Abstract
Objectives:
Analyzing expression patterns of Krüppel-like factor (KLF) transcription factors in normal and osteoarthritis (OA) human cartilage, and determining functions and mechanisms of KLF4 and KLF2 in joint homeostasis and OA pathogenesis.
Methods:
Experimental approaches included human joint tissues cells, transgenic mice, and mouse OA model with viral KLF4 gene delivery to demonstrate therapeutic benefit in structure and pain improvement. Mechanistic studies applied global gene expression analysis and chromatin immunoprecipitation sequencing (ChIP-seq).
Results:
Several KLF genes were significantly decreased in OA cartilage. Among them, KLF4 and KLF2 were strong inducers of cartilage collagen genes and Proteoglycan-4. Cartilage-specific deletion of Klf2 in mature mice aggravated severity of experimental OA. Transduction of human chondrocytes with Adenovirus (Ad) expressing KLF4 or KLF2 enhanced expression of major cartilage extracellular matrix (ECM) genes and SRY-box transcription factor-9, and suppressed mediators of inflammation and ECM-degrading enzymes. Ad-KLF4 and Ad-KLF2 enhanced similar protective functions in meniscus cells and synoviocytes, and promoted chondrocytic differentiation of human mesenchymal stem cells. Viral KLF4 delivery into mouse knees reduced severity of OA-associated changes in cartilage, meniscus and synovium, and improved pain behaviors. ChIP-seq analysis suggested that KLF4 directly bound cartilage signature genes. Ras-related protein-1 signaling was the most enriched pathway in KLF4-transduced cells, and its signaling axis was involved in upregulating cartilage ECM genes by KLF4 and KLF2.
Conclusions:
KLF4 and KLF2 may be central transcription factors that increase protective and regenerative functions in joint tissue cells, suggesting that KLF gene transfer or molecules upregulating KLFs are therapeutic candidates for OA.
Keywords: Osteoarthritis, Chondrocytes, Inflammation
INTRODUCTION
Osteoarthritis (OA) is the most common joint disease.1 Despite substantial progress in identifying mechanisms of OA pathogenesis and molecular targets for intervention2, there have thus far not been any successful clinical trials and there are no approved pharmacological treatments to prevent the disease onset or progression. A potential explanation is that more important molecular mechanisms than those previously targeted are involved in OA.
Degradation and loss of articular cartilage is a major factor of OA pathogenesis.2 Cartilage extracellular matrix (ECM) molecules, including type-2 and type-11 collagen (COL2A1 and COL11A2), aggrecan (ACAN) and cartilage oligomeric matrix protein (COMP), are regulated by SRY-box transcription factor-9 (SOX9) cooperating with SOX5 and SOX6.3–7 Proteoglycan-4 (PRG4) also known as lubricin is dominantly expressed in the superficial zone of articular cartilage, and is essential for homeostasis of articular joints to prevent damage to the articular surface.8 While several transcription factors are reported to regulate PRG49–11, PRG4 is not subject to regulation by SOX9, which is a different regulatory mechanism compared to other cartilage ECM genes described earlier.5 12 A transcription factor that upregulates all these cartilage signature genes would be a promising therapeutic for cartilage engineering and in treatment of OA.
Activation of catabolic and inflammatory events is another key mechanism in OA.2 13 14 Therefore, suppressing catabolic and inflammatory genes, including a disintegrin and metalloproteinase with thrombospondin motifs-5 (ADAMTS5), matrix metalloproteinase-3 (MMP3), MMP13, interleukin-6 (IL6), prostaglandin-endoperoxide synthase-2 (PTGS2), and nitric oxide synthase-2 (NOS2), will also be important for therapeutic intervention in OA.
We previously performed RNA-seq analysis of normal and OA human knee cartilage and found that expression of several members in the Krüppel-like factor (KLF) family of transcription factors was suppressed in OA.15 The KLF family includes 17 zinc-finger transcription factors that are typically categorized into several groups based on similarities in structure and transcriptional activity: (1) KLF3, −8 and −12 serving as transcriptional repressors; (2) KLF1, −2, −4, −5, −6 and −7 functioning predominantly as transcriptional activators; (3) KLF9, −10, −11, −13, −14 and −16 having repressor activity; KLF15 and KLF17 have not been classified because their interaction motifs have yet to be determined.16 While the zinc-finger domains are highly conserved among the KLFs, the non-DNA-binding regions are highly divergent, and modular activation and repression domains have been suggested to regulate transcriptional activity.16 KLFs are involved in various biological and pathological mechanisms, including differentiation, apoptosis and tumorigenesis.16 However, there is limited knowledge about potential roles of KLFs in cartilage and OA pathogenesis.
Here, we analyzed functions of KLF4 and KLF2 in cells from human joint tissues and OA models in vivo and determined mechanisms by which KLF4 and KLF2 protected joint tissues from OA-associated damage.
RESULTS
Expression of KLF family genes in cartilage and regulation of cartilage ECM genes by KLFs
We utilized our previous RNA-seq dataset of human knee cartilage tissues15, to determine which KLF genes were highly expressed in normal cartilage and differentially expressed between normal and OA cartilage (supplementary figure 1; supplementary table 1). KLF1, KLF14 and KLF17 were excluded from further analyses because of their low expression levels. KLF genes with log2(counts per million reads mapped [CPM]) >5 in normal cartilage were considered to be expressed. Among those KLFs, KLF2, KLF4, KLF9, KLF10 and KLF15 were significantly decreased in OA cartilage, which suggested that these five KLFs might be associated with the compromised maintenance of cartilage homeostasis in OA.
We transduced TC28a2 human chondrocyte cells17 with the five KLFs (figure 1A; supplementary figure 2). Overexpression of KLF2 and KLF4 upregulated expression of COL2A1 and COL11A2. Notably, these two KLFs also increased expression of PRG4. Based on these findings, we focused subsequent studies on KLF4 and KLF2.
Figure 1. Regulation of cartilage collagen genes and PRG4 by KLFs, and expression of KLF4 and KLF2 in human and mouse cartilage tissues.
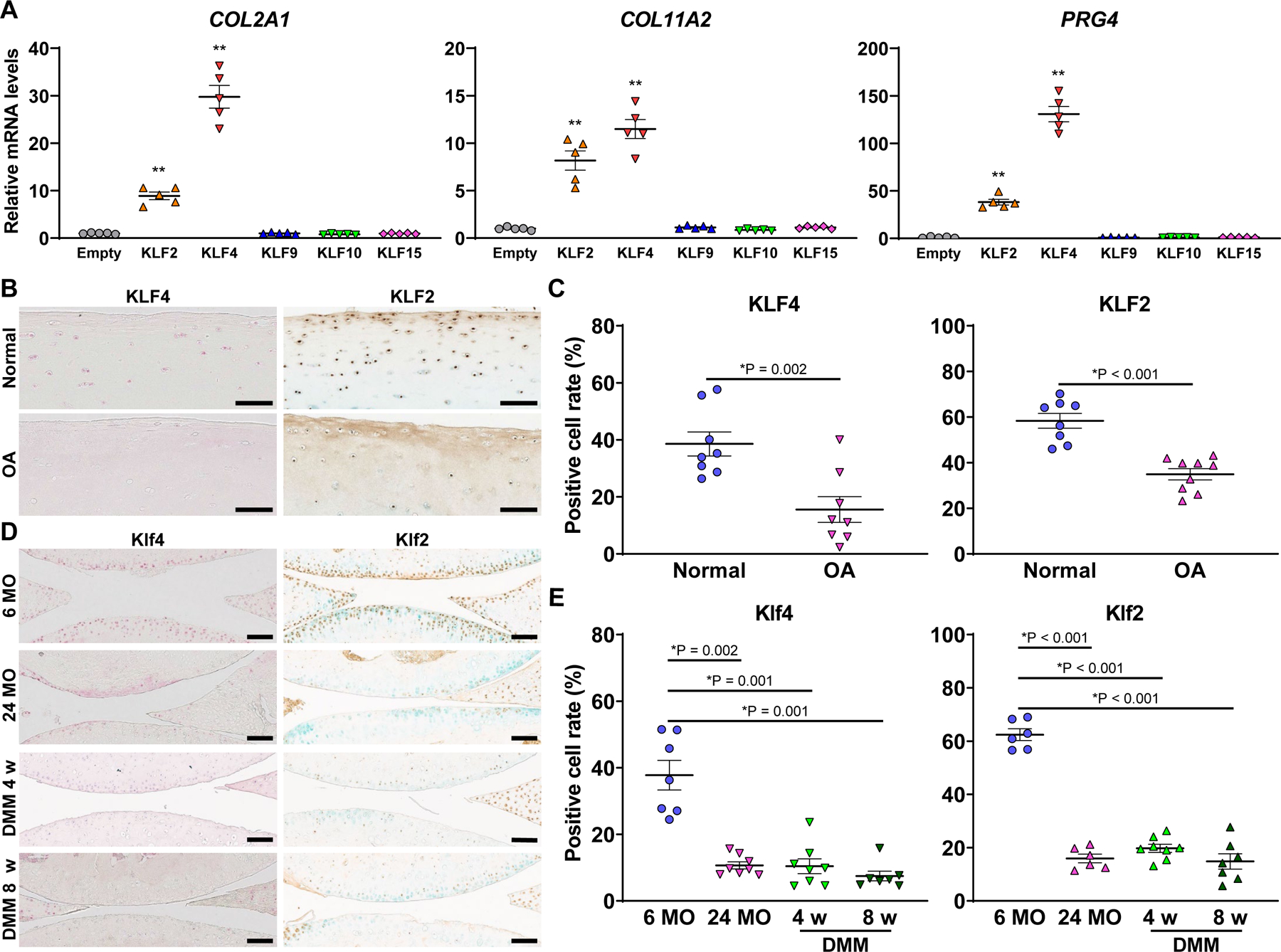
(A) TC28a2 cells were transfected with an empty vector, or expression vectors for either KLF2, KLF4, KLF9, KLF10 or KLF15, and RNA was collected 48 hours after transfection (n=5 from five independent experiments). mRNA levels relative to empty vector are shown. *P<0.05, **P<0.01 versus empty vector, Dunnett’s test. (B and C) Immunohistochemistry (IHC) for KLF4 and KLF2 in human knee cartilage. For KLF4, normal cartilage samples were from 8 donors (5 males and 3 females; age 22–48, mean 33±3), while osteoarthritis (OA)-affected cartilage samples were from 8 donors (4 males and 4 females; age 29–90, mean 65±7). For KLF2, normal cartilage samples were from 8 donors (5 males and 3 females; age 18–53, mean 38 ± 4), while OA-affected cartilages were from 9 donors (3 males and 6 females; age 51–90, mean 70±5). *P<0.05, Welch’s t-test in (C). (D and E) IHC for Klf4 and Klf2 in mouse knee cartilage (n=6–8 per condition). While 6 MO samples were from knees of 6-month-old mice, 24 MO indicates samples from knees of 24-month-old mice. DMM 4 w and DMM 8 w samples were from knees of 6-month-old mice 4 and 8 weeks after destabilization of the medial meniscus (DMM) surgery. *P<0.05 versus 6 MO, Dunnett’s T3 test in (E). All quantitative data are expressed as means±SE, and results of omnibus tests for multiple comparisons are shown in supplementary table 2. Scale bars, 100 µm.
