Abstract
The molecular basis of Escherichia coli traversal of the blood-brain barrier in the development of E. coli meningitis is not well understood. We have previously shown that a novel Ibe10 protein found in cerebrospinal fluid isolates of E. coli is necessary for invasion of the brain microvascular endothelial cells (BMEC) that constitute the blood-brain barrier both in vitro and in a newborn rat model of hematogenous meningitis. Here we identified a novel Ibe10 binding molecule/receptor (Ibe10R) on both bovine BMEC (HBMEC) and human BMEC (HBMEC) that is responsible for invasion by E. coli. Ibe10R, an approximately 55-kDa protein, was purified from BBMEC by Ibe10-Ni-Sepharose affinity chromatography. Bovine Ibe10R, as well as polyclonal antibodies to Ibe10R, blocked E. coli invasion of BBMEC very effectively. The N-terminal amino acid sequence of Ibe10R showed 75% homology to serum albumin. However, the amino acid sequence of an Ibe10R fragment generated by limited enzymatic digestion did not reveal homology to any other proteins, suggesting that Ibe10R represents a novel albumin-like protein. Immunocytochemical analysis of BBMEC using anti-Ibe10R antibody suggested that only a subset of cultured BBMEC express Ibe10R on their surface. Enrichment of Ibe10R-positive BBMEC by fluorescence-activated cell sorting with anti-Ibe10R antibody resulted in enhanced invasion by E. coli. The anti-Ibe10R antibody raised against bovine Ibe10R also blocked E. coli invasion of HBMEC very effectively. Interestingly, anti-Ibe10R antibody affinity chromatography of HBMEC membrane proteins revealed a smaller protein with an approximate molecular mass of 45 kDa. These results suggest that the Ibe10 of E. coli interacts with a novel BMEC surface protein, Ibe10R, for invasion of both BBMEC and HBMEC.
Many pathogenic bacteria colonize human hosts through interactions between specific bacterial surface adhesins and respective binding sites on the mammalian cell surface (2, 3, 19, 20). Identification of the host cell surface receptors exploited by these bacteria is important for delineation of issues like host range and tissue tropism of different infections. Unraveling of the mechanisms of these interactions may help define domains on both bacterial and host tissues which are critical for establishing disease and may provide a basis for the development of novel therapeutic or preventive strategies.
Escherichia coli is one of the most common gram-negative bacteria that cause meningitis during the neonatal period. The mortality and morbidity associated with this disease have remained significant despite advances in antimicrobial chemotherapy (4, 18). This is attributed mainly to inadequate knowledge of the pathogenesis and pathophysiology of this disease. E. coli meningitis develops as a result of hematogenous spread, but it is not clear how circulating bacteria cross the blood-brain barrier, a lining comprised of brain microvascular endothelial cells (BMEC). The invasion by E. coli preceded by adherence to BMEC is thought to be a critical step in the pathogenesis of E. coli meningitis. E. coli expresses several surface structures that can potentially interact with host cells, such as lipopolysaccharide, K1 capsule, fimbriae, and outer membrane proteins. S-fimbriae, the filamentous protein appendages that bind to terminal NeuAcα2,3-Gal sequences present on glycoproteins, have been implicated as one of the microbial factors involved in the pathogenesis of neonatal meningitis (6, 8, 10). We have shown that S-fimbria expression enhances the binding of E. coli to BMEC, which is mediated by a lectin-like activity of the SfaS adhesin specific for NeuAcα2,3-Gal residues (15), and also to sulfated glycolipids via the major fimbrillin protein SfaA (11). We have previously identified the S-fimbria binding sialoglycoproteins present on BMEC containing NeuAcα2,3-Gal residues (14); however, binding via S-fimbriae was not accompanied by invasion of BMEC, indicating that S-fimbriae may be the prime attachment-promoting factor for E. coli.
In search of structures contributing to E. coli invasion of BMEC, we have observed that expression of OmpA, one of the major outer membrane proteins, enhances the invasion of BMEC by E. coli (12). This event occurs by OmpA interaction with GlcNAc1-4GlcNAc epitopes present on N-linked oligosaccharides of BMEC surface glycoproteins (13). However, OmpA is highly conserved and present in both nonclinical and clinical isolates of E. coli, suggesting that there are other structures specific to meningitis-causing E. coli that contribute to the invasion of BMEC.
In order to identify other E. coli structures that contribute to the invasion of BMEC, we have used transposon TnphoA mutagenesis to generate a collection of noninvasive mutants. One of the noninvasive mutants, 10A-23, with a single TnphoA insertion and without any changes in other phenotypic and genotypic characteristics, was found to be significantly less invasive of BMEC in vitro and of the central nervous system in the newborn rat model of hematogenous E. coli meningitis (5). The sequence of flanking regions of TnphoA revealed an open reading frame encoding a novel 8.3-kDa protein (Ibe10) with potential multiple transmembrane domains. The purified recombinant Ibe10 protein significantly inhibited E. coli invasion of BMEC. In addition, the invasive determinant encoded by ibe10 appears to be common in cerebrospinal fluid (CSF) isolates of E. coli K1 (e.g., C5 and RS218), while laboratory strains of E. coli K-12 (e.g., DH5α and HB101), as well as noninvasive E. coli (e.g., E412), lack ibe10 (1, 5). However, it is unclear how Ibe10 interacts with BMEC, resulting in enhanced E. coli invasion of BMEC. In this study, we set out to identify the BMEC cell surface molecule(s) that interacts with Ibe10 and is responsible for E. coli invasion of BMEC.
MATERIALS AND METHODS
Bacterial strains and chemicals.
E. coli RS218 is a clinical isolate from the CSF of a newborn infant with meningitis. E44 is a spontaneous rifampin-resistant mutant of RS218 (12), and 10A-23 is a noninvasive strain derived from E44 by TnphoA mutagenesis with a disrupted ibe10 locus (5). E. coli M15 was used as a host strain for transformation with pQIB10. Transformed bacteria were grown in Luria broth at 37°C with ampicillin (50 μg/ml) and kanamycin (50 μg/ml). Restriction endonucleases and other enzymes were purchased from New England Biolabs (Beverly, Mass.) unless otherwise stated. Protein A-Sepharose columns, the Fab fragment generation kit, and sulfo-NHS-LC biotin were obtained from Pierce Co. (Rockford, Ill.). All other chemicals were obtained from Sigma (St. Louis, Mo.) unless otherwise stated.
