Abstract
Streptococcus mutans, the principal etiologic agent of dental caries in humans, possesses a variety of virulence traits that enable it to establish itself in the oral cavity and initiate disease. A 185-kDa cell surface-localized protein known variously as antigen I/II, antigen B, PAc, and P1 has been postulated to be a virulence factor in S. mutans. We showed previously that P1 expression is necessary for in vitro adherence of S. mutans to salivary agglutinin-coated hydroxyapatite as well as for fluid-phase aggregation. Since adherence of the organism is a necessary first step toward colonization of the tooth surface, we sought to determine what effect deletion of the gene for P1, spaP, has on the colonization and subsequent cariogenicity of this organism in vivo. Germ-free Fischer rats fed a diet containing 5% sucrose were infected with either S. mutans NG8 or an NG8-derived spaP mutant strain, PC3370, which had been constructed by allelic exchange mutagenesis. At 1-week intervals for 6 weeks after infection, total organisms recovered from mandibles were enumerated. At week 6, caries lesions also were scored. A significantly lower number of enamel and dentinal carious lesions was observed for the mutant-infected rats, although there was no difference between parent and mutant in the number of organisms recovered from teeth through 6 weeks postinfection. Coinfection of animals with both parent and mutant strains resulted in an increasing predominance of the mutant strain being recovered over time, suggesting that P1 is not a necessary prerequisite for colonization. These data do, however, suggest a role for P1 in the virulence of S. mutans, as reflected by a decrease in the cariogenicity of bacteria lacking this surface protein.
The oral pathogen Streptococcus mutans colonizes the hard surfaces in the human oral cavity and is considered to be the principal etiologic agent of dental caries (16). Key to understanding how S. mutans colonizes the oral cavity is discerning how the various molecular components comprising the bacterial cell surface interact with the acquired dental pellicle. There is much evidence in the literature suggesting that a number of cell surface molecules play important roles in the adherence (and cohesion) process and that the presence or absence of dietary sugar determines which of these molecules is operative (23, 24). It is widely accepted that adherence is mediated by mechanisms involving bacterial extracellular polymers (e.g., glucans) synthesized from sucrose when that common sugar is present. In the absence of sucrose, a 185-kDa cell wall-associated adhesin belonging to a family of oral streptococcal polypeptides called antigen I/II (22) has been suggested to play a major role in colonization (8, 9). The gene for this adhesin in S. mutans was cloned by Lee et al. (13) and called spaP and then by Okahashi et al. (20), who called it pac. Subsequent restriction fragment length polymorphism (RFLP) analysis revealed only minor differences between these adhesin genes (3). To be consistent with our earlier reports, we refer to the gene as spaP and to its product as P1 in this paper.
Early experiments to test the hypothesis that the spaP gene product is involved in non-sucrose-mediated adherence involved the construction of P1-negative mutants by insertional inactivation of spaP (14). In vitro experiments with hydroxyapatite beads coated with either parotid gland saliva or salivary agglutinin demonstrated nonadherence by spaP mutant strain 834 (2, 14) as well as by naturally occurring non-P1-containing strains of S. mutans (5). Also, both native P1 and recombinant P1 competitively inhibited binding by whole cells in these in vitro assays (5). Likewise, Koga et al. (12) demonstrated a lack of adherence by two pac mutants to saliva-coated hydroxapatite and a lack of saliva-induced aggregation of these strains compared to the pac+ parent strain. Collectively, these findings suggested an important role for the spaP gene product in the primary colonization of teeth in the absence of glucan.
Previously, the virulence properties of P1 mutant strain 834 and its parent strain were tested in an in vivo system in which each strain was used to infect desalivated specific-pathogen-free rats fed a high (56%)-sucrose diet (2). Both parent and mutant strains caused similar levels of smooth-surface caries. These findings were confounded by our simultaneous discoveries that mutant 834, which retained an active promoter, expressed the N-terminal 612 amino acids of the 1,561-amino-acid P1 molecule (3) and that an alanine-rich salivary agglutinin-binding domain was associated with this retained fragment (6). In addition, because of the high-sucrose diet, the glucan-independent phase of colonization was likely obscured in this animal study. Therefore, to determine the role of P1 in virulence, we undertook a new study using germ-free rats fed a low (5%)-sucrose diet and infected with a newly constructed mutant of S. mutans NG8 engineered to be completely devoid of spaP.
