Abstract
Inherited retinal diseases (IRDs) are a genetically and phenotypically heterogeneous group of genetic eye disorders. There are more than 300 disease entities, and together this group of disorders affects millions of people globally and is a frequent cause of blindness or low-vision certification. However, each type is rare or ultra-rare. Characteristically, the impaired vision in IRDs is due to retinal photoreceptor dysfunction and loss resulting from mutation in a gene that codes for a retinal protein. Historically, IRDs have been considered incurable and individuals living with these blinding conditions could be offered only supportive care. However, the treatment landscape for IRDs is beginning to evolve. Progress is being made, driven by improvements in understanding of genotype–phenotype relationships, through advances in molecular genetic testing and retinal imaging. Alongside this expanding knowledge of IRDs, the current era of precision medicine is fueling a growth in targeted therapies. This has resulted in the first treatment for an IRD being approved. Several other therapies are currently in development in the IRD space, including RNA-based therapies, gene-based therapies (such as augmentation therapy and gene editing), cell therapy, visual prosthetics, and optogenetics. RNA-based therapies are a novel approach within precision medicine that have demonstrated success, particularly in rare diseases. Three antisense oligonucleotides (AONs) are currently in development for the treatment of specific IRD subtypes. These RNA-based therapies bring several key advantages in the setting of IRDs, and the potential to bring meaningful vision benefit to individuals living with inherited blinding disorders. This review will examine the increasing breadth and relevance of RNA-based therapies in clinical medicine, explore the key features that make AONs suitable for treating genetic eye diseases, and provide an overview of the three-leading investigational AONs in clinical trials.
Keywords: antisense oligonucleotides, genetic eye diseases, inherited retinal diseases, QR-1123, RNA therapies, sepofarsen, ultevursen
Introduction
Globally, an estimated 43.4 million people are blind and another 295 million have moderate-to-severe visual impairment.1 Around one-third of cases of blindness or severe visual impairment have a genetic basis, either as part of a multifactorial etiology, for example, as in age-related macular degeneration, diabetic retinopathy, or glaucoma, or as the direct result of genetic mutations, such as in inherited retinal diseases (IRDs), inherited corneal dystrophies, or optic neuropathies.2,3
IRDs are a genetically and phenotypically heterogeneous group of genetic eye disorders. There are more than 300 disease entities, and together this group of disorders affects at least 1 in 1400 individuals, or around 5.5 million people worldwide.4–9 However, each type is rare or ultra-rare. Despite the lack of global data documenting the overall disease burden of IRDs in terms of loss of sight, studies in the United Kingdom and Australia show that they account for around 6% of blindness or low-vision certifications,10 12% among children (making it the second most common diagnosis), and 20–23% among individuals of working age (making it the most common diagnosis). Cost of illness (blindness) studies in the Republic of Ireland and in the United Kingdom estimate total costs of £42.6 and £523.3 million, respectively, in 2019.11 Well-being costs accounted for approximately one-third of these sums, illustrating the significant impact sight loss has on the individual with an IRD.
The impaired vision in IRDs is due to retinal photoreceptor dysfunction or loss resulting from mutation in a gene that codes for a retinal protein.8,12,13 The precise phenotype of each IRD depends on the role of the affected protein, which may include phototransduction, photoreceptor morphogenesis and maintenance, photoreceptor ciliary transport, or the retinoid cycle.13 Other body systems may also be implicated if the aberrant protein synthesis impacts tissues other than the retina: IRDs that affect other systems, such as hearing, are known as syndromic disorders.14
Historically, IRDs have been considered incurable and individuals living with these blinding conditions could only be offered supportive care to treat associated complications, such as cataracts and macular edema, or in some cases with nutritional adjustments.15,16 However, the treatment landscape for IRDs is changing. Progress is being made, driven by improvements in understanding of genotype–phenotype relationships, through advances in molecular genetic testing and retinal imaging.13 Alongside this expanding knowledge of IRDs, the current era of precision medicine is fueling a growth in targeted therapies. This has resulted in the first treatment for an IRD being approved in 2017 in the United States and in 2018 in Europe.17,18 Voretigene neparvovec-rzyl (Luxturna®; Spark Therapeutics Inc.) is a gene therapy for adult and pediatric patients with vision loss due to IRD caused by biallelic mutations in the retinoid isomerohydrolase (RPE65) gene.18 This approval represents a landmark change for individuals living with an IRD. However, voretigene neparvovec-rzyl is applicable only for patients with RPE65-related IRD, and only in those with sufficient viable retinal cells.17,18 Consequently, there remains a pressing medical need for treatment options for people with a genetic eye disease outside of this small subset of patients. Several other therapies are in development in the IRD space, building on the success of voretigene neparvovec-rzyl, including DNA-based therapies (such as augmentation therapy and gene editing), RNA-based therapies, cell therapy, visual prosthetics, and optogenetics.19,20
RNA-based therapies are a novel approach within precision medicine that have demonstrated success, particularly in rare diseases;21 and a number of therapies are currently in development for the treatment of IRDs. This review will examine the increasing breadth and relevance of RNA-based therapies in clinical medicine and, more specifically, in ophthalmology, explore the key features that make such therapies suitable for treating genetic eye diseases, and provide an overview of investigational RNA-based therapies for IRDs that have reached clinical trials.
RNA-based therapies
Ribonucleic acid (RNA) is produced from deoxyribonucleic acid (DNA) via the transcription process. There are different types of RNAs, some of them called coding RNAs (e.g. mRNAs) are involved in protein synthesis while some others, the non-coding RNAs (ncRNAs), do not code for proteins but may modulate gene expression.22–24 The ncRNAs, such as microRNAs (miRNAs), long non-coding RNAs (lncRNA), silencing RNAs (siRNAs), etc., are involved in a variety of biological functions. For example, they regulate gene expression and translation, RNA processing, and translation levels; they protect genomes from foreign nucleic acids; and they can guide DNA synthesis or genome rearrangement.22–24 Within the context of genetic diseases involving protein disruption, coding RNAs have been considered a potential therapeutic target for more than four decades; the correction at the RNA level of changes induced by a pathogenic variant in the DNA could restore normal protein production or stop pathogenic protein production.22,23 A class of non-coding RNAs regrouped under a competing endogenous RNA (ceRNA) network can target coding and non-coding RNA molecules and indirectly regulate each other by competing for them. Thus, leading to an additional post-transcriptional regulatory layer accomplished by non-coding RNAs. However, ceRNAs are also involved in the pathogenesis of several diseases.25 Further understanding this ceRNA network may provide a base to identify potential therapeutic targets or new prognostic biomarkers.26,27
There are several categories of RNA-based therapies, including single-stranded antisense oligonucleotides (known as AONs or ASOs), double-stranded small interfering RNAs, mRNAs, and aptamers.21,28,29 RNA-targeting small molecule drugs are not considered RNA-based therapies per se because these are not analogs of nucleic acid polymers.30 Not including vaccines, 13 RNA-based therapies have already been approved across several therapeutic areas, but none of them for IRDs (Table 1).21,29,31,32 These include one aptamer (pegaptanib; for age-related macular degeneration) and three small interfering RNAs (patisiran, givosiran, and lumasiran; all for hepatic conditions). However, the majority of approvals are among the family of AONs; these comprise fomivirsen (to treat cytomegalovirus-induced retinitis) and eight therapies for hereditary conditions, including familial hypercholesterolemia, Duchenne muscular dystrophy, spinal muscular atrophy, hereditary transthyretin amyloidosis, and familial chylomicronemia syndrome.21,29,32,33 In addition, more than 400 drug development programs for RNA-based therapies are underway across a variety of ocular and non-ocular areas, but predominantly for oncology.21 Within this pipeline, several RNA-based therapies are being investigated for IRDs, all of which are AONs. Because AONs can target a patient-specific mutation, there is also precedent for ‘N-of-1’ use of patient-specific AONs.34
Table 1.
Summary of approved RNA-based therapies.a21,29,31,32
| RNA therapy class | Name | Target organ | Indication | Administration route | Year of approval | |
|---|---|---|---|---|---|---|
| FDA | EMA | |||||
| AON | Fomivirsenc | Eye | CMV-induced retinitis | Intravitreal | 1998 | 1999 |
| Mipomersenb | Liver | Familial hypercholesterolemia | SC | 2013 | ||
| Eteplirsen | Skeletal muscle | Duchenne muscular dystrophy | IV | 2016 | ||
| Nusinersen | Spinal cord | Spinal muscular atrophy | Intrathecal | 2016 | 2017 | |
| Inotersen | Liver | Hereditary transthyretin-mediated amyloidosis | SC | 2018 | 2018 | |
| Volanesorsen | Blood lipids | Familial chylomicronemia syndrome | SC | 2019 | ||
| Golodirsen | Skeletal muscle | Duchenne muscular dystrophy | IV | 2019 | ||
| Viltolarsen | Skeletal muscle | Duchenne muscular dystrophy | IV | 2020 | ||
| Casimersen | Skeletal muscle | Duchenne muscular dystrophy | IV | 2021 | ||
| Aptamer | Pegaptanib | Eye | Age-related macular degeneration | Intravitreal | 2004 | 2006b |
| Small interfering RNA | Patisiran | Liver | Hereditary transthyretin amyloidosis | IV | 2018 | 2018 |
| Givosiran | Liver | Acute hepatic porphyria | SC | 2019 | 2020 | |
| Lumasiran | Liver | Primary hyperoxaluria type 1 | SC | 2020 | 2020 | |
Source: EMA and FDA websites.
Approval status as of August 23, 2021.
AON: antisense oligonucleotide; CMV: cytomegalovirus; EMA: European Medicines Agency; FDA: Food and Drug Administration; IV: intravenous; SC: subcutaneous.
Excluding RNA-based vaccines.
Discontinued due to lack of commercial interest.
Withdrawn from use due to reduced clinical need.
