Abstract
Objective
This research aims to develop a mathematical model that relates the structural features of noscapine with anti-tumor activity, to explains the mode of binding between noscapine compounds and the target receptor tubulin by docking analysis. By considering the results of docking analysis and predictions of pharmacokinetic properties/drug likeness, we designed novel noscapine compounds as anti-tumor agents against pancreatic cancer.
Methods
We used an in silico quantitative structure–activity relationship (QSAR) approach, molecular docking analysis and online tools for pharmacokinetics and drug likeness prediction to develop novel compounds.
Results
A QSAR model with good validations parameters and quality of fit (R2 = 0.9731, Q2CV = 0.9434, R2adj = 0.9647 and R2test set = 0.8343) was built utilizing 70% of the dataset as a training set and the remaining 30% as an external validation to ascertain its predictive capability. Three novel compounds were designed: D3, D4 and D6 with binding scores of −11.2, −10.2 and 10.6 kcal/mol, respectively, exhibiting high affinity towards the tubulin receptor than the template (parent compound) and the co-crystallized ligand (E∗) with a binding score of 9.2 kcal/mol.
Conclusion
The QSAR approach and molecular docking analysis is an important approach for modern drug discovery. Pharmacokinetics studies of the selected novel compounds revealed good drug properties and can be used as candidate compounds for the development of anti-tumor agents for pancreatic cancer.
Keywords: Anti-tumor, Bioisosterism, Docking, Model, Pharmacokinetics, Tubulin
الملخص
أهداف البحث
يسعى هذا البحث إلى تطوير نموذج رياضي يربط بين ميزات نوسكابين الهيكلية ونشاطه المضاد للأورام، ويشرح طريقة الارتباط بين مركبات نوسكابين والمستقبل المستهدف (توبولين) من خلال عملية الإرساء، وتصميم مركبات نوسكابين جديدة بناء على المعرفة والمعلومات التي تم الحصول عليها من النموذج وعملية الإرساء، والتنبؤ بخصائص الحرائك الدوائية/التشابه الدوائي لهذه المركبات الجديدة كعامل مضاد للورم ضد ورم البنكرياس.
طرق البحث
تم استخدام نهج علاقة البنية-الفعالية الكمية، وعملية الإرساء الجزيئي، وأدوات قائمة على الإنترنت للتنبؤ بالحرائك الدوائية وتشابه الأدوية للمركبات الجديدة.
النتائج
تم بناء نموذج "علاقة البنية-الفعالية الكمية" مع معلمات تحقق جيدة وجودة ملاءمة باستخدام ٧٠٪ من البيانات كمجموعة تدريب واستخدمت النسبة المتبقية ٣٠٪ في التحقق من صحة النموذج خارجيًا للتأكد من قدرته التنبؤية كمجموعة اختبار. أظهرت ثلاثة مركبات جديدة (مصممة) دي٣ و دي٤ و دي٦ بدرجة ربط أفضل تبلغ −١١.٢ و −١٠.٢ و ١٠.٦ كيلوكالوري/مول على التوالي تقاربا أعلى تجاه مستقبلات توبولين مقارنة بالقالب (المركب الأصلي) و اللجين المتبلور المشترك بدرجة ربط ٩.٢.
الاستنتاجات
يعتبر نهج علاقة البنية-الفعالية الكمية وعملية الإرساء الجزيئي نهجا مهما في البحث عن الأدوية الحديثة، والذي تم استخدامه في هذا العمل. أظهرت دراسات الحرائك الدوائية للمركبات الجديدة المختارة خصائص دوائية أفضل ويمكن استخدامها كمركب ناجح في البحث عن الأدوية المضادة للأورام وتحديدا ورم البنكرياس.
الكلمات المفتاحية: مضاد الورم, الخواص الحيوية, الإرساء الجزيئي, نموذج, الحرائك الدوائية, توبولين
Introduction
Cancer has been become the most common cause of death globally. Pancreatic cancer has received significant attention from many medicinal chemists and pharmaceutical companies because of its devastating effect, impact and high mortality rate. Despite efforts to identify, diagnose and treat pancreatic cancer, the disease remains a significant global challenge. The difficulties in managing and treating this cancer type because of the late manifestation of its symptoms, relapse and reoccurrence. These factors render most current chemotherapy approaches ineffective32
A recent study described the proper coordination of synthetic and mitotic phases involved in the cell division cycle that is responsible for the duplication and separation of duplicate chromosomal DNA via the checkpoint pathways in the microtubules in a cell; the microtubules represent polymers of tubulin. Malignancy arises due to dysregulation in any of these checkpoints.10
The inability to identify appropriate new drugs and poor success rates in clinical trials has created significant challenges for drug discovery and design processes. Despite significant cost and efforts, the majority of drugs evaluated in clinical trials did not reach the market due to poor pharmacokinetics parameters or unacceptable side effects. In addition to the potency of a drug, its pharmacokinetics and toxicity properties also dictate the success and effectiveness of a drug in clinical trials.35
The recent development of quantitative structure-activity relationship (QSAR) analysis provides us with a cost-effective, easy and powerful tool for drug searches. QSAR is based on the principle that compounds with similar structures may show similar activities.20 Being a ligand-based drug design (LBDD) method, QSAR relates molecular properties (encoded as descriptors) to the qualitative activities of compounds using mathematical equations. The model reliably interprets the contribution of some structural features of a compound within a given dataset and can also predict the potency of a chemical compound.37
The optimal binding of a new drug to a therapeutic target needs to be established and ascertained to ensure that it can get to the target site at a reasonable concentration, exert physiological effects and be eliminated in a reasonable timeframe. The binding of a drug to its target can be studied by docking analysis. Information obtained from docking analysis can help to avoid late failure during the drug development process.23 Docking refers to the computational determination of binding affinity between molecules (the protein structure and the ligand) and attempts to identify the best match between the two molecules.40 Given the two selected molecules, docking analysis can predict (1) whether the two molecules interact or not, (2) the binding affinity, (3) the three-dimensional structure of the complex, (4) the optimal orientation and conformation of the interacting molecules in space, and (5) the stability of the complex.23
Therefore, balanced interactions between toxicity, pharmacokinetics and potency is important if we are to ensure the success of a drug. It is paramount that we identify suitable parameters relating to the adsorption, distribution, metabolism, excretion and toxicity (ADMET) of drugs during pre-clinical trials.35
ADMET studies, as a crucial aspect of the drug developmental process, is carried out by conducting drug metabolism and pharmacokinetics (DMPK) studies.25 These studies assist help to define the viability of a drug candidate based on information obtained from ADMET properties/data.24
Early ADMET profiling of a drug candidate during preclinical studies can reduce the potential risk during clinical development. As drug development failure results from potency or safety issues, there is a necessity to perform preclinical ADMET characterization of a drug candidate to identify drugs with potency and those with major limitations.4,9
A recent study showed that pancreatic cancer may be treated by combining mitogen activated protein kinase (MEK) inhibitors and hydroxychloroquine to block Kirsten Rat Saromma viral oncogene homolog (KRAS). As mutated KRAS is believed to be involved and be partly responsible for this type of cancer, MEK or ERK inhibitors do not appear to work with the majority of pancreatic cancer patients even after blocking the MEK or ERK pathways; these attempts are often associated with increased autophagy in many patients and a poor response. Other studies have shown a more significant response when this treatment is combined with hydroxychloroquine.6
Over recent years, a new approach for cancer treatment has evolved that involves targeted and personalized therapy; however, this approach is often ineffective despite its high cost. This clearly indicates the need to identify new methods to treat cancer.32
Scientific research has demonstrated the expression of tubulin in tumors in different locations in the human body. Class III β-tubulin (TUB β3) is the most dominant tubulin associated with advanced tumors.38 Protein studies have shown that tubulin plays a vital role in cell behavior and represents a structural unit of the microtubules.
