ABSTRACT
Hepatitis B virus (HBV) infection is a major health burden worldwide, and currently there is no cure. The persistence of HBV covalently closed circular DNA (cccDNA) is the major obstacle for antiviral trement. HBV core protein (HBc) has emerged as a promising antiviral target, as it plays important roles in critical steps of the viral life cycle. However, whether HBc could regulate HBV cccDNA transcription remains under debate. In this study, different approaches were used to address this question. In synthesized HBV cccDNA and HBVcircle transfection assays, lack of HBc showed no effect on transcription of HBV RNA as well as HBV surface antigen (HBsAg) production in a hepatoma cell line and primary human hepatocytes. Reconstitution of HBc did not alter the expression of cccDNA-derived HBV markers. Similar results were obtained from an in vivo mouse model harboring cccDNA. Chromatin immunoprecipitation (ChIP) or ChIP sequencing assays revealed transcription regulation of HBc-deficient cccDNA chromatin similar to that of wild-type cccDNA. Furthermore, treatment with capsid assembly modulators (CAMs) dramatically reduced extracellular HBV DNA but could not alter viral RNA and HBsAg. Our results demonstrate that HBc neither affects histone modifications and transcription factor binding of cccDNA nor directly influences cccDNA transcription. Although CAMs could reduce HBc binding to cccDNA, they do not suppress cccDNA transcriptional activity. Thus, therapeutics targeting capsid or HBc should not be expected to sufficiently reduce cccDNA transcription.
IMPORTANCE Hepatitis B virus (HBV) core protein (HBc) has emerged as a promising antiviral target. However, whether HBc can regulate HBV covalently closed circular DNA (cccDNA) transcription remains elusive. This study illustrated that HBc has no effect on epigenetic regulation of cccDNA, and it does not participate in cccDNA transcription. Given that HBc is dispensable for cccDNA transcription, novel cccDNA-targeting therapeutics are needed for an HBV cure.
KEYWORDS: capsid inhibitor, cccDNA, core protein, hepatitis B virus, transcriptional regulation
INTRODUCTION
Hepatitis B virus (HBV) creates a major health burden, with around 257 million chronically infected individuals worldwide (1). Those people are at high risk of developing cirrhosis and hepatocellular carcinoma (2, 3). Although vaccinations and treatments are available, there is no cure for HBV infection. HBV establishes covalently closed circular DNA (cccDNA) in the nucleus of an infected hepatocyte. This cccDNA is organized as a minichromosome and is resistant to currently available antivirals. Hence, complete eradication of HBV from the infected cells remains unattainable (4).
Currently, two types of therapeutics are approved for chronic HBV infection treatment, including interferon alpha (IFN-α) and nucleos(t)ide reverse transcriptase inhibitors (NRTIs) (5). Both treatments are effective to some extent but have limitations (6). IFN-α is the only approved immunomodulatory drug. However, the response rate remains low and side effects are often difficult to tolerate (7, 8). NRTIs target the reverse transcriptase activity of viral polymerase and limit virus replication. However, these treatments show little or no effect on HBV cccDNA and thus cannot clear virus infection. Therefore, there is an urgent need to develop novel therapies with targets that can lead to an HBV cure.
Recently, HBV core protein (HBc) has emerged as a promising antiviral target, as it plays important roles in critical steps of the viral life cycle including nucleocapsid formation and pregenomic RNA (pgRNA) packaging (5). Several capsid assembly modulators (CAMs) are currently under clinical development and have demonstrated effective antiviral abilities, to a certain extent (5). However, important questions regarding additional mechanisms of CAMs, particularly the role of HBc in cccDNA transcriptional regulation, remain largely unclarified. HBc is a component of the cccDNA minichromosome and is preferentially associated with CpG island 2 of the HBV genome that overlaps enhancers I and II (9, 10). By using a transient-transfection model, it has been demonstrated that the HBc carboxyl-terminal domain (CTD) plays a role in regulating HBV transcription (11). In duck hepatitis B virus (DHBV) infection, a DHBV mutant deficient in HBc expression leads to similar amounts of cccDNA formation but significantly less viral RNA, suggesting a potential role of HBc in cccDNA transcription regulation (12). In contrast, in HBV infection cell culture models, HBc-deficient HBV and wild-type (WT) HBV establish equal amounts of cccDNA and HBV RNA (13, 14). These results indicate that the newly synthesized HBc is not required for the maintenance or transcriptional activities of cccDNA. However, the major drawback of those studies is that the role of HBc derived from incoming virions could not be examined, as a recent study reported that virion-delivered HBc can associate with cccDNA for at least 6 weeks in cell cultures (15). Therefore, the question whether HBc participates in cccDNA transcriptional regulation remains open.
In this study, to avoid the influence of HBc carried by the incoming virion, we employed transfection of synthesized HBV cccDNA-like molecules and adeno-associated-virus (AAV)-mediated HBV cccDNA formation to study cccDNA transcription. Additionally, by using point mutation to create HBc deficiency and by CAM treatment, we revealed that HBc is not required for cccDNA transcriptional regulation.
RESULTS
HBc is not required for cccDNA transcription in cccDNA transfection models.
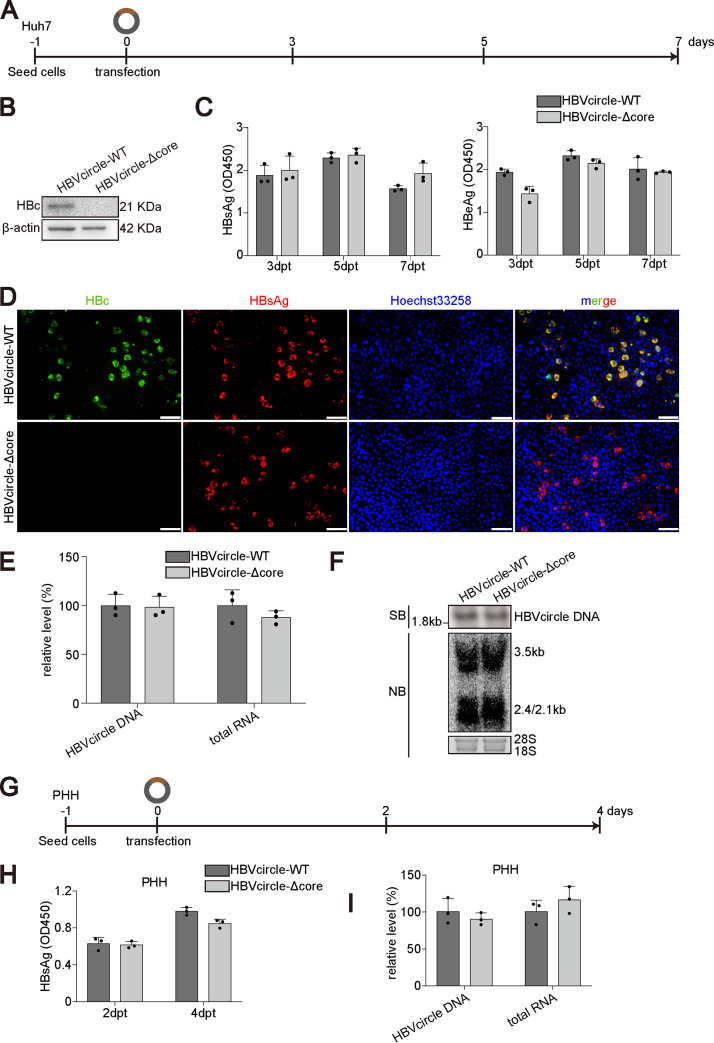
One major concern about previous studies performed on hepatocytes infected with mutant HBV virions deficient in HBc expression is that the role of HBc derived from incoming virions could not be excluded (13–15). To this end, we took advantage of HBVcircle, a close mimic of HBV cccDNA which supports cccDNA-dependent transcription (16). As shown in Fig. 1A, HBVcircle with a wild-type HBV genome (HBVcircle-WT) or an HBc-deficient HBV genome (HBVcircle-Δcore) was produced and transfected into hepatocytes. As expected, only HBVcircle-WT but not HBVcircle-Δcore resulted in HBc expression (Fig. 1B). Interestingly, the two HBVcircles led to similar amounts of secreted HBeAg and HBsAg (Fig. 1C). Immunostaining results confirmed that while HBVcircle-Δcore transfection could not produce HBc, the expression of HBsAg was comparable to that of HBVcircle-WT at the single-cell level (Fig. 1D). Intracellularly, quantitative PCR (qPCR) revealed that equal amounts of HBVcircle-WT and HBVcircle-Δcore were transfected and similar amounts of HBV RNA were transcribed (Fig. 1E). These results were further confirmed by Southern and Northern blotting (Fig. 1F). Since hepatoma cells could not entirely represent the physiological situation due to impaired signaling, we then conducted the same experiment in primary human hepatocytes (PHHs) (Fig. 1G) (17). Similarly, HBVcircle-WT and HBVcircle-Δcore resulted in equal amounts of HBsAg (Fig. 1H) and intracellular HBV RNA (Fig. 1I).
FIG 1.
Comparison of cccDNA transcription with and without HBc in HBVcircle transfection model. (A) Schematic representation of the experimental setting. Huh7 cells were transfected with HBVcircle-WT or HBVcircle-Δcore in 48-well plates, and the culture supernatants were collected at 3 days, 5 days, and 7 days posttransfection (dpt). (B) The expression of HBc protein was detected by Western blotting. (C) The levels of HBeAg and HBsAg in supernatants were measured by ELISA. OD450, optical density at 450 nm. (D) The levels of HBsAg and HBc protein in HBVcircle-transfected Huh7 cells were evaluated by immunofluorescence staining. Bar, 50 μm. (E) The relative levels of HBV RNA and HBVcircle DNA were determined by qPCR. (F) HBVcircle DNA and RNA were detected by Southern and Northern blotting, respectively. (G) Schematic representation of the experimental setting, PHHs were transfected with HBVcircle-WT or HBVcircle-Δcore in 48-well plates, and the culture supernatants were collected at 2 days and 4 days posttransfection (dpt). (H) The levels of HBsAg in supernatants were evaluated by ELISA after 1:5 dilution. (I) The relative levels of intracellular HBV RNA and HBVcircle DNA were determined by qPCR. Data were presented as mean ± SD. SB, Southern bloting; NB, Northern bloting.
