Abstract
The B-cell receptor (BCR) consists of surface-bound immunoglobulin (Ig) and a heterodimeric signaling unit comprised of CD79A and CD79B. Upon cognate antigen recognition, the receptor initiates important signals for B-cell development and function. The receptor also conveys antigen-independent survival signals termed tonic signaling. While the requirement of a CD79A/CD79B heterodimer for BCR complex assembly and surface expression is well established based on mice models, few studies have investigated this in human mature B cells. Here, we found that human tonsillar B cells with high surface expression of IgM or IgG had potentiated BCR signaling compared to BCRlow cells, and high IgM expression in germinal center B cells was associated with reduced apoptosis. We explored the mechanism for IgM surface expression by CRISPR/Cas9-induced deletion of CD79A or CD79B in four B-lymphoma cell lines. Deletion of either CD79 protein caused loss of surface IgM in all cell lines and reduced fitness in three. From two cell lines, we generated stable CD79A or CD79B knockout clones, and demonstrated that loss of CD79A or CD79B caused a block in N-glycan maturation and accumulation of immature proteins, compatible with retention of BCR components in the endoplasmic reticulum. Rescue experiments with CD79B WT restored surface expression of CD79A and IgM with mature glycosylation, whereas a naturally-occurring CD79B G137S mutant disrupting CD79A/CD79B heterodimerization did not. Our study highlights that CD79A and CD79B are required for surface IgM expression in human B cells and illuminates the importance of IgM expression level for signaling and fitness.
Introduction
The development and function of B cells depend on signals from the B-cell receptor (BCR). The antigen-binding part of the BCR consists of two identical membrane-bound Ig heavy chains (IgH), each linked to identical Ig light chains. The Ig molecule pairs with a signaling unit comprised of a transmembrane heterodimer of CD79A (Igα) and CD79B (Igβ). Upon antigen binding, BCR signaling is initiated by phosphorylation of the ITAMs of CD79A and CD79B by the SRC-family kinase LYN (1). Signaling is further relayed by recruitment and phosphorylation of SYK and activation of downstream signaling including PLC-γ, BLNK and BTK. Deletion of any of the BCR components in mice, including IgH, CD79A or CD79B, results in a block in B-cell development in the bone marrow and absence of mature B cells (2–4). Signaling through CD79A or CD79B is needed in this developmental checkpoint as replacement of the tyrosines in the ITAMs of CD79A and CD79B in mice leads to a block in the B-cell development (5). In mature mouse B cells, the presence of BCR on the cell surface provides a low-level constitutive antigen-independent survival signal called tonic signaling. This was first shown in conditional knockout (KO) mice where deletion of Igh or Cd79a in mature B cells depleted the mature B-cell pool with a half-life of 3–6 days (6, 7). Mature BCR-negative B cells can be saved by constitutive active PI3K signaling or deletion of the phosphatase Pten, a negative regulator of AKT signaling and a key target of PI3K signaling (8). Primary B cells with high BCR expression have increased survival and elevated tonic signaling compared to cells with low BCR expression(9).
The BCR similarly has an essential role for human B cells. Immunodeficient patients with mutation in IGHM, CD79A or CD79B do not have mature B cells (10–13), confirming the developmental checkpoint seen in mice. B-cell malignancies arising from mature B cells usually retain BCR expression on the surface (14) and are addicted to tonic (15), chronic active (16, 17) or antigen-induced BCR signaling (18–20), hence making this pathway an important therapeutic target. CD79A and CD79B were identified as essential in prior CRISPR/Cas9 screens in lymphoma cell lines representing diffuse large B-cell lymphoma (DLBCL) (21–23), the most common aggressive form of B-cell non-Hodgkin lymphoma (B-NHL). The activated B-cell like (ABC) subtype of DLBCL is characterized by chronic activation of NF-κB, induced by BCR engament by self-antigens (17) or activating mutations in genes involved in the BCR pathway (16). In younger ABC DLBCL patients, especially the MCD genetic subtype harbouring mutations in CD79B and/or MYD88, show high response rates to the BTK inhibitor Ibrutinib (24). Mantle cell lymphoma (MCL), a rarer form of aggressive B-NHL which also show activation of the BCR pathway and constitutive NF-κB signaling, have high response-rate to Ibrutinib (25, 26) and next-generation BTK inhibitors acalabrutinib and zanubrutinib (27).
The Germinal centre like (GCB) subtype of DLBCL depends on tonic BCR signaling relayed via PI3K-mediated activation of AKT and inhibition of FOXO1, a positive regulator of apoptosis (15, 28). In cell lines representing GCB DLBCL, Y188F mutation in the ITAM of CD79A or KO of AKT is toxic, and BCR KO cells can be rescued by forced activation of AKT (15). Furthermore, silencing of FOXO1 in B-cell lymphomas renders the cells insensitive to BCR signaling inhibitors (28). Burkitt lymphoma, which is a MYC-dependent malignancy, are outcompeted after induction of shRNA targeting CD79A, suggesting a dependency on tonic signaling (29). However, in a MYC-driven lymphoma mouse model, BCR loss did not kill the cells, but the competitiveness of BCR-negative cells was reduced due to rewiring of the metabolism and increased acitivty of GSK3β (30). Recently, the requirement of the CD79A/CD79B heterodimer for survival of human B cells was questioned, as the Burkitt lymphoma cell line Ramos could be rescued upon reintroduction of CD79B WT in BCR KO cells (31). In this cell line, CD79B homodimers could associate with CD19 and provide an alternative survival signal when CD79A or IgM was lost (31).
Although it is generally accepted that CD79A and CD79B are needed for BCR assembly and expression, few studies have investigated this in detail using human B cells. Overexpression of BCR components in Drosophila S2 cells have shown that CD79A and CD79B are covalently bound by a disulfide bridge in the extracellular domain (32) and this CD79A/CD79B heterodimer is non-covalently linked to the Ig molecule through aromatic proline interactions in the transmembrane domain (33, 34). In a murine cell line, the CD79A/CD79B heterodimer is observed in a 1:1 stoichiometry to plasma-membrane Ig (35). Findings in the Burkitt lymphoma cell line Ramos suggest that the entire BCR complex must be assembled in the endoplasmic reticulum (ER) before transport to the cell membrane (36), a mechanism controlled by chaperone proteins of the ER quality control system that retains improperly folded proteins or improperly assembled protein complexes (37). For instance, in the absence of CD79A in a murine B cell line, Igμ heavy chain is retained in the ER through interactions of polar amino acids in the transmembrane domain with the ER chaperone calnexin (38). Transport of protein complexes from ER through Golgi stacks and trafficking to the plasma membrane depends on N-glycosylation in a well-regulated, multistep process (39). The progression of the BCR complex can be measured by the glycosylation pattern as immature, pre-Golgi proteins are decorated with a high-mannose N-acetyl glucosamine core whereas mature, post-Golgi proteins have more complex N-glycans (40).
Here, we aimed to systematically investigate the requirement of CD79A and CD79B for IgM membrane expression and B-cell fitness. We used human tonsillar B cells to investigate the association between the level of membrane expression of IgM and protection against apoptosis. For mechanistic studies, we focused on human MCL cell lines as model system, as lymphoma cells from MCL usually overexpress IgM as compared to normal B cells, which likely contribute to the pathogenesis of this lymphoma type (41). CRISPR/Cas9 editing was used to generate mutants lacking CD79A or CD79B, followed by monitoring of KO cells over time in a competition assay. CD79A and CD79B KO clones were generated to study their interdependence for protein maturation and for maturation of IgM.
Materials and Methods
Human samples and cell lines
Tonsils were obtained from patients undergoing tonsillectomy with written informed consent in accordance with the Declaration of Helsinki and the Regional Committees for Medical and Health Research Ethics, Region Eastern Norway (REK#2010/1147a). The tonsils were processed to single-cell suspension by mincing and then stored in liquid nitrogen. Before experiments, the cells were thawed and rested in RPMI w/10% FCS at 37°C for at least 15 min before counting and further processing. The B-cell lines Z-138 (ATCC CRL-3001) and Granta-519 (DSMZ ACC 342) were cultured in RPMI/10% FCS + 2 μM L-glutamine. MINO (DSMZ ACC 687) and SUDHL-6 (DMSZ ACC 572) were cultured in RPMI/10% FCS. Cell line authentication was done by PCR-single-locus technology for 21 independent PCR systems (Eurofins, Denmark).
CRISPR/Cas9 editing and generation of CD79A or CD79B KOs
Stable Cas9-expressing cell lines were made as previously described (42). Briefly, Z-138, MINO, SUDHL-6 and Granta-519 were transduced with retroviral particles expressing humanized wild type Cas9 (MSCV_Cas9_puro was a gift from Christopher Vakoc (Addgene plasmid # 65655)) (43), and then selected by puromycin at concentrations determined by a cytotoxicity assay (Invitrogen). sgRNAs targeting CD79A and CD79B were designed using the online tool CHOPCHOP (44). The three sgRNAs with the highest scores were delivered into Cas9-expressing Granta-519 and MINO cells by electroporation using the ECM 830 Square Wave Electroporation System (BTX). To check the genome editing efficiency, surface staining of CD79A and CD79B was performed 72 h after electroporation and the targeted loci were amplified by PCR and subjected to Sanger sequencing. The results were analyzed using the TIDE online software (45). The sgRNAs with the highest editing efficiencies (CD79A: GCACCACCGCGCAGAACAGG and CD79B: GACCTTTGGGATTCCGGTAC) were selected for functional analyses. Single cell clones were sorted by FACS (Sony SH800S CellSorter) and expanded for several weeks, before repeating surface staining and Sanger sequencing to enable selection of potential KO clones. Stable clones were obtained for Granta-519 (CD79A KO clone II-G6– homozygous 85-bp deletion, CD79B KO clone E4 – homozygous 1-bp insertion) and Z-138 (CD79B KO clone B3 – homozygous 2-bp deletion and clone B1 – complex alteration with three alleles; 1-bp deletion, 14-bp deletion and 22-bp deletion (only used in Supplemental Figure 2)). In addition, clones with in-frame 6-bp deletions in CD79B were obtained for MINO (clone B and D with identical alterations; 1-bp insertion and 6-bp deletion) and Z-138 (clone C7 and C9 with identical homozygous 6-bp deletions). Clones with a 6-bp deletion maintained surface IgM expression.
