Abstract
Background:
Asthma is the most common chronic condition in children and the third leading cause of hospitalization in pediatrics. The genome-wide association study catalog reports 140 studies with genome wide significance. A polygenic risk score (PRS) with predictive value across ancestries has not been evaluated for this important trait.
Objective:
We aim to train and validate a PRS relying on genetic determinants for asthma to provide predictions for disease occurrence in pediatric cohorts of diverse ancestries.
Methods:
We applied a Bayesian regression framework method using the Trans-National Asthma Genetic Consortium (TAGC) GWAS summary statistics to derive a multiancestral PRS score, used one eMERGE cohort as a Training set, used a second independent eMERGE cohort to Validate the score, and used the UK Biobank data to Replicate the findings. PheWAS was performed using the PRS to identify shared genetic etiology with other phenotypes.
Results:
The multiancestral asthma PRS was associated with asthma in the two pediatric Validation data sets. Overall, the multiancestral asthma PRS has an area under the curve (AUC) of 0.70 (0.69–0.72) in the pediatric Validation 1 and AUC of 0.66 (0.65–0.66) in the pediatric Validation 2 datasets. We found significant discrimination across pediatric sub-cohorts of European (AUC, 0.60 and 0.66), African (AUC, 0.61 and 0.66), admixed American (AUC, 0.64 and 0.70), Southeast Asian (AUC, 0.65), and East Asian (AUC, 0.73) ancestry. Pediatric participants with the top 5% PRS had a 2.80 – 5.82 increased odds of asthma compared to the bottom 5% across the Training, Validation 1, and Validation 2 cohorts when adjusted for ancestry. PheWAS analysis confirmed the strong association of the identified PRS with asthma (odds ratio (OR) 2.71, PFDR =3.71×10−65) and related phenotypes.
Conclusions:
A multiancestral PRS for asthma based on Bayesian posterior genomic effect sizes identifies increased odds of pediatric asthma.
Clinical implication:
This PRS will be used to identify children at increased risk for asthma across a multisite multiancestral prospective intervention cohort study.
Keywords: Genetics, Asthma, GWAS, Polygenic risk score (PRS), PheWAS
Capsule summary:
Clinical tools to identify risk of asthma in pediatric groups exist. In contrast to previously reported PRS for asthma, we present a PRS that performs well across multiancestral groups.
Introduction
Asthma is an inflammatory disease of the airway with symptoms including coughing, wheezing, chest tightness, and shortness of breath caused by airflow obstruction and hyperresponsiveness (1, 2). Asthma affects seven million children across the United States of America, yielding a prevalence equaling 10% of children, but this prevalence can vary by sex, ancestry, and age (1). Patients with asthma experience increased morbidity with respiratory infection, days absent from school and work, emergency department visits, hospitalizations, and even death. While asthma is a significant economic and health burden, proactive treatment and specific interventions can prevent severe disease requiring emergency or inpatient management (3, 4). Given this, a major focus is primary prevention (5). There have been substantial efforts to develop asthma prediction models based on clinical factors (6, 7), but these tools rely on early life phenotypes that may not have been assessed. Thus, development of additional predictive tools are warranted.
The etiology of asthma is multifactorial with contributions from environmental and genetic factors (4, 8, 9). Twin studies estimate a heritability of 60–80% of asthma susceptibility attributable to genetic factors while also highlighting the etiological contributions of shared environment (10). At the time of development of this multiancestral asthma PRS, there were more than 140 studies in the GWAS catalog with 2,167 variants reported with genome wide significant results; 50 of the reported studies resulted from studies of participants not of full European ancestry (11). These GWAS identify genetic loci that increase risk for asthma with a common genetic variant-based narrow sense heritability of 14.9% (12); however, any one risk locus does not provide sufficient risk discrimination to be of practical clinical use. By accounting for genotypes at risk variants in proportion to the effect size of each genetic locus, polygenic risk scores (PRS) have the potential to be tools for incorporating genetic risk into clinical decision support (13).
A PRS for asthma has been developed, however the data used to generate the PRS was limited to individuals of European descent. The limited diversity of participants used to generate the PRS is a problem because different demographic groups may have different underlying genetic etiology. For example, although pediatric and adult-onset asthma have numerous shared risk loci, genetic studies have also identified distinct risk loci in children (14). Further, the risk of asthma is different across ancestral groups, with children of African descent having higher frequencies than children of European descent (15–20), so it is important that for a PRS to be maximally clinically informative, the PRS should be developed and validated using training datasets that are similar to the populations to whom the PRS will be applied. Thus, a PRS developed using primarily adult studies or based on limited ancestral diversity may not be optimal to predict pediatric asthma. To address the current limitations, our goal was to develop a high quality PRS for pediatric asthma using data from a diverse cohort representing multiple ancestries at all stages of the process. To address this goal, we utilized an existing large scale genome wide association derived from diverse populations and a Bayesian framework to derive a multiancestral PRS score and tested and validated this score in multiple cohorts and phenotype definitions. With the development of this pediatric asthma PRS, we are now poised to test the clinical utility.
Our intent is to evaluate the clinical utility of this PRS in a prospective study that will include individuals across ancestral groups. It is routine in clinical settings to ask about an individual’s race and ethnicity. Yet, how an individual self identifies is not equivocal to their genetic ancestral group. Further, many individuals have admixed ancestry. Thus, we developed a multiancestral PRS that could be used for all individuals.
Methods
Study overview
The study design is presented in Figure 1. We divided the combined eMERGE I, II, III imputed data set from over 105,000 samples (welcome to eMerge (emerge-network.org)) into two non-overlapping independent data sets (eMERGE Training dataset and eMERGE “Validation 1” dataset 2) based upon date of participation in eMERGE. The UK Biobank cohort was used as a “Validation 2” dataset. For the Training eMERGE dataset, patients with asthma and controls were identified based on a validated algorithm using structured data (International Classification of Diseases (ICD) version 9 (ICD-9 493.xx) or version 10 (ICD-10 J45, J46) codes)) for asthma as well as unstructured data as previously described (21). Participants in the Validation eMERGE dataset were classified as cases by having two or more ICD-9/10 codes for asthma and as controls by having no history of asthma or atopy. The minor differences in case definitions were based on improvements in clinical data extraction over the past fifteen years of eMERGE. For Validation 2 dataset in the UK Biobank, we used a previously established algorithm to identify participants with asthma, developed by UK Biobank team (Resource ID 4124).
Figure 1. Study Design.
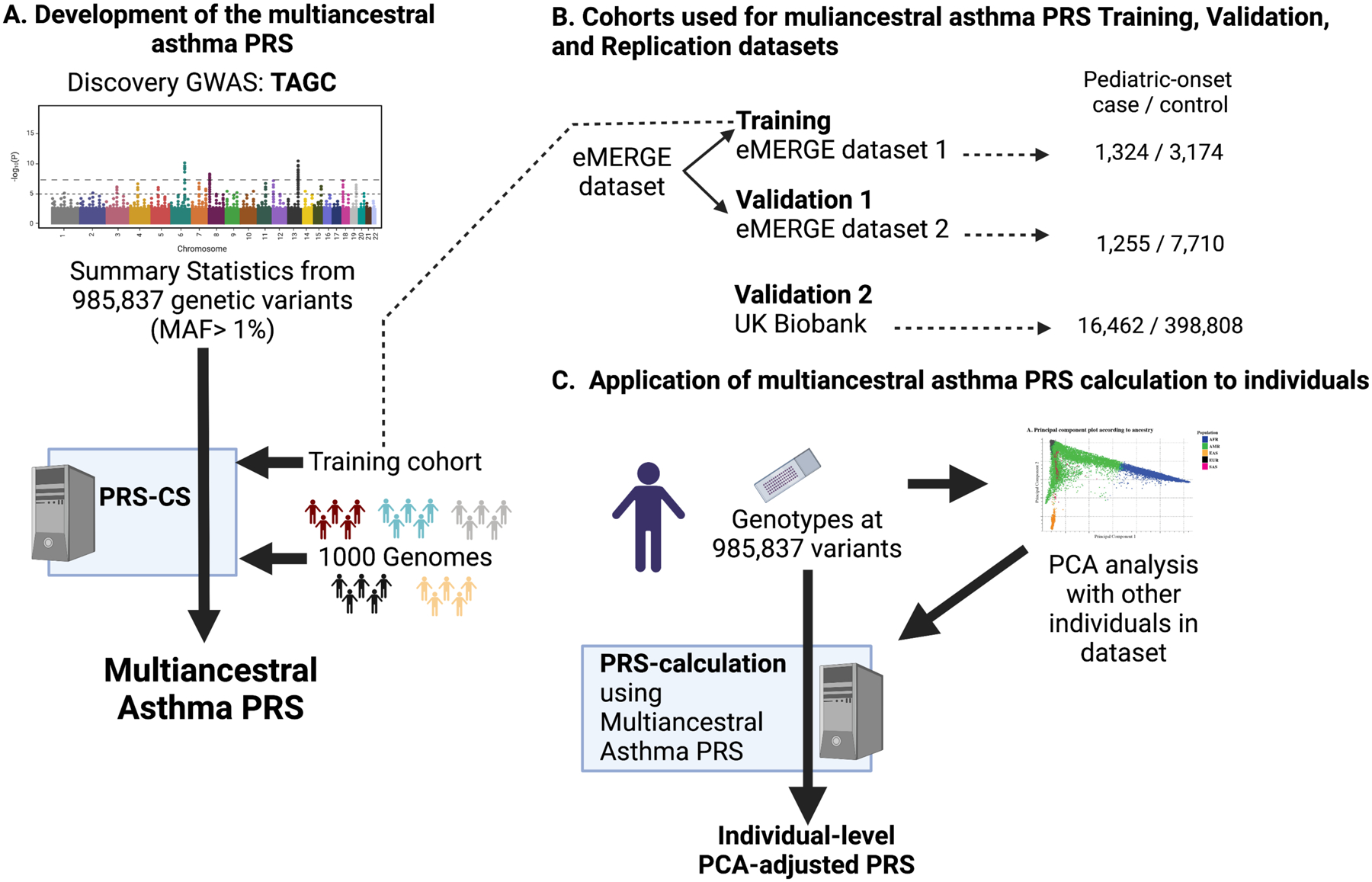
(A) Summary statistics from 985,837 genetic variants with minor allele frequencies (MAF) greater than 1% in the overall (cases and controls combined) Trans-National Asthma Genetic Consortium (TAGC) Discovery genome-wide association studies (GWAS) were used to develop the multiancestral asthma PRS in the context of the linkage disequilibrium of the reference 1000 genomes reference panel and the Training dataset. (B) The eMERGE dataset was split into two independent datasets (Training and Validation 1) and the UK Biobank was used as a Validation 2 dataset. Subjects with confirmed pediatric-onset asthma and controls were used for PRS Training, Validation 1, and Validation 2. C) Individual genotypes from each subject in each dataset was used to perform a principal component analysis (PCA). For each individual, genotypes at each of the 985,837 variants included in the multiancestral asthma PRS were used to calculate a PCA-adjusted PRS. The adjusted multiancestral PRS was applied to each individual in the Training, Validation 1, and Validation 2 cohorts in preparation for it to be similarly applied to individuals recruited into an IRB-approved prospective intervention study. Figure created in BioRender.
