Abstract
Individuals with chronic obstructive pulmonary disease (COPD) are more susceptible to exacerbation crisis triggered by secondary lung infections due to the dysfunction of antiviral signaling, principally via suppression of IFN-γ. Although the probiotic is known for controlling pulmonary inflammation in COPD, the influence of the Lactobacillus rhamnosus (Lr) on antiviral signaling in bronchial epithelium exposed to cigarette smoke extract (CSE) and viruses, remains unknown. Thus, the present study investigated the Lr effect on the antiviral signaling and the secretion of inflammatory mediators from bronchial epithelial cells (16HBE cells) exposed to CSE and SARS-CoV-2. The 16HBE cells were cultured, treated with Lr, stimulated with CSE, and infected with SARS-CoV-2. The cellular viability was evaluated using the MTT assay and cytotoxicity measured by lactate dehydrogenase (LDH) activity. The viral load, TLR2, TLR3, TLR4, TLR7, TLR8, MAVS, MyD88, and TRIF were quantified using specific PCR. The pro-inflammatory mediators were measured by a multiplex biometric immunoassay, and angiotensin converting enzyme 2 (ACE2) activity, NF-κB, RIG-I, MAD5, and IRF3 were measured using specific ELISA kits. Lr decreased viral load, ACE2, pro-inflammatory mediators, TLR2, TLR4, NF-κB, TLR3, TLR7, and TLR8 as well as TRIF and MyD88 expression in CSE and SARS-CoV-2 -exposed 16HBE cells. Otherwise, RIG-I, MAD5, IRF3, IFN-γ, and the MAVS expression were restored in 16HBE cells exposed to CSE and SARS-CoV-2 and treated with Lr. Lr induces antiviral signaling associated to IFN-γ secreting viral sensors and attenuates cytokine storm associated to NF-κB in bronchial epithelial cells, supporting its emerging role in prevention of COPD exacerbation.
Keywords: COVID-19, Smoking, Bronchial epithelium, Antiviral signaling, Cytokine storm, Probiotic
Introduction
Inhalation of cigarette smoke is closely linked to the pathophysiology of chronic obstructive pulmonary disease (COPD) [1]. In this scenario, structural cells such as airway epithelial cells have a key role in both initial and later phase of inflammatory response in COPD [2]. In fact, the integrity of airway epithelial cells is tightly regulated by immunological response against pathogens or tissue damage [3]. Therefore, airway epithelial cells can be activated by harmful particles, and thus these cells secrete exaggerated concentrations of pro-inflammatory cytokines that amplify the migration of inflammatory cells towards the lung tissue [4].
It is well known that individuals with COPD present a defective immune response, and it has a critical role in secondary respiratory infection, such as viral infection [5, 6]. Particularly, cytoplasmic receptors capable of identifying the viral RNA, such as RIG-I (retinoic acid-inducible gene I) and MDA5 (melanoma differentiation-associated protein 5), through MAVS (mitochondrial antiviral-signaling protein) and IRF3 (interferon regulatory factor) activation, present a lower expression in airway from COPD subjects, resulting in a lower secretion of IFN-γ associated to antiviral response [7–10]. Oppositely, some studies indicate that the activation of endosomal viral sensors, such as TLR3 (Toll-like receptor 3), induce a marked secretion of pro-inflammatory cytokines [11, 12] from epithelial airway cells via signaling TRIF (TIR-domain-containing adapter-inducing interferon-β)/NF-κB (nuclear factor kappa B) [13]. In addition, both TLR7 (Toll-like receptor 7) and TLR8 (Toll-like receptor 8) also activate NF-κB signaling via MyD88 (myeloid differentiation primary response 88) to secrete pro-inflammatory cytokines [14, 15]. Therefore, COPD individuals present a lung microenvironmental in which the antiviral immune response is compromised, and the pro-inflammatory response is exacerbated.
The Coronavirus disease 2019, or COVID-19, is an acute viral illness caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in which the main features are epithelial barrier destruction, microvascular dilatation, edema, and immune cell infiltration [16, 17]. This condition amplifies the inflammatory response in COPD leading to a synergistic deterioration in lung function and prolonged hospitalization [18, 19].
It is well known that viral load is critical to determine the degree of infection and the severity of SARS [20]. The angiotensin-2 converting enzyme (ACE2) is highly expressed in airway epithelium [21], and it is crucial in facilitating the entry of SARS-CoV-2 into the respiratory system of host [22, 23]. COPD individuals present an increased activity of ACE2 in airway epithelial cells [24], and for this reason are included into the risk group for COVID-19. In fact, some authors have demonstrated that COPD individuals and infected with SARS-CoV-2 present an exacerbated secretion of cytokines from airway epithelial cells [25, 26].
Due to limitations in the treatment of COPD exacerbation, innovative actions are needed [27]. In this scenario, the beneficial bacterial strain, called probiotics, have been recommended to relieve the symptoms of asthma, COPD, and virus-induced pneumonia due to the anti-inflammatory action and restoration of balance immune response [28–31]. Nevertheless, the probiotic effect on antiviral response of airway epithelium in COPD exacerbation remains not elucidated.
Thus, the present study investigated whether L. rhamnosus to reinstate antiviral signaling as well as attenuate the pro-inflammatory storm in a lineage of human bronchial epithelial cells exposed to CSE and infected with SARS-CoV-2.
Material and Methods
SARS-CoV-2
SARS-CoV-2 was kindly provided by Laboratory of Virology at Department of Immunology of State University of Rio de Janeiro, Rio de Janeiro, Brazil. SARS-CoV-2 was employed in the experiments at a multiplicity of infection (MOI) of 0.1. The study was approved by the Research Ethics Committee of the Federal University of São Paulo, under opinion number 2161190520.
Experimental Groups
All experimental assays were performed in human bronchial epithelial cells which were divided in 8 experimental groups, as follows: (1) control: cells not stimulated; (2) Lr — cells exposed to Lactobacillus rhamnosus; (3) CSE — cells stimulated with cigarette smoke extract; (4) Lr + CSE: cells treated with Lactobacillus rhamnosus and stimulated with CSE; (5) SARS-CoV-2: cells infected with SARS-CoV-2; (6) Lr + SARS-CoV-2: cells treated with Lactobacillus rhamnosus and infected with SARS-CoV-2; (7) CSE + SARS-CoV-2: cells exposed to CSE and infected with SARS-CoV-2; and (8) Lr + CSE + SARS-CoV-2: cells treated with Lactobacillus rhamnosus and stimulated with CSE and infected with SARS-CoV-2.
Human Bronchial Epithelial Cells (16HBE) and Culture Conditions
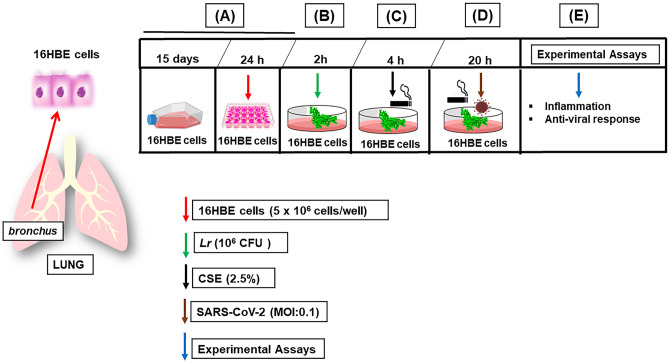
The human bronchial epithelium cell line (16HBE (ATCC® CRL-9609™)) was isolated from normal human bronchial epithelium obtained from autopsy of healthy individuals. 16HBE cells was cultured for 15 days in T25 flasks at a density of 1 × 106 cells per flask with DMEM medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37 °C with 5% CO2 in a humidified incubator. 16HBE cells were centrifuged and resuspended in fresh complete medium and in 24-well plates at a concentration of 5 × 106 cells per well, 24 h prior to experimental assay. After that, the cells were incubated with Lr (1 × 106 CFU) for 2 h. Two hours later of Lr addition, the cells were stimulated with 2.5% of CSE during 4 h (Lr + CSE group). The CSE was made from 1 unfiltered cigarette which was burned in 10 mL of culture medium. A vacuum pump was used at a pressure of −11 Kpa so that the cigarette smoke could be incorporated into the culture medium. Two hours later of Lr addition, 16HBE cells were infected with SARS-CoV-2 at 0.1 of multiplicity of infection index (MOI) during 20 h (Lr + SARS-CoV-2 group). To mimic in vitro the COPD exacerbation induced by COVID-19, 16HBE cells were stimulated with 2.5% of CSE during 4 h; elapsed 4 h of CSE addition, the cells were infected with SARS-CoV-2 at 0.1 of multiplicity of infection index (MOI) during 20 h (CSE + SARS-CoV-2 group). The 16HBE cells remained exposed to CSE associated to SARS-CoV-2 during 20 h to induce the COPD exacerbation. Elapsed 20 h of SARS-CoV-2 addition, the experimental assays were performed. Two hours later of Lr addition, 16HBE cells were stimulated with 2.5% of CSE during 4 h and infected with SARS-CoV-2 (MOI 0.1) during 20 h (Lr + CSE + SARS-CoV-2 group). 16HBE cells not stimulated were used as control group. The Lr group consisted of 16HBE cells stimulated only with 1 × 106 CFU of Lr for 2 h. The SARS-CoV-2 group consisted of 16HBE cells infected with SARS-CoV-2 for 20 h. The experimental design is illustrated in Fig. 1.
Fig. 1.
