the cultivation Platycodon grandiflorus. Northern Horticulture. 2010:132–135. [Google Scholar]
- 39.Noh P., Kim W.J., Yang S., Choi G., Moon B.C. PCR-based rapid diagnostic tools for the authentication of medicinal mistletoe species. Phytomedicine. 2021;91 doi: 10.1016/j.phymed.2021.153667. [DOI] [PubMed] [Google Scholar]
- 40.Long C., Bin W., Li Y., Guo J., Zhang T., Yu Z., et al. RAPD analysis of the genetic relationship of Viscum coloratum (Kom.) nakai. Mol Plant Breed. 2019;17:175–180. [Google Scholar]
- 41.Grazina L., Costa J., Amaral J.S., Mafra I. High-resolution melting analysis as a tool for plant species authentication. Methods Mol Biol. 2021;2264:55–73. doi: 10.1007/978-1-0716-1201-9_5. [DOI] [PubMed] [Google Scholar]
- 42.Sun W., Li J.J., Xiong C., Zhao B., Chen S. The potential power of Bar-HRM technology in herbal medicine identification. Front Plant Sci. 2016;7:367. doi: 10.3389/fpls.2016.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song M., Li J., Xiong C., Liu H., Liang J. Applying high-resolution melting (HRM) technology to identify five commonly used Artemisia species. Sci Rep. 2016;6 doi: 10.1038/srep34133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim W.J., Yang S., Choi G., Park I., Noh P., Kim M.J., et al. Accurate and rapid identification of longan Arillus and Litchi semen by a multiplex PCR assay. Plants. 2020;9:948. doi: 10.3390/plants9080948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang W., Cui S., Cheng X.L., Wei F., Ma S. An optimized TaqMan real-time PCR method for authentication of ASINI CORII COLLA (donkey-hide gelatin) J Pharm Biomed Anal. 2019;170:196–203. doi: 10.1016/j.jpba.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 46.Jiang L.L., Wong K.L., Wong Y.L., Chen W.T., Li M., Lau C.B.S., et al. Helicase-dependent amplification is effective in distinguishing Asian ginseng from American ginseng. Food Control. 2014;43:199–205. [Google Scholar]
- 47.Zheng X., An W., Yao H., Xu J., Chen S. Rapid authentication of the poisonous plant Gelsemium elegans by combining filter-paper-based DNA extraction and RPA-LFD detection. Engineering. 2021;7:14–16. [Google Scholar]
- 48.Liu L., Chao Z., Yang Z., Pan S., Wu B., Preng K. Identification of Bupleurum plants-derived crude drugs by oligonucleotide DNA chip based on ITS region polymorphism. Chin Pharmaceut J. 2010;45:821–824. [Google Scholar]
- 49.Li S., Zhang C., Wang S., Liu Q., Feng H., Ma X., et al. Electrochemical microfluidics techniques for heavy metal ion detection. Analyst. 2018;143:4230–4246. doi: 10.1039/c8an01067f. [DOI] [PubMed] [Google Scholar]
- 50.Fourou H., Zazoua A., Braiek M., Jaffrezic-Renault N. An enzyme biosensor based on beta-galactosidase inhibition for electrochemical detection of cadmium (II) and chromium (VI) Int J Environ An Ch. 2016;96:872–885. [Google Scholar]
- 51.Kim Y., Choi H., Shin W.H., Oh J.M., Koo S.M., Kim Y., et al. Development of colorimetric whole-cell biosensor for detection of heavy metals in environment for public health. Int J Environ Res Publ Health. 2021;18 doi: 10.3390/ijerph182312721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tu J., Zhou Z., Liu Y., Li T., Lu S., Xiao L., et al. Nanochannel-based sensor for the detection of lead ions in traditional Chinese medicine. RSC Adv. 2021;11:3751–3758. doi: 10.1039/d0ra10157e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang Y., Xu K., Zhao B., Zhang M., Gong C., Wan H., et al. A novel electronic nose for the detection and classification of pesticide residue on apples. RSC Adv. 2021;11:20874–20883. doi: 10.1039/d1ra03069h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aly N., Mosallam E., Ahmed N., El-Gendy K. Development and validation of a novel enzyme-linked immunosorbent assay for monitoring ethylene thiourea in soil and vegetable samples. Food Chem. 2020;325 doi: 10.1016/j.foodchem.2020.126931. [DOI] [PubMed] [Google Scholar]
- 55.Watanabe E., Miyake S., Yogo Y. Review of enzyme-linked immunosorbent assays (ELISAs) for analyses of neonicotinoid insecticides in agro-environments. J Agric Food Chem. 2013;61:12459–12472. doi: 10.1021/jf403801h. [DOI] [PubMed] [Google Scholar]
- 56.Wu K.H., Huang W.C., Chang S.C., Shyu R.H. Colloidal silver-based lateral flow immunoassay for detection of profenofos pesticide residue in vegetables. RSC Adv. 2022;12:13035–13044. doi: 10.1039/d2ra01654k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L., Zeng S., Chen T., Qu H. Direct analysis in real time mass spectrometry, a process analytical technology tool for real-time process monitoring in botanical drug manufacturing. J Pharm Biomed Anal. 2014;91:202–209. doi: 10.1016/j.jpba.2013.12.034. [DOI] [PubMed] [Google Scholar]
- 58.Feng W.W., Zhang Y., Tang J.F., Zhang C.E., Dong Q., Li R.Y., et al. Combination of chemical fingerprinting with bioassay, a preferable approach for quality control of Safflower Injection. Anal Chim Acta. 2018;1003:56–63. doi: 10.1016/j.aca.2017.11.069. [DOI] [PubMed] [Google Scholar]
- 59.Yan D., Li J., Xiong Y., Zhang C., Luo J., Han Y., et al. Promotion of quality standard of herbal medicine by constituent removing and adding. Sci Rep. 2014;4:3668. doi: 10.1038/srep03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sathish J.G., Sethu S., Bielsky M.C., Haan Ld, French N.S., Govindappa K., et al. Challenges and approaches for the development of safer immunomodulatory biologics. Nat Rev Drug Discov. 2013;12:306–324. doi: 10.1038/nrd3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Z., Wang Y., Cheng Y. Mass spectrometry-sensitive probes coupled with direct analysis in real time for simultaneous sensing of chemical and biological properties of botanical drugs. Anal Chem. 2019;91:9001–9009. doi: 10.1021/acs.analchem.9b01251. [DOI] [PubMed] [Google Scholar]
