Abstract
Stable isotope labeling techniques have been widely applied in the field of metabolomics and proteomics. Before the measured mass spectral data can be used for quantitative analysis, it must be accurately corrected for isotope natural abundance and tracer isotopic impurity. Despite the increasing popularity of dual-isotope tracing strategy such as 13C-15N or 13C-2H, there are no accurate tools for correcting isotope natural abundance for such experiments in a resolution-dependent manner. Here, we present AccuCor2 as an R-based tool to perform the correction for 13C-15N or 13C-2H labeling experiments. Our method uses a newly designed algorithm to construct the correction matrices that link labeling pattern and measured mass fractions, then use non-negative least-squares to solve the labeling patterns. Our results show that the dual-isotope experiments often require a mass resolution that is high enough to resolve 13C and 15N or 13C and 2H. Otherwise, the labeling pattern is not solvable. However, this mass resolution may not be sufficiently high to resolve other non-tracer elements such as oxygen or sulfur from the tracer elements. Therefore, we design AccuCor2 to perform the correction based on the actual mass resolution of the measurements. Using both simulated and experimental data, we show that AccuCor2 performs accurate and resolution-dependent correction for dual-isotope tracer data.
INTRODUCTION
Stable isotope tracers have been widely used to study the metabolism of carbohydrates, protein, lipids, and metabolic co-factors [1]. Tracers can be used to determine the origins of specific metabolites by quantifying the incorporation of the isotope atoms from the labeled substrates [2-4]. Tracers can also be infused in vivo in a non-perturbative way to measure the turnover rates of circulating metabolites [5, 6]. Another application of stable isotope tracing is metabolic flux analysis, which provides a quantitative evaluation of the rates of biochemical reactions in the entire metabolic network [7-9]. The most commonly used tracing isotopes are 13C, 2H, and 15N. 13C tracers are frequently used to study the biosynthesis of metabolites and quantify the carbon flow in the metabolic network [10]. However, there are also many metabolic reactions such as transamination, (de)hydration, and redox reactions that do not involve the formation or cleavage of carbon─carbon bonds [11]. In these cases, the 13C tracers are not informative and tracers that include 2H or 15N should be used instead. 2H tracers are commonly used to study isomerase reactions and dehydrogenase reactions that involve NADH or NADPH as co-factors [12, 13]. Meanwhile, 15N tracers are widely used to investigate the metabolism of amino acids and nucleotides [14, 15]. Modern mass spectrometry provides a high mass resolution that enables unambiguous detection of various stable isotopes simultaneously. As a result, tracers containing dual isotopes (e.g., 13C-15N and 13C-2H) have been increasingly deployed in biological experiments. For example, L-[13C6,15N4] arginine was used to investigate autophagy and melanoma growth [16]. L-[1-13C, 15N] leucine was used to study the whole-body amino acid metabolism [17]. Methyl[2H3]-13C-methionine was used to study protein methylation and breakdown [18]. In these applications, the biological interpretation of the results depends on the accurate determination of the isotopic tracer incorporation in the metabolites.
It should be noted that the measured mass isotopologue distribution does not directly reflect the isotope incorporation from tracers. This is because of the natural abundance (NA) of isotopes. For example, the unlabeled metabolite NAD+ (C21H27N7O14P2) would have 19% being M + 1 because of the NA of 13C. Furthermore, 2.8% of unlabeled NAD+ could be M + 2 because of the NA of 18O. These NAs must be accounted for to reveal the actual tracer isotope incorporation, which is done through a process called isotope natural abundance correction (INAC).
Many efforts have been made to advance the methodology of INAC. Biemann [19] proposed the first INAC algorithm, which subtracts the isotopologue fractions in a stepwise manner (i.e., NA contributions from m + 0 were corrected first, followed by contributions from m + 1, m + 2, etc.). Although the stepwise approach is mathematically correct, this method is prone to experimental errors since the measurement error in one mass fraction may be propagated across all other mass fractions and lead to a biased result. This issue was solved by Brauman [20], Wittmann and Heinzle [21], and Van Winden et al. [22] using a least-squares approach where all measurement errors were weighted equally. This method first uses probability theory to generate a correction matrix that links the isotope labeling pattern and the measured mass fractions affected by NA. Then, a least-squares approach is applied to solve the isotope labeling pattern. Millard et al. [23] further improved this method by applying a non-negative least-squares method to solve the labeling pattern and developed a well-established software IsoCor. IsoCor is an efficient tool that can perform both tracer impurity correction and INAC for many different tracer elements on large datasets. While IsoCor is accurate for low-resolution mass spectrometry data, it may have an over-correction problem when handling high-resolution data. This is because under high resolution, some isotopologues may be resolved from the tracer isotopologues and therefore, the corresponding fraction should not be subtracted. To address this issue, Su et al. [24] developed the first resolution-dependent INAC tool called AccuCor. AccuCor uses resolution information to evaluate the impact of all non-tracer isotopes and only corrects the fractions that affect the tracer isotopologue channels. Millard et al. [25] and Du et al. [26] further refined this method by calculating the correction limits for non-tracer isotope combinations and developed IsoCor2 and ElemCor, respectively. As the use of dual-isotope tracers becomes increasingly popular, several tools have been developed to perform INAC on multi-isotope experiments [27-29]. However, these tools assume that all the non-tracer isotopologues can be resolved from tracer isotopologues with an ultra-high-resolution instrument, which is not necessarily true and would be discussed later.
In this study, we developed AccuCor2 as the first tool to perform a resolution-dependent INAC in data from dual-isotope tracers. We found that such correction problems require the two tracer isotopes to be resolved under sufficiently high mass resolution. The non-tracer isotopes are often not fully resolved, and AccuCor2 can account for them in a resolution-dependent manner. We also compared the performance of AccuCor2 with two other available tools IsoCorrectoR [28] and PICor [29], using both simulated and experimental data. The result shows both IsoCorrectoR and PICor under-corrected isotopologues caused by non-tracer isotope NA, whereas AccuCor2 correctly solved this problem.
