Abstract
Liver abscess etiology in feedlot steers involves the escape of bacteria from the digestive tract to form a polymicrobial abscess within or on the external surface of the liver. However, little is known about the effects of feedlot finishing systems on the microbial composition of the liver abscess purulent material. Liver abscesses were collected at the time of harvest from steers originating from a single feedlot managed in either a traditional program (which included tylosin phosphate supplementation) or a natural program (without tylosin phosphate supplementation). The purulent material of liver abscesses from traditionally managed steers (N = 53 abscesses) and that of naturally managed steers (N = 62 abscesses) was characterized using the V4 region of the 16S rRNA gene. Two phyla and three genera were found in greater than 1% relative abundance across all abscesses. The genus Fusobacterium was identified in all liver abscess samples and accounted for 64% of sequencing reads. Bacteroides and Porphyromonas genera accounted for 33% and 1% of reads, respectively. Trueperella was more likely to be found in the liver abscesses of naturally managed steers than traditionally managed steers (P = 0.022). Over 99% of the genus-level bacterial sequences observed across all liver abscesses belonged to Gram-negative genera. Bacteria known to colonize both the rumen and hindgut were identified within liver abscesses. No differences in alpha diversity or beta diversity were detected between liver abscess communities (between the two management programs or individual pens) when tested as richness, Shannon Diversity Index, or weighted UniFrac distances (P > 0.05). These results were consistent with previous identification of Fusobacterium necrophorum as the primary bacteriologic agent within liver abscesses and emphasized the relationship between the gastrointestinal microbiota and liver abscess formation. Though the microbiota of the liver abscess purulent material was similar between steers fed an antibiotic-free diet and those fed an antibiotic-containing diet from the same feedlot, divergence was detected in liver abscess communities with some being dominated by Fusobacterium and others being dominated by Bacteroides.
Keywords: antibiotic, feedlot, liver abscess, steers, tylosin, 16S
Liver abscess microbial communities from feedlot steers receiving tylosin phosphate or no tylosin phosphate had similar microbial communities dominated by the genus Fusobacterium. Bacteroides and Porphyromonas were also identified within liver abscesses, and Trueperella was observed more frequently in liver abscesses from cattle not supplemented with tylosin phosphate.
Introduction
Occurrence of liver abscesses in feedlot steers is associated with a reduction in live performance, hot carcass weight, and visceral value (Brink et al., 1990; Brown and Lawrence, 2010). Prevalence of liver abscesses among commercial cattle has increased from the earliest reports of 5.3% (Smith, 1940) to 17.8% reported by the most recent National Beef Quality Audit (Eastwood et al., 2017). While administering in-feed antibiotics to livestock is scrutinized (Aarestrup, 1999; Haskell et al., 2018; Chen et al., 2019), tylosin phosphate supplementation remains the most economical method to reduce liver abscess prevalence (Brown et al., 1975; Nagaraja and Lechtenberg, 2007). Historically, Fusobacterium necrophorum has been implicated as the primary etiologic agent in the liver abscess formation (Nagaraja and Lechtenberg, 2007). Although the use of culture-based microbiological techniques has consistently demonstrated F. necrophorum presence within polymicrobial abscesses, these results may oversimplify the microbial community of liver abscess purulent material as many unculturable microbes are not assessed (Nagaraja and Chengappa, 1998). In recent years, a limited number of studies have utilized high-throughput sequencing to assess the microbiota within liver abscesses (Amachawadi et al., 2016, 2021; Weinroth et al., 2017; Stotz et al., 2021). Differences in liver abscess microbiota have been attributed to cattle source (ranch of origin, backgrounding location, etc.), breed composition, feedlot location, and inclusion of tylosin phosphate in the diet (Amachawadi et al., 2017; Weinroth et al., 2017). Natural feedlot finishing programs exclude tylosin phosphate and the use of growth technologies (steroidal implants and beta-adrenergic agonists). Additionally, natural programs that have specifications for cattle management before feedlot entry often require cattle to have never been given an antibiotic. These specifications create microbial selection pressures even before the cattle arrive at the feedlot. Therefore, the objective of this study was to use 16S rRNA gene sequencing to compare the liver microbiota of steers managed within a single feedlot under a traditional program with that of steers managed in a natural program (without tylosin phosphate supplementation, steroidal implants, beta-adrenergic agonists, or antimicrobial treatments).
Materials and Methods
Animal care and use
All cattle were harvested in a commercial processing facility and samples were collected postmortem under commercial conditions. All procedures were reviewed and approved by Colorado State University Institutional Animal Care and Use Committee via waiver (IACUC waivers #2018-888 and #2019-773). Observations and sample collections did not change normal commercial practices.
Cattle population
Fourteen pens of yearling steers with an average of 281 steers per pen (range of 212 to 323 steers per pen) were identified for observation in a commercial feedlot in the High Plains region. Pens enrolled in a traditional management program (N = 7) and pens enrolled in a natural management program (N = 7) arrived at the feedlot over a 45-d period from late August through early October 2018. Traditional and natural pens were selected in pairs by the arrival date to minimize temporal effects. Steers were observed in the management program in which they were placed in accordance with management prior to feedlot arrival. Upon arrival, all steers were sorted and vaccinated according to standard feedlot protocol. Traditionally managed cattle received standard hormonal implants containing trenbolone acetate and estradiol.
All steers were fed using a step-up feeding program that included receiving, intermediate, and finishing diets. Diet ingredients and nutrient analysis are summarized in Supplementary Table S1. Traditionally managed steers were fed an additional diet including ractopamine hydrochloride (Optaflexx; Elanco Animal Health; Greenfield, IN) during the final 28 to 42 d of the finishing period. Monensin (Rumensin; Elanco Animal Health) and tylosin phosphate (Tylan; Elanco Animal Health) were also fed to traditionally managed steers. Naturally managed cattle were not administered growth-promoting technologies or antibiotics. When treated for illness with an antibiotic, naturally managed cattle were removed from natural pens and, consequently, removed from the study population.
Liver abscess collection
Cattle were transported to a commercial processing facility for harvest from February through April of 2019. Traditionally managed cattle were fed for an average of 179 d (SD = 12.6), and naturally managed cattle were fed for an average of 214 d (SD = 14.4). Identities of feedlot pens were maintained through the harvest process to allow for the collection of liver abscesses from cattle from each pen. Livers identified as inedible (because of abscess, adherence to internal tissues, cirrhosis, flukes, telangiectasias, or contamination) were removed from the production line and evaluated by trained personnel for abscess presence. Livers were palpated to identify abscesses that harbored purulent material. Abscesses and surrounding liver tissue were extracted with sterile scalpels. When the liver abscess capsule was compromised during tissue removal, the entire abscess sample was discarded. Abscesses from individual livers were placed in sterile collection bags (VWR; Radnor, PA), sealed, and transported in insulated containers to the Center for Meat Safety and Quality at Colorado State University (Fort Collins, CO) for further processing.
Liver abscess processing
On the same date as sample collection, liver abscess samples were processed and prepared for storage. Abscess capsules or the external surface of the liver tissue were flame sterilized using 100% ethanol. Following sterilization, abscess capsules were opened with a sterile scalpel, and purulent material was extracted and transferred to sterile 50-mL conical tubes (VWR) using sterile tongue depressors. Aliquots of liver abscess purulent material were stored at −80 °C until the time of DNA extraction.
