Abstract
Little is known about how variables, such as carcass mass, affect the succession pattern of microbes in soils during decomposition. To investigate the effects of carcass mass on the soil microbial community, soils associated with swine (Sus scrofa domesticus) carcasses of four different masses were sampled until the fifteenth day of decomposition during the month of June in a pasture near Lincoln, Nebraska. Soils underneath swine of 1 kg, 20 kg, 40 kg, and 50 kg masses were investigated in triplicate, as well as control sites not associated with a carcass. Soil microbial communities were characterized by sequencing the archaeal, bacterial (16S), and eukaryotic (18S) rRNA genes in soil samples. We conclude that time of decomposition was a significant influence on the microbial community, but carcass mass was not. The gravesoil associated with 1 kg mass carcasses differs most compared to the gravesoil associated with other carcass masses. We also identify the fifteen most abundant bacterial and eukaryotic taxa, and discuss changes in their abundance as carcass decomposition progressed. Finally, we show significant decreases in alpha diversity for carcasses of differing mass in pre-carcass rupture (days 0,1,2,4,5, and 6 postmortem) versus post-carcass rupture (days 9 and 15 postmortem) microbial communities.
Keywords: postmortem microbiology, decomposition, bacteria, nematode, pathology
Postmortem microbial communities are crucial and dynamic contributors to corpse decomposition. The activity of these decomposer microorganisms drives many postmortem changes, such as bloating [1] and ethanol production [2]. The structure of these microbial communities changes as a corpse decomposes because available nutrients are consumed [3]. Postmortem microbial communities have received much interest lately because they change in a predictable way, particularly the microorganisms on the skin [3,4] and in carcass-associated soils [3]. These developmental shifts are analogous to those associated with insects [5] and have great potential to be developed as a means to estimate postmortem interval [3,4]. We are particularly interested in utilizing soil microbial communities associated with decomposition, also known as gravesoils, because they host a clock-like succession of microbes [3] and are easily accessible at crime-scenes in outdoor scenarios.
The development of soil microorganisms as physical evidence requires us to answer several fundamental questions about the relationships between corpses, decomposition, and soil microbial communities. It is known that microbial activity in gravesoils increases rapidly and significantly during the early stages of decomposition [6,7] and that this activity is influenced by several variables including soil texture, temperature, moisture, vegetation, and pH [8,7,9–11]. Microbial gravesoil activity is primarily driven by bacteria during the early stages of decomposition [3,12], followed by increased activity of eukaryotes such as nematodes [3] and fungi [13,14] during later stages of decomposition. Yet one variable has received little experimental attention: the mass of the corpse.
Corpse mass is an important variable to understand because it can affect the rate of decomposition. However, this relationship is still under investigation. Many studies have utilized swine carcasses since the decomposition rate and arthropod colonization in Sus scrofa domesticus corpses mimics that in humans [15–17]. One of the first studies, which was not replicated, reported that larger mass corpses decay faster than smaller mass corpses [18]. Most of the later studies concluded that smaller corpse masses decay faster [19–23]. The exact functional nature of the decay rate in these studies however is not fully agreed upon [20–23]. Also, the effect of corpse mass and decay rate on the host-associated invertebrate and microbial community is not well understood. Hewadikaram and Goff found that corpse mass did not affect arthropod taxa composition or its succession over time [18]. Simmons et al. [20] concluded that corpses of different masses only decayed at different rates if insects were present. Finally, only a few small studies with tiny carcass masses have investigated the relationship between carrion carcass mass and insect types [24,25].
To our knowledge, no studies have yet investigated the effect of carcass mass on the associated microbial communities. In this paper, we focus on the samples of Spicka et al. [21], which is a statistically well designed study of swine decomposition during a Nebraska summer using four different mass carcasses in triplicate. Spicka et al. [21] observed that larger swine carcasses (20 kg – 50 kg) released a greater concentration of ninhydrin-reactive nitrogen into gravesoil than neonatal (1 kg) carcasses. An additional mass effect was observed where the largest carcasses (40 kg – 50 kg) released a significant amount of total nitrogen more rapidly then 20 kg carcasses. This release of nutrients, along with the recent observation that soil microorganisms contribute directly to the breakdown of carcass materials [12], leads to our hypothesis that carcass mass will influence the structure of associated soil microbial communities.
To investigate the effect of carcass mass on the structure of postmortem microbial communities in gravesoil, we sequenced the archaeal, bacterial, and eukaryotic microbial communities of soils collected by Spicka et al. [21]. We used the universal and taxonomically-informative 16S rRNA gene and 18S rRNA gene to analyze the structure of the microbial communities associated with the control soil and with carcasses of mass 1 kg, 20 kg, 40 kg, and 50 kg.
Materials and Methods
Carcasses and Decomposition Site
Swine (Sus scrofa domesticus) carcasses of different masses (~1 kg, 20 kg, 40 kg, and 50 kg) were killed by blunt force trauma to the skull with a bolt gun, and placed on a weighing frame (2.5 cm2 polypropylene mesh bound to a 85 cm × 40 cm PVC frame: Fig. 1) directly on the surface of a grassland soil near Mead, Nebraska, USA in the summer within 60 minutes of death [21]. The grassland soil was a deep, silty, clay loam with a texture of 15.1% sand, 53.6% silt, and 31.3% clay. The soil surface of the decomposition site was flat so that decomposition fluids released from a carcass would collect around the carcass, but was not influenced by slope. Coyotes (Canis latrans) and turkey vultures (Cathartes aura) were the primary scavengers in the area, however no scavenger activity was observed at this site for five years [21]. Insect activity was not restricted in the current experiment.
Fig 1.

The gross decomposition of swine (Sus scrofa domesticus) carcasses of contrasting mass (~1 kg, 20 kg, 40 kg, and 50 kg) on the soil surface of a pasture near Mead, Nebraska where postmortem interval was measured as days (d) and Accumulated Degree Days (ADD).
Soil Collection and Storage
Gravesoils and control soils (soils not associated with carcasses) were collected as described in Spicka et al. [21]. Gravesoil and control plots were at least 5 m apart. Soil samples were collected from underneath each carcass (0 cm - 5 cm depth) while it was lifted to measure mass loss. Soils were collected from an unsampled location each time using a 2.54 cm diameter KHS soil probe (M&M Supply Company, Clear Lake, Iowa, USA). Probe surfaces were cleaned with ethanol between each sample collection. There was no need to clear plant detritus from the soil surface before each sampling, as it was sparse. Soil samples were collected from the initial time of placement and at 24-hour intervals for 1, 2, 4–6, 9 and 15 days postmortem. Day 3 and day 8 were skipped due to severe thunderstorms. Three carcasses of each weight were placed at once, resulting in a total of 12 carcasses. Daily temperature ranged from 13.7 °C to 32.9 °C. Accumulated Degree Days (ADDs) were calculated as in Arnold [26] using a base temperature of 0 °C [27]. All soils were stored at −20 °C until DNA extraction.
