Abstract
Studies of the Vibrio cholerae population, using molecular typing techniques, have shown the existence of several pathogenic clones, mainly sixth-pandemic, seventh-pandemic, and U.S. Gulf Coast clones. However, the relationship of the pathogenic clones to environmental V. cholerae isolates remains unclear. A previous study to determine the phylogeny of V. cholerae by sequencing the asd (aspartate semialdehyde dehydrogenase) gene of V. cholerae showed that the sixth-pandemic, seventh-pandemic, and U.S. Gulf Coast clones had very different asd sequences which fell into separate lineages in the V. cholerae population. As gene trees drawn from a single gene may not reflect the true topology of the population, we sequenced the mdh (malate dehydrogenase) and hlyA (hemolysin A) genes from representatives of environmental and clinical isolates of V. cholerae and found that the mdh and hlyA sequences from the three pathogenic clones were identical, except for the previously reported 11-bp deletion in hlyA in the sixth-pandemic clone. Identical sequences were obtained, despite average nucleotide differences in the mdh and hlyA genes of 1.52 and 3.25%, respectively, among all the isolates, suggesting that the three pathogenic clones are closely related. To extend these observations, segments of the recA and dnaE genes were sequenced from a selection of the pathogenic isolates, where the sequences were either identical or substantially different between the clones. The results show that the three pathogenic clones are very closely related and that there has been a high level of recombination in their evolution.
Vibrio cholerae is a gram-negative bacterium which comprises part of the autochthonous microflora of aquatic environments, often found in close association with a variety of algae and crustaceans (13, 14, 22, 24). Of medical importance, however, is that certain members of the species have evolved mechanisms to become pathogenic to humans, with the potential to cause the severe life-threatening diarrheal disease cholera. A characteristic of the disease is its ability to emerge as explosive outbreaks in human populations. Since epidemiological records of cholera were initiated, the outbreaks have been divided into seven pandemics, with the fifth, sixth, and seventh pandemics caused by strains which carry the O1 antigen (39).
At the end of 1992, a strain of V. cholerae with a novel antigen emerged as a major cause of cholera around the Bay of Bengal in India and Bangladesh (11, 41). Prior to this, non-O1 V. cholerae strains were known to be responsible only for sporadic cases of gastroenteritis and for extraintestinal infections (36). The new form of the antigen was designated O139, and the strain is known as V. cholerae O139 Bengal, after its initial appearance in the Indian subcontinent. V. cholerae O139 Bengal rapidly spread through the immunologically naive populations in neighboring Asian countries. Genetic studies indicate it is closely related to the O1 seventh-pandemic clone and presumably arose by lateral transfer of genes for lipopolysaccharide biosynthesis from an O139 strain to an organism of the seventh-pandemic clone (4, 5).
The techniques traditionally used to assess the relationship between V. cholerae isolates were based mainly on the biochemical characteristics. Recently, various molecular biology-based techniques have been used to study the relationships among clinical and environmental isolates. They include multilocus enzyme electrophoresis (MLEE) (10, 16, 45), pulse-field gel electrophoresis (PFGE) (9), ribotyping (25, 40), and randomly amplified polymorphic DNA (RAPD) (43, 46), which have differentiated isolates of the V. cholerae population into different electrophoretic types (ETs or zymovars), PFGE types, ribotypes, and RAPD fingerprint types, respectively. Application of these molecular epidemiological techniques has shown the existence within the V. cholerae population of several pathogenic clones, primarily isolates considered to be remnants of the sixth pandemic, isolates from the seventh pandemic, and isolates from the U.S. Gulf Coast region of North America.
Another molecular technique which has been used to study the relationships of the pathogenic clones to the predominantly nontoxigenic, environmental isolates of V. cholerae is comparative nucleotide sequence analysis. This technique provides particularly valuable data for population genetic studies, aimed at determining the genetic structures of populations of bacteria and understanding the evolutionary processes that affect rates of nucleotide and amino acid substitutions. Karaolis et al. (26) analyzed the sequence variation in the asd gene from 45 isolates of V. cholerae. No variation was found within the sixth-pandemic, seventh-pandemic, or U.S. Gulf Coast clones, but the asd sequences of the three clones were not closely related.
A single locus may not be representative of a given genome, and as MLEE and other data are discordant with the conclusion from the asd sequences, we sequenced the mdh (malate dehydrogenase [MDH]) gene and a segment of the hlyA (hemolysin) gene from 32 isolates of V. cholerae. In contrast to findings for the asd gene, we found no variation in the mdh gene and hlyA gene (except for the 11-bp deletion in the sixth-pandemic clone) within or between isolates of the pathogenic clones, suggesting that the clones are very closely related. This observation was supported by the limited sequencing of segments of the recA and dnaE genes, which also showed that the level of recombination is high for V. cholerae.
MATERIALS AND METHODS
Bacterial isolates.
In this study, we examined a total of 33 V. cholerae isolates, comprising 13 O1 clinical isolates of the sixth-pandemic clone (M642, M644, M648, M967, and 569B), along with isolates from pre-seventh-pandemic outbreaks (M543, M640, M645, and M802), from the seventh-pandemic outbreak (M663 and M793), and from outbreaks in the U.S. Gulf Coast region (M794 and M796); 2 O1 nontoxigenic environmental isolates (M535 and M536); 2 O139 Bengal isolates (M539 and M831); and 16 environmental, nontoxigenic, non-O1, non-O139 isolates (M548, M549, M550, M551, M552, M553, M554, M555, M556, M557, M558, M559, M560, M561, M562, and M563). These isolates are from diverse geographical locations and were previously described by Karaolis et al. (26). Strain M967 (#75) is a sixth-pandemic isolate from Japan (1921), and 569B is a remnant of the sixth pandemic from India (1940). Vibrio mimicus M547 was selected for use as an outgroup.
DNA methods.
Isolates were stored at −70°C and subcultured onto nutrient agar, from which a single colony was selected and chromosomal DNA was extracted as previously described (3). PCR was performed in reaction mixtures containing 50 mM KCl, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, bovine serum albumin (200 μg/ml), 100 μM each deoxyribonucleoside triphosphate, 0.3 μM each primer, purified chromosomal DNA (∼10 ng/μl), and Taq polymerase (0.02 U/μl). Amplification was performed in an FTS-960 thermal cycler (Corbett Research) with the following program: denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 50 to 60°C for 15 s, and 72°C for 30 s, and a final cycle of 72°C for 10 min. Amplified products were resolved by 1% agarose gel electrophoresis with 0.5× Tris-borate-EDTA as the running buffer and visualized by ethidium bromide staining followed by UV transillumination. PCR primers used in this study are listed in Table 1. PCR amplicons were purified by using the Promega Wizard PCR purification system and sequenced by the dye terminator method at the Sydney University and Prince Alfred Hospital Macromolecular Analysis Centre, using a model 877 integrated thermal cycler and model 377 automated DNA sequencer (Applied Biosystems).
TABLE 1.
