Abstract
Purpose
Seroepidemiology and genomic surveillance are valuable tools to investigate infection transmission during a pandemic. North East (NE) India is a strategically important region being the gateway connecting the country with Southeast Asia. Here, we examined the spread of SARS-CoV-2 in NE India during the first and second waves of COVID-19 using serological and whole genome sequencing approaches.
Methods
qRT-PCR analysis was performed on a selected population (n = 16,295) from June 2020 to July 2021, and metadata was collected. Immunoassays were studied (n = 2026) at three-time points (August 2020, February 2021, and June 2021) and in a cohort (n = 35) for a year. SARS-CoV-2 whole genomes (n = 914) were sequenced and analyzed with those obtained from the databases.
Results
Test positivity rates (TPR) in the first and second waves were 6.34% and 6.64% in Assam, respectively, and a similar pattern was observed in other NE states. Seropositivity in the three time points was 10.63%, 40.3%, and 46.33%, respectively, and neutralizing antibody prevalence was 90.91%, 52.14%, and 69.30%, respectively. Persistence of pan-IgG-N SARS-CoV-2 antibody for over a year was observed among three subjects in the cohort group. Normal variants dominated the first wave, while B.1.617.2 and AY-sublineages dominated the second wave in the region. The prevalence of the variants co-related well with high TPR and seropositivity rate in the region and identified mostly among vaccinated individuals.
Conclusion
The COVID-19 first wave in the region witnessed low transmission with the evolution of diverse variants. Seropositivity increased during the study period with over half of the individuals carrying neutralizing antibodies against SARS-CoV-2. High infection and seroprevalence in NE India during the second wave were associated with the dominant emergence of variants of concern.
Keywords: COVID-19, Test positivity rate, Seroprevalence, Neutralizing antibody, Whole genome sequencing
1. Introduction
SARS-CoV-2 has rapidly infected people across the world. The real burden of the COVID-19 pandemic remains unclear, though established to be higher than the reported cases, due to various inherent factors [1]. Seroepidemiology and genome surveillance have helped understand the true burden, transmission and evolution of SARS-CoV-2 [[2], [3], [4]]. Genome surveillance can supplement serosurvey in informing effective public health responses and policy decisions.
North East (NE) India is a unique region with significant geographical and ethnic diversity. It shares international border with five countries and is strategically important as India's gateway to Southeast Asia. COVID-19 gradually spread throughout the NE region since its first confirmed case was reported in March 2020 [5]. This region is crucial in India's effort to trace, test, isolate and treat COVID-19. Strict surveillance of SARS-CoV-2 in the region is important to restrict transnational and interethnic SARS-CoV-2 transmission. As per the Indian SARS-CoV-2 Genomics Consortium (INSACOG), only 2.36% of genome sequences was reported from NE India indicating a critical dearth of surveillance in this region [6]. Moreover, very limited seroepidemiological studies have been reported from the population of this region [1].
This observational study investigates COVID-19 infectivity, seroepidemiology and genome surveillance during first wave (June–September 2020), intermittent phase (February 2021) and second wave (May–September 2021) in NE India. Furthermore, a longitudinal seroepidemiological follow-up was conducted for a year to assess anti-SARS-CoV-2 antibodies and their neutralizing efficacy.
2. Materials and methods
This observational study was approved by the Institutional Human Ethics Committee, CSIR–North East Institute of Science and Technology (IHEC/NEIST20-21/201). The study was conducted among the population of Jorhat district, Assam. The study design and methodological approaches used are represented in Supplementary Fig. S1. Before enrollment, written informed consent was obtained, and all subjects participated voluntarily. Subject metadata were obtained using a standard questionnaire. Blood (5 mL) and naso-oropharyngeal swabs were collected and handled as per guidelines issued by the Ministry of Health and Family Welfare (MoHFW), Government of India. Swabs collected during first wave (June–September 2020) (n = 12,879) and second wave (May–July 2021) (n = 3416) were tested using quantitative reverse transcription polymerase chain reaction (qRT-PCR) kits approved by the Indian Council of Medical Research (ICMR). All individuals, irrespective of