(This work was presented in part at the 97th General Meeting of the American Society for Microbiology, Miami, Fla., 4 to 8 May 1997.)
MATERIALS AND METHODS
Bacterial strains, culture conditions, and plasmids.
S. mutans NG8 and the NG8-derived spaP mutant strain, PC3370, were routinely cultured aerobically at 37°C in Todd-Hewitt broth (BBL, Cockeysville, Md.) supplemented with 0.3% yeast extract (THYE broth) or on THYE agar in a candle jar. THYE media were supplemented with tetracycline (15 μg/ml) as required. A comparison of the plating efficiencies of the parent and mutant strains was performed by inoculating 2 ml of THYE broth with single colonies of each strain, followed by growth for 16 h and enumeration of CFU by serial dilution and plating. Colonies on triplicate plates of each strain were counted, and the mean ± standard error of the mean (SEM) CFU per milliliter for each were determined.
Escherichia coli JM109 and INVαF′ (InVitrogen, La Jolla, Calif.) were cultured aerobically at 37°C with vigorous shaking in Luria-Bertani (LB) broth supplemented with 15 μg of tetracycline or 50 μg of kanamycin per ml as required.
Plasmid pCR1000 (InVitrogen) was used for cloning of PCR-amplified DNA fragments in accordance with the manufacturer’s instructions.
PCRs.
PCRs were performed with primers having the sequences 5′-CCGGATCCGTGTCAGGTACTATTGTCA-3′ and 5′-GGCTGCAGACGCCTTCGCCTTGTTTAG-3′ to amplify a 480-bp DNA sequence (bp 31 to 511 of the spaP sequence with GenBank accession no. X17390) upstream of the putative spaP promoter (underlined bases indicate engineered restriction sites for BamHI and PstI, respectively) and primers having the sequences 5′-GGAAGCTTTGACAGCATAGACATTACA-3′ and 5′-CGGGATCCAAGGCAGTGCGAAGTACCT-3′ to amplify a 275-bp DNA sequence downstream (including the translational stop codon) of spaP (bp 4895 to 5170 of pac [20], a gene homologous to spaP and encoding Pac [P1] in S. mutans MT8148 [GenBank accession no. X14490]) (underlined bases indicate engineered restriction sites for HindIII and BamHI, respectively). PCRs were performed with a Biometra (Tampa, Fla.) UNO Thermoblock thermocycler, S. mutans NG5 chromosomal DNA as the template, and Taq polymerase (Promega, Madison, Wis.) for 30 cycles under the following conditions: denaturation at 96°C for 30 s, annealing at 56°C for 1 min, and extension at 72°C for 2 min. A final extension cycle was carried out for 5 min at 72°C.
DNA preparations and sequencing.
Plasmid DNA from E. coli recombinant strains and chromosomal DNA from S. mutans were prepared as described previously (7). All vector constructions were verified by restriction analysis and/or DNA sequencing, which was performed at the DNA Sequencing Core Laboratory of the University of Florida Interdisciplinary Center for Biotechnology Research. Primers were prepared at the DNA Synthesis Core Laboratory of the Interdisciplinary Center for Biotechnology Research.
Construction of the spaP mutant.