AONs are short lengths of chemically stabilized single-stranded RNA or DNA that modify the expression of a given nucleotide sequence by binding to target pre-mRNA or mRNA via base pairing.28,32,33,35 They act on the processing of the mutant RNAs to suppress the defects that result in abnormal synthesis or function of proteins, and thereby slow down or reverse the course of an associated disease.28,36 The precise mechanism of action depends on the AON; for example, they can be engineered for pre-mRNA splicing correction, exon skipping, or mRNA knockdown.28,32,33,35
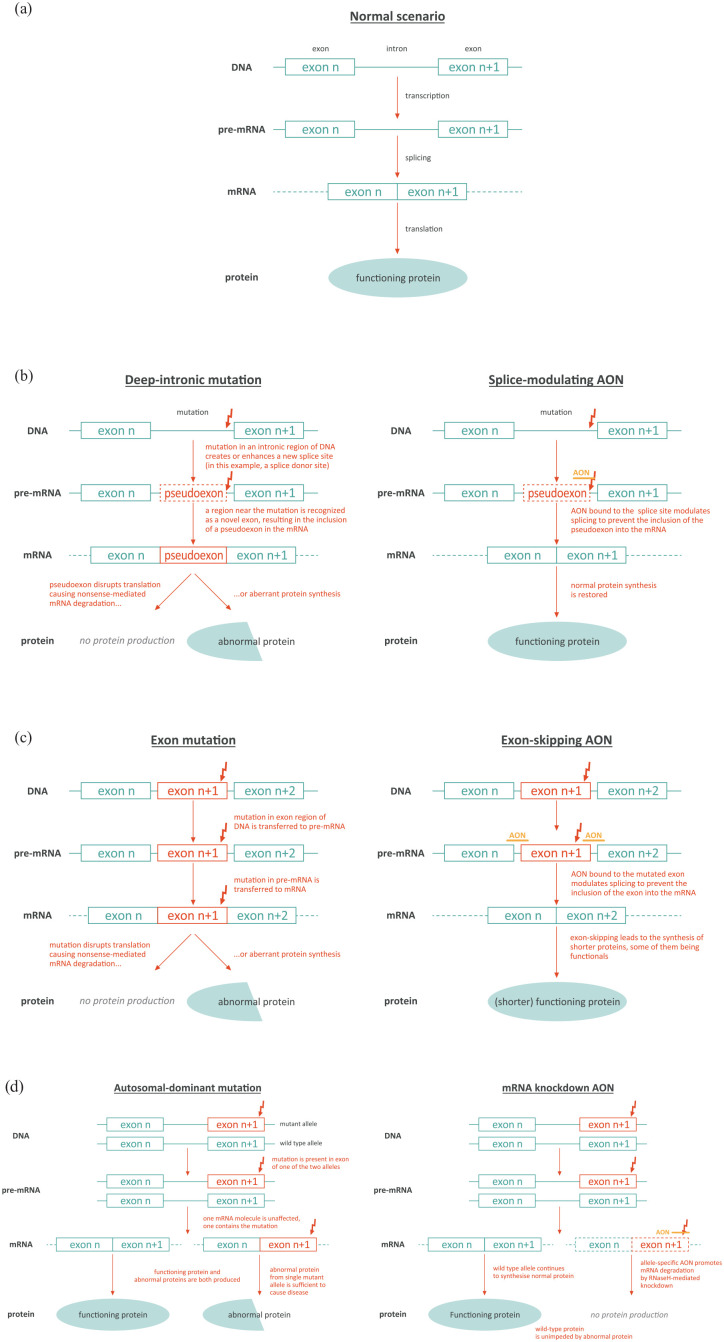
Under normal circumstances, genes are transcribed into pre-mRNA transcripts, comprising exons holding the genetic code required to synthesize a protein, separated by non-coding introns. Splicing removes the introns from the pre-mRNA to generate the mature mRNA that is then translated into the protein (Figure 1(a)).33,35 Splicing defects can arise, for example, due to deep-intronic mutations that generate a new splice donor or acceptor site in the intron, which leads to retention of a part of that intron during splicing, and thereby its inclusion in the mature mRNA (i.e. insertion of a pseudoexon).33,35 Such is the case in CEP290-associated IRD, as described later in this article. The addition of this pseudoexon leads to either nonsense-mediated mRNA decay and therefore an absence of protein production, or synthesis of an abnormal protein.33 By binding to the deep-intronic mutant region, AONs can modulate splicing and prevent inclusion of the pseudoexon and the resultant disruption of protein synthesis (Figure 1(b)).33 AONs can also be designed to bind to splice sites or regulatory elements on a target exon to prevent recruitment of splicing factors, causing skipping of that exon during RNA splicing.28,33 This design stops mutation-containing exons being incorporated into the mature mRNA (Figure 1(c)). This strategy is relevant for USH2A mutations discussed below.
Figure 1.
Mechanisms of action of AONs in genetic eye disease. (a) Wild-type scenario. (b) AON correcting deep-intronic mutation. (c) AON inducing exon skipping. (d) AON producing mRNA knockdown (adapted from Collin 2017).33
In some autosomal-dominant conditions, such as autosomal-dominant retinitis pigmentosa (adRP) detailed below, protein production from a single mutant allele is sufficient to manifest disease (i.e. dominant negative effect), despite the presence of a normal allele that produces normal protein. In these circumstances, AONs can be used to trigger allele-selective degradation or ‘knockdown’ of mutant mRNA transcripts, so that the wild-type allele can continue to produce wild-type protein unhindered.33 In this approach, the AON is designed to recognize the mutant allele and activate the enzyme RNase H (which is involved in the degradation of transcripts) at the level of the (pre-)mRNA, to reduce the number of mutant mRNA transcripts, and therefore the amount of mutant target protein (Figure 1(d)).32 Further applications of AONs include base editing of the RNA to directly change individual sites (e.g. to correct G > A mutations) and repeat-targeting to prevent the toxic effects caused by expanded, non-coding trinucleotide sequences in RNA.37,38
Characteristics of AONs in genetic eye diseases
RNA therapies, such as AONs, can be considered a form of genetic therapy as they are highly specific and target the underlying genetic cause of a disease.5,28,32,39 RNA-based therapy and DNA-based therapy (gene therapy), are two different approaches.32 They differ in several ways including mechanism of action, permanency of effect, and delivery to the target cells.32,39 Table 2 summarizes the key differences between AONs and DNA-based therapy in the setting of genetic eye diseases and these are discussed below.
Table 2.
Key differentiating features of RNA- and DNA-based therapies.
| RNA AONs | DNA-based therapies |
|---|---|
| Act at the level of the RNA; do not alter DNA32,35
No DSB generation |
Target the DNA; some can directly alter the genome;5,39,40
Induces DSB |
| Long half-life; require repeat dosing (in IRDs typically once or twice per year)36,41 | Potential for single treatment/dosing (per
eye); Long-term expression of nuclease |
| Non-permanent; treatment can be discontinued36,41 | Long-term durability still being investigated42–44 |
| Naked, no vectors needed; can target diseases with large, affected genes45 | Require viral vectors for delivery; usually limited to diseases with small (trans)gene size36,46 immune response against adeno-associated virus leads to the production of neutralizing antibodies |
| Can be administered via routine intravitreal injection, using local anesthesia47,48 | Usually require subretinal administration; surgery involves vitrectomy and usually requires general anesthesia18,49 |
| Intravitreal administration allows exposure to central and peripheral areas of retina50 | Subretinal administration targets usually sub-macular area or the retinal area with available target cells45 |
RNA: ribonucleic acid; DNA: deoxyribonucleic acid; DSB: double-strand breaks; IRD: inherited retinal disease.
Act at the RNA level and do not alter the genome
AONs make transient alterations to the RNA without altering the genome.32 At a maximum, splice-modulating AONs restore normal levels of target mRNA and do not bring a risk of overexpression of target protein that may cause cellular toxicity.36,50 DNA-based therapies act at gene level and could make changes or additions to the DNA.32 The principal forms of DNA-based therapy are gene augmentation therapy which uses vectors (such as adeno-associated viruses; AAVs) to insert a normal, non-mutated cDNA copy of the optimized coding sequence of the target gene into the host cells; and gene editing therapy, which corrects defects in the genetic sequence of the mutated gene using clustered regularly interspaced short palindromic repeats (CRISPR) technology.5,39,51 Wild-type AAVs integrate into the host DNA. When recombinant AAV vectors are used, the genomic DNA is not modified and the introduced genetic sequence is very rarely integrated into the host DNA; instead, it typically coexists as a so-called episome in the nucleus of the target cell.5,39,40 This is the case with the gene augmentation therapy voretigene neparvovec-rzyl, which uses AAV serotype 2 (AAV2) to carry a functional cDNA copy of the RPE65 gene into the retinal pigment epithelial cells in individuals who have reduced or absent RPE65 protein due to biallelic RPE65 mutations.5,18 With lentivirus vectors, random integration into the genome can occur, which may be associated with a risk of insertional mutagenesis potentially affecting a tumor suppressor gene, leading to cancer.5,52 However, the risk of malignant transformation is probably low with insertion in non-dividing cells, such as the neurosensory retina. Gene editing, by definition, makes changes directly to the genomic DNA of the mutated gene.5,51,40 The risks associated with this approach have not yet been fully elucidated.
