Abstract
The epidemic of infectious diseases caused by contagious pathogens is a life-threatening hazard to the entire human population worldwide. A timely and accurate diagnosis is the critical link in the fight against infectious diseases. Aptamer-based biosensors, the so-called aptasensors, employ nucleic acid aptamers as bio-receptors for the recognition of target pathogens of interest. This review focuses on the design strategies as well as state-of-the-art technologies of aptasensor-based diagnostics for infectious pathogens (mainly bacteria and viruses), covering the utilization of three major signal transducers, the employment of aptamers as recognition moieties, the construction of versatile biosensing platforms (mostly micro and nanomaterial-based), innovated reporting mechanisms, and signal enhancement approaches. Advanced point-of-care testing (POCT) for infectious disease diagnostics are also discussed highlighting some representative ready-to-use devices to address the urgent needs of currently prevalent coronavirus disease 2019 (COVID-19). Pressing issues in aptamer-based technology and some future perspectives of aptasensors are provided for the implementation of aptasensor-based diagnostics into practical application.
Graphical Abstract
Keywords: Aptasensors, Infectious pathogens, Nanomaterial-based biosensors, Point-of-care testing
Introduction
The outbreak and epidemic of infectious diseases have been continuously posing serious threats to global human health since the last century [1]. According to the latest situation report released by the World Health Organization (WHO), the ongoing pandemic of coronavirus disease 2019 (COVID-19) has caused more than 6 million deaths worldwide, and currently, the Omicron variant remains the dominant variant circulating globally [2]. The emergence of unknown pathogens, as well as the reemergence and surging variants of known pathogens, could bring enormous challenges to the diagnosis, treatment, and control of infectious diseases. Highly contagious pathogens can be transmitted between animals and human bodies easily through contaminated air, water, food, etc. A timely and accurate diagnosis is the critical link in the fight against infectious diseases, which could function as the basis of an effective treatment and the foundation of prophylaxis protocol formulation.
Conventional pathogenic detection techniques in laboratory and clinic mainly include microbiological method, molecular biology genetic method, and antibody-based immunological method. The microbiological method, which involves the isolation, culture, and microscopy visualization of pathogens from clinical samples, is highly specific but time-consuming and laborious. In addition, traditional culture-based assay is poorly adapted to the detection of some fastidious pathogens [3]. Molecular biology genetic technique contains a nucleic acid amplification method to identify the genome of pathogens, i.e., polymerase chain reaction (PCR) detection. PCR detection possesses superior sensitivity and specificity toward a wide range of pathogens, including those microorganism species that are hard to culture when using the microbiological method. However, the extremely high sensitivity attributed to amplification can also lead to false positive or negative results [4]. Hence, sophisticated equipment, highly trained operators, and repeated measurements are usually needed to ensure the accuracy of the test results. The immunological method relies on the specific binding of antibodies to the antigens corresponding to specific pathogens [5]. It is a traditional clinical pathogen detection method including enzyme-linked immunosorbent assay (ELISA) [6, 7], fluorescence and luminescence immunoassay [8, 9], and immunoblotting [10]. Immunoassay-based technique is sensitive and easy to perform in a basic clinical laboratory but demands particular antibody-related production, storage, and handling procedures. The specificity of antibodies might also be compromised due to pathogen evolution [11]. Therefore, continuous efforts should be devoted to the development of rapid, sensitive, reliable, and cost-effective techniques for pathogen detection, especially those in response to a pandemic.
Biosensor-based diagnostic represents an advanced technology in pursuit of rapid detection of various analytes including pathogens [12, 13]. A typical biosensor is composed of immobilized bio-sensitive materials as recognition moieties and physical or chemical transducers that translate the recognition information into measurable signals. Commonly employed bio-recognition elements involve antibodies, nucleic acid derivatives, peptides, enzymes, and whole cells [14, 15]. Aptamer-based biosensors, the so-called aptasensors, employ aptamers as the recognition moieties or bio-receptors to recognize and bind with targets of interest, followed by the translation and output of the recognition information into identifiable signals via versatile transducers [16, 17]. According to signal transduction strategies (sensing mechanisms), they can be mainly classified into acoustic, electrochemical, and optical aptasensors.
This review article focuses on the design strategies as well as state-of-the-art technologies of aptasensor-based diagnostics for infectious pathogens (mostly bacteria and viruses). We intend to highlight the establishment of pathogen diagnostics and point-of-care testing (POCT) methodologies developed in recent years. Current technical barriers of aptamer-based technology are also discussed in the last section of the article, followed by representative solutions and future perspectives toward the implementation of aptamer-based biosensing technology into practical application.
Selection of aptamers against pathogen-related targets
Systematic evolution of ligands by exponential enrichment (SELEX) method was first described in 1990, and the obtained nucleic acid molecules which showed high affinity and specificity toward their targets were named “aptamers” [18–20]. Essentially, SELEX is a technique that simulates natural evolution in vitro to screen the “fittest” aptamers from a random oligonucleotide library. A typical SELEX process involves three main steps: incubation, separation, and amplification [21]. Target molecules are mixed and incubated with an oligonucleotide library containing 1012–1016 DNA or RNA single strands [22]. The unbound sequences can be removed from bound sequences via membrane filtration, magnetic bead-based separation, affinity chromatography, capillary electrophoresis [23–25], etc. Following separation, the bound sequences are eluted from the targets and amplified by PCR (DNA SELEX) or reverse transcription PCR (RNA SELEX) to generate an enriched pool of selected oligonucleotides for a successive round of screening. After 8–15 rounds, the obtained aptamers with high affinity are cloned and sequenced for identification, structure characterization, and binding analysis. Negative selection or counter selection can be employed to eliminate the non-specific aptamers that bind to the matrix or target analogs [26].
Pathogenic microorganisms are biological macromolecules with complex structures. The targets of aptamers for the detection of pathogens can be a specific purified protein or receptor on the surface of pathogen microorganisms [27], bacterial virulence factors [28, 29], the whole cells [30, 31], etc. For the protein-based SELEX, purified proteins with relatively simple structures would ensure their binding stability with aptamers in each selection round to improve the efficiency of screening. However, some proteins might not be able to maintain their conformation as identical as those in their native state, and the lack of glycosylation pattern, association lipids, or carbohydrates could influence the affinity of the selected aptamers to their natural targets [22, 27]. In addition, some target proteins are not easy to obtain, especially when the pathogen is unknown. Whole-cell SELEX is developed for the selection of aptamers against the whole bacterial cells and viral particles [30, 31]. This approach allows effective screening of aptamers without prior knowledge of the detailed structural information of target molecules. However, complicated target molecules with multiple binding sites, the presence of non-target cells and dead cells in the screening system, or non-consistent cell-culture conditions might lead to compromised specificity of the screened aptamers [32]. Homologous microorganisms with similar structures or cell surfaces to target cells could be used in the counter selection round of SELEX to eliminate the non-specific aptamers. In addition, efficient aptamer library regeneration and binding confirmation assays are also critical for the achievement of a successful cell-SELEX [33].
To date, a considerable number of aptamers have been selected against versatile pathogenic species. The implementation of these aptamers for the diagnosis and treatment of infectious diseases have been vastly studied [17, 34–36]. Recently, we published a perspective review article on the state-of-the-art of aptamer-based therapeutic strategies for the treatment of common infectious diseases [21]. Aptamers against membrane-fusion-related proteins, key enzymes, and whole microbial cells could function as effective inhibitors or drug delivery vehicles to inhibit infection. Besides their therapeutic application, the research on aptamer-based testing kits and biosensors is also blooming in recent years, especially motivated by the current situation of the COVID-19 pandemic [37, 38]. Several clinical trials of aptasensors have been carried out, including detection or diagnostic for carcinoma, COVID-19, oxytocin, and anti-HIV drugs (searched at “ClinicalTrials.gov” database, Table 1) [39].
Table 1.
Clinical trials for aptasensors
| Row | Study title | Conditions | Status | Locations |
|---|---|---|---|---|
| 1 | Saliva-Based COVID-19 DNA Aptamer Test | COVID-19 | Recruiting | Udayana University Hospital, Badung, Bali, Indonesia |
| 2 | Clinical Proof-of-Concept of a Tenofovir (TFV) Aptamer-Based Biosensor | HIV/AIDS (tenofovir) |
Active, not recruiting Early phase 1 |
Clinical Research Center, Eastern Virginia Medical School, Norfolk, VA, USA |
| 3 | Identify Proteomic Biomarkers for Outcome Prediction of Lipiodol TACE Treatment (Lipiodol TACE) | Hepatocellular carcinoma | Not yet recruiting | UT Southwestern Medical Center, Dallas, TX, USA |
| 4 | Molecular Biosensors for Detection of Bladder Cancer | Bladder cancer | Recruiting | University of California Irvine, Orange, CA, USA |
| 5 | The Clinical Application of 68 Ga Labeled ssDNA Aptamer Sgc8 in Healthy Volunteers and Colorectal Patients | Colorectal cancer |
Recruiting status unknown Early phase 1 |
Xijing Hospital Nuclear Medicine Department, Xi’an, Shaanxi, China |
| 6 | Non-Invasive, Highly Specific Detection of Oxytocin in Biological Fluids | Pregnancy (oxytocin) | Completed | Lucile Packard Children’s Hospital, Palo Alto, CA, USA |
Aptasensors for pathogen detection
In a typical aptasensor, selected aptamers are immobilized on substrates or materials, which, upon interaction with pathogens of interest, would adopt unique three-dimensional conformational changes that lead to identifiable signal output of the transducers [40]. Those signals can be detected and analyzed by acoustic, electrochemical, or optical-based measurements, depending on different signal transduction techniques. In this section, versatile aptasensors are systematically introduced according to classifications of signal transduction technique, and the brief details of the selected diagnostic aptamers are summarized in Table 2 for reference.
Table 2.
Summarized details of the selected diagnostic aptamers
| Type of aptasensor | Target | Aptamer selection method | Name of aptamer (type) | Sequence (from 5′ to 3′) | Ref |
|---|---|---|---|---|---|
| Acoustic aptasensors | |||||
| QCM aptasensor | E. coli O157:H7 | Whole-cell SELEX | S1 (DNA) | TGGTCGTGGTGAGGTGCGTGTATGGGTGGTGGATGAGTGTGTGGC | [49] |
| AIV H5N1 | Protein-based SELEX | Surface protein-specific aptamer (DNA) | GTGTGCATGGATAGCACGTAACGGTGTAGTAGTAACGTGCGGGTAGGAAGAAAGGGAAATAGTTGTCGTGTTG | [50] | |
| Salmonella | Whole-cell SELEX | S8-7 (DNA) | CTGATGTGTGGGTAGGTGTCGTTGATTTCTTCTGGTGGGG | [51] | |
| Brucella melitensis | Whole-cell SELEX | Brucella melitensis binding aptamer (DNA) | GAGAGTAAAGGCCATCGGCGGCCATTTATGTTGTACCC | [52] | |
| Lysozyme | Protein-based SELEX | Lysozyme-specific aptamer (DNA) | TTTTTTATCAGGGCTAAAGAGTGC | [53] | |
| Thrombin | Protein-based SELEX | 15-Mer thrombin aptamer (DNA) | GGTTGGTGTGGTTGG | [54] | |
| S. typhimurium | QCM-based cell SELEX | B5 (DNA) | CAGTCCAGGACAGATTCGCGAGCCCACTCCAAACACGACCAACTCACGCTCTATCAACATCGCTATCCACGTGGATTTCATTCAGCGATT | [55] | |
| SAW aptasensor | α-thrombin; HIV-1 Rev peptide | Protein-based SELEX | Thrombin aptamer (DNA); Rev peptide aptamer (RNA) | GGTGGTGGTGGTGTT; GGGUGUCUUGGAGUGCUGAUCGGACACC | [59] |
| Endotoxin | SELEX | Endotoxin aptamer (DNA) | CTTCTGCCCGCCTCCTTCCTAGCCGGATCGCGCTGGCCAGATGATATAAAGGGTCAGCCCCCCAGGAGACGAGATAGGCGGACACT | [62] | |
| Electrochemical aptasensors | |||||
| Labeled electrochemical aptasensors | M. tb HspX | Protein-based SELEX | H63 SL-2 M6 (DNA) | AGGGCTTTTTTTTTTTTTAGTTCGTTTG | [71] |
| M. tb antigen MPT64 | Protein-based SELEX | MBA I (DNA) | TGGGAGCTGATGTCGCATGGGTTTTGATCACATGA | [72] | |
| P. aeruginosa | Whole-cell SELEX | P. aeruginosa aptamer (DNA) | CCCCCGTTGCTTTCGCTTTTCCTTTCGCTTTTGTTCGTTTCGTCCCTGCTTCCTTTCTTG | [73] | |
| Norovirus | Protein-based SELEX | Capsid-specific aptamer (DNA) | AGTATACCGTATTACCTGCAGCCATGTTTTGTAGGTGTAATAGGTCATGTTAGGGTTTCTGCGATATCTCGGAGATCTTGC | [75] | |
| AIV | Magnetic bead-based SELEX | AIV nucleoprotein aptamer NP5 (DNA) | TATTGTCTATTCATTCCTGTTTCGGTTCTATCTCTTTTTT | [77] | |
| Label-free electrochemical aptasensors | S. Typhimurium | Outer-membrane protein-based SELEX | S. Typhimurium aptamer (DNA) | TTTGGTCCTTGTCTTATGTCCAGAATGCGAGGAAAGTCTATAGCAGAGGAGATGTGTGAACCGAGTAAATTTCTCCTACTGGGATAGGTGGATTAT | [79] |
| S. Typhimurium | Outer-membrane protein-based SELEX | S. Typhimurium aptamer (DNA) | TTTGGTCCTTGTCTTATGTCCAGAATGCGAGGAAAGTCTATAGCAGAGGAGATGTGTGAACCGAGTAAATTTCTCCTACTGGGATAGGTGGATTAT | [80] | |
| Aflatoxin M1 | Protein-based SELEX | AFM1-aptamer (DNA) | ACTGCTAGAGATTTTCCACAT | [81] | |
| SARS-CoV-2 | SARS-CoV-2 RBD-based SELEX | CoV2-RBD-1C (DNA) | CAGCACCGACCTTGTGCTTTGGGAGTGCTGGTCCAAGGGCGTTAATGGACA | [83] | |
| Human adenovirus; SARS-CoV-2 | Whole-virus SELEX | HAdV-Seq4; SARS2-AR10 (DNA) | GGCTGCAGCTGAAGCACTGGTTTTGAGTCAAACCCAGACGATGGA (HAdV-Seq4); CCCGACCAGCCACCATCAGCAACTCTTCCGCGTCCATCCCTGCTG (SARS2-AR10) | [84] | |
| S. Typhimurium | Outer-membrane protein-based SELEX | Salmonella-specific aptamer (DNA) | TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG | [85] | |
| E. coli | Whole-cell SELEX | Anti-E. coli aptamer (DNA) | GCAATGGTACGGTACTTCCACTTAGGTCGAGGTTAGTTTGTCTTGCTGGCGCATCCACTGAGCGCAAAAGTGCACGCTACTTTGCTAA | [86] | |
| HIV-1 | HIV-1 Tat protein-based SELEX | Split aptamer (RNA) | UCGGUCGAUCGCUUCAUAA; GAAGCUUGAUCCCGAA | [90] | |
| Optical aptasensors | |||||
| Colorimetric aptasensors | S. enteritidis | Whole-cell SELEX | crn-1 and crn-2 (DNA) | AAGGGCTGGCTGGGATGGACCCTCCCGAAACGAGCTGTCTCTTAACGGAAGCTAATCTGCCTCACTCCACGGACCCCACT (crn-1); AAGGGCTGGCTGGGATGGATGTAAGAAGGGAGGAAAGGACCTAAGACCTGCTATATTGCGATCACTCCACGGACCCCACT (crn-2) | [93] |
| S. typhimurium | Whole-cell SELEX | apt 1 and apt 2 (DNA) | AGTAATGCCCGGTAGTTATTCAAAGATGAGTAGGAAAAGA (apt 1); AAAAAAAAAAAAAGTAATGCCCGGTAGTTATTCAAAGATGAGTAGGAAAAGA (apt 2) | [95] | |
| C. jejuni | Whole-cell SELEX | ONS-23 (DNA) | GCAAGATCCCGAGATATCGTGCTGGGGGGTGGTTTGTTTGGGTCGGTTGTTTTGGTTGGGCTGCAGGTAATACGTATACT | [96] | |
| Vp | Whole-cell SELEX and post-SELEX optimization | A4 (DNA) | CAACGAAACAGTGACTCGTTG | [97] | |
| L. monocytogenes | Whole-cell SELEX | Aptamer (DNA) | TATCCATGGGGCGGAGATGAGGGGGAGGAGGGCGGGTACCCGGTTGAT | [98] | |
| S. typhimurium | Whole-cell SELEX | Apt (DNA) | GCGCTCGGCCTCCTCTGCCATCTCATTCGCGAGCGC | [99] | |
| Shigella flexneri | Whole cell-SELEX | Shigella flexneri binding aptamer (DNA) | CCGGACTAGGGCTGGTTAGCTTCAATACTGCTGGGCGAGG | [100] | |
| C. sakazakii | Whole-cell SELEX | CS4 (DNA) | GTGGTCGGGGTGGTGGGTGGGAGGGCGACTTCATCTGCGC | [101] | |
| Dengue virus | Viral protein-based SELEX | DENV aptamer (DNA) | CCCGCACCGGGCAGGACGTCCGGGGTCCTCGGGGGGCGGG | [102] | |
| Avian influenza H5N2 | GO-SELEX | J3APT and JH4APT (DNA) | CGTACGGAATTCGCTAGCTGATGGTGTGGCGGGGGGCG(J3APT); CGTACGGAATTCGCTAGCGGTGGCTCTAGGGCCTATC (JH4APT) | [105] | |
| Fluorescent aptasensors | P. aeruginosa | Whole-cell SELEX | F23 (DNA) | CCCCCGTTGCTTTCGCTTTTCCTTTCGCTTTTGTTCGTTTCGTCCCTGCTTCCTTTCTTG | [109, 110] |
| S. aureus | Whole-cell SELEX | Apt (DNA) | GCAATGGTACGGTACTTCCTCGGCACGTTCTCAGTAGCGCTCGCTGGTCATCCCACAGCTACGTCAAAAGTGCACGCTACTTTGCTAA | [111, 112] | |
| E. coli ATCC 8739 | Whole-cell SELEX | Aptamer 8739 (DNA) | TGAAATGTTGGGACACTAGGTGGCATAGAGCCGCAAAAGTGCACGCTACTTTGCTAA | [115] | |
| SARS-CoV-2 | SARS-CoV-2 NP-based SELEX | Np-A48 (DNA) | GCTGGATGTCGCTTACGACAATATTCCTTAGGGGCACCGCTACATTGACACATCCAGC | [118] | |
| E. coli/pUC19 | Whole-cell SELEX | E. coli-specific aptamer (DNA) | GCAATGGTACGGTACTTCCCCATGAGTGTTGTGAAATGTTGGGACACTAGGTGGCATAGAGCCGCAAAAGTGCACGCTACTTTGCTAA | [119] | |
| SARS-CoV2 | SARS-CoV-2 NP-based SELEX | Apt 58 and 61 (DNA) | GCTGGATGTCACCGGATTGTCGGACATCGGATTGTCTGAGTCATATGACACATCCAGC (Apt 58); GCTGGATGTTGACCTTTACAGATCGGATTCTGTGGGGCGTTAAACTGACACATCCAG C (Apt 61) | [120] | |
| Influenza A H1N1 virus | Microfluidic SELEX | H1N1-specific aptamer (DNA) | TTTTTTTTGGCAGGAAGACAAACAGCCAGCGTGACAGCGACGCGTAGGGACCGGCATCCGCGGGTGGTCTGTGGTGCTGT | [121] | |
| Acinetobacter baumannii | Whole-cell SELEX | AB-specific aptamer (DNA) | CAGCACCACAGACCACATATCACATGCTGTCGCCTTGCGATATCAATTCCAGTGATGTTTGTCTTCCTGCC | [122] | |
| SERS aptasensors | Influenza virus | Recombinant hemagglutinin-based SELEX | RHA0385 (DNA) | TTGGGGTTATTTTGGGAGGGCGGGGGTT | [128] |
| E. coli O157:H7 | Whole-cell SELEX | Apt-1 and Apt-2 (DNA) | CCGGACGCTTATGCCTTGCCATCTACAGAGCAGGTGTGACGG (Apt-1); -CCGGACGCTTATGCCTTGCCATCTACAGAGCAGGTGTGACGG (Apt-2) | [129] | |
| Vp | Whole-cell SELEX | apt-1 and apt-2 (DNA) | TCTAAAAATGGGCAAAGAAACAGTGACTCGTTGAGATACT (apt-1); TCTAAAAATGGGCAAAGAAACAGTGACTCGTTGAGATACT (apt-2) | [131] | |
| Vp | Whole-cell SELEX | A3 (DNA) | TCTAAAAATGGGCAAAGAAACAGTGACTCGTTGAGATACT | [132, 133] | |
| E. coli and S. aureus | Whole-cell SELEX | E1 and Sa1 (DNA) | GCAATGGTACGGTACTTCCTCGGCACGTTCTCAGTAGCGCTCGCTGGTCATCCCACAGCTACGTCAAAAGTGCACGCTACTTTGCTAA (E1); GCAATGGTACGGTACTTCCACTTAGGTCGAGGTTAGTTTGTCTTGCTGGCGCATCCACTGAGCGCAAAAGTGCACGCTACTTTGCTAA (Sa1) | [134] | |
| SPR aptasensors | Whole H5Nx avian influenza viruses | Multi-GO-SELEX | IF10 and IF22 (DNA) | CGTACGGTCGACGCTAGCTAACGGTGTGCCCCGGGGGTACAGCGCACTCACGTGGAGCTCGGATCC (IF10); CGTACGGICGACGCTASCTAAATGGGCGTGGGAATGACTCTACGGGGCCACGTGGAGCTCGGATCC (IF22) | [137] |
| Norovirus | Norovirus capsid protein-based SELEX | Aptamer I and II (DNA) | GCTAGCGAATTCCGTACGAAGGGCGAATTCCACATTGGGCTGCAGCCCGGGGGATCC (aptamer I); CGTACGGAATTC GCTAGCACGGGGCTTAAGGAATACAGATGTACTACCGAGCTCATGAGGATCCGAGCTCCACGTG (aptamer II) | [138] | |
| S. typhimurium | Outer-membrane protein-based SELEX | APT33 and 45 (DNA) | TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG (APT33); GAGGAAAGTCTATAGCAGAGGAGATGTGTGAACCGAGTAA (APT45) | [139] | |
| S. typhimurium | Outer-membrane protein-based SELEX | APT33 (DNA) | TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG | [141] | |
| Other aptasensors | |||||
| Microcantilever aptasensor | HCV helicase | Protein-based SELEX | Helicase aptamer (RNA) | GGGAGAGCGGAAGCGUGCUGGGCCACAUUGUGAGGGGCUCAGGUGGAUCGCAUGGCCGUGUCCAUAACCCAGAGGUCGAUGGAUCCU | [144] |
| Gravity-based aptasensor | Influenza virus H3N2 | Hemagglutinin (HA) protein-based SELEX | HA12-16 (RNA) | GCUUGACGGAGAUCAAGGGCGAGUCUCAUACCAAGUUGAUGGGG | [145] |
| Aptamer-assisted proximity ligation assay | SARS-CoV-2 | N-protein based SELEX | N48 and N58 (DNA) |
GCTGGATGTCGCTTACGACAATATTCCTTAGGGGCACCGCTACATTGACACATCCAGC (N48); GCTGGATGTCACCGGATTGTCGGACATCGGATTGTCTGAGTCATATGACACATCCAGC (N58) |
[146] |
| Thermophoretic aptasensor | SARS-CoV-2 | S protein-based SELEX | CoV2-RBD-4C (DNA) | ATCCAGAGTGACGCAGCATTTCATCGGGTCCAAAAGGGGCTGCTCGGGATTGCGGATATGGACACGT | [147] |
Acoustic aptasensors
Acoustic sensors are often considered “mass sensitive,” because mass effect is one of the crucial contributions to the response of the sensor [41–43]. Mass-sensitive transducers are employed in acoustic sensors in which the changes on the transducer surface can be detected by measuring the changes of the resonant behavior. Acoustic biosensors are bioassay techniques known for label-free detection, ease of operation, and real-time measurement [44, 45]. In this section, the basic working principles of commonly developed acoustic sensors are briefly introduced, followed by the examples of their application in pathogen detection.