- 62.Ma H., Li J., An M., Gao X.M., Chang Y.X. A powerful on line ABTS(+)-CE-DAD method to screen and quantify major antioxidants for quality control of Shuxuening injection. Sci Rep. 2018;8:5441. doi: 10.1038/s41598-018-23748-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X., Chen X., Li J., Evans O.B., Wang H., Yang X., et al. Thrombin-based discovery strategy of bioactive-chemical quality marker combination for pollen of Typha orientalis by metabolomics coupled with chemometrics. Phytomedicine. 2020;75 doi: 10.1016/j.phymed.2020.153246. [DOI] [PubMed] [Google Scholar]
- 64.Jabbari S., Dabirmanesh B., Arab S.S., Amanlou M., Daneshjou S., Gholami S., et al. A novel enzyme based SPR-biosensor to detect bromocriptine as an ergoline derivative drug. Sensor Actuator B Chem. 2017;240:519–527. [Google Scholar]
- 65.Tao Y., Zhang Y., Wang Y., Chen Y. Hollow fiber based affinity selection combined with high performance liquid chromatography-mass spectroscopy for rapid screening lipase inhibitors from lotus leaf. Anal Chim Acta. 2013;785:75–81. doi: 10.1016/j.aca.2013.04.058. [DOI] [PubMed] [Google Scholar]
- 66.Wang H., Huang Y., Zhao X., Gong W., Wang Y., Cheng Y. A novel aggregation-induced emission based fluorescent probe for an angiotensin converting enzyme (ACE) assay and inhibitor screening. Chem Commun. 2014;50:15075–15078. doi: 10.1039/c4cc07161a. [DOI] [PubMed] [Google Scholar]
- 67.Zhang N., Li J., Liu B., Wang H., Zhang D., Li Z. A facile "turn-on" fluorescent aptasensor for simultaneous detection of dual mycotoxins in traditional Chinese medicine based on graphene oxide and FRET. Toxicon. 2022;206:42–50. doi: 10.1016/j.toxicon.2021.12.006. [DOI] [PubMed] [Google Scholar]
- 68.Zhao Y.W., Wang H.X., Jia G.C., Li Z. Application of aptamer-based biosensor for rapid detection of pathogenic Escherichia coli. Sensors (Basel) 2018;18:2518. doi: 10.3390/s18082518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gong X., Shao J., Guo S., Pan J., Fan X. Determination of inhibitory activity of Salvia miltiorrhiza extracts on xanthine oxidase with a paper-based analytical device. J Pharm Anal. 2021;11:603–610. doi: 10.1016/j.jpha.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo S., Shao J., Gong X. Paper-based analytical devices prepared with polycaprolactone printing and their application in the activity determination of mulberry extracts. J Pharm Biomed Anal. 2018;161:28–34. doi: 10.1016/j.jpba.2018.08.022. [DOI] [PubMed] [Google Scholar]
- 71.Guo S., Lin X., Wang Y., Gong X. Fabrication of paper-based enzyme immobilized microarray by 3D-printing technique for screening alpha-glucosidase inhibitors in mulberry leaves and lotus leaves. Chin Med. 2019;14:13. doi: 10.1186/s13020-019-0236-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z.H., Ai N., Yu L.X., Qian Z.Z., Cheng Y.Y. A multiple biomarker assay for quality assessment of botanical drugs using a versatile microfluidic chip. Sci Rep. 2017;7 doi: 10.1038/s41598-017-12453-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong Y., Ru C., Wang S., Li Z., Cheng Y. An online, non-destructive method for simultaneously detecting chemical, biological, and physical properties of herbal injections using hyperspectral imaging with artificial intelligence. Spectrochim Acta Mol Biomol Spectrosc. 2022;264 doi: 10.1016/j.saa.2021.120250. [DOI] [PubMed] [Google Scholar]
- 74.Ding G., Li B., Han Y., Liu A., Zhang J., Peng J., et al. A rapid integrated bioactivity evaluation system based on near-infrared spectroscopy for quality control of Flos Chrysanthemi. J Pharm Biomed Anal. 2016;131:391–399. doi: 10.1016/j.jpba.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 75.Fraser K., Lane G.A., Otter D.E., Harrison S.J., Quek S.Y., Hemar Y., et al. Monitoring tea fermentation/manufacturing by direct analysis in real time (DART) mass spectrometry. Food Chem. 2013;141:2060–2065. doi: 10.1016/j.foodchem.2013.05.054. [DOI] [PubMed] [Google Scholar]
- 76.Zhu X., Rehman K.U., Wang B., Shahzad M. Modern soft-sensing modeling methods for fermentation processes. Sensors. 2020;20:1771. doi: 10.3390/s20061771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andryukov B.G., Lyapun I.N., Matosova E.V., Somova L.M. Biosensor technologies in medicine: from detection of biochemical markers to research into molecular targets (review) Sovrem Tekhnologii Med. 2021;12:70–83. doi: 10.17691/stm2020.12.6.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bhavadharini B., Kavimughil M., Malini B., Vallath A., Prajapati H.K., Sunil C.K. Recent advances in biosensors for detection of chemical contaminants in food—a review. Food Anal Methods. 2022;15:1545–1564. [Google Scholar]
- 79.Vigneshvar S., Sudhakumari C.C., Senthilkumaran B., Prakash H. Recent advances in biosensor technology for potential applications―an overview. Front Bioeng Biotechnol. 2016;4:11. doi: 10.3389/fbioe.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ahmadi A., Danesh N.M., Ramezani M., Alibolandia M., Lavaee P., Emrani A.S., et al. A rapid and simple ratiometric fluorescent sensor for patulin detection based on a stabilized DNA duplex probe containing less amount of aptamer-involved base pairs. Talanta. 2019;204:641–646. doi: 10.1016/j.talanta.2019.06.057. [DOI] [PubMed] [Google Scholar]
- 81.Caglayan M.O., Üstündağ Z. Detection of zearalenone in an aptamer assay using attenuated internal reflection ellipsometry and it's cereal sample applications. Food Chem Toxicol. 2020;136 doi: 10.1016/j.fct.2019.111081. [DOI] [PubMed] [Google Scholar]