Long-lasting effect but non-permanent nature
AONs are similar to drug therapies in that their duration of effect depends on their half-life and clearance rate.41 This means that repeated administration is required, and this could have implications for patient safety and convenience and for healthcare resource utilization costs. For AONs used in the eye, repeat treatment is relatively infrequent; currently twice per year.36 This is because, unlike other target cells, retinal cells are post-mitotic, enabling AONs to have a long-lasting therapeutic effect.53 Also, the chemical modification of the AONs, such as the inclusion of phosphorothioate (PS) backbone, the attachment of the 2′-O-methoxyethyl or 2′-O-methyl group to the sugar residues of the PS backbone, and the formulation of emulsification drops, ensures a long half-life, preserves the AONs from early degradation, and therefore ensures prolonged response.33 Conversely, the transient nature of both RNA and AONs means the AON effects are somewhat reversible.36,41 It may, therefore, be possible to adjust the strength of the effects by modifying subsequent dosing (if required), and potential side effects might be transient.41,45
DNA-based therapies are assumed to be a one-time therapy. The durability of DNA-based therapies in ophthalmological settings has not yet been categorically shown; long-term data for voretigene neparvovec-rzyl extend only up to 4 years post-treatment to date.42 Other DNA-based therapies aimed at RPE65-associated Leber congenital amaurosis (LCA) have shown variable durability of effect, in some cases, reporting subsequent declining visual efficacy alongside persisting degenerative loss of photoreceptors.43,44,54 If these therapies prove to be permanent when targeting non-dividing cells, this could represent an advantage in patient burden in terms of requiring only one-off surgery, albeit with one eye treated at a time. This single intervention will require thorough assessment to identify the appropriate dosing prior to surgery, especially taking into account individual variation, to achieve maximum benefit while avoiding any permanent side effects.
Do not require vectors for delivery
AONs can be administered as naked molecules as the chemical modifications and the optimized formulation ameliorate their cell uptake. As such, vectors are not needed and this approach is not hampered by limitations of vector capacity or vector-associated intraocular inflammation.45,55,56 Their relatively small size means that AONs may provide therapeutic benefit and restore protein function in diseases in which gene sequences are too large to be delivered via AAV2 (e.g. ABCA4 for Stargardt disease and MYO7A or USH2A associated with Usher syndrome) or other acceptable viral vectors used in gene augmentation therapy.52,57 It should be noted that dual-vector strategies with the potential to overcome this limitation are currently in preclinical development.58 Both approaches offer alternatives and more potential therapeutic options for the patients.
Administered via intravitreal injection
AONs can be administered via an intravitreal injection.28,59 This method is low-risk, fast, and convenient and can be performed in a clinic or office setting with topical anesthesia depending on patient age and acceptance.47,48 Intravitreal administration is known to be less invasive than subretinal delivery which is required for many AAV gene augmentation therapies. It has a low rate of procedural complications, including intraocular inflammation which could represent an advantage for the patients.39,46,49,52,53,60 It must be noted that cataracts occur with intravitreal treatments.60 Subretinal delivery requires, in most cases, a general anesthetic and is relatively invasive, entailing removal of vitreous via pars plana vitrectomy, retinotomy, temporary retinal/macular detachment, and then injection of the therapy into the subretinal space.18,49 Attempts to deliver AAV gene augmentation through intravitreal route have so far not proven successful and have led to intraocular inflammation.61 Individuals receiving subretinal voretigene neparvovec-rzyl require the retina to be > 100 microns thick at the posterior pole.17
Broad distribution within the retina
The optimal mode of delivery for an IRD treatment has not been fully defined. Intravitreal administration of a treatment allows broad distribution within the retina to reach a larger number of retinal cells.45,52 Furthermore, preclinical and early clinical data show that intravitreally administered AONs can be used to target the peripheral and central regions of the retina50 and can reach all layers of the retina.36 Therapies delivered to the subretinal space target mainly the macular region of the total retinal area.45 More research is needed to understand whether there are any potential advantages of targeting the entire retina, and especially the periphery, in terms of whether this confers any potential for early intervention for AONs.
Low risk of systemic side effects
AONs used to treat genetic eye diseases are associated with limited off-target effects.28,36 This low risk of systemic side effects also applies to DNA-based therapies. This is due to several unique features of the eye. First, the small size of the eye means that only small doses are needed.5,19,51 Local delivery also enhances drug bioavailability which supports the use of low doses.5,59 Second, the eye is a closed compartment, so intravitreal and subretinal delivery methods have a lower rate of systemic side effects versus systemic administration, with blood–retinal barriers helping to prevent systemic spread.5,59 Third, the eye is highly compartmentalized, so treatment can be targeted to specific regions (e.g. subretinal space or vitreous cavity), thus reducing off-target exposure.5,19,51 However, despite delivery into the vitreous cavity, there is evidence of exposure of AONs anteriorly, even as far as the cornea.62 Finally, as certain sub-compartments of the eye are the sites of immune deviation (referred to as immune privileged), treatments can be administered into the eye with reduced risk of provoking inflammation and immune-mediated damage.5,19,51
RNA-based therapies in development for IRDs
There are currently no approved RNA-based therapies for IRDs. Three investigational RNA-based therapies for IRDs are in phase I/II and II/III clinical trials: sepofarsen, ultevursen, and QR-1123 (Tables 3 and 4). All are AONs, developed by ProQR Therapeutics and Ionis Pharmaceuticals [in the case of P23H autosomal dominant retinitis pigmentosa treatment (QR-1123), and which is licensed to ProQR Therapeutics]. The ongoing studies will build on knowledge gained from both animal and preclinical research including studies using retinal organoids36 and aim to validate results from these translational models within the clinical trial setting for each respective IRD.
Table 3.
| IRD | In vitro/in vivo | Model | Treatment | Proof of concept |
|---|---|---|---|---|
| CEP290-associated IRD | In vitro | Homozygous and compound heterozygous LCA primary fibroblasts | AON | AON (QR-110) treatment restored CEP290 wild-type (exon 26–27) mRNA levels (dose-dependent), reduced exon X-containing transcripts (exon X-27), and translated into a detectable increase in full-length CEP290 protein levels |
| CEP290-associated IRD | In vitro | Patient iPSC-derived 3D retinal organoids |
AON | AON (QR-110) reduced aberrant splicing, increased wild-type CEP290, restored ciliogenesis, and showed no off-target pharmacology |
| CEP290-associated IRD | In vivo | C57BL/6 mice | AON | Presence of AON (QR-110) in all retinal layers, including the RPE, which was more prominent in the ganglion cell layer, inner nuclear layer, and outer limiting membrane |
| CEP290-associated IRD | In vivo | Dutch-belted rabbits | AON | Confirmed findings of the mice model. Rapid uptake of AON (QR-110) by retinal cells, with an estimated retinal half-life of 58 days. |
| USH2A-associated Usher syndrome | In vitro | WERI-Rb1 cells | AON | AON (QR-421a) induced a concentration-dependent increase in USH2A exon 13 skipping in WERI-Rb1 cells |
| USH2A-associated Usher syndrome | In vitro | Patient-derived photoreceptor progenitor cells (PPCs) | AON | AON (QR-421a) induced a concentration-dependent increase in USH2A exon 13 skipping in iPSC-derived PPCs |
| USH2A-associated Usher syndrome | In vivo | Zebrafish ush2a-exon13 | PMO | PMO-induced skipping of ush2a exon 13 in the mutant zebrafish model restored usherin protein expression and visual function |
| USH2A-associated Usher syndrome | In vivo | Humanized exon 13 mutant mouse model (Ush2a-Δ12 / Ush2a-KO) | AON | AON (mQR-421a) induced Ush2a exon 12 skipping (dose-dependent) that lasted at least 203 days. Highest skipping level detected at 56 days. Retinal uptake in all retinal layers including the pharmacological target site (photoreceptors) |
Table 4.
Summary of clinical trials of RNA therapies in IRDs.a
| IRD | Trial | Phase | Sponsor | Type of RNA therapy | Design and objectives | Intervention | Population | Status |
|---|---|---|---|---|---|---|---|---|
| CEP290-associated IRD | PQ-110-001 (NCT03140969) |
Ib/II | ProQR Therapeutics | AON | Open-label, multiple-dose study to evaluate the efficacy, safety, tolerability, and systemic exposure; 24-month follow-up | Sepofarsen IVT to one eye every 3 months, up to four doses; up to three dose levels |
N = 11 Age ⩾ 6 years with LCA due to the CEP290 c.2991 + 1655A > G mutation |
Completion in 2019 |
| CEP290-associated IRD |
Insight extension
/ PQ-110-002 (NCT03913130) |
Ib/II | ProQR Therapeutics | AON | Extension study of PQ-110-001 (NCT03140969) examining long-term safety and efficacy, 24-month follow-up | Sepofarsen IVT continued dosing of previously treated eye plus treatment of contralateral eye; every 6 months |
N = 9 Age ⩾ 6 years with LCA due to the CEP290 c.2991 + 1655A > G mutation |
Ongoing; completion expected May 2022 |
| CEP290-associated IRD |
Illuminate / PQ-110-003 (NCT03913143) |
II/III | ProQR Therapeutics | AON | Double-masked, randomized, controlled, multiple-dose study to evaluate the efficacy, safety, tolerability, and systemic exposure; 24-month follow-up | Sepofarsen IVT to one eye; every 6 months; two dose levels; crossover to contralateral eye after 12 months of treatment |
N = 36 Age ⩾ 8 years with LCA due to the CEP290 c.2991 + 1655A > G mutation |
Ongoing; completion expected 2023 |
| CEP290-associated IRD |
Brighten / PQ-110-005 (NCT04855045) |
II/III | ProQR Therapeutics | AON | Open-label, dose escalation followed by double-masked, randomized study evaluating safety and tolerability; 24-month follow-up | Sepofarsen IVT to one eye; every 6 months; three dose levels |
N = 15 Age < 8 years with LCA due to the CEP290 c.2991 + 1655A > G mutation |
Recruiting; completion expected 2023 |
| USH2A-associated Usher syndrome |
Stellar / PQ-421a-001 (NCT03780257) |
Ib/II | ProQR Therapeutics | AON | Dose-escalation, masked, randomized, controlled study to evaluate safety and tolerability; 24-month follow-up | Ultevursen IVT to one eye, single dose; three dose levels |
N = 18 Age ⩾ 18 years with Usher syndrome due to USH2A exon 13 mutation |
Recruiting; completion expected 2022 |
| USH2A-associated Usher syndrome |
Helia extension / PQ-421a-002 (NCT05085964) |
Ib/II | ProQR Therapeutics | AON | Extension study of PQ-421a-001 (NCT03780257) examining long-term safety and efficacy, 24-month follow-up | Ultevursen IVT continued dosing of previously treated eye plus treatment of contralateral eye; every 6 months | Up to 100 participants Age ⩾ 18 years with Usher syndrome due to USH2A exon 13 mutation |
Ongoing |
| USH2A-associated Usher syndrome | Sirius / PQ-421a-003 (NCT05158296) | II/III | ProQR Therapeutics | AON | Double-masked, randomized, controlled, multiple-dose study to evaluate the efficacy, safety and tolerability; 27-month follow-up | Ultevursen IVT to one eye, single dose; two dose levels; crossover to contralateral eye after 12 months of treatment |
N = 81 Age ⩾ 12 years with Usher syndrome due to USH2A exon 13 mutation |
Ongoing; completion expected 2024 |
| P23H adRP |
Aurora / PQ-1123-001 (NCT04123626) |
Ib/II | ProQR Therapeutics | AON | Dose-escalation, open-label or masked, randomized, controlled study to evaluate safety, tolerability, and efficacy; 12-month follow-up | QR-1123 IVT to one eye; single dose or repeat doses every 3 months |
N = 35 Age ⩾ 18 years with adRP due to P23H mutation in RHO |
Completion in 2021 |
adRP: autosomal-dominant retinitis pigmentosa; AON: antisense oligonucleotide; IVT: intravitreal; LCA: Leber congenital amaurosis.