Quartz crystal microbalance (QCM) aptasensors
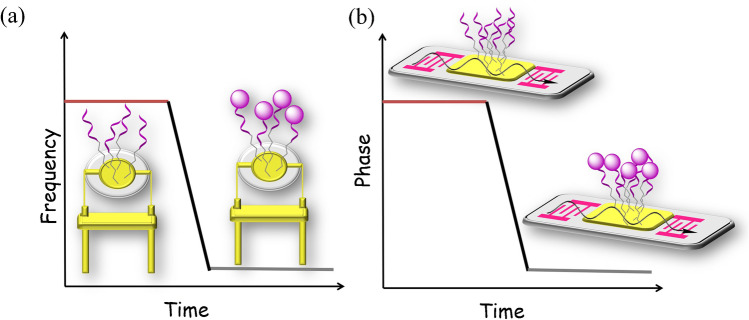
QCM sensors have been extensively studied for its high sensitivity, low noise level, and ease of fabrication. QCM utilizes the piezoelectric effect of quartz crystal to convert the change of surface quality into frequency fluctuation in output signals [46, 47]. The vibration frequency of quartz crystal under alternating electric field is closely related to its surface quality. Increase of surface loading due to substance adsorption could change the vibration frequency and induce a phase shift. In QCM-based diagnostic devices for infectious diseases, the receptors employed for target recognition include antibodies and antigens, nucleic acid probes, and molecularly imprinted polymers [48]. In a typical QCM aptasensor, aptamers are immobilized on gold-coated quartz crystals (Fig. 1a), and their interaction with target molecules can be measured by the corresponding decrease of frequency.
Fig. 1.
Schematic illustration of basic (a) QCM and (b) SAW aptasensors for target binding and signal measurement
A whole-bacterium SELEX technique was developed for the selection of aptamers specifically bound to Escherichia coli (E. coli) O157:H7 [49]. Aptamer sequence S1 was then modified with a biotin moiety and immobilized onto the surface of a streptavidin fabricated QCM electrode. The resulting QCM aptasensor showed a detection limit of 1.46 × 103 CFU (colony-forming unit) mL−1 for E. coli with a response time of 50 min. Wang et al. fabricated a QCM sensor with aptamer-ssDNA crosslinked polymeric hydrogel for rapid and sensitive detection of Avian influenza viruses (AIV) H5N1 [50]. The “smart” AIV-responsive hydrogel was constructed by introducing the selected aptamer against AIV H5N1 surface protein and a crosslinked ssDNA to the polymer backbones, followed by immobilization onto the gold surface of QCM sensor. Upon exposure to the virus, the binding between aptamer and H5N1 virus caused the dissolution of the linkage and led to the swelling of the hydrogel, which could be monitored by the QCM sensor as decreased frequency of the output signals. The detection limit reached 0.0128 HAU (hemagglutinating unit) in 30 min, showing no interference from non-target AIV subtypes, which was superior to the anti-H5 antibody immobilized QCM biosensor in terms of the detection limit and detection time. To develop sensors applicable for the detection of bacterial cells in food samples, an integrated system was reported combing an aptamer-based magnetic separation system for target enrichment and QCM analysis for real-time monitoring [51]. Salmonella specific aptamer immobilized magnetic beads were conjugated to gold-coated QCM electrode, which could efficiently capture the Salmonella cells at 100 CFU mL−1 in milk in less than 10 min to induce frequency changes of the QCM sensor. Treatment of the crystal surface with NaOH solution could regenerate the sensing system for reuse. By employing a similar pre-concentration strategy, Brucella melitensis bacteria in milk and milk products could be quantitatively detected with high selectivity, and the detection limit was determined to be 103 cells (Fig. 2a) [52]. Those nanoparticles remained high pre-concentration efficiency after recycling for 8 times. Aptamer-magnetic system has also been applied to the detection of lysozyme, and the QCM chip sensor showed an observed detection limit of 17.9 ± 0.6 ng/mL with high selectivity [53]. Another interesting example developed a label-free aptasensor for thrombin based on target-triggered release of cargo molecules from gold nanocages [54]. An array of gold nanocages were loaded with cargo molecules in their interiors, and DNA probes were immobilized on the surface for hybridization with thrombin-specific aptamers as the gatekeeper. In the presence of thrombin, surface aptamers disassociated from the nanocages, resulting in the release of interior cargo molecules. The loss of cargo molecules was monitored by QCM. The use of polyamidoamine as cargo molecules achieved an optimized detection limit of 7.7 pM.
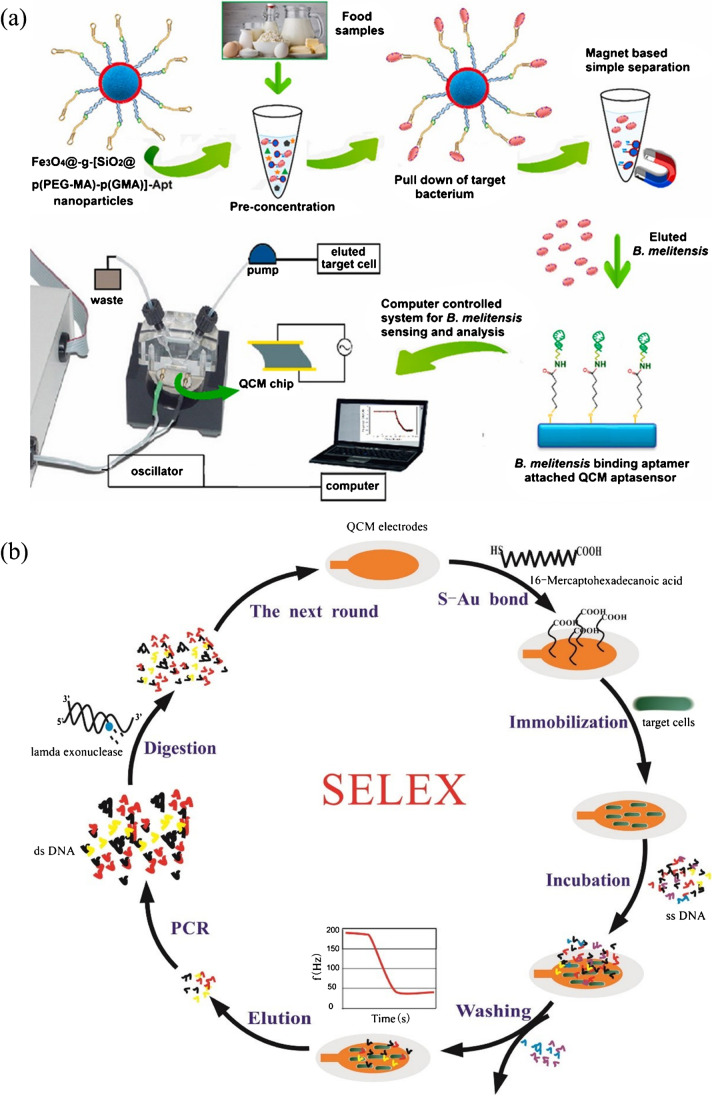
Fig. 2.
(a) Aptamer-magnetic bead pre-concentration and detection of Brucella melitensis bacteria in food samples with a QCM aptasensor. Reproduced with permission from [52], Copyright 2019 Elsevier. (b) QCM-based SELEX for S. typhimurium. Reproduced with permission from [55], Copyright 2017 Elsevier
QCM can also be applied for aptamer selection to increase the success rate of SELEX. In one example, QCM was used to simultaneously track the affinity of DNA pool in each selection round to Salmonella typhimurium (S. typhimurium), and the candidate pool was cloned and sequenced when the frequency change reached a maximum value after several rounds of selection and counter-selection (Fig. 2b) [55]. Aptamer B5 was chosen for the fabrication of a QCM aptasensor, exhibiting a detection limit of 103 CFU mL−1 within 1 h. This study successfully demonstrated the feasibility of a more effective aptamer selection by QCM-based SELEX, especially for the fabrication of a QCM aptasensor.
Surface acoustic wave (SAW) aptasensors
SAW sensors are also comprised of a piezoelectric substrate, together with an interdigital transducer (IDT) that generate and detect acoustic wave signals on chip surface [56, 57]. The measurement principle is based on the propagation of surface acoustic waves. While passing through the chip surface, the phase and amplitude of acoustic waves vary with surface quality and viscosity, which can be detected as decrease in resonance frequency or phase shift between input and output signals (Fig. 1b).
Shear horizontal SAW (SH-SAW) sensors, also known as Love-wave sensors, are special SAW sensors with higher sensitivity by utilizing shear horizontal waves guided through the layer on the sensor surface to minimize the acoustic losses into substrate [58]. Schlensog et al. reported a Love-wave biosensor array for specific detection of human α-thrombin and HIV-1 Rev peptide [59]. Compared with a bulk acoustic wave sensor (detection limit = 3.3 ng/cm2) [60], the sensitivity of the Love-wave sensor (detection limit < 80 pg/cm2) was significantly lower. In addition, the sensor could be easily regenerated by simple washing steps. Endotoxin is a general term of toxic substances in gram-negative bacteria, which is responsible for symptoms like fever, microcirculation disturbance, septic shock, and disseminated intravascular coagulation [61]. A label-free and highly sensitive SH-SAW aptasensor was developed for endotoxin detection [62]. Instead of using Au-based electrode materials, single-layered graphene films were employed, accompanied by chemical vapor deposition technique for device fabrication. Aptamers specifically bind to endotoxin were chemically bonded to the surface, and the sensing platform exhibited a detection limit of 3.53 ng/mL, with excellent specificity in discriminating the endotoxin and the aflatoxin obtained from Pseudomonas aeruginosa (P. aeruginosa) (Fig. 3).
Fig. 3.
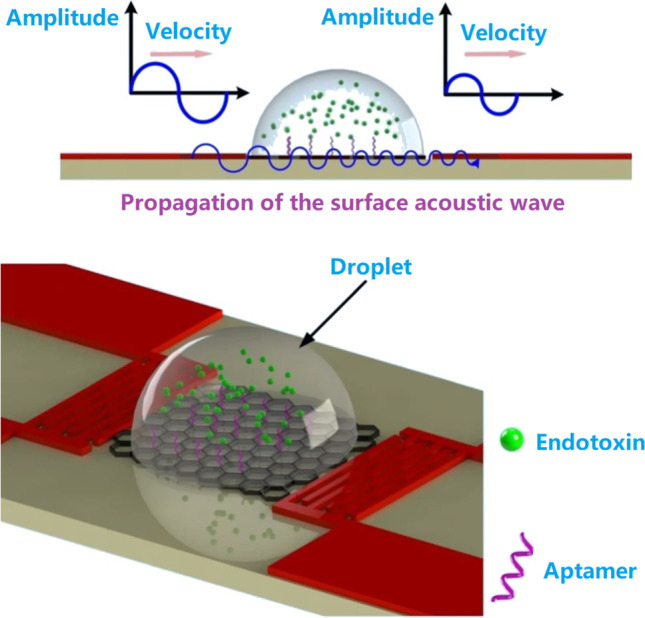
Principles and schematic illustration of endotoxin detection on the SH-SAW aptasensor. Reproduced with permission from [62] under a CC BY license, Copyright 2020 Springer Nature
Electrochemical aptasensors
Among the wide variety of biosensing techniques, sensors based on the changes of electric properties are attractive options owing to their high sensitivity, convenient operation, and ease of miniaturization for use in portable devices. Electrochemical biosensors are capable of transducing target recognition events at the electrochemical interface into detectable electrochemical signals [63, 64]. To date, a considerable amount of research work has explored versatile electrochemical strategies for the diagnosis of infectious diseases and healthcare monitoring [65–67]. Aptamers have been integrated on electrode surfaces as recognition elements [68, 69]. In this section, we focus on recent developed strategies of electrochemical aptasensors and their application in pathogen detection for diagnostic purposes.
Labeled electrochemical aptasensors
In traditional electrochemical aptasensors, electroactive species such as enzymes, ferrocene (Fc), and methylene blue (MB) are commonly incorporated as labels, because the binding of aptamers with pathogenic microorganisms cannot generate electrochemical signal by themselves. Aptamers labeled with electroactive species are immobilized onto the electrode surface in a typical electrochemical aptasensor (Fig. 4a). In some other cases, redox probes are added to the solution as indicators for the recognition events (Fig. 4b–c). Those recognition events could change the electron transfer efficiencies of the redox labels, resulting in the changes of the corresponding electrochemical signals (potential, current, conductivity, or impedance) [70].
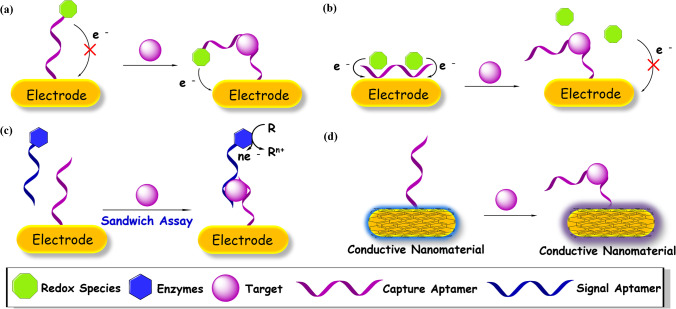
Fig. 4.
Schematic illustration of some representative electrochemical aptasensors. (a) Working principle of an aptasensor with a redox probe tagged on the aptamer for signal “OFF–ON” detection. (b) Working principle of an aptasensor to release the redox probe upon target binding for a signal “ON–OFF” detection. (c) Working principle of an aptasensor with a signal aptamer carrying enzymes to report the recognition information via a sandwich assay. (d) Working principle of a label-free aptasensor to detect target through the electrochemical variation of the conductive material coated on the electrode
A highly sensitive structural switching electrochemical aptasensor was developed based on a DNA aptamer tagged with electroactive MB for HspX, a Mycobacterium tuberculosis (M. tb) antigen [71]. Upon target binding, the electron transfer between MB and electrode was interfered by the conformation change of aptamer, leading to a sharp decrease in current. The sensor exhibited a response time of less than 30 min with a detection limit of 10 pg HspX. MPT64 is a 24-kDa protein only secreted by M. tb, which is usually employed as a target molecule for M. tb detection. An electrochemical aptasensor was constructed for ultrasensitive detection of MPT64 in human serum [72]. The gold electrode was immobilized with capture aptamers (MBA I) to capture the MPT64 antigen, and coil-like fullerene-doped polyaniline (C60-PAn) redox nanoprobes were decorated with gold nanoparticles (GNPs or AuNPs) and labeled with signal aptamers (MBA II) to form the tracer label. In the presence of the targets, the sandwich reaction between capture aptamers and the tracer label resulted in obvious changes of the detection signal by differential pulse voltammetry (DPV) measurement, and the signal could be further enhanced by the electrocatalytic activity of C60-PAn toward ascorbic acid (AA) (Fig. 5a). The sensor exhibited a detection limit of 20 fg/mL, showing excellent specificity and sensitivity for MPT64 detection in real serum samples of tuberculosis (TB) patients. Shahrokhian et al. developed a sensitive diagnostic device for P. aeruginosa whole-cell detection based on aptamers immobilized on the surface of engineered zeolitic imidazolate Framework-8 (ZIFs-8) [73]. Ferrocene − graphene oxide (Fc-GO) was employed as the electroactive indicator. In the absence of P. aeruginosa, Fc-GO adsorbed on the aptasensor through interaction with aptamers, while the presence of target bacteria resulted in the removal of Fc-GO, giving a signal “OFF” of the DPV signal. The sensing platform was able to detect P. aeruginosa with a detection limit of 1 CFU mL−1. Excellent recovery rate from spiked human urine samples revealed the potential of the proposed device for application in clinical analysis.
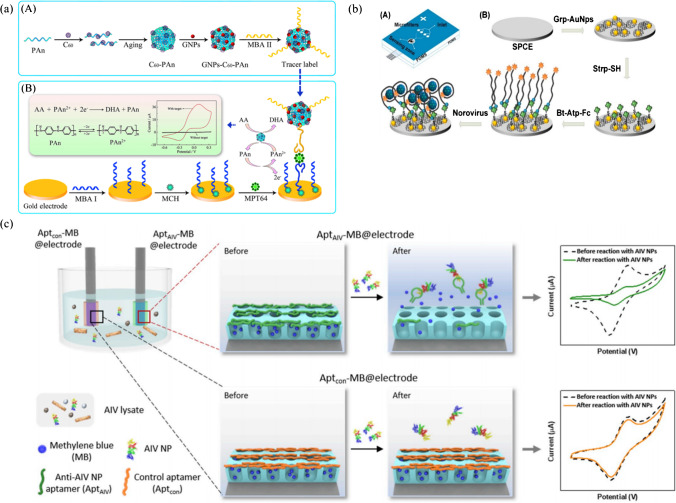
Fig. 5.
(a) Preparation procedure of the tracer label and schematic diagram of the electrochemical aptasensor for the detection of MPT64 via sandwich reaction. Reproduced with permission from [72] Copyright 2017 Elsevier. (b) Structure of PDMS microfluidic chip and electrode functionalization and detection of norovirus. Reproduced with permission from [75], Copyright 2017 Elsevier. (c) Self-calibrating dual-electrode-based electrochemical aptasensing platform for AIV detection. Reproduced with permission from [77], Copyright 2020 Elsevier
Microfluidics and electrochemistry have a synergistic relationship, and the integration of microtechnology with electrochemistry has boosted the development toward the miniaturization and portability of novel electrochemical systems and eventually next-generation POCT microsystems [74]. An electrochemical aptasensor integrated with an all-polydimethylsiloxane (PDMS) microfluidic platform was constructed, aiming at on-site sample processing and detection of norovirus in clinical samples [75]. Norovirus-infected clinical samples were filtered and enriched by the microfluidic chip containing packed silica microbead zones. Carbon electrode was modified with graphene-AuNPs composite, followed by functionalization with Fc tagged viral capsid-specific aptamers. The binding of aptamers with norovirus resulted in a decrease in the electrochemical signal from Fc (Fig. 5b). As shown by DPV analysis, the detection limit was determined to be 100 pM for norovirus. The aptasensor was also utilized to detect norovirus in spiked blood samples for real sample application assessment, exhibiting high sensitivity and selectivity in the presence of peptidoglycan as an interferon.
It was found that aptamer attachment geometry on the electrode could dramatically alter the performance of electrochemical aptasensors. By switching the 3′ and 5′ terminus for electrode binding and reporter tagging, the orientation of aptamers strongly affected the output signals, suggesting that the attachment geometry of aptamers is a worthwhile parameter to optimize in the design of new electrochemical aptasensors [76]. Besides the essential components of electrochemical aptasensors, namely, aptamers, electrode, and electroactive species, the existence of external interference factors in the supporting buffer such as salts, ions, and cell lysates may lead to electrochemical signal errors. Pre-treatment and purification of the samples, optimization of detection conditions, and control tests are necessary to minimize the interferences. To address the problem in a more effortless approach, Lee et al. invented a self-calibrating dual-electrode-based electrochemical aptasensing platform for reliable and stable detection of avian influenza viruses (AIV) [77]. Both electrodes were fabricated using tungsten rods, followed by modification with 3D nanostructured porous silica film on the surface. MB molecules were trapped into the pores and capped with the corresponding aptamers: one with anti-AIV nucleoprotein (NP) aptamers (AptAIV) for target binding and the other with control aptamers (Aptcon) to correct the false responses generated by non-specific aptamer detachment and MB release and provide a corrected baseline for the output signals (Fig. 5c). Compared with a conventional single-electrode platform (RSD: 30.13%), the dual-electrode platform exhibited superior output stability (relative standard deviation, RSD: 5.86%) for AIV nucleoprotein samples with no need of further purification and washing steps. This work presented a universal strategy for the design of more reliable electrochemical aptasensors.
