ABSTRACT
Hepatoviruses are atypical hepatotropic picornaviruses that are released from infected cells without lysis in small membranous vesicles. These exosome-like, quasi-enveloped virions (eHAV) are infectious and the only form of hepatitis A virus (HAV) found circulating in blood during acute infection. eHAV is released through multivesicular endosomes in a process dependent on endosomal sorting complexes required for transport (ESCRT). Capsid protein interactions with the ESCRT-associated Bro1 domain proteins, ALG-2-interacting protein X (ALIX) and His domain-containing protein tyrosine phosphatase (HD-PTP), which are both recruited to the pX domain of 1D (VP1pX), are critical for this process. Previous proteomics studies suggest pX also binds the HECT domain, NEDD4 family E3 ubiquitin ligase, ITCH. Here, we confirm this interaction and show ITCH binds directly to the carboxy-terminal half of pX from both human and bat hepatoviruses independently of ALIX. A small chemical compound (compound 5) designed to disrupt interactions between WW domains of NEDD4 ligases and substrate molecules blocked ITCH binding to pX and demonstrated substantial antiviral activity against HAV. CRISPR deletion or small interfering RNA (siRNA) knockdown of ITCH expression inhibited the release of a self-assembling nanocage protein fused to pX and also impaired the release of eHAV from infected cells. The release could be rescued by overexpression of wild-type ITCH, but not a catalytically inactive ITCH mutant. Despite this, we found no evidence that ITCH ubiquitylates pX or that eHAV release is strongly dependent upon Lys residues in pX. These data indicate ITCH plays an important role in the ESCRT-dependent release of quasi-enveloped hepatovirus, although the substrate molecule targeted for ubiquitylation remains to be determined.
IMPORTANCE Mechanisms underlying the cellular release of quasi-enveloped hepatoviruses are only partially understood, yet play a crucial role in the pathogenesis of this common agent of viral hepatitis. Multiple NEDD4 family E3 ubiquitin ligases, including ITCH, have been reported to promote the budding of conventional enveloped viruses but are not known to function in the release of HAV or other picornaviruses from infected cells. Here, we show that the unique C-terminal pX extension of the VP1 capsid protein of HAV interacts directly with ITCH and that ITCH promotes eHAV release in a manner analogous to its role in budding of some conventional enveloped viruses. The catalytic activity of ITCH is required for efficient eHAV release and may potentially function to ubiquitylate the viral capsid or activate ESCRT components.
KEYWORDS: ESCRT, extracellular vesicle, hepatitis A, nonlytic viral release, picornavirus, quasi-envelope, ubiquitin
INTRODUCTION
Hepatoviruses are one of several genera within the Picornaviridae that have been shown to be released from cells in a nonlytic manner in small extracellular vesicles (EVs) (1–4). These quasi-enveloped hepatovirus virions (eHAV) contain 1 to 3 viral capsids and are infectious, despite the absence of any virus-encoded protein on their surface (1, 5). They are the only form of the virus detected in the blood during acute hepatitis A in humans (1). The quasi-envelope membrane of eHAV cloaks it from the immune system and contributes to the stealth-like nature of early HAV infection (1, 6). More conventional, “naked” virus particles (nHAV) are shed in feces and account for person-to-person spread of the virus. Studies in nonhuman primates and mice indicate that these fecally shed virions are replicated in the liver and are stripped of membranes by bile salts following their passage from the liver to the gut through the biliary system (7–9).
Quasi-enveloped and naked hepatitis A virions enter cells via distinct pathways that converge in the late endosome/lysosome (10). Whereas cell surface receptors for the naked particle remain poorly defined, phosphatidylserine receptors facilitate clathrin-mediated endocytosis of eHAV, leading to its transport to the lysosome where lysosomal acid lipase (LAL) and Niemann-Pick disease C1 protein (NPC1) degrade the quasi-envelope (10–12). This results in the capsid becoming accessible to endosomal gangliosides, such as GD1a, that bind the capsid and are required for cytoplasmic delivery of the genomes of both naked and quasi-enveloped viruses (13). Virus strains that are highly adapted to growth in cell culture may induce apoptosis and are cytopathic (14), whereas wild-type virus is noncytopathic, and liver injury results entirely from innate and adaptive immune responses (8, 15, 16).
The assembly and release of quasi-enveloped eHAV are only partly understood. These virions are released from infected cells via a mechanism dependent upon the cellular endosomal sorting system required for transport (ESCRT) (1). ALG-2-interacting protein X (ALIX, otherwise known as PDCD6-interacting protein), an ESCRT-associated protein that functions in the release of conventional enveloped viruses, is required for efficient eHAV release (1, 5). Several lines of evidence suggest eHAV forms by outward budding of preassembled capsids on the cytosolic surface of multivesicular endosomes (MVEs), leading to their enclosure within intralumenal vesicles that are released to the extracellular environment following fusion of the limiting membrane of the MVE with the plasma membrane of the cell. Multiple host proteins associated with the eHAV envelope have a lysosomal or endosomal origin, and His domain-containing protein tyrosine phosphatase (HD-PTP, otherwise known as tyrosine-protein phosphatase nonreceptor type 23), an ALIX paralog that functions in a specialized endosome-specific ESCRT pathway, is required for eHAV release (5, 17, 18). Other ESCRT-associated proteins identified within extracellular eHAV vesicles include the ESCRT-III components, charged multivesicular body protein 4A (CHMP4A), CHMP1A, CHMP1B, and Ist1 homolog 1 (Ist1) (5).
Recent studies provide insight into how ALIX and HD-PTP are recruited to the HAV capsid to mediate the association with ESCRT required for eHAV release. The capsid protein VP2 contains tandem YPX3L “late domains,” similar to peptide motifs in structural proteins of enveloped viruses, that bind ALIX and recruit ESCRT (19, 20). Mutant viruses in which these VP2 YPX3L motifs were ablated failed to be released from cells (20). However, the residues forming this motif are minimally accessible on the exterior surface of the fully formed, extracellular naked virus capsid (21), rendering these results difficult to interpret. More recent studies show that an unusual 8-kDa C-terminal extension of the VP1 capsid protein known as “pX” (Fig. 1A) also interacts with ALIX and that this interaction is crucial for eHAV release (17, 22). The pX sequence plays a crucial role in polyprotein processing and HAV capsid assembly (23). Capsids present within quasi-enveloped virions contain an unprocessed 38.5-kDa VP1pX protein, but pX is rapidly cleaved from the capsid upon disruption of the quasi-envelope, and it is not present in extracellular naked particles (1).
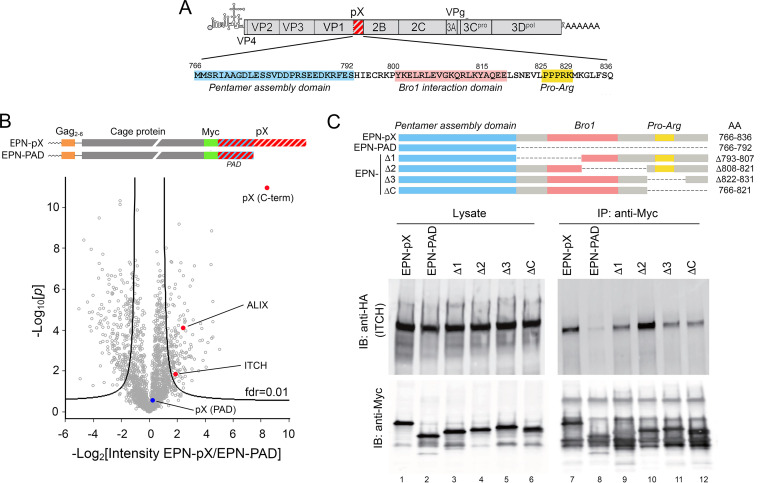
FIG 1.
The C terminus of the hepatovirus pX polypeptide forms a complex with the HECT domain E3 ubiquitin ligase ITCH. (A) HAV genome organization showing the location of pX at the C terminus of VP1; at the bottom is the amino acid sequence of pX (residues numbered according to position in the polyprotein), with the pentamer assembly domain (PAD) (23), Bro1 protein (ALIX and HD-PTP) interaction domain (17), and Pro-Arg motifs highlighted. (B) Volcano plot showing the previously reported results of a label-free quantitative proteomics comparison of cellular proteins associating with nanocage assemblies displaying full-length pX (EPN-pX) versus nanocages displaying only the pentamer association domain (EPN-PAD) of pX (17). Domain structure of the EPN-pX and EPN-PAD nanocage proteins is shown at the top. fdr, false-discovery rate. pX peptides derived from the C-terminal pX sequence (“C-term,” highly enriched) and peptides derived from the PAD domain (no enrichment) are highlighted. (C) Anti-Myc coimmunoprecipitation of HA-ITCH with Myc-tagged EPN-pX, EPN-PAD, and other EPN-pX deletion mutants coexpressed in 293T cells. IB, immunoblot; IP, immunoprecipitation.