Carcass Decomposition
The mass loss of carcasses followed a sigmoidal curve [21] typically associated with the breakdown of carrion [28]. Adult flies were observed on all carcasses within seconds of placement and larval masses were established on all replicates. Peak volume of larval mass was apparently a function of carcass mass; it took more time for larger carcasses to support peak maggot volume (Fig. 1). However, the majority of migration was completed by 9 days postmortem (144 ADD) in all replicates. These carcasses did not tend to undergo an abdominal rupture that is often observed with carrion. Rather, fly larvae feeding from the head toward the posterior end typically consumed the carcasses in the current study (Fig. 1).
Microbiome Analysis
DNA extraction, PCR amplification were conducted as described in Metcalf et al. [3] and following Earth Microbiome Project standard protocols (http://www.earthmicrobiome.org). Archaeal and bacterial 16S rRNA gene amplicons were sequenced using the Illumina HiSeq 2000 (100 basepair reads) and microbial eukaryotic 18S rRNA amplicons were sequenced using the Illumina MiSeq (150 basepair reads). Sequence processing and data analyses were conducted as described in Metcalf et al. [3], except that updated taxonomy databases were used, specifically Greengenes version 13_5 (http://greengenes.secondgenome.com, [29]) for open-reference OTU picking of 16S rRNA sequences, and SILVA version 111 [30] for closed-reference OTU picking of 18S rRNA sequences. Additionally, primer and adapters were removed from the end of the 18S read, resulting in read lengths of approximately 120 basepairs.
For 16S sequences, taxa that were not classified in the Domains Bacteria or Archaea were removed. For 18S sequences, we focused on the microbial community by filtering out taxa classified in groups Craniata, Chloroplastida, Mollusca, and Arthropoda. After these filtering steps, our 16S and 18S data sets included 9,953,274 sequence reads (mean 82,943 reads per sample) and 567,129 (mean 4,975 reads per sample), respectively. The average number of reads per sample was substantially lower for the 18S data set because of the lower depth of sequencing on the MiSeq platform and because some samples contained a high relative abundance of chloroplast, insect, and host DNA reads, which were filtered out. We rarified the 16S data set to 14,000 sequences per sample and the 18S data set to 430 sequences per sample, which allowed us to include most samples in analyses.
To confirm our rarefying results, and to maximize the statistical power of our data set, we also ran analyses using Cumulative Sum Scaling (CSS) as an alternate normalization technique to rarefying [31]. We only used weighted UniFrac [32] analysis on the CSS transformed data, as rarefying is a more appropriate technique for unweighted UniFrac [33]. Before analysis with CSS, we removed very low depth samples (below 940 and 850 sequences/sample for 16S and 18S datasets) and extreme outliers [34]. This is because low depth samples have a higher proportion of contaminants [35]. We also ensured that the CSS-transformed results did not display clustering based on sample sequencing depth.
Using the QIIME pipeline [36], we explored relative taxon abundances and patterns of community dissimilarity with phylogeny-based UniFrac unweighted and weighted distances. We report p-values and type I sequential sums-of-squares error (R2) for the strength and statistical significance of sample grouping based on unweighted and weighted UniFrac distances using a non-parametric analysis of variance (PERMANOVA) statistical test [37] with the adonis function in the ‘vegan’ [38] statistical package for R [39]. We also report Bonferroni-corrected p-values for the distance boxplots using the nonparametric two-sided Student’s t-test (999 permutations). Error bars are based on the standard deviation of the UniFrac distance distributions. We report differentially abundant taxa using the Kruskal-Wallis statistical test.
Results
The decomposition of a carcass had a significant effect on the structure of gravesoil microbial communities; all gravesoils were significantly (PERMANOVA p < 0.001) different compared to control soils by the end of the trial. However, we observed no sustained significant differences between soil microbial communities associated with carcasses of contrasting mass (Fig. 2). The 1 kg mass was the most different of the masses, although not quite significant (Fig. 2, Table 1). This non-significant finding was supported using an alternate normalization technique (Table 1Sa). Spicka et al. found significant differences in the amount of Ninhydrin-reactive Nitrogen (NRN) released by corpses of contrasting masses. Specifically, Spicka et al. found that the 1kg mass had a greater concentration of NRN per unit carcass (NRNc) compared to other masses, and the 20kg mass also briefly had greater NRNc compared to the 40kg and 50kg masses. When NRN differences were controlled for, carcass mass became even less significant (Table 2a). Although mass was not a significant factor in determining the 16S microbial community, time was (Table 1, Table 1Sa).
Fig 2.
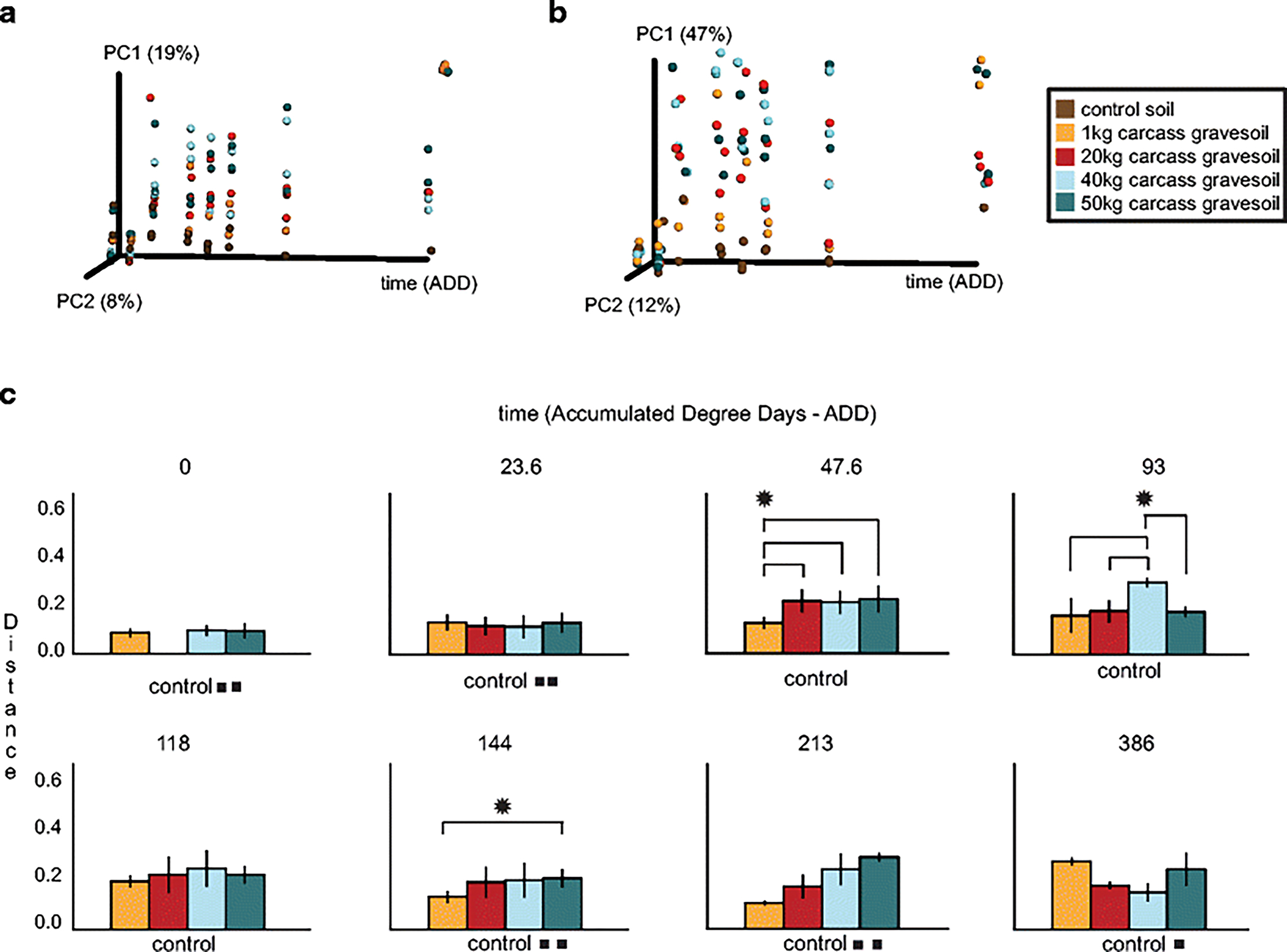
Ordination and bar plots to visualize differences between the structure of gravesoil microbial communities during the decomposition of swine (Sus scrofa domesticus) carcasses on the soil surface of a pasture near Mead, Nebraska, USA during the summer. (a) 16S rarefied unweighted UniFrac Principal Coordinates Analysis (PCoA), (b) 16S rarefied weighted UniFrac PCoA, and (c) 16S rarefied weighted UniFrac distance comparison bar plots for each mass compared to the control soil on each accumulated degree day (ADD). * indicates a significant nonparametric-t test difference with a Bonferroni-corrected p < 0.05. For example, at ADD 144, the weighted UniFrac distance from control soil to 50kg carcass gravesoil is significantly different compared to the distance from control soil to 1kg carcass gravesoil. We use the control soil as a baseline. In cases where less than three samples were in the analysis due to quality concerns, the number of squares (∎) indicates how many samples were analyzed. Results for weighted and unweighted UniFrac analyses were nearly identical.