Decreased expression of KLF4 and KLF2 in cartilage with OA and aging
Among human primary joint cells and bone marrow-derived mesenchymal stem cells (BMSCs), chondrocytes had the highest expression levels of KLF4 and KLF2 (supplementary figure 3). Then, we examined whether there were zone-specific expression patterns of KLF4 and KLF2 in articular cartilage, utilizing our DNA microarray dataset for three zones (superficial, middle and deep) of human knee cartilage tissues.18 Expression levels of KLF4 and KLF2 were not significantly different among the zones (supplementary figure 4). Six-month-old mouse knee joints were analyzed with immunohistochemistry (IHC), and there were no differences in positive cell rates of Klf4 and Klf2 between articular cartilage and subchondral bone, and between weight-bearing and non-weight-bearing regions of articular cartilage (supplementary figure 5).
Next, we examined how expression of KLF4 and KLF2 would change with OA and aging in human and mouse cartilage. Positive cell rates of both KLF4 and KLF2 were decreased in human OA cartilage compared to those in normal cartilage (figure 1B and C). IHC of Klf4 and Klf2 was also performed in knee joints from 6- and 24-month-old mice, and mice 4 or 8 weeks after OA induction by surgical destabilization of the medial meniscus (DMM)19 (figure 1D and E). Positive cell rates of both Klf4 and Klf2 in 24-month-old mice and in mice after DMM surgery were decreased when compared to 6-month-old control mice. KLF4 and KLF2 in human normal chondrocytes were down-regulated by proinflammatory cytokine IL-1β (supplementary figure 6), which is compatible with previous studies.20 21 Meanwhile, treatment of human OA chondrocytes with TGF-β3 upregulated KLF4 and KLF2 (supplementary figure 7). Collectively, these findings indicated OA- and aging-associated suppression of KLF4 and KLF2 in cartilage, and that mediators of joint tissue catabolism and inflammation were potential mechanisms of this suppression.
Klf2 functions in joint homeostasis and OA pathogenesis
When KLF2 and KLF4 were knocked down using small interfering RNAs (siRNAs) in human normal chondrocytes, expression levels of COL2A1, COL11A2, PRG4 and SOX9 were suppressed, while IL6, MMP3, PTGS2, ADAMTS5 and MMP13 were upregulated (supplementary figure 8).
We obtained Klf2flox/flox mice22 and Aggrecan-CreERT2 knock-in mice23, and created Aggrecan-CreERT2;Klf2flox/flox (conditional knock-out, cKO) mice to analyze roles of Klf2 in maintenance of mature articular cartilage and in OA pathogenesis. Klf2 deletion in articular cartilage of skeletally mature mice was confirmed using knee cartilage of 12-week-old cKO mice after tamoxifen injection (figure 2A). We performed DMM surgery to induce OA in cKO and littermate control mice and harvested knee joints 8 weeks after surgery for histological analysis (figure 2B). Severity of OA in cKO mice was significantly higher than in control mice, as shown by the Osteoarthritis Research Society International (OARSI) scores24 (figure 2C and D). In cKO mice, meniscus histopathological scores25, synovitis scores26 and bone scores9 were also higher than in control mice (figure 2E and F; supplementary figure 9). These results demonstrated that Klf2 deletion in articular cartilage increased severity of OA.
Figure 2. Postnatal deletion of Klf2 in cartilage using Aggrecan-CreERT2 mice.
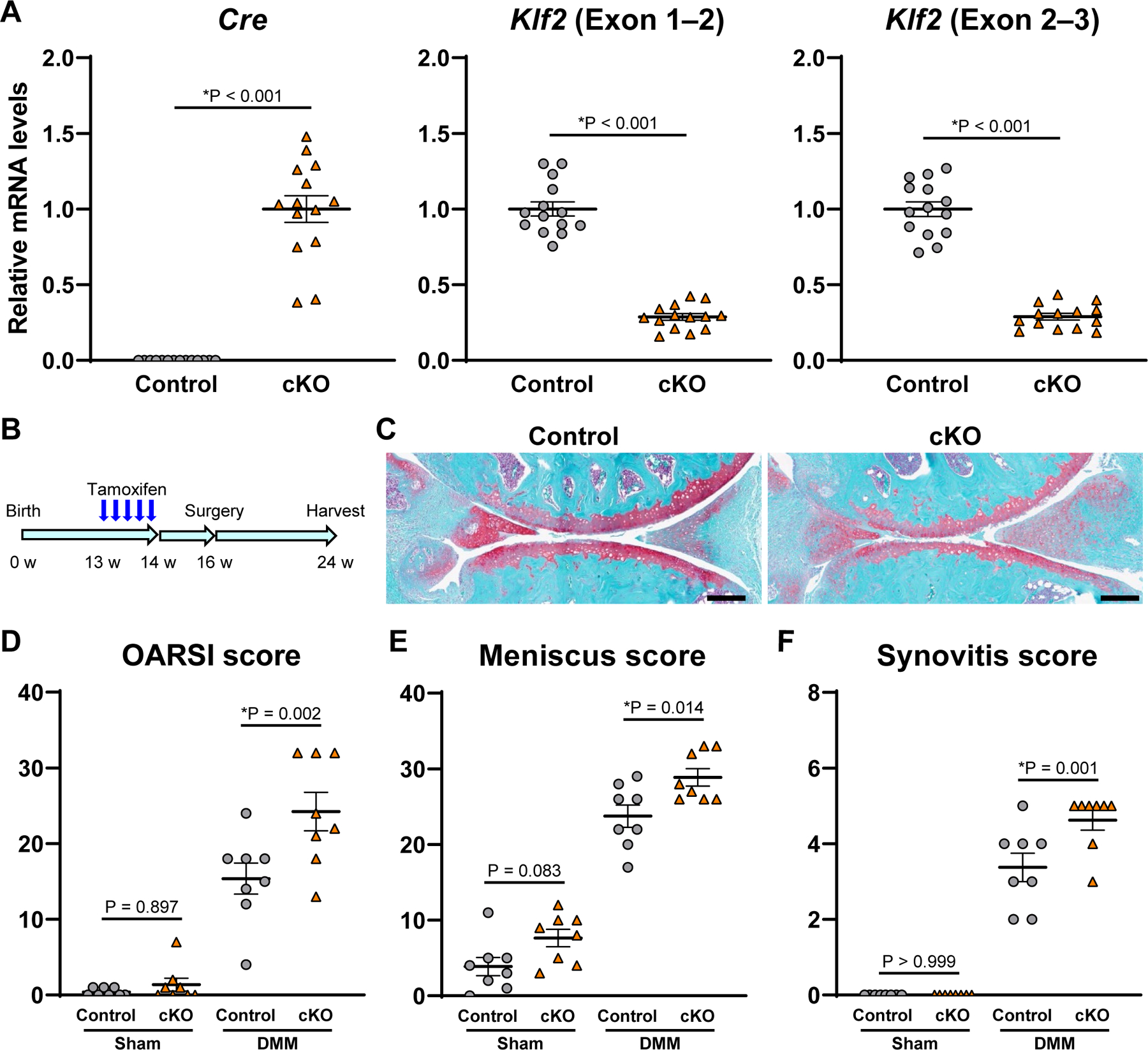
(A) RNA was collected from knee cartilage of 12-week-old Aggrecan-CreERT2;Klf2flox/flox (cKO) and littermate control mice 2 weeks after tamoxifen injection (n=14 per group). mRNA levels are relative to cKO for Cre, and relative to control for Klf2. *P<0.05, unpaired Welch’s t-test. (B) Sixteen-week-old cKO and littermate control mice underwent DMM or sham surgery 2 weeks after tamoxifen injection. The knees were harvested at 8 weeks postoperatively for histological analysis (n=8 per group). (C) Representative Safranin-O staining images of control and cKO mice with DMM surgery. Scale bars, 200 µm. (D) Summed Osteoarthritis Research Society International (OARSI) scores for the medial femoral condyle and tibial plateau. (E) Meniscus histopathological scores. (F) Synovitis scores. For (D–F), *P<0.05, Sidak’s multiple comparison test. All quantitative data are expressed as means±SE, and results of two-way ANOVA test are shown in supplementary table 2.
KLF4 and KLF2 regulation of cartilage signature genes
To investigate functions of KLF4 and KLF2 in regulation of joint tissue homeostasis, we used adenovirus (Ad) to overexpress KLF4 and KLF2 in several different cell types. In human OA chondrocytes, Ad-KLF4 increased expression of COL2A1, COL11A2, COMP, PRG4 and SOX9 (figure 3A; supplementary figure 10A), and Ad-KLF2 upregulated COL2A1, COL11A2, PRG4 and SOX9 (figure 3B; supplementary figure 10B). Human meniscal cells transfected with Ad-KLF4 showed higher mRNA levels of COL2A1, COL11A2, COMP and SOX9 (figure 3C; supplementary figure 10C). Transduction of KLF4 also upregulated scleraxis (SCX) and tenascin-B (TNXB), which are reported to be highly expressed in the meniscus.27 In Ad-KLF2-transduced meniscal cells, expression of COL2A1, COL11A2, SOX9 and TNXB was increased (figure 3D; supplementary figure 10D).
