Abstract
Artificial intelligence (AI) has been a very active research topic over the last years and thoracic imaging has particularly benefited from the development of AI and in particular deep learning. We have now entered a phase of adopting AI into clinical practice. The objective of this article was to review the current applications and perspectives of AI in thoracic oncology. For pulmonary nodule detection, computer-aided detection (CADe) tools have been commercially available since the early 2000s. The more recent rise of deep learning and the availability of large annotated lung nodule datasets have allowed the development of new CADe tools with fewer false-positive results per examination. Classical machine learning and deep-learning methods were also used for pulmonary nodule segmentation allowing nodule volumetry and pulmonary nodule characterization. For pulmonary nodule characterization, radiomics and deep-learning approaches were used. Data from the National Lung Cancer Screening Trial (NLST) allowed the development of several computer-aided diagnostic (CADx) tools for diagnosing lung cancer on chest computed tomography. Finally, AI has been used as a means to perform virtual biopsies and to predict response to treatment or survival. Thus, many detection, characterization and stratification tools have been proposed, some of which are commercially available.
Keywords: Artificial intelligence, Deep learning, Diagnostic imaging, Multidetector computed tomography, Lung neoplasms
Introduction
The implementation of artificial intelligence (AI)-based tools in clinical practice is the next step in the digitization of medical imaging. With the breakthrough of deep learning, AI has become particularly popular in recent years. After the initial hype, during which the peak of inflated expectations caused some to fear the replacement of radiologists by artificial intelligence, we have now entered an adoption phase. Thoracic imaging has particularly benefited from the development of AI and deep learning, and is the most frequently targeted area for AI software [1, 2]. Multiple AI-based tools have been developed, notably for lung segmentation, nodule detection and characterization, but also for quantification, characterization and follow-up of interstitial lung disease, bronchial disease and COVID-19 [3–7]. The objective of this article was to review the current applications and perspectives of AI in thoracic oncology.
From radiomics to deep learning
Although using very different algorithms, the invention of radiomics preceded the use of deep learning in medical imaging by only a few years. The term “radiomics” was corresponds to a field of medical imaging that aims to extract features invisible to the human eye from medical images; for tasks such as characterization or prediction [8]. Several feature types can be extracted, the most common being histogram characteristics, texture parameters and shape parameters [9]. The number of extracted features is variable but can reach up to more than 2000, leading to high dimensionality [10]. Therefore, even if most of them can be correlated with the target, an intermediate step of feature selection is often conducted to keep only a small number of relevant features and this is usually based on machine learning algorithms for dimensionality reduction. The feature selection step is performed in conjunction with or separately from the learning step that combines the selected features to create a radiomic signature that correlates with the clinical outcome. Machine learning algorithms used in radiomics are referred to as classical machine learning algorithms as opposed to deep learning. In contrast to radiomics, deep-learning features are not predefined and are learned together with the clinical problem. The most common types of deep-learning algorithms for image analysis are convolutional neural networks (CNNs). CNNs correspond to deep neural networks based on a sequence of convolutional operations. Deep learning with CNNs gained momentum in 2012 when Krizhevsky et al. won the ImageNet Large-Scale Visual Recognition Challenge by a large margin, with a CNN called AlexNet [11]. For medical image analysis, the application of deep learning began and grew rapidly in 2015 and 2016 [12]. Deep learning has been applied in ultrasound, X-rays, computed tomography (CT) and magnetic resonance imaging [13–18]. Deep learning is now considered as the state-of-the-art method in medical image analysis. Pros and cons of radiomics and deep learning are summarized in Table 1. It has been shown to outperform traditional machine learning methods and radiomics for many applications, but it requires more data for training, and this can be a limiting factor. Computer-aided diagnosis (CAD) tools consist of software that uses algorithms derived from AI to provide indicators and assist radiologists. CAD systems are subdivided in computer-aided detection (CADe) tools and computer-aided diagnosis (CADx) tools.
Table 1.
Pros and cons of radiomics and deep learning
| Pros | Cons |
|---|---|
| Radiomics | |
| Explainable | Generalizability issues |
| Require less data for training | Radiomics signature to assess the same problem are often based on different parameters |
| Deep learning | |
| Often achieve better results | Black box |
| Better generalizability | Require more data for training |
Lung nodule detection
By definition, lung nodules are focal opacities, well or poorly defined, measuring 3–30 mm in diameter [19]. The majority of pulmonary nodules are solid, co-existing with a subsolid category; which includes pure ground glass and part-solid nodules. The mean prevalence of lung nodules varies between incidentally detected series and screening trials, ranging from 13% (range: 2–24%) to 33% (17–53%), respectively [20]. In the National Lung Screening Trial (NLST), 27.3% of 26,309 participants who underwent low-dose chest CT had at least one non-calcified lung nodule measuring 4 mm in diameter or larger and which was considered screen positive [21]. In the Dutch–Belgian lung-cancer screening trial [Nederlands–Leuvens Longkanker Screenings Onderzoek (NELSON)], 19.7% of the 6,309 individuals screened had at least one non-calcified pulmonary nodule measuring 5–10 mm on their first screening chest CT [22]. Assisting in the detection of lung nodules is an important task in thoracic imaging, particularly in the context of mass screening.
The first CADe tools for the detection of pulmonary nodules on chest CT were developed in the 2000s (Fig. 1). These tools were based on classical machine learning methods and already had excellent sensitivity. In an ancillary study from the NELSON trial, Zhao et al. found that a commercially available CADe tool had a sensitivity of 96.7%, whereas double reading had only 78.1% sensitivity [23]. Nonetheless, these CADe tools are known to encounter a high number of false positives and insufficient performances for the detection of subsolid nodules (Fig. 2). Evaluating one of these early CADe tools, Bae et al. reported a sensitivity of 95.6% for the detection of pulmonary nodules nonetheless with a number of false positives per examination of 6.9 and 4.0 for detection sensitivity thresholds of 3 and 5 mm, respectively [24]. Regarding the detection of subsolid nodules, Benzakoun et al. showed that only 50% were detected with a sensitivity adjusted for 3-mm nodule detection, at the average cost of 17 CAD marks per CT [25].