- 82.He D., Wu Z., Cui B., Jin Z., Xu E. A fluorometric method for aptamer-based simultaneous determination of two kinds of the fusarium mycotoxins zearalenone and fumonisin B1 making use of gold nanorods and upconversion nanoparticles. Mikrochim Acta. 2020;187:254. doi: 10.1007/s00604-020-04236-4. [DOI] [PubMed] [Google Scholar]
- 83.Jiang M., Chen C., He J., Zhang H., Xu Z. Fluorescence assay for three organophosphorus pesticides in agricultural products based on magnetic-assisted fluorescence labeling aptamer probe. Food Chem. 2020;307 doi: 10.1016/j.foodchem.2019.125534. [DOI] [PubMed] [Google Scholar]
- 84.Khan I.M., Zhao S., Niazi S., Mohsin A., Shoaib M., Duan N., et al. Silver nanoclusters based FRET aptasensor for sensitive and selective fluorescent detection of T-2 toxin. Sensor Actuator B Chem. 2018;277:328–335. [Google Scholar]
- 85.Khan R., Sherazi T.A., Catanante G., Rasheed S., Marty J.L., Hayat A. Switchable fluorescence sensor toward PAT via CA-MWCNTs quenched aptamer-tagged carboxyfluorescein. Food Chem. 2020;312 doi: 10.1016/j.foodchem.2019.126048. [DOI] [PubMed] [Google Scholar]
- 86.Mahmoud M., Laufer S., Deigner H.P. Visual aptamer-based capillary assay for ethanolamine using magnetic particles and strand displacement. Mikrochim Acta. 2019;186:690. doi: 10.1007/s00604-019-3795-9. [DOI] [PubMed] [Google Scholar]
- 87.Wu S., Liu L., Duan N., Li Q., Zhou Y., Wang Z. An aptamer-based lateral flow test strip for rapid detection of zearalenone in corn samples. J Agric Food Chem. 2018;66:1949–1954. doi: 10.1021/acs.jafc.7b05326. [DOI] [PubMed] [Google Scholar]
- 88.Xing K.Y., Peng J., Shan S., Liu D., Huang Y.N., Lai W. Green enzyme-linked immunosorbent assay based on the single-stranded binding protein-assisted aptamer for the detection of mycotoxin. Anal Chem. 2020;92:8422–8426. doi: 10.1021/acs.analchem.0c01073. [DOI] [PubMed] [Google Scholar]
- 89.Zhang M., Wang Y., Yuan S., Sun X., Huo B., Bai J., et al. Competitive fluorometric assay for the food toxin T-2 by using DNA-modified silver nanoclusters, aptamer-modified magnetic beads, and exponential isothermal amplification. Mikrochim Acta. 2019;186:219. doi: 10.1007/s00604-019-3322-z. [DOI] [PubMed] [Google Scholar]
- 90.Wang H., Zhao Y., Bie S., Suo T., Jia G., Liu B., et al. Development of an electrochemical biosensor for rapid and effective detection of pathogenic Eoscherichia coli in licorice extract. Appl Sci. 2019;9:295. [Google Scholar]
- 91.Gu Z., Zhao M., Sheng Y., Bentolila L.A., Tang Y. Detection of mercury ion by infrared fluorescent protein and its hydrogel-based paper assay. Anal Chem. 2011;83:2324–2329. doi: 10.1021/ac103236g. [DOI] [PubMed] [Google Scholar]
- 92.He Y., Wua W.B., Fu J.Z. Rapid fabrication of paper-based microfluidic analytical devices with desktop stereolithography 3D printer. RSC Adv. 2015;5:2694–2701. [Google Scholar]
- 93.Mitchell H.T., Noxon I.C., Chaplan C.A., Carlton S.J., Liu C.H., Ganaja K.A., et al. Reagent pencils: a new technique for solvent-free deposition of reagents onto paper-based microfluidic devices. Lab Chip. 2015;15:2213–2220. doi: 10.1039/c5lc00297d. [DOI] [PubMed] [Google Scholar]
- 94.Ahmed S., Bui M.-P.N., Abbas A. Paper-based chemical and biological sensors: engineering aspects. Biosens Bioelectron. 2016;77:249–263. doi: 10.1016/j.bios.2015.09.038. [DOI] [PubMed] [Google Scholar]
- 95.Fan X., Zhao X., Jin Y., Shen X., Liu C. Network toxicology and its application to traditional Chinese medicine. Zhongguo Zhongyao Zazhi. 2011;36:2920–2922. [PubMed] [Google Scholar]
- 96.Liu Y., Tan Y., Huang J., Wu C., Fan X., Stalin A., et al. Revealing the mechanism of Huazhi Rougan granule in the treatment of nonalcoholic fatty liver through intestinal flora based on 16S rRNA, metagenomic sequencing and network pharmacology. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.875700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li X.Y., Jin X., Li Y.Z., Gao D.D., Liu R., Liu C.X. Network toxicology and LC‒MS-based metabolomics: new approaches for mechanism of action of toxic components in traditional Chinese medicines. Chin Herbal Med. 2019;11:357–363. [Google Scholar]
- 98.He T., Liu C., Li M., Wang M., Liu N., Zhang D., et al. Integrating non-targeted metabolomics and toxicology networks to study the mechanism of Esculentoside A-induced hepatotoxicity in rats. J Biochem Mol Toxicol. 2021;35:1–15. doi: 10.1002/jbt.22761. [DOI] [PubMed] [Google Scholar]
- 99.Eduardoa N., Juana A.P., Josefinaa A.S., Martinb T.J.M., Cirilob P., Victoriac T.M., et al. Cardiovascular activity of a methanolic extract of Digitalis purpurea spp. heywoodii. J Ethnopharmacol. 2000;71:437–442. doi: 10.1016/s0378-8741(00)00175-6. [DOI] [PubMed] [Google Scholar]
- 100.Sacks D., Baxter B., Campbell B.C.V., Carpenter J.S., Cognard C., Dippel D., et al. Multisociety consensus quality improvement revised consensus statement for endovascular therapy of acute ischemic stroke. Int J Stroke. 2018;13:612–632. doi: 10.1177/1747493018778713. [DOI] [PubMed] [Google Scholar]
- 101.Bissig K.D., Han W., Barzi M., Kovalchuk N., Ding L., Fan X., et al. P450-humanized and human liver chimeric mouse models for studying xenobiotic metabolism and toxicity. Drug Metab Dispos. 2018;46:1734–1744. doi: 10.1124/dmd.118.083303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Susukida T., Aoki S., Shirayanagi T., Yamada Y., Kuwahara S., Ito K. HLA transgenic mice: application in reproducing idiosyncratic drug toxicity. Drug Metab Rev. 2020;52:540–567. doi: 10.1080/03602532.2020.1800725. [DOI] [PubMed] [Google Scholar]