We carried out a label-free quantitative proteomics analysis of host cell proteins associated with the C terminus of the pX sequence that identified ALIX, HD-PTP, and IST1 (17). Each of these ESCRT-associated proteins is required for efficient eHAV release (1, 5, 17). We also found the E3 ubiquitin-protein ligase Itchy homolog (ITCH) in the pX-protein complex (Fig. 1B) (17). ITCH is a member of the NEDD4 HECT domain E3 ubiquitin ligase family that has been shown to regulate ESCRT recruitment during budding of multiple enveloped viruses (24, 25). Here, we confirm the direct interaction of ITCH with pX protein and show that ITCH plays an important role in ESCRT-dependent release of quasi-enveloped hepatovirus.
RESULTS
The ubiquitin ligase ITCH associates with the C terminus of pX. In previous studies assessing the role of pX in eHAV release (17), we fused the pX sequence to the C terminus of a 231-amino-acid engineered protein nanocage (EPN) protein that self-assembles into a 60-copy, 25-nm wire cage dodecahedron (Fig. 1B) (26). The pX fusion did not interfere with assembly of the nanocage. Fully assembled EPN-pX nanocages were released from transfected cells in small EVs by an ESCRT-dependent process (17). These results mirrored earlier studies demonstrating ESCRT-dependent release of EPN fused at its C terminus to the p6Gag protein of human immunodeficiency virus type 1 (HIV-1), which contains a YPX3L late domain interacting with ALIX, as well as a PTAP late domain interacting with TSG101 (ESCRT-0) (27). HAV replication is slow and inefficient, and nanocage display of pX proved to be a useful strategy for characterizing the interaction of pX with ESCRT (17). The EPN-pX construct contains a Myc tag (Fig. 1B), allowing us to carry out a label-free quantitative (LFQ) proteomics study comparing proteins present in anti-Myc precipitates of 293T cells expressing either EPN-pX or EPN-pentamer assembly domain (PAD), which contains the N-terminal pX sequence required for capsid pentamer assembly but lacks the C-terminal 44-amino-acid sequence of pX (Fig. 1B) and does not mediate ESCRT-dependent release of EPN (17). As we reported previously, EPN-pX precipitates were enriched in ALIX, HD-PTP, and other ESCRT-associated proteins (IST1 and VTA). Also present in the complex was the HECT domain E3 ligase, ITCH (Fig. 1B). To confirm that pX associates with ITCH, as suggested by this proteomics study, we coexpressed hemagglutinin (HA)-tagged ITCH and EPN-pX (or EPN-PAD) in 293T cells. ITCH was efficiently coimmunoprecipitated with EPN-pX using an anti-Myc antibody, but not with EPN-PAD (Fig. 1C). Surprisingly, ITCH continued to coimmunoprecipitate with each of a series of deletion mutants that span the C-terminal pX sequence. Moreover, coimmunoprecipitation was reproducibly increased by a deletion (Δ2 mutant) spanning residues 808 to 821 (HAV polyprotein numbering) (Fig. 1C), which includes part of the pX-Bro1 protein interaction domain mapped in our prior studies (17). These results confirm that ITCH associates with C terminal pX sequence downstream of the PAD domain, but they indicate that the association is complex and likely involves more than one segment of the pX sequence.
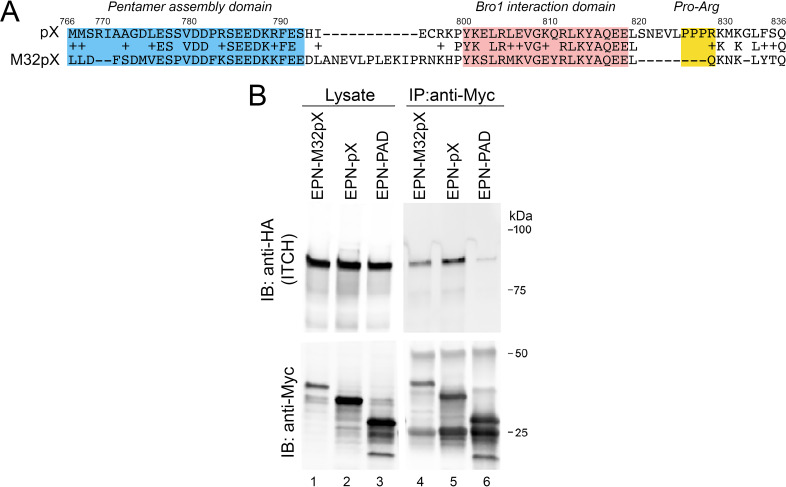
Multiple hepatovirus species have been recovered in recent years from a wide variety of mammalian host species (28, 29). Although phylogenetically distinct, these viruses appear to be hepatotropic and share numerous features in common with human HAV, including the tandem ALIX-interacting YPX3L late domains in VP2 mentioned above. The pX polypeptide of M32 virus, which was recovered from the African fruit bat, Eidolon helvum (28), binds ALIX and drives extracellular nanocage release when fused to the C terminus of EPN (EPN-M32pX) (17), despite having only 39% amino acid identity with the pX of human virus (Fig. 2A). To determine whether it also interacts with ITCH, we coexpressed HA-tagged ITCH and EPN-M32pX in 293T cells. Similar to EPN-pX, ITCH was efficiently coimmunoprecipitated with EPN-M32pX (Fig. 2B). This suggests that the recruitment of a protein complex containing ITCH is a conserved attribute of hepatovirus pX proteins.
FIG 2.
ITCH forms a complex with bat hepatovirus pX. (A) Global Needleman-Wunch amino acid sequence alignment of pX from human hepatovirus A virus (HAV; HM175 virus; GenBank accession no. NP_041007) and M32 virus, which was recovered from the African straw-colored fruit bat Eidolon helvum (hepatovirus H; GenPept accession no. ALL35271). (B) Coimmunoprecipitation of EPN-M32pX, EPN-pX, and EPN-PAD from lysates of transfected 293T cells.
Mutational analysis of the pX-ITCH interaction.
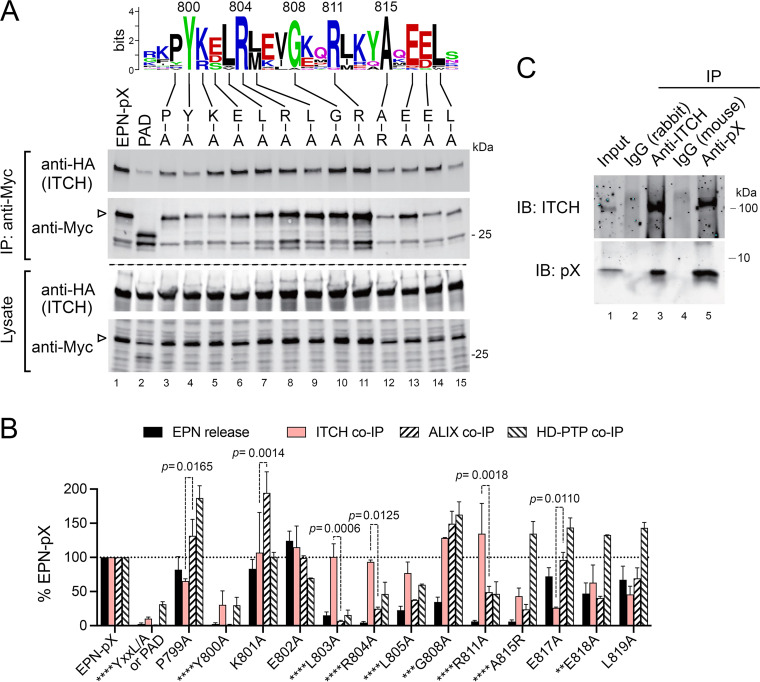
ITCH contains four modular class I WW domains that bind well-defined PPxY late domains in proteins targeted for ubiquitylation (30). Importantly, neither the EPN-pX nor EPN-M32pX sequences that coimmunoprecipitate with ITCH contain such a motif (Fig. 2A). A Pro-Arg motif (PPPR) like that bound by class III WW domains (31) does exist near the C terminus of the primate hepatovirus pX sequences, but this is absent in M32pX. Moreover, deleting the Pro-Arg motif caused only a minor reduction in EPN-pX coimmunoprecipitation with ITCH (Fig. 1C, Δ3 mutant). One possible explanation for these results is that ITCH interacts with pX through a combination of direct binding and an indirect association bridged by ALIX, which has a canonical PPxY motif and has been shown previously to interact with ITCH (32). To assess this possibility, we measured the coimmunoprecipitation of ITCH with a series of EPN-pX mutants in which single amino acid substitutions were made at pX residues between Pro799 and Leu819 that are highly conserved among multiple hepatovirus species (Fig. 3A, see WebLogo) (17). We compared these results with previous results from experiments assessing the impact of these mutations on EPN-pX release and EPN-pX coimmunoprecipitation with the Bro1 proteins, ALIX and HD-PTP (Fig. 3B) (17). Although no single amino acid substitution completely ablated EPN-pX coimmunoprecipitation with ITCH, these experiments revealed crucial roles for Pro799 and Tyr800, as well as residues between Ala815 and Leu819 (Fig. 3A and B). Some of these residues, especially Tyr800, are also critical for pX binding to ALIX and its paralog, HD-PTP (17). However, whereas Ala substitutions between Leu-803 and Leu-805 had little impact on ITCH coimmunoprecipitation, they substantially reduced or ablated ALIX and HD-PTP binding to pX (17) (Fig. 3B). On the other hand, Ala substitution at E817 significantly reduced coimmunoprecipitation with ITCH but had no impact on ALIX while substantially increasing HD-PTP coimmunoprecipitation with EPN-pX.