Table 1.
16S rarefied unweighted UniFrac Type 1 sequential sums of squares PERMANOVA. The model y ∼ ADD_time + mass was fit to control for differences in the number of replicates at each time point before assessing the effect of carcass mass on gravesoil microbial communities. The FDR procedure is Bonferroni correction.
| time (ADD) | mass | |||
|---|---|---|---|---|
|
| ||||
| Comparison | R2 | FDR p | R2 | FDR p |
|
| ||||
| 1kg v. 20kg | 0.086 | 0.01 | 0.053 | 0.084 |
| 1kg v. 40kg | 0.1 | 0.006 | 0.042 | 0.28 |
| 1kg v. 50kg | 0.16 | 0.006 | 0.039 | 0.3 |
| 20k v. 40kg | 0.05 | 0.066 | 0.025 | 1 |
| 20 kg v. 50 kg | 0.064 | 0.006 | 0.033 | 0.38 |
| 40 kg v. 50 kg | 0.078 | 0.006 | 0.021 | 1 |
| control v. all masses | 0.052** | 0.034* | ||
p < 0.001
p<0.01
Table 2.
18S rarefied unweighted UniFrac Type 1 sequential sums of squares PERMANOVA. The model y ∼ ADD_time + mass was fit to control for differences in the number of replicates at each time point before assessing the effect of carcass mass on gravesoil microbial communities. The FDR procedure is Bonferroni correction.
| time (ADD) | mass | |||
|---|---|---|---|---|
|
| ||||
| Comparison | R2 | FDR p | R2 | FDR p |
|
| ||||
| 1kg v. 20 kg | 0.072 | 0.006 | 0.041 | 0.066 |
| 1kg v. 40 kg | 0.068 | 0.006 | 0.055 | 0.012 |
| 1kg v. 50 kg | 0.06 | 0.048 | 0.06 | 0.018 |
| 20k v. 40 kg | 0.095 | 0.006 | 0.025 | 1 |
| 20kg v. 50kg | 0.087 | 0.006 | 0.033 | 0.54 |
| 40kg v. 50kg | 0.099 | 0.006 | 0.029 | 1 |
| control v. all masses | 0.049** | 0.03** | ||
p < 0.001
Archaeal and bacterial groups changed significantly in relative abundance during decomposition. For example, bacterial family “Candidatus Chthoniobacteraceae” dominated all soils during the early stages of decomposition but the abundance of these bacteria decreased as carcasses decomposed (Fig. 3). The abundance of bacteria from taxa Gaiellaceae, Acidobacteria, and Rhodoplanes also decreased during decomposition (Bonferroni p < 0.01). This was also true for the archaeal taxa “Candidatus Nitrososphaera”. However, several bacterial taxa significantly increased during decomposition, including those from taxa Planococcaceae, Sporosarcina sp., Ignatzschineria sp., and Chitinophagaceae (Bonferroni p < 0.01).
Fig 3.
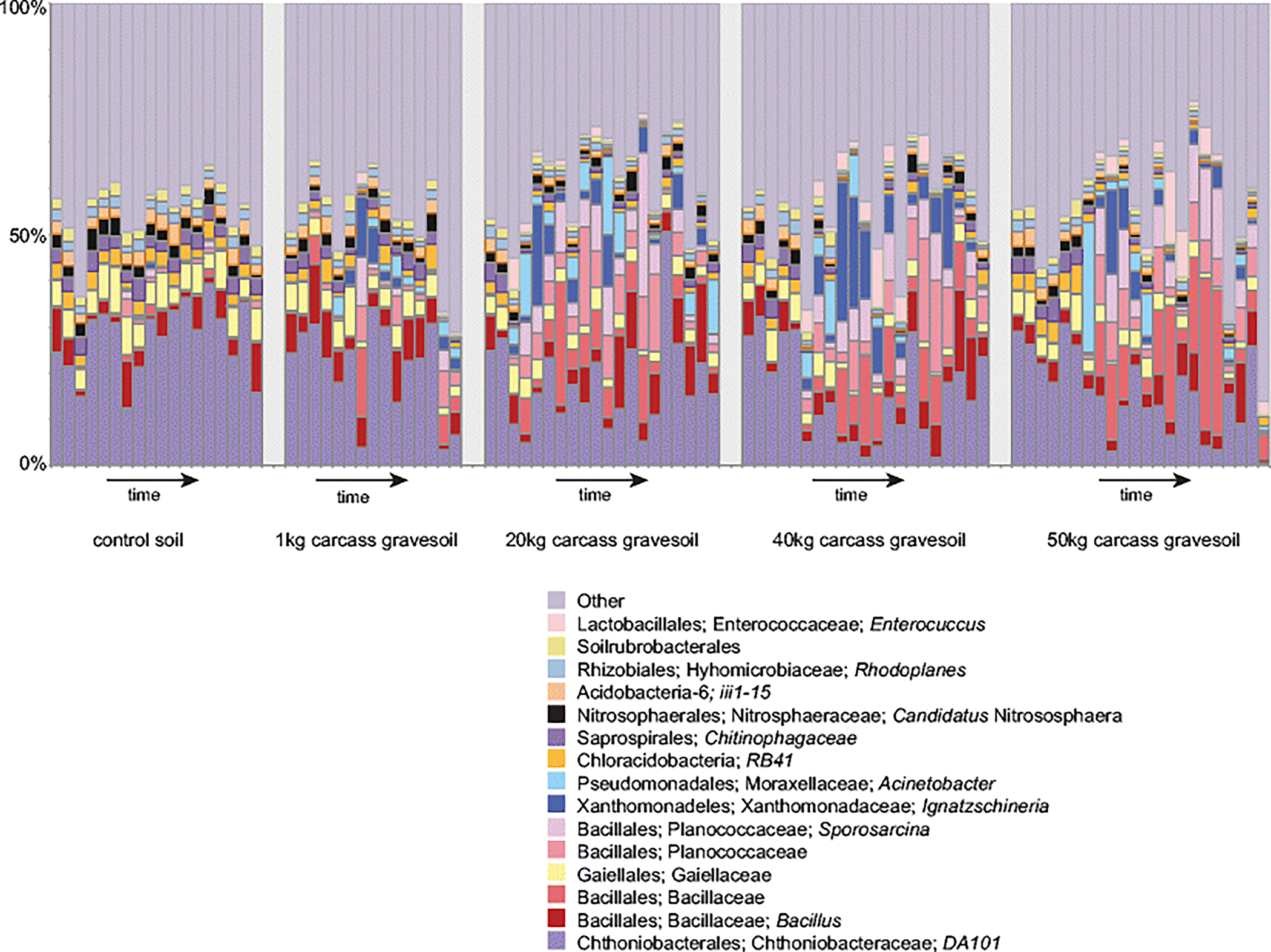
Sample relative abundance of control and gravesoil microbial (16S) communities during the decomposition of swine (Sus scrofa domesticus) carcasses on the soil surface of a pasture near Mead, Nebraska, USA during the summer. One bar represents each sample. Only the 15 highest relative abundance taxa are shown, starting at the order level. Genus “Candidatus Nitrososphaera”, is the only archaeal taxa of high abundance in the data set. Additional archaeal and bacterial taxa in each sample are combined into a single ‘other’ category. Archaea occupy approximately 0.06% of the ‘other’ category.