MATERIALS AND METHODS
Theory of INAC
INAC is the process whereby isotope labeling patterns are solved from the measured mass fractions. The isotope labeling pattern (denoted by L) and the measured mass fractions (denoted by M) can be expressed as column vectors. These two vectors are related by the correction matrix (denoted by CM): CM × L = M [21, 22]. The ith row and jth column element of the correction matrix CMij represents the probability of the jth labeled fraction contributing to the ith measured mass fraction due to the isotope NA and the isotopic impurity of the tracer [21, 22]. The INAC algorithm that is suitable for high-resolution mass spectrometry data will construct the correction matrix based on the chemical formula of the ion, the mass resolution of the measurements, and the isotopic purity of the tracer [24-26]. Once the correction matrix is built, the labeling pattern can be solved by non-negative least-squares algorithms such as Hanson–Lawson or L-BFGS-B [23, 24]. Here, we will demonstrate the construction of the correction matrix for dual-isotope data.
The form of the correction matrix is determined by the forms of the isotope labeling pattern vector L and measured mass fractions vector M. Here, we use serine ([M-H]− C3H6NO3) as an example. If serine is only labeled with 13C isotope in the experiment, the labeling pattern vector L = [13C0, 13C1, 13C2, 13C3]⊤ and the measured mass fractions vector M = [M + 0, M + 1, M + 2, M + 3]⊤. The CM connecting these two vectors would be a 4 × 4 matrix. In contrast, if serine is labeled with both 13C and 15N, L and M vectors and their relationship can be expressed as Eq. (1).
| (1) |
In Eq. (1), the element 13Cm 15Nn in the L vector denotes the fraction with m-13C and n-15N labeling. The element M+CiNj in the M vector denotes the observed mass fraction that corresponds to the incorporation of i 13C and j 15N atoms. As we shall see in “Results”, in the dual-isotope labeling experiment, it is necessary to be able to resolve mass fractions of the same nominal mass such as M+C1N0 and M+C0N1. If the measurements are done under unit mass resolution, and the M + 1 (m/z 105 for serine) fraction is obtained instead of the resolved M+C1N0 (m/z 105.03867 for serine) and M+C0N1 (m/z 105.03235 for serine) fractions, the correction may fail to generate the accurate labeling pattern. For serine, which has three C and one N atoms, the correction matrix CM is an 8 × 8 matrix.
Construction of the correction matrix
CM can be factorized into three matrices: the tracer isotopic purity matrix IPM, the tracer elements matrix TM, and the non-tracer elements matrix NTM: CM = NTM × TM × IPM. We begin the demonstration with the construction of TM because this matrix usually has the greatest impact on CM. For convenience, we always use the following convention to arrange the terms in the L and M vectors: ascending numbers of 13C followed by ascending numbers of 2H or 15N (Eq. (1)). Therefore, the TM for 13C-15N labeling of serine can be constructed (Eq. (2)).
| (2) |
As shown in Eq. (2), for a metabolite with m carbon atoms and n nitrogen atoms, TM is a square matrix with (m + 1) × (n + 1) row and columns. In fact, TM is composed of (n + 1) × (n + 1) blocks where each block is an (m + 1) × (m + 1) submatrix. For example, the element in row 7 column 6 (element [7, 6]) is within block [2, 2] and position [3, 2] in the submatrix. This element represents the probability of the 13C1 15N1-labeled fraction that is appearing as M + C2N1 due to the carbon and nitrogen NA. Since the 13C1 15N1-labeled fraction of serine already has one 13C from the tracer, only two carbon atoms are natural carbon. Similarly, since the 13C1 15N1-labeled fraction of serine has one 15N from the tracer, no nitrogen atom is natural. We use the term to represent the probability of having one 13C out of two natural carbon atoms: , in which pC = 0.011 represents the NA of 13C. Therefore, the element [7, 6] is . More generally, for the carbon─nitrogen TM, the element [(i − 1) × (m + 1) + f, (j − 1) × (m + 1) + g], which is in the ith row and jth column of the blocks and the fth row and gth column of the submatrix has the value of . The element [7,6] has the block indices of i = 2, j = 2 and submatrix indices f = 3, g = 2, and this element has the value of . Because when f < g or i < j, TM is a lower-triangular matrix, and each nonzero submatrix is also a lower-triangular matrix. Because of this triangularity, TM is always well-conditioned.
Similar to TM, we can also construct the isotopic purity matrix IPM for 13C-15N-labeled serine (Eq. (3)).
| (3) |
For a metabolite with m carbon atoms and n nitrogen atoms, IPM is a square matrix composed of (n + 1) × (n + 1) blocks where each block is an (m + 1) × (m + 1) submatrix. The element in row 5 column 7 (element [5, 7]) is within block [2, 2] and position [1, 3] in the submatrix. This element represents the probability of the 13C2 15N1-labeled fraction that is appearing as M + C0N1 due to the tracer isotopic impurities. We use the term to represent the probability of having two natural carbon atoms out of two tracer carbon atoms: , in which pIPC = 0.99 represents the isotopic purity of 13C in the tracer. Therefore, the element [5, 7] is . More generally, for the carbon─nitrogen IPM, the element [(i − 1) × (m + 1) + f, (j − 1) × (m + 1) + g], which is in the ith row and jth column of the blocks and the fth row and gth column of the submatrix has the value of . The element [5, 7] has the block indices of i = 2, j = 2 and submatrix indices f = 1, g = 3, and this element has the value of . Because when g < f or j < i, IPM is an upper triangular matrix, and each nonzero submatrix is also an upper triangular matrix. Again, because of this triangularity, IPM is always well-conditioned.