DNA extraction and sequencing
DNA extraction and library preparation were performed at Colorado State University consistent with the recommendations of Weinroth et al. (2022). A randomly selected subset of the liver abscess purulent material aliquots (N = 10 per pen) was thawed for 16 h at 4 °C before extraction. Purulent material aliquots were individually sampled with sterile swabs (Becton, Dickinson and Company; Franklin Lakes, NJ) and randomized and loaded into 96-well plates by cutting the inoculated swab tip into the plate well with flame-sterilized scissors. Cross contamination was prevented by covering all inactive wells. Sixteen negative controls and two positive controls (ZymoBIOMICS Microbial Community Standard 6300, Zymo Research; Irvine, CA) were included. Ten technical replicates were also included to evaluate consistency across extraction plates; 10 randomly selected liver abscess purulent samples were swabbed in duplicate, and duplicate swabs were loaded on separate plates. Loaded plates were stored at −20 °C until the time of DNA extraction.
Upon thawing the loaded plates, DNA was extracted using the DNEasy PowerSoil HTP 96 Kit (Qiagen; Hilden, Germany) following the manufacturer’s protocol. Extracted DNA was amplified with barcoded primers targeting the V4 region of the 16S rRNA gene. Primer constructs included the Illumina MiSeq adaptor (Illumina; San Diego, CA), Golay barcode, spacer, and primer. Earth Microbiome Project (EMP) primers 515F and 806R were used for amplification (Caporaso et al., 2011, 2012; Apprill et al., 2015; Parada et al., 2016).
Amplification was conducted in duplicate by PCR using an Eppendorf Vapo.Protect MasterCycler Pro-S thermocycler (Eppendorf; Hauppauge, NY). For the initial PCR, 25 μL of reaction mix was prepared by combining 1 μL of template DNA, 1 μL of each barcoded primer (10 μM), 12 μL of molecular-grade water, and 10 μL of Platinum Hot Start PCR Master Mix (Thermo Fisher Scientific; Waltham, MA). Conditions for PCR followed EMP protocol and included initial denaturation at 94 °C for 3 min; 30 cycles of denaturation (94 °C, 45 s), annealing (50 °C, 60 s) and elongation (72 °C, 90 s); and a final 10-min extension at 72 °C. Products of PCR were visually evaluated for effective amplification by agarose gel electrophoresis with expected band size of approximately 300 to 350 base pairs. Similarly, negative controls were visually evaluated for lack of banding pattern. The second PCR process was completed using the same reaction conditions as described above; however, 50 μL of reaction mix was prepared by combining 2 μL template DNA, 2 μL of each barcoded primer (10 μM), 24 μL of molecular-grade water, and 20 μL of PCR Master Mix. The main goal of the second amplification was to generate sufficient quantities of amplicons for next-generation sequencing. Agarose gel evaluation of the product was performed as described above.
Duplicate PCR products were combined for each sample. The concentration of amplicon products was determined by Quant-IT PicoGreen dsDNA Assay Kit (Thermo Fisher Scientific) read on a Fluoroskan (Thermo Fisher Scientific) plate reader. Pico assay concentration results were qualitatively verified by comparison to agarose gel banding patterns. Amplicons from all wells of a single 96-well plate (including samples, technical replicates, and controls) were combined before cleaning. To combine amplicons, 300 ng of DNA from each sample was added to a single tube for each plate (referred to as “plate pool”). No more than 50 μL from a single sample was added to the plate pool to maintain the integrity of the negative control. Plate pools were cleaned using MinElute PCR Purification Kit (Qiagen) following manufacturer’s protocols. The cleaned plate pools were evaluated for DNA concentration by NanoDrop Lite spectrophotometer (Thermo Fisher Scientific) and combined in equimolar concentrations to form the final sequencing library.
The amplicon library was diluted to a loading concentration of 8 pM and combined with 15% PhiX control library. Paired-end sequencing (2 × 250 bp) was performed using the 500 cycle MiSeq Reagent Kit v2 (Illumina; San Diego, CA) on the Illumina MiSeq platform at the Next Generation Sequencing Core Laboratory at Colorado State University.
Bioinformatics and statistical analysis
All amplicon sequence data were bioinformatically processed in QIIME2 version 2019.4 (Bolyen et al., 2018). Imported and demultiplexed paired-end sequences were denoised with DADA2 (Callahan et al., 2016) with both forward and reverse reads trimmed to 250 bp. Taxonomy was assigned with the q2-feature-classifier plugin (Bokulich et al., 2018) using a pretrained naive Bayes Greengenes 13_8 classifier (DeSantis et al., 2006; McDonald et al., 2012), a pretrained naive Bayes Silva 132 classifier, and a Silva 132 (Quast et al., 2013) classifier trained specifically for the primer set used for amplification. The pretrained Silva classifier was selected for downstream analysis due to a fewer amplicon sequence variants (ASV) being identified as unclassified. Reads classified as mitochondria or chloroplasts were removed from the data set; controls and technical replicates were also removed. ASV was placed in the Greengenes 13_8 phylogenetic tree by SEPP methodology using q2-fragment-insertion (Janssen et al., 2018). Adequate sampling depth was justified by constructing a rarefaction curve with diversity metrics. Because alpha diversity rarefaction curves showed little diversity added with a sampling depth above 3,000 sequences per sample, diversity analysis was standardized by subsampling without replacement (Weiss et al., 2017) to 10,049 sequences per sample, which allowed retention of most samples in the analyses.
Alpha diversity was measured by richness (the number of observed ASV) and Shannon Diversity Index (Shannon, 1948). Differences in richness and Shannon Diversity were evaluated between management programs and pen assignments with Kruskal–Wallis testing (Kruskal and Wallis, 1952) and visualized in R version 3.4.1 (R Core Team, 2017) using ggplot2 (Wickham, 2009). Beta diversity was measured with unweighted UniFrac (Lozupone and Knight, 2005) and weighted UniFrac (Lozupone et al., 2007). K-means clustering was used to group samples based on the unweighted UniFrac distance matrix (Lloyd, 1982; MacQueen, 1967). Differences in beta diversity between management programs, pen assignments, and K-means clusters were evaluated with PERMANOVA testing (Anderson, 2017) using the vegan package of R (Oksanen et al., 2022). Taxa differential abundance was evaluated by ANCOM testing at both the phylum and genus level (Mandal et al., 2015). Significance for differential abundance was evaluated as a W value indicating log-fold change against a model-determined threshold based on a bimodal distribution. Rarefied abundance data was exported from QIIME2 as relative abundance and visualized in R version 3.4.1 (R Core Team, 2017).
The sequencing depth of each negative control was evaluated to ensure cleanliness of extraction and library preparation; the number of reads generated by each control well before and after denoising was recorded. The sequencing depth of positive controls was similarly recorded. Additionally, the taxa relative abundance of each positive control was exported and visualized in R version 3.4.1 (R Core Team, 2017) using ggplot2 (Wickham, 2009) and compared to the known composition of the mock community for qualitative evaluation. Technical replicates were qualitatively evaluated in pairs by taxa relative abundance to ensure consistency of taxa relative abundance between separate extraction plates.
For each individual liver abscess microbial community, the presence or absence of Fusobacterium, Bacteroides, and Trueperella was determined from rarefied taxonomy tables. Bacterial presence was compared between management programs by logistic regression in R version 3.4.1 (R Core Team, 2017) using lme4 (Bates et al., 2015) and emmeans (Searle et al., 1980). A mixed effects model was fit using management program of the live steer as a fixed effect and pen assignment of the live steer as a random effect. A predetermined alpha level of 0.05 was used for all comparisons in the observational study.