The centrosome can be monitored as an indicator for cancer progression; this structure is a dynamic element that organizes the machinery responsible for cell division. The centrosome is made up of a pair of centrioles from which the spindle and astral microtubules originate. Cancer cells often feature extra centrosomes and chromosomal instability. These numerical and structural changes in the centrosome represent a useful indicator and hallmark for human cancer and chromosomal disorders. Human tissue contains various tubulin isoforms in a specific ratio and in many combinations; this complexity highlights metabolic variation in every tissue. Changes or alterations in the number and ratio of these isoforms may create a mechanism for cancer progression and inversion.
Recent knowledge gained from the study of tubulin and its functions have identified potential treatment options for cancer that could increase the survival rate of cancer patients with diverse tumor localizations.21 The progression of pancreatic cancer is associated with tubulin expression, metastasis and resistance to chemotherapy. Therefore, the inhibition of TUB β3 may limit the growth of pancreatic tumors and reduce the risk of metastasis. Tubulin-targeted therapy combined with immunotherapy appears to be a promising approach. Agents acting as anti-polymerization and anti-depolymerization agents for microtubules have led to increased immune response in the body.13
Therefore, optimization of tubulin-targeted agents, along with early ADMET evaluations and predictions using the quantum computational approach, will assist in the drug development process and allow the selection of good drug candidates. This will benefit the process of drug discovery development by avoiding failure in the late stages and will undoubtedly yield fast, novel and effective chemotherapy options for oncology research and the treatment of cancer patients. Furthermore, noscapine has been shown to eliminate human tumors by binding to tubulins, thus leading to distortion of the microtubule spindle and incapacitate the assembly of chromosomes at the metaphase plate. Noscapine can also arrest cells in mitosis with distorted and fragmented nuclei and with apoptotic morphologies (Ye et al., 1998). Hence, identifying the binding site of noscapine with tubulin and optimizing binding sites to enhance binding affinity is an important area in cancer research. These novel compounds with enhanced affinity to tubulin represent a significant breakthrough in the search for better drug candidates in cancer research.
In this research, we developed anti-tumor compounds with high potency and improved pharmacokinetics properties that will meet the requirements of a lead compound for clinical trials. This was carried out using QSAR studies and docking analysis; we also performed ADMET predictions of the compounds tested to determine anti-tumor activity. Subtle manipulation of some compounds within the dataset led to the design of novel compounds with increased docking scores (a high affinity towards the target receptor) and good ADMET predictions to avoid failure in clinical trials. Noscapine compounds are considered as a viable option for microtubule-targeted agents (MTAs) because of their relatively safe pharmacokinetic profiles, including high gastro-intestinal absorption, blood-brain barrier permeability, central nervous system permeability, total clearance and toxicity.28
Studies have shown that tumor aggressiveness, invasion, metastasis, infinite growth and chemotherapeutic resistance are all associated to the over-expression of tubulin, particularly the beta-isotype (β-isotype).1
Noscapine is an anti-mitotic agent that binds to tubulin and effectively causes cell cycle arrest in the G2-M phase and results in the production of irregular mitotic spindles and adjusts the structure of microtubules to hinder proliferation by altering the dynamics of spindle microtubules in tumors, thus causing cell death.1
Therefore, effectively interrupting and destabilizing the metaphase-anaphase stage in a tumor's cell cycle can prevent growth and metastasis and cause apoptosis. Using noscapine compounds to target tubulin represents a safer approach to eliminate malignant cells and provides a promising approach to treat tumors.
Materials and Methods
QSAR Modelling methodology
Data sourcing
Reddy and colleagues synthesized 30 imidazo[2,1,b]thiazole-coupled natural noscapine derivatives and tested their activities against four different human cancer cell lines by in vitro assays.32
In vitro assays in anti-tumor drug studies incorporates the use of isolated tumor cell(s) in an artificial environment that is entirely different from its natural setting. Such studies seek to identify the short-term response of a tumor to specific drugs and determine primary resistance; however, drugs that acquire specific resistance due to the tumor microenvironment cannot be studied.33 Human immortalized cancer cell lines isolated from a cancer patient do not provide a tumor microenvironment; however, in vivo assays provide a native microenvironment where the tumor can originate and reside. The microenvironment can significantly contribute to the behavior and response of a tumor to a drug. Furthermore, the presence of different populations such as endothelial, inflammatory and stromal cells may lead to acquired resistance or may improve the effect of the drug in terms of suppressing tumor growth, aggressiveness, inversion and metastasis.5 An example of an in vivo model is the human xenograft studies performed by inoculating immortalized human cancer cells or the use of the Genetically Engineered Mouse Model (GEMM).