FIG 3.
EPN-pX binds ITCH in an ALIX-independent manner. (A) Immunoblot of HA-tagged ITCH coimmunoprecipitating (co-IP) with Myc-tagged EPN-pX, EPN-PAD (Fig. 1C), and scanning Ala or Arg substitution EPN-pX mutants from lysates of cotransfected 293T cells. At the top is a WebLogo showing conserved pX residues in the export domain that previous studies show interacts directly with the Bro1 proteins ALIX and HD-PTP (17). (B) Relative impact of the mutations shown in panel A on HA-ITCH co-IP with EPN-pX (Myc), quantified from two independent experiments. Shown for comparison are EPN pX release and HA-ALIX and HA-HD-PTP co-IP with EPN-pX reported previously (17). YKEL/A mutant was a negative control for Bro1 protein co-IP and contains Ala substitutions at each of the residues between Tyr800 and Leu803. P values for HA-ITCH versus HA-ALIX co-IP are shown in the graph. Asterisks preceding mutant name indicate significance for EPN-pX release as follows: ****, P < 0.0001; ***, P = 0.0004; and **, P = 0.0050. Significance was determined by two-way ANOVA with Sidak’s multiple-comparison test. (C) Co-IP of ITCH, expressed by in vitro translation of synthetic RNA, and recombinant pX protein expressed in E. coli. Immunoprecipitation (IP) was done with rabbit anti-ITCH and mouse monoclonal anti-pX antibodies with species-specific isotype controls as shown. IB, immunoblot.
Taken collectively, these data provide strong evidence that ITCH interacts with pX independently of ALIX or HD-PTP. This conclusion is supported also by experiments demonstrating that HA-ITCH produced in a cell-free translation reaction coimmunoprecipitates with purified recombinant, bacterially expressed pX protein (Fig. 3C, lane 4). Conversely, recombinant pX protein was also present in anti-ITCH precipitates of the cell-free translation product (Fig. 3C, lane 2). These data argue strongly for a direct interaction of ITCH with pX.
ITCH promotes ESCRT-dependent release of EPN-pX and eHAV. ITCH plays important regulatory roles in ESCRT-dependent budding of multiple canonical enveloped viruses (24, 25, 33). However, it has not been suggested previously to contribute to the nonlytic release of hepatoviruses or other naked viruses. We thus assessed its importance to the cellular release of EPN-pX from transfected Huh-7.5 cells in EVs. Since the EPN protein contains a Myc tag, EVs containing the nanocage are readily detected by immunoblotting following their centrifugation through a sucrose cushion. We used CRISPR/Cas9 gene editing to knockout ITCH expression in Huh-7.5 cells (Fig. 4) and demonstrated that this significantly reduced the release of EPN-pX (Fig. 4A and B). We also assessed the impact of ITCH knockout on release of EPN-pX containing the E817A mutation. We selected the E817A mutant for further analysis because of the opposing effects it demonstrated on EPN-pX coimmunoprecipitation with ITCH (decreased by ~75%) versus coimmunoprecipitation with ALIX and HD-PTP (no effect or increased by ~50%) (Fig. 3B). The mutation had little effect by itself on EPN-pX release (Fig. 3B and 4A and B). Consistent with its decreased coimmunoprecipitation with ITCH, there was no difference in the efficiency of EPN-E817A release from ITCH knockout versus control cells transduced with a nontargeting single guide RNA (sgRNA) (Fig. 4A and B). Surprisingly, however, the E817A mutation significantly increased EPN-pX release from ITCH knockout cells. We speculate that this could be related to increased coimmunoprecipitation of EPN-E817A with HD-PTP (Fig. 3B).
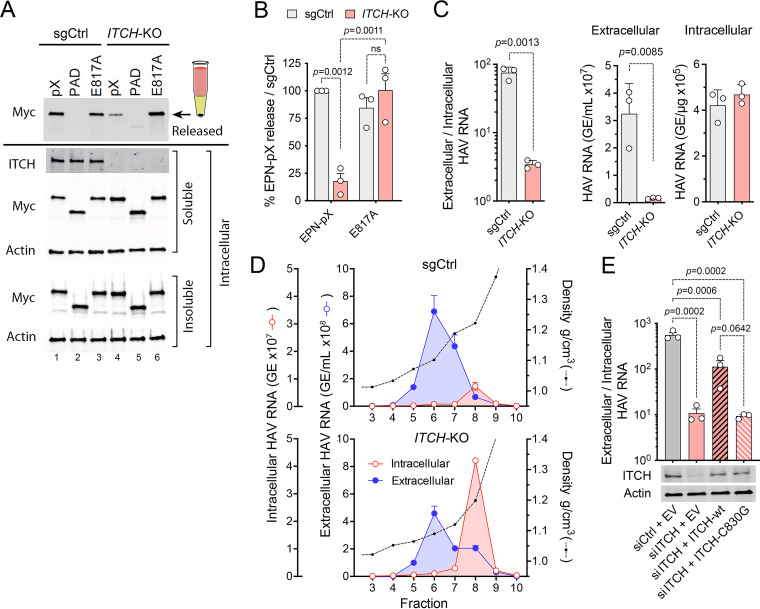
FIG 4.
ITCH depletion impairs the release of EPN-pX and quasi-enveloped eHAV. (A) EPN-pX release from ITCH-KO and sgCtrl cells transfected with EPN-pX. (A, Top) Myc immunoblot of released extracellular EPN-pX protein recovered following centrifugation through a 20% sucrose cushion. (A, Bottom) Immunoblots of soluble intracellular ITCH and soluble and insoluble intracellular EPN-pX. Actin is included in blots as a loading control. (B) Quantitation of EPN-pX and EPN-E817A (Fig. 3) release from ITCH-KO cells relative to release from sgCtrl cells in 3 independent experiments. Percent release was normalized to EPN expression based on the quantitation of soluble intracellular Myc. (C) Intracellular and extracellular HAV RNA abundance in ITCH-KO and sgCtrl cells 24 h postinfection with 18f virus, determined by RT-PCR. Data shown are means ± SD from a representative experiment. The relative abundance of RNA in extracellular fluids (genome equivalents [GE] per mL) versus intracellular viral RNA (GE/μg total RNA) is shown on the right. (D) HAV RNA in fractions of isopycnic iodixanol minigradients loaded with supernatant fluids or lysates of sgCtrl and ITCH-KO cells 5 days postinfection with 18f virus. (E) eHAV release from infected cells depleted of ITCH by transfection of ITCH-specific siRNA and transfected with vectors expressing either wild-type (WT) or catalytically inactive (C830G) ITCH. Also shown are immunoblots of ITCH and actin (loading control). EV, empty vector. Data shown are from 3 technical replicates in a representative experiment. Statistical comparisons were done by two-way ANOVA.