According to planned, completed and ongoing trials listed on www.clinicaltrials.gov as of 29 April 2022.
Sepofarsen (formerly named QR-110)
Sepofarsen is being developed as a treatment for CEP290-associated IRD, also commonly referred to as LCA10. LCA comprises a group of heterogeneous disease subtypes classified according to the associated gene defect.13 At least 25 genes that code for a diverse range of retinal functions have been linked to 17 phenotypes of LCA9,13 and more than 400 mutations have been implicated in these known phenotypes.14 Additional subtypes exist but the gene defects are yet to be identified.13 Typically, CEP290-associated IRD is inherited as an autosomal recessive trait.14,64 CEP290-associated IRD is the most common and severe subtype of LCA; it accounts for 15–30% of cases13,65–67 and has an estimated prevalence of < 1 patient per 100,000 individuals.55 This subtype is linked to mutations in the centrosomal protein 290 (CEP290) gene, which encodes the CEP290 protein localized in the transition zone of the connecting cilium of photoreceptors68,69 located between the inner and outer segments of rods and cones.69,70 This ciliary transition zone transports proteins to and from the outer segment of the photoreceptor where phototransduction occurs to convert light into electrical signals and vision.69,70 CEP290 is required for normal ciliary function and photoreceptor survival.36,64,69
The most common disease-causing mutation in CEP290-associated IRD is c.2991 + 1655A > G (also known as p.Cys998*).13,67,71,72,73 This is present in at least one allele in 21–77% of individuals with CEP290-associated IRD.65–68,73 The c.2991 + 1655A > G point mutation activates a cryptic splice site in intron 26, resulting in the inclusion of a pseudoexon between exons 26 and 27 in the CEP290 mRNA. Within this aberrant exon is a stop codon that prevents expression of a functional protein, either by resulting in the degradation of the mRNA or in a truncated CEP290 protein.35,65 As a consequence of the absence of functional CEP290 protein, protein transport through the cilium is hampered and the outer segment of the photoreceptor degenerates and shortens in length,64 the latter manifesting as a reduction in thickness in the outer nuclear layer in the peripheral to perifoveal region of the retina. Retained photoreceptor nuclei in the fovea with abnormal segments are also observed on optical coherence tomography (OCT).64 Loss of integrity of the retinal layers is correlated with reduced visual function.70 Electroretinography (ERG) responses are also undetectable, indicative of photoreceptor dysfunction.14,68 Interestingly, the photoreceptor degeneration progresses gradually over several decades; in the first decades of life despite widespread loss of rod photoreceptors, the foveal outer nuclear layer may be preserved.13,68,70 This suggests that a window of opportunity for early treatment may exist during which cone rescue could be possible.5,45,74
Symptoms of CEP290-associated IRD (which are also common to all LCA and EORD/EOSRD subtypes) include severe, congenital or early vision loss, nystagmus, and sluggishly reactive pupils.14 Sight varies from bare light perception to 20/200 in most of the cases, although there are also subjects with milder progression and less dramatically reduced visual acuity until adulthood.68,71,75 At presentation, 64% of individuals with CEP290-associated IRD have vision classified as counting fingers or worse in the better-seeing eye.73
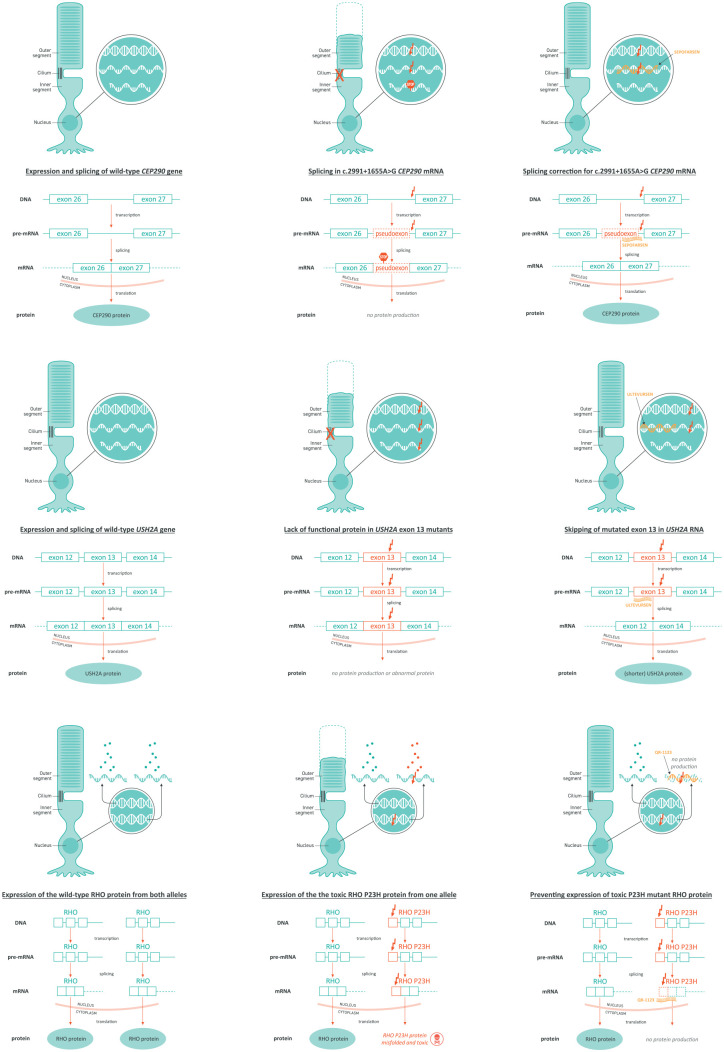
Sepofarsen is a 17-mer single-stranded, fully phosphorothioated, and 2′-O-methyl-modified AON that has been designed to modulate RNA splicing by blocking the cryptic splice site, thus restoring normal splicing and normal CEP290 protein synthesis (Figure 2(a)).36 In preclinical studies, sepofarsen restored normal CEP290 mRNA and protein expression in fibroblasts and retinal organoids derived from individuals with c.2991 + 1655A > G mutation in the CEP290 gene, and in animal models, this AON was able to reach all retinal layers.36 The positive preclinical data from optic cup studies with sepofarsen have been substantiated by positive early clinical data, confirming these models to be a good predictor of clinical outcomes.
Figure 2.
Mechanisms of action of (a) sepofarsen, (b) Ultevursen, and (c) QR-1123 detailing wild-type condition, mutant condition, and mechanism of action of AON.
The first-in-human, 12-month, phase Ib/II dose-escalation study of sepofarsen (PQ-110-001; NCT03140969) assessed the safety and efficacy of two doses of this AON in 11 individuals with CEP290-associated IRD aged ⩾ 6 years in the worse-seeing eye.57,76,77 Six participants received loading/maintenance doses of 160 µg/80 µg and five received 320 µg/160 µg. The final 12-month pooled data reported clinically meaningful improvements from baseline to Month 12 in treated eyes versus untreated eyes in best-corrected visual acuity (BCVA) (p < 0.05), red full-field stimulus test (FST) (p < 0.01), and blue FST (p < 0.02). There were also signals of improvement in functional vision as shown in composite mobility course score.57,77 The most common adverse events were cataracts; all participants who showed visual decline during cataract development regained pre-cataract visual acuity after lens replacement, with no procedural complications.57 There were two cases of mild cystoid macular edema and two of retinal thinning (all with the 320 µg/160 µg regimen).57 No study discontinuations occurred,77 and no inflammation was observed. Based on efficacy observations at the first two dose levels, the 500 µg/270 µg dose was not initiated, and following safety assessment, the 320 µg/160 µg dose was discontinued.57,77
The Insight Study (PQ-110-002/NCT03913130) is an ongoing extension of the above study in which the same participants are receiving treatment of the contralateral eye and continued treatment of the first treated eye with sepofarsen 160 µg/80 µg every 6 months. Initial results from four patients showed the safety profile of sepofarsen dosed in the contralateral eye to be consistent with that observed in the phase Ib/II trial. Improvements in BCVA of ⩾ 0.8 logMAR were observed in two patients which was similar to the efficacy in the first treated eye. All four patients showed similar improvements in FST to those observed in the first treated eye.78
Additional ongoing clinical trials of sepofarsen in CEP290-associated IRD include the phase II/III Illuminate study (PQ-110-003/NCT03913143) in adults and children aged ⩾ 8 years, and the phase II/III dose-escalation Brighten study (PQ-110-005/NCT04855045) in children aged < 8 years (Table 4). The phase II/III Illuminate study (PQ-110-003/NCT03913143) did not meet the primary endpoint nor notable secondary endpoints – and despite having good responders in the active treatment arms, no additional benefit was observed in either treatment arm versus sham in prespecified analyses. However, post hoc analyses reveal multiple pointers of a beneficial effect when comparing sepofarsen against sham group if the contralateral eyes of each group are adjusted for.79 Sepofarsen showed to be generally well tolerated with a consistent safety profile as observed in the phase Ib/II trial (PQ-110-001; NCT03140969). Additional analyses are being conducted to understand the conflicting data observed in the sepofarsen trials.