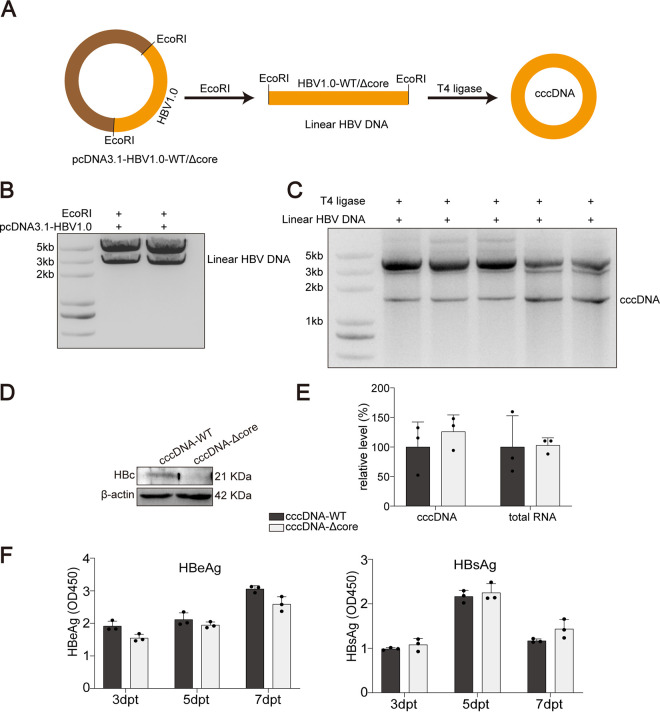
As HBVcircle was generated through plasmid recombination in a genetically engineered Escherichia coli strain, a small attR site was introduced in the HBV cccDNA sequence at the recombination site (16). To exclude the effect of this insertion, authentic HBV cccDNA was produced through circularization of the linear HBV genome cut from the HBV construct as previously described (Fig. 2A) (11). The production and isolation of cccDNA were determined (Fig. 2B and C). Wild-type HBV cccDNA (cccDNA-WT) or HBc-deficient HBV cccDNA (cccDNA-Δcore) was transfected into hepatocytes. Western blotting confirmed the phenotype of HBc deficiency of cccDNA-Δcore (Fig. 2D). In agreement with the results from HBVcircle, comparable transcriptional activities of these two cccDNA molecules were observed (Fig. 2E). In addition, similar amounts of HBeAg and HBsAg were produced by cccDNA-WT and cccDNA-Δcore (Fig. 2F).
FIG 2.
Comparison of cccDNA transcription with and without HBc in cccDNA transfection model. (A) Plasmids with HBV1.0-fold genome were digested by restriction enzyme EcoRI to produce linear HBV DNA. (B and C) For obtaining cccDNA in vitro, linear HBV DNAs were self-linked by using T4 ligase. DNA gel electrophoresis was used to detect and isolate linear HBV DNA (B) and cccDNA (C). (D) Huh7 cells were transfected with cccDNA-WT or cccDNA-Δcore. The expression of HBc was tested by Western blotting. (E) The relative levels of HBV RNA and HBV cccDNA were determined by qPCR. (F) The levels of HBeAg and HBsAg in supernatants were measured by ELISA. Data were presented as mean ± SD.
Together, these results suggested that a complete loss of HBc did not hamper the existing cccDNA transcription.
Overexpression or reconstitution of HBc has no impact on cccDNA transcription.
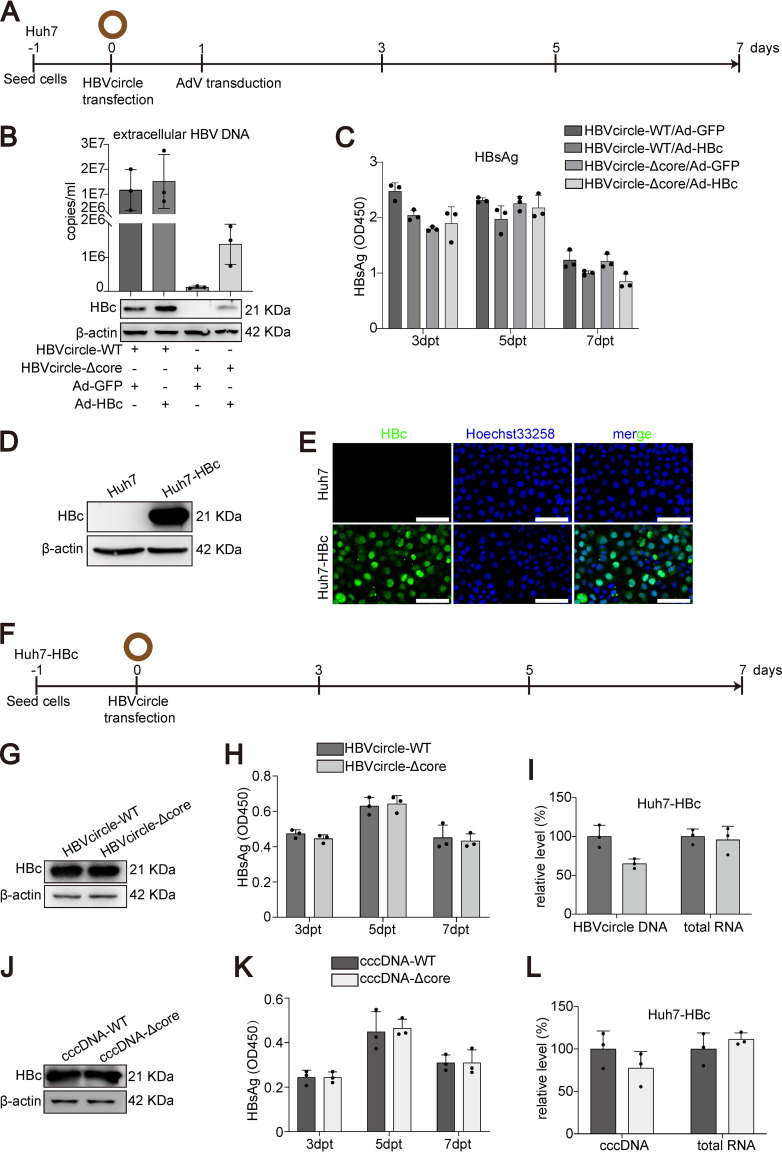
To further dissect the possible role of HBc in cccDNA transcription, we conducted HBc overexpression or reconstitution experiments. Huh7 cells were first transfected with HBVcircle-WT or HBVcircle-Δcore, followed by adenoviral vector (AdV) transduction to overexpress HBc, or green fluorescent protein (GFP) as a control (Fig. 3A). As shown in Fig. 3B, only HBVcircle-WT but not HBVcircle-Δcore could result in the production of HBV particles, as demonstrated by HBV DNA in the cell culture supernatant. Interestingly, overexpression of HBc mediated by AdV partially rescued HBV DNA secretion, supporting the essential role of HBc in HBV production (Fig. 3B). However, the secretion of HBsAg showed no difference among four groups (Fig. 3C). These data further questioned a role of HBc in cccDNA transcription.
FIG 3.
Effect of HBc overexpression or reconstitution on cccDNA transfection models. (A) Schematic representation of the experimental setting. Huh7 cells were transfected with HBVcircle-WT or HBVcircle-Δcore in 24-well plates, the cells were transduced with AdV (MOI = 1) for 24 h, and the culture supernatants were collected at 3 days, 5 days, and 7 days posttransfection (dpt). (B) HBV DNA from supernatants was determined by qPCR, and the expression of HBc was detected by Western blotting. (C) The level of HBsAg in cell supernatants was measured by ELISA. (D) The expression of HBc was detected by Western blotting in Huh7 and Huh7-HBc cells. (E) HBc immunostaining was performed. HBc is shown in green, cell nuclei are stained with Hoechst 33258 in blue. Bar, 50 μm. (F) Schematic representation of the experimental setting. Huh7-HBc cells were transfected with HBVcircle-WT or HBVcircle-Δcore in 24-well plates, and the culture supernatants were collected at 3 days, 5 days, and 7 days posttransfection. (G) The expression of HBc in HBVcircle-transfected Huh7-HBc cells was tested by Western blotting. (H) The level of secreted HBsAg was evaluated by ELISA. (I) Intracellular HBV RNA and HBVcircle DNA were measured by qPCR. (J) cccDNA-WT or cccDNA-Δcore was transfected into Huh7-HBc cells. Detection of HBc expression was performed by Western blotting. (K) HBsAg from supernatants of Huh7-HBc cells transfected with cccDNA-WT or cccDNA-Δcore at 3, 5, and 7 dpt was measured by ELISA. (L) The relative levels of HBV RNA and HBV cccDNA normalized to GAPDH mRNA or PRNP DNA, respectively, at 7 dpt were determined by qPCR. Data were presented as mean ± SD.
To obtain a more sustained HBc expression covering all the cells, Huh7-HBc, a stable cell line expressing HBc, was established. The expression of HBc was determined by Western blotting (Fig. 3D) and immunostaining (Fig. 3E). Huh7-HBc cells were transfected with HBVcircle-WT or HBVcircle-Δcore, and different HBV replication markers were evaluated (Fig. 3F). Western blotting showed that HBc was overexpressed in Huh7-HBc cells at a much higher level than HBVcircle-WT-derived HBc, as no obvious difference was observed between HBVcircle-WT and HBVcircle-Δcore transfection (Fig. 3G). Kinetics of HBsAg expression revealed that the two groups expressed similar amounts of viral antigens over time (Fig. 3H). When equal amounts of HBVcircle were used to transfect Huh7-HBc cells, comparable levels of HBV RNA were detected (Fig. 3I). In keeping with that, consistent results were obtained from cccDNA-WT- and cccDNA-Δcore-transfected Huh7-HBc cells (Fig. 3J to L).