Purification of recombinant Ibe10 protein.
The expression and purification of recombinant histidine-tagged Ibe10 protein (rIbe10) was carried out in accordance with the instructions of the manufacturer (Qiagen) as described in detail previously (5). Briefly, the BamHI-HindIII fragment (250 bp) from pCIB10A, which encodes Ibe10, was eluted from 2% agarose gel after digestion and then ligated to the same restriction sites of a pQE30 His-tag expression vector (Qiagen). This construct, pQIB10, encodes a 9-kDa Ibe10 protein with a histidine tag at the N terminus. Transformation of E. coli M15 with pQIB10 was performed by electroporation in 0.1-cm cuvettes using an E. coli gene pulser (Bio-Rad Laboratories, Richmond, Calif.). Transformants were identified by ampicillin and kanamycin selection. Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside at 30°C. The rIbe10 protein containing the histidine tag was purified on nickel-nitrilotriacetic acid (Ni-NTA) resin in 6 M guanidine-HCl. The eluted proteins containing 8 M urea were refolded by sequential dialysis against decreasing concentrations of urea in 25 mM Tris-HCl (pH 8.0)–0.2 M NaCl–1 mM EDTA–10 mM β-mercaptoethanol.
Infection experiments.
The bovine BMEC (BBMEC) and human BMEC (HBMEC) used for invasion assays in this study were isolated as described previously (15–17). For infection experiments, BMEC were plated in a 24-well plate coated with collagen and invasion assays were performed as previously described (12, 13). Approximately 107 bacteria were added to a confluent monolayer of BMEC (bacterium-BMEC ratio of 100:1) in experimental medium containing M199 and Ham F-12 (1:1) with 5% heat-inactivated fetal calf serum (FCS), 2 mM l-glutamine, and 1 mM sodium pyruvate. The plates were incubated for 1.5 h at 37°C in 5% CO2 without shaking. Infection was stopped by rinsing of the cells four times with RPMI medium, followed by additional incubation in experimental medium containing 100-μg/ml gentamicin for 1 h to kill extracellular bacteria. The monolayers were again washed four times and lysed with 0.5% Triton X-100. The released intracellular bacteria were enumerated by plating on sheep blood agar. The assays were carried out at least three times in triplicate. Bacterial viability was not affected by 0.5% Triton X-100 treatment under the experimental conditions employed.
rIbe10 affinity chromatography.
The rIbe10 protein (10 mg) in Tris buffer (50 mM, pH 7.3) containing NaCl (0.2 M), EDTA (1 mM), β-mercaptoethanol (10 mM), and urea (0.5 M) was added to the Ni-NTA-Sepharose (5 ml), which was pre-equilibrated with Tris–0.2 M NaCl and incubated for 1 h at 4°C with gentle rotation. After the incubation, the Sepharose was centrifuged at 500 × g for 5 min and the supernatant containing uncoupled rIbe10 was removed. The gel was washed with Tris-NaCl several times, packed into a column, and stored at 4°C until use. Whole membrane proteins from BBMEC were isolated by cell lysis, Triton X-100 solubilization of membrane proteins, and ultracentrifugation as described earlier (14). Approximately 10 mg of BBMEC membrane proteins was passed through the 10-ml bed volume of a wheat germ agglutinin (WGA)-Sepharose column over a period of 2 h at 4°C (14). The column was then washed with 2 bed volumes of Tris-buffered saline (TBS), and the washes were combined with the original runthrough solution. The column was further washed with TBS, and the bound proteins were eluted with 200 mM N-acetylglucosamine in TBS, dialyzed extensively against TBS, and concentrated. The WGA runthrough proteins were preincubated with Ni-NTA-Sepharose overnight at 4°C on a rotator. On the next day, the mixture was centrifuged to remove the Ni-NTA-Sepharose, and the supernatant was collected. The Sepharose was washed twice with Tris buffer and combined with the original protein solution. The treated BBMEC membrane proteins were then passed through the rIbe10-Ni-Sepharose column over a period of 1 h at 4°C. The column was washed again with 10 bed volumes of Tris-NaCl buffer, and the bound proteins were eluted with 100 mM glycine-HCl, pH 2.5. The eluted fraction was extensively dialyzed against TBS overnight with several changes of buffer in a dialysis bag (8,000 molecular weight cutoff) and concentrated by Centricon tubes and lyophilization.
Anti-Ibe10R antibody affinity chromatography of BBMEC membrane proteins.
Polyclonal antiserum to the rIbe10-Ni-Sepharose-bound BBMEC proteins (Ibe10R) was generated in rabbits and coupled to CNBr-activated Sepharose by the methods described earlier (14). For anti-Ibe10R antibody affinity chromatography, BBMEC membrane proteins (2 mg) were initially passed through the 10-ml bed volumes of WGA-Sepharose columns to remove N-glycosylated proteins several times. The WGA runthrough proteins were subjected to anti-Ibe10R antibody-Sepharose chromatography (14). In addition, both BBMEC and HBMEC surface proteins were biotinylated and membrane proteins were prepared as described previously (14) and subjected to anti-Ibe10R antibody Sepharose chromatography. The bound proteins were released with glycine-HCl (pH 2.5), dialyzed against phosphate-buffered saline (PBS), and concentrated, and the protein content was estimated.
Two-dimensional gel electrophoresis.
Ibe10R was subjected to two-dimensional electrophoresis for N-terminal amino acid sequencing. For the first dimension, 10 μg of Ibe10R was separated by electrophoresis in a minicapillary containing urea and Bio-ampholyte (pI 3 to 10) in accordance with the manufacturer’s (Bio-Rad) protocol. The gel was removed and placed onto the second-dimension sodium dodecyl sulfate (SDS)–10% polyacrylamide gel horizontally. After separation, the proteins were transferred to a nitrocellulose membrane and immunoblotted with anti-Ibe10R antibody. A duplicate gel was briefly stained with Coomassie brilliant blue R-250; the band corresponding to anti-Ibe10R antibody-reactive protein was excised and subjected to N-terminal amino acid sequencing.
Immunocytochemical staining.