Human CD79B-2A-GFP cloning
A population of naive and memory B cells was isolated from tonsils by bead-depletion of CD3+ T cells and CD38+ GC B cells as previously described (46). Approximately 1 million naive/memory B cells were pelleted, total mRNA was extracted (RNeasy Plus kit, Qiagen) and cDNA was synthesized (Quantitect kit, Qiagen). CD79B coding sequence was amplified using 10 ng of cDNA and the following primers: CD79B_FWD: CAC CAT GGC CAG GCT GGC GTT GTC and CD79B_REV: CTC TCT TGG CTC TCT CCT GGC CTG GGT GCT CAC. The PCR conditions were the following 30 cycles (30” 94°C, 30” 53°C and 2’ 68°C) using Pfx polymerase (ThermoFisher Scientific, Waltham, MA, USA) and generated a 690 bp product which contains the natural start and a replacement of the STOP codon. In parallel, a GFP fragment was amplified using a 2A-GFP containing template (CSK-2A-GFP) (47) with these primers: 2Af_UNIVERSAL AGA GCC AAG AGA GGC AGC GGC G and pWS_GFPXI CTC GAG TTA CTT GTA CAG CTC GTC CAT GCC GAG, to generate a GFP fragment containing a 2A coding sequence on its 5’-end. The two fragments were purified, mixed and amplified using the CD79B_FWD and pWS_GFPXI primers in these conditions: 20X (1’ 94°C, 1’ 53°C and 1’30” 68°C) with Pfx. The amplicon was subsequently purified and subcloned into pENTR/D-TOPO vector (ThermoFisher Scientific) and sent to sequencing (MWG Eurofins, Erlangen, Germany). Validated sequences were further recombined into the retroviral vector pMP71-Gateway vector (48) using Gateway recombinase system (ThermoFisher Scientific). CD79B mutants were created by Pfu-Turbo-based site direct mutagenesis procedure using pENTR-CD79B-2A-GFP as a template and the following primers: G137S: GTC TAC CAG GGC TGC AGC ACA GAG CTG CGAG and CTC GCA GCT CTG TGC TGC AGC CCT GGT AGAC, Y196N: GAA GAT CAC ACC AAC GAG GGC CTG GAC and GTC CAG GCC CTC GTT GGT GTG ATC TTC, Y196C: GAA GAT CAC ACC TGC GAG GGC CTG GAC and GTC CAG GCC CTC GCA GGT GTG ATC TTC, Y196F: GGA AGA TCA CAC CTT CGA GGG CCT GGAC and GTC CAG GCC CTC GAA GGT GTG ATC TTCC. All mutants were sequence verified and further recombined into pMP71-gateway vector.
Retroviral transduction
CD79B retroviral particles were prepared in HEK-Phoenix cells as previously described (48). Granta-519 CD79B KO cells were transduced by one cycle of spinoculation. Non-treated culture plates (ThermoFisher Scientific) were coated over night with Retronectin (Takara, Shiga, Japan) at 4°C and blocked with PBS containing FCS at 2% final concentration for 30 min at RT. Wells were then washed and 1 ml of cells at 0.5 million/ml was added and incubated for 10 min. Then 1 ml of retroviral supernatant was mixed, and the plates were spun at 750 × g for 1 hour at 32°C. Cells were harvested after 18 hours, washed in PBS and grown in complete medium for another 48 hours. Transgene presence was monitored by flow cytometry analysis of the GFP signal.
Phenotyping by flow cytometry
Tonsillar single cell suspensions or B-lymphoma cell lines were stained with antibodies on ice for 30 min before analysis on a FACSCanto II (BD). The following antibodies from BioLegend were used: CD20-PacBlue (clone 2H7), CD3-BV570 (clone UCHT1), CD38-PECy7 (clone HIT2), CD79A-PECy7 (clone HM47) and IgM-PacBlue (clone MHM-88). The following antibodies from BD were used: CD27-APC (clone M-T271), CD27-BV605 and -FITC (clone L128), CD3-V500 and -Ax700 (clone UCHT1), CD79B-PerCPCy5.5 and -APC (clone SN8/3A2–2E7), IgD-PerCPCy5.5 (clone IA6–2) and IgG-APC-H7 (clone G18–145). In addition, we used CD3-PE (clone UCHT1) and poly rabbit IgD (cat. No. R5112) from Dako, CD79A-PE (clone 706931) from R&D, poly goat IgA-PE (Cat no. 2052–09) from Southern Biotech, and poly goat IgG- and IgM-FITC (Cat. No. AHI1308 and AHI 1608) from Invitrogen.
Signaling by phospho-specific flow cytometry (phospho-flow)
To induce BCR signaling, cell suspensions from tonsils were distributed at 107 cells/ml and rested for an additional 15–30 min, then stimulated with 10 μg/ml anti-IgM F(ab’)2 or IgG F(ab’)2 (Invitrogen cat. AHI1601 (discontinued) and cat. no. AHI1301 or Jackson Immuno Research cat. no. 309–006-043) for 4 min. Cell lines were stimulated with 20 μg/ml anti-IgM for 15 min as previously described (41, 49). Fixation was done by adding paraformaldehyde to a 1.6% final concentration for 5 min at room temperature. In experiments where signaling was induced by FITC-label ed anti-IgM F(ab’)2 (5 min; Invitrogen cat. no. AHI1608 (discontinued) or Jackson Immuno Research cat. No. 309–096-043) or anti-IgG F(ab’)2 (5 min; Cat. No. AHI1308), the unstimulated cells were stained for 15 min, room temperature, after fix. Cells were washed with PBS and then permeabilized with ice-cold methanol (final concentration > 90%) and stored in −20 °C or −80 °C. At the day of flow cytometry analysis, cells were rehydrated by washing twice in PBS. For barcoding, cells were resuspended in PBS and incubated with different concentrations of Pacific Blue dye for 30 min in the dark. Antibody staining (P-BLNK-Ax647 (clone J117–1278), p-BTK-Ax647 (clone 24a/BTK), p-BTK-Ax488 (clone N-35–88), p-LCK-Ax488 and -PE (clone 4/LCK-Y505), p-PLC-γ -Ax647 and -Ax488 (clone K86–689.37), p-SFK-Ax647 (clone 4/LCK-Y505), p-SYK-Ax647, -PE, and -Ax488 (clone 17a/P-ZAP70), CD20-PerCPCy5.5 (clone H1) from BD and p-ERK-Ax647 (clone 197G2) from Cell Signaling Technologies) was performed at room temperature for 15–30 min before washing and running the samples on a FACS LSRII or a FACS Canto II (BD). Cytobank Software (www.cytobank.org) was used for data analysis, and relative phosphorylation changes were calculated using arcsinh transformation of median fluorescence intensity (MFI) of the cell population of interest.
Apoptosis detection
Tonsillar cell suspensions were thawed, rested for 30 min and distributed at107 cells/ml. Cells were either left unstained or stained for 5 min with anti-IgM-FITC, then left untreated of treated with FASL. As a live/dead cell stain, Pacific Blue dye was added at a final concentration of 0.1 ng/μl for the last 10 min of culture. After 3 h, cells were fixed by adding PFA (1.6%, 5 min, room temperature). The cells that were left unstained prior to culture, were now stained with anti-IgM-FITC (15 min, room temperature), followed by permeabilization with ice cold methanol. Cells were stored at −20 °C until the day of flow cytometry analysis. Antibody staining (cleaved caspase-3-Ax647, clone C92–605 from BD) was performed at room temperature for 15–30 min before washing and running the samples on a FACS LSRII (BD).
CD79B−IgM− and CD79B+IgM+ MINO cells were sorted by FACS (BD FACSAria) on day 3 after transfection with CD79A or CD79B sgRNA, then cultured in separate wells for up to 7 days. 24 hours after sorting, cells were incubated with Alexa750 NHS Ester (1 μg/ml) or LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Invitrogen) for 10 minutes as live/dead cell stain before fixation in PFA, washing, permeabilization and staining with anti-cleaved caspase-3-PE (clone C92–605) from BD and anti-phospho-GSK3β-Ax647 (clone D85E12) from Cell Signaling.
Western Blot analysis and Endo H treatment
Protein lysates were generated by lysis with SDS buffer containing protease and phosphatase inhibitors. Endoglycosidase H (Endo H) treatment was carried out by combining 10 μg glycoprotein with 1 μl of 10X Glycoprotein Denaturing Buffer (100°C, 10 min) before treatment with GlycoBuffer and Endo H for 1 h as previously described (50). The samples were run on Criterion 15 % Tris-HCL, 12% or 7.5% TGX gels from Bio-Rad and analyzed on Chemidoc MP (Bio-Rad) applied for imaging. The following antibodies were used: anti-CD79A (clone: D1X5C) and anti-CD79B (clone: D7V2F) from Cell Signaling Technologies, anti-CD79B (clone: CB3–1) from eBioscience, anti-IgM rabbit F(ab’)2 (cat. no. 309–006-043) from Jackson Immunoresearch, anti-Histone H3 rabbit polyclonal from Millipore (cat. no. 05–928) and anti-GAPDH rabbit polyclonal from Genetex (cat. no. GTX100118). Image processing was performed in ImageLab (Bio-Rad). Global background-adjusted volume of bands was quantified, and the lane normalization factor was calculated by normalizing all housekeeping control bands to the strongest housekeeping signal on the gel. Each band of interest was then normalized by the lane normalization factor and intensities are shown relative to expression of total protein by original cells (lane 1).
Real-time PCR
RNA was isolated using the Qiagen RNeasy Plus kit and cDNA was synthesized using the Qiagen Quantitect kit. qPCR was carried out using TaqMan™ Universal PCR Master Mix on a 7500 Real Time PCR System with probes against CD79A (Hs00998119_m1), CD79B (Hs00236881_m1) and IGHM (Hs00941538_g1), using GAPDH (Hs99999905_m1) and PGK1 (Hs9999906_m1) as endogenous control (Applied Biosciences). Relative gene expression was calculated by the comparative CT method.
Statistical analysis
Statistical analyses were performed in GraphPad Prism and data were considered statistically significant when p < 0.05. One-way ANOVA, one-sample t-test or two-sample t-test were applied to determine the level of statistical significance. In Figure 1B, linear regression and Pearson correlation were performed to investigate the relationship between signaling and surface Ig expression.
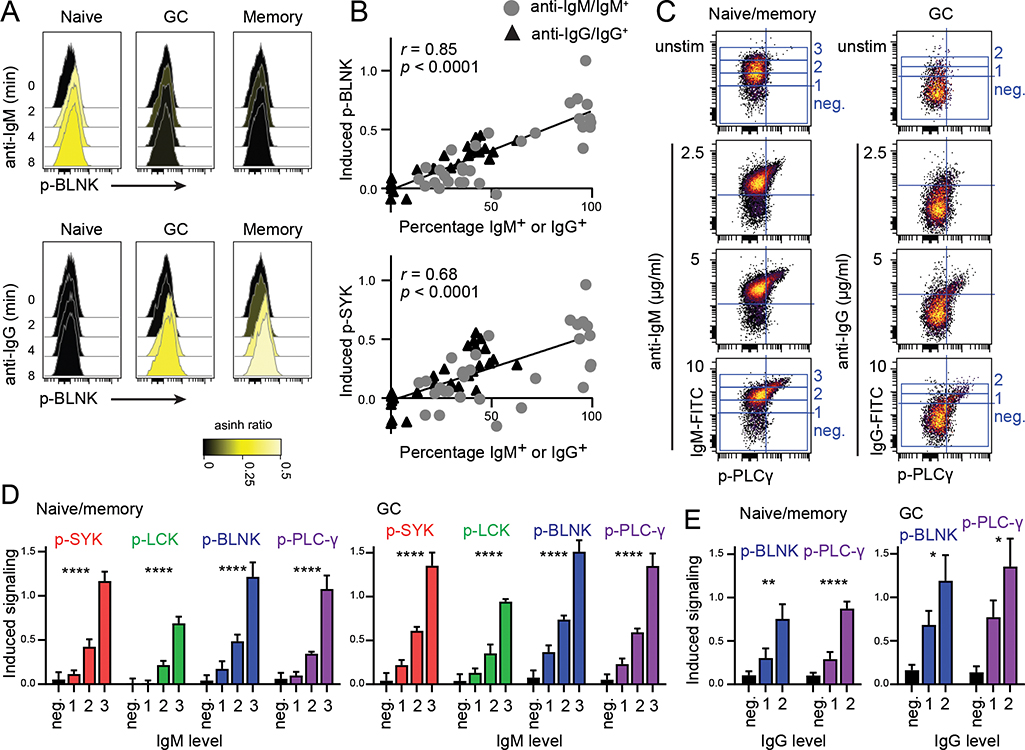
Figure 1. BCR signaling strength correlates with level of BCR expression in primary human B cells.