Ultevursen (formerly named QR-421a)
Ultevursen is being developed for Usher syndrome and non-syndromic RP (nsRP) associated with biallelic USH2A mutations. USH2A mutations are the most common cause of autosomal recessive RP (implicated in 7–23% of cases), and the two most frequent mutations in USH2A are located in exon 13.63
There are three main clinical types of Usher syndrome (numbered 1–3) varying in severity of vision and sensorineural hearing loss, age of onset, and involvement of vestibular dysfunction. Usher syndrome has been linked to mutations in 15 genes,8,14 and the pattern of inheritance is autosomal recessive.14 USH2A mutations are linked to Usher syndrome type 2.63,80–82 In this subtype, hearing impairment is moderate to severe and vision is affected from the second decade of life.81
Mutations within the USH2A gene disrupt the production of usherin protein which is localized in the connecting cilia of photoreceptors and stereocilia of cochlear hair cells (the sensory cells of the auditory system).83 This protein is thought to be important for the structural maintenance of photoreceptors and normal development of cochlear hair cells.84 Disrupted usherin protein production causes progressive photoreceptor degeneration beginning with the rods and eventually also involving the cones.81,84 In USH2A-associated nsRP severe visual impairment occurs by the age of 50 years.85 Patients with Usher syndrome have an early decline in vision, beginning with night blindness and constricted visual field, followed by the loss of central vision and severe visual impairment.81,85 Congenital hearing impairment is seen in this syndromic form.81,84
Ultevursen is a 21-mer single-stranded, fully phosphorothioated, and 2′-O-(2-methoxyethyl)-modified AON that binds to USH2A pre-mRNA and modulates splicing by exon skipping to exclude exon 13 from the mature mRNA (Figure 2(b)).63 This mechanism restores synthesis of a shorter but functional usherin protein.63,86
Preclinical proof-of-concept in vitro and in vivo studies have shown that the dose-dependent exon skipping induced by ultevursen is sustained at least 3 months, resulting in the restoration of usherin protein expression and functional ERG responses.86
The Stellar study (PQ-421a-001/NCT03780257) is a first-in-human phase Ib/II single-dose, dose-escalation trial assessing the safety and efficacy of ultevursen by intravitreal injection in adults with biallelic USH2A pathogenic variants, with at least one mutation occurring in exon 13 (Table 4).80 Interim data from unilateral treatment in 20 participants showed ultevursen to be well tolerated.80 There were no serious adverse events or inflammation, one case of worsening of pre-existing cataract, and one case of progression of pre-existing cystoid macula edema. Ultevursen demonstrated encouraging levels of BCVA stabilization (i.e. there was no decline versus baseline), compared with a deterioration in the control eyes, in keeping with the natural history of the disease.80 Treatment led to clinically significant improvements in retinal sensitivity as measured by the mean change from baseline in the number of loci that improved by ⩾ 7 dB on static perimetry in treated versus untreated eyes.80 These findings were supported by objective retinal structural optical coherence tomography imaging.80 The response to ultevursen was similar in all dose cohorts.
Participants from the Stellar study have been given the opportunity to enroll into the Helia extension study (PQ-421a-002/NCT05085964) for continued dosing and follow-up. This study will provide long-term safety, tolerability, and efficacy data of ultevursen. Following reported results in the Stellar study, a double-masked, randomized, sham controlled, 24-month, multiple-dose study, Sirius (PQ-421a-003; NCT05158296) has been initiated to further evaluate efficacy and safety of ultevursen.
QR-1123
QR-1123 is under evaluation as a treatment for adRP associated with the c.68 C > A mutation in RHO leading to a proline-to-histidine (P23 H; p.Pro23His) substitution. Around 15–25% cases of RP are inherited in an autosomal-dominant manner,14,87 and there are several subtypes of adRP.87 At least 22 genes may be involved in adRP,9 although the most commonly implicated is the RHO gene, which is responsible for 20–30% of cases of adRP.87,88 RHO codes for the rhodopsin protein,87 a photosensitive protein when intimately bound to 11-cis-retinal, present in rods and essential for rod-based phototransduction. In turn, the most common mutation in RHO is c.68 C > A; p.Pro23His.88,89 This mutation is seen almost exclusively in the United States, affecting an estimated 2500–3000 individuals89 and results in a P23H substitution in rhodopsin protein.87 The mutated protein is misfolded, leading to progressive photoreceptor degeneration.89 Like other forms of RP, adRP is characterized by rod–cone dystrophy with characteristic photoreceptor degeneration and intraretinal pigment migration predominantly in the mid-peripheral retina.90 Retinal degeneration progresses from the mid-periphery to the macula and fovea, manifesting as night blindness, followed by progressive, concentric constriction of the visual fields, leading to tunnel vision and eventually blindness.91
QR-1123 is an allele-specific, RNase H1-activating AON designed to treat P23H adRP by suppressing the formation of the toxic, mutant protein through knockdown of P23H rhodopsin mRNA. This selective inhibition of the mutant protein with dominant negative effect, while retaining the expression of the normal variant,92 enables the restoration of photoreceptor function (Figure 2(c)).89
Preclinical proof-of-concept studies of QR-1123 have demonstrated selective reduction of P23H rhodopsin expression and prevention of retinal degeneration in adRP animal and human cell models.89 The first-in-human phase I/II single-dose, dose-escalation study, Aurora (PQ-1123-001; NCT04123626), is ongoing (Table 4). This study will examine the safety and target engagement of QR-1123 intravitreal injection in adults with adRP due to P23H mutation in RHO.
Discussion
Genetic eye diseases are a leading cause of blindness and moderate-to-severe visual impairment that have, to date, been considered untreatable. However, there has been a surge in research interest in IRDs driven by advanced technologies and precision medicine capabilities, bringing new hope for improved outcomes for individuals living with IRD. This follows the approval of the first interventional therapy for an IRD, and a growing number of innovative, investigational therapies following behind. Among these pipeline products, three AONs targeting specific subtypes of LCA, Usher syndrome, nsRP, and adRP are leading the way.
AONs are well suited for treating the eye, which is a relatively immune-privileged closed compartment and have limited off-target effects. The most common adverse events of sepofarsen are cataracts.57,77–79 Instances were manageable and vision restored to pre-cataract levels after lens replacement surgery.57 The mechanism behind these cataract events is as yet unknown, but cataracts are reported with intravitreal treatments.60 They also feature in the natural course of CEP290-associated IRD, occurring spontaneously in 18–63% of cases in natural history studies, and such instances have been managed successfully with lens removal or replacement.65,73,75
The AON platform shares a common technology that encompasses a variety of mechanisms, potentially extending its use across multiple IRDs with different mechanisms (e.g. splice correction, exon skipping, repeat-targeting, exon inclusion, and allele-specific knockdown). These AONs could be useful in both peripheral and central IRDs, and may have the potential to treat a disease at its early stages, which may be particularly beneficial in diseases, such as CEP290-associated IRD.45 There is also a potential to broaden the application of this RNA platform technology to a range of genetic eye disorders including macular dystrophies (e.g. Stargardt disease) and beyond IRDs (e.g. Fuchs endothelial corneal dystrophy), and possibly other therapeutic areas, in the future.
Generally, AONs are developed to be specific to one mutation in one gene or a defined genetic sequence (such as the exon 13 of the USH2A gene). Therefore, the complex array of genetic mutations within each type and subtype of IRD brings an inherent challenge to the development of RNA-based therapies for genetic eye diseases.20 There are more than 270 different genes known to cause IRDs,9 and a large number of mutations may be involved within these genes.20 On this backdrop of broad genetic heterogeneity, identification and subsequent clinical development of AONs suitable for every individual with an IRD is a time and resource intensive process. The future success of AONs will require more work to understand the presence and significance of retinal-specific splice variants and alternative transcripts for all IRD genes in the humans, which may differ from animal models and non-retinal tissues. Observing clear signs of efficacy in preclinical testing does not always translate to efficacy in human trials, for a variety of reasons.93 The temporary nature of the therapeutic effect of AONs should also be considered as an important factor for development, potentially bringing an additional therapeutic burden versus a one-administration treatment but also conferring additional flexibility in terms of adjusting the treatment effects. Finally, as a novel therapy, the long-term efficacy and safety of AONs remain to be fully understood, although this will be addressed through the planned clinical research programs for these therapies.