Immunocytochemical staining of BMEC with the anti-Ibe10R antibody was carried out in eight-well chamber slides as previously described (14), except that the BMEC were fixed with 2% paraformaldehyde in PBS for 15 min at room temperature. The fixed BMEC monolayers were washed with PBS five times and incubated with 1% normal goat serum to block the nonspecific binding sites. The monolayers were further incubated with protein A-purified anti-Ibe10R antibody (1:500), followed by peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG; 1:1,000). In some experiments, Triton X-100 was added to the solutions (final concentration, 0.1%) to facilitate the permeation of the cells by the antibody.
Partial proteolytic digestion.
Ibe10R and bovine serum albumin (BSA) (50 μg of each) were lyophilized and resuspended separately in 50 mM ammonium bicarbonate containing 1 mM CaCl2, pH 8.0. The proteins were mixed with 1:1,000 (wt/wt) endoproteinase Glu-C (proteinase type XXXV) from Staphylococcus aureus V8 and incubated at room temperature overnight (7). The digestion was quenched with SDS sample buffer, and the proteins were separated on an SDS–12% polyacrylamide gel in duplicate. After electrophoresis, the proteins were transferred to nitrocellulose sheets and one blot was blocked with 5% nonfat milk in PBS for 1 h at room temperature. The blot was then incubated with anti-Ibe10R antibody (1:500) for 2 h, followed by goat anti-rabbit IgG coupled to horseradish peroxidase (1:1,000), and washed five times with PBS–0.05% Tween 20. The blots were developed with diaminobenzidine and hydrogen peroxide as substrates to identify the Ibe10R band. The duplicate blot was briefly stained with Coomassie brilliant blue R-250 stain, and the protein band corresponding to the anti-Ibe10R antibody-reactive fragment was excised and subjected to N-terminal protein sequencing.
Solid-phase binding assay.
The binding of E. coli strains and rIbe10 protein to Ibe10R immobilized on microtiter plates (Immunlon 3; Dynatech Laboratories) was determined by enzyme-linked immunosorbent assay (14). E. coli and rIbe10 protein were biotinylated as described earlier (11). Ibe10R (0.4 μg of protein/well) was allowed to bind to the microtiter plate wells in PBS (50 μl) overnight at 4°C. The solution containing the unbound protein was aspirated, washed with PBS three times, and incubated with 5% BSA for 1 h at room temperature to block nonspecific binding sites. Either biotinylated bacteria (2 × 103 CFU/well) or rIbe10 protein (5 μg/well) was added to wells in 5% BSA, and the plates were incubated for 2 h at 4°C with occasional shaking. The wells were washed four times with PBS and incubated with streptavidin-peroxidase (1:2,000 in 5% BSA) for 30 min at room temperature. Bound bacteria and rIbe10 were quantified as previously described (14).
Fluorescence-activated cell sorting of BBMEC.
Confluent monolayers of BBMEC were washed three times with RPMI medium and detached from the surface by using 1 mM EDTA in RPMI medium. The detached cells were resuspended in RPMI medium containing 10% FCS. Cells (2.0 × 105) were incubated with polyclonal anti-Ibe10R antibody (1:500 dilution) for 15 min on ice. The cells were then washed with RPMI medium containing 2% FCS and counterstained with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG (diluted 1:5,000 in RPMI medium–10% FCS) on ice for 15 min. The cells were washed and resuspended in PBS for fluorescence-activated cell sorter (FACS) analysis. Anti-OmpA polyclonal antibody was used as a negative control. Stained cells were analyzed by flow microfluorimetry on a FACScan flow cytometer (Becton Dickinson & Co., Mountain View, Calif.) using the LYSYS II program. For sorting, 5 × 106 cells were stained as described above and then resuspended in RPMI medium containing 30% FCS. The top 8% of the positive cells and the bottom 10% of the negative cells were collected, and the serum was diluted to 10% before plating into T25 flasks. The cells were then transferred to a T75 flask and, after reaching confluence, plated in a 24-well tissue culture plate for infection experiments as described above. These cells were also plated in an eight-well chamber slide for immunocytochemical staining.
RESULTS
E. coli invasin Ibe10 binds to a 55-kDa BBMEC protein.
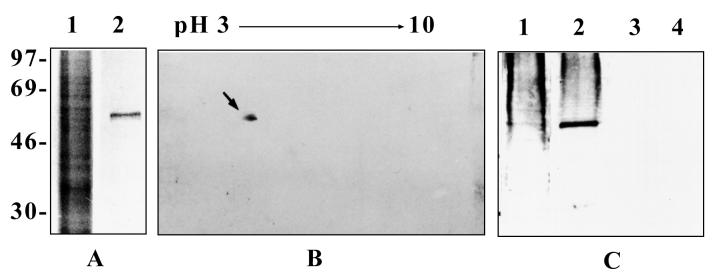
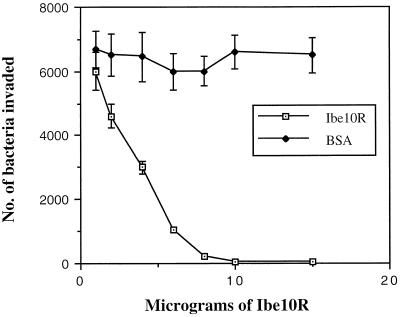
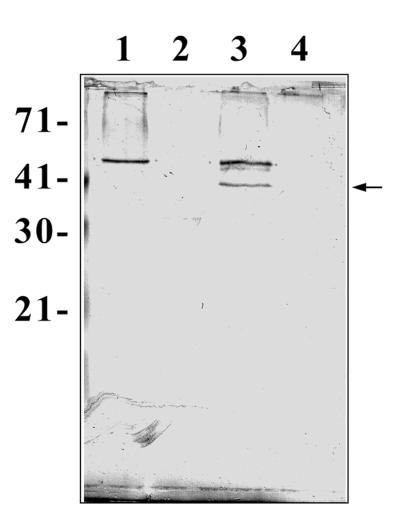
To identify the surface structures interacting with the novel E. coli protein Ibe10, BBMEC membrane proteins were subjected to rIbe10-Ni-Sepharose affinity chromatography. Since rIbe10 contains a histidine tag, the protein was coupled to Ni-Sepharose to immobilize the rIbe10 protein. We have previously identified two carbohydrate epitopes, NeuAcα2,3-galactose for S-fimbria-mediated binding and GlcNAcβ1,4GlcNAc for OmpA-mediated invasion, present on N-linked oligosaccharides of BMEC glycoproteins that interact with E. coli (13). To eliminate the interaction of these glycoproteins with an rIbe10-Ni-Sepharose column, the BBMEC membrane proteins were initially subjected to WGA-Sepharose chromatography several times to remove N-glycosylated proteins. The WGA-Sepharose runthrough proteins were then subjected to rIbe10-Ni-Sepharose chromatography, and the BBMEC membrane proteins bound to the affinity column were eluted with glycine-HCl buffer. The eluted fraction showed a major protein with an apparent molecular mass of 55 kDa (Fig. 1A). This fraction did not show any low-molecular-mass proteins, e.g., 9 kDa for rIbe10, suggesting that lowering the pH of the elution buffer did not cause any leaching of the bound rIbe10 protein. However, use of the rIbe10 affinity column more than two times resulted in leaching of the rIbe10 protein from the column. Thus, we used these columns only twice to isolate the rIbe10-bound molecules. Isoelectric focusing of rIbe10-bound BBMEC protein revealed a pI of 4 to 5, and further separation by two-dimensional gel electrophoresis showed a major band along with several minor faint bands (Fig. 1B). The 55-kDa BBMEC protein (designated Ibe10R) effectively inhibited the invasion of BBMEC by E. coli in a dose-dependent manner (Fig. 2), whereas the control protein BSA showed no such inhibition. Neither Ibe10R nor BSA affected bacterial viability under the experimental conditions employed. Approximately 50% inhibition was achieved with 4 to 5 μg of affinity-purified Ibe10R per well (500-μl volume). These results suggest that a 55-kDa BBMEC protein interacts E. coli invasion Ibe10 for E. coli invasion of BBMEC.