A. BCR signaling was induced with anti-IgM F(ab’)2 or anti-IgG F(ab’)2 in single-cell suspensions from human tonsils and analyzed by phospho-flow. Gating of B-cell subsets is shown in Supplemental Fig. 1A. B: In separate experiments using the same tonsils, the percentage of IgM+, IgD+, IgG+ and IgA+ B cells were calculated from surface phenotyping (Supplemental Fig. 1B, 1C). Induced signaling (arcsinh ratio of median relative to unstimulated cells) is plotted against percentage of IgM+ and IgG+ cells for each population in each tonsil. Line indicates simple linear regression of all data points. Pearson r and significance is indicated. C-E. For simultaneous measurement of surface Ig level and signaling readout, signaling was induced by FITC-labeled anti-IgM F(ab’)2 or anti-IgG F(ab’)2. In unstimulated cells, FITC-labeled anti-Ig F(ab’)2 was added after PFA fixation. C. Anti-IgM-induced signaling in naive/memory B cells and IgG-induced signaling in GC B cells is shown for one representative tonsil. D. Surface IgM-negative cells and three populations of IgM-positive cells were gated in unstimulated and stimulated cells (gating shown in C). Induced signaling was measured in GC B cells or naïve/memory B cells within specific IgM levels relative to the same Ig level in unstimulated cells (arcsinh ratio of median). Mean and SD, n = 4. *indicates significance after ANOVA tested for the three IgM+ populations. ****p < 0.0001. E. Similarly, surface IgG-negative and two levels of IgG-positive cells were gated and signaling were measured within specific IgG levels. Mean and SD, n = 4. *indicates significance after t-test of the two IgG+ populations. *p < 0.05, **p < 0.01, ****p < 0.0001.
Results
Surface BCR expression level is associated with BCR signaling strength
To investigate the relationship between BCR expression and BCR signaling strength, we started by mapping the IgH isotype (IgM, IgD, IgG, IgA) and BCR expression level in tonsillar B-cell subsets. Naive, germinal center (GC) and memory B cells were distinguished using CD20, CD38, CD27 and IgD (Supplemental Fig. 1A). All naive B cells expressed IgM and/or IgD whereas GC and memory B cells had a mix of IgM+, IgD+, IgG+ and IgA+ B cells (Supplemental Fig. 1B, 1C). GC B cells had a large fraction of BCR-negative cells as expected (Supplemental Fig. 1B, 1C). The expression level of CD79B and IgM/IgG showed a linear relationship (Supplemental Fig. 1D, 1E). Similarly, IgD levels were also associated with CD79B levels, but a population of IgDlowIgMhigh cells interrupted the linearity of this relationship (Supplemental Fig. 1D–1G). The maximum expression level of IgM, IgG, IgD, IgA and CD79B was similar across the different B-cell subsets (Supplemental Fig. 1H, 1I). In separate experiments, using the same ten tonsils, we measured BCR-signaling by phospho-flow in the three B-cell populations (Fig. 1A). On a population level, anti-IgM/IgG-induced signaling varied between the subsets but exhibited good correlation between percentage surface IgG/IgM expression and anti-IgM/IgG-induced signaling strength (Pearson r = 0.86 for p-BLNK and r = 0.67 for p-SYK; Fig. 1B). To directly link BCR-signaling strength to surface IgM or IgG expression level, we induced signaling by activating tonsillar cells with FITC-labeled anti-IgM F(ab’)2 or anti-IgG F(ab’)2, whereas unstimulated cells were fixed before staining with the FITC-labelled antibodies (Fig. 1C). Different levels of IgM or IgG expression were gated, and BCR-induced signaling was then quantified for specific levels of IgM/IgG relative to the same level in unstimulated B cells (Fig. 1C). This approach revealed a strong association between surface IgM or IgG and induced phosphorylation of the downstream signaling proteins p-SYK, p-BLNK, p-LCK and p-PLCγ in naïve/memory as well as in GC B cells (Fig. 1D, 1E). A potential limitation in these experiments is that the total level of signaling proteins were not measured. However, due to the short activation time, changes are not expected. Thus, we conclude that BCR expression level determined the BCR signaling strength in human mature B cells.
Surface BCR level affects spontaneous and FASL-induced apoptosis
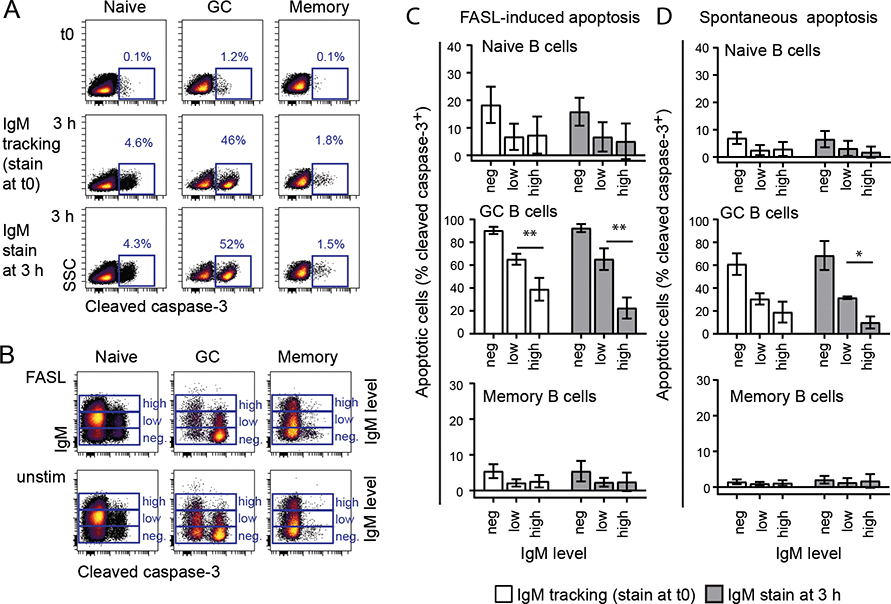
To further examine how the surface expression level of BCR affects B-cell function, we measured apoptosis in human tonsillar B cells. GC B cells rapidly underwent spontaneous apoptosis when cultured in vitro for 3 hours as measured by expression of cleaved caspase-3 (Fig. 2A). To link IgM expression to apoptosis, we used the same approach as for tracking of BCR-induced signaling and briefly stained tonsillar B cells with FITC-labeled anti-IgM F(ab’)2 at time zero for tracking of IgMlow and IgMhigh B cells. The IgM-FITC signal and IgD expression were stable for at least 6 hours (Supplemental Fig. 1J). Of importance, the anti-IgM-FITC induced BCR signaling (Fig. 1), but did not induce apoptosis as unstimulated cells and IgM-stained cells displayed the same percentage of apoptotic cells at 3 h (Fig. 2A). To further rule out that anti-IgM-FITC binding to IgM molecules at the start of the culture induced apoptosis; we also stained cells at the end of the culture. These two approaches showed that high surface IgM expression protected GC B cells from FASL-induced apoptosis, as cleaved caspase-3 was significantly lower in IgMhigh cells compared to IgMlow cells (Fig. 2B, 2C). Spontaneous apoptosis was also significantly lower in IgMhigh cells than IgMlow cells when the IgM staining was performed at the end of the culture (Fig. 2C). IgM-negative GC B cells displayed the highest frequency of apoptotic cells (Fig. 2B, 2C). Naive and memory B cells had very low level of spontaneous or FASL-induced apoptosis during this short-term culture and no significant difference was found related to difference in IgM expression level (Fig. 2). These experiments demonstrate a link between IgM levels and apoptosis in GC B cells.
Figure 2. High BCR surface expression is associated with low spontaneous and FASL-induced apoptosis.
Single cell suspensions from human tonsils were thawed and cultured for 3 hours with or without FASL. Dead cells were excluded by a live/dead marker and apoptosis was measured by anti-cleaved caspase-3 antibody using flow cytometry. Cells were either stained with anti-IgM F(ab’)2 after fix at the end of the 3-hour culture or prior to culturing; 5 min stain and wash before culturing (“IgM tracking”). Gating of B-cell subsets were performed as in Figure 1. A. IgM-tracking did not affect spontaneous apoptosis in unstimulated naive, memory or GC B cells. B-D. B cells were divided in three populations based on IgM level and percentage of cleaved caspase-3+ apoptotic cells were gated as in A. Mean ± SD, n = 3. *indicates significance in a t-test between high and low IgM expression. *p < 0.05, **p < 0.01.
Loss of CD79A or CD79B results in loss of surface IgM expression and reduced B-cell fitness
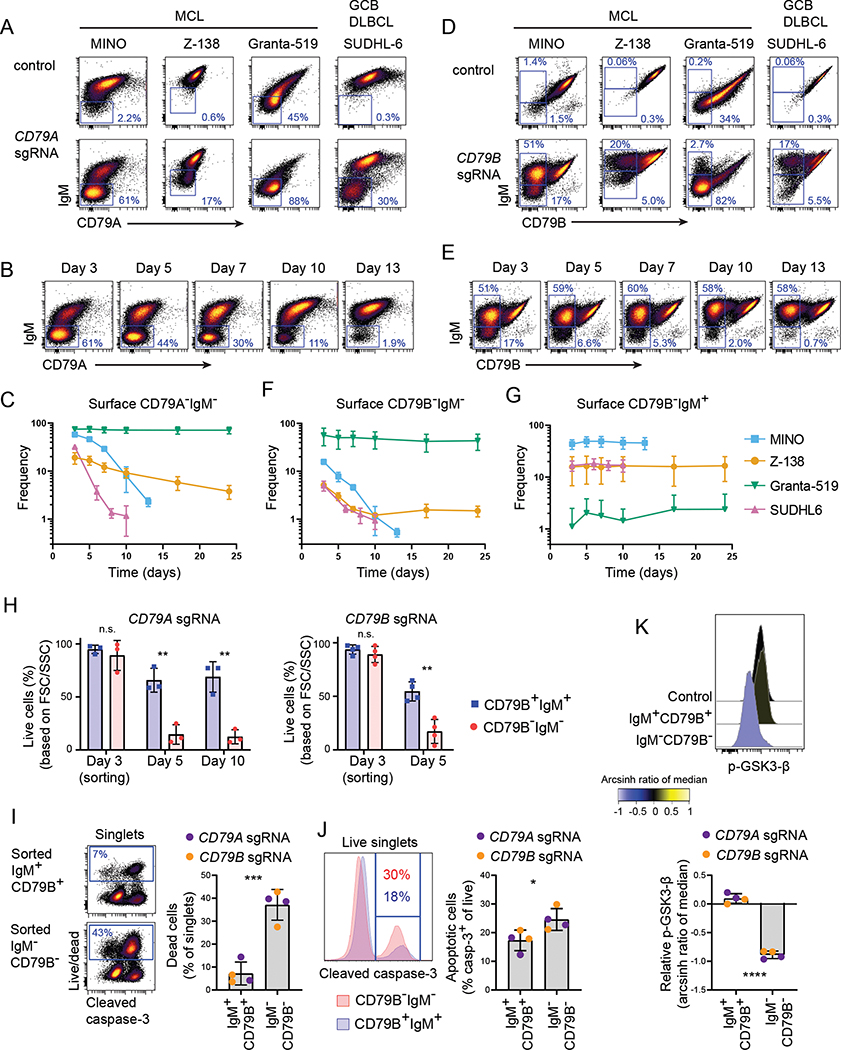
Since IgM surface levels correlated with level of apoptosis in primary GC B cells, we further investigated the role of CD79A and CD79B for IgM expression and survival using CRISPR/Cas9 in cell line models. After CD79A-CRISPR editing in three IgM+ MCL cell lines (MINO, Z-138, Granta-519) and a GCB DLBCL line (SUDHL-6), the cells that lost surface CD79A expression also lost surface IgM and CD79B expression (Fig. 3A and Supplemental Fig. 2A, 2B). We did not observe CD79A-negative cells with CD79B expression as reported in previous studies (31). The CD79A-edited cultures were immunophenotyped regularly for up to 24 days and in most cell lines, the triple negative population was outcompeted by unedited original cells (Fig. 3B, 3C). In MINO and SUDHL-6, the CD79A−IgM− population rapidly declined, whereas Z-138 displayed a slower reduction of the edited population. CRISPR/Cas-9 editing of CD79B also led to a triple negative population that was rapidly outcompeted in MINO, SUDHL-6 and Z-138 (Fig. 3D–3F, Supplemental Fig. 2A, 2B). We also observed a CD79B−IgM+ population that remained stable over time (Fig. 3D, 3E, 3G). We generated single cell clones from this population and CD79B−IgM+ MINO clones (C7 and C9) had a homozygous 6 base-pair deletion in CD79B (Supplemental Fig. 2C), which did not reduce the total levels of CD79B or IgM protein (Supplemental Fig. 2D–2F, 2H). Z-138 clones (B and D) with 6 base-pair deletion in one allele and a 1 base-pair deletion in the other had reduced levels of CD79B and slightly lower IgM (Supplemental Fig. 2G, 2H). As CD79B protein was still present, we did not further investigate these clones and focused on CD79A−CD79B−IgM− triple negative cells.