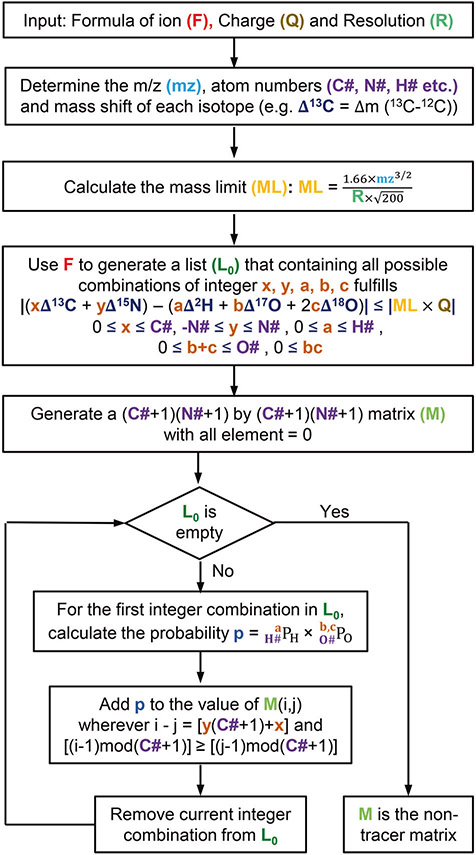
Unlike the construction of TM and IPM described above, the construction of NTM is affected by the mass resolution of the measurement [24]. For serine that is 13C-15N-labeled, it takes a minimum nominal mass resolution of 50,800 (defined at m/z 200 on Orbitrap analyzer) [24] to resolve 18O1 and 13C2 peaks but a minimum nominal mass resolution of 140,000 to resolve 17O1 and 13C1 peaks on the mass spectrum. Therefore, if data were measured at a mass resolution of 100,000, NTM should include the term of 17O1 but exclude the term of 18O1 to avoid over-correction. In our previous work, NTM was further factorized. The contributions from each non-tracer element such as oxygen or sulfur were described by separate matrices. These matrices were multiplied together to obtain the full NTM. Later, Millard et al. [25] pointed out that the combination of certain non-tracer elements may be harder to resolve than individual elements. For example, a minimum resolution of 50,800 and 42,500 is required to resolve 18O1 versus 13C2 and 2H1 versus 13C1 of serine, respectively. However, it takes a minimal resolution of 260,000 to resolve 18O1 + 2H1 versus 13C3 of serine. As a result, a single NTM should be constructed instead of a product of matrices for individual elements. The latest version of AccuCor has adopted such an algorithm for improved accuracy. For AccuCor2, we also construct a single NTM to account for the NA of the non-tracer elements. The algorithm for constructing NTM is shown in Fig. 1. First, the algorithm uses the input data to calculate the mass limit (ML) of resolvable peaks. Second, the algorithm generates a list of atom combinations that satisfy the following two criteria: (1) for each element, the atom number in the list does not exceed the atom number in the chemical formula; (2) the difference between the mass shift caused by the tracer atoms and the non-tracer atoms is within the ML. Last, the probability of each atom combination in the list is calculated and added to the corresponding position of NTM. The construction completes when all entries in the list are accounted.
Fig. 1. NTM construction algorithm for 13C-15N labeling experiments.
In this example, carbon and nitrogen are the tracer elements, and we assume, for simplicity, that the non-tracer elements only include hydrogen and oxygen. The algorithm takes the chemical formula of the ion, the charge and the mass resolution as the input information. The output is the resolution dependent NTM. Algorithm for 13C-2H labeling experiments follows the same structure.
| (4) |
As shown in Eq. (4), for a metabolite with m carbon and n nitrogen atoms, NTM is a square matrix composed of (n + 1) × (n + 1) blocks where each block is an (m + 1) × (m + 1) submatrix. The element in row 4 column 1 (element [4, 1]) represents the probability of the 13C0 15N0-labeled fraction contributing to the M + C3N0 m/z channel due to the non-tracer elements. Under 100,000 resolution, the only non-tracer element combination that makes such mass shift is 2H1 + 18O1, which has the probability of . The term represents the probability of having one 2H atom out of six natural hydrogen atoms. The term represents the probability of having zero 17O and one 18O atom out of three natural oxygen atoms. Note that NTM is not strictly lower-triangular since element [4, 5] is nonzero. This represents the probability of 13C0 15N1-labeled serine contributing to the M + C3N0 channel, which reflects the fact that 15N1 + 2H2 and 12C3 are not resolved for serine under 100,000 resolution.
Materials
LCMS-grade methanol (A456), acetonitrile (A955), acetic acid (A35), and water (ACROS 61515) were purchased from Fisher Chemicals (Pittsburgh, PA). 13C3-15N1 L-serine (99% isotopic purity on both 13C and 15N, CNLM-474-H-PK) was purchased from Cambridge Isotope Laboratories (Tewksbury, MA). L-serine (S4500) was purchased from MilliporeSigma (Burlington, MA).
Sample preparation
One hundred micrometer of L-serine, 13C3-15N1 L-serine, or 1:1 mixture were dissolved in 40:40:20 (acetonitrile:methanol:water) and centrifuged for 10 min at 4 °C. The supernatant was collected for LC–MS analysis.
Liquid chromatography–mass spectrometry
The LC–MS method is the same as previously published [30]. Conditions were optimized on an HPLC–ESI–MS system fitted with a Vanquish Horizon UHPLC and a Thermo Q Exactive Plus MS. The MS scans were obtained in negative ionization mode with a nominal mass resolution of 70,000 (defined at m/z 200), in addition to an automatic gain control target of 3 × 106 and m/z scan range of 72–1000. Metabolite data were obtained using the MAVEN software package [31] with each labeled isotope fraction (mass accuracy window: 16 ppm).
Simulation of mass fractions
Simulations of NAD+ mass fractions were done using Thermo Xcalibur Qual Browser software with 750,000 nominal mass resolution (defined at m/z 200).
Isotope natural abundance correction (INAC)
AccuCor2 works for data acquired on Obitrap mass analyzers. AccuCor2 calculates the ML for correction based on the nominal mass resolution for data acquisition and the m/z values of the metabolites [24].
| (5) |
AccuCor2 supports the isotopes of 13C, 2H, 15N, 17O, 18O, 33S, 34S, 29Si, 30Si, 37Cl, and 81Br. The isotope abundance data are taken from Isotopic Compositions of the Elements 1997 [32], and the users are allowed to change the isotope abundance values. The natural isotope abundance correction code was written in R [33]. The labeling pattern solving was done using the Hanson–Lawson non-negative least-squares method, which was implemented in the NNLS package [34], to avoid negative fractions. We used PICor [29] and IsoCorrectoR [28] to compare the performance of INAC. For the correction of NAD labeling, the root-mean-square deviation (RMSD) is calculated using the sum of squared residue between the corrected values and the theoretical values of the labeling pattern. For serine and the sodium acetate adduct of serine, we only calculated the RMSD for Sample 3 which is the 100% 13C3,15N1. The AccuCor2 code is freely available for both 13C-2H, and 13C-15N labeling studies (https://github.com/wangyujue23/AccuCor2).