The dominant soil eukaryotes were fungi and nematodes (Fig. 4). As observed in the bacterial communities the structure of these decomposer communities shifted significantly with time compared to the control soils (Table 2, Table 1Sb.). Similar to archaeal and bacterial communities, the eukaryotic microbial communities associated with different carcass masses shifted similarly over time regardless of mass (Fig. 5, Table 2). Again the 1 kg mass gravesoil was the most different of the masses, and was significantly different compared to the 50 kg mass (Table 2, Table 1Sb). However, this difference disappeared when NRN was controlled for (Table 2Sb). The top fifteen relative abundance gravesoil eukaryote communities comprised a large fraction of the total sequences, with the greatest shifts observed as increases in the abundance of nematodes in the family Rhabditidae and slime mold Fonticula alba (Bonferroni p < 0.01).
Fig 4.
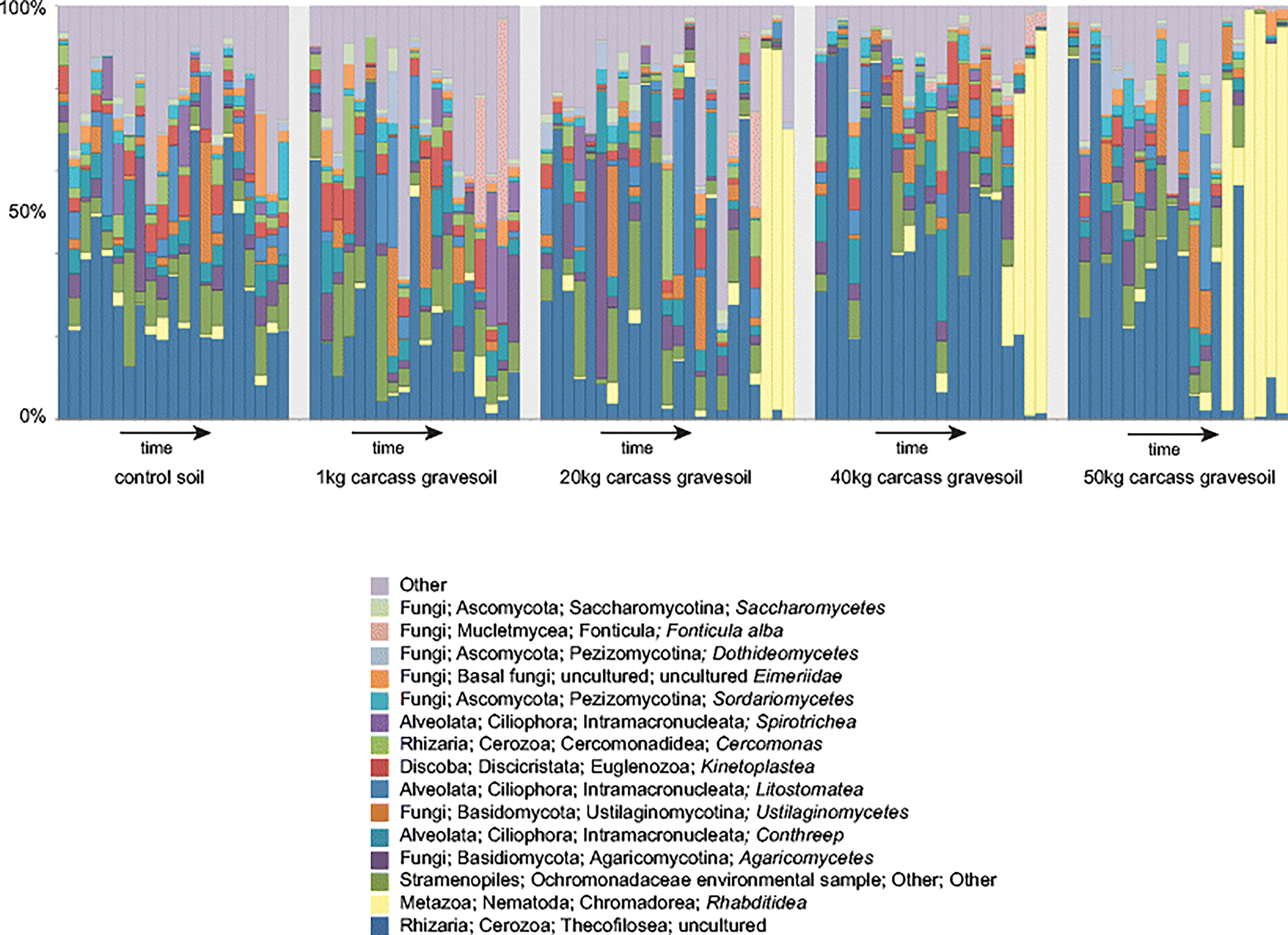
Relative abundance of control and gravesoil microbial (18S) communities during the decomposition of swine (Sus scrofa domesticus) carcasses. One bar represents each sample. Only the 15 highest relative abundance taxa are shown starting at the class level, additional taxa in each sample are combined into a single ‘other’ category. The apparent lack of nematode bloom in the 1 kg samples at the later time points is because the later time point samples were filtered out due to quality concerns.
Fig 5.
