Abstract
Purpose
Cone and rod photoreceptors in the retina convert light to electrical signals which are transmitted to the visual cortex of the brain. Abnormal photoreceptor development and degeneration results in blindness. So far, the mechanism that controls photoreceptor specification and its subsequent fate bifurcation remain elusive.
Methods
To trace and enrich the human photoreceptor lineage, we first engineered H9 human embryonic stem cell (hESC) reporter line by fusing EGFP to endogenous BLIMP1 using CRISPR/CAS9 gene-editing technology, and then used the cell line to generate 3D retinal organoids. Following EGFP-based cell sorting, single-cell RNA-sequencing was conducted via 10x Genomics Chromium system, and the data were analyzed using Seurat. Immunofluorescence combined with lentivirus-mediated knockdown and overexpression experiments were used as validation approaches.
Results
Single-cell transcriptomic profiling revealed that retinal progenitor cells were temporally programmed to differentiate to cone and rod sequentially. We identified PHLDA1 as a novel regulator of photoreceptor specification. PHLDA1 mediated the effects of IGF1 through IGF1R, and inhibited AKT phosphorylation during photoreceptor development.
Conclusions
Our data established a transcriptomic cell atlas of the human photoreceptor lineage, and identified IGF1-PHLDA1 axis to regulate human photoreceptor development.
Keywords: photoreceptor development, PHLDA1, single-cell RNA sequencing, retinal organoid
Photoreceptors are highly specialized retinal sensory neurons that transduce environmental light stimuli into electrical potentials that form the primary retinal image.1,2 Cone photoreceptors enable color and high-acuity vision,3,4 whereas rod photoreceptors mediate vision in darker conditions.5 Both photoreceptor types are derived from multipotent retinal progenitor cells (RPCs) that undergo terminal neurogenic divisions to become photoreceptor precursors and further differentiate to mature cones or rods.6 When and how the photoreceptors are fate-determined is of great interest to the field of neuron development and regeneration.
Several developmental models of photoreceptor fate specification have been formulated based on genetic and lineage tracing studies.6–8 In the “competence model,” RPCs follow intrinsic temporal order of competency states. At each stage, the RPCs produce particular retinal cell types at that time.7 In the “transcriptional dominance” model, all differentiated photoreceptors originate from a common postmitotic precursor that can differentiate into either cones or rods.9,10 In this model, photoreceptor subtype is dictated by the dominant expression of subtype-specific genes in a given precursor, such as NRL.6,9 Alternatively, in the “binary fate choice model,” lineage diverges early in RPC development and cell subpopulations exhibit distinct transcriptional profiles at different developmental stages.11 In accord with the “transcriptional dominance” model, recent large-scale single-cell RNA sequencing (scRNA-seq) studies of retinal development have revealed a subpopulation of multipotent photoreceptor precursors segregated from RPC clusters that then bifurcate into separate cone and rod lineages.12–14 However, the molecular mechanisms underlying the development of photoreceptor lineage remains largely unknown.
Both intrinsic and extrinsic cues guide the fate determination of photoreceptor precursors.7,11 Multiple transcriptional factors (TFs), such as OTX2, BLIMP1, and CRX, are expressed in most photoreceptor precursors and dictate key steps of photoreceptor differentiation. For example, OTX2 is expressed in nascent and mature photoreceptors and bipolar cells.15,16 Conditional knockout of Otx2 results in complete loss of cones and rods, as well as a loss of bipolar cells in mice.17 The transcriptional repressor Blimp1 (Prdm1) is expressed solely by Otx2+ cells and promotes nascent Otx2+ cell differentiated to photoreceptor cells by repressing bipolar fate.18 The expression of Blimp1 is downregulated and becomes undetectable between P6 and P10 in mouse, and mis-expression of Blimp1 in mature photoreceptors appears to be toxic.19 CRX is required for the growth of outer segments and is important for terminal differentiation of rods and cones but does not determine the photoreceptor cell fate.20
In addition, numerous extrinsic factors can influence the differentiation of specific photoreceptor subtypes by modulating intrinsic factors, such as insulin-like growth factor 1 (IGF1), retinoic acid (RA), taurine, ciliary neurotrophic factor (CNTF), and leukemia inhibitory factor (LIF).21,22 For example, IGF1, as a neurotropic factor, has been demonstrated to enhance retinal lamination at the early stages of differentiation and photoreceptor production at the later stages of differentiation.23 Studies also revealed the positive effect of IGF1 on rod differentiation through specific PKC isoforms.24 Nevertheless, precisely how IGF1 signaling influences photoreceptor cell fate determination and integrates into the transcriptional network of photoreceptor differentiation remains largely unknown.
With the development of state-of-the-art single-cell transcriptional profiling and lineage tracing technologies,25 it has become possible to identify signals mediating cell fate determination for photoreceptors. Several studies have reported tracing photoreceptors by fusing the GFP to photoreceptor marker genes, such as CRX26 and NRL.27 However, these reporter cell lines were not ideal models for tracing the photoreceptors’ development trajectory, as CRX+ cells can differentiate to bipolar in the human fetal retina,11,28 NRL only labels rods but not cones. Compared with other photoreceptor marker genes, BLIMP1 is exclusively expressed in photoreceptor precursors.19 We hypothesized that the BLIMP1 is an ideal marker to trace and enrich the photoreceptor lineage, and help to reveal the molecular mechanisms of photoreceptor differentiation.
In this study, we construct a fluorescence-labeled embryonic stem cell (ESC) line to trace photoreceptor lineages by fusing enhanced green fluorescence protein (EGFP) with BLIMP1, and generate a cell atlas of human photoreceptor developmental trajectory by using single-cell RNA sequencing (scRNA-seq) technology. We observed the separate temporal emergence of rod and cone specified from neurogenic RPCs in which genetic programs were altered to differentially bias cell fate. Further, we identified PHLDA1 as a novel regulator of photoreceptor specification. PHLDA1 mediated the effects of IGF1 and it can inhibit AKT phosphorylation during photoreceptor development. Collectively, these data identify the IGF1-PHLDA1-pAKT axis as a signaling pathway regulating human photoreceptor development, with potential utility for vision repair and therapeutics.
Materials and Methods
Establishment of a BLIMP1 Reporter Human Embryonic Stem Cell Line
To track photoreceptor development, retinal organoids were derived from a human embryonic stem cell (hESC) H9 line in which the EGFP gene was introduced at the endogenous BLIMP1 locus using CRISPR/CAS9 (Biocytogen). Briefly, a BLIMP1-sgRNA (AAATGGTTTCCCCTCACCTC) was designed to target a 567-bp region between Exon 7 and the 3′UTR of BLIMP1, and expression was validated using UCATM (Biocytogen). A target vector containing the knock-in gene, P2A-EGFP-5′TR-JT-puroDeltaTK-3′TR-JT, was generated and co-transfected with the BLIMP1-sgRNA into H9 hESCs. Transfected hESCs were selected with puromycin starting on the fourth day after transfection, and colonies were chosen 10 days later. The colonies were further screened by PCR genotyping and karyotype testing (see Supplementary Fig. S1E).