We used a range of different human cancer cell lines: HeLa (cervical), Mia Paca-2 (pancreatic), SK-N-SH (neuroblastoma) and DU145 (prostate cancer). In this research, we selected only the activities (inhibitory concentration at 50% (IC50)) of noscapine compounds against pancreatic cancer cell lines; these were reported in μM. The activities were then converted/transformed into their corresponding logarithm scale using Equation (1).2
| (1) |
Structure generation and optimization
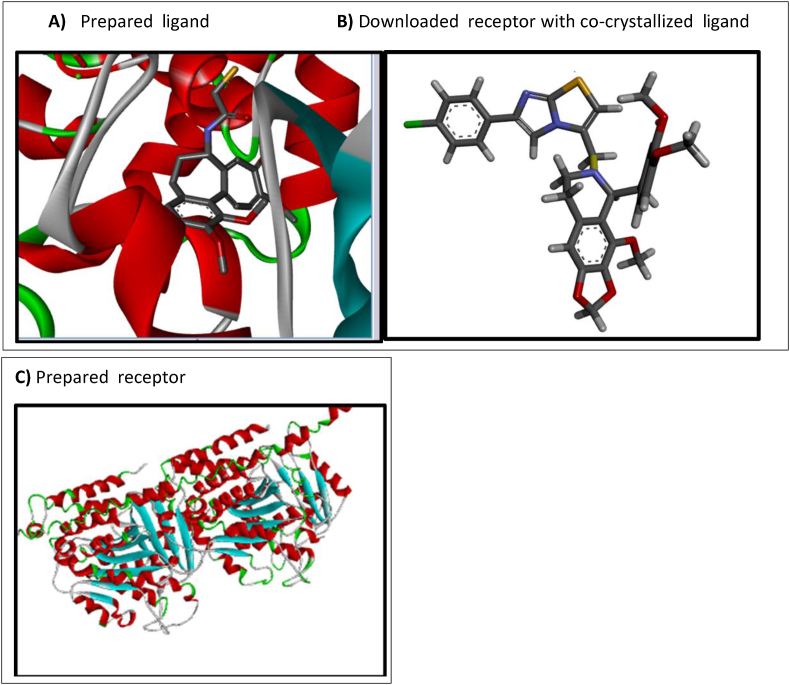
Two-dimensional (2D) structures of the noscapine derivatives were created using Chem draw ultra-software version 12.0. These 2D-structures were converted to three-dimensional (3D) structures by Spartan 14 software and an equilibrium geometry search was carried out.14 This equilibrium geometry structure with minimized energy corresponds to the structure and behaviors of the compound as it exists in nature.3 Identifying the equilibrium structure and minimized energy was performed by Density Functional Theory (DFT) using Becke's three-parameter hybrid function and LYP correlation function with the 6-31G∗ basis set.14 The optimized structures were saved in a protein data bank (pdb) file format. Figure 1A shows the three-dimensional structure of the ligand in pdp file format.15
Figure 1.
Ligand structure, co-crystallized receptor and the prepared receptor.11
Molecular descriptors, data pretreatment and division
Independent variables (molecular descriptors) were computed using the Pharmaceutical Data Exploration Laboratory (PaDEL) descriptors tool kit for all of the calculated structures with minimized energy. The PaDEL tool kit generated different types of descriptors (one-dimensional, two-dimensional and three dimensional).15 Data pretreatment software was use to pretreat the generated data, including the removal of uninformative data, redundancy and constant descriptors.2 The treated data were then divided into a training and test set by application of the Kennard–Stone algorithm, in which 70% constituted the training set and 30% the test set.16
Model generation
The mathematical models were generated by applying multi-linear regression (MLR) analysis on the training set with genetic function approximation techniques using material studio software. Using this approach, the biological activities (pIC50) of the compounds were taken as response variables and the predictors/descriptors were used as independent variables.16
Model validation
For internal validation, the generated models were validated internally with the following statistical tools; coefficient of determination (R2), adjusted coefficient of determination (R2adj), cross validation coefficient (Q2cv) and other validation tools/techniques. The mathematical equations are described in Equation (2).
| (2) |
In Equation (2), Ypred, Yexp and Ym/trn represent the predicted, experimental and mean experimental activities of the training set.
To authenticate the stability of a model apart from the coefficient of correlation R2, the adjusted coefficient of determination R2adj was also computed as shown in Equation (3).
| (3) |
In Equation (3), P represents the number of descriptors in the model and n represents the number of compounds in the training set.
Leave-one-out cross validation Q2cv was used to ascertain the predictive power of the generated models using Equation (4).
| (4) |
In Equation (4), Ypred, Yexp and Ym/trn represent the predicted, experimental and mean experimental activities of the training set.
For external validation, we further validated and ascertained the robustness and predictive power of the coefficient of determination of a model; this was calculated using the test set with Equation (5).
| (5) |
In Equation (5), Ypred, Yexp and Ym/tst represent the predicted, experimental and mean experimental activities of the test set.
Randomization test, mean effect and multi-co-linearity evaluation
Another important validation tool commonly employed to determine the robustness of a QSAR model is the Y-randomization test. This is carried out by adjusting dependent variables with respect to the independent variables which were not adjusted. This test was performed on a training set where the R2 and Q2 generated should be lower than the actual R2 and Q2; this would indicate that the model is reliable, robust and not based on chance correlation. The CR2p was also calculated; this should be greater than or equal to 0.515,27; the formula is shown in Equation (6).
| (6) |
In Equation (6), CR2p represents the coefficient of determination for Y-randomization, R represents the coefficient of correlation for Y-randomization and RR represents the average ‘R’ of random models.
Effective participation and contribution of each descriptor in a given model, also known as the mean effect, was calculated by Equation (7).
| (7) |
In Equation (7), MEj represents the mean effect of the descriptor j, βj represent the coefficient of the descriptor j, dij represents the value of the selected descriptors of each compound and m represents the number of descriptors in a generated model; n represents the number of molecules that made up the training set. The calculated ME value shows the significance and direction of the contribution of each descriptor in a model.2
To identify multi-co-linearity between the descriptors used in a model, we calculated the variance inflation factor (VIF) for each descriptor. This revealed whether these descriptors correlate with another descriptor or not. If the VIF value is equal to 1 then there is no correlation between one descriptor and the other; if it ranges from 1 to 5, there is a chance of accepting the model but if greater than 10, the model has to be rejected because the descriptor is highly correlated with another.15,29 VIF was calculated by Equation (8).
| (8) |
In Equation (8), VIFi represents the variance inflation factor of a descriptor i and R2ij represents the correlation coefficient of a multiple regression between the descriptor i and the rest j in the developed model.
Applicability domain of the generated model
The theoretical region within a chemical space is known as the applicability domain (AD). This is a William's plot delineated by standardized residual value (differences between predicted and observed activities) and the leverage value of the compounds in a given data set. AD identifies if there are outliers or influencer molecules and checks if a new chemical lies within the applicability domain by calculating its leverage value.29 The leverage value hi of a given molecule was calculated by Equation (9).
| (9) |
In Equation (9), Xi represents the training compounds matrix i, X represents the n × k descriptor matrix of the training set compound and XT represents the transpose matrix of X used in generating the model
h∗ is a threshold or warning leverage value determined by Equation (10).
| (10) |
In Equation (10), n and p represent the number of training compounds and descriptors used in a generated model, respectively.