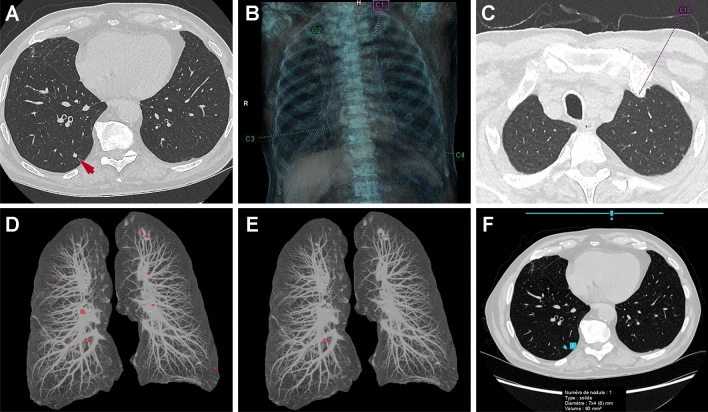
Fig. 1.
Detection of a pulmonary solid nodule. A Axial chest CT image shows a 7 × 4 mm solid nodule (arrow) in the right lower lobe. B, C A first computer-aided detection (CADe) tool based on classical machine learning method correctly detects the nodule at the cost of four false positives (an example of a false positive is pointed in C). D, E A second CADe tool based on classical machine learning method also correctly detects the nodule at the cost of numerous false positives when the sensitivity is adjusted for 3-mm nodule detection (D) whereas there are no false positives when the threshold is set at 6-mm (E). F A deep learning-based CADe tool also correctly identifies the nodule with no false positives
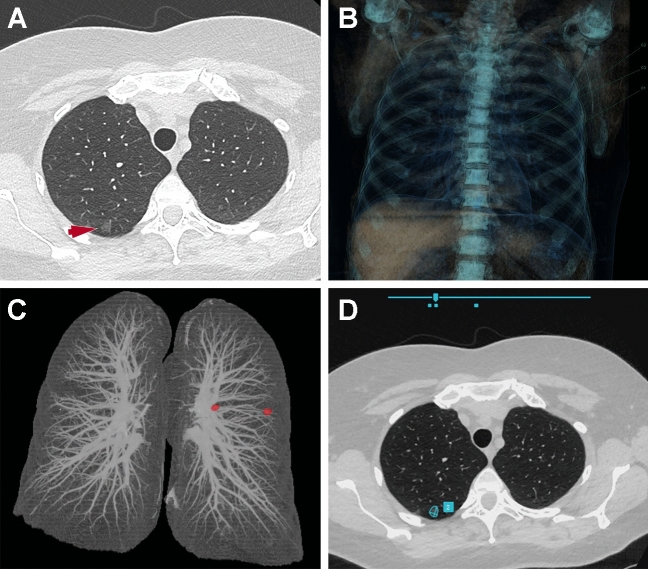
Fig. 2.
Detection of a pulmonary ground-glass nodule. A Axial chest CT image shows a 10 × 8 mm ground-glass nodule (arrow) in the right upper lobe. The ground-glass nodule is not detected by two different computer-aided detection tools based on classical machine learning methods (B and C) but is correctly detected by the one based on deep learning (D)
In recent years, several CADe tools based on deep learning have been developed. Either two-dimensional or three-dimensional CNN were used. These tools have been developed either on proprietary data or on publicly available databases. Several public databases have been released including the NLST dataset and the Lung Image Database Consortium Image collection (LIDC) [26]. The main advantages of public databases are to allow a larger number of data scientists to work on the subject and to allow a performance comparison between the proposed algorithms. Of the 1018 chest CTs from the LIDC dataset, 888 CTs had 1,186 nodule annotations that were used by the Lung Nodule Analysis 2016 (LUNA16) challenge in which participants were invited to develop a CADe tool to automatically detect pulmonary nodules on CT [27]. The proposed algorithms achieved a sensitivity ranging between 79.3 and 98.3% at the respective costs of 1 and 8 false positives per CT. In a recent study, Masood et al. reported a deep learning-based CADe tool which was also developed from the LIDC dataset; reaching 98.1% sensitivity for nodules ≥ 3 mm, with 2.21 false positives per CT [28]. Other challenges for lung nodule detection have been organized, for example: the data challenge held at the annual congress of the French Society of Radiology in 2019 (Journées Francophones de Radiologie 2019). This challenge included 1237 chest CT examinations and participants were asked to develop AI models that detected lung nodules; calculating their volume after segmentation and classifying them as either: probably benign (volume < 100 mm3) or probably malignant (volume ≥ 100 mm3) nodules [29, 30].
Several deep learning-based tools are already commercially available. These tools are expected to increase the adoption of CADe tools for lung nodule detection. Their sensitivity is generally greater than that of radiologists, but even if the number of false-positive per CT has decreased compared with previous CADe tools it remains a limiting factor. Li et al. evaluated a commercially available CADe tool on 346 patients in a lung-cancer screening program and found a greater detection rate compared to that of double reading (86.2% vs. 79.2%), however, the number of false-positives per CT examination was also significantly greater (1.53 vs. 0.13; P < 0.001) [31]. For lung-cancer screening, these deep learning-based CADe systems could be used as a second, concurrent or first reader, or even as a stand-alone solution if their performance is confirmed as superior to that of expert radiologists [32]. It is important to note that dose reduction and iterative reconstruction parameters impact AI software in its ability to detect lung nodules. In a study using chest phantoms, Schwyzer et al. showed that the detection of ground-glass nodules decreased significantly when the radiation dose was decreased and it was also influenced by the level of iterative reconstruction [33].