age, gender, symptomatic, asymptomatic, vaccinated and non-vaccinated, who are resident of Jorhat, Assam and whose samples were tested in the COVID-19 Testing Laboratory, CSIR-NEIST, Jorhat were included in the present study as per the guidelines of the MoHFW. To study the seropositivity in the onset of first wave, end of first wave and second wave of infection, a repeated cross-sectional serological investigation was conducted. A total of 2026 participants were included, and blood samples were collected in August 2020 (n = 724), February 2021 (n = 866), and June 2021 (n = 436). Immunoassays specific to SARS-CoV-2 IgG-N were performed using Elecsys Anti-SARS-CoV-2 kit (Roche Diagnostics, Germany), and both IgG-S and IgM-S were determined using VoxPress New Corona Virus (COVID-19) IgG/IgM Rapid Test kit (Voxtur Bio Ltd, India) according to the manufacturers' protocol. Samples seropositive for IgG-N, IgG-S and/or IgM-S were analyzed for SARS-CoV-2 neutralizing antibody using cPass SARS-CoV-2 Neutralization Antibody Detection Kit (GenScript, USA). A longitudinal follow-up of a cohort (n = 35) among the participants enrolled for the serological study was conducted for a year to monitor anti-SARS-CoV-2 antibodies and their neutralizing efficacies. The follow-up individuals were selected based on the criteria that the individual must have participated in all the three time points of the above serological investigation and continued to volunteer to the end of the follow up. From the collected swab samples, whole genome sequencing was performed on 914 qRT-PCR positive samples (first wave: n = 591; second wave: n = 323) that had Ct ≤ 35 for any of the three confirmatory genes, viz. ORF1ab, RdRp and N, on a Nanopore MinION Mk1c platform (Oxford Nanopore Technologies, UK) using a multiplex tiling PCR approach (NEBNext ARTIC SARS-CoV-2 Companion Kit, New England Biolabs, USA) [7,8]. Wuhan-Hu-1 SARS-CoV-2 genome (MN908947.3) was used as reference, and variant calling and consensus genome assembly were performed using ARTIC FieldBioinformatics pipeline v1.1.0 (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html). Mutation calling and lineage assignment were carried out using NextClade v.154 (https://clades.nextstrain.org/) and Pangolin Lineage Assigner v3.1.16 (https://pangolin.cog-uk.io). The genome-based phylogenetic tree was constructed using IQ-TREE v1.6.12 and Fig Tree v1.4.4. COVID-19 and vaccination data of NE states were retrieved from the public COVID19INDIA dashboard (https://www.covid19india.org/). COVID-19 data of the neighboring countries were sourced from Our World in Data (https://ourworldindata.org/coronavirus). Serology data of India and the neighboring countries were sourced from Serotracker (https://serotracker.com/en/Explore). SARS-CoV-2 variant datasets of NE states and their neighboring countries were retrieved from INSACOG (http://clingen.igib.res.in/covid19genomes/) and GISAID (https://www.gisaid.org/). The genome sequences generated in this study were deposited to GISAID and available upon registration. Statistical analyses were performed using Sigma Stat software (San Jose, CA, USA). One way ANOVA and Student's T-test were performed to evaluate significant differences among the study groups at significant level of 0.05.
3. Results
3.1. COVID-19 test positivity rate during first and second waves
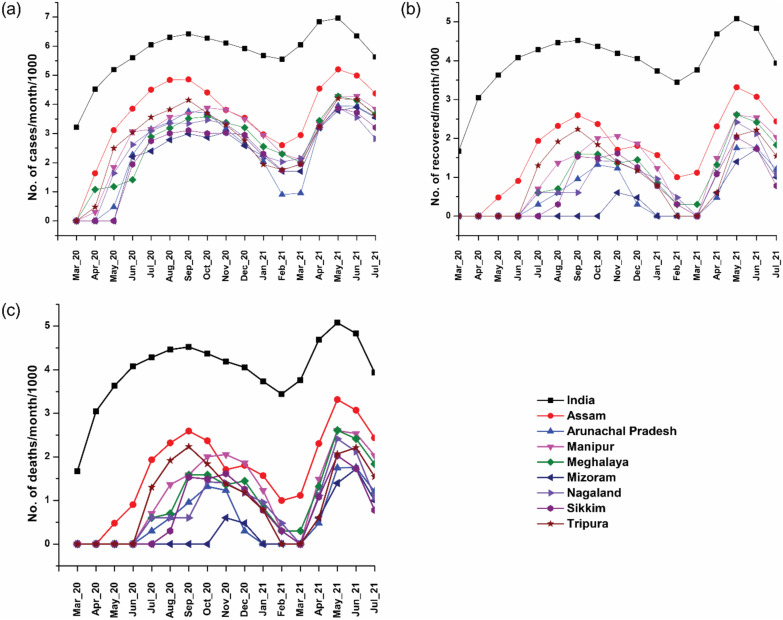
The first COVID-19 infection was reported in Assam on March 31, 2020 and registered the highest daily positive cases (n = 2354) on August 18, 2020 (Fig. 1 ). The second wave started in early April 2021, and the highest daily registered cases were 5704 on May 27, 2021. The SARS-CoV-2 test positivity rate (TPR) in Assam during first wave and second wave was 6.34% and 6.64%, respectively. The outbreak of second wave was also considerably high in other NE states like Manipur, Meghalaya, Mizoram and Sikkim (Fig. 1). During first wave, out of 12,879 samples from Jorhat district, Assam, 637 tested positive (TPR: 4.95%). However, out of 3416 samples in the second wave, 248 tested positive (TPR: 7.26%). The TPR increased with age in first wave indicating higher infection in older subjects (Table S1). However, a significant increase (p < 0.01) in TPR was observed in children <10 years in the second wave compared to those seen in first wave. Similarly, significant increase in TPR was also observed in subjects with 11–30 years (p < 0.01) and 31–60 years (p < 0.001) in the second wave compared to those seen in first wave. On the other hand, no significant (p = 0.085) increase in TPR was observed in subjects with >60 years in the second wave compared to those seen in first wave [9].