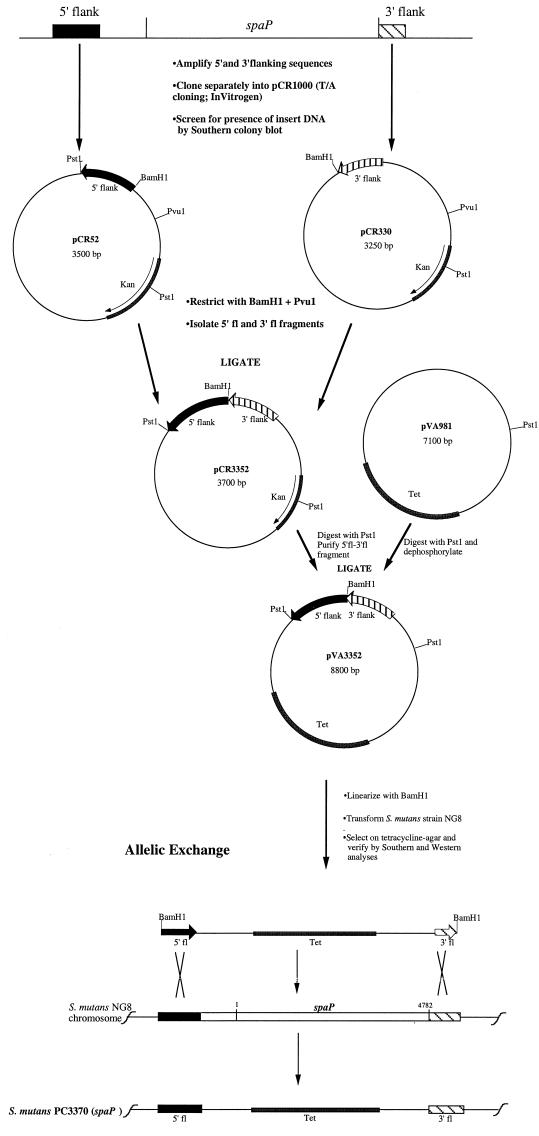
spaP mutant strain PC3370 was constructed as outlined in Fig. 1. PCR-amplified streptococcal DNA fragments (described above) into which appropriate restriction enzyme recognition sequences had been engineered for cloning were ligated separately into the cloning vector pCR1000 to yield pCR52 and pCR330. Each cloned fragment was excised from the host vector so that, upon ligation with each other, the host plasmid was reconstructed and the two amplified DNA fragments were oriented in tandem to produce pCR3352. E. coli INVαF′ was used to propagate each plasmid construct. The PCR-amplified products were excised from the constructs as single fragments by use of PstI and ligated into the PstI site of pVA981 (25). The resulting recombinant plasmid, pVA3352, was propagated in E. coli JM109, restricted with BamHI to linearize it at a unique site engineered to lie between the amplified sequences, and used to naturally transform (21) S. mutans NG8. The spaP mutants of NG8 generated via allelic exchange were selected on THYE agar containing 10% sucrose and 15 μg of tetracycline per ml. Mutant PC3370 was selected for further evaluation as described below.
FIG. 1.
Schematic representation of the construction of spaP mutant strain PC3370. fl, flanking.
Analysis of the spaP mutant.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), Western immunoblotting, radioimmunoassay (RIA), adherence to agglutinin-coated hydroxyapatite beads, and aggregation analyses of parent and mutant strains were all performed as described previously (4, 5). The API 20 Strep (bioMérieux Vitek, Inc., Hazelwood, Mo.) test was performed in accordance with the manufacturer’s instructions. Antibodies used for Western and RIA analyses were mouse anti-P1 monoclonal antibodies 4-10A8c and 1-6F6b, respectively (1). Southern analysis was performed on chromosomal DNA from parent and mutant strains blotted onto nylon membranes by use of the Photogene nonradioactive detection system (Gibco BRL, Bethesda, Md.) in accordance with the manufacturer’s instructions. Blots were probed with biotinylated pVA981 and a 763-bp internal fragment from the 3′ end of spaP (bp 4019 to 4782 of the spaP gene sequence [10]).
Infection and analysis of germ-free rats.
For caries assessments, groups of five 19-day-old weanling germ-free Fischer rats [CDF(344) GN/Crl BR] fed a low (5%)-sucrose diet (Diet 305 [18]) were inoculated orally for 3 consecutive days with saturated swabs of parent strain NG8 or spaP mutant strain PC3370 diluted to 3 × 108 CFU/ml or a mixture of both strains (1 × 108 CFU/ml each). After 6 weeks, whole mandibles were removed from each rat, sonicated as described above for bacterial enumeration, and stored in 95% ethanol for 24 h. Then, excess tissue was manually cleaned from around the teeth, which were stained overnight in 0.4% murexide for caries scoring (11).
To determine the time course of colonization, we performed an experiment in which three groups of 20 animals fed Diet 305 were infected with strain NG8, strain PC3370, or both. At 1-week intervals for 5 weeks following inoculation, three animals from each group were sacrificed, and S. mutans was enumerated from mandibular molars extracted with a sterile rongeur as follows. Left and right molars were placed in separate tubes on ice and containing 3 ml of 67 mM sodium phosphate buffer (pH 7.2) and sonicated (Branson Instruments Co., Plainview, N.Y.) for 10 s on power setting 4. One hundred-microliter aliquots of 10−4, 10−5, and 10−6 dilutions of each sonicated sample were plated in duplicate on Todd-Hewitt agar. In the mixed-infection experiment, 100 colonies derived from these platings were patched separately onto THYE agar with and without added tetracycline to determine the ratio of NG8- to PC3370-infected cells present in each sample. At 6 weeks postinfection, caries lesions were scored in the remaining five animals from each group.