Figure 3. Regulation of chondrogenic and anabolic genes by KLF4 and KLF2 in human OA chondrocytes, meniscal cells and BMSC pellets.
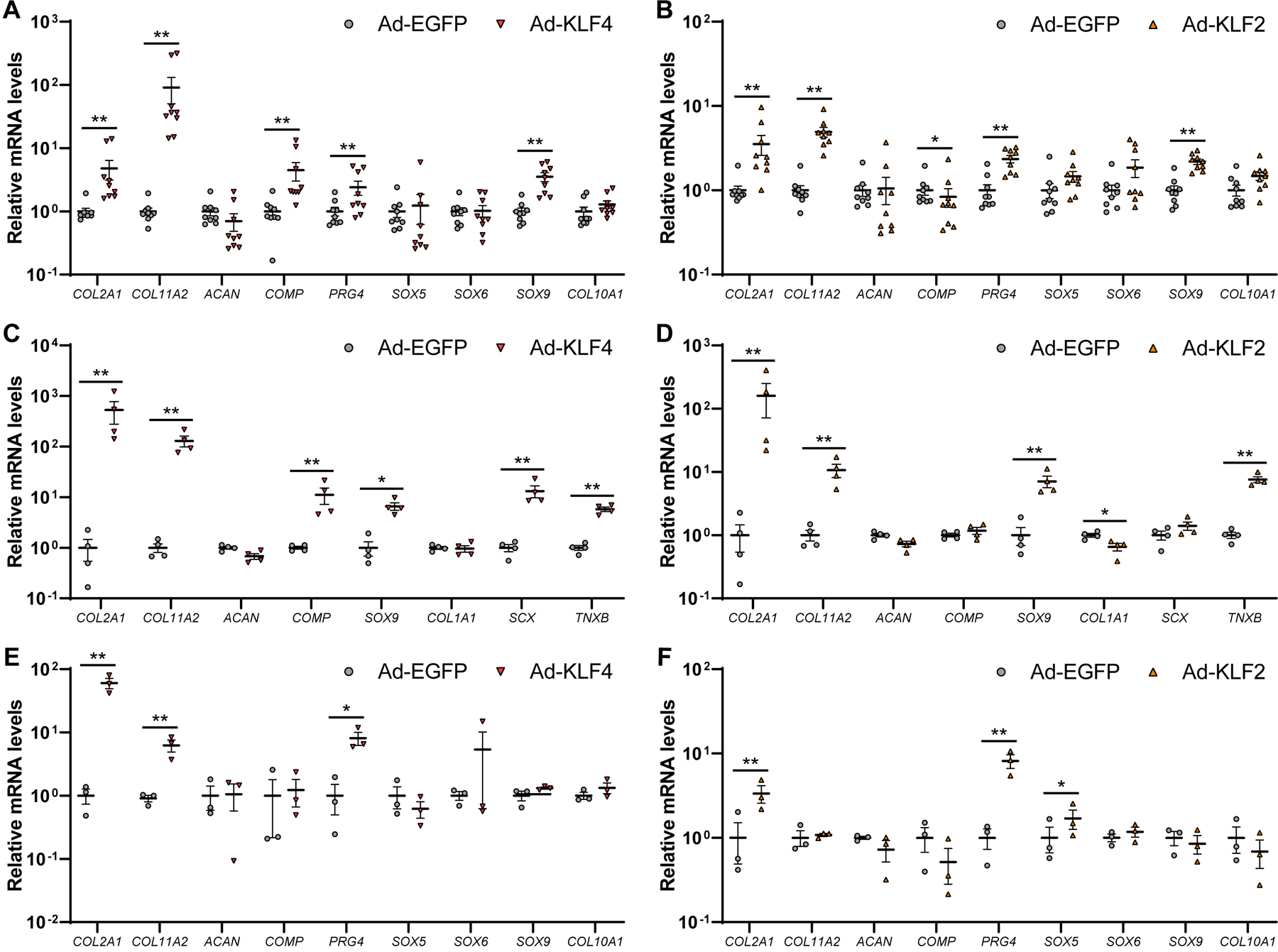
(A and B) Human OA chondrocytes were transfected with adenovirus (Ad-KLF4 or Ad-EGFP for (A), and Ad-KLF2 or Ad-EGFP for (B)), and RNA was collected 48 hours after transfection (n=9 donors). (C and D) Human meniscal cells were transfected with adenovirus (Ad-KLF4 or Ad-EGFP for (C), and Ad-KLF2 or Ad-EGFP for (D)), and RNA was collected 48 hours after transfection (n=4 donors). (E and F) Human bone marrow-derived mesenchymal stem cells (BMSCs) were transfected with adenovirus (Ad-KLF4 or Ad-EGFP for (E), and Ad-KLF2 or Ad-EGFP for (F)), and were cultured in pellets. RNA was collected 2 weeks after pellet culture (n=3 donors). mRNA levels are expressed as means±SE, relative to Ad-EGFP. *P<0.05, **P<0.01, paired t-test.
We further studied KLF4 and KLF2 in chondrogenesis of human BMSCs. In BMSCs under monolayer culture, Ad-KLF4 increased COL2A1, COL11A2, ACAN, COMP, PRG4, SOX5 and SOX9 (supplementary figure 11), while Ad-KLF2 upregulated expression of COL2A1, COL11A2 and PRG4 (supplementary figure 12). During 2-week pellet culture of BMSCs, KLF4 upregulated COL2A1, COL11A2 and PRG4 (figure 3E; supplementary figure 13A), and transduction of KLF2 increased COL2A1, PRG4 and SOX5 (figure 3F; supplementary figure 13B). Collectively, KLF4 and KLF2 upregulated various chondrogenic and anabolic genes in human chondrocytes, meniscal cells and BMSCs.
Suppression of inflammatory and catabolic genes by KLF4 and KLF2
We examined regulation of genes related to inflammation and ECM degradation by the KLFs. When OA chondrocytes were treated with IL-1β, Ad-KLF4 down-regulated IL6, MMP3, NOS2, PTGS2, ADAMTS5 and MMP13, and Ad-KLF2 decreased expression levels of IL6, MMP3, NOS2, ADAMTS5 and MMP13 (figure 4A; supplementary figure 14). In IL-1β stimulated meniscal cells, IL6, MMP3, PTGS2, ADAMTS5 and MMP13 were down-regulated by KLF4- or KLF2-transduction (figure 4B; supplementary figure 15). Similarly in synoviocytes, Ad-KLF4 and Ad-KLF2 suppressed expression levels of IL6, MMP3, PTGS2, ADAMTS5 and MMP13 under IL-1β stimulation (figure 4C; supplementary figure 16C). These findings suggested that KLF4 and KLF2 suppressed catabolic and inflammatory genes in joint tissue cells.
Figure 4. Regulation of inflammatory and catabolic genes by KLF4 and KLF2 in human OA chondrocytes, meniscal cells and synoviocytes on IL-1β stimulation.
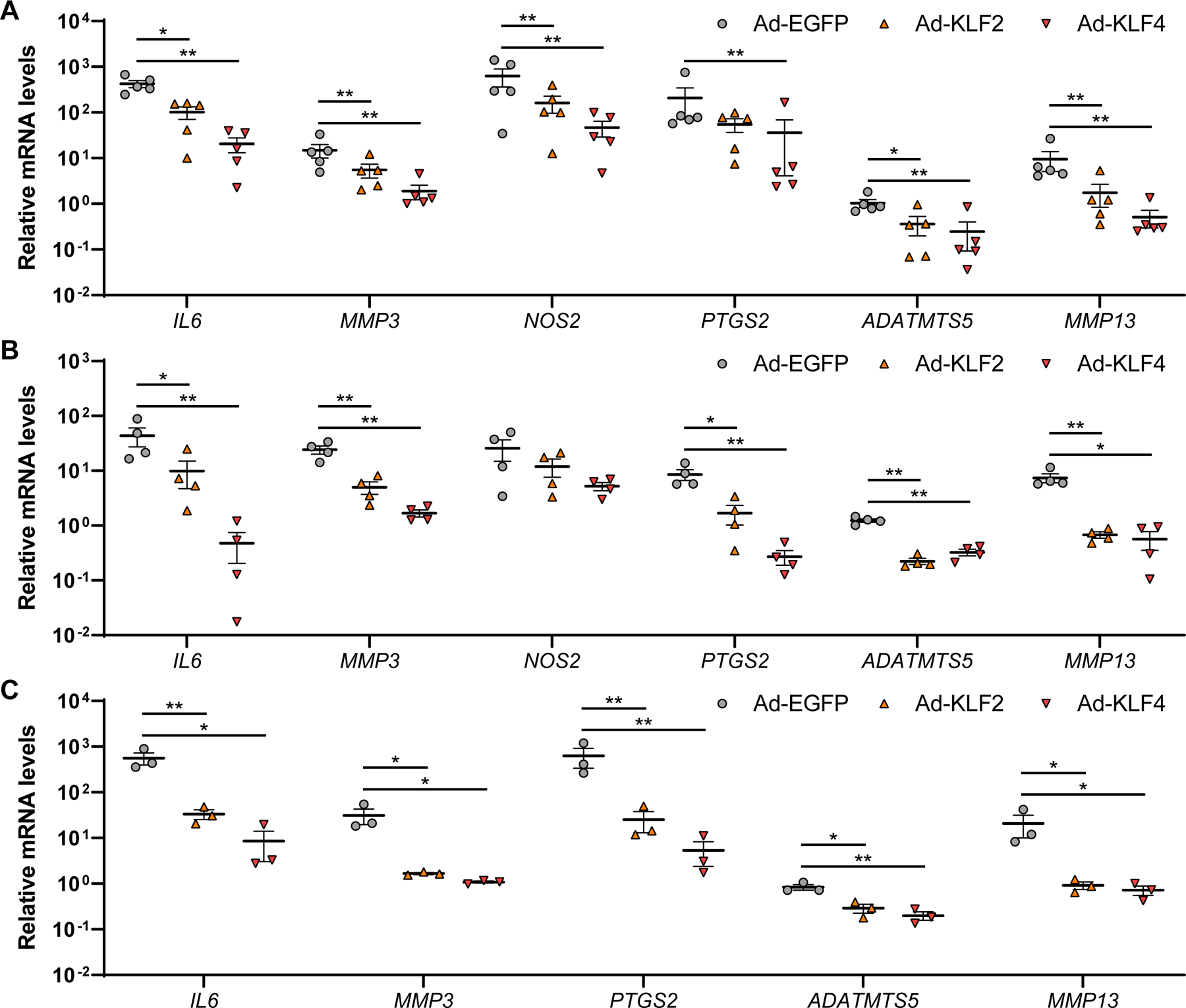
Cells were transfected with Ad-EGFP, Ad-KLF2 or Ad-KLF4, and RNA was collected 48 hours after adenoviral transfection and 6 hours after treatment with 10 ng/ml of interleukin-1β (IL-1β). (A) n=5 donors were used for human OA chondrocytes. (B) n=4 donors were used for human meniscal cells. (C) n=3 donors were used for human synoviocytes. mRNA levels are expressed as means±SE, relative to Ad-EGFP without IL-1β as controls. *P<0.05, **P<0.01 versus Ad-EGFP, Dunnett’s test. Results of one-way mixed-effects ANOVA test are shown in supplementary table 2.