Fig. 1.
Trend of COVID-19 (a) positivity (b) recovery (c) death in India and states of North East India during first wave and second wave. The data were sourced from www.covid19india.org (retrieved on July 17, 2021).
3.2. Seroprevalence during first and second waves
In the repeated cross-sectional serological investigation, the anti-SARS-CoV-2 IgG-N, IgG-S, and IgG-M antibodies were found to be 10.63%, 8.83%, and 1%, respectively, during August 2020, while the seropositivity in February 2021 and June 2021 were 40.3%, 16.85%, and 3.49%, and 46.33%, 15.82%, and 3.66%, respectively (Table 1 ). The frequencies of neutralizing antibody in these periods were 90.91%, 52.14%, and 69.30%, respectively. Seropositivity increased while the frequency of neutralizing antibodies decreased both in the intermittent phase and in second wave compared to the first wave (Table S2). Seropositivity and neutralizing antibody decreased with age in August 2020, indicating higher infection in older subjects, while higher infection was observed in younger subjects in June 2021. Results showed that there is a positive correlation between COVID-19 infectivity and seropositivity and negative correlation between COVID-19 infectivity and neutralizing antibody, but statistically not significant. The less neutralizing activity of the antibodies supports the current finding of breakthrough infection by variant of concern as the developed antibody could not neutralize the SARS-CoV-2 variants emerging in the first wave.
Table 1.
Seropositivity of the participants against SARS-CoV-2a.
| August 2020 (n = 724) |
February 2021 (n = 866) |
June 2021 (n = 436) |
||||
|---|---|---|---|---|---|---|
| Parameterb | Positive result [n (%)] | Adjusted Positivityc[% (95% CI)] | Positive result [n (%)] | Adjusted Positivityc[% (95% CI)] | Positive result [n (%)] | Adjusted Positivityc[% (95% CI)] |
| IgG-N | 77 (10.63%) | 10.61% (8.37–12.85) | 349 (40.3%) | 40.21% (36.95–43.48) | 202 (46.33%) | 46.23% (41.55–50.91) |
| IgG-S | 64 (8.83%) | – | 146 (16.85%) | – | 69 (15.82%) | – |
| IgM-S | 7 (1%) | – | 31 (3.49%) | – | 16 (3.66%) | – |
| Neutralizing antibodyd | 70 (90.91%) | 90.91% (97.33–84.49) | 182 (52.14%) | 52.14% (57.39–46.91) | 140 (69.30%) | 69.30% (75.67–62.95) |
Assessments were done at three different time points, namely August 2020, February 2021, and June 2021.
IgG-N: anti-N protein IgG; IgG-S: anti-S protein IgG; IgM-S: anti-S protein IgM; Neutralizing antibody: neutralizing antibody against SARS-CoV-2.
Adjusted positivity was calculated considering 99.8% sensitivity and 100% specificity for the IgG-N kit (Roche) and 100% sensitivity and 100% specificity for the neutralizing antibody kit (Genscript). Kits used for IgG-S and IgM-S were ICMR-approved, but, there were no information about the sensitivity and specificity.
Only samples that were seropositive for IgG-N, IgG-S and/or IgM-S were considered for the neutralizing antibody assay.
