Abstract
The development of human microbiome has collectively correlated the sophisticated interactions between Fusobacterium nucleatum and colorectal cancers (CRCs). However, the treatment of CRC via disruption of gastrointestinal flora remains less explored. Aiming at the up-regulated activity of nitroreductase in F. nucleatum–infected tumors, here, we developed the nitroreductase-instructed supramolecular self-assembly. The designed assembly precursors underwent enzymatic transformation to form assemblies, which agglutinated F. nucleatum and eradicated the targeted bacteria. These assemblies with anti–F. nucleatum activity could further alleviate the bacteria-induced drug resistance effect, thus sensitizing CRC cells against chemo-drugs. Eventually, in mice bearing F. nucleatum–infected CRC, the local introduction of nitroreductase-instructed assemblies could efficiently inhibit the tumor growth. Overall, this study incorporated nitroreductase to broaden the toolbox of enzyme-instructed supramolecular self-assembly. The local introduction of nitroreductase-instructed assemblies could target F. nucleatum to eliminate its contribution to CRC drug resistance and ameliorate chemotherapy outcomes.
Supramolecular self-assembly is an abiotic strategy to interrupt bacteria-mammalian cell interactions and enhance CRC therapy.
INTRODUCTION
Colorectal cancers (CRCs) have emerged as frequently diagnosed diseases and the second leading cause of cancer death globally (1). Along with the expanding knowledge in cancer biology, the array of CRC treatments could largely extend the overall survival for those nonmetastasized cases (2). In contrast, those advanced and metastatic CRC often fail the whole treatment, suffering from recurrence and drug resistance (3). Bacterial infections likely result in the difficulty in CRC treatment. Accumulating evidence from CRC patients and animal experiments has correlated the intestinal microbiota with CRC (4). Fusobacterium nucleatum is one of the most prevalent bacterial strains in CRC. The increase of F. nucleatum not only elicits the high-grade dysplasia and established CRC but also promotes the metastasis and drug resistance (5–9).
Consequently, CRC treatments that target the microbiota are evolving, including selective elimination of oncogenic microbes, lipopolysaccharide–promoted immunotherapy, and targeted bacteriophage therapy (10, 11). A treatment involving the antibiotic metronidazole (MTZ) directly reduced F. nucleatum and tumor growth, implying a potential CRC treatment via antimicrobial interventions (8). However, the use of antibiotics should be restricted due to potential microbial imbalance or dysbiosis (12). Because bacteriophages are often highly specific to eliminate single species of bacteria while sparing others, a proper workflow to isolate bacteriophage against F. nucleatum enabled the phage-guided CRC treatment, which can target F. nucleatum–burdened CRC to lyse F. nucleatum, proliferate Clostridium butyricum, and release anticancer drugs simultaneously (13). Alternatively, peptide-based supramolecular self-assembly serves as a typical abiotic method to inhibit bacteria via agglutination both in vitro and in vivo (14, 15). Specifically, the minimal nature of diphenylalanine motif made this scaffold both the shortest peptide-based agent with antimicrobial effect so far and an important skeleton to construct the alternative antimicrobial materials (16). Our previous work designed a tetrapeptide that conjugated with a reactive oxygen species labile trigger to mimic neutrophil extracellular traps. The in situ formation of assemblies at inflammatory loci succeeded in inhibiting methicillin-resistant bacteria in vivo (17). Therefore, the introduction of peptidic supramolecular self-assembly to inhibit F. nucleatum should provide an opportunity to enhance CRC treatments.