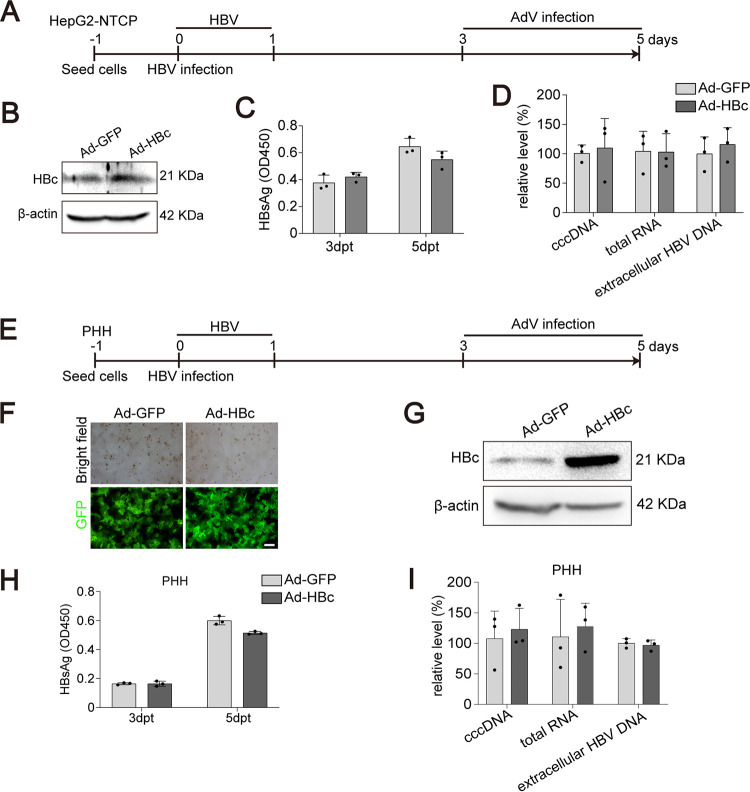
We further tested the role of additional exogenous HBc in HBV-infected cell cultures. HepG2-NTCP cells were first infected with HBV. After 3 days to allow cccDNA establishment, cells were transduced with AdV expressing HBc or GFP (Fig. 4A). Western blotting demonstrated successful HBc overexpression (Fig. 4B). As expected, additional HBc neither affected HBsAg (Fig. 4C) nor influenced intracellular HBV RNA and extracellular HBV DNA (Fig. 4D). Similar results were obtained from HBV-infected PHHs (Fig. 4E to I).
FIG 4.
Effect of HBc overexpression in HBV infection models. (A) Schematic representation of the experimental setting. HepG2-NTCP cells were infected with HBV (MOI = 100). Three days postinfection, the cells were transduced with AdV (MOI = 1) for 48 h. The culture supernatants were collected at 3 days and 5 days postinfection. (B) The expression of HBc was evaluated by Western blotting. (C) The level of extracellular HBsAg was measured by ELISA. (D) The relative levels of intracellular HBV RNA, HBV cccDNA, and extracellular HBV DNA were determined by qPCR. (E) Schematic representation of the experimental setting. PHHs were infected with HBV (MOI = 100). Three days postinfection, the cells were transduced with AdV (MOI = 1) for 48 h. The culture supernatants were collected at 3 days and 5 days postinfection. (F) Transduction efficiency of adenovirus was evaluated by fluorescence intensity of GFP. Bar, 50 μm. (G) The expression of HBc in adenovirus-transduced cells was detected by Western blotting. (H) The level of secreted HBsAg was evaluated by ELISA. (I) Intracellular HBV RNA, HBV cccDNA, and extracellular HBV DNA were measured by qPCR. Data were presented as mean ± SD.
Taken together, overexpression or reconstitution of HBc does not change cccDNA-derived HBV markers, further elaborating that HBc is dispensable for cccDNA transcriptional regulation.
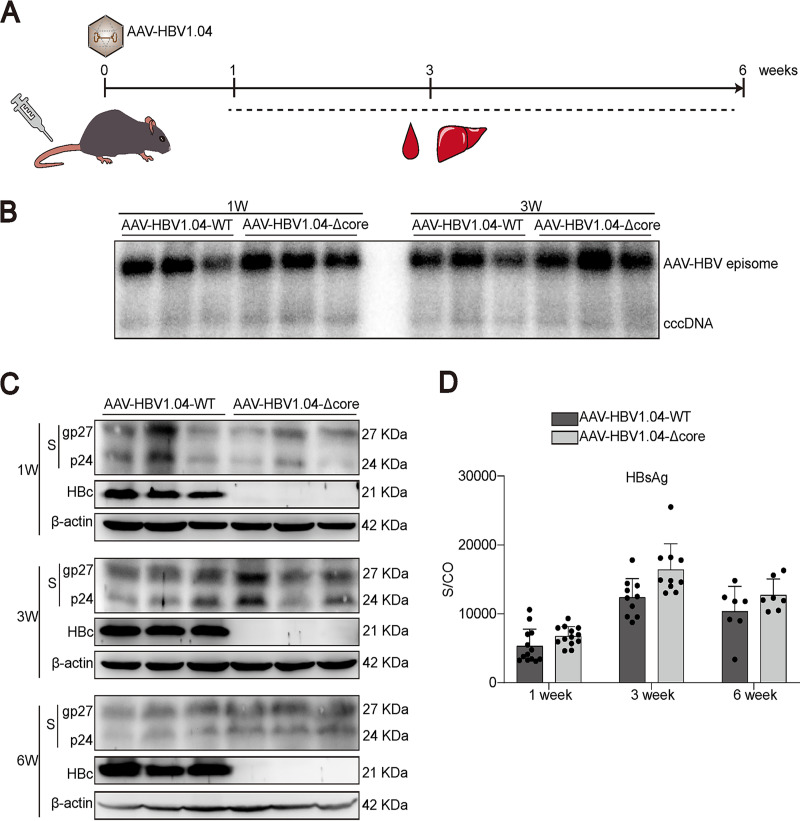
HBc is not required for cccDNA transcription in vivo.
Considering the possible differences between cells cultured in vitro and the hepatocytes in vivo, we continued to evaluate the role of HBc using a novel mouse model harboring HBV cccDNA (18). Briefly, an AAV vector carrying a wild-type or HBc-deficient HBV1.04-fold genome (AAV-HBV1.04-WT and AAV-HBV1.04-Δcore, respectively) was constructed. Owing to the discontinuity of the S gene on the linear HBV genome, AAV-HBV1.04 itself cannot express HBsAg. After transduction of AAV-HBV1.04 into mouse, cccDNA is formed through recombination and HBsAg can be produced through transcription of cccDNA (18). Thus, HBsAg in this model can serve as a marker for cccDNA activity.
We injected mice with the recombinant AAV-HBV1.04-WT or AAV-HBV1.04-Δcore through the tail vein and collected different samples at the indicated time points (Fig. 5A). Southern blotting demonstrated that amounts of HBV cccDNA comparable between the two groups were established in the livers (Fig. 5B). Western blotting results showed that while AAV-HBV1.04-Δcore-transduced mice could not produce HBc in the liver, the expression of HBsAg was comparable with that in AAV-HBV1.04-WT-transduced mice (Fig. 5C). Consistent with this observation, the two groups showed similar serum HBsAg kinetics (Fig. 5D).
FIG 5.
Comparison of cccDNA transcription with and without HBc in vivo. (A) Female C57BL/6 mice (6 to 8 weeks old) were transduced with AAV-HBV1.04 at 1 × 1011 viral genome (vg) equivalents through tail vein injection. Samples were collected at the indicated time points. (B) HBV cccDNA in mouse liver was analyzed by Southern blotting. (C) The expression of HBc and HBsAg in mouse liver was evaluated by Western blotting. (D) The kinetics of HBsAg in mouse serum were detected. Data were presented as mean ± SD.
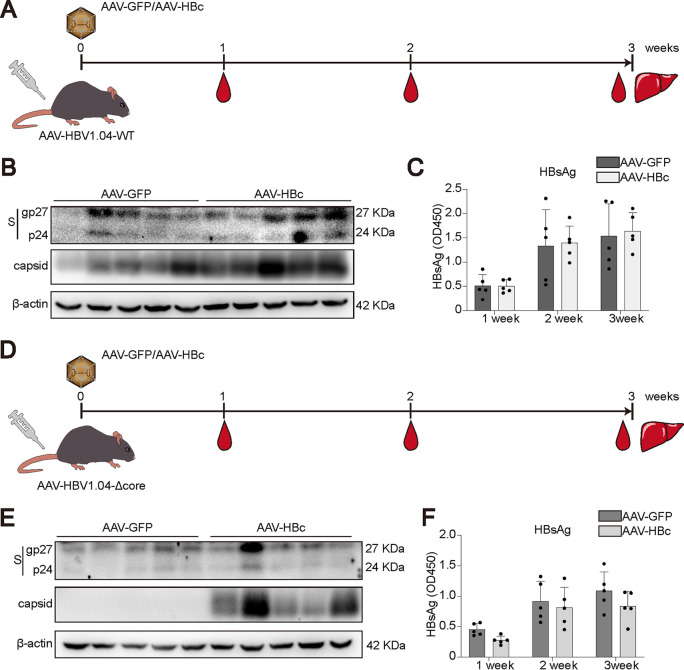
To further determine the role of HBc in vivo, AAV-GFP or AAV-HBc was injected together with AAV-HBV1.04-WT (Fig. 6A). Western blotting showed that although the AAV-HBc group generated more capsids, the expression of HBsAg was not altered by HBc (Fig. 6B). The kinetics of HBsAg confirmed that the secretion levels of HBsAg were comparable between the AAV-GFP and AAV-HBc groups (Fig. 6C). Similar experiments were conducted with AAV-HBV1.04-Δcore-transduced mice (Fig. 6D). As expected, HBV capsid could be detected only in the AAV-HBc group (Fig. 6E). In agreement with results for AAV-HBV1.04-WT-transduced mice, AAV-HBc did not affect the expression of HBsAg (Fig. 6E and F). In conclusion, all these data support the concept that HBc is not required for cccDNA transcription.
FIG 6.
Effect of HBc overexpression or reconstitution on cccDNA transcription in vivo. (A) AAV-HBV1.04-WT was cotransduced into C57BL/6 mice with AAV-GFP or AAV-HBc at 1 × 1011 viral genome (vg) equivalents through tail vein injection, and samples were collected at the indicated time points. (B) Liver samples were collected and subjected to Western blotting for cellular HBsAg and capsid detection. (C) The level of HBsAg in mouse serum was tested by ELISA after 1:1,000 dilution. (D) AAV-HBV1.04-Δcore was cotransduced into C57BL/6 mice with AAV-GFP or AAV-HBc at 1 × 1011 viral genome (vg) equivalents through tail vein injection, and samples were collected at the indicated time points. (E) The expression of HBsAg and capsid was detected by Western blotting. (F) The level of HBsAg in mouse serum was evaluated by ELISA after 1:1,000 dilution. Data were presented as mean ± SD.
HBc does not change major transcription regulators of cccDNA.