Study population of eMERGE and UK Biobank and phenotype ascertainment
The Electronic Medical Records and Genomics (eMERGE) Network is a National Institutes of Health (NIH)-organized and funded consortium of United States medical research institutions (https://www.genome.gov/Funded-Programs-Projects/Electronic-Medical-Records-and-Genomics-Network-eMERGE). Post-imputation whole genome genotyping data for participants from the eMERGE network were made available for this study (dbGAP (phs000888.v1.p1). The associated BAM, xml, and vcf files are available on the eMERGE Commons web portal, accessible to sites as well as outside investigators who apply for access (see eMERGE Network in Web Resources). The imputation process and genotype quality control in eMERGE followed guidelines that have been published previously (22). Briefly, all subjects and variants had missingness less than 2%. Individual level genotype data was derived from 78 array batches across 12 academic medical centers. Each batch was imputed using the Michigan Imputation Server, which provides a missing single-nucleotide variant genotype imputation service using the minimac3 imputation algorithm with the Haplotype Reference Consortium genotype reference set. Only one genetic dataset was retained for each participant (22).
The UK Biobank is a large long-term biobank study in the United Kingdom developed to support the investigation of the respective contributions of genetic predisposition and environmental exposure to the development of diseases (23) including asthma (24, 25). The UK Biobank post imputed data was obtained through application ID: 47377.
All participants in the eMERGE and UK Biobank cohorts provided written informed consent prior to study inclusion. The institutional review board of each contributing institution approved the eMERGE study. The North West Multicenter Research Ethics Committee and Patient Information Advisory Group approved the UK Biobank study. All analyses were conducted using deidentified data.
Prior to analyses, participant level quality control was employed. Self-reported race and ethnicity was not used to identify ancestry. We used genetically-defined ancestry for ancestry-specific analyses. Principal component analysis (PCA) of genome-wide genetic variants was used to establish ancestry (Table 1). The FRAPOSA software package was used to perform PCA and assign all individuals into five major super-populations (European (EUR), African (AFR), Admixed American (AMR), East Asian (EAS) and South Asian (SAS)) (26). We used the Phase 3 release of the 1000 Genomes data as a reference that consists of 2,504 individuals from five super-populations as shown in Supplemental Table 1 (27). The major steps in the FRAPOSA algorithm include computing principal components of the reference dataset using the matched variants only and projecting computed principal components to the target data using an optimized implementation of the Online Augmentation, Decomposition, and Procrustes (OADP) transformation. Next, the algorithm predicts the ancestry membership by using the K(20)-nearest-neighbor method (19). The pairwise correlation between self-reported race and genetic ancestry was 99% in UK Biobank and 97% in the emerge data set, if we exclude those who self-identified as having a mixed or Hispanic race/ethnicity. All participants with sex inconsistencies were removed, additionally the dataset was pruned to remove participants to prevent duplicated individuals, twins, and first-degree relatives using PLINK’s implementation of KING robust kinship coefficients (28). In the KING pipeline, after the kinship (relationship) matrix is calculated using high quality markers for all individuals, kinship-based pruning of samples is performed in which the program by default, randomly excludes one member of each pair of samples and print all independent individuals for downstream analysis.
Table 1. Pediatric study population.
A total of 70,290 cases and 467,247 controls across eMERGE and the UK Biobank in five super-populations [EUR (European), AMR (Admixed American), AFR (African), EAS (East Asians), SAS (South Asian)] were evaluated. Pediatric-onset was defined as diagnosis before age 18 years old. Please see Supplemental Table 4 for a list of all participants, including those without pediatric-onset asthma.
| Cohort | Ancestry | Case/Control | Female/Male | Mean age in years |
|---|---|---|---|---|
| Training | All | 1,324/3,174 | 2,058/2,440 | 11.83 (5.32) |
| European | 322/1,736 | 941/1,117 | 11.82 (5.20) | |
| African | 768/727 | 697/798 | 12.55 (5.17) | |
| Admixed | 203/642 | 367/478 | 12.14 (5.15) | |
| Eastern Asian* | 21/40 | 34/27 | 11.68 (5.11) | |
| Southern Asian* | 10/29 | 19/20 | 10.95 (6.01) | |
| Validation 1 | All | 1,255/7,710 | 3,988/4,977 | 10.37 (5.97) |
| European | 386/3,982 | 1,883/2,485 | 10.90 (5.75) | |
| African | 588/1,352 | 905/1,035 | 10.04 (6.25) | |
| Admixed | 270/2,180 | 1,105/1,345 | 11.13 (5.50) | |
| Eastern Asian* | 9/114 | 62/61 | 9.69 (6.17) | |
| Southern Asian* | 2/82 | 33/51 | 10.07 (6.18) | |
| Validation 2 | All | 16,462/398,808 | 219,728/195,542 | 8.40 (4.69) |
| European | 15,586/376,234 | 207,785/184,035 | 8.10 (4.82)I | |
| African | 310/7,508 | 4,385/3,433 | 8.21 (4.70)I | |
| Admixed | 205/5,020 | 2,717/2,508 | 8.16 (5.06)I | |
| Eastern Asian | 49/1,622 | 877/794 | 7.96 (4.12)I | |
| Southern Asian | 312/8,424 | 3,964/4,772 | 9.54 (4.74)I |
Mean age of onset for Validation 2 cases at the time of diagnosis (see Methods)
Due to low sample size, subgroups identified with asterisks only included in combined adjusted PRS analysis; ancestry-specific results from these subgroups are not presented.
Results were evaluated in all individuals as well as participants who were enrolled in eMERGE as a child (age ≤ 18). It was not possible to identify asthma age of onset for all subjects in eMERGE who were over 18. Because the UK Biobank enrolled only adult participants, pediatric-onset asthma was identified through an assessment of the date of diagnosis in the context of the subject’s current age (UK Biobank field identifiers 21003, 22147, 3786). For both eMERGE and the UK Biobank, we are confident in the identification of subjects with pediatric-onset asthma, while the true adult-onset asthma with no past medical history of asthma in childhood was unable to be accurately determined using electronic medical records.
Discovery GWAS for identifying variants to include in PRS analysis
The PRS was built using the GWAS results from a 2018 study published by the Trans-National Asthma Genetic Consortium (TAGC), which assessed 23,948 patients with adult and pediatric-onset asthma and 118,538 controls (29). Supplemental Table 2 describes the studies included in TAGC. The summary statistics from 2,001,280 autosomal genetic variants that passed quality control filters were accessed through the GWAS catalog (https://www.ebi.ac.uk/gwas/home – assessed on January 8, 2021) and were used to calculate the PRS. 985,837 autosomal genetic variants with minor allele frequencies greater than one percent in the combined Training and Validation datasets (with cases and controls combined) were identified. These 985,837 common markers were found in the 1000 genome reference panel, the TAGC, Training, and Validation datasets with genotyping rates of 99.9% in the Training and Validation 1 datasets. 983,520 (99.7%) of these markers were present in the Validation 2 dataset with a total genotyping rate of 98%. The complete list of the selected variants, the effect alleles, allele frequencies across ancestral groups, and posterior effect sizes (see below) are included in Supplemental Table 3.