3.3. SARS-CoV-2 variants during first and second wave
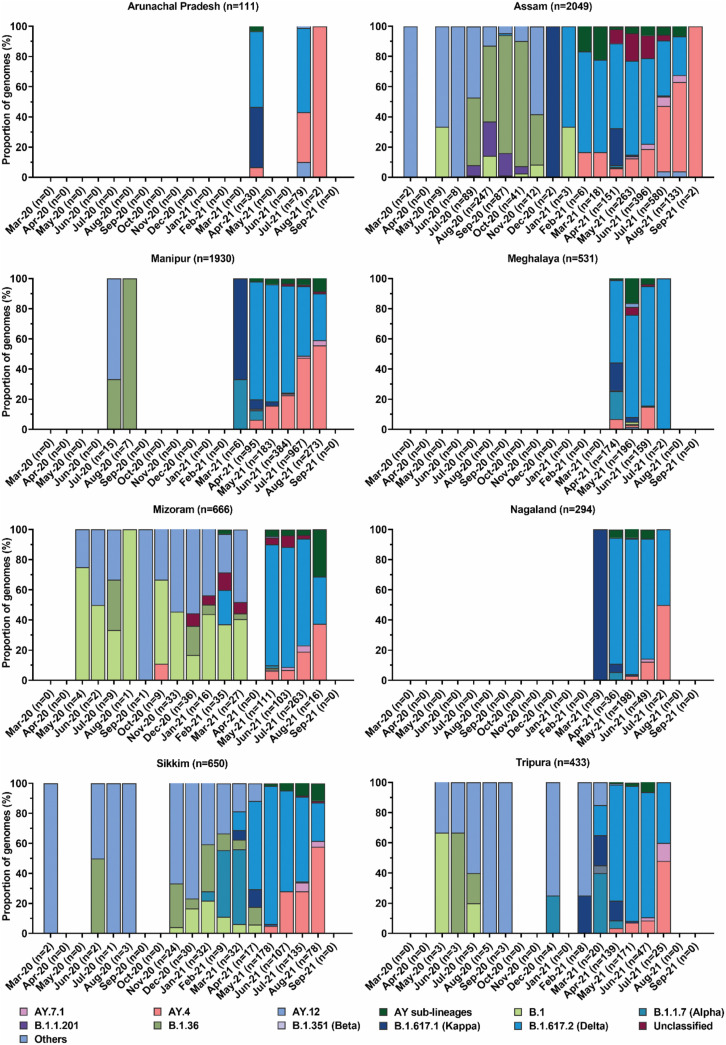
Out of total 815 samples that passed genome read QC and lineage assignment, 39 variants were detected, including B.1.617.2 (n = 158), AY-sublineages (n = 112), and B.1.617.1 (n = 1). Thirty two normal variants, predominated by B.1.36 (n = 329), B.1.1.201 (n = 105) and B.1 (n = 36), were detected in first wave, while second wave was identified with 9 variants predominated by B.1.617.2 (n = 157), AY-sublineages (n = 112) and B.1.617.1 (n = 1). The maximum-likelihood time-resolved phylogenetic tree of the detected variants is shown in Fig. S2. Among 162 individuals with vaccination details, B.1.617.2 and AY-sublineages were identified mostly in vaccinated individuals (n = 94, 58.02%) with highest infections among single-dose vaccinated individuals (n = 51, 54.26%), followed by breakthrough infection (n = 38, 40.43%). Total 3373 mutations were observed across the genomes (Table S3, Fig. S3, Fig. S4), wherein 865 mutations (synonymous: 451; missense: 395; non-sense: 19) were exclusively observed in first wave, while 1731 mutations (synonymous: 447; missense: 1284) were observed exclusively in second wave. The mutation pattern and frequency among the variants shifted between the two waves and were predominantly associated with non-synonymous mutations in S, N, ORF1a, ORF1b, ORF3a, ORF7a, and ORF7b during the second wave. Deletion mutations were predominantly observed only in the second wave among B.1.617.2. NE India reported numerous incidences of variants of concern (VOC), especially B.1.617.2 and AY-sublineages (Fig. 2, Fig. 3 ). Meta-analysis of variant datasets for the NE states revealed that VOC emerged in the region as early as September 2020 when B.1.617.2 was first detected in Assam. The data showed the displacement of normal variants in first wave by B.1.617.2 and AY-sublineages in the second wave, indicating intra-lineage mutation.
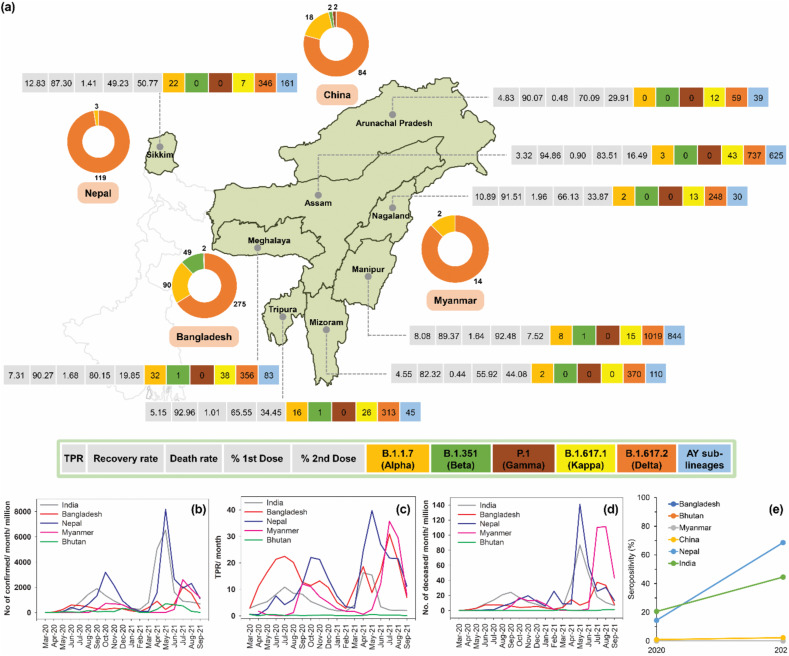
Fig. 2.