FIG. 1.
Polyacrylamide gel electrophoresis and Western blotting of Ibe10R. (A) rIbe10-Ni-Sepharose-bound BBMEC membrane proteins were separated by SDS–10% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue R-250. Lanes: 1, whole BBMEC membrane proteins (20 μg); and 2, rIbe10-Ni-Sepharose-bound proteins (Ibe10R, 2 μg). The values on the left are molecular sizes in kilodaltons. (B) Ibe10R was subjected to minicapillary isoelectric focusing using Bio-ampholyte (pH 3 to 10) for the first dimension and to SDS–10% polyacrylamide gel electrophoresis for the second dimension. The gel was transblotted to a nitrocellulose sheet and immunoblotted with anti-Ibe10R antibody. The arrow indicates the protein that reacted with the antibody. (C) Affinity-purified Ibe10R was separated on an SDS–10% polyacrylamide gel and immunoblotted with anti-Ibe10R antibody. Lanes: 1, whole BBMEC membrane proteins (10 μg); 2, rIbe10 affinity-purified Ibe10R (1.5 μg); 3, BSA (4 μg); 4, WGA-Sepharose-bound BBMEC membrane proteins (10 μg).
FIG. 2.
Inhibition of E. coli invasion of BBMEC by Ibe10R. E. coli was incubated with various concentrations of either Ibe10R or BSA for 1 h on ice before being added to confluent BBMEC monolayers for invasion assays as described in Materials and Methods. All values represent the means of triplicate determinations; error bars indicate standard deviations.
Antibody to Ibe10R inhibits E. coli invasion of BBMEC.
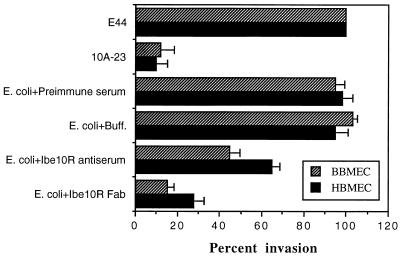
Polyclonal antiserum raised against Ibe10R reacted with a 55-kDa protein in both the whole BBMEC membrane proteins and the rIbe10 affinity-purified fraction (Fig. 1C, lanes 1 and 2). In contrast, the anti-Ibe10R antibody showed no reactivity with proteins in the WGA-bound fraction of BBMEC membrane proteins (Fig. 1C, lane 4). In addition, Ibe10R did not react with WGA on Western blots, suggesting that Ibe10R is not an N-glycosylated protein (data not shown). The anti-Ibe10R antibody was purified by protein A columns, and Fab fragments were prepared to eliminate nonspecific interaction of Fc the portion with BBMEC. As shown in Fig. 3, anti-Ibe10R serum (40 μl) and the Fab fragments (100 μg) significantly blocked invasion by E. coli, whereas neither preimmune serum nor the buffer control had any blocking effect (6,613 ± 915 CFU/well for E. coli with preimmune serum versus 3,850 ± 450 CFU/well for anti-Ibe10R serum and 945 ± 209 CFU/well for anti-Ibe10R Fab fragments [P < 0.001]). The anti-Ibe10R antibody Fab fragments also showed dose-dependent inhibition of invasion of both BBMEC and HBMEC by E. coli (data not shown). These results suggest that the anti-Ibe10R antibody interacts with BBMEC protein probably at the same binding site as Ibe10, thus preventing the interaction of E. coli with BBMEC for invasion. These data are also in agreement with those of the inhibition obtained with Ibe10R (Fig. 2).
FIG. 3.
Inhibition of E. coli invasion of BBMEC by anti-Ibe10R antibody. Confluent monolayers of BBMEC and HBMEC were incubated with either preimmune serum, PBS (Buff.), anti-Ibe10R serum, or anti-Ibe10R antibody Fab fragments for 1 h at 37°C before addition of bacteria. Invasion assays were carried out as described in Materials and Methods. E. coli E44 is a spontaneous rifampin-resistant mutant of RS218 expressing Ibe10. 10A-23 was derived from E44 by TnphoA mutagenesis and has an inactivated ibe10 locus. The invasion results were expressed as percentages of E44 invasion (which was taken as 100%). Each value represents the mean of at least four experiments done in triplicate, and the error bars indicate the standard deviations.
Anti-Ibe10R antibody recognizes BBMEC surface protein.