Polygenic Risk Scores (PRS)
PRS were calculated using PRS-CS, a Bayesian polygenic prediction method that infers posterior effect sizes of genetic variants using GWAS summary statistics in the context of linkage disequilibrium between variants as assessed on an external reference panel (i.e., the Phase 3 release of the 1000 Genomes data) (30). The genome-wide association study upon which the PRS is derived is a multiancestral metanalysis, and the effect size for all genetic variants in the study are inverse-variance weighted with fixed effects accounting for all ancestral populations. Continuous shrinkage priors that were implemented in this pipeline allowed for marker-specific adaptive shrinkage: the amount of shrinkage applied to each genetic marker is adaptive to the strength of its association signal in GWAS. The pipeline can accommodate diverse underlying genetic architectures. Linkage disequilibrium (LD) and an LD matrix were determined and built based on the highest number of ancestry representation in the discovery set, which in our case was European. The Training process (Figure 1) used the Discovery GWAS summary statistics (multi-ancestral TAGC Discovery GWAS), the reference population (individual-level 1000 genomes genotype data), and the individual-level genotype and phenotype data of target population (Training data set in order to tune the hyper-parameters of the prediction model using CS (auto mode)) so that the pipeline automatically learned the sparseness of the genetic architecture from data and adjusted for the LD structure accordingly (24).
We adjusted for confounding effects due to population stratification with a linear regression model using the ten principal components of ancestry in all participants (31). After calculating a principal component adjusted PRS, age and sex were used as covariates using a logistic regression fitting model implemented in R version 4.1.0 (19). The residuals from this model were used to create an ancestry corrected PRS distribution. The distribution of unadjusted compared to the ancestry-adjusted PRS scores across the five ancestral groups in the Training and Validation cohorts are presented in Supplemental Figure 1.
The PRS prediction accuracy and performance was assessed by using Area Under the Receiver Operating Curve (AUROC), odds ratio (OR) per one standard deviation, and by variance explained (R2 based on the Pseudo R2 calculation based on the McFadden method as applied in Stata) in logistic regression after accounting for covariates (10 principal components, age, and sex). The median of the adjusted percentile distribution between cases and controls after ancestry standardization (i.e., mean PRS of zero and SD of one in each group) was assessed. As our long-term goal is to evaluate this PRS clinically, we selected the top 5% as a threshold for high risk. This threshold was selected to identify those at highest genetic risk while minimizing the number of people receiving a “high risk” result who would not develop asthma (Supplemental Figure 2). To measure the discrimination of the multiancestral asthma PRS, we report the top 5% of this distribution as a high polygenic score and report the increased odds of asthma by comparing the top 5% compared to both the bottom 5% and bottom 95%. After fitting the regression model, the marginal effect of sex and ancestry were also evaluated using the delta method implemented in Stata. These marginal effects measure the impact of unit change in one variable on the prediction of asthma while all other variables of the adjusted multiancestral asthma PRS are constant.
Our primary outcome is predictive value in the pediatric cohorts based on the plan to use this PRS in a prospective study focused on children. In the supplemental tables, we also report outcomes in the overall cohort because it has additional power and allows us to compare performance in the subset of pediatric individuals.
We also benchmarked the performance of the multiancestral PRS using two previously published PRS algorithms (32, 33). Notably, the number of genetic variants included and the populations used in the previously published PRS are different.
PheWAS analyses
To evaluate pleiotropic effects of the multiancestral PRS for asthma against other traits, a phenome-wide association study (PheWAS) was performed using the R PheWAS software package in the Training and Validation cohorts (34). Briefly, ICD9 codes were translated into PheWAS codes according to PheWAS map (34). Cases and controls were identified based on at least two occurrences of the PheWAS code on different days in the cases and no instances in the controls (34). For each PheWAS code, the asthma PRS score was included in a logistic regression model adjusted for age, sex and the ten principal components. The Odds Ratio (OR) is based on regression analyses using each phenotype as the dependent variable and adjusted PRS (a quantitative value calculated for each individual) as an independent variable. A false discovery rate (FDR) of 0.05 using the Benjamini–Hochberg procedure was implemented to account for multiple testing.
Results
The number of participants in this study in each ancestral group that passed quality control steps (see Methods) are broken down by age and sex and presented in Table 1. In total, we analyzed 70,290 participants with asthma and 467,247 controls across the multiancestral Training, Validation 1, and Validation 2 datasets. Each dataset includes participants from each of the five super populations as defined by principal component analysis of independent genetic variants (Table 1, Supplemental Table 4).
An ancestry-specific asthma PRS for asthma is not optimal for clinical implementation, as individuals may align with multiple ancestries. Thus, we evaluated a single multiancestral asthma PRS which accounted for the underlying ancestral differences in the PRS scores (Figure 1). The ancestry-harmonization of the PRS distribution was performed to account for the density and range of each ancestry-specific distribution (Supplemental Figure 1). After adjustment for ancestry, the multiancestral AUC for the Validation pediatric cohorts was 0.70 (0.69–0.72) and AUC of 0.66 (0.65–0.66) in the pediatric Validation 2 cohort (Figure 2, Table 2, Supplemental Table 4). The discrimination of the PRS between the top 5th percentile to the bottom 5th percentile was measured in the Training (OR=2.80, 95% Confidence Interval (CI) (1.87–4.12)), Validation 1 (OR=3.31, 95% CI (2.29–4.78) and Validation 2 (OR=5.82, 95% CI (5.19–6.53)), datasets as shown in Table 2 for pediatric cohorts and Supplemental Table 5 for the full datasets. A comparison of the PRS percentile distribution between cases and controls is presented in Figure 3. The risk prediction per each decile in the Training, Validation 1, and Validation 2 cohorts are included in Supplementary Figure 2.
Figure 2. Multiancestral polygenic risk score (PRS) performance.
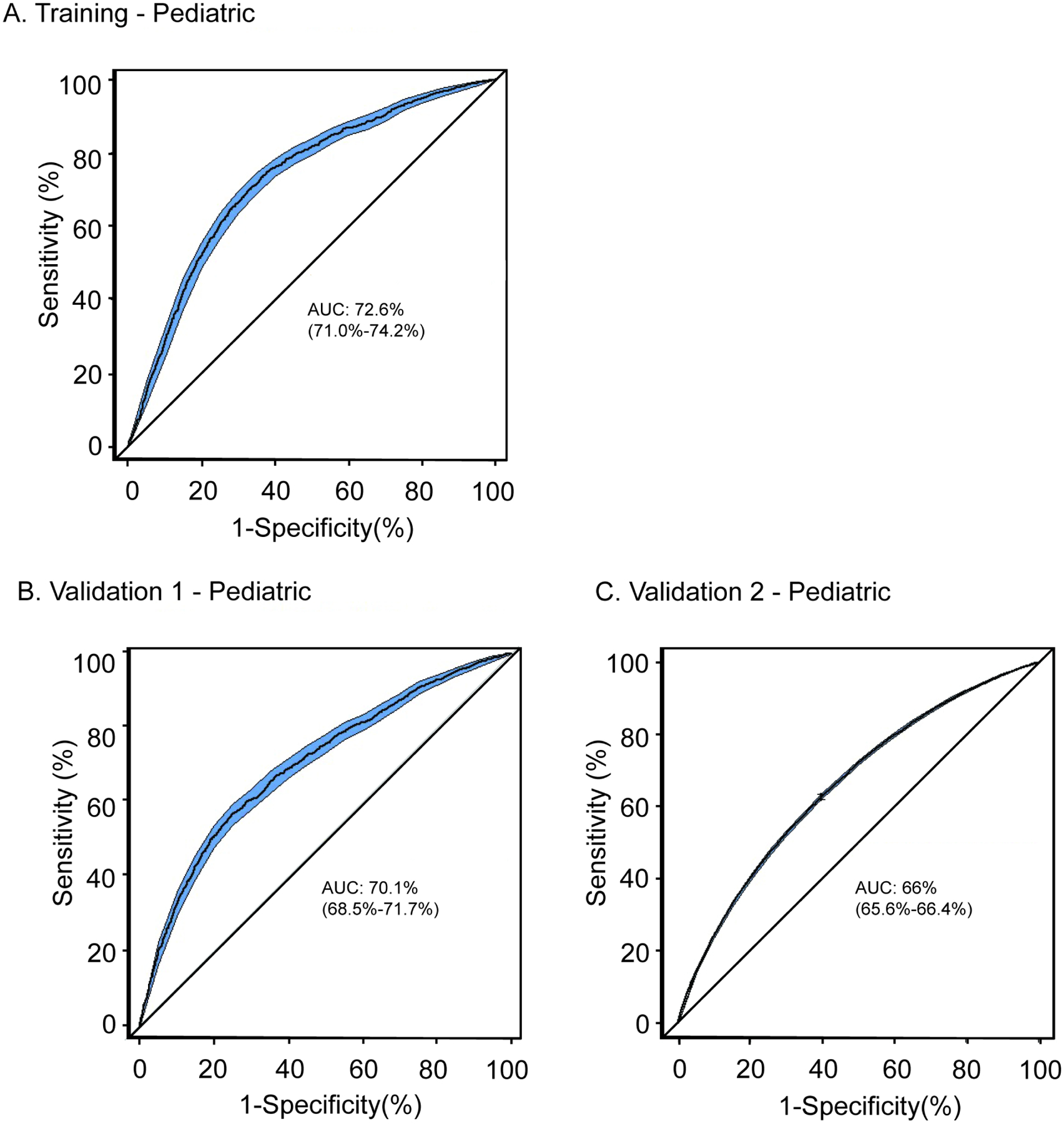
Overall adjusted PRS performance are shown for the Training pediatric cohort (A), Validation 1 pediatric cohort (B), and the Validation 2 pediatric cohort (C). The area under the curve (AUC) and 95% percentile of the confidence interval, are shown adjusted for age, sex and 10 ancestral principal components.