COVID-19 spread in various states of North East region of India and in the neighboring countries sharing international border with the region. (a) Demographics of test positivity rate (TPR), recovery rate, death rate, vaccination rate and variants detected for the states of North East India, and variants identified in the neighboring countries during the study period. The data related to COVID-19 test, recovery, death and vaccination were sourced from www.covid19india.org (retrieved on July 17, 2021). Only the variants of concern and variant of interest are shown. The variant data was retrieved from the Indian SARS-CoV-2 Genomics Consortium (INSACOG) (http://clingen.igib.res.in/covid19genomes/), and GISAID databases (https://www.gisaid.org/) (retrieved on October 15, 2021). The variant dataset for the state of Assam also included the variants identified from the whole genome sequencing conducted in the present study. (b) COVID-19 confirmed cases, (c) TPR, and (d) death cases in the neighboring countries of North East region of India. Data were sourced from Our World in Data (https://ourworldindata.org/coronavirus) (retrieved on October 15, 2021). (e) Seropositivity rate of India and neighboring countries. Data were sourced from Serotracker (https://serotracker.com/en/Explore) (retrieved on October 24, 2021). Seropositivity rate for India, China and Nepal are shown as the average of three different time points of each year. Seropositivity data was not available for Bangladesh, Bhutan and Myanmar.
Fig. 3.
Evolution of SARS-CoV-2 variants at different time points of their detection in the states of North East India. The variant data was sourced from the Indian SARS-CoV-2 Genomics Consortium (INSACOG) (http://clingen.igib.res.in/covid19genomes/), and GISAID databases (https://www.gisaid.org/) (retrieved on October 15, 2021). The variant dataset for the state of Assam also included the variants identified from the whole genome sequencing conducted in the present study.
3.4. Serological study of cohort
In the longitudinal follow-up of cohort (n = 35), pan-IgG-N antibody increased from 8.57% in August 2020 to 28.57%, 31.42%, 45.71% and 45.71% in February 2021, June 2021, July 2021 and August 2021, respectively, correlating well with the increase in COVID-19 incidences (Table 2 ). Three subjects tested seropositive in August 10, 2020 and persisted for over a year till August 14, 2021 (Fig. S5). The neutralizing activity increased 2.33-fold from August 2020 to February 2021 and over 4-fold from February 2021 to July 2021 to its maximum (Table 2). Interestingly, IgG-N antibody remained low compared to neutralizing activity after June 2021. The increase in neutralizing activity within a short span of time could be due to the effect of vaccination (Table S4). Vaccine breakthrough was observed in only one single individual vaccinated with Covishield who was asymptomatic seropositive during first wave. Moreover, it was observed that individuals infected with SARS-CoV-2 prior to vaccination showed higher neutralizing activity irrespective of lifestyle and co-morbidity.
Table 2.
IgG-N reactive antibody and neutralizing activity against SARS-CoV-2 among the cohort of 35 subjectsa.
| Parameters |
2020 (n = 35) |
2021 (n = 35) |
||||||
|---|---|---|---|---|---|---|---|---|
| Sampling Date | 10 August | 10 February | 10 June | 10 July | 16 July | 24 July | 31 July | 14 August |
| IgG-N antibody [n (%)] | 3 (8.57%) | 10 (28.57%) | 11 (31.42%) | 16 (45.71%) | 16 (45.71%) | 16 (45.71%) | 16 (45.71%) | 16 (45.71%) |
| Neutralizing antibody [n (%)] | 3 (8.57%) | 7 (20%) | 11 (31.42%) | 29 (82.85%) | 31 (88.57) | 25 (71.41%) | 24 (68.57%) | 26 (74.28%) |
| Vaccinated, Single dose [n (%)] | – | – | 20 (57.1%) | 15 (42.85%) | – | – | – | – |
| Vaccinated, Double dose [n (%)] | – | – | 10 (28.57%) | 4 (11.42%) | – | – | – | – |
| qRT-PCR positive [n (%)] | 5 (14.2%) | – | 1 (2.85%) | 1 (2.85%) | 1 (2.85%) | – | – | 2 (5.71%) |
| Vaccine breakthroughb[n (%)] | – | – | – | – | – | – | – | 1 (2.85%) |
Cohort group includes 29 male and 6 female participants aged between 25 and 55 years with an average age of 35.77 years.
Vaccine breakthrough is defined as one who got infected after receiving both doses and completed at least 14 days of follow-up after the second dose.