Figure 3. Loss of CD79A or CD79B cause loss of IgM surface expression and can confer reduced B cell fitness.
Cas9-expressing B-cell lines were transfected with CD79A sgRNA (A-C) or CD79B sgRNA (D-G) and phenotyped from day 3 and up to three weeks. Live singlets are shown in all dot plots. A: Representative plots of Cas9 control cells and CD79A-edited cells at day 3 after transduction. Gates are drawn on CD79A−IgM− cells. B: CD79A-edited MINO cells analyzed at day 3–13 after transduction. C: Percentage of surface CD79A−IgM− cells from CD79A-edited cultures. D: Representative plots of Cas9 control cells and CD79B-edited cells at day 3 after transfection. Gates are drawn on CD79B−IgM− cells (lower gate) and CD79B−IgM+ cells (upper gate). E: CD79B-edited MINO cells analyzed at day 3–13 after transfection. F-G: Percentage of surface CD79B−IgM− cells and surface CD79B−IgM+ from CD79B-edited cultures. Mean ± SD; MINO: n = 4; SUDHL-6, Z-138 and Granta-519: n = 3. H-K: CD79B−IgM− cells and CD79B+IgM+ cells were sorted three days after transfection and cultured for up to 10 days. Too few CD79B-edited cells survived until day 10 and could therefore not be phenotyped after day 5. Percentage of live cells based on FSC and SSC is shown (H). 24 hours after sorting, cells were stained with a live/dead marker (I) before fix/perm and staining with anti-cleaved caspase-3 (J) and anti-p-GSK3β antibodies (K). Graphs show percentage of dead cells out of all singlets (I), percentage of apoptotic cells gated as cleaved caspase-3+ cells out of live singlets (J), and phospho-GSK3β relative to control cells in live, non-apoptotic cells (K). Representative plots shown for CD79B sgRNA-edited cells (n = 2). CD79A sgRNA-edited cells (n = 2) is plotted in the same graphs and statistics run on both combined (I-K). * indicates significance in a t-test between CD79B+IgM+ cells and CD79B−IgM− cells. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To determine if the CD79A−CD79B−IgM− triple negative cells were outcompeted or died by apoptosis, we sorted CD79B−IgM− and CD79B+IgM+ MINO cells 3 days after CD79A/CD79B-CRISPR editing and cultured the two populations separately. More than 90% of the sorted CD79A sgRNA-edited cells and >80% of the CD79B sgRNA-edited cells maintained a triple negative phenotype (on day 10 and day 5, respectively; data not shown). The sorting selected for live cells and directly after the sort, the viability was equal in CD79B−IgM− and CD79B+IgM+ MINO cells (Fig. 3H). During culturing, the viability became significantly lower in CD79B−IgM− compared to CD79B+IgM+ cells. The day after sorting, we detected high cell death and apoptosis in the CD79B−IgM− cells. We also observed lower p-GSK3β in CD79B−IgM− cells compared to CD79B+IgM+ and Cas9 control cells. Reduced p-GSK3β has previously been associated with reduced fitness in lymphoma cells (30).
The survival and fitness of Granta-519 cells were not affected by loss of CD79A or CD79B and surface IgM as the frequency of edited cells remained unchanged after 24 days in culture (Fig. 3C–3F). Unlike the other cell lines, Granta-519 is EBV-positive and may express EBV genes such as LMP2A which provides a constitutive surrogate for BCR signaling using LYN and SYK kinases (51) or LMP1 that activates TRAF2,3 and the NF-κB pathway, rendering these cells less dependent on tonic BCR signaling. This might also explain the wide range of IgM surface expression in original Granta-519 cells. These cells displayed a large population of IgM− cells prior to CRISPR/Cas9-editing, contrasting the other cells lines displaying mostly surface IgM+/high cells (Fig. 3A).
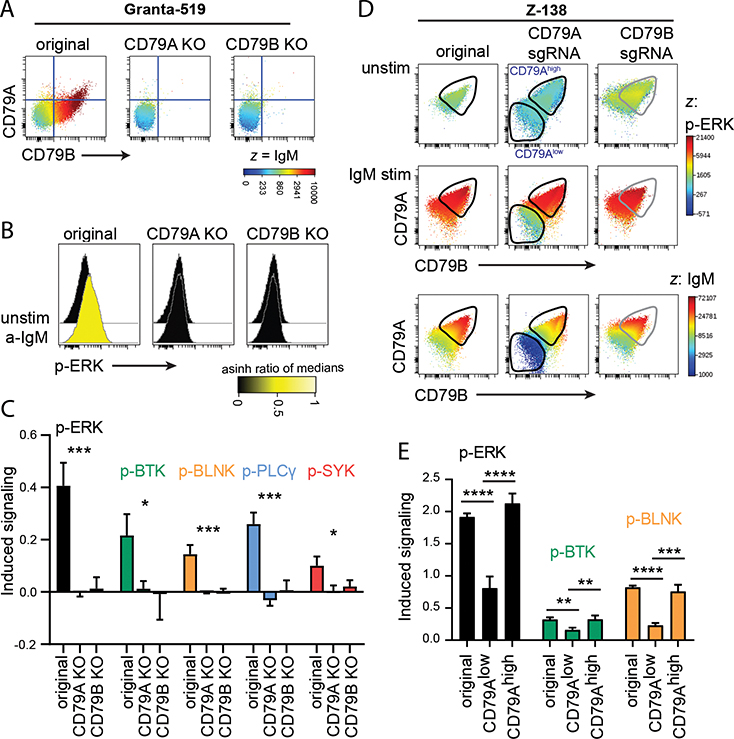
BCR-induced signaling is lost in CD79A KO and CD79B KO cells
Stable CD79A and CD79B KO clones were established from the cultures of edited Granta-519 cells (Supplemental Fig. 2C). The clones maintained a triple negative phenotype, lacking surface expression of CD79A, CD79B and IgM (Fig. 4A). To confirm that the clones had no functional BCR, we compared anti-IgM-induced signaling in KO cells to the original Granta-519 cells and did not detect any BCR signaling in the KO cells (Fig. 4 B, 4C).
Figure 4: B cells lacking surface BCR are incapable of BCR-induced signaling.
A. Surface expression of CD79A, CD79B and IgM is shown in Granta-519 original, CD79A KO (clone II-G6) and CD79B KO (clone E4). B-C. Granta-519 original, CD79A KO and CD79B KO clones were stimulated with anti-IgM F(ab’)2. Phosphorylation was measured by intracellular phospho-flow. One representative experiment (B) or phosphorylation of several proteins (C).
D-E. Z-138 was transfected with CD79A or CD79B sgRNA. Cells were frozen after 10 days, then thawed and cultured for 9 days. The original cell line and the two sgRNA-transfected cell lines were stimulated with FITC-labeled anti-IgM F(ab’)2 for simultaneous induction of BCR signaling and detection of IgM surface level. Phosphorylation and intracellular levels of CD79A and CD79B were measured by phospho-flow. D: Unstimulated and stimulated cells are plotted and gated on CD79A+ unedited cells and CD79Alow edited cells. CD79B-edited cells are shifted towards CD79Blow cells, however, the small population of CD79B−IgM− cells are not distinguished from CD79B−IgM+ cells that signal as original cells. E: Phosphorylation of several proteins in original Z-138 and CD79A-edited cells gated on CD79A+ unedited cells and CD79Alow edited cells. Values are shown relative to unstimulated cells, using arcsinh transformed data of median. Mean ± SD, n = 3. *indicates significance after ANOVA (C) with Tukey’s multiple comparison test (E). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
In Z-138 cells that had undergone CRISPR/Cas-9-editing of CD79A or CD79B, IgM-induced signaling was measured in bulk populations, using antibodies against CD79A, CD79B and IgM to identify original and edited cells (Fig. 4D). In CD79Alow cells, reduced BCR signaling and low/no IgM expression were observed, confirming the loss of a functional BCR in the edited cells (Fig. 4D, 4E). The population of CD79B−IgM− negative cells in CD79B-edited cells was lost at the time point when these experiments were performed, preventing similar analysis of BCR signaling in these cells.