Treatment with Lactobacillus rhamnosus and 16HBE cell exposure to CSE and SARS-CoV-2. A 16HBE cells was cultured for 15 days in T25 flasks at a density of 1 × 106 cells per flask with DMEM medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin at 37 °C with 5% CO2 in a humidified incubator. 16HBE cells were centrifuged and resuspended in fresh complete medium and in 24-well plates at a concentration of 5 × 106 cells per well, 24 h prior to add Lr, or cigarette smoke extract (CSE), or SARS-CoV-2, or CSE + SARS-CoV-2, or Lr + CSE + SARS-CoV-2. B After that, the cells were incubated with Lr (1 × 106 CFU) for 2 h. C Two hours later of Lr addition, the cells were stimulated with 2.5% of CSE during 4 h. D Elapsed 4 h of CSE addition, the cells were infected with SARS-CoV-2 at 0.1 of multiplicity of infection index (MOI) during 20 h. 16HBE cells remained exposed to CSE associated to SARS-CoV-2 for 20 h to induce the COPD exacerbation. E Elapsed 20 h of SARS-CoV-2 addition, the experimental assays were performed
Cytotoxicity Assay by MTT Assay
To assess the cytotoxicity to CSE, MTT assay was carried out. Briefly, 5 × 104 viable 16HBE cells were placed into clear 96-well flat-bottom plates (Corning USA) in MEM medium supplemented with 10% FBS (Invitrogen) and 1% penicillin/streptomycin and immediately after different concentrations (1.5%, 2.5%, 3.5%, 5%) of CSE. Following 24 h after incubation in a humidified atmosphere of a CO2 incubator (5% CO2, 37 °C), 10 μL/well of MTT (5 mg/mL) was added to the cells (both in the control and CSE), which was incubated for 4 h. After this time, 100 μL of 10% sodium dodecyl sulfate solution in deionized water was added to the cells and incubated overnight. The absorbance was measured at 595 nm in a benchtop multimode reader SpectraMax i3 (Molecular Devices, San Jose, CA, USA).
Cell Viability
The 16HBE cells viability test was evaluated using the MTT [3-(4.5-dimethylthiazol-2-yl)-2.5-diphenyltetrazolium bromide] method in bronchial epithelial cells in all experimental groups. The cells were initially cultivated with 2 × 104 cells in 200 µL/well in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin in a 96-well plate in an incubator at 37 °C and 5% CO2 for 24 h. Then, the cells were stimulated with 2.5% of cigarette smoke extract and infected with SARS-CoV-2 at 0.1 of multiplicity of infection index (MOI) remaining for 24 h in a humidified oven at 37 °C in an atmosphere of 5% CO2. After the experimental time, the medium of the wells with cells stimulated with the compounds was replaced by 100 µL/well of medium containing MTT at a concentration of 0.5 mg/mL. The plate with cells in the presence of MTT was left in the oven for 4 h, then 100 µL of SDS was added to each well, and the plate remained incubated overnight in the oven. After this process, the formazan crystal concentration was quantified by spectroscopy using a microplate reader (ELISA Reader — Spectra Count — Packards Instrument, Offenburg — Germany) with a wavelength of 570 nm.
LDH Activity
Lactate dehydrogenase (LDH) activity was measured in the culture medium after 24 h as an index of cytotoxicity, employing an LDH kit (Bayer Diagnostics VR, France). The optical density of the samples was measured by an adjusted microplate reader at 490 nm. Enzyme activity was expressed as the extra-cellular LDH activity percentage of the total activity in the wells.
Measurement of ACE2
Supernatant samples from 16HBE cells were processed to assess the concentration of angiotensin converting enzyme 2 (ACE2) using ELISA kit for humans. Briefly, 16HBE cell supernatant was used in a quantitative sandwich enzyme immunoassay technique. The intensity of the measurement is proportional to the amount of ACE2 evaluated in specific lysate kits in the initial step. Values are expressed as pg/mL deduced from standard runs in parallel with recombinant ACE2.
Viral RNA Extraction
16HBE cells were washed and harvested by trypsinization and centrifugation, 24,000 rpm for 1 min at room temperature. To each tube, 600 mL of pre-chilled denaturing solution (Promega, Southampton, UK) was added, and the mixture homogenized at total RNA was extracted from the homogenate with phenol:chloroform:isoamyl alcohol (99:24:1, pH 4.7) (Promega), precipitated from the upper phase with an equal volume of isopropanol at −2 °C for 30 min. The RNA was recovered by centrifugation at 10,000 g for 20 min at 41 °C, washed with ice-cold 75% v/v ethanol and air dried in an RNase-free environment for 15 min. RNA was dissolved in nuclease-free water, and RNA concentration and purity were estimated by optical density determination. The 260/280 ratio was usually 41.6, indicating suitable purity of RNA.
Real-time Polymerase Chain Reaction (PCR) for Detection of Viral Load
Allplex™ 2019-nCoV Assay (Seegene, Korea), which targets envelope gene of SARS-CoV-2, was used for SARS-CoV-2 RNA detection according to the manufacturer’s instructions. Briefly, 8 μL of extracted RNA was added to 5 μL of 5 × real-time one-step buffer, 5 μL of 2019-nCoV MuDT Oligo Mix (2019-nCoV-MOM), 2 μL of real-time one-step enzyme, and 5 μL of RNase free water. The CFX-96 real-time thermal cycler (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used for amplification. The conditions consisted of 1 cycle of 20 min at 50 °C, 1 min at 95 °C and followed by 45 cycles of 15 s at 94 °C and 30 s at 58 °C. The result was analyzed using Seegene Viewer (Seegene, Korea), in which a cycle threshold value (Ct-value) < 40 for all three target genes was defined as a positive result.
Gene Expression of TLR2, TLR3, TLR4, TLR7, TLR8, TRIF, MyD88, and MAVS by Real-time Quantitative PCR
Gene expression of TLR2 [32], TLR3 [33], TLR4 [32], TLR7 [32], TLR8 [32], TRIF [34], MyD88 [34], and MAVS [35] was quantified by real-time reverse transcription-polymerase chain reaction using the Promega SV Total RNA Isolation System kit, according to the manufacturer’s instructions. Values were normalized by the expression of GAPDH [35] expressed by arbitrary units. The sequence of primers can be found in Table 1.
Table 1.
List of primers sequence used in this study
| Primers sequence | ||
|---|---|---|
| Gene | Forward | Reverse |
| TLR2 | AACTTACTGGGAAATCCTTAC | AAAAATCTCCAGCAGTAAAAT |
| TLR3 | AGTGCCGTCTATTTGCCACACA | AACAGTGCACTTGGTGGTGGAG |
| TLR4 | CGAGGAAGAGAAGACACCAGT | CATCATCCTCACTGCTTCTGT |
| TLR7 | AAACTCCTTGGGGCTAGATG | AGGGTGAGGTTCGTGGTGTT |
| MAVS | ATAAGTCCDGAGGGCACCTTT | GTGACTACCAGCACCCCTGT |
| TRIF | CCTCCTCCTCCTCCTCCTC | GCGTGGAGGATCACAAAGTT |
| MYD88 | TGGCACCTGTGTCTGGTCTA | ACATTCCTTGCTCTGCAGGT |
| GAPDH | GCACCGTCAAGGCTGAGAAC | ATGGTGGTGAAGACGCCAGT |
TLR Toll-like receptor, MAVS mitochondrial antiviral signaling, TRIF TIR-domain containing adapter inducing IFN-β, MyD88 myeloid differentiation primary response gene 88, GAPDH glyceraldehyde-3-phosphate dehydrogenase
Immunoassay
Supernatant samples from 16HBE cells were processed for a multiplex biometric immunoassay, using fluorescently dyed microspheres conjugated to monoclonal antibodies specific for a target protein, was used to measure 11 cytokines (TNF-α, IL-1β, IL-6, IL-8, IL-33, TSLP, MCP-1, GM-CSF, and IFN-γ), the viral sensors RIG-I, MDA5, and IRF3, and the transcription factor NF-Κb, according to the manufacturer’s instructions (Bio-Plex Human Cytokine Assay; Bio-Rad Inc., Hercules, CA, USA). The protein concentration of each cytokine, chemokine, receptor, and transcription factor was determined using a multiplex array reader from Luminex™ Instrumentation System (Bio-Plex Workstation from Bio-Rad Laboratories, Hercules, CA, USA). The analyte concentrations were calculated using software provided by the manufacturer (Bio-Plex Manager Software).
Statistical Analysis
Statistical analyses were conducted using the GraphPad Prism v.6.0 software. For the treatment of the obtained data, we used the analysis of variance (ANOVA) followed by the Tukey post-test. The results indicated by “p < 0.05” were considered significant, with a 95% confidence interval.