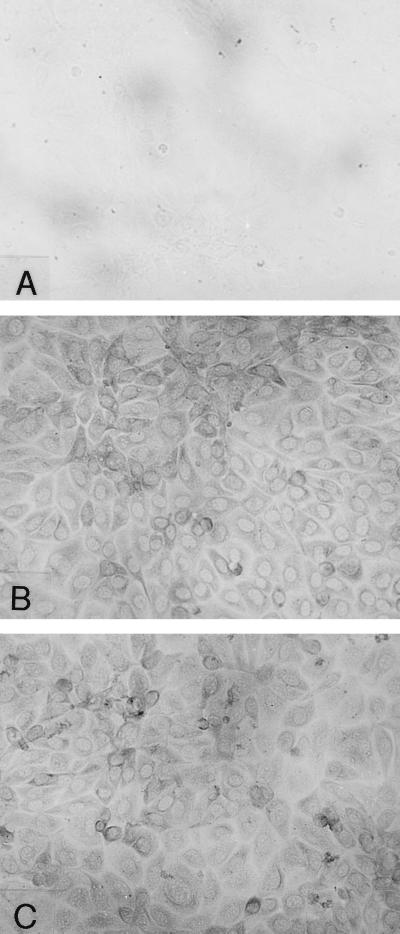
Since E. coli enters through the cell surface of BBMEC, not through the intercellular junctions (14), Ibe10R is most likely to be a cell surface molecule. To verify whether Ibe10 recognizes BMEC surface protein, BBMEC membrane proteins were biotinylated and subjected to either anti-Ibe10R antibody affinity or rIbe10-Ni-Sepharose chromatography. Both anti-Ibe10R–Sepharose (Fig. 4A, lane 2)- and rIbe10-Ni-Sepharose (lane 3)-bound fractions showed a major 55-kDa protein when probed with streptavidin-peroxidase. These proteins were also reactive with the anti-Ibe10R antibody (Fig. 4B, lanes 2 and 3), suggesting that the antibody recognized a BBMEC surface protein. The protein bands other than that of the 55-kDa protein were due to nonspecific interaction of the secondary antibody with biotinylated proteins. The presence of Ibe10R on the BBMEC surface was also established by immunocytochemical analysis (Fig. 5). The anti-Ibe10R antibody showed enhanced staining of BBMEC membrane proteins (panel B), while there was no discernible staining with the unrelated control antibody (panel A). The use of Triton X-100 to permeabilize BBMEC did not change the staining pattern (panel C), indicating that the reactivity was primarily on the surface of BBMEC. Interestingly, only a small number of cells showed stronger reactivity with the anti-Ibe10R antibody, suggesting that a subpopulation of BBMEC expresses Ibe10R.
FIG. 4.
Western blotting of biotinylated BBMEC membrane proteins. BBMEC surface proteins were biotinylated, and the membrane proteins were prepared as described in Materials and Methods. The proteins were passed over either an rIbe10-Ni-Sepharose column or an anti-Ibe10R antibody–Sepharose column. The bound proteins were eluted, dialyzed, and concentrated. The proteins were separated by SDS–10% polyacrylamide gel electrophoresis, transblotted to nitrocellulose sheets, and probed with either streptavidin-peroxidase (A) or anti-Ibe10R antibody (B). Lanes: 1, biotinylated BMEC membrane proteins (20 μg); 2, biotinylated membrane proteins bound to anti-Ibe10R antibody–Sepharose (4 μg); 3, biotinylated membrane proteins bound to an rIbe10-Ni-Sepharose column (4 μg). Molecular size markers are indicated on the left in kilodaltons.
FIG. 5.
Surface localization of Ibe10R by immunocytochemical analysis. Confluent BBMEC monolayers grown in eight-well chamber slides were fixed with 2% paraformaldehyde and processed for immunocytochemical analysis as described in Materials and Methods. Panels: A, control unrelated antibody; B, anti-Ibe10R antibody; C, anti-Ibe10R antibody in the presence of 0.1% Triton X-100. Original magnification, ×200.
rIbe10 binds to affinity-purified Ibe10R on a solid phase.
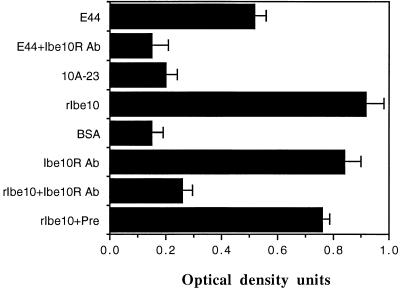
Although Ibe10R was isolated by using rIbe10 affinity chromatography, it is necessary to verify its reactivity with Ibe10 when present as a native molecule in E. coli. Thus, we examined the binding of biotinylated wild-type (E44) and Ibe10 mutant (10A-23) E. coli strains to an Ibe10R-coated solid surface by using an immunosorbent assay (14). We also used biotinylated rIbe10 and BSA as positive and negative controls, respectively. As expected, E. coli expressing Ibe10 bound significantly better than Ibe10 mutant E. coli (Fig. 6). This binding of E44 to Ibe10R was also significantly inhibited by the anti-Ibe10R antibody. Purified rIbe10 bound to Ibe10R very efficiently, similar to the anti-Ibe10R antibody and more efficiently than native E. coli, whereas the control protein BSA did not bind. Preincubation of Ibe10R-coated wells with unlabeled anti-Ibe10R antibody blocked the binding of rIbe10 to Ibe10R by more than 75%, suggesting that rIbe10 and the anti-Ibe10R antibody compete for the same binding site(s). These results are in agreement with that of the inhibition of E. coli invasion of BBMEC by Ibe10R, suggesting that the binding domains of both rIbe10 and Ibe10R were unaltered, even after isolation.
FIG. 6.
Binding of E. coli and rIbe10 to immobilized Ibe10R. rIbe10-Ni-Sepharose-bound Ibe10R was used to coat an Immunlon 3 enzyme-linked immunosorbent assay plate as described in Materials and Methods. Biotinylated E. coli, rIbe10, anti-Ibe10R Fab fragments, or BSA was incubated with immobilized Ibe10R in the plate. In some experiments, the Ibe10R was preincubated with unlabeled anti-Ibe10R Fab fragments or preimmune serum (Pre) prior to the addition of biotinylated bacteria or proteins. The bound bacteria or proteins were identified by probing with streptavidin-peroxidase. All values represent the means of triplicate determinations, and the error bars indicate the standard deviations. Ab, antibody.
Ibe10R represents a novel BBMEC protein.