Descriptive statistics were used to summarize the data. A mixed-model approach was used to assess differences in caries scores and microbiological counts between strains (15). Tukey’s method was used to adjust for multiple comparisons, and a square root transformation was used on the count data. A two-way analysis of variance was used to analyze the time course experiment.
RESULTS AND DISCUSSION
Characterization of the spaP mutant.
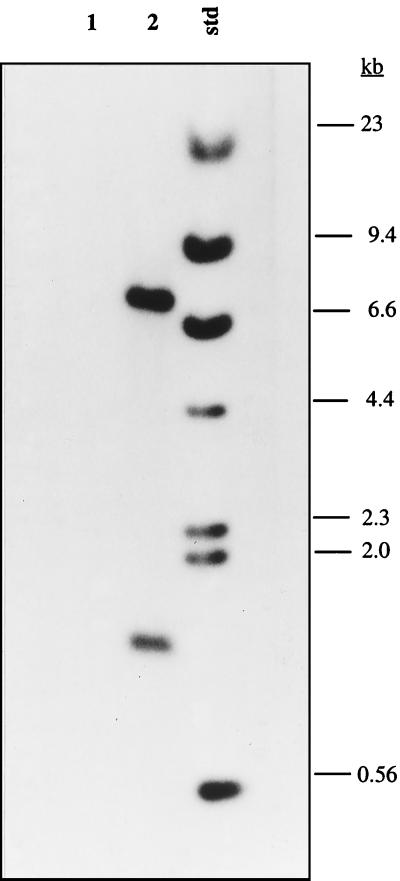
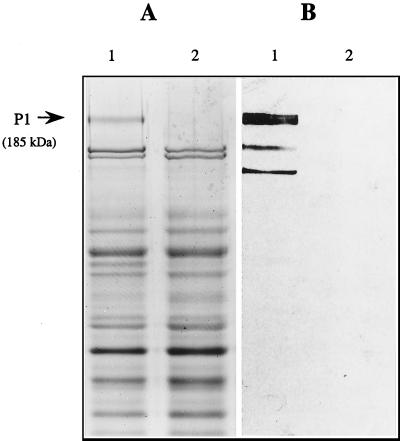
Transformation of parent strain S. mutans NG8 with 1 μg of BamHI-linearized pVA3352 yielded 65 Tetr colonies. All transformants were initially screened for the lack of surface expression of P1 by an RIA with an anti-P1 monoclonal antibody. All but one transformant showed no reactivity with the antibody (data not shown). One P1-negative transformant, PC3370, was chosen for further analysis. The generation time for parent and mutant strains in THYE broth was 80 min. Biochemical and fermentation profiles for parent and mutant strains were identical, as determined with the API 20 Strep test (data not shown). Southern analysis of BamHI-restricted chromosomal DNA from mutant strain PC3370 (Fig. 2, lane 1) showed no hybridization with a labeled internal fragment of spaP, while the parent strain hybridized with the two predicted fragments (Fig. 2, lane 2). SDS-PAGE (Fig. 3A) analysis of SDS extracts of mutant and parent cells demonstrated nearly identical protein profiles, except that bands corresponding to P1 were not present in the mutant. Western immunoblotting with an anti-P1 monoclonal antibody confirmed the loss of P1-reactive polypeptides in the mutant (Fig. 3B). Additionally, there was virtually no in vitro adherence (3.2% ± 2.1%) of radiolabeled mutant PC3370 cells to agglutinin-coated hydroxyapatite, whereas 55.8% ± 2.0% of NG8 cells adhered. Taken together, these data provide ample evidence that spaP was deleted from the genome of mutant PC3370, that P1 expression was eliminated, and that in vitro adherence properties were dramatically affected.
FIG. 2.