Although chest radiography is not effective in lung-cancer screening, assisting in the detection of lung nodules on radiographs remains a task of major importance in daily practice. Several CADe tools have been developed for detecting major chest X-rays abnormalities, of those which include pulmonary nodules (Fig. 3). Yoo et al. evaluated a commercially available deep learning-based CADe on three chest X-rays from the NLST trial [34]. The sensitivity and specificity for detecting pulmonary nodules were not statistically different from those of radiologists. (86.2% vs. 87.7%; P = 0.80 and 85.0% vs. 86.7%; P = 0.42). However, when they only considered malignant nodules from the first round of the NLST, AI had a similar sensitivity to that of radiologists despite a lower specificity (94.1% vs. 94.1%; P > 0.99 and 83.3% vs. 91.3%; P < 0.001 respectively). Using another commercially available deep learning-based CADe tool for lung nodule detection on chest X-ray, Sim et al. showed that the mean sensitivity of radiologists improved significantly from 65.1 to 70.3% (P < 0.001), and the number of false-positive results per radiograph decreased significantly from 0.20 to 0.18 (P < 0.001) when radiologists had access to AI results [35]. In their study, sensitivity of the CADe tool alone was 67.3% for 0.20 false-positive per radiograph [35].
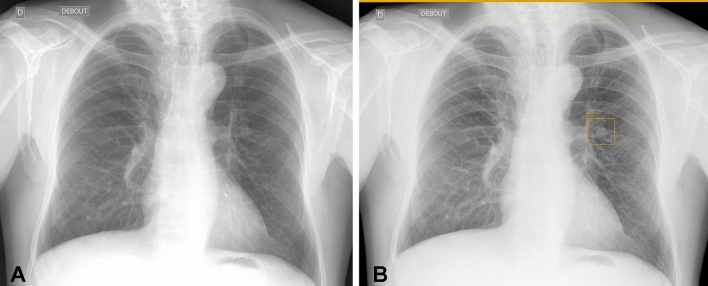
Fig. 3.
Lung nodule detection on chest X-ray. Lung nodule seen on chest X-ray (A) is correctly detected by the deep learning-based computer-aided detection tool (box with orange borders in B)
Lung nodule segmentation and characterization
In daily practice, the strategy for characterizing pulmonary nodules is based primarily on their size, morphology and evolution over time. For incidentally detected nodules, the most commonly used guidelines are those of the Fleischner Society [36] and British Thoracic Society (BTS) [20]. The Fleischner guidelines are based on the mean diameter and type (solid or subsolid) of the nodule. By contrast, the BTS guidelines recommend volumetry rather than 2D- measurements of the nodule and include clinical information through the Brock model [20]. The Brock model, which was developed using data from the Pan-Canadian Early Lung Cancer Detection Study, is designed to assess the likelihood of malignancy of pulmonary nodules [37]. It is based on demographic characteristics, the presence of emphysema, as well as the size, shape and location of the nodule [37].
For lung-cancer screening, the American College of Radiology established the Lung-RADS classification (Lung CT Screening Reporting and Data System) [38], which is based on the diameter measurement similar to that used in the NLST. Whereas in Europe, the NELSON trial used another approach based on volumetry and the calculation of volume doubling times [22]. In line with the European position statement guidelines [39], three French scientific societies recommended the use of volumetry when evaluating lung nodules [40].
The first major application of AI tools in the characterization of pulmonary nodules is therefore the segmentation of lung nodules. Accurate segmentation of pulmonary nodules is essential as it allows automated volume calculation. These measurements are used to decide the management of the nodule according to the guidelines previously mentioned and also to calculate the volume doubling time when a subsequent follow-up CT examination is performed. Some AI tools dedicated to lung-cancer screening can not only automatically detect and measure lung nodules but also suggest Lung-RADS categorization. This is done by combining the mean nodule diameter measurement and the nodule type (solid or not) (Fig. 4). Lancaster et al. recently evaluated this type of deep-learning tool in 283 participants from the Moscow lung-cancer screening program who had at least one solid lung nodule [41]. CT examinations were analyzed independently by five experienced thoracic radiologists. They were asked to determine whether each participant’s largest nodule measured < 100 mm3 (nodule classified as negative) or ≥ 100 mm3 (nodule classified as indeterminate/positive). A consensus among radiologists was reached to determine the ground truth. The threshold of 100 mm3 for a solid nodule was chosen in accordance with the NELSON-Plus protocol for pulmonary nodules detected by low-dose CT [42]. The authors found that the AI tool had fewer negative misclassifications (i.e., nodules ≥ 100 mm3 misclassified as nodules < 100 mm3) than four out of five experienced radiologists, however it resulted in more positive misclassifications (53 vs. 6 to 25). In another study, Jacobs et al. showed that using a dedicated viewer with automatic segmentation instead of a PACS-like viewer increased the interobserver agreement (Fleiss kappa = 0.58 vs. 0.66) and significantly decreased reading time (86 vs. 160 s; P < 0.001) [43]. Several morphology-based segmentation methods have been evaluated for lung nodule segmentation such as region growing and graph cut, however, deep-learning methods can also be used [44, 45]. Rocha et al. evaluated three different approaches for lung nodule segmentation: a conventional approach using the Sliding Band Filter and two deep-learning approaches, one using the very popular U-Net architecture and the other using the SegU-Net architecture [46]. The models were trained on 2653 nodules from the LIDC dataset. Deep learning methods clearly outperformed the conventional approach with Dice similarity coefficients of 0.830 for the U-net and 0.823 for the SegU-Net compared to 0.663 for the Sliding Band Filter [46]. Wang et al. proposed a different CNN architecture for the same task [47]. Their model was also trained on the LIDC dataset and they obtained performances in the same ranges as Rocha et al. In their study, the Dice similarity coefficient ranged from 0.799 to 0.823 in the test dataset and external test datasets [47].
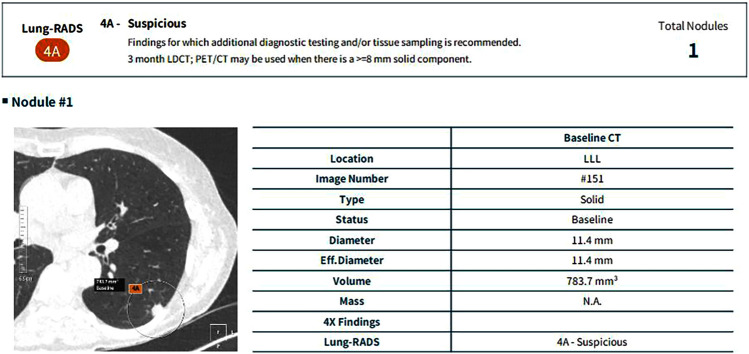
Fig. 4.