Label-free electrochemical aptasensors
Labeled aptasensors are highly sensitive due to the signal amplification resulted from enzymatic reactions or the electroactive labels. Nevertheless, the modification of aptamer with tagged molecule is time- and effort-consuming, and moreover, labeling might affect the binding affinity of aptamers toward their analytes [78]. Label-free strategy has become an attractive alternative in electrochemical sensing technology, especially with the development of versatile nanomaterials and surface modification technique of electrodes in recent years (Fig. 4d). A label-free impedimetric biosensor for Salmonella Typhimurium (S. Typhimurium) detection was developed by combination of S. typhimurium specific aptamers and conductive polypyrrole-based polymers [79]. Impedimetric measurements was facilitated by the variation of the electrical properties of the polymeric surface resulted from aptamer-pathogen interaction. The aptasensor exhibited a detection limit of 3 CFU mL−1. Another impedimetric label-free aptasensor for S. typhimurium was reported using an aptamer-immobilized diazonium-supporting layer to fabricate the screen-printed carbon electrodes (SPEs) [80]. The electrochemical immobilization of the diazonium-grafting layer allowed the formation of a denser aptamer layer, which resulted in high sensitivity of the aptasensor with a detection limit of 6 CFU mL−1. Both examples performed real sample assessment in spiked apple juice to prove these aptasensors as viable approaches for rapid detection of pathogens in food. Istamboulié et al. also used diazonium activated SPEs to immobilize a hexaethylene glycol-modified 21-mer oligonucleotide aptamer for the determination of aflatoxin M1 (AFM1) through electrochemical impedance spectroscopy detection [81]. AFM1 is the hydroxylated metabolite of aflatoxin B1 (AFB1), one of the most toxic aflatoxins that has been designated as a primary carcinogenic compound by the International Agency for Research on Cancer (IARC) [82]. The binding interaction of aptamers with AFM1 could induce an increase in electron-transfer resistance for the determination of AFM1 with a detection limit of 1.15 ng/L. AFM1 ranging from 20 to 1000 ng/kg in milk could be detected after a simple filtration through a 0.2-mm polytetrafluoroethylene (PTFE) membrane. An aptamer targeting the receptor-binding domain (RBD) in the spike protein (S protein) of the SARS-CoV-2 was immobilized on AuNPs/SPEs platform, and the aptasensor yielded a limit of detection of 1.30 pM (66 pg/mL) for SARS-CoV-2 S protein as revealed by electrochemical impedance spectroscopy after 40-min incubation [83]. A whole-virus in vitro selection approach was applied to achieve high selectivity against active human adenovirus and SARS-CoV-2 over the inactive ones. The selected aptamers (HAdV-Seq4, SARS2-AR10) were then immobilized onto the inner wall of a solid-state nanopore for direct detection of the viruses, and steady-state current–voltage measurements indicated a sensitivity down to 1 PFU/mL (plaque forming unit per mL) for human adenovirus and 1 × 104 copies/ml for SARS-CoV-2 [84]. The incorporation of nanowires into the construction of electrochemical biosensors is promising attributed to their small size, high aspect ratios, and excellent electronic properties. A facile fabrication technique for sub-100-nm suspended carbon nanowire sensors was presented as an innovative platform for chemiresistive biosensing [85]. Aptamers modified with amine moieties were immobilized by carbodiimide crosslinker chemistry with the carboxylic groups on the surface of carbon nanowire. The platform was then integrated with a microfluidic chip to form a lab-on-a-chip device for label-free detection of S. Typhimurium. The sensor showed highly specific and sensitive detection of target bacteria in 5-min assay time with a detection limit of 10 CFU mL−1.
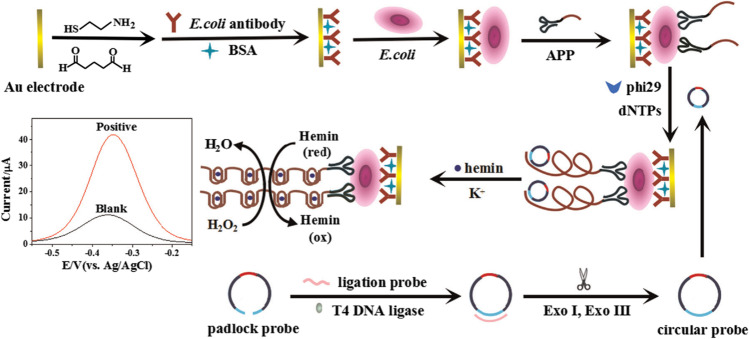
Besides modification of electrodes, some other techniques with signal enhancement features have been adopted for the construction of label-free aptasensors. Guo et al., for the first time, integrated rolling circle amplification (RCA) coupled peroxidase-mimicking DNAzyme amplification technique into electrochemical assay of E. coli [86]. An aptamer-primer probe containing anti-E. coli aptamer and a primer sequence complementary to a circular probe including two G-quadruplex units was designed for target recognition and triggering of the RCA-based polymerase elongation. Upon binding with E. coli, numerous G-quadruplex oligomers were formed on the electrode due to RCA coupled DNAzyme amplification. The oligomers folded into G-quadruplex/hemin complexes in the presence of K+ and hemin, generating extremely strong catalytic activity toward H2O2 to give an obvious current increase in DPV measurements. The proposed aptasensor exhibited a detection limit of 8 CFU mL−1 as a simple, rapid platform for E. coli detection (Fig. 6).
Fig. 6.
Electrochemical assay of E. coli using RCA and DNAzyme amplification technique. Reproduced with permission from [86], Copyright 2016 Elsevier
Field effect transistor (FET) biosensor is another type of electrochemical sensing platform that make use of FET as the signal transducer to provide a label-free and ultrasensitive detection technique for various targets [87, 88]. Aptamers can be immobilized on the sensing channel of FET, and their recognition behaviors that cause the variation of the channel conductance can be recorded and further processed by an electrical measurement system as output signals [89]. The interaction between split RNA aptamer with HIV-1 Tat, the clinically important target, was investigated via a multi-wall carbon nanotube-modified biosensing FET [90]. Electrical measurements showed that the immobilization of aptamer on the multi-wall carbon nanotube gave a 34.4-mV gate voltage shift, and in the presence of HIV-1 Tat, the current flow decreased with a concomitant gate voltage shift of 23.5 mV. The aptasensor exhibited a sensitivity of 600 pM for HIV-1 Tat with negligible interferences from other tested HIV-1 proteins Nef and p24.
Optical aptasensors
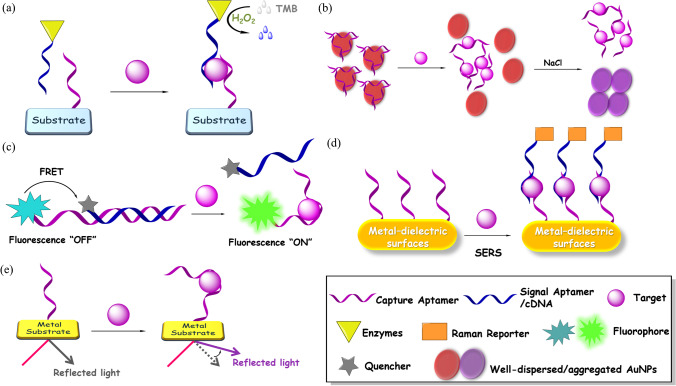
Optical biosensors possess great advantages over traditional analytical techniques for their high sensitivity, accessibility, small size, and cost-effectiveness [91]. In addition, they can be easily miniaturized and present potential for chip integration [92]. Aptamer-based biosensors can be mainly classified into colorimetric, fluorescent, surface-enhanced Raman scattering (SERS), and surface plasmon resonance (SPR) aptasensors depending on the corresponding applied optics (Fig. 7). Basic working principles and a selection of recently reported examples for infectious pathogen detection are summarized in this section.
Fig. 7.
Schematic illustration of representative optical aptasensors. (a) Working principle of an ELONA aptasensor. Colorless substrate TMB is oxidized by enzymes like HRP to generate a blue color. (b) Working principle of a AuNPs-based colorimetric aptasensor. Released AuNPs aggregate in the presence salt accompanied by a color change from red to purple. (c) Working principle of a fluorescent aptasensor. The fluorescence of the fluorophore is recovered due to inhibition of FRET upon target binding. (d) Working principle of a SERS aptasensor. Raman intensity is amplified via the formation of a sandwich style complex. (e) Working principle of a SPR aptasensor. Aptamer-target binding leads to the variation of refractive index
Colorimetric aptasensors
Colorimetric detection allows direct analysis of samples by visual observation with naked eyes. This “instrumental-free” approach is cost-effective and user-friendly, which can be applied in basic clinical laboratories. The most widely adopted colorimetric technique is ELISA, which is a standard method in the diagnosis of various microbial or viral infections [6, 7]. Specific antibodies are immobilized on the surface of solid substrate to capture target antigens, followed by complexation with another antibody tagged enzymes. Those enzymes can promote the oxidation of a colorless substrate to its colored product. Therefore, the corresponding immune response can be determined by colorimetric output signals. As alternatives to antibodies, aptamers have been utilized as the bio-recognition elements in enzyme-linked oligonucleotide assay (ELONA) (Fig. 7a), which is also known as enzyme-linked aptamer assay (ELAA) or enzyme linked aptamer sorbent assay (ELASA).
Two highly specific aptamers against Salmonella enteritidis (S. enteritidis) were selected via cell-SELEX and employed for the development of a sandwich type aptamer-based colorimetric capillary detection platform [93]. Poly enzyme-horseradish peroxidase (HRP) was used to promote the H2O2-mediated oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB) for color generation. Capturing aptamers were covalently immobilized to the inner surface of capillary to bind with target cells, followed by the attachment of signaling aptamers. The detection limit was determined to be 103 CFU mL−1 by absorbance measurements.
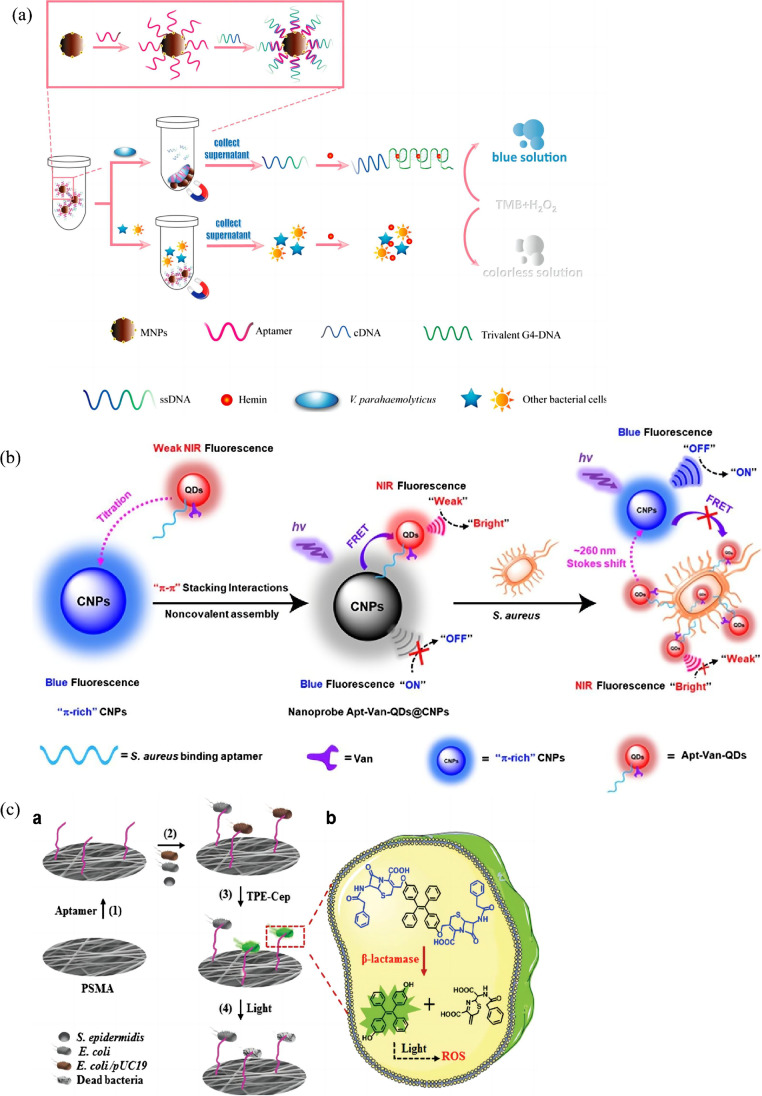
Although highly efficient, HRP is a natural enzyme with some limitations such as high-cost, inherent instability, and sensitivity to environmental interferences [94]. Therefore, various artificial enzyme peroxidase mimetics have been developed as substitutions to HRP. Wu et al. reported the preparation of ZnFe2O4-reduced graphene oxide (ZnFe2O4/rGO) nanostructures as an effective enzyme mimetics for the detection of S. typhimurium [95]. Aptamer (on microplate)-target-aptamer-ZnFe2O4/rGO sandwich complexes were formed via aptamer-target recognition, exhibiting a detection limit of 11 CFU mL−1 in buffer solution. Au@Pd nanoparticles could also function as enzyme mimetics for colorimetric determination of Campylobacter jejuni (C. jejuni) in milk samples [96]. Originally, a large number of aptamers specific for C. jejuni were floated in the solution. Au@Pd nanoparticles added to the solution were covered with free aptamers via electrostatic interactions, which hindered the reactivity of the catalyst for TMB oxidation. Aptamers bound with target cells were released from the nanoparticles to recover the peroxidase-like activity of the catalyst, inducing an obvious color change to blue. The intensity of blue color was quantified by absorbance spectroscopy, and the detection limit for C. jejuni was 100 CFU mL−1 in milk. Sun et al. established a colorimetric aptasensor based on G-quadruplex DNAzyme for the determination of Vibrio parahaemolyticus (V. parahaemolyticus, Vp), a widespread foodborne pathogen causing food poisoning derived from seafood [97]. Vp aptamers were immobilized on magnetic nanoparticles (MNPs) as capture probes, and label-free ssDNA containing both the complementary DNA (cDNA) sequence of Vp aptamer and a CatG4 sequence was partly hybridized with the aptamer. Upon exposure to Vp, ssDNA dissociated from the aptamer to form trivalent DNAzyme via the interaction of CatG4 with hemin. TMB solution containing H2O2 was then added to the supernatant for colorimetric output signals (Fig. 8a). The detection limit was determined to be as low as 10 CFU mL−1.
Fig. 8.
(a) Schematic illustration of the colorimetric aptasensor for Vp. Reproduced with permission from [97], Copyright 2019 American Chemical Society. (b) Dual-recognition ratiometric fluorescent nanosensor for S. aureus. Reproduced with permission from [112], Copyright 2020 American Chemical Society. (c) Schematic illustration of bacterial capture and photodynamic destruction on fibrous strips. Reproduced with permission from [119], Copyright 2020 Royal Society of Chemistry
RCA was employed as a signal enhancement approach in enzyme-linked aptasensor for colorimetric detection of Listeria monocytogenes (L. monocytogenes) [98]. L. monocytogenes-specific aptamer bound to biotin probe 1 (BP1) was immobilized on the surface of a microplate. In the presence of target bacteria, BP1 was released to hybridize with the RCA probe that was complementary to BP1 for the initiation of the RCA reaction. The RCA process produced copies of the ssDNA (cssDNA) to form complexes with biotin-probe 3 (BP3), followed by the addition of streptavidin labeled HRP (SA-HRP) to generate SA-HRP ~ BP3 ~ cssDNA complexes, which was responsible for the oxidation of the enzyme substrate for colorimetric detection. The competition-based assay showed a limit of detection of 4.6 × 102 CFU mL−1 in pure culture, which was three orders of magnitude higher than that without RCA signal enhancement. Real sample analysis of spiked fresh lettuce showed a detection limit of 6.1 × 103 CFU g−1.
AuNPs have been intensively applied in colorimetric assays, serving as color indicators. When aptamers adsorb onto the surface of AuNPs via electrostatic interactions, the addition of salt cannot induce the aggregation of AuNPs. Upon disassociation of the aptamers, the released bare nanoparticles could form salt-induced aggregates to result in a color change from red to purple, which can be easily differentiated by naked eye (Fig. 7b). By applying this phenomenon, an aptamer-based assay was described for the detection of S. typhimurium [99]. The absorbance at 550 nm increased linearly with the logarithm of the bacteria concentration ranging from 100 to 109 CFU mL−1, exhibiting a detection limit of 16 CFU mL−1. Similar strategy was employed for the determination of Shigella flexneri in food samples [100]. The sensing platform was capable of simple, convenient, and on-site detection at a concentration as low as 80 CFU mL−1 within 20 min. A two-stage colorimetric sensing platform was established for the detection of Cronobacter sakazakii (C. sakazakii) in powdered infant formula [101]. Aptamers were originally bound with target bacteria, and followed centrifugation, the supernatant containing unbound aptamers was incubated with AuNP solution. By addition of salt solution, the color and spectral change were observed by naked eye or with a spectrometer. Compared with conventional single stage method, this platform could minimize the interference factors that might inhibit salt-induced AuNPs aggregation. C. sakazakii in powdered infant formula at a concentration of 7.1 × 103 CFU mL−1 could be determined by naked eye within 30 min. A hybrid nanomaterial-based immunosensor was developed for the detection of Dengue virus [102]. Super paramagnetic nanoparticles γ-Fe2O3 named surface-active maghemite nanoparticles (SAMNs) were modified with 3-mercaptopropionic acid (MPA) to form a self-assembled monolayer with thiol groups to bind with AuNPs, followed by covalent linkage of aptamers with thiolated ends. Colorimetric tests were performed at each stage of material fabrication, and a color change from red to dark purple was observed for AuNPs and SAMNs@MPA@AuNPs@aptamer complex, respectively, due to the increase of the local refractive index on the surface of AuNPs. A pool containing four dengue serotypes was added to the complex for conjugation with aptamers, inducing further color change to green owing to the reduction of the surface area of AuNPs caused by aptamer-target binding. Interfering analysis was carried out to rule out possible false positive diagnoses from other viruses of the same genus such as zika virus (ZIKV) and yellow fever virus (YFV).
Lateral flow assay is a strip paper-based platform with great convenience for rapid POCT due to the low development costs and ease of production [103, 104]. Kim et al. applied a cognate pair of aptamers screened by graphene oxide SELEX (GO-SELEX) on lateral flow strips for sandwich-type detection of avian influenza H5N2 whole virus particles for the first time [105]. Aptamer J3APT was used as a capturing aptamer for test line, and a secondary aptamer JH4APT labeled with AuNPs was used as a reporter. The detection limit for H5N2 virus (H5N2/K08-404) was estimated to be 1.27 × 105 EID50 mL−1 and 2.09 × 105 EID50 mL−1 in buffer and spiked duck’s feces, respectively. The assay exhibited remarkable specificity with no significant signals for other subtype species due to the cognate pair of aptamers.
Fluorescent aptasensors
A wide variety of fluorophores and fluorescent quenchers can be tagged onto oligonucleotides during aptamer synthesis, providing versatile labeled fluorescent aptasensors [106, 107]. The recognition information between aptamers and target molecules is monitored in real time through changes of fluorescence intensity as output signals. The competition between target molecules with quencher-tagged complementary DNA (cDNA) sequences to bind with fluorophore tagged aptamers is a commonly employed format in the design of fluorescent aptasensors. Originally, aptamers hybridize with their cDNA sequences, and the fluorescence of the fluorophore that serves as the energy donor can be quenched by the energy accepting quencher through Förster resonance energy transfer (FRET) upon light excitation. The separation of the fluorophore from adjacent quencher due to target binding and cDNA disassociation interferes with the FRET process, leading to the recovery of fluorescent emission as detectable signals (Fig. 7c) [108].
Based on the abovementioned DNA hybridization strategy, a FRET-based fluorescent aptasensor was engineered for the detection of P. aeruginosa [109]. The hybridized complex of P. aeruginosa-specific aptamer and 5-carboxyfluorescein labeled cDNA (FAM-cDNA) was adsorbed onto the surface of graphene oxide quantum dots (GOQDs) through the single-stranded flanking region in the aptamer, so that the fluorescence of FAM was quenched by GOQDs owing to the FRET. Aptamer-target binding led to the disassociation of FAM-cDNA from GOQDs, and the fluorescence was recovered due to the inhibition of fluorescence quenching. The aptasensor showed a limit of detection of 100 CFU mL−1 with a detection time of 2 h toward P. aeruginosa. By employing MNPs, the same research group developed another fluorometric assay for the detection of P. aeruginosa [110]. This aptasensor was based on the hybridization of aptamer-modified MNPs and fluorescein-labeled complementary DNA (FAM-cDNA). Magnetic separation was employed for the enrichment of aptamer-target complexes, and the number of bacteria could be quantified by the fluorescence intensity of the released FAM-cDNA in the supernatant. The detection process took less than 1.5 h with a detection limit as low as 1 CFU mL−1. Cui et al. constructed an ultrasensitive sensing platform for Staphylococcus aureus (S. aureus) based on self-assembled fluorescent carbon dots (CDs) and Fe3O4 nanoparticles [111]. S. aureus-specific aptamers were immobilized on the surface of Fe3O4 to form a nano-dimer with CDs modified cDNA, and the fluorescence of CDs was quenched via the FRET process, in which CDs acted as energy donors and Fe3O4 as acceptors. In the presence of S. aureus, the fluorescence of CDs was regenerated, and the detection limit was determined to be 8 CFU mL−1. A dual-recognition ratiometric fluorescent nanosensor was reported, exhibiting a remarkably large stoke shift for accurate tracking of pathogenic bacteria at the single-cell level [112]. In this system, blue fluorescent π-rich electronic carbon nanoparticles (CNPs) acted as the energy donor, and upon titration of the near-infrared (NIR) fluorescent QD tagged with aptamer and broad-spectrum glycopeptide antibiotic vancomycin (Apt-Van-QDs), efficient energy transfer happened from the blue fluorescent energy donor to NIR Apt-Van-QDs (energy acceptor), leading to the quenching of the emission at ∼465 nm and the emergence of the emission at ∼725 nm. The FRET process was disrupted in the presence of target bacteria S. aureus, and the emission of complex Apt-Van-QDs@CNPs switched to the blue channel with a stoke shift of ∼260 nm (Fig. 8b). The nanoprobe showed an ultrahigh specificity and single-cell level sensitivity (detection limit = 1.0 CFU/mL) for ratiometric fluorescence detection of S. aureus.
Lanthanide-doped upconversion nanoparticles (UCNPs) possess great potential as alternatives to traditional organic fluorophores for their excellent photo-stability, low cytotoxicity, and high signal-to-noise ratio [113, 114]. A novel detection platform for E. coli ATCC 8739 was developed based on the FRET between UCNPs (as the donor) and AuNPs (as the acceptor) [115]. Dissociation of UCNPs-cDNA from AuNPs-aptamers resulted in the recovery of upconversion fluorescence. The biosensor showed a detection limit of 3 CFU mL−1.