Southern analysis of BamHI-restricted chromosomal DNA from spaP mutant strain PC3370 (lane 1) and parent strain NG8 (lane 2). The membrane was probed with a biotinylated internal fragment of spaP (see Materials and Methods). Lane std contains biotinylated HindIII-digested lambda DNA markers.
FIG. 3.
SDS-PAGE and Western immunoblot analysis of whole cells of parent strain NG8 (lanes 1) and spaP mutant strain PC3370 (lanes 2) boiled in nonreducing SDS sample buffer. After electrophoresis, proteins were stained with Coomassie brilliant blue R-250 (A) or transferred to nitrocellulose membranes and reacted with a 1:1,000 dilution of anti-P1 monoclonal antibody 4-10A8c (B).
Virulence in gnotobiotic rats.
Caries scores from four separate experiments in which rats fed the low (5%)-sucrose diet were infected with either parent strain NG8 or spaP mutant strain PC3370 or coinfected with both strains are compiled in Table 1. The data clearly indicate a role for the spaP gene product in the virulence of S. mutans. There were significantly fewer enamel and dentinal lesions on all molar surfaces in rats infected with mutant PC3370 than in those infected with NG8 (P ≤ 0.009), except for both moderate and deep proximal dentinal lesions, for which there were either no or too few lesions to compare. The number of streptococci recovered from week-6 plaque samples was not significantly different (P = 0.3471) in animals infected with NG8 (26.9 × 106 ± 8.8 × 106 CFU/ml) and those infected with PC3370 (21.4 × 106 ± 6.8 × 106 CFU/ml), demonstrating that the mutant strain clearly was able to colonize the oral cavity of the rats to the same extent as the parent strain. Therefore, P1 may function as an adhesin but is not a necessary requirement for adherence leading to colonization in this model. Other mechanisms that contribute to colonization, such as glucan-mediated adherence, must be considered here, since sucrose was provided in the rat diet, albeit at a low concentration. In this regard, it has been shown that as little as 0.1% dietary sucrose is a sufficient substrate for the accumulation of S. mutans plaque in rats infected with a single strain (19). Therefore, it is possible that the reduction in caries scores seen in mutant-infected animals might have been greater in the absence of sucrose, since glucan might have contributed to the adherence of these organisms and therefore likely obscured the actual ability of the mutant to colonize the oral cavities of the rats in a sucrose-independent manner. Unfortunately, some sucrose in the diet is necessary for colonization by wild-type S. mutans to occur in this particular animal model.
TABLE 1.
Caries scores for S. mutans NG8 and PC3370 in an experimental gnotobiotic rat model
| Infecting strain | No. of rats | Rat wt (g) | Mean
caries scorea
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buccal
|
Sulcal
|
Proximal
|
||||||||||||
| E | Ds | Dm | Dx | E | Ds | Dm | Dx | E | Ds | Dm | Dx | |||
| NG8 (parent) | 20 | 159 ± 25 | 17.7 ± 0.49 | 15.0 ± 0.67 | 12.1 ± 0.57 | 7.84 ± 0.55 | 22.7 ± 0.53 | 19.7 ± 0.64 | 12.4 ± 0.49 | 6.73 ± 0.56 | 5.68 ± 0.54 | 1.63 ± 0.44 | 0.10 ± 0.07 | 0.0 |
| PC3370 (spaP mutant) | 20 | 158 ± 33 | 14.0b ± 0.59 | 12.0b ± 0.67 | 8.09b ± 0.43 | 5.09b ± 0.47 | 19.8b ± 0.52 | 17.0b ± 0.55 | 10.1c ± 0.66 | 4.33d ± 0.59 | 3.33e ± 0.57 | 0.28f ± 0.16 | 0.00 | 0.0 |
| NG8 + PC3370 (mixed) | 5 | 144 ± 12 | 16.0 ± 0.71 | 14.2 ± 0.86 | 11.0 ± 1.10 | 7.40 ± 1.08 | 20.4 ± 0.75 | 17.4 ± 0.51 | 12.8 ± 1.18 | 6.80 ± 1.02 | 5.60 ± 0.75 | 2.80g ± 0.49 | 0.40 ± 0.40 | 0.0 |
Values are means ± standard errors of the means. Mean caries scores are the extent of caries activity on molar surfaces involving enamel (E), slight dentinal (Ds), moderate dentinal (Dm), and extensive dentinal (Dx) lesions.