Automated Lung-RADS classification. Automated lung nodule detection, volumetry and Lung-RADS classification using a deep-learning-based computer-aided detection tool in a patient who underwent an ultra-low-dose chest CT examination as part of a lung-cancer screening program
Another potential application of AI in the management of pulmonary nodules is the prediction of risk of malignancy based on the study of the nodule’s global characteristics rather than just its size. Several studies have been conducted to predict the risk of lung nodule malignancy using either a radiomics or a deep-learning approach [48–51]. Hawkins et al. used a radiomic approach on a subset of NLST participants who had lung nodules (198 participants, 170 of whom had lung cancers) to predict which nodules would later become cancers [48]. Of the 219 three-dimensional radiomic features they extracted, 23 stable features were selected to build a model capable of predicting which nodules would become cancerous at one and two years with accuracies of 80% (area under the curve [AUC] = 0.83) and 79% (AUC = 0.75), respectively [48]. In a retrospective study of 290 patients from two institutions, Beig et al. found that adding radiomic features of the perinodular space to radiomic features of the intranodular space improved the AUC from 0.75 to 0.80 for distinguishing lung adenocarcinomas from granulomas on non-contrast chest CT [49]. In their study, a total of 1776 features were extracted from each solitary pulmonary nodule, of which 12 were retained for each model. One of the concerns regarding the use of the radiomic approach is the lack of consistency between studies, notably for the characterization of pulmonary nodules [49]. Indeed, radiomic features retained by the machine learning models vary from one study to another. Emaminejad et al. evaluated the effects of several CT acquisitions and reconstruction parameters on radiomic features [52]. In a cohort of 89 lung-cancer screening participants with a solid lung nodule, they found that the 226 radiomic features included, lacked interconditional reproducibility [52]. Furthermore, slice thickness, dose and reconstruction kernel had a major impact on feature reproducibility. In a study of 99 nodules in 170 patients who underwent full-dose and ultra-low-dose unenhanced chest CT, Autrusseau et al. evaluated the consistency of a radiomic model for predicting the relative risk of lung nodule malignancy [53]. They used a research version of a CADx tool that had been trained on the NLST dataset and only found a good but not perfect agreement (kappa = 0.60) for classifying nodules as “benign,” “indeterminate,” or “malignant” on full-dose and ultra-low-dose chest CT. These studies show that the impact of dose is an important issue for the generalizability of radiomic-based CADe tools in the setting of lung-cancer screening. In the NLST, low-dose chest CT examinations were performed at an effective dose of 1.5 mSv, but ultra-low-dose protocols with sub-mSv radiation doses are increasingly being used [54], especially for lung-cancer screening. In addition, further dose reduction is expected in the coming years with the advent of photon-counting CT systems [55]. As shown by Emaminejad et al. reconstruction parameters also significantly impact the results of radiomic-based approaches [52]. The impact of the recently introduced deep-learning image reconstruction algorithms on radiomics models has not yet been evaluated. These new reconstruction algorithms are known to reduce image noise and improve lesion detectability [56]. Deep learning models are also influenced by image parameters. Hoang-Thi et al. reported that the choice of the reconstruction kernel had an impact on the performance of deep-learning models trained to segment diffuse lung diseases [57]. They showed that deep-learning models perform better on images reconstructed with the same kernel which was used to reconstruct the images in the training dataset [52]. They also showed that combining the mediastinal and pulmonary kernels in the training dataset improves the performance of the models.
Several studies have evaluated the use of deep-learning methods to predict the risk of lung nodule malignancy. For instance, Massion et al. used the NLST dataset (14,761 benign and 932 malignant nodules) to develop their model, which they then tested on 2 independent external datasets [50]. They obtained AUCs of 0.835 and 0.919 to predict malignancy on external datasets, compared with 0.781 and 0.819, respectively, for the Mayo model, which is a commonly used clinical risk model for incidental nodules. These results are better than those previously reported using a radiomics approach. According to the authors, a deep-learning model score above 65% indicates the need for tissue sampling [50]. However, in the two independent external datasets on which they tested their model, this threshold would have resulted in an interventional procedure being performed in up to 21% of benign nodules. Ardila et al. also developed a deep-learning model on the NLST dataset, but instead of focusing on the characterization of pre-identified nodules, they aimed to build an end-to-end approach, performing both localization and lung-cancer risk categorization [51]. Their model was tested on a subset of 6716 individuals (86 positive for cancer) from the NLST dataset and on an independent dataset of 1139 individuals (27 positive for cancer), on which it obtained AUCs of 0.944 and 0.955, respectively [51]. Their model outperformed radiologists with absolute reductions of 11% in false positives and 5% in false negatives [51]. Their model was also trained to input the prior CT examination in addition to the current one. In this setting, the model performance was on-par with that of the radiologists [51]. Despite impressive results, these two studies show that current CADx systems do not yet outperform the clinical strategy which is based on close follow-up of suspected pulmonary nodules.
Lung cancer characterization and stratification
Several studies have evaluated whether AI could go beyond cancer detection and attempt to predict histological type or even the probability of treatment response or prognosis.