As another distance-dependent fluorescence assay technology, protein-induced fluorescence enhancement (PIFE) offers a promising alternative to FRET for higher spatial resolution and specificity to probe protein-nucleic acid interactions [108, 116]. The response distance for PIFE is within a 0–30 Å range, shorter than that in FRET (10–100 Å). Cy3 is preferentially to lock into its trans conformational state in close proximity to a protein, resulting in significant fluorescence enhancement [117]. Based on PIFE, a Cy3-labeled aptamer was used to monitor the protein binding with the nucleocapsid proteins (NP) of SARS-CoV-2 [118]. The detection limit of the assay was calculated to be 2.5 ng for N protein.
Electrospun fibrous strips integrated with capture aptamers and aggregation-induced emission (AIE) probes were utilized for fluorescent trace sensing and photodynamic destruction of antibiotic-resistant E. coli [119]. AIE probes were synthesized from the linkage of two cephalosporin moieties to hydroxyl tetraphenylethene (TPE), followed by the conjugation onto electrospun fibers. The fluorescence emission of the probe was turned on only in the presence of b-lactamase, a critical marker for screening antibiotic-resistant bacteria. The apparent color change from blue to green was given as visual readout of bacterial levels, and the detection limit was determined to be 60 CFU mL−1 toward E. coli/pUC19. In the meantime, captured bacteria could be efficiently destroyed by the reactive oxygen species (ROS) generated by the AIE probes under room light illumination (Fig. 8c).
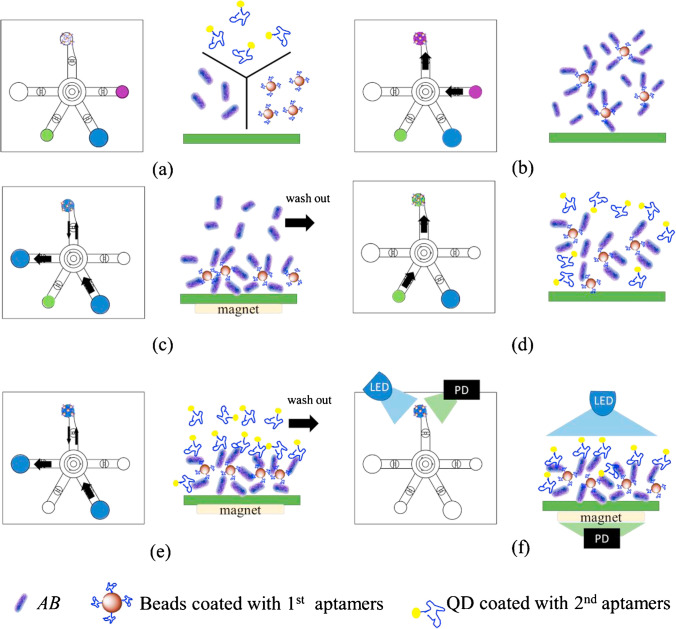
Microfluidic platforms have also been integrated for the fluorescent assay of infectious pathogens. A microfluidic chip with femtoliter-sized wells was fabricated for sensitive fluorescence-based detection of SARS-CoV2 NP [120]. An aptamer/antibody sandwich method was applied, forming β-galactosidase (β-Gal)-linked antibody/NP/aptamer immunocomplexes on magnetic beads. The magnetic beads and β-Gal substrate were injected into the wells of the microfluidic chip, and the fluorescence images were monitored by a conventional inverted fluorescent microscope, showing a detection limit of 33.28 pg/mL for NP. Lee’s research group demonstrated a digital microfluidic platform utilizing ELISA-like assay for influenza A H1N1 virus detection [121]. Magnetic beads served as three-dimensional substrates for H1N1-specific aptamers to capture the virus, while an HRP-conjugated antibody was used to form sandwich complexes with activate tyramide-tetramethylrhodamine (TTMR) to generate amplified fluorescent signals. Droplets containing magnetic beads were driven by electromagnetic force to automate the entire detection process within 40 min. The experimental detection limit was determined to be 0.032 HAU, which was of enough sensitivity for clinical diagnosis of influenza A H1N1 virus. The same group developed another integrated microfluidic device was for the diagnosis of Acinetobacter baumannii (AB), a lethal multidrug-resistant pathogen mainly responsible for nosocomial infections [122]. Replacing antibodies with a pair of aptamers, the dual-aptamer assay was constructed by attaching one aptamer to the magnetic beads for the collection bacterial cells, while a secondary aptamer tagged with quantum dot (QD) functioned as a quantitative tool (see Fig. 9 for detailed assay). The electromagnetically driven microfluidic system featuring light-emitting diode (LED)-induced fluorescence modules exhibited a limit of detection of 100 CFU/reaction within a detection time of 30 min. The whole detection process could be carried out automatically without human intervention or external equipment.
Fig. 9.
Dual aptamer assay for the detection of AB. (a) Bacterial samples and reagents were loaded into the corresponding chambers. (b) Magnetic beads and bacteria were pumped into the reaction chamber by a micropump and mixed by a micromixer. (c) Unbound materials were washed away with wash buffer while applying an external electromagnetic field. (d) Bead-bacteria complexes and QD were mixed by a micromixer. (e) Excessive QD was washed away with wash buffer while applying an external electromagnetic field. (f) Fluorescent signals were excited by LED and detected by photodiodes (PD). Reproduced with permission from [122], Copyright 2020 Elsevier
SERS aptasensors
SERS effect refers to the phenomenon of an exceptional increase in Raman cross-section of adsorbed molecules than that of the normal Raman scattering signal due to the enhancement of electromagnetic field on nanostructured metal–dielectric surfaces [123]. SRES overcomes the limitation of the low sensitivity of normal Raman spectrum and represents a powerful tool to analyze the change of adsorption state of molecules on nanostructured metal surfaces [124]. The combination of SERS-based strategies with aptamer-based capture biomolecules holds significant potential for rapid, real-time, and on-site pathogen detection [125–127].
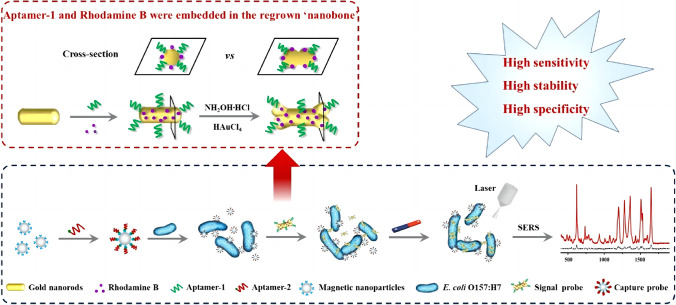
SERS aptasensing strategy is a label-based approach. Typically, aptamers are immobilized onto the surface of metallic nanoparticles as capturing probes to bind with target molecules, followed by the formation of a sandwich style complex with the secondary aptamers labeled with Raman reporters such as 4-mercaptobenzoic acid (4-MBA) and 5,5-dithiobis-(2-nitrobenzoic acid) (DTNB). The recognition information could be monitored through the changes of the Raman spectra of the labels (Fig. 7d). Employing a sandwich strategy, a SERS aptasensor was achieved for highly sensitive detection of influenza virus [128]. Aptamer RHA0385 was utilized as the primary aptamer to attach onto metal particles of a SERS substrate for the recognition of H1, H3, and H5 influenza virus subtypes, and a secondary aptamer was labeled with Raman-active molecules for signal detection upon binding with target viruses. The sensor showed a limit of detection of 10−4 HAU per sample, which was significantly lower than the values of other commonly used techniques for influenza virus detection. Applying a similar sandwich assay, a SERS-based aptasensor for quantitative detection of E. coli O157:H7 was developed, exhibiting a detection limit of 3 CFU mL−1 [129]. The high sensitivity resulted from applying a combination of MNPs-based capture probe and gold nanobones (NBs)-based signal probe (GNRApt‑1+RhB) prepared from gold nanorods (GNRs) comediated by an aptamer (Apt-1) and signal molecule rhodamine B (RhB) (Fig. 10). The capture probe could specifically bind with target bacteria via the affinity of Apt-2, followed by the formation of a sandwich style complex with the signal probe to produce a greatly enhanced Raman intensity, which was derived from the strong electromagnetic field distribution with the locations at the apex of both ends of the GNRApt‑1+RhB.
Fig. 10.
Schematic illustration of the SRES aptasensor for the detection of E. coli O157:H7. Reproduced with permission from [129], Copyright 2020 American Chemical Society
The substrate of SERS-based aptasensor is critical for aptamer adsorption and sensitivity optimization through both electromagnetic and chemical mechanisms [130]. Toward the aim of signal enhancement, novel SERS substrates such as graphene oxide-wrapped Fe3O4@Au nanostructures [131] and gold-coated polydimethylsiloxane film [132] were developed for the enhancement of Raman scattering to determine Vp. For sensitive detection of Vp, a hetero-sandwich-based SERS method was described in conjunction with the RCA signal amplification strategy [133]. Antibodies were immobilized on the microplate to capture the target bacteria, followed by binding with silver-coated AuNPs fabricated by aptamer-included ssDNA probes. The DNA probes could also serve as primers of the RCA process to initiate the assembly of metallic nanoparticles, creating excess “hot-spot” sites for Raman measurement. The achieved sensor provided a detection limit of 1 CFU mL−1 for Vp and was successfully applied to determine Vp in spiked food samples showing satisfactory sensitivity and specificity. A dual-recognition platform for the simultaneous detection of two pathogenic bacteria, E. coli and S. aureus, was constructed based on the dual recognition by vancomycin and aptamers [134]. Vancomycin, a broad-spectrum glycopeptide antibiotic, was incorporated on Fe3O4@Au nanoparticles as a universal bacteria capturer to efficiently enrich target bacteria, while aptamer-modified AuNPs with two different types of SERS tags were employed for the specific and sensitive quantification of target bacteria. The platform exhibited a detection limit of 20 and 50 cells mL−1 for S. aureus and E. coli, respectively.
SPR aptasensors
SPR sensor is developed upon the basis of surface plasmon resonance absorption. The resonance frequency or resonance angle of surface plasmons changes with the variation of refractive index of the adjacent medium bound on the metal surface, so that the measurement of SPR frequency or angle could indicate the reaction of biomolecules adsorbed on the substrate surface [135]. As for a SPR aptasensor, aptamers are adsorbed on the metal surface. Target binding increases the thickness of the surface, leading to the change of the refractive index (Fig. 7e). Similar to QCM and SAW sensors, SPR sensor is also mass sensitive. From this perspective, SPR sensor can also be classified as acoustic sensors. Our classification here is based on the working principle, that ultimately, surface plasmon resonance is an optical phenomenon. SPR sensor provides a label-free detection platform for a wide variety of target molecules including pathogens [136].
The sensitivity and selectivity of SPR-based sensors are generally lower than some other labeled optical techniques such as fluorescent sensors, and these shortcomings have been significantly improved in recent years by the incorporation of AuNPs and the sandwich assay involving a pair of recognition units. Nguyen et al. screened and characterized a series of aptamers against whole H5Nx avian influenza viruses using a multi GO-SELEX method [137]. Aptamers IF10 and IF22, which could bind H5N1 virus simultaneously at different site of the same virus, were applied to develop a sandwich-type AuNPs-based SPR aptasensor. The angel shift of the sensor using a 2nd aptamer conjugated with AuNPs was enhanced by 50-fold, compared to those using single aptamer or 2nd aptamer without modification. By applying a pair of aptamers with a sandwich-type signal enhancing technique, the sensor exhibited high sensitivity toward H5N1 whole virus with a detection limit of 200 EID50/mL. Employing a similar strategy, norovirus capsid protein detection at attomolar concentration was achieved by using a gold nanorod enhanced surface sandwich assay [138]. The sensitivity was found to be 105 times higher than that of sandwich platform without any gold nanorod particles. Wang et al. investigated the binding reactions between aptamers and outer membrane proteins on the surface of S. typhimurium at single-cell level using SPR and atomic force microscopy (AFM) [139]. Two DNA aptamers were used for the label-free detections of S. typhimurium, and the high-throughput biosensor exhibited a detection limit of 3 × 104 CFU mL−1. AFM topography and recognition images of the bacteria was obtained by attaching aptamer APT33 to AFM tip and on modified Au (111) surface. This study provided some fundamental insight for a better understanding of the binding activities between aptamers and bacteria on a biosensor surface.
In the last few years, various plasmonic nanoparticle-based localized surface plasmon resonance (LSPR) biosensors have been established and applied in biotechnological and clinical assay for their inherent label-free nature, portability, low cost, and real-time sensing capacity [140]. A portable plasmonic biosensor-based device was developed for ultrasensitive and selective detection of S. typhimurium in pork meat samples [141]. AuNP monolayer with a regulated diameter of 20 nm was deposited on a transparent glass substrate to produce longitudinal wavelength extinction shifts via a LSPR signal. Aptamers were conjugated onto the chip by a simple dipping adsorption method. The plasmonic-active biodevice exhibited quantitative detection of the bacteria with an upper detection limit of 104 CFU/mL in pure culture as well as in artificially contaminated pork meat samples within 30–35 min. No pre-enrichment step was needed for spiked pork meat samples.
Instead of being the target, pathogenic bacteria were employed as recognition moieties to detect lysozyme via a SPR-based system [142]. Micrococcus lysodeikticus (M. lysodeikticus) whole cells were adsorbed on graphene oxide (GO)-coated SPR interfaces. As M. lysodeikticus is a typical enzymatic substrate for lysozyme, the cell walls of bacteria were destroyed upon exposure to the lysozymes in serum, followed by cell detachment from the GO surface to result in obvious changes in the SPR signal. The sensor showed a detection limit (0.05 μg mL−1) suitable for clinical use in undiluted serum.
Other aptasensors
Microcantilever array sensing technology provides another analytical platform featuring label-free detection, high scalability, low sample volumes, and short response time. Generally, a micron-sized rectangular cantilever beam is coated with receptor molecules on one side, and the binding with target molecules could induce the dynamic response change of microcantilevers [143]. Piezoelectric microcantilevers, operated in vibration mode (oscillation), were demonstrated to detect hepatitis C virus (HCV) helicase using RNA aptamers as receptor molecules. The engineered RNA aptamers provided sufficient surface stress for the ligand-binding to RNA aptamers on the microcantilever surface, which enabled the label-free detection of proteins at low concentrations (100 pg/mL) [144].
A gravity-based method was applied for rapid visual detection of flu viruses. RNA aptamers specific for different strains of human influenza virus were assembled onto gold nanoparticles that subsequently formed a gold nanoshell (AuNS) around the viral envelope. This AuNS could be visualized by transmission electron microscopy or sedimentation with a low-cost centrifuge to detect 3 × 108 virus particles with naked eye [145].
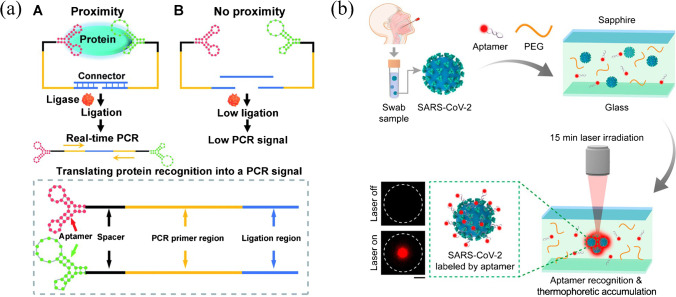
An aptamer-assisted proximity ligation assay (Apt-PLA) was reported as a generalizable method for the detection of serum COVID-19-associated antigens (Fig. 11a) [146]. The binding of two aptamer probes to the same SARS-CoV-2 N protein could bring the ligation DNA region into close proximity, thereby initiating ligation-dependent qPCR amplification to generate a significant qPCR signal. A limit of detection of 30.9 pg mL−1 for N protein in spiked human serum was obtained within a workflow of 2 h.
Fig. 11.
(a) Scheme of aptamer-assisted proximity ligation assay for COVID-19 antigens. Reproduced with permission from [146] under a CC BY license, Copyright 2020 The Royal Society of Chemistry. (b) Schematic of the one-step aptamer-based thermophoretic assay for rapid detection of SARS-CoV-2 viral particles. Reproduced with permission from [147], Copyright 2021 American Chemical Society
Aptamers can also be adapted to thermophoretic sensors to provide a homogeneous approach for quantitative analysis of biomolecules in a temperature gradient. The binding of aptamers with their target would give rise to differences in thermophoretic movement between the aptamer-bound complex and non-binding aptamer [147]. A one-step thermophoretic assay was described for rapid and quantitative detection of pseudotyped SARS-CoV-2 viral particles (Fig. 11b). Aptamers specific for the S protein of SARS-CoV-2 were labeled with fluorescent Cy5 dyes, and under a temperature gradient induced by localized infrared laser heating, high-affinity binding of aptamers to the S protein and PEG-enhanced thermophoretic accumulation resulted in an enhanced fluorescence intensity, indicating the level of S protein on viral particles. A limit of detection of 170 particles μL−1 (26 fM of the spike protein) was achieved in 15 min.
Aptasensors for POCT
A timely and reliable diagnosis is critical for the treatment and control of infectious diseases. POCT, or bedside testing, allows a quick and accurate analysis of the patient with minimum human intervention in hospitals, resource-constrained clinics, and communities [11, 148, 149]. The “ASSURED” criteria, i.e., Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free, and Deliverable to end-users, can be used as a guideline to aid the selection of diagnostic tests in resource-constrained settings [150]. As a vast number of aptasensors have been established, the integration of aptamers as molecular recognition elements into lab-on-a-chip devices to construct POCT facilities seems a promising approach for rapid diagnosis of infectious diseases [151, 152]. The employment of aptamers in POCT would enable discrimination and real-time monitoring of disease-causing pathogens for subsequent personalized therapy. Some LSPR, paper-based lateral flow assay, electrochemical and microfluidic aptasensing platforms listed in the “Aptasensors for pathogen detection” section have showcased their potential for the development of POCT devices as proof-of-concept models [75, 85, 105, 118, 120–122, 141]. In this section, we would like to highlight some ready-to-use devices, focusing on addressing the urgent needs of currently prevalent COVID-19, and the brief details of the selected aptamers are summarized in Table 3 for reference.
Table 3.
Summarized details of the selected aptamers for POCT
| POCT platform | Target | Aptamer selection method | Name of aptamer (type) | Sequence (from 5′ to 3′) | Ref |
|---|---|---|---|---|---|
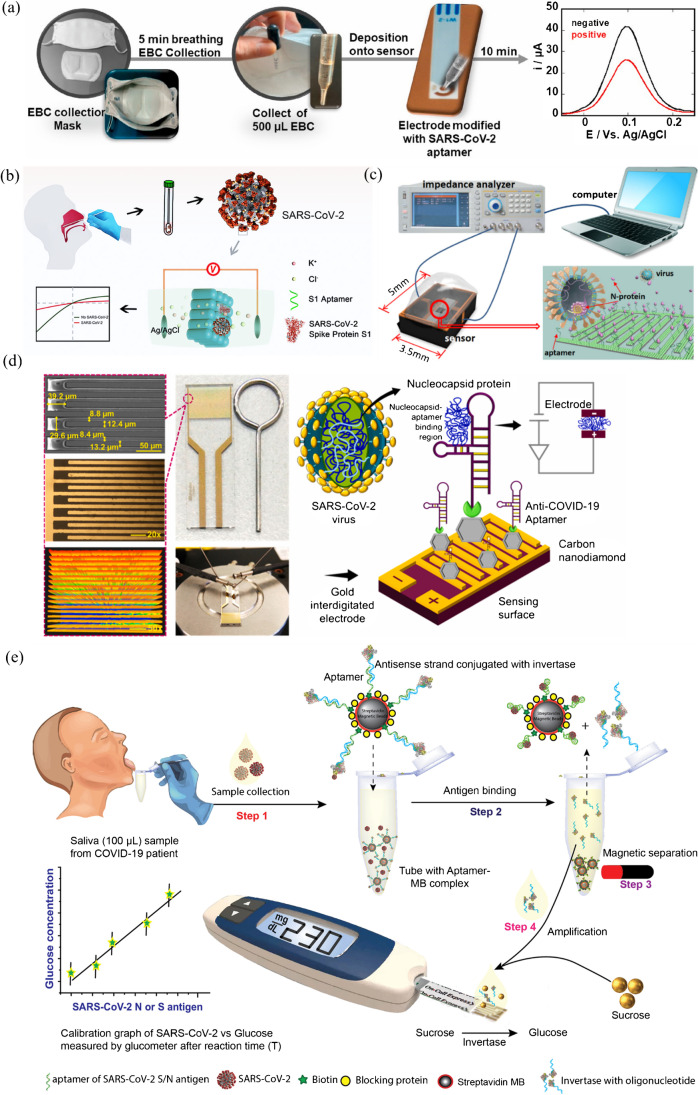
| Mask-based electrochemical platform | SARS-CoV-2 | S1 protein-based SELEX | CFA0688T (DNA) | BasePairBio Inc | [158] |
| Nanochannel-based electrochemical platform | SARS-CoV-2 | S1 protein-based SELEX | XN-268 s (DNA) | CACGCATAACCGAGCTGGGGTGGGGTAGTGGTATGGAGCGTCAGTTGTTATGCGTG | [159] |
| Microelectrode array chip-based platform | SARS-CoV-2 | N protein-based SELEX | A48 (DNA) | GCTGGATGTCGCTTACGACAATATTCCTTAGGGGCACCGCTACATTGACACATCCAGC | [160] |
| Gold interdigitated electrode-based platform | SARS-CoV-2 | N protein-based SELEX | NP aptamer (DNA) | GCAATGGTACGGTACTTCCGGATGCGGAAACTGGCTAATTGGTGAGGCTGGGGCGGT | [161] |
| Glucometer-based platform | SARS-CoV-2 | N protein-based SELEX | NP aptamer (DNA) | GCAATGGTACGGTACTTCCGGATGCGGAAACTGGCTAATTGGT GAGGCTGGGGCGGT | [162] |
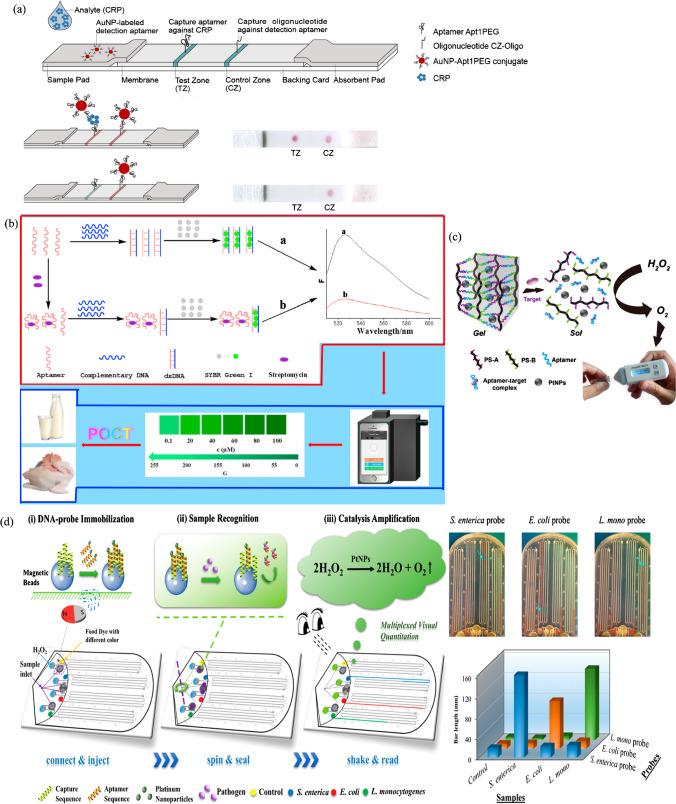
| Lateral flow assay-based platform | C-reactive protein | Microfluidic technology-assisted SELEX | Apt1 (DNA) | GGCAGGAAGACAAACACGATGGGGGGGTATGATTTGATGTGGTTGTTGCATGATCGTGGTCTGTGGTGCTGT | [177] |
| Smartphone-based platform | Streptomycin | SELEX | APT (DNA) | TAGGGAATTCGTCGACGGATCCGGGGTCTGGTGTTCTGCTTTGTTCTGTCGGGTCGTCTGCAGGTCGACGCATGCGCCG | [179] |
| Smartphone-based platform | Mycotoxin (ochratoxin A) | SELEX | 1.12.2 (DNA) | GATCGGGTGTGGGTGGCGTAAAGGGAGCATCGGACA | [180] |
| Pressuremeter-based platform; microfluidic paper-based platform | Cocaine and ochratoxin A | SELEX | Aptamer (DNA) | GGGAGACAAGGATAAATCCTTCAATGAAGTGGGTCTCCC (cocaine); GATCGGGTGTGGGTGGCGTAAAGGGAGCATCGGACA (ochratoxin A) | [183, 184] |
| Multiplexed bar-chart chip-based platform | S. enterica, E. coli, and L. monocytogenes | Cell-SELEX | Aptamer (DNA) | TATGGCGGCGTCACC CGACGGGGACTTGACATTATGACAG (S. enterica); CCGGACGCTTATGCC TTGCCATCTACAGAG CAGGTGTGACGG (E. coli); TACTATCGCGGAGACAGCGCGGGAGGCACCGGGGA (L. monocytogenes) | [185] |
COVID-19
Currently, two standard screening methods for SARS-CoV-2 are reverse transcription PCR (RT-PCR) and serological tests (detection of antibodies) [153, 154]. The molecular diagnosis of COVID-19 is surrounded by some unneglectable issues, including false negative results from the detection of viral RNA, temporal variations of viral loads, selection and treatment of specimens, and limiting factors in detecting viral proteins [155]. There are urgent needs and tremendous opportunities for the development of alternative diagnostic techniques, especially for POCT methods [156]. To date, a considerate number of aptamers have been selected for diagnostic and therapeutic purposes toward COVID-19, which are ideal candidates to be functionalized as recognition moieties in corporation with versatile POCT techniques [157].