To assess the role of ITCH in a more physiological context, we similarly examined the impact of ITCH knockout on release of the 18f-NLuc reporter virus from infected cells. 18f-NLuc release was reduced significantly by ITCH knockout, as was the ratio of extracellular to intracellular viral RNA 24 h after infection (Fig. 4C). These results were confirmed by isopycnic gradient analysis of extracellular fluids and lysates of 18f virus-infected cell cultures, which revealed an ~6.5-fold reduction in extracellular eHAV (banding at a density of ~1.09 gm/cm3) relative to intracellular viral RNA (banding at the density of nHAV) in the knockout cells (Fig. 4D). Virus release was also impaired in Huh-7.5 cells depleted of ITCH by prior transfection of siRNA (Fig. 4E). Importantly, the defect in eHAV release in RNA interference (RNAi)-treated cells was partially corrected by reconstituting expression of wild-type ITCH, but not by expressing a catalytically inactive mutant (C830G) (Fig. 4E). Collectively, these data show that ITCH is required for optimal ESCRT-dependent release of both the EPN-pX nanocage and quasi-enveloped eHAV, mirroring the role that ITCH and other NEDD4 family E3 ligases play in budding of canonical enveloped viruses.
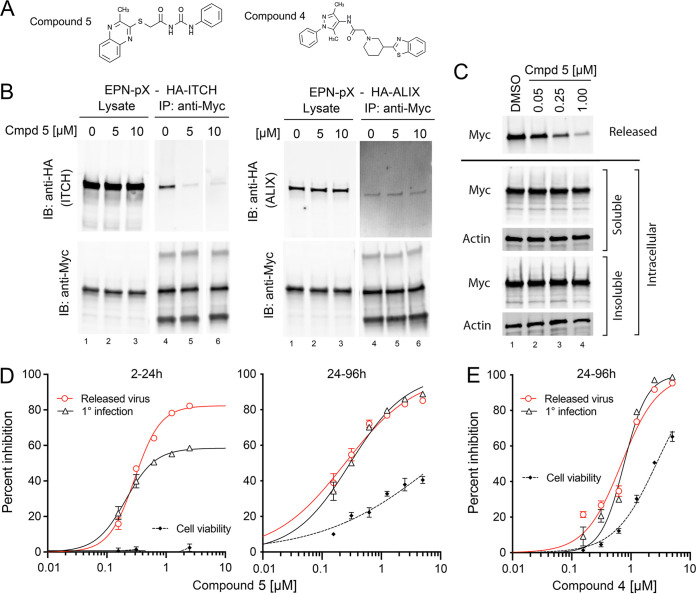
eHAV replication and release are blocked by small-molecule NEDD4 inhibitors. Several small-molecule inhibitors of NEDD4 E3 ligases have been designed to disrupt the PPxY-NEDD4 WW domain interface (30, 34) (Fig. 5A). One such compound (compound 5) ablated EPN-pX coimmunoprecipitation with ITCH when added to cell lysates (Fig. 5B, left). This effect was specific, as the compound did not inhibit coimmunoprecipitation of EPN-pX with ALIX (Fig. 5B, right). Compound 5 also substantially reduced EPN-pX release from transfected cells (Fig. 5C). When administered 2 h after infection of cells with the 18f-NLuc reporter virus under single-cycle conditions (35), compound 5 inhibited both replication (NLuc expression), as well as the release of infectious virus, which was quantified by NLuc expression in cells infected with supernatant fluids in the absence of compound (Fig. 5D, left). Although both measures were inhibited with similar concentration achieving 50% maximal inhibition values (IC50, 207 to 295 nM) (Table 1), virus release was inhibited to a greater extent than replication (82% versus 58%, P < 0.001 by the extra sum-of-squares F test) (Fig. 5D, left). Over multiple rounds of replication (compound 5 present from 24 to 96 h postinfection), both replication and release were similarly inhibited (Fig. 5D, right). The IC50 for reporter virus replication was 30- to 40-fold less than that causing cytotoxicity (Fig. 5D; Table 1) and substantially lower than that required to inhibit arenavirus or filovirus budding (24, 34). A related small molecule, compound 4 (30, 34), demonstrated similar but less potent inhibition of HAV infection (Fig. 5E). These data are consistent with NEDD4 family ligases like ITCH playing a role in virus release, but also suggest a previously unrecognized role for WW domain protein-protein interactions in HAV replication (30). We also tested TAK-243 (MLN7843) (36), a small-molecule inhibitor of UAE, the E1 ubiquitin-activating enzyme, but found it too cytotoxic to draw any conclusions (Table 1).
FIG 5.
HAV replication and eHAV release are inhibited by small molecules designed to disrupt the PPxY-NEDD4 WW domain interface. (A) Chemical structures of compound 5 and compound 4 from Han et al. (34). (B) Compound 5 (Cmpd 5) disrupts EPN-pX coimmunoprecipitation with HA-ITCH (left) but not HA-ALIX (right) when added to lysates of cotransfected 293T cells. 0 μM, DMSO vehicle only. (C) Compound 5 inhibits release of EPN-pX from transfected 293T cells. The compound was added 2 h after transfection; release was assessed at 24 h. See legend to Fig. 3B for details. (D) Compound 5 inhibition of 18f-NLuc reporter virus replication (1° infection) and release. Compound 5 was added to media from 2 to 24 h postinfection (single-cycle replication) (left) or 24 to 96 h postinfection (multiple replication cycles) (right), with cell lysate harvested at the end of treatment for the NLuc assay. Virus present in supernatant fluids was quantified by measuring NLuc expressed by it in naive cells in the absence of compound, as described in the Materials and Methods. Data were fit to a 4-parameter, variable-slope regression model. (E) Similar treatment of 18f-NLuc-infected cells with compound 4 from 24 to 96 h postinfection. See Table 1 for estimated IC50 and 50% cytotoxic concentration (CC50) values.
TABLE 1.
Suppression of HAV replication and virus release by ubiquitylation inhibitors
| HAV type | Compound | Exposure (hpi)a | Replication IC50 (95% CI [nM]) | Virus release (95% CI [nM]) | CC50 (95% CI [μM]) |
|---|---|---|---|---|---|
| 18f-NLuc | Cmpd 5 | 2–24 | 207 (177–255) | 295 (261–337) | >2.5 |
| Cmpd 5 | 24–96 | 245 (197–305) | 298 (261–340) | 8.7 (5.6–13.7) | |
| Cmpd 4 | 24–96 | 765 (680–860) | 690 (589–808) | 2.7 (2.5–2.9) | |
| TAK-243 | 24–96 | 20.9 (19.2–22.9) | 25.9 (23.4–28.6) | 39.3 (37.9–40.8) |
hpi, hours postinfection.
Protein ubiquitylation and eHAV release.
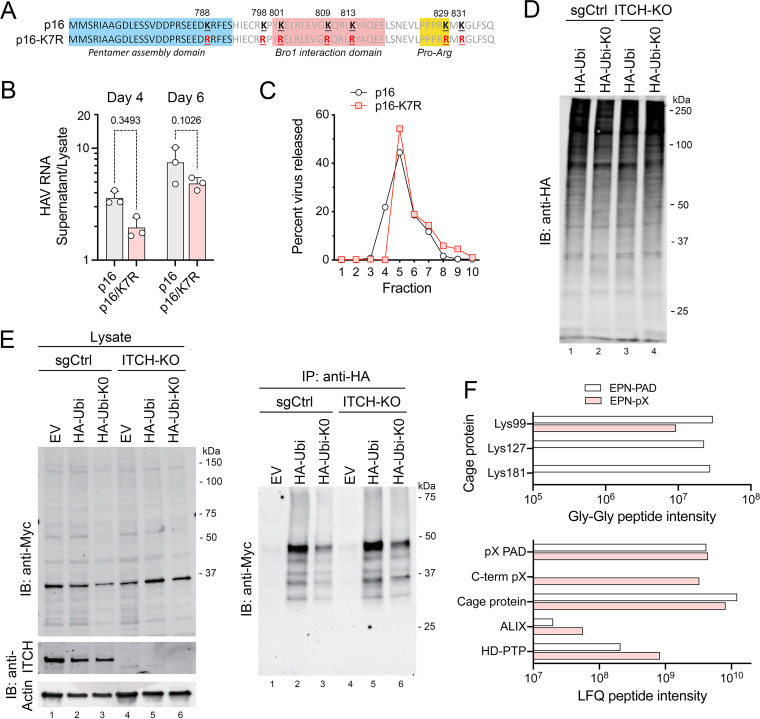
Although ITCH is recruited to pX and facilitates the release of quasi-enveloped virus and EPN-pX nanocages, we found no evidence that ITCH ubiquitylates pX. The pX sequence is rich in lysines, with lysines representing 6 or 7 of the 71 residues in pX in human viruses (Fig. 6A). With the exception of Lys788, which is within the pentamer association domain and is variably Lys or Arg among hepatovirus A viruses, these Lys residues, including 3 within the Bro1 interaction domain, are highly conserved. To assess their functional importance, we generated a mutant from HM175/p16 virus in which all 7 Lys residues in pX were substituted with Arg (p16-K7R). This mutant replicated well, and although there was a clear trend toward less efficient release than the parental p16 virus, the difference did not achieve statistical significance (Fig. 6B). Extracellular p16-K3R virus particles, like the p16 virus itself, banded uniformly in iodixanol gradients at the density of quasi-enveloped virions (Fig. 6C). These data suggest ubiquitylation of pX lysine residues is unlikely to be required for efficient release of virus.