RESULTS
Requirement on the mass resolution for dual-isotope tracer experiment
Early isotope tracer experiments that use a single isotope were typically done on unit resolution mass spectrometers. The dual-isotope tracer experiments, however, often have a minimum requirement on the mass resolution of the instrument. Take 13C-15N serine as an example, there are eight possible labeled fractions of serine: 13C0-3-15N0-1. The labeling pattern of serine can be correctly solved if the sample is measured on a high-resolution instrument (see “Materials and methods: Theory of INAC”). Under low mass resolution, different labeled fractions with the same nominal mass are combined (e.g., M + C1N0 and M + C0N1 are combined as M + 1), resulting in fewer mass fraction measurements than the labeled fractions. Therefore, when measured on a unit resolution mass spectrometer, serine has only five mass fractions M + 0−M + 4. It is obviously impossible to solve eight labeling fractions based on the measurements of only five mass fractions. Therefore, the labeling fractions of the metabolite cannot be determined.
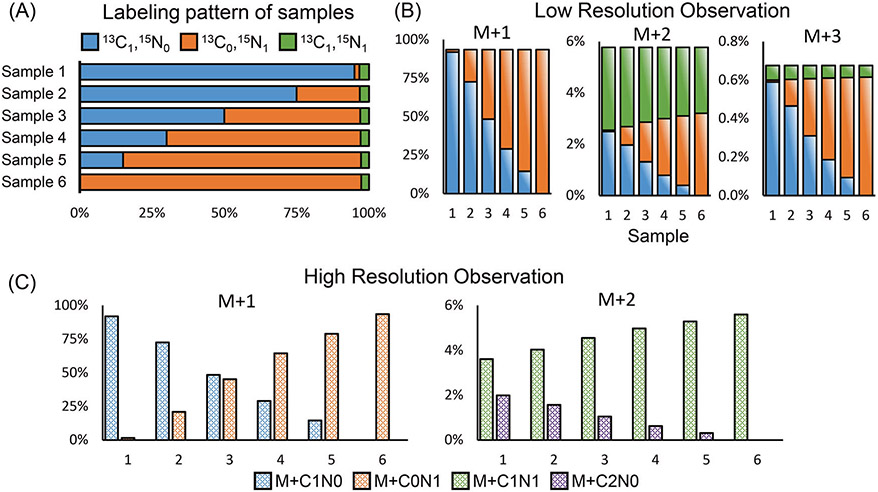
Even if the choice of the tracer limits the number of possible labeling fractions to be no more than the number of measured mass fractions, the correction may still fail. Suppose serine can be labeled in only three forms: 13C1-15N0, 13C0-15N1, and 13C1-15N1. Under unit mass resolution, the 13C1-15N0 and 13C0-15N1 serine are both measured as M + 1 and the 13C1-15N1 fraction is measured as M + 2. Due to the isotope NA, the 13C1-15N0 and 13C0-15N1 serine also contribute to the M + 2 and M + 3 fractions; the 13C1-15N1 serine also contributes to the M + 3 fraction. Therefore, it would appear to be possible to determine three labeling fractions 13C1-15N0, 13C0-15N1, and 13C1-15N1 from the measurement of three mass fractions M + 1, M + 2, and M + 3. However, Fig. 2 provides a set of examples to demonstrate that such correction cannot be done. We calculated six combinations of labeling fractions (Fig. 2A, Samples 1–6), which have the 13C1-15N0 fraction ranging from 0 to 95%. These samples have exactly the same M + 1, M + 2, and M + 3 fractions when measured under unit mass resolution (Fig. 2B, Supplementary Table 1). Therefore, the labeling fractions cannot be calculated from the measured mass fractions. In fact, there are infinitely many combinations that could fit these mass measurements. The mathematical reason behind this phenomenon is that low resolution causes certain columns of the correction matrix, such as those corresponding to M + C1N0 and M + C0N1, to be combined as M + 1. These combinations make the correction matrix singular. With a singular correction matrix, the mass fraction vector M becomes insensitive to the changes in L. Therefore, given a measurement of M, the labeling pattern vector L cannot be uniquely determined.
Fig. 2. Dual-isotope tracer experiment requires high mass resolution.
A Serine isotopologue mixtures (13C1-15N0, 13C0-15N1 and 13C1-15N1) were measured under low and high resolution MS. B Low resolution do not distinguish 13C and 15N, leading to the same observation among all samples. C Consequently, the labeling pattern is unsolvable under low resolution. In contrast, high resolution can distinguish 13C and 15N isotopologues, leading to unique solution of labeling pattern.
In contrast, when these samples were measured under a mass resolution that is high enough to resolve 13C and 15N, the labeling fraction of 13C1-15N0, 13C0-15N1, and 13C1-15N1 would be measured as M + C1N0, M + C0N1, and M + C1N1, respectively (Fig. 2C). 13C1-15N0 would also contribute to M + C2N0 due to NA, which can be resolved from M + C1N1 under high mass resolution. In this case, all six samples give different mass fraction measurements so that their respective labeling patterns can be solved accurately. For serine ([M-H]−, m/z 104.03532), the nominal mass resolution (for Orbitrap instrument, defined at m/z 200) [24, 35] should be >19,700 to resolve 13C and 15N. In general, the mass resolution should be high enough to resolve all the labeling fractions of interest. This requirement should be taken into consideration when designing a dual-isotope tracer experiment.