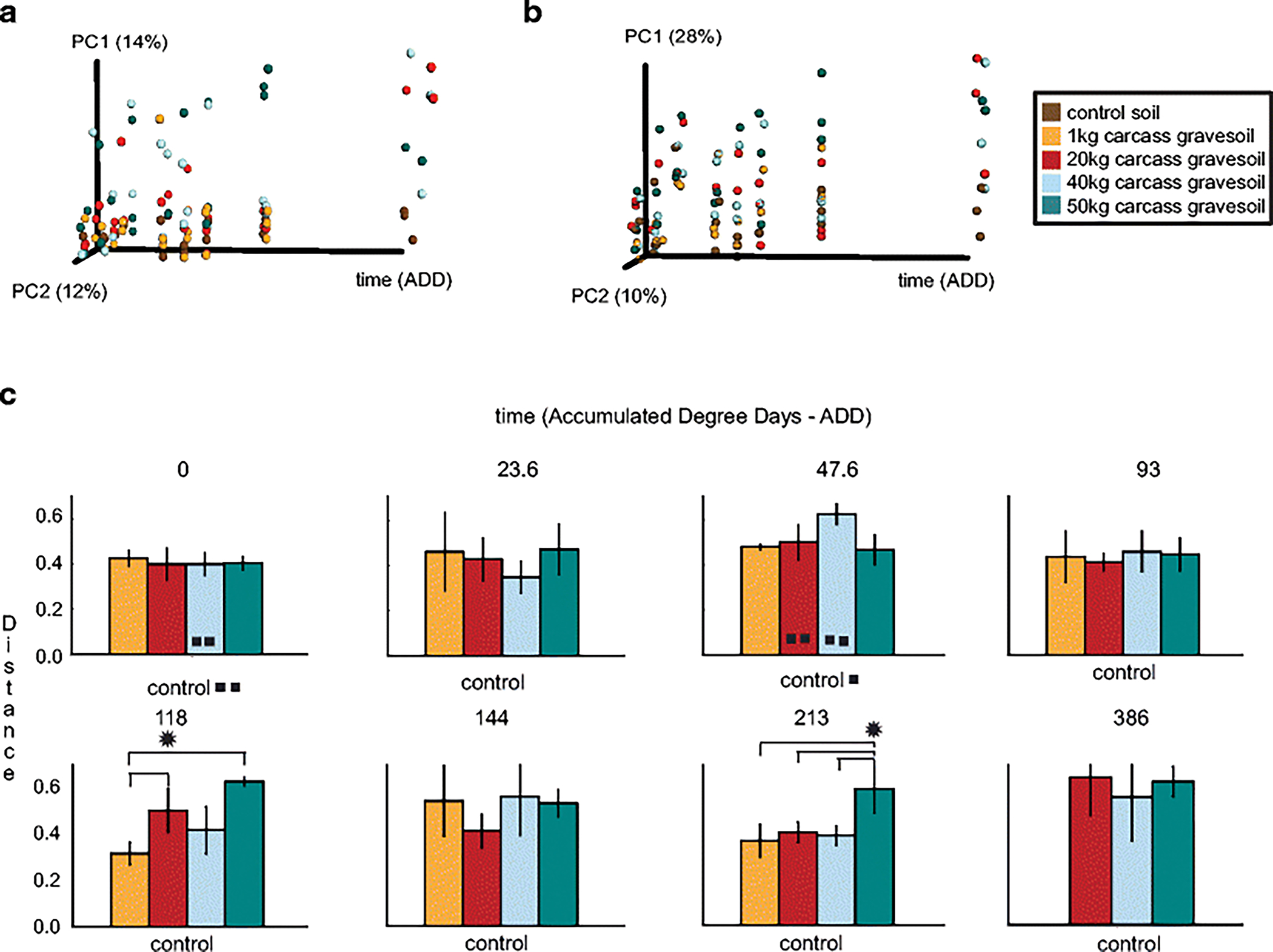
Ordination and bar plots to visualize differences between the structure of gravesoil microbial communities during the decomposition of swine (Sus scrofa domesticus) carcasses on the soil surface of a pasture near Mead, Nebraska, USA during the summer. (a) 18S rarefied unweighted UniFrac Principal Coordinates Analysis (PCoA), (b) 18S Cumulative Sum Scaling (CSS) weighted UniFrac PCoA, and (c) 18S CSS weighted UniFrac distance comparison bar plots for each mass compared to the control soil on each accumulated degree day (ADD). * indicates a significant nonparametric-t test difference with a Bonferroni-corrected p < 0.05. In cases where less than three samples were in the analysis due to quality concerns, the number of squares (∎) indicates how many samples were retained. Results for weighted and unweighted UniFrac analyses were nearly identical.
For both 16S microbes and 18S eukaryotes, we observed a significant decrease in the alpha diversity, or a measure of how many species are in each sample, of gravesoils during decomposition compared to control soils (Fig. 6, Fig. S1). Pre vs. post rupture differences in alpha diversity were present but harder to detect, especially when the masses were analyze separately (Fig. S1), possibly due to only three replicates per mass. For all analyses, resolution of more subtle effects would require more replicates for increased statistical power.
Fig 6.
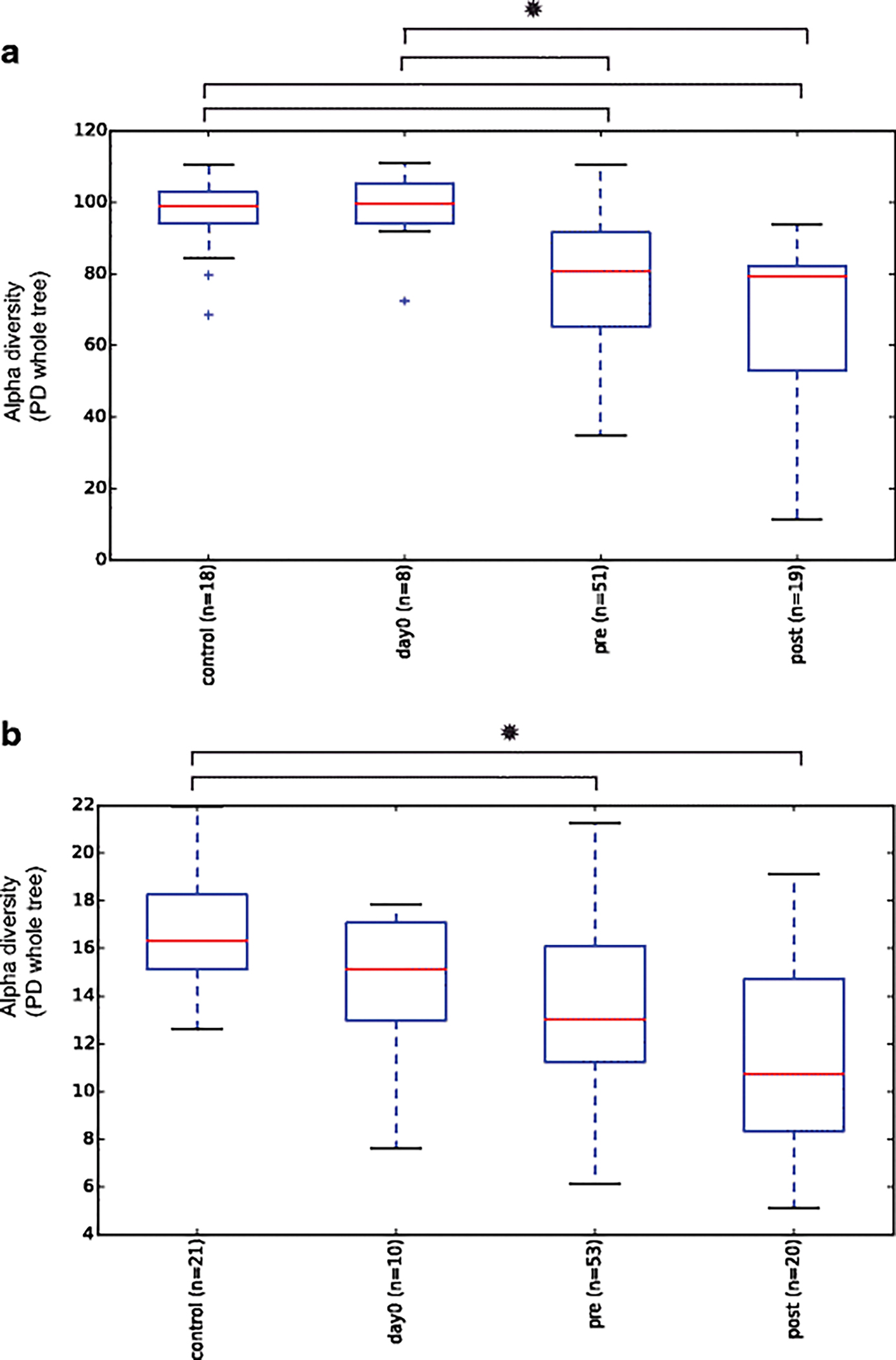
Phylogenetic distance (PD) alpha diversity boxplots. This includes control, day0, pre-carcass rupture, and post-carcass rupture samples for all gravesoil masses. * indicates significant differences between boxplots (p < 0.05). (a) 16S (b) 18S
Discussion
Our data show that soil microbial communities associated with carcasses greatly differ from control soils during decomposition, but are robust to carcass mass. The 1 kg carcasses were marginally statistically significant compared to other masses, particularly the 50 kg mass. However, all other carcass masses (20 kg, 40 kg, and 50 kg) did not display significant differences in their microbial communities throughout decomposition. This finding suggests that microbial clocks to estimate the postmortem interval may be robust to human cadaver mass, at least between 20 kg – 50 kg. When NRN was controlled for the 1kg mass moved far away from borderline statistically significant. While the pH of the soil increases during decomposition [40,3], we observed it to have a smaller effect compared to NRN, as assessed by PERMANOVA R2 (25% and 50% less on average in 16S and 18S data respectively).
The current findings are similar to those of other recent investigations into the postmortem microbiome. Other studies have identified the smallest mass carcasses as exhibiting the most variable decomposition patterns [21,22]. Our results also agree that the major variables influencing the structure of the microbial communities is the death of the host [41], and the time since death [3,4]. Postmortem microbial communities shift when a carcass is decomposing, probably due to rupture, increased resource availability, or the proliferation of insects. It has been show that the cadaver microbial community can influence insect activity and vice versa [42–45]. Added to this is likely an example of ‘resource selects community’ [46] where the different stages of decomposition offer different nutrients, e.g. pre and post-rupture. We observed a decrease in alpha diversity during decomposition, but the result was only strongly significant when all mass classes were combined (Fig. 5, Fig. S1). Metcalf and colleagues found a stronger decrease in alpha diversity of gravesoils during decomposition [3] possibly because their samples were collected for a longer time period and the sample size was larger providing better power.
A striking similarity between the current results and those reported by Metcalf et al. [3] was the increase in nematode abundance. The nematodes in this study were of the same family (Rhabditidae) as those reported by Metcalf et al [3]. This flush of nematodes is probably due to the increase in the abundance of the postmortem bacteria, their primary food source. Nematodes have long been used for environmental monitoring [47] and we find it very interesting that similar nematode taxa have been observed with decomposing mice [3] and swine in two different soil types. We recommend that the forensic value of nematodes be explored in more detail.