Our understanding of the role of RNA-based therapies in relation to emerging DNA-based therapies will need to be defined. Alongside the pipeline AONs, at least 12 investigational gene augmentation therapies are in development for IRDs and other ophthalmic conditions, which currently include subtypes of LCA, RP, Bietti crystalline dystrophy, and Leber hereditary optic neuropathy. In addition, gene editing therapy is also being evaluated in the IRD space. EDIT-101 (AGN-151587; Editas Medicine) is an in vivo gene editing therapy based on CRISPR. An early-stage trial of this strategy is ongoing with preliminary data from the initial cohorts showing a favorable safety profile, transient EDIT-101 shedding in body fluids with low levels of detection, and signs suggesting biologic activity in some participants.94,95 As our knowledge of DNA- and RNA-based therapy approaches and their side effects grows, we may better understand at which stages of disease each may be applied. We anticipate that the future could bring a multimodal therapeutic strategy to treating certain individuals with IRDs. For instance, definitive DNA-based therapy risk reward may be appropriate for advanced stages of an IRD, whereas RNA-based therapy may offer a better trade-off in early-stage disease due to widespread accessibility to the peripheral retina and lower procedural risks. Therefore, in early disease, as DNA-based therapy would typically not be applicable, there may be a rationale for trialing RNA-based therapy soon after diagnosis. In end-stage disease, combination of DNA-based and RNA-based therapies might be applicable. However, more research is needed to better understand the roles of these targeted therapies across the family of IRDs.
Overall, RNA-based therapies have the potential to become a standard of care for genetic eye conditions. The close structure of oligonucleotides and the several categories of RNA-based therapies targeting different type of mutations could help accelerate the development of new therapies addressing unmet medical needs with urgency. Furthermore, the inclusion of new chemical modifications could further improve stability, half-life, biodisponibility and ultimately the efficacy, safety, and patient comfort.
Of critical importance, to take full advantage of advances in therapy in genetic eye diseases, it is important to improve genetic identification of eligible patients and streamline patient referral pathways to ensure access to treatments or clinical trials, as appropriate. This involves raising awareness of IRDs at every stage of the care pathway with general ophthalmologists, healthcare professionals, genetic counselors, and family practitioners. Improving access to, and use of, genetic testing is fundamental to these objectives as more than one-third of individuals with IRDs currently do not receive these critical investigations.96
Conclusion
In summary, the treatment paradigm for genetic eye diseases is in the midst of great change, driven by advances in knowledge of the diseases and innovations in therapeutic technologies. RNA-based therapies are an innovative approach within precision medicine that offers several key advantages in the setting of IRDs, and the potential to bring meaningful vision benefit to individuals living with these inherited blinding disorders. Three leading AONs are in clinical trials in specific subtypes of IRDs. Early data from ongoing trials of these therapies are promising and full results are eagerly awaited.
Acknowledgments
Medical writing support for this article was provided by Lyndsey Wood, BSc, of ApotheCom, and Janne Turunen, Luca Guerriero, and Agathe Plichta of ProQR Therapeutics.
Footnotes
ORCID iDs: Aniz Girach  https://orcid.org/0000-0003-0641-3028
https://orcid.org/0000-0003-0641-3028
Katarina Stingl  https://orcid.org/0000-0002-8132-911X
https://orcid.org/0000-0002-8132-911X
Contributor Information
Aniz Girach, ProQR Therapeutics, Zernikedreef 9, 2333 CK Leiden, the Netherlands.
Isabelle Audo, Centre Hospitalier National d’Ophtalmologie des Quinze-Vingts, Centre de référence maladies rares REFERET and INSERM-DHOS CIC 1423, CHNO des Quinze-Vingts, Paris, France; Institute of Ophthalmology, University College London, London, UK; Sorbonne Université, INSERM, CNRS, Institut de la Vision, Paris, France.
David G. Birch, Retina Foundation of the Southwest, Dallas, TX, USA
Rachel M. Huckfeldt, Department of Ophthalmology, Harvard Medical School, Massachusetts Eye and Ear Infirmary, Boston, MA, USA
Byron L. Lam, Bascom Palmer Eye Institute, University of Miami Miller School of Medicine, Miami, FL, USA
Bart P. Leroy, Department of Ophthalmology & Center for Medical Genetics, Ghent University Hospital & Ghent University, Ghent, Belgium Division of Ophthalmology & Center for Cellular & Molecular Therapeutics, The Children’s Hospital of Philadelphia, Philadelphia, PA, USA.
Michel Michaelides, UCL Institute of Ophthalmology, University College London and Moorfields Eye Hospital, London, UK.
Stephen R. Russell, The University of Iowa Institute for Vision Research, University of Iowa, Iowa City, IA, USA
Juliana M.F. Sallum, Department of Ophthalmology, Universidade Federal de São Paulo, São Paulo, Brazil Instituto de Genética Ocular, São Paulo, Brazil.
Katarina Stingl, Center for Ophthalmology, University Eye Hospital, University of Tübingen, Tübingen, Germany; Center for Rare Eye Diseases, University of Tübingen, Tübingen, Germany.
Stephen H. Tsang, Jonas Children’s Vision Care and Bernard and Shirlee Brown Glaucoma Laboratory, Columbia Stem Cell Initiative, Vagelos College of Physicians and Surgeons, Columbia University, New York, NY, USA Edward S. Harkness Eye Institute, New York-Presbyterian Hospital, New York, NY, USA.
Paul Yang, Casey Eye Institute, Oregon Health & Science University, Portland, OR, USA.
Declarations
Ethics approval and consent to participate: Not applicable.
Consent for publication: Not applicable.
Author contributions: Aniz Girach: Conceptualization; Data curation; Formal analysis; Investigation; Methodology; Writing – original draft; Writing – review & editing.
Isabelle Audo: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing.
David G. Birch: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing
Rachel M. Huckfeldt: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing.
Byron L. Lam: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing.
Bart P. Leroy: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing.
Michel Michaelides: Conceptualization; Data curation; Writing – original draft; Writing – review & editing.
Stephen R Russell: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing.
Juliana M.F. Sallum: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing.
Katarina Stingl: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing.
Stephen H. Tsang: Conceptualization; Data curation; Writing – original draft; Writing – review & editing.
Paul Yang: Conceptualization; Data curation; Investigation; Writing – original draft; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Editorial support for this article was funded by ProQR Therapeutics.
The authors declared the following potential conflicts of interest with respect to the research, authorship, and/or publication of this article: K.S., I.A., B.P.L., and M.M. are the members of ERN-EYE (www.ern-eye.eu). A.G. is an employee of ProQR Therapeutics. I.A. is a consultant/advisor for Novartis, Sparing Vision, Janssen, Roche, and ProQR Therapeutics; she is supported by the Foundation Fighting Blindness, Retina France, and UNADEV. D.G.B. reports support from the National Eye Institute EY09076 and Foundation Fighting Blindness; he has served as a consultant for ProQR Therapeutics, AGTC, Nacuity, Editas, and Biogen and has received clinical trial support from AGTC, ProQR Therapeutics, NightstaRx, and 4D Therapeutics. R.M.H. is a consultant for AGTC, Annexon, ProQR, Regeneron, and Vida Ventures and an advisor for Intergalactic Therapeutics; she receives grant funding from FFB and clinical trial support from AGTC, Biogen, MeiraGTx/Janssen, ProQR, and Spark. B.L.L. reports grant funding from ProQR Therapeutics, Biogen, AGTC, Allergan, and Spark Therapeutics; he has served as a consultant for ProQR Therapeutics, Biogen, and Allergan. B.P.L. is a Senior Clinical Investigator of the Research Foundation – Flanders, Belgium (Grant No. 1.8.038.11 N); he reports trial support from Second Sight Medical Products, consultancy fees from Bayer, REGENXBIO, and Vedere Bio, and trial support and consultancy fees and travel support from GenSight Therapeutics, IVERIC Bio, Novartis Pharma, Spark Therapeutics, and ProQR Therapeutics; he reports no personal financial gain, with all consultancy fees paid into Ghent University Hospital research accounts to support research. M.M. is supported by a grant from the National Institute for Health Research Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology. S.R.R. reports grant funding from Spark Therapeutics and ProQR Therapeutics, is a consultant for Novartis and co-founded an artificial intelligence-based retinal imaging company, Digital Diagnostics Incorporated (formerly IDx, LLC). J.M.F.S. has nothing to disclose. K.S. is or has served as a consultant for ProQR Therapeutics, ViGeneron, Novartis, Santen and Nayan, with consultancy fees paid to the Center for Ophthalmology, University of Tuebingen to support research. S.H.T. receives salary from the National Institute of Health (Health U01 EY030580, U54OD020351, R24EY028758, R24EY027285, R01EY018213, R01EY024698, R01EY026682), New York State (SDHDOH01-C32590GG-3450000), Research to Prevent Blindness and Alcon Research Institute, and serves as a consultant for SAB, EmendoBio, Nanoscope, and Rejuvitas. P.Y. is supported by grant from the Foundation Fighting Blindness TRAP1 Award (TA-NMT-0521-0803-OHSU-TRAP), the National Institutes of Health (Bethesda, MD) (P30 EY010572), and by unrestricted departmental funding from Research to Prevent Blindness (New York, NY); he serves as a consultant for 4D Molecular Therapeutics, Adverum, AGTC, Annexon Bio, EcoR1, ExpertConnect, Guidepoint, Janssen, MeiraGTx, Nanoscope Therapeutics, Otonomy, ProQR and Vedere and receives research support from 4D Molecular Therapeutics, Acucela, AGTC, Biogen, Editas, Foundation Fighting Blindness, Iveric bio, ProQR Therapeutics, Reneuron, Sanofi, and Spark.
Availability of data and materials: Not applicable.