Molecular docking methodology
Identification of active sites in the tubulin receptor
The crystalline structure of the receptor was downloaded from https://www.rcsb.org with the protein data bank (pdb) entry data code 1sa0. The receptor was visualized with discovery studio software and found to be co-crystallized with a ligand molecule (Shown in Figure 1B) that facilitated the identification of active sites based on the ligand–complex interaction.7 The amino acid residues identified as the active sites were SER178, VAL177, GLY146, ALA122, SER140, THR145, ASN206; some of these were involved in conventional hydrogen bonds while other bonds were donor–donor, acceptor–acceptor, pi-sigma, pi–pi stacked and amine pi-stacked.
Molecular docking analysis
The downloaded receptor from RCSB PDB in pdb file format was prepared using Discovery Studio in which residues, such as ligands, water molecules and other traces associated with the receptor protein were removed.22 The ligand (the optimized compound, shown in Figure 1A) in pdb file format and the prepared receptor (Figure 1C) were imported in to PyRx docking software to carry out the docking simulation.19
Design of a novel compounds
The design of new compounds involved enhancing and improving the affinity of noscapine analogs towards the receptor with the aim of increasing drug likeness. Compounds 20 and 30 shown in Figure 5, Figure 6 were chosen because of their higher activities, as evidenced by a PIC50 of 5.2 and 5.4, respectively (as seen in Table 1) against pancreatic tumor cells; these were also within the applicability domain. This was carried out by subtle manipulation of these two compounds, by either addition, deletion or substitution of their substituents based on the knowledge of bioisosterism (a strategy adopted by medicinal chemists for the ration design of new drug candidates applied on a target compound as a special process of molecular modification). This led to the design of six different novel compounds.8,30
Figure 5.

Structure of compound 20 used as a first template for design.
Figure 6.

Structure of compound 30 used as asecond template for design.
Table 1.
Observed and predicted activities, residuals and docking score of 30 noscapine compound against pancreatic (Mia PaCa-2) cancer (see also the Supplementary table).
| S/no. | Observed pIC50 | Predicted pIC50 | Residuals | Docking score (kcal/mol) |
|---|---|---|---|---|
| 1a | 5.37675 | 5.160952 | 0.215799 | −9.5 |
| 2 | 4.245651 | 4.318454 | −0.0728 | −8.1 |
| 3 | 4.272458 | 4.319147 | −0.04669 | −9.9 |
| 4 | 4.36957 | 4.376663 | −0.00709 | −9.5 |
| 5 | 4.328827 | 4.308319 | 0.020508 | −7.7 |
| 6 | 4.300162 | 4.319354 | −0.01919 | −9.2 |
| 7 | 4.272458 | 4.241228 | 0.031231 | −8.8 |
| 8 | 4.260427 | 4.190956 | 0.069472 | −8.9 |
| 9 | 4.301029 | 4.4257 | −0.12467 | −9.2 |
| 10 | 4.313363 | 4.303806 | 0.009558 | −9 |
| 11 | 4.328827 | 4.448648 | −0.11982 | −9.1 |
| 12 | 4.339134 | 4.29763 | 0.041504 | −9.4 |
| 13 | 4.312471 | 4.240756 | 0.071715 | −8.3 |
| 14 | 4.298432 | 4.212189 | 0.086243 | −9.1 |
| 15 | 4.347753 | 4.394798 | −0.04705 | −9.4 |
| 16 | 4.318758 | 4.341063 | −0.0223 | −8.6 |
| 17 | 5.408935 | 5.509189 | −0.10025 | −8.4 |
| 18 | 5.37675 | 5.333544 | 0.043206 | −9.2 |
| 19 | 4.349692 | 4.271134 | 0.078559 | −9.7 |
| 20 | 5.16115 | 5.074242 | 0.086909 | −9.6 |
| 21a | 4.356547 | 4.56621 | −0.20966 | −9.4 |
| 22a | 4.37059 | 4.446317 | −0.07573 | −8.8 |
| 23 | 4.343901 | 4.404393 | −0.06049 | −7.8 |
| 24a | 4.365522 | 4.281083 | 0.084439 | −9.5 |
| 25a | 4.389339 | 4.532786 | −0.14345 | −9.6 |
| 26a | 4.336299 | 4.190657 | 0.145642 | −9.4 |
| 27a | 4.375717 | 4.436243 | −0.06053 | −9 |
| 28 | 4.312471 | 4.306833 | 0.005638 | −8.7 |
| 29a | 4.333482 | 4.369651 | −0.03617 | −9.3 |
| 30 | 5.443697 | 5.367885 | 0.075813 | −9.4 |
Denote test set.
SWISSADME (http://www.swissadme.ch/index.php) a free online web tool, along with pkCSM, an online web server (http://structure.bioc.cam.ac.uk/pkcsm), were used to predict the pharmacokinetics, drug-likeness and medicinal chemistry of these compounds (Antoine et al., 2016).17,18,36
Result and discussion
QSAR model
The quantitative structure–activity relationship (QSAR) model was generated by employing the biological activity as a response or dependent variable and molecular descriptors as the independent variable. This QSAR model quantitatively relates the activity of noscapine to its chemical structure and forms the basis for predicting the biological response of a novel compound that falls within the same chemical domain.
QSAR model generation
The QSAR model was generated based on the Genetic Algorithm–Multiple Linear Regression approach using material studio software. Five different models were generated for the data set, from which the best model was selected based on a high squared correlation coefficient R2, a low residual value and a high coefficient of determination value for the test set, R2test set. Equation (10) shows the selected model. The predicted pIC50 value and the residual values of the data set generated by the selected mathematical model are shown in Table 1.
| pIC50 = 3.117190356∗(MATS8p) + 10.848890534∗(SpMin1_Bhv) − 378.303268598∗(VE2_D) + 0.119179001∗(RDF70p) − 0.015170243∗(RDF75i) − 17.661807511 (10) |
| R2 = 0.9731, Q2CV = 0.9434, R2adj = 0.9647, R2test set = 0.8343 |
The QSAR model generated was robust, reliable and powerfully predictive, as determined by the validation test results indicated below.
| R2 = 0.9731, Q2CV = 0.9434, R2adj = 0.9647, R2test set = 0.8343 |
This made the model good and powerfully predictive as it passed the validation tests shown in Table 3.
Table 3.