To expand on this research, we also recommend more detailed study into the dynamics between carcass mass, decomposition, and microbial communities with more replicates and over a longer time period to confirm the apparent lack of relationship between carcass mass and the time required for a shift in microbial community structure. Also, additional time points would allow for the use of regression models to estimate PMI. Similarly, we recommend that the decomposition of corpses greater than 50 kg should be investigated in detail to determine if additional trends can be identified, i.e. are corpses greater than 50 kg associated with different gravesoil microbial communities? Also, further investigation would be ideally done on human rather than swine corpses; however, it is difficult to find human donors, and donors are usually older than crime scene victims. While Sus scrofa is accepted as a model system most similar to humans because they have similar decay rates, body mass, and skin structure among other factors [48], some differences have been found between the two, for example four times as much stearic acid in swine fat compared to human [49].
The current data contribute to postmortem microbiology, a branch of forensic medicine designed to serve as a useful adjunct to autopsy [50]. The identification of postmortem microorganisms can be used to confirm the presence of a suspected antemortem infection, identify an infection when the cause of death is unknown, and assess the efficacy of antibiotics in treating an infection [51]. Recently, we demonstrated that corpses host a large and diverse microbial community at death [3,52,41,53,54]. The structure of this microbial community shifts significantly and predictably as a corpse decomposes, and can become less diverse as it decomposes into an increasingly specialized habitat [3,4,55]. These are exciting developments for forensic medicine because they likely foreshadow an expanded use of microorganisms as physical evidence. Indeed, we can envision the development of a postmortem microbiology to aid in establishing cause of death, associating people with objects and locations [56], and estimating postmortem interval [3,4]. These would be significant developments toward the development of a comprehensive forensic microbiology. The current data add to this fundamental understanding by showing that postmortem microbial communities can be similar regardless of initial carcass mass, which has the potential to simplify initial postmortem analysis. However, we caution that more replicates, time points, and mass types should be investigated; and this experiment was done with swine, therefore results could differ with human cadavers.
Supplementary Material
Fig 1S: Phylogenetic distance (PD) alpha diversity boxplots. This includes day 0, pre-carcass rupture (days 1, 2, 4, 5 and 6), and post-carcass rupture (days 9 and 15) gravesoils for carcasses of masses 1 kg, 20 kg, 40 kg, and 50 kg. For example, label pre_1, signifies alpha diversity of pre-rupture 1kg carcass gravesoils. Squares (∎) indicate siginificant differences from the control soils. * indicates possible (p < 0.08) weak significant difference between boxplots. Shannon diversity yielded similar results. (a) 16S (b) 18S.
Table 1S. CSS-normalized weighted UniFrac Type 1 sequential sums of squares PERMANOVA. The model y ~ ADD_time + mass was fit to control for differences in the number of replicates at each time point before assessing the effect of carcass mass on gravesoil microbial communities. The FDR procedure is Bonferroni correction. (a) 16S (b) 18S
Table 2S. Rarefied unweighted UniFrac Type 1 sequential sums of squares PERMANOVA. The model y ~ ADD_time + NRN + mass was fit to control for differences in the number of replicates at each time point, and the amount of released NRN, before assessing the effect of carcass mass on gravesoil microbial communities. The FDR procedure is Bonferroni correction. (a) 16S (b) 18S
Acknowledgments
This research was funded by the Office of Justice Programs National Institute of Justice Award# NIJ-2011-DN-BX-K533. S.W. was funded by the National Human Genome Research Institute Grant# 3 R01 HG004872-03S2, and the National Institute of Health Grant# 5 U01 HG004866-04. Research capacity at Chaminade University of Honolulu was supported by NIH-BRIC P20MD006084.