Electrochemical aptasensors are advantageous for low cost, rapid response, readily amenable for miniaturization, convenient for real-time and on-filed detection, which are desired qualities for the development of POCT devices [68, 69]. A mask-based diagnostic platform integrating an electrochemical aptasensor for SARS-CoV-2 was developed as a POCT method for COVID-19 screening [158]. SARS-CoV-2 aptamers targeting the S1 protein were immobilized onto SPEs with ferrocenemethanol as a redox mediator. Exhaled breath condensate (EBC) was readily collected by a mask-based sampling device, followed by screening with the biosensor which detected SARS-CoV-2 viral particles down to 10 PFU mL−1 in cultured SARS-CoV-2 suspensions. Results about the infectivity state of the patient could be obtained in 10 min (Fig. 12a). However, the sampling process was directly correlated to viral load in breath and breathing of the patient, and the sampling step needs further modification to overcome some false negative issues. In another example, a SARS-CoV-2 spike S1 protein specific aptamer was screened via SELEX and immobilized into the sub-nanometer channels of porous anodic alumina structures [159]. Target binding resulted in increased steric hindrance and decreased charge density, which affected the ion transport through the nanochannels along with a rapid electrochemical response (Fig. 12b). The aptasensor exhibited an ultrahigh detection sensitivity to S1 protein (down to 1 fM level) and was successfully applied to detect the SARS-CoV-2 viral particles in clinical pharyngeal swab specimens. Based on a microelectrode array (MEA) chip modified with SARS-CoV-2 NP-specific aptamers, a microfluidics-coupled capacitive sensor was developed for real-time, label-free SARS-CoV-2 screening and diagnosis (Fig. 12c) [160]. The limit of detection reached an ultralow level of femtogram per milliliter attributed to the ultrasensitive solid − liquid interface capacitance indicator, and by employing a microfluidic enrichment method, the response time of the sensor was shortened to only 15 s. Aiming at developing portable biosensors, a 10-μm gap-sized gold interdigitated electrode (AuIDE) aptasensing platform was presented for ultrasensitive detection of SARS-CoV-2 (Fig. 12d) [161]. Its sensitivity was enhanced by depositing carbon nanodiamond on the sensing surface. The aptasensing was evaluated by electrochemical impedance spectroscopy measurements, and the detection limit was evidenced as 0.389 fM for NP in spiked human serum.
Fig. 12.
Selected examples of candidate POCT platforms for SARS-CoV-2. (a) Exhaled breath condensate-based diagnostic strategy for SARS-CoV-2 infectivity. Reproduced with permission from [158], Copyright 2021 Elsevier. (b) Aptamer-functionalized nanochannels for one-step detection of SARS-CoV-2. Reproduced with permission from [159], Copyright 2021 American Chemical Society. (c) Conceptual illustration of the sensor, measurement system, and NP capturing. Reproduced with permission from [160], Copyright 2022 American Chemical Society. (d) Diamond enhanced AuIDE for the detection of SARS-CoV-2 NP. Reproduced with permission from [161], Copyright 2022 Elsevier. (e) Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Reproduced with permission from [162], Copyright 2021 Elsevier
Commercially available glucometers can be repurposed to detect a variety of non-glucose-based targets including infectious pathogens. These meters are portable, inexpensive, and user-friendly. Singh et al. reported a point-of-care salivary antigen testing for SARS-CoV-2 viral antigen with an off-the-shelf glucometer [162]. An aptamer-based competitive assay was conducted based on aptamer-oligo-invertase complex preassembled on magnetic beads. In the presence of the cognate antigen, antigen-sensitive switch occurred, and after magnetic separation, the released enzyme could hydrolyze sucrose into glucose, which allowed signal readout with a glucometer (Fig. 12e). This assay could clinically discriminate infected and non-infected individuals with low-cost reagents ($3.20/test) and an unmodified glucometer ($29). As for the glucometer, issues on optimization of aptamer binding conditions, enzyme selection, and increasing shelf life need to be addressed prior to clinical approval.
The ongoing pandemic of COVID-19 has roused a surging increase in the development of aptasensors for the detection of SARS-CoV-2. Besides the abovementioned examples, there have also been a considerable number of excellent examples presented during the last 2 years, mainly employing electrochemical [163–167], photoelectrochemical [168, 169], and optical [170–174] aptasensing strategies. Many of these aptasensors were capable of detecting viral proteins/particles with similar or better sensitivities than those of the conventional assays within an hour or even in real time. Device miniaturization and incorporation with lateral flow assays, microfluidic platforms, or smartphones would further promote the implementation of these aptasensors into POCT utilization [11, 149, 175].
Other selected examples of anti-infective POCT devices
Lateral flow assays are well-acknowledged potent POCT approaches, especially in resource-limited settings owing to the advantages of ease of use, low cost, test rapidity, disposable format, and low sample volume required [176]. The high inter-batch variability and relatively low accuracy due to the use antibodies could be addressed by employing synthetic aptamers as recognition molecules. A ready-to-use lateral flow assay was developed for detecting inflammation/infection biomarker C-reactive protein (CRP) using microarray technology as a prescreening platform (Fig. 13a) [177]. CRP-specific aptamer was immobilized on microarray chips in sandwich format for lateral flow assay, and the cutoff for CRP detection was 10 mg/L for the determination of infections/inflammation (≥ 10 mg/L CRP) in human patient samples.
Fig. 13.
(a) Schematic illustration of an aptamer-based lateral flow assay for detection of CRP. Reproduced with permission from [177], Copyright 2020 American Chemical Society. (b) A POCT method for on-site and visual detection of streptomycin by smartphones. Reproduced with permission from [179], Copyright 2018 Elsevier. (c) Target-responsive hydrogel pressure-based assay for POCT. Reproduced with permission from [183], Copyright 2017 American Chemical Society. (d) A multiplexed bar-chart chip for visual quantitative detection of multiple pathogens. Reproduced with permission from [185], Copyright 2018 American Chemical Society
Smartphones is well-suited for POCT and personalized healthcare for their high-resolution cameras, advanced computing and data processing capabilities, and most importantly, global accessibility to users [178]. Optical signal output can be captured and analyzed readily with self-built cameras and programs installed on smartphones. A POCT method for on-site and visual detection of streptomycin was established based on aptamer recognition and digital image colorimetry by smartphone [179]. The fluorescence intensity of anthocyanins dye SYBR Green I was employed as indicator for the concentration of streptomycin, and the RGB values of acquired images were analyzed by the Touch Color APP installed in smartphone (Fig. 13b). The limit of detection of this method was 94 nM, and the recovery rates reached 94.1–110% for determining streptomycin in chicken and milk samples. Smartphone-based system was also established for the real-time detection of mycotoxin by combining nucleic acid aptamer recognition and shape coding microhydrogel techniques, exhibiting a detection limit of 0.1 ng/mL [180].
Yang’s research group developed a target-responsive hydrogel system encapsulated Au core/Pt shell nanoparticle (Au@PtNP) as a universal platform for quantitative POCT. The hydrogels were constructed with DNA-grafted linear polyacrylamide and cross-linking DNA aptamers for selective target recognition. The presence of target molecules would trigger the release of the preloaded Pt nanoparticles that possessed excellent catalytic capability for decomposing H2O2 to O2. The generation of O2 in sealed environment could be measured by a portable volumetric bar-chart chip [181, 182] or a handheld pressuremeter [183] (Fig. 13c). In addition, glucoamylase was also used as alternatives to Pt for cascade enzymatic reactions for signal amplification and visualization with microfluidic paper-based analytic devices [184]. This strategy was employed for portable and highly sensitive detection of a series of targets, including cocaine and ochratoxin A. Similar pressure amplification technique was utilized for an instrument-free detection of multiple pathogens by integrating a portable multiplexed bar-chart chip with nanoparticle-mediated magnetic aptasensors (Fig. 13d) [185]. This user-friendly device allowed visual quantitative detection of multiple pathogens simultaneously without cross-interference. Three major foodborne pathogens in apple juice were specifically quantified with limits of detection of about 10 CFU/mL.
The remold of commercially available handheld meters such as glucometer [162, 186], pressuremeter [183, 185], and thermometer [187] represents promising approach for the development of POCT diagnostic devices [188]. These household meters possess merits of wide availability, easy operation, good portability, rapid response, and low cost. The transformation of the detection of targets into readable signals, i.e., changes of glucose, temperature, or pressure, is the major technical challenge which requires enormous investment of manpower and resources at the research and development stage. Due to the inherent poor sensitivity of these handheld meters, signal amplification strategies are crucial to detect subtle signal variation in response to biomarkers at low abundance.
Conclusions, challenges, and perspectives
Under the current circumstances of global epidemic, a vast number of aptasensors aiming at rapid, ultrasensitive, highly specific, cost-effective, and user-friendly detection of infectious pathogens have been established, integrating innovated reporting mechanisms, signal enhancers, nanomaterial-based sensing platforms, and automated detection technologies. Herein, we have discussed the design strategies and advanced techniques of aptasensors, covering the utilization of three major signal transducers, the employment of aptamers as recognition moieties, and the construction of versatile sensing platforms. The latest development of POCT devices for anti-infective purposes has also been highlighted. Real sample analyses of spiked food, drinks, urine, and blood are always performed, some of which were tested on patients to demonstrate the capability of these sensors as potential candidates for pathogen detection and clinical diagnosis of infectious diseases.
Despite being compared as promising alternatives to antibodies, the market share of aptasensor-based diagnostic products lags far behind than that of the antibodies. Antibody-based immunologic diagnostics currently dominates the market of in vitro diagnostics (IVD) [189]. There are a limited number of clinical trials on aptasensors for infectious disease diagnostics (Table 1). Current situation is that the market recognition of aptamers is relatively low compared with that of antibodies, and few companies are willing to invest on the commercialization of aptamer-based diagnostic devices. Several exposed technical barriers of aptamer-based technology have hamstrung the implementation of aptasensors into clinical utilization: (i) highly sensitive and specific anti-infective aptamers are still insufficient compared with the rich database of antibodies. The main reason lies in the relatively low screening efficiency and success rate of conventional SELEX process. Hence, many of the analytically aimed studies are based on limited number of well-characterized model aptamers [190]; (ii) the inherent structural flexibility of oligonucleotides reduces the repeatability of aptamer-target binding. The binding is also highly sensitive to exterior interferences such as pH, temperature, and species of buffer. Sometimes the high binding affinity cannot be reproduced without a standard protocol; (iii) most of the in vitro SELEX were carried out in saline buffer or cell culture medium, and those selected aptamers may not retain their high affinity and specificity toward pathogens in their natural forms, concerning the complexity of real sample analysis and diverse interfering factors [21]; (iv) DNA/RNA aptamers, especially unmodified ones, are intrinsically susceptible to nuclease degradation. Their metabolic instability and other pharmacokinetic problems such as rapid renal filtration may have adverse effect on their binding performance in physiological environment [191].
As for aptasensors, despite that a vast number of papers have been published in this field, the majority of them are proof-of-concept models that lack the potential for practical application in clinic. Robust and affordable POCT devices are scarce due to several critical issues: (i) the reliability of the sensor still needs to be improved. Although some aptamers are characterized as highly sensitive and specific to infectious agents during selection, whether those features could be retained after fabrication should be carefully evaluated. Infectious pathogens, especially bacteria and viruses, are prone to mutation, and aptamers selected against certain strain of pathogens might give false negative result to other mutated strains; (ii) the stability and reproducibility of the sensors are crucial factors to be considered. For instance, fluorescent dyes serving as reporting probes are susceptible to photobleaching, and those nanomaterial-based platforms may suffer from batch-to-batch difference in their performance [107]; (iii) the fabrication procedures of many reported aptasensors are relatively tedious, which is unfavorable for industrial commercial production; (iv) the overall cost of an aptasensor manifests insignificant advantage over that of antibody, concerning the complicated production process, and the use of precious metal-based materials in some cases.
The SELEX procedure, cornerstone of aptamer technology, has been fast-developing in the last few years to address the aforementioned deficiencies of aptamers [192, 193]. Cell-SELEX has been extensively applied for the identification of aptamers for infectious pathogens to broaden the scope of targets, avoid target purification, and retain the natural conformation of molecules on cell surfaces [30–33]. Slow off-rate modified aptamers (SOMAmers), selected from library with functionalized nucleotides combined with a strategy of slow off-rate selection, possess enhanced (sub-nM) affinity to various protein targets [194, 195]. In addition, SOMAmers-based platform is capable of measuring hundreds of protein analytes in one single sample, providing a new tool for biomarker identification [196]. Microfluidic chip technique has been incorporated into SELEX to provide a rapid, highly efficient, and automated screening method in which the sample and reagent consumption can be reduced to microliter grade, and each selection round only takes about 1 h [197]. The utilization of high-throughput sequencing during each SELEX round allows supervision of the whole enrichment process for a timely adjustment of the screening procedures, hence reducing the screening blindness and accelerating the screening process [198]. To circumvent the susceptibility of aptamers to nuclease digestion and renal filtration, versatile post-SELEX chemical modifications have been explored, including PEGylation, sugar modification, phosphodiester linkage modification, and truncation [16, 97]. In silico approaches carried out during selection and modification process are expected to reduce the time and cost associated with aptamer development. There are methods enabling the estimation of aptamer-target binding affinity, performing thermodynamics and kinetic analysis, and optimizing aptamer sequence modification [199, 200].
Although superior specificity is highly appreciated in aptamer selection, aptamers with broad-spectrum recognition of targeted pathogens can be especially useful in the context of infectious disease diagnostics and therapy. The molecular diagnosis of virus infection by aptasensors primarily relies on the detection of viral proteins. Aptamers that selected against highly conserved residues of an evolving viral protein would present a broad-spectrum recognition of the variants, hence possessing more reliable target binding and higher genetic barrier to resistance in contrast to highly specific aptamers [201, 202]. However, some viral proteins share certain amount of sequence identity with other viral species (such as the envelope, nucleocapsid, membrane, and spike proteins of SARS-CoV-2, SARS-CoV, and MERS-CoV [203]), and the choice of epitopes specific to each virus need to be cautious to reduce cross-reactivity and eliminate false positive results [155]. The choice of broad-spectrum or highly selective aptamer depends on specific application scenarios, whether the intention is to make generalized disease diagnostics or to differentiate variants of pathogens for precise medication.
Besides optimization of the screening process, innovative ideas have been presented to ameliorate the performance of various aptasensing platforms. The development of nanomaterial-based aptasensors has become a hotspot in aptamer-based diagnostic research filed [204]. Attributed to the outstanding optical, electrical, or magnetic properties of versatile nanomaterials, the sensitivity and specificity of aptasensors could be significantly enhanced [205]. The distinct color difference between well-dispersed and aggregated AuNPs has been vastly employed for the preparation of colorimetric aptasensors, and generally, owing to the large surface-to-volume ratio of the nanomaterial, aptamers can be coated onto the surfaces of AuNPs for signal amplification [206–208]. Carbon nanomaterials such as carbon nanotubes, carbon nanowires, and graphenes that possess hydrophobic surfaces can be easily adsorbed by aptamers, and their excellent conductivity enables readily functionalization for electrochemical aptasensors, especially label-free detection systems [209–211]. As alternatives to conventional organic fluorophores, quantum dots and UCNPs feature enhanced photo-stability and high signal-to-noise ratio, which are particularly suitable for the development of fluorescent aptasensors [115, 212, 213]. MNPs such as Fe3O4 are advantageous for facile separation, extraction, target cell enrichment, and signal amplification owing to their ease of production, environmentally friendliness, and good biocompatibility [214, 215]. Hybrid nano-conjugated systems have also been fabricated for the integration of the unique properties of single nanomaterial species [216–218]. Future efforts should be focused on the development of robust, multi-functional, low-cost, readily to large-scale production nanomaterial-based sensing platforms.
Multiplexed detection refers to the detection of multiple targets simultaneously in a single assay. The features of enhanced diagnostic efficiency, higher throughput, reduced cost, and improved operational convenience are highly desired, especially in clinic diagnostics and POCT [219–222]. Aptamers specific for different targets can be labeled with distinguishable signal tags and integrated onto one single platform for multiplexed biosensing. Toward the development of highly efficient, on-site, and cost-effective simultaneous screening methods, multiplexed aptasensors have been designed for the detection of antibiotics, mycotoxins, and pathogens [185, 223, 224]. For example, an aptamer-immobilized LSPR-based sensor was used to detect and identify three different bacterial species (Lactobacillus acidophilus, S. typhimurium, and P. aeruginosa) in a multiplexed mode [225]. Quantitative detection of S. typhimurium and S. aureus simultaneously has also been realized using GNPs modified with different Raman molecules, while aptamers functioned as the signal and capture probes [226]. Aptamer-functionalized quantum dots and upconverting nanoparticles, which can be excited at different wavelength to minimize cross-talk between the luminescent signals, were successfully employed for the detection of S. typhimurium and S. aureus using a dual excitation strategy [227]. Recently, a microfluidic paper-based colorimetric multiplexed aptasensor was developed for E. coli O157:H7 and S. Typhimurium, and image analysis was used to read and quantify the colorimetric signal and measure target concentration [228]. Despite the fast development of multiplexed aptasensors for pathogen detection in the last few years, there exist some challenges for simultaneous detection techniques, such as limited number of distinguishable signal tags, multiple signal overlap, mutual interference, and cross-reactivity between different targets [223]. Meanwhile, complicated fabrication procedures of the sensing platforms will compromise their practical significance and application prospect. How to strike a balance between delicate design and simple configuration is an important issue to be explored.
POCT is currently a subsector of accelerated development in IVD industry. The outbreaks of influenza, Ebola, the field of sexually transmitted disease control, and the recent pandemic of COVID-19 have brought in unprecedented market demand for rapid diagnosis of infectious diseases [229–231]. As discussed in the last section, the incorporation of anti-infective aptamers with lateral flow assays, microfluidic platforms, and smartphones have manifested great potential in rapid, user-friendly pathogen testing and patient diagnosis in resource-constrained facilities [152, 232]. With the development of tailor-made aptamers and the advancement of cutting-edge devices, aptamer-based POCT is anticipated for a prosperous future in infectious disease diagnostics. Tremendous efforts from researchers and bio-pharmaceutical companies have been devoted to promote the development of aptamer-based technology over the past two decades. We believe the pressing needs in the diagnosis of infectious diseases will motivate continuous innovations and breakthroughs from diverse aspects for the translation of aptasensors into clinical practices.
Dedication
This paper is dedicated to the Institute of Analysis, Guangdong Academy of Sciences (China National Analytical Center, Guangzhou) on the occasion of its 50th anniversary.
Funding
This work was supported by the Guangdong Academy of Sciences (2020GDASYL-20200103046, 2020GDASYL-20200103045).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xin Zhao, Email: zhaoxin2017@foxmail.com.
Zifeng Yang, Email: jeffyah@163.com.