- 103.Song B., Aoki S., Liu C., Susukida T., Ito K. An animal model of abacavir-induced HLA-mediated liver injury. Toxicol Sci. 2018;162:713–723. doi: 10.1093/toxsci/kfy001. [DOI] [PubMed] [Google Scholar]
- 104.Liu C., Chen S., Xiao X., Zhang T., Hou W., Liao M. A new concept on quality marker of Chinese materia medica: quality control for Chinese medicinal products. Chin Tradit Herb Drugs. 2016;47:1443–1457. [Google Scholar]
- 105.Cully M. Zebrafish earn their drug discovery stripes. Nat Rev Drug Discov. 2019;18:811–813. doi: 10.1038/d41573-019-00165-x. [DOI] [PubMed] [Google Scholar]
- 106.Patton E.E., Zon L.I., Langenau D.M. Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials. Nat Rev Drug Discov. 2021;20:611–628. doi: 10.1038/s41573-021-00210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang C., Qian D.D., Yu T., Yang H., Li P., Li H.J. Multi-parametric cellular imaging coupled with multi-component quantitative profiling for screening of hepatotoxic equivalent markers from Psoraleae Fructus. Phytomedicine. 2021;93 doi: 10.1016/j.phymed.2021.153518. [DOI] [PubMed] [Google Scholar]
- 108.Yan Z., Huang X., Xie Y., Song M., Zhu K., Ding S. Macrolides induce severe cardiotoxicity and developmental toxicity in zebrafish embryos. Sci Total Environ. 2019;649:1414–1421. doi: 10.1016/j.scitotenv.2018.07.432. [DOI] [PubMed] [Google Scholar]
- 109.Sharma M., Gupta P., Garabadu D. Bacopa monnieri attenuates glutamate-induced nociception and brain mitochondrial toxicity in Zebrafish. Metab Brain Dis. 2022;37:383–396. doi: 10.1007/s11011-021-00874-6. [DOI] [PubMed] [Google Scholar]
- 110.Gao S., Chen W., Zhang N., Xu C., Jing H., Zhang W., et al. A high-throughput assay for the prediction of chemical toxicity by automated phenotypic profiling of Caenorhabditis elegans. J Vis Exp. 2019;145 doi: 10.3791/59082. [DOI] [PubMed] [Google Scholar]
- 111.Camacho J., Conti Ad, Pogribny I.P., Sprando R.L., Hunt P.R. Assessment of the effects of organic vs. inorganic arsenic and mercury in Caenorhabditis elegans. Curr Res Toxicol. 2022;3 doi: 10.1016/j.crtox.2022.100071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Martínez-Fernández C., Bergamino M., Schiavi A., Brena D., Ventura N., Honnen S., et al. Insights into cisplatin-induced neurotoxicity and mitochondrial dysfunction in Caenorhabditis elegans. Dis Model Mech. 2022;15 doi: 10.1242/dmm.049161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang Y., Cheng Q., Su Q., Yu X., Shen T., Yang X., et al. Aesculin offers increased resistance against oxidative stress and protective effects against Aβ-induced neurotoxicity in Caenorhabditis elegans. Eur J Pharmacol. 2022;917 doi: 10.1016/j.ejphar.2022.174755. [DOI] [PubMed] [Google Scholar]
- 114.Karlsson O., Hanrieder J. Imaging mass spectrometry in drug development and toxicology. Arch Toxicol. 2016;91:2283–2294. doi: 10.1007/s00204-016-1905-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Im K., Mareninov S., Diaz M.F.P., Yong W.H. An introduction to performing immunofluorescence staining. Methods Mol Biol. 2019;1897:299–311. doi: 10.1007/978-1-4939-8935-5_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fu M., Wang K., Xue J., Li Y., Bian M., Zhu Q. A dual-response fluorescent probe for N2H4 and viscosity in living cells and zebrafish to evaluate liver injury. Org Biomol Chem. 2022;20:3359–3364. doi: 10.1039/d2ob00260d. [DOI] [PubMed] [Google Scholar]
- 117.Nolan J.P., Lauer S., Prossnitz E.R., Sklar L.A. Flow cytometry: a versatile tool for all phases of drug discovery. Drug Discov Today. 1999;4:173–180. doi: 10.1016/S1359-6446(99)01320-3. [DOI] [PubMed] [Google Scholar]
- 118.Heisler J., Elvir L., Barnouti F., Charles E., Wolkow T.D., Pyati R. Morphological effects of natural products on Schizosaccharomyces pombe measured by imaging flow cytometry. Nat Prod Bioprospect. 2014;4:27–35. doi: 10.1007/s13659-014-0004-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Qiao J., Zhao Y., Liu Y., Zhang S., Zhao W., Liu S., et al. Neuroprotective effect of Ginsenoside Re against neurotoxin-induced Parkinson's disease models via induction of Nrf2. Mol Med Rep. 2022;25:215. doi: 10.3892/mmr.2022.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu Z., Huang R., Roberts R., Tong W. Toxicogenomics: a 2020 vision. Trends Pharmacol Sci. 2019;40:92–103. doi: 10.1016/j.tips.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xiao F., Qiu J., Zhao Y. Exploring the potential toxicological mechanisms of vine tea on the liver based on network toxicology and transcriptomics. Front Pharmacol. 2022;13 doi: 10.3389/fphar.2022.855926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meissner F., Geddes-McAlister J., Mann M., Bantscheff M. The emerging role of mass spectrometry-based proteomics in drug discovery. Nat Rev Drug Discov. 2022;21:637–654. doi: 10.1038/s41573-022-00409-3. [DOI] [PubMed] [Google Scholar]
- 123.Li X., Liu Q., Zhang S., Yang W., Zhou Y. Biolabel-led research pattern reveals serum profile in rats after treatment with herba Lysimachiae: combined analysis of metabonomics and proteomics. Biomed Chromatogr. 2022;36:e5385. doi: 10.1002/bmc.5385. [DOI] [PubMed] [Google Scholar]
- 124.Sun S., Wang M., Yuan Y., Wang S., Ding H., Liang C., et al. A new strategy for the rapid identification and validation of direct toxicity targets of psoralen-induced hepatotoxicity. Toxicol Lett. 2022;363:11–26. doi: 10.1016/j.toxlet.2022.05.002. [DOI] [PubMed] [Google Scholar]
- 125.Wishart D.S. Metabolomics for investigating physiological and pathophysiological processes. Physiol Rev. 2019;99:1819–1875. doi: 10.1152/physrev.00035.2018. [DOI] [PubMed] [Google Scholar]