Accepted QSAR Validation tool.41
| Validation tools | Interpretation | Acceptable value | Developed model value | Remarks |
|---|---|---|---|---|
| R2 | Co-efficient of determination | ≥0.6 | 0.9731 | Pass |
| Q2CV | Cross validation co-efficient | ˃0.5 | 0.9434 | Pass |
| R2adj | Adjusted co-efficient of determination | ˃0.5 | 0.9647 | Pass |
| R2−Q2cv | Different between R2 and Q2cv | ≤0.03 | 0.0297 | Pass |
| Next/test set | Minimum number of external test set | ≥5 | 8.0000 | Pass |
| R2test set | Co-efficient of determination of external and test set | ≥0.5 | 0.8343 | Pass |
The two-dimensional (2-D) and three-dimensional (3-D) descriptors used in the model were as follows: Moran autocorrelation − lag 8/weighted by polarizabilities (MATS8p) 2D, smallest absolute eigen value of Burden modified matrix − n 1/weighted by relative van der Waals volumes (SpMin1_Bhv) 2D, average coefficient sum of the last eigen vector from detour matrix (VE2_Dt) 2D, radial distribution function − 070/weighted by relative polarizabilities (RDF70p) 3D and radial distribution function − 075/weighted by relative first ionization potential (RDF75i) 3D (Table 2).
Table 2.
Descriptors, definitions and descriptor types.
| Descriptor | Descriptor definition | Descriptor type |
|---|---|---|
| MATS8p | Moran autocorrelation − lag 8/weighted by polarizabilities | 2D |
| SpMin1_Bhv | Smallest absolute eigenvalue of Burden modified matrix − n 1/weighted by relative van der Waals volumes | 2D |
| VE2_Dt | Average coefficient sum of the last eigen vector from detour matrix | 2D |
| RDF70p | Radial distribution function −070/weighted by relative polarizabilities | 3D |
| RDF75i | Radial distribution function −075/weighted by relative first ionization potential | 3D |
The Y- randomization test value shows that the Model was not based on chance correlation. This is because of the lower average randomized values of R2 and Q2 shown in Table 4. Also, the cRp2 value was greater than 0.5.
Table 4.
Y-Randomization test, descriptor variance inflation factor (VIF) and mean effect (M/E).
| Model | R | R2 | Q2 | Descriptor | VIF | M/E |
|---|---|---|---|---|---|---|
| Original | 0.885843 | 0.784718 | 0.565892 | MATS8p | 2.229932 | 0.002544 |
| Random 1 | 0.294292 | 0.086608 | −0.36191 | SpMin1_Bhv | 1.203092 | 0.869475 |
| Random 2 | 0.372131 | 0.138482 | −0.62327 | VE2_DzZ | 3.071484 | −0.03337 |
| Random 3 | 0.590231 | 0.348373 | 0.089259 | RDF70p | 2.453743 | 0.152919 |
| Random 4 | 0.440807 | 0.194311 | −0.16057 | RDF75i | 6.255071 | 0.008427 |
| Random 5 | 0.502612 | 0.252619 | −0.22613 | |||
| Random 6 | 0.536386 | 0.287709 | −0.22521 | |||
| Random 7 | 0.428398 | 0.183525 | −0.75932 | |||
| Random 8 | 0.433775 | 0.188161 | −0.28028 | |||
| Random 9 | 0.449071 | 0.201664 | −0.22311 | |||
| Random 10 | 0.630951 | 0.398099 | −0.21973 | |||
| Random models parameters | ||||||
| Average r: | 0.467865 | |||||
| Average r2: | 0.227955 | |||||
| Average Q2: | −0.29903 | |||||
| cRp2: | 0.66634 | |||||
| cRp2: | 0.66634 | |||||
The descriptors showed no multi-co-linearity and the VIF value in Table 4 was within the acceptable range. This signified that the generated model was acceptable.
The effective contribution of each descriptor, known as the mean effect, is shown in the last column of Table 4. This shows the contribution of each descriptor in predicting the response activity; a negative value means that the descriptor contributes negatively (the presence of the descriptor lowers the activity, while its absence increases the activity). A positive value shows a positive contribution.
The plot of the predicted activities against the observed activities in Figure 2 shows a linear correlation between the predicted and observed activities, thus proving the strength of the model in the response variables. The plot of standardized residuals against observed residuals, shows a distribution for the compound on both sides of zero (Figure 3); this ensured the absence of systematic error in the developed model.
Figure 2.
Plot of predicted activities against observed activities.
Figure 3.
Plot of standardized residuals against observed activities.
The William's plot (Figure 4) identified four influential compounds among the test set; this is due to their high leverage value greater than the threshold (h∗ = 0.82) value. This means that the compounds are outside the applicability domain and should be disregarded when designing novel noscapine compounds.
Figure 4.
Plot of standardized residuals against leverages; an applicability domain/Williams plot.
Docking results
Docking analysis was conducted between novel noscapine compounds and the downloaded tubulin receptor with protein data bank code 1sa0. The results of interactions known as binding scores are shown in Table 5, Table 6, and represent the affinity of a compound to its receptor and can also explain as readiness and strength of the interactions.26 Docking studies revealed strong interactions between all the docked ligands and receptors. We observed a range of bonding interactions, including hydrophobic, electrostatic bonds, amide and pie–pie interactions within the ligand–receptor complex. Re-docking of the co-crystallized ligand with the receptor validated the docking protocol used in this study.
Table 5.
Novel ligands from template 20 and their binding energies.
| Compound | R1 | R2 | R3 | Binding energy (kcal/mol) |
|---|---|---|---|---|
| D1 | CH3 | NH2 | Cl | −9.2 |
| D2 | H3CO | Cl | CH3 | −8.0 |
| D3 | NH2 | CH3 | Cl | −11.2 |
Table 6.
Novel ligands from template 30 with their binding energies.
| Compound | R1 | R2 | R3 | R4 | Binding energy (kcal/mol) |
|---|---|---|---|---|---|
| D4 | H3CO | OCH3 | CH3 | Br | −10.2 |
| D5 | Cl | Cl | CO2H | CH3 | −8.9 |
| D6 | CH3 | NH3 | Cl | NH2 | −10.8 |
| E∗ | C0-crystallized ligand | −9.2 | |||
E∗ indicates the ligand co-crystallizes in the pocket of the downloaded receptor.
D1, D2, and D3 compounds with binding energies of −9.2, −8.0 and −11.2, respectively; those of D4, D5 and D6 compounds were −10.2, −9.8 and −10.8, respectively. This data indicates a lower energy of binding when compared to the co-crystallized ligand E∗ except for D2 and D5. D1 had a binding energy that was equivalent to E∗. With this discovery compounds D3, D4 and D6 were found to be better compounds with activity against pancreatic tumors (Figure 7, Figure 8, Figure 9, Figure 10).