Resolution-dependent correction for the non-tracer elements
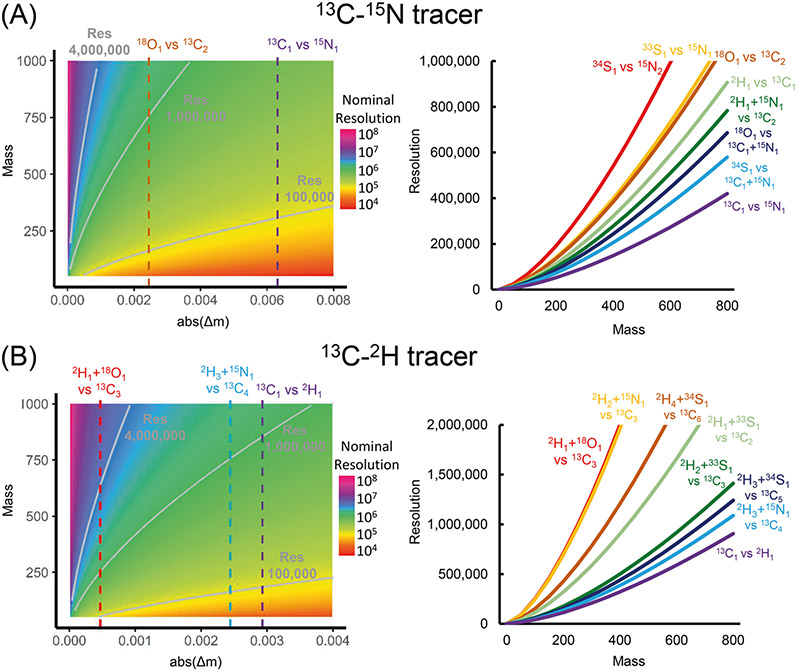
Dual-isotope tracer experiments require a minimum mass resolution under which the tracer isotopes can be resolved. However, this resolution may not be high enough to resolve other non-tracer elements, which should also be corrected. As shown by Su et al. [24], the minimum resolution for isotopologue separation depends on both the absolute mass difference (Δm) and m/z (represented by m in the formula).
| (5) |
Equation (5) suggests that as the mass difference Δm goes lower, a higher mass resolution is required to resolve the corresponding isotopologues. Figure 3 provides a visual illustration of the relationship among the resolution requirement, the mass difference, and the mass of the ion. For serine ([M-H]−, m/z 104.03532) that is 13C- and 15N-labeled, it only takes a minimum resolution of 19,700 to resolve the 13C1-15N0 and 13C0-15N1 isotopologues. However, it takes a minimum resolution of 50,800 to resolve 13C2 and 18O1 (Δm = 0.00245). Similarly, for serine that is 13C- and 2H-labeled, it takes a minimum resolution of 42,500 to resolve the 13C1-2H0 and 13C0-2H1 isotopologues. However, it takes a minimum resolution of 260,000 to resolve 2H1-18O1 and 13C3 (Δm = 0.00048) and 50,400 to resolve 2H3-15N1 and 13C4 (Δm = 0.00247).
Fig. 3. Effect of resolution on isotopologue separation.
The minimum nominal mass resolution (defined at m/z 200 on Orbitrap analyzer) for separating isotopologues according to the mass of ion and mass difference (Δm) for experiments using 13C-15N (A) and 13C-2H (B) tracer experiments. The most commonly observed isotopologue pairs are shown in different colors. All isotopologues shown were measured with −1 charge.
Moreover, as the mass of the ion goes up, the required resolution also goes up (Fig. 3). For acetyl-CoA ([M-H]−, C23H37N7O17P3S) that is 13C-15N-labeled, it takes a minimum resolution of 427,000 to resolve the 13C1-15N0 and 13C0-15N1 isotopologues, which is achievable on modern Orbitrap instruments. However, it takes a minimum resolution of 697,000 to resolve 13C1-15N1 and 18O1 (Δm = 0.00387); 1,100,000 to resolve 13C2 and 18O1 (Δm = 0.00245); and 1,550,000 to resolve 34S1 and 15N2 (Δm = 0.00174). The mass resolution for resolving these isotopologues is not attainable on many current mainstream instruments. Fortunately, as long as the tracer isotopes are resolved, the non-tracer correction matrix is always well-conditioned so that the correction can be done even when the non-tracer isotopes are not resolved.
To accurately account for the contribution from the NA of the non-tracer elements, NTM should be constructed based on the actual mass resolution of the experiment (see “Materials and methods: Theory of INAC”). AccuCor2 will calculate whether a specific isotopologue is resolved from any of the tracer isotopologues based on the mass resolution of the measurement. If the resolution is not high enough so that the non-tracer NA contributes to a mass channel of a labeling fraction, AccuCor2 will add associated probability to NTM and correct for it. If the resolution is high enough to resolve the non-tracer isotopologue from the labeling fraction, AccuCor2 will leave this fraction off from NTM to avoid over-correction.