Results and Discussion
Summary of samples processed
In total, 140 unique liver abscess purulent samples were processed. Seventy liver abscesses from naturally managed cattle (N = 10 from each of 7 pens) and 70 liver abscesses from traditionally managed cattle (N = 10 from each of 7 pens) were selected for analysis.
DNA sequencing data
A total of 3,437,552 sequence reads were generated by Illumina MiSeq sequencing for liver abscess purulent material samples, technical replicates, and controls. Liver abscess purulent material samples and replicates (N = 150) averaged 21,037 sequences per sample (range: 18 to 43,192; SD = 11,673). Negative controls (N = 16) averaged 146 sequences per sample (range: 7 to 547; SD = 189). Positive controls (N = 2) averaged 34,636 sequences per sample (range: 31,915 to 37,356; SD = 3,847).
After denoising, filtering for sequencing depth, and removing controls, replicates, chloroplasts, and mitochondria, a total of 1,055,145 reads mapped to 69 unique features were included for analysis. Rarefying to 10,049 sequences per sample resulted in retaining 53 of 70 liver abscess purulent material samples from steers managed in the traditional program and 62 of 70 liver abscess purulent material samples from steers managed in the natural program. After denoising, negative controls (N = 16) averaged 32 sequences per sample (range: 0 to 311; SD = 77) and positive controls (N = 2) averaged 24,943 sequences per sample (range: 22,272 to 27,614; SD = 3,777).
Core microbial composition
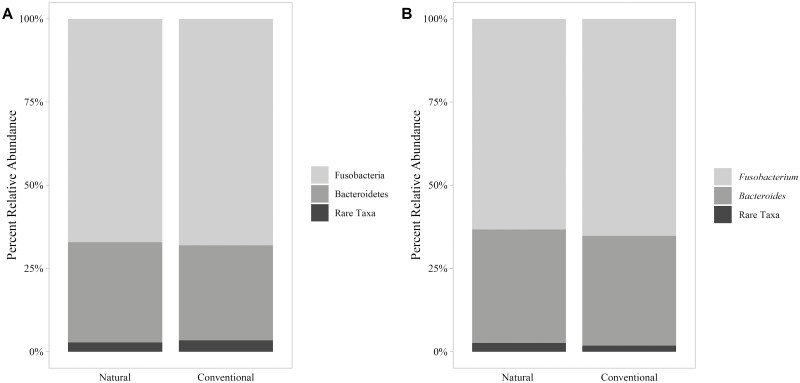
Taxonomic classification with the pretrained naive Bayes Silva 132 classifier identified two phyla and three genera of bacteria in greater than 1% relative abundance of all reads (Figure 1A and B). The dominant phyla observed included Fusobacteria (64% of reads) and Bacteroidetes (34% of reads). This represented a much simpler microbial community than was reported by Weinroth et al. (2017) and Amachawadi et al. (2021) based on the number of taxa observed. The referenced studies used mean sequencing depths that were a magnitude of order greater than the sequencing depth of the present study; however, alpha diversity rarefaction curves constructed for the current study plateaued at approximately 3,000 sequences per sample. Regardless of sequencing depth, other factors could have affected richness of the microbial communities. For instance, the purulent material analyzed in this observational study originated from steers housed in a single feedlot over the same time period while the purulent material analyzed by Weinroth et al. (2017) originated from cattle from five different feedlots, and Amachawadi et al. (2021) evaluated liver abscess representing 22 feedlots. In addition, the length of time between the initial abscess occurrence and harvest could have affected succession of abscess microbial communities. Others have observed decreased microbial richness as selection pressure is applied to a microbial community (Chaillou et al., 2015). If the abscesses evaluated in this study were chronologically older than those of previous evaluations, action of the immune system could have contributed to decreased alpha diversity. Though difficult to identify the time of abscess occurrence in the live animal, further investigation into the age of the abscess relative to the complexity of its microbial community could help explain observed differences in complexity of liver abscess communities.
Figure 1.
Average taxonomic composition (% relative abundance) of the bacterial communities within liver abscesses from steers in a single feedlot under natural (N = 62) and traditional (N = 53) management programs averaged by management program at the phylum-level (A) and genus-level (B).
The genera identified in greater than 1% relative abundance of all reads included Fusobacterium (64% of reads; 100% of samples), Bacteroides (33% of reads; 93% of samples), and Porphyromonas (1% of reads; 7% of samples). Bacteroides is a Gram-negative bacterium found in the gastrointestinal tracts of cattle (Miura et al., 1980; Wetzels et al., 2017; Ozbayram et al., 2018) and has been previously associated with liver abscesses (Simon and Stovell, 1971; Kanoe et al., 1979; Scanlan and Hathcock, 1983; Nagaraja and Lechtenberg, 2007). A previous study demonstrated an increase in the relative abundance of Bacteroides on the rumen epithelium during acidosis challenge (Wetzels et al., 2017). Porphyromonas is a Gram-negative ruminal bacterium closely related to Bacteroides (Summanen et al., 2005) that has been found in bovine liver abscesses (Scanlan and Hathcock, 1983; Nagaraja and Lechtenberg, 2007; Weinroth et al., 2017). Driven by Fusobacterium, Bacteroides¸ and Porphyromonas, over 99% of the bacterial sequences observed across all liver abscesses belonged to genera previously classified as Gram-negative by Langworth (1977), Hofstad (1984), and Bostanci and Belibasakis (2012).
Taxa of interest
The genus Fusobacterium was identified in all liver samples from both traditionally and naturally managed steers (Table 1). Fusobacterium was the sole microbial genus in only two of the 115 purulent material samples (2%); all other samples (98%) were identified as mixed cultures of Fusobacterium with other bacteria. Though bovine liver abscesses have often been described as polymicrobial infections, F. necrophorum has historically been considered the primary etiologic agent of liver abscesses in fed cattle (Newsom, 1938; Jensen et al., 1954; Calkins and Dewey, 1968; Scanlan and Hathcock, 1983; Nagaraja et al., 1996; Nagaraja and Chengappa, 1998; Nagaraja and Lechtenberg, 2007). In agreement with the current study, Weinroth et al. (2017) found Fusobacterium in all liver abscess samples collected from fed cattle using molecular techniques. Similarly, Nagaraja et al. (1999) and Amachawadi and Nagaraja (2016) found Fusobacterium in all liver abscess samples collected from feedlot cattle using culture-based microbiological techniques. Historically, others have identified Fusobacterium within nearly all liver abscess samples collected from commercial processing facilities using culture-based microbiological techniques (Newsom, 1938; Simon and Stovell, 1971).