Significantly different from value for NG8 (P = 0.0001).
Significantly different from value for NG8 (P = 0.0094).
Significantly different from value for NG8 (P = 0.0015).
Significantly different from value for NG8 (P = 0.0004).
Significantly different from value for NG8 (P = 0.0018).
Significantly different from value for PC3370 (P = 0.0018).
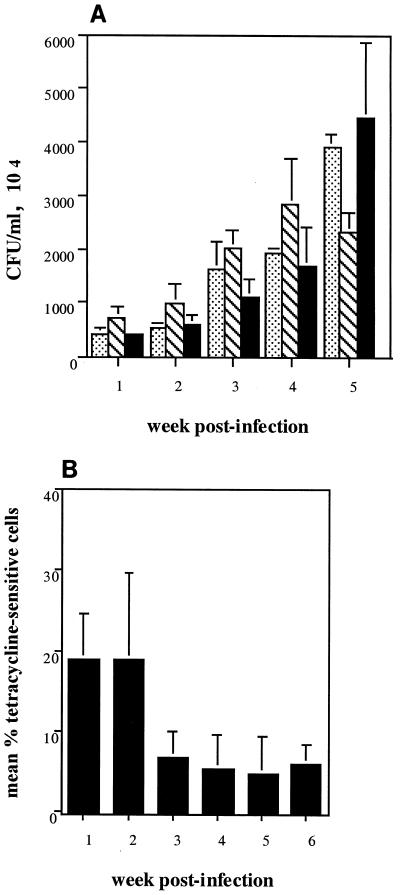
We speculated that the reduction in caries scores seen in mutant-infected animals might be explained by a slower colonization rate due to the lack of the major surface adhesin. Figure 4A shows the results of one experiment in which groups of 20 rats were infected with either the parent or the mutant strain to determine if there were differences in the recoverable numbers of parent versus mutant organisms at 1-week intervals through the course of the experiment. As determined by a two-way analysis of variance, the average number of organisms did not vary significantly by strain (P = 0.2466); however, differences in means were detected over time (P = 0.0001), and a strain-time interaction was significant (P = 0.0093). In this type of analysis, a significant strain-time interaction indicates that the relationship between strain means is not uniform over time. Therefore, while the overall trend appears to indicate that mutant strain PC3370 colonized the oral cavities of the rats in the same time frame as parent strain NG8, this result is inconclusive, perhaps due to our small sample size.
FIG. 4.
Time course of colonization. (A) Groups of 20 rats were infected with parent strain NG8 ( ) or mutant strain PC3370 (▧) or coinfected with both strains (■), and the number of cells recovered from mandibles of three animals at each time point after infection was enumerated. (B) The percentage of tetracycline-sensitive cells (NG8) recovered from mandibles of coinfected animals was determined as described in Materials and Methods. Error bars indicate standard errors of the means.
The total numbers of organisms recovered at all times from animals coinfected with a mixture of both the parent and the mutant strains were not significantly different (P = 0.3471) from those obtained from animals infected with either parent strain NG8 or mutant strain PC3370 (Fig. 4A). Interestingly, when the percentages of recoverable CFU of NG8 and PC3370 from coinfected animals were compared to determine if the expression of P1 conferred a competitive advantage, the percentage of cultivable NG8 cells (Tets) was only 19.3% ± 5.5% of the total population after 1 week and steadily decreased to 6.0% ± 2.3% at week 6 (Fig. 4B). It is unclear whether the difference in the numbers of recovered bacteria represents a difference in the efficiency of colonization between the two strains or instead reflects a trend toward less efficient recovery of parent strain NG8 from plaque over longer time periods in coinfected animals. At week 6, total bacteria were enumerated after sonication of whole rat mandibles (as opposed to individual teeth, which were examined during weeks 1 to 5 of the experiment). This difference in technique presumably accounts for the artificially lower level of organisms (ca. 2 × 106 CFU/ml for all groups) recovered at week 6 in this experiment.
Efficiencies of plating of parent and mutant strains on nonselective media were nearly identical; mean CFU per milliliter from overnight cultures were 2.62 × 109 for NG8 and 2.96 × 109 for PC3370. Caries scores for the coinfected animals (Table 1) were mostly intermediate between those for PC3370-infected animals and those for NG8-infected animals but did not differ significantly from those for animals infected with a single strain (Table 1), except in the number of slight proximal dentinal lesions. For the latter, there was a significant difference in comparison with mutant-infected animals (P = 0.0018).