Virtual biopsy using AI may be of interest in frail patients for whom percutaneous biopsy is considered risky, especially before treatment with stereotactic radiotherapy, or to select the best surgical time for subsolid nodules. Indeed, subsolid nodules may correspond either to atypical adenomatous hyperplasia and adenocarcinoma in situ, both considered pre-invasive lesions, or to minimally invasive adenocarcinoma and invasive adenocarcinoma. Pre-invasive lesions and minimally invasive adenocarcinomas can be followed-up clinically whereas invasive adenocarcinoma requires surgery. Although they all present as subsolid nodules, certain morphological characteristics are known to differ between these lesions [58]. Fan et al. developed a radiomic signature in 160 pathologically confirmed lung adenocarcinomas to differentiate invasive adenocarcinomas from non-invasive lesions manifesting as ground-glass nodules. Out of 355 radiomic features extracted, two were retained to build their radiomic signature, which had an AUC ranging from 0.917 to 0.971 in four independent datasets [59]. Wang et al. evaluated a deep-learning approach to address the same question [60]. They used a dataset of 886 ground-glass nodules and trained three different deep-learning algorithms. They obtained lower AUCs ranging from 0.827 to 0.941.
AI has also been used to non-invasively search for certain mutations that can impact therapeutic choices. For instance, epidermal growth factor receptor (EGFR) mutation is strongly predictive of therapy response to anti-EGFR tyrosine kinase inhibitor therapy and programmed death-ligand 1 (PD-L1) expression on tumors is a predictive biomarker for sensitivity to immunotherapy. EGFR-positive adenocarcinomas are more frequently found in women without centrilobular emphysema and are more frequently of smaller size with spiculated margins and with no associated lymphadenopathy [61]. Jia et al. developed and tested a radiomic signature to predict EGFR mutations in a cohort of 503 patients with lung adenocarcinoma who had received surgery [62]. They extracted 440 features of which 94 were retained and combined using a random forest classifier. They obtained an AUC of 0.802 to predict the presence of the mutation and the AUC was further improved to 0.828 by adding sex and smoking history, although the significance of this improvement was not assessed. For the same task, Wang et al. evaluated a deep-learning approach in a larger multicentric study [63]. They used data from 18,232 patients with lung cancer with EGFR gene sequencing taken from nine cohorts in China and the USA including a prospective cohort of 891 patients. Their model to predict EGFR mutation, yielded an AUC ranging from 0.748 to 0.813 in the different cohorts (0.756 in the prospective cohort) [60]. When combing imaging and clinical data, the AUC raised to the range of 0.763–0.834 (0.788 in the prospective cohort) [63]. To predict immunotherapy response, several approaches have been tested. Sun et al. used a radiomic model to predict tumor infiltration by CD8 cells in patients treated by anti PD-1 or anti PD-L1 immunotherapy [64]. They used data from 135 patients with five types of cancer including lung cancer. Eight features including five radiomic features were retained to compose the radiomic signature which had an AUC of 0.67 predicting the abundance of CD8 cells [61]. This radiomic signature was associated with improved overall survival. By contrast, Trebeschi et al. constructed a radiomic signature to directly predict immunotherapy response in patients with advanced melanoma and non-small cell lung cancer [65]. They obtained an AUC of 0.66 to predict individual response for each lesion within the test dataset. By combining predictions made on individual lesions, they obtained significant performances in the prediction of overall survival for both tumor types (AUC = 0.76) [65].
Finally, in a stratification study, Hosny et al. used a deep-learning approach for mortality risk stratification in patients with non-small cell lung cancer [66]. They used data from seven independent datasets across five institutions, including publicly available datasets totaling 1194 patients. They trained three-dimensional CNN to predict 2-year overall survival from pre-treatment chest CT examinations in patients treated with radiotherapy or surgery. They obtained an AUC of 0.70 for radiotherapy and 0.71 for surgery [66]. These models outperformed random forest models based on clinical information (age, sex, and TNM stage) which had an AUC of 0.55 and 0.58 for radiotherapy and surgery, respectively. By exploring the features captured by the CNN, they identified that the region that contributed most to the predictions was the interface between the tumor and stroma (parenchyma or pleura).
All these results show that images contain information that can help the characterization and stratification of lung cancer. However, the results of these CADx tools remain currently insufficient to replace biopsy in eligible patients or to prevent patients from receiving potentially effective treatment.
Conclusion
Lung cancer imaging is a very active research topic in AI. Many detection, characterization and stratification tools have been proposed and some of which are commercially available. The prospect of massive lung-cancer screening programs represents a major challenge in terms of patient volume and medical resources. AI-based tools will likely play an important role in limiting the medico-economic impact and allowing the largest possible population to benefit from efficient screening (i.e., with a minimal number of false negatives and false positives). To date, commercially available tools remain primarily CADe-type detection software.
Author contributions
Conceptualization: All authors. Methodology: GC, PS. Formal analysis and investigation: All authors. Writing—original draft preparation: GC, CM-M, PS. Writing—review and editing: All authors. Funding acquisition: N.A. Resources: GC. Supervision: PS.
Funding
This work did not receive specific funding.
Declarations
Conflict of interest
The authors have no conflicts of interest to disclose in relation with this article.
Ethical approval
Not applicable for review article.
Informed consent
Not applicable for review article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Guillaume Chassagnon, Email: guillaume.chassagnon@aphp.fr.
Constance De Margerie-Mellon, Email: constance.de-margerie@aphp.fr.
Maria Vakalopoulou, Email: maria.vakalopoulou@ecp.fr.
Rafael Marini, Email: marinirfsilva@gmail.com.
Trieu-Nghi Hoang-Thi, Email: htrieunghi@yahoo.fr.
Marie-Pierre Revel, Email: marie-pierre.revel@aphp.fr.
Philippe Soyer, Email: philippe.soyer@aphp.fr.