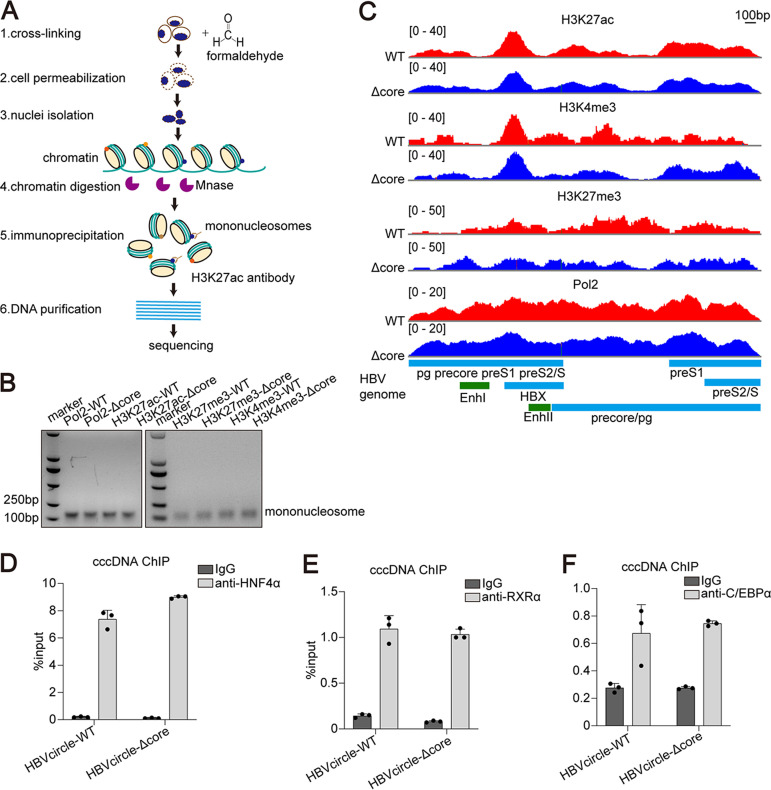
Histones with various modifications are associated with transcriptionally active or repressive HBV cccDNA minichromosome to regulate its transcription. We next investigated whether HBc could affect histone modifications in HBV cccDNA. Huh7 cells were transfected with either HBVcircle-WT or HBVcircle-Δcore, and the cells were harvested at day 3 for HBV cccDNA chromatin immunoprecipitation (ChIP) followed by massive parallel sequencing (Fig. 7A). The HBVcircle was digested into mononucleosomes (Fig. 7B). We selected two reported histone posttranslational modifications associated with active transcription, H3K4me3 and H3K27ac, and one known modification associated with gene silencing, H3K27me3 (19). In addition, RNA polymerase II (Pol2), which is the major enzyme engaging in cccDNA transcription, was also analyzed for its distribution in HBVcircle (4). As shown in Fig. 7C, the mapping of H3K27ac and H3K4me3 showed very similar patterns in HBVcircle-WT and HBVcircle-Δcore: they were both enriched at the transcription start site of X (between enhancer I and enhancer II) and pre-S1 genes. In contrast, H3K27me3 showed no typical enrichment in both HBVcircles (Fig. 7C). The enrichment of RNA polymerase II was found on the entire sequences of HBVcircle-WT and HBVcircle-Δcore with a higher signal at the transcription start sites of X and the pre-S1 gene (Fig. 7C). These results suggested that HBVcircle-Δcore shared similar histone modification patterns and RNA polymerase II-mediated transcription activities with HBVcircle-WT.
FIG 7.
ChIP-Seq and ChIP-PCR analysis of HBVcircle-transfected cells. (A) Schematic of cccDNA ChIP-Seq assay. Histone modifications are located at specific sites within nucleosomes in cccDNA and cellular chromatin. Huh7 cells transfected with HBVcircle DNA were digested with micrococcal nuclease (1,500 U/mL), and resulting mononucleosomes were released from the cell nucleus. Several specific antibodies, such as H3K27ac antibody, were applied to enrich nucleosomes by ChIP assay. The associated DNA extracted with a genome extraction kit was sequenced by next-generation sequencing. (B) Nuclei isolated from HBVcircle-transfected Huh7 cells were incubated with MNase (1,500 U/mL). Sample DNA was extracted, separated by agarose gel electrophoresis, and visualized by ethidium bromide staining. (C) Distributions of nucleosomes and posttranslational modifications along the HBV genome are shown. Relative read density for each track is represented by height on the y axis. HBV transcripts and enhancer elements are represented on the x axis. (D to F) ChIP-PCR assays were performed with antibodies to HNF4α (D), RXRα (E), and C/EBPα (F) or nonspecific IgG antibody control. HBV cccDNA-specific primers were used for qPCR amplification. Data were presented as mean ± SD.
In addition, the associations of cccDNA with three known transcription factors which are important for HBV cccDNA activities were evaluated by ChIP PCR. As the results show, HNF4α (Fig. 7D), RXRα (Fig. 7E), and C/EBPα (Fig. 7F) shared similar abundances on both HBVcircle-WT and HBVcircle-Δcore.
Overall, our ChIP sequencing (ChIP-Seq) and ChIP PCR data indicate that the HBc-deficient cccDNA chromatin showed histone modifications and a transcription factor profile similar to those of wild-type cccDNA. In addition, HBc is not required for the binding of transcription factors and RNA polymerase II on cccDNA.
CAMs could not alter cccDNA transcription.
CAMs are a novel drug class that was initially developed to target virus assembly and may have a potential impact on the functional cure (5). Recently, interesting questions regarding their alternative antiviral mechanisms are under active discussion. One such proposed mechanism is that CAMs can inhibit cccDNA transcription (20).
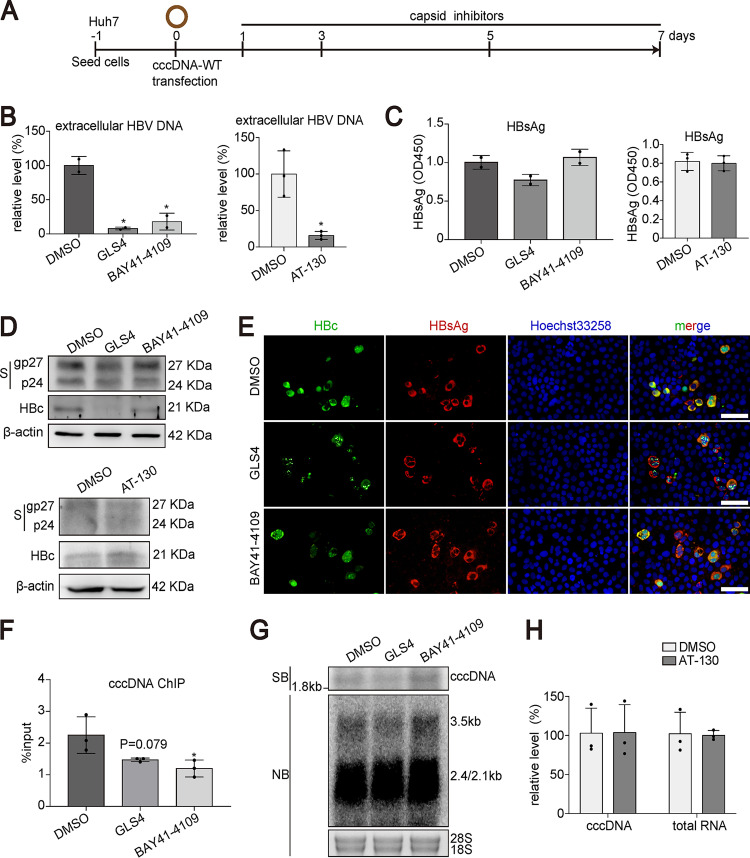
To address this question, we transfected Huh7 cells with cccDNA-WT and treated the cells with three CAMs, GLS4, BAY41-4109, and AT-130 (Fig. 8A). As expected, CAM treatment significantly reduced HBV DNA levels generated from cccDNA-WT (Fig. 8B). Both enzyme-linked immunosorbent assay (ELISA) and Western blotting demonstrated that CAM treatment had no effect on HBsAg (Fig. 8C and D). Immunostaining revealed altered HBc signal after GLS4 or BAY41-4109 treatment (Fig. 8E). However, HBsAg production was not altered by GLS4 or BAY41-4109 in cccDNA-WT-transfected cells (Fig. 8E). We next performed cccDNA ChIP to determine whether CAMs could influence the amount of HBc on cccDNA. As shown in Fig. 8F, both GLS4 and BAY41-4109 treatment led to reduced cccDNA signal in the ChIP assay using antibody specific for HBc. However, a smaller amount of HBc on cccDNA did not affect cccDNA transcription or cccDNA stability within the experimental time as demonstrated by Southern and Northern blotting (Fig. 8G). Along the same line, AT-130 treatment did not affect cccDNA transcription (Fig. 8H).
FIG 8.
Effect of CAMs on cccDNA transcription in cccDNA transfection model. (A) Schematic representation of the experimental setting. Huh7 cells were transfected with cccDNA-WT in 48-well plates. One day posttransfection, the cells were treated with BAY41-4109 (5 μM), GLS4 (0.2 μM), AT-130 (5 μM), or DMSO and harvested at 7 days posttransfection. (B) Extracellular HBV DNA from supernatants at 7 dpt was determined by qPCR. (C) The level of secreted HBsAg at 7 dpt was evaluated by ELISA. (D) The expression of intracellular HBsAg and HBc was detected by Western blotting. (E) The levels of HBsAg and HBc proteins were evaluated by immunofluorescence staining. Bar, 50 μm. (F) ChIP PCR assays were performed with antibody against HBc. HBV cccDNA-specific primers were used for qPCR. (G) HBV cccDNA and RNA were subjected to Southern blotting and Northern blotting, respectively. (H) Intracellular HBV RNA and HBV cccDNA were measured by qPCR. Data were presented as mean ± SD. *, P < 0.05; SB, Southern bloting; NB, Northern bloting.