Therapeutic effects of KLF4 gene delivery using adeno-associated virus
To directly confirm therapeutic effects against OA, mice with DMM or sham surgery received intraarticular injection of adeno-associated virus (AAV) expressing KLF4 (figure 5A; supplementary figure 17A), AAV-EGFP (supplementary figure 17B) or phosphate-buffered saline (PBS) at 2 and 4 weeks after surgery. Knee joints were harvested at 8 weeks postoperatively, as illustrated in figure 5B. We performed von Frey test28 29 preoperatively, and at 4 and 8 weeks after surgery to evaluate mechanical allodynia (figure 5C; supplementary figures 18 and 19). AAV-KLF4 injected mice with DMM surgery showed significantly decreased numbers of paw withdrawals at 4 and 8 weeks. Histological analyses revealed that AAV-KLF4 injection significantly improved the OARSI scores, meniscus histopathological scores, synovitis scores and bone scores (figure 5D–G; supplementary figure 20). These results clearly demonstrated that KLF4 reduced OA-associated joint damage and mechanical allodynia.
Figure 5. Therapeutic effects of OA by KLF4 gene delivery.
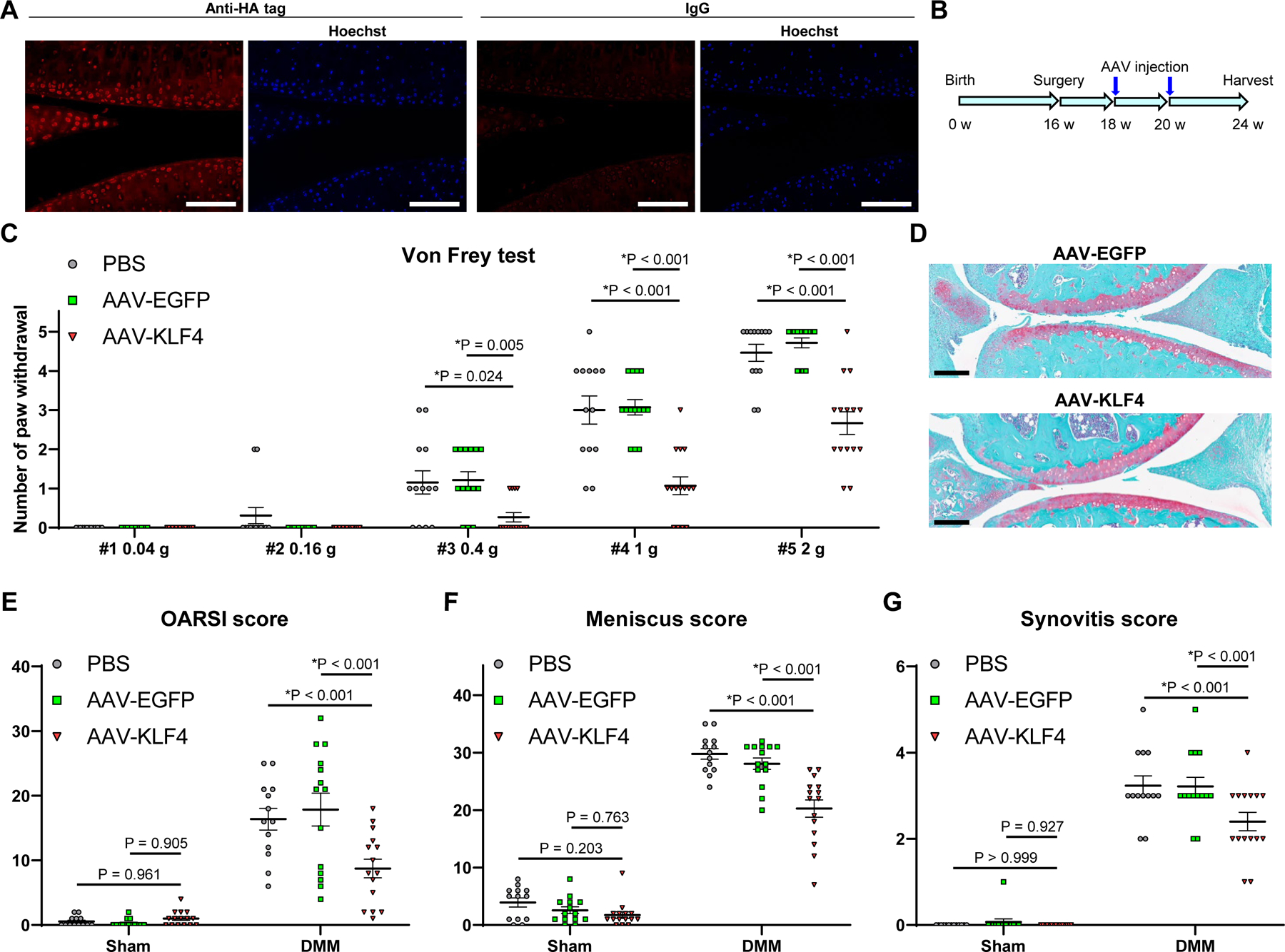
(A) Immunofluorescence of HA tag with nuclear staining by Hoechst 33342 in 12-week-old mouse knee joints one week after injection of adeno-associated virus (AAV) expressing KLF4 and HA tag. Representative images from n=6 are shown. Scale bars, 100 µm. (B) Sixteen-week-old mice underwent DMM or sham surgery, and AAV-KLF4 (n=15), AAV-EGFP (n=14) or PBS (n=13) was injected into knee joints at 2 and 4 weeks after surgery. Knees were harvested at 8 weeks postoperatively for histological analysis. (C) Von Frey test in mice at 8 weeks after DMM surgery. Numbers of paw withdrawals from 5 stimuli per filament per mouse are shown. *P<0.05 versus AAV-KLF4, Dunn’s test. (D) Representative Safranin-O staining images of AAV-EGFP- and AAV-KLF4-injected mice with DMM surgery. Scale bars, 200 µm. (E) Summed OARSI scores for the medial femoral condyle and tibial plateau. (F) Meniscus histopathological scores. (G) Synovitis scores. For (E–G), *P<0.05 versus AAV-KLF4, Dunnett’s test. All quantitative data are expressed as means±SE, and results of omnibus tests for multiple comparisons are shown in supplementary table 2.
Global analysis of KLF4-regulated genes
To study genes regulated by KLF4 comprehensively and to elucidate regulatory mechanisms of KLF4, we performed global expression profiling by digital RNA with pertUrbation of Genes (DRUG-seq) analysis30 of Ad-KLF4-, Ad-EGFP- and non-transduced TC28a2 cells with and without IL-1β stimulation (supplementary figure 21). For differential expression analysis, we compared cells that were treated with various combinations of IL-1β, Ad-EGFP and Ad-KLF4. Lists of all significantly upregulated genes (URGs) and down-regulated genes (DRGs) in each comparison are shown in supplementary tables 3–10, where genes with a false discovery rate (FDR) of <0.05 and a |log2(fold change [FC])| >1 were considered to be significantly differentially expressed genes.
Ad-KLF4 significantly upregulated cartilage ECM genes such as COL2A1, COL11A2, ACAN, COMP and PRG4 (Fig. 6A and supplementary table 7). Notably, a number of genes which are anabolic or protective against OA were also significantly upregulated, including tissue inhibitor of metalloproteinases-1 (TIMP1), TIMP2, TIMP3, cluster of differentiation-24 (CD24), COL6A1, COL6A2, COL9A2, COL9A3, nuclear factor of activated T cells-1 (NFATC1), NFATC2, forkhead box O1 (FOXO1), FOXO3 and fibroblast growth factor-18 (FGF18)9 31–38 (supplementary table 7).
Figure 6. Digital RNA with pertUrbation of Genes (DRUG-seq) analysis of Ad-EGFP or Ad-KLF4 transfected TC28a2 cells.