Table 2. Adjusted multiancestral polygenic risk score performance in three independent multiancestral pediatric cohorts.
The overall PRS multiancestry performance with 95% confidence intervals after covariate and ancestry adjustment is presented. The odds ratio of having asthma in patients within the top 5th percentile of the PRS distribution compared to the remaining 95% are shown with 95% confidence intervals. AUC: area under the curve; OR: odds ratio; PRS: polygenic risk score; SD: standard deviation.
| Transancestral PRS performance In Pediatric cohorts | AUC | OR per 1 SDI | Pseudo R2 | ORII Top 5% vs 95% | ORIII Top 5% vs bottom 5% |
|---|---|---|---|---|---|
| Training | 0.73 (0.71–0.74) | 1.21 (1.12–1.30) | 0.11 | 1.84 (1.42–2.39) | 2.80 (1.87–4.12) |
| Validation 1 | 0.70 (0.69–0.72) | 1.22 (1.15–1.30) | 0.08 | 2.16 (1.74–2.67) | 3.31 (2.29–4.78) |
| Validation 2 | 0.66 (0.65–0.66) | 1.59 (1.57–1.62) | 0.04 | 2.37 (2.25–2.49) | 5.82 (5.19–6.53) |
The odds ratio (95% CI) per one standard deviation (P<0.0001).
The odds ratio (95% CI) when comparing the top 5% of standardized adjusted PRS distribution against remaining 95% (P<0.0001).
The odds ratio (95% CI) when comparing the top 5% of standardized adjusted PRS distribution against bottom 5% (p<0.0001)
Figure 3. PRS percentile comparison between pediatric patients with asthma and controls.
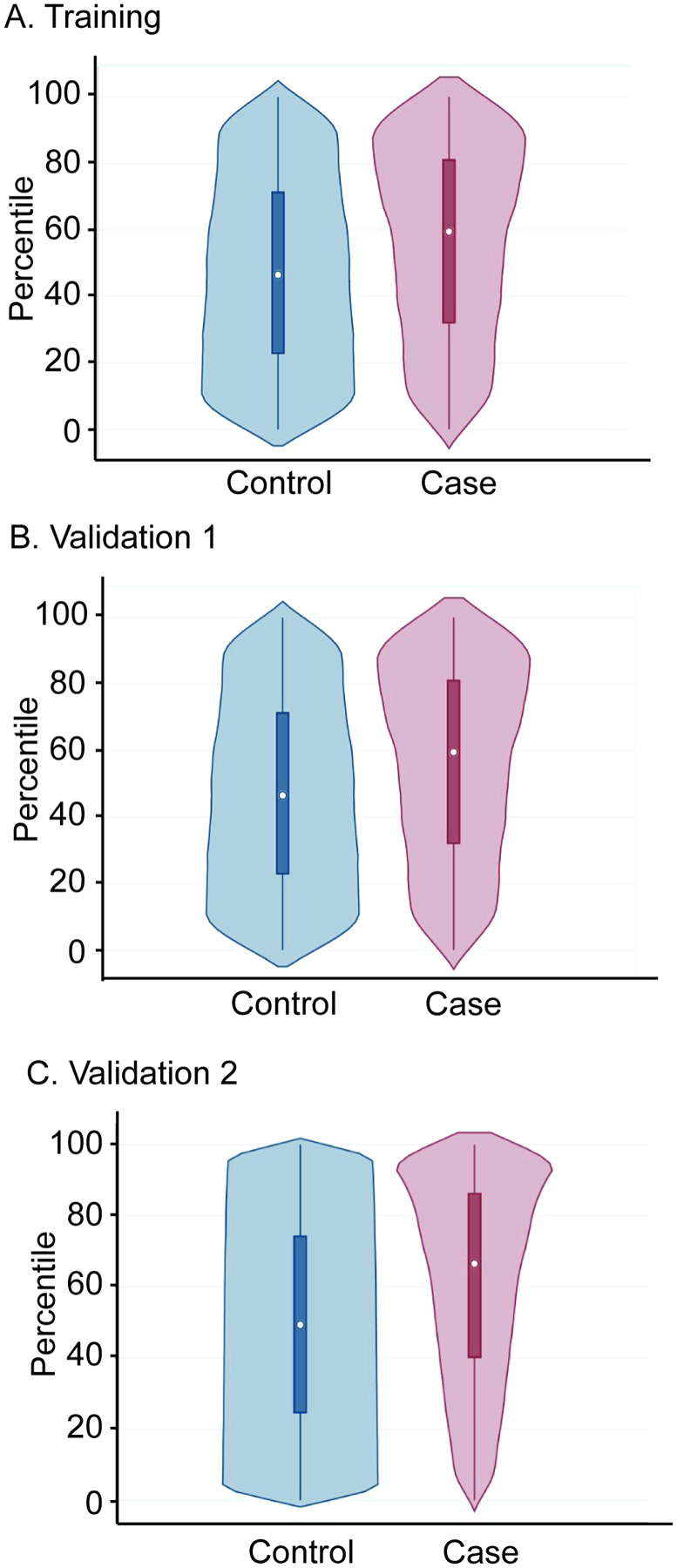
The violin plots of the median ancestry standardized PRS distributions between cases and controls in pediatric Training (A), Validation 1 (B), and Validation 2 cohorts. In the boxplot insert, the dot within each box indicates the median score. In the Training cohort (A), the median standardized score of cases was 60% vs 47% in controls (p<0.0001). In the Validation 1 cohort (B), the median standardized score of cases was 58% vs 48% in controls (p<0.0001). In the Validation 2 cohort (C), 67% in cases versus 49% in controls (p<0.0001). The top and bottom of the boxes indicate the interquartile range (75th and 25th percentiles), respectively.
To confirm that using multiancestral priors did not reduce the performance of the PRS, we calculated PRS for European cohorts using posterior effect sizes after training using only the European studies in TAGC (Supplemental Table 6) with trends towards better performance with multiancestral priors. As shown in Table 3 and Supplemental Table 7, the multiancestral PRS performance was consistent across all ancestries, with better performance in the Validation pediatric cohorts. The pediatric-only Training dataset demonstrated significant discrimination using the covariate-adjusted multiancestral PRS (European AUC 0.67 (0.64–0.70), 1.27 OR per SD; African AUC 0.57 (0.54–0.60), 1.13 OR per SD; and Admixed American AUC 0.68 (0.64–0.72), 1.61 OR per SD). These results were replicated in the pediatric Validation 1 cohorts, (European AUC 0.60 (0.57–0.63), 1.20 OR per SD; African AUC 0.61 (0.58–0.63), 1.27 OR per SD; and Admixed American AUC 0.64 (0.61–0.68), 1.25 OR per SD) (Table 3 (pediatric), Supplemental Table 7 (overall)). The pediatric Validation 2 cohort was used to further replicate the multiancestral PRS and provided the opportunity to measure the performance of the multiancestral asthma PRS in Eastern and Southern Asian cohorts (European AUC 0.66 (0.65–0.67), 1.57 OR per SD; African AUC 0.66 (0.63–0.69), 1.43 OR per SD; Admixed American AUC 0.70 (0.67–0.74), 1.63 OR per SD); Eastern Asian AUC 0.73 (0.66–0.80), 1.32 OR per SD; and Southern Asian AUC 0.65 (0.62–0.68), 1.32 OR per SD).
Table 3: Multiancestral asthma PRS performance in three independent cohorts.
The standardized PRS distribution was evaluated in a logistic regression model adjusted for age and sex as well as the 10 principal components that informed the five super populations. The Area under the curve (AUC) and 95% confidence interval (CI), odds ratio (OR) per one standard deviation (SD) and the total variation explained (pseudo R2) are shown.
| Pediatric Cohorts | AUC (Full model)I | pseudo R2 | OR per one SDII |
|---|---|---|---|
| European ancestry | |||
| Training | 0.67 (0.64–0.70) | 0.05 | 1.27 (1.12–1.44) |
| Validation 1 | 0.60 (0.57–0.63) | 0.02 | 1.20 (1.08–1.34) |
| Validation 2 | 0.66 (0.65–0.67) | 0.04 | 1.57 (1.55–1.60) |
| African ancestry | |||
| Training | 0.57 (0.54–0.60) | 0.01 | 1.13 (1.01–1.26)III |
| Validation 1 | 0.61 (0.58–0.63) | 0.02 | 1.27 (1.14–1.42) |
| Validation 2 | 0.66 (0.63–0.69) | 0.03 | 1.43 (1.24–1.65) |
| Admixed American ancestry | |||
| Training | 0.68 (0.64–0.72) | 0.07 | 1.61 (1.28–2.02) |
| Validation 1 | 0.64 (0.61–0.68) | 0.03 | 1.25 (1.06–1.47) |
| Validation 2 | 0.70 (0.67–0.74) | 0.07 | 1.63 (1.36–1.95) |
| Eastern Asian ancestry | |||
| Validation 2 | 0.73 (0.66–0.80) | 0.08 | 1.32 (0.99–1.77)III |
| Southern Asian ancestry | |||
| Validation 2 | 0.65 (0.62–0.68) | 0.03 | 1.32 (1.18–1.48) |
AUC Full model includes age, sex and 10 principal components
The odds ratio per one standard deviation of PRS distribution (logistic regression P<0.0001)
For the African Ancestry eMERGE datasets 1- Pediatric and Eastern Asian UK Biobank-Pediatric cohorts, P=0.03 and P=0.07 respectively.