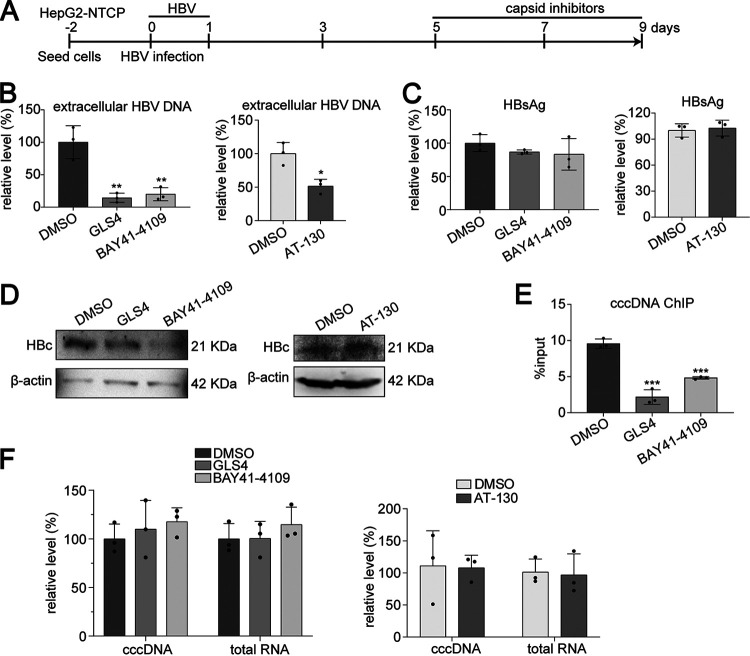
We further investigated the effects of CAMs in the HBV-infected cell culture model. Five days after HBV infection, HepG2-NTCP cells were treated with GLS4, BAY41-4109, or AT-130, and different markers were assessed (Fig. 9A). Consistent with previous observation, CAMs significantly reduced HBV DNA (Fig. 9B) but not HBsAg (Fig. 9C) levels. HBc was determined by Western blotting (Fig. 9D). Although the amounts of HBc on cccDNA were significantly suppressed by GLS4 or BAY41-4109 (Fig. 9E), quantification of HBV RNA further proved that transcription of cccDNA was not affected (Fig. 9F).
FIG 9.
Effect of CAMs on cccDNA transcription in HBV infection model. (A) HepG2-NTCP cells were infected with HBV (MOI = 100). Five days postinfection, cells were treated with BAY41-4109 (5 μM), GLS4 (0.2 μM), AT-130 (5 μM), or DMSO and harvested at 9 days postinfection. (B and C) Extracellular HBV DNA (B) and HBsAg (C) from supernatants at 9 days postinfection were detected by qPCR and ELISA, respectively. (D) Intracellular HBc was detected by Western blotting. (E) ChIP PCR assays were performed with antibody against HBc. HBV cccDNA-specific primers were used for qPCR. (F) Intracellular HBV cccDNA and RNA were measured by qPCR. Data were presented as mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Altogether, our data strongly support the idea that although CAMs can reduce the HBc amount on cccDNA, they cannot influence cccDNA transcription.
DISCUSSION
Our current understanding of the HBV life cycle, especially cccDNA transcriptional regulation, is still largely vague. Previous studies reported controversial roles of HBc in cccDNA transcription. Important knowledge has been gained using the HBV-Δcore mutant virus infection model. However, essential limitations in such models do exist. During the production of HBV-Δcore mutant virus, complementary HBc has to be provided to enable the mutant virion assembly. Thus, the potential effect of HBc from the incoming virion on cccDNA cannot be teased out, which has been a hurdle for a long time preventing us from explicitly defining the role of HBc. In this study, we used synthesized HBV cccDNA or HBVcircle transfection models and AAV-mediated HBV cccDNA mouse models to dissect the function of HBc in cccDNA transcription. By comparing HBV replication markers from HBc-deficient cccDNA with those from wild-type cccDNA, we demonstrated that HBc is not required for cccDNA transcriptional regulation. In addition, overexpression of HBc did not alter wild-type or HBc-deficient cccDNA transcription. Furthermore, ChIP revealed similar epigenetic regulation of cccDNA with and without HBc. Finally, CAM treatment reduced the HBc amount on cccDNA, but there was no impact on cccDNA transcription. In sum, our data support the notion that HBV core protein is not required for cccDNA transcriptional regulation.
As the structural protein that is able to self-assemble to form the viral capsid, HBc is a polypeptide with an assembly domain followed by a linker region and an arginine-rich CTD. Besides its function as the building block of the HBV nucleocapsid, HBc is known to play important functions in multiple stages of the HBV life cycle. HBc regulates transport and nuclear release of the viral genome (21). It binds to the CpG islands of HBV cccDNA (10). The CTD of HBc is required for viral pgRNA encapsidation and productive viral positive-strand DNA synthesis (22, 23). Furthermore, the persistence of cccDNA may involve recycling of nucleocapsids into the nucleus, during which HBc is also indispensable (24).
Reports on subcellular localization of HBc in chronically infected patients demonstrated both cytoplasmic and nuclear distribution (25–27). An in vitro study suggested that nuclear localization of the HBV core protein was negatively regulated by phosphorylation during the cell cycle (28). Given that the CTD of HBc contains an arginine-rich region which binds to nucleic acids due to its positive charge, CTD is considered a nucleic acid chaperone (22, 23). In addition, the HBc CTD facilitated annealing and unwinding of DNA and enhanced the in vitro cleavage activity of the RNA substrate by a hammerhead ribozyme (29). Based on its nucleic acid binding ability, HBc was believed to act as a regulator of transcription. HBc has the ability to bind a large number of human gene promoters and thus may disrupt their expression (30). HBc was reported to enhance the transcription activation of cAMP response element (CRE) via the CRE/CREB/CBP pathway (31). HBc may also function as a transcriptional repressor. Transcriptional repression of the human p53 gene by HBc in human liver cells was observed previously (32). HBc may also protect hepatocytes from tumor necrosis factor-alpha-related apoptosis-inducing ligand (TRAIL)-induced apoptosis by blocking DR5 expression through repressing its promoter (33). In addition, HBc has been shown to downregulate MxA, an interferon-stimulated gene, by interacting with its promoter in hepatocytes (34). However, most of these studies were based on overexpression of HBc in hepatoma cell lines. Whether the conclusions have physiological relevance is questionable.
Recently, the effect of HBc on HBV cccDNA has been gaining more attention. Electron microscopy and ChIP studies revealed that HBc is a component of the cccDNA minichromosome (9, 35). HBc binds to cccDNA and reduces the nucleosome spacing of the cccDNA-histone complex, which may regulate HBV transcription by altering the nucleosomal arrangement of the HBV genome (9). By analyzing liver biopsy samples of 22 chronic hepatitis B patients, Guo et al. demonstrated that HBc preferentially bound to the CpG islands in cccDNA (10). In the overexpression model, HBc was reported to be responsible for the recruitment of histone acetyltransferases to cccDNA minichromosome and to subsequently regulate cccDNA transcription (11). In contrast, HBc is not required for HBV transcription in the monomeric linear HBV genome transfection assay (36). The discordant results may be caused by different experimental conditions. In HBV-infected cell culture models, HBc-deficient and wild-type HBV virions resulted in comparable cccDNA, HBV RNA, and HBsAg amounts (13, 14). These studies implied that de novo-synthesized HBc plays a minor role in transcriptional regulation of cccDNA. However, the role of HBc from the incoming virion remains uncertain. In the present study, by using in vitro and in vivo systems that support cccDNA-initiated HBV transcription, we demonstrated that HBc could affect neither epigenetic status nor transcriptional activity of cccDNA.
A previous study on DHBV showed that a mutant DHBV deficient in HBc expression results in significantly less viral RNA, suggesting a role of HBc in DHBV cccDNA transcription regulation (12). This observation contrasts with HBV infection. These contradictory findings on the role of HBc in DHBV and HBV cccDNA transcription regulation may be caused by the distinct structures and biological functions of the avian and human HBV core in the viral life cycle (20). DHBV HBc is approximately 80 amino acids longer than HBV HBc, and the function of the DHBV HBc N terminus is different from that of the corresponding domain of the HBV HBc (37). As DHBV lacks X protein, it had been speculated that DHBV HBc may have an HBx-like function (20). A key function of HBx is to redirect the DNA damage binding protein 1-CUL4 E3 ubiquitin ligase to target SMC5/6 for degradation and thus relieve the suppression and activate the transcription of cccDNA (38, 39). Whether DHBV cccDNA transcription activates through a similar mechanism and whether DHBV HBc can act like HBx protein are currently unknown.
The association of HBc with cccDNA is very stable in experimental models (15). In addition, HBc has the potential to recruit APOBEC3A to cccDNA for its degradation (40, 41). These observations suggest that HBc may be involved in the maintenance or stabilization of the cccDNA minichromosome. Although HBc expression is not required for cccDNA stability or transcription within 9 weeks of in vitro infection, the virion-derived HBc can associate with cccDNA in the absence of de novo synthesis of viral proteins (14, 15). In our study, the long-term stability of HBV cccDNA with or without HBc was not determined. As our AAV-HBV1.04 mouse model can support HBV cccDNA formation and long-term maintenance, it will provide a unique platform for answering this question (18).
In our study, transfection of cccDNA-like molecules may result in higher copy numbers of DNAs and potential induction of innate immune responses. Furthermore, it is unknown whether these cccDNA-like molecules respond to HBc in the same way as authentic cccDNA. It is a technical challenge to produce cccDNA without HBc in HBV infection models. At least, additional exogenous HBc showed no effect on cccDNA formed during HBV infection, as we demonstrated here.
Our observations are of particular relevance as several drugs targeting HBc are being clinically evaluated and will likely become available in the near future. Concerning the effects of CAMs on the nuclear function of HBc, one study showed that prolonged treatment of HBV-infected HepaRG cells with two CAMs, JNJ-827 and JNJ-890, can reduce HBV RNA and HBsAg production (42). However, whether these two compounds have additional effects other than targeting HBc is not clear. Though HBV core protein has been shown to be associated with the cccDNA minichromosome, in accordance with our results, CAM treatment did not reduce the preexisting cccDNA copy numbers or transcription in HBV-infected humanized mice (43). As CAMs are able to suppress virion production and subsequently also inhibit spreading of or new infection by HBV, it is anticipated that the long-term effect of these capsid inhibitors may result in reduced viral cccDNA and RNA in patients. Nevertheless, to achieve a higher rate of HBV cure, novel therapeutics targeting cccDNA are needed.
MATERIALS AND METHODS
Plasmid constructs.
HBc-null plasmids (pcDNA3.1-HBV1.0-Δcore and pAAV-HBV1.04-Δcore) derived from parental plasmids (pcDNA3.1-HBV1.0 and pAAV-HBV1.04, respectively) were generated by site-directed mutagenesis using the oligonucleotide CTTTGGGGCATAGACATCGACCCTTATAAAGAATTTG, corresponding to HBV nucleotides 1894 to 1930. In that way, the core AUG was altered to AUA by mismatch at the A site.
Cell culture and transient transfection.