4. Discussion
Considering the unique regional importance of NE India and the global need to generate and exchange genomic and serological profiles of SARS-CoV-2 transmission and evolution, we have conducted this study of NE India. Our analysis showed that the COVID-19 incidence across NE states was relatively low compared to the trend observed in the overall Indian population (Fig. 1). The higher incidence in the overall Indian population compared to the NE states could be due to extremely higher local incidences in some parts of India, such as Maharashtra, Delhi, and Karnataka. When we normalized the COVID-19 incidence data per 1000 cases, we observed interesting variations in number of waves and peak intensities among the NE states (Tables S5 and S6). During the first wave, the increase in infection was steady and gradual across NE India, which might have led to the emergence of new variants while the virus adapted to the hosts. Skewed distribution observed during the first wave indicated rapid and effective control of the COVID-19 transmission in the region that might have resulted from strict public health strategies implemented by the state governments. The observed rapid rise in infection during the second wave might have resulted from the emergence of B.1.617.2 and AY-sublineages, with high transmission and immune escape properties [10]. This was further elevated with relaxation in COVID-19 preventive measures by the respective state governments.
Our repeated cross-sectional serological study observed that seropositivity increased while the frequency of neutralizing antibody decreased both in the intermittent phase and in the second wave compared to those seen in the first wave. The observed seropositivity of 10.61% during first wave was in concordance with our earlier finding [11,12]. A possible reason for the observation was the emergence of B.1.617.2 and AY variants that were reported to possess reduced sensitivity to antibody neutralization [13]. Moreover, the increase in seropositivity and neutralizing antibody in older subjects in the second wave might suggest the additional influence of age-selective mass vaccination programmes. Several explanations have been put forward as possible factors for the generation of multi-waves and their characteristics in pandemics. Some of these are co-infection [14], pandemic fatigue [15], sub-optimal prevention and control measures during intermittent endemic periods [16], climatic variations [17] and genetic drift of the pathogen associated with diffusion and herd immunity [18,19].
In India, the first wave of COVID-19 lasted from August 2020 to January 2021 and the second wave persisted from March 2021 to June 2021. The vaccination drive started at the end of January 2021. Since, most of the study participants were vaccinated from April 2021 onwards, the antibody examination in the longitudinal cohort was conducted relatively more frequently post July 2021 to assess the changes in the antibody titre after vaccination. Moreover, it has been reported that the antibody level reaches its peak by 3 months [20]. Our longitudinal cohort study recorded that seropositivity could last more than a year, which corroborated earlier findings that some individuals could sustain SARS-CoV-2 antibodies for a relatively long period for prolonged protection [21]. It was observed that SARS-CoV-2 infection before vaccination resulted in higher neutralizing activity irrespective of lifestyle and health condition. This result assented to earlier findings that a single dose of mRNA vaccine could enhance effective immune response in individuals with prior COVID-19 infection [22]. The observed decrease in neutralizing antibodies in the vaccinated individuals could be assumed as a possible shifting of humoral immunity towards the generation of memory B cells for longer uses [23,24].
The whole genome sequencing allowed us to detect and compare the representative diversity of SARS-CoV-2 in the NE states. Most importantly, our genome sequencing work sought to confirm the first occurrence of VOC in NE India. Our findings confirmed the earliest occurrence of B.1.617.2 in the NE state of Assam in September 2020, within one month of its first identification in India in August 2020. This indicated that B.1.617.2 transmission was swift and prompt from mainland India to NE India. Upon its first occurrence in Assam, the rapid transmission of B.1.617.2 and its AY-sublineages was observed. We also observed that the occurrence of B.1.617.2 and AY-sublineages was mainly associated with partial vaccination. This observation was consistent with earlier reports [25,26]. It was reported that the secondary attack rate of B.1.617.2 in UK was higher than B.1.1.7 in partially immunized population [27]. The linkage between the SARS-CoV-2 variants was mainly characterized by unique substitution or deletion in the spike protein. Changes in L452R, E484Q and P681R amino acids were identified in B.1.617.1, while B.1.617.2 was identified with changes in L452R and P681R amino acids [28]. The changes in these three amino acids were directly linked with the strong host binding affinity of SARS-CoV-2 and its capability to escape neutralizing antibodies [29]. Overall, the genomic characterization of SARS-CoV-2 from NE India increased the availability of geographical-based genomes of the virus and illustrated the emerging trend of genomic disparities among the variants.
During the second wave, most neighboring countries of NE India had a relatively small number of cases (Fig. 2). Bhutan was an exception, where it was experiencing one of its largest surges despite vaccination of 62% of the population with at least one dose [30]. The case surge in Bangladesh corresponded with widespread detection of B.1.351 in February and March 2021 and B.1.617.2 in subsequent months [31]. The case surge in Nepal and India, particularly in the NE region, assented to the increasing incidences of B.1.617.2. The case surge in Bhutan and Myanmar, though it started after a rapid case decline in India and its NE region, coincided with the prevalence of B.1.617.2. Overall, the second wave in the countries neighboring India's NE region followed similar pattern of the rise and fall of cases and was driven by B.1.617.2. Considering the fragile and porous nature of these international borders, undetected cross-border spillage of SARS-CoV-2 is highly likely, and further investigation is warranted to ascertain pertinent transmission of the infection across the border.