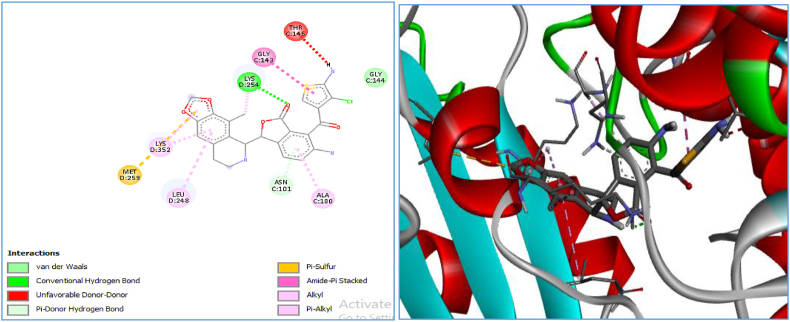
Figure 7.
2-D and 3-D visualization of compound D3 in the active pocket of the receptor.
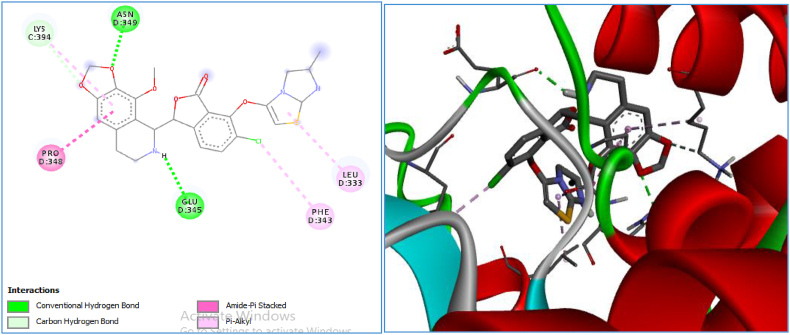
Figure 8.
2-D and 3-D visualization of compound D6 in the active pocket of the receptor.
Figure 9.
2-D and 3-D visualization of compound D4 in the active pocket of the receptor.
Figure 10.
2-D and 3-D visualization of the re-docked co-crystalline in the active pocket of the receptor.
Three compounds (D3, D6 and D4) were selected out of the six newly designed novel compounds by virtue of their high affinity towards the receptor. Table 7 shows the types of interactions and amino acid residues involved in the interactions with the receptor. Carbon hydrogen bonds, conventional hydrogen bonds and van der Waals are the predominant interactions among the designed ligands and are common to those of the co-crystalline ligand, E∗. The higher binding score shown by these novel compounds may be attributed to the presence of van der Waals, pi-donor, amide-pi stacked and pi–alkyl interactions.
Table 7.
Binding scores, types of interactions and amino acid residues associated with the novel compounds with that of the co-crystalline ligand.
| Compound | Binding score (kcal/mol) | Type of interactions | Amino acids residues involved |
|---|---|---|---|
| D3 | −11.2 | Van der Waal, conventional hydrogen bond, unfavorable donor–donor, Pi–donor hydrogen bond, Pi–sulfur, amide–Pi stacked alkyl, and Pi–alkyl | MET 259, LYS352, LEU 248, ASN 101, ALA 180, GLY 143 and THR 145 |
| D6 | −10.8 | Van der Waal, conventional hydrogen bond, carbon hydrogen bond, amide–Pi stacked and Pi–alkyl | LYS 394, ASN 349, PRO348, GLU34, PHE343, and LEU 333 |
| D4 | −10.2 | Van der Waal, carbon hydrogen bond, conventional hydrogen bond, Pi–donor hydrogen bond, amide–Pi stacked, alkyl and Pi–alkyl | LEU 255, LYS 254, GLN 11, ALA 12, GLY 10, SER 178, GLU 183, ALA 180, ASN 101 and LYS 352 |
| Ea | −9.2 | Salt bridge, attractive charge, conventional hydrogen and carbon–hydrogen bond | ASN 206, TYR 224, SER 178, GLY,142, GLY 143, GLY 146, THR 145, ALA 12, SER 140, GLY 144 and LYS 254 |
Co-crystallized ligand downloaded with the receptor.
The different amino acid residues that are involved in these interactions were LYS, ASN, LEU, GLY and SER, as indicated in Table 7. The D3 compound predicts a higher binding score; this may be attributed to the presence of the THR145 residue that added an unfavorable donor–donor interaction and MET259 acid residues that formed pi–sulfur interactions; these interactions were absent in the other ligand–receptor interactions.
Pharmacokinetics and ADMET predictions of some selected novel noscapine compounds are shown in the first column of Table 8. Compounds D3, D4, D6 showed high human intestinal absorption values of 94.799, 91.997 and 95.957, respectively. The gastro-intestinal solubility (over 90%) of these compounds determine the handling and formulation of the drug and also ensure delivery of the target. Therefore, these novel noscapine compounds have absorbance values between 91.997 and 95.957%; consequently, these values passed the minimum recommended values of 30%, thus indicating good human intestinal absorption. Blood–brain barrier (BBB) permeability (log BB) was predicted to be −1.066–1.391 and −1.406, respectively; while central nervous system permeability (log PS) was predicted to be −2.433, −3.183 and −3.192, respectively, as shown in the second and third columns of Table 8. The recommended limits for blood–brain barrier (BBB) and central nervous system permeability are >0.3 to <−1 Log BB and >−2 to <−3 Log PS, respectively (Carpenter & Krishner, 2014).15 For these selected novel noscapine compounds, the Log BB was >−1, thus implying that the compounds are better distributed to the brain with a Log PS >−2; thus, these compounds are predicted to better penetration of the central nervous system. Cytochrome P450 (CYP450) is an isoenzyme that is involved in the metabolism of drugs and the biotransformation of drugs in the body. Several isoenzymes play a major role in drug metabolism (1A2, 2C9, 2C19, 2D6 and 3A4), as indicated in Table 8. Members of the 3A4 family was found to act as both a substrate and an inhibitor of the selected novel noscapine compounds; these represent an important family with regards to the metabolism of the noscapine drug candidates. Total clearance was used use to show the relationship between the rate of elimination of a drug and its concentration in the body. The selected compounds showed total clearance values within the permissible limit of a drug molecule in the body. Two of the designed compounds (D4 and D6) were found to be non-toxic while D3 was found to be mildly toxic because it inhibited C II (ether-a-go-go related gene), inhibitor II but did not inhibit type I. Both D4 and D6 are non-toxic and do not act as an inhibitor of hERG I and II. hERG (ether-a-go-go related gene) is an anti-target; blockade by a drug candidate leads to QT prolongation (thus delaying ventricular repolarization) and cardio-toxicity12 The ADMET properties of these compounds revealed their good pharmacokinetic profiles in most of the predictions.