FIG 6.
Protein ubiquitylation and eHAV release. (A) pX sequence in p16 and mutant p16-K7R virus with Arg substitutions of all 7 pX Lys residues. (B) Extracellular release of p16-K7R virus relative to the parent p16 virus, calculated from the ratio of HAV RNA in extracellular fluids versus cell lysate generated by each viral RNA at 4 and 6 days posttransfection. P values were calculated by two-way ANOVA with Sidak’s multiple-comparison test. (C) Percentage of extracellular HAV RNA banding in fractions of isopycnic minigradients loaded with supernatant fluids from cells transfected 5 days previously with p16 or p16-K7R genomic RNA. Extracellular p16-K7R virus bands at the same density as eHAV. (D) Immunoblot showing global protein ubiquitylation in ITCH-KO and control (sgCtrl) cells transfected with EPN-pX and vectors expressing either HA-tagged ubiquitin (HA-Ubi) or an HA-ubiquitin mutant lacking Lys residues (HA-Ubi-K0). Cells were treated with MG132 for 2 h prior to harvest. (E) EPN-pX ubiquitylation in cells transfected with HA-Ubi, HA-Ubi-KO, or empty vector (EV). Immunoblots show EPN-pX (Myc, ~33 kDa) and ITCH in cell lysates (left) and EPN-pX in anti-HA precipitates (right). (F, Top) Normalized intensities of nanocage protein peptides with Gly-Gly ubiquitin remnants identified in the EPN-pX and EPN-PAD proteomics samples. (F, Bottom) Normalized LFQ intensities of all peptides from selected proteins present in EPN-pX and EPN-PAD samples. C-term pX, C-terminal sequence unique to EPN-pX and not present in EPN-PAD.
To gain further insight into whether ITCH might ubiquitylate EPN-pX, we cotransfected Huh-7.5 cells with EPN-pX and vectors expressing HA-tagged ubiquitin (HA-Ubi) or a ubiquitin mutant containing no lysines (HA-Ubi-K0) (Fig. 6D). An ~48-kDa protein reacting with anti-Myc and consistent with ubiquitylated EPN-pX was immunoprecipitated by anti-HA from lysates of cells expressing either HA-Ubi or HA-Ubi-K0 (Fig. 6E). This 48-kDa protein was increased in abundance in similarly transfected ITCH-KO cells, indicating that ITCH does not mediate this apparent ubiquitylation of EPN-pX. To determine which residues in EPN-pX might be ubiquitylated, we searched our previously reported proteomics data (ProteomeXchange identifier PXD022107) (17) for Gly-Gly ubiquitin remnants present among tryptic peptides recovered from EPN-pX and EPN-PAD precipitates. This revealed sites of lysine ubiquitylation at Lys99, Lys127, and Lys181 of the nanocage protein (Fig. 6F). These ubiquitin remnants were more common among peptides derived from EPN-PAD, which does not bind ITCH (Fig. 1C), than EPN-pX. No Gly-Gly remnants were detected among pX peptides.
DISCUSSION
The hepatovirus C-terminal VP1 pX extension is unique among picornaviruses, and its function beyond directing the assembly of 14S capsid pentamer intermediates has been enigmatic (23, 37, 38). Recent studies by us and others have revealed that pX contains crucial determinants of quasi-envelopment, acting as an adaptor that recruits ESCRT to the viral capsid by binding two distinct Bro1-domain proteins, ALIX and HD-PTP, thereby forming a complex containing multiple ESCRT-related proteins (17, 22). This adaptor function is central to the biology of the virus and conserved among pX sequences of hepatoviruses infecting mammalian species separated by tens of millions of years of evolution (17). Here, we show that pX also binds the E3 ubiquitin ligase ITCH and that this interaction is important for the quasi-envelopment and release of eHAV from infected cells.
pX is cleaved from VP1 and rapidly lost from the capsid following dissolution of the quasi-envelope (1). The importance of this cleavage event in the viral life cycle is uncertain, but both trypsin and cathepsin L have been suggested to contribute to it (39, 40). Although our data show that ITCH is recruited to the C-terminal half of pX (Fig. 1C), exactly how it interacts with pX remains uncertain. Deletions in the C-terminal pX sequence (Fig. 1, Δ3 and ΔC) and some single amino acid substitutions (Fig. 3A) reduced coimmunoprecipitation with ITCH, but no single mutation other than removal of the sequence C ternminal to the pentamer association domain eliminated coimmunoprecipitation. Reproducible increases in coimmunoprecipitation were observed with a pX mutant deficient in ALIX and HD-PTP protein binding (EPN-Δ2 mutant) (Fig. 1C). This suggests the possibility that the ITCH interface may partially overlap and potentially compete with ALIX and HD-PTP for binding. Unfortunately, nothing is known about the structure of pX and the conformation it adopts within the quasi-enveloped virion. It is possible that it is presented differently on the viral capsid during quasi-envelopment and on the EPN-pX particles we studied here. The pX polypeptide is not present in the crystallographic model of the naked HAV capsid (21), and cryo-electron microscopy of purified EPN-pX nanocages revealed no clearly discernible density for pX (17). Nonetheless, in both contexts, pX is critical for optimal ESCRT-dependent release.
Fusing pX to the C terminus of the nanocage protein has proven to be a useful approach to studying its role in the release of quasi-enveloped virus (17). With respect to the involvement of ITCH, it allowed us to demonstrate the impact of compound 5 on ESCRT-dependent release (Fig. 5C) in the absence of the confounding inhibitory effect it has on intracellular virus replication (Fig. 5D). There is no membrane interaction domain within pX. Our previous results show that the ESCRT-dependent release of EPN-pX from transfected cells requires the inclusion of a specific membrane interaction signal upstream of the cage sequence (17). This was shown also for the release of the nanocage protein when fused to the p6Gag sequence of the human immunodeficiency virus (HIV), which contains ESCRT-interacting late domains (27). In the case of EPN-pX, this membrane interaction signal can be provided by either an exogenous myristoylation signal or the N-terminal 9 residues of the HAV VP4 polypeptide (17). VP4 has been suggested to integrate into membranes and form pores (41), but what drives the membrane association of assembled capsids during the process of quasi-envelopment remains to be determined.
NEDD4 family E3 ubiquitin ligases, including ITCH, are known to promote the budding of a wide range of conventional enveloped viruses (34). How they do this is not well understood. NEDD4L overexpression is capable of driving the budding and release of an HIV-1 mutant lacking both the TSG101- and ALIX-interacting late domains of p6Gag (33), showing that these ligases can act independently of viral late domains in mediating release. Arenavirus and filovirus budding is partially dependent upon ITCH (24, 25, 42). In both cases, the ligase associates with viral structural proteins, and the catalytic activity of ITCH is important for budding. We found efficient quasi-envelopment and release of hepatoviruses similarly require the catalytic activity of ITCH (Fig. 4C to E). The complex pX forms with ITCH can be disrupted by inhibitors targeting the WW domains of ITCH (30), and these compounds have substantial antiviral activity against HAV (Fig. 5B and E). Taken collectively, these results suggest that the ligase plays an important role in the hepatovirus life cycle and represents an aspect of eHAV release shared with several conventional enveloped viruses.