- 126.Ma K.F., Zhang X.G., Jia H.Y. CYP1A2 polymorphism in Chinese patients with acute liver injury induced by Polygonum multiflorum. Genet Mol Res. 2014;13:5637–5643. doi: 10.4238/2014.July.25.19. [DOI] [PubMed] [Google Scholar]
- 127.Li C.Y., Tu C., Gao D., Wang R.L., Zhang H.Z., Niu M., et al. Metabolomic study on idiosyncratic liver injury induced by different extracts of Polygonum multiflorum in rats integrated with pattern recognition and enriched pathways analysis. Front Pharmacol. 2016;7:483. doi: 10.3389/fphar.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu L., Wang Y., Ma Z., Tang X., Gao Y. Urine metabolomics study on potential hepatoxic biomarkers identification in rats induced by aurantio-obtusin. Front Pharmacol. 2020;11:1237. doi: 10.3389/fphar.2020.01237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harvey A.L., Edrada-Ebel R., Quinn R.J. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov. 2015;14:111–129. doi: 10.1038/nrd4510. [DOI] [PubMed] [Google Scholar]
- 130.Li F.S., Weng J.K. Demystifying traditional herbal medicine with modern approach. Native Plants. 2017;3 doi: 10.1038/nplants.2017.109. [DOI] [PubMed] [Google Scholar]
- 131.Luo S., Guo L., Sheng C., Zhao Y., Chen L., Li C., et al. Rapid identification and isolation of neuraminidase inhibitors from mockstrawberry (Duchesnea indica Andr.) based on ligand fishing combined with HR-ESI-Q-TOF-MS. Acta Pharm Sin B. 2020;10:1846–1855. doi: 10.1016/j.apsb.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gu Y., Chen X., Wang Y., Liu Y., Zheng L., Li X., et al. Development of 3-mercaptopropyltrimethoxysilane (MPTS)-modified bone marrow mononuclear cell membrane chromatography for screening anti-osteoporosis components from Scutellariae Radix. Acta Pharm Sin B. 2020;10:1856–1865. doi: 10.1016/j.apsb.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen X., Wu Y., Chen C., Gu Y., Zhu C., Wang S., et al. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm Sin B. 2021;11:222–236. doi: 10.1016/j.apsb.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mejias M., Gallego J., Naranjo-Suarez S., Ramirez M., Pell N., Manzano A., et al. CPEB4 increases expression of PFKFB3 to induce glycolysis and activate mouse and human hepatic stellate cells, promoting liver fibrosis. Gastroenterology. 2020;159:273–288. doi: 10.1053/j.gastro.2020.03.008. [DOI] [PubMed] [Google Scholar]
- 135.Mangan M.S.J., Olhava E.J., Roush W.R., Seidel H.M., Glick G.D., Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17:588–606. doi: 10.1038/nrd.2018.97. [DOI] [PubMed] [Google Scholar]
- 136.Riccardi C., Napolitano E., Platella C., Musumeci D., Montesarchio D. G-quadruplex-based aptamers targeting human thrombin: discovery, chemical modifications and antithrombotic effects. Pharmacol Ther. 2021;217 doi: 10.1016/j.pharmthera.2020.107649. [DOI] [PubMed] [Google Scholar]
- 137.Kurbanoglu S., Erkmen C., Uslu B. Frontiers in electrochemical enzyme based biosensors for food and drug analysis. Trends Anal Chem. 2020;124 [Google Scholar]
- 138.Wang Z., Zhang Y., Zeng Y., Li X., Chen Z., Luo J., et al. Discovery of TAS2R14 agonists from Platycodon grandiflorum using virtual screening and affinity screening based on a novel TAS2R14-functionalized HEMT sensor combined with UPLC–MS analysis. J Agric Food Chem. 2018;66:11663–11671. doi: 10.1021/acs.jafc.8b04455. [DOI] [PubMed] [Google Scholar]
- 139.Dong C., Ly C., Dunlap L.E., Vargas M.V., Sun J., Hwang I.W., et al. Psychedelic-inspired drug discovery using an engineered biosensor. Cell. 2021;184:2779–2792 e18. doi: 10.1016/j.cell.2021.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lima H.R.S., Silva JSd, Farias EAdO., Teixeira P.R.S., Eiras C., Nunes eLCC. Electrochemical sensors and biosensors for the analysis of antineoplastic drugs. Biosens Bioelectron. 2018;108:27–37. doi: 10.1016/j.bios.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 141.Gong W., Li J., Chu Z., Yang D., Subhan S., Li J., et al. A low-cost high-entropy porous CrO/CrN/C biosensor for highly sensitive simultaneous detection of dopamine and uric acid. Microchem J. 2022;175 [Google Scholar]
- 142.Chen H., Liu Y., Feng S., Cao Y., Wu T., Liu Z. Cotton thread-based multi-channel photothermal biosensor for simultaneous detection of multiple microRNAs. Biosens Bioelectron. 2022;200 doi: 10.1016/j.bios.2021.113913. [DOI] [PubMed] [Google Scholar]
- 143.Driehuis E., Kretzschmar K., Clevers H. Establishment of patient-derived cancer organoids for drug-screening applications. Nat Protoc. 2020;15:3380–3409. doi: 10.1038/s41596-020-0379-4. [DOI] [PubMed] [Google Scholar]
- 144.Schuster B., Junkin M., Kashaf S.S., Romero-Calvo I., Kirby K., Matthews J., et al. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat Commun. 2020;11:5271. doi: 10.1038/s41467-020-19058-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mohapatra S., Kar R.K., Biswal P.K., Bindhani S. Approaches of 3D printing in current drug delivery. Sensors International. 2022;3 [Google Scholar]
- 146.Mohammed A.A., Algahtani M.S., Ahmad M.Z., Ahmad J., Kotta S. 3D printing in medicine: technology overview and drug delivery applications. Annals of 3D Printed Medicine. 2021;4 [Google Scholar]
- 147.Chen K.Y., Zeng J.J., Lin G.T. Fabrication of 5-fluorouracil-loaded tablets with hyperbranched polyester by digital light processing 3D printing technology. Eur Polym J. 2022;171 [Google Scholar]
- 148.Baskar G., Palaniyandi T., Viswanathan S., Rajendran B.K., Ravi M., Sivaji A. Development of patient derived organoids for cancer drug screening applications. Acta Histochem. 2022;124 doi: 10.1016/j.acthis.2022.151895. [DOI] [PubMed] [Google Scholar]