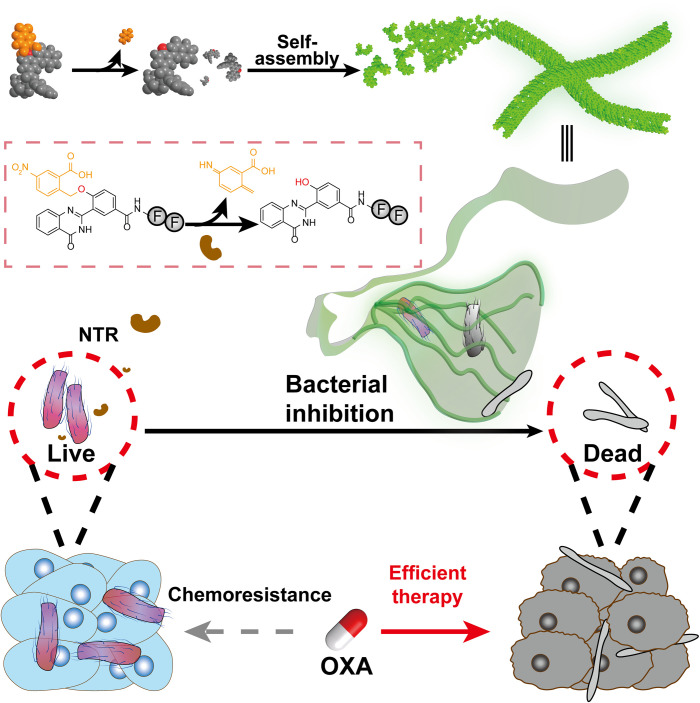
Because F. nucleatum relatively overexpressed nitroreductase (NTR) (18), we designed short peptide–based precursors, which underwent NTR-instructed self-assembly to form nanofibers to target F. nucleatum. On the basis of the general principles for enzyme-instructed supramolecular self-assembly (EISA) strategy (19, 20), the precursor contained two major components: (i) an NTR-responsive moiety and (ii) an assembly-prone short peptide. NTR efficiently converted the precursors into hydrogelators, which self-assembled into nanofibers both in vitro and in biological milieu. The formed nanofibers trapped F. nucleatum, resulting in the bacterial morphology change and growth inhibition. In addition, the treatment with the nanofibers attenuated the F. nucleatum–assisted chemoresistance for CRC cells against chemo-drugs, e.g., oxaliplatin (OXA). Furthermore, the in vivo application of NTR-instructed self-assembly improved the chemotherapy of CRC tumors via the elimination of intratumoral F. nucleatum. Overall, the NTR-instructed assemblies would provide an antibiotic-free strategy to inhibit specific bacterial strains for alternative treatment regime against CRC (Fig. 1).
Fig. 1. NTR-instructed assemblies to regulate microbiome for enhanced chemotherapy against CRCs.
Upon the enzymatic reduction, the precursors underwent a self-elimination reaction to produce hydrogelators, which self-assembled into supramolecular nanofibers. The nanofibers inhibited intratumoral bacteria and enhanced the subsequent chemotherapy.
RESULTS
NTR-catalyzed self-assembly and hydrogelation
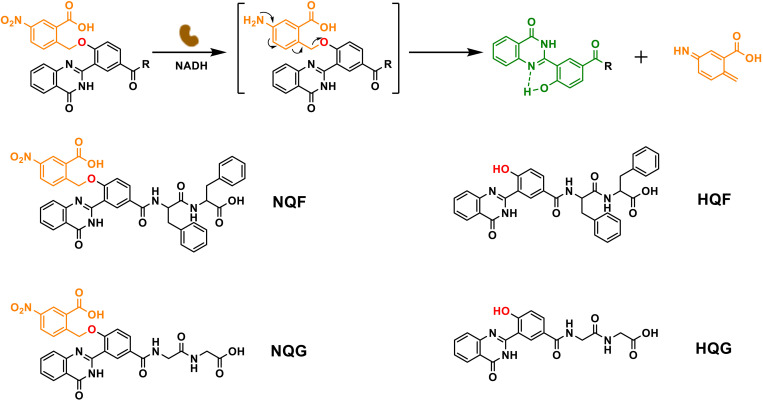
The quinazolinone core was derived with a 4-hydroxybenzoic acid. The phenol group was caged with a typical self-immolative residue, p-nitrobenzyl alcohol, which was readily removed upon NTR-catalyzed reduction with NADH (reduced form of nicotinamide adenine dinucleotide) (Fig. 2). The liberated phenol tended to form an intramolecular hydrogen bond, which planarized the quinazolinone derivative and emitted green fluorescence simultaneously (21). Certain short peptides (e.g., Phe-Phe, FF; Gly-Gly, GG) were coupled to 4 on its carboxylic group via a sequential N,N’-Diisopropylcarbodiimide/N-hydroxysuccinimide activation and amide exchange reaction to yield NQF and NQG, respectively. (The complete synthesis route was shown in the Supplementary Materials.)