Data presented in this report strongly suggest that the lack of P1 reduces the virulence of S. mutans in terms of carious lesion formation; however, our data also suggest that P1 is not a strict requirement for the successful colonization of rodent dental surfaces, since the P1-negative mutant was capable of colonizing in the absence or presence of wild-type cells. Recent findings by Love et al. (17), who used some of our S. mutans strains, may provide a possible explanation for the apparent disparity in the role of P1 in cariogenicity versus colonization. These authors reported an affinity for type 1 collagen by antigen I/II polypeptides from S. gordonii (SspA and SspB) and S. mutans (P1). Wild-type strains of each species, including strain NG8, invaded dentinal tubules, while mutant strains of each species, including S. mutans 834 (spaP), demonstrated markedly reduced invasive abilities. For example, invasion by an S. gordonii mutant (sspA sspB) was reduced by 66%, and that by S. mutans 834 was reduced by 79%. When the S. gordonii mutant was complemented with spaP and antigen I/II (P1) was expressed on the cell surface, normal invasive abilities were restored.
While the data presented here do not demonstrate a necessary role for P1 in colonization of gnotobiotic rats, the involvement of P1 in the cariogenicity of S. mutans is significant and reproducible. Furthermore, the data presented by Love et al. (17) clearly establish antigen I/II polypeptides as dental invasins. Studies of dentinal tubule invasion with the improved spaP mutant PC3370 described in this report should further clarify the function of surface antigen P1 of S. mutans in virulence.
ACKNOWLEDGMENTS
We thank Cecily C. Harmon for expertise with the gnotobiotic rat model and Charlotte J. Hammond for help with the rat microbiologic analysis. Statistical analyses were provided by Sue McGorray of the Biostatistics Department at the University of Florida. We also thank Jeffrey D. Hillman for helpful discussions.
This study was supported by NIDR grants DE08007 (to A.S.B.) and DE09081 and DE08182 (to S.M.M.).
REFERENCES
- 1.Ayakawa G Y, Boushell L W, Crowley P J, Erdos G W, McArthur W P, Bleiweis A S. Isolation and characterization of monoclonal antibodies specific for antigen P1, a major surface protein of mutans streptococci. Infect Immun. 1987;55:2759–2767. doi: 10.1128/iai.55.11.2759-2767.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowen W H, Schilling K M, Giertsen E, Pearson S, Lee S F, Bleiweis A S, Beeman D. Role of a cell surface-associated protein in adherence and dental caries. Infect Immun. 1991;59:4606–4609. doi: 10.1128/iai.59.12.4606-4609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady L J, Crowley P J, Ma J K-C, Kelly C, Lee S F, Lehner T, Bleiweis A S. Restriction fragment length polymorphisms and sequence variation within the spaP gene of Streptococcus mutansserotype c. Infect Immun. 1991;59:1803–1810. doi: 10.1128/iai.59.5.1803-1810.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brady L J, Piacentini D A, Crowley P J, Bleiweis A S. Identification of monoclonal antibody-binding domains within antigen P1 of Streptococcus mutansand cross-reactivity with related surface antigens of oral streptococci. Infect Immun. 1991;59:4425–4435. doi: 10.1128/iai.59.12.4425-4435.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brady L J, Piacentini D A, Crowley P J, Oyston P C F, Bleiweis A S. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect Immun. 1992;60:1008–1017. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crowley P J, Brady L J, Piacentini D A, Bleiweis A S. Identification of a salivary agglutinin-binding domain within cell surface adhesin P1 of Streptococcus mutans. Infect Immun. 1993;61:1547–1552. doi: 10.1128/iai.61.4.1547-1552.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutierrez J A, Crowley P J, Brown D P, Hillman J D, Youngman P, Bleiweis A S. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenkinson H F, Demuth D R. Structure, function and immunogenicity of streptococcal antigen I/II polypeptides. Mol Microbiol. 1997;23:183–190. doi: 10.1046/j.1365-2958.1997.2021577.x. [DOI] [PubMed] [Google Scholar]