References
- 1. GBD Study Group. Trends in prevalence of blindness and distance and near vision impairment over 30 years: an analysis for the Global Burden of Disease Study. Lancet Glob Health 2021; 9: e130–e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Méjécase C, Malka S, Guan Z, et al. Practical guide to genetic screening for inherited eye diseases. Ther Adv Ophthalmol 2020; 12: 2515841420954592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Green JS, Bear JC, Johnson GJ. The burden of genetically determined eye disease. Br J Ophthalmol 1986; 70: 696–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. American Academy of Ophthalmology, Clinical statement: recommendations on clinical assessment of patients with inherited retinal degenerations, 2016, https://www.aao.org/clinical-statement/recommendations-on-clinical-assessment-of-patients
- 5. Hu ML, Edwards TL, O’Hare F, et al. Gene therapy for inherited retinal diseases: progress and possibilities. Clin Exp Optom 2021; 104: 444–454. [DOI] [PubMed] [Google Scholar]
- 6. Hanany M, Rivolta C, Sharon D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc Natl Acad Sci U S A 2020; 117: 2710–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsang S, Sharma T. Atlas of inherited retinal diseases. Berlin: Springer, 2018. [Google Scholar]
- 8. Georgiou M, Fujinami K, Michaelides M. Inherited retinal diseases: therapeutics, clinical trials and end points-A review. Clin Exp Ophthalmol 2021; 49: 270–288. [DOI] [PubMed] [Google Scholar]
- 9. RetNet. Summaries of genes and loci causing retinal diseases, https://sph.uth.edu/retnet/sum-dis.htm#A-genes (accessed 12 May 2021).
- 10. Bunce C, Xing W, Wormald R. Causes of blind and partial sight certifications in England and Wales: April 2007-March 2008. Eye (Lond) 2010; 24: 1692–1699. [DOI] [PubMed] [Google Scholar]
- 11. Galvin O, Chi G, Brady L, et al. The impact of inherited retinal diseases in the Republic of Ireland (ROI) and the United Kingdom (UK) from a cost-of-illness perspective. Clin Ophthalmol 2020; 14: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farrar GJ, Carrigan M, Dockery A, et al. Toward an elucidation of the molecular genetics of inherited retinal degenerations. Hum Mol Genet 2017; 26: R2–R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kumaran N, Moore AT, Weleber RG, et al. Leber congenital amaurosis/early-onset severe retinal dystrophy: clinical features, molecular genetics and therapeutic interventions. Br J Ophthalmol 2017; 101: 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sahel JA, Marazova K, Audo I. Clinical characteristics and current therapies for inherited retinal degenerations. Cold Spring Harb Perspect Med 2014; 5: a017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berson EL, Rosner B, Sandberg MA, et al. A randomized trial of vitamin A and vitamin E supplementation for retinitis pigmentosa. Arch Ophthalmol 1993; 111: 761–772. [DOI] [PubMed] [Google Scholar]
- 16. Sacchetti M, Mantelli F, Merlo D, et al. Systematic review of randomized clinical trials on safety and efficacy of pharmacological and nonpharmacological treatments for retinitis pigmentosa. J Ophthalmol 2015; 2015: 737053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Annex I summary of Product Characteristics - European Medicines Agency (no date). https://www.ema.europa.eu/en/documents/product-information/luxturna-epar-product-information_en.pdf (accessed 24 October 2022).
- 18. HIGHLIGHTS OF PRESCRIBING INFORMATION - LUXTURNA (voretigene neparvovec-rzyl) intraocular suspension for subretinal injection (no date). https://www.fda.gov/media/109906/download (acessed 24 October 2022).
- 19. Dalkara D, Goureau O, Marazova K, et al. Let there be light: gene and cell therapy for blindness. Hum Gene Ther 2016; 27: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duncan JL, Pierce EA, Laster AM, et al. Inherited retinal degenerations: current landscape and knowledge gaps. Transl Vis Sci Technol 2018; 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang F, Zuroske T, Watts JK. RNA therapeutics on the rise. Nat Rev Drug Discov 2020; 19: 441–442. [DOI] [PubMed] [Google Scholar]
- 22. Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014; 157: 77–94. [DOI] [PubMed] [Google Scholar]
- 23. Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov 2017; 16: 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Crooke ST, Witztum JL, Bennett CF, et al. RNA-targeted therapeutics. Cell Metab 2018; 27: 714–739. [DOI] [PubMed] [Google Scholar]
- 25. Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014; 505: 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang R, Wu J, Zheng Z, et al. The construction and analysis of ceRNA network and patterns of immune infiltration in mesothelioma with bone metastasis. Front Bioeng Biotechnol 2019; 7: 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen J, Song Y, Li M, et al. Comprehensive analysis of ceRNA networks reveals prognostic lncRNAs related to immune infiltration in colorectal cancer. BMC Cancer 2021; 21: 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xue K, MacLaren RE. Antisense oligonucleotide therapeutics in clinical trials for the treatment of inherited retinal diseases. Expert Opin Investig Drugs 2020; 29: 1163–1170. [DOI] [PubMed] [Google Scholar]
- 29. Kim YK. RNA therapy: current status and future potential. Chonnam Med J 2020; 56: 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu AM, Choi YH, Tu MJ. RNA drugs and RNA targets for small molecules: principles, progress, and challenges. Pharmacol Rev 2020; 72: 862–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roberts TC, Langer R, Wood MJA. Advances in oligonucleotide drug delivery. Nat Rev Drug Discov 2020; 19: 673–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuijper EC, Bergsma AJ, Pijnappel WWMP, et al. Opportunities and challenges for antisense oligonucleotide therapies. J Inherit Metab Dis 2021; 44: 72–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collin RW, Garanto A. Applications of antisense oligonucleotides for the treatment of inherited retinal diseases. Curr Opin Ophthalmol 2017; 28: 260–266. [DOI] [PubMed] [Google Scholar]
- 34. Kim J, Hu C, Moufawad El, Achkar C, et al. Patient-customized oligonucleotide therapy for a rare genetic disease. N Engl J Med 2019; 381: 1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bacchi N, Casarosa S, Denti MA. Splicing-correcting therapeutic approaches for retinal dystrophies: where endogenous gene regulation and specificity matter. Invest Ophthalmol Vis Sci 2014; 55: 3285–3294. [DOI] [PubMed] [Google Scholar]
- 36. Dulla K, Aguila M, Lane A, et al. Splice-modulating oligonucleotide QR-110 restores CEP290 mRNA and function in human c. 2991+1655A>G LCA10 models. Mol Ther Nucleic Acids 2018; 12: 730–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zarouchlioti C, Sanchez-Pintado B, Hafford Tear NJ, et al. Antisense therapy for a common corneal dystrophy ameliorates TCF4 repeat expansion-mediated toxicity. Am J Hum Genet 2018; 102: 528–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Porto EM, Komor AC, Slaymaker IM, et al. Base editing: advances and therapeutic opportunities. Nat Rev Drug Discov 2020; 19: 839–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vázquez-Domínguez I, Garanto A, Collin RWJ. Molecular therapies for inherited retinal diseases-current standing, opportunities and challenges. Genes (Basel) 2019; 10: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sanjurjo-Soriano C, Kalatzis V. Guiding lights in genome editing for inherited retinal disorders: implications for gene and cell therapy. Neural Plast 2018; 2018: 5056279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Levin AA. Treating disease at the RNA level with oligonucleotides. N Engl J Med 2019; 380: 57–70. [DOI] [PubMed] [Google Scholar]
- 42. Maguire AM, Russell S, Chung DC, et al. Durability of voretigene neparvovec for biallelic RPE65-mediated inherited retinal disease: phase 3 results at 3 and 4 years. Ophthalmology 2021; 128: 1460–1468. [DOI] [PubMed] [Google Scholar]
- 43. Jacobson SG, Cideciyan AV, Roman AJ, et al. Improvement and decline in vision with gene therapy in childhood blindness. N Engl J Med 2015; 372: 192020150503–192020151926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bainbridge JW, Mehat MS, Sundaram V, et al. Long-term effect of gene therapy on Leber’s congenital amaurosis. N Engl J Med 2015; 372: 188720150504–188720151897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Garanto A, Chung DC, Duijkers L, et al. In vitro and in vivo rescue of aberrant splicing in CEP290-associated LCA by antisense oligonucleotide delivery. Hum Mol Genet 2016; 25: 2552–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maguire AM, Russell S, Wellman JA, et al. Efficacy, safety, and durability of voretigene neparvovec-rzyl in RPE65 mutation-associated inherited retinal dystrophy: results of phase 1 and 3 trials. Ophthalmology 2019; 126: 1273–1285. [DOI] [PubMed] [Google Scholar]
- 47. Avery RL, Bakri SJ, Blumenkranz MS, et al. Intravitreal injection technique and monitoring: updated guidelines of an expert panel. Retina 2014; 34(Suppl. 12): S1–S18. [DOI] [PubMed] [Google Scholar]
- 48. Fagan XJ, Al-Qureshi S. Intravitreal injections: a review of the evidence for best practice. Clin Exp Ophthalmol 2013; 41: 500–507. [DOI] [PubMed] [Google Scholar]
- 49. Moore NA, Morral N, Ciulla TA, et al. Gene therapy for inherited retinal and optic nerve degenerations. Expert Opin Biol Ther 2018; 18: 37–49. [DOI] [PubMed] [Google Scholar]
- 50. Gerard X, Perrault I, Munnich A, et al. Intravitreal injection of splice-switching oligonucleotides to manipulate splicing in retinal cells. Mol Ther Nucleic Acids 2015; 4: e250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee JH, Wang JH, Chen J, et al. Gene therapy for visual loss: opportunities and concerns. Prog Retin Eye Res 2019; 68: 31–53. [DOI] [PubMed] [Google Scholar]