- 149.Kondo J., Inoue M. Application of cancer organoid model for drug screening and personalized therapy. Cells. 2019;8:470. doi: 10.3390/cells8050470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chadwick M., Yang C., Liu L., Gamboa C.M., Jara K., Lee H., et al. Rapid processing and drug evaluation in glioblastoma patient-derived organoid models with 4D bioprinted arrays. iScience. 2020;23 doi: 10.1016/j.isci.2020.101365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen L., Dai Z., Ge C., Huang D., Zhou X., Pan K., et al. Specific metabolic response of patient-derived organoids to curcumin of colorectal cancer. J Chromatogr, B: Anal Technol Biomed Life Sci. 2022;1203 doi: 10.1016/j.jchromb.2022.123260. [DOI] [PubMed] [Google Scholar]
- 152.Clapp N., Amour A., Rowan W.C., Candarlioglu P.L. Organ-on-chip applications in drug discovery: an end user perspective. Biochem Soc Trans. 2021;49:1881–1890. doi: 10.1042/BST20210840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ren L., Zhou X., Nasiri R., Fang J., Jiang X., Wang C., et al. Combined effects of electric stimulation and microgrooves in cardiac tissue-on-a-chip for drug screening. Small Methods. 2020;4 doi: 10.1002/smtd.202000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Delalat B., Cozzi C., Ghaemi S.R., Polito G., Kriel F.H., Michl T.D., et al. Microengineered bioartificial liver chip for drug toxicity screening. Adv Funct Mater. 2018;28 [Google Scholar]
- 155.Romano G., Marino I.R. Organoids and organs-on-chips: systems for disease modeling, drug screening and identification of environmental risk factors for human illnesses. Drugs Future. 2020;45:553. [Google Scholar]
- 156.Azizipour N., Avazpour R., Rosenzweig D.H., Sawan M., Ajji A. Evolution of biochip technology: a review from lab-on-a-chip to organ-on-a-chip. Micromachines. 2020;11:599. doi: 10.3390/mi11060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ma C., Peng Y., Li H., Chen W. Organ-on-a-chip: a new paradigm for drug development. Trends Pharmacol Sci. 2021;42:119–133. doi: 10.1016/j.tips.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ma Z., Li B., Peng J., Gao D. Recent development of drug delivery systems through microfluidics: from synthesis to evaluation. Pharmaceutics. 2022;14:434. doi: 10.3390/pharmaceutics14020434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Huang X., Tang J., Hu L., Bian R., Liu M., Cao W., et al. Arrayed microfluidic chip for detection of circulating tumor cells and evaluation of drug potency. Anal Biochem. 2019;564–565:64–71. doi: 10.1016/j.ab.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 160.Liu Y., Sun L., Zhang H., Shang L., Zhao Y. Microfluidics for drug development: from synthesis to evaluation. Chem Rev. 2021;121:7468–7529. doi: 10.1021/acs.chemrev.0c01289. [DOI] [PubMed] [Google Scholar]
- 161.Bein A., Shin W., Jalili-Firoozinezhad S., Park M.H., Sontheimer-Phelps A., Tovaglieri A., et al. Microfluidic organ-on-a-chip models of human intestine. Cell Mol Gastroenterol Hepatol. 2018;5:659–668. doi: 10.1016/j.jcmgh.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yang X., Wang Y., Byrne R., Schneider G., Yang S. Concepts of artificial intelligence for computer-assisted drug discovery. Chem Rev. 2019;119:10520–10594. doi: 10.1021/acs.chemrev.8b00728. [DOI] [PubMed] [Google Scholar]
- 163.Patel V., Shah M. Artificial intelligence and machine learning in drug discovery and drug development. Intelligent Medicine. 2021;2:134–140. [Google Scholar]
- 164.Gupta R., Srivastava D., Sahu M., Tiwari S., Ambasta R.K., Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers. 2021;25:1315–1360. doi: 10.1007/s11030-021-10217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xu Z., Wang X., Zeng S., Ren X., Yan Y., Gong Z. Applying artificial intelligence for cancer immunotherapy. Acta Pharm Sin B. 2021;11:3393–3405. doi: 10.1016/j.apsb.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Polykovskiy D., Zhebrak A., Vetrov D., Ivanenkov Y., Aladinskiy V., Mamoshina P., et al. Entangled conditional adversarial autoencoder for de novo drug discovery. Mol Pharm. 2018;15:4398–4405. doi: 10.1021/acs.molpharmaceut.8b00839. [DOI] [PubMed] [Google Scholar]
- 167.Kong W.J., Zhang S.S., Zhao Y.L., Wu M.Q., Chen P., Wu X.R., et al. Combination of chemical fingerprint and bioactivity evaluation to explore the antibacterial components of Salvia miltiorrhizae. Sci Rep. 2017;7:8112. doi: 10.1038/s41598-017-08377-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Meusnier I., Singer G.A., Landry J.F., Hickey D.A., Hebert P.D., Hajibabaei M. A universal DNA mini-barcode for biodiversity analysis. BMC Genom. 2008;9:214. doi: 10.1186/1471-2164-9-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Little D.P. Authentication of Ginkgo biloba herbal dietary supplements using DNA barcoding. Genome. 2014;57:513–516. doi: 10.1139/gen-2014-0130. [DOI] [PubMed] [Google Scholar]
- 170.Lo Y.T., Shaw P.C. DNA barcoding in concentrated Chinese medicine granules using adaptor ligation-mediated polymerase chain reaction. J Pharm Biomed Anal. 2018;149:512–516. doi: 10.1016/j.jpba.2017.11.048. [DOI] [PubMed] [Google Scholar]
- 171.Liu Y., Wang X.Y., Wei X.M., Gao Z.T., Han J.P. Rapid authentication of Ginkgo biloba herbal products using the recombinase polymerase amplification assay. Sci Rep. 2018;8:8002. doi: 10.1038/s41598-018-26402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Taberlet P., Prud'Homme S.M., Campione E., Roy J., Miquel C., Shehzad W., et al. Soil sampling and isolation of extracellular DNA from large amount of starting material suitable for metabarcoding studies. Mol Ecol. 2012;21:1816–1820. doi: 10.1111/j.1365-294X.2011.05317.x. [DOI] [PubMed] [Google Scholar]