Similar to the situation with NEDD4 ligases and budding of arenaviruses, filoviruses, and lentiviruses (24, 25, 33), the substrate upon which ITCH acts to promote eHAV release remains to be identified. The recruitment of ITCH to the carboxy-terminal half of pX would position the ligase for possible ubiquitylation of the hepatovirus capsid. This could potentially provide a signal for the recruitment of ESCRT-related proteins, which contain multiple ubiquitin-binding domains (43). Such a signal could enhance the likelihood that a capsid is recruited to ESCRT and bud into the MVE and thus possibly also influence the number of capsids enclosed within each newly formed intralumenal vesicle. Our results do not suggest that ITCH ubiquitylates pX or that ubiquitylation of pX itself is required for eHAV release, as virus lacking any Lys residue in pX continues to be efficiently released as quasi-enveloped virions (Fig. 6B). However, it is difficult to exclude a requirement for ubiquitylation elsewhere on the surface of the assembled capsid or the possibility of noncanonical ubiquitylation of pX residues other than lysine (44). Alternatively, it is possible that one or more ESCRT components may be activated by ubiquitylation, as suggested previously in the budding of lentiviruses and arenaviruses (24, 33).
MATERIALS AND METHODS
Cells and virus.
H1 HeLa and human hepatocyte-derived Huh-7.5 cells were cultured as described previously (45). HEK293FT (293T) cells were procured from the ATCC (CRL-3216). All cells were mycoplasma free. Viruses were derived from modified infectious molecular clones of low (HM175/p16)- or high (HM175/18f)-passage cell culture-adapted HAV pT7-p16.2 (GenBank accession no. KP879217) and pT7-18f.2 (KP879216) (14, 46). The 18f-NLuc reporter virus expressing nanoluciferase has been described previously (47).
Plasmids and oligonucleotides.
Plasmids encoding the EPN protein nanocage with an N-terminal HIV-1 myristoylation signal (26, 27) and fused at the C terminus to wild-type or mutated pX sequences of human or bat HAV (17) have been described previously and have the expression vectors for ALIX (48) and wild-type and mutant ITCH (49). pRK5-HA-ubiquitin-WT (catalog no. 17608) and pRK5-HA-ubiquitin-K0 (catalog no. 17603) were from Addgene. SMARTPool ON-TARGETplus and nontargeting control 2 siRNAs were purchased from Dharmacon.
Antibodies.
Murine monoclonal antibody to pX (clone 59.7.1) was produced against bacterially expressed 6 × His-tagged pX protein as described (17) (RRID AB_2868525). Other antibodies used in this study include mouse anti-Myc, clone 4A6 (EMD Millipore; catalog no. 05-724; RRID AB_568800); rabbit anti-HA tag, clone C29F4 (Cell Signaling; catalog no. 3724; RRID AB_1549585); rat anti-HA, clone 3F10 (Roche; catalog no. 11867423001; RRID AB_390918); RRID AB_673819; rabbit anti-ALIX, clone E6P9B (Cell Signaling; catalog no. 92880; RRID AB_2800192); rabbit anti-ITCH, clone D8Q6D (Cell Signaling; catalog no. 12117; RRID AB_2797822); rabbit anti-actin (Sigma-Aldrich; catalog no. A2066; RRID AB_476693); and mouse anti-actin, clone C-2 (Santa Cruz; catalog no. sc-843; RRID AB_626630).
Proteomics analysis of the carboxy-terminal pX interactome.
Label-free quantitative proteomics methods and results are described in detail elsewhere (17). In brief, lysates were prepared from cultures of 293T cells transfected with pEPN-pX or pEPN-PAD, immunoprecipitated with anti-Myc antibody, and subjected to SDS-PAGE. Proteins were extracted from segments cut from the gel, subjected to tryptic digestion, and analyzed by liquid chromatography-mass spectrometry (LC/MS). A total of 2,192 cellular proteins were identified in the two sample sets, with 87 proteins enriched more than 4-fold in samples from pEPN-pX- than pEPN-PAD-transfected cells (17). Mass spectrometry data are available from the ProteomeXchange Consortium with the data set identifier PXD022107.
Chemical reagents.
Compound 4 (Amb123203) and compound 5 (Amb21795397) were gifts from Christopher Ziegler of the University of Vermont and purchased from Ambinter (Orléans, France). TAK-243 (MLN7243) was purchased from MedChemExpress (catalog no. HY-100487). Iodixanol (Opti-Prep density gradient medium) was from MilliporeSigma (catalog no. D1556).
siRNA protein depletion.
RNAi depletion of cellular proteins was carried out as described previously (47). In brief, cells were transfected with 50 nM ON-TARGETplus siRNA (set of 4) or ON-TARGETplus nontargeting control siRNA (control 2; “siCtrl”) (Dharmacon), using the Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. Four different siRNA sequences were used, and the siRNA providing the most efficient knockdown was selected for use in subsequent experiments.
CRISPR/Cas9 gene editing.
CRISPR/Cas9 genetic knockouts were generated as previously described (47) using precloned lentivirus vectors purchased from Applied Biological Materials (Richmond, Canada). Dual sgRNA/CRISPR/Cas9-expressing lentivirus was produced by cotransfection of specific and nontargeting sgRNA-expressing vectors (Applied Biological Materials) and 3rd-generation packaging system mix (Applied Biological Materials) into 293T cells using the Fugene HD transfection reagent (Promega). Supernatant fluids were harvested at 72 h, passed through a 0.22-μm syringe filter, and used subsequently to transduce 293T cells. Puromycin (5 mg/mL) was added to the medium 72 h following transfection for antibiotic selection.
DNA transfection.
DNA transfections were carried out with Fugene HD according to the manufacturer’s recommended protocol. Briefly, 293T cells (5 × 105 per well in 6-well plates) were transfected with 500 ng of EPN vectors. For coimmunoprecipitation experiments, 2.5 × 106 293T cells, seeded previously in 10-cm dishes, were cotransfected with 500 ng EPN vector and 5 μg empty or HA-tagged ALIX, ITCH, Ubi, or Ubi-K0 expression vectors. The Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) was used for transfection of 50 nM SMARTPool ON-TARGETplus or control 2 siRNAs (Dharmacon) according to the manufacturer’s recommended procedure.
Isopycnic gradient centrifugation of virus and measurement of viral RNA.
The buoyant densities of extracellular and intracellular virus particles were determined by centrifugation in isopycnic iodixanol step gradients as previously described (1, 20). Fractions were collected manually from the top of the gradient. RNA was extracted from fractions using the RNeasy kit (Qiagen), followed by two-step reverse transcriptase quantitative PCR (RT-qPCR) quantitation of HAV RNA.
Chemical inhibition of viral release.
Stock solutions of compound 4 and compound 5 (34) were prepared in dimethyl sulfoxide (DMSO) at 10 mM. 18f-NLuc nHAV (~1,000 genome equivalents [GE] per cell) was allowed to adsorb to Huh-7.5 cells for 2 h at 37°C, after which cells were refed with maintenance medium. Compound 5 was added to the medium at concentrations ranging from 0.3125 to 2.5 μM immediately after virus inoculation, and cells and supernatant fluids were harvested 22 h later. Alternatively, compounds (0.3125 to 5 μM) were added 24 h after infection, and cells were incubated for an additional 72 h. At harvest, cell pellets were lysed to measure NLuc activity (1° infection). Supernatant fluids were inoculated onto naive cells in the absence of compound to assess the amount of released virus. The supernatant inoculum was removed from these cells after 2 h adsorption, and the cells were washed with fresh media and then cultured for an additional 22 h until they were lysed for NLuc assay (released virus). The cellular toxicity of the compounds was assessed by ATP assay (CellTiter Glo; Promega).
Nanocage release assays.
The release of EPN-pX from transfected 293T cells was assayed as previously described (17). Supernatant fluids were collected 24 h after transfection with nanocage vectors, clarified by centrifugation at 8,000 rpm for 10 min, and then centrifuged through a 200-μL 20% sucrose cushion in an Eppendorf 5424R centrifuge at full speed for 90 min. Samples removed from the bottom of the cushion were resuspended in 50 μL Laemmli sample buffer and subjected to SDS-PAGE followed by immunoblotting with anti-Myc antibody and visualization with an Odyssey infrared fluorescence scanner (LI-COR Biosciences). To assess the nonreleased EPN-pX fraction, cells were lysed in 200 μL lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, and 1× complete protease inhibitor cocktail [MilliporeSigma]) on ice for 5 min. Soluble and insoluble proteins were separated by centrifugation at 12,000 rpm for 5 min in an Eppendorf 5424R centrifuge and subjected to immunoblotting for Myc as described above. The efficiency of release was calculated as the percentage of EPN protein pelleted from culture supernatant against total EPN protein expressed (normalized intracellular EPN protein plus supernatant fluid EPN protein) based on quantitation of immunoblots, using the Odyssey imaging system.