5. Conclusion
This study demonstrates that each approach can derive different epidemiological profiles of transmission and evolution of SARS-CoV-2 in a given population. The states of NE India have similar trend of COVID-19 test positivity in both the infection waves, and the first wave in the region witnesses low transmission with the evolution of diverse variants. The prevalence of various variants across NE India implies the potential to spread any communicable diseases through and beyond this region. Seropositivity increased during the study period with over half of the study individuals carrying neutralizing antibodies against SARS-CoV-2. High SARS-CoV-2 infection rate and seroprevalence in the region during the second wave were associated with the dominant emergence of variants of concern B.1.617.2 and its AY-sublineages, which were identified mostly among vaccinated individuals. Additional studies are warranted to understand how the virus will evolve post-vaccination, which will dictate the outcome of any possible future wave of COVID-19.
Funding
Council of Scientific and Industrial Research (CSIR) [grant numbers MLP0158, MLP1019].
Role of the funding source
The funding source had no role in study design; collection, analysis, and interpretation of data; manuscript writing; and the decision to submit the article for publication.
Conflicts of interest
The authors declare no conflicts of interest related to this article.
Ethics approval
IHEC/NEIST20-21/201.
CRediT author statement
G Narahari Sastry, Jatin Kalita: Conceptualization, Study design, Project administration, Supervision, Funding acquisition, Writing - Review & Editing; Romi Wahengbam, Pankaj Bharali, Prasenjit Manna, Tridip Phukan: Study design, Methodology, Software, Investigation, Formal analysis, Validation, Resources, Writing - Original Draft, Writing - Review & Editing, Visualization; Moirangthem Goutam Singh, Gayatri Gogoi, Yasmin Begam Tapadar, Himakshi Sarma, Ravi Kumar Sahu, Prachurjya Dutta: Investigation; Anil Kumar Singh: Investigation, Formal analysis, Visualization, Writing - Original Draft, Writing - Review & Editing; Rituraj Konwar: Investigation, Writing - Original Draft, Writing - Review & Editing; Selvaraman Nagamani, Hridoy Jyoti Mahanta: Software, Formal analysis; Natarajan Velmurugan, Channakeshavaiah Chikkaputtaiah, Sawlang Borsingh Wann: Writing - Original Draft.
Acknowledgments
We would like to acknowledge doctors, technical staffs, and nurses of the Clinical Centre, CSIR-NEIST, and CSIR-NEIST COVID-19 Warriors for their support in subject recruitment, sample collection and vaccination. The Indian SARS-CoV-2 Genomics Consortium (INSACOG) is thanked for supporting the SARS-CoV-2 genome surveillance. We acknowledge the Council of Scientific and Industrial Research (CSIR), India for funding the research. qRT-PCR testing kits were provided by the COVID-19 depot, ICMR-RMRCNE, Dibrugarh, Assam, India.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijmmb.2022.10.011.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Murhekar M.V., Bhatnagar T., Selvaraju S., Saravanakumar V., Thangaraj J.W.V., Shah N., et al. SARS-CoV-2 antibody seroprevalence in India, August-September, 2020: findings from the second nationwide household serosurvey. Lancet Global Health. 2021;9(3):e257–e266. doi: 10.1016/S2214-109X(20)30544-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genome sequencing by INSACOG shows variants of concern and a novel variant in India . 2021. Ministry of Health and Family Welfare, Govt. of India.https://pib.gov.in/PressReleaseIframePage.aspx?PRID=1707177 Available at. [Google Scholar]
- 3.Indian SARS-CoV-2 Genomics Consortium (INSACOG) 2021. Ministry of Health and Family Welfare, Department of Biotechnology (DBT), Council of Scientific & Industrial Research (CSIR) and Indian Council of Medical Research (ICMR)https://dbtindia.gov.in/insacog Available at. [Google Scholar]
- 4.Tessema S.K., Inzaule S.C., Christoffels A., Kebede Y., de Oliveira T., Ouma A.E.O., et al. Accelerating genomics-based surveillance for COVID-19 response in Africa. Lancet Microbe. 2020;1(6):e227–e228. doi: 10.1016/S2666-5247(20)30117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.COVID19 INDIA. 2021. https://www.covid19india.org/ Available at. [Google Scholar]
- 6.Indian COVID-19 Genome Surveillance . 2021. Department of Biotechnology (DBT), Govt. of India.http://clingen.igib.res.in/covid19genomes/ Available at. [Google Scholar]
- 7.ARTIC network SARS-CoV-2. 2021. https://artic.network/ncov-2019 Available at. [Google Scholar]