We compared the multiancestral asthma PRS from this study to the two previously published PRS (32, 33) in our two European Training and Validation datasets. The number of genetic variants in these PRS were limited (15, 22) and were developed based upon genetic studies of European ancestry while the current PRS was developed based on a multiancestral GWAS. The AUCs in the full, covariate-adjusted models were lower for the Belsky et al. study (AUC=0.59, 95% CI: 0.57–0.61) and Dijk et al. study (AUC=0.60, 95% CI: : 0.59–0.62) compared to the multiancestral PRS in our Training and Validation Europeans cohorts. Further, the multiancestral PRS outperformed the previous European-derived PRS in non-European ancestries (Supplemental Figure 3).
In the full model logistic regression analyses of the PRS, female sex was associated with reduced odds of asthma in pediatric cohorts across all ancestries (Training - Pediatric: OR=0.74, 95% CI 0.65–0.86, p<0.0001, Validation 1 - Pediatric: OR=0.69, 95% CI 0.61–0.79, p<0.0001, Validation 2 - Pediatric: 0.67, 95% CI 0.65–0.69, p<0.0001). This finding is consistent with pediatric-onset asthma being more common in males than females (35, 36). To assess its confounding effect, we calculated the marginal effects of sex for prediction probability of asthma after fitting the logistic regression in Validation 2 cohort as a combined cohort and in each ancestry separately (Supplemental Figure 4). Indeed, the better predictive probability of asthma from the overall PRS model in males compared to females is consistent with the regression analysis. Similarly, we evaluated the marginal effects of ancestry on the multiancestral PRS and found that there was substantial overlap consistent with ancestral normalization.
A phenome-wide association study (PheWAS) was performed in the combined full Training and Validation cohorts to evaluate potential pleiotropic effects of the multiancestral asthma PRS in this study with other traits. As expected, this approach confirmed the strong association of the multiancestral asthma PRS with asthma (OR 2.71, 95% CI 2.04–3.03, PFDR=3.71×10−65) (Table 4). This exploratory analysis also identified more than 300 pleiotropic association effects (false discovery rate (FDR)-corrected p<0.05) including positive associations with asthma severity and exacerbation, emphysema, and pulmonary insufficiency, as well as diabetes, eosinophilic esophagitis, food allergy and white blood cell disorders (Figure 4, Table 4, Supplemental Table 8). Notably, the asthma PRS was more strongly associated with asthma than with the related phenotype allergic rhinitis (OR 1.24, 95% CI 1.10–1.40, PFDR=3.21×10−4) (Supplemental Tables 8 and 9), supporting the phenotype-specificity of the asthma PRS. This approach also detected novel negative associations with traits such as hyperlipidemia and hypercholesterolemia (OR=0.62, 0.95% CI (0.55–0.69), PFDR=6.55×10−17) (Figure 4, Table 4, Supplemental Table 8).
Table 4.
Selected results of PheWAS applying Multiancestral Asthma PRS to Training and Validation 1 cohorts. CI: confidence interval; OR: odds ratio; PheWAS: phenome-wide association study; PRS: polygenic risk score.
| DescriptionI | PheWAS-code | Case | Control | ORII | 95% CI | PFDR |
|---|---|---|---|---|---|---|
| Positively associated with Asthma PRS | ||||||
| Asthma | 495 | 12,963 | 70,020 | 2.71 | 2.41– 3.02 | 3.17×10−65 |
| Asthma with exacerbation | 495.2 | 3,043 | 70,020 | 4.72 | 3.74 – 5.95 | 3.39×10−39 |
| Emphysema | 508 | 8,276 | 73,217 | 1.78 | 1.56 – 2.05 | 1.22×10−16 |
| Chronic obstructive asthma | 495.1 | 1,433 | 70,020 | 3.23 | 2.35 – 4.45 | 4.90×10−13 |
| Type 1 diabetes | 250.1 | 4,283 | 69,292 | 1.91 | 1.58 –2.30 | 1.16×10−11 |
| Chronic airway obstruction | 496 | 8,056 | 70,020 | 1.56 | 1.36 –1.80 | 5.57×10−10 |
| Respiratory failure | 509 | 6,029 | 73,217 | 1.61 | 1.37 – 1.88 | 3.05×10−9 |
| Wheezing | 512.1 | 2,234 | 47,495 | 2.17 | 1.68 – 2.82 | 4.37×10−9 |
| Diabetes mellitus | 250 | 19,764 | 69,292 | 1.34 | 1.21 – 1.48 | 9.36×10−9 |
| Eosinophilic esophagitis | 530.15 | 607 | 62,044 | 3.85 | 2.38 –6.20 | 3.40×10−8 |
| Type 2 diabetes | 250.2 | 19,137 | 69,292 | 1.32 | 1.19 – 1.46 | 9.78×10−8 |
| Diseases of white blood cells | 288 | 4,694 | 77,026 | 1.61 | 1.35 –1.91 | 1.14×10−7 |
| Allergic reaction to food | 930 | 1,688 | 65,842 | 2.25 | 1.66 – 3.04 | 1.49×10−7 |
| Negatively associated with Asthma PRS | ||||||
| Postmenopausal disorders | 627 | 11,313 | 75,725 | 0.55 | 0.48 – 0.63 | 8.00×10−18 |
| Peripheral enthesopathies | 726 | 16,894 | 66,608 | 0.64 | 0.57 –1.71 | 6.31×10−17 |
| Hypercholesterolemia | 272.11 | 19,899 | 51,696 | 0.62 | 0.55 –0.69 | 6.55×10−17 |
| Hematuria | 593 | 7,632 | 68,213 | 0.55 | 0.48 – 0.64 | 2.50×10−16 |
| Benign neoplasm of skin | 216 | 11,233 | 79,947 | 0.63 | 0.56 – 0.71 | 1.86×10−14 |
| Disorders of lipid metabolism | 272 | 41,493 | 51,696 | 0.73 | 0.67 –0.80 | 1.04×10−11 |
| Hyperlipidemia | 272.1 | 41,299 | 51,696 | 0.73 | 0.67 – 0.80 | 1.11×10−11 |
| Disorders of synovium | 727 | 9,291 | 66,608 | 0.64 | 0.56 –1.73 | 3.65×10−11 |
| Cataract | 366 | 14,306 | 81,833 | 0.67 | 0.60 –0.76 | 5.14×10−11 |
| Carbohydrate transport disorder | 271 | 1,410 | 97,301 | 0.36 | 0.27 – 0.49 | 1.34×10−10 |
| Disaccharide malabsorption | 271.3 | 1,302 | 97,301 | 0.35 | 0.25 – 0.48 | 1.40×10−10 |
| Elevated prostate specific antigen | 796 | 2,746 | 84,778 | 0.53 | 0.41 – 0.67 | 2.92×10−7 |
Selected results at false discovery rate PFDR<0.05. The complete lists of traits are included in Supplemental Table 5.
OR<1 indicates negative association of trait with asthma PRS
Figure 4. A plot of PheWAS analysis of the asthma PRS within the combined Training and Validation cohorts.