The N-terminal amino acid sequence of purified Ibe10R was determined twice, i.e., after electroeluting the protein (i) from an SDS-polyacrylamide gel as previously described (14) and (ii) from a nitrocellulose membrane after two-dimensional gel electrophoresis. The N-terminal amino acid sequence of Ibe10R is NTHLSGIAFDDLG. A search of protein databases (PIR and Swiss-Prot) revealed that the N-terminal portion of Ibe10R has 75% homology to the serum albumin precursor of various species. However, serum albumin neither reacted to anti-Ibe10R antibody (Fig. 1C, lane 3) nor showed any ability to inhibit invasion by E. coli (Fig. 2). In contrast, Ibe10R showed significant inhibition of E. coli invasion of BBMEC in a dose-dependent manner. These results suggest that the Ibe10R protein is an albumin-like protein that interacts with Ibe10 invasin. To further confirm that Ibe10R differs from albumin, partial enzymatic cleavage using endoproteinase Glu-C from S. aureus V8 was performed. This enzyme cleaves the amide bond between glutamic acid and other amino acid towards the CO side. As shown in Fig. 7, the enzyme generated a large fragment with an approximate molecular mass of 35 to 38 kDa that was also reactive with the anti-Ibe10R antibody on a Western blot. SDS-polyacrylamide gel electrophoresis of the endoproteinase Glu-C-digested Ibe10R protein showed several other lower-molecular-mass fragments (approximately 6 kDa and less) that did not react with the anti-Ibe10R antibody (data not shown). Similarly, BSA was also subjected to endoproteinase digestion, and the resulting products did not react with the anti-Ibe10R antibody (Fig. 7, lane 4). The N-terminal amino acid sequence of the 35- to 38-kDa fragment is MRGSHKHGGAGIFTQ. This sequence did not show homology to any known proteins from a protein database. These results suggest that Ibe10R represents a novel albumin-like protein present on BBMEC and interacts with E. coli invasin Ibe10.
FIG. 7.
Enzymatic cleavage of Ibe10R with endoproteinase Glu-C. Ibe10R was partially digested with endoproteinase Glu-C overnight at room temperature, and the reaction was stopped with SDS buffer. The proteins were separated by SDS–12% polyacrylamide gel electrophoresis, transferred to a nitrocellulose sheet, and immunoblotted with anti-Ibe10R antibody. Lanes: 1, untreated Ibe10R; 2, endoproteinase enzyme alone; 3, Ibe10R treated with endoproteinase Glu-C; 4, BSA treated with endoproteinase Glu-C. Molecular size markers are on the left in kilodaltons, and the arrow indicates the Ibe10R fragment submitted for N-terminal amino acid sequencing.
Anti-Ibe10R antibody recognizes a smaller protein on HBMEC.
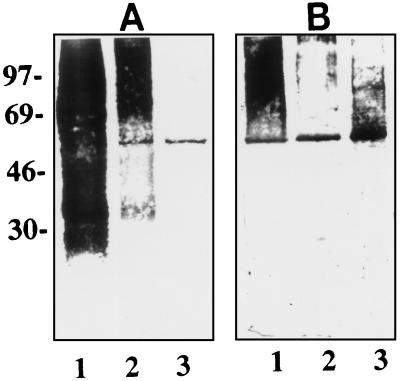
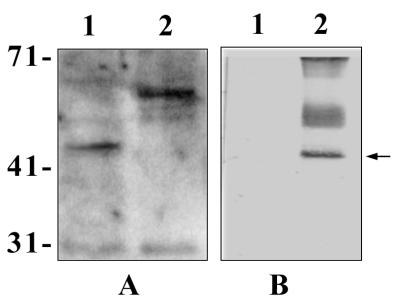
Since E. coli causes meningitis in humans, we examined the presence of Ibe10R in HBMEC. As shown in Fig. 3, the anti-Ibe10R antibody significantly blocked the invasion of HBMEC by E. coli, but its inhibition was 15 to 20% less than that of BBMEC invasion. Western blot analysis (Fig. 8A) revealed that the anti-Ibe10R antibody reacted to a smaller protein (45 kDa, lane 1) in the HBMEC membrane fraction than that observed with BBMEC (55 kDa, lane 2). In addition, the anti-Ibe10R antibody affinity-purified surface biotinylated HBMEC membrane fraction also yielded a 45-kDa protein (Fig. 8B, lane 2). The protein band with an approximate molecular mass of 30 kDa in Fig. 8A was also observed in immunoblots without the primary antibody, probably due to nonspecific interaction of the secondary antibody with BMEC proteins. The diffuse protein bands around 50 and 71 kDa in Fig. 8B was due to nonspecific interaction of biotinylated BMEC proteins with the Sepharose gel (data not shown). These findings suggest that a 45-kDa HBMEC protein that reacted with the anti-Ibe10R antibody is involved in the invasion of HBMEC by E. coli. However, we could not carry out invasion assays by using purified human Ibe10R because of the difficulty in obtaining sufficient quantities of proteins from HBMEC. Immunocytochemical analysis of HBMEC with the anti-Ibe10R antibody showed staining similar to that of BBMEC (data not shown), indicating that human Ibe10R is also a surface protein. The basis of the molecular size differences between bovine Ibe10R and human Ibe10R awaits further characterization.
FIG. 8.
Detection of HBMEC membrane proteins reactive to anti-Ibe10R antibody. (A) Anti-Ibe10R antibody immunoblotting of HBMEC membrane proteins (20 μg, lane 1) and BBMEC membrane proteins (20 μg, lane 2). The blot was processed with an ECL detection kit (Amersham). (B) Either unbiotinylated (lane 1) or biotinylated (lane 2) HBMEC membrane proteins (8 μg of each) were subjected to anti-Ibe10R antibody affinity chromatography as described in the legend to Fig. 4, and the bound proteins were eluted, separated on SDS–10% polyacrylamide gels, and probed with streptavidin-peroxidase. The values on the left are molecular sizes in kilodaltons.
Fluorescence-activated cell sorting of BBMEC using anti-Ibe10R antibody.