PCR primers used in this study
| Primer | Gene or purpose | Oligonucleotide sequence (5′-3′) | 5′ start positiona | Degen-eracy | Anneal-ing temp (°C) |
|---|---|---|---|---|---|
| 502 | mdh | GCNGGYGGYATYGGYCARGC | +25 | 128 | 50 |
| 457 | mdh | CCYTCIACRTAIGSACAYTC | +827 | 16 | 50 |
| 516 | Used in IPCR | CAGAACCAGCAGGAAGACGG | +85 | 1 | 50 |
| 517 | Used in IPCR | AAGCACTGCAAGGCGAATCT | +719 | 1 | 50 |
| 540 | mdh | GTTTGACGGTCGGATACACC | −67 | 1 | 60 |
| 541 | mdh | AGAGCGGTATTTTCCAATGC | +972 | 1 | 60 |
| 584 | mdh | AAAGTYGCWGTHMTYGGYGC | +4 | 96 | 45 |
| 585 | mdh | BMWGCYGCYTGRCCCATAGA | +696 | 96 | 45 |
| 644 | hlyA | GCCAAAACTCAATCGTTCCG | +2 | 1 | 60 |
| 645 | hlyA | TGTAAAGCTAACCGCTTGCG | +1078 | 1 | 60 |
| 884 | recA | TGGACGAGAATAAACAGAAGGC | +2 | 1 | 60 |
| 885 | recA | AACCTCTTTGCATTCAGCCC | +1069 | 1 | 60 |
| 713 | dnaE | GATTTCTCTATGGTGGATGG | +39 | 1 | 60 |
| 714 | dnaE | ATTCCAGCGGATCAAGGTCG | +1130 | 1 | 60 |
Relative to the adenosine of the translational start codon ATG.
Sequencing of the mdh gene of V. cholerae M793.
Degenerate primers 502 and 457 were based on highly conserved regions in MDH of closely related species (Photobacterium spp. [accession no. P37226 {52}], Escherichia coli [accession no. M24777 {50}], and Salmonella enterica [accession no. P25077 {31}]) and used in a PCR with chromosomal DNA from strain M793. The expected fragment of 742 bp was excised from a 1% low-melting-temperature agarose gel with 1× Tris-acetate-EDTA as the running buffer, purified by using the Promega Wizard PCR purification system, and ligated into the cloning vector pGEM-T (Promega) for dye-labeled primer sequencing.
Primers 516 and 517, based on the partial mdh sequence of V. cholerae M793, were used in an inverse PCR (IPCR) (21) to amplify the flanking regions. The primers amplified a fragment of approximately 800 bp from the template DNA derived by digestion with PstI, which was cloned and sequenced as described above. Amplification of IPCR fragments from templates obtained from digestions with AccI, StyI, NcoI, SalI, SphI, XhoI, AflII, BclI, BglII, BssHII, EcoRI, SacI, and XbaI was unsuccessful.
Sequencing of the mdh, hlyA, recA, and dnaE genes from selected V. cholerae strains.
Primers 540 and 541, based on the flanking sequences of the mdh gene of strain M793, were used to amplify and sequence a 1,039-bp fragment containing the entire 936-bp coding region of the mdh gene from purified chromosomal DNA. These primers failed to amplify from V. mimicus M547 and V. cholerae M552; however, PCR amplicons and partial mdh sequences were obtained from these isolates by using degenerate primers 584 and 585.
Primers 644 and 645, based on the hlyA sequence of V. cholerae 017 (accession no. Y00557 [1]), were used to amplify and sequence a 1,038-bp segment of the 2,226-bp coding sequence of the hlyA gene. Primers 644 and 645 failed to amplify from strains M547 and M552, despite the lower stringency of annealing conditions.
Primers 884 and 885, based on the recA sequence of V. cholerae 017 (accession no. X71969 [49]), were used to amplify and sequence a 1,041-bp segment of the 1,061-bp coding sequence of the recA gene. Primers 713 and 714, based on the dnaE sequence of V. cholerae C6706 (accession no. U30472 [19]), were used to amplify and sequence a 1,067-bp segment of the 3,477-bp coding region of dnaE.
Computer analysis of the sequences.
The nucleotide sequences of the mdh, hlyA, recA, and dnaE genes were edited and assembled with the TED (20) and GAP4 (47) programs. Sequences were aligned with the CLUSTALW program, and phylogenetic analysis was performed with PHYLIP (17) and MULTICOMP (42). These programs are accessed through the Australian National Genomic Information Service at the University of Sydney.
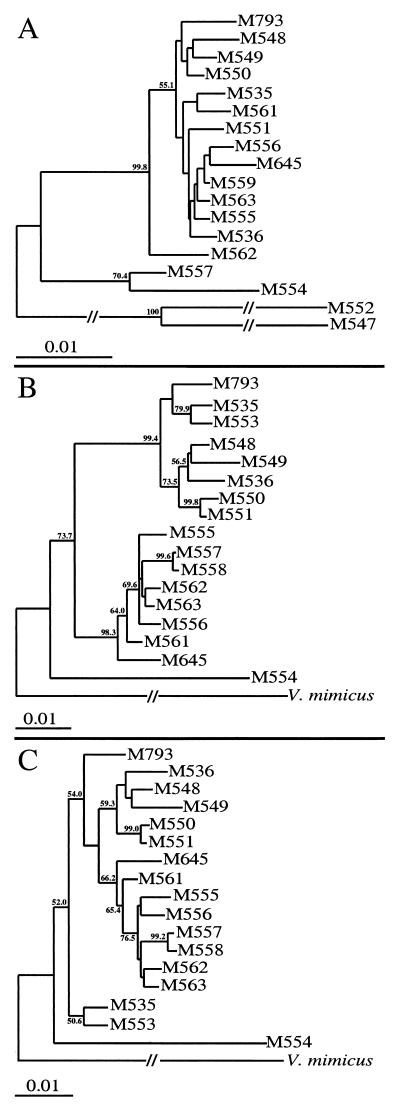
Phylogenetic trees were constructed by the neighbor-joining method (44) for the mdh and hlyA genes (Fig. 2). The mdh gene tree was rooted with the partial mdh sequence from V. mimicus M547. The hlyA gene tree was rooted with the vmhA sequence of V. mimicus (accession no. U68271 [28]), which shows 76% nucleotide identity to hlyA from pathogenic isolates and is clearly the same gene with a different name.
FIG. 2.
Phylogenetic tree for the mdh gene (A) and for the hlyA gene with (B) and without (C) the 60-bp region of recombination. The mdh tree was rooted with the partial mdh sequence of strain M547. The hlyA trees were rooted with the vmhA gene sequence of V. mimicus. Bootstrap values are percentages of 1,000 computer-generated trees and are shown at the nodes. Values of less than 50 are not shown.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequences determined in this study are AF117833 to AF117883.
RESULTS
Sequence of the V. cholerae mdh gene.
At the time this study was initiated, no suitable housekeeping genes from V. cholerae were available in the databases. The mdh gene, coding for the metabolic enzyme MDH, was selected because it has been used previously for population studies, which would enable comparison of mdh variation between species (7). Also, the mdh gene trees constructed for E. coli and S. enterica isolates were shown to be congruent with phylogenetic relationships inferred from MLEE.
The mdh gene from strain M793 was sequenced by degenerate PCR and IPCR. The mdh gene shows high levels of similarity to the mdh genes of other bacteria: 72.33% identity to mdhA of a Photobacterium sp. (81.4% amino acid identity; accession no. P37226) and 72.44% identity to mdh of E. coli (79.8% amino acid identity; accession no. M24777). Surprisingly, the V. cholerae mdh gene shows only 70.5% identity to the mdh gene from a psychrophilic Vibrio sp. (37), which shows greater similarity to the mdhA gene of a Photobacterium sp. (74% identity). If the difference in the mdh genes of V. cholerae and the Vibrio sp. isolate reflects the overall genetic differences in their genomes, the taxonomical classification of these isolates requires reassessment.