Protein coimmunoprecipitation.
Cell lysates were prepared from 10-cm cell culture dishes with lysis buffer (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 250 mM NaCl, 10% glycerol, 5 mM EDTA, 1× complete protease inhibitor, 1× PhoStop, and 0.5% NP-40) and clarified by low-speed centrifugation prior to immunoprecipitation using Pierce protein A/G agarose beads (Thermo Scientific).
In vitro translation of ITCH.
ITCH protein was synthesized using the TnT quick coupled transcription/translation system (Promega). Briefly, 1 μg of circular plasmid DNA was combined with TnT reaction buffer and amino acid mixture to a total volume of 50 μL and incubated for 1 h at 30°C. pX protein was added, and the mixture was placed at room temperature for 30 min, followed by addition of antibody and immunoprecipitation.
Statistical analysis.
Unless indicated otherwise, significance was assessed by unpaired t test or analysis of variance (ANOVA). Calculations were carried out with GraphPad Prism 9.4.0 software.
Data availability.
Mass spectrometry data have been deposited with the ProteomeXchange Consortium and are available with the data set identifier PXD022107.
ACKNOWLEDGMENTS
We gratefully acknowledge Hugh Pelham of the MRC Laboratory of Molecular Biology, Cambridge, United Kingdom, for ITCH expression vectors, Ted Dawson of the Johns Hopkins University for ubiquitin expression vectors, and Christopher Ziegler of the University of Vermont for the gift of compounds 4 and 5.
This work was supported in part by the following grants from the U.S. National Institutes of Health: R01-AI103083, R01-AI131685, and R01-AI150095 to S.M.L. and R01-GM133107 and R21-AG071229 to X.C.
Contributor Information
Stanley M. Lemon, Email: smlemon@med.unc.edu.
J.-H. James Ou, University of Southern California.
REFERENCES
- 1.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, Williams W, Kha N, Cruz C, Hancock BM, Nguyen DP, Sayen MR, Hilton BJ, Doran KS, Segall AM, Wolkowicz R, Cornell CT, Whitton JL, Gottlieb RA, Feuer R. 2014. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog 10:e1004045. 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, Jiang P, Wimmer E, Altan-Bonnet G, Altan-Bonnet N. 2015. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160:619–630. 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Grein SG, Defourny KAY, Rabouw HH, Galiveti CR, Langereis MA, Wauben MHM, Arkesteijn GJA, van Kuppeveld FJM, Nolte-'t Hoen ENM. 2019. Picornavirus infection induces temporal release of multiple extracellular vesicle subsets that differ in molecular composition and infectious potential. PLoS Pathog 15:e1007594. 10.1371/journal.ppat.1007594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKnight KL, Xie L, Gonzalez-Lopez O, Rivera-Serrano EE, Chen X, Lemon SM. 2017. Protein composition of the hepatitis A virus quasi-envelope. Proc Natl Acad Sci USA 114:6587–6592. 10.1073/pnas.1619519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemon SM, Ott JJ, Van Damme P, Shouval D. 2018. Type A viral hepatitis: a summary and update on the molecular virology, epidemiology, pathogenesis and prevention. J Hepatol 68:167–184. 10.1016/j.jhep.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 7.Schulman AN, Dienstag JL, Jackson DR, Hoofnagle JH, Gerety RJ, Purcell RH, Barker LF. 1976. Hepatitis A antigen particles in liver, bile, and stool of chimpanzees. J Infect Dis 134:80–84. 10.1093/infdis/134.1.80. [DOI] [PubMed] [Google Scholar]
- 8.Hirai-Yuki A, Hensley L, McGivern DR, Gonzalez-Lopez O, Das A, Feng H, Sun L, Wilson JE, Hu F, Feng Z, Lovell W, Misumi I, Ting JP, Montgomery S, Cullen J, Whitmire JK, Lemon SM. 2016. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science 353:1541–1545. 10.1126/science.aaf8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirai-Yuki A, Hensley L, Whitmire JK, Lemon SM. 2016. Biliary secretion of quasi-enveloped human hepatitis A virus. mBio 7:e01998-16. 10.1128/mBio.01998-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera-Serrano EE, Gonzalez-Lopez O, Das A, Lemon SM. 2019. Cellular entry and uncoating of naked and quasi-enveloped human hepatoviruses. Elife 8:e43983. 10.7554/eLife.43983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A, Hirai-Yuki A, Gonzalez-Lopez O, Rhein B, Moller-Tank S, Brouillette R, Hensley L, Misumi I, Lovell W, Cullen JM, Whitmire JK, Maury W, Lemon SM. 2017. TIM1 (HAVCR1) is not essential for cellular entry of either quasi-enveloped or naked hepatitis A virions. mBio 8. 10.1128/mBio.00969-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das A, Maury W, Lemon SM. 2019. TIM1 (HAVCR1): an essential “receptor” or an “accessory attachment factor” for hepatitis A virus? J Virol 93:e01793-18. 10.1128/JVI.01793-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das A, Barrientos RC, Shiota T, Madigan V, Misumi I, McKnight KL, Sun L, Li Z, Meganck RM, Li Y, Kaluzna E, Asokan A, Whitmire JK, Kapustina M, Zhang Q, Lemon SM. 2020. Gangliosides are essential endosomal receptors for quasi-enveloped and naked hepatitis A virus. Nat Microbiol 5:1069–1078. 10.1038/s41564-020-0727-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemon SM, Murphy PC, Shields PA, Ping LH, Feinstone SM, Cromeans T, Jansen RW. 1991. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: evidence for genetic recombination. J Virol 65:2056–2065. 10.1128/JVI.65.4.2056-2065.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun L, Li Y, Misumi I, González-López O, Hensley L, Cullen JM, McGivern DR, Matsuda M, Suzuki R, Sen GC, Hirai-Yuki A, Whitmire JK, Lemon SM. 2021. IRF3-mediated pathogenicity in a murine model of human hepatitis A. PLoS Pathog 17:e1009960. 10.1371/journal.ppat.1009960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim J, Chang DY, Lee HW, Lee H, Kim JH, Sung PS, Kim KH, Hong SH, Kang W, Lee J, Shin SY, Yu HT, You S, Choi YS, Oh I, Lee DH, Lee DH, Jung MK, Suh KS, Hwang S, Kim W, Park SH, Kim HJ, Shin EC. 2018. Innate-like cytotoxic function of bystander-activated CD8(+) T cells is associated with liver injury in acute hepatitis A. Immunity 48:161–173.e5. 10.1016/j.immuni.2017.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Shirasaki T, Feng H, Duyvesteyn HME, Fusco WG, McKnight KL, Xie L, Boyce M, Kumar S, Barouch-Bentov R, González-López O, McNamara R, Wang L, Hertel-Wulff A, Chen X, Einav S, Duncan JA, Kapustina M, Fry EE, Stuart DI, Lemon SM. 2022. Nonlytic cellular release of hepatitis A virus requires dual capsid recruitment of the ESCRT-associated Bro1 domain proteins HD-PTP and ALIX. PLoS Pathog 18:e1010543. 10.1371/journal.ppat.1010543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tabernero L, Woodman P. 2018. Dissecting the role of His domain protein tyrosine phosphatase/PTPN23 and ESCRTs in sorting activated epidermal growth factor receptor to the multivesicular body. Biochem Soc Trans 46:1037–1046. 10.1042/BST20170443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee S, Joshi A, Nagashima K, Freed EO, Hurley JH. 2007. Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol 14:194–199. 10.1038/nsmb1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez-Lopez O, Rivera-Serrano EE, Hu F, Hensley L, McKnight KL, Ren J, Stuart DI, Fry EE, Lemon SM. 2018. Redundant late domain functions of tandem VP2 YPX3L motifs in nonlytic cellular egress of quasi-enveloped hepatitis A virus. J Virol 92:e01308-18. 10.1128/JVI.01308-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Ren J, Gao Q, Hu Z, Sun Y, Li X, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2015. Hepatitis A virus and the origins of picornaviruses. Nature 517:85–88. 10.1038/nature13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W, Ma P, Deng L, Liu Z, Wang X, Liu X, Long G. 2020. Hepatitis A virus structural protein pX interacts with ALIX and promotes the secretion of virions and foreign proteins through exosome-like vesicles. J Extracell Vesicles 9:1716513. 