- 173.Xin T., Xu Z., Jia J., Leon C., Hu S., Lin Y., et al. Biomonitoring for traditional herbal medicinal products using DNA metabarcoding and single molecule, real-time sequencing. Acta Pharm Sin B. 2018;8:488–497. doi: 10.1016/j.apsb.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Liu Y., Wang X., Wang L., Chen X., Pang X., Han J. A nucleotide signature for the identification of American Ginseng and its products. Front Plant Sci. 2016;7:319. doi: 10.3389/fpls.2016.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ririe K.M., Rasmussen R.P., Wittwer C.T. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal Biochem. 1997;245:154–160. doi: 10.1006/abio.1996.9916. [DOI] [PubMed] [Google Scholar]
- 176.Herrmann M.G., Durtschi J.D., Bromley L.K., Wittwer C.T., Voelkerding K.V. Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem. 2006;52:494–503. doi: 10.1373/clinchem.2005.063438. [DOI] [PubMed] [Google Scholar]
- 177.Gao Z., Liu Y., Wang X., Wei X., Han J. DNA mini-barcoding: a derived barcoding method for herbal molecular identification. Front Plant Sci. 2019;10:987. doi: 10.3389/fpls.2019.00987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Vlasov Y.G., Legin A.V., Rudnitskaya A.M., DiNatale C., DAmico A. Multisensor system with an array of chemical sensors and artificial neural networks (electronic tongue) for quantitative analysis of multicomponent aqueous solutions. Russ J Appl Chem. 1996;69:848–853. [Google Scholar]
- 179.Vlasov Y., Legin A., Rudnitskaya A., Natale C.D., D'Amico A. Nonspecific sensor arrays ("electronic tongue") for chemical analysis of liquids (IUPAC technical report) Pure Appl Chem. 2005;77:1965–1983. [Google Scholar]
- 180.Łaba´nska M., Ciosek-Skibi´nska P., Wróblewski W. Critical evaluation of laboratory potentiometric electronic tongues for pharmaceutical analysis-an overview. Sensors. 2019;19:5376. doi: 10.3390/s19245376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Woertz K., Tissen C., Kleinebudde P., Breitkreutz J. Taste sensing systems (electronic tongues) for pharmaceutical applications. Int J Pharm. 2011;417:256–271. doi: 10.1016/j.ijpharm.2010.11.028. [DOI] [PubMed] [Google Scholar]
- 182.Wasilewski T., Migon D., Ge˛bicki J., Kamysz W. Critical review of electronic nose and tongue instruments prospects in pharmaceutical analysis. Anal Chim Acta. 2019;1077:14–29. doi: 10.1016/j.aca.2019.05.024. [DOI] [PubMed] [Google Scholar]
- 183.Guhmann M., Preis M., Gerber F., Pöllinger N., Breitkreutz J., Weitschies W. Design, development and in-vitro evaluation of diclofenac taste-masked orodispersible tablet formulations. Drug Dev Ind Pharm. 2015;41:540–551. doi: 10.3109/03639045.2014.884122. [DOI] [PubMed] [Google Scholar]
- 184.Zidan A.S., Aljaeid B.M., Mokhtar M., Shehata T.M. Taste-masked orodispersible tablets of cyclosporine self-nanoemulsion lyophilized with dry silica. Pharmaceut Dev Technol. 2015;20:652–661. doi: 10.3109/10837450.2014.908307. [DOI] [PubMed] [Google Scholar]
- 185.Amelian A., Wasilewska K., Wesoły M., Ciosek-Skibińska P., Winnicka K. Taste-masking assessment of orally disintegrating tablets and lyophilisates with cetirizine dihydrochloride microparticles. Saudi Pharmaceut J. 2017;25:1144–1150. doi: 10.1016/j.jsps.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Guhmann M., Preis M., Gerber F., Pöllinger N., Breitkreutz J., Weitschies W. Development of oral taste masked diclofenac formulations using a taste sensing system. Int J Pharm. 2012;438:81–90. doi: 10.1016/j.ijpharm.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 187.Tokuyama E., Matsunaga C., Yoshida K., Mifsud J.C., Irie T., Yoshida M., et al. Famotidine orally disintegrating tablets: bitterness comparison of original and generic products. Chem Pharm Bull (Tokyo) 2009;57:382–387. doi: 10.1248/cpb.57.382. [DOI] [PubMed] [Google Scholar]
- 188.Ahmad M.N., Ismail Z., Chew O.S., Islam A.S., Shakaff A.Y.M. Development of multichannel artificial lipid-polymer membrane sensor for phytomedicine application. Sensors. 2006;6:1333–1344. [Google Scholar]
- 189.Woertz K., Tissen C., Kleinebudde P., Breitkreutz J. A comparative study on two electronic tongues for pharmaceutical formulation development. J Pharm Biomed Anal. 2011;55:272–281. doi: 10.1016/j.jpba.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 190.Douroumis M.M.D. An in-vitro-in-vivo taste assessment of bitter drug: comparative electronic tongues study. J Pharm Pharmacol. 2015;67:43–55. doi: 10.1111/jphp.12319. [DOI] [PubMed] [Google Scholar]
- 191.Pein M., Gondongwe X.D., Habara M., Winzenburg G. Interlaboratory testing of Insent e-tongues. Int J Pharm. 2014;469:228–237. doi: 10.1016/j.ijpharm.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 192.Pein M., Kirsanov D., Ciosek P., Valle Md, Yaroshenko I., Wesoły M., et al. Independent comparison study of six different electronic tongues applied for pharmaceutical analysis. J Pharm Biomed Anal. 2015;114:321–329. doi: 10.1016/j.jpba.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 193.Alam J., Cook J.L. Reporter genes application to the study of mammalian gene transcription. Anal Biochem. 1990;188:245–254. doi: 10.1016/0003-2697(90)90601-5. [DOI] [PubMed] [Google Scholar]
- 194.Naylor L.H. Reporter gene technology: the future looks bright. Biochem Pharmacol. 1999;58:749–757. doi: 10.1016/s0006-2952(99)00096-9. [DOI] [PubMed] [Google Scholar]