The mdh gene of V. cholerae is 936 bp in length, coding for 311 amino acids. Compared to other bacterial mdh sequences, the V. cholerae MDH is one amino acid shorter than the 312-residue MDH of other, closely related species (37, 52).
Within the sequence obtained from the 0.8-kb IPCR fragment, a partial open reading frame was identified 327 bp upstream of mdh and in the opposite orientation (data not shown). The open reading frame identified shows 70.7% identity to the argR gene of E. coli (accession no. M17532 [30]). The argR gene is also found upstream of and in the opposite orientation from mdh in the E. coli and Haemophilus influenzae genomes, which suggests that the gene order around the mdh locus has been conserved in these bacteria.
Nucleotide sequence variation in V. cholerae mdh.
The nucleotide sequence of the 936-bp coding region of mdh was determined for 32 isolates and a partial mdh sequence obtained from strain M552. Among the 32 complete mdh sequences, 16 variants were identified. Interestingly, 14 of the 15 pathogenic isolates used in this study have identical mdh sequences, the exception being the pre-seventh-pandemic strain M645. Other isolates with identical mdh sequences were environmental isolates M535 and M553, M557 and M558, and M559 and M560. For comparative analysis, the 16 unique mdh sequences were used.
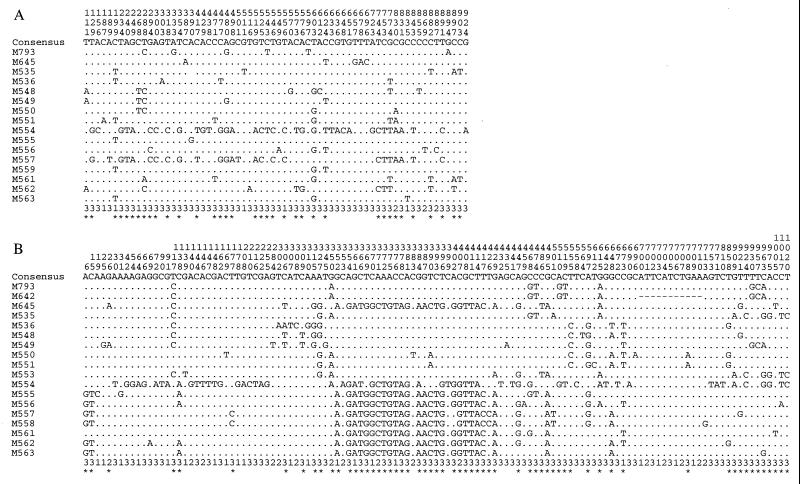
There were 64 polymorphic sites within the mdh sequences analyzed (Fig. 1), with the majority of the substitutions occurring at the 3′ end of the gene (codons 151 to 311). Similarly, the polymorphisms detected in mdh of E. coli and S. enterica were mostly within the 3′ region of the gene (7). Of the 64 nucleotide substitutions, 57 occurred at the third base of a codon, 1 was at the second base, and 6 were at the first base. Four nonsynonymous substitutions, representing 1.28% of the 311 codons, were found. Of the 64 polymorphic sites, 36 were phylogenetically informative (at least two bases present in two or more of the 16 sequences).
FIG. 1.
Polymorphic sites within the mdh (A) and hlyA (B) genes of V. cholerae isolates. (A) Polymorphic sites within unique mdh sequences. M793 represents the sixth-pandemic, seventh-pandemic, and U.S. Gulf Coast clones, M535 also represents M553, M557, also represents M558, and M559 also represents M560. (B) Polymorphic sites within the 1,038-bp segment of the hlyA gene. M793 represents the seventh-pandemic and U.S. Gulf Coast clones, M642 represents the sixth-pandemic clone, and M555 also represents M559 and M560. Numbering of the polymorphic sites (vertical format) are from the first position of the sequence segment. The position within the codon for each polymorphic site is shown below the sequences. Asterisks indicate polymorphic sites which are informative.
The average pairwise difference for the 16 mdh sequence variants was 1.52%, with a maximum of 4.49% observed between strains M553 and M554 (Table 2). This is similar to the average pairwise difference for 21 unique sequences of asd (1.41%). Average nucleotide differences in mdh variants from E. coli and S. enterica populations were 1.1 and 4.5%, respectively, (7).
TABLE 2.
Nucleotide differences between mdh and hlyA genes among isolatesa
| M793 | M645 | M535 | M536 | M548 | M549 | M550 | M551 | M553 | M554 | M555 | M556 | M557 | M558 | M559 | M560 | M561 | M562 | M563 | M552 | M547 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M793 | 1.18 | 1.18 | 1.07 | 1.28 | 0.75 | 0.85 | 1.28 | 1.18 | 4.17 | 0.85 | 1.07 | 3.10 | 3.10 | 0.95 | 0.95 | 1.28 | 1.50 | 0.96 | 12.23 | 10.60 | |
| M645 | 3.95 | 1.07 | 0.96 | 1.39 | 0.85 | 0.96 | 1.18 | 1.07 | 4.27 | 0.75 | 0.96 | 3.21 | 3.21 | 0.64 | 0.64 | 1.28 | 1.60 | 0.85 | 10.60 | 11.93 | |
| M535 | 1.16 | 4.05 | 0.75 | 1.39 | 1.07 | 0.96 | 0.96 | 0.00 | 4.49 | 0.53 | 0.96 | 3.21 | 3.21 | 0.64 | 0.64 | 0.64 | 1.28 | 0.64 | 12.08 | 10.45 | |
| M536 | 2.02 | 3.95 | 2.12 | 1.07 | 0.96 | 0.85 | 0.64 | 0.75 | 4.06 | 0.43 | 1.07 | 2.78 | 2.78 | 0.53 | 0.53 | 0.96 | 1.28 | 0.53 | 10.45 | 11.77 | |
| M548 | 1.83 | 3.47 | 1.93 | 0.96 | 0.75 | 0.64 | 1.07 | 1.39 | 4.38 | 1.07 | 1.28 | 3.31 | 3.31 | 0.96 | 0.96 | 1.07 | 1.28 | 0.96 | 10.91 | 12.23 | |
| M549 | 1.64 | 4.05 | 2.41 | 1.64 | 1.16 | 0.53 | 1.18 | 1.07 | 4.06 | 0.75 | 0.96 | 2.99 | 2.99 | 0.64 | 0.64 | 1.18 | 1.18 | 0.85 | 10.75 | 11.77 | |
| M550 | 1.93 | 4.24 | 2.02 | 1.35 | 1.25 | 2.02 | 0.64 | 0.96 | 3.95 | 0.64 | 0.85 | 2.88 | 2.88 | 0.53 | 0.53 | 0.85 | 1.28 | 0.53 | 10.45 | 11.93 | |
| M551 | 1.73 | 4.05 | 1.83 | 1.16 | 1.06 | 1.83 | 0.39 | 0.96 | 3.95 | 0.64 | 1.07 | 2.88 | 2.88 | 0.53 | 0.53 | 0.75 | 1.50 | 0.53 | 10.60 | 12.08 | |
| M553 | 1.73 | 3.47 | 0.77 | 2.31 | 2.12 | 2.60 | 2.22 | 2.02 | 4.49 | 0.53 | 1.18 | 3.21 | 3.21 | 0.64 | 0.64 | 0.64 | 1.28 | 0.64 | 10.45 | 12.08 | |
| M554 | 5.78 | 5.59 | 5.39 | 7.13 | 6.94 | 7.23 | 6.84 | 6.65 | 5.59 | 4.17 | 3.95 | 1.71 | 1.71 | 3.85 | 3.85 | 4.27 | 3.95 | 4.06 | 10.14 | 11.01 | |
| M555 | 3.76 | 1.73 | 4.05 | 4.24 | 4.05 | 4.43 | 4.05 | 3.85 | 4.24 | 5.39 | 0.85 | 2.88 | 2.88 | 0.32 | 0.32 | 0.96 | 1.28 | 0.32 | 10.45 | 12.08 | |
| M556 | 4.05 | 1.45 | 4.24 | 4.14 | 3.95 | 4.34 | 3.95 | 3.76 | 3.85 | 5.20 | 0.87 | 3.10 | 3.10 | 0.53 | 0.53 | 1.18 | 1.71 | 0.75 | 10.45 | 11.47 | |
| M557 | 4.14 | 1.54 | 4.34 | 4.14 | 3.66 | 4.43 | 3.85 | 3.66 | 4.05 | 5.78 | 1.16 | 1.06 | 0.00 | 2.99 | 2.99 | 3.21 | 3.10 | 2.99 | 11.06 | 12.23 | |