The Huh7, Huh7-HBc, HepG2-NTCP, and HEK293T cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Lonsera, Uruguay), 1% penicillin-streptomycin (P/S; Gibco, USA), at 37°C in a 5% CO2 incubator. Primary human hepatocytes (PHHs) were purchased from Liver Biotechnology (Shenzhen) Co., Ltd. To generate a stable Huh7-HBc cell line, HBc coding sequence (CDS) was cloned into lentiviral vector pWPI-BSD. To produce lentiviral particles, HEK293T cells cultured in T-25 flasks were cotransfected with 5 μg lentivirus plasmid, 3.75 μg psPAX2 plasmid, and 1.25 μg pMD2.G plasmid using PEI MAX 40K according to the manufacturer’s instructions. The supernatants containing lentivirus were collected 72 h posttransfection. Huh7 cells were transduced by lentivirus and selected in the presence of 10 μg/mL blasticidin S hydrochloride (Solarbio, China). Lipofectamine 2000 transfection reagent (Invitrogen, USA) and FuGENE HD transfection reagent (Promega, USA) were used for HBVcircle and cccDNA transfection, respectively, according to the manufacturer’s instructions.
HBVcircle and cccDNA synthesis in vitro.
For HBVcircle production in vitro, minicircle DNA technology (Roche) was used according to the manufacturer’s protocols (16). Original constructs were gifted by Roche R&D Center (China) Ltd. with a material transfer agreement.
To obtain cccDNA in vitro, linear HBV DNA was acquired by EcoRI restriction digestion from pcDNA3.1-HBV1.0 and self-linked by using T4 ligase at 1 ng/μL. Circularized HBV DNA was purified by agarose gel electrophoresis and DNA gel extraction.
Adeno-associated viral vector preparation.
Adeno-associated viral vector production was performed as described above. In brief, plasmids (pAAV-HBV1.04, pAAV2/8-RC, and pHelper in a 1:1:1 molar ratio) were cotransfected into HEK293T cells seeded in a 150-mm plate, and 72 h posttransfection, the cells and supernatants were harvested and filtered with an 0.22-μm filter. Then, the cells and supernatants were concentrated with an Amicon Ultra-15 centrifugal filter unit, and virus yield was quantified by quantitative PCR (qPCR) using targeting HBV genome primers. Primer sequences are listed in Table 1.
TABLE 1.
Primer sequences for qPCR
| Oligonucleotide name | Sequence order (5′–3′) |
|---|---|
| HBV total RNA fwd | CCGTCTGTGCCTTCTCATCTGC |
| HBV total RNA rev | ACCAATTTATGCCTACAGCCTCC |
| cccDNA 92 fwd | GCCTATTGATTGGAAAGTATGT |
| cccDNA 2251 rev | AGCTGAGGCGGTATCTA |
| Human GAPDH fwd | GCACAAGAGGAAGAGAGAGACC |
| Human GAPDH rev | AGGGGAGATTCAGTGTGGTG |
| Human PRNP fwd | TGCTGGGAAGTGCCATGAG |
| Human PRNP rev | CGGTGCATGTTTTCACGATAGTA |
| AAV-GFP-F | CCCGACAACCACTACCTGAG |
| AAV-GFP-R | GTCCATGCCGAGAGTGATCC |
| AAV-HBc-F | TTGGTGTCTTTCGGAGTGTG |
| AAV-HBc-R | AGGGGCATTTGGTGGTCTAT |
Drugs and inhibitors.
BAY41-4109 racemate (HY-100029A), GLS4 (HY-108917), and AT-130 (HY-100028) were purchased from MedChemExpress (MCE; USA).
Virus infection and transduction experiment.
For HBV infection, HepG2-NTCP cells and PHHs were cultured in medium with 2.5% dimethyl sulfoxide (DMSO) for 2 days. The cells were infected with 100 HBV genomes (multiplicity of infection [MOI] = 100) per cell diluted in medium with 4% polyethylene glycol 8000 (PEG 8000). One day postinfection, cells were washed three times with phosphate-buffered saline (PBS) and treated further as indicated for the following experiment.
For AAV transduction, AML12 cells were seeded in 6-well plates and transduced with 50,000 viral genomes (vg) per cell in the presence of 4% PEG 8000 for 24 h. One day postransduction, cells were washed three times with PBS and cultured further as indicated for each experiment.
For adenoviral vector transduction, Huh7 cells were infected with 1 viral genome (multiplicity of infection [MOI] = 1) per cell diluted in DMEM supplemented with 10% fetal bovine serum (FBS) (Lonsera, Uruguay), 1% penicillin-streptomycin (P/S) (Gibco, USA), for 24 h. One day postinfection, cells were washed three times with PBS and treated further as indicated for the following experiment.
Mouse experiments.
C57BL/6 mice were purchased from China Three Gorges University Laboratory Animal Center (Yichang, China). Six- to 8-week-old female mice were used in the experiments. The wild type of AAV-HBV1.04 (AAV-HBV1.04-WT), the core-null form of AAV-HBV1.04 (AAV-HBV1.04-Δcore), AAV-CMV-GFP (AAV-GFP), and AAV-CMV-HBc (AAV-HBc) were delivered into mice through tail vein injection, with 1 × 1011 viral genomes diluted in 200 μL PBS per mouse. The blood sample (orbital blood) and liver tissue were harvested at indicated time points. Serum samples were obtained after centrifugation at 3,500 × g for 15 min at 4°C to examine HBsAg. Mouse liver was used for the detection of HBV cccDNA, HBc, capsid, and intracellular HBsAg. The protocol was approved by the ethics committee of the animal facility, Wuhan University.
Detection of HBV markers.
The levels of HBeAg and HBsAg from the cell supernatants and serum were evaluated with an enzyme-linked immunosorbent assay (ELISA) kit (Kehua Bio-Engineering Co., Shanghai, China) according to the manufacturer’s instructions. Serum HBsAg levels were measured at Renmin Hospital of Wuhan University (Wuhan, China) by electrochemiluminescent immunoassay (ELCIA) using the Elecsys HBsAg II kits (Roche, Penzberg, Germany) according to the manufacturer’s instructions, and the sample results were presented in the form of a relative cutoff index (sample/cutoff [S/CO]). Extracellular HBV DNA was tested by qPCR using an HBV DNA quantitative fluorescence diagnostic kit (Sansure Biotech, Changsha, China). Total DNA from transfected cells was extracted using a TIANamp genomic DNA kit (Tiangen Biotech Co., Beijing, China). Total RNA from transfected cells was purified using an RNApure tissue and cell kit (Cowin Biotech, Beijing, China) and transcribed into cDNA with ReverTra Ace qPCR RT master mix (Toyobo Biotech Co., Osaka, Japan). HBV total RNA and cccDNA were detected using a LightCycler 480 (Roche, Penzberg, Germany), and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA or prion protein (PRNP) DNA, respectively (44). Primer sequences are listed in Table 1.
Western blotting.
Protein samples from cells and mouse liver were extracted with protein lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 1% NP-40, 0.1% SDS, protease inhibitor) and radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma-Aldrich, USA) with protease inhibitor, respectively, and the protein concentration of the cells and liver tissues was determined by using a Bradford assay kit (Bio-Rad, USA) and a bicinchoninic acid (BCA) assay kit (Biosharp, China), respectively. Thirty micrograms of total protein was separated by SDS-PAGE using Mini-Protean Tetra (Bio-Rad, USA) and transferred into a polyvinylidene difluoride (PVDF) membrane (Millipore, USA) through a mini-Trans-Blot system (Bio-Rad, USA). At room temperature, the membrane was blocked in 5% nonfat milk for 1 h, and the membrane was incubated with the primary antibody overnight at 4°C. After incubation with secondary antibody for 1 h at 37°C, protein band signals were detected using the Immobilon Western chemiluminescent horseradish peroxidase (HRP) substrate (Millipore, USA) with a Luminescent image analyzer (Syngene, England). The information for antibodies is shown in Table 2.
TABLE 2.
Antibodies
| Antibody | Company/reference | Catalog no. | Host | Dilution(s) |
|---|---|---|---|---|
| HBc | Self-made (18) | Rabbit | 1:1,000 (Western blot); 1:20 (ChIP assay) | |
| HBc | Gene Technology | GB058629 | Rabbit | 1:50 (immunostaining) |
| HBsAg | Novus Biologicals | NB100-62652 | Rabbit | 1:2,000 (Western blot) |
| HBs (S1) | Kindly provided by Xinwen Chen (49) | Mouse | 1:50 (immunostaining) | |
| β-Actin | Cell Signaling Technology | 4970 | Rabbit | 1:10,000 (Western blot) |
| RNA polymerase II | Kindly provided by Kaiwei Liang (50) | Rabbit | 1:30 (HBVcircle ChIP-Seq) | |
| H3K27ac | Kindly provided by Kaiwei Liang | Rabbit | 1:20 (HBVcircle ChIP-Seq) | |
| H3K4me3 | Cell Signaling Technology | 9751S | Rabbit | 1:30 (HBVcircle ChIP-Seq) |
| H3K27me3 | Cell Signaling Technology | 9733S | Rabbit | 1:30 (HBVcircle ChIP-Seq) |
| HNF4α | Santa Cruz Biotechnology | sc-374229 | Mouse | 1:50 (ChIP assay) |
| RXRα | Cell Signaling Technology | 5388S | Rabbit | 1:50 (ChIP assay) |
| C/EBPα | Santa Cruz Biotechnology | sc-365318 X | Mouse | 1:50 (ChIP assay) |
| Rabbit IgG | Cell Signaling Technology | 2729 | Rabbit | 5 μg (ChIP assay) |
| Mouse IgG | Proteintech | B900620 | Mouse | 5 μg (ChIP assay) |
Detection of HBV capsid.
Detection of HBV nucleocapsid in liver tissue was performed as described previously (45). Briefly, 20 mg liver tissue was minced with a Tissue Cell-Destroyer (DS1000; Novastar, China), homogenized in capsid lysis buffer (10 mM Tris-HCl [pH 7.5], 50 mM NaCl, 1 mM EDTA, 8% sucrose, 1% NP-40, protease inhibitor), and incubated on ice for 10 min. After centrifugation at 15,800 × g for 10 min at 4°C, the supernatant was supplemented with 1.2 μL 1 M MgCl2, 4 μL 10-mg/mL DNase I, and 3 μL 100-mg/mL RNase A at 37°C for 20 min. Then, the supernatant was harvested by centrifugation at 15,800 × g for 10 min at 4°C, subjected to 1.6% (wt/vol) agarose gel electrophoresis, and transferred to a PVDF membrane by the capillary siphon method. At room temperature, the membrane was blocked in 5% nonfat milk for 1 h and incubated with the primary antibody overnight at 4°C. After incubation with secondary antibody for 1 h at 37°C, protein band signals were detected using the Immobilon Western chemiluminescent HRP substrate (Millipore, USA) by a Luminescent image analyzer (Syngene, England). The information on antibodies is shown in Table 2.