Trueperella pyogenes has been identified as the second most frequently cultured microbe from liver abscess purulent material (Berg and Scanlan, 1982; Nagaraja et al., 1996). T. pyogenes is an opportunistic, Gram-positive bacterium known to inhabit ruminant gastrointestinal tracts (Ribeiro et al., 2015). In the current study, the genus Trueperella was found more frequently within liver abscesses from naturally managed cattle (14 of 62; 23%) than traditionally managed cattle (3 of 53; 6%; Table 1). ANCOM testing identified Trueperella as the only genus that was differentially abundant within liver abscesses of steers managed in traditional and natural programs (W = 6). This result is likely associated with traditionally managed cattle receiving an ionophore and tylosin phosphate, a macrolide antibiotic that acts as a bacteriostat against Gram-positive bacteria (Hof, 1994). However, in a previous study, T. pyogenes was isolated more often in liver abscesses of cattle fed tylosin compared to cattle not fed tylosin (Nagaraja et al., 1999). Weinroth et al. (2017) identified T. pyogenes in all liver abscesses sampled by molecular techniques (using greater sequencing depth than the current study), regardless of cattle management program. Given the variability of previous results, it is likely that the presence of Trueperella within liver abscesses of feedlot cattle is not solely dependent on the inclusion of tylosin in the diet. In the current study, Trueperella comprised only 0.13% of all sequences identified, indicating limited presence of Trueperella within liver abscesses of the observed population.
Table 1.
Adjusted probability1 of an individual abscess from a steer managed in a natural2 or traditional3 program to harbor Fusobacterium, Bacteroides, or Trueperella
| Bacterial genus | Natural | Traditional | P-value4 | ||
|---|---|---|---|---|---|
| Probability of presence, % | SE | Probability of presence, % | SE | ||
| Fusobacterium5 | 100.00 | NA | 100.00 | NA | NA |
| Bacteroides | 95.56 | 3.422 | 94.71 | 4.326 | 0.850 |
| Trueperella | 23.82 | 8.676 | 4.71 | 3.316 | 0.022 |
Adjusted probability generated by transforming the odds estimated by logistic regression to a probability scale.
Program included no growth-promoting technologies or antimicrobials.
Program included hormonal implants, tylosin phosphate, monensin, ractopamine hydrochloride, and treatment with antimicrobials when necessary.
P-value for the test of the log odds ratio between natural and traditional management programs.
No logistic regression analysis performed given Fusobacterium presence in all liver abscesses; unadjusted prevalence presented.
Rare taxa
Bacteria known to inhabit the ruminant gastrointestinal tract were found as rare taxa within liver abscess microbial communities. Relative abundances of rare taxa are summarized in Supplementary Table S2. Families Ruminococcaceae and Prevotellaceae were identified in 26% and 3% of all liver abscesses, respectively. Ruminococcus and Prevotella genera are commonly identified as constituents of the rumen microbiota (Firkins and Yu, 2015; Henderson et al., 2015; Ozbayram et al., 2018; Holman and Gzyl, 2019). The genus Treponema was identified in 9% of all liver abscesses and has been found in the rumen, specifically on the rumen epithelium (Stanton and Canale-Parola, 1980; Liu et al., 2016). Campylobacter was found within 10 liver abscesses and composed 0.28% of all reads. Campylobacter has been previously identified in bovine liver abscesses (Weinroth et al., 2017) and has been found more prevalently on rumen epithelium as a commensal organism than in rumen contents or feces (Liu et al., 2016; Pacifico et al., 2021). Generally, bacteria known to inhabit the microbial communities of the rumen epithelium were found more frequently within liver abscesses than bacteria known to strictly inhabit the rumen contents. This supports the proposed etiology of bovine liver abscesses with respect to the rumenitis-liver abscess complex described by Jensen et al. (1954) and the hypothesis that bacteria escape the rumen by damage to epithelial tissues (Nagaraja et al., 1996).
However, many bacteria found in the rumen are also found in the lower gastrointestinal tract and feces. Ruminococcaceae and Bacteroidaceae have been found in greater relative abundance in the feces compared to the rumen (Ozbayram et al., 2018; Holman and Gzyl, 2019). Turicibacter, a genus previously identified in fecal microbial communities (Liu et al., 2016), was identified in 13% of all liver abscesses. Additionally, Romboutsia and Clostridium sensu stricto 1, bacteria known to colonize the hindgut in ruminants as early as immediately after birth, were identified in 10% and 11% of all liver abscesses, respectively (Alipour et al., 2018).
Rare genera identified in the liver abscess samples included bacteria of the Ruminococcaceae, Provotellaceae, Clostridiaceae 1, Spirochaetaceae, Erysipelotrichaceae, and Peptostreptococcaceae families; the same families were also identified in fecal samples from the same population of steers (Fuerniss et al., unpublished data). While greater taxonomic resolution is needed to identify potential homology between taxa present within both feces and liver abscess purulent material, the results suggest that escape of bacteria from the hind gut could be a factor in the formation of polymicrobial liver abscesses. In theory, intestinal epithelial damage from acidosis could allow passage of microbes into the portal blood (Oba and Wertz-Lutz, 2011). Gressley et al. (2011) suggested that hindgut epithelium might be more susceptible to damage caused by fermentation products compared with ruminal epithelium due to the lack of salivary flow, limited protozoa, and fewer epithelial layers. Thoefner et al. (2004) induced ruminal acidosis in dairy heifers and observed mild signs of inflammation in the cecum and upper colon upon postmortem examination. Acidosis in feedlot cattle fed high-concentrate diets could contribute to liver abscess formation by compromising the barrier function of the epithelium of the hind gut and promoting microbial translocation to the liver.
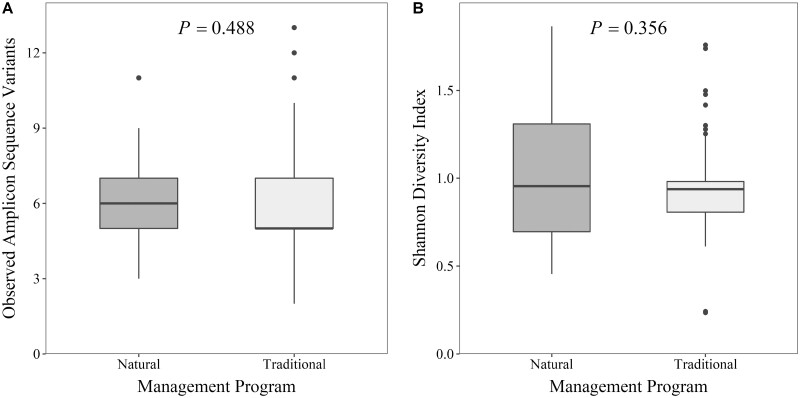
Alpha diversity within liver abscess microbial communities
Alpha diversity did not differ between liver abscess purulent material from steers managed in traditional and natural programs when measured as richness (P = 0.488; Figure 2A) or Shannon Diversity Index (P = 0.356; Figure 2B). Numerically, a slightly greater mean number of ASV and slightly greater mean Shannon Diversity Index value were found within liver abscesses microbiota from naturally managed steers in comparison to those of traditionally managed steers. Richness of liver abscess communities between pens was also similar (P = 0.855) with a range of mean richness values per pen between 5.13 and 7.22 ASV. Similarly, Shannon Diversity Index of liver abscess communities was also similar (P = 0.495) between pens with a range of mean Shannon Diversity Index values per pen between 0.83 and 1.30.
Figure 2.
Alpha diversity, depicted as richness (A) and Shannon Diversity Index (B), of the bacterial communities within liver abscess purulent material from steers in a single feedlot within natural (N = 62) and traditional (N = 53) management programs.