- 9.Jenkinson H F, Lamont R J. Streptococcal adhesion and colonization. Crit Rev Oral Biol Med. 1997;8:175–200. doi: 10.1177/10454411970080020601. [DOI] [PubMed] [Google Scholar]
- 10.Kelly C P, Evans P, Bermeier L, Lee S F, Progulske-Fox A, Harris A C, Aitken A, Bleiweis A S, Lehner T. Sequence analysis of the cloned streptococcal surface antigen I/II. FEBS Lett. 1989;258:127–132. doi: 10.1016/0014-5793(89)81632-1. [DOI] [PubMed] [Google Scholar]
- 11.Keyes P H. Dental caries in the molar teeth of rats. II. A method for diagnosing and scoring several types of lesions simultaneously. J Dent Res. 1958;37:1088–1099. doi: 10.1177/00220345580370060901. [DOI] [PubMed] [Google Scholar]
- 12.Koga T, Okahashi N, Takahashi I, Kanamoto T, Asakawa H, Iwaki M. Surface hydrophobicity, adherence, and aggregation of cell surface protein antigen mutants of Streptococcus mutansserotype c. Infect Immun. 1990;58:289–296. doi: 10.1128/iai.58.2.289-296.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S F, Progulske-Fox A, Bleiweis A S. Molecular cloning and expression of a Streptococcus mutans major surface protein antigen, P1 (I/II), in Escherichia coli. Infect Immun. 1988;56:2114–2119. doi: 10.1128/iai.56.8.2114-2119.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee S F, Progulske-Fox A, Erdos G W, Piacentini D A, Ayakawa G Y, Crowley P J, Bleiweis A S. Construction and characterization of isogenic mutants of Streptococcus mutansdeficient in major surface protein antigen P1 (I/II) Infect Immun. 1989;57:3306–3313. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Littell R C, Milliken G A, Stroup W, Wolfinger R D. SAS system for mixed models. Cary, N.C: SAS Institute Inc.; 1996. A setting for mixed models applications: randomized blocks designs; pp. 1–29. [Google Scholar]
- 16.Loesche W. Role of Streptococcus mutansin human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love R M, McMillan M D, Jenkinson H F. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect Immun. 1997;65:5157–5164. doi: 10.1128/iai.65.12.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michalek S M, McGhee J R, Navia J M. Virulence of Streptococcus mutans. A sensitive method for evaluating cariogenicity in young gnotobiotic rats. Infect Immun. 1975;12:69–75. doi: 10.1128/iai.12.1.69-75.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michalek S M, McGhee J R, Shiota T S, Devenyns D. Low sucrose levels promote extensive Streptococcus mutans-induced dental caries. Infect Immun. 1977;16:712–714. doi: 10.1128/iai.16.2.712-714.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okahashi N, Sasakawa C, Yoshikawa M, Hamada S, Koga T. Cloning of a surface protein antigen gene from serotype c Streptococcus mutans. Mol Microbiol. 1989;3:221–228. doi: 10.1111/j.1365-2958.1989.tb01811.x. [DOI] [PubMed] [Google Scholar]
- 21.Perry D, Kuramitsu H K. Genetic transformation of Streptococcus mutans. Infect Immun. 1981;32:1295–1297. doi: 10.1128/iai.32.3.1295-1297.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell M W, Lehner T. Characterization of antigens extracted from cells and culture fluids of Streptococcus mutans serotype c. Arch Oral Biol. 1978;23:7–15. doi: 10.1016/0003-9969(78)90047-x. [DOI] [PubMed] [Google Scholar]
- 23.Schilling K M, Bowen W H. Glucans synthesized in situ in experimental salivary pellicle function as specific binding sites for Streptococcus mutans. Infect Immun. 1992;60:284–295. doi: 10.1128/iai.60.1.284-295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanzer J M. Virulence of mutants defective in glucosyltransferase dextran-mediated aggregation or dextranase activity. In: Mergenhagen S A, Rosan B, editors. Molecular basis of oral microbial adhesion. Washington, D.C: American Society for Microbiology; 1985. pp. 204–211. [Google Scholar]
- 25.Tobian J A, Cline M L, Macrina F. Characterization and expression of a cloned tetracycline resistance determinant from the chromosome of Streptococcus mutans. J Bacteriol. 1984;160:556–563. doi: 10.1128/jb.160.2.556-563.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]