References
- 1.Tadavarthi Y, Vey B, Krupinski E, Prater A, Gichoya J, Safdar N, et al. The state of radiology AI: considerations for purchase decisions and current market offerings. Radiol Artif Intell. 2020;2:e200004. doi: 10.1148/ryai.2020200004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chassagnon G, Vakalopoulou M, Paragios N, Revel M-P. Artificial intelligence applications for thoracic imaging. Eur J Radiol. 2020;123:108774. doi: 10.1016/j.ejrad.2019.108774. [DOI] [PubMed] [Google Scholar]
- 3.Chassagnon G, Vakalopoulou M, Régent A, Zacharaki EI, Aviram G, Martin C, et al. Deep learning-based approach for automated assessment of interstitial lung disease in systemic sclerosis on CT images. Radiol Artif Intell. 2020;2:e190006. doi: 10.1148/ryai.2020190006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chassagnon G, Vakalopoulou M, Régent A, Sahasrabudhe M, Marini R, Hoang-Thi T-N, et al. Elastic registration-driven deep learning for longitudinal assessment of systemic sclerosis interstitial lung disease at CT. Radiology. 2021;298:189–198. doi: 10.1148/radiol.2020200319. [DOI] [PubMed] [Google Scholar]
- 5.Chassagnon G, Vakalopoulou M, Battistella E, Christodoulidis S, Hoang-Thi T-N, Dangeard S, et al. AI-driven quantification, staging and outcome prediction of COVID-19 pneumonia. Med Image Anal. 2021;67:101860. doi: 10.1016/j.media.2020.101860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campredon A, Battistella E, Martin C, Durieu I, Mely L, Marguet C, et al. Using chest CT scan and unsupervised machine learning for predicting and evaluating response to lumacaftor-ivacaftor in people with cystic fibrosis. Eur Respir J. 2021 doi: 10.1183/13993003.01344-2021. [DOI] [PubMed] [Google Scholar]
- 7.Chassagnon G, Zacharaki EI, Bommart S, Burgel P-R, Chiron R, Dangeard S, et al. Quantification of cystic fibrosis lung disease with radiomics-based CT scores. Radiol Cardiothorac Imaging. 2020;2:e200022. doi: 10.1148/ryct.2020200022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chassagnon G, Vakalopolou M, Paragios N, Revel M-P. Deep learning: definition and perspectives for thoracic imaging. Eur Radiol. 2020;30:2021–2030. doi: 10.1007/s00330-019-06564-3. [DOI] [PubMed] [Google Scholar]
- 9.Nakaura T, Higaki T, Awai K, Ikeda O, Yamashita Y. A primer for understanding radiology articles about machine learning and deep learning. Diagn Interv Imaging. 2020;101:765–770. doi: 10.1016/j.diii.2020.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Abunahel BM, Pontre B, Kumar H, Petrov MS. Pancreas image mining: a systematic review of radiomics. Eur Radiol. 2021;31:3447–3467. doi: 10.1007/s00330-020-07376-6. [DOI] [PubMed] [Google Scholar]
- 11.Shi Z. Learning. In: Shi Z, editor. Intelligence science. New York: Elsevier; 2021. pp. 267–330. [Google Scholar]
- 12.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 13.Blanc-Durand P, Schiratti J-B, Schutte K, Jehanno P, Herent P, Pigneur F, et al. Abdominal musculature segmentation and surface prediction from CT using deep learning for sarcopenia assessment. Diagn Interv Imaging. 2020;101:789–794. doi: 10.1016/j.diii.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 14.Gao X, Wang X. Performance of deep learning for differentiating pancreatic diseases on contrast-enhanced magnetic resonance imaging: a preliminary study. Diagn Interv Imaging. 2020;101:91–100. doi: 10.1016/j.diii.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Gogin N, Viti M, Nicodème L, Ohana M, Talbot H, Gencer U, et al. Automatic coronary artery calcium scoring from unenhanced-ECG-gated CT using deep learning. Diagn Interv Imaging. 2021;102:683–690. doi: 10.1016/j.diii.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Dupuis M, Delbos L, Veil R, Adamsbaum C. External validation of a commercially available deep learning algorithm for fracture detection in children. Diagn Interv Imaging. 2022;103:151–159. doi: 10.1016/j.diii.2021.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Roca P, Attye A, Colas L, Tucholka A, Rubini P, Cackowski S, et al. Artificial intelligence to predict clinical disability in patients with multiple sclerosis using FLAIR MRI. Diagn Interv Imaging. 2020;101:795–802. doi: 10.1016/j.diii.2020.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Evain E, Raynaud C, Ciofolo-Veit C, Popoff A, Caramella T, Kbaier P, et al. Breast nodule classification with two-dimensional ultrasound using Mask-RCNN ensemble aggregation. Diagn Interv Imaging. 2021;102:653–658. doi: 10.1016/j.diii.2021.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Hansell DM, Bankier AA, MacMahon H, McLoud TC, Müller NL, Remy J. Fleischner society: glossary of terms for thoracic imaging. Radiology. 2008;246:697–722. doi: 10.1148/radiol.2462070712. [DOI] [PubMed] [Google Scholar]
- 20.Callister MEJ, Baldwin DR, Akram AR, Barnard S, Cane P, Draffan J, et al. British thoracic society guidelines for the investigation and management of pulmonary nodules. Thorax. 2015;70:1–54. doi: 10.1136/thoraxjnl-2015-207168. [DOI] [PubMed] [Google Scholar]
- 21.National Lung Screening Trial Research Team. Aberle DR, Adams AM, Berg CD, Black WC, Clapp JD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. New Engl J Med. 2011;365:395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Koning HJ, van der Aalst CM, de Jong PA, Scholten ET, Nackaerts K, Heuvelmans MA, et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. New Engl J Med. 2020;382:503–513. doi: 10.1056/NEJMoa1911793. [DOI] [PubMed] [Google Scholar]
- 23.Zhao Y, de Bock GH, Vliegenthart R, van Klaveren RJ, Wang Y, Bogoni L, et al. Performance of computer-aided detection of pulmonary nodules in low-dose CT: comparison with double reading by nodule volume. Eur Radiol. 2012;22:2076–2084. doi: 10.1007/s00330-012-2437-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bae KT, Kim J-S, Na Y-H, Kim KG, Kim J-H. Pulmonary nodules: automated detection on CT images with morphologic matching algorithm–preliminary results. Radiology. 2005;236:286–293. doi: 10.1148/radiol.2361041286. [DOI] [PubMed] [Google Scholar]