Alpha diversity of the liver abscess purulent material observed in this study was considerably lesser than values previously observed for rumen or fecal samples (Shanks et al., 2011; Yang et al., 2016; Azad et al., 2019). However, similarly low levels of alpha diversity were found in bovine liver abscesses by Amachawadi et al. (2021) and Stotz et al. (2021). Since abscesses develop from a specific insult to the body from an etiological agent or agents, dominance by the etiological agent is expected in abscess microbial communities. Microbial dominance and diversity are antagonistic which supports lesser alpha diversity for abscess communities. Still, the relative abundances of minor genera (anything other than Fusobacterium or Bacteroides) observed in this study was less than that of previous studies.
Beta diversity between liver abscess microbial communities
Using UniFrac phylogenetic distances to compare microbiota, differences between purulent material microbial communities were not observed when compared by management program for unweighted (P = 0.169) and weighted (P = 0.799) metrics. However, differences in microbiota were observed between individual pens by unweighted UniFrac (P = 0.013). Variation between pens was expected due to the intrinsic pen-specific factors including ranch of origin, contemporary environment, and feeding behaviors. Unweighted UniFrac is sensitive to presence and absence of rare taxa (Lozupone and Knight, 2005) and indicated that the observed differences in diversity of liver abscess microbiota between pens were driven by the presence or absence of unique taxa that could be associated with pen-specific factors such as cattle source, prefeedlot management, and pen dynamics. However, differences in microbiota were not observed between individual pens by weighted UniFrac (P = 0.067). Weighted UniFrac analysis (Lozupone et al., 2007) accounts for the ecological mass-ratio hypothesis which implies that community dynamics are largely influenced by dominant species and are insensitive to the abundance of rare taxa (Grime, 1998). Applied to the context of the findings of this study, the abundance of Fusobacterium and Bacteroides (combined average relative abundance of 98%) characterize major differences in liver abscess microbial communities. Together, unweighted and weighted UniFrac results suggest more variation in abscess communities between pens than between feedlot management programs, especially regarding rare taxa. As such, pen-specific factors such as cattle source, prefeedlot management, intake patterns, and health challenges should be considered in future studies, especially in large-pen, population-based designs.
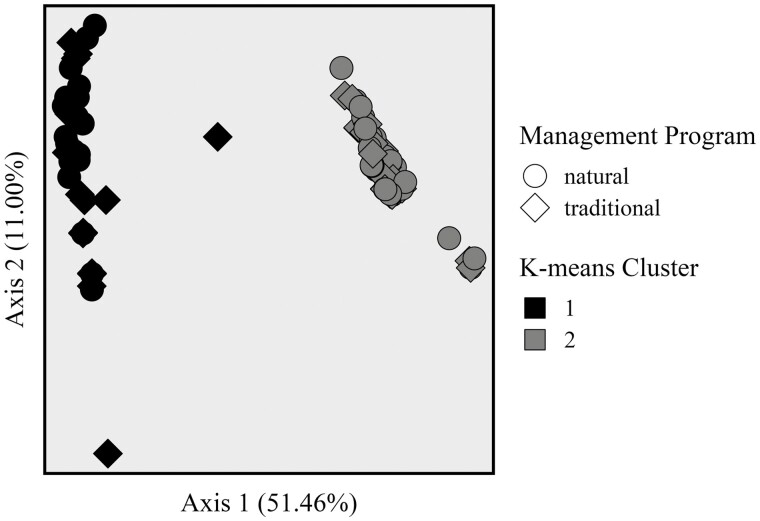
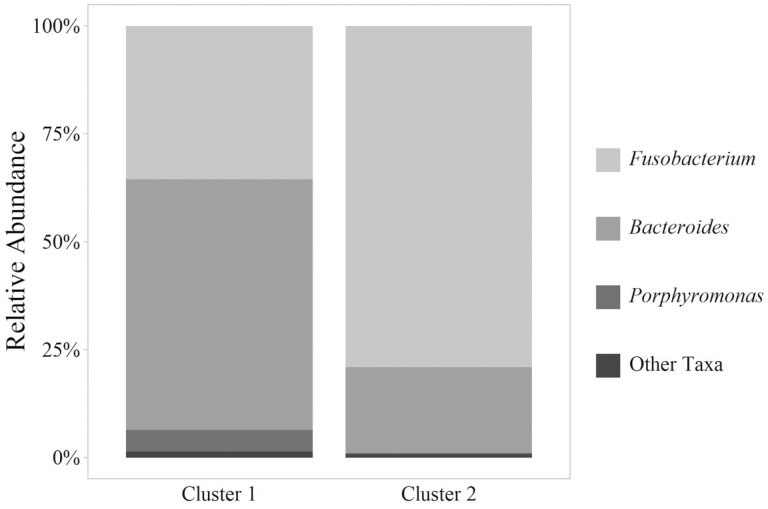
To investigate the difference between abscesses regardless of management program, K-means clusters were evaluated. Two distinct K-means clusters were observed (Figure 3). K-means cluster 1 included 20 abscess communities from naturally managed cattle and 20 microbial communities from traditionally managed cattle. K-means cluster 2 included 42 abscess communities from naturally managed cattle and 33 microbial communities from traditionally managed cattle. No interaction was observed between K-means cluster and management program (P = 0.079) by PERMANOVA testing of unweighted UniFrac distances. The relative abundances of genera within each cluster are visualized in Figure 4, and rare genera are summarized in Supplementary Table S3. Fusobacterium relative abundance was greater in K-means cluster 2 (ANCOM W = 37; 79% vs. 39% relative abundance) while Bacteroides relative abundance was greater in K-means cluster 1 (ANCOM W = 37; 58% vs. 20% relative abundance). While Porphyromonas relative abundance was not statistically different between K-means clusters, mean relative abundance in K-means cluster 1 was 5% and mean relative abundance in K-means cluster 2 was 0%. While generally more rare taxa were observed in K-means cluster 1, alpha diversity measured as Shannon Diversity Index was not different between K-means clusters (P = 0.307).
Figure 3.
Principal coordinate analysis of liver abscess communities from steers managed naturally (no growth-promoting technologies or antimicrobials) or traditionally (with hormonal implants, tylosin phosphate, monensin, ractopamine hydrochloride, and treatment with antimicrobials when necessary). Microbiota of liver abscess samples from naturally managed cattle was similar to microbiota of liver abscess samples from traditionally managed cattle based on PERMANOVA analysis of unweighted UniFrac distances (P = 0.169). Microbiota of liver abscess samples in K-means group 1 differed from microbiota of liver abscess samples in K-means group 2 based on PERMANOVA analysis of unweighted UniFrac distances (P = 0.001).
Figure 4.
Average taxonomic composition (% relative abundance) of the bacterial communities within liver abscesses from steers in a single feedlot by K-means cluster assignment (Figure 3). K-means cluster 1 included 40 samples, and K-means cluster 2 included 75 samples.
Divergence of liver abscess communities, particularly those dominated by Fusobacteria and Bacteroidetes, was recently reported by Stotz et al. (2021) and Pinnell et al. (2022). The findings of the present study are consistent with that of previous authors who found greater relative abundance of Bacteroides within liver abscess communities than previously reported. The work of Pinnell et al. (2022) suggested that Bacteroides-dominated abscess communities could be linked to more distal portions of the gastrointestinal tract (the cecum through rectum) than Fusobacterium-dominated abscess communities. The rare taxa observed in numerically greater relative abundance in Bacteroides-dominated K-means cluster 1 are consistent with the genera discriminant of Bacteroides-dominated liver abscesses reported by Pinnell et al. (2022). While the results of this study and previous reports suggested a role of the hind gut in liver abscess occurrence, further research is needed with microbiota comparisons between body sites of individual animals to definitively link gastrointestinal microbiota to liver abscess communities.