- 195.Guan F., Lam W., Hu R., Kim Y.K., Han H., Cheng Y.C. Majority of Chinese medicine herb category "Qing Re Yao" have multiple mechanisms of anti-inflammatory activity. Sci Rep. 2018;8:7416. doi: 10.1038/s41598-018-25813-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lam W., Ren Y., Guan F., Jiang Z., Cheng W., Xu C.H., et al. Mechanism based quality control (MBQC) of herbal products: a case study YIV-906 (PHY906) Front Pharmacol. 2018;9:1324. doi: 10.3389/fphar.2018.01324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chao J., Dai Y., Cheng H.Y., Lam W., Cheng Y.C., Li K., et al. Improving the concentrations of the active components in the herbal tea ingredient, Uraria crinita: the effect of post-harvest oven-drying processing. Sci Rep. 2017;7 doi: 10.1038/srep38763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dai Y., Lia Q., Tong J., Verpoorte R., Zhao S.J., Qin X.M., et al. Quality marker identification based on standard decoction of differently processed materials of Ephedrae Herba. J Ethnopharmacol. 2019;237:47–54. doi: 10.1016/j.jep.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 199.Dai Y., Jin R., Verpoorte R., Lam W., Cheng Y.C., Xiao Y., et al. Natural deep eutectic characteristics of honey improve the bioactivity and safety of traditional medicines. J Ethnopharmacol. 2020;250 doi: 10.1016/j.jep.2019.112460. [DOI] [PubMed] [Google Scholar]
- 200.Ai Y., Zhang F., Wang C., Xie R., Liang Q. Recent progress in lab-on-a-chip for pharmaceutical analysis and pharmacological/toxicological test. Trends Anal Chem. 2019;117:215–230. [Google Scholar]
- 201.Nigon V.M., Félix M.A. History of research on C. elegans and other free-living nematodes as model organisms. Worm. 2017;2017:1–84. doi: 10.1895/wormbook.1.181.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Blount Z.D. The unexhausted potential of E. coli. Elife. 2015;2015 doi: 10.7554/eLife.05826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Subramanian S. Zebrafish as a model organism–can a fish mimic human? J Basic Clin Physiol Pharmacol. 2021 doi: 10.1515/jbcpp-2021-0113. Available from: [DOI] [PubMed] [Google Scholar]
- 204.Howe KClark MDTorroja CFTorrance JBerthelot CMuffato M., et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Weyand A.C., Shavit J.A. Zebrafish as a model system for the study of hemostasis and thrombosis. Curr Opin Hematol. 2014;21:418–422. doi: 10.1097/MOH.0000000000000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Genschow E., Spielmann H., Scholz G., Seiler A., Brown N., Piersma A., et al. The ECVAM international validation study on in vitro embryotoxicity tests: results of the definitive phase and evaluation of prediction models. Altern Lab Anim. 2002;30:151–176. doi: 10.1177/026119290203000204. [DOI] [PubMed] [Google Scholar]
- 207.Eimon P.M., Rubinstein A.L. The use of in vivo zebrafish assays in drug toxicity screening. Expet Opin Drug Metabol Toxicol. 2009;5:393–401. doi: 10.1517/17425250902882128. [DOI] [PubMed] [Google Scholar]
- 208.Ma X., Chen Y., Jiang S., Zhao X. A bioassay-based approach for the batch-to-batch consistency evaluation of Xuesaitong injection on a Zebrafish Thrombosis model. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.623533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Qi Y., Zhao X., Liu H., Wang Y., Zhao C., Zhao T., et al. Identification of a quality marker (Q-marker) of Danhong injection by the zebrafish thrombosis model. Molecules. 2017;22:1443. doi: 10.3390/molecules22091443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Gao P., Li S., Liu K., Sun C., Song S., Li L. Antiplatelet aggregation and antithrombotic benefits of terpenes and flavones from hawthorn leaf extract isolated using the activity-guided method. Food Funct. 2019;10:859–866. doi: 10.1039/c8fo01862f. [DOI] [PubMed] [Google Scholar]
- 211.Cuong V.T., Chen W., Yang P., Fei J. Establishment of quality evaluation indicator system for Ganoderma lucidum by enhancing the antioxidant capacity of Caenorhabditis elegans. Chin J Cell Biol. 2016;38:499–509. [Google Scholar]
- 212.Yang C.G., Xu Z.R., Wang J.H. Manipulation of droplets in microfluidic systems. Trends Anal Chem. 2010;29:141–157. [Google Scholar]
- 213.Yin B., Qian C., Wang S., Wan X., Zhou T. A microfluidic chip-based MRS immunosensor for biomarker detection via enzyme-mediated nanoparticle assembly. Front Chem. 2021;9 doi: 10.3389/fchem.2021.688442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Deshpande S., Dekker C. On-chip microfluidic production of cell-sized liposomes. Nat Protoc. 2018;13:856–874. doi: 10.1038/nprot.2017.160. [DOI] [PubMed] [Google Scholar]
- 215.Yu F., Choudhury D. Microfluidic bioprinting for organ-on-a-chip models. Drug Discov Today. 2019;24:1248–1257. doi: 10.1016/j.drudis.2019.03.025. [DOI] [PubMed] [Google Scholar]
- 216.Ho N.T., Desai D., Zaman M.H. Rapid and specific drug quality testing assay for artemisinin and its derivatives using a luminescent reaction and novel microfluidic technology. Am J Trop Med Hyg. 2015;92:24–30. doi: 10.4269/ajtmh.14-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sezgin S., Hassan R., Zühlke S., Kuepfer L., Hengstler J.G., Spiteller M., et al. Spatio-temporal visualization of the distribution of acetaminophen as well as its metabolites and adducts in mouse livers by MALDI MSI. Arch Toxicol. 2018;92:2963–2977. doi: 10.1007/s00204-018-2271-3. [DOI] [PubMed] [Google Scholar]
- 218.Herland A., Maoz B.M., Das D., Somayaji M.R., Prantil-Baun R., Novak R., et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng. 2020;4:421–436. doi: 10.1038/s41551-019-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Chen S., Song J., Sun C., Xu J., Zhu Y., Verpoorte R., et al. Herbal genomics: examining the biology of traditional medicines. Science. 2015;347:S27–S29. [Google Scholar]