Table 8.
Adsorption, distribution, metabolism, excretion and toxicity of the novel compounds.
| Absorption | Distribution | Metabolism |
Excretion | Toxicity | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Substrate inhibitors | ||||||||||||
| CYP | ||||||||||||
| Compound | Intestinal absorption (human) | BBB permeability | CNS perm. | 2D6 | 3A4 | 1A2 | 2C19 | 2C9 | 2D6 | 3A4 | Total | AMES |
| (log BB) | (log PS) | Clearance | Toxicity | |||||||||
| D3 | 94.799 | −1.066 | −2.433 | No | Yes | No | Yes | No | No | Yes | 0.69 | YES |
| D4 | 91.997 | −1.391 | −3.183 | No | Yes | No | No | No | No | Yes | 0.72 | No |
| D6 | 95.957 | −1.406 | −3.192 | No | Yes | No | No | Yes | No | Yes | 1.195 | No |
The selected novel noscapine compounds respect Lipinski's rule of five with only one violation; the molecular weight of all three compounds were greater than 500 kDa but were still within the permissible limit for drug molecules to be orally bioavailable, as shown in Table 9. The remaining rules (Number of hydrogen bond donors ≤ 5, Number of hydrogen bond acceptors ≤ 10, and Calculated Log p ≤ 5) were all respected. All of the selected novel compounds (D3, D4 and D6) had a good bioavailability score of 0.55, thus confirming the drug-likeness properties of these selected and reported compounds, shown in Table 9. As such, these compounds are orally bioavailable with good pharmacokinetic properties and represent lead compounds for developing anti-tumor drugs. Lead drugs need to be small and less hydrophobic so that they can be optimized further to become drug candidates31; minor manipulations of our newly designed compounds could lead to promising noscapine based anti-tumor drugs.
Table 9.
Molecular weight (MW), number of H-bond acceptors, number of H-bond donors, MLOGP, number of Lipinski violations, bioavailability scores and drug-likeness.
| Molecule | MW | No. of H-bond acceptors | No. of H-bond donors | MLOGP | No. of Lipinski's violations | Bioavailability score | Drug-likeness |
|---|---|---|---|---|---|---|---|
| D3 | 512.975 | 7 | 2 | 2.5 | 1 | 0.55 | Yes |
| D4 | 527.986 | 8 | 1 | 2.98 | 1 | 0.55 | Yes |
| D6 | 572 | 8 | 1 | 2.57 | 1 | 0.55 | Yes |
Conclusion
Quantitative structure–activity relationship (QSAR) and molecular docking studies were carried out on 30 noscapine analogs as potential anti-tumor agents for pancreatic cancer. We also developed predictive and robust models. Based on our results, novel noscapine compounds were designed; we studied the pharmacokinetics of these drugs and performed docking analysis to determine potency against tumor cells (pancreatic cancer). In this study, we targeted the tubulin receptor as it is believed to be associated with different types of cancer in humans. Noscapine binds to tubulin and causes arrest at the G2-M checkpoint in the cell cycle (the mitotic checkpoint), thus preventing cell growth, delaying division and inducing apoptosis. These newly designed novel compounds exhibit improved potency, safe pharmacokinetics profiles and low binding scores to tubulin, thus confirming that noscapine might be used as lead candidate for developing anti-tumor drugs against pancreatic cancer.
Source of funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
The section is not applicable to this research work.
Authors contributions
BN contributed throughout the research work. AU provided directives and technical advice. ITB, MTI and ABU assisted with technical activities. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
Acknowledgment
We would like to thank Assistant Professor Yunus M. Marhe, Dr. Saminu Dagari, M I Shago and Auwal Salisu Isa for their support and encouragement during the course of this research work.
Footnotes
Peer review under responsibility of Taibah University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jtumed.2022.07.013.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Albahde H.A.M., Abdrakhimov B., Li G., Zhou X., Zhou D., Xu H., Qian H., Wang W. The role of microtubules in Pancreatics cancer: therapeutic progress. Front Oncol. 2021;11(640863):1–14. doi: 10.3389/fonc.2021.640863. Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdullahi M., Uzairu A., Shallangwa G.A., Arthur D.E., Umar B.A., Ibrahim M.T. Virtual molecular docking study of some novel carboxamide series as new anti-tubercular agents. Eur J Chem. 2020;11:30–36. [Google Scholar]
- 3.Baldi A. Computational approaches for drug design and discovery: an overview. Sys Rev Pharm. 2010;1(1):99–105. doi: 10.4103/0975-8453.59519. [DOI] [Google Scholar]
- 4.BioPharma . Thermofisher Scientific; 2020. The role of of ADME and toxicology studies in drug discovery and development. [Google Scholar]
- 5.Chen L.X., Ni X.L., Zhang H., Wu M., Liu J., Xu S., Yang L.L., Fu Z.S., Wu J. Preparation, characterisation, in vitro and in vivo antitumour effect of thalidomide nanoparticles on lung cancer. Int J Nanomed. 2018;8(3):2463–2476. doi: 10.2147/IJN.S159327. Dove press. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien S. University of Texas MD Anderson Centre; 2021. Targeting KRAS mutation for effective cancer treatment. [Google Scholar]
- 7.Cui W., Aouidate A., Wang S. Yu Q., Li Y., Yuan S. Discovering anti-cancer drugs via computational methods. Front Pharmacol. 2020;11:733. doi: 10.3389/fpar.2020.00733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuozzo A., Daina A., Perez M.A.S., Michielin O., Zoete V. SwissBioisostere 2021: updated structural, bioactivity and physicochemical data delivered by a reshaped web interface. Nucleic Acids Res. 2021;50:D1382–D1390. doi: 10.1093/nar/gkab1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2016;7 doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong A., Durkin A., Lee H. The potential of combining tubulin-targeting anticancer therapeutics and immune therapy. Int J Mol Sci. 2019;20(3) doi: 10.3390/ijms20030586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadni H., Elhallaoui M. 3D-QSAR, docking and ADMET properties of aurone analogues as antimalarial agents. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e03580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanser T., Steinmetz F.P., Plante J., Rippman F., Krier M. Avoiding hERG-liability in drug design via synergetic combination of different QSAR methodologies and data sources: a case study in an industrial setting. J Cheminformatic. 2019;11:9. doi: 10.1186/s13321-019-0334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heinemann S. Microtubules, Leukemia and cough syrup. Blood. 2006;107:22216–22217. [Google Scholar]