- 52. Ziccardi L, Cordeddu V, Gaddini L, et al. Gene therapy in retinal dystrophies. Int J Mol Sci 2019; 20: 5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chiu W, Lin TY, Chang YC, et al. An update on gene therapy for inherited retinal dystrophy: experience in Leber congenital amaurosis clinical trials. Int J Mol Sci 2021; 26: 4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang X, Yu C, Tzekov RT, et al. The effect of human gene therapy for RPE65-associated Leber’s congenital amaurosis on visual function: a systematic review and meta-analysis. Orphanet J Rare Dis 2020; 15: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Timmers AM, Newmark JA, Turunen HT, et al. Ocular inflammatory response to intravitreal injection of adeno-associated virus vector: relative contribution of genome and capsid. Hum Gene Ther 2020; 31: 80–89. [DOI] [PubMed] [Google Scholar]
- 56. Bucher K, Rodríguez-Bocanegra E, Dauletbekov D, et al. Immune responses to retinal gene therapy using adeno–associated viral vectors – Implications for treatment success and safety. Prog Retin Eye Res 2021; 83: 100915. [DOI] [PubMed] [Google Scholar]
- 57. Russell SR, Drack AV, Cideciyan AV, et al. Intravitreal antisense oligonucleotide sepofarsen in Leber congenital amaurosis type 10: a phase 1b/2 trial. Nat Med 2022; 28: 1014–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McClements ME, Barnard AR, Singh MS, et al. An AAV dual vector strategy ameliorates the stargardt phenotype in adult Abca4(-/-) mice. Hum Gene Ther 2019; 30: 59020181224–59020181600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gupta A, Kafetzis KN, Tagalakis AD, et al. RNA therapeutics in ophthalmology – translation to clinical trials. Exp Eye Res 2021; 205: 108482. [DOI] [PubMed] [Google Scholar]
- 60. Leroy BP, Birch DG, Duncan JL, et al. Leber congenital amaurosis due to CEP290 mutations: severe vision impairment with a high unmet medical need: a review. Retina 2021; 41: 898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cukras C, Wiley HE, Jeffrey BG, et al. Retinal AAV8-RS1 gene therapy for X-linked retinoschisis: initial findings from a phase I/IIa trial by intravitreal delivery. Mol Ther 2018; 26: 2282–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cideciyan AV, Jacobson SG. Leber Congenital Amaurosis (LCA): potential for Improvement of Vision. Invest Ophthalmol Vis Sci 2019; 60: 1680–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Dulla K, Slijkerman R, van Diepen HC, et al. Antisense oligonucleotide-based treatment of retinitis pigmentosa caused by USH2A exon 13 mutations. Mol Ther 2021; 29: 2441–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. den Hollander AI, Roepman R, Koenekoop RK, et al. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog Retin Eye Res 2008; 27: 391–419. [DOI] [PubMed] [Google Scholar]
- 65. Chau VQ, Hu J, Gong X, et al. Delivery of antisense oligonucleotides to the cornea. Nucleic Acid Ther 2020; 30: 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kondkar AA, Abu-Amero KK. Leber congenital amaurosis: current genetic basis, scope for genetic testing and personalized medicine. Exp Eye Res 2019; 189: 107834. [DOI] [PubMed] [Google Scholar]
- 67. den Hollander AI, Koenekoop RK, Yzer S, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet 2006; 79: 556–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Coppieters F, Casteels I, Meire F, et al. Genetic screening of LCA in Belgium: predominance of CEP290 and identification of potential modifier alleles in AHI1 of CEP290-related phenotypes. Hum Mutat 2010; 31: E1709–E1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Perrault I, Delphin N, Hanein S, et al. Spectrum of NPHP6/CEP290 mutations in Leber congenital amaurosis and delineation of the associated phenotype. Hum Mutat 2007; 28: 416. [DOI] [PubMed] [Google Scholar]
- 70. Sheck L, Davies WIL, Moradi P, et al. Leber congenital amaurosis associated with mutations in CEP290, clinical phenotype, and natural history in preparation for trials of novel therapies. Ophthalmology 2018; 125: 894–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shimada H, Lu Q, Insinna-Kettenhofen C, et al. In vitro modeling using ciliopathy-patient-derived cells reveals distinct cilia dysfunctions caused by CEP290 mutations. Cell Rep 2017; 20: 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Feldhaus B, Weisschuh N, Nasser F, et al. CEP290 mutation spectrum and delineation of the associated phenotype in a large German cohort: a monocentric study. Am J Ophthalmol 2020; 211: 142–150. [DOI] [PubMed] [Google Scholar]
- 73. McAnany JJ, Genead MA, Walia S, et al. Visual acuity changes in patients with Leber congenital amaurosis and mutations in CEP290. JAMA Ophthalmol 2013; 131: 178–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Garafalo AV, Cideciyan AV, Heon E, et al. Progress in treating inherited retinal diseases: early subretinal gene therapy clinical trials and candidates for future initiatives. Prog Retin Eye Res 2020; 77: 100827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yzer S, Hollander AI, Lopez I, et al. Ocular and extra-ocular features of patients with Leber congenital amaurosis and mutations in CEP290. Mol Vis 2012; 18: 412–425. [PMC free article] [PubMed] [Google Scholar]
- 76. Cideciyan AV, Jacobson SG, Drack AV, et al. Effect of an intravitreal antisense oligonucleotide on vision in Leber congenital amaurosis due to a photoreceptor cilium defect. Nat Med 2019; 25: 225–228. [DOI] [PubMed] [Google Scholar]
- 77. Russell SR, Drack AV, Cideciyan AV, et al. Results of a phase 1b/2 trial of intravitreal (IVT) sepofarsen (QR-110) antisense oligonucleotide in Leber congenital amaurosis 10 (LCA10) due to p.Cys998X mutation in the CEP290 gene. ARVO 2020; 61: 866. [Google Scholar]
- 78. Leroy B, Russell SR, Drack AV, et al. Safety and efficacy of sepofarsen in the second treated eye in the Phase 1b/2 extension trial in Leber congenital amaurosis due to mutations in the CEP290 gene (Insight Trial). EURETINA, 2021, https://euretina.org/resource/abstract_2021_safety-and-efficacy-of-sepofarsen-in-the-second-treated-eye-in-the-phase-1b-2-extension-trial-in-leber-congenital-amaurosis-due-to-mutations-in-the-cep290-gene-insight-trial/
- 79. Leroy BP, Stingl K, Audo I, et al. Efficacy and safety of sepofarsen, an intravitreal RNA antisense oligonucleotide, for the treatment of CEP290-associated Leber congenital amaurosis (LCA10): a randomized, double-masked, sham-controlled, Phase 3 study (ILLUMINATE). ARVO 2022; 63: 4536. [Google Scholar]
- 80. Birch DG, Audo I, Jayasundera KT, et al. Phase 1b/2 interim results of QR-421a RNA therapy in retinitis pigmentosa due to mutations in the USH2A gene (Stellar trial). EURETINA, 2021, https://euretina.org/resource/abstract_2021_phase-1b-2-interim-results-of-qr-421a-rna-therapy-in-retinitis-pigmentosa-due-to-mutations-in-the-ush2a-gene-stellar-trial/
- 81. Toualbi L, Toms M, Moosajee M. USH2A-retinopathy: from genetics to therapeutics. Exp Eye Res 2020; 201: 108330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Seyedahmadi BJ, Rivolta C, Keene JA, et al. Comprehensive screening of the USH2A gene in Usher syndrome type II and non-syndromic recessive retinitis pigmentosa. Exp Eye Res 2004; 79: 167–173. [DOI] [PubMed] [Google Scholar]
- 83. Cosgrove D, Zallocchi M. Usher protein functions in hair cells and photoreceptors. Int J Biochem Cell Biol 2014; 46: 80–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liu X, Bulgakov OV, Darrow KN, et al. Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc Natl Acad Sci U S A 2007; 104: 4413–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Pierrache LH, Hartel BP, van Wijk E, et al. Visual prognosis in USH2A-associated retinitis pigmentosa is worse for patients with usher syndrome type IIa than for those with nonsyndromic retinitis pigmentosa. Ophthalmology 2016; 123: 1151–1160. [DOI] [PubMed] [Google Scholar]
- 86. van Diepen H, Dulla K, Chan H, et al. QR-421a, an antisense oligonucleotide, for the treatment of retinitis pigmentosa due to USH2A exon 13 mutations. ARVO 2019; 60: 3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tsang S, Sharma T. Autosomal dominant retinitis pigmentosa. In: Tsang S, Sharma T. (eds) Atlas of inherited retinal diseases. Berlin: Springer, 2018, pp. 70–77. [Google Scholar]
- 88. Daiger SP, Bowne SJ, Sullivan LS. Genes and mutations causing autosomal dominant retinitis pigmentosa. Cold Spring Harb Perspect Med 2014; 5: a017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Biasutto P, Adamson P, Dulla K, et al. Allele specific knock-down of human P23H rhodopsin mRNA and prevention of retinal degeneration in humanized P23H rhodopsin knock-in mouse, following treatment with an intravitreal GAPmer antisense oligonucleotide (QR–1123). Invest Ophthalmol Vis Sci 2019; 60: 5719. [Google Scholar]
- 90. Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis 2006; 1: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ferrari S, Di Iorio E, Barbaro V, et al. Retinitis pigmentosa: genes and disease mechanisms. Curr Genomics 2011; 12: 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Woods KN, Pfeffer J. Conformational perturbation, allosteric modulation of cellular signaling pathways, and disease in P23H rhodopsin. Sci Rep 2020; 10: 265720200216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Leung LSB, Voleti VB, Lin JH, et al. Genetic pathways in retinal degenerations and targets for therapy. In: Traboulsi MD. (ed.) Genetic diseases of the eye. Oxford: Oxford University Press, 2012, pp. 356–372. [Google Scholar]
- 94. Penessi M, Swan K, Casey P. BRILLIANCE: a phase 1/2 single ascending dose study of EDIT-101, an in vivo CRISPR gene editing therapy in CEP290-related retinal degeneration, 2021, https://editasmedicine.gcs-web.com/static-files/22b32d3d-e38f-4e90-899c-a2e701872745
- 95. Jaskolka MC, El-Husayni S, Duke B, et al. Exploratory safety profile of EDIT-101, a first-in-human in vivo CRISPR gene editing therapy for CEP290-related retinal degeneration. Invest Ophthalmol Vis Sci 2022; 63: 2836. [Google Scholar]
- 96. Deloitte Access Economics. The socioeconomic impact of inherited retinal dystrophies (IRDs) in the United Kingdom. London: Deloitte Access Economics, 2019. [Google Scholar]