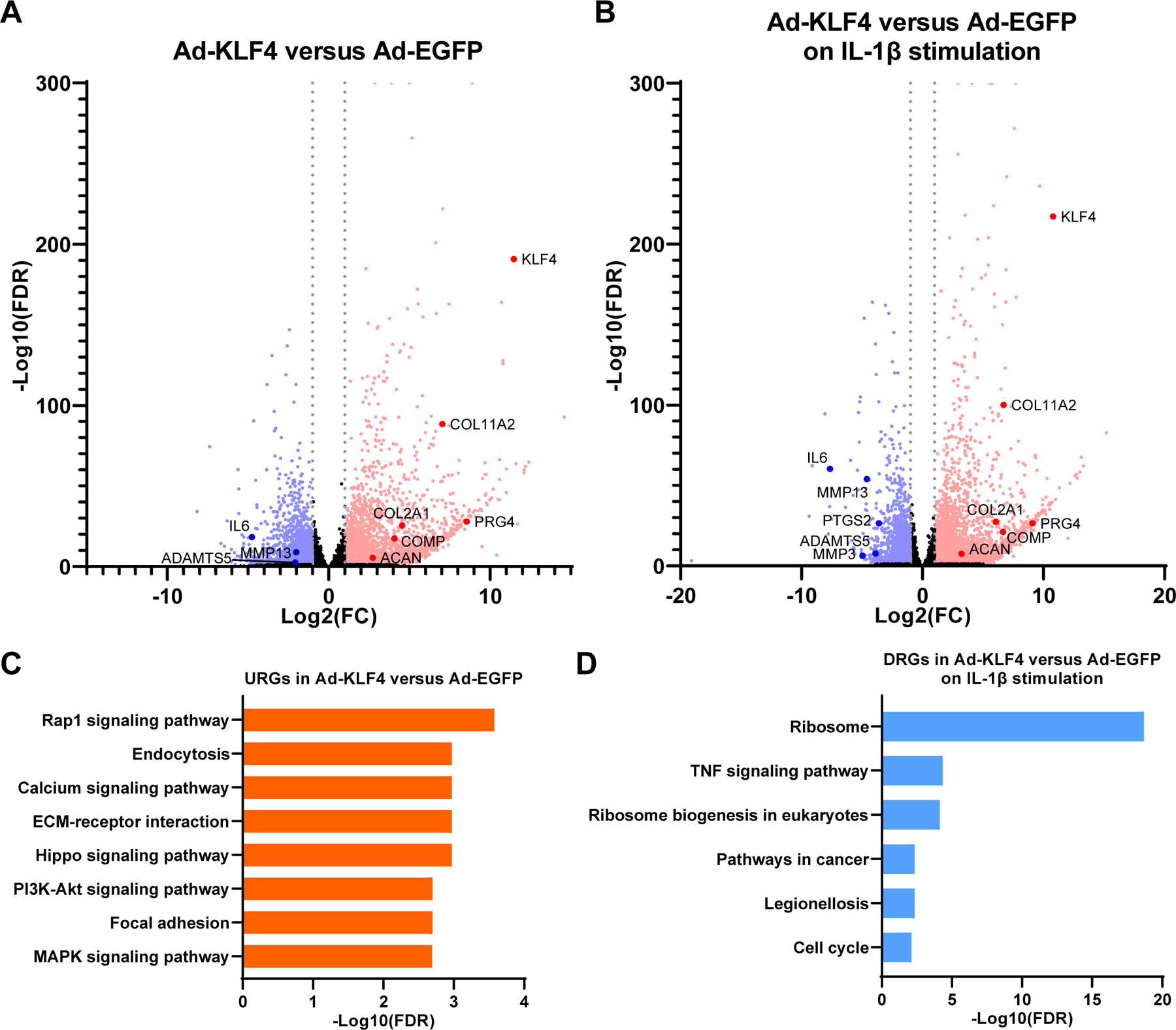
In samples with IL-1β stimulation, cells were treated with 10 ng/ml of IL-1β for 6 hours before RNA collection. n=7 per condition from seven independent experiments were analyzed. (A) Volcano plot analysis to identify differentially expressed genes (DEGs) in Ad-KLF4 versus Ad-EGFP transfected samples. (B) Volcano plot analysis to identify DEGs in Ad-KLF4 versus Ad-EGFP transfected samples on IL-1β stimulation. For (A and B), grey dotted lines indicate |log2(fold change [FC])|=1. Significantly upregulated genes (URGs) are shown as red dots, while significantly down-regulated genes (DRGs) are indicated as blue dots; black dots represent non-significant DEGs. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis for URGs in Ad-KLF4 versus Ad-EGFP transfected samples. The top eight enriched pathways are shown. (D) KEGG pathway analysis for DRGs in Ad-KLF4 versus Ad-EGFP transfected samples on IL-1β stimulation. All significantly enriched pathways are shown. FDR, false discovery rate.
In the presence of IL-1β, Ad-KLF4 significantly suppressed IL6, MMP3, PTGS2, ADAMTS5, MMP13, and also other genes associated with OA pathogenesis, including fibronectin-1 (FN1), periostin (POSTN), cellular communication network factor-2 (CCN2), leukemia inhibitory factor (LIF), gremlin-1 (GREM1), SOX4, IL-1 receptor type 1 (IL1R1), IL1B, IL11, IL33, ADAMTS1, ADAMTS12 and MMP114 39–49 (figure 6B; supplementary table 10). These findings suggested that KLF4 suppressed key mediators of OA pathogenesis.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses50 were performed using URGs and DRGs. Significantly enriched pathways are shown in supplementary tables 11–16. In the URGs by KLF4, “ECM-receptor interaction”, where cartilage ECM genes such as COL2A1, COL11A2 and COMP are annotated, was significantly enriched (figure 6C; supplementary table 13). “Rap1 signaling pathway” was the top pathway, and several signaling pathways including “Calcium signaling pathway” and “MAPK signaling pathway” were highly ranked, suggesting their regulation by KLF4. Among the DRGs by KLF4 on IL-1β stimulation, “TNF signaling pathway” was the second most enriched pathway (figure 6D; supplementary table 16).
Genome-wide analysis of KLF4-DNA association in KLF4-transduced cells
Chromatin immunoprecipitation sequencing (ChIP-seq) was performed using TC28a2 cells transfected with a vector expressing KLF4 with HA tag (HA-KLF4) or an empty vector (supplementary table 17). Non-specific peaks were removed, and only specific peaks were used for subsequent analyses (supplementary figure 22). In gene ontology analysis, terms related to chondrocyte differentiation were significantly enriched, such as “growth plate cartilage chondrocyte differentiation”, and “regulation of chondrocyte development” (supplementary table 18). Next, de novo motif analysis was performed for the obtained ChIP-seq peaks. A KLF motif (figure 7A) was top-ranked among the significantly recovered motifs (supplementary figure 23), and this motif was highly enriched at the predicted centers of the peaks (figure 7B). These findings supported direct binding of KLF4 to DNA as well as integrity of the ChIP-seq dataset. Then, we examined peak signals of the ChIP-seq dataset around cartilage signature genes, as well as the top four URGs by KLF4 in the DRUG-seq analysis (supplementary table 7). Large numbers of peaks were detected in the putative regulatory regions of SOX9, COL2A1, COL11A2, PRG4, ACAN and COMP, and most of these peak regions contained the recovered KLF motif, while similar findings were found for the top URGs (figure 7C and D; supplementary figures 24–31). These data suggested that KLF4 directly bound to DNA around major chondrogenic genes, indicating that KLF4 was an important regulator of chondrogenesis.
Figure 7. Chromatin immunoprecipitation sequencing (ChIP-seq) for genome-wide analysis of KLF4-DNA association in KLF4 overexpressed TC28a2 cells.
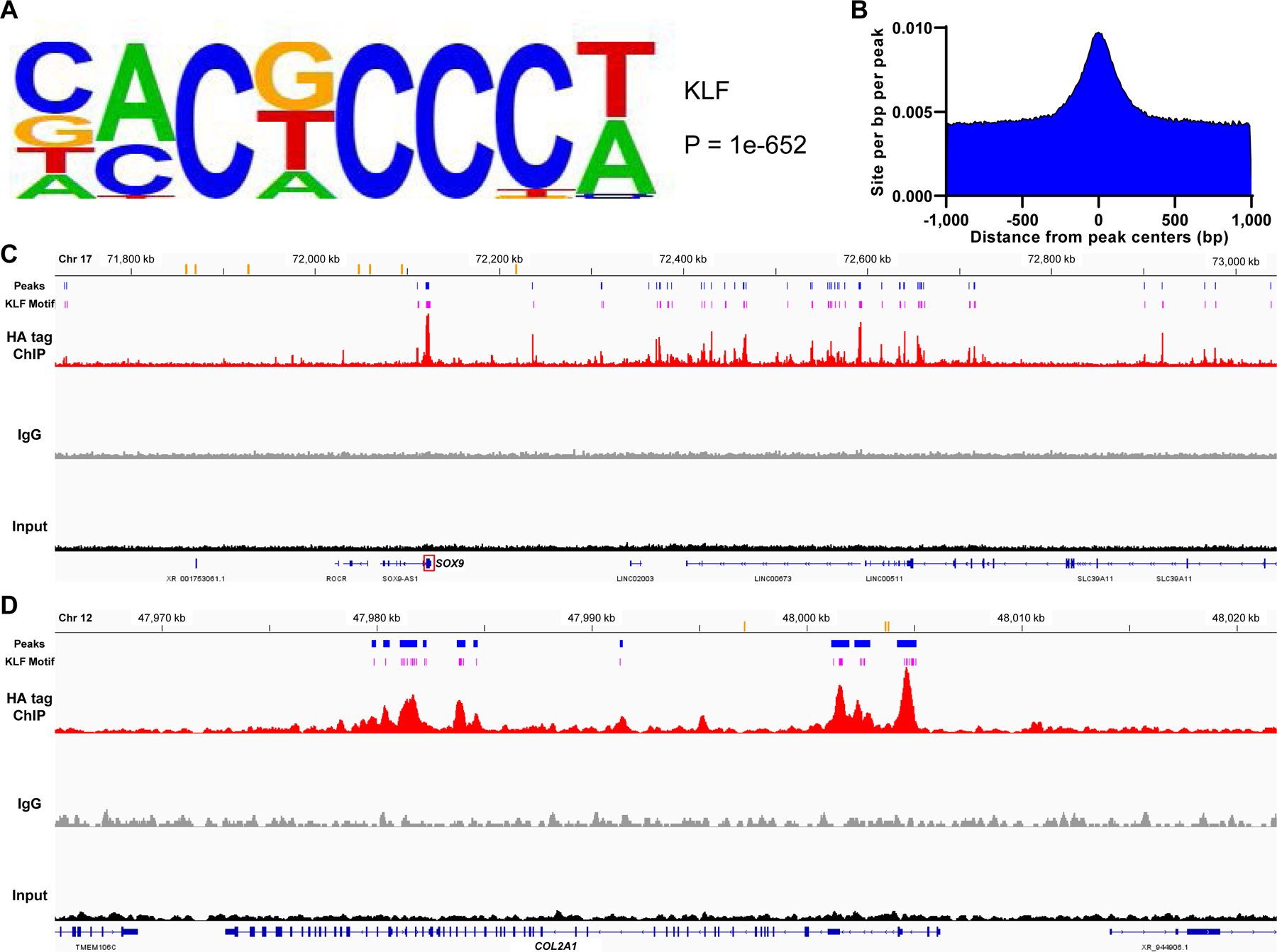
(A) A KLF motif recovered from ChIP-seq peaks. The motif logo displays nucleotide frequencies (scaled relative to the information content) at each position. (B) Distribution of the KLF motif within a 1,000 base pair (bp) window from the peak centers with a 10 bp step size. (C and D) Visualization of aligned reads for HA tag ChIP-seq, IgG and input samples around the SOX9 gene (C) and around the COL2A1 gene (D). Orange bars indicate previously identified enhancers for each gene. Blue bars indicate peak regions that were putative regulatory domains of SOX9 or COL2A1 and common between the replicates of ChIP samples. Pink bars show the predicted sites of the recovered KLF motif which were located within the peak regions. Chr, chromosome. kb, kilobase.