Summary
In this observational study, Fusobacterium and Bacteroides appeared to dominate the microbial communities of liver abscess purulent material and could be linked to different portions of the gastrointestinal tract. The liver abscess microbial communities characterized in this study were much simpler than in previously published studies, emphasizing the need for further investigation. Though management program did not influence the diversity of the microbiota of the liver abscess, Trueperella was found more frequently within liver abscesses from steers managed in the natural program (without exposure to tylosin phosphate). Rare taxa identified suggest a link between the microbiota of the gastrointestinal tract and the microbiota of the liver abscess purulent material. Regardless of the program, steers managed within the same feedlot had similar liver abscess microbiota. Further research is needed to evaluate the routes of entry of gastrointestinal-associated bacteria into the liver. Particularly, the microbiota of the rumen and hindgut epithelial tissues should be evaluated in greater taxonomic depth and compared to bacteria found within the liver abscess.
Supplementary Material
Acknowledgments
This work utilized the Summit supercomputer, which is supported by the National Science Foundation (awards ACI-1532235 and ACI-1532236), the University of Colorado Boulder, and Colorado State University. The Summit supercomputer is a joint effort of the University of Colorado Boulder and Colorado State University.
Glossary
Abbreviations:
- ASV
amplicon sequence variant
Contributor Information
Luke K Fuerniss, Department of Animal Sciences, Colorado State University, Fort Collins, Colorado, USA.
Haley E Davis, Department of Animal Sciences, Colorado State University, Fort Collins, Colorado, USA.
Aeriel D Belk, Department of Animal Sciences, Colorado State University, Fort Collins, Colorado, USA.
Jessica L Metcalf, Department of Animal Sciences, Colorado State University, Fort Collins, Colorado, USA.
Terry E Engle, Department of Animal Sciences, Colorado State University, Fort Collins, Colorado, USA.
John A Scanga, Department of Animal Sciences, Colorado State University, Fort Collins, Colorado, USA.
Franklyn B Garry, Department of Clinical Sciences, Colorado State University, Fort Collins, Colorado, USA.
Tony C Bryant, Five Rivers Cattle Feeding, LLC, Johnstown, Colorado, USA.
Jennifer N Martin, Department of Animal Sciences, Colorado State University, Fort Collins, Colorado, USA.
Conflict of Interest Statement
J.A. Scanga serves as an affiliate faculty member of Colorado State University and is employed by Meyer Natural Foods (Loveland, Colorado). Meyer Natural Foods was not involved in this research. All other authors declare no conflict of interest.
Literature Cited
- Aarestrup, F. M. 1999. Association between the consumption of antimicrobial agents in animal husbandry and the occurrence of resistant bacteria among food animals. Int. J. Antimicrob. Agents 12:279–285. doi: 10.1016/s0924-8579(99)90059-6 [DOI] [PubMed] [Google Scholar]
- Alipour, M. J., Jalanka J., Pessa-Morikawa T., Kokkonen T., Satokari R., Hynönen U., Iivanainen A., and Niku M... 2018. The composition of the perinatal intestinal microbiota in cattle. Sci. Rep. 8:10437. doi: 10.1038/s41598-018-28733-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amachawadi, R. G., and Nagaraja T. G... 2016. Liver abscesses in cattle: a review of incidence in Holsteins and of bacteriology and vaccine approaches to control in feedlot cattle. J. Anim. Sci. 94:1620–1632. doi: 10.2527/jas.2015-0261 [DOI] [PubMed] [Google Scholar]
- Amachawadi, R. G., Thomas M., Nagaraja T. G., and Scaria J... 2016. Genome sequences of Salmonella enterica subsp. enterica serovar Lubbock strains isolated from liver abscesses of feedlot cattle. Genome Announc. 4:e00319–e00316. doi: 10.1128/genomeA.00319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amachawadi, R. G., Purvis T. J., Lubbers B. V., Homm J. W., Maxwell C. L., and Nagaraja T. G... 2017. Bacterial flora of liver abscesses in crossbred beef cattle and Holstein steers fed finishing diets with or without tylosin. J. Anim. Sci. 95:3425–3434. doi: 10.2527/jas.2016.1198 [DOI] [PubMed] [Google Scholar]
- Amachawadi, R. G., Tom W. A., Hays M. P., Fernando S. C., Hardwidge P. R., and Nagaraja T. G... 2021. Bacterial community analysis of purulent material from liver abscesses of crossbred cattle and Holstein steers fed finishing diets with or without tylosin. J. Anim. Sci. 99:1–10. doi: 10.1093/jas/skab076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M. J. 2017. Permutational multivariate analysis of vari’ance (PERMANOVA). In: Wiley StatsRef: Statistics Reference Online. Chichester, UK John Wiley & Sons, Ltd.; p. 1–15. doi: 10.1002/9781118445112.stat07841 [DOI] [Google Scholar]
- Apprill, A., McNally S., Parsons R., and Weber L... 2015. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75:129–137. doi: 10.3354/ame01753 [DOI] [Google Scholar]
- Azad, E., Derakhshani H., Forster R. J., Gruninger R. J., Acharya S., McAllister T. A., and Khafipour E... 2019. Characterization of the rumen and fecal microbiome in bloated and non-bloated cattle grazing alfalfa pastures and subjected to bloat prevention strategies. Sci. Rep. 9:4272. doi: 10.1038/s41598-019-41017-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates, D., Mächler M., Bolker B., and Walker S... 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67:1–48. doi: 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Berg, J. N., and Scanlan C. M... 1982. Studies of Fusobacterium necrophorum from bovine hepatic abscesses: biotypes, quantitation, virulence, and antibiotic susceptibility. Am. J. Vet. Res. 43:1580–1586. [PubMed] [Google Scholar]
- Bokulich, N. A., Kaehler B. D., Rideout J. R., Dillon M., Bolyen E., Knight R., Huttley G. A., and Caporaso J. G... 2018. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 6:90. doi: 10.1186/s40168-018-0470-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen, E., Rideout J. R., Dillon M. R., Bokulich N. A., Abnet C., Al-Ghalith G. A., Alexander H., Alm E. J., Arumugam M., Asnicar F., et al. 2018. QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr. 6:e27295–v2.. doi: 10.7287/peerj.preprints.27295v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostanci, N., and Belibasakis G. N... 2012. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiol. Lett. 333:1–9. doi: 10.1111/j.1574-6968.2012.02579.x [DOI] [PubMed] [Google Scholar]
- Brink, D. R., Lowry S. R., Stock R. A., and Parrott J. C... 1990. Severity of liver abscesses and efficiency of feed utilization of feedlot cattle. J. Anim. Sci. 68:1201–1207. doi: 10.2527/1990.6851201x [DOI] [PubMed] [Google Scholar]
- Brown, T. R., and Lawrence T. E... 2010. Association of liver abnormalities with carcass grading performance and value. J. Anim. Sci. 88:4037–4043. doi: 10.2527/jas.2010-3219 [DOI] [PubMed] [Google Scholar]
- Brown, H., Bing R. F., Grueter H. P., McAskill J. W., Cooley C. O., and Rathmacher R. P... 1975. Tylosin and chlortetracycline for the prevention of liver abscesses, improved weight gains and feed efficiency in feedlot cattle. J. Anim. Sci. 40:207–213. doi: 10.2527/jas1975.402207x [DOI] [PubMed] [Google Scholar]