| M558 | 4.14 | 1.73 | 4.34 | 4.14 | 3.85 | 4.43 | 3.85 | 3.66 | 4.05 | 5.78 | 1.16 | 1.06 | 0.19 | 2.99 | 2.99 | 3.21 | 3.10 | 2.99 | 11.06 | 12.23 | |
| M559 | 3.76 | 1.73 | 4.05 | 4.24 | 4.05 | 4.43 | 4.05 | 3.85 | 4.24 | 5.39 | 0.00 | 0.87 | 1.16 | 1.16 | 0.00 | 0.85 | 1.39 | 0.21 | 10.45 | 11.62 | |
| M560 | 3.76 | 1.73 | 4.05 | 4.24 | 4.05 | 4.43 | 4.05 | 3.85 | 4.24 | 5.39 | 0.00 | 0.87 | 1.16 | 1.16 | 0.00 | 0.85 | 1.39 | 0.21 | 10.45 | 11.62 | |
| M561 | 3.56 | 0.96 | 3.85 | 3.85 | 3.66 | 4.05 | 3.66 | 3.47 | 3.66 | 5.01 | 0.96 | 0.87 | 1.16 | 1.16 | 0.96 | 0.96 | 1.18 | 0.85 | 10.45 | 11.93 | |
| M562 | 4.14 | 1.54 | 4.43 | 3.95 | 3.85 | 4.43 | 3.66 | 3.47 | 4.05 | 5.39 | 0.96 | 0.67 | 0.96 | 0.96 | 0.96 | 0.96 | 0.77 | 1.39 | 10.29 | 11.77 | |
| M563 | 3.76 | 1.45 | 3.95 | 4.05 | 3.76 | 4.24 | 3.76 | 3.56 | 3.76 | 5.11 | 0.67 | 0.77 | 0.87 | 0.87 | 0.67 | 0.67 | 0.87 | 0.48 | 10.29 | 11.77 | |
| M552 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 10.45 | |
| Vm | 23.27 | 22.35 | 22.93 | 23.22 | 22.93 | 23.70 | 23.31 | 23.41 | 23.03 | 23.70 | 22.54 | 22.45 | 22.93 | 23.12 | 22.54 | 22.54 | 22.64 | 22.45 | 22.45 | NA |
Percent nucleotide differences in the mdh (upper right) and hlyA (lower left) genes of V. cholerae isolates. M793 represents the sixth-pandemic, seventh-pandemic, and U.S. Gulf Coast clones; Vm is the vmhA sequence of V. mimicus (accession no. U68271). NA, not applicable.
The mdh sequences of the pathogenic isolates (except M645) were all identical, suggesting that the pathogenic clones are very closely related. The sequence is most similar to that of the environmental isolate M549, with seven synonymous substitutions and a pairwise difference of 0.75%. The pre-seventh-pandemic isolate M645 differed from the other pathogenic isolates at 1.18% of the nucleotide sites and was most similar to environmental isolates M559 and M560 (0.64% difference).
Partial mdh sequences from V. mimicus M547 and from strain M552 revealed that the average pairwise difference between the typical V. cholerae isolates and M547 is 10.52%, whereas M552 differed from the other V. cholerae isolates at 11.87% and from M547 at 10.45% of the nucleotide sites. The level of divergence of strain M552 from the other V. cholerae isolates and from V. mimicus suggests that it may represent a different species.
hlyA.
The hlyA gene codes for the hemolysin which is traditionally used to differentiate between the two biotypes of V. cholerae. It was selected for this study because it is ubiquitous within V. cholerae (8), suggesting that it probably plays a role in the survival of V. cholerae in its natural environment. The loss of hemolytic function among isolates of the sixth pandemic and the gradual loss in isolates from the seventh pandemic would indicate that it is not essential for human pathogenesis (2).
The nucleotide sequence of a 1,038-bp segment of hlyA, extending from codons 12 to 358 and representing 46.6% of the 2,226-bp coding region of hlyA, was determined for 32 V. cholerae isolates. This segment could not be amplified from V. cholerae M552 and V. mimicus M547, even when less stringent conditions were used. From the 32 isolates studied, 17 different hlyA sequences were identified. Similar to the findings for the mdh gene, 14 of the 15 pathogenic isolates have the same hlyA sequence (except for the 11-bp deletion in sixth-pandemic isolates [1]). A different hlyA sequence is also shared by the environmental isolates M555, M559, and M560.
Analysis of the 17 hlyA sequence variants revealed 121 polymorphic nucleotide sites (Fig. 1): 84 at the third base of a codon, 16 at the second base, and 21 at the first base. We found 61 polymorphic sites to be phylogenetically informative, with detection of 24 nonsynonymous substitutions, which represents 6.94% of the 346 residues studied. The average pairwise percent difference within the V. cholerae isolates studied was 3.21%, with a maximum difference of 7.23% observed between isolates M549 and M554 (Table 2). The level of variation in hlyA is more than twice that observed in asd and mdh. The hlyA sequence from the pathogenic isolates is most closely related to the hlyA sequence of environmental O1 isolate M535, with a difference of 1.16% of the nucleotides.
Evidence for recombination.
Application of the Stephens test for nonrandom clustering of polymorphic nucleotide sites (48) revealed no detectable cases of intragenic recombination over the 936-bp coding region of mdh. However, a significant partition of 60 bp (bases 354 to 414) (Fig. 1) supported by 16 sites was detected in the hlyA sequences, which separates all of the pathogenic strains (except M645) and the environmental isolates M535, M536, M548, M549, M550, M551, and M553 from the other isolates studied (P < 0.00001). Of the 16 sites, 4 were nonsynonymous, resulting in three amino acid substitutions. As several environmental isolates were affected by the recombination event, which is obscured by subsequent mutation within or proximal to the recombinant region, the recombination event probably occurred significantly before the emergence of the V. cholerae pathogenic clones. Omitting this region, the average pairwise difference between isolates falls to 2.25%, still higher than that for asd or mdh.