References
- 1.Gill-King H (1997) Chemical and Ultrastructural Aspects of Decomposition. In: Haglund WD and Sorg MH e (ed) Forensic Taphonomy: The Postmortem Fate of Human Remains. CRC Press, Boca Raton, FL., [Google Scholar]
- 2.Correy JEL (1978) Possible sources of ethanol ante- and post-mortem: its relationships to the biochemistry and microbiology of decomposition. Journal of Applied Bacteriology 44:1–56 [DOI] [PubMed] [Google Scholar]
- 3.Metcalf JL, Wegener Parfrey L, Gonzalez A, Lauber CL, Knights D, Ackermann G, Humphrey GC, Gebert MJ, Van Treuren W, Berg-Lyons D, Keepers K, Guo Y, Bullard J, Fierer N, Carter DO, Knight R (2013) A microbial clock provides an accurate estimate of the postmortem interval in a mouse model system. eLife 2:e01104. doi: 10.7554/eLife.01104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pechal JL, Crippen TL, Benbow ME, Tarone AM, Dowd S, Tomberlin JK (2014) The potential use of bacterial community succession in forensics as described by high throughput metagenomic sequencing. Int J Leg Med 128:193–205 [DOI] [PubMed] [Google Scholar]
- 5.Haskell NH and Catts EP, editors. (2008) Entomology and Death: A Procedural Guide. Second edn. East Park Printing [Google Scholar]
- 6.Putman RJ (1978) Patterns of carbon dioxide evolution from decaying carrion. 1. Decomposition of small mammal carrion in temperate systems. Oikos 31:47–57 [Google Scholar]
- 7.Carter DO, Yellowlees D, Tibbett M (2008) Temperature affects microbial decomposition of cadavers (Rattus rattus) in contrasting soils. Appl Soil Ecol 40:129–137 [Google Scholar]
- 8.Carter DO, Yellowlees D, Tibbett M (2010) Moisture can be the dominant environmental parameter governing cadaver decomposition in soil. Forensic Sci Int 200:60–66 [DOI] [PubMed] [Google Scholar]
- 9.Ibekwe AM, Kennedy AC, Frohne PS, Papiernik SK, Yang CH, Crowley DE (2002) Microbial diversity along a transect of agronomic zones. FEMS microbiology ecology 39 (3):183–191. doi: 10.1111/j.1574-6941.2002.tb00921.x [DOI] [PubMed] [Google Scholar]
- 10.Kuske CR, Ticknor LO, Miller ME, Dunbar JM, Davis JA, Barns SM, Belnap J (2002) Comparison of soil bacterial communities in rhizospheres of three plant species and the interspaces in an arid grassland. Applied and environmental microbiology 68 (4):1854–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haslam TC, Tibbett M (2009) Soils of contrasting pH affect the decomposition of buried mammalian (Ovis aries) skeletal muscle tissue. Journal of forensic sciences 54 (4):900–904. doi: 10.1111/j.1556-4029.2009.01070.x [DOI] [PubMed] [Google Scholar]
- 12.Lauber CL, Metcalf JL, Keepers K, Ackermann G, Carter DO, Knight R (2014) Vertebrate decomposition is accelerated by soil microbes. Appl Environ Microbiol 80:4920–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tibbett M, Carter DO (2003) Mushrooms and taphonomy: the fungi that mark woodland graves. Mycologist 17:20–24 [Google Scholar]
- 14.Sagara N, Yamanaka T, Tibbett M (2008) Soil fungi associated with graves and latrines: toward a forensic mycology. In: Tibbett M, Carter DO (eds) Soil Analysis in Forensic Taphonomy: Chemical and Biological Effects of Buried Human Remains. CRC Press, Boca Raton, FL, USA, pp 67–108 [Google Scholar]
- 15.Anderson GS (1995) The use of insects in death investigations: an analysis of cases in British Columbia over a five year period. Canadian Society of Forensic Science Journal 28:277–292 [Google Scholar]
- 16.Anderson GS, VanLaerhoven SL (1996) Initial studies on insect succession on carrion in southwestern British Columbia. Journal Forensic Science 41:617–625 [PubMed] [Google Scholar]
- 17.Campobasso CP, Introna F (2001) The forensic entomologist in the context of the forensic pathologist’s role. Forensic Sci Int 120 (1–2):132–139 [DOI] [PubMed] [Google Scholar]
- 18.Hewadikaram KA, Goff ML (1991) Effect of carcass size on rate of decomposition and arthropod succession patterns. American Journal of Forensic Medicine and Pathology 12:235–240 [DOI] [PubMed] [Google Scholar]
- 19.Komar D, Beattie O (1998) Effects of carcass size on decay rates of shade and sun exposed carrion. Can Soc Forensic Sci J 31:35–43 [Google Scholar]
- 20.Simmons T, Adlam RE, Moffatt C (2010) Debugging decomposition data--comparative taphonomic studies and the influence of insects and carcass size on decomposition rate. Journal of forensic sciences 55 (1):8–13. doi: 10.1111/j.1556-4029.2009.01206.x [DOI] [PubMed] [Google Scholar]
- 21.Spicka A, Johnson R, Bushing J, Higley LG, and Carter DO (2011) Carcass mass can influence rate of decomposition and release of ninhydrin-reactive nitrogen. Forensic Science International 209:80–85 [DOI] [PubMed] [Google Scholar]
- 22.Sutherland A, Myburgh J, Steyn M, Becker PJ (2013) The effect of body size on the rate of decomposition in a temperate region of South Africa. Forensic Sci Int 231 (1–3):257–262. doi: 10.1016/j.forsciint.2013.05.035 [DOI] [PubMed] [Google Scholar]
- 23.Matuszewski S, Konwerski S, Fratczak K, Szafalowicz M (2014) Effect of body mass and clothing on decomposition of pig carcasses. Int J Legal Med 128 (6):1039–1048. doi: 10.1007/s00414-014-0965-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuusela S, Hanski I (1982) The Structure of Carrion Fly Communities - the Size and the Type of Carrion. Holarctic Ecol 5 (4):337–348 [Google Scholar]
- 25.Kneidel KA (1984) Influence of Carcass Taxon and Size on Species Composition of Carrion-Breeding Diptera. Am Midl Nat 111 (1):57–63. doi:Doi 10.2307/2425542 [DOI] [Google Scholar]
- 26.Arnold CY (1959) The determination and significance of the base temperature in a linear heat unit system. Proc Am Soc Hortic Sci 74:430–445 [Google Scholar]
- 27.Vass AA, Bass WM, Wolt JD, Foss JE, Ammons JT (1992) Time since death determinations of human cadavers using soil solution. J Forensic Sci 37:1236–1253 [PubMed] [Google Scholar]
- 28.Carter DO, Yellowlees D, Tibbett M (2007) Cadaver decomposition in terrestrial ecosystems. Die Naturwissenschaften 94 (1):12–24. doi: 10.1007/s00114-006-0159-1 [DOI] [PubMed] [Google Scholar]
- 29.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P (2012) An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME journal 6 (3):610–618. doi: 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research 41 (Database issue):D590–596. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulson JN, Stine OC, Bravo HC, Pop M (2013) Differential abundance analysis for microbial marker-gene surveys. Nature methods 10 (12):1200–1202. doi: 10.1038/nmeth.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lozupone CA, Hamady M, Kelley ST, Knight R (2007) Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Applied and environmental microbiology 73 (5):1576–1585. doi: 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology 71 (12):8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, Oundo J, Tamboura B, Mai V, Astrovskaya I, Corrada Bravo H, Rance R, Stares M, Levine MM, Panchalingam S, Kotloff K, Ikumapayi UN, Ebruke C, Adeyemi M, Ahmed D, Ahmed F, Alam MT, Amin R, Siddiqui S, Ochieng JB, Ouma E, Juma J, Mailu E, Omore R, Morris JG, Breiman RF, Saha D, Parkhill J, Nataro JP, Stine OC (2014) Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome biology 15 (6):R76. doi: 10.1186/gb-2014-15-6-r76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW (2014) Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC biology 12:87. doi: 10.1186/s12915-014-0087-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzalez A, Goodrich JK, Gordon JL, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone C, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widman J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecology 26:32–46 [Google Scholar]
- 38.Jari Oksanen FGB, Kindt Roeland, Legendre Pierre, Minchin Peter R., O’Hara RB, Simpson Gavin L., Solymos Peter, Henry M Stevens H and Wagner Helene (2015) vegan: Community Ecology Package.