- 25.Benzakoun J, Bommart S, Coste J, Chassagnon G, Lederlin M, Boussouar S, et al. Computer-aided diagnosis (CAD) of subsolid nodules: evaluation of a commercial CAD system. Eur J Radiol. 2016;85:1728–1734. doi: 10.1016/j.ejrad.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Armato SG, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, et al. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans: the LIDC/IDRI thoracic CT database of lung nodules. Med Phys. 2011;38:915–931. doi: 10.1118/1.3528204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Setio AAA, Traverso A, de Bel T, Berens MSN, van den Bogaard C, Cerello P, et al. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal. 2017;42:1–13. doi: 10.1016/j.media.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 28.Masood A, Sheng B, Yang P, Li P, Li H, Kim J, et al. Automated decision support system for lung cancer detection and classification via enhanced RFCN with multilayer fusion RPN. IEEE Trans Ind Inf. 2020;16:7791. doi: 10.1109/TII.2020.2972918. [DOI] [Google Scholar]
- 29.Lassau N, Bousaid I, Chouzenoux E, Lamarque JP, Charmettant B, Azoulay M, et al. Three artificial intelligence data challenges based on CT and MRI. Diagn Interv Imaging. 2020;101:783–788. doi: 10.1016/j.diii.2020.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Blanc D, Racine V, Khalil A, Deloche M, Broyelle J-A, Hammouamri I, et al. Artificial intelligence solution to classify pulmonary nodules on CT. Diagn Interv Imaging. 2020;101:803–810. doi: 10.1016/j.diii.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Li L, Liu Z, Huang H, Lin M, Luo D. Evaluating the performance of a deep learning-based computer-aided diagnosis (DL-CAD) system for detecting and characterizing lung nodules: comparison with the performance of double reading by radiologists. Thorac Cancer. 2019;10:183–192. doi: 10.1111/1759-7714.12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.On behalf of the European Society of Radiology (ESR) and the European Respiratory Society (ERS) Kauczor H-U, Baird A-M, Blum TG, Bonomo L, Bostantzoglou C, et al. ESR/ERS statement paper on lung cancer screening. Eur Radiol. 2020;30:3277–3294. doi: 10.1007/s00330-020-06727-7. [DOI] [PubMed] [Google Scholar]
- 33.Schwyzer M, Messerli M, Eberhard M, Skawran S, Martini K, Frauenfelder T. Impact of dose reduction and iterative reconstruction algorithm on the detectability of pulmonary nodules by artificial intelligence. Diagn Interv Imaging. 2022;103:273–280. doi: 10.1016/j.diii.2021.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Yoo H, Kim KH, Singh R, Digumarthy SR, Kalra MK. Validation of a deep learning algorithm for the detection of malignant pulmonary nodules in chest radiographs. JAMA Netw Open. 2020;3:e2017135. doi: 10.1001/jamanetworkopen.2020.17135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sim Y, Chung MJ, Kotter E, Yune S, Kim M, Do S, et al. Deep convolutional neural network–based software improves radiologist detection of malignant lung nodules on chest radiographs. Radiology. 2020;294:199–209. doi: 10.1148/radiol.2019182465. [DOI] [PubMed] [Google Scholar]
- 36.MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, et al. Guidelines for management of incidental pulmonary nodules detected on CT images: from the Fleischner Society 2017. Radiology. 2017;284:228–243. doi: 10.1148/radiol.2017161659. [DOI] [PubMed] [Google Scholar]
- 37.McWilliams A, Tammemagi MC, Mayo JR, Roberts H, Liu G, Soghrati K, et al. Probability of cancer in pulmonary nodules detected on first screening CT. New Engl J Med. 2013;369:910–919. doi: 10.1056/NEJMoa1214726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.American College of Radiology. Lung CT screening reporting and data system (Lung-RADS) 2014. Available from: https://www.acr.org/Quality-Safety/Resources/LungRADS. Accessed 11 Nov 2022.
- 39.Oudkerk M, Devaraj A, Vliegenthart R, Henzler T, Prosch H, Heussel CP, et al. European position statement on lung cancer screening. Lancet Oncol. 2017;18:e754–e766. doi: 10.1016/S1470-2045(17)30861-6. [DOI] [PubMed] [Google Scholar]
- 40.Couraud S, Ferretti G, Milleron B, Cortot A, Girard N, Gounant V, et al. Intergroupe francophone de cancérologie thoracique, Société de pneumologie de langue française, and Société d’imagerie thoracique statement paper on lung cancer screening. Diagn Interv Imaging. 2021;102:199–211. doi: 10.1016/j.diii.2021.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Lancaster HL, Zheng S, Aleshina OO, Yu D, Yu Chernina V, Heuvelmans MA, et al. Outstanding negative prediction performance of solid pulmonary nodule volume AI for ultra-LDCT baseline lung cancer screening risk stratification. Lung Cancer. 2022;165:133–140. doi: 10.1016/j.lungcan.2022.01.002. [DOI] [PubMed] [Google Scholar]
- 42.Oudkerk M, Liu S, Heuvelmans MA, Walter JE, Field JK. Lung cancer LDCT screening and mortality reduction: evidence, pitfalls and future perspectives. Nat Rev Clin Oncol. 2021;18:135–151. doi: 10.1038/s41571-020-00432-6. [DOI] [PubMed] [Google Scholar]
- 43.Jacobs C, Schreuder A, van Riel SJ, Scholten ET, Wittenberg R, Wille MMW, et al. Assisted versus manual Interpretation of low-dose CT scans for lung cancer screening: impact on lung-RADS agreement. Radiol Imaging Cancer. 2021;3:e200160. doi: 10.1148/rycan.2021200160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farag AA, El Munim HEA, Graham JH, Farag AA. A novel approach for lung nodules segmentation in chest CT using level sets. IEEE Trans on Image Process. 2013;22:5202–5213. doi: 10.1109/TIP.2013.2282899. [DOI] [PubMed] [Google Scholar]