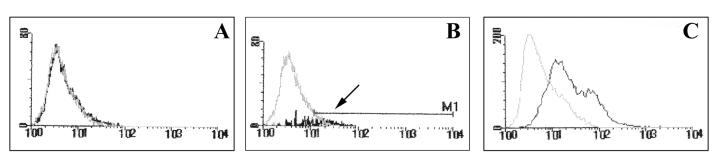
As described above, immunocytochemical analysis using the anti-Ibe10R antibody revealed that only a fraction of the BBMEC population showed enhanced positive staining compared to that obtained with an unrelated antibody, suggesting that the Ibe10R protein is expressed only by a subset of endothelial cells. To examine this possibility, BBMEC were sorted by using a FACS and the anti-Ibe10R antibody. As shown in Fig. 9, approximately 7 to 10% of the total BBMEC population was reactive to the anti-Ibe10R antibody (panel B). These positive cells were separated and grown to confluence before the invasion assays were performed. The separated Ibe10R-negative cells (10% of the bottom fraction) were also grown and used as a negative control. Ibe10R-positive cells showed 10-fold higher invasion by Ibe10+ E. coli (E44) than by Ibe10R-negative cells (Table 1). In contrast, an Ibe10-negative mutant, E. coli 10A-23, showed considerably less invasion of both Ibe10R-positive and -negative BMEC. In agreement with these results, immunocytochemical analysis using the anti-Ibe10R antibody showed considerably more positive anti-Ibe10R antibody-sorted cells than Ibe10R-negative cells (data not shown). The Ibe10R-positive cells were analyzed again by FACS, which revealed 50 to 60% positivity for the anti-Ibe10R antibody (Fig. 9C), in agreement with the immunocytochemical and invasion results. These results indicate that a certain population of BBMEC exhibits Ibe10R protein expression in culture and might be the target for E. coli invasion.
FIG. 9.
FACS sorting of BBMEC using anti-Ibe10 antibody. BBMEC were FACS sorted using anti-Ibe10R antibody as described in Materials and Methods. Panels: A, control antibody; B, anti-Ibe10R antibody; C, anti-Ibe10R staining of Ibe10R+ cells. The arrow indicates the Ibe10R+ cells that were separated and grown for use in invasion assays as well as in the experiment whose results are shown in panel C.
TABLE 1.
E. coli invasion of FACS-sorted Ibe10R+ and Ibe10R− BMEC
| BMEC | Invasion of BMEC (CFU/well) by strain:
|
|
|---|---|---|
| E44 (Ibe10R+) | 10A-23 (Ibe10−) | |
| Ibe10R+ | 10,125 ± 500a | 795 ± 250 |
| Ibe10R− | 1,250 ± 255 | 1,270 ± 355 |
P < 0.01 by two-tailed t test compared to Ibe10− cells.
DISCUSSION
We have previously shown that E. coli crossing of the blood-brain barrier involves interactions of bacterial ligands with specific receptors on BMEC. Specific surface molecules on both bacterial and host cells are likely to contribute to the tissue tropism of this disease. Several specific traits of E. coli contributing to invasion of BMEC have been identified, and one such determinant is the Ibe10 invasin. This Ibe10 invasin is commonly found in CSF isolates of E. coli but not in laboratory strains or other noninvasive clinical isolates (1, 5). We propose that study of the interaction of this invasin with host cell surface molecules would help unravel the mechanisms involved in the pathogenesis of this serious infection.
In the present study, we identified an approximately 55-kDa protein (Ibe10R) on the surface of BBMEC that interacts with E. coli invasin Ibe10. Although the N-terminal amino acid sequence of Ibe10R from BBMEC showed 75% sequence homology to the serum albumin precursor, the internal sequence did not show any sequence homology to other known proteins, suggesting that it is a novel albumin-like protein present on BBMEC. Ibe10R also differed from albumin in the lack of reactivity of the anti-Ibe10R antibody with albumin and the inability of albumin to affect the invasion of BBMEC by E. coli. In contrast, Ibe10R and the anti-Ibe10R antibody blocked the invasion very effectively. Onozuka et al. have reported an albumin-like 70-kDa protein associated with seizure activities (9). Ibe10R has 75% sequence homology with the N-terminal sequence and a pI (4 to 5) similar to that of the seizure activity-related protein but differs in molecular size. The complete characterization of this seizure activity-related protein is not available, precluding further comparison between Ibe10R and the seizure activity-related protein. Interestingly, a 45-kDa Ibe10R molecule on the HBMEC surface reacted with the anti-Ibe10R antibody raised against bovine Ibe10R. Moreover, the anti-Ibe10R antibody also showed an inhibitory effect on the E. coli invasion of HBMEC, suggesting that the bovine and human Ibe10R proteins have similar functional epitopes. The basis of the molecular size difference between the human and bovine Ibe10Rs and also the question of whether the human Ibe10R protein, like bovine Ibe10R, has any N-terminal sequence homology to the serum albumin precursor remain to be determined.
The ability of the anti-Ibe10R antibody to react with both BBMEC and HBMEC, as shown by immunocytochemical analysis, indicates that the epitope(s) of both the bovine and human Ibe10R proteins interacting with Ibe10 invasin is surface exposed. In addition, the unchanged pattern of the reactivity of the anti-Ibe10R antibody to the surface of both BMEC in the presence of a detergent suggests that Ibe10R is located primarily on the cell surface membrane. FACS analysis of BBMEC using the Ibe10R antibody yielded only 8 to 10% positive BBMEC, indicating that the immunoreactivity was confined to a particular fraction of the BBMEC population. This fraction of FACS-sorted cells showed greater susceptibility to E. coli invasion than did unsorted BBMEC, suggesting that enrichment of Ibe10R cells allows greater expression of the invasion phenotype.
Our previous studies have shown that invasion of BMEC by E. coli is decreased by 95% in the absence of either the OmpA or Ibe10 protein of E. coli, indicating the requirement of both proteins for efficient entry (5, 12). OmpA was shown to interact with GlcNAcβ1,4GlcNAc epitopes present on N-linked oligosaccharides of BMEC glycoproteins, and the disaccharide receptor analogues (e.g., chito-oligomers) blocked the E. coli invasion of BBMEC (13). In contrast, the novel E. coli adhesin Ibe10 interacts with a non-N-glycosylated novel Ibe10R protein that may differ from the OmpA binding protein that is necessary for E. coli invasion. Taken together, these results suggest that it is possible that OmpA interaction with GlcNAcβ1,4GlcNAc epitopes allows E. coli to have more-intimate contact with BMEC and allows the Ibe10 invasin to interact with Ibe10R on BMEC for invasion by E. coli. Alternatively, the presence of both OmpA and Ibe10 receptors on BMEC may enhance the efficiency of the invasion of BMEC by E. coli. Work is in progress to identify the genes encoding these receptor molecules from an HBMEC cDNA library and to determine their role in E. coli invasion of BMEC.