Immunofluorescence assay.
To analyze the expression of HBc and HBsAg, an immunofluorescence assay was performed as described previously (46). In brief, cells were washed with precooled PBS and fixed with 4% paraformaldehyde (Invitrogen, USA) for 20 min on ice. Fixed cells were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) for 10 min at room temperature and then blocked in PBS containing 1% goat serum for 1 h at room temperature. Then, the cells were inoculated with primary antibodies diluted with 1% blocking serum overnight at 4°C. After cells were washed with PBS, the secondary antibodies were added into cells for 1 h in the dark at room temperature. Cells were inoculated with Hoechst 33258 (Invitrogen, USA) for 5 min at room temperature, and images were captured with an Olympus CKX53 FL microscope (Olympus, Japan). The information on antibodies is listed in Table 2.
Southern blotting.
DNA from cells or homogenized liver tissues was extracted by the Hirt procedure as described previously (47). Briefly, a DNA sample (cells, 20 μg; liver tissues, 40 μg) was subjected to 1.2% agarose gel electrophoresis and transferred onto an Amersham Hybond-N+ membrane (GE Healthcare). Then, the membrane was cross-linked by UV, followed by hybridization with HBV probes prepared with the random primer DNA labeling kit, ver. 2 (TaKaRa Bio, Japan). Finally, the hybridization signal was captured with a Typhoon FLA 9500 imager (GE Healthcare Lifesciences, USA).
Northern blotting.
Total cellular RNA was extracted by TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions, and total RNA (20 μg) was subjected to denaturizing 1% (wt/vol) agarose gel electrophoresis with 2% formaldehyde reagents and transferred to Hybond XL nylon membranes by the capillary siphon method. Then, the membrane was cross-linked by UV, followed by hybridization with HBV probes prepared with the random primer DNA labeling kit, ver. 2 (TaKaRa Bio, Japan). Finally, the images were acquired by a Typhoon FLA 9500 imager (GE Healthcare Lifesciences, USA).
ChIP PCR.
The ChIP assay was performed as described previously (48). Briefly, the cells were cross-linked by 1% formaldehyde (Invitrogen, USA) for 10 min at room temperature, and a 1/20 volume of 2.5 M glycine (Sigma-Aldrich, St. Louis, MO, USA) was added to quench formaldehyde. Next, the cells were incubated in ChIP lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 1% NP-40, 0.1% SDS, protease inhibitor) on ice for 30 min. The lysates were added into protein A/G-conjugated agarose beads after protein A/G-conjugated agarose beads were incubated with indicated antibodies in a rotator overnight at 4°C. Then, the beads were collected and washed three times, immunocomplexes were eluted in elution buffer (1% SDS, 100 mM NaHCO3) and de-cross-linked at 65°C for 5 to 6 h, and the DNA was purified by a TIANamp genomic DNA kit (Tiangen Biotech Co., Beijing, China). The purified DNA was detected by qPCR amplification with primers specific for cccDNA. The information on antibodies is listed in Table 2.
ChIP-Seq assay.
The cccDNA ChIP-Seq assay was performed according to previously described procedures, with slight modifications (19). Huh7 cells transfected with HBVcircle were cross-linked by 1% formaldehyde (Invitrogen, USA) for 10 min at room temperature, and fixation was quenched by adding 125 mM (final concentration) glycine for 5 min. Next, the cells were lysed in nucleus isolation buffer (1× PBS with 0.1% Triton X-100, 0.1% NP-40, 1 mM dithiothreitol [DTT], 10 mM sodium butyrate, 1× protease inhibitor [Roche]) for 10 min at 4°C. Nuclei were pelleted after centrifugation at 9,000 × g for 3 min at 4°C and resuspended and digested in micrococcal nuclease (MNase) digestion buffer (50 mM Tris [pH 7.5], 4 mM MgCl2, 1 mM CaCl2, 10% [vol/vol] glycerol, 10 mM sodium butyrate, 1× protease inhibitor) with 1,500 U/mL MNase (New England Biolabs, Ipswich, MA) at 37°C for 1 h, and digestion was quenched with 10 mM EGTA on ice. Then, the digested nuclei were harvested by centrifugation at 9,000 × g for 3 min at 4°C and resuspended in ChIP lysis buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, 1% NP-40, 0.1% SDS, 10 mM sodium butyrate, protease inhibitor) with occasional mixing for 2 h at 4°C. The samples were centrifuged at 6,500 × g for 5 min, and the supernatants were collected for immunoprecipitation. For ChIP, the supernatants from the last step above were added into antibody-preincubated protein A/G-conjugated agarose beads, and beads were collected and washed five times. Immunocomplexes were eluted in elution buffer (1% SDS, 100 mM NaHCO3) and de-cross-linked at 65°C for 5 to 6 h, and the DNA fragments were purified by a TIANamp genomic DNA kit (Tiangen Biotech Co., Beijing, China). For sequencing, the DNA fragments were sent to a sequencing company (Annoroad Genomics Company, China). The information on antibodies used for the ChIP-Seq assay is presented in Table 2.
Bioinformatic analysis.
Paired-end sequencing reads in fastq format were mapped to the HBVcircle genome with Bowtie2 in no-discordant and no-mixed mode. Sequencing reads from transfected cells were aligned to the HBVcircle genome, and the alignment output files (SAM format) were converted into the files (BAM format) by using SAMtools. ChIP-Seq density profiles were generated by operating the alignment output files (BAM format) in the IGV genome browser.
Statistical analysis.
Statistical analysis was performed with GraphPad Prism 8.0 software (GraphPad Software, USA). The data were presented as mean ± standard deviation (SD). The statistical analyses were carried out using Student’s unpaired two-tailed t test. P values of <0.05 were considered significant: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Data availability.
Raw and processed ChIP-Seq data were deposited in the Gene Expression Omnibus under accession no. GSE199653.
ACKNOWLEDGMENTS
We thank the staff at the Research Center for Medicine and Structural Biology of Wuhan University for technical assistance.
Conceptualization, supervision: Yuchen Xia; investigation, data curation: Youquan Zhong, Chuanjian Wu, Zaichao Xu, Yan Teng, Li Zhao, Kaitao Zhao, Jingjing Wang, Wen Wang, Qiong Zhan, Chengliang Zhu; formal analysis: Youquan Zhong, Chuanjian Wu, Zaichao Xu, Yan Teng, Li Zhao, Kaitao Zhao, Jingjing Wang, Wen Wang, Xinwen Chen, Kaiwei Liang, Xiaoming Cheng, Yuchen Xia; data interpretation: Youquan Zhong, Chuanjian Wu, Zaichao Xu, Li Zhao, Yuchen Xia; funding acquisition: Yuchen Xia; writing—original draft: Youquan Zhong, Yuchen Xia; writing—review and editing: all authors.
There is no conflict of interest.
This work was supported by the National Natural Science Foundation of China (project no. 81971936), Hubei Province’s Outstanding Medical Academic Leader Program, the Gilead Sciences Research Scholars Program in Liver Disease Asia, the Foundation for Innovative Research Groups of the Hubei Natural Science Foundation (project no. 2020CFA015), the Foundation for Innovative Research Groups of Hubei Health Commission (project no. WJ2021C002), and Basic-Clinical Medicine Translation Joint Platform Fund of Zhongnan Hospital Wuhan University.
Contributor Information
Yuchen Xia, Email: yuchenxia@whu.edu.cn.
J.-H. James Ou, University of Southern California.