- 8.Tyson J.R., James P., Stoddart D., Sparks N., Wickenhagen A., Hall G., et al. Improvements to the ARTIC multiplex PCR method for SARS-CoV-2 genome sequencing using nanopore. bioRxiv. 2020;2020 doi: 10.1101/2020.09.04.283077. 09.04.283077. [DOI] [Google Scholar]
- 9.Mahanta H.J., Sastry G.N. COVID-19 impact on socio-economic and health interventions: A gaps and peaks analysis using clustering approach. J Statistics Manag Syst. 2022;25:2123–2153. doi: 10.1080/09720510.2022.2117335. [DOI] [Google Scholar]
- 10.Mlcochova P., Kemp S.A., Dhar M.S., Papa G., Meng B., Ferreira I.A.T.M., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naushin S., Sardana V., Ujjainiya R., Bhatheja N., Kutum R., Bhaskar A.K., et al. Insights from a Pan India Sero-Epidemiological survey (Phenome-India Cohort) for SARS-CoV2. Elife. 2021;10 doi: 10.7554/eLife.66537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh P., Ujjainiya R., Prakash S., Naushin S., Sardana V., Bhatheja N., et al. A machine learning-based approach to determine infection status in recipients of BBV152 (Covaxin) whole-virion inactivated SARS-CoV-2 vaccine for serological surveys. Comput Biol Med. 2022;146:1054419. doi: 10.1016/j.compbiomed.2022.105419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 14.Merler S., Poletti P., Ajelli M., Caprile B., Manfredi P. Coinfection can trigger multiple pandemic waves. J Theor Biol. 2008;254(2):499–507. doi: 10.1016/j.jtbi.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Tammemi A.B., Tarhini Z., Akour A. A swaying between successive pandemic waves and pandemic fatigue: where does Jordan stand? Ann Med Surg (Lond) 2021;65 doi: 10.1016/j.amsu.2021.102298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cacciapaglia G., Cot C., Sannino F. Multiwave pandemic dynamics explained: how to tame the next wave of infectious diseases. Sci Rep. 2021;11(1) doi: 10.1038/s41598-021-85875-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pietro R.D., Basile M., Antolini L., Alberti S. Genetic drift versus regional spreading dynamics of COVID-19. Front Genet. 2021;12 doi: 10.3389/fgene.2021.663371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y., Li J., Li H., Lei P., Shen G., Yang C. Persistence of SARS-CoV-2-specific antibodies in COVID-19 patients. Int Immunopharm. 2021;90 doi: 10.1016/j.intimp.2020.107271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao K., Yang H., Liu B., Pang X., Du J., Liu M., et al. Antibodies can last for more than 1 year after SARS-CoV-2 infection: a follow-up study from survivors of COVID-19. Front Med. 2021;8(684864) doi: 10.3389/fmed.2021.684864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds C.J., Pade C., Gibbons J.M., Butler D.K., Otter A.D., Menacho K., et al. Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose. Science. 2021;372:1418–1423. doi: 10.1126/science.abh1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goel R.R., Apostolidis S.A., Painter M.M., Mathe D., Pattekar A., Kuthuru O., et al. Distinct antibody and memory B cell responses in SARS-CoV-2 naïve and recovered individuals following mRNA vaccination. Sci Immunol. 2021;6(58) doi: 10.1126/sciimmunol.abi6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner J.S., O'Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596(7870):109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challen R., Dyson L., Overton C.E., Guzman-Rincon L.M., Hill E.M., Stage H.B., et al. Early epidemiological signatures of novel SARS-CoV-2 variants: establishment of B.1.617.2 in England. medRxiv. 2021;2021 doi: 10.1101/2021.06.05.21258365. 06.05.21258365. [DOI] [Google Scholar]
- 26.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W., Shaman J. COVID-19 pandemic dynamics in India, the SARS-CoV-2 Delta variant, and implications for vaccination. medRxiv. 2021;2021 doi: 10.1101/2021.06.21.21259268. 06.21.21259268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yadav P.D., Mohandas S., Shete A.M., Nyayanit D.A., Gupta N., Patil D.Y., et al. SARS CoV-2 variant B.1.617.1 is highly pathogenic in hamsters than B.1 variant. bioRxiv. 2021;2021 doi: 10.1101/2021.05.05.442760. 05.05.442760. [DOI] [Google Scholar]
- 29.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310. doi: 10.1016/j.cell.2020.08.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallapaty S. India's neighbours race to sequence genomes as COVID surges. Nature. 2021;593(7860):485–486. doi: 10.1038/d41586-021-01287-2. [DOI] [PubMed] [Google Scholar]
- 31.Saha S., Tanmoy A.M., Hooda Y., Tanni A.A., Goswami S., Sium S.M.A., et al. COVID-19 rise in Bangladesh correlates with increasing detection of B.1.351 variant. BMJ Glob Health. 2021;6(5) doi: 10.1136/bmjgh-2021–6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.