Phylogenetic analysis.
Phylogenetic trees constructed from the mdh and hlyA sequences (Fig. 2), with and without the regions involved in recombination, show few examples of congruence between the two trees. Low bootstrap values were obtained for most of the nodes in the mdh gene tree, possibly due to recombinational events in mdh which were not detected by the Stephens test. The recombination in hlyA distributed the set of strains into two distinct clusters, which is evident even with the 60-bp segment omitted.
Strain M554 is the most divergent V. cholerae strain, as was expected from the pairwise comparisons, although in the mdh tree, strains M557 and M558 cluster with strain M554, whereas in the hlyA tree, they cluster with the other isolates. The clinical pre-seventh-pandemic isolate M645 is found in different clusters than the other pathogenic isolates in both gene trees and has a significant sequence divergence from them in the mdh and hlyA genes, with 1.18 and 3.92% nucleotide differences, respectively.
Nucleotide sequence variation in recA and dnaE.
There is no variation within the mdh and hlyA genes for 14 pathogenic isolates of V. cholerae, whereas three distinct sequences are found for asd. To extend these observations, two more genes were selected for sequencing. At the time these experiments were done, sequences of housekeeping genes from biosynthetic pathways more traditionally used for such studies were not available in the databases. Segments of the recA (coding for the RecA protein involved in homologous recombination) and dnaE (coding for the α subunit of the DNA polymerase III holoenzyme) genes were selected because they encode proteins involved in housekeeping roles and therefore are not expected to be under diversifying selection. Sequences were obtained from four isolates of the sixth-pandemic clone (M642, M648, M967, and 569B), four pre-seventh-pandemic outbreak isolates (M645, M802, M543, and M640), two seventh-pandemic isolates (M793 and M663), two U.S. Gulf Coast isolates (M794 and M796), and two non-O1, non-O139 environmental isolates (M549 and M553) which are closely related to the pathogenic isolates in the mdh and hlyA genes.
recA.
A 1,041-bp fragment of the recA gene, from positions 25 to 1065 (residues 9 to 354), representing 98.11% of the 1,061-bp coding region, was sequenced for the 14 selected V. cholerae isolates. All the pre-seventh-pandemic (except M645), seventh-pandemic, and U.S. Gulf Coast isolates had the same recA sequence, which is identical to the published recA sequence (accession no. U10162 [32]), in agreement with the differences noted for previously published recA sequences (accession no. X71969 [49] and X61384). The recA sequence of the sixth-pandemic clone differs from that of the seventh-pandemic and U.S. Gulf Coast clones at 48 nucleotide sites, or 4.59% the 1,041-bp segment (Table 3). Of the 48 substitutions, 3 were nonsynonymous. The recA sequence from isolates of the sixth-pandemic clone in this study, one of which is strain 569B, differed from the GenBank sequence of recA from strain 569B (accession no. L42384), which is identical to the recA sequence (U10162) from a U.S. Gulf Coast isolate. We believe that GenBank entry L42384 is recA of a seventh-pandemic strain.
TABLE 3.
Nucleotide differences between recA and dnaE genes among isolatesa
| M793 | M642 | M794 | M645 | M549 | M553 | |
|---|---|---|---|---|---|---|
| M793 | 4.59 | 0.00 | 2.49 | 4.02 | 2.39 | |
| M642 | 0.00 | 4.59 | 3.63 | 2.10 | 3.44 | |
| M794 | 1.97 | 1.97 | 2.49 | 4.02 | 2.39 | |
| M645 | 1.97 | 1.97 | 2.25 | 2.87 | 0.67 | |
| M549 | 1.59 | 1.59 | 0.56 | 1.87 | 2.58 | |
| M553 | 0.28 | 0.28 | 1.87 | 1.87 | 1.50 |
Percent nucleotide differences in the recA (upper right) and dnaE (lower left) genes of V. cholerae isolates. M793, M642, and M794 represent seventh-pandemic, sixth-pandemic, and U.S. Gulf Coast clones, respectively.
The recA sequence difference between the sixth- and seventh-pandemic clones was greater than that observed between the clones in the asd locus. Visual inspection of the polymorphic sites (Fig. 3) within the recA sequences shows that the sixth-pandemic clone and the environmental isolate M549 differ substantially from the other recA sequences between bases 765 and 1011 at 15 sites, indicative of a recombination event. This conclusion is supported by statistical analysis of the data, using the Stephens test, which detected a significant partition, supported by the 15 sites, between (i) strain M549 and the sixth-pandemic isolates and (ii) the other V. cholerae isolates studied (P < 0.00001).
FIG. 3.
Polymorphic sites within the 1,041-bp fragment of recA (A) and the 1,067-bp fragment of dnaE (B) of selected V. cholerae isolates. (A) M793 represents the seventh-pandemic and U.S. Gulf Coast clones, and M642 represents the sixth-pandemic clone. (B) M793 represents the sixth- and seventh-pandemic clones, and M794 represents the U.S. Gulf Coast clone. See the legend to Fig. 1 for further details.
dnaE.
A 1,067-bp fragment of the dnaE gene, from positions 1 to 1128 (residues 21 to 376), representing 30.69% of the 3,477-bp coding region, was sequenced for the 14 selected V. cholerae isolates. The sequences revealed that isolates of the sixth- and seventh-pandemic clones and from pre-seventh-pandemic outbreaks (except M645) all have the same dnaE sequence, which differs from that of the U.S. Gulf Coast clone at 21 synonymous sites, which represents 1.97% of the 1,067-bp sequence (Table 3). There was one site of conflict between the sequence from the seventh-pandemic clone and the published dnaE sequence of strain C6706 (accession no. U30472 [19]), at position 1027, which results in an amino acid substitution from Val to Ile at residue 343. However, as the dnaE sequences from 10 isolates in this study were identical, we are confident that our sequence is correct.
The dnaE sequence of the U.S. Gulf Coast clone was most similar to that of the environmental isolate M549, differing at only six nucleotide sites, which represents 0.56% of the region sequenced. Visual inspection of the polymorphic sites (Fig. 3) suggests that the U.S. Gulf Coast clone and M549 may have undergone recombination between bases 135 and 645, an inference supported by application of the Stephens test (P < 0.02594). The dnaE sequence from the sixth- and seventh-pandemic clones is most similar to that of environmental isolate M553, with a pairwise difference of 0.28%.
DISCUSSION
The species V. cholerae shows considerable variation in ETs (16, 45), ribotypes (25, 29, 40), PFGE types (9), RAPD fingerprint types (43, 46), and insertion sequence fingerprint types (6). MLEE studies, which analyze the electrophoretic mobility differences in multiple housekeeping enzymes to determine strain relationships, have differentiated several ETs among pathogenic isolates, being remnants of the sixth-pandemic isolates seventh-pandemic (including O139 Bengal) isolates, isolates from the Latin American outbreak and from outbreaks in the U.S. Gulf Coast region (10), and clinical isolates from Australia (15) and the Amazon rainforest (12). The pathogenic isolates all exhibit similar electrophoretic profiles, differing at only 1 to 3 of the 12 to 16 loci analyzed, implying a closer relationship among these pathogenic ETs than to most of the ETs represented by the predominantly nonpathogenic, environmental isolates, which display diverse electrophoretic patterns and serotypes.