10.1080/20013078.2020.1716513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen L, Benichou D, Martin A. 2002. Analysis of deletion mutants indicates that the 2A polypeptide of hepatitis A virus participates in virion morphogenesis. J Virol 76:7495–7505. 10.1128/jvi.76.15.7495-7505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziegler CM, Dang L, Eisenhauer P, Kelly JA, King BR, Klaus JP, Manuelyan I, Mattice EB, Shirley DJ, Weir ME, Bruce EA, Ballif BA, Botten J. 2019. NEDD4 family ubiquitin ligases associate with LCMV Z's PPXY domain and are required for virus budding, but not via direct ubiquitination of Z. PLoS Pathog 15:e1008100. 10.1371/journal.ppat.1008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Z, Sagum CA, Bedford MT, Sidhu SS, Sudol M, Harty RN. 2016. ITCH E3 ubiquitin ligase interacts with Ebola virus VP40 to regulate budding. J Virol 90:9163–9171. 10.1128/JVI.01078-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsia Y, Bale JB, Gonen S, Shi D, Sheffler W, Fong KK, Nattermann U, Xu C, Huang PS, Ravichandran R, Yi S, Davis TN, Gonen T, King NP, Baker D. 2016. Design of a hyperstable 60-subunit protein dodecahedron. Nature 535:136–139. 10.1038/nature18010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Votteler J, Ogohara C, Yi S, Hsia Y, Nattermann U, Belnap DM, King NP, Sundquist WI. 2016. Designed proteins induce the formation of nanocage-containing extracellular vesicles. Nature 540:292–295. 10.1038/nature20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drexler JF, Corman VM, Lukashev AN, van den Brand JMA, Gmyl AP, Brünink S, Rasche A, Seggewiβ N, Feng H, Leijten LM, Vallo P, Kuiken T, Dotzauer A, Ulrich RG, Lemon SM, Drosten C, Hepatovirus Ecology Consortium . 2015. Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci USA 112:15190–15195. 10.1073/pnas.1516992112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Oliveira Carneiro I, Sander AL, Silva N, Moreira-Soto A, Normann A, Flehmig B, Lukashev AN, Dotzauer A, Wieseke N, Franke CR, Drexler JF. 2018. A novel marsupial hepatitis A virus corroborates complex evolutionary patterns shaping the genus Hepatovirus. J Virol 92:e00082-18. 10.1128/JVI.00082-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shepley-McTaggart A, Fan H, Sudol M, Harty RN. 2020. Viruses go modular. J Biol Chem 295:4604–4616. 10.1074/jbc.REV119.012414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, Taylor L, Yeung B, Vassilovski G, Amin M, Chen F, Matskova L, Winberg G, Ernberg I, Linding R, O'Donnell P, Starostine A, Keller W, Metalnikov P, Stark C, Pawson T. 2005. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol 25:7092–7106. 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CP, Liu GT, Kung HN, Liu PT, Liao YT, Chow LP, Chang LS, Chang YH, Chang CW, Shu WC, Angers A, Farina A, Lin SF, Tsai CH, Bouamr F, Chen MR. 2016. The ubiquitin ligase Itch and ubiquitination regulate BFRF1-mediated nuclear envelope modification for Epstein-Barr virus maturation. J Virol 90:8994–9007. 10.1128/JVI.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung HY, Morita E, von Schwedler U, Müller B, Kräusslich HG, Sundquist WI. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol 82:4884–4897. 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han Z, Lu J, Liu Y, Davis B, Lee MS, Olson MA, Ruthel G, Freedman BD, Schnell MJ, Wrobel JE, Reitz AB, Harty RN. 2014. Small-molecule probes targeting the viral PPxY-host Nedd4 interface block egress of a broad range of RNA viruses. J Virol 88:7294–7306. 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whetter LE, Day SP, Elroy-Stein O, Brown EA, Lemon SM. 1994. Low efficiency of the 5' nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J Virol 68:5253–5263. 10.1128/JVI.68.8.5253-5263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyer ML, Milhollen MA, Ciavarri J, Fleming P, Traore T, Sappal D, Huck J, Shi J, Gavin J, Brownell J, Yang Y, Stringer B, Griffin R, Bruzzese F, Soucy T, Duffy J, Rabino C, Riceberg J, Hoar K, Lublinsky A, Menon S, Sintchak M, Bump N, Pulukuri SM, Langston S, Tirrell S, Kuranda M, Veiby P, Newcomb J, Li P, Wu JT, Powe J, Dick LR, Greenspan P, Galvin K, Manfredi M, Claiborne C, Amidon BS, Bence NF. 2018. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat Med 24:186–193. 10.1038/nm.4474. [DOI] [PubMed] [Google Scholar]
- 37.Harmon SA, Emerson SU, Huang YK, Summers DF, Ehrenfeld E. 1995. Hepatitis A viruses with deletions in the 2A gene are infectious in cultured cells and marmosets. J Virol 69:5576–5581. 10.1128/JVI.69.9.5576-5581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Probst C, Jecht M, Gauss-Muller V. 1999. Intrinsic signals for the assembly of hepatitis A virus particles. Role of structural proteins VP4 and 2A. J Biol Chem 274:4527–4531. 10.1074/jbc.274.8.4527. [DOI] [PubMed] [Google Scholar]
- 39.Morace G, Kusov Y, Dzagurov G, Beneduce F, Gauss-Muller V. 2008. The unique role of domain 2A of the hepatitis A virus precursor polypeptide P1-2A in viral morphogenesis. BMB Rep 41:678–683. 10.5483/bmbrep.2008.41.9.678. [DOI] [PubMed] [Google Scholar]
- 40.Seggewiß N, Paulmann D, Dotzauer A. 2016. Lysosomes serve as a platform for hepatitis A virus particle maturation and nonlytic release. Arch Virol 161:43–52. 10.1007/s00705-015-2634-5. [DOI] [PubMed] [Google Scholar]
- 41.Shukla A, Padhi AK, Gomes J, Banerjee M. 2014. The VP4 peptide of hepatitis A virus ruptures membranes through formation of discrete pores. J Virol 88:12409–12421. 10.1128/JVI.01896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baillet N, Krieger S, Carnec X, Mateo M, Journeaux A, Merabet O, Caro V, Tangy F, Vidalain PO, Baize S. 2019. E3 ligase ITCH interacts with the Z matrix protein of Lassa and Mopeia viruses and is required for the release of infectious particles. Viruses 12:49. 10.3390/v12010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frankel EB, Audhya A. 2018. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin Cell Dev Biol 74:4–10. 10.1016/j.semcdb.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClellan AJ, Laugesen SH, Ellgaard L. 2019. Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol 9:190147. 10.1098/rsob.190147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng H, Lenarcic EM, Yamane D, Wauthier E, Mo J, Guo H, McGivern DR, Gonzalez-Lopez O, Misumi I, Reid LM, Whitmire JK, Ting JP, Duncan JA, Moorman NJ, Lemon SM. 2017. NLRX1 promotes immediate IRF1-directed antiviral responses by limiting dsRNA-activated translational inhibition mediated by PKR. Nat Immunol 18:1299–1309. 10.1038/ni.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jansen RW, Newbold JE, Lemon SM. 1988. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology 163:299–307. 10.1016/0042-6822(88)90270-x. [DOI] [PubMed] [Google Scholar]
- 47.Yamane D, Feng H, Rivera-Serrano EE, Selitsky SR, Hirai-Yuki A, Das A, McKnight KL, Misumi I, Hensley L, Lovell W, Gonzalez-Lopez O, Suzuki R, Matsuda M, Nakanishi H, Ohto-Nakanishi T, Hishiki T, Wauthier E, Oikawa T, Morita K, Reid LM, Sethupathy P, Kohara M, Whitmire JK, Lemon SM. 2019. Constitutive expression of interferon regulatory factor 1 drives intrinsic hepatocyte resistance to multiple RNA viruses. Nat Microbiol 4:1096–1104. 10.1038/s41564-019-0425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhai Q, Landesman MB, Robinson H, Sundquist WI, Hill CP. 2011. Structure of the Bro1 domain protein BROX and functional analyses of the ALIX Bro1 domain in HIV-1 budding. PLoS One 6:e27466. 10.1371/journal.pone.0027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mund T, Pelham HR. 2009. Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep 10:501–507. 10.1038/embor.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Mass spectrometry data have been deposited with the ProteomeXchange Consortium and are available with the data set identifier PXD022107.