- Calkins, H. E., and Dewey M. L... 1968. Quantitative analysis of the microflora of a bovine liver abscess. J. Bacteriol. 96:1439–1440. doi: 10.1128/jb.96.4.1439-1440.1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B. J., McMurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P... 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581–583. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., Fierer N., and Knight R... 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108:4516–4522. doi: 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Huntley J., Fierer N., Owens S. M., Betley J., Fraser L., Bauer M., et al. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6:1621–1624. doi: 10.1038/ismej.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillou, S., Chaulot-Talmon A., Caekebeke H., Cardinal M., Christieans S., Denis C., Desmonts M. H., Dousset X., Feurer C., Hamon E., et al. 2015. Origin and ecological selection of core and food-specific bacterial communities associated with meat and seafood spoilage. ISME. 9:1105–1118. doi: 10.1038/ismej.2014.202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M., Qiu T., Sun Y., Song Y., Wang X., and Gao M... 2019. Diversity of tetracycline- and erythromycin-resistant bacteria in aerosols and manures from four types of animal farms in China. Environ. Sci. Pollut. Res. 2019:1–10. doi: 10.1007/s11356-019-05672-3 [DOI] [PubMed] [Google Scholar]
- DeSantis, T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., and Andersen G. L... 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood, L. C., Boykin C. A., Harris M. K., Arnold A. N., Hale D. S., Kerth C. R., Griffin D. B., Savell J. W., Belk K. E., Woerner D. R., et al. 2017. National Beef Quality Audit-2016: transportation, mobility, and harvest-floor assessments of targeted characteristics that affect quality and value of cattle, carcasses, and by-products. Transl. Anim. Sci. 1:229–238. doi: 10.2527/tas2017.0029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firkins, J. L., and Yu Z... 2015. Ruminant nutrition symposium: how to use data on the rumen microbiome to improve our understanding of ruminant nutrition. J. Anim. Sci. 93:1450–1470. doi: 10.2527/jas.2014-8754 [DOI] [PubMed] [Google Scholar]
- Gressley, T. F., Hall M. B., and Armentano L. E... 2011. Ruminant nutrititon symposium: productivity, digestion, and health responses to hindgut acidosis in ruminants. J. Anim. Sci. 89:1120–1130. doi: 10.2527/jas.2010-3460 [DOI] [PubMed] [Google Scholar]
- Grime, J. P. 1998. Benefits of plant diversity to ecosystems: immediate, filter and founder effects. J. Ecol. 86:902–910. doi: 10.1046/j.1365-2745.1998.00306.x [DOI] [Google Scholar]
- Haskell, K. J., Schriever S. R., Fonoimoana K. D., Haws B., Hair B. B., Wienclaw T. M., Holmstead J. G., Barboza A. B., Berges E. T., Heaton M. J., et al. 2018. Antibiotic resistance is lower in Staphylococcus aureus isolated from antibiotic-free raw meat as compared to conventional raw meat. K. Becker, editor. PLoS One 13(12):e0206712. doi: 10.1371/journal.pone.0206712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, G., Cox F., Ganesh S., Jonker A., Young W., Janssen P. H., Abecia L., Angarita E., Aravena P., Arenas G. N., et al. 2015. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 5:14567. doi: 10.1038/srep14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hof, H. 1994. Macrolides, a group of antibiotics with a broad spectrum of activity. Immun. Infekt. 22:66–71. [PubMed] [Google Scholar]
- Hofstad, T. 1984. Pathogenicity of anaerobic gram-negative rods: possible mechanisms. Rev. Infect. Dis. 6:189–199. doi: 10.1093/clinids/6.2.189 [DOI] [PubMed] [Google Scholar]
- Holman, D. B., and Gzyl K. E... 2019. A meta-analysis of the bovine gastrointestinal tract microbiota. FEMS Microbiol. Ecol. 95(.6 ). doi: 10.1093/femsec/fiz072 [DOI] [PubMed] [Google Scholar]
- Janssen, S., McDonald D., Gonzalez A., Navas-Molina J. A., Jiang L., Xu Z. Z., Winker K., Kado D. M., Orwoll E., Manary M., et al. 2018. Phylogenetic placement of exact amplicon sequences improves associations with clinical information. N. Chia, editor. mSystems. 3:e00021–e00018. doi: 10.1128/mSystems.00021-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, R., Deane H. M., Cooper L. J., Miller V. A., and Graham W. R... 1954. The rumenitis-liver abscess complex in beef cattle. Am. J. Vet. Res. 15:202–216. [PubMed] [Google Scholar]
- Kanoe, M., Izuchi Y., Kemi M., Toda M., and Hara Y... 1979. Hepatic abscess in fattened dairy steers. Japanese J. Vet. Sci. 41:73–76. doi: 10.1292/jvms1939.41.73 [DOI] [PubMed] [Google Scholar]
- Kruskal, W. H., and Wallis W. A... 1952. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47:583. doi: 10.2307/2280779 [DOI] [Google Scholar]
- Langworth, B. F. 1977. Fusobacterium necrophorum: its characteristics and role as an animal pathogen. Bacteriol. Rev 41:373–390. doi: 10.1128/br.41.2.373-390.1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Zhang M., Zhang R., Zhu W., and Mao S... 2016. Comparative studies of the composition of bacterial microbiota associated with the ruminal content, ruminal epithelium and in the faeces of lactating dairy cows. Microb. Biotechnol. 9:257–268. doi: 10.1111/1751-7915.12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, S. P. 1982. Least squares quantization in PCM. IEEE Trans. Inf. Theory 28(2):129–137. doi: 10.1109/TIT.1982.1056489 [DOI] [Google Scholar]
- Lozupone, C., and Knight R... 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C. A., Hamady M., Kelley S. T., and Knight R... 2007. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73:1576–1585. doi: 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen, J. B. 1967. Some methods for classification and analysis of multivariate observations. In Le Cam L. M. and Neyman J., editors. Proceedings of the fifth Berkeley symposium on mathematical statistics and probability; California: University of California Press; p. 281–297. [Google Scholar]
- Mandal, S., Van Treuren W., White R. A., Eggesbø M., Knight R., and Peddada S. D... 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Heal. Dis. 26:27663. doi: 10.3402/mehd.v26.27663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., Andersen G. L., Knight R., and Hugenholtz P... 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. doi: 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura, H., Horiguchi M., and Matsumoto T... 1980. Nutritional interdependence among rumen bacteria, Bacteroides amylophilus, Megasphaera elsdenii, and Ruminococcus albus. Appl. Environ. Microbiol. 40:294–300. doi: 10.1128/aem.40.2.294-300.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja, T. G., and Chengappa M. M... 1998. Liver abscesses in feedlot cattle: a review. J. Anim. Sci. 76:287–298. doi: 10.2527/1998.761287x [DOI] [PubMed] [Google Scholar]
- Nagaraja, T. G., and Lechtenberg K. F... 2007. Liver abscesses in feedlot cattle. Vet. Clin. North Am. Food Anim. Pract. 23:351–369. doi: 10.1016/j.cvfa.2007.05.002 [DOI] [PubMed] [Google Scholar]
- Nagaraja, T. G., Laudert S. B., and Parrott J. C... 1996. Liver abscesses in feedlot cattle. Part I. Causes, pathogenesis, pathology, and diagnosis. Compend. Cont. Educ. Pr. Vet. 18:S230–S241. [Google Scholar]
- Nagaraja, T. G., Beharka A. B., Chengappa M. M., Carroll L. H., Raun A. P., Laudert S. B., and Parrott J. C... 1999. Bacterial flora of liver abscesses in feedlot cattle fed tylosin or no tylosin. J. Anim. Sci. 77:973–978. doi: 10.2527/1999.774973x [DOI] [PubMed] [Google Scholar]
- Newsom, I. E. 1938. A bacteriologic study of liver abscesses in cattle. J. Infect. Dis. 63:232–238. doi: 10.1093/infdis/63.3.232 [DOI] [Google Scholar]
- Oba, M., and Wertz-Lutz A. E... 2011. Ruminant nutrition Symposium: acidosis: new insights into the persistent problem. J. Anim. Sci. 89:1090–1091. doi: 10.2527/jas.2010-3727 [DOI] [PubMed] [Google Scholar]
- Oksanen, J., Simpson G. L., Blanchet F. G., Kindt R., Legendre P., Minchin P. R., O’Hara R. B., Solymos P., Stevens M. H. H., Szoecs E., et al. 2022. vegan: Community Ecology Package. R package version 2.6-2. Available from https://CRAN.R-project.org/package=vegan.