In summary, we demonstrated that E. coli invasin Ibe10 interacts with endothelial cells via a 55-kDa BBMEC Ibe10R and a 45-kDa HBMEC Ibe10R for E. coli invasion. Partial characterization by N-terminal and internal amino acid sequencing of Ibe10R from BBMEC reveals that it represents a novel albumin-like protein present on the surface of BBMEC whose enrichment enhances the invasion of BBMEC by E. coli.
ACKNOWLEDGMENTS
We thank M. F. Stins for providing brain endothelial cells and H. Shimada for immunocytochemistry photographs. We also thank Felix Burotto of the Division of Bone Marrow Transplantation for FACS analysis.
This work was supported by the American Heart Association Grants-in-Aid program affiliated with the greater Los Angeles area (N.V.P.) and Public Health Service grants R29-AI40567 (N.V.P.), R29-AI40635 (S.H.), and R01-NS26310 (K.S.K.).
REFERENCES
- 1.Bingen E, Bonacorsi S, Brahimi N, Denamur E, Elion J. Virulence patterns of Escherichia coli K1 strains associated with neonatal meningitis. J Clin Microbiol. 1997;35:2981–2982. doi: 10.1128/jcm.35.11.2981-2982.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Vries F P, Cole R, Dankert J, Frosch M, van Putten J P M. Neisseria meningitidis producing the Opc adhesin binds epithelial cell proteoglycan receptors. Mol Microbiol. 1998;27:1203–1212. doi: 10.1046/j.1365-2958.1998.00763.x. [DOI] [PubMed] [Google Scholar]
- 3.Finlay B B, Cossart P. Exploitation of mammalian host cell function by bacterial pathogens and the references therein. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 4.Gladstone I M, Ehrenkranz R A, Edberg S C, Baltimore R S. A ten-year review of neonatal sepsis and comparison with previous fifty-year experience. Pediatr Infect Dis J. 1990;9:819–825. doi: 10.1097/00006454-199011000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Huang S-H, Wass C, Fu Q, Prasadarao N V, Stins M, Kim K S. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect Immun. 1995;63:4470–4475. doi: 10.1128/iai.63.11.4470-4475.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korhonen T K, Valtonen M V, Parkkinen J, Väisänen-Rhen V, Finne J, Ørskov F, Ørskov I, Svenson S B, Mäkelä P H. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect Immun. 1985;48:486–491. doi: 10.1128/iai.48.2.486-491.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsudaira P. Limited N-terminal sequence analysis. Methods Enzymol. 1990;182:602. doi: 10.1016/0076-6879(90)82047-6. [DOI] [PubMed] [Google Scholar]
- 8.Moch T, Hoschutzky H, Hacker J, Kroncke K D, Jann K. Isolation and characterization of the α-sialyl-β-galactosyl specific adhesin from fimbriated E. coli. Proc Natl Acad Sci USA. 1987;84:3462–3466. doi: 10.1073/pnas.84.10.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onozuka M, Imai S, Isobe T, Yen C T, Watanabe K. Purification and characterization of a novel 70 kDa brain protein associated with seizure activities. Neurochem Res. 1995;20:901–905. doi: 10.1007/BF00970735. [DOI] [PubMed] [Google Scholar]
- 10.Parkkinen J, Korhonen T K, Pere A, Hacker J, Soiinilla S. Binding sites of the rat brain for E. coli S-fimbriae associated with neonatal meningitis. J Clin Investig. 1988;81:860–865. doi: 10.1172/JCI113395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasadarao N V, Wass C A, Hacker J, Jann K, Kim K S. Adhesion of S-fimbriated E. coli to brain glycolipids mediated by sfaA gene-encoded protein of S-fimbriae. J Biol Chem. 1993;268:10356–10363. [PubMed] [Google Scholar]
- 12.Prasadarao N V, Wass C A, Weiser J N, Stins M F, Huang S H, Kim K S. Outer membrane protein of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prasadarao N V, Wass C A, Kim K S. Endothelial cell GlcNAcβ1-4GlcNAc epitopes for outer membrane protein A enhance the traversal of Escherichia coli across the blood-brain barrier. Infect Immun. 1996;64:154–160. doi: 10.1128/iai.64.1.154-160.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasadarao N V, Wass C A, Kim K S. Identification and characterization of S fimbria-binding sialoglycoproteins on brain microvascular endothelial cells. Infect Immun. 1997;65:2852–2860. doi: 10.1128/iai.65.7.2852-2860.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stins M F, Prasadarao N V, Ibric L, Wass C A, Luckett P, Kim K S. Binding characteristics of S-fimbriated E. coli to isolated brain microvascular endothelial cells. Am J Pathol. 1994;145:1228–1236. [PMC free article] [PubMed] [Google Scholar]
- 16.Stins M F, Prasadarao N V, Gilles F, Kim K S. Transfection of human brain microvascular endothelial cells with SV40-large T antigen: development of an immortalized cell line with blood-brain barrier characteristics. Mol Biol Cell. 1994;5:245. [Google Scholar]
- 17.Stins M F, Gilles F, Kim K S. Selective expression of adhesion molecules on human brain microvascular endothelial cells. J Neuroimmunol. 1997;76:81–90. doi: 10.1016/s0165-5728(97)00036-2. [DOI] [PubMed] [Google Scholar]
- 18.Unhanand M, Mustafa M M, McCracken G H, Nelsen J D. Gram-negative enteric bacillary meningitis: a twenty year experience. J Pediatr. 1993;122:15–21. doi: 10.1016/s0022-3476(05)83480-8. [DOI] [PubMed] [Google Scholar]
- 19.Virji M, Makepeace K, Ferguson D J P, Watt S M. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol Microbiol. 1996;22:941–950. doi: 10.1046/j.1365-2958.1996.01551.x. [DOI] [PubMed] [Google Scholar]
- 20.Watari M, Funato S, Sasakawa C. Interaction of Ipa proteins of Shigella flexneri with α5β integrin promotes entry of the bacteria into mammalian cells. J Exp Med. 1996;183:991–999. doi: 10.1084/jem.183.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]