Loss of CD79A or CD79B interferes with protein maturation
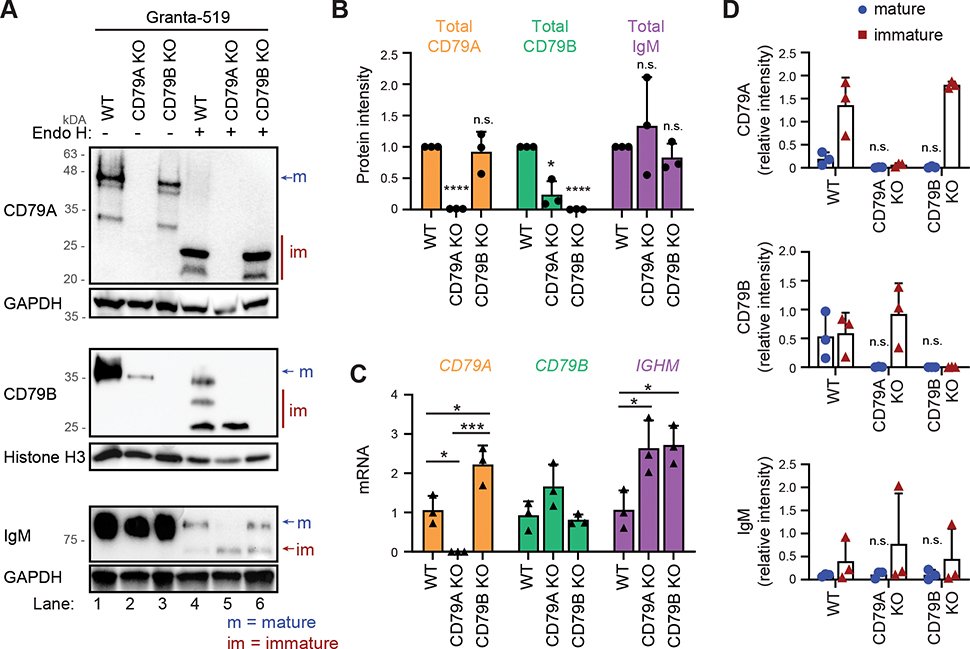
To investigate the mechanisms underlying loss of surface BCR expression upon deletion of CD79A or CD79B, we investigated gene transcription and protein expression in original cells and CRISPR-edited cells (Fig. 5, Supplemental Fig. 3). Protein lysates from Granta-519 CD79A KO cells did not express CD79A protein (Fig. 5A lane 2, 5B) and CD79A mRNA was not detected in these cells (Fig. 5C). CD79B KO cells did not express CD79B (Fig. 5A lane 3, 5B), but CD79B mRNA was detected at the same level as in original cells (Fig. 5C). The total protein level of IgM was not altered in the two KO models (Fig. 5A lane 1–3, 5B), but IGHM mRNA was increased, possibly through a compensatory mechanism (Fig. 5C). In contrast, the total protein level of CD79B was significantly reduced in CD79A KO cells (Fig. 5A lane 2, 5B) although the CD79B mRNA expression level was not altered (Fig. 5C). This suggests that the protein stability of CD79B depends on CD79A. CD79A mRNA was, similarly to IGHM, increased in CD79B KO cells (Fig. 5C), but total CD79A protein was not altered (Fig. 5A lane 3, 5B).
Figure 5: Deletion of CD79A blocks the maturation of CD79B and vice versa in Granta-519.
A: Protein lysates from original Granta-519 cells, CD79A KO (clone II-G6) and CD79B KO (clone E4) were left untreated or treated with Endo H for 1 hour before western blot analysis. One representative of three blots is shown and all blots are shown in Supplemental Fig. 3A. B: The intensity of the bands in lane 1–3 (total protein level) were quantified and plotted relative to total protein expression in original cells (lane 1). Mean ± SD (n = 3). * indicates significance after one sample t-test comparing sample means of CD79A KO and CD79B KO to 1. C: mRNA level of CD79A, CD79B and IGHM in original Granta-519 cells, CD79A KO and CD79B KO were measured by real-time RT PCR. Mean ± SD (n = 3). * indicates significance after ANOVA and Tukey’s multiple comparison test. D: The intensity of the mature and immature fraction of the bands in lane 4–6 of CD79A, CD79B and IgM were quantified and plotted relative to protein expression in original cells (lane 1). Mean ± SD (n = 3). *indicates significance after ANOVA and Dunnet’s multiple comparison test of mature fraction in CD79A KO and CD79B KO against original. *p < 0.05, ***p < 0.001, ****p < 0.0001
The loss of surface BCR in CD79A/CD79B KO cells could be due to a block in protein maturation and transport of proteins from the ER to the surface. To test this in our KO models, we used the enzyme endoglycosidase H (Endo H) to cleave off N-linked glycans from immature proteins. The complex N-linked glycans of mature protein is Endo H resistant, whereas immature protein will have the glycans cleaved off upon Endo H treatment. Western blot analysis can then be applied to separate the longer mature protein from the shorter immature protein (40). Whereas original Granta-519 cells expressed both mature and immature fraction of CD79B (Fig. 5A lane 4, 5D), CD79A KO cells only expressed the immature fraction of CD79B (Fig. 5A lane 5, 5D). The mature fraction of CD79A is barely detectable in Granta-519 (Fig. 5A lane 4), but the weak band of mature CD79A is missing in CD79B KO cells (Fig. 5A lane, 5D), indicating that CD79A protein cannot mature without CD79B and vice versa. Original Granta-519 cells had variable and low level of mature IgM protein (Fig. 5A lane 4, 5D), in agreement with the low fraction of cells with surface expression observed by flow cytometry (Fig. 3A). We did not observe an effect on IgM protein maturation in Granta-519 CD79A KO and CD79B KO cells (Fig. 5A lane 4–6, 5D).
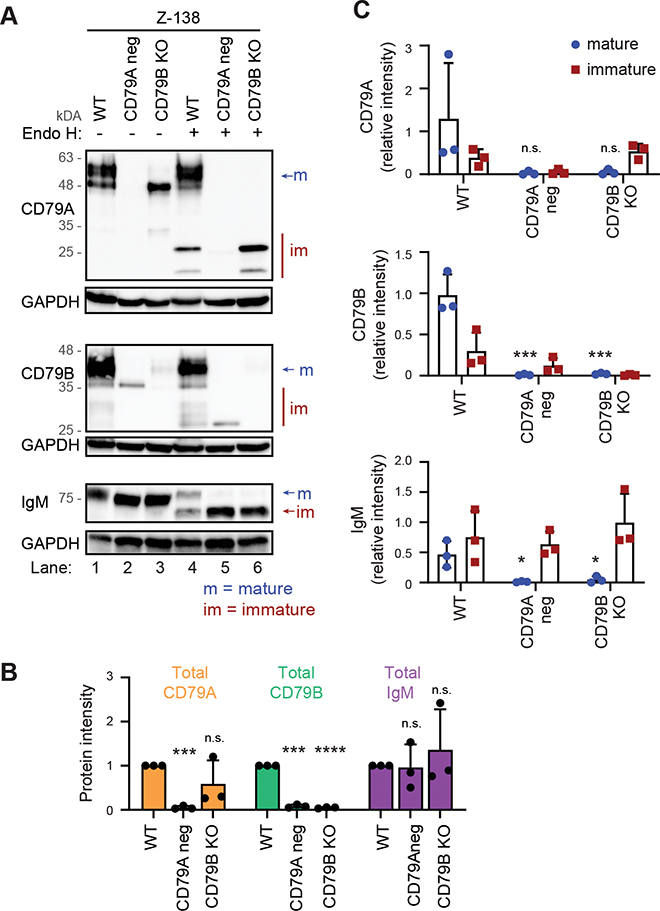
Due to the low surface expression and mature fraction of IgM in Granta-519, we used Z-138 as a second model system. We were able to obtain a stable Z-138 CD79B KO clone whereas CD79A KO cells did not survive in culture. Instead, CD79A-negative cells were sorted from CD79A-edited cultures before cell lysis. CD79A protein was lost in CD79A-negative cells (Fig. 6A lane 2) and CD79B protein was lost in CD79B KO cells (Fig. 6A lane 3), whereas total IgM protein was not altered by CD79A/CD79B KO (Fig. 6A lane 1–3, 6B). As in Granta-519, the total protein level of CD79B was significantly reduced in CD79A-negative cells whereas the total protein level of CD79A was less affected in CD79B KO cells (Fig. 6A lane 1–3, 6B). Original Z-138 cells expressed both mature and immature fractions of IgM, but the mature fraction of IgM was lost in CD79A-negative cells and in CD79B KO cells (Fig. 6A lane 4–6, 6C). Similarly, the mature fraction of CD79A was lost in CD79B KO cells and the mature fraction of CD79B was lost in CD79A negative cells (Fig. 6A lane 4–6, 6C). These results support a model where retention of immature CD79A/CD79B and IgM in the ER is the main mechanism for reduced surface BCR expression in CD79A/CD79B KO cells.
Figure 6. Deletion of CD79A or CD79B blocks the maturation of CD79A, CD79B and IgM in Z-138.
Protein lysates from original Z-138 cells, FACS-sorted CD79A-negative and CD79B KO cells (clone B3) were left untreated or treated without Endo H for 1 hour before western blot analysis. A One representative of three blots. All blots are shown in Supplemental Fig. 3B. B-C The intensity of the bands in lane 1–3 (total protein level; B) and the mature and immature fraction (lane 4–6; C) of CD79A, CD79B and IgM were quantified and plotted relative to total protein expression in original cells (lane 1). Mean ± SD (n = 3). *in B indicates significance after one sample t-test comparing sample means of CD79A KO and CD79B KO to 1. * in C indicates significance after ANOVA and Dunnet’s multiple comparison test of mature fraction in CD79A negative and CD79B KO against original. *p < 0.05, ***p < 0.001, ****p < 0.0001
CD79A and IgM protein maturation and surface expression in CD79B KO cells can be rescued by reintroducing CD79B WT
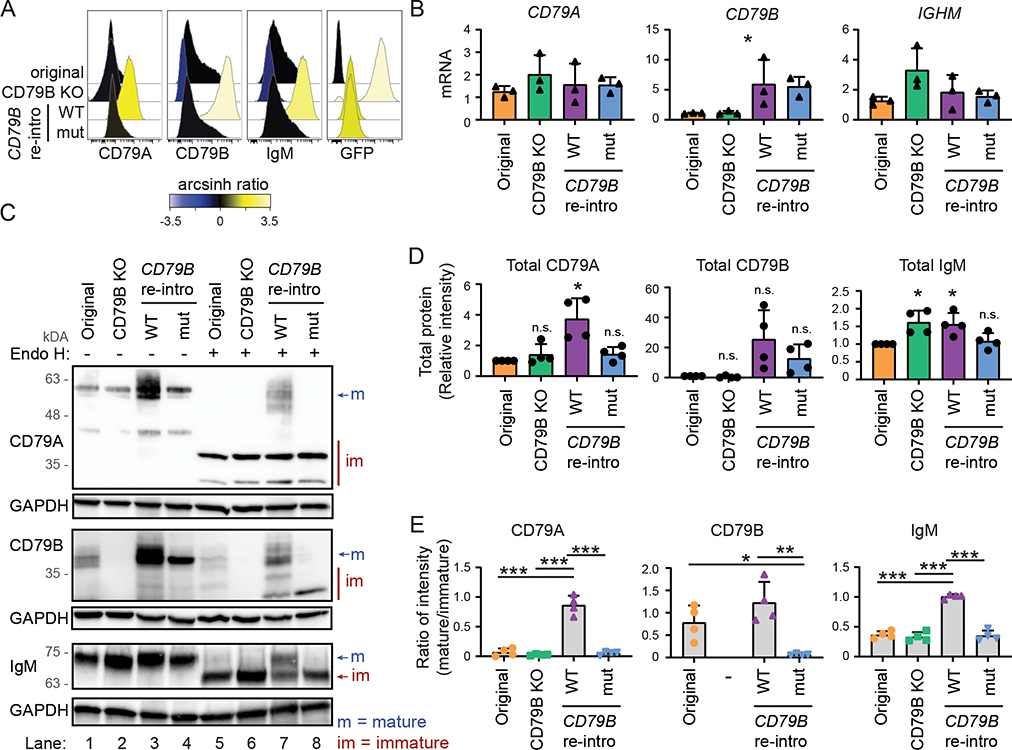
We next performed a rescue experiment with reintroduction of wild type CD79B or overexpression of a validated dominant negative mutant, G137S (12), in Granta-519 CD79B KO cells. The G137S mutation is next to the cysteine required for the disulfide bond with CD79A (C136) and was detected in a patient with agammaglobulinemia where the mutation resulted in inefficient heterodimerization of the CD79 molecules, low IgM expression and a strong reduction of mature peripheral blood B cells (12). In order to assess equivalent production of the rescue proteins, we linked CD79B to GFP using a 2A ribosome skipping peptide (47). Reintroduction of wild type CD79B resulted in strong surface expression of CD79A, CD79B and IgM, contrasting CD79B G137S which led to low surface expression of the three BCR components (Fig. 7A). Reintroduction of CD79B variants with point mutations of Y196 led to even higher surface expression of CD79A, CD79B and IgM, both when comparing the total GFP+ population (Supplemental Fig. 4A, 4B) and cells with equivalent GFP levels (Supplemental Fig. 4C), consistent with previous observations (16, 50). Importantly, the low surface expression levels observed with CD79B G137S was not due to lower retroviral transduction efficiency since all CD79B constructs showed similar GFP expression level (Fig. 7A). In addition, CD79B mRNA expression levels were similar (Fig. 7B), whereas the total level of CD79B protein was higher for CD79B WT than CD79B G137S (Fig. 7C lane 3–4), suggesting that this mutant has reduced protein stability. However, the most striking difference was in protein maturation of CD79B (Fig. 7E, Supplemental Fig. 4D). Whereas cells with wild type CD79B expressed both mature and immature CD79B at the same ratio as original Granta-519 cells, the cells expressing CD79B G137S only expressed the immature fraction of CD79B (Fig. 7C lane 5, 7 and 8, 7E). CD79A mRNA expression was not affected by reintroduction of CD79B (Fig. 7B), but the total protein level of CD79A was increased (Fig. 7C lane 3, 7D) and the mature fraction was strongly increased in cells with reintroduced CD79B WT (Fig. 7C lane 7, 7E). Similarly, IGHM mRNA expression was not affected (Fig. 7B), but a strong skewing towards more mature IgM protein (Fig. 7C lane 7, 7E) and a small increase in IgM total protein was observed after reintroduction of CD79B WT (Fig. 7C lane 3, 7D).