AccuCor2 performs accurate NA correction from dual-isotope tracer data
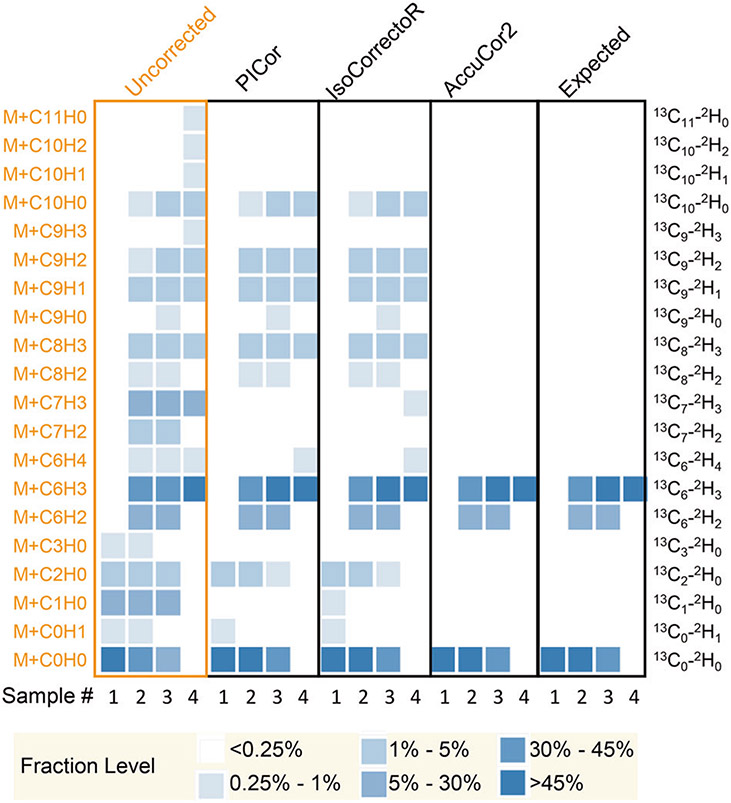
To test the performance of AccuCor2, we first used a simulated dataset of 13C-2H-labeled NAD+ ([M-H]−, C21H26N7O14P2) (Fig. 4). The simulation assumed a mass resolution of 750,000, which is sufficient to resolve 13C and 2H. The simulation also assumed the following labeling patterns: sample 1: 100% non-labeled; sample 2: 50% non-labeled + 10% 13C6-2H2 + 40% 13C6-2H3; sample 3: 36% non-labeled + 14% 13C6-2H2 + 50% 13C6-2H3; sample 4: 100% 13C6-2H3. These isotopologues were chosen because they are possible products when 13C6-2H4 nicotinamide tracer is used [36]. The simulated dataset shows that besides the expected labeled fractions such as M + C0H0, M + C6H2, and M + C6H3, there are many unexpected isotopologues in the uncorrected data. For example, in sample 4, M + C8H3 is 3.4% of the total intensity and M + C9H1 is 2.1% of the total intensity (Fig. 4). Carbon NA alone does not explain the intensity of these signals. The observed signals are in fact mainly contributed by 13C6-2H3-18O1 and 13C6-2H3-15N1, respectively. The mass difference between 13C6-2H3-18O1 and 13C8-2H3 NAD+ is 0.00245, so that they are not resolved when the resolution is below 816,000. The mass difference between 13C6-2H3-15N1 and 13C9-2H1 NAD+ is 0.00048, so that they are not resolved when the resolution is below 4,200,000. AccuCor2 recognized these fractions that arose due to NA of the non-tracer elements and corrected them successfully. To quantitatively evaluate the performance of different tools, we also calculated the RMSD to show how far the corrected results deviate from the expected labeling pattern. The corrected results from AccuCor2 are mostly identical to the expected labeling pattern of the samples (RMSD = 7.7 × 10−7 ± 8.9 × 10−7). PICor and IsoCorrectoR, on the other hand, failed to correct these fractions and the corrected results are heavily deviated from the expected data (RMSD = 1.1 × 10−2 ± 5.9 × 10−3 and 1.2 × 10−2 ± 6.0×10−3, respectively). We believe that this is because these tools essentially assume infinite mass resolution and ignore non-tracer elements and therefore lead to under-correction of the data (Fig. 4).
Fig. 4. Performance of PICor, IsoCorrectoR, and AccuCor2 in simulated data.
Uncorrected data are generated using NAD+ MS spectra simulated by Thermo Xcalibur Qual Browser at 750,000 resolution. The original labeling patterns are sample 1: 100% non-labeled; sample 2: 50% nonlabeled + 10% 13C6-2H2 + 40% 13C6-2H3; sample 3: 36% nonlabeled + 14% 13C6-2H2 + 50% 13C6-2H3; sample 4: 100% 13C6-2H3.
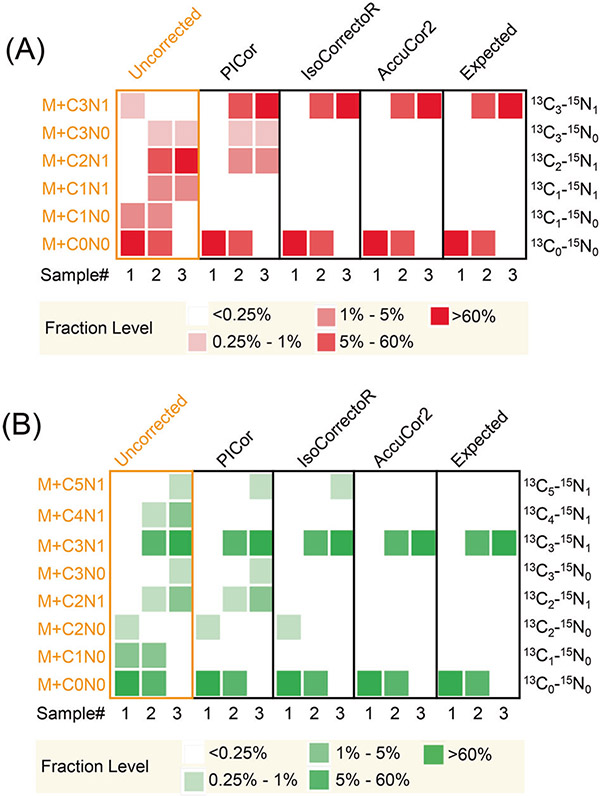
In addition to the simulated data, we have also tested the performance of AccuCor2 using experimental data (Fig. 5). In this experiment, we used [13C3-15N1] serine and unlabeled serine to make the following samples: sample 1: 100% non-labeled; sample 2: 50% non-labeled + 50% 13C3-15N1; sample 3: 100% 13C3-15N1. The measurement was performed at 70,000 resolution. For serine (Fig. 5A), this resolution is sufficient to resolve all the non-tracer elements. The only unanticipated measured mass fractions are M + 2C1N and M + 3C0N due to the isotopic impurity of the tracer (99% isotopic purity on both 13C and 15N). AccuCor2 and IsoCorrectoR both return the expected results (RMSD = 9.5 × 10−5 ± 4.2 × 10−5 and 9.5 × 10−5 ± 4.2 × 10−5, respectively for Sample 3), whereas PICor left the 13C2-15N1 and 13C3-15N0 fractions uncorrected (RMSD = 1.6 × 10−2 ± 5.6 × 10−5 for Sample 3) because PICor does not handle tracer isotopic impurity (Fig. 5A). In this experiment, serine is also observed as a sodium acetate adduct ([M+NaAc-H]−, C5H9NO5Na) [30]. We observed signals that correspond to M + C5N1 serine + NaAc (Fig. 5B). Since the NaAc is from the LC mobile phase, which is unlabeled, this fraction should have been generated because of isotope NA. In fact, serine + NaAc has the m/z of 186.04, which means a minimum resolution of 120,000 is needed to resolve 13C2 and 18O1. The observed 13C5-15N1 fraction is in fact 13C3-15N1-18O1 fraction. The correction results show that AccuCor2 recognized this fraction and corrected it (RMSD = 0.0 ± 0.0 for Sample 3). In contrast, PICor and IsoCorrectoR failed to correct this fraction (RMSD = 1.1 × 10−2 ± 5.6 × 10−4 and 3.2 × 10−3 ± 2.8 × 10−4, respectively, for Sample 3). Our tests using simulated and experimental data demonstrate that the resolution-dependent correction, as implemented in AccuCor2, is important for the accuracy of INAC for dual-isotope tracer experiments.