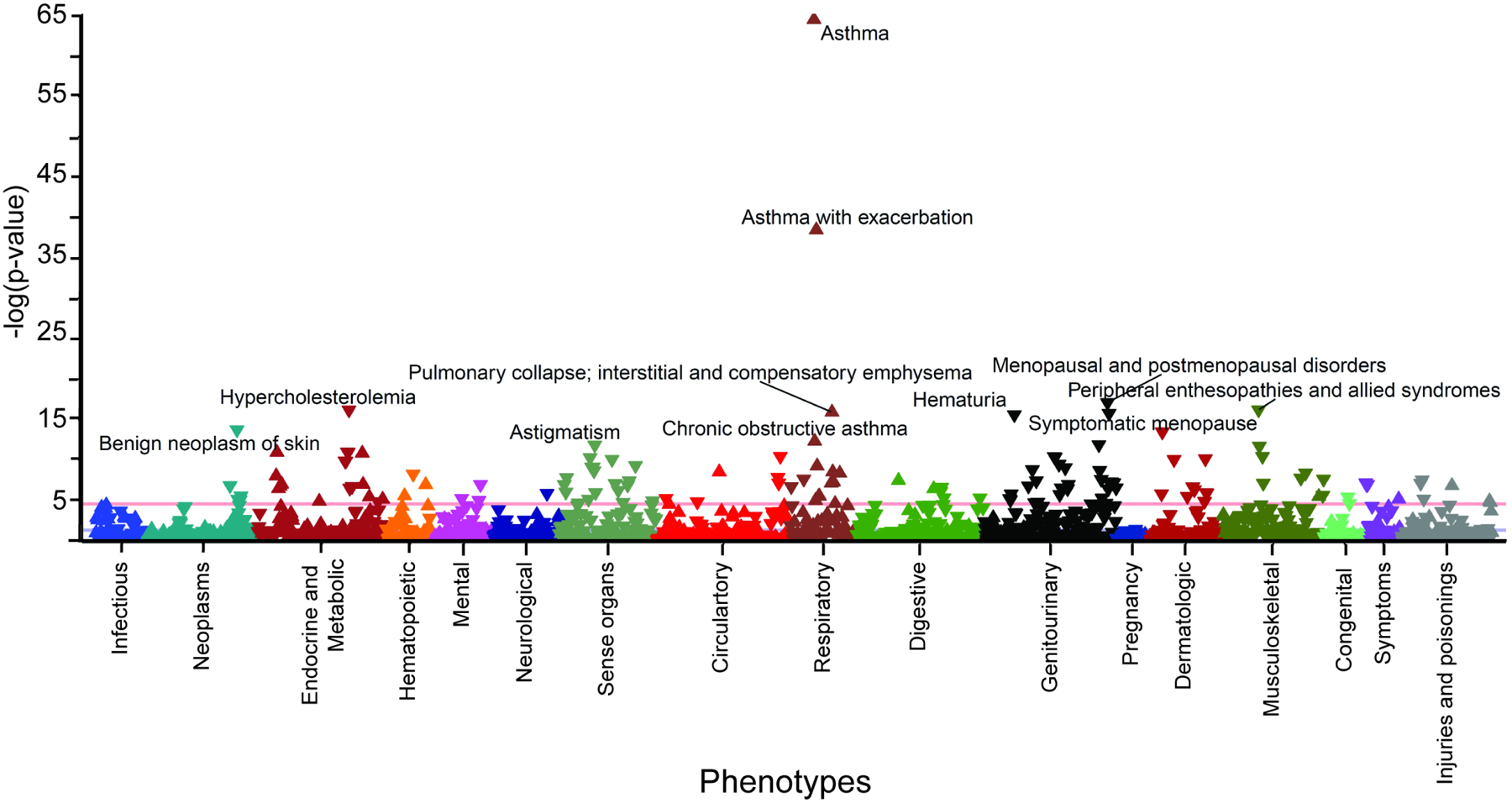
Manhattan plots of phenome-wide association analyses with phecodes (X-axis) and FDR-corrected PheMap phenotype probability (y-axis). The red line indicates the Bonferroni level of significance (5.0 × 10−5).
Discussion
Pediatric asthma affects ~10% of the children in the United States of America. There is no cure, underscoring the importance of prevention and early identification. In this study, we developed a PRS for asthma using an ancestrally diverse group of individuals and trained and validated the PRS’s performance using multiple independent cohorts, which included pediatric-onset as well as any age of onset. We demonstrate that our asthma PRS has good discriminatory performance in people of diverse ancestries and especially children, is more discriminating than prior scores and reveals potential pleiotropic effects. Taken together, these results support the value of our multiancestral PRS.
The PRS performed well across three independent datasets and in five different ancestral groups. Because of the large number of participants assembled, we were able to evaluate the performance of the PRS overall as well as in documented pediatric cases. This evaluation revealed that our asthma PRS performed better in children than in the overall cohort of subjects with combined pediatric and adult-onset asthma. These findings are consistent with previous genetic variant-based heritability estimates supporting a larger genetic contribution in children compared to adults (14). While we confidently identified individuals with pediatric-onset asthma, a limitation of this study was our inability to identify individuals with adult-onset asthma (i.e. some adults with asthma could have developed disease as a child).
Notably, our multiancestral asthma PRS performed better based upon AUC than prior asthma PRSs, especially when considering non-European populations. Based on assessment of the PGS catalog (https://www.pgscatalog.org/) in August 2021, there are currently two published studies focusing on PRS development for asthma alone (32, 33). These studies were limited primarily to European ancestral groups and PRS development was based on p-value thresholding. In complex diseases with many modest genetic effects such as asthma, the p-value thresholding methodology can underperform due to the omission of many variants with weaker phenotype association (30). In contrast, the Bayesian approach used in this study incorporates genome-wide variants after considering the underlying linkage disequilibrium population sub-structure and generates a posterior effect size for all variants included in the study. The benefits of the Bayesian strategy relative to other PRS approaches that use effect-size weighted additive model include (30): 1) all association data from a GWAS are used – including information that is usually not included in approaches that start with robust genetic associations. 2) the approach incorporates the differences in linkage disequilibrium and genetic architecture between ancestral groups, and 3) the use of continuous shrinkage priors allows the model to consider robust genetic signals with large effect sizes as well as small effects with less significant association signals. This suggests that the Bayesian methodology is superior for the development of a PRS. However, as we compared different methodology and different population (multi-ancestral) a broader comparison of these methods for the development of multi-ancestral pediatric asthma PRSs is justified.
The improved performance of our PRS may also be due to both the Bayesian approach and the multiancestral approach used. As asthma risk varies by ancestry, including individuals from diverse ancestral groups will be essential to ensure the clinical use of PRSs does not exacerbate health disparities (37). Other approaches to develop transancestral and multiancestral PRS differ by how they select and weigh genetic risk variants and how they integrating genetic data with other clinical and environmental data (38–42). While we used PRS-CS, other models use best linear unbiased prediction (BLUP) and least absolute shrinkage and section operator approaches (LASSO) to estimate genetic effect sizes in joint models of multiple variants and predictions are performed simultaneously (43–46). There are also numerous methods that use different approaches to account for differences in linkage disequilibrium in individuals of different ancestry (47, 48). As statistical methods are developed to improve multiancestral PRS, multiancestral asthma PRS should be continuously refined with future publications of genetic association studies of asthma in larger, admixed populations. It is possible that similarly powered ancestry-specific analyses would identify additional loci with more impactful effect sizes; however, the results of the multiancestral study might prove to be more broadly useful when applied to a heterogenous population, such as the type of people who go to a primary care setting. Further, a major strength of our multiancestral PRS is that a single PRS is applicable to all ancestral groups. Thus, clinicians and researchers will not be required to a priori assign an individual to an ancestral group.
While TAGC might be the largest and most diverse meta-analysis from the perspective of genetic ancestry, there are several limitations to consider in the context of the multi-ancestral asthma PRS. Specifically, if there are differences in the genetic etiology by age of onset of asthma, the TAGC is not composed of a majority of pediatric cases. Further the degree to which pediatric onset asthma cases are represented in the meta-analysis differs greatly based on the race-ethnicity of the component studies. Thus, while our PRS performed well for pediatric asthma, continued refinement of the PRS is warranted with special focus on pediatric cases and capturing more ancestral diversity in the discovery data.
To understand how underlying asthma risk may relate to a variety of conditions, we performed a PheWAS analysis to examine positive or negative association of other conditions with the asthma PRS. Notably, we measured a stronger effect size for asthma with exacerbation (PheWAS code 495.2; OR = 4.72) and chronic asthma (PheWAS code 495.1; OR=3.23) than asthma (PheWAS code 495; OR = 2.71) (Table 4). In the case of this PheWAS, the Odds Ratio (OR) is based on regression analyses using each phenotype as the dependent variable and adjusted PRS as an independent variable. These findings suggests that the asthma PRS may be useful not only for prevention but also to help clinicians select treatment strategies for children already diagnosed with asthma. These findings require replication, as we do not have sufficiently uniform measurements in the subjects in the Training, Validation 1, and Validation 2 cohorts to identify if PRS in patients is associated with disease severity. However, these findings provide rationale for a controlled prospective study to test the association of asthma severity with PRS. Not surprisingly, other atopic conditions such as eosinophilic esophagitis and food allergy were positively associated with the asthma PRS, as these conditions have been noted to have increased rates in patients with asthma (49, 50). However, we also found positive associations with type I and type II diabetes. Intriguingly, several studies have reported a higher-than-expected co-occurrence of asthma and type I diabetes, supporting a partially shared genetic etiology (51–53). We also found a surprising negative association between our asthma PRS and hypercholesterolemia/hyperlipidemia. In contrast to our findings, previous meta analyses have reported that asthma is associated with worse lipid profiles at a phenotypic level (54). One possible explanation for this discrepancy is that the use of inhaled corticosteroids (a primary treatment of asthma) is associated with a worse lipid profile in adults (55). It is also possible that environmental factors, such as pollution, are driving increased risks for asthma as well as poor lipid profiles (56–58). While the PheWAS analysis suggests potential pleiotropic effects (both increased and decreased risk of other diseases), additional work is required to clarify these relationships.
This study is an initial step towards developing a multiancestral PRS to be used in the eMERGE IV network (https://www.genome.gov/Funded-Programs-Projects/Electronic-Medical-Records-and-Genomics-Network-eMERGE) prospective intervention cohort study beginning in 2022. 5000 children (underrepresented, non-European preferred) will be enrolled across the 10 eMERGE clinical sites. Multiancestral PRS for 4 phenotypes (asthma, obesity, type 1 diabetes, type 2 diabetes) will be calculated and returned to participants’ parents and primary care providers. Parents and primary care providers of children with a high risk asthma PRS (top 5th percentile) will also receive guideline informed health recommendations (59, 60). We seek to understand how primary care providers, patients, and patient families change their behavior in reaction to a top 5th percentile asthma PRS. The prospective study will collect family history information and clinical factors to display along with an asthma high risk PRS that providers can use to calculate a Pediatric Asthma Risk Score (PARS). Recent studies have validated PARS as a tool to predict asthma development in young children based upon family history, eczema before age 3, wheezing apart from colds before age 3, African American ancestry, and sensitization to two or more food or aero allergens (7).