References
- 1.Bloom DE, Cadarette D. Infectious disease threats in the twenty-first century: strengthening the global response. Front Immunol. 2019;10:549. doi: 10.3389/fimmu.2019.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO COVID-19 situation dashboard. https://covid19.who.int/. Accessed 04–07–2022
- 3.Fournier P-E, Drancourt M, Colson P, Rolain J-M, Scola BL, Raoult D. Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol. 2013;11(8):574–585. doi: 10.1038/nrmicro3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maurer JJ. Rapid detection and limitations of molecular techniques. Annu Rev Food Sci Technol. 2011;2(1):259–279. doi: 10.1146/annurev.food.080708.100730. [DOI] [PubMed] [Google Scholar]
- 5.Andreotti PE, Ludwig GV, Peruski AH, Tuite JJ, Morse SS, Peruski LF. Immunoassay of infectious agents. Biotechniques. 2003;35(4):850–859. doi: 10.2144/03354ss02. [DOI] [PubMed] [Google Scholar]
- 6.Tsurusawa N, Chang JYH, Namba M, Makioka D, Yamura S, Iha K, Kyosei Y, Watabe S, Yoshimura T, Ito E. Modified ELISA for ultrasensitive diagnosis. J Clinic Med. 2021;10(21):5197. doi: 10.3390/jcm10215197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Q, Lu D, Zhang G, Zhang D, Shi X. Recent improvements in enzyme-linked immunosorbent assays based on nanomaterials. Talanta. 2021;223:121722. doi: 10.1016/j.talanta.2020.121722. [DOI] [PubMed] [Google Scholar]
- 8.Chang XH, Zhang J, Wu LH, Peng YK, Yang XY, Li XL, Ma AJ, Ma JC, Chen GQ. Research progress of near-infrared fluorescence immunoassay. Micromachines. 2019;10(6):422. doi: 10.3390/mi10060422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rongen HAH, Hoetelmans RMW, Bult A, Vanbennekom WP. Chemiluminescence and immunoassays. J Pharm Biomed Anal. 1994;12(4):433–462. doi: 10.1016/0731-7085(94)80027-8. [DOI] [PubMed] [Google Scholar]
- 10.Towbin H, Gordon J. Immunobloting and dot immunobinding – current status and outlook. J Immunol Meth. 1984;72(2):313–340. doi: 10.1016/0022-1759(84)90001-2. [DOI] [PubMed] [Google Scholar]
- 11.Sharafeldin M, Davis JJ. Point of care sensors for infectious pathogens. Anal Chem. 2021;93(1):184–197. doi: 10.1021/acs.analchem.0c04677. [DOI] [PubMed] [Google Scholar]
- 12.Nesakumar N, Lakshmanakumar M, Srinivasan S, Jbb AJ, Rayappan JBB. Principles and recent advances in biosensors for pathogens detection. ChemistrySelect. 2021;6(37):10063–10091. doi: 10.1002/slct.202101062. [DOI] [Google Scholar]
- 13.Wang Q, Wang J, Huang Y, Du YC, Zhang Y, Cui YX, Kong DM. Development of the DNA-based biosensors for high performance in detection of molecular biomarkers: more rapid, sensitive, and universal. Biosens Bioelectron. 2022;197:113739. doi: 10.1016/j.bios.2021.113739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sande MG, Rodrigues JL, Ferreira D, Silva CJ, Rodrigues LR. Novel biorecognition elements against pathogens in the design of state-of-the-art diagnostics. Biosensors-Basel. 2021;11(11):418. doi: 10.3390/bios11110418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Q, Wu C, Cai H, Hu N, Zhou J, Wang P. Cell-based biosensors and their application in biomedicine. Chem Rev. 2014;114(12):6423–6461. doi: 10.1021/cr2003129. [DOI] [PubMed] [Google Scholar]
- 16.Ni S, Zhuo Z, Pan Y, Yu Y, Li F, Liu J, Wang L, Wu X, Li D, Wan Y, Zhang L, Yang Z, Zhang BT, Lu A, Zhang G. Recent progress in aptamer discoveries and modifications for therapeutic applications. ACS Appl Mater Interfaces. 2021;13(8):9500–9519. doi: 10.1021/acsami.0c05750. [DOI] [PubMed] [Google Scholar]
- 17.Davydova A, Vorobjeva M, Pyshnyi D, Altman S, Vlassov V, Venyaminova A. Aptamers against pathogenic microorganisms. Crit Rev Microbiol. 2016;42(6):847–865. doi: 10.3109/1040841X.2015.1070115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment - RNA ligands to Bateriophage-T4 DNA-polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 19.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 20.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature. 1990;344(6265):467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 21.Chen XF, Zhao X, Yang Z. Aptamer-based antibacterial and antiviral therapy against infectious diseases. J Med Chem. 2021;64(24):17601–17626. doi: 10.1021/acs.jmedchem.1c01567. [DOI] [PubMed] [Google Scholar]
- 22.Platella C, Riccardi C, Montesarchio D, Roviello GN, Musumeci D (2017) G-quadruplex-based aptamers against protein targets in therapy and diagnostics. Biochim Biophys Acta Gen Subj 1861(5 Pt B):1429–1447. 10.1016/j.bbagen.2016.11.027 [DOI] [PMC free article] [PubMed]
- 23.Gopinath SCB. Methods developed for SELEX. Anal Bioanal Chem. 2007;387(1):171–182. doi: 10.1007/s00216-006-0826-2. [DOI] [PubMed] [Google Scholar]
- 24.Saito S. SELEX-based DNA aptamer selection: a perspective from the advancement of separation techniques. Anal Sci. 2021;37(1):17–26. doi: 10.2116/analsci.20SAR18. [DOI] [PubMed] [Google Scholar]
- 25.Yuce M, Ullah N, Budak H. Trends in aptamer selection methods and applications. Analyst. 2015;140(16):5379–5399. doi: 10.1039/c5an00954e. [DOI] [PubMed] [Google Scholar]
- 26.Xi ZJ, Huang RR, Deng Y, He NY. Progress in selection and biomedical applications of aptamers. J Biomed Nanotechnol. 2014;10(10):3043–3062. doi: 10.1166/jbn.2014.1979. [DOI] [PubMed] [Google Scholar]
- 27.Dua P, Kim S, Lee D-k. Nucleic acid aptamers targeting cell-surface proteins. Methods. 2011;54(2):215–225. doi: 10.1016/j.ymeth.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Wang K, Gan L, Jiang L, Zhang X, Yang X, Chen M, Lan X. Neutralization of staphylococcal enterotoxin B by an aptamer antagonist. Antimicrob Agents Chemother. 2015;59(4):2072–2077. doi: 10.1128/AAC.04414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lahousse M, Park HC, Lee SC, Ha NR, Jung IP, Schlesinger SR, Shackelford K, Yoon MY, Kim SK. Inhibition of anthrax lethal factor by ssDNA aptamers. Arch Biochem Biophys. 2018;646:16–23. doi: 10.1016/j.abb.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shangguan D, Bing T, Zhang N (2015) Cell-SELEX: aptamer selection against whole cells. In: Tan W, Fang X (eds) Aptamers Selected by Cell-SELEX for Theranostics. Springer Berlin Heidelberg, Berlin, Heidelberg, 13–33. 10.1007/978-3-662-46226-3_2
- 31.Kaur H (2018) Recent developments in cell-SELEX technology for aptamer selection. Biochim Biophys Acta Gen Subj 1862(10):2323–2329. 10.1016/j.bbagen.2018.07.029 [DOI] [PubMed]
- 32.Duan Y, Zhang C, Wang Y, Chen G. Research progress of whole-cell-SELEX selection and the application of cell-targeting aptamer. Mol Biol Rep. 2022 doi: 10.1007/s11033-022-07317-0. [DOI] [PubMed] [Google Scholar]
- 33.Rahimizadeh K, AlShamaileh H, Fratini M, Chakravarthy M, Stephen M, Shigdar S, Veedu RN (2017) Development of cell-specific aptamers: recent advances and insight into the selection procedures. Molecules 22(12). 10.3390/molecules22122070 [DOI] [PMC free article] [PubMed]
- 34.Pan Q, Luo F, Liu M, Zhang XL. Oligonucleotide aptamers: promising and powerful diagnostic and therapeutic tools for infectious diseases. J Infect. 2018;77(2):83–98. doi: 10.1016/j.jinf.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan Q, Liu X, Zu Y. Oligonucleotide aptamers for pathogen detection and infectious disease control. Theranostics. 2021;11(18):9133–9161. doi: 10.7150/thno.61804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afrasiabi S, Pourhajibagher M, Raoofian R, Tabarzad M, Bahador A. Therapeutic applications of nucleic acid aptamers in microbial infections. J Biomed Sci. 2020;27(1):6. doi: 10.1186/s12929-019-0611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torabi R, Ranjbar R, Halaji M, Heiat M. Aptamers, the bivalent agents as probes and therapies for coronavirus infections: a systematic review. Mol Cell Probes. 2020;53:101636. doi: 10.1016/j.mcp.2020.101636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandal M, Dutta N, Dutta G. Aptamer-based biosensors and their implications in COVID-19 diagnosis. Anal Methods. 2021;13(45):5400–5417. doi: 10.1039/d1ay01519b. [DOI] [PubMed] [Google Scholar]
- 39.ClinicalTrials.gov database. https://clinicaltrials.gov/ct2/home. Accessed 04-11-2022
- 40.Mascini M, Palchetti I, Tombelli S. Nucleic acid and peptide aptamers: fundamentals and bioanalytical aspects. Angew Chem Int Ed. 2012;51(6):1316–1332. doi: 10.1002/anie.201006630. [DOI] [PubMed] [Google Scholar]
- 41.Lucklum R, Hauptmann P. Acoustic microsensors—the challenge behind microgravimetry. Anal Bioanal Chem. 2006;384(3):667–682. doi: 10.1007/s00216-005-0236-x. [DOI] [PubMed] [Google Scholar]
- 42.Oprea A, Weimar U. Gas sensors based on mass-sensitive transducers Part 1: Transducers and receptors—basic understanding. Anal Bioanal Chem. 2019;411(9):1761–1787. doi: 10.1007/s00216-019-01630-7. [DOI] [PubMed] [Google Scholar]
- 43.Fogel R, Limson J, Seshia Ashwin A. Acoustic biosensors. Essays Biochem. 2016;60(1):101–110. doi: 10.1042/ebc20150011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grammoustianou A, Gizeli E (2018) Chapter 9 - Acoustic wave–based immunoassays. In: Vashist SK, Luong JHT (eds) Handbook of Immunoassay Technologies. Academic Press, 203–239. 10.1016/B978-0-12-811762-0.00009-8
- 45.Zhang J, Zhang X, Wei X, Xue Y, Wan H, Wang P. Recent advances in acoustic wave biosensors for the detection of disease-related biomarkers: a review. Anal Chim Acta. 2021;1164:338321. doi: 10.1016/j.aca.2021.338321. [DOI] [PubMed] [Google Scholar]
- 46.Migoń D, Wasilewski T, Suchy D. Application of QCM in peptide and protein-based drug product development. Molecules. 2020;25(17):3950. doi: 10.3390/molecules25173950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skládal P. Piezoelectric biosensors. Trends. Anal Chem. 2016;79:127–133. doi: 10.1016/j.trac.2015.12.009. [DOI] [Google Scholar]
- 48.Lim HJ, Saha T, Tey BT, Tan WS, Ooi CW. Quartz crystal microbalance-based biosensors as rapid diagnostic devices for infectious diseases. Biosens Bioelectron. 2020;168:112513. doi: 10.1016/j.bios.2020.112513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X, Chen F, Wang R, Li Y. Whole-bacterium SELEX of DNA aptamers for rapid detection of E. coli O157:H7 using a QCM sensor. J Biotechnol. 2018;266:39–49. doi: 10.1016/j.jbiotec.2017.12.011. [DOI] [PubMed] [Google Scholar]
- 50.Wang R, Li Y. Hydrogel based QCM aptasensor for detection of avian influenza virus. Biosens Bioelectron. 2013;42:148–155. doi: 10.1016/j.bios.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 51.Ozalp VC, Bayramoglu G, Erdem Z, Arica MY. Pathogen detection in complex samples by quartz crystal microbalance sensor coupled to aptamer functionalized core-shell type magnetic separation. Anal Chim Acta. 2015;853:533–540. doi: 10.1016/j.aca.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 52.Bayramoglu G, Ozalp VC, Oztekin M, Arica MY. Rapid and label-free detection of brucella melitensis in milk and milk products using an aptasensor. Talanta. 2019;200:263–271. doi: 10.1016/j.talanta.2019.03.048. [DOI] [PubMed] [Google Scholar]
- 53.Bayramoglu G, Kilic M, Yakup Arica M. Selective isolation and sensitive detection of lysozyme using aptamer based magnetic adsorbent and a new quartz crystal microbalance system. Food Chem. 2022;382:132353. doi: 10.1016/j.foodchem.2022.132353. [DOI] [PubMed] [Google Scholar]
- 54.Xi X, Niyonshuti II, Yu N, Yao L, Fu Y, Chen J, Li Y. Label-free quartz crystal microbalance biosensor based on aptamer-capped gold nanocages loaded with polyamidoamine for thrombin detection. ACS Appl Nano Mater. 2021;4(10):10047–10054. doi: 10.1021/acsanm.1c01350. [DOI] [Google Scholar]
- 55.Wang L, Wang R, Chen F, Jiang T, Wang H, Slavik M, Wei H, Li Y. QCM-based aptamer selection and detection of Salmonella typhimurium. Food Chem. 2017;221:776–782. doi: 10.1016/j.foodchem.2016.11.104. [DOI] [PubMed] [Google Scholar]
- 56.Mandal D, Banerjee S. Surface acoustic wave (SAW) sensors: physics, materials, and applications. Sensors. 2022;22(3):820. doi: 10.3390/s22030820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lange K, Rapp BE, Rapp M. Surface acoustic wave biosensors: a review. Anal Bioanal Chem. 2008;391(5):1509–1519. doi: 10.1007/s00216-008-1911-5. [DOI] [PubMed] [Google Scholar]
- 58.Li Z, Jones Y, Hossenlopp J, Cernosek R, Josse F. Analysis of liquid-phase chemical detection using guided shear horizontal-surface acoustic wave sensors. Anal Chem. 2005;77(14):4595–4603. doi: 10.1021/ac0504621. [DOI] [PubMed] [Google Scholar]
- 59.Schlensog MD, Gronewold TMA, Tewes M, Famulok M, Quandt E. A love-wave biosensor using nucleic acids as ligands. Sens Actuat B Chem. 2004;101(3):308–315. doi: 10.1016/j.snb.2004.03.015. [DOI] [Google Scholar]
- 60.Liss M, Petersen B, Wolf H, Prohaska E. An aptamer-based quartz crystal protein biosensor. Anal Chem. 2002;74(17):4488–4495. doi: 10.1021/ac011294p. [DOI] [PubMed] [Google Scholar]
- 61.Dullah EC, Ongkudon CM. Current trends in endotoxin detection and analysis of endotoxin-protein interactions. Crit Rev Biotechnol. 2017;37(2):251–261. doi: 10.3109/07388551.2016.1141393. [DOI] [PubMed] [Google Scholar]
- 62.Ji J, Pang Y, Li D, Huang Z, Zhang Z, Xue N, Xu Y, Mu X (2020) An aptamer-based shear horizontal surface acoustic wave biosensor with a CVD-grown single-layered graphene film for high-sensitivity detection of a label-free endotoxin. Microsyst Nanoeng 6(1). 10.1038/s41378-019-0118-6 [DOI] [PMC free article] [PubMed]
- 63.Jia X, Dong S, Wang E. Engineering the bioelectrochemical interface using functional nanomaterials and microchip technique toward sensitive and portable electrochemical biosensors. Biosens Bioelectron. 2016;76:80–90. doi: 10.1016/j.bios.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 64.Bakirhan NK, Topal BD, Ozcelikay G, Karadurmus L, Ozkan SA. Current advances in electrochemical biosensors and nanobiosensors. Crit Rev Anal Chem. 2022;52(3):519–534. doi: 10.1080/10408347.2020.1809339. [DOI] [PubMed] [Google Scholar]
- 65.Cesewski E, Johnson BN. Electrochemical biosensors for pathogen detection. Biosens Bioelectron. 2020;159:112214. doi: 10.1016/j.bios.2020.112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoon J, Cho H-Y, Shin M, Choi HK, Lee T, Choi J-W. Flexible electrochemical biosensors for healthcare monitoring. J Mater Chem B. 2020;8(33):7303–7318. doi: 10.1039/D0TB01325K. [DOI] [PubMed] [Google Scholar]
- 67.Goud KY, Reddy KK, Khorshed A, Kumar VS, Mishra RK, Oraby M, Ibrahim AH, Kim H, Gobi KV. Electrochemical diagnostics of infectious viral diseases: trends and challenges. Biosens Bioelectron. 2021;180:113112. doi: 10.1016/j.bios.2021.113112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Radi A, Abd-Ellatief MR (2021) Electrochemical aptasensors: current status and future perspectives. Diagnostics 11(1). 10.3390/diagnostics11010104 [DOI] [PMC free article] [PubMed]
- 69.Labib M, Berezovski MV. Electrochemical aptasensors for microbial and viral pathogens. Adv Biochem Eng Biotechnol. 2014;140:155–181. doi: 10.1007/10_2013_229. [DOI] [PubMed] [Google Scholar]
- 70.Sadeghi AS, Ansari N, Ramezani M, Abnous K, Mohsenzadeh M, Taghdisi SM, Alibolandi M. Optical and electrochemical aptasensors for the detection of amphenicols. Biosens Bioelectron. 2018;118:137–152. doi: 10.1016/j.bios.2018.07.045. [DOI] [PubMed] [Google Scholar]
- 71.Das R, Dhiman A, Mishra SK, Haldar S, Sharma N, Bansal A, Ahmad Y, Kumar A, Tyagi JS, Sharma TK. Structural switching electrochemical DNA aptasensor for the rapid diagnosis of tuberculous meningitis. Int J Nanomedicine. 2019;14:2103–2113. doi: 10.2147/IJN.S189127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bai L, Chen Y, Bai Y, Chen Y, Zhou J, Huang A. Fullerene-doped polyaniline as new redox nanoprobe and catalyst in electrochemical aptasensor for ultrasensitive detection of mycobacterium tuberculosis MPT64 antigen in human serum. Biomaterials. 2017;133:11–19. doi: 10.1016/j.biomaterials.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 73.Shahrokhian S, Ranjbar S. Development of a sensitive diagnostic device based on zeolitic imidazolate frameworks-8 using ferrocene–graphene oxide as electroactive indicator for Pseudomonas aeruginosa detection. ACS Sustain Chem Eng. 2019;7(15):12760–12769. doi: 10.1021/acssuschemeng.9b01314. [DOI] [Google Scholar]
- 74.Fernández-la-Villa A, Pozo-Ayuso DF, Castaño-Álvarez M. Microfluidics and electrochemistry: an emerging tandem for next-generation analytical microsystems. Curr Opin Electrochem. 2019;15:175–185. doi: 10.1016/j.coelec.2019.05.014. [DOI] [Google Scholar]
- 75.Chand R, Neethirajan S. Microfluidic platform integrated with graphene-gold nano-composite aptasensor for one-step detection of norovirus. Biosens Bioelectron. 2017;98:47–53. doi: 10.1016/j.bios.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 76.Chamorro-Garcia A, Ortega G, Mariottini D, Green J, Ricci F, Plaxco KW. Switching the aptamer attachment geometry can dramatically alter the signalling and performance of electrochemical aptamer-based sensors. Chem Commun. 2021;57(88):11693–11696. doi: 10.1039/D1CC04557A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee I, Kim SE, Lee J, Woo DH, Lee S, Pyo H, Song CS, Lee J. A self-calibrating electrochemical aptasensing platform: correcting external interference errors for the reliable and stable detection of avian influenza viruses. Biosens Bioelectron. 2020;152:112010. doi: 10.1016/j.bios.2020.112010. [DOI] [PubMed] [Google Scholar]
- 78.Hai X, Li Y, Zhu C, Song W, Cao J, Bi S. DNA-based label-free electrochemical biosensors: from principles to applications. Trends Anal Chem. 2020;133:116098. doi: 10.1016/j.trac.2020.116098. [DOI] [Google Scholar]
- 79.Sheikhzadeh E, Chamsaz M, Turner APF, Jager EWH, Beni V. Label-free impedimetric biosensor for salmonella typhimurium detection based on poly [pyrrole-co-3-carboxyl-pyrrole] copolymer supported aptamer. Biosens Bioelectron. 2016;80:194–200. doi: 10.1016/j.bios.2016.01.057. [DOI] [PubMed] [Google Scholar]
- 80.Bagheryan Z, Raoof JB, Golabi M, Turner APF, Beni V. Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Biosens Bioelectron. 2016;80:566–573. doi: 10.1016/j.bios.2016.02.024. [DOI] [PubMed] [Google Scholar]
- 81.Istamboulie G, Paniel N, Zara L, Reguillo Granados L, Barthelmebs L, Noguer T. Development of an impedimetric aptasensor for the determination of aflatoxin M1 in milk. Talanta. 2016;146:464–469. doi: 10.1016/j.talanta.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 82.Marchese S, Polo A, Ariano A, Velotto S, Costantini S, Severino L (2018) Aflatoxin B1 and M1: biological properties and their involvement in cancer development. Toxins 10(6). 10.3390/toxins10060214 [DOI] [PMC free article] [PubMed]
- 83.Abrego-Martinez JC, Jafari M, Chergui S, Pavel C, Che D, Siaj M. Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy. Biosens Bioelectron. 2022;195:113595. doi: 10.1016/j.bios.2021.113595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peinetti AS, Lake RJ, Cong W, Cooper L, Wu Y, Ma Y, Pawel GT, Toimil-Molares ME, Trautmann C, Rong L, Mariñas B, Azzaroni O, Lu Y (2021) Direct detection of human adenovirus or SARS-CoV-2 with ability to inform infectivity using DNA aptamer-nanopore sensors. Sci Adv 7 (39):eabh2848. 10.1126/sciadv.abh2848 [DOI] [PMC free article] [PubMed]