Fig. 5. Performance of PICor, IsoCorrectoR, and AccuCor2 in experimental data.
Raw data are serine (A) and sodium acetate adduct of serine (B) (C5H10NO5Na) measured under 70,000 nominal mass resolution (defined at m/z 200 on Orbitrap analyzer). The original labeling patterns are sample 1: 100% non-labeled; sample 2: 50% non-labeled + 50% 13C3-15N1; sample 3: 100% 13C3-15N1. Each sample on the plot is the average of four replicates.
DISCUSSION
AccuCor2 is the first resolution-dependent method to perform INAC on dual-isotope tracer experiments. As shown in “Results”, the reason AccuCor2 performs better INAC than other tools is because AccuCor2 does not assume all the non-tracer isotopologues are resolvable from the tracer isotopologues. Instead, AccuCor2 first use the resolution information to determine whether a specific non-tracer isotopologue is resolvable from the tracer isotopologues and then only subtract the fractions of non-resolvable isotopologues from the measured mass fraction. Previous tools for dual-INAC take the assumption that the non-tracer elements are fully resolved due to the use of ultra-high-resolution instrument. However, as we calculated in “Results”, in order to resolve 2H2-15N1 and 13C3 in NAD+, a minimum nominal mass resolution of 4,200,000 is required. This requirement is far beyond the capability of current ultra-high-resolution LC–MS instruments. Therefore, the resolution-dependent feature of AccuCor2 is critical for accurate INAC.
It is important to note that accurate input data are crucial for INAC. The input data should contain all the detectable labeled isotopologues and the isotopologues that are not resolved from the labeled ones. When picking and integrating the peaks for metabolites, the mass tolerance window should be made narrow enough to exclude the resolved mass peaks. In reality, this is rarely an issue because most people use narrow mass tolerance window of 5 or 10 ppm for peak picking, which is smaller than the resolvable ML. For serine ([M-H]−, m/z 104.03532) measured under the nominal mass resolution of 70,000, the resolvable ML is 0.00178, which is 17 ppm. On instruments with mass resolution >200,000, the mass tolerance window need to be carefully adjusted to avoid resolved mass peaks. On the other hand, if two mass peaks are not fully resolved, the m/z of the centroid peak is the intensity-weighted average m/z of the two peaks. This may cause some labeled isotopologue peaks to shift from the calculated m/z. We suggest the users to carefully inspect the raw data to ensure the accuracy of peak picking.
Several limitations of this study are noted. First, although sharing major features with AccuCor, AccuCor2 is specifically designed to handle data generated from dual-isotope experiments. For single-isotope experiments, it is not efficient to use AccuCor2 since additional input parameters are required. In the future, we plan to combine the functions of AccuCor and AccuCor2 into a single R package and make it available in CRAN. Second, AccuCor2 only handle data generated from 13C-2H and 13C-15N tracers, which suits the most needs of dual-isotope experiments. In the future, however, the use of other combinations of isotopes such as 2H-15N and 13C-18O might become popular. To perform INAC on these combinations of isotopes, similar algorithms as shown in Fig. 1 can be designed. Last, the current version of AccuCor2 only works for Orbitrap mass analyzers. We plan to add support for TOF analyzers in the updated versions.
In summary, we developed AccuCor2 as a new tool to perform INAC of data from dual-isotope tracer experiments in a resolution-dependent manner. Our results show that the dual-isotope tracer experiment should use a mass resolution that is high enough to resolve all the labeling fractions of interest. Otherwise, the labeling pattern may not be solvable. The non-tracer elements may require an even higher mass resolution to fully resolve, but this can be handled by the resolution-dependent correction in AccuCor2. AccuCor2 is freely available in open-source format and would enable more accurate calculation of metabolite labeling patterns from MS.
Supplementary Material
FUNDING
This research is supported, in part, by NIH Grants P30CA072720-5923 (XS).
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study do not require ethical approval.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41374-021-00631-4.
DATA AVAILABILITY
The datasets are available from GitHub repository, https://github.com/wangyujue23/AccuCor2.
CODE AVAILABILITY
The codes are available from GitHub repository, https://github.com/wangyujue23/AccuCor2.