- 14.Ibrahim M.T., Uzairu A., Shallangwa G.A., Uba S. Computer-aided molecular modeling studies of some 2, 3-dihydro-[1, 4] dioxino [2, 3-f] quinazoline derivatives as EGFR WT inhibitors Beni-Suef University. J Basic Appl Sci. 2020;9:1–10. [Google Scholar]
- 15.Ibrahim M.T., Uzairu A., Uba S., Shallangwa G.A. Quantitative structure-activity relationship, molecular docking, drug-likeness, and pharmacokinetic studies of some non-small cell lung cancer therapeutic agents. Beni-Suef Univ J Basic Appl Sci. 2020;9:1–14. [Google Scholar]
- 16.Ibrahim M.T., Uzairu A., Shallangwa G.A., Uba S. Structure-based design and activity modeling of novel epidermal growth factor receptor kinase inhibitors; an in silico approach. Sci Afr. 2020 [Google Scholar]
- 17.Ibrahim M.T., Uzairu A., Shallangwa G.A., Uba S. Structure-based design of some quinazoline derivatives as epidermal growth factor receptor inhibitors Egyptian. J Med Hum Genet. 2020;21:1–12. [Google Scholar]
- 18.Ismail Y.S., Uzairu A. In silico QSAR and molecular docking studies of sulfur containing shikonin oxime derivatives as anti-cancer agent for colon cancer. Rad Inf Dis. 2019;6(2019):108–121. [Google Scholar]
- 19.Isyaku Y., Uzairu A., Uba s., Ibrahim M.T., Umar A. QSAR, molecular docking, and design of novel 4-(N,N-diarylmethyl amines) Furan2(5H)-one derivatives as insecticides against Aphis craccivora. Bull Natl Res Cent. 2020 doi: 10.1186/s42269-020-00297-w. [DOI] [Google Scholar]
- 20.Johnson M.A., Maggiora G.M. Wiley; New York: 1990. Concepts and applications of molecular similarity. [Google Scholar]
- 21.Kamal A., Balakrishna M., Nyak V.L., Shaik T.B., Faazil S., Nimbarte V.D. Design and synthesis of imidazo [2,1-b] thiazole chalcone conjugates: microtubule destabilizing agents. ChemMedChem. 2014:276–2780. doi: 10.1002/cmdc.201402310. [DOI] [PubMed] [Google Scholar]
- 22.Kausar S., Falcao O.A. An automated framework for QSAR model building. J Cheminf. 2018;10(1) doi: 10.1186/s13321-017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavallaris M. Microtubules and resistance to tubulin-binding agents. Nat Rev Cancer. 2010;10(3):194–204. doi: 10.1038/nrc2803. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy T. Managing the drug discovery/development. J Drug Discov. 1997;2(10):436–444. doi: 10.1016/s1359644(97)01099-4. [DOI] [Google Scholar]
- 25.Madison E, Ogilvie B. What is ADME and how does it fit into drug development? Test system and method, 2020.
- 26.Mahmud A.W., Shallangwa G.A., Uzairu A. QSAR and molecular docking studies of 1,3-dioxoisoindoline-4-aminoquinolines as Potent antiplasmodium hybrid compounds. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e03449. Elsevier Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhammad U., Uzairu A., Authur E.D. Review on: quantitative structure activity relationship (QSAR) modelling. J Anal Pharm Res. 2018 MedCrave. Mini review. [Google Scholar]
- 28.Oliva A.M., Prosta E.A., Rodriguesz-Salarichs J., Benanai I.y., Jeminenez-Barbero J., Bargsten K., Canales A., Steinmetz O.M., Diaz F.T. Structural basis of noscapine activation for tubulin binding. J Med Chem. 2020;63:8495–8501. doi: 10.1021/acs.jmedchem.0c00855. https://dx.doi/10.1021/acs/jmedchem.0c00855 [DOI] [PubMed] [Google Scholar]
- 29.Olasupo S.B., Uzairu A., Shallangwa G., Uba S. QSAR analysis and molecular docking simulation of norepinephrine transporter (NET) inhibitors as anti-Psychotic therapeutic agents. Heliyon. 2019;5:e02640. doi: 10.1016/j.heliyon.2019.e02640. Elsevier research article Heliyon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantami A.G., LaVoie J.E. Bioisosterism: a rational approach in drug design. Chem Rev. 1996;96:3147–3176. doi: 10.1021/cr950066q. [DOI] [PubMed] [Google Scholar]
- 31.Polinski A. 3rd ed. 2008. Practice of medicinal chemistry. (Chapter 12-lead-likeness and drug-likeness). [DOI] [Google Scholar]
- 32.Reddy P.K.N., Krishna Kommalapati Vamsi, Siva Krishna Vagolu, Sriram Dharmarajan, Devi Tangutur Anjana, Kantevari Srinivas. Imidazo[2,1-b]thiazole-coupled natural Noscapine Derivatives as anticancer agents. J ACS Omega. 2019;4:19382–19398. doi: 10.1021/acsomega.9b02789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosa R., Montaleone F., Zambrano N., Bianco R. In vitro and in vivo models for analysis of resistance to anticancer molecular therapies. Curr Med Chem. 2014;21(14):1596–1606. doi: 10.2174/09298673113209990226. Benthan science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saeed M.A., Abdou M.I., Salem A.A., Ghatter A.M., Attreh N., Alneyadi S.S. Anticancer activity and molecular docking of some pyrano[3,2-c]quinolone analogues. Open J Med Chem. 2020;10:1–14. ISSBN online: 2164-313x. [Google Scholar]
- 36.Sliwoski G., Kothiwale S., Meiler J., Lowe W.E. Computational methods in drug discovery. Pharmacol Rev. 2014;66:334–395. doi: 10.1124/pr.112.007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svetnik V., Liaw A., Tong C., Culberson J.C., Sheridan R.P., Feuston B.P. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 38.Tepp K., Mado K., Varikmaa M., Klepinin A., Timohhina N., Shevchuk I., Chekulayev V., Kuznetsov A.V., Guzun R., Kaambre T. The role of tubulin in the mitochondrial metabolism and arrangement in muscle cells. J Bioenerg Biomembr. 2014;46(5):421–434. doi: 10.1007/s10863-014-9579-3. [DOI] [PubMed] [Google Scholar]
- 40.Thomas C. Al Azhar College of Pharmacy; Thodupuzhar: 2020. Drug design and molecular docking (video Webinar)-Department of Pharmaceutical Sciences, CPAS, Ettunanoor ,Kerala. [Google Scholar]
- 41.Veersamy R.,V., Harish R., Abhishek J., Shalini S., Christapher P.V., Ram K.A. Validation of QSAR models – strategies and importance. Int J Drug Des Discov. 2011;2(3):511–519. doi: 10.1128/AAC.39.4.964. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.