Retinal Organoid Differentiation
These hESCs were cultured in Essential 8 (E8) medium (Invitrogen) on Vitronectin (VTN-N)-coated plates. To initiate retinal differentiation, hESC colonies were dissociated into small cell clusters with dispase (2 mg/mL) and allowed to aggregate as the medium was gradually switched from E8 to neural induction medium (NIM: DMEM/F12 [1:1], 1% N2 supplement, MEM nonessential amino acids, penicillin-streptomycin, and 2 mg/mL heparin sulfate) over 4 days. Starting on day 6, recombinant human BMP4 (50 ng/mL) was added to increase the efficiency of retinal differentiation. Half of the NIM volume was replaced every third day with fresh NIM also containing 50 ng/mL BMP4. Cell aggregates were then reseeded onto VTN-N-coated plates on day 7 with NIM medium containing 10% fetal bovine serum (FBS) and BMP4. On day 8, 10% FBS was removed and half of the serum-free NIM was changed. On day 16, neural rosettes were dislodged from plates manually and henceforth maintained in retinal differentiation medium (RDM: DMEM/F12 [3:1], 2% B27 supplement, MEM nonessential amino acids, and penicillin-streptomycin) to allow the formation of retinal organoids. From day 30, RDM medium was supplemented with 10% FBS, 100 µM taurine, 2 mM GlutaMAX, and 0.5 µM retinoic acid for long-term retinal organoid culture.
Retinal Organoid Cell Dissociation for scRNA-seq
Retinal organoids (4 or 5) were randomly selected at each indicated developmental time point, dissociated into single-cell suspensions by treatment with Accutase for 30 minutes at 37°C, filtered through a 35-µm cell strainer, screened for EGFP+ expression using a Flow Cell Sorter (BD FACSAria), and resuspended in PBS containing 0.04% bovine serum albumin. The scRNA-seq libraries were prepared using the Single-cell Gene Expression 3′ V2 or 3′ V3 kit (10x Genomics, Pleasanton, CA, USA) following the manufacturer's protocol. Briefly, single cells were partitioned on gel beads in EMulsion (GEMs) within a Chromium instrument followed by cell lysis, reverse transcription, cDNA amplification, and library construction. Libraries were then sequenced on the Illumina HiSeq 2500 platform.
scRNA-seq Analysis
Pre-Processing of Droplet-Based scRNA-seq Data
The raw scRNA-seq data were demultiplexed and aligned to the human reference genome (GRCh38) using Cell Ranger (version 3.1) with default parameters. The expression level of each transcript was determined using the UMI and assigned to cells based on cell barcodes. The filtered gene expression matrices produced by Cell Ranger were then used for downstream analyses.
Processing and Cell-Type Annotation of Human Fetal Retina scRNA-seq Data
Age-matched human fetal retina data were downloaded from the Gene Expression Omnibus (GEO, dataset GSE138002) and pre-processed. Cell-type annotation was then modified using Seurat (version 4.0.2)48 and the processed dataset was used to predict cell types in scRNA-seq data from retinal organoids. Briefly, scRNA-seq data from 9 and 11 to 18 gestational weeks were normalized and integrated. Principal component analysis (PCA) was performed on integrated data to reduce dimensionality, and a k-nearest neighbor graph (k = 30) was constructed based on Euclidean distance in the significantly PC space. Distinct cell clusters were identified using the Louvain-Jaccard graph-based method and labeled based on marker genes and cell type annotation from a previous study.13
Labeling of Organoid Datasets
To predict the cell type from retinal organoid scRNA-seq data, we transferred the cell type metadata from the fetal retina dataset to the retinal organoid dataset using the Seurat FindTransferAnchors and TransferData functions, and embedded cells into the UMAP structure of the reference using the MapQuery function.48
Identification of the Photoreceptor/Bipolar Cell Precursor Subcluster
Sub-clustering analysis was performed on photoreceptor/bipolar cell precursors using the Seurat FindSubCluster function to identify potential cone, rod, and bipolar cell precursors. Differentially expressed genes (DEGs) among the three subclusters were identified using the FindAllMarkers function, and genes with false discovery rate (FDR)-correct P < 0.05 and > 0.5 average log2-fold-change were considered marker genes for the subcluster.
Identification of Differentially Expressed Genes in the scRNA-seq Dataset
Model-based analysis of single-cell transcriptomics (MAST)49 was used to identify DEGs for each cell type. Identified DEGs were then tested against an asymptotic chi-square null distribution. Genes with FDR-correct P < 0.05 are considered differentially expressed.
Gene Ontology Analysis of DEGs
Gene Ontology (GO) analysis was performed using clusterProfiler (version 4.0)50 and visualized using the ggplot2 R package (https://github.com/tidyverse/ggplot2). GO terms or pathways were considered enriched at P < 0.01.
Developmental Trajectory Analysis
Photoreceptor developmental trajectories were reconstructed using Monocle2.31 Briefly, the UMI matrix was used as input and the marker genes previously generated in Seurat with default parameters were used to calculate the trajectory.
Co-Expression Analysis
Co-expression modules within the scRNA-seq dataset were inferred using a regression per-target approach implemented in GENIE3.51 Briefly, gene co-expression networks were constructed from the UMI matrix using GENIE3 and visualized using Cytoscape (version 3.7.1).
Retinal Organoids Sectioning and Immunofluorescence
Retinal organoids were fixed in 4% paraformaldehyde for 20 minutes at 4°C, washed 3 times in 1X phosphate buffered saline (PBS), cryoprotected in 30% sucrose solution at 4°C overnight, embedded in OCT, and cryosectioned at 5 µm. Frozen sections were incubated sequentially in blocking buffer (10% FBS, 2% donkey serum, and 0.2% Triton X-100 in PBS) for 1 hour, primary antibody overnight at 4°C, and in secondary antibodies (1:1000) for 45 minutes, then counterstained with DAPI, washed, and mounted. Primary antibodies used in this study are available in Supplementary Table S3. Immunolabeled sections were imaged using a fluorescence microscope (Zeiss) and analyzed for fluorescence intensity distribution using Adobe Photoshop. For each image, the normalized fluorescence intensity was calculated as the mean gray value of each immunostaining channel divided by the mean DAPI or mCherry gray value.
BrdU Labeling
Retinal organoids were cultured in RDM+3 medium in the presence of BrdU (5 µg/mL; MCE cat #HY-15910) for 24 hours. BrdU-treated organoids were incubated with 2 N HCl for 10 minutes at room temperature and then with 0.1 M sodium borate (pH 8.5) for 10 minutes at room temperature. After rinsing with TBS three times, immunostaining was performed as described above.
TUNEL Staining
Fluorescent terminal dUTP nick end labeling (TUNEL; Serologicals Corporation, Norcross, GA, USA) was used to detect the presence of DNA strand breaks in the retinal organoids’ sections, following the manufacturer's instructions. Only cells with evident nuclei were scored as TUNEL-positive.
RNA Fluorescence In Situ Hybridization
RNA in situ hybridization was conducted using digoxigenin-labeled antisense riboprobes. Probe templates were generated from organoid cDNAs by RNA extraction and reverse transcription using Superscript III (Thermo Fisher). Antisense probes were then generated by PCR using a reverse primer with a T7 sequence adaptor. DIG rUTP was used for riboprobe in vitro transcription.
The fixed sections were treated with 10 µg/mL Proteinase K and ethanolamine-acetic anhydride solution for 10 minutes each, and then washed sequentially in DEPC-PBS, 0.85% NaCl, 75% ETOH, and 95% ET OH for dehydration. Dehydrated sections were incubated in hybridization solution (50% deionized formamide, 10% dextran sulfate, 1 mg/mL rRNA, and 1x Denhardt's solution) containing digoxigenin-labeled antisense riboprobes (0.25 ng/µL) at 68°C overnight. Sections were then incubated with horseradish peroxidase (HRP)-labeled anti-DIG (1:1000), followed by tyramide amplification for probe visualization.