- 39.Team RC (2014) R: A language and environment for statistical computing. Vienna, Austria [Google Scholar]
- 40.Meyer J, Anderson B, Carter DO (2013) Seasonal variation of carcass decomposition and gravesoil chemistry in a cold (Dfa) climate. Journal of forensic sciences 58 (5):1175–1182. doi: 10.1111/1556-4029.12169 [DOI] [PubMed] [Google Scholar]
- 41.Pechal JL, Crippen TL, Tarone AM, Lewis AJ, Tomberlin JK, Benbow ME (2013) Microbial community functional change during vertebrate carrion decomposition. PloS one 8 (11):e79035. doi: 10.1371/journal.pone.0079035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeBlanc HN, Logan JG (2010) Exploiting Insect Olfaction in Forensic Entomology. Current Concepts in Forensic Entomology:205–221. doi:Doi 10.1007/978-1-4020-9684-6_11 [DOI] [Google Scholar]
- 43.Tomberlin JK, Benbow ME, Tarone AM, Mohr RM (2011) Basic research in evolution and ecology enhances forensics. Trends in ecology & evolution 26 (2):53–55. doi:Doi 10.1016/J.Tree.2010.12.001 [DOI] [PubMed] [Google Scholar]
- 44.Ma Q, Fonseca A, Liu WQ, Fields AT, Pimsler ML, Spindola AF, Tarone AM, Crippen TL, Tomberlin JK, Wood TK (2012) Proteus mirabilis interkingdom swarming signals attract blow flies. Isme Journal 6 (7):1356–1366. doi:Doi 10.1038/Ismej.2011.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomberlin JK, Crippen TL, Tarone AM, Singh B, Adams K, Rezenom YH, Benbow ME, Flores M, Longnecker M, Pechal JL, Russell DH, Beier RC, Wood TK (2012) Interkingdom responses of flies to bacteria mediated by fly physiology and bacterial quorum sensing. Anim Behav 84 (6):1449–1456. doi:Doi 10.1016/J.Anbehav.2012.09.013 [DOI] [Google Scholar]
- 46.De Wit R, Bouvier T (2006) ‘Everything is everywhere, but the environment selects’; what did Baas Becking and Beijerinck really say? Environmental Microbiology 8:755–758 [DOI] [PubMed] [Google Scholar]
- 47.Bongers T, Ferris H (1999) Nematode community structure as a bioindicator in environmental sampling. Trends Ecol Evol 14:224–228 [DOI] [PubMed] [Google Scholar]
- 48.Tibbett M, Carter DO (2008) Soil analysis in forensic taphonomy : chemical and biological effects of buried human remains. CRC Press, Boca Raton [Google Scholar]
- 49.Gunstone FD (1967) An Introduction to the Chemistry and Biochemistry of Fatty Acids and their Glycerides. Chapman Hall, London, UK [Google Scholar]
- 50.Ridgway EJ, Harvey DJ (2010) Microbiology of the autopsy. In: Burton JL, Rutty G (eds) The Hospital Autopsy: A Manual of Fundamental Autopsy Practice. CRC Press, Boca Raton, FL, pp 227–245 [Google Scholar]
- 51.Caplan MJ, Koontz FP (2001) Cumitech 35 Postmortem Microbiology. ASM Press, Washington DC [Google Scholar]
- 52.Lauber CL, Metcalf JL, Keepers K, Ackermann G, Carter DO, Knight R (2014) Vertebrate decomposition is accelerated by soil microbes. Applied and environmental microbiology 80 (16):4920–4929. doi: 10.1128/AEM.00957-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyde ER, Haarmann DP, Lynne AM, Bucheli SR, Petrosino JF (2013) The living dead: bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PloS one 8 (10):e77733. doi: 10.1371/journal.pone.0077733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maujean G, Guinet T, Fanton L, Malicier D (2013) The interest of postmortem bacteriology in putrefied bodies. Journal of forensic sciences 58 (4):1069–1070. doi: 10.1111/1556-4029.12155 [DOI] [PubMed] [Google Scholar]
- 55.Hyde ER, Haarmann DP, Lynne AM, Bucheli SR, Pertrosino JF (2013) The living dead: bacterial community structure of a cadaver at the onset and end of the bloat stage of decomposition. PLOS One 8:e77733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R (2010) Forensic identification using skin bacterial communities. Proceedings of the National Academy of Sciences of the United States of America 107 (14):6477–6481. doi: 10.1073/pnas.1000162107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig 1S: Phylogenetic distance (PD) alpha diversity boxplots. This includes day 0, pre-carcass rupture (days 1, 2, 4, 5 and 6), and post-carcass rupture (days 9 and 15) gravesoils for carcasses of masses 1 kg, 20 kg, 40 kg, and 50 kg. For example, label pre_1, signifies alpha diversity of pre-rupture 1kg carcass gravesoils. Squares (∎) indicate siginificant differences from the control soils. * indicates possible (p < 0.08) weak significant difference between boxplots. Shannon diversity yielded similar results. (a) 16S (b) 18S.
Table 1S. CSS-normalized weighted UniFrac Type 1 sequential sums of squares PERMANOVA. The model y ~ ADD_time + mass was fit to control for differences in the number of replicates at each time point before assessing the effect of carcass mass on gravesoil microbial communities. The FDR procedure is Bonferroni correction. (a) 16S (b) 18S
Table 2S. Rarefied unweighted UniFrac Type 1 sequential sums of squares PERMANOVA. The model y ~ ADD_time + NRN + mass was fit to control for differences in the number of replicates at each time point, and the amount of released NRN, before assessing the effect of carcass mass on gravesoil microbial communities. The FDR procedure is Bonferroni correction. (a) 16S (b) 18S