Regulation of chondrogenesis by KLFs via PKA-RAP1-MEK-CREB signaling axis
In addition to the function of KLF4 as a transcription factor to directly upregulate the cartilage signature genes, we focused on relation between Ras-related protein-1 (RAP1) signaling and KLFs, since “Rap1 signaling pathway” was the top-ranked enriched pathway in the KEGG pathway analysis (figure 6C; supplementary tables 13 and 15). It is reported that activation of RAP1 promotes chondrogenesis.51 While KLF4 and KLF2 did not increase expression levels of RAP1A and RAP1B in OA chondrocytes (supplementary figure 32) or in TC28a2 cells (supplementary table 19), KLF4 directly interacts with RAP1 protein.52 Our results showed that treatment of KLF4- and KLF2-transfected TC28a2 cells with Rap1 inhibitor GGTI-298 diminished or eliminated upregulation of cartilage ECM genes, COL2A1, COL11A2, ACAN, COMP and PRG4 (figure 8A; supplementary figure 33).
Figure 8. KLF4 regulation of cartilage ECM genes through the RAP1-related signaling axis.
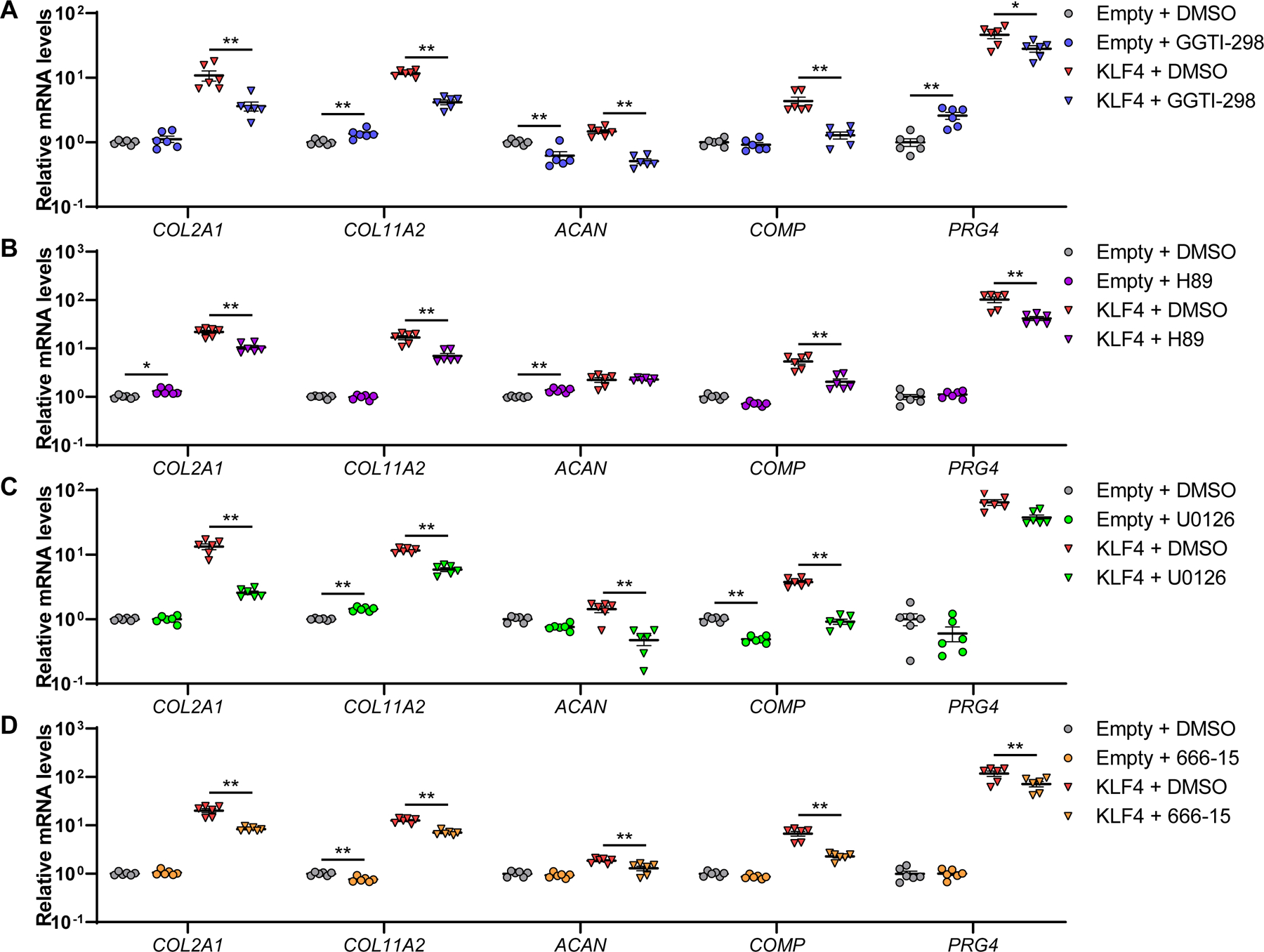
TC28a2 cells were transfected with an expression vector encoding KLF4 or an empty vector. Then, cells were treated with GGTI-298 (A), H89 (B), U0126 (C) or 666–15 (D), or dimethyl sulfoxide (DMSO). RNA was collected 48 hours after transfection (n=6 from six independent experiments). mRNA levels are expressed as means±SE, relative to empty vector-transfected samples with DMSO as controls. *P<0.05, **P<0.01, Sidak’s multiple comparison test. Results of two-way mixed-effects ANOVA test are shown in supplementary table 2.
RAP1 is a small GTPase53 and a member of the PKA (protein kinase A)-RAP1-MEK (mitogen-activated protein kinase kinase)-CREB (cyclic-AMP [cAMP] response element-binding protein) signaling axis.54–56 Fluid flow shear stress induces expression of Prg4 in articular cartilage via activation of this signaling pathway11, and treatment of TC28a2 cells with the cAMP inducer forskolin increased expression levels of cartilage ECM genes (supplementary figure 34). We thus hypothesized that KLF4 and KLF2 might upregulate cartilage ECM genes via the PKA-RAP1-MEK-CREB signaling axis. To test this hypothesis, we treated KLF4- and KLF2-transfected TC28a2 cells with inhibitors of several members in the signaling axis; PKA inhibitor H89, MEK inhibitor U0126, and CREB inhibitor 666–15. Upregulation of the cartilage ECM genes was attenuated or cancelled by each of these inhibitors (figure 8B–D; supplementary figure 35). Collectively, these findings demonstrated that KLF4 and KLF2 upregulated major cartilage ECM genes, not only by direct regulation as a transcription factor but also via the PKA-RAP1-MEK-CREB signaling axis (supplementary figure 36).
DISCUSSION
This is the first detailed analysis of KLFs in the context of cartilage biology, joint homeostasis and OA pathogenesis. We focused on KLF4 and KLF2 as they were the strongest inducers of cartilage collagen genes and PRG4, and as their expression was suppressed in OA cartilage. KLF4 and KLF2 are closely related members within the KLF family.16 57
KLF4 and KLF2 down-regulated mediators of inflammation and ECM-degrading enzymes in human joint tissue cells, which are compatible with previous studies showing that they suppressed NF-κB activity.58 59 Previous reports show KLF4 binding to the promoter proximal region of COL2A1 gene60 and KLF4 colocalization and interaction with SOX9 in chondrocytes.61 The present study suggested that KLF4 and KLF2 were important regulators of chondrogenesis in joint tissue cells because they upregulated cartilage signature genes with different regulatory mechanisms; SOX9, a master regulator of chondrogenesis, SOX9-regulated genes such as COL2A1, COL11A2, ACAN and COMP3–7, and PRG4 which is not subject to regulation by SOX9.5 12 ChIP-seq analysis supported the model of KLF4 as a central transcription factor of chondrogenesis. KLF4 bound to large numbers of the putative regulatory regions of major chondrogenic genes, and many of the bound sites included the KLF motif. All these findings indicated direct regulation of the cartilage signature genes by KLF4. We also revealed a novel link of KLF4 with RAP1 signaling. The PKA-RAP1-MEK-CREB signaling axis is involved in modulation of various types of genes54–56, and Prg4 expression in articular cartilage is induced by fluid flow shear stress via this signaling pathway.11 Our results showed that the KLF4- or KLF2-effects on cartilage ECM genes were dependent on each molecule in the PKA-RAP1-MEK-CREB axis.