- Ozbayram, E. G., Ince O., Ince B., Harms H., and Kleinsteuber S... 2018. Comparison of rumen and manure microbiomes and implications for the inoculation of anaerobic digesters. Microorganisms. 6(.1 ):15. doi: 10.3390/microorganisms6010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacífico, C., Petri R. M., Ricci S., Mickdam E., Wetzels S. U., Neubauer V., and Zebeli Q... 2021. Unveiling the obvine epimural microbiota composition and putative function. Microorganisms. 9:342. doi: 10.3390/microorganisms9020342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada, A. E., Needham D. M., and Fuhrman J. A... 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18:1403–1414. doi: 10.1111/1462-2920.13023 [DOI] [PubMed] [Google Scholar]
- Pinnell, L. J., Whitlow C. W., Huebner K. L., Bryant T. C., Martin J. N., Belk K. E., and Morley P. S... 2022. Not all liver abscesses are created equal: the impact of tylosin and antibiotic alternatives on bovine liver abscess microbial communities and a first look at bacteroidetes-dominated communities. Front. Microbiol. 13:882419. doi: 10.3389/fmicb.2022.882419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glöckner F. O... 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. doi: 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2017. R: a language and environment for statistical computing. Available from: https://www.R-project.org/.
- Ribeiro, M. G., Risseti R. M., Bolaños C. A. D., Caffaro K. A., De Morais A. C. B., Lara G. H. B., Zamprogna T. O., Paes A. C., Listoni F. J. P., and Franco M. M. J... 2015. Trueperella pyogenes multispecies infections in domestic animals: a retrospective study of 144 cases (2002 to 2012). Vet. Q 35:82–87. doi: 10.1080/01652176.2015.1022667 [DOI] [PubMed] [Google Scholar]
- Scanlan, C. M., and Hathcock T. L... 1983. Bovine rumenitis-liver abscess complex: a bacteriological review. Cornell Vet. 73:288–297. [PubMed] [Google Scholar]
- Shanks, O. C., Kelty C. A., Archibeque S., Jenkins M., Newton R. J., McLellan S. L., Huse S. M., and Sogin M. L... 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl. Environ. Microbiol. 77:2992–3001. doi: 10.1128/AEM.02988-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, C. E. 1948. A mathematical theory of communication. Bell Syst. Tech. J. 27:623–656. doi: 10.1002/j.1538-7305.1948.tb00917.x [DOI] [Google Scholar]
- Simon, P. C., and Stovell P. L... 1971. Isolation of Sphaerophorus necrophorus from bovine hepatic abscesses in British Columbia. Can. J. Comp. Med. 35:103–106. [PMC free article] [PubMed] [Google Scholar]
- Smith, H. R. 1940. Beef liver condemnations. J. Anim. Sci. 1940:272–276. doi: 10.2527/jas1940.19401272x [DOI] [Google Scholar]
- Stanton, T. B., and Canale-Parola E... 1980. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch. Microbiol. 127:145–156. doi: 10.1007/BF00428018 [DOI] [PubMed] [Google Scholar]
- Stotz, M. K., Henry D. D., and Crossland W. L... 2021. Characterization of bacterial DNA identified in abscessed and non-abscessed bovine hepatic tissue at the time of harvest. J. Anim. Sci. 99:1–11. doi: 10.1093/jas/skab076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summanen, P. H., Durmaz B., Väisänen M. -L., Liu C., Molitoris D., Eerola E., Helander I. M., and Finegold S. M... 2005. Porphyromonas somerae sp. nov., a pathogen isolated from humans and distinct from porphyromonas levii. J. Clin. Microbiol. 43:4455–4459. doi: 10.1128/JCM.43.9.4455-4459.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoefner, M. B., Pollitt C. C., Van Eps A. W., Milinovich G. J., Trott D. J., Wattle O., and Andersen P. H... 2004. Acute bovine laminitis: a new induction model using alimentary oligofructose overload. J. Dairy Sci. 87:2932–2940. doi: 10.3168/jds.S0022-0302(04)73424-4 [DOI] [PubMed] [Google Scholar]
- Weinroth, M. D., Carlson C. R., Martin J. N., Metcalf J. L., Morley P. S., and Belk K. E... 2017. Rapid Communication: 16S ribosomal ribonucleic acid characterization of liver abscesses in feedlot cattle from three states in the United States. J. Anim. Sci. 95:4520–4525. doi: 10.2527/jas2017.1743 [DOI] [PubMed] [Google Scholar]
- Weinroth, M. D., Belk A. D., Dean C., Noyes N., Dittoe D. K., Rothrock M. J., Ricke S. C., Myer P. R., Henniger M. T., Ramírez G. A., et al. 2022. Considerations and best practices in animal science 16S ribosomal RNA gene sequencing microbiome studies. J. Anim. Sci. 100:1–18. doi: 10.1093/JAS/SKAB346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, S., Xu Z. Z., Peddada S., Amir A., Bittinger K., Gonzalez A., Lozupone C., Zaneveld J. R., Vázquez-Baeza Y., Birmingham A., et al. 2017. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5:27. doi: 10.1186/s40168-017-0237-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzels, S. U., Mann E., Pourazad P., Qumar M., Pinior B., Metzler-Zebeli B. U., Wagner M., Schmitz-Esser S., and Zebeli Q... 2017. Epimural bacterial community structure in the rumen of Holstein cows with different responses to a long-term subacute ruminal acidosis diet challenge. J. Dairy Sci. 100:1829–1844. doi: 10.3168/jds.2016-11620 [DOI] [PubMed] [Google Scholar]
- Wickham, H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. doi: 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- Yang, X., Noyes N. R., Doster E., Martin J. N., Linke L. M., Magnuson R. J., Yang H., Geornaras I., Woerner D. R., Jones K. L., et al. 2016. Use of metagenomic shotgun sequencing technology to detect foodborne pathogens within the microbiome of the beef production chain. Appl. Environ. Microbiol. 82:2433–2443. doi: 10.1128/AEM.00078-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.