Figure 7: BCR surface expression and protein maturation in CD79B KO cells can be rescued by reintroducing CD79B.
Wild type CD79B and mutant CD79B G137S were re-introduced into Granta-519 CD79B KO cells (clone E4) by retroviral transduction. A: Surface protein expression was measured by flow cytometry. One representative out of three experiments is shown. All blots are shown in Supplemental Fig. 4D. B: mRNA level of CD79A, CD79B and IGHM were measured by real-time RT PCR. Mean ± SD (n = 3). *indicates significance after ANOVA, Tukey’s multiple comparison test did not show significance. C: Protein lysates were left untreated or treated with Endo H for 1 hour before western blot analysis. One representative out of three experiments is shown. D: The intensity of the bands in lane 1–4 (total protein level) were quantified and plotted relative to total protein expression in original cells (lane 1). Mean ± SD, (n = 3). * indicates significance after one-sample t-test comparing sample means of CD79B KO, reintroduction of CD79B WT or CD79B G137S to 1. E: The intensity of the mature and immature fraction of the bands in lane 5–8 of CD79A, CD79B and IgM were quantified and plotted as ratio between mature and immature fraction. Mean ± SD, (n = 4). *indicates significance after ANOVA and Tukey’s multiple comparison test *p < 0.05, **p < 0.01, ***p < 0.001.
Collectively, these data demonstrate that CD79B protein governs the maturation and surface expression of both CD79A and IgM. The model that emerges from these results is that heterodimerization of CD79A and CD79B is required for proper protein maturation.
Discussion
The requirement for CD79A and CD79B for BCR expression and B-cell fitness is well established in mice, but few studies have investigated this using human B cells. Prior CRISPR/Cas9 screens in human DLBCL cell lines identified CD79A and CD79B as essential but did not investigate consequences for BCR assembly and trafficking to the cell surface (21–23). In human MCL cell lines, we herein demonstrated that surface expression and glycan maturation of IgM and CD79 are lacking after CD79A or CD79B KO, indicating retention of BCR complex components in the ER. The loss of surface IgM had important consequences for B-cell survival. In three out of four cell lines, CRISPR/Cas9 editing of either CD79 gene reduced the fitness relative to original cells and the edited cells were outcompeted by non-sgRNA-transfected cells over time. Cell sorting of MINO cells that had lost surface expression of IgM after CD79A or CD79B editing demonstrated that these cells rapidly underwent apoptosis. The consequence of IgM surface level for survival was further shown in primary human GC B cells as high surface density of IgM protected against spontaneous- and FasL-induced apoptosis.
Loss of BCR surface expression following CD79 KO was previously shown in a screen using shRNA in DLBCL cell lines where the degree of CD79A or CD79B knockdown correlated with the level of surface BCR (16). Assembly of the BCR complex occurs in the ER (36). Without dimerization, CD79A and CD79B are blocked from transport to the Golgi apparatus and this mechanism depends on conserved motifs in the transmembrane domain that contains charged amino acids (34, 52). The glycosylation pattern of CD79A, CD79B and IgM in CD79A or CD79B KO cells and in CD79B G137S-reconstituted cells in our study fit with a model where these proteins are retained in the ER when CD79A/CD79B heterodimer formation is not possible (40). Reintroduction of wild type as well as CD79B Y196 variants in Granta-519 CD79B KO cells led to high surface expression of CD79A and IgM, suggesting that CD79B is the rate-limiting factor for the level of surface BCR expression in these cells. The same conclusion has previously been drawn for normal mouse B cells (50). The BCR surface expression in Granta-519 was not regulated at the transcriptional level since reintroduction of CD79B WT increased the surface BCR expression without increasing CD79A and IGHM mRNA level and high levels of mutated CD79B G137S mRNA did not affect CD79A or IGHM mRNA levels. Instead, pools of immature CD79A and IgM protein were matured as the level of mature CD79B increased, shown by the skewing in the mature/immature ratio of CD79A and IgM. In primary mouse B cells, which do not have a large pool of immature IgM, overexpression of CD79B WT led to a small increase in surface BCR expression and a large increase in immature CD79B protein (50). The increased CD79A and IgM maturation in Granta-519 depended on CD79A/CD79B heterodimerization as the CD79B G137S variant did not rescue the protein maturation of CD79A and IgM. In addition, the total protein level of CD79A and IgM increased in cells overexpressing CD79B, suggesting that mature CD79B stabilizes the other two proteins and prevents protein degradation, in agreement with earlier reports (37). In contrast, CD79A was suggested to be the rate-liming factor for BCR expression in the Burkitt lymphoma cell line Ramos (36). Using the same cell line, He et al. showed that CD79B can be expressed as homodimers on the surface in the absence of CD79A and IgM (31). Crystal structures of human and murine CD79B suggest that the protein can form homodimers (33). However, this was not seen in the three MCL cell lines and the GCB DLBCL cell line used in our study, as CD79B was not expressed on the surface of CD79A KO cells. The mechanism for how CD79B can exist as homodimers and be expressed at the plasma membrane in the absence of other BCR components in Burkitt lymphoma but not in DLBCL and MCL, remains unknown.
Glycosylation defects and retention of IgM in the ER is a mechanism for reduced BCR expression in CLL (53) which can be rescued by IL-4 (54, 55). Furthermore, surface IgM in CLL cells can differ in the N-glycosylation pattern in the Igμ constant region; one being similar to normal B cells with mature complex glycans while the other form is high in mannose, characteristic of immature Igμ chains in the ER (56). Similar to CLL cells, tolerant B cells express normal levels of IGHM mRNA and surface IgD, but IgM is selectively blocked in the ER after repeatedly stimulation by self-antigen (40). B cells expressing mutated MYD88 downregulate their IgM similarly to the trafficking block of IgM induced by chronic exposure to self-antigen (50). This can be overcome by CD79B Y196 ITAM mutations causing elevated surface BCR expression (50). Co-occuring mutations in CD79B and MYD88 is characteristic of the genetic subtype MCD/C5 of DLBCL (57, 58), demonstrating that malignant B cells can acquire mutations to escape IgM retention or maturation block that would normally cause low BCR expression.
CD79A or CD79B KO reduced the fitness of MCL cell lines MINO and Z-138. MINO was rapidly outcompeted by unedited cells due to rapid induction of apoptosis upon loss of surface IgM, whereas Z-138 had a slower decline. It has previously been shown that some MCL cell lines, including MINO, are sensitive to BTK-inhibitors due to constitutive BCR activation and canonical NF-κB signaling (26). Z-138 represent another subset of MCL cell lines that are insensitive to BTK-inhibitors and demonstrate activation of the non-canonical NF-κB pathway (26). This difference in NF-κB pathway activation can explain why Z-138 is less sensitive to loss of surface BCR expression. Granta-519 was resistent to BCR loss, likely due to EBV (51), showing that lymphoma cells can replace the survival signal that should arise from a functional BCR. In Ramos cells, CD79B formed homodimers in the absence of other BCR complex components and constituted an alternative survival pathway (31). In DLBCL cell lines, KO of the Ig heavy chain reduced the fitness of DLBCL cell lines, but the decline of the cells varied greatly (15). Reduced proliferation was the cause of the decline in most cells, but apoptosis was also seen in some cells. Collectively, this demonstrates that malignant B cells can acquire mutations or express viral proteins that provides better fitness, compared to healthy B cells.
In some lymphoma cells, IgM expression has been shown to have other roles that providing survival signals. In Ramos, the production of IgM was crucial for the metabolic fitness of B cells (59). Without expression of immunoglubulins, ER homeostasis was not maintained and the cells died (59). In a myc-driven mouse lymphoma model, BCR depletion did not affect lymphoma growth, but BCR− cells with reduced p-GSK3β were outcompeted by their BCR+ counterparts (30). GSK3 is a metabolic sensor downstream of BCR signaling. The kinase promotes the survival of naïve cells by restricting cell mass accumulation, but is inhibited by phosphorylation in GC B cells to allow for growth, proliferation and increased metabolic activity (60). The BCR− cells could be rescued by GSK3β inhibition, showing that BCR-induced inhibition of GSK3β was essential for metabolic fitness when nutrients were scarce. BCR− MINO cells displayed reduced p-GSK3β, but they also had increased cell death.
Emerging evidence demonstrate that also healthy B cells depend on surface BCR for survival and metabolic fitness. We found that high BCR expression protected against apoptosis in GC B cells. Consistent with this, it has been shown in both mouse and human that BCRlow cells have reduced signaling compared to BCRhigh cells (9, 61). All GC B cells, independent of BCR expression level, displayed increased apoptosis compared to naïve and memory B cells. We observed more than 50% apoptosis after 3 hours in culture, consistent with a study showing that 50% of GC B cells die within six hours in vivo (62). In the GC reaction, B cells go through rounds of somatic hypermutation in the dark zone to increase the affinity of the BCR. Downregulation of the BCR is important in this step as the GC B cell need to re-express and test the affinity of their newly mutated BCR. A block in CD79B downregulation reduces the number of GC B cells and affinity maturation in transgenic mice (63). This downregulation of the BCR explains the large percentage of GC B cells lacking surface BCR expression in our study. A functional BCR must be re-expressed on the surface for dark zone B cells to reach the light zone, and dark zone B cells with damaging mutations in the BCR do not reach the light zone (64). Light zone B cells are programmed to die unless rescued by T cells whereas dark zone cells depend on tonic signaling as apoptotic cells in this zone are highly enriched for Ig genes with deleterious mutations lacking surface BCR (62). Yet, the role of antigen-induced BCR signaling in GC B cells is not fully understood. It has been suggested BCR signaling is suppressed in GC B cells and that T-cell help is the main selective mechanism in the germinal center (65). In our approach, detecting maximal BCR-induced signaling capacity, we observed similar BCR signaling capacity in human GC B cells compared to naïve and memory B cells. Newer studies suggest that BCR signals do play an important role during the germinal center reaction and that the antigen-induced BCR signaling is rewired to inactivate FOXO1 through PI3K-AKT in GC B cells (66).