A previous group suggested that their pediatric asthma PRS did not provide any discriminatory value above clinical risk factors (33). Yet, assessing clinically predictive factors can be challenging due to the lack of consistent capturing of such data in cohorts with sufficient statistical power. Even if the PRS and the clinical risk prediction substantially overlap in who is identified at risk, the development of such a PRS would still be of value. This is because not all children undergo allergic sensitization testing before age 3 and clinical presentations such as eczema and wheezing without a cold may not be recognized by parents. Thus, alternative strategies for risk stratification are needed. Notably, there are already preventive measures which can be prioritized if a child is identified as high risk (61). For example, once a child is identified as high risk for asthma, families can be counseled to limit smoke exposure, identify and avoid known allergens, prevent viral infection, and limit dust and mold exposure (4, 62, 63). However, we recognize that the use of genetics alone to predict asthma has inherent limitations, as both genes and environment contribute to asthma risk.
A long-term goal beyond eMERGE IV will be to create and validate a combined/integrated predictive model that includes genetic, family history, clinical and environmental risk factors. Data collected during the eMERGE IV prospective study will provide essential elements toward our long term goal. In addition to genotype data, family history information and relevant clinical factors we will also have geocodes to develop a combined/integrated predictive model.
In conclusion, we present the development and validation of a pediatric asthma PRS that performs effectively across ancestries in three independent cohorts and identifies novel pleiotropic relationships. In the future, this PRS will be used in the context of additional demographic and clinical risk factors as part of a genome informed risk assessment to help families of children at high risk for asthma take preventive steps to avoid disease.
Supplementary Material
Financial Disclosure
R01 HG010730, R01 NS099068, R01 GM055479, U01 AI130830, R01 AI141569, and U01 AI150748 to MTW; R01 DK107502, R01 AI148276, U19 AI070235, U01 HG011172, and P30 AR070549 to LCK; R01 AR073228, R01 AI024717, and CCHMC ARC Award 53632 to MTW and LCK; R01 HG010166, R01 HL145422, R25 GM129808, R01 AI127392, UG3 OD023282, U19 AI070235, U54 AI117804, R01 NS096053, R01 DK107502, R01 HD089458, R01 HL132153, R01 AI139126, R01 HL135114, R01 HD099775, U01 HG011172 to LJM; R01 HL132344 and R01 HG011411 to TBM.
The eMERGE Network was initiated and funded by NHGRI through the following grants:
Phase IV: U01 HG011172 (Cincinnati Children’s Hospital Medical Center); U01 HG011175 (Children’s Hospital of Philadelphia); U01 HG008680 (Columbia University); U01 HG011176 (Icahn School of Medicine at Mount Sinai); U01 HG008685 (Mass General Brigham); U01 HG006379 (Mayo Clinic); U01 HG011169 (Northwestern University); U01 HG011167 (University of Alabama at Birmingham); U01 HG008657 (University of Washington); U01 HG011181 (Vanderbilt University Medical Center); U01 HG011166 (Vanderbilt University Medical Center serving as the Coordinating Center).
Phase III: U01 HG8657 (Kaiser Permanente Washington/University of Washington); U01 HG8685 (Brigham and Women’s Hospital); U01 HG8672 (Vanderbilt University Medical Center); U01 HG8666 (Cincinnati Children’s Hospital Medical Center); U01 HG6379 (Mayo Clinic); U01 HG8679 (Geisinger Clinic); U01 HG8680 (Columbia University Health Sciences); U01 HG8684 (Children’s Hospital of Philadelphia); U01 HG8673 (Northwestern University); U01 HG8701 (Vanderbilt University Medical Center serving as the Coordinating Center); U01 HG8676 (Partners Healthcare/Broad Institute); and U01 HG8664 (Baylor College of Medicine).
Phase II: U01 HG006828 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital); U01 HG006830 (Children’s Hospital of Philadelphia); U01 HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01 HG006382 (Geisinger Clinic); U01 HG006375 (Group Health Cooperative/University of Washington); U01 HG006379 (Mayo Clinic); U01 HG006380 (Icahn School of Medicine at Mount Sinai); U01 HG006388 (Northwestern University); U01 HG006378 (Vanderbilt University Medical Center); and U01 HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center).
Genotyping Center support U01 HG004438 (CIDR) and U01 HG004424 (the Broad Institute).
Phase I: U01 HG004610 (Group Health Cooperative/University of Washington); U01 HG004608 (Marshfield Clinic Research Foundation and Vanderbilt University Medical Center); U01 HG04599 (Mayo Clinic); U01 HG004609 (Northwestern University); U01 HG04603 (Vanderbilt University Medical Center, also serving as the Administrative Coordinating Center); U01 HG004438 (CIDR) and U01 HG004424 (the Broad Institute) serving as Genotyping Centers.
Abbreviations
- AFR
African
- AMR
Admixed American
- AUC
Area under the curve
- CI
Confidence interval
- EAS
East Asian
- eMERGE
Electronic Medical Records and Genomics
- EUR
European
- FDR
False discovery rate
- GWAS
Genome-wide association study
- ICD
International Classification of Diseases
- NIH
National Institutes of Health
- OR
Odds ratio
- PARS
Pediatric Asthma Risk Score
- PheWAS
Phenome-wide association study
- PRS
Polygenic risk score
- SAS
South Asian
- TAGC
Trans-National Asthma Genetic Consortium
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests:
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research has been conducted using data from UK Biobank, a major biomedical database www.ukbiobank.ac.uk
References
- 1.Holgate ST, Wenzel S, Postma DS, Weiss ST, Renz H, Sly PD. Asthma. Nat Rev Dis Primers 2015;1:15025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuruvilla ME, Lee FE, Lee GB. Understanding Asthma Phenotypes, Endotypes, and Mechanisms of Disease. Clin Rev Allergy Immunol. 2019;56(2):219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsahai JM, Hansbro PM, Wark PAB. Mechanisms and Management of Asthma Exacerbations. Am J Respir Crit Care Med. 2019;199(4):423–32. [DOI] [PubMed] [Google Scholar]
- 4.Castillo JR, Peters SP, Busse WW. Asthma Exacerbations: Pathogenesis, Prevention, and Treatment. J Allergy Clin Immunol Pract. 2017;5(4):918–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiksten J, Toppila-Salmi S, Makela M. Primary Prevention of Airway Allergy. Curr Treat Options Allergy. 2018;5(4):347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castro-Rodriguez JA, Holberg CJ, Wright AL, Martinez FD. A clinical index to define risk of asthma in young children with recurrent wheezing. Am J Respir Crit Care Med. 2000;162(4 Pt 1):1403–6. [DOI] [PubMed] [Google Scholar]
- 7.Biagini Myers JM, Schauberger E, He H, Martin LJ, Kroner J, Hill GM, et al. A Pediatric Asthma Risk Score to better predict asthma development in young children. J Allergy Clin Immunol. 2019;143(5):1803–10 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoettler N, Rodriguez E, Weidinger S, Ober C. Advances in asthma and allergic disease genetics: Is bigger always better? J Allergy Clin Immunol. 2019;144(6):1495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheth KK, Lemanske RF Jr. Pathogenesis of asthma. Pediatrician. 1991;18(4):257–68. [PubMed] [Google Scholar]
- 10.Thomsen SF. Exploring the origins of asthma: Lessons from twin studies. Eur Clin Respir J. 2014;1(Suppl 1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buniello A, MacArthur JAL, Cerezo M, Harris LW, Hayhurst J, Malangone C, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47(D1):D1005–D12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira MA, Vonk JM, Baurecht H, Marenholz I, Tian C, Hoffman JD, et al. Shared genetic origin of asthma, hay fever and eczema elucidates allergic disease biology. Nat Genet. 2017;49(12):1752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19(9):581–90. [DOI] [PubMed] [Google Scholar]
- 14.Pividori M, Schoettler N, Nicolae DL, Ober C, Im HK. Shared and distinct genetic risk factors for childhood-onset and adult-onset asthma: genome-wide and transcriptome-wide studies. Lancet Respir Med. 2019;7(6):509–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vergara C, Murray T, Rafaels N, Lewis R, Campbell M, Foster C, et al. African ancestry is a risk factor for asthma and high total IgE levels in African admixed populations. Genet Epidemiol. 2013;37(4):393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flores C, Ma SF, Pino-Yanes M, Wade MS, Perez-Mendez L, Kittles RA, et al. African ancestry is associated with asthma risk in African Americans. PLoS One. 2012;7(1):e26807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brim SN, Rudd RA, Funk RH, Callahan DB. Asthma prevalence among US children in underrepresented minority populations: American Indian/Alaska Native, Chinese, Filipino, and Asian Indian. Pediatrics. 2008;122(1):e217–22. [DOI] [PubMed] [Google Scholar]
- 18.Pino-Yanes M, Thakur N, Gignoux CR, Galanter JM, Roth LA, Eng C, et al. Genetic ancestry influences asthma susceptibility and lung function among Latinos. J Allergy Clin Immunol. 2015;135(1):228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fishe J, Zheng Y, Lyu T, Bian J, Hu H. Environmental effects on acute exacerbations of respiratory diseases: A real-world big data study. Sci Total Environ. 2022;806(Pt 1):150352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur S, Rosenstreich D, Cleven KL, Spivack S, Grizzanti J, Reznik M, et al. Severe asthma in adult, inner-city predominantly African-American and latinx population: demographic, clinical and phenotypic characteristics. J Asthma. 2021:1–11. [DOI] [PubMed] [Google Scholar]