Results
Cell Viability
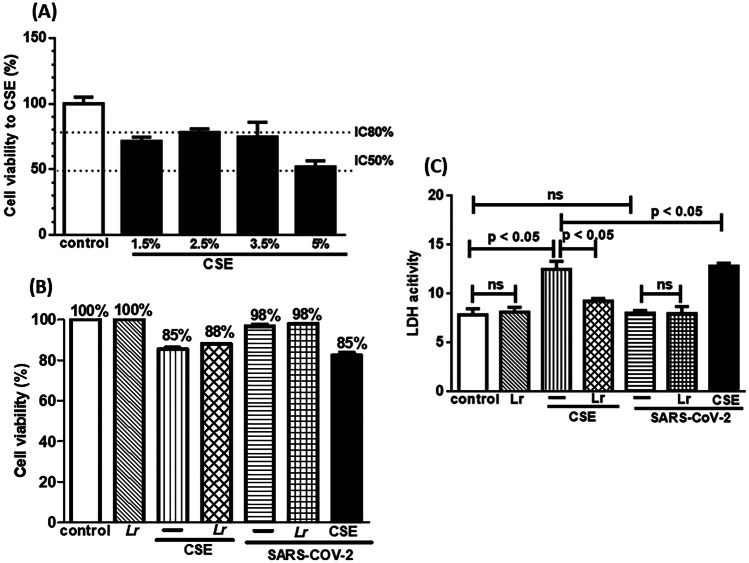
Figure 2 illustrates the cytotoxicity to CSE (Fig. 2A), the cellular viability (Fig. 2B), and the membrane integrity (Fig. 2C). To evaluate the cytotoxic effect of CSE, 16HBE cells were exposed to different concentrations of CSE (1.5%, 2.5%, 3.5%, and 5%). Among the studied concentrations, 2.5% of CSE was chosen due to fact that there is 80% (IC80%) of viable cells compared with 5% CSE concentration, in which the cell viability reached 50% (IC50%), compared with the control group (Fig. 2A). Figure 2B shows that cell viability to control, Lr, SARS-CoV-2, Lr + SARS-CoV-2 groups was not significantly different, compared to the control group. On the contrary, the cell viability represented in CSE, Lr + CSE, CSE + SARS-CoV-2, and Lr + CSE + SARS-CoV-2 groups was statistically different in comparison to the control group. It is worth noting that the SARS-CoV-2 infection with MOI 0.1 used in the present manuscript did not induce a significant change in the cell viability. Therefore, the reduction of cell viability against CSE, Lr + CSE, CSE + SARS-CoV-2, and Lr + CSE + SARS-CoV-2 was totally dependent on CSE. Figure 2C corroborates with these findings, once that the evaluation of membrane integrity through LDH activity in control, Lr, SARS-CoV-2, and Lr + SARS-CoV-2 was not also significantly different, compared to the control. Otherwise, the LDH activity was increased in cells exposed to CSE as well as in CSE + SARS-CoV-2 group. Whereas SARS-CoV-2 group did not present change in LDH activity, so surely the rise of LDH activity in CSE + SARS-coV-2 group was also due to the cell exposure to CSE. In fact, it is well known that individual with COPD or airway cells exposed to CSE present high levels of LDH activity [36, 37]. Still in Fig. 2C, the treatment with Lr does not change the LDH activity in Lr + SARS-CoV-2 group, compared to SARS-CoV-2 group. On the contrary, Lr-treated cells and exposed to CSE (Lr + CSE group) present a significant reduction of LDH activity, compared to CSE group. At the same sense, treatment with Lr reduced the LDH activity in Lr + CSE + SARS-CoV-2 group to similar levels those found in Lr + CSE group. These data indicate that the low levels of LDH activity in Lr + CSE + SARS-CoV-2 group cells are due to Lr effect on exposure to CSE. There is no difference between the control and Lr groups. An important data illustrated in Fig. 2C shows that the multiplicity of infection (MOI) used in the present manuscript does not interfere on the LDH activity because the viral particles with 0.1 MOI are not able to induce death cell, a situation in which there is a substantial rise of LDH activity [38, 39]. The pro-inflammatory storm and the defective antiviral response are present with MOI 0.1 but it is unable to induce significant cell death in conditions performed in the present manuscript. There really are some authors who have demonstrated that MOI higher than 1 [40, 41] is usual in studies that focus on the effect of SARS-CoV-2 on mechanisms such as cell death or in an infection level in which cytokine storm is uncontrollable and not treatable. In this scenario, there is a significant increase of LDH activity, as a marker of cell membrane damage. Our experiments show that the bronchial epithelial cells are infected, since that occurred an increase of viral load in the presence of SARS-CoV-2 (Fig. 3A); however, there is no alteration of LDH activity because the cell viability was not significantly altered in the presence of virus (Fig. 2B). In fact, the measurement of LDH activity in our study was due to exposure of cells to cigarette smoke, once it is well known that cigarette smoke induces cell death by injuring the cell membrane. It was evidenced when the cell viability was reduced in the presence of CSE. However, the reduction of cell viability after CSE did not compromise the experimental assays.
Fig. 2.
Cell viability and membrane integrity of 16HBE cells. Cell viability to cigarette smoke extract (CSE) at 1.5%, 2.5%, 3.5%, and 5% (A). Cell viability (B) and catalytic activity of LDH (C) in 16HBE cells exposed to Lr (1 × 106 CFU), CSE (2.5%), Lr + CSE or SARS-CoV-2 (MOI: 0.1), or CSE + SARS-CoV-2 (B). Cell viability was determined using the MTT assay with an absorbance of 570 nm, and the LDH activity was expressed as the extra-cellular LDH activity percentage of the total activity in the wells. Data are presented as means ± SE from triplicate samples for each treatment versus control. Ns, non-significant difference
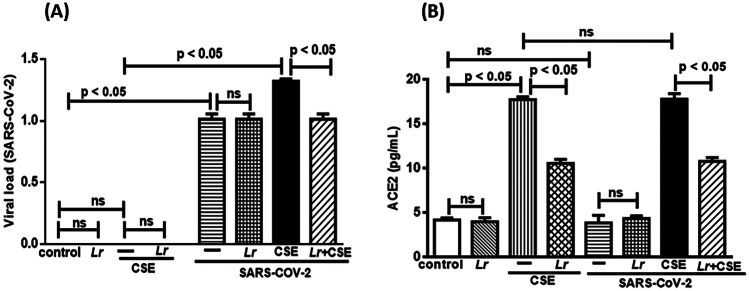
Fig. 3.
Lactobacillus rhamnosus on viral load and ACE2 level. Viral charge (A) and ACE2 concentration (B) in 16HBE cells treated with Lr (1 × 106 CFU) for 2 h, and then exposed to CSE (2.5%) associated to SARS-CoV-2 (MOI: 0.1) during 20 h. The viral load of SARS-CoV-2 was measured by real-time polymerase chain reaction (PCR), and the ACE2 level was detected using an ELISA kit for humans. Data are presented as means ± SE from triplicate samples for each treatment versus control. ns, non-significant difference
Effect of L. rhamnosus on Viral Load and ACE2 Activity
It is well known that the rise of SARS-CoV-2 viral load is related to increased activity of ACE2 in COPD individuals, and consequently to severity of lung inflammation during COVID-19. According to these findings, Fig. 3A illustrates that viral load in Lr, CSE, Lr + CSE groups was not changed, compared to the control group. Obviously, it is due to the fact that cells from control, Lr, CSE, and Lr + CSE group were not exposed to SARS-CoV-2. On the contrary, there was a significant increase of viral load in cells infected with SARS-CoV-2 (SARS-CoV-2 group), compared to the control group. These data evidence that the rise of the load viral is totally dependent on SARS-CoV-2 infection, and that no experimental artifact or stimulus performed in the present manuscript is capable of increasing the viral load, besides the virus itself. However, cells exposed to CSE and infected with SARS-CoV-2 (CSE + SARS-CoV-2 group) had a significantly higher viral load than that found in SARS-CoV-2 group. This result can be interpreted as an indirect effect of CSE, since the increase in both the expression and the activity of ACE2 in COPD individuals is well described [25]. Whereas that the ACE2 is the main receptor for the SARS-CoV-2 entry into the host cell is reasonable, suggests that the exposure to CSE increases the viral load in a way dependent on ACE2 rise. Conversely, Fig. 3A shows that Lr induced a significant reduction of viral load in Lr + CSE + SARS-CoV-2 group, compared to CSE + SARS-CoV-2. These findings demonstrate that Lr-induced viral load reduction was depedent on CSE presence, once that Lr did not change the viral load of SARS-CoV-2-exposed cells, compared to SARS-CoV-2 group. There is no difference between the control and Lr groups. Figure 3 gives support to the findings of the load viral related to the CSE exposure, since Fig. 3B illustrates an increased activity of ACE2 in CSE group, compared to the control. Conversely, the ACE2 level was reduced in Lr + CSE group in comparision to CSE. In addition, Fig. 3B shows that the ACE2 level was not changed in cells from SARS-CoV-2 group, compared to the control group, as well as the ACE2 level in SARS-CoV-2 group was not statistically different of Lr + SARS-CoV-2 group. Still in Fig. 3B, there is no exacerbation of ACE2 level in cells from CSE + SARS-CoV-2 group, compared to the CSE group. These results evidence that the rise of ACE2 level is due to exposure to CSE. Lastly, the ACE2 levels found in Lr + CSE + SARS-CoV-2 group presented values similar to Lr + CSE group, evidencing that Lr acts on CSE stimulus only without interfering with SARS-CoV-2 to lower the ACE2 levels. Regarding to ACE2 activity, there is no significant difference between the control and Lr groups.
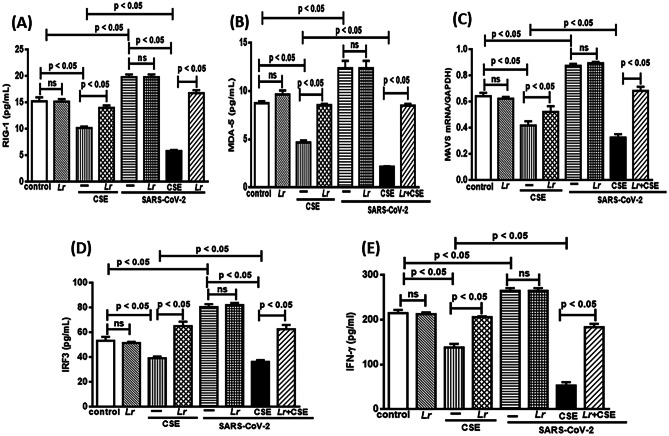
Effect of L. rhamnosus on Cytoplasmatic Antiviral Sensor: RIG-I, MAD-5, MAVS, IRF3, and IFN-γ
The signaling of immune response suppression against viral infection with lower levels of IFN-γ is a hallmark of COPD. Based on that, we investigated the antiviral response-associated innate immunity sensors RIG-1, MAD-5, MAVS, IRF3, and IFN-γ in 16HBE cells from the experimental groups control, Lr, CSE, Lr + CSE, SARS-CoV-2, Lr + SARS-CoV-2, CSE + SARS-CoV-2, and Lr + CSE + SARS-CoV-2. As shown in Fig. 4, there is a significant decrease of RIG-1 (4A) and MDA-5 (Fig. 4B) in CSE-exposed 16HBE cells (CSE group), compared to the control group. These findings show that CSE induces a defectuous antiviral response. Otherwise, Lr markedly restored RIG-1 and MDA-5 in cells from Lr + CSE group, compared with CSE group. Figure 4 also illustrates a marked increase of RIG-1 (4A) and MDA-5 (Fig. 4B) in SARS-CoV-2-exposed cells (SARS-CoV-2 group), compared to the control group. Otherwise, the Lr did not change the increase RIG-1 (Fig. 4A) and MDA-5 (Fig. 4B) in cells from Lr + SARS-CoV-2 group in comparison with cells infected with SARS-CoV-2 (SARS-CoV-2 group). Still in Fig. 4 is represented a drastic fall of RIG-1 (Fig. 4A) and MDA-5 (Fig. 4B) in cells from CSE + SARS-CoV-2 group, compared to CSE group. In counterpart, our results show that Lr-treated 16HBE cells and exposed to CSE and infected with SARS-CoV-2 (Lr + CSE + SARS-CoV-2) had a restoration of both the RIG-I and MAD-5 in comparison with CSE + SARS-CoV-2 group. It is well known that both RIG-I and MAD5 engage the adaptor protein MAVS on the mitochondrial outer membrane to induce the IFN-γ secretion via IRF3 signaling. For this reason, we investigated the MAVS expression as well as the protein concentration of IRF3 and IFN-γ in the following experimental groups: control, Lr, CSE, Lr + CSE, SARS-CoV-2, Lr + SARS-CoV-2, CSE + SARS-CoV-2, and Lr + CSE + SARS-CoV-2. Figure 4C illustrates a significant downregulation of MAVS expression and Fig. 4D, E show lower levels of IRF3 as well as IFN-γ, respectively, in cells from CSE group, compared to the control group. Meanwhile, Lr-treated cells and exposed to CSE (Lr + CSE group) had marked restoration of MAVS expression (Fig. 4C) and increase of both IRF3 (Fig. 4D) and the IFN-γ (Fig. 4E), compared to CSE group. On the contrary, the MAVS expression (Fig. 4C) and IRF3 level (Fig. 4D) as well as IFN-γ concentration (Fig. 4E) was markedly accentuated in cells from SARS-CoV-2 group, compared to control. In addition, Lr did not change MAVS, IRF3 and IFN-γ in cells from Lr + SARS-CoV-2 group in comparison with SARS-CoV-2 group. Finally, Fig. 4 illustrates an exacerbated fall of antiviral sensors MAVS (Fig. 4C), IRF3 (Fig. 4D), and IFN-γ (Fig. 4E) in cells from CSE + SARS-CoV-2 group, compared to CSE group. Conversely, Lr markedly upregulated the MAVS expression (Fig. 4C) and the level of IRF3 (Fig. 4D) and the IFN-γ protein concentration (Fig. 4E) in Lr + CSE + SARS-CoV-2, compared to CSE + SARS-CoV-2 group. There is no significant difference between control and Lr groups.
Fig. 4.
Lactobacillus rhamnosus on concentration of RIG-1, MDA-5, MAVS, IRF3, and IFN-γ. Protein concentration of RIG-1 (A), MDA-5 (B), MAVS (C), IRF3 (D), and IFN-γ (E) in 16HBE cells treated with Lr (1 × 106 CFU) for 2 h, and then exposed to CSE (2.5%) associated to SARS-CoV-2 (MOI 0.1) during 20 h. The RIG-1, MDA-5, and IFN-γ concentration was performed using the ELISA kit, according to “Material and Methods.” Data are presented as means ± SE from triplicate samples for each treatment versus control. ns: non-significant difference
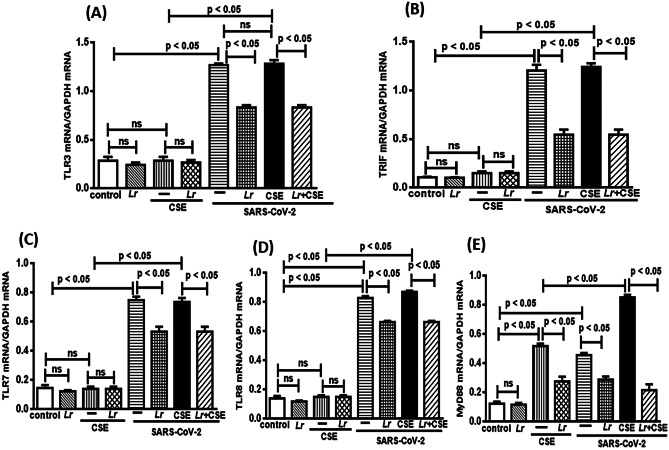
Effect of L. rhamnosus on Endosomal Viral Sensors: TLR3/TRIF and TLR7/TLR8/MyD88
The TLR3, TLR7, and TLR8 are crucial for virus recognition through cell signaling involving TRIF and MyD88, with subsequent activation of NF-κB transcription factor to secrete pro-inflammatory mediators. Thus, Fig. 5 illustrates the mRNA expression of TLR3 (Fig. 5A), TRIF (Fig. 5B), TLR7 (Fig. 5C), TLR8 (Fig. 5D), and MyD88 (Fig. 5E) in cells from control, Lr, CSE, Lr + CSE, SARS-CoV-2, Lr + SARS-CoV-2, CSE + SARS-CoV-2, and Lr + CSE + SARS-CoV-2 groups. Our results show that CSE (CSE group) did not change the expression of TLR3, TLR7, and TLR8, except for MyD88, in comparison with cells from the control group. At the same sense, the Lr did not work on the expression of TLR3 (Fig. 5A), TRIF (Fig. 5B), TLR7 (Fig. 5C), and TLR8 (Fig. 5D) in cells from Lr + CSE group, compared to CSE. Although it is a viral receptor, MyD88 is also known as activation factor for TLR2 and TLR4 which are receptors also sensitive to CSE [42]. Therefore, Fig. 5E illustrates a significant increase of MyD88 expression in cells exposed to CSE (CSE group), compared to the control. Otherwise, the MyD88 expression was markedly reduced in cells from Lr + CSE group in comparison with CSE group. In counterpart, the expression of these pro-inflammatory viral sensors was markedly increased in cells from SARS-CoV-2 group, compared to the control group. It is due to the fact that the signaling via TLR3/TRIF as well as signaling via TLR7/TLR8/MyD88 is involved in viral recognition. On the contrary, Lr induced significant fall in expression of TLR3 (Fig. 5A), TRIF (Fig. 5B), TLR7 (Fig. 5C), TLR8 (Fig. 5D), and MyD88 (Fig. 5E) in cells from Lr + SARS-CoV-2 group, compared to the SARS-CoV-2 group. Figure 5 also illustrates a significant exacerbation of the pro-inflammatory viral sensor’s expression in cells from CSE + SARS-CoV-2 group, compared to CSE group. However, the expression of TLR3 (Fig. 5A), TRIF (Fig. 5B), TLR7 (Fig. 5C) and TLR8 (Fig. 5D) in cells from CSE + SARS-CoV-2 group was similar with that found in SARS-CoV-2 group. Whereas CSE does not influence the expression of viral sensors TLR3, TRIF, TLR7, and TLR8 (CSE group), the Fig. 5 shows that the rise in expression these viral sensors, except MyD88 (5E), depends on presence of SARS-CoV-2. In addition, the downregulation of TLR3 (Fig. 5A), TRIF (Fig. 5B), TLR7 (Fig. 5C), and TLR8 (Fig. 5D) after Lr (Lr + CSE + SARS-CoV-2) had similar values with those found in Lr + SARS-CoV-2 group. These data indicate that, in this condition, the Lr exclusively acts on the increased expression of TLR3/TRIF as well as TLR7 and TLR8 induced by viral infection. Regarding the MyD88 expression, cells from CSE + SARS-CoV-2 group had a significant upregulation, compared to the CSE group. Herein, the MyD88 expression in cells from CSE + SARS-CoV-2 group was significantly higher than that found in CSE group or in SARS-CoV-2 group alone, indicating that MyD88 has an important role against both stimuli. On the contrary, Lr induced a drastic reduction of MyD88 expression in cells from Lr + CSE + SARS-CoV-2 group, compared to CSE + SARS-CoV-2. There is no significant difference between control and Lr groups.
Fig. 5.
Lactobacillus rhamnosus on gene expression of TLR3, TRIF, TLR7, TLR8, and MyD88. Gene expression of TLR3 (A), TRIF (B), TLR7 (C), TLR8 (D), and MyD88 (E) in 16HBE cells treated with Lr (1 × 106 CFU) for 2 h, and then exposed to CSE (2.5%) associated to SARS-CoV-2 (MOI 0.1) during 20 h. Gene expression was performed using a real-time polymerase chain reaction (qPCR). Data are presented as means ± SE from triplicate samples for each treatment versus control. ns, non-significant difference
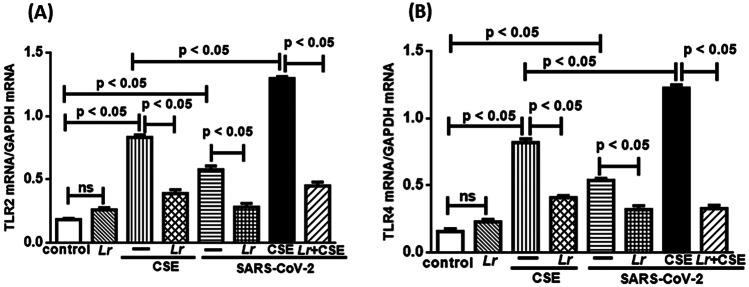
Effect of L. rhamnosus on mRNA Expression for TLR2 and TLR4
The TLR2/TLR4 activation-induced MyD88/NF-κB signaling, which results in an intense secretion of pro-inflammatory cytokines, can be activated in presence of viruses. For this reason, we investigated the mRNA expression of both TLR2 and TLR4 in cells from control, Lr, CSE, Lr + CSE, SARS-CoV-2, Lr + SARS-CoV-2, CSE + SARS-CoV-2, and Lr + CSE + SARS-CoV-2 group. The results illustrated in Fig. 6 show that expression of both TLR2 (Fig. 6A) and TLR4 (Fig. 6B) was significantly upregulated in CSE group as well as SARS-CoV-2 group, compared to the control. On the contrary, Lr-treated cells and exposed to CSE (Lr + CSE group) or infected with SARS-CoV-2 (Lr + SARS-CoV-2 group) had expression levels of TLR2 (Fig. 6A) and TLR4 (Fig. 6B) lower than compared to the CSE group or SARS-CoV-2 group, respectively. Still in Fig. 6, our results show that the TLR2 (Fig. 6A) and TLR4 (Fig. 6B) expressions were markedly incremented in cells from CSE + SARS-CoV-2 group, compared to CSE group. In fact, the expression of these receptors in CSE + SARS-CoV-2 group was higher than in CSE group or SARS-CoV-2 group alone, evidencing that infection with SARS-CoV-2 exacerbates the upregulated expression of TLR2 as well as TLR4 previously induced by exposure to CSE. On the other hand, Lr downregulated expression of both the TLR2 (Fig. 6A) and TLR4 (Fig. 6B) in cells from Lr + CSE + SARS-CoV-2, compared to CSE + SARS-CoV-2 group. There is no significant difference between control and Lr groups.
Fig. 6.
Lactobacillus rhamnosus on gene expression of TLR2 and TLR4. Gene expression of TLR2 (A) and TLR4 (B) in 16HBE cells treated with Lr (1 × 106 CFU), and then exposed to CSE (2.5%) associated to SARS-CoV-2 (MOI 0.1) for 20 h. Gene expression was performed using a real-time time polymerase chain reaction (qPCR). Data are presented as means ± SE from triplicate samples for each treatment versus control. ns, non-significant difference
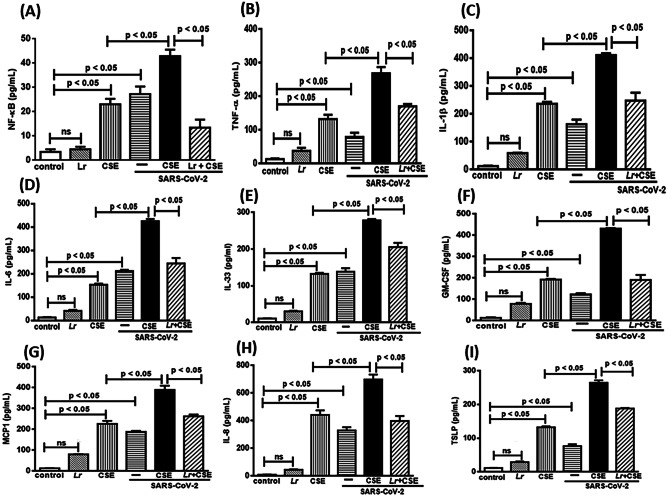
Effect of L. rhamnosus on Pro-inflammatoryResponse: NF-κB and Cytokine Storm
The SARS-CoV-2 infection has a critical role in exacerbation of COPD due to secretion of NF-κB-related-pro-inflammatory mediators in lung milieu [43]. As shown in Fig. 7A, there is a significant increase of NF-κB concentration in cells exposed to CSE (CSE group) or infected with SARS-CoV-2 (SARS-CoV-2 group), compared to the control group. The CSE presence in association with SARS-CoV-2 (CSE + SARS-CoV-2 group) induced an exacerbated increase of NF-κB concentration in cells, compared to the CSE group. On the other hand, the treatment with Lr attenuated the exacerbated concentration of NF-κB in cells from Lr + CSE + SARS-CoV-2 group, compared to CSE + SARS-CoV-2 group. In accordance with these findings, our results show increased secretion of TNF-α (Fig. 7B), IL-1β (Fig. 7C), IL-6 (Fig. 7D), IL-8 (Fig. 7E), IL-33 (Fig. 7F), GM-CSF (Fig. 7G), MCP1 (Fig. 7H), and TSLP (Fig. 7I) in cells exposed to CSE (group CSE) or infected with SARS-CoV-2 (SARS-CoV-2 group), compared to the control. Otherwise, Lr attenuated the secretion of pro-inflammatory mediators in cells from the Lr + CSE group and Lr + SARS-CoV-2 in comparison with CSE group and SARS-CoV-2 group, respectively. Figure 7 also illustrates that the pro-inflammatory mediators TNF-α (Fig. 7B), IL-1β (Fig. 7C), IL-6 (Fig. 7D), IL-8 (Fig. 7E), IL-33 (Fig. 7F), GM-CSF (Fig. 7G), MCP1 (Fig. 7H), and TSLP (Fig. 7I) secreted from cell exposure to CSE and infected with SARS-CoV-2 (CSE + SARS-CoV-2 group) had concentrations higher than those found in CSE group or SARS-CoV-2 group. It means that infection with SARS-CoV-2 in bronchial epithelial cells exacerbates the pro-inflammatory response previously induced by exposure to CSE. On the other hand, our results show that Lr-treated cells and exposed to CSE and infected with SARS-CoV-2 (Lr + CSE + SARS-CoV-2 group) had a markedly reduced secretion of pro-inflammatory mediators compared to CSE + SARS-CoV-2 group. There is no significant difference between the control and Lr groups.
Fig. 7.
Lactobacillus rhamnosus on NF-κB and cytokine storm. The NF-κB (A) and the cytokines TNF-α (B), IL-1β (C), IL-6 (D), IL-8 (E), IL-33 (F), GM-CSEF (G), MCP1 (H), and TSLP (I) were detected in 16HBE cells treated with Lr (1 × 106 CFU) for 2 h, and then exposed to CSE (2.5%) associated to SARS-CoV-2 (MOI 0.1) during 20 h. The NF-κB and cytokines were measured using ELISA kits, according to “Material and Methods.” Data are presented as means ± SE from triplicate samples for each treatment versus control. ns, non-significant difference
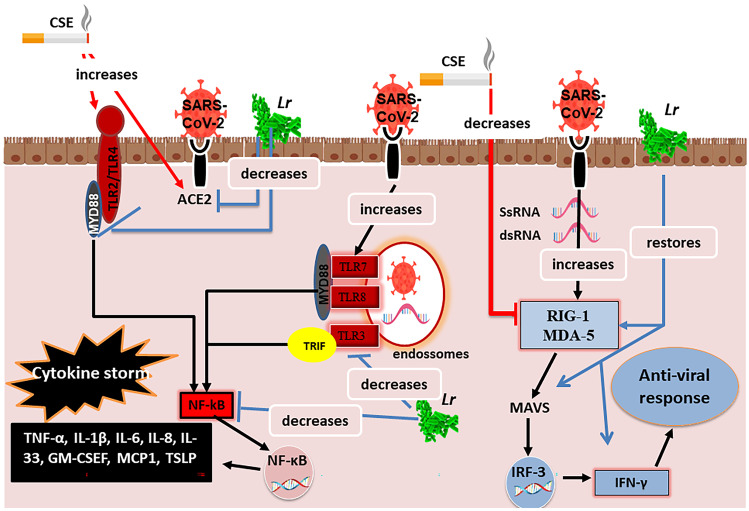
Proposed Molecular Mechanism for the Effect of L. rhamnosus on the Antiviral Signaling and the Cytokine Storm in Human Bronchial Epithelial Cells Exposed to CSE and Infected with SARS-CoV-2
Figure 8 illustrates that cigarette smoke extract (CSE) increase the levels of angiotensin-converser enzyme 2 (ACE2). This condition facilitates the entry of SARS-CoV-2 into the airway epithelium, once the ACE2 was identified as a functional receptor of the SARS-CoV-2. Therefore, individuals with COPD are more susceptible to secondary lung infections, such as COVID-19. CSE also increases the TLR2 and TLR4 expression culminating in the secretion of pro-inflammatory mediators via MyD88/NF-κB activation. Once SARS-CoV-2 entries into airway epithelium, it is recognized in endosome by TLR3 activating NF-κB via TRIF, while TLR7 and TLR8 stimulate the NF-κB signaling via MyD88. Therefore, the COVID-19 exacerbates the COPD due to cytokine storm released in lung milieu. In COPD, the innate immune response is defective due to downregulation of viral sensors RIG-I and MDA-5 with consequent decreased expression of MAVS as well as secretion of antiviral IFN-γ. This scenario reflects the inefficiency of the lung antiviral response against SARS-CoV-2. On the other hand, Lr restores antiviral response associated with IFN-γ-secreting viral sensors and attenuates the NF-κB-associated cytokine storm in human bronchial epithelial cells, supporting its emerging role in preventing COPD exacerbation.
Fig. 8.
Proposed molecular mechanism for the effect of the Lactobacillus rhamnosus on the antiviral signaling and the cytokine storm in human bronchial epithelial cells exposed to cigarette smoke and infected with SARS-CoV-2
Discussion
The present study evidences for the first time that probiotic Lactobacillus rhamnosus (Lr) attenuates both the pro-inflammatory response and the immune dysfunction in an in vitro model of SARS-CoV-2-induced COPD exacerbation. In fact, our findings evidence that Lr controls the cytokine storm as well as restores antiviral signaling in 16HBE cells exposed to CSE and infected with SARS-CoV-2. These results gain highlights because exacerbation is defined as a worsening of COPD symptoms, resulting in the need for additional pharmacological treatment [44, 45]. Some authors have shown that steroids are able to attenuate the intense secretion of pro-inflammatory mediators from bronchoalveolar lavage fluid of individuals infected with COVID-19, and thus it reduces the hospitalization in intensive care units [46]. However, individuals with COPD do not have a good response to steroids [47–49].
In this concern, new anti-inflammatory therapies as probiotics have deserved highlights due to beneficial effect on lung diseases, such as asthma, ARDS, COPD, and pneumonia [50–53]. Even in that preliminary, the present study shows that Lr restores the antiviral signaling and reduces the pro-inflammatory response, therefore presenting a possibility of being used in association with pharmacological therapy.
Regarding the choice of the probiotic Lactobacillus rhamnosus (Lr) to carry out the experimental assays of the present study, this decision was made based on results published by our group that describe the beneficial effect of Lr on cell signaling of the lung inflammatory response and the airway remodeling observed in murine models of COPD and ARDS [54, 55]. Thus, the present manuscript expands the knowledge of the action mechanism of Lr on COPD, once it investigated the Lr effect on antiviral signaling and cytokine storm in cells exposed to CSE and infected with SARS-CoV-2. The idea of this in vitro model was to evaluate the Lr effect in a condition that mimics, at least in human bronchial epithelial cells, the COVID-19-induced exacerbation of COPD.
Both asthma and COPD has been targeting anti-inflammatory and immunomodulatory effect of probiotics, including the Lactobacillus, through signaling Treg/IL-10, [56–59]. However, some authors have demonstrated that probiotic present also the ability to reinstate the antiviral signaling acting on TLR3, TLR8, and MyD88, by modulating the production of pro-inflammatory and anti-inflammatory cytokines [60–62]. Likewise, some authors have revealed that probiotic is able to sustain the antiviral response associated with IFN-γ production by inducing the IRF3, IRF5, and IRF7 [63, 64]. However, studies focused in Lr effect on antiviral signaling and cytokine storm in human bronchial epithelial cells exposed to CSE and infected with SARS-CoV-2 were not still described.
Our results show that Lr restores the antiviral signaling associated to the RIG-I and MAD-5 receptors activation-induced MAVS signaling which induces secretion of IFN-γ via IRF3, once that Lr increased concentrations of RIG-I, MAD-5, and IFN-γ as well as MAVS mRNA expression to levels close to control in 16HBE cells exposed to CSE and infected. Otherwise, Lr reduces the pro-inflammatory response associated to TLR3 activation-induced TRIF signaling as well as decreases the pro-inflammatory response associated to TLR7/TLR8 activation-induced MyD88 signaling in 16HBE cells exposed to CSE and SARS-CoV-2. It occurred due to fact that Lr downregulated mRNA expression of TLR3 and TRIF as well as mRNA expression TLR3, TLR7, and MyD88. These results strongly suggest that Lr modulates the antiviral sensors located in bronchial epithelium, so that pro-inflammatory response is attenuated and antiviral signaling is restored after probiotic treatment. In addition, our findings explore that the bronchial epithelial cells have an important role in virus-induced COPD exacerbation acting as target of beneficial effect of probiotic.
It is well known that the transcriptional factor NF-κB is involved in COPD and COVID-19, due to fact that in both diseases the NF-κB activation leads the pro-inflammatory mediator’s secretion [65]. However, the cellular signaling that activates NF-κB is different from COPD to COVID-19, once that the signaling responsible by secreting pro-inflammatory mediators in COVID-19 activates NF-κB via TRIF (TLR3) and MyD88 (TLR7 and TLR8), while in COPD the NF-κB is activated via TLR2/TLR4 and MyD88. For this reason, the results suggest that the exacerbated increase of NF-κB levels in 16HBE cells is due to synergic effect induced by exposing to CSE and infection with SARS-CoV-2. Otherwise, our findings show that Lr downregulated NF-κB concentration in CSE-exposed 16HBE cells and infected; however, in the present study it is not possible to identify exactly which NF-κB signaling the Lr interfered. Despite it, our results show that the downregulatory effect of Lr on NF-κB concentration, reflected in the reduction of the pro-inflammatory mediators secreted from 16HBE cells, is exposed to CSE and SARS-CoV-2.
In the present manuscript, the Lr reduced the exacerbated expression both TLR2 and TLR4 in 16HBE cells exposed to CSE and infected. It corroborates with some authors that showed a reduction of pro-inflammatory cytokine secretion due to lower levels of TLR4 from macrophages exposed to LPS [66]. In fact, it is well known that the protector effect of probiotic on the immune response activation is linked to the downregulation of TLR2 and TLR4 [67]. However, the SARS-CoV-2 can also interact with TLR2 and TLR4, but differently of Lr, to induce a secretion of pro-inflammatory cytokines via NF-κB signaling [68, 69]. Taking into account that the 16HBE cells received Lr before infection with SARS-CoV-2, it is possible to suggest that probiotics bind to TLR2 and TLR4 first that SARS-CoV-2, and thus the Lr impairs the recognization of SARS-CoV-2 through of TLR2 an TLR4 located in the cell membrane, which result in the reduction of cytokins storm. Recently, some authors have showed that binding of probiotic bacteria to the epithelial surface can cause steric hindrance and block the virus’s attachment to the host cell receptor [70]. Nevertheless, more studies are required to clarify if the interaction of Lr with TLR2/TLR4 can truly interfere in the secretion of SARS-CoV-2-induced pro-inflammatory mediators.
There is a close relation between viral load and severity of COVID-19 [71]. The viral load able to cause lung inflammation depends on the entry of SARS-CoV-2 into host cells through angiotensin converser enzyme-2 (ACE2) [72, 73]. Therefore, the binding of S1 subunit of the Spike protein of SARS-CoV-2 to the ACE2 receptor triggers the cleavage of ACE2 at the ectodomain sites [74] and a soluble form that retains its catalytic activity is produced [64]. It was evidenced when some authors showed that ACE2 inhibitors can decrease viral entry in vitro and in vivo, demonstrating their key role in determining SARS-CoV-2 infectivity and their potential use as a target for antiviral therapies [75]. In addition, some authors have evidenced elevated levels of ACE2 in the bronchial epithelium and lung tissue of COPD patients [76–78]. Therefore, COPD patients infected with SARS-CoV-2 are more susceptible to higher viral load in the lung. The present study shows an exacerbation of load viral when 16HBE cells were exposed to CSE and infected with SARS-CoV-2. Likewise, exposure to CSE markedly increased the ACE2 activity in 16HBE cells, while the SARS-CoV-2 infection did not change ACE2. These results suggest that the increased viral load in CSE + SARS-CoV-2 group is due to CSE effect on ACE2 activity. On the contrary, our results show that Lr reduced both the viral load and ACE2 activity in 16HBE cells exposed to CSE and infected. It suggests that Lr influences the viral load into airway epithelial cells by a mechanism involving the reduction of ACE2 activity. From use of the probiotic therapy opens a new window of opportunities for controlling COVID-19 induced-COPD exacerbation.
It is worthy to note that, the in vitro model of SARS-CoV-2-induced COPD exacerbation in human bronchial epithelial cells adopted in the present study mimics the impairment of antiviral signaling and the increase of pro-inflammatory mediators’ secretion. Thus, it is possible to assume that the Lr effect was investigated in a similar situation to that the bronchial epithelium of COPD patients with COVID-19. In addition, the Lr effect on the bronchial epithelium suggests that the beneficial effect of Lr on COPD exacerbation is not necessarily dependent on the axis gut-lung. Based on the data, it is reasonable to suggest that probiotic Lr can be intranasally administered for treatment of virus-dependent COPD exacerbation.
Regarding the action mechanism of Lr on immune response in 16HBE cells exposed to CSE and infected with SARS-CoV-2, it is curious that Lr interferes with antiviral sensors of innate immunity located in the inner bronchial epithelial cells. Although there is no data about interaction of Lr with airway epithelial cells, it is able to induce antiviral signaling against SARS-CoV-2. Therefore, further studies should be performed to better investigate this point.
Despite the results obtained in the present manuscript, there are limitations in the present study that must be considered when translating the results of Lr therapy, obtained in an in vitro model, for clinical use. Therefore, we recognize that in vitro studies with anti-inflammatory therapies to control the immune response do not fill some gaps when compared to in vivo assays. However, the present study evaluated the Lr effect on human bronchial epithelial cells, and it is well known that the airway epithelium is closely related to lung diseases because the airway epithelial cells are the first barrier against external stimuli, including exposure to cigarette smoke and pathogens, such as bacterial and virus [79]. For this reason, the findings of the present study help to understand which are the molecular targets in airway epithelial cells that can be influenced by Lr in order to attenuate COPD exacerbation induced by SARS-CoV-2 infection.
In conclusion, the present manuscript, reports for the first time, that the Lr reinstate the antiviral signaling and attenuates the cytokine storm secreted from human bronchial epithelial cells exposed to CSE and infected with SARS-CoV-2, which seems to be an important mechanism of Lr for controlling COVID-19 induced-COPD exacerbation.
Author Contribution
F. O. and F. A.: conceptualization; F. O., R. A. S, P. C. R, W. S., C. O.: methodology; F. O. and F. A.: investigation; F. Aimbire and F. O.: resources; F. O.: writing — original draft; F. Aimbire: writing — review and editing; F. Aimbire: supervision; F. A. and F. O.: funding acquisition. All the authors read and approved the final manuscript.
Funding
This work is funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Grant. 88887.506517/2020–00.
Availability of Data and Materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Salvi S. Tobacco smoking and environmental risk factors for chronic obstructive pulmonary disease. Clin Chest Med. 2014;35:17–27. doi: 10.1016/j.ccm.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Pace E, Ferraro M, Di Vincenzo S, et al. Cigarette smoke increases BLT2 receptor functions in bronchial epithelial cells: in vitro and ex vivo evidence. Immunology. 2013;139(2):245–255. doi: 10.1111/imm.12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y, Xu J, Meng Y, et al. Role of inflammatory cells in airway remodeling in COPD. Int J Chron Obstruct Dis. 2014;13:3341–3348. doi: 10.2147/COPD.S176122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwak S, Choi YS, Na HG, et al. Glyoxal and methylglyoxal as e-cigarette vapor ingredients-induced pro-inflammatory cytokine and mucins expression in human nasal epithelial cells. Am J Rhinol Allergy. 2020;35(2):213–220. doi: 10.1177/1945892420946968. [DOI] [PubMed] [Google Scholar]
- 5.Sikkel MB, Quint JK, Mallia P, et al. Respiratory syncytial virus persistence in chronic obstructive pulmonary disease. Pediatr Infect Dis J. 2008;27(10 Suppl):S63–70. doi: 10.1097/INF.0b013e3181684d67. [DOI] [PubMed] [Google Scholar]
- 6.McManus TE, Coyle PV, Kidney JC. Childhood respiratory infections and hospital admissions for COPD. Respir Med. 2006;100(3):512–518. doi: 10.1016/j.rmed.2005.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menzel M, Ramu S, Calvén J, et al. Oxidative stress attenuates TLR3 responsiveness and impairs anti-viral mechanisms in bronchial epithelial cells from COPD and asthma patients. Front Immunol. 2019;10:2765. doi: 10.3389/fimmu.2019.02765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu W, Zhang W, More S, et al. Cigarette smoke attenuates the RIG-I-initiated innate antiviral response to influenza infection in two murine models. Am J Physiol Lung Cell Mol Physiol. 2014;307(11):L848–L858. doi: 10.1152/ajplung.00158.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.García-Valero J, Olloquequi J, Montes JF, et al. Deficient pulmonary IFN-β expression in COPD patients. PLoS ONE. 2019;14(6):e0217803. doi: 10.1371/journal.pone.0217803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Modestou MA, Manzel LJ, El-Mahdy S, et al. Inhibition of IFN-gamma-dependent antiviral airway epithelial defense by cigarette smoke. Respir Res. 2010;11(1):64. doi: 10.1186/1465-9921-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahmutovic-Persson I, Johansson M, Brandelius A, et al. Capacity of capsazepinoids to relax human small airways and inhibit TLR3-induced TSLP and IFNβ production in diseased bronchial epithelial cells. Int Immunopharmacol. 2012;13(3):292–300. doi: 10.1016/j.intimp.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 12.Gasparello J, D'Aversa E, Papi C, et al. Sulforaphane inhibits the expression of interleukin-6 and interleukin-8 induced in bronchial epithelial IB3–1 cells by exposure to the SARS-CoV-2 Spike protein. Phytomedicine. 2021;87:153583. doi: 10.1016/j.phymed.2021.153583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillot L, Le Goffic R, Bloch S, et al. Involvement of Toll-like receptor 3 in the immune response of lung epithelial cells to double-stranded RNA and influenza A virus. J Biol Chem. 2005;280(7):5571–5580. doi: 10.1074/jbc.M410592200. [DOI] [PubMed] [Google Scholar]
- 14.Moreno-Eutimio MA, López-Macías C, Pastelin-Palacios R. Bioinformatic analysis and identification of single-stranded RNA sequences recognized by TLR7/8 in the SARS-CoV-2, SARS-CoV, and MERS-CoV genomes. Microbes Infect. 2020;22(4–5):226–229. doi: 10.1016/j.micinf.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bortolotti D, Gentili V, Rizzo S, et al. TLR3 and TLR7 RNA sensor activation during SARS-CoV-2 infection. Microorganisms. 2021;9(9):1820. doi: 10.3390/microorganisms9091820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimenez-Guardeño JM, Nieto-Torres JL, DeDiego ML, et al. The PDZ-binding motif of severe acute respiratory syndrome coronavirus envelope protein is a determinant of viral pathogenesis. PLoS Pathog. 2014;10(8):e1004320. doi: 10.1371/journal.ppat.1004320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Z, Zhao Y, Zhou Y, et al. Single-cell analysis identified lung progenitor cells in COVID-19 patients. Cell Prolif. 2020;53(12):e12931. doi: 10.1111/cpr.12931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo-Parke H, Linden D, Weldon S, et al. Mechanisms of virus-induced airway immunity dysfunction in disease pathogenesis, progression and exacerbation of COPD. Front Immunol. 2020;11:1205. doi: 10.3389/fimmu.2020.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coronavirus Brazil COVID-19 (2020) Ministry of Health. https://covid.saude.gov.br. Accessed 2020–05–08
- 20.Fajnzylber J, Regan J, Coxen K, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020 doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwata M, Silva Enciso JE, Greenberg BH. Selective and specific regulation of ectodomain shedding of angiotensin-converting enzyme 2 by tumor necrosis factor alpha-converting enzyme. Am J Physiol Cell Physiol. 2009;297(5):C1318–C1329. doi: 10.1152/ajpcell.00036.2009. [DOI] [PubMed] [Google Scholar]
- 22.Barton MI, MacGowan SA, Kutuzov MA, et al. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetiCSE. Elife. 2021;10:e70658. doi: 10.7554/eLife.70658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davies NG, Abbott S, Barnard RC, et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021 doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin J, Kasper B, Petersen F, et al. Association of cigarette smoking, COPD, and lung cancer with expression of SARS-CoV-2 entry genes in human airway epithelial cells. Front Med (Lausanne) 2020;7:619453. doi: 10.3389/fmed.2020.619453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung JM, Yang CX, Tam A, et al. ACE-2 expression in the small airway epithelia of smokers and COPD patients: implications for COVID-19. Eur Respir J. 2020;55(5):2000688. doi: 10.1183/13993003.00688-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vardavas CI, Nikitara K (2020) COVID-19 and smoking: a systematic review of the evidence. Tob Induc Dis 18:20. 10.18332/tid/119324 [DOI] [PMC free article] [PubMed]
- 27.Vollenweider DJ, Jarrett H, Steurer-Stey CA, et al. Antibiotics for exacerbations of chronic obstructive pulmonary disease. Cochrane Database Syst Rev. 2012;12:CD010257. doi: 10.1002/14651858.CD010257. [DOI] [PubMed] [Google Scholar]
- 28.Spacova I, Van Beeck W, Seys S, et al. Lactobacillus rhamnosus probiotic prevents airway function deterioration and promotes gut microbiome resilience in a murine asthma model. Gut Microbes. 2020;11(6):1729–1744. doi: 10.1080/19490976.2020.1766345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mortaz E, Adcock IM, Ricciardolo FL, et al. Anti-inflammatory effects of Lactobacillus rhamnosus and Bifidobacterium breve on cigarette smoke activated human macrophages. PLoS ONE. 2015;10(8):e0136455. doi: 10.1371/journal.pone.0136455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zelaya H, Tada A, Vizoso-Pinto MG, et al. Nasal priming with immunobiotic Lactobacillus rhamnosus modulates inflammation-coagulation interactions and reduces influenza virus-associated pulmonary damage. Inflamm Res. 2015;64(8):589–602. doi: 10.1007/s00011-015-0837-6. [DOI] [PubMed] [Google Scholar]
- 31.Shi CW, Zeng Y, Yang GL, et al. Effect of Lactobacillus rhamnosus on the development of B cells in gut-associated lymphoid tissue of BALB/c mice. J Cell Mol Med. 2020;24(15):8883–8886. doi: 10.1111/jcmm.15574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y, Cai B, Xu M, et al. Gene silencing of Toll-like receptor 2 inhibits proliferation of human liver cancer cells and secretion of inflammatory cytokines. PLoS ONE. 2012;7(7):e38890. doi: 10.1371/journal.pone.0038890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheyhidin I, Nabi G, Hasim A, et al. Overexpression of TLR3, TLR4, TLR7 and TLR9 in esophageal squamous cell carcinoma. World J Gastroenterol. 2011;17(32):3745–3751. doi: 10.3748/wjg.v17.i32.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajebrahimi B, Bagheri M, Hassanshahi G, et al. The adapter proteins of TLRs, TRIF and MYD88, are upregulated in depressed individuals. Int J Psychiatry Clin Pract. 2014;18(1):41–44. doi: 10.3109/13651501.2013.859708. [DOI] [PubMed] [Google Scholar]
- 35.Xing F, Matsumiya T, Onomoto K, et al. Foreign RNA induces the degradation of mitochondrial antiviral signaling protein (MAVS): the role of intracellular antiviral factors. PLoS ONE. 2012;7(9):e45136. doi: 10.1371/journal.pone.0045136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alharbi KS, Fuloria NK, Fuloria S, et al. Nuclear factor-kappa B and its role in inflammatory lung disease. Chem Biol Interact. 2021;345:109568. doi: 10.1016/j.cbi.2021.109568. [DOI] [PubMed] [Google Scholar]
- 37.Zhang MY, Jiang YX, Yang YC, et al. Cigarette smoke extract induces pyroptosis in human bronchial epithelial cells through the ROS/NLRP3/caspase-1 pathway. Life Sci. 2021;269:119090. doi: 10.1016/j.lfs.2021.119090. [DOI] [PubMed] [Google Scholar]
- 38.Gamage AM, Tan KS, Chan WOY, et al. Infection of human nasal epithelial cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020;16(12):e1009130. doi: 10.1371/journal.ppat.1009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pedrosa CDSG, Goto-Silva L, Temerozo JR, et al. Non-permissive SARS-CoV-2 infection in human neurospheres. BioRxiv. 2021 doi: 10.1101/2020.09.11.293951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gamage AM, Tan KS, Chan WOY, et al. Infection of human nasal epithelial cells with SARS-CoV-2 and a 382- nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020;16(12):e1009130. doi: 10.1371/journal.ppat.1009130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodrigues TS, de Sá KSG, Ishimoto AY, et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. DS. J Exp Med. 2021;218(3):e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Budulac SE, Boezen HM, Hiemstra PS, et al. Toll-like receptor (TLR2 and TLR4) polymorphisms and chronic obstructive pulmonary disease. PLoS ONE. 2012;7(8):e43124. doi: 10.1371/journal.pone.0043124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Do-Umehara HC, Chen C, Zhang Q, et al. Epithelial cell-specific loss of function of Miz1 causes a spontaneous COPD-like phenotype and up-regulates Ace2 expression in mice. Sci Adv. 2020;6(33):eabb7238. doi: 10.1126/sciadv.abb7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neufeldt CJ, Cerikan B, Cortese M, et al. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. Commun Biol. 2022;5(1):45. doi: 10.1038/s42003-021-02983-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beeh KM, Beier J, Candler H, et al. Effect of ELOM-080 on exacerbations and symptoms in COPD patients with a chronic bronchitis phenotype - a post-hoc analysis of a randomized, double-blind, placebo-controlled clinical trial. Int J Chron Obstruct Pulmon Dis. 2016;11:2877–2884. doi: 10.2147/COPD.S117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomazini BM, Maia IS, Cavalcanti AB, COALITION COVID-19 Brazil III Investigators et al. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: The CoDEX Randomized Clinical Trial. JAMA. 2020;324(13):1307–1316. doi: 10.1001/jama.2020.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crim C, Calverley PM, Anderson JA, et al. Pneumonia risk in COPD patients receiving inhaled corticosteroids alone or in combination: TORCH study results. Eur Respir J. 2009;34:641–647. doi: 10.1183/09031936.00193908. [DOI] [PubMed] [Google Scholar]
- 48.Kew K, Seniukovich A. Inhaled steroids and risk of pneumonia for chronic obstructive pulmonary disease. Cochrane Database Syst Rev. 2014;10:CD010115. doi: 10.1002/14651858.CD010115.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ernst P, Gonzalez AV, Brassard P, et al. Inhaled corticosteroid use in chronic obstructive pulmonary disease and the risk of hospitalization for pneumonia. Am J Respir Crit Care Med. 2007;176:162–166. doi: 10.1164/rccm.200611-1630OC. [DOI] [PubMed] [Google Scholar]
- 50.Wu CT, Lin FH, Lee YT, et al. Effect of Lactobacillus rhamnosus GG immunopathologic changes in chronic mouse asthma model. J Microbiol Immunol Infect. 2019;52(6):911–919. doi: 10.1016/j.jmii.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Zhao L, Mao Y, Yu H, et al. The preventive effects of Lactobacillus casei on acute lung injury induced by lipopolysaccharide. Indian J Microbiol. 2021;61(3):1–13. doi: 10.1007/s12088-021-00949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jamalkandi SA, Ahmadi A, Ahrari I, et al. Oral and nasal probiotic administration for the prevention and alleviation of allergic diseases, asthma and chronic obstructive pulmonary disease. Nutr Res Rev. 2021;34(1):1–16. doi: 10.1017/S0954422420000116. [DOI] [PubMed] [Google Scholar]
- 53.Ji T, Zhu X, Shang F, et al. Preventive effect of probiotics on ventilator-associated pneumonia: a meta-analysis of 2428 patients. Ann Pharmacother. 2021;55(8):949–962. doi: 10.1177/1060028020983021. [DOI] [PubMed] [Google Scholar]
- 54.Carvalho JL, Miranda M, Fialho AK, et al. Oral feeding with probiotic Lactobacillus rhamnosus attenuates cigarette smoke-induced COPD in C57Bl/6 mice: relevance to inflammatory markers in human bronchial epithelial cells. PLoS ONE. 2020;15(4):e0225560. doi: 10.1371/journal.pone.0225560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olimpio F, da Silva JRM, Vieira RP, et al. Lacticaseibacillus rhamnosus modulates the inflammatory response and the subsequent lung damage in a murine model of acute lung inflammation. Clinics (Sao Paulo) 2022;15(77):100021. doi: 10.1016/j.clinsp.2022.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sagar S, Morgan ME, Chen S, et al. Bifidobacterium breve and Lactobacillus rhamnosus treatment is as effective as budesonide at reducing inflammation in a murine model for chronic asthma. Respir Res. 2014;15(1):46. doi: 10.1186/1465-9921-15-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Fang Z, Liu X, et al. Lactobacillus reuteri attenuated allergic inflammation induced by HDM in the mouse and modulated gut microbes. PLoS ONE. 2020;15(4):e0231865. doi: 10.1371/journal.pone.0231865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin SW, Lee GH, Jang MJ, et al. Lactic acid bacteria ameliorate diesel exhaust particulate matter-exacerbated allergic inflammation in a murine model of asthma. Life (Basel) 2020;10(11):260. doi: 10.3390/life10110260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jang SO, Kim HJ, Kim YJ, et al. Asthma prevention by Lactobacillus rhamnosus in a mouse model is associated with CD4(+)CD25(+)Foxp3(+) T cells. Allergy Asthma Immunol Res. 2012;4(3):150–156. doi: 10.4168/aair.2012.4.3.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiba E, Tomosada Y, Vizoso-Pinto MG, et al. Immunobiotic Lactobacillus rhamnosus improves resistance of infant mice against respiratory syncytial virus infection. Int Immunopharmacol. 2013;17(2):373–382. doi: 10.1016/j.intimp.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 61.Tomosada Y, Chiba E, Zelaya H, et al. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol. 2013;14:40. doi: 10.1186/1471-2172-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.DeMuri GP, Lehtoranta LM, Eickhoff JC, et al. Ex vivo peripheral blood mononuclear cell response to R848 in children after supplementation with the probiotic Lactobacillus acidophilus NCFM/Bifidobacterium lactis Bi-07. Benef Microbes. 2021;12(1):85–93. doi: 10.3920/BM2020.0068. [DOI] [PubMed] [Google Scholar]
- 63.Lu W, Fang Z, Liu X, et al. The potential role of probiotics in protection against influenza a virus infection in mice. Foods. 2021;10(4):902. doi: 10.3390/foods10040902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Groeger D, Schiavi E, Grant R, et al. Intranasal Bifidobacterium longum protects against viral-induced lung inflammation and injury in a murine model of lethal influenza infection. EBioMedicine. 2020;60:102981. doi: 10.1016/j.ebiom.2020.102981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Wen M, Liu X, et al. Granules ameliorate pulmonary inflammation in the rat model of chronic obstructive pulmonary disease via TLR2/4-mediated NF-κB pathway. BMC Complement Med Ther. 2020;20(1):170. doi: 10.1186/s12906-020-02964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mortaz E, Adcock IM, Ricciardolo FL, et al. Anti-Inflammatory effects of Lactobacillus rahmnosus and Bifidobacterium breve on cigarette smoke activated human macrophages. PLoS ONE. 2015;10(8):e0136455. doi: 10.1371/journal.pone.0136455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khan S, Shafiei MS, Longoria C, et al. SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-κB pathway. Elife. 2021;10:e68563. doi: 10.7554/eLife.68563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khailova L, Petrie B, Baird CH, Dominguez Rieg JA, Wischmeyer PE. Lactobacillus rhamnosus GG and Bifidobacterium longum attenuate lung injury and inflammatory response in experimental sepsis. PLoS ONE. 2014;9(5):e97861. doi: 10.1371/journal.pone.0097861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aboudounya MM, Heads RJ. COVID-19 and Toll-like receptor 4 (TLR4): SARS-CoV-2 may bind and activate TLR4 to increase ACE2 expression, facilitating entry and causing hyperinflammation. Mediators Inflamm. 2021 doi: 10.1155/2021/8874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singh K, Rao A. A potential immunomodulator in COVID-19 infection management. Nutr Res. 2021;87:1–12. doi: 10.1016/j.nutres.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Guo F, Liu K, et al. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008;136(1–2):8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bayati A, Kumar R, Francis V, et al. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J Biol Chem. 2021;296:100306. doi: 10.1016/j.jbc.2021.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bermejo-Martin JF, González-Rivera M, Almansa R, et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit Care. 2020;24(1):691. doi: 10.1186/s13054-020-03398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heurich A, Hofmann-Winkler H, Gierer S, et al. TMPRSS2 and ADAM17 cleave ACE2 diferentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein. J Virol. 2014;88(2):1293–1307. doi: 10.1128/JVI.02202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lambert DW, Yarski M, Warner FJ, et al. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2) J Biol Chem. 2005;280(34):30113–30119. doi: 10.1074/jbc.M505111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haga S, Nagata N, Okamura T, et al. TACE antagonists blocking ACE2 shedding caused by the spike protein of SARS-CoV are candidate antiviral compounds. Antiviral Res. 2010;85(3):551–555. doi: 10.1016/j.antiviral.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith JC, Sausville EL, Girish V, et al. Cigarette smoke exposure and inflammatory signaling increase the expression of the SARS-CoV-2 receptor ACE-2 in the respiratory tract. Dev Cell. 2020;53:514–529. doi: 10.1016/j.devcel.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Russo P, Bonassi S, Giacconi R, et al. COVID-19 and smoking. Is nicotine the hidden link Eur Respir J. 2020;55:2001116. doi: 10.1183/13993003.01116-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Adil MS, Verma A, Rudraraju M, et al. Akt-independent effects of triciribine on ACE2 expression in human lung epithelial cells: potential benefits in restricting SARS-CoV2 infection. J Cell Physiol. 2021;236(9):6597–6606. doi: 10.1002/jcp.30343. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.