Here, combining CRISPR/Cas9 deletion of CD79A or CD79B with studies of glycan maturation in MCL cell lines gave mechanistic insight in protein maturation and BCR complex assembly in human B cells. Our study highlights the central role of the survival signal provided by BCR-signaling for primary human GC B cells and in EBV negative MCL cell lines. Activation through the BCR guides decisions about life, death, and B-cell function, and our results showed that the IgM surface level contributes to these decisions. The presence of IgM on the surface of these B cells equally depended on CD79A and CD79B, and if one of these proteins was deleted, it blocked glycan maturation of the other CD79 protein as well as IgM, with subsequent blocked transportation of the IgM complex to the plasma membrane. We have confirmed the requirement of the CD79A/CD79B heterodimer for surface expression of IgM in human B cells as previously comprehensively demonstrated in mice.
Supplementary Material
Key points.
High surface expression of IgM protects against apoptosis in human GC B cells
CD79A/B depletion resulted in loss of IgM surface expression and reduced fitness
Glycan maturation of BCR complex components was interrupted upon CD79A or CD79B KO
Acknowledgement
We are grateful for services provided by the Flow Cytometry Core Facility at Oslo University Hospital.
Funding:
This work was supported by the Norwegian Cancer Society (16294- Career Development Grant (KH), 163260 (JHM), and 182694 (EBS)), South-Eastern Norway Regional Health Authority (2014075, EBS), the Foundation KG Jebsen (MED019, EBS and JHM), and National Institutes of Health (R01 CA226833, JMI).
References
- 1.Sanchez M, Misulovin Z, Burkhardt AL, Mahajan S, Costa T, Franke R, Bolen JB, and Nussenzweig M. 1993. Signal transduction by immunoglobulin is mediated through Ig alpha and Ig beta. J Exp Med 178: 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pelanda R, Braun U, Hobeika E, Nussenzweig MC, and Reth M. 2002. B Cell Progenitors Are Arrested in Maturation but Have Intact VDJ Recombination in the Absence of Ig-α and Ig-β. The Journal of Immunology 169: 865–872. [DOI] [PubMed] [Google Scholar]
- 3.Kitamura D, Roes J, Kühn R, and Rajewsky K. 1991. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350: 423–426. [DOI] [PubMed] [Google Scholar]
- 4.Gong S, and Nussenzweig MC. 1996. Regulation of an early developmental checkpoint in the B cell pathway by Ig beta. Science 272: 411–414. [DOI] [PubMed] [Google Scholar]
- 5.Gazumyan A, Reichlin A, and Nussenzweig MC. 2006. Ig beta tyrosine residues contribute to the control of B cell receptor signaling by regulating receptor internalization. J Exp Med 203: 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lam KP, Kühn R, and Rajewsky K. 1997. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell 90: 1073–1083. [DOI] [PubMed] [Google Scholar]
- 7.Kraus M, Alimzhanov MB, Rajewsky N, and Rajewsky K. 2004. Survival of Resting Mature B Lymphocytes Depends on BCR Signaling via the Igα/β Heterodimer. Cell 117: 787–800. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, and Rajewsky K. 2009. PI3 Kinase Signals BCR-Dependent Mature B Cell Survival. Cell 139: 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yasuda S, Zhou Y, Wang Y, Yamamura M, and Wang JY. 2017. A model integrating tonic and antigen-triggered BCR signals to predict the survival of primary B cells. Sci Rep 7: 14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yel L, Minegishi Y, Coustan-Smith E, Buckley RH, Trübel H, Pachman LM, Kitchingman GR, Campana D, Rohrer J, and Conley ME. 1996. Mutations in the mu heavy-chain gene in patients with agammaglobulinemia. N Engl J Med 335: 1486–1493. [DOI] [PubMed] [Google Scholar]
- 11.Minegishi Y, Coustan-Smith E, Rapalus L, Ersoy F, Campana D, and Conley ME. 1999. Mutations in Igalpha (CD79a) result in a complete block in B-cell development. J Clin Invest 104: 1115–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobbs AK, Yang T, Farmer D, Kager L, Parolini O, and Conley ME. 2007. Cutting edge: a hypomorphic mutation in Igbeta (CD79b) in a patient with immunodeficiency and a leaky defect in B cell development. J Immunol 179: 2055–2059. [DOI] [PubMed] [Google Scholar]
- 13.Ferrari S, Lougaris V, Caraffi S, Zuntini R, Yang J, Soresina A, Meini A, Cazzola G, Rossi C, Reth M, and Plebani A. 2007. Mutations of the Igbeta gene cause agammaglobulinemia in man. J Exp Med 204: 2047–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuppers R 2005. Mechanisms of B-cell lymphoma pathogenesis. Nat Rev Cancer 5: 251–262. [DOI] [PubMed] [Google Scholar]
- 15.Havranek O, Xu J, Köhrer S, Wang Z, Becker L, Comer JM, Henderson J, Ma W, Man Chun Ma J, Westin JR, Ghosh D, Shinners N, Sun L, Yi AF, Karri AR, Burger JA, Zal T, and Davis RE. 2017. Tonic B-cell receptor signaling in diffuse large B-cell lymphoma. Blood 130: 995–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, Xu W, Shaffer AL, Wright G, Xiao W, Powell J, Jiang JK, Thomas CJ, Rosenwald A, Ott G, Muller-Hermelink HK, Gascoyne RD, Connors JM, Johnson NA, Rimsza LM, Campo E, Jaffe ES, Wilson WH, Delabie J, Smeland EB, Fisher RI, Braziel RM, Tubbs RR, Cook JR, Weisenburger DD, Chan WC, Pierce SK, and Staudt LM. 2010. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 463: 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young RM, Wu T, Schmitz R, Dawood M, Xiao W, Phelan JD, Xu W, Menard L, Meffre E, Chan WC, Jaffe ES, Gascoyne RD, Campo E, Rosenwald A, Ott G, Delabie J, Rimsza LM, and Staudt LM. 2015. Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc Natl Acad Sci U S A 112: 13447–13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevenson FK, Krysov S, Davies AJ, Steele AJ, and Packham G. 2011. B-cell receptor signaling in chronic lymphocytic leukemia. Blood 118: 4313–4320. [DOI] [PubMed] [Google Scholar]
- 19.Coelho V, Krysov S, Ghaemmaghami AM, Emara M, Potter KN, Johnson P, Packham G, Martinez-Pomares L, and Stevenson FK. 2010. Glycosylation of surface Ig creates a functional bridge between human follicular lymphoma and microenvironmental lectins. Proceedings of the National Academy of Sciences 107: 18587–18592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dühren-von Minden M, Übelhart R, Schneider D, Wossning T, Bach MP, Buchner M, Hofmann D, Surova E, Follo M, Köhler F, Wardemann H, Zirlik K, Veelken H, and Jumaa H. 2012. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature 489: 309–312. [DOI] [PubMed] [Google Scholar]
- 21.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, Leppa S, Pasanen A, Meriranta L, Karjalainen-Lindsberg ML, Nørgaard P, Pedersen M, Gang AO, Høgdall E, Heavican TB, Lone W, Iqbal J, Qin Q, Li G, Kim SY, Healy J, Richards KL, Fedoriw Y, Bernal-Mizrachi L, Koff JL, Staton AD, Flowers CR, Paltiel O, Goldschmidt N, Calaminici M, Clear A, Gribben J, Nguyen E, Czader MB, Ondrejka SL, Collie A, Hsi ED, Tse E, Au-Yeung RKH, Kwong YL, Srivastava G, Choi WWL, Evens AM, Pilichowska M, Sengar M, Reddy N, Li S, Chadburn A, Gordon LI, Jaffe ES, Levy S, Rempel R, Tzeng T, Happ LE, Dave T, Rajagopalan D, Datta J, Dunson DB, and Dave SS. 2017. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 171: 481–494.e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phelan JD, Young RM, Webster DE, Roulland S, Wright GW, Kasbekar M, Shaffer AL 3rd, Ceribelli M, Wang JQ, Schmitz R, Nakagawa M, Bachy E, Huang DW, Ji Y, Chen L, Yang Y, Zhao H, Yu X, Xu W, Palisoc MM, Valadez RR, Davies-Hill T, Wilson WH, Chan WC, Jaffe ES, Gascoyne RD, Campo E, Rosenwald A, Ott G, Delabie J, Rimsza LM, Rodriguez FJ, Estephan F, Holdhoff M, Kruhlak MJ, Hewitt SM, Thomas CJ, Pittaluga S, Oellerich T, and Staudt LM. 2018. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 560: 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, Wang JQ, Schmitz R, Morin RD, Tang J, Jiang A, Bagaev A, Plotnikova O, Kotlov N, Johnson CA, Wilson WH, Scott DW, and Staudt LM. 2020. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell 37: 551–568.e514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson WH, Wright GW, Huang DW, Hodkinson B, Balasubramanian S, Fan Y, Vermeulen J, Shreeve M, and Staudt LM. 2021. Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell 39: 1643–1653.e1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saba NS, Liu D, Herman SE, Underbayev C, Tian X, Behrend D, Weniger MA, Skarzynski M, Gyamfi J, Fontan L, Melnick A, Grant C, Roschewski M, Navarro A, Beà S, Pittaluga S, Dunleavy K, Wilson WH, and Wiestner A. 2016. Pathogenic role of B-cell receptor signaling and canonical NF-κB activation in mantle cell lymphoma. Blood 128: 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahal R, Frick M, Romero R, Korn JM, Kridel R, Chan FC, Meissner B, Bhang HE, Ruddy D, Kauffmann A, Farsidjani A, Derti A, Rakiec D, Naylor T, Pfister E, Kovats S, Kim S, Dietze K, Dörken B, Steidl C, Tzankov A, Hummel M, Monahan J, Morrissey MP, Fritsch C, Sellers WR, Cooke VG, Gascoyne RD, Lenz G, and Stegmeier F. 2014. Pharmacological and genomic profiling identifies NF-κB-targeted treatment strategies for mantle cell lymphoma. Nat Med 20: 87–92. [DOI] [PubMed] [Google Scholar]
- 27.Bond DA, and Maddocks KJ. 2020. Current Role and Emerging Evidence for Bruton Tyrosine Kinase Inhibitors in the Treatment of Mantle Cell Lymphoma. Hematol Oncol Clin North Am 34: 903–921. [DOI] [PubMed] [Google Scholar]
- 28.Szydlowski M, Kiliszek P, Sewastianik T, Jablonska E, Bialopiotrowicz E, Gorniak P, Polak A, Markowicz S, Nowak E, Grygorowicz MA, Prochorec-Sobieszek M, Szumera-Cieckiewicz A, Malenda A, Lech-Maranda E, Warzocha K, and Juszczynski P. 2016. FOXO1 activation is an effector of SYK and AKT inhibition in tonic BCR signal-dependent diffuse large B-cell lymphomas. Blood 127: 739–748. [DOI] [PubMed] [Google Scholar]