REFERENCES
- 1.Revill PA, Chisari FV, Block JM, Dandri M, Gehring AJ, Guo H, Hu J, Kramvis A, Lampertico P, Janssen HLA, Levrero M, Li W, Liang TJ, Lim SG, Lu F, Penicaud MC, Tavis JE, Thimme R, Zoulim F, Members of the ICE-HBV Working Groups, ICE-HBV Stakeholders Group Chairs, ICE-HBV Senior Advisors . 2019. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol 4:545–558. 10.1016/S2468-1253(19)30119-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polaris Observatory Collaborators. 2018. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: a modelling study. Lancet Gastroenterol Hepatol 3:383–403. 10.1016/S2468-1253(18)30056-6. [DOI] [PubMed] [Google Scholar]
- 3.Zhao K, Liu A, Xia Y. 2020. Insights into hepatitis B virus DNA integration-55 years after virus discovery. Innovation (Camb) 1:100034. 10.1016/j.xinn.2020.100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia Y, Guo H. 2020. Hepatitis B virus cccDNA: formation, regulation and therapeutic potential. Antiviral Res 180:104824. 10.1016/j.antiviral.2020.104824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia Y, Liang TJ. 2019. Development of direct-acting antiviral and host-targeting agents for treatment of hepatitis B virus infection. Gastroenterology 156:311–324. 10.1053/j.gastro.2018.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang LSY, Covert E, Wilson E, Kottilil S. 2018. Chronic hepatitis B infection: a review. JAMA 319:1802–1813. 10.1001/jama.2018.3795. [DOI] [PubMed] [Google Scholar]
- 7.Ghany MG. 2017. Current treatment guidelines of chronic hepatitis B: the role of nucleos(t)ide analogues and peginterferon. Best Pract Res Clin Gastroenterol 31:299–309. 10.1016/j.bpg.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Xia Y, Protzer U. 2017. Control of hepatitis B virus by cytokines. Viruses 9:18. 10.3390/v9010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. 2001. Structural organization of the hepatitis B virus minichromosome. J Mol Biol 307:183–196. 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 10.Guo YH, Li YN, Zhao JR, Zhang J, Yan Z. 2011. HBc binds to the CpG islands of HBV cccDNA and promotes an epigenetic permissive state. Epigenetics 6:720–726. 10.4161/epi.6.6.15815. [DOI] [PubMed] [Google Scholar]
- 11.Chong CK, Cheng CYS, Tsoi SYJ, Huang FY, Liu F, Seto WK, Lai CL, Yuen MF, Wong DK. 2017. Role of hepatitis B core protein in HBV transcription and recruitment of histone acetyltransferases to cccDNA minichromosome. Antiviral Res 144:1–7. 10.1016/j.antiviral.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Schultz U, Summers J, Staeheli P, Chisari FV. 1999. Elimination of duck hepatitis B virus RNA-containing capsids in duck interferon-alpha-treated hepatocytes. J Virol 73:5459–5465. 10.1128/JVI.73.7.5459-5465.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Y, Gao Z, Xu G, Peng B, Liu C, Yan H, Yao Q, Sun G, Liu Y, Tang D, Song Z, He W, Sun Y, Guo JT, Li W. 2016. DNA polymerase kappa is a key cellular factor for the formation of covalently closed circular dna of hepatitis B virus. PLoS Pathog 12:e1005893. 10.1371/journal.ppat.1005893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tu T, Zehnder B, Qu B, Urban S. 2021. De novo synthesis of hepatitis B virus nucleocapsids is dispensable for the maintenance and transcriptional regulation of cccDNA. JHEP Rep 3:100195. 10.1016/j.jhepr.2020.100195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucifora J, Pastor F, Charles E, Pons C, Auclair H, Fusil F, Rivoire M, Cosset FL, Durantel D, Salvetti A. 2021. Evidence for long-term association of virion-delivered HBV core protein with cccDNA independently of viral protein production. JHEP Rep 3:100330. 10.1016/j.jhepr.2021.100330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan Z, Zeng J, Yu Y, Xiang K, Hu H, Zhou X, Gu L, Wang L, Zhao J, Young JAT, Gao L. 2017. HBVcircle: a novel tool to investigate hepatitis B virus covalently closed circular DNA. J Hepatol 66:1149–1157. 10.1016/j.jhep.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Xia Y, Carpentier A, Cheng X, Block PD, Zhao Y, Zhang Z, Protzer U, Liang TJ. 2017. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J Hepatol 66:494–503. 10.1016/j.jhep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Z, Zhao L, Zhong Y, Zhu C, Zhao K, Teng Y, Cheng X, Chen Q, Xia Y. 2022. A novel mouse model harboring hepatitis B virus covalently closed circular DNA. Cell Mol Gastroenterol Hepatol 13:1001–1017. 10.1016/j.jcmgh.2021.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tropberger P, Mercier A, Robinson M, Zhong W, Ganem DE, Holdorf M. 2015. Mapping of histone modifications in episomal HBV cccDNA uncovers an unusual chromatin organization amenable to epigenetic manipulation. Proc Natl Acad Sci USA 112:E5715–E5724. 10.1073/pnas.1518090112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viswanathan U, Mani N, Hu Z, Ban H, Du Y, Hu J, Chang J, Guo JT. 2020. Targeting the multifunctional HBV core protein as a potential cure for chronic hepatitis B. Antiviral Res 182:104917. 10.1016/j.antiviral.2020.104917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blondot ML, Bruss V, Kann M. 2016. Intracellular transport and egress of hepatitis B virus. J Hepatol 64:S49–S59. 10.1016/j.jhep.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 22.Lewellyn EB, Loeb DD. 2011. The arginine clusters of the carboxy-terminal domain of the core protein of hepatitis B virus make pleiotropic contributions to genome replication. J Virol 85:1298–1309. 10.1128/JVI.01957-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nassal M. 1992. The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J Virol 66:4107–4116. 10.1128/JVI.66.7.4107-4116.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nassal M. 2015. HBV cccDNA: viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64:1972–1984. 10.1136/gutjnl-2015-309809. [DOI] [PubMed] [Google Scholar]
- 25.Petit MA, Capel F, Pillot J. 1985. Demonstration of a firm association between hepatitis B surface antigen proteins bearing polymerized human albumin binding sites and core-specific determinants in serum hepatitis B viral particles. Mol Immunol 22:1279–1287. 10.1016/0161-5890(85)90047-1. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Lu W, Zheng Y, Wang W, Bai L, Chen L, Feng Y, Zhang Z, Yuan Z. 2016. In situ analysis of intrahepatic virological events in chronic hepatitis B virus infection. J Clin Invest 126:1079–1092. 10.1172/JCI83339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu CM, Liaw YF. 1987. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology 92:220–225. 10.1016/0016-5085(87)90863-8. [DOI] [PubMed] [Google Scholar]
- 28.Yeh CT, Wong SW, Fung YK, Ou JH. 1993. Cell cycle regulation of nuclear localization of hepatitis B virus core protein. Proc Natl Acad Sci USA 90:6459–6463. 10.1073/pnas.90.14.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu TH, Liou AT, Su PY, Wu HN, Shih C. 2014. Nucleic acid chaperone activity associated with the arginine-rich domain of human hepatitis B virus core protein. J Virol 88:2530–2543. 10.1128/JVI.03235-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Kang W, Lei X, Li Y, Xiang A, Liu Y, Zhao J, Zhang J, Yan Z. 2012. Hepatitis B viral core protein disrupts human host gene expression by binding to promoter regions. BMC Genomics 13:563. 10.1186/1471-2164-13-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiang A, Ren F, Lei X, Zhang J, Guo R, Lu Z, Guo Y. 2015. The hepatitis B virus (HBV) core protein enhances the transcription activation of CRE via the CRE/CREB/CBP pathway. Antiviral Res 120:7–15. 10.1016/j.antiviral.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 32.Kwon JA, Rho HM. 2003. Transcriptional repression of the human p53 gene by hepatitis B viral core protein (HBc) in human liver cells. Biol Chem 384:203–212. 10.1515/BC.2003.022. [DOI] [PubMed] [Google Scholar]
- 33.Du J, Liang X, Liu Y, Qu Z, Gao L, Han L, Liu S, Cui M, Shi Y, Zhang Z, Yu L, Cao L, Ma C, Zhang L, Chen Y, Sun W. 2009. Hepatitis B virus core protein inhibits TRAIL-induced apoptosis of hepatocytes by blocking DR5 expression. Cell Death Differ 16:219–229. 10.1038/cdd.2008.144. [DOI] [PubMed] [Google Scholar]
- 34.Fernandez M, Quiroga JA, Carreno V. 2003. Hepatitis B virus downregulates the human interferon-inducible MxA promoter through direct interaction of precore/core proteins. J Gen Virol 84:2073–2082. 10.1099/vir.0.18966-0. [DOI] [PubMed] [Google Scholar]
- 35.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130:823–837. 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Mao R, Yan R, Cai D, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Huang Y, Guo H, Zhang J. 2014. Transcription of hepatitis B virus covalently closed circular DNA is regulated by CpG methylation during chronic infection. PLoS One 9:e110442. 10.1371/journal.pone.0110442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Weizsacker F, Wieland S, Blum HE. 1995. Identification of two separable modules in the duck hepatitis B virus core protein. J Virol 69:2704–2707. 10.1128/JVI.69.4.2704-2707.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Decorsiere A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP, Hantz O, Strubin M. 2016. Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 531:386–389. 10.1038/nature17170. [DOI] [PubMed] [Google Scholar]
- 39.Murphy CM, Xu Y, Li F, Nio K, Reszka-Blanco N, Li X, Wu Y, Yu Y, Xiong Y, Su L. 2016. Hepatitis B virus X protein promotes degradation of SMC5/6 to enhance HBV replication. Cell Rep 16:2846–2854. 10.1016/j.celrep.2016.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, Remouchamps C, Chou WM, Thasler WE, Huser N, Durantel D, Liang TJ, Munk C, Heim MH, Browning JL, Dejardin E, Dandri M, Schindler M, Heikenwalder M, Protzer U. 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343:1221–1228. 10.1126/science.1243462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia Y, Stadler D, Lucifora J, Reisinger F, Webb D, Hösel M, Michler T, Wisskirchen K, Cheng X, Zhang K, Chou WM, Wettengel JM, Malo A, Bohne F, Hoffmann D, Eyer F, Thimme R, Falk CS, Thasler WE, Heikenwalder M, Protzer U. 2016. Interferon-γ and tumor necrosis factor-α produced by T cells reduce the HBV persistence form, cccDNA, without cytolysis. Gastroenterology 150:194–205. 10.1053/j.gastro.2015.09.026. [DOI] [PubMed] [Google Scholar]
- 42.Lahlali T, Berke JM, Vergauwen K, Foca A, Vandyck K, Pauwels F, Zoulim F, Durantel D. 2018. Novel potent capsid assembly modulators regulate multiple steps of the hepatitis B virus life cycle. Antimicrob Agents Chemother 62:e00835-18. 10.1128/AAC.00835-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klumpp K, Shimada T, Allweiss L, Volz T, Lutgehetmann M, Hartman G, Flores OA, Lam AM, Dandri M. 2018. Efficacy of NVR 3-778, alone and in combination with pegylated interferon, vs entecavir in uPA/SCID mice with humanized livers and HBV infection. Gastroenterology 154:652–662.e8. 10.1053/j.gastro.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 44.Xia Y, Stadler D, Ko C, Protzer U. 2017. Analyses of HBV cccDNA quantification and modification. Methods Mol Biol 1540:59–72. 10.1007/978-1-4939-6700-1_6. [DOI] [PubMed] [Google Scholar]
- 45.Zhang S, Guo JT, Wu JZ, Yang G. 2013. Identification and characterization of multiple TRIM proteins that inhibit hepatitis B virus transcription. PLoS One 8:e70001. 10.1371/journal.pone.0070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teng Y, Xu Z, Zhao K, Zhong Y, Wang J, Zhao L, Zheng Z, Hou W, Zhu C, Chen X, Protzer U, Li Y, Xia Y. 2021. Novel function of SART1 in HNF4alpha transcriptional regulation contributes to its antiviral role during HBV infection. J Hepatol 75:1072–1082. 10.1016/j.jhep.2021.06.038. [DOI] [PubMed] [Google Scholar]
- 47.Cai D, Nie H, Yan R, Guo JT, Block TM, Guo H. 2013. A southern blot assay for detection of hepatitis B virus covalently closed circular DNA from cell cultures. Methods Mol Biol 1030:151–161. 10.1007/978-1-62703-484-5_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee TI, Johnstone SE, Young RA. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc 1:729–748. 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao L, Wu C, Shi H, Gong Z, Zhang E, Wang H, Zhao K, Liu S, Li S, Gao X, Wang Y, Pei R, Lu M, Chen X. 2014. Coexistence of hepatitis B virus quasispecies enhances viral replication and the ability to induce host antibody and cellular immune responses. J Virol 88:8656–8666. 10.1128/JVI.01123-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang K, Woodfin AR, Slaughter BD, Unruh JR, Box AC, Rickels RA, Gao X, Haug JS, Jaspersen SL, Shilatifard A. 2015. Mitotic transcriptional activation: clearance of actively engaged Pol II via transcriptional elongation control in mitosis. Mol Cell 60:435–445. 10.1016/j.molcel.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw and processed ChIP-Seq data were deposited in the Gene Expression Omnibus under accession no. GSE199653.