One method to confirm and clarify the phylogenies inferred from the MLEE studies is comparative nucleotide sequence analysis of housekeeping genes, which detects synonymous as well as nonsynonymous substitutions in chromosomally located genes for pairwise comparisons and construction of phylogenetic trees.
Nucleotide sequence variation in mdh, hlyA, recA, and dnaE.
The mdh gene and a segment of the hlyA gene were analyzed for 32 V. cholerae isolates. Identical mdh and hylA sequences were found in isolates of the sixth-pandemic clone (except for an 11-bp deletion in hlyA) and isolates from pre-seventh-pandemic (except M645), seventh pandemic, O139 Bengal, and U.S. Gulf Coast outbreaks. The mdh and hlyA sequences from the pathogenic isolates were distinct from the sequences obtained from the environmental nontoxigenic, non-O1, non-O139 isolates, where the average pairwise nucleotide differences between variants were 1.52 and 3.21%, respectively. The pathogenic isolates (except M645) generally have identical recA and dnaE genes, although for recA, the sixth-pandemic clone differed substantially from the other pathogenic isolates, while for dnaE, the U.S. Gulf Coast clone had a sequence which differed substantially from that of the other pathogenic strains.
The frequent occurrence of identical housekeeping genes among the pathogenic isolates suggests that the pathogenic clones are indeed closely related, despite three asd sequence variants being present among these clones, as it seems highly unlikely that identical mdh, hlyA, recA, and dnaE genes could have been transferred into independent lineages, even by means such as hitchhiking with genes acquired as part of their adaptation to pathogenesis.
The pre-seventh-pandemic El Tor outbreak strains M543, M640, and M802, but not M645, can be considered at this stage to be precursors to the seventh-pandemic isolates, as for the five genes studied there are no differences between them and the seventh-pandemic isolates. Strain M645 appears to be an anomalous clinical isolate, as it differed from the other pathogenic isolates in all of the five genes studied and was located within different clusters than the other pathogenic isolates in the mdh and hlyA gene trees.
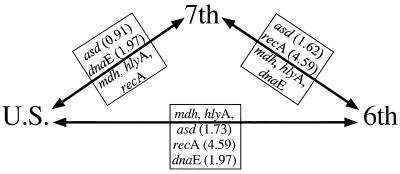
For each of the genes asd, mdh, hlyA, recA, and dnaE, there is no variation within the sixth-pandemic, seventh-pandemic, or U.S. Gulf Coast clones. These clones are clearly closely related, as for each pair they were identical at two or three of the five genes analyzed (Fig. 4). The significant nucleotide differences at the other loci must be due to recombination, as it seems inconceivable that mutation alone could give such levels of divergence while other genes did not diverge at all. The genes all encode proteins involved in housekeeping functions, with no reason to expect differences in the level of selection to account for the disparity in sequence variation in the different genes. Only the 11-bp deletion in the hlyA gene of the sixth-pandemic clone is attributed to mutation, and this mutation in the sixth-pandemic clone may well have been established by selection, as hlyA-negative forms appeared soon after the major expansion of the seventh-pandemic clone. Thus, among the 4,082-bp sequence in the four genes, there are no differences in the three pathogenic clones attributed to random genetic drift of neutral mutation. This finding indicates a much closer relationship than can be inferred from MLEE data.
FIG. 4.
Representation of the genetic identities of four housekeeping genes between the pathogenic clones of V. cholerae. Genes within the triangle are identical to the adjacent pathogenic clones. Genes outside the triangle are different between adjacent pathogenic clones; percent nucleotide differences are in parentheses.
MLEE studies show that the seventh-pandemic and U.S. Gulf Coast clones differ in the leucine aminopeptidase, DA1 (NADPH diaphorase), and NSE (carboxylesterase) loci, and the sixth- and seventh-pandemic clones differ in the 6-phosphogluconate dehydrogenase and glucose phosphate isomerase loci (16, 45). Whether the differences are due to recombination or arise by mutation cannot be determined by MLEE, but they are very obvious from the sequences. In light of our observations and conclusions from the sequence data, the difference in mobilities of the enzymes between the clones is most likely due to recombination, but this can be confirmed only by sequence analysis of the genes.
Recombination in V. cholerae.
The recombination discussed above is not expected to be due to diversifying selection, suggesting a high level of recombination for V. cholerae. We applied the algorithm used by Maynard Smith et al. to detect levels of association between alleles at different loci (33) to the MLEE data set for 260 V. cholerae strains (45). The index of association (IA) is a generalized measure of linkage disequilibrium, with an expected value of zero if the association between loci is random. For all V. cholerae isolates, IA equals 1.57 with a standard error of 0.09, indicating there is a nonrandom distribution of alleles, which is evidence for a clonal population structure. However, this value is biased by the overrepresentation of isolates of the sixth and seventh pandemics of cholera, and when only ETs are considered, IA falls to −0.092 ± 0.17, a value consistent with a nonclonal or weakly clonal population.
A comparison of the phylogenetic trees for mdh, hlyA, and asd shows a lack of congruence between the trees, which is concordant with the conclusion from the statistical test on the isozyme data. The only exception to the lack of congruence are two pairs of strains, M559-M560 and M557-M558, each pair possessing either identical or very similar mdh, hlyA, and asd sequences.
The mdh and hlyA gene trees are more congruent with each other than with the asd gene tree, where the difference in phylogenies is due to recombination. For example, strain M555 has the same hlyA sequence as strains M559 and M560 and a similar mdh sequence (0.32% pairwise difference) but differs in its asd sequence from strains M559 and M560 by 4.16 and 4.06% of the nucleotides, respectively. In addition, strains M535 and M553 have identical mdh sequences and similar hlyA sequences (0.77% pairwise difference) but differ in the asd locus by 2.13%.
The asd locus has been affected by gene transfer among the pathogenic clones also, since these clonal lineages diverge significantly at this locus (26). Across the mdh and hlyA gene trees, the pathogenic clones cluster with the environmental isolates M535 and M548, but in the asd gene tree, it is only the seventh-pandemic clone which clusters with these two environmental isolates. The sixth-pandemic and U.S. Gulf Coast clones are found within different lineages in the asd gene tree, where the noncongruence in the genealogies of these pathogenic clones is most likely a result of independent gene transfers which occurred after their divergence from a common ancestor.
It is interesting that the asd locus has undergone two (or three) recombination events among the pathogenic isolates, whereas there has been only one event involving the whole gene in each of the dnaE and recA genes and no recombination in the mdh and hlyA genes in the pathogenic lineage. This is consistent with a high but random level of recombination involving large (greater than gene size) fragments. However, the fact that there is evidence for intragenic recombination in asd suggests that it is particularly subject to recombination. We have no explanation for the high rate of recombination observed for asd.
The emergence of cholera.
Characteristics of the disease cholera, such as long-term immunity of the host, lack of a carrier state, and no known animal host, suggest that it was probably rare or nonexistent in the Paleolithic period, when the relative isolation in which the small hunter-gatherer societies existed could not have supported the continual propagation of such infectious diseases (18). Cholera most probably emerged after the Neolithic revolution, which occurred first in the Middle East some 10,000 years ago, where the invention and/or adoption of agricultural practices by nomadic groups enabled higher densities of humans to subsist. With the establishment of villages and their water supplies, the change from the nomadic to the sedentary lifestyle of human populations provided an opportunity for an environmental V. cholerae bacterium to acquire the necessary virulence mechanisms to survive and multiply in the specific niche of the intestines of humans. Diarrhea induced by extracellular proteins, mainly the cholera toxin, provided the means by which the organism could be released back to the environment, to await infection of the next host.