- 29.Corso J, Pan KT, Walter R, Doebele C, Mohr S, Bohnenberger H, Ströbel P, Lenz C, Slabicki M, Hüllein J, Comoglio F, Rieger MA, Zenz T, Wienands J, Engelke M, Serve H, Urlaub H, and Oellerich T. 2016. Elucidation of tonic and activated B-cell receptor signaling in Burkitt’s lymphoma provides insights into regulation of cell survival. Proc Natl Acad Sci U S A 113: 5688–5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varano G, Raffel S, Sormani M, Zanardi F, Lonardi S, Zasada C, Perucho L, Petrocelli V, Haake A, Lee AK, Bugatti M, Paul U, Van Anken E, Pasqualucci L, Rabadan R, Siebert R, Kempa S, Ponzoni M, Facchetti F, Rajewsky K, and Casola S. 2017. The B-cell receptor controls fitness of MYC-driven lymphoma cells via GSK3β inhibition. Nature 546: 302–306. [DOI] [PubMed] [Google Scholar]
- 31.He X, Kläsener K, Iype JM, Becker M, Maity PC, Cavallari M, Nielsen PJ, Yang J, and Reth M. 2018. Continuous signaling of CD79b and CD19 is required for the fitness of Burkitt lymphoma B cells. Embo j 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegers GM, Yang J, Duerr CU, Nielsen PJ, Reth M, and Schamel WW. 2006. Identification of disulfide bonds in the Ig-alpha/Ig-beta component of the B cell antigen receptor using the Drosophila S2 cell reconstitution system. Int Immunol 18: 1385–1396. [DOI] [PubMed] [Google Scholar]
- 33.Radaev S, Zou Z, Tolar P, Nguyen K, Nguyen A, Krueger PD, Stutzman N, Pierce S, and Sun PD. 2010. Structural and functional studies of Igalphabeta and its assembly with the B cell antigen receptor. Structure 18: 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottwick C, He X, Hofmann A, Vesper N, Reth M, and Yang J. 2019. A symmetric geometry of transmembrane domains inside the B cell antigen receptor complex. Proc Natl Acad Sci U S A 116: 13468–13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tolar P, Sohn HW, and Pierce SK. 2005. The initiation of antigen-induced B cell antigen receptor signaling viewed in living cells by fluorescence resonance energy transfer. Nat Immunol 6: 1168–1176. [DOI] [PubMed] [Google Scholar]
- 36.Brouns GS, de Vries E, and Borst J. 1995. Assembly and intracellular transport of the human B cell antigen receptor complex. Int Immunol 7: 359–368. [DOI] [PubMed] [Google Scholar]
- 37.Foy SP, and Matsuuchi L. 2001. Association of B lymphocyte antigen receptor polypeptides with multiple chaperone proteins. Immunol Lett 78: 149–160. [DOI] [PubMed] [Google Scholar]
- 38.Grupp SA, Mitchell RN, Schreiber KL, McKean DJ, and Abbas AK. 1995. Molecular mechanisms that control expression of the B lymphocyte antigen receptor complex. J Exp Med 181: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Helenius A, and Aebi M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 73: 1019–1049. [DOI] [PubMed] [Google Scholar]
- 40.Bell SE, and Goodnow CC. 1994. A selective defect in IgM antigen receptor synthesis and transport causes loss of cell surface IgM expression on tolerant B lymphocytes. Embo j 13: 816–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Myklebust JH, Brody J, Kohrt HE, Kolstad A, Czerwinski DK, Wälchli S, Green MR, Trøen G, Liestøl K, Beiske K, Houot R, Delabie J, Alizadeh AA, Irish JM, and Levy R. 2017. Distinct patterns of B-cell receptor signaling in non-Hodgkin lymphomas identified by single-cell profiling. Blood 129: 759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai B, Myklebust JH, and Wälchli S. 2020. Gene Editing in B-Lymphoma Cell Lines Using CRISPR/Cas9 Technology. Methods Mol Biol 2115: 445–454. [DOI] [PubMed] [Google Scholar]
- 43.Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, and Vakoc CR. 2015. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33: 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montague TG, Cruz JM, Gagnon JA, Church GM, and Valen E. 2014. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42: W401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brinkman EK, Chen T, Amendola M, and van Steensel B. 2014. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42: e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bollum LK, Huse K, Oksvold MP, Bai B, Hilden VI, Forfang L, Yoon SO, Wälchli S, Smeland EB, and Myklebust JH. 2017. BMP-7 induces apoptosis in human germinal center B cells and is influenced by TGF-β receptor type I ALK5. PLoS One 12: e0177188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inderberg EM, Mensali N, Oksvold MP, Fallang LE, Fane A, Skorstad G, Stenvik GE, Progida C, Bakke O, Kvalheim G, Myklebust JH, and Walchli S. 2018. Human c-SRC kinase (CSK) overexpression makes T cells dummy. Cancer Immunol Immunother 67: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wälchli S, Løset GÅ, Kumari S, Nergård Johansen J, Yang W, Sandlie I, and Olweus J. 2011. A Practical Approach to T-Cell Receptor Cloning and Expression. PLOS ONE 6: e27930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Irish JM, Czerwinski DK, Nolan GP, and Levy R. 2006. Kinetics of B Cell Receptor Signaling in Human B Cell Subsets Mapped by Phosphospecific Flow Cytometry. The Journal of Immunology 177: 1581–1589. [DOI] [PubMed] [Google Scholar]
- 50.Wang JQ, Jeelall YS, Humburg P, Batchelor EL, Kaya SM, Yoo HM, Goodnow CC, and Horikawa K. 2017. Synergistic cooperation and crosstalk between MYD88(L265P) and mutations that dysregulate CD79B and surface IgM. J Exp Med 214: 2759–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fish K, Comoglio F, Shaffer AL 3rd, Ji Y, Pan KT, Scheich S, Oellerich A, Doebele C, Ikeda M, Schaller SJ, Nguyen H, Muppidi J, Wright GW, Urlaub H, Serve H, Staudt LM, Longnecker R, and Oellerich T. 2020. Rewiring of B cell receptor signaling by Epstein-Barr virus LMP2A. Proc Natl Acad Sci U S A 117: 26318–26327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dylke J, Lopes J, Dang-Lawson M, Machtaler S, and Matsuuchi L. 2007. Role of the extracellular and transmembrane domain of Ig-alpha/beta in assembly of the B cell antigen receptor (BCR). Immunol Lett 112: 47–57. [DOI] [PubMed] [Google Scholar]
- 53.Vuillier F, Dumas G, Magnac C, Prevost MC, Lalanne AI, Oppezzo P, Melanitou E, Dighiero G, and Payelle-Brogard B. 2005. Lower levels of surface B-cell-receptor expression in chronic lymphocytic leukemia are associated with glycosylation and folding defects of the mu and CD79a chains. Blood 105: 2933–2940. [DOI] [PubMed] [Google Scholar]
- 54.Guo B, Zhang L, Chiorazzi N, and Rothstein TL. 2016. IL-4 rescues surface IgM expression in chronic lymphocytic leukemia. Blood 128: 553–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aguilar-Hernandez MM, Blunt MD, Dobson R, Yeomans A, Thirdborough S, Larrayoz M, Smith LD, Linley A, Strefford JC, Davies A, Johnson PM, Savelyeva N, Cragg MS, Forconi F, Packham G, Stevenson FK, and Steele AJ. 2016. IL-4 enhances expression and function of surface IgM in CLL cells. Blood 127: 3015–3025. [DOI] [PubMed] [Google Scholar]
- 56.Krysov S, Potter KN, Mockridge CI, Coelho V, Wheatley I, Packham G, and Stevenson FK. 2010. Surface IgM of CLL cells displays unusual glycans indicative of engagement of antigen in vivo. Blood 115: 4198–4205. [DOI] [PubMed] [Google Scholar]
- 57.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, Lawrence MS, Roemer MGM, Li AJ, Ziepert M, Staiger AM, Wala JA, Ducar MD, Leshchiner I, Rheinbay E, Taylor-Weiner A, Coughlin CA, Hess JM, Pedamallu CS, Livitz D, Rosebrock D, Rosenberg M, Tracy AA, Horn H, van Hummelen P, Feldman AL, Link BK, Novak AJ, Cerhan JR, Habermann TM, Siebert R, Rosenwald A, Thorner AR, Meyerson ML, Golub TR, Beroukhim R, Wulf GG, Ott G, Rodig SJ, Monti S, Neuberg DS, Loeffler M, Pfreundschuh M, Trümper L, Getz G, and Shipp MA. 2018. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 24: 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, Roulland S, Kasbekar M, Young RM, Shaffer AL, Hodson DJ, Xiao W, Yu X, Yang Y, Zhao H, Xu W, Liu X, Zhou B, Du W, Chan WC, Jaffe ES, Gascoyne RD, Connors JM, Campo E, Lopez-Guillermo A, Rosenwald A, Ott G, Delabie J, Rimsza LM, Tay Kuang Wei K, Zelenetz AD, Leonard JP, Bartlett NL, Tran B, Shetty J, Zhao Y, Soppet DR, Pittaluga S, Wilson WH, and Staudt LM. 2018. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N Engl J Med 378: 1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jumaa H, Caganova M, McAllister EJ, Hoenig L, He X, Saltukoglu D, Brenker K, Köhler M, Leben R, Hauser AE, Niesner R, Rajewsky K, Reth M, and Jellusova J. 2020. Immunoglobulin expression in the endoplasmic reticulum shapes the metabolic fitness of B lymphocytes. Life Sci Alliance 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jellusova J, Cato MH, Apgar JR, Ramezani-Rad P, Leung CR, Chen C, Richardson AD, Conner EM, Benschop RJ, Woodgett JR, and Rickert RC. 2017. Gsk3 is a metabolic checkpoint regulator in B cells. Nat Immunol 18: 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ashouri JF, and Weiss A. 2017. Endogenous Nur77 Is a Specific Indicator of Antigen Receptor Signaling in Human T and B Cells. J Immunol 198: 657–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mayer CT, Gazumyan A, Kara EE, Gitlin AD, Golijanin J, Viant C, Pai J, Oliveira TY, Wang Q, Escolano A, Medina-Ramirez M, Sanders RW, and Nussenzweig MC. 2017. The microanatomic segregation of selection by apoptosis in the germinal center. Science 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Todo K, Koga O, Nishikawa M, and Hikida M. 2015. Modulation of Igβ is essential for the B cell selection in germinal center. Sci Rep 5: 10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stewart I, Radtke D, Phillips B, McGowan SJ, and Bannard O. 2018. Germinal Center B Cells Replace Their Antigen Receptors in Dark Zones and Fail Light Zone Entry when Immunoglobulin Gene Mutations are Damaging. Immunity 49: 477–489.e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khalil AM, Cambier JC, and Shlomchik MJ. 2012. B Cell Receptor Signal Transduction in the GC Is Short-Circuited by High Phosphatase Activity. Science 336: 1178–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo W, Weisel F, and Shlomchik MJ. 2018. B Cell Receptor and CD40 Signaling Are Rewired for Synergistic Induction of the c-Myc Transcription Factor in Germinal Center B Cells. Immunity 48: 313–326.e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.