Because KLF4 upregulated more chondrogenic and anabolic genes, and down-regulated more inflammatory and catabolic genes than KLF2 in our in vitro experiments, we assumed KLF4 to be more promising as a therapeutic target for OA. Intraarticular Ad-KLF2 had previously shown to protect against cartilage damage induced by monoiodoacetate.21 Therefore, we tested intraarticular injection of AAV-KLF4 in the mouse DMM model. AAV-KLF4 improved pain behaviors, and reduced severity of OA histopathological changes in cartilage, meniscus and synovium. These results indicated that KLF4 had therapeutic and protective effects against OA-associated tissue damage and pain.
Several limitations should be noted in the present study. First, all mouse experiments were done with only male animals, and sex differences were not addressed. Second, aging-associated spontaneous OA model was not performed in our study. Third, only experiments using Aggrecan-CreERT2;Klf2flox/flox mice were performed, because Klf4 deletion in articular cartilage of Aggrecan-CreERT2;Klf4flox/flox mice was not sufficient. Finally, because KLF4-overexpressed cells were used in our ChIP-seq analysis, more sites could be detected as peaks than in ChIP-seq of endogenous KLF4.
In conclusion, this study identifies KLF4 and KLF2 as new transcription factors that are important regulators of the chondrocyte phenotype, and that also have effects on other joint tissues by upregulating ECM components and by suppressing ECM-degrading enzymes and inflammatory mediators. KLF4 and KLF2 thus have a spectrum of biological activities with promise in treatment of joint diseases such as OA.
MATERIALS AND METHODS
Experimental procedures are provided in the supplementary materials and methods.
Supplementary Material
Key messages:
What is already known about this subject?
We previously performed RNA-seq analysis of normal and OA human knee cartilage and found that several members in the Krüppel-like factor (KLF) family of transcription factors are suppressed in OA.
What does this study add?
The suppression of KLF4 and KLF2 seen in OA does have functional consequences for the phenotype of cells in joint tissues, leading to increased OA severity.
KLF4 and KLF2 upregulate major cartilage extracellular matrix and chondrogenic genes, and they also suppress mediators of inflammation and ECM-degrading enzymes.
Delivery of KLF4 gene into mouse knee joints reduces severity of experimental OA-associated changes in cartilage, meniscus and synovium and improves pain behaviors.
How might this impact on clinical practice or future developments?
KLF4 and KLF2 are central transcription factors regulating protective functions in joints, and are promising therapeutic targets for joint diseases such as OA.
Acknowledgments:
We thank Gogce Crynen, PhD for her expert advice on statistical analysis.
Funding:
National Institutes of Health grants R01AG049617 and R01AG056144 (M.K.L.).
Footnotes
Competing interests: The authors declare that they have no competing interests.
Ethics statements
Patient consent for publication: Obtained.
Ethics approval: All human tissues were obtained with approval by the Scripps Human Subjects Committee. All animal studies were performed with approval by the Scripps Institutional Animal Care and Use Committee.
Data availability statement:
The GEO accession numbers for all the datasets utilized in the present study are available in the supplementary materials and methods.
REFERENCES
- 1.Lawrence RC, Felson DT, Helmick CG, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum 2008;58(1):26–35. doi: 10.1002/art.23176 [published Online First: 2008/01/01] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeser RF, Goldring SR, Scanzello CR, et al. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum 2012;64(6):1697–707. doi: 10.1002/art.34453 [published Online First: 2012/03/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smits P, Li P, Mandel J, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell 2001;1(2):277–90. doi: 10.1016/s1534-5807(01)00003-x [published Online First: 2001/11/13] [DOI] [PubMed] [Google Scholar]
- 4.Liu CJ, Zhang Y, Xu K, et al. Transcriptional activation of cartilage oligomeric matrix protein by Sox9, Sox5, and Sox6 transcription factors and CBP/p300 coactivators. Front Biosci 2007;12:3899–910. doi: 10.2741/2359 [published Online First: 2007/05/09] [DOI] [PubMed] [Google Scholar]
- 5.Oh CD, Maity SN, Lu JF, et al. Identification of SOX9 interaction sites in the genome of chondrocytes. PloS one 2010;5(4):e10113. doi: 10.1371/journal.pone.0010113 [published Online First: 2010/04/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kronenberg HM. Developmental regulation of the growth plate. Nature 2003;423(6937):332–6. doi: 10.1038/nature01657 [published Online First: 2003/05/16] [DOI] [PubMed] [Google Scholar]
- 7.Song H, Park KH. Regulation and function of SOX9 during cartilage development and regeneration. Semin Cancer Biol 2020;67(Pt 1):12–23. doi: 10.1016/j.semcancer.2020.04.008 [published Online First: 20200504] [DOI] [PubMed] [Google Scholar]
- 8.Jay GD, Waller KA. The biology of lubricin: near frictionless joint motion. Matrix Biol 2014;39:17–24. doi: 10.1016/j.matbio.2014.08.008 [published Online First: 2014/08/31] [DOI] [PubMed] [Google Scholar]
- 9.Matsuzaki T, Alvarez-Garcia O, Mokuda S, et al. FoxO transcription factors modulate autophagy and proteoglycan 4 in cartilage homeostasis and osteoarthritis. Sci Transl Med 2018;10(428) doi: 10.1126/scitranslmed.aan0746 [published Online First: 2018/02/16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang CH, Gao Y, Jadhav U, et al. Creb5 establishes the competence for Prg4 expression in articular cartilage. Commun Biol 2021;4(1):332. doi: 10.1038/s42003-021-01857-0 [published Online First: 2021/03/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogawa H, Kozhemyakina E, Hung HH, et al. Mechanical motion promotes expression of Prg4 in articular cartilage via multiple CREB-dependent, fluid flow shear stress-induced signaling pathways. Genes Dev 2014;28(2):127–39. doi: 10.1101/gad.231969.113 [published Online First: 2014/01/23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry SP, Liang S, Akdemir KC, et al. The postnatal role of Sox9 in cartilage. J Bone Miner Res 2012;27(12):2511–25. doi: 10.1002/jbmr.1696 [published Online First: 2012/07/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lepetsos P, Papavassiliou AG. ROS/oxidative stress signaling in osteoarthritis. Biochim Biophys Acta 2016;1862(4):576–91. doi: 10.1016/j.bbadis.2016.01.003 [published Online First: 2016/01/16] [DOI] [PubMed] [Google Scholar]
- 14.Kumavat R, Kumar V, Malhotra R, et al. Biomarkers of Joint Damage in Osteoarthritis: Current Status and Future Directions. Mediators Inflamm 2021;2021:5574582. doi: 10.1155/2021/5574582 [published Online First: 2021/03/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fisch KM, Gamini R, Alvarez-Garcia O, et al. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis Cartilage 2018;26(11):1531–38. doi: 10.1016/j.joca.2018.07.012 [published Online First: 2018/08/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev 2010;90(4):1337–81. doi: 10.1152/physrev.00058.2009 [published Online First: 2010/10/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kokenyesi R, Tan L, Robbins JR, et al. Proteoglycan production by immortalized human chondrocyte cell lines cultured under conditions that promote expression of the differentiated phenotype. Arch Biochem Biophys 2000;383(1):79–90. doi: 10.1006/abbi.2000.2044 [published Online First: 2000/11/30] [DOI] [PubMed] [Google Scholar]
- 18.Grogan SP, Duffy SF, Pauli C, et al. Zone-specific gene expression patterns in articular cartilage. Arthritis Rheum 2013;65(2):418–28. doi: 10.1002/art.37760 [published Online First: 2012/11/06] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glasson SS, Blanchet TJ, Morris EA. The surgical destabilization of the medial meniscus (DMM) model of osteoarthritis in the 129/SvEv mouse. Osteoarthritis Cartilage 2007;15(9):1061–9. doi: 10.1016/j.joca.2007.03.006 [published Online First: 2007/05/02] [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Guo H, Li L, et al. Long Non-Coding RNA (lncRNA) Small Nucleolar RNA Host Gene 15 (SNHG15) Alleviates Osteoarthritis Progression by Regulation of Extracellular Matrix Homeostasis. Med Sci Monit 2020;26:e923868. doi: 10.12659/MSM.923868 [published Online First: 2020/07/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao X, Jiang S, Du Z, et al. KLF2 Protects against Osteoarthritis by Repressing Oxidative Response through Activation of Nrf2/ARE Signaling In Vitro and In Vivo. Oxid Med Cell Longev 2019;2019:8564681. doi: 10.1155/2019/8564681 [published Online First: 2019/12/13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JS, Yu Q, Shin JT, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell 2006;11(6):845–57. doi: 10.1016/j.devcel.2006.09.006 [published Online First: 2006/12/05] [DOI] [PubMed] [Google Scholar]
- 23.Henry SP, Jang CW, Deng JM, et al. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis 2009;47(12):805–14. doi: 10.1002/dvg.20564 [published Online First: 2009/10/16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pritzker KP, Gay S, Jimenez SA, et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage 2006;14(1):13–29. doi: 10.1016/j.joca.2005.07.014 [published Online First: 2005/10/26] [DOI] [PubMed] [Google Scholar]
- 25.Kwok J, Onuma H, Olmer M, et al. Histopathological analyses of murine menisci: implications for joint aging and osteoarthritis. Osteoarthritis Cartilage 2016;24(4):709–18. doi: 10.1016/j.joca.2015.11.006 [published Online First: 2015/11/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krenn V, Morawietz L, Haupl T, et al. Grading of chronic synovitis--a histopathological grading system for molecular and diagnostic pathology. Pathol Res Pract 2002;198(5):317–25. doi: 10.1078/0344-0338-5710261 [published Online First: 2002/07/03] [DOI] [PubMed] [Google Scholar]