- 85.Thiha A, Ibrahim F, Muniandy S, Dinshaw IJ, Teh SJ, Thong KL, Leo BF, Madou M. All-carbon suspended nanowire densors as a rapid highly-sensitive label-free chemiresistive biosensing platform. Biosens Bioelectron. 2018;107:145–152. doi: 10.1016/j.bios.2018.02.024. [DOI] [PubMed] [Google Scholar]
- 86.Guo Y, Wang Y, Liu S, Yu J, Wang H, Wang Y, Huang J. Label-free and highly sensitive electrochemical detection of E. coli based on rolling circle amplifications coupled peroxidase-mimicking DNAzyme amplification. Biosens Bioelectron. 2016;75:315–319. doi: 10.1016/j.bios.2015.08.031. [DOI] [PubMed] [Google Scholar]
- 87.Sang SB, Wang YJ, Feng QL, Wei Y, Ji JL, Zhang WD. Progress of new label-free techniques for biosensors: a review. Crit Rev Biotechnol. 2016;36(3):465–481. doi: 10.3109/07388551.2014.991270. [DOI] [PubMed] [Google Scholar]
- 88.Sedki M, Chen Y, Mulchandani A (2020) Non-carbon 2D materials-based field-effect transistor biosensors: recent advances, challenges, and future perspectives. Sensors 20(17). 10.3390/s20174811 [DOI] [PMC free article] [PubMed]
- 89.Vu CA, Chen WY (2020) Predicting future prospects of aptamers in field-effect transistor biosensors. Molecules 25(3). 10.3390/molecules25030680 [DOI] [PMC free article] [PubMed]
- 90.Fatin MF, Rahim Ruslinda A, Gopinath SCB, Arshad MKM. High-performance interactive analysis of split aptamer and HIV-1 Tat on multiwall carbon nanotube-modified field-effect transistor. Int J Biol Macromol. 2019;125:414–422. doi: 10.1016/j.ijbiomac.2018.12.066. [DOI] [PubMed] [Google Scholar]
- 91.Damborský P, Švitel J, Katrlík J. Optical biosensors. Essays Biochem. 2016;60(1):91–100. doi: 10.1042/ebc20150010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chen C, Wang J. Optical biosensors: an exhaustive and comprehensive review. Analyst. 2020;145(5):1605–1628. doi: 10.1039/C9AN01998G. [DOI] [PubMed] [Google Scholar]
- 93.Bayrac C, Eyidogan F, Avni Oktem H. DNA aptamer-based colorimetric detection platform for Salmonella enteritidis. Biosens Bioelectron. 2017;98:22–28. doi: 10.1016/j.bios.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 94.Xie J, Zhang X, Wang H, Zheng H, Huang Y, Xie J. Analytical and environmental applications of nanoparticles as enzyme mimetics. Trends Anal Chem. 2012;39:114–129. doi: 10.1016/j.trac.2012.03.021. [DOI] [Google Scholar]
- 95.Wu S, Duan N, Qiu Y, Li J, Wang Z. Colorimetric aptasensor for the detection of salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int J Food Microbiol. 2017;261:42–48. doi: 10.1016/j.ijfoodmicro.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 96.Dehghani Z, Hosseini M, Mohammadnejad J, Bakhshi B, Rezayan AH. Colorimetric aptasensor for campylobacter jejuni cells by exploiting the peroxidase like activity of Au@Pd nanoparticles. Microchim Acta. 2018;185(10):448. doi: 10.1007/s00604-018-2976-2. [DOI] [PubMed] [Google Scholar]
- 97.Sun Y, Duan N, Ma P, Liang Y, Zhu X, Wang Z. Colorimetric aptasensor based on truncated aptamer and trivalent DNAzyme for Vibrio parahemolyticus determination. J Agric Food Chem. 2019;67(8):2313–2320. doi: 10.1021/acs.jafc.8b06893. [DOI] [PubMed] [Google Scholar]
- 98.Zhan Z, Li H, Liu J, Xie G, Xiao F, Wu X, Aguilar ZP, Xu H. A competitive enzyme linked aptasensor with rolling circle amplification (ELARCA) assay for colorimetric detection of listeria monocytogenes. Food Control. 2020;107:106806. doi: 10.1016/j.foodcont.2019.106806. [DOI] [Google Scholar]
- 99.Yi J, Wu P, li G, Xiao W, Li L, He Y, He Y, Ding P, Chen C (2019) A composite prepared from carboxymethyl chitosan and aptamer-modified gold nanoparticles for the colorimetric determination of Salmonella typhimurium. Microchim Acta 186.10.1007/s00604-019-3827-5 [DOI] [PubMed]
- 100.Feng J, Shen Q, Wu J, Dai Z, Wang Y. Naked-eyes setection of shigella flexneri in food samples based on a novel gold nanoparticle-based colorimetric aptasensor. Food Control. 2019;98:333–341. doi: 10.1016/j.foodcont.2018.11.048. [DOI] [Google Scholar]
- 101.Kim H-S, Kim Y-J, Chon J-W, Kim D-H, Yim J-H, Kim H, Seo K-H. Two-stage label-free aptasensing platform for rapid detection of Cronobacter sakazakii in powdered infant formula. Sens Actuat B Chem. 2017;239:94–99. doi: 10.1016/j.snb.2016.07.173. [DOI] [Google Scholar]
- 102.Basso CR, Crulhas BP, Magro M, Vianello F, Pedrosa VA. A new immunoassay of hybrid nanomater conjugated to aptamers for the detection of dengue virus. Talanta. 2019;197:482–490. doi: 10.1016/j.talanta.2019.01.058. [DOI] [PubMed] [Google Scholar]
- 103.Jauset-Rubio M, El-Shahawi MS, Bashammakh AS, Alyoubi AO, O′Sullivan CK. Advances in aptamers-based lateral flow assays. Trends Anal Chem. 2017;97:385–398. doi: 10.1016/j.trac.2017.10.010. [DOI] [Google Scholar]
- 104.Nguyen VT, Song S, Park S, Joo C. Recent advances in high-sensitivity detection methods for paper-based lateral-flow assay. Biosens Bioelectron. 2020;152:112015. doi: 10.1016/j.bios.2020.112015. [DOI] [PubMed] [Google Scholar]
- 105.Kim SH, Lee J, Lee BH, Song C-S, Gu MB. Specific detection of avian influenza H5N2 whole virus particles on lateral flow strips using a pair of sandwich-type aptamers. Biosens Bioelectron. 2019;134:123–129. doi: 10.1016/j.bios.2019.03.061. [DOI] [PubMed] [Google Scholar]
- 106.Zhao X, Dai X, Zhao S, Cui X, Gong T, Song Z, Meng H, Zhang X, Yu B. Aptamer-based fluorescent sensors for the detection of cancer biomarkers. Spectrochim Acta A Mol Biomol Spectrosc. 2021;247:119038. doi: 10.1016/j.saa.2020.119038. [DOI] [PubMed] [Google Scholar]
- 107.Li Y, Su R, Li H, Guo J, Hildebrandt N, Sun C. Fluorescent aptasensors: design strategies and applications in analyzing chemical contamination of food. Anal Chem. 2021 doi: 10.1021/acs.analchem.1c04294. [DOI] [PubMed] [Google Scholar]
- 108.Wu L, Huang C, Emery BP, Sedgwick AC, Bull SD, He X-P, Tian H, Yoon J, Sessler JL, James TD. Förster resonance energy transfer (FRET)-based small-molecule sensors and imaging agents. Chem Soc Rev. 2020;49(15):5110–5139. doi: 10.1039/C9CS00318E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gao R, Zhong Z, Gao X, Jia L. Graphene oxide quantum dots assisted construction of fluorescent aptasensor for rapid detection of Pseudomonas aeruginosa in food samples. J Agric Food Chem. 2018;66(41):10898–10905. doi: 10.1021/acs.jafc.8b02164. [DOI] [PubMed] [Google Scholar]
- 110.Zhong Z, Gao X, Gao R, Jia L. Selective capture and sensitive fluorometric determination of Pseudomonas aeruginosa by using aptamer modified magnetic nanoparticles. Microchim Acta. 2018;185(8):377. doi: 10.1007/s00604-018-2914-3. [DOI] [PubMed] [Google Scholar]
- 111.Cui F, Sun J, de Dieu HJ, Yang X, Ji J, Zhang Y, Lei H, Li Z, Zheng J, Fan M, Sun X. Ultrasensitive fluorometric angling determination of staphylococcus aureus in vitro and fluorescence imaging in vivo using carbon dots with full-color emission. Anal Chem. 2019;91(22):14681–14690. doi: 10.1021/acs.analchem.9b03916. [DOI] [PubMed] [Google Scholar]
- 112.Shen Y, Wu T, Zhang Y, Ling N, Zheng L, Zhang SL, Sun Y, Wang X, Ye Y. Engineering of a dual-recognition ratiometric fluorescent nanosensor with a remarkably large stokes shift for accurate tracking of pathogenic bacteria at the single-cell level. Anal Chem. 2020;92(19):13396–13404. doi: 10.1021/acs.analchem.0c02762. [DOI] [PubMed] [Google Scholar]
- 113.Zheng W, Huang P, Tu DT, Ma E, Zhu HM, Chen XY. Lanthanide-doped upconversion nano-bioprobes: electronic structures, optical properties, and biodetection. Chem Soc Rev. 2015;44(6):1379–1415. doi: 10.1039/c4cs00178h. [DOI] [PubMed] [Google Scholar]
- 114.Li Y, Chen C, Liu FF, Liu JL (2022) Engineered lanthanide-doped upconversion nanoparticles for biosensing and bioimaging application. Microchimica Acta 189(3). 10.1007/s00604-022-05180-1 [DOI] [PubMed]
- 115.Jin B, Wang S, Lin M, Jin Y, Zhang S, Cui X, Gong Y, Li A, Xu F, Lu TJ. Upconversion nanoparticles based FRET aptasensor for rapid and ultrasenstive bacteria detection. Biosens Bioelectron. 2017;90:525–533. doi: 10.1016/j.bios.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 116.Hwang H, Myong S. Protein induced fluorescence enhancement (PIFE) for probing pro-tein-nucleic acid interactions. Chem Soc Rev. 2014;43(4):1221–1229. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Levitus M, Ranjit S. Cyanine dyes in biophysical research: the photophysics of polymethine fluorescent dyes in biomolecular environments. Q Rev Biophys. 2011;44(1):123–151. doi: 10.1017/S0033583510000247. [DOI] [PubMed] [Google Scholar]
- 118.Lee JM, Kim CR, Kim S, Min J, Lee MH, Lee S. Mix-and-read, one-minute SARS-CoV-2 diagnostic assay: development of PIFE-based aptasensor. Chem Commun (Camb) 2021;57(79):10222–10225. doi: 10.1039/d1cc04066a. [DOI] [PubMed] [Google Scholar]
- 119.Zhao L, Liu Y, Zhang Z, Wei J, Xie S, Li X. Fibrous testing papers for fluorescence trace sensing and photodynamic destruction of antibiotic-resistant bacteria. J Mater Chem B. 2020;8(13):2709–2718. doi: 10.1039/d0tb00002g. [DOI] [PubMed] [Google Scholar]
- 120.Ge C, Feng J, Zhang J, Hu K, Wang D, Zha L, Hu X, Li R. Aptamer/antibody sandwich method for digital detection of SARS-CoV2 nucleocapsid protein. Talanta. 2022;236:122847. doi: 10.1016/j.talanta.2021.122847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lu PH, Ma YD, Fu CY, Lee GB. Structure-free digital microfluidic platform for detection of influenza A virus by using magnetic beads and electromagnetic forces. Lab Chip. 2020;20(4):789–797. doi: 10.1039/c9lc01126a. [DOI] [PubMed] [Google Scholar]
- 122.Su C-H, Tsai M-H, Lin C-Y, Ma Y-D, Wang C-H, Chung Y-D, Lee G-B. Dual aptamer assay for detection of Acinetobacter baumannii on an electromagnetically-driven microfluidic platform. Biosens Bioelectron. 2020;159:112148. doi: 10.1016/j.bios.2020.112148. [DOI] [PubMed] [Google Scholar]
- 123.Wu D-Y, Li J-F, Ren B, Tian Z-Q. Electrochemical surface-enhanced raman spectroscopy of nanostructures. Chem Soc Rev. 2008;37(5):1025–1041. doi: 10.1039/B707872M. [DOI] [PubMed] [Google Scholar]
- 124.Fan M, Andrade GFS, Brolo AG. A review on recent advances in the applications of surface-enhanced raman scattering in analytical chemistry. Anal Chim Acta. 2020;1097:1–29. doi: 10.1016/j.aca.2019.11.049. [DOI] [PubMed] [Google Scholar]
- 125.Xu K, Zhou R, Takei K, Hong M. Toward flexible surface-enhanced raman scattering (SERS) sensors for point-of-care diagnostics. Adv Sci. 2019;6(16):1900925. doi: 10.1002/advs.201900925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang HX, Zhao YW, Li Z, Liu BS, Zhang D (2019) Development and application of aptamer-based surface-enhanced raman spectroscopy sensors in quantitative analysis and biotherapy. Sensors 19(17). 10.3390/s19173806 [DOI] [PMC free article] [PubMed]
- 127.Muhammad M, Huang Q. A REVIEW OF APTAMER-BASED SERS biosensors: design strategies and applications. Talanta. 2021;227:122188. doi: 10.1016/j.talanta.2021.122188. [DOI] [PubMed] [Google Scholar]
- 128.Kukushkin VI, Ivanov NM, Novoseltseva AA, Gambaryan AS, Yaminsky IV, Kopylov AM, Zavyalova EG. Highly sensitive detection of influenza virus with SERS aptasensor. PLoS One. 2019;14(4):e0216247. doi: 10.1371/journal.pone.0216247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou S, Lu C, Li Y, Xue L, Zhao C, Tian G, Bao Y, Tang L, Lin J, Zheng J. Gold nanobones enhanced ultrasensitive surface-enhanced raman scattering aptasensor for detecting Escherichia coli O157:H7. ACS Sens. 2020;5(2):588–596. doi: 10.1021/acssensors.9b02600. [DOI] [PubMed] [Google Scholar]
- 130.Krajczewski J, Ambroziak R, Kudelski A. Substrates for surface-enhanced raman scattering formed on nanostructured non-metallic materials: preparation and characterization. Nanomaterials. 2021;11(1):75. doi: 10.3390/nano11010075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Duan N, Shen M, Wu S, Zhao C, Ma X, Wang Z. Graphene oxide wrapped Fe3O4@Au nanostructures as substrates for aptamer-based detection of vibrio parahaemolyticus by surface-enhanced raman spectroscopy. Microchim Acta. 2017;184(8):2653–2660. doi: 10.1007/s00604-017-2298-9. [DOI] [Google Scholar]
- 132.Wu S, Duan N, Shen M, Wang J, Wang Z. Surface-enhanced raman spectroscopic single step detection of Vibrio parahaemolyticus using gold coated polydimethylsiloxane as the active substrate and aptamer modified gold nanoparticles. Microchim Acta. 2019;186(7):401. doi: 10.1007/s00604-019-3499-1. [DOI] [PubMed] [Google Scholar]
- 133.Yao L, Ye Y, Teng J, Xue F, Pan D, Li B, Chen W. In vitro isothermal nucleic acid amplification assisted surface-enhanced raman spectroscopic for ultrasensitive detection of Vibrio parahaemolyticus. Anal Chem. 2017;89(18):9775–9780. doi: 10.1021/acs.analchem.7b01717. [DOI] [PubMed] [Google Scholar]
- 134.Zhang C, Wang C, Xiao R, Tang L, Huang J, Wu D, Liu S, Wang Y, Zhang D, Wang S, Chen X. Sensitive and specific detection of clinical bacteria via vancomycin-modified Fe3O4@Au nanoparticles and aptamer-functionalized SERS Tags. J Mater Chem B. 2018;6(22):3751–3761. doi: 10.1039/c8tb00504d. [DOI] [PubMed] [Google Scholar]
- 135.Singh P. SPR biosensors: historical perspectives and current challenges. Sens Actuat B Chem. 2016;229:110–130. doi: 10.1016/j.snb.2016.01.118. [DOI] [Google Scholar]
- 136.Park JH, Cho YW, Kim TH (2022) Recent advances in surface plasmon resonance sensors for sensitive optical detection of pathogens. Biosensors (Basel) 12(3). 10.3390/bios12030180 [DOI] [PMC free article] [PubMed]
- 137.Nguyen VT, Seo HB, Kim BC, Kim SK, Song CS, Gu MB. Highly sensitive sandwich-type SPR based detection of whole H5Nx viruses using a pair of aptamers. Biosens Bioelectron. 2016;86:293–300. doi: 10.1016/j.bios.2016.06.064. [DOI] [PubMed] [Google Scholar]
- 138.Kim S, Lee S, Lee HJ. An aptamer-aptamer sandwich assay with nanorod-enhanced surface plasmon resonance for attomolar concentration of norovirus capsid protein. Sens Actuat B Chem. 2018;273:1029–1036. doi: 10.1016/j.snb.2018.06.108. [DOI] [Google Scholar]
- 139.Wang B, Park B, Xu B, Kwon Y. Label-free biosensing of Salmonella enterica serovars at single-cell level. J Nanobiotechnology. 2017;15(1):40. doi: 10.1186/s12951-017-0273-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim DM, Park JS, Jung SW, Yeom J, Yoo SM (2021) Biosensing applications using nanostructure-based localized surface plasmon resonance sensors. Sensors (Basel) 21(9). 10.3390/s21093191 [DOI] [PMC free article] [PubMed]
- 141.Oh SY, Heo NS, Shukla S, Cho HJ, Vilian ATE, Kim J, Lee SY, Han YK, Yoo SM, Huh YS. Development of gold nanoparticle-aptamer-based LSPR sensing chips for the rapid detection of Salmonella typhimurium in pork meat. Sci Rep. 2017;7(1):10130. doi: 10.1038/s41598-017-10188-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Vasilescu A, Gaspar S, Gheorghiu M, David S, Dinca V, Peteu S, Wang Q, Li M, Boukherroub R, Szunerits S. Plasmon resonance based sensing of lysozyme in serum on Micrococcus lysodeikticus-modified graphene oxide surfaces. Biosens Bioelectron. 2017;89(Pt 1):525–531. doi: 10.1016/j.bios.2016.03.040. [DOI] [PubMed] [Google Scholar]
- 143.Li C, Ma X, Guan Y, Tang J, Zhang B. Microcantilever array biosensor for simultaneous detection of carcinoembryonic antigens and alpha-fetoprotein based on real-time monitoring of the profile of cantilever. ACS Sens. 2019;4(11):3034–3041. doi: 10.1021/acssensors.9b01604. [DOI] [PubMed] [Google Scholar]
- 144.Hwang KS, Lee SM, Eom K, Lee JH, Lee YS, Park JH, Yoon DS, Kim TS (2007) Nanomechanical microcantilever operated in vibration modes with use of RNA aptamer as receptor molecules for label-free detection of HCV helicase. Biosens Bioelectron 23(4):459–465. 10.1016/j.bios.2007.05.006 [DOI] [PubMed]
- 145.Le TT, Adamiak B, Benton DJ, Johnson CJ, Sharma S, Fenton R, McCauley JW, Iqbal M, Cass AE. Aptamer-based biosensors for the rapid visual detection of flu viruses. Chem Commun (Camb) 2014;50(98):15533–15536. doi: 10.1039/c4cc07888h. [DOI] [PubMed] [Google Scholar]
- 146.Liu R, He L, Hu Y, Luo Z, Zhang J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem Sci. 2020;11(44):12157–12164. doi: 10.1039/d0sc03920a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Deng J, Tian F, Liu C, Liu Y, Zhao S, Fu T, Sun J, Tan W. Rapid one-step detection of viral particles using an aptamer-based thermophoretic assay. J Am Chem Soc. 2021;143(19):7261–7266. doi: 10.1021/jacs.1c02929. [DOI] [PubMed] [Google Scholar]
- 148.Vashist SK, Luppa PB, Yeo LY, Ozcan A, Luong JHT. Emerging technologies for next-generation point-of-care testing. Trends Biotechnol. 2015;33(11):692–705. doi: 10.1016/j.tibtech.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 149.Zarei M. Pathogens meet point-of-care diagnostics. Biosens Bioelectron. 2018;106:193–203. doi: 10.1016/j.bios.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 150.Kosack CS, Page A-L, Klatser PR. A guide to aid the selection of diagnostic tests. Bull World Health Organ. 2017;95(9):639–645. doi: 10.2471/BLT.16.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gopinath SC, Lakshmipriya T, Chen Y, Phang WM, Hashim U. Aptamer-based 'point-of-care testing'. Biotechnol Adv. 2016;34(3):198–208. doi: 10.1016/j.biotechadv.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 152.Citartan M, Tang TH. Recent developments of aptasensors expedient for point-of-care (POC) diagnostics. Talanta. 2019;199:556–566. doi: 10.1016/j.talanta.2019.02.066. [DOI] [PubMed] [Google Scholar]
- 153.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 154.Ji T, Liu Z, Wang G, Guo X, Akbar Khan S, Lai C, Chen H, Huang S, Xia S, Chen B, Jia H, Chen Y, Zhou Q. Detection of COVID-19: a review of the current literature and future perspectives. Biosens Bioelectron. 2020;166:112455. doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Feng W, Newbigging AM, Le C, Pang B, Peng H, Cao Y, Wu J, Abbas G, Song J, Wang DB, Cui M, Tao J, Tyrrell DL, Zhang XE, Zhang H, Le XC. Molecular diagnosis of COVID-19: challenges and research needs. Anal Chem. 2020;92(15):10196–10209. doi: 10.1021/acs.analchem.0c02060. [DOI] [PubMed] [Google Scholar]
- 156.Erdem O, Es I, Saylan Y, Inci F. Unifying the efforts of medicine, chemistry, and engineering in biosensing technologies to tackle the challenges of the COVID-19 pandemic. Anal Chem. 2021 doi: 10.1021/acs.analchem.1c04454. [DOI] [PubMed] [Google Scholar]
- 157.Zhang W, Liu N, Zhang J. Functional nucleic acids as modular components against SARS-CoV-2: from diagnosis to therapeutics. Biosens Bioelectron. 2022;201:113944. doi: 10.1016/j.bios.2021.113944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Daniels J, Wadekar S, DeCubellis K, Jackson GW, Chiu AS, Pagneux Q, Saada H, Engelmann I, Ogiez J, Loze-Warot D, Boukherroub R, Szunerits S. A mask-based diagnostic platform for point-of-care screening of Covid-19. Biosens Bioelectron. 2021;192:113486. doi: 10.1016/j.bios.2021.113486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shi L, Wang L, Ma X, Fang X, Xiang L, Yi Y, Li J, Luo Z, Li G. Aptamer-functionalized nanochannels for one-step detection of SARS-CoV-2 in samples from COVID-19 patients. Anal Chem. 2021;93(49):16646–16654. doi: 10.1021/acs.analchem.1c04156. [DOI] [PubMed] [Google Scholar]