- 45.Ye X, Beddoe G, Slabaugh G. Automatic graph cut segmentation of lesions in CT using mean shift superpixels. Intern J Biomed Imaging. 2010;2010:1–14. doi: 10.1155/2010/983963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rocha J, Cunha A, Mendonça AM. Conventional filtering versus U-net based models for pulmonary nodule segmentation in CT images. J Med Syst. 2020;44:81. doi: 10.1007/s10916-020-1541-9. [DOI] [PubMed] [Google Scholar]
- 47.Wang S, Zhou M, Liu Z, Liu Z, Gu D, Zang Y, et al. Central focused convolutional neural networks: developing a data-driven model for lung nodule segmentation. Med Image Anal. 2017;40:172–183. doi: 10.1016/j.media.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawkins S, Wang H, Liu Y, Garcia A, Stringfield O, Krewer H, et al. Predicting malignant nodules from screening CT scans. J Thorac Oncol. 2016;11:2120–2128. doi: 10.1016/j.jtho.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beig N, Khorrami M, Alilou M, Prasanna P, Braman N, Orooji M, et al. Perinodular and intranodular radiomic features on lung CT images distinguish adenocarcinomas from granulomas. Radiology. 2019;290:783–792. doi: 10.1148/radiol.2018180910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massion PP, Antic S, Ather S, Arteta C, Brabec J, Chen H, et al. Assessing the accuracy of a deep learning method to risk stratify indeterminate pulmonary nodules. Am J Respir Crit Care Med. 2020;202:241–249. doi: 10.1164/rccm.201903-0505OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25:954–961. doi: 10.1038/s41591-019-0447-x. [DOI] [PubMed] [Google Scholar]
- 52.Emaminejad N, Wahi-Anwar MW, Kim GHJ, Hsu W, Brown M, McNitt-Gray M. Reproducibility of lung nodule radiomic features: Multivariable and univariable investigations that account for interactions between CT acquisition and reconstruction parameters. Med Phys. 2021;48:2906–2919. doi: 10.1002/mp.14830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Autrusseau P-A, Labani A, De Marini P, Leyendecker P, Hintzpeter C, Ortlieb A-C, et al. Radiomics in the evaluation of lung nodules: intrapatient concordance between full-dose and ultra-low-dose chest computed tomography. Diagn Interv Imaging. 2021;102:233–239. doi: 10.1016/j.diii.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 54.Martini K, Moon JW, Revel MP, Dangeard S, Ruan C, Chassagnon G. Optimization of acquisition parameters for reduced-dose thoracic CT: a phantom study. Diagn Interv Imaging. 2020;101:269–279. doi: 10.1016/j.diii.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 55.Si-Mohamed S, Boccalini S, Rodesch P-A, Dessouky R, Lahoud E, Broussaud T, et al. Feasibility of lung imaging with a large field-of-view spectral photon-counting CT system. Diagn Interv Imaging. 2021;102:305–312. doi: 10.1016/j.diii.2021.01.001. [DOI] [PubMed] [Google Scholar]
- 56.Greffier J, Frandon J, Si-Mohamed S, Dabli D, Hamard A, Belaouni A, et al. Comparison of two deep learning image reconstruction algorithms in chest CT images: a task-based image quality assessment on phantom data. Diagn Interv Imaging. 2022;103:21–30. doi: 10.1016/j.diii.2021.08.001. [DOI] [PubMed] [Google Scholar]
- 57.Hoang-Thi T-N, Vakalopoulou M, Christodoulidis S, Paragios N, Revel M-P, Chassagnon G. Deep learning for lung disease segmentation on CT: which reconstruction kernel should be used? Diagn Interv Imaging. 2021;102:691–695. doi: 10.1016/j.diii.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 58.Cao L, Wang Z, Gong T, Wang J, Liu J, Jin L, et al. Discriminating between bronchiolar adenoma, adenocarcinoma in situ and minimally invasive adenocarcinoma of the lung with CT. Diagn Interv Imaging. 2020;101:831–837. doi: 10.1016/j.diii.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 59.Fan L, Fang M, Li Z, Tu W, Wang S, Chen W, et al. Radiomics signature: a biomarker for the preoperative discrimination of lung invasive adenocarcinoma manifesting as a ground-glass nodule. Eur Radiol. 2019;29:889–897. doi: 10.1007/s00330-018-5530-z. [DOI] [PubMed] [Google Scholar]
- 60.Wang X, Li Q, Cai J, Wang W, Xu P, Zhang Y, et al. Predicting the invasiveness of lung adenocarcinomas appearing as ground-glass nodule on CT scan using multi-task learning and deep radiomics. Transl Lung Cancer Res. 2020;9:1397–1406. doi: 10.21037/tlcr-20-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.AlGharras A, Kovacina B, Tian Z, Alexander JW, Semionov A, van Kempen LC, et al. Imaging-based surrogate markers of epidermal growth factor receptor mutation in lung adenocarcinoma: a local perspective. Can Assoc Radiol J. 2020;71:208–216. doi: 10.1177/0846537119888387. [DOI] [PubMed] [Google Scholar]
- 62.Jia T-Y, Xiong J-F, Li X-Y, Yu W, Xu Z-Y, Cai X-W, et al. Identifying EGFR mutations in lung adenocarcinoma by noninvasive imaging using radiomics features and random forest modeling. Eur Radiol. 2019;29:4742–4750. doi: 10.1007/s00330-019-06024-y. [DOI] [PubMed] [Google Scholar]
- 63.Wang S, Yu H, Gan Y, Wu Z, Li E, Li X, et al. Mining whole-lung information by artificial intelligence for predicting EGFR genotype and targeted therapy response in lung cancer: a multicohort study. Lancet Digit Health. 2022;4:e309–e319. doi: 10.1016/S2589-7500(22)00024-3. [DOI] [PubMed] [Google Scholar]
- 64.Sun R, Limkin EJ, Vakalopoulou M, Dercle L, Champiat S, Han SR, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 65.Trebeschi S, Drago SG, Birkbak NJ, Kurilova I, Cǎlin AM, DelliPizzi A, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann Oncol. 2019;30:998–1004. doi: 10.1093/annonc/mdz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hosny A, Parmar C, Coroller TP, Grossmann P, Zeleznik R, Kumar A, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med. 2018;15:e1002711. doi: 10.1371/journal.pmed.1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]