Fig. 2. Molecular design and enzymatic transformation.
The NTR-catalyzed transformation from nitro-quinazolinone (NQ) derivatives to hydroxyl-quinazolinone (HQ) derivatives.
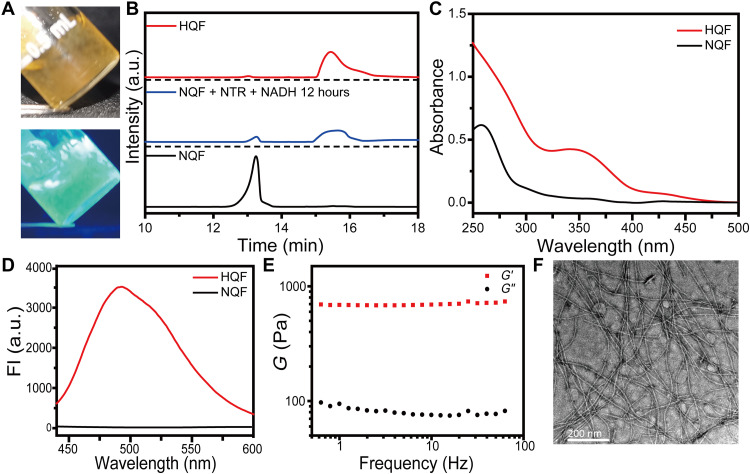
Then, NQF or NQG were treated with NTR (10 μg/ml; 1 U/ml) and 50 mM NADH for hydrogelation test. NQF (5 mM) was sufficient to form a self-supported hydrogel after enzymatic transformation. The hydrogel emitted bright green fluorescence under ultraviolet (UV) irradiation (Fig. 3A). The underlined NTR-catalyzed transformation was monitored by high-performance liquid chromatography (HPLC) and matrix-assisted laser desorption/ionization–time-of-flight (MALDI-TOF). As shown in Fig. 3B, after the addition of NTR/NADH, the peak of NQF (retention time: 13.24 min) disappeared with the emergence of a new peak (retention time: 15.44 min) whose mass spectrum indicated the generation of HQF (fig. S1). Accordingly, the UV-visible (UV-vis) spectrum gave a new absorption peak at 339 nm after reduction (Fig. 3C). With the excitation at 405 nm, the assemblies of HQF gave an emission at 493 nm, while there was no emission of NQF solution (Fig. 3D). The rheological test showed that the dynamic storage modulus (G′) of the hydrogel was almost one order magnitude higher than the loss modulus (G″). Meanwhile, both G′ and G″ were independent with a broad range of frequency, confirming the formation of a stable hydrogel of 5 mM HQF (Fig. 3E and fig. S2). Within the hydrogel, there were abundant amounts of supramolecular nanofibers (Fig. 3F). Lower concentration of NQF (e.g., 1 to 2 mM) formed soft hydrogels and displayed weaker green fluorescence, while the critical assembly concentration (CAC) of HQF was around 100 μM where nearly no green fluorescence could be observed (fig. S3). In contrast, after the treatment of NTR/NADH, NQG remained a solution without green fluorescence emission, despite the full conversion to HQG (fig. S4, A to C). Nanofibers were absent, and no hydrogel formed even in 10 mM HQG (fig. S4, D to G). Therefore, both the peptide FF (22) and the intramolecular hydrogen bond within HQ promoted the intermolecular π-π stacking and the subsequent supramolecular self-assembly, which can be well correlated with the fluorescence emission.
Fig. 3. NTR-instructed self-assembly.
(A) Optical images of a stable hydrogel of HQF, which emitted green fluorescence with UV light. (B) HPLC trace of the reