Abstract
Selective breeding is a classic technique that enables an experimenter to modify a heritable target trait as desired. Direct selective breeding for extreme sleep and circadian phenotypes in flies successfully alters these behaviors, and sleep and circadian perturbations emerge as correlated responses to selection for other traits in mice, rats, and dogs. The application of sequencing technologies to the process of selective breeding identifies the genetic network impacting the selected trait in a holistic way. Breeding techniques preserve the extreme phenotypes generated during selective breeding, generating community resources for further functional testing. Selective breeding is thus a unique strategy that can explore the phenotypic limits of sleep and circadian behavior, discover correlated responses of traits having shared genetic architecture with the target trait, identify naturally-occurring genomic variants and gene expression changes that affect trait variability, and pinpoint genes with conserved roles.
Keywords: selective breeding, artificial selection, sleep, circadian rhythms, Drosophila, mice, rats, dogs
Statement of Significance.
Selective breeding makes the improvement of animals and crops possible. Recent work couples this classic breeding strategy with sequencing technology to trace the genomic and gene expression changes underlying observed changes in sleep and circadian behavior. Selective breeding emerges as a comprehensive approach for the identification of genetic networks and the detection of conserved genomic variants for sleep.
Introduction
For thousands of years human beings have used selective breeding to improve the characteristics of domesticated animals and crops [1]. Selective breeding has a prominent role in the history of experimental biology and genetics: the results of selective breeding experiments underlie Darwin’s theory of natural selection [2]. Selective breeding involves the active participation of a breeder or experimenter with the goal of altering a specific phenotype. To alter a target trait, an experimenter uses a three-step process. First, the experimenter measures the target trait in an outbred population. Second, the experimenter chooses males and females with the most extreme (highest or lowest) values of the target trait as the parents for the next generation and allows them to mate. Third, the experimenter measures the target trait in the resulting progeny to assess the response to selection. Selection continues in this fashion each generation. Usually the experimenter chooses a consistent proportion of the individuals measured each generation as the breeding parents, referred to as the intensity of selection [1]. Selective breeding differs from evolved changes that occur when populations are exposed to different environments in nature, such as the eye loss, reduced pigmentation, and reduced sleep observed in the cave-dwelling Mexican tetra (Astyanax mexicanus) compared to surface populations [3]. Selective breeding is also distinct from laboratory evolution, where an experimenter exposes the population to certain conditions (i.e. high temperatures) but does not choose which animals will breed in successive generations [4]. Instead, selective breeding relies on the intent of an experimenter to “push” the values of a trait to extremes not normally observed in nature by choosing a small percentage of animals at either end of the phenotypic distribution for breeding.
Virtually any genetically variable trait is amenable to selective breeding, and in fact traits that respond to selective breeding confirm the presence of an underlying genetic basis [1]. The majority of sleep parameters studied to date, such as sleep duration, numbers of naps, average nap length, sleep latency, and day-to-day variability in sleep [5–17] are heritable, as are behaviors related to the circadian regulation of sleep such as circadian period, chronotype, and rhythmicity index [8, 18–21]. Furthermore, sleep characteristics related to human sleep disorders, such as insomnia symptoms, excessive daytime sleepiness, daytime napping, and sleep-disordered breathing [22–26] all have a heritable basis. Consequently, these heritable phenotypes should respond to selective breeding to the extent that they can be modeled in nonhuman animals.
Modern selective breeding experiments have a distinct advantage over earlier experiments. Using next-generation sequencing, it is now possible to trace the allele frequency changes (i.e. the change in the proportion of a given allelic variant in the population) and gene expression changes that respond to selective breeding. Doing so has led to new insights into the origins of genetic variation in sleep and will ultimately lead to an understanding of its function. In this perspective, I discuss what selective breeding experiments have discovered about sleep and circadian behaviors including some of the advantages and challenges, and outline a strategy for the future application of this technique.
Selective Breeding to Discover the Limits of What is Possible in Nature
Selective breeding enables the experimenter to explore the phenotypic limits of the target trait. Like experimental mutagenesis and CRISPR strategies, selective breeding modifies the underlying genome. However, selective breeding alters the allele frequencies of naturally-occurring polymorphic variants in the population rather than engineering synthetic alleles with altered function [1]. In this way, selective breeding capitalizes on the genetic variation already present in a population [1], providing a more realistic model of phenotypic potential.
Sleep and circadian parameters respond rapidly and dynamically to selective breeding (Table 1). Selection for 30 generations on a combination of increased sleep latency, reduced sleep bout duration, and increased activity produced flies with insomnia-like behavior as well as extremely short average sleep duration—less than 100 min in a 24-hour day [27], well below the average 923 min seen in natural populations [5]. Likewise, selection for long and short night sleep duration for 13 generations in flies produced divergent populations with a 9.97-hour difference in this parameter (Figure 1A) [28]. The extreme reduction in sleep duration achieved by these two studies was similar to that of engineered mutations in single genes on sleep duration [29–33], demonstrating that the combined effects of naturally occurring genomic variants on sleep can be large. Selective breeding modified traits under circadian regulation as well. Selective breeding changed the timing of adult fly eclosion (i.e. the emergence of the adult fly from the pupal stage) to specific early and late hourly windows during the day [34]. Furthermore, selective breeding produced robust nocturnal and diurnal activity patterns in flies [35]. One intriguing application of selective breeding is to produce animals sensitive or resistant to the effects of certain drugs, enabling a detailed understanding of their efficacy. For example, selective breeding based on the response of rats to γ-hydroxybutyric acid (GHB) created an animal model sensitive to the sedating effects of the drug [36]. One oft-noted challenge of selective breeding is the many generations required to produce populations with extreme phenotypes, making it a more favored strategy for species with short generation times. This need not be an intractable limitation, however, in some instances, the selective breeding has already been accomplished; the key is to recognize the utility of the resulting population in the study of sleep. For example, brachycephalic dog breeds—dogs selectively bred for a shortened muzzle and flattened face—have increased sleep disturbances and decreased sleep latency compared to other dog breeds, making them a readily available model for sleep-disordered breathing [37]. Selective breeding is thus a powerful strategy to effect change in sleep and circadian-related behaviors, demonstrating the limits of what is possible in nature.
Table 1.
Sleep- and circadian-related characters that respond to selective breeding.
| Organism | Selected trait | Starting population | Selection intensity (%)b | No. generations of selection | High Trait Mean |
Low Trait Mean |
Trait(s) with correlated response | Ref. |
|---|---|---|---|---|---|---|---|---|
| D. melanogaster | insomnia-like behaviora | Canton-S | – | 30–65 | – | 100 min. sleep in 24 h | APS learning; No. falls; DA level; triglycerides, free fatty acids, cholesterol, lifespan, desiccation, starvation | [27] |
| D. melanogaster | nocturnal/diurnal behavior | Outbred pooled from 272 isofemale lines from 33 regions in Europe and Africa | 8.3 (male flies only) | 10 | 1.20c | 0.32c | Acrophase of morning/evening activity, night and day sleep duration, circadian period, eclosion phase, phase delay of per signals; female lifespan and no. of progenye |
[35] |
| D. melanogaster | Early/late adult emergence | Outbred from B populations of M. Rose |
– | 75 | 17:00–21:00 | 5:00–9:00 | Circadian period of emergence, circadian period of rest/activity, mRNA expression parameters in canonical clock genesf, egg to puparium duration, egg to adult duration, fecundity, median longevity | [34, 38, 39] |
| D. melanogaster | Window of emergence | Outbred from B populations of M. Rose |
– | 90 | ZT 10-11 | ZT 01-02 | Mid-life fecundity, female lifespan | [40] |
| D. melanogaster | Long/short night sleep duration | Outbred created by mating 10 DGRP lines for 21 generations | 25 | 13 | 685.0 and 678.5 min. | 111.9 and 54.8 min. | Day sleep duration, night avg. bout length, sleep latency, day bout number, day sleep CVE, night avg. bout length CVE, sleep latency CVE, day bout number CVEd | [28] |
aInsomnia-like behavior includes reduced sleep time, increased sleep latency, reduced sleep bout duration, and elevated levels of waking activity. APS, aversive phototaxic suppression; DA, dopamine level.
bHere selection intensity refers to the proportion of flies selected as parents for the next generation.
cNumbers refer to ND ratio, which is the ratio of the activity during the 12-hr dark phase to the activity during the 12-hr light phase.
d CV E, coefficient of environmental variation.
eFemale lifespan and numbers of progeny were tested after selection was relaxed.
fDifferences observed included phase and mean of per mRNA expression, phase of tim mRNA expression, phase and amplitude of Clk mRNA expression, amplitude and mean of cry mRNA expression, phase and amplitude of vri mRNA expression, phase, amplitude, mean of PDF mRNA expression.
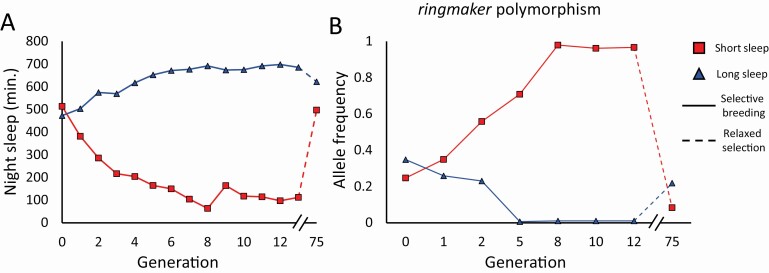
Figure 1.
Response to selective breeding for night sleep followed by relaxed selection. Flies were selectively bred for 13 generations, followed by relaxed selection for 62 generations (ending at generation 75). (A) Night sleep duration. (B) Allele frequency for a polymorphism in ringmaker. The y-axis indicates the proportion of alleles that are “A” in each population, as opposed to the opposite allele, “G”. Plots created by combining data from references [28, 41], used under CC BY 4.0.
Selective Breeding Reveals the Genetic Basis of Sleep and Circadian Behavior
It has recently been noted that the term sleep actually refers to a set of complex phenotypes, each with a polygenic basis [17, 42]. Consequently, genome-wide association studies and systems genetics approaches have detected hundreds of genes affecting different aspects of sleep and circadian behavior [5–7, 9–17, 19–26]. Furthermore, mutational screens in mice and flies revealed that a striking 14%–16% of mutations tested have quantitative effects on some aspect of sleep [42, 43] and 0.1%–0.2% of those tested have Mendelian (2–3 standard deviations) effects on sleep [29, 44]. As sleep is polygenic, it is likely that a single mutation does not act in isolation. At least two mutagenesis studies detected background modifiers to sleep-altering mutations in a single gene, for example [29, 45]. Adding to this complexity is the effect of circadian rhythms, which are thought to regulate sleep [46]. While the canonical molecular circadian clock genes are known, modifiers across the genome can alter circadian behavior [19–21, 47–49]. Selective breeding coupled with the measure of underlying molecular endophenotypes offers a solution to this challenge as outlined below.
As stated previously, selective breeding alters the allele frequencies of naturally occurring polymorphic variants within a population [1]. Accordingly, following the trajectories of allele frequency change per generation during the selection process via whole-genome sequencing facilitates the discovery of genomic variants that affect the target trait. This was done during selection for long and short night sleep duration in flies as exemplified in Figure 1B. The figure shows how allele frequency trajectories diverge between long- and short-sleeper populations for a polymorphism within an intron of the gene ringmaker [28]. This strategy uncovered 126 divergent polymorphisms tagging 80 genes [28]. Some of these genes had known functions in sleep and circadian behavior as well as in classic developmental pathways [EGFR and MAPK (pointed), Wnt (frizzled, dally, and shaggy), and Hippo (scribble)] [28]. Adding information from known gene–gene and protein–protein interaction databases suggested a network of predicted interactions among some of the genes [28]. This is a key advantage of selective breeding over mutagenesis or candidate gene studies: the ability to track genomic modifications over time reveals the entire suite of candidate polymorphisms and their modifiers impacting sleep rather than identifying genes of interest one at a time.
While selective breeding can reveal changes in the genome that are important for sleep, random genetic drift acts simultaneously to alter the allele frequencies of polymorphisms not relevant to sleep, increasing the potential for false positives. Hundreds of thousands of allele frequency changes are possible in an unselected control population over several generations of selection due to drift [28]. A critical step, therefore, is to simulate the effects of random genetic drift to distinguish it from the signal of selection [28, 41]. Likelihood simulations of both the unselected and selected populations provide an empirical threshold eliminating less promising variants [41]. Increasing the size of the measured population is another common way to mitigate the effects of drift but may be difficult for sleep studies, which rely on phenotypic measurements in individual animals. Similarly, replicate populations offer a means to avoid false positives due to drift. For a given candidate variant, an experimenter may conservatively require statistical significance in all replicates of selected populations. An additional alternative is that statistical significance for a given variant is lacking in unselected control populations and/or that the allele frequency of the variant diverges between populations selected for high and low values of the trait (Figure 1B) [28]. Furthermore, the polymorphic variants identified via sequence comparisons often map to noncoding regions of the genome [28]. A strategy to demonstrate the causality of these noncoding polymorphic variants in sleep is critical to understanding the maintenance of genetic variation in sleep (see below). Giving consideration to these factors will mitigate erroneous interpretation of variant function.
Like allele frequency trajectories, the comparison of the transcriptome between diverged selection populations or with an unselected control population [27, 35] reveals the entire gene network underlying the phenotypic changes brought about by selective breeding. RNA-Seq of fly heads revealed differential expression among flies with nocturnal, normal, and diurnal activity [35]. The expression of one core circadian clock gene, PAR-domain protein 1, increased in diurnal flies, but differentially expressed genes were largely novel or had known functions upstream and downstream of the canonical clock circuit [35]. Diurnal flies had increased levels of photoreceptor genes functioning upstream of the clock such as Rhodopsin 3, Turandot A, and Turandot C while nocturnal flies had higher levels of Rhodopsin 2 and Photoreceptor dehydrogenase [35]. Nocturnal flies also had increased expression of known downstream targets of the clock, such as takeout and Pigment dispersing factor [35]. Microarray profiling of heads from flies with insomnia-like phenotypes identified 1,350 differentially-expressed genes compared to Canton-S control flies [27]. The enriched genes were involved in sensory perception of external stimuli such as light and radiation; metabolism, particularly lipid metabolism; cellular signaling; neuronal activity; and locomotor behavior [27]. The authors speculated that differential expression of sensory genes in particular in the insomnia-like flies may be related to a state of hyperarousal [27]. In addition, flies selectively bred for eclosion in specific time windows had altered mean, amplitude and phase of expression in canonical clock genes [38]. Each of these studies measured gene expression at the end of the selective breeding process in selected populations and controls, finding altered gene networks underlying sleep and circadian processes.
An alternative to this approach is to assess the transcriptome in selected populations and controls during each generation of selective breeding. In addition to a per-gene differential expression analysis, Gaussian Process models identified nonlinear trends in the data and inferred gene–gene interactions facilitating changes in sleep (C. Souto-Maior and S. Harbison, unpublished data). Transcriptomic analyses thus reveal both the individual components as well as the combinatorial networks responsible for genetic variation in sleep.
Selective Breeding Identifies Variants Under Natural Selection
Given that sleep is both complex and polygenic, elucidating the functional role of all genes involved in sleep presents a formidable task. Is there a way to determine which genes are the most crucial to sleep? Selective breeding gives us the opportunity to answer this question, provided that natural selection operates on the target trait. A procedure known as relaxed selection can be used to determine which genomic variants for the target trait are under natural selection [50]. These variants are the most critical to the maintenance of genetic variation for sleep, and thus the top candidates for functional studies. Consider the example of night sleep duration [28, 41]. Traits affected by natural selection will presumably be maintained at an optimal level in an outbred population at the start of selective breeding [1]. Selective breeding subsequently alters this optimum, resulting in a divergence in night sleep duration (Figure 1A) [28, 41]. If the experimenter then allows random mating within the population to occur, referred to as relaxing selection, natural selection will return the trait values back towards the optimum level, as shown by the dashed lines in Figure 1A [1, 51]. This phenotypic reversal is a demonstration that natural selection acts on the target trait, in this example night sleep duration. Likewise, allele frequency changes that diverge under selective breeding and then reverse course under relaxed selection strongly implicate these polymorphisms in the maintenance of genetic variation for sleep, as shown for the ringmaker variant in Figure 1B [41]. Notice the more pronounced response to relaxed selection in the short-sleeping population as against the long-sleeping population, which suggests that natural selection acts strongly against extreme short night sleep. This observation also explains why genes with severe short-sleeping mutants accrued genomic modifiers over time that increased sleep duration in flies [29, 45]. Thus, relaxed selection enables the prioritization of candidate variants for sleep.
A response to selective breeding that is much greater in one direction than the other is another indication that natural selection acts on the trait [52]. Selective breeding for nocturnal and diurnal behavior in flies produced a strong asymmetrical response in the direction of increased diurnal behavior [35]. After 15 generations of relaxed selection, the differences in activity patterns between the diurnal population and the control were gone, suggesting that natural selection acts against diurnal activity patterns in flies [35]. The authors observed increased female lifespan in the nocturnal population as well as greater numbers of progeny produced by nocturnal females, which suggests increased fitness in nocturnal flies (but see the section on correlated responses to selection below). The observation of asymmetry suggests the action of natural selection on the target trait.
One should not expect natural selection to act on every phenotype examined. Some traits show little or inconsistent responses to relaxed selection. Some examples in the literature include studies of wing shape [53], sex comb bristle number [54], and thorax length [55] in flies, in which one or more replicate populations did not change appreciably after selective breeding ceased. A lack of response may indicate optimal fitness in the population, or it may reflect a loss of genetic variation due to inbreeding [54, 55]. Still, in cases where natural selection is operating the relaxed selection procedure is an unbiased way to identify key variants maintaining genetic variation in sleep and circadian behavior.
Selective Breeding Identifies Traits Having a Shared Genetic Architecture with Sleep
Selective breeding can establish the genetic relationship between sleep and other complex traits. If the target trait has a shared genetic architecture with a secondary trait, the secondary trait may also respond to selective breeding for the target trait, a phenomenon known as a correlated response to selection [1]. Correlated responses to selection may or may not be desirable in the selected population. Table 1 shows traits selected for sleep and circadian phenotypes where a correlated response to selection was observed in another trait. Related sleep and circadian phenotypes respond to breeding for a target sleep trait, which might be anticipated, but correlated responses also include metabolic traits, life-history and fitness traits, and other behaviors such as learning and memory [27, 28, 34, 35, 38–40]. A correlated response to selection implies a shared genetic architecture between sleep and other traits.
Interestingly, animals selected for other phenotypes may also have altered sleep due to selective breeding (Table 2). For example, flies selected for increased starvation resistance also have increased day and night sleep duration [56, 57], while flies selected for increased desiccation resistance do not [57]. Mice selected for immobility during the tail suspension test, a model of depression, had correspondingly longer slow-wave sleep and REM sleep [58]. Rats bred for sensitivity to diisopropyl fluorophosphate (DFP) had increased REM and advanced circadian phase [59, 60]. Additionally, young rats selectively bred for high alcohol preference had longer sleep–wake stages than controls [61]. These studies reveal a previously under-appreciated shared genetic architecture between sleep and the selected trait.
Table 2.
Sleep- and circadian-related characters that emerged as a correlated response to selection.
| Organism | Selected trait | Starting population | Selection intensity (%)a | No. Generations of selection | High Trait |
Low Trait |
Trait(s) with correlated response | Ref. |
|---|---|---|---|---|---|---|---|---|
| D. melanogaster | Starvation resistance | Wild-collected Terhune Orchards, Princeton, NJ, USA | 15 | 80 | 18 days | – | Day and night sleep duration | [56] |
| Mus musculus | Immobility in tail suspension testb | CD1 mice | Fixed criteria: High immobility (>115 s); low immobility (<35 s) | 14 | 100% >115s | 100% < 35s |
Light SWS, REM sleep latency, wakefulness, sucrose consumption, seric corticosterone and brain 5HT levels, 8-OH-DPAT-induced hypothermia, DRN firing inhibition, response to antidepressants | [58] |
| Mus musculus | Stress reactivity (corticosterone level after restraint stress) | CD-1 outbred mice | 8 | 7 | Median 13.0 male/26.9 female (ng/ml) | Median 1.5 male/7.2 female (ng/ml) | Percentage of time in wake, REM, and NREM; transition frequency between wake and sleep states; EEG frequency power | [62] |
| Rattus norvegicus | Sensitivity to diisopropyl fluorophosphate (DFP)d | Sprague–Dawley | – | 9 | – | – | Increased REM, reduced REM onset, advanced circadian phase, body temperature, drinking behavior | [59, 60] |
| Rattus norvegicus | Intake of 10% alcohol per body weight | Sprague–Dawley, Wistar, Long-Evans | – | 8–51 | 0.48 ± 0.25 in males; 0.97 ± 0.34 in femalesc | 0.18 ± 0.13 in males; 0.29 ± 0.26 in femalesc | Increased time in sleep–wake stages | [61, 63] |
| Rattus norvegicus | Righting reflex after intraperitoneal injection of GHB 1 g/kg | Ratio of sleep duration/onset of loss of righting reflex ≥ 8 sensitive; ≤ 2 resistant |
10 | 13.2 in males; 16.5 in females | 0.2 in males; 0.5 in females | [36] | ||
| Canis familiaris | Brachycephaly | – | – | – | – | – | Sleep-disordered breathing index; nadir in O2 saturation | [37] |
aHere selection intensity refers to the proportion of flies selected as parents for the next generation.
bImmobility in the tail suspension test is a model for depression. DRN, dorsal raphe nucleus.
cml alcohol consumed per 100 g of body weight.
dSensitivity was defined from a composite score of changes in body temperature, drinking behavior, and body weight.
While a correlated response to selection suggests common genes between the target trait and the correlated trait, selective breeding may alter underlying complex processes involving many genes. It is therefore critical to note that the genetic correlation between the two traits will be less than one in the vast majority of cases. Thus, some care must be taken in the interpretation of a correlated response as there will not be a one-to-one relationship between the genes influencing the target trait and those influencing the correlated trait. Furthermore, a correlated response observed in one replicate population may not occur in a second replicate population or experiment [64].
The relationship between sleep and lipid stores is one example of this situation. Selection for insomnia-like symptoms in flies greatly reduced sleep duration but produced increases in triglycerides, free fatty acid, and cholesterol stores, as well as increased starvation resistance [27]. However, selection for increased starvation resistance in flies also increased lipid stores but increased rather than decreased sleep duration. One explanation for these disparate results is that the correlated response is a consequence of inbreeding or random genetic drift, not selective breeding. Masek et al. demonstrated that this was the case for sleep and lipid stores. First, they noted that starvation-resistant flies ate more as larvae than unselected controls. By restricting feeding in starvation resistant populations, lipid stores decreased, yet sleep remained unchanged [56]. Crosses of starvation-resistant populations to unselected controls demonstrated that there was no correlation between sleep and the amount of food consumed [56]. These experiments demonstrate that sleep duration and starvation resistance have a shared genetic architecture, but not via lipid stores.
A similar issue emerges in the genetic relationship between night sleep duration and lifespan. A recent evaluation of lifespan in the Sleep Inbred Panel (SIP) showed increased lifespan and reduced mortality rate in short sleepers as compared to long sleepers [65]. This is in contrast to the similar lifespan observed in long and short sleepers of the progenitor populations from which the SIP was derived [28]. Differences in methodology for the lifespan measurements, including parameters known to affect lifespan such as mating status and diet composition, may have unmasked this relationship between lifespan and short sleep [65], suggesting a context-dependent relationship between the two traits.
Thus, it is important to confirm the relationship between the target trait and the trait having the correlated response. Crossing divergent populations will enable the experimenter to determine whether traits are inherited together [56, 58, 66] as replicate populations with similar phenotypic results can be constructed from disparate underlying processes [28].
Selective Breeding Facilitates Development of Community Resources
If selective breeding coupled with relaxed selection reveals the action of natural selection as outlined above, then the extreme phenotypes achieved with selective breeding are volatile. The selective breeding program must either be maintained continuously, or the experimenter must take action to preserve the phenotypic changes; otherwise, natural selection will act to return the trait to optimal levels. One way to preserve the extreme phenotypes achieved with selective breeding is to create isogenized or inbred lines from the selected populations. For example, Pegoraro et al. used Drosophila balancer chromosomes to create a single isogenized line for each of the nocturnal, diurnal, and control fly populations they created, which maintains the extreme phenotypes in a stable, usable form [35]. Likewise, the Sleep Inbred Panel (SIP) is a group of 39 long-sleeping and short-sleeping inbred lines created by 20 generations of full-sib inbreeding of fly populations selected for long and short night sleep duration [67]. These lines form important laboratory and community resources for the further study of sleep as outlined below [35, 67].
First, these resources enable more in-depth exploration of phenotypes potentially correlated with extreme sleep or circadian behavior. Several phenotypes have been explored as potential correlates with extreme long and short sleep duration in the SIP. As mentioned previously, lifespan was longer in short-sleeping SIP flies than in long sleepers, and their age-specific mortality rate was lower [65]. Body weight was lower in both sexes, and triglycerides and glucose content was lower for males of SIP short sleeper lines [65]. In addition, both long and short sleeper lines of the SIP exhibited impaired learning [68]. Wildtype flies housed in same-sex groups for several days show increased sleep, predominantly during the daytime, compared to flies isolated during the same period of time [69, 70]. Males of the SIP responded to this paradigm largely as expected [68, 71]; however, both long- and short-sleeping females were more variable in their response to social exposure [68]. These examples demonstrate how extreme lines can identify traits potentially having a genetic correlation with sleep and circadian behavior.
Second, underlying molecular pathways contributing to the extreme phenotypes can be studied in greater detail using stabilized lines. For example, lines isogenized for nocturnal and diurnal behavior in flies had phase shifts in PER protein in clock neurons associated with the changes in behavior [35]. Likewise, immunological staining of the brains of SIP short and long sleepers showed the differential effects of social exposure on brain morphology [68].
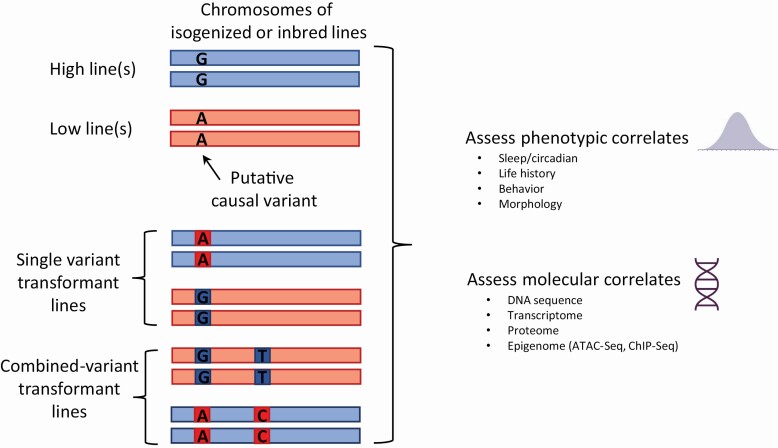
Third, stable lines with extreme phenotypes produced by selective breeding provide a means to demonstrate the causality of highly conserved candidate variants (Figure 2). CRISPR scarless allelic replacement [72, 73] of candidate alleles in a long-sleeping line with their short-sleeping counterparts enables a variant-by-variant comparison of sleep differences between the modified line and the long-sleeping line in the same genetic background. If the variant is causal, a full systems genetics comparison between the modified line and the long-line is warranted, which may include RNA-Seq, proteomics, metabolomics, and an assessment of chromatin structural modifications to reveal the underlying changes induced by a single variant change. Transgenic approaches could then be applied to assess whether the variant directly affects sleep through known sleep-active neurons [74]. As mentioned previously, sleep is highly polygenic; thus, some interaction among candidate variants should occur. Crossing modified lines enables the assessment of combinatorial effects among polymorphisms (Figure 2) [75]; alternatively, polycistronic CRISPR approaches facilitate the assessment of combinatorial effects simultaneously [76]. This framework will enable the construction of the network of naturally-occurring polymorphisms that maintain variation in the target sleep trait.
Figure 2.
Isogenized/inbred lines are a resource to gain functional understanding of the role of sleep-associated variants.
Future Directions
Selective breeding in nonhuman models offers certain advantages over conventional mutagenesis and GWAS studies. Selective breeding can be used to develop extreme high and low phenotypes for heritable traits, within the limits of the underlying genetic variation. Like GWAS, selective breeding enables the discovery of the suite of genomic variants and corresponding genes that modify the target trait. However, the relaxed selection procedure unique to selective breeding is an unbiased way of picking variants to pursue mechanistically as it reveals the variants under natural selection. Preserving extreme phenotypes via breeding creates community resources for further study. Importantly, these resources can be used to confirm the causality of candidate variants by perturbing extreme phenotypes while maintaining the integrity of the genetic background. A further systems understanding of the trait can be gleaned by measuring additional phenotypic and molecular correlates. Some outstanding challenges in this area are to (1) develop new mathematical approaches to the analyses of multiomic, multigenerational data to derive causal genetic networks; and (2) to generate methods that will more rapidly verify genomic variants and elucidate their function. Advancements in these areas will link this classic technique with 21st-century biology.
Acknowledgements
I would like to thank N. Singh, C. Souto-Maior, and A. Zimmerman for helpful comments on this manuscript.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, the National Heart, Lung, and Blood Institute.
Disclosure Statement
Financial disclosure: The Sleep Inbred Panel lines are commercially available from the Bloomington Drosophila Stock Center (Bloomington, IN). The author does not receive any compensation for the sale or distribution of these lines.
Non-financial disclosure: The author declares no non-financial conflicts of interest.
Data Availability
No new data were generated or analyzed in support of this article.
References
- 1. Falconer DS, et al. Introduction to Quantitative Genetics. 4th ed. Edinburgh Gate, Harlow: Addison Wesley Longman Limited; 1996. [Google Scholar]
- 2. Darwin C. The Origin of Species. New York: Barnes & Noble Books; 2004. [Google Scholar]
- 3. Moran RL, et al. Hybridization underlies localized trait evolution in cavefish. iScience 2022;25(2):103778. doi: 10.1016/j.isci.2022.103778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kawecki TJ, et al. Experimental evolution. Trends Ecol Evol. 2012;27(10):547–560. doi: 10.1016/j.tree.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 5. Harbison ST, et al. Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics. 2013;14:281. doi: 10.1186/1471-2164-14-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wu KJ, et al. Genotype influences day-to-day variability in sleep in Drosophila melanogaster. Sleep 2018:41(2). doi: 10.1093/sleep/zs1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Smith BR, et al. Dissecting the genetic basis of variation in Drosophila sleep using a multiparental QTL mapping resource. Genes (Basel) 2020;11(3):294. doi: 10.3390/genes11030294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keenan BT, et al. High-throughput sleep phenotyping produces robust and heritable traits in diversity outbred mice and their founder strains. Sleep 2020;43(5). doi: 10.1093/sleep/zsz278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Amin N, et al. Genetic variants in RBFOX3 are associated with sleep latency. Eur J Hum Genet. 2016;24(10):1488–1495. doi: 10.1038/ejhg.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dashti HS, et al. Genome-wide association study identifies genetic loci for self-reported habitual sleep duration supported by accelerometer-derived estimates. Nat Commun. 2019;10(1):1100. doi: 10.1038/s41467-019-08917-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gottlieb DJ, et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol Psychiatry. 2015;20(10):1232–1239. doi: 10.1038/mp.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doherty A, et al. GWAS identifies 14 loci for device-measured physical activity and sleep duration. Nat Commun. 2018;9(1):5257. doi: 10.1038/s41467-018-07743-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones SE, et al. Genetic studies of accelerometer-based sleep measures yield new insights into human sleep behaviour. Nat Commun. 2019;10(1):1585. doi: 10.1038/s41467-019-09576-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khoury S, et al. Multi-ethnic GWAS and meta-analysis of sleep quality identify MPP6 as a novel gene that functions in sleep center neurons. Sleep 2021;44(3). doi: 10.1093/sleep/zsaa211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lane JM, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49(2):274–281. doi: 10.1038/ng.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marinelli M, et al. Heritability and genome-wide association analyses of sleep duration in children: The EAGLE Consortium. Sleep 2016;39(10):1859–1869. doi: 10.5665/sleep.6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Diessler S, et al. A systems genetics resource and analysis of sleep regulation in the mouse. PLoS Biol. 2018;16(8):e2005750. doi: 10.1371/journal.pbio.2005750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emery PT, et al. Investigation of natural genetic variation in the circadian system of Drosophila melanogaster. II. Biometrical analyses of locomotor activity rhythms recorded in constant darkness. Chronobiol Int. 1995;12(2):77–86. doi: 10.3109/07420529509064503. [DOI] [PubMed] [Google Scholar]
- 19. Jones SE, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12(8):e1006125. doi: 10.1371/journal.pgen.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones SE, et al. Genome-wide association analyses of chronotype in 697,828 individuals provides insights into circadian rhythms. Nat Commun. 2019;10(1):343. doi: 10.1038/s41467-018-08259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harbison ST, et al. Mackay TFC. Genome-wide association study of circadian behavior in Drosophila melanogaster. Behav Genet. 2019;49(1):60–82. doi: 10.1007/s10519-018-9932-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Byrne EM, et al. A genome-wide association study of sleep habits and insomnia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B(5):439–451. doi: 10.1002/ajmg.b.32168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dashti HS, et al. Genetic determinants of daytime napping and effects on cardiometabolic health. Nat Commun. 2021;12(1):900. doi: 10.1038/s41467-020-20585-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hammerschlag AR, et al. Genome-wide association analysis of insomnia complaints identifies risk genes and genetic overlap with psychiatric and metabolic traits. Nat Genet. 2017;49(11):1584–1592. doi: 10.1038/ng.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jansen PR, et al. Genome-wide analysis of insomnia in 1,331,010 individuals identifies new risk loci and functional pathways. Nat Genet. 2019;51(3):394–403. doi: 10.1038/s41588-018-0333-3. [DOI] [PubMed] [Google Scholar]
- 26. Lane JM, et al. Biological and clinical insights from genetics of insomnia symptoms. Nat Genet. 2019;51(3):387–393. doi: 10.1038/s41588-019-0361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seugnet L, et al. Identifying sleep regulatory genes using a Drosophila model of insomnia. J Neurosci. 2009;29:7148–7157. doi: 10.1523/JNEUROSCI.5629-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harbison ST, et al. Selection for long and short sleep duration in Drosophila melanogaster reveals the complex genetic network underlying natural variation in sleep. PLoS Genet. 2017;13(e1007098):e10007098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cirelli C, et al. Reduced sleep in Drosophila Shaker mutants. Nature 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 30. Rogulja D, et al. Control of sleep by Cyclin A and its regulator. Science 2012;335:1617–1621. doi: 10.1126/science.1212476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stavropoulos N, et al. insomniac and Cullin-3 regulate sleep and wakefulness in Drosophila. Neuron 2011;72:964–976. doi: 10.1016/j.neuron.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koh K, et al. Identification of SLEEPLESS, a sleep-promoting factor. Science 2008;321:372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shi M, et al. Identification of Redeye, a new sleep-regulating protein whose expression is modulated by sleep amount. Elife 2014;3:e01473. doi: 10.7554/eLife.01473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kumar S, et al. Selection for early and late adult emergence alters the rate of pre-adult development in Drosophila melanogaster. BMC Dev Biol. 2006;6:57. doi: 10.1186/1471-213X-6-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pegoraro M, et al. The genetic basis of diurnal preference in Drosophila melanogaster. BMC Genomics. 2020;21(1):596. doi: 10.1186/s12864-020-07020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lobina C, et al. Completion by the 10th generation of the bidirectional selective breeding of GHB-sensitive and GHB-resistant rats. Brain Res Brain Res Protoc 2004;13(1):53–56. doi: 10.1016/j.brainresprot.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 37. Hendricks JC, et al. The English bulldog: a natural model of sleep-disordered breathing. J Appl Physiol (1985) 1987;63(4):1344–1350. [DOI] [PubMed] [Google Scholar]
- 38. Nikhil KL, et al. Molecular correlates of circadian clocks in fruit fly Drosophila melanogaster populations exhibiting early and late emergence chronotypes. J Biol Rhythms. 2016;31(2):125–141. doi: 10.1177/0748730415627933. [DOI] [PubMed] [Google Scholar]
- 39. Nikhil KL, et al. Life-history traits of Drosophila melanogaster populations exhibiting early and late eclosion chronotypes. BMC Evol Biol. 2016;16:46. doi: 10.1186/s12862-016-0622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Varma V, et al. Selection for narrow gate of emergence results in correlated sex-specific changes in life history of Drosophila melanogaster. Biol Open 2014;3(7):606–613. doi: 10.1242/bio.20147906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Souto-Maior C, et al. Natural selection on sleep duration in Drosophila melanogaster. Sci Rep. 2020;10:20652. doi: 10.1038/s41598-020-77680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jan M, et al. Recent advances in understanding the genetics of sleep. F1000Res 2020;9:214. doi: 10.12688/f1000research.22028.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harbison ST, et al. Quantitative genetic analysis of sleep in Drosophila melanogaster. Genetics 2008;178:2341–2360. doi: 10.1534/genetics.107.081232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Miyoshi C, et al. Methodology and theoretical basis of forward genetic screening for sleep/wakefulness in mice. Proc Natl Acad Sci USA. 2019;116(32):16062–16067. doi: 10.1073/pnas.1906774116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yuan Q, et al. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 46. Dubowy C, et al. Circadian rhythms and sleep in Drosophila melanogaster. Genetics 2017;205(4):1373–1397. doi: 10.1534/genetics.115.185157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lane JM, et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat Commun. 2016;7:10889. doi: 10.1038/ncomms10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hu Y, et al. GWAS of 89,283 individuals identifies genetic variants associated with self-reporting of being a morning person. Nat Commun. 2016;7:10448. doi: 10.1038/ncomms10448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shimomura K, et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11(6):959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- 50. Dobzhansky T, et al. Artificial and natural selection for two behavioral traits in Drosophila pseudoobscura. Proc Natl Acad Sci USA. 1969;62(1):75–80. doi: 10.1073/pnas.62.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mackay TF. Transposable element-induced response to artificial selection in Drosophila melanogaster. Genetics 1985;111(2):351–374. doi: 10.1093/genetics/111.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Frankham R, et al. Reproductive fitness and artificial selection in animal breeding: culling on fitness prevents a decline in reproductive fitness in lines of Drosophila melanogaster selected for increased inebriation time. Theor Appl Genet. 1988;76(6):909–914. doi: 10.1007/BF00273680. [DOI] [PubMed] [Google Scholar]
- 53. Bolstad GH, et al. Complex constraints on allometry revealed by artificial selection on the wing of Drosophila melanogaster. Proc Natl Acad Sci USA. 2015;112(43):13284–13289. doi: 10.1073/pnas.1505357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ahuja A, et al. Variation and evolution of male sex combs in Drosophila: nature of selection response and theories of genetic variation for sexual traits. Genetics 2008;179(1):503–509. doi: 10.1534/genetics.107.086363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Robertson FW. Selection response and the properties of genetic variation. Cold Spring Harb Symp Quant Biol. 1955;20:166–177. doi: 10.1101/sqb.1955.020.01.017. [DOI] [PubMed] [Google Scholar]
- 56. Masek P, et al. Altered regulation of sleep and feeding contributes to starvation resistance in Drosophila melanogaster. J Exp Biol. 2014;217(Pt 17):3122–3132. doi: 10.1242/jeb.103309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Slocumb ME, et al. Enhanced sleep is an evolutionarily adaptive response to starvation stress in Drosophila. PLoS One. 2015;10(7):e0131275. doi: 10.1371/journal.pone.0131275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. El Yacoubi M, et al. Behavioral, neurochemical, and electrophysiological characterization of a genetic mouse model of depression. Proc Natl Acad Sci USA. 2003;100(10):6227–6232. doi: 10.1073/pnas.1034823100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Overstreet DH. The Flinders sensitive line rats: a genetic animal model of depression. Neurosci Biobehav Rev. 1993;17(1):51–68. doi: 10.1016/s0149-7634(05)80230-1. [DOI] [PubMed] [Google Scholar]
- 60. Overstreet DH, et al. Selective breeding for sensitivity to the anticholinesterase DFP. Psychopharmacology (Berl) 1979;65(1):15–20. doi: 10.1007/BF00491972. [DOI] [PubMed] [Google Scholar]
- 61. Hilakivi I, et al. Strain difference in early postnatal sleep-wake behaviour between Alko Alcohol and Wistar rats. Acta Physiol Scand. 1995;154(1):75–80. doi: 10.1111/j.1748-1716.1995.tb09888.x. [DOI] [PubMed] [Google Scholar]
- 62. Fenzl T, et al. Sleep disturbances in highly stress reactive mice: modeling endophenotypes of major depression. BMC Neurosci. 2011;12:29. doi: 10.1186/1471-2202-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eriksson K. Genetic selection for voluntary alcohol consumption in the albino rat. Science 1968;159(3816):739–741. doi: 10.1126/science.159.3816.739. [DOI] [PubMed] [Google Scholar]
- 64. Morozova TV, et al. Phenotypic and transcriptional response to selection for alcohol sensitivity in Drosophila melanogaster. Genome Biol. 2007;8:R231. doi: 10.1186/gb-2007-8-10-r231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Thompson JB, et al. Sleep-length differences are associated with altered longevity in the fruit fly Drosophila melanogaster. Biol Open 2020;9(9):bio054361. doi: 10.1242/bio.054361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. El Yacoubi M, et al. Genetic association between helpless trait and depression-related phenotypes: evidence from crossbreeding studies with H/Rouen and NH/Rouen mice. Int J Neuropsychopharmacol. 2012;15(3):363–374. doi: 10.1017/S1461145711000605. [DOI] [PubMed] [Google Scholar]
- 67. Serrano Negron YL, et al. The sleep inbred panel, a collection of inbred Drosophila melanogaster with extreme long and short sleep duration. G3 (Bethesda) 2018;8(9):2865–2873. doi: 10.1534/g3.118.200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kumar S, et al. Short-term memory deficits in the Sleep Inbred Panel. Clocks & Sleep 2019;1:471–488. doi: 10.3390/clockssleep1040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Donlea JM, et al. Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 2009;324(5923):105–108. doi: 10.1126/science.1166657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ganguly-Fitzgerald I, et al. Waking experience affects sleep need in Drosophila. Science 2006;313:1775–1781. doi: 10.1126/science.1130408. [DOI] [PubMed] [Google Scholar]
- 71. Li W, et al. Chronic social isolation signals starvation and reduces sleep in Drosophila. Nature 2021;597(7875):239–244. doi: 10.1038/s41586-021-03837-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Gratz SJ, et al. CRISPR-Cas9 genome editing in Drosophila. Curr Protoc Mol Biol. 2015;111:31 32 31–31 32 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lamb AM, et al. Tools and strategies for scarless allele replacement in Drosophila using CRISPR/Cas9. Fly (Austin) 2017;11(1):53–64. doi: 10.1080/19336934.2016.1220463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bringmann H. Sleep-active neurons: conserved motors of sleep. Genetics 2018;208(4):1279–1289. doi: 10.1534/genetics.117.300521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu ET, et al. Of mice and CRISPR: The post-CRISPR future of the mouse as a model system for the human condition. EMBO Rep. 2017;18(2):187–193. doi: 10.15252/embr.201643717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Port F, et al. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods. 2016;13(10):852–854. doi: 10.1038/nmeth.3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analyzed in support of this article.