- 21.Almoguera B, Vazquez L, Mentch F, Connolly J, Pacheco JA, Sundaresan AS, et al. Identification of Four Novel Loci in Asthma in European American and African American Populations. Am J Respir Crit Care Med. 2017;195(4):456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanaway IB, Hall TO, Rosenthal EA, Palmer M, Naranbhai V, Knevel R, et al. The eMERGE genotype set of 83,717 subjects imputed to ~40 million variants genome wide and association with the herpes zoster medical record phenotype. Genet Epidemiol. 2019;43(1):63–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Z, Lee PH, Chaffin MD, Chung W, Loh PR, Lu Q, et al. A genome-wide cross-trait analysis from UK Biobank highlights the shared genetic architecture of asthma and allergic diseases. Nat Genet. 2018;50(6):857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z, Zhu X, Liu CL, Shi H, Shen S, Yang Y, et al. Shared genetics of asthma and mental health disorders: a large-scale genome-wide cross-trait analysis. Eur Respir J. 2019;54(6). [DOI] [PubMed] [Google Scholar]
- 26.Zhang D, Dey R, Lee S. Fast and robust ancestry prediction using principal component analysis. Bioinformatics. 2020;36(11):3439–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demenais F, Margaritte-Jeannin P, Barnes KC, Cookson WOC, Altmuller J, Ang W, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat Genet. 2018;50(1):42–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge T, Chen CY, Ni Y, Feng YA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10(1):1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khera AV, Chaffin M, Zekavat SM, Collins RL, Roselli C, Natarajan P, et al. Whole-Genome Sequencing to Characterize Monogenic and Polygenic Contributions in Patients Hospitalized With Early-Onset Myocardial Infarction. Circulation. 2019;139(13):1593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belsky DW, Sears MR, Hancox RJ, Harrington H, Houts R, Moffitt TE, et al. Polygenic risk and the development and course of asthma: an analysis of data from a four-decade longitudinal study. Lancet Respir Med. 2013;1(6):453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dijk FN, Folkersma C, Gruzieva O, Kumar A, Wijga AH, Gehring U, et al. Genetic risk scores do not improve asthma prediction in childhood. J Allergy Clin Immunol. 2019;144(3):857–60 e7. [DOI] [PubMed] [Google Scholar]
- 34.Carroll RJ, Bastarache L, Denny JC. R PheWAS: data analysis and plotting tools for phenome-wide association studies in the R environment. Bioinformatics. 2014;30(16):2375–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frohlich M, Pinart M, Keller T, Reich A, Cabieses B, Hohmann C, et al. Is there a sex-shift in prevalence of allergic rhinitis and comorbid asthma from childhood to adulthood? A meta-analysis. Clin Transl Allergy. 2017;7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinart M, Keller T, Reich A, Frohlich M, Cabieses B, Hohmann C, et al. Sex-Related Allergic Rhinitis Prevalence Switch from Childhood to Adulthood: A Systematic Review and Meta-Analysis. Int Arch Allergy Immunol. 2017;172(4):224–35. [DOI] [PubMed] [Google Scholar]
- 37.Oh SS, Galanter J, Thakur N, Pino-Yanes M, Barcelo NE, White MJ, et al. Diversity in Clinical and Biomedical Research: A Promise Yet to Be Fulfilled. PLoS Med. 2015;12(12):e1001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudolph A, Song M, Brook MN, Milne RL, Mavaddat N, Michailidou K, et al. Joint associations of a polygenic risk score and environmental risk factors for breast cancer in the Breast Cancer Association Consortium. Int J Epidemiol. 2018;47(2):526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50(9):1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mars N, Koskela JT, Ripatti P, Kiiskinen TTJ, Havulinna AS, Lindbohm JV, et al. Polygenic and clinical risk scores and their impact on age at onset and prediction of cardiometabolic diseases and common cancers. Nat Med. 2020;26(4):549–57. [DOI] [PubMed] [Google Scholar]
- 41.Mak TSH, Porsch RM, Choi SW, Zhou X, Sham PC. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol. 2017;41(6):469–80. [DOI] [PubMed] [Google Scholar]
- 42.Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S, et al. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97(4):576–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Speed D, Balding DJ. MultiBLUP: improved SNP-based prediction for complex traits. Genome Res. 2014;24(9):1550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Carbonetto P, Stephens M. Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet. 2013;9(2):e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi J, Park JH, Duan J, Berndt ST, Moy W, Yu K, et al. Winner’s Curse Correction and Variable Thresholding Improve Performance of Polygenic Risk Modeling Based on Genome-Wide Association Study Summary-Level Data. PLoS Genet. 2016;12(12):e1006493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lello L, Avery SG, Tellier L, Vazquez AI, de Los Campos G, Hsu SDH. Accurate Genomic Prediction of Human Height. Genetics. 2018;210(2):477–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amariuta T, Ishigaki K, Sugishita H, Ohta T, Koido M, Dey KK, et al. Improving the trans-ancestry portability of polygenic risk scores by prioritizing variants in predicted cell-type-specific regulatory elements. Nat Genet. 2020;52(12):1346–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marquez-Luna C, Loh PR, South Asian Type 2 Diabetes C, Consortium STD, Price AL. Multiethnic polygenic risk scores improve risk prediction in diverse populations. Genet Epidemiol. 2017;41(8):811–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez-Cervera J, Arias A, Redondo-Gonzalez O, Cano-Mollinedo MM, Terreehorst I, Lucendo AJ. Association between atopic manifestations and eosinophilic esophagitis: A systematic review and meta-analysis. Ann Allergy Asthma Immunol. 2017;118(5):582–90 e2. [DOI] [PubMed] [Google Scholar]
- 50.Foong RX, du Toit G, Fox AT. Asthma, Food Allergy, and How They Relate to Each Other. Front Pediatr. 2017;5:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metsala J, Lundqvist A, Virta LJ, Kaila M, Gissler M, Virtanen SM, et al. The association between asthma and type 1 diabetes: a paediatric case-cohort study in Finland, years 1981–2009. Int J Epidemiol. 2018;47(2):409–16. [DOI] [PubMed] [Google Scholar]
- 52.Hsiao YT, Cheng WC, Liao WC, Lin CL, Shen TC, Chen WC, et al. Type 1 Diabetes and Increased Risk of Subsequent Asthma: A Nationwide Population-Based Cohort Study. Medicine (Baltimore). 2015;94(36):e1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smew AI, Lundholm C, Savendahl L, Lichtenstein P, Almqvist C. Familial Coaggregation of Asthma and Type 1 Diabetes in Children. JAMA Netw Open. 2020;3(3):e200834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peng J, Huang Y. Meta-analysis of the association between asthma and serum levels of high-density lipoprotein cholesterol and low-density lipoprotein cholesterol. Ann Allergy Asthma Immunol. 2017;118(1):61–5. [DOI] [PubMed] [Google Scholar]
- 55.Fessler MB, Massing MW, Spruell B, Jaramillo R, Draper DW, Madenspacher JH, et al. Novel relationship of serum cholesterol with asthma and wheeze in the United States. J Allergy Clin Immunol. 2009;124(5):967–74 e1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGuinn LA, Schneider A, McGarrah RW, Ward-Caviness C, Neas LM, Di Q, et al. Association of long-term PM2.5 exposure with traditional and novel lipid measures related to cardiovascular disease risk. Environ Int. 2019;122:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Keet CA, Keller JP, Peng RD. Long-Term Coarse Particulate Matter Exposure Is Associated with Asthma among Children in Medicaid. Am J Respir Crit Care Med. 2018;197(6):737–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brunst KJ, Ryan PH, Brokamp C, Bernstein D, Reponen T, Lockey J, et al. Timing and Duration of Traffic-related Air Pollution Exposure and the Risk for Childhood Wheeze and Asthma. Am J Respir Crit Care Med. 2015;192(4):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Expert Panel Working Group of the National Heart L, Blood Institute a, coordinated National Asthma E, Prevention Program Coordinating C, Cloutier MM, Baptist AP, et al. 2020 Focused Updates to the Asthma Management Guidelines: A Report from the National Asthma Education and Prevention Program Coordinating Committee Expert Panel Working Group. J Allergy Clin Immunol. 2020;146(6):1217–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cloutier MM, Teach SJ, Lemanske RF Jr., Blake KV. The 2020 Focused Updates to the NIH Asthma Management Guidelines: Key Points for Pediatricians. Pediatrics. 2021;147(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tackett AP, Farrow M, Kopel SJ, Coutinho MT, Koinis-Mitchell D, McQuaid EL. Racial/ethnic differences in pediatric asthma management: the importance of asthma knowledge, symptom assessment, and family-provider collaboration. J Asthma. 2020:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elenius V, Jartti T. Vaccines: could asthma in young children be a preventable disease? Pediatr Allergy Immunol. 2016;27(7):682–6. [DOI] [PubMed] [Google Scholar]
- 63.Szefler SJ. Advances in pediatric asthma in 2012: moving toward asthma prevention. J Allergy Clin Immunol. 2013;131(1):36–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