- 160.Qi H, Hu Z, Yang Z, Zhang J, Wu JJ, Cheng C, Wang C, Zheng L. Capacitive aptasensor coupled with microfluidic enrichment for real-time detection of trace SARS-CoV-2 nucleocapsid protein. Anal Chem. 2022;94(6):2812–2819. doi: 10.1021/acs.analchem.1c04296. [DOI] [PubMed] [Google Scholar]
- 161.Ramanathan S, Gopinath SCB, Ismail ZH, Md Arshad MK, Poopalan P. Aptasensing nucleocapsid protein on nanodiamond assembled gold interdigitated electrodes for impedimetric SARS-CoV-2 infectious disease assessment. Biosens Bioelectron. 2022;197:113735. doi: 10.1016/j.bios.2021.113735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Singh NK, Ray P, Carlin AF, Magallanes C, Morgan SC, Laurent LC, Aronoff-Spencer ES, Hall DA. Hitting the diagnostic sweet spot: point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. Biosens Bioelectron. 2021;180:113111. doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hwang MT, Park I, Heiranian M, Taqieddin A, You S, Faramarzi V, Pak AA, van der Zande AM, Aluru NR, Bashir R. Ultrasensitive detection of dopamine, IL-6 and SARS-CoV-2 proteins on crumpled graphene FET biosensor. Adv Mater Technol. 2021;6(11):2100712. doi: 10.1002/admt.202100712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Adeel M, Asif K, Canzonieri V, Barai HR, Rahman MM, Daniele S, Rizzolio F. Controlled, partially exfoliated, self-supported functionalized flexible graphitic carbon foil for ultrasensitive detection of SARS-CoV-2 spike protein. Sens Actuators B Chem. 2022;359:131591. doi: 10.1016/j.snb.2022.131591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Amouzadeh Tabrizi M, Acedo P (2022) An electrochemical impedance spectroscopy-based aptasensor for the determination of SARS-CoV-2-RBD using a carbon nanofiber-gold nanocomposite modified screen-printed electrode. Biosensors (Basel) 12(3). 10.3390/bios12030142 [DOI] [PMC free article] [PubMed]
- 166.Curti F, Fortunati S, Knoll W, Giannetto M, Corradini R, Bertucci A, Careri M. A folding-based electrochemical aptasensor for the single-step detection of the SARS-CoV-2 spike protein. ACS Appl Mater Interfaces. 2022 doi: 10.1021/acsami.2c02405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Han C, Li W, Li Q, Xing W, Luo H, Ji H, Fang X, Luo Z, Zhang L. CRISPR/Cas12a-derived electrochemical aptasensor for ultrasensitive detection of COVID-19 nucleocapsid protein. Biosens Bioelectron. 2022;200:113922. doi: 10.1016/j.bios.2021.113922. [DOI] [PubMed] [Google Scholar]
- 168.AmouzadehTabrizi M, Nazari L, Acedo P. A photo-electrochemical aptasensor for the determination of severe acute respiratory syndrome coronavirus 2 receptor-binding domain by using graphitic carbon nitride-cadmium sulfide quantum dots nanocomposite. Sens Actuators B Chem. 2021;345:130377. doi: 10.1016/j.snb.2021.130377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jiang ZW, Zhao TT, Li CM, Li YF, Huang CZ. 2D MOF-based photoelectrochemical aptasensor for SARS-CoV-2 spike glycoprotein detection. ACS Appl Mater Interfaces. 2021;13(42):49754–49761. doi: 10.1021/acsami.1c17574. [DOI] [PubMed] [Google Scholar]
- 170.Cennamo N, Pasquardini L, Arcadio F, Lunelli L, Vanzetti L, Carafa V, Altucci L, Zeni L. SARS-CoV-2 spike protein detection through a plasmonic D-shaped plastic optical fiber aptasensor. Talanta. 2021;233:122532. doi: 10.1016/j.talanta.2021.122532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Chen H, Park SG, Choi N, Kwon HJ, Kang T, Lee MK, Choo J. Sensitive detection of SARS-CoV-2 using a SERS-based aptasensor. ACS Sens. 2021;6(6):2378–2385. doi: 10.1021/acssensors.1c00596. [DOI] [PubMed] [Google Scholar]
- 172.Wang Y, Zhang Y, Chen J, Wang M, Zhang T, Luo W, Li Y, Wu Y, Zeng B, Zhang K, Deng R, Li W. Detection of SARS-CoV-2 and its mutated variants via CRISPR-Cas13-based transcription amplification. Anal Chem. 2021;93(7):3393–3402. doi: 10.1021/acs.analchem.0c04303. [DOI] [PubMed] [Google Scholar]
- 173.Chen H, Park SK, Joung Y, Kang T, Lee MK, Choo J. SERS-based dual-mode DNA aptasensors for rapid classification of SARS-CoV-2 and influenza A/H1N1 infection. Sens Actuators B Chem. 2022;355:131324. doi: 10.1016/j.snb.2021.131324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang Y, Xue T, Wang M, Ledesma-Amaro R, Lu Y, Hu X, Zhang T, Yang M, Li Y, Xiang J, Deng R, Ying B, Li W. CRISPR-Cas13a cascade-based viral RNA assay for detecting SARS-CoV-2 and its mutations in clinical samples. Sens Actuators B Chem. 2022;362:131765. doi: 10.1016/j.snb.2022.131765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang W, He Y, Feng Z, Zhang J. Recent advances of functional nucleic acid-based sensors for point-of-care detection of SARS-CoV-2. Microchim Acta. 2022;189(3):128. doi: 10.1007/s00604-022-05242-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Reid R, Chatterjee B, Das SJ, Ghosh S, Sharma TK. Application of aptamers as molecular recognition elements in lateral flow assays. Anal Biochem. 2020;593:113574. doi: 10.1016/j.ab.2020.113574. [DOI] [PubMed] [Google Scholar]
- 177.Phung NL, Walter JG, Jonczyk R, Seiler LK, Scheper T, Blume C. Development of an aptamer-based lateral flow assay for the detection of c-reactive protein using microarray technology as a prescreening platform. ACS Comb Sci. 2020;22(11):617–629. doi: 10.1021/acscombsci.0c00080. [DOI] [PubMed] [Google Scholar]
- 178.Xiao M, Tian F, Liu X, Zhou Q, Pan J, Luo Z, Yang M, Yi C (2022) Virus detection: from state-of-the-art laboratories to smartphone-based point-of-care testing. Adv Sci (Weinh):e2105904. 10.1002/advs.202105904 [DOI] [PMC free article] [PubMed]
- 179.Lin B, Yu Y, Cao Y, Guo M, Zhu D, Dai J, Zheng M. Point-of-care testing for streptomycin based on aptamer recognizing and digital image colorimetry by smartphone. Biosens Bioelectron. 2018;100:482–489. doi: 10.1016/j.bios.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 180.Ji W, Zhang Z, Tian Y, Yang Z, Cao Z, Zhang L, Qi Y, Chang J, Zhang S, Wang H. Shape coding microhydrogel for a real-time mycotoxin detection system based on smartphones. ACS Appl Mater Interfaces. 2019;11(8):8584–8590. doi: 10.1021/acsami.8b21851. [DOI] [PubMed] [Google Scholar]
- 181.Zhu Z, Guan Z, Jia S, Lei Z, Lin S, Zhang H, Ma Y, Tian ZQ, Yang CJ. Au@Pt nanoparticle encapsulated target-responsive hydrogel with volumetric bar-chart chip readout for quantitative point-of-care testing. Angew Chem Int Ed. 2014;53(46):12503–12507. doi: 10.1002/anie.201405995. [DOI] [PubMed] [Google Scholar]
- 182.Ma Y, Mao Y, Huang D, He Z, Yan J, Tian T, Shi Y, Song Y, Li X, Zhu Z, Zhou L, Yang CJ. Portable visual quantitative detection of aflatoxin B1 using a target-responsive hydrogel and a distance-readout microfluidic chip. Lab Chip. 2016;16(16):3097–3104. doi: 10.1039/c6lc00474a. [DOI] [PubMed] [Google Scholar]
- 183.Liu D, Jia S, Zhang H, Ma Y, Guan Z, Li J, Zhu Z, Ji T, Yang CJ. Integrating target-responsive hydrogel with pressuremeter readout enables simple, sensitive, user-friendly, quantitative point-of-care testing. ACS Appl Mater Interfaces. 2017;9(27):22252–22258. doi: 10.1021/acsami.7b05531. [DOI] [PubMed] [Google Scholar]
- 184.Tian T, Wei X, Jia S, Zhang R, Li J, Zhu Z, Zhang H, Ma Y, Lin Z, Yang CJ. Integration of target responsive hydrogel with cascaded enzymatic reactions and microfluidic paper-based analytic devices (microPADs) for point-of-care testing (POCT) Biosens Bioelectron. 2016;77:537–542. doi: 10.1016/j.bios.2015.09.049. [DOI] [PubMed] [Google Scholar]
- 185.Wei X, Zhou W, Sanjay ST, Zhang J, Jin Q, Xu F, Dominguez DC, Li X. Multiplexed instrument-free bar-chart spinchip integrated with nanoparticle-mediated magnetic aptasensors for visual quantitative detection of multiple pathogens. Anal Chem. 2018;90(16):9888–9896. doi: 10.1021/acs.analchem.8b02055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu R, Hu Y, He Y, Lan T, Zhang J. Translating daily COVID-19 screening into a simple glucose test: a proof of concept study. Chem Sci. 2021;12(26):9022–9030. doi: 10.1039/d1sc00512j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhou S, Zhao Y, Mecklenburg M, Yang D, Xie B. A novel thermometric biosensor for fast surveillance of β-lactamase activity in milk. Biosens Bioelectron. 2013;49(15):99–104. doi: 10.1016/j.bios.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 188.Zhang J, Lan T, Lu Y. Translating in vitro diagnostics from centralized laboratories to point-of-care locations using commercially-available handheld meters. Trends Analyt Chem. 2020;124:115782. doi: 10.1016/j.trac.2019.115782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.WHO (2021) The selection and use of essential in vitro diagnostics. WHO Technical Report Series. https://www.who.int/publications/i/item/9789240019102. Accessed 04–07–2022
- 190.Lautner G, Balogh Z, Bardóczy V, Mészáros T, Gyurcsányi RE. Aptamer-based biochips for label-free detection of plant virus coat proteins by SPR imaging. Analyst. 2010;135(5):918–926. doi: 10.1039/B922829B. [DOI] [PubMed] [Google Scholar]
- 191.Kovacevic KD, Gilbert JC, Jilma B. Pharmacokinetics, pharmacodynamics and safety of aptamers. Adv Drug Deliver Rev. 2018;134:36–50. doi: 10.1016/j.addr.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 192.Yu H, Alkhamis O, Canoura J, Liu Y, Xiao Y. Advances and challenges in small-molecule DNA aptamer isolation, characterization, and sensor development. Angew Chem Int Ed. 2021;60(31):16800–16823. doi: 10.1002/anie.202008663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Yan J, Xiong H, Cai S, Wen N, He Q, Liu Y, Peng D, Liu Z. Advances in aptamer screening technologies. Talanta. 2019;200:124–144. doi: 10.1016/j.talanta.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 194.Eid C, Palko JW, Katilius E, Santiago JG. Rapid slow off-rate modified aptamer (SOMAmer)-based detection of c-reactive protein using isotachophoresis and an ionic spacer. Anal Chem. 2015;87(13):6736–6743. doi: 10.1021/acs.analchem.5b00886. [DOI] [PubMed] [Google Scholar]
- 195.Duo J, Chiriac C, Huang RYC, Mehl J, Chen G, Tymiak A, Sabbatini P, Pillutla R, Zhang Y. Slow off-rate modified aptamer (SOMAmer) as a novel reagent in immunoassay development for accurate soluble glypican-3 quantification in clinical samples. Anal Chem. 2018;90(8):5162–5170. doi: 10.1021/acs.analchem.7b05277. [DOI] [PubMed] [Google Scholar]
- 196.Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, Kim N, Saccomano NA, Wilcox SK, Zichi D, Sanders GM. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: a SOMAmer-based, streamlined multiplex proteomic assay. PLoS One. 2011;6(10):e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Dembowski SK, Bowser MT. Microfluidic methods for aptamer selection and characterization. Analyst. 2018;143(1):21–32. doi: 10.1039/c7an01046j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nguyen Quang N, Perret G, Ducongè F. Applications of high-throughput sequencing for in vitro selection and characterization of aptamers. Pharmaceuticals. 2016;9:76. doi: 10.3390/ph9040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hamada M. In silico approaches to RNA aptamer design. Biochimie. 2018;145:8–14. doi: 10.1016/j.biochi.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 200.Thevendran R, Navien TN, Meng X, Wen K, Lin Q, Sarah S, Tang T-H, Citartan M. Mathematical approaches in estimating aptamer-target binding affinity. Anal Biochem. 2020;600:113742. doi: 10.1016/j.ab.2020.113742. [DOI] [PubMed] [Google Scholar]
- 201.Lange MJ, Nguyen PDM, Callaway MK, Johnson MC, Burke DH. RNA-protein interactions govern antiviral specificity and encapsidation of broad spectrum anti-HIV reverse transcriptase aptamers. Nucleic Acids Res. 2017;45(10):6087–6097. doi: 10.1093/nar/gkx155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Alam KK, Chang JL, Lange MJ, Nguyen PDM, Sawyer AW, Burke DH. Poly-target selection identifies broad-spectrum RNA aptamers. Mol Ther Nucleic Acids. 2018;13:605–619. doi: 10.1016/j.omtn.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ahmed SF, Quadeer AA, McKay MR (2020) Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses 12(3). 10.3390/v12030254 [DOI] [PMC free article] [PubMed]
- 204.Sharifi S, Vahed SZ, Ahmadian E, Dizaj SM, Eftekhari A, Khalilov R, Ahmadi M, Hamidi-Asl E, Labib M. Detection of pathogenic bacteria via nanomaterials-modified aptasensors. Biosens Bioelectron. 2020;150:111933. doi: 10.1016/j.bios.2019.111933. [DOI] [PubMed] [Google Scholar]
- 205.Kurup CP, Mohd-Naim NF, Ahmed MU. Recent trends in nanomaterial-based signal amplification in electrochemical aptasensors. Crit Rev Biotechnol. 2021;42(5):794–812. doi: 10.1080/07388551.2021.1960792. [DOI] [PubMed] [Google Scholar]
- 206.Zahra QuA, Luo Z, Ali R, Khan MI, Li F, Qiu B. Advances in gold nanoparticles-based colorimetric aptasensors for the detection of antibiotics: an overview of the past decade. Nanomaterials. 2021;11(4):840. doi: 10.3390/nano11040840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yu L, Song Z, Peng J, Yang M, Zhi H, He H. Progress of gold nanomaterials for colorimetric sensing based on different strategies. Trends Anal Chem. 2020;127:115880. doi: 10.1016/j.trac.2020.115880. [DOI] [Google Scholar]
- 208.Gopinath SCB, Lakshmipriya T, Awazu K. Colorimetric detection of controlled assembly and disassembly of aptamers on unmodified gold nanoparticles. Biosens Bioelectron. 2014;51:115–123. doi: 10.1016/j.bios.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Evtugyn G, Porfireva A, Shamagsumova R, Hianik T. Advances in electrochemical aptasensors based on carbon nanomaterials. Chemosensors. 2020;8(4):96. doi: 10.3390/chemosensors8040096. [DOI] [Google Scholar]
- 210.Yang Y, Yang X, Yang Y, Yuan Q. Aptamer-functionalized carbon nanomaterials electrochemical sensors for detecting cancer relevant biomolecules. Carbon. 2018;129:380–395. doi: 10.1016/j.carbon.2017.12.013. [DOI] [Google Scholar]
- 211.Vasilescu A, Hayat A, Gáspár S, Marty J-L. Advantages of carbon nanomaterials in electrochemical aptasensors for food analysis. Electroanalysis. 2018;30(1):2–19. doi: 10.1002/elan.201700578. [DOI] [Google Scholar]
- 212.Wen L, Qiu LP, Wu YX, Hu XX, Zhang XB (2017) Aptamer-modified semiconductor quantum dots for biosensing applications. Sensors 17(8). 10.3390/s17081736 [DOI] [PMC free article] [PubMed]
- 213.Li HH, Ahmad W, Rong YW, Chen QS, Zuo M, Ouyang Q, Guo ZM. Designing an aptamer based magnetic and upconversion nanoparticles conjugated fluorescence sensor for screening Escherichia coli in food. Food Control. 2020;107:106761. doi: 10.1016/j.foodcont.2019.106761. [DOI] [Google Scholar]
- 214.Reddy LH, Arias JL, Nicolas J, Couvreur P. Magnetic nanoparticles: design and characterization, toxicity and biocompatibility, pharmaceutical and biomedical applications. Chem Rev. 2012;112(11):5818–5878. doi: 10.1021/cr300068p. [DOI] [PubMed] [Google Scholar]
- 215.Modh H, Scheper T, Walter JG. Aptamer-modified magnetic beads in biosensing. Sensors. 2018;18(4):1041. doi: 10.3390/s18041041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gou D, Xie G, Li Y, Zhang X, Chen H. Voltammetric immunoassay for Mycobacterium tuberculosis secretory protein MPT64 based on a synergistic amplification strategy using rolling circle amplification and a gold electrode modified with graphene oxide, Fe3O4 and Pt nanoparticles. Microchim Acta. 2018;185(9):436. doi: 10.1007/s00604-018-2972-6. [DOI] [PubMed] [Google Scholar]
- 217.Yu X, Song HL, Huang J, Chen YX, Dai M, Lin XC, Xie ZH. An aptamer@AuNP-modified POSS-polyethylenimine hybrid affinity monolith with a high aptamer coverage density for sensitive and selective recognition of ochratoxin A. J Mater Chem B. 2018;6(13):1965–1972. doi: 10.1039/C7TB03319B. [DOI] [PubMed] [Google Scholar]
- 218.Ding CP, Zhang CL, Cheng SS, Xian YZ. Multivalent aptamer functionalized Ag2S nanodots/hybrid cell membrane-coated magnetic nanobioprobe for the ultrasensitive isolation and detection of circulating tumor cells. Adv Funct Mater. 2020;30(16):1909781. doi: 10.1002/adfm.201909781. [DOI] [Google Scholar]
- 219.Huang L, Tian S, Zhao W, Liu K, Ma X, Guo J. Multiplexed detection of biomarkers in lateral-flow immunoassays. Analyst. 2020;145(8):2828–2840. doi: 10.1039/c9an02485a. [DOI] [PubMed] [Google Scholar]
- 220.Han X, Liu Y, Yin J, Yue M, Mu Y. Microfluidic Devices for Multiplexed Detection of Foodborne Pathogens. Food Res Int. 2021;143:110246. doi: 10.1016/j.foodres.2021.110246. [DOI] [PubMed] [Google Scholar]
- 221.Berry ME, Kearns H, Graham D, Faulds K. Surface enhanced raman scattering for the multiplexed detection of pathogenic microorganisms: towards point-of-use applications. Analyst. 2021;146(20):6084–6101. doi: 10.1039/d1an00865j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Laing S, Gracie K, Faulds K. Multiplex in vitro detection using SERS. Chem Soc Rev. 2016;45(7):1901–1918. doi: 10.1039/c5cs00644a. [DOI] [PubMed] [Google Scholar]
- 223.Zhang K, Li H, Wang W, Cao J, Gan N, Han H. Application of multiplexed aptasensors in food contaminants detection. ACS Sens. 2020;5(12):3721–3738. doi: 10.1021/acssensors.0c01740. [DOI] [PubMed] [Google Scholar]
- 224.Grabowska I, Hepel M, Kurzatkowska-Adaszynska K. Advances in design strategies of multiplex electrochemical aptasensors. Sensors (Basel) 2022;22(1):161. doi: 10.3390/s22010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Yoo SM, Kim DK, Lee SY. Aptamer-functionalized localized surface plasmon resonance sensor for the multiplexed detection of different bacterial species. Talanta. 2015;132:112–117. doi: 10.1016/j.talanta.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 226.Zhang H, Ma X, Liu Y, Duan N, Wu S, Wang Z, Xu B. Gold nanoparticles enhanced SERS aptasensor for the simultaneous detection of Salmonella typhimurium and Staphylococcus aureus. Biosens Bioelectron. 2015;74:872–877. doi: 10.1016/j.bios.2015.07.033. [DOI] [PubMed] [Google Scholar]
- 227.Kurt H, Yuce M, Hussain B, Budak H. Dual-excitation upconverting nanoparticle and quantum dot aptasensor for multiplexed food pathogen detection. Biosens Bioelectron. 2016;81:280–286. doi: 10.1016/j.bios.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 228.Somvanshi SB, Ulloa AM, Zhao M, Liang Q, Barui AK, Lucas A, Jadhav KM, Allebach JP, Stanciu LA. Microfluidic paper-based aptasensor devices for multiplexed detection of pathogenic bacteria. Biosens Bioelectron. 2022;207:114214. doi: 10.1016/j.bios.2022.114214. [DOI] [PubMed] [Google Scholar]
- 229.Kost GJ. Geospatial hotspots need point-of-care strategies to stop highly infectious outbreaks: ebola and coronavirus. Arch Pathol Lab Med. 2020;144(10):1166–1190. doi: 10.5858/arpa.2020-0172-RA. [DOI] [PubMed] [Google Scholar]
- 230.Zhang Z, Ma P, Ahmed R, Wang J, Akin D, Soto F, Liu B-F, Li P, Demirci U. Advanced point-of-care testing technologies for human acute respiratory virus detection. Adv Mater. 2022;34(1):2103646. doi: 10.1002/adma.202103646. [DOI] [PubMed] [Google Scholar]
- 231.Van Der Pol B (2020) Making the most of point-of-care testing for sexually transmitted diseases. Clin Infect Dis70(9):1824–1825. 10.1093/cid/ciz523 [DOI] [PubMed]
- 232.Dhiman A, Kalra P, Bansal V, Bruno JG, Sharma TK. Aptamer-based point-of-care diagnostic platforms. Sens Actuat B Chem. 2017;246:535–553. doi: 10.1016/j.snb.2017.02.060. [DOI] [Google Scholar]