The genes encoding the cholera toxin comprise part of the genome of the phage encoding cholera toxin, CTXφ (51), which uses the toxin coregulated pilus (TCP) as a receptor, with the genes encoding the TCP being found within a potentially mobile element, the V. cholerae pathogenicity island (VPI) (27). It has been suggested that the adaptation to pathogenesis of V. cholerae involved a sequential process (35), initially requiring the expression of the TCP for CTXφ transduction. For the sixth- and seventh-pandemic clones, whether this process of acquisition occurred before or after their divergence from a common ancestor remains unclear, as the two clones differ in the chromosomal location and copy number of the CTX (cholera toxin) element; the sixth-pandemic clone containing two separate copies compared to the one to three tandem copies of the CTX element found in the seventh-pandemic clone (34). They also exhibit sequence divergence in the ctxB (38) and tcpA (23) genes, which may reflect diversifying selection pressures or indicate independent acquisitions of the CTX and VPI elements.
The emergence of a common ancestor of the present pathogenic clones of V. cholerae probably occurred relatively recently, as no variation was detected within the mdh gene or in the hlyA segment (except for the 11-bp deletion in the sixth-pandemic clone) of the pathogenic isolates over a 57-year time period. Similarly, no variation was detected in the recA and dnaE genes of the pathogenic clones, although recombination in the recA gene of the sixth-pandemic clone, in the dnaE gene of the U.S. Gulf Coast clone, and in the asd gene of these clonal lineages suggests that recombination is frequent in V. cholerae, higher than the mutation rate in these pathogenic clones.
The lack of mutational changes and the high frequency of recombination in the loci studied make it difficult for clear relationships to be determined for the pathogenic clones. From MLEE data, it appears that the U.S. Gulf Coast clone diverged before the emergence of the sixth- and seventh-pandemic clones, suggesting that the U.S. Gulf Coast isolates are remnants from one of the previous pandemics that swept across North America. The 11-bp deletion in hlyA of the sixth-pandemic clone and the fact that some of the characteristics which distinguish the classical and El Tor biotypes involve loss of function (e.g., Vogues-Proskauer reaction and hemagglutination) indicate that it diverged from a common ancestor with the seventh-pandemic clone which had these properties intact. The high rate of recombination and the existence of pathogenic strains from outbreaks between 1937 and 1954 which are very closely related to isolates of the seventh pandemic has major implications for our understanding of how new pandemics emerge. Recombination could be seen as a mechanism whereby recombinant phenotypes are generated from existing pathogenic isolates which, given the right selection pressures, emerge as new pandemics of cholera.
ACKNOWLEDGMENTS
This project was supported by grants from the Australian Research Council and the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Alm R A, Stroeher U H, Manning P A. Extracellular proteins of Vibrio cholerae: nucleotide sequence of the structural gene (hlyA) for the haemolysin of the haemolytic El Tor strain 017 and characterization of the hlyA mutation in the non-haemolytic classical strain 569B. Mol Microbiol. 1988;2:481–488. doi: 10.1111/j.1365-2958.1988.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 2.Barrett T J, Blake P A. Epidemiological usefulness of changes in hemolytic activity of Vibrio cholerae biotype El Tor during the seventh pandemic. J Clin Microbiol. 1981;13:126–129. doi: 10.1128/jcm.13.1.126-129.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastin D A, Romana L K, Reeves P R. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol Microbiol. 1991;5:2223–2231. doi: 10.1111/j.1365-2958.1991.tb02152.x. [DOI] [PubMed] [Google Scholar]
- 4.Berche P, Poyart C, Abachin E, Lelievre H, Vandepitte J, Dodin A, Fournier J M. The novel epidemic strain O139 is closely related to the pandemic strain O1 of Vibrio cholerae. J Infect Dis. 1994;170:701–704. doi: 10.1093/infdis/170.3.701. [DOI] [PubMed] [Google Scholar]
- 5.Bik E M, Bunschoten A E, Gouw R D, Mooi F R. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 1995;14:209–216. doi: 10.1002/j.1460-2075.1995.tb06993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bik E M, Gouw R D, Mooi F R. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J Clin Microbiol. 1996;34:1453–1461. doi: 10.1128/jcm.34.6.1453-1461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd E F, Nelson K, Wang F S, Whittam T S, Selander R K. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown M H, Manning P A. Haemolysin genes of Vibrio cholerae: presence of homologous DNA in non-haemolytic O1 and haemolytic non-O1 strains. FEMS Microbiol Lett. 1985;30:197–201. [Google Scholar]
- 9.Cameron D N, Khambaty F M, Wachsmuth I K, Tauxe R V, Barrett T J. Molecular characterization of Vibrio cholerae O1 strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1685–1690. doi: 10.1128/jcm.32.7.1685-1690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen F, Evins G M, Cook W L, Almeida R, Hargrett-Bean N, Wachsmuth K. Genetic diversity among toxigenic and nontoxigenic Vibrio cholerae O1 isolated from the Western Hemisphere. Epidemiol Infect. 1991;107:225–233. doi: 10.1017/s0950268800048846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cholera Working Group, International Centre for Diarrhoeal Diseases Research, Bangladesh. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet. 1993;342:387–390. [PubMed] [Google Scholar]
- 12.Coelho A, Andrade J R, Vicente A C, Salles C A. New variant of Vibrio cholerae O1 from clinical isolates in Amazonia. J Clin Microbiol. 1995;33:114–118. doi: 10.1128/jcm.33.1.114-118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colwell R R, Huq A. Environmental reservoir of Vibrio cholerae. The causative agent of cholera. Ann N Y Acad Sci. 1994;740:44–54. doi: 10.1111/j.1749-6632.1994.tb19852.x. [DOI] [PubMed] [Google Scholar]
- 14.Colwell R R, Spira W M. The ecology of Vibrio cholerae. In: Barua D, Greenough III W B, editors. Cholera. New York, N.Y: Plenum Medical Book Co.; 1992. pp. 107–127. [Google Scholar]
- 15.Desmarchelier P M, Momen H, Salles C A. A zymovar analysis of Vibrio cholerae isolated in Australia. Trans R Soc Trop Med Hyg. 1988;82:914–917. doi: 10.1016/0035-9203(88)90041-7. [DOI] [PubMed] [Google Scholar]
- 16.Evins G M, Cameron D N, Wells J G, Greene K D, Popovic T, Giono-Cerezo S, Wachsmuth I K, Tauxe R V. The emerging diversity of the electrophoretic types of Vibrio cholerae in the Western Hemisphere. J Infect Dis. 1995;172:173–179. doi: 10.1093/infdis/172.1.173. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP package, version 3.5. Seattle, Wash: University of Washington; 1993. http://evolution.genetics.washington.edu/phylip.html. . ( http://evolution.genetics.washington.edu/phylip.html.) ) [Google Scholar]