Lentivirus Transduction and Validation
Lentiviral vectors encoding shRNA targeting PHLDA1, shRNA targeting INSR, or Scramble shRNA were purchased from Vectorbuilder. The human RB cell line Y-79 was used to validate the knockdown effect of shRNA. Briefly, Y-79 cells were infected with shPHLDA1, shINSR, or Scramble shRNA lentivirus for 3 days, followed by RNA extraction and quantitative real-time PCR (qRT-PCR) measurement of PHLDA1 or INSR expression using the human ACTB gene as the internal reference. The shRNA target sequences and qRT-PCR primer sequences are provided in Supplementary Table S2. For lentiviral infection of retinal organoids, 1 million lentiviral transduction units per retinal organoid was added to medium containing 5 µg/mL polybrene (Sigma). After incubation for 12 hours, lentivirus particles were removed by changing the medium, and infected organoids were cultured for 2 to 3 weeks before observation.
Statistical Analyses
All experiments were performed at least three times using independently isolated and treated organoids. Results are expressed as mean ± SEM. Two treatment group means were compared by unpaired 2-tailed Student's t-test and three or more group means by 1-way ANOVA with post hoc Dunnett's tests for pair-wise comparisons. All statistical analyses were performed using GraphPad Prism version 7.04. A *P < 0.05 was considered significant for all tests.
Results
Single-cell Transcriptomic Profiling of Human Photoreceptor Lineage
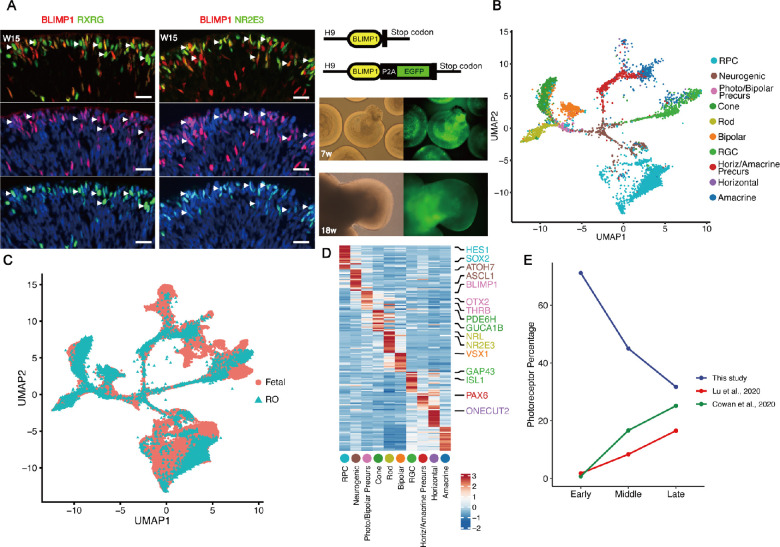
Previously, mouse RPCs expressing the transcriptional repressor Blimp1 (Prdm1) were found to become photoreceptors rather than bipolar cells.18 Consistently, we observed that BLIMP1 was widely expressed across almost all stages of the photoreceptor lineage in human retinal organoids as evidenced by immunofluorescent colocalization with the neurogenic RPC marker OLIG2 (Supplementary Fig. S1A), the cone marker RXRG, and the rod marker NR2E3 (Fig. 1A). Further, by using a public dataset of human retinal single-cell spatiotemporal transcriptome profiles,13 we observed that BLIMP1 was expressed in photoreceptor cell lineages at different developmental stages (Supplementary Figs. S1B-D). Therefore, we hypothesized that BLIMP1 could be used as a marker for tracing the developmental lineage of photoreceptors in retinal organoids, and generated retinal organoids in vitro from a human embryonic stem cell (hESC) line in which endogenous BLIMP1 was fused to enhanced green fluorescent protein (EGFP; Fig. 1A). As expected, EGFP expression paralleled endogenous BLIMP1 expression (Supplementary Figs. S1E, S1F), validating this in vitro cell lineage tracing technique.
Figure 1.
Tracing of photoreceptor lineages in hESC-derived retinal organoids. (A) Left: Immunofluorescence staining of retinal organoids showing that a subset of BLIMP1+ cells co-expressed either the cone marker RXRG or the rod marker NR2E3. Arrowheads indicate colocalization of BLIMP1 with RXRG or NR2E3. Right (upper panel): Schematic diagram showing the targeting strategy at the BLIMP1 insertion site. The EGFP cDNA sequence was fused in-frame into BLIMP1 beyond the stop codon. Right (lower panels): Fluorescence and light microscopic images of BLIMP1-EGFP hESC-derived retinal organoids at W7 and W18 in vitro. Scale bars = 20 µm. (B) UMAP plot of the single-cell RNA-seq data of human retinal organoid after integrating with the fetal retina RNA-seq data from GSE138002, recolored, and showing the major cell clusters. RPC, multipotent retinal progenitor cell; neurogenic, neurogenic RPC; Photo/Bipolar Precurs, photoreceptor/bipolar cell precursor; Horiz/Amacrine Precurs, horizontal/amacrine cell precursor. (C) The integrated UMAP plot colored according to origin. (D) Heatmap of representative markers of each cell type. (E) Line chart comparing the proportion (%) of photoreceptor lineage cells at early developmental stages (week 7 in this study, weeks 6 in Cowan et al. 2020, weeks 9 in Lu et al. 2020), middle developmental stages (week 12 in this study, weeks 12 in Cowan et al. 2020, weeks 14 in Lu et al. 2020), and late developmental stages (week 18 in this study, weeks 18 in Cowan et al. 2020, weeks 18 in Lu et al. 2020) for our enrichment assay, retinal organoids, and fetal retina.
Further, EGFP+ cells from retinal organoids were isolated by FACS after 7, 12, and 18 weeks in culture, and subjected to scRNA-seq (Supplementary Fig. S1G). In total, 10,455 cells were profiled to a mean depth of 8064 (SD = 6275) – unique molecular identifiers (UMIs) and 2844 (SD = 1316) genes (Supplementary Fig. S1H, Supplementary Table S1). Construction of a single-cell transcriptomic cell atlas revealed 10 distinct cell clusters (Figs. 1B, 1C) distinguished by differential expression of known marker genes, including proliferative RPCs (HES1+/SOX2+), neurogenic RPCs (ATOH7+/ASCL1+), photoreceptor/bipolar cell precursors (BLIMP1+/OTX2+), cone (PDE6H+/GUCA1B+), rod (NRL+/NR2E3+), bipolar cell (VSX1+), and horizontal/amacrine cell precursors (PAX6+/ONECUT2+) etc. (Fig. 1D). Cells of the photoreceptor lineage were significantly enriched in this study (Fig. 1E), enabling further dissection of the molecular events related to the lineage specification.
PHLDA1 Was Upregulated in Photoreceptor Precursors
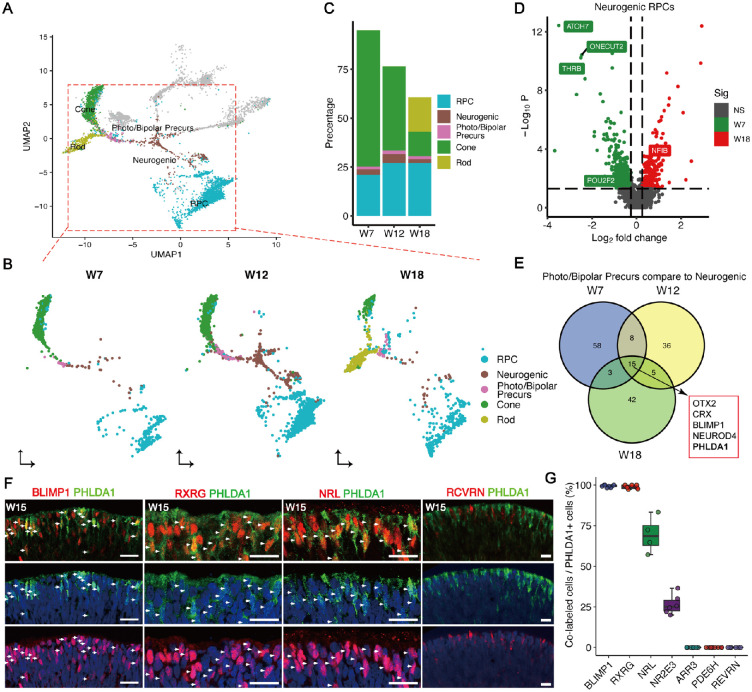
We next examined the temporal windows of cell-fate specification of photoreceptor subtypes, and observed that cones emerge at W7, whereas rods appear after W12 (Figs. 2A-C). The timing of photoreceptor subtypes specification in vivo was similar to that demonstrated by another scRNA-seq dataset of retinal organoids13 (Supplementary Figs. S2A, S2B). Further, by comparing neurogenic RPCs at in vitro durations corresponding to the emergence of cone and rod precursors, we found that cone fate-related genes such as THRB, ATOH7,12 and POU2F229 showed higher expression in early neurogenic RPCs, whereas the rod-related gene NFIB30 had higher expression in late neurogenic RPCs (Fig. 2D, Supplementary Fig. S2C). Upregulated genes in photoreceptor precursors compared to neurogenic RPCs at different developmental stages were further characterized. Interestingly, intersection of the marker genes at W7, W12, and W18 led to the identification of the photoreceptor cell-fate commitment genes, such as OTX2 and BLIMP1 (Fig. 2E). We also found a novel gene, PHLDA1, which is highly expressed in photoreceptor precursors (see Fig. 2E, Supplementary Fig. S2D). Consistent with the transcriptomic result, immunostaining data showed almost every PHLDA1+ cell expressed BLIMP1 and rarely expressed the mature pan-photoreceptor marker RCVRN (Figs. 2F, 2G). A subset of PHLDA1+ cells was positive for RXRG staining and NRL staining (early rod genes), but no expression of PDE6H and ARR3 (mature cone genes) was colocalized with PHLDA1 (see Figs. 2F, 2G, Supplementary Fig. S2E), suggesting an important role of PHLDA1 in photoreceptor precursors. Therefore, it is intriguing to investigate the uncharacterized function of PHLDA1 in photoreceptor development, especially its specific functions in the specification of photoreceptor precursors.
Figure 2.
Temporal specification of photoreceptor subtypes in retinal organoids. (A) UMAP plot highlighting the cell types in photoreceptor cell development trajectory. (B) Cell type distribution of cell types in photoreceptor cell development trajectory at three points. (C) Stacked plots showing the proportions (%) of each photoreceptor lineage at the indicated development time points. (D) Volcano plot displaying fate determining factors differentially expressed between neurogenic RPCs at early stage (W7) and late stage (W18). (E) Venn plot showing the common upregulated genes in photoreceptor precursors across three stages. (F) Co-expression of PHLDA1 with BLIMP1, RCVRN, RXRG, or NRL shown by immunofluorescence. Scale bars = 20 µm. Arrowheads indicate co-localization of PHLDA1 with others markers of photoreceptors. (G) Histogram showing the proportions (%) of PHLDA1+ cells co-expressing either BLIMP1, RXRG, NRL, NR2E3, PDE6H, ARR3, or RCVRN.
PHLDA1 Implicated in Temporal Specification of Photoreceptors From RPCs
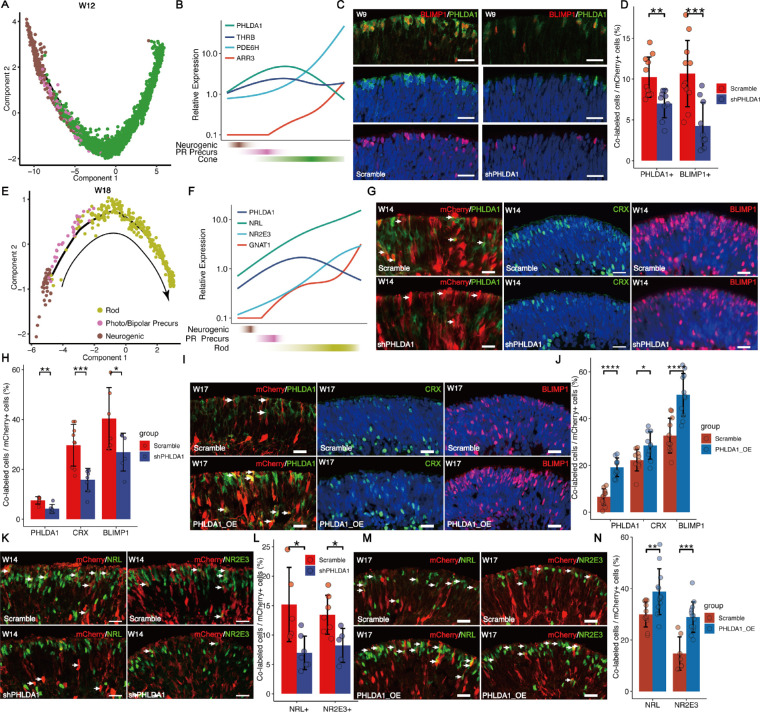
We further characterized the developmental trajectory of cone development using Monocle2.31 Pseudotime analysis showed that photoreceptor precursors emerged after neurogenic RPCs and before cones (Fig. 3A, Supplementary Fig. S3A, S3B). With the organoid development ongoing, the expression of PHLDA1 was initially upregulated and then downregulated, and the expression of phototransduction-related genes, such as PDE6H and ARR3, was upregulated with the terminal cone differentiation (Fig. 3B). We also found the same dynamic gene expression changes during cone development in another scRNA-seq dataset from human retinal organoids13 (Supplementary Fig. S3C). We validated the dynamic expression of PHLDA1 during the photoreceptor differentiation via immunostaining, and observed that PHLDA1 was colocalized with pan-photoreceptor marker OTX2 and BLIMP1, but rarely with neurogenic RPCs marker ASCL132 and mature photoreceptor marker RHO33 (Supplementary Figs. S3K, S3L). To further investigate the function of PHLDA1 in the lineage specification of cone precursors, we performed knockdown experiments by transfecting W7 organoids with lentivirus expressing PHLDA1-targeted shRNA together with the fluorescence marker mCherry (Supplementary Fig. S4A), and assessed the organoids 1 week after transfection. We confirmed that shPHLDA1 lentivirus significantly reduced PHLDA1 mRNA compared to a scrambled control in the Y-79 cell line (Supplementary Fig. S4B) and reduced PHLDA1+ cells in retinal organoids (Figs. 3C, 3D). Strikingly, we observed that PHLDA1 knockdown significantly reduced the number of BLIMP1+ cells in retinal organoids (see Figs. 3C, 3D). These results suggested that PHLDA1 promotes the cone generation.
Figure 3.
PHLDA1 promotes cone and rod specification. (A) Trajectory analysis of the cone lineage showing progression from neurogenic RPC to photoreceptor precursors and cone. (B) Line chart showing the dynamic expression of PHLDA1, THRB, ARR3, and PDE6H. (C, D) The number of PHLDA1+ and BLIMP1+ cells were significantly downregulated in the shPHLDA1 (KD) group compared to the Scramble shRNA group. Scale bars = 20 µm. (E) Trajectory analysis of the rod lineage showing progression from neurogenic RPC to rod precursors and rod. (F) Line chart showing the dynamic expression of PHLDA1, NRL, NR2E3, and GNAT1. (G, H) The number of PHLDA1+, CRX+, and BLIMP1+ cells was significantly downregulated in the shPHLDA1 (KD) group compared to the Scramble shRNA group. Scale bars = 20 µm. Arrowheads indicate PHLDA1+ cells expressing scramble or PHLDA1 shRNA. (I, J) The number of PHLDA1+, CRX+, and BLIMP1+ cells was significantly increased in the PHLDA1 overexpression (OE) group compared to the Scramble. Scale bars = 20 µm. Arrowheads indicate the cells transfected by lentivirus (mCherry+). (K, L) The number of NRL+ and NR2E3+ cells was significantly downregulated in the shPHLDA1 (KD) group compared to the Scramble shRNA group. Scale bars = 20 µm. Arrowheads indicate NR2E3+ cells transfected by lentivirus (mCherry+). (M, N) The number of NRL+ and NR2E3+ cells was significantly increased in the PHLDA1 overexpression (OE) group compared to the Scramble group. Scale bars = 20 µm. Arrowheads indicate NR2E3+ cells transfected by lentivirus (mCherry+).
Next, we characterized the developmental trajectory of rod lineages (Fig. 3E, Supplementary Figs. S3D, S3E). As similar to cones, the expression of PHLDA1 was downregulated and phototransduction-related genes (e.g. NR2E3 and GNAT1) concurrently upregulated during terminal differentiation of rods (Fig. 3F, Supplementary Figs. S3I, S3J). Similar expression patterns of these genes were shown by another scRNA-seq dataset (Supplementary Fig. S3F). To directly test the effects of PHLDA1 on the differentiation of photoreceptor subtypes, we co-transduced retinal organoids of 13 weeks with lentivirus expressing PHLDA1-targeted shRNA together with mCherry, and organoids were assessed 1 week after transfection. Knockdown of PHLDA1 in organoids significantly reduced the number of BLIMP1+, NRL+, and NR2E3+ cells (Figs. 3G, 3H, 3K, 3L, Supplementary Figs. S4C, S4D), whereas overexpression of PHLDA1 increased the number of BLIMP1+, NRL+, and NR2E3+ cells (Figs. 3I, 3J, 3M, 3N). By counting the number of RPCs (VSX2+ and MKI67+), we observed a complementary increase of RPCs along with the decrease of photoreceptors after PHLDA1 knockdown (Figs. 3G, 3H, Supplementary Figs. S4E, S4F). In addition, PHLDA1 overexpression decreased the number of RPCs (Supplementary Figs. S3G, S3H.). These results suggested that PHLDA1 promotes the specification of photoreceptors. We further performed BrdU pulse-labeling and found that knockdown of PHLDA1 significantly increased the number of BrdU+ cells (Supplementary Figs. S4I, S4J). Although we cannot rule out that the increase in BrdU incorporation results from cell-cycle shortening, we suggest knockdown of PHLDA1 increased the number of proliferative RPCs, at least in part due to the reduced specification of RPC to photoreceptors. Further, we investigate whether knockdown of PHLDA1 affects the photoreceptor apoptosis. Although knockdown of PHLDA1 significantly increased the TUNEL+ cells, from 0.93% to 1.63% (Supplementary Figs. S4K, S4L), the increased number of TUNEL+ cells was much less than the decreased number of photoreceptor cells (Figs. 3C, 3D, Supplementary Figs. S4K, S4L), suggesting that the reduction in the number of photoreceptor precursors caused by PHLDA1 knockdown was mainly due to abnormal cell specification rather than cell death.
PHLDA1 Inhibits AKT Phosphorylation in Photoreceptors
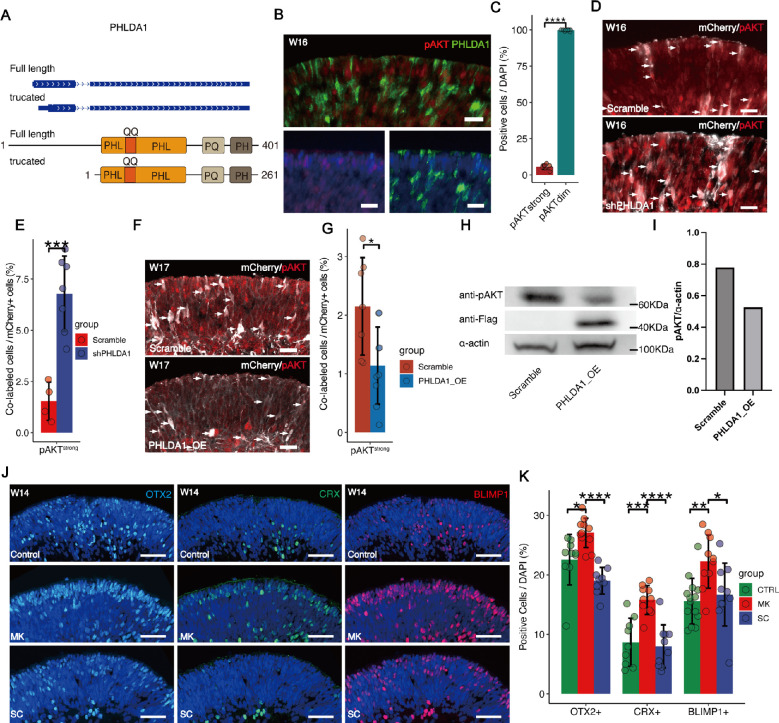
We then examined alternative splicing in the PHLDA1 gene by performing single-cell full-length sequencing based on the Pacbio platform. We found that a truncated PHLDA1 mRNA, which encoded a truncated isoform, was mainly expressed in retinal organoids. Analysis of PHLDA1 amino acids sequencing identified that both isoforms are enriched in polyglutamine (QQ), proline-glutamine (PQ), and proline-histidine tracts with a PHL domain (Fig. 4A). Previous studies reported that PHL domain was involved in inhibition of AKT activation by binding to phosphatidylinositol lipids in cancer cells.34 Concordantly, we found that pAKT was weakly expressed in PHLDA1+ cells of human retinal organoids (Figs. 4B, 4C). Furthermore, PHLDA1 knockdown upregulated pAKT (Figs. 4D, 4E), and pAKT was significantly reduced by PHLDA1 overexpression in retinal organoids (Figs. 4F, 4G) and Y79 cells (Figs. 4H, 4I), indicating that PHLDA1 inhibited pAKT in photoreceptors.
Figure 4.
PHLDA1 inhibits phosphorylation of AKT. (A) Genome browser tracks showing the structure of PHLDA1 mRNA and schematic representation of the structure of the PHLDA1 protein. (B, C) Immunofluorescence staining showing that strong pAKT expression was rarely colocalized with PHLDA1 expression. Scale bars = 20 µm. (D, E) Immunofluorescence of pAKT in retinal organoids transfected with shPHLDA1. Expression was markedly increased by PHLDA1 KD compared to cells transfected with Scramble shRNA. Scale bars = 20 µm. Arrowheads indicate the cells transfected by lentivirus (mCherry+). (F, G) Immunofluorescence of pAKT in retinal organoids transfected with lentivirus to overexpress PHLDA1 (OE). Scale bars = 20 µm. Arrowheads indicate the cells transfected by lentivirus (mCherry+). (H, I) Western blot analysis of pAKT in Y79 with PHLDA1 overexpression (OE). The exogenously overexpressed PHLDA1 was detected by using the FLAG antibody. (J, K) Effects of AKT activation and inhibition on photoreceptor differentiation. W12 retinal organoids were treated with the AKT agonist SC79 (SC, 10 µM) or antagonist MK2206 (MK, 1 µM) for 2 weeks. AKT inhibition significantly increased the proportion of photoreceptors. Scale bars = 20 µm.
Further, we tested the effects of the canonical AKT agonist SC7935 and antagonist MK220636 on photoreceptor development. Treatment with MK2206 for 2 weeks increased the number of OTX2+, CRX+, and BLIMP1+ cells (Figs. 4J, 4K), confirming its function to facilitate photoreceptor differentiation. In contrast, SC79 reduced OTX2+ and CRX+ cell numbers, although the effect did not reach statistical significance (Figs. 4J, 4K). In summary, these data implicated the reduced activity of the AKT pathway downstream of PHLDA1 may specify human photoreceptors from neurogenic RPCs.
PHLDA1 May Mediate the Effects of IGF1 on Promoting Specification of Photoreceptors from RPCs
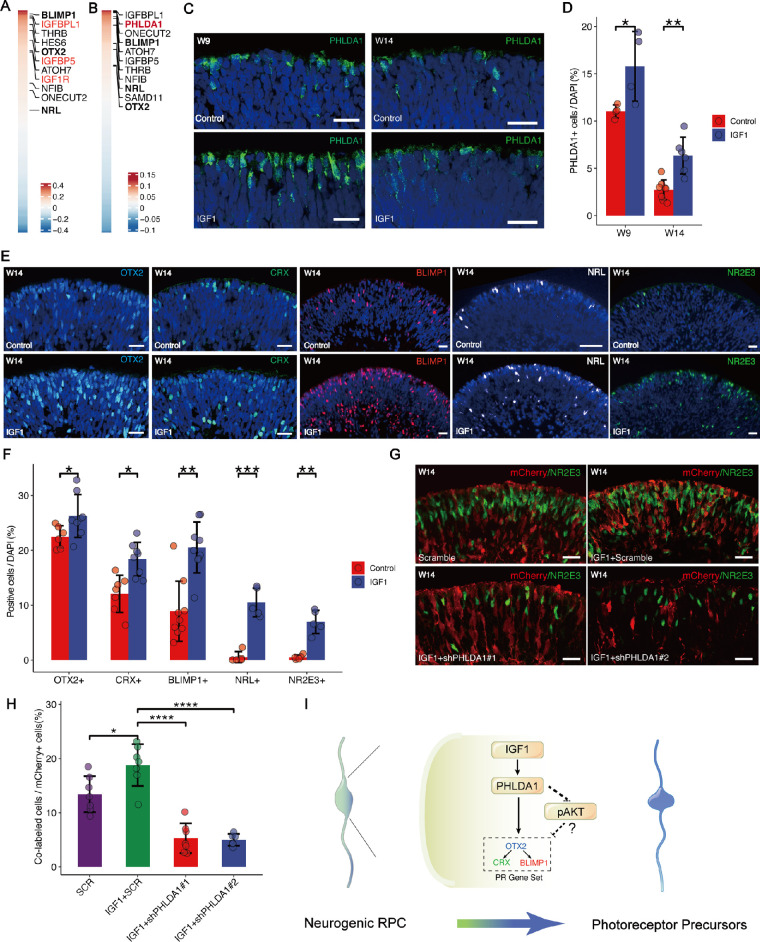
To further investigate the molecular mechanism underlying specification of photoreceptors promoted by PHLDA1, we performed the correlation analysis on cells of photoreceptors. As expected, we found that PHLDA1 was highly correlated with regulators of photoreceptor cell-fate specification (BLIMP1 and OTX2), of cones (THRB, ATOH7, and ONECUT2) and rods (NFIB and NRL; Fig. 5A). Surprisingly, we observed PHLDA1 was correlated with IGF1-related genes, such as IGFBPL1, IGFBP5, and IGF1R (see Fig. 5A), and colocalization of PHLDA1 with IGF1R protein was confirmed by immunofluorescence (Supplementary Figs. S5A, S5B). Similarly, correlation analysis of IGF1R revealed that IGF1R was highly correlated with regulators of photoreceptor cell-fate specification (Fig. 5B), suggesting that IGF1R and PHLDA1 act cooperatively to promote the differentiation of photoreceptors.
Figure 5.
IGF1-PHLDA1 promotes rod specification. (A) Heatmap of genes positively correlated with PHLDA1 expression. The top 10 genes are highlighted in the box. (B) Heatmap of genes positively correlated with IGF1R expression. The top 10 genes are highlighted in the box. (C, D) PHLDA1+ cell number in organoids was significantly upregulated by IGF1 (50 ng/mL) treatment from W7 to W9 and W13 to W14 compared to untreated controls. Scale bars = 20 µm. (E, F) The number of OTX2+, CRX+, BLIMP1+, NRL+, and NR2E3+ cells in organoids were significantly upregulated by IGF1 (50 ng/mL) treatment from W13 to W14 compared to untreated controls. Scale bars = 20 µm. (G, H) IGF1-induced NR2E3+ cell number in organoids was reversed by shRNA-mediated PHLDA1 knockdown (KD) from week W12 to W13 compared to Scramble controls. Scale bars = 20 µm. (I) Schematic illustration of the potential mechanism by which the IGF1-PHLDA1 axis promotes photoreceptor cell specification. PHLDA1, upregulated by IGF1, promotes photoreceptor specification in neurogenic RPCs by inactivating AKT.
By supplementing the IGF1 to the culture media, we found that the number of PHLDA1+ was significantly increased within the organoids of both early and late stages (Figs. 5C, 5D), and the number of OTX2+, CRX+, BLIMP1+, NRL+, and NR2E3+ (Figs. 5E, 5F) cells increased as well. We also found that the number of BrdU+ cells increased after treatment with IGF1 (Supplementary Figs. S5C, S5D), indicating an increase in proliferative RPCs. In line with PHLDA1 overexpression, IGF1 promoted the generation of photoreceptors and upregulated the expression of PHLDA1 (see Figs. 5C, 5D). The knockdown of PHLDA1 blocked the effect of IGF1 on promoting photoreceptor differentiation (Figs. 5G, 5H, Supplementary Fig. S5E.). Together, these results suggest that IGF1 can promote the proliferation of RPCs, and may also promote photoreceptor specification through PHLDA1.
It was reported that IGF1 can act through IGF receptor (IGF1R), insulin receptor (INSR), and the hybrid receptor.37 We then inhibit the IGF1R by treatment with the specific IGF1R inhibitor picropodophyllin (PPP) and knockdown INSR using lentivirus expressing INSR-targeted shRNA together with mCherry (Supplementary Figs. S5F, S5G). The IGF1-induced PHLDA1 expression and -increased NR2E3+ cells number were reversed by PPP (Supplementary Figs. S5H, S5I) but not by shRNA-mediated INSR knockdown (Supplementary Figs. S5J, S5K), suggesting that IGF1R, instead of INSR, mediates the effects of IGF1-PHLDA1 on photoreceptor differentiation.
Discussion
Here, based on a strategy to enrich the photoreceptor lineage, we established a cell atlas during the human photoreceptor development, and successfully characterized the developmental progression of human photoreceptor precursors, identified the IGF1-PHLDA1-AKT signaling pathway to regulate photoreceptor differentiation (see Supplementary Fig. 5I.). This study will provide potential targets for future studies on the mechanisms underlying photoreceptor development and regeneration.
PHLDA1 has been found to act as a downstream regulator of IGF1 signaling pathway in multi cancer cell lines and inhibit AKT activity through its PH domain competing with AKT for phosphatidylinositol binding.34 Our data showed that PHLDA1 knockdown significantly upregulated pAKT and reduced the number of cells expressing the photoreceptor lineage marker, such as OTX2, CRX, and BLIMP1, and then we further confirmed that PHLDA1 overexpression reduced the phosphorylation of AKT and increased the number of photoreceptor cells. Previous studies have characterized PHLDA1 as a mediator of apoptosis, proliferation, and differentiation dependent on cell type and content,34 and AKT plays key roles in cell proliferation and differentitaion.38 We speculate that PHLDA1 promotes the RPCs’ exit from the cell cycle and differentiation from neurogenic RPC to photoreceptor via AKT deactivation. BrdU-labeling after PHLDA1 knockdown revealed that PHLDA1 promotes RPCs exit the cell cycle and differentiation into photoceptor cells. Consistent with our hypothesis, AKT has also been reported to stabilize SOX2 through phosphorylation39 and activate the neurogenic program in hippocampal neuronal precursor cells.40 SOX2 plays an important role in retinal development, knockout of Sox2 in postnatal mice moderately increased rod density, suggesting that SOX2 negatively regulates rod differentiation.41 Thus, these suggested that the IGF1-PHLDA1-AKT cascade may regulate photoreceptor differentiation by reducing SOX2 in RPCs and leading to the RPCs exit of the cell cycle.
Our data showed that IGF1-PHLDA1 promotes retinal cell survival during RPC differentiation, and PHLDA1 inhibits pAKT. Consistently, studies by others reported the role PHLDA1 in inhibiting Pakt.42 As activation of PI3K/AKT signaling can promote neuronal survival,43 we speculate that the effect of PHLDA1 to promote retinal cell survival was not mediated by PI3K/AKT signaling. Previous studies demonstrated that PHLDA1 has pro- and anti-apoptotic roles, dependent on the cell type and state.44 The mechanism to promote the survival of retinal neurons remains to be elucidated by future studies.
Previously, IGF1/IGF1R signaling pathway has been reported to promote the proliferation of mouse RPCs in vitro, predominantly relying on the phosphorylation of MAPK/ERK signaling pathway.45 IGF1 was also shown to induce rod differentiation through PKC signaling pathway.24 Consistently, our data showed that IGF1-IGF1R signaling stimulates the proliferation of RPCs and promotes the differentiation of both cones and rods. As the expression of BLIMP1 commences as cells are exiting mitosis,11 we speculated that changes in photoreceptor cell numbers are not due to the proliferation of BLIMP1+ cells but rather the differentiation of RPCs. In addition, our data showed that PHLDA1 inhibits cell proliferation, suggesting its role in mediating the cell cycle exiting during photoreceptor specification. Together, this study provided novel insights into the molecular mechanisms of IGF1-mediated neuron differentiation.
In addition, this study provided novel approaches to enrich the photoreceptors ex vivo. Loss and degeneration of photoreceptors is the leading cause of blindness. Retinal organoid-derived photoreceptor can integrate into retina and form axons and synaptic terminals. Previous studies showed that the photoreceptor cells from neonatal mouse retina display more efficient integration into the host retina than cells from either embryonic retina, or older than P11.46 Furthermore, forced expression of IGF1 led to significantly increased levels of cell integration, which may result from the potential of IGF1 on photoreceptor differentiation.47 The IGF1-PHLDA1-AKT axis proposed by our study will provide novel targets to enrich the photoreceptors and improve the therapeutic efficiency in the future.
This study has several limitations. We observed multiple roles of PHLDA1 to regulate the photoreceptor specification, RPC proliferation, and survival, but it is still under investigation how PHLDA1 integrates into the genetic programs underlying photoreceptor differentiation. In addition, as the number of assessment time points was limited, the precise functions of PHLDA1 in mediating photoreceptor differentiation need to be fully addressed by studying retinal organoids over the differentiation time course.
Supplementary Material
Acknowledgments
Funded by the National Natural Science Foundation of China (32171445 and 81721003); National Key R&D Program of China (2017YFC1001300 and 2018YFA0108300); Clinical Innovation Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR0201001); Research Unit Of Ocular Development And Regeneration, Chinese Academy Of Medical Sciences (2019-I2M-5-005); Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2017BT01S138); and the Science and Technology Program of Guangzhou; the Young Teacher Training Project of Sun Yat-sen University (20ykpy142).
Data Availability. All data supporting the findings of this study are included in the article and supplementary information files. The scRNA-seq data have been deposited in the Gene Expression Omnibus (GEO) under series number GSE181737.
Author Contributions: Y.H., X.M., Q.L., and Y.L. conceived and designed the study. X.M., Y.X., X.H., K.C., X.C., W.D., Y.L., and P.M. performed experiments. Y.H., Y.X., S.Y., S.Z., and X.M. analyzed the data and performed statistical analyses. Y.H., X.M., Y.X., and Y.L. interpreted the data and wrote the manuscript in discussion with all authors.
Disclosure: Y. Xiao, None; X. Mao, None; X. Hu, None; S. Yuan, None; X. Chen, None; W. Dai, None; S. Zhang, None; Y. Li, None; M. Chen, None; P. Mao, None; Y. Liu, None; Q. Liu, None; Y. Hu, None
References
- 1. Zeng H, Sanes JR.. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci. 2017; 18: 530–546. [DOI] [PubMed] [Google Scholar]
- 2. Baden T, Euler T, Berens P.. Understanding the retinal basis of vision across species. Nat Rev Neurosci. 2020; 21: 5–20. [DOI] [PubMed] [Google Scholar]
- 3. Mustafi D, Engel AH, Palczewski K.. Structure of cone photoreceptors. Prog Retin Eye Res. 2009; 28: 289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ingram NT, Sampath AP, Fain GL.. Why are rods more sensitive than cones? J Physiol. 2016; 594: 5415–5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamb TD. Why rods and cones? Eye (Lond). 2016; 30: 179–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swaroop A, Kim D, Forrest D.. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010; 11: 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat Rev Neurosci. 2014; 15: 615–627. [DOI] [PubMed] [Google Scholar]
- 8. Livesey FJ, Cepko CL.. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001; 2: 109–118. [DOI] [PubMed] [Google Scholar]
- 9. Ng L, Lu A, Swaroop A, Sharlin DS, Swaroop A, Forrest D.. Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J Neurosci. 2011; 31: 11118–11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim JW, Yang HJ, Oel AP, et al.. Recruitment of Rod Photoreceptors from Short-Wavelength-Sensitive Cones during the Evolution of Nocturnal Vision in Mammals. Dev Cell. 2016; 37: 520–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cepko CL. The Determination of Rod and Cone Photoreceptor Fate. Annu Rev Vis Sci. 2015; 1: 211–234. [DOI] [PubMed] [Google Scholar]
- 12. Lu Y, Shiau F, Yi W, et al.. Single-Cell Analysis of Human Retina Identifies Evolutionarily Conserved and Species-Specific Mechanisms Controlling Development. Dev Cell. 2020; 53: 473–491.e479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cowan CS, Renner M, De Gennaro M, et al.. Cell Types of the Human Retina and Its Organoids at Single-Cell Resolution. Cell. 2020; 182: 1623–1640.e1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clark BS, Stein-O'Brien GL, Shiau F, et al.. Single-Cell RNA-Seq Analysis of Retinal Development Identifies NFI Factors as Regulating Mitotic Exit and Late-Born Cell Specification. Neuron. 2019; 102: 1111–1126.e1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamamoto H, Kon T, Omori Y, Furukawa T.. Functional and Evolutionary Diversification of Otx2 and Crx in Vertebrate Retinal Photoreceptor and Bipolar Cell Development. Cell Rep. 2020; 30: 658–671.e655. [DOI] [PubMed] [Google Scholar]
- 16. Torero Ibad R, Mazhar B, Vincent C, et al.. OTX2 Non-Cell Autonomous Activity Regulates Inner Retinal Function. eNeuro. 2020; 7(5): 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koike C, Nishida A, Ueno S, et al.. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007; 27: 8318–8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brzezinski JAT, Lamba DA, Reh TA.. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development. 2010; 137: 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goodson NB, Park KU, Silver JS, Chiodo VA, Hauswirth WW, Brzezinski JAT. Prdm1 overexpression causes a photoreceptor fate-shift in nascent, but not mature, bipolar cells. Dev Biol. 2020; 464: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Furukawa T, Morrow EM, Li T, Davis FC, Cepko CL.. Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat Genet. 1999; 23: 466–470. [DOI] [PubMed] [Google Scholar]
- 21. Rhee KD, Goureau O, Chen S, Yang XJ.. Cytokine-induced activation of signal transducer and activator of transcription in photoreceptor precursors regulates rod differentiation in the developing mouse retina. J Neurosci. 2004; 24: 9779–9788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Graham DR, Overbeek PA, Ash JD.. Leukemia inhibitory factor blocks expression of Crx and Nrl transcription factors to inhibit photoreceptor differentiation. Invest Ophthalmol Vis Sci. 2005; 46: 2601–2610. [DOI] [PubMed] [Google Scholar]
- 23. Zerti D, Molina MM, Dorgau B, et al.. IGFBPs mediate IGF-1's functions in retinal lamination and photoreceptor development during pluripotent stem cell differentiation to retinal organoids. Stem Cells. 2021; 39: 458–466. [DOI] [PubMed] [Google Scholar]
- 24. Pinzon-Guzman C, Zhang SS, Barnstable CJ.. Specific protein kinase C isoforms are required for rod photoreceptor differentiation. J Neurosci. 2011; 31: 18606–18617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nie J, Hashino E.. Organoid technologies meet genome engineering. EMBO Rep. 2017; 18: 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Collin J, Zerti D, Queen R, et al.. CRX Expression in Pluripotent Stem Cell-Derived Photoreceptors Marks a Transplantable Subpopulation of Early Cones. Stem Cells. 2019; 37: 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phillips MJ, Capowski EE, Petersen A, et al.. Generation of a rod-specific NRL reporter line in human pluripotent stem cells. Sci Rep. 2018; 8: 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Glubrecht DD, Kim JH, Russell L, Bamforth JS, Godbout R.. Differential CRX and OTX2 expression in human retina and retinoblastoma. J Neurochem. 2009; 111: 250–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Javed A, Mattar P, Lu S, et al.. Pou2f1 and Pou2f2 cooperate to control the timing of cone photoreceptor production in the developing mouse retina. Development. 2020; 147(18): dev188730. [DOI] [PubMed] [Google Scholar]
- 30. Xie H, Zhang W, Zhang M, et al.. Chromatin accessibility analysis reveals regulatory dynamics of developing human retina and hiPSC-derived retinal organoids. Sci Adv .2020; 6: eaay5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Qiu X, Mao Q, Tang Y, et al.. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017; 14: 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brzezinski JAT, Kim EJ, Johnson JE, Reh TA.. Ascl1 expression defines a subpopulation of lineage-restricted progenitors in the mammalian retina. Development. 2011; 138: 3519–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin ZB, Okamoto S, Osakada F, et al.. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS One. 2011; 6: e17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagai MA. Pleckstrin homology-like domain, family A, member 1 (PHLDA1) and cancer. Biomed Rep. 2016; 4: 275–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jo H, Mondal S, Tan D, et al.. Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proc Natl Acad Sci USA. 2012; 109: 10581–10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng Y, Zhang Y, Zhang L, et al.. MK-2206, a novel allosteric inhibitor of Akt, synergizes with gefitinib against malignant glioma via modulating both autophagy and apoptosis. Mol Cancer Ther .2012; 11: 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ziegler AN, Levison SW, Wood TL.. Insulin and IGF receptor signalling in neural-stem-cell homeostasis. Nat Rev Endocrinol. 2015; 11: 161–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu JS, Cui W.. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016; 143: 3050–3060. [DOI] [PubMed] [Google Scholar]
- 39. Jeong CH, Cho YY, Kim MO, et al.. Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells. 2010; 28: 2141–2150. [DOI] [PubMed] [Google Scholar]
- 40. Mir S, Cai W, Carlson SW, Saatman KE, Andres DA.. IGF-1 mediated Neurogenesis Involves a Novel RIT1/Akt/Sox2 Cascade. Sci Rep. 2017; 7: 3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Surzenko N, Crowl T, Bachleda A, Langer L, Pevny L.. SOX2 maintains the quiescent progenitor cell state of postnatal retinal Muller glia. Development. 2013; 140: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen Y, Takikawa M, Tsutsumi S, et al.. PHLDA1, another PHLDA family protein that inhibits Akt. Cancer Sci. 2018; 109: 3532–3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saito A, Narasimhan P, Hayashi T, Okuno S, Ferrand-Drake M, Chan PH.. Neuroprotective role of a proline-rich Akt substrate in apoptotic neuronal cell death after stroke: relationships with nerve growth factor. J Neurosci. 2004; 24: 1584–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yousof T, Byun JH, Chen J, Austin RC.. Pleckstrin Homology-Like Domain, Family A, Member 1 (PHLDA1): A Multifaceted Cell Survival Factor that Drives Metabolic Disease. Engineering [published online ahead of print July 2, 2022], 10.1016/j.eng.2022.05.014. [DOI] [Google Scholar]
- 45. Wang Y, Zhang D, Zhang Y, et al.. Insulin-like growth factor-1 regulation of retinal progenitor cell proliferation and differentiation. Cell Cycle. 2018; 17: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. MacLaren RE, Pearson RA, MacNeil A, et al.. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006; 444: 203–207. [DOI] [PubMed] [Google Scholar]
- 47. West EL, Pearson RA, Duran Y, et al.. Manipulation of the recipient retinal environment by ectopic expression of neurotrophic growth factors can improve transplanted photoreceptor integration and survival. Cell Transplant. 2012; 21: 871–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hao Y, Hao S, Andersen-Nissen E, et al.. Integrated analysis of multimodal single-cell data. Cell. 2021; 184: 3573–3587.e3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finak G, McDavid A, Yajima M, et al.. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015; 16: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu T, Hu E, Xu S, et al.. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb). 2021; 2: 100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aibar S, Gonzalez-Blas CB, Moerman T, et al.. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017; 14: 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.