- 27.Lee KI, Gamini R, Olmer M, et al. Mohawk is a transcription factor that promotes meniscus cell phenotype and tissue repair and reduces osteoarthritis severity. Sci Transl Med 2020;12(567) doi: 10.1126/scitranslmed.aan7967 [published Online First: 2020/10/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deuis JR, Dvorakova LS, Vetter I. Methods Used to Evaluate Pain Behaviors in Rodents. Front Mol Neurosci 2017;10:284. doi: 10.3389/fnmol.2017.00284 [published Online First: 2017/09/22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran PB, Miller RE, Ishihara S, et al. Spinal microglial activation in a murine surgical model of knee osteoarthritis. Osteoarthritis Cartilage 2017;25(5):718–26. doi: 10.1016/j.joca.2016.09.007 [published Online First: 2016/09/21] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye C, Ho DJ, Neri M, et al. DRUG-seq for miniaturized high-throughput transcriptome profiling in drug discovery. Nat Commun 2018;9(1):4307. doi: 10.1038/s41467-018-06500-x [published Online First: 2018/10/20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao X, Zhang J, Jing X, et al. Fibroblast growth factor 18 exerts anti-osteoarthritic effects through PI3K-AKT signaling and mitochondrial fusion and fission. Pharmacol Res 2019;139:314–24. doi: 10.1016/j.phrs.2018.09.026 [published Online First: 2018/10/03] [DOI] [PubMed] [Google Scholar]
- 32.Greenblatt MB, Ritter SY, Wright J, et al. NFATc1 and NFATc2 repress spontaneous osteoarthritis. Proceedings of the National Academy of Sciences of the United States of America 2013;110(49):19914–9. doi: 10.1073/pnas.1320036110 [published Online First: 2013/11/20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qu H, Li J, Wu LD, et al. Trichostatin A increases the TIMP-1/MMP ratio to protect against osteoarthritis in an animal model of the disease. Mol Med Rep 2016;14(3):2423–30. doi: 10.3892/mmr.2016.5523 [published Online First: 2016/07/20] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi M, Shi S, Li T, et al. TIMP2 deficient mice develop accelerated osteoarthritis via promotion of angiogenesis upon destabilization of the medial meniscus. Biochem Biophys Res Commun 2012;423(2):366–72. doi: 10.1016/j.bbrc.2012.05.132 [published Online First: 2012/06/06] [DOI] [PubMed] [Google Scholar]
- 35.Nakamura H, Vo P, Kanakis I, et al. Aggrecanase-selective tissue inhibitor of metalloproteinase-3 (TIMP3) protects articular cartilage in a surgical mouse model of osteoarthritis. Sci Rep 2020;10(1):9288. doi: 10.1038/s41598-020-66233-0 [published Online First: 2020/06/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee J, Smeriglio P, Dragoo J, et al. CD24 enrichment protects while its loss increases susceptibility of juvenile chondrocytes towards inflammation. Arthritis Res Ther 2016;18(1):292. doi: 10.1186/s13075-016-1183-y [published Online First: 2016/12/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jakkula E, Melkoniemi M, Kiviranta I, et al. The role of sequence variations within the genes encoding collagen II, IX and XI in non-syndromic, early-onset osteoarthritis. Osteoarthritis Cartilage 2005;13(6):497–507. doi: 10.1016/j.joca.2005.02.005 [published Online First: 2005/06/01] [DOI] [PubMed] [Google Scholar]
- 38.Alexopoulos LG, Youn I, Bonaldo P, et al. Developmental and osteoarthritic changes in Col6a1-knockout mice: biomechanics of type VI collagen in the cartilage pericellular matrix. Arthritis Rheum 2009;60(3):771–9. doi: 10.1002/art.24293 [published Online First: 2009/02/28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuo CL, Liu ST, Chang YL, et al. Zac1 regulates IL-11 expression in osteoarthritis. Oncotarget 2018;9(65):32478–95. doi: 10.18632/oncotarget.25980 [published Online First: 2018/09/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attur M, Duan X, Cai L, et al. Periostin loss-of-function protects mice from post-traumatic and age-related osteoarthritis. Arthritis Res Ther 2021;23(1):104. doi: 10.1186/s13075-021-02477-z [published Online First: 2021/04/10] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroder A, Nazet U, Muschter D, et al. Impact of Mechanical Load on the Expression Profile of Synovial Fibroblasts from Patients with and without Osteoarthritis. Int J Mol Sci 2019;20(3) doi: 10.3390/ijms20030585 [published Online First: 2019/02/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tu M, Yao Y, Qiao FH, et al. The pathogenic role of connective tissue growth factor in osteoarthritis. Biosci Rep 2019;39(7) doi: 10.1042/BSR20191374 [published Online First: 2019/07/03] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang Y, Xiao Q, Hu Z, et al. Tissue levels of leukemia inhibitory factor vary by osteoarthritis grade. Orthopedics 2014;37(5):e460–4. doi: 10.3928/01477447-20140430-57 [published Online First: 2014/05/09] [DOI] [PubMed] [Google Scholar]
- 44.Takahata Y, Nakamura E, Hata K, et al. Sox4 is involved in osteoarthritic cartilage deterioration through induction of ADAMTS4 and ADAMTS5. FASEB J 2019;33(1):619–30. doi: 10.1096/fj.201800259R [published Online First: 2018/07/18] [DOI] [PubMed] [Google Scholar]
- 45.Chang SH, Mori D, Kobayashi H, et al. Excessive mechanical loading promotes osteoarthritis through the gremlin-1-NF-kappaB pathway. Nat Commun 2019;10(1):1442. doi: 10.1038/s41467-019-09491-5 [published Online First: 2019/03/31] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu CJ, Kong W, Xu K, et al. ADAMTS-12 associates with and degrades cartilage oligomeric matrix protein. The Journal of biological chemistry 2006;281(23):15800–8. doi: 10.1074/jbc.M513433200 [published Online First: 2006/04/14] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang CY, Chanalaris A, Troeberg L. ADAMTS and ADAM metalloproteinases in osteoarthritis - looking beyond the ‘usual suspects’. Osteoarthritis Cartilage 2017;25(7):1000–09. doi: 10.1016/j.joca.2017.02.791 [published Online First: 2017/02/22] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mehta S, Akhtar S, Porter RM, et al. Interleukin-1 receptor antagonist (IL-1Ra) is more effective in suppressing cytokine-induced catabolism in cartilage-synovium co-culture than in cartilage monoculture. Arthritis Res Ther 2019;21(1):238. doi: 10.1186/s13075-019-2003-y [published Online First: 2019/11/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Z, Song Y, Yi Y, et al. Blockade of IL-33 signalling attenuates osteoarthritis. Clin Transl Immunology 2020;9(10):e1185. doi: 10.1002/cti2.1187 [published Online First: 2020/11/03] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic acids research 2000;28(1):27–30. doi: 10.1093/nar/28.1.27 [published Online First: 1999/12/11] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maruyama T, Jiang M, Abbott A, et al. Rap1b Is an Effector of Axin2 Regulating Crosstalk of Signaling Pathways During Skeletal Development. J Bone Miner Res 2017;32(9):1816–28. doi: 10.1002/jbmr.3171 [published Online First: 2017/05/19] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anuja K, Kar M, Chowdhury AR, et al. Role of telomeric RAP1 in radiation sensitivity modulation and its interaction with CSC marker KLF4 in colorectal cancer. Int J Radiat Biol 2020;96(6):790–802. doi: 10.1080/09553002.2020.1721609 [published Online First: 2020/01/28] [DOI] [PubMed] [Google Scholar]
- 53.Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol 2011;21(10):615–23. doi: 10.1016/j.tcb.2011.07.001 [published Online First: 2011/08/09] [DOI] [PubMed] [Google Scholar]
- 54.Cohen JD, Tham KY, Mastrandrea NJ, et al. cAMP-dependent cytosolic mislocalization of p27(kip)-cyclin D1 during quinol-thioether-induced tuberous sclerosis renal cell carcinoma. Toxicol Sci 2011;122(2):361–71. doi: 10.1093/toxsci/kfr118 [published Online First: 2011/06/23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grewal SS, Fass DM, Yao H, et al. Calcium and cAMP signals differentially regulate cAMP-responsive element-binding protein function via a Rap1-extracellular signal-regulated kinase pathway. The Journal of biological chemistry 2000;275(44):34433–41. doi: 10.1074/jbc.M004728200 [published Online First: 2000/08/22] [DOI] [PubMed] [Google Scholar]
- 56.Otani T, Mizokami A, Hayashi Y, et al. Signaling pathway for adiponectin expression in adipocytes by osteocalcin. Cell Signal 2015;27(3):532–44. doi: 10.1016/j.cellsig.2014.12.018 [published Online First: 2015/01/07] [DOI] [PubMed] [Google Scholar]
- 57.Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome biology 2003;4(2):206. doi: 10.1186/gb-2003-4-2-206 [published Online First: 2003/03/07] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang X, Zhang Q, Gao Z, et al. Down-Regulation of MiR-150 Alleviates Inflammatory Injury Induced by Interleukin 1 via Targeting Kruppel-Like Factor 2 in Human Chondrogenic Cells. Cell Physiol Biochem 2018;47(6):2579–88. doi: 10.1159/000491654 [published Online First: 2018/07/12] [DOI] [PubMed] [Google Scholar]
- 59.Chang SF, Huang KC, Chang HI, et al. 2 dyn/cm(2) shear force upregulates kruppel-like factor 4 expression in human chondrocytes to inhibit the interleukin-1beta-activated nuclear factor-kappaB. J Cell Physiol 2018;234(1):958–68. doi: 10.1002/jcp.26924 [published Online First: 2018/08/23] [DOI] [PubMed] [Google Scholar]
- 60.Zhang X, Nham GTH, Ito K, et al. Gene expression of type II collagen is regulated by direct interaction with Kruppel-like factor 4 and AT-rich interactive domain 5B. Gene 2021;773:145381. doi: 10.1016/j.gene.2020.145381 [published Online First: 2021/01/01] [DOI] [PubMed] [Google Scholar]
- 61.Yu SM, Kim SJ. Kruppel-like factor 4 (KLF-4) plays a crucial role in simvastatin (SVT)-induced differentiation of rabbit articular chondrocytes. Biochem Biophys Res Commun 2018;501(3):814–19. doi: 10.1016/j.bbrc.2018.05.094 [published Online First: 2018/05/19] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GEO accession numbers for all the datasets utilized in the present study are available in the supplementary materials and methods.