- 18.Fenner F. The effects of changing social organisation on the infectious diseases of man. In: Boyden S V, editor. The impact of civilisation on the biology of man. Canberra, Australia: Australian National University Press; 1970. pp. 49–76. [Google Scholar]
- 19.Franco A A, Yeh P-E, Johnson J A, Barry E M, Guerra H, Maurer R, Morris J G., Jr Cloning and characterization of dnaE, encoding the catalytic subunit of replicative DNA polymerase III, from Vibrio cholerae strain C6706. Gene. 1996;175:281–283. doi: 10.1016/0378-1119(96)00155-2. [DOI] [PubMed] [Google Scholar]
- 20.Gleeson T J, Staden R. An X Windows and UNIX implementation of our sequence analysis package. Comput Appl Biosci. 1991;7:398. doi: 10.1093/bioinformatics/7.3.398. [DOI] [PubMed] [Google Scholar]
- 21.Hartl D L, Ochman H. Inverse polymerase chain reaction. Methods Mol Biol. 1994;31:187–196. doi: 10.1385/0-89603-258-2:187. [DOI] [PubMed] [Google Scholar]
- 22.Huq A, Small E B, West P A, Huq M I, Rahman R, Colwell R R. Ecological relationships between Vibrio cholerae and planktonic crustacean copepods. Appl Environ Microbiol. 1983;45:275–283. doi: 10.1128/aem.45.1.275-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iredell J R, Manning P A. Biotype-specific tcpA genes in Vibrio cholerae. FEMS Microbiol Lett. 1994;121:47–54. doi: 10.1111/j.1574-6968.1994.tb07074.x. [DOI] [PubMed] [Google Scholar]
- 24.Kaper J, Lockman H, Colwell R R, Joseph S W. Ecology, serology, and enterotoxin production of Vibrio cholerae in Chesapeake Bay. Appl Environ Microbiol. 1979;37:91–103. doi: 10.1128/aem.37.1.91-103.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karaolis D K, Lan R, Reeves P R. Molecular evolution of the seventh-pandemic clone of Vibrio cholerae and its relationship to other pandemic and epidemic V. cholerae isolates. J Bacteriol. 1994;176:6199–6206. doi: 10.1128/jb.176.20.6199-6206.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaolis D K, Lan R, Reeves P R. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim G T, Lee J Y, Huh S H, Yu J H, Kong I S. Nucleotide sequence of the vmhA gene encoding hemolysin from Vibrio mimicus. Biochim Biophys Acta. 1997;1360:102–104. doi: 10.1016/s0925-4439(97)00008-2. [DOI] [PubMed] [Google Scholar]
- 29.Koblavi S, Grimont F, Grimont P A. Clonal diversity of Vibrio cholerae O1 evidenced by rRNA gene restriction patterns. Res Microbiol. 1990;141:645–657. doi: 10.1016/0923-2508(90)90059-y. [DOI] [PubMed] [Google Scholar]
- 30.Lim D B, Oppenheim J D, Eckhardt T, Maas W K. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc Natl Acad Sci USA. 1987;84:6697–6701. doi: 10.1073/pnas.84.19.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu C D, Abdelal A T. Complete sequence of the Salmonella typhimurium gene encoding malate dehydrogenase. Gene. 1993;123:143–144. doi: 10.1016/0378-1119(93)90554-g. [DOI] [PubMed] [Google Scholar]
- 32.Margraf R L, Roca A I, Cox M M. The deduced Vibrio cholerae RecA amino-acid sequence. Gene. 1995;152:135–136. doi: 10.1016/0378-1119(94)00686-m. [DOI] [PubMed] [Google Scholar]
- 33.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 35.Mekalanos J J, Rubin E J, Waldor M K. Cholera—molecular basis for emergence and pathogenesis. FEMS Immunol Med Microbiol. 1997;18:241–248. doi: 10.1111/j.1574-695X.1997.tb01052.x. [DOI] [PubMed] [Google Scholar]
- 36.Morris J G., Jr Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol Rev. 1990;12:179–191. doi: 10.1093/oxfordjournals.epirev.a036052. [DOI] [PubMed] [Google Scholar]
- 37.Ohkuma M, Ohtoko K, Takada N, Hamamoto T, Usami R, Kudo T, Horikoshi K. Characterization of malate dehydrogenase from deep-sea psychrophilic Vibrio sp. strain no. 5710 and cloning of its gene. FEMS Microbiol Lett. 1996;137:247–252. doi: 10.1111/j.1574-6968.1996.tb08113.x. [DOI] [PubMed] [Google Scholar]
- 38.Olsvik O, Wahlberg J, Petterson B, Uhlen M, Popovic T, Wachsmuth I K, Fields P I. Use of automated sequencing of polymerase chain reaction-generated amplicons to identify three types of cholera toxin subunit B in Vibrio cholerae O1 strains. J Clin Microbiol. 1993;31:22–25. doi: 10.1128/jcm.31.1.22-25.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pollitzer R. History of the disease. In: Pollitzer R, editor. Cholera. Geneva, Switzerland: World Health Organization; 1959. pp. 11–50. [Google Scholar]
- 40.Popovic T, Bopp C, Olsvik O, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramamurthy T, Garg S, Sharma R, Bhattacharya S K, Nair G B, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. Emergence of a novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993;341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 42.Reeves P R, Farnell L, Lan R. MULTICOMP: a program for preparing sequence data for phylogenetic analysis. CABIOS. 1994;10:281–284. doi: 10.1093/bioinformatics/10.3.281. [DOI] [PubMed] [Google Scholar]
- 43.Rivera I G, Chowdhury M A, Huq A, Jacobs D, Martins M T, Colwell R R. Enterobacterial repetitive intergenic consensus sequences and the PCR to generate fingerprints of genomic DNAs from Vibrio cholerae O1, O139, and non-O1 strains. Appl Environ Microbiol. 1995;61:2898–2904. doi: 10.1128/aem.61.8.2898-2904.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 45.Salles C A, Momen H. Identification of Vibrio cholerae by enzyme electrophoresis. Trans R Soc Trop Med Hyg. 1991;85:544–547. doi: 10.1016/0035-9203(91)90251-s. [DOI] [PubMed] [Google Scholar]
- 46.Shangkuan Y H, Tsao C M, Lin H C. Comparison of Vibrio cholerae O1 isolates by polymerase chain reaction fingerprinting and ribotyping. J Med Microbiol. 1997;46:941–948. doi: 10.1099/00222615-46-11-941. [DOI] [PubMed] [Google Scholar]
- 47.Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982;10:2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephens J C. Statistical methods of DNA sequence analysis: detection of intragenic recombination or gene conversion. Mol Biol Evol. 1985;2:539–556. doi: 10.1093/oxfordjournals.molbev.a040371. [DOI] [PubMed] [Google Scholar]
- 49.Stroeher U H, Lech A J, Manning P A. Gene sequence of recA+ and construction of recA mutants of Vibrio cholerae. Mol Gen Genet. 1994;244:295–302. doi: 10.1007/BF00285457. [DOI] [PubMed] [Google Scholar]
- 50.Vogel R F, Entian K D, Mecke D. Cloning and sequence of the mdh structural gene of Escherichia coli coding for malate dehydrogenase. Arch Microbiol. 1987;149:36–42. doi: 10.1007/BF00423133. [DOI] [PubMed] [Google Scholar]
- 51.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 52.Welch T J, Bartlett D H. Cloning, sequencing and overexpression of the gene encoding malate dehydrogenase from the deep-sea bacterium Photobacterium species strain SS9. Biochim Biophys Acta. 1997;1350:41–46. doi: 10.1016/s0167-4781(96)00200-x. [DOI] [PubMed] [Google Scholar]