REFERENCES
- 1.Kim IY, Suh SH, Lee IK, Wolfe RR. Applications of stable, nonradioactive isotope tracers in in vivo human metabolic research. Exp Mol Med. 2016;48:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell. 2015;162:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills EL, Pierce KA, Jedrychowski MP, Garrity R, Winther S, Vidoni S, et al. Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature. 2018;560:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.TeSlaa T, Bartman CR, Jankowski CSR, Zhang Z, Xu X, Xing X, et al. The source of glycolytic intermediates in mammalian tissues. Cell Metab. 2021;33:367–78.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Kwon H, Su X, Wondisford FE. Glycerol not lactate is the major net carbon source for gluconeogenesis in mice during both short and prolonged fasting. Mol Metab. 2020;31:36–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiechert W, Möllney M, Isermann N, Wurzel M, De Graaf AA. Bidirectional reaction steps in metabolic networks: III. Explicit solution and analysis of isotopomer labeling systems. Biotechnol Bioeng. 1999;66:69–85. [PubMed] [Google Scholar]
- 8.Antoniewicz MR. Methods and advances in metabolic flux analysis: a mini-review. J Ind Microbiol Biotechnol. 2015;42:317–25. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, Wondisford FE, Song C, Zhang T, Su X. Metabolic flux analysis—linking isotope labeling and metabolic fluxes. Metabolites. 2020;10:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang C, Chen L, Rabinowitz JD. Metabolomics and isotope tracing. Cell. 2018;173:822–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Martien J, Gilbertson C, Ma J, Amador-Noguez D, Park JO. Metabolic flux analysis and fluxomics-driven determination of reaction free energy using multiple isotopes. Curr Opin Biotechnol. 2020;64:151–60. [DOI] [PubMed] [Google Scholar]
- 12.Liu L, Shah S, Fan J, Park JO, Wellen KE, Rabinowitz JD. Malic enzyme tracers reveal hypoxia-induced switch in adipocyte NADPH pathway usage. Nat Chem Biol. 2016;12:345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L, Zhang Z, Hoshino A, Zheng HD, Morley M, Arany Z, et al. NADPH production by the oxidative pentose-phosphate pathway supports folate metabolism. Nat Metab. 2019;1:404–15. [PMC free article] [PubMed] [Google Scholar]
- 14.Neinast MD, Jang C, Hui S, Murashige DS, Chu Q, Morscher RJ, et al. Quantitative analysis of the whole-body metabolic fate of branched-chain amino acids. Cell Metab. 2019;29:417–29.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spinelli JB, Yoon H, Ringel AE, Jeanfavre S, Clish CB, Haigis MC. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science. 2017;358:941–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poillet-Perez L, Xie X, Zhan L, Yang Y, Sharp DW, Hu ZS, et al. Autophagy maintains tumour growth through circulating arginine. Nature. 2018;563:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu YM, Burke JF, Vogt JA, Chambers L, Young VR. Splanchnic and whole body L-[1-13C,15N]leucine kinetics in relation to enteral and parenteral amino acid supply. Am J Physiol Endocrinol Metab. 1992;262:E687–94. [DOI] [PubMed] [Google Scholar]
- 18.Crossland H, Smith K, Atherton PJ, Wilkinson DJ. A novel stable isotope tracer method to simultaneously quantify skeletal muscle protein synthesis and breakdown. Metab Open. 2020;5:100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Biemann K. Mass spectrometry: organic chemical applications. New York: McGraw-Hill; 1962. [Google Scholar]
- 20.Brauman JI Least. Squares analysis and simplification of multi-isotope mass spectra. Anal Chem. 1966;38:607–10. [Google Scholar]
- 21.Wittmann C, Heinzle E. Mass spectrometry for metabolic flux analysis. Biotechnol Bioeng. 1999;62:739–50. [DOI] [PubMed] [Google Scholar]
- 22.Van Winden WA, Wittmann C, Heinzle E, Heijnen JJ. Correcting mass isotopomer distributions for naturally occurring isotopes. Biotechnol Bioeng. 2002;80:477–9. [DOI] [PubMed] [Google Scholar]
- 23.Millard P, Letisse F, Sokol S, Portais JC. IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics. 2012;28:1294–6. [DOI] [PubMed] [Google Scholar]
- 24.Su X, Lu W, Rabinowitz JD. Metabolite spectral accuracy on orbitraps. Anal Chem. 2017;89:5940–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Millard P, Delépine B, Guionnet M, Heuillet M, Bellvert F, Létisse F. IsoCor: isotope correction for high-resolution MS labeling experiments. Bioinformatics. 2019;35:4484–7. [DOI] [PubMed] [Google Scholar]
- 26.Du D, Tan L, Wang Y, Peng B, Weinstein JN, Wondisford FE, et al. ElemCor: accurate data analysis and enrichment calculation for high-resolution LC-MS stable isotope labeling experiments. BMC Bioinform. 2019;20:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carreer W, Flight R, Moseley H. A computational framework for high-throughput isotopic natural abundance correction of omics-level ultra-high resolution FT-MS datasets. Metabolites. 2013;3:853–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinrich P, Kohler C, Ellmann L, Kuerner P, Spang R, Oefner PJ, et al. Correcting for natural isotope abundance and tracer impurity in MS-, MS/MS- and high-resolution-multiple-tracer-data from stable isotope labeling experiments with IsoCorrectoR. Sci Rep. 2018;8:17910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietze J, van Pijkeren A, Ziegler M, Kwiatkowski M, Heiland I. Natural isotope correction improves analysis of protein modification dynamics. bioRxiv. 2020. 10.1101/2020.10.31.361725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su X, Chiles E, Maimouni S, Wondisford FE, Zong W-X, Song C. In-source CID ramping and covariant ion analysis of hydrophilic interaction chromatography metabolomics. Anal Chem. 2020;92:4829–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Melamud E, Vastag L, Rabinowitz JD. Metabolomic analysis and visualization engine for LC - MS data. Anal Chem. 2010;82:9818–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosman KJR, Taylor PDP. Isotopic compositions of the elements 1997. J Phys Chem Ref Data. 1998;27:1275–87. [Google Scholar]
- 33.Team R Development Core. A language and environment for statistical computing, vol. 2. R Foundation for Statistical Computing; 2018. [Google Scholar]
- 34.Lawson CL, H. R. Solving least squares problems. New York: Prentice Hall; 1974. [Google Scholar]
- 35.Zubarev RA, Makarov A. Orbitrap mass spectrometry. Anal Chem. 2013;85:5288–96. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Su X, Quinn WJ, Hui S, Krukenberg K, Frederick DW, et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 2018;27:1067–80.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets are available from GitHub repository, https://github.com/wangyujue23/AccuCor2.
The codes are available from GitHub repository, https://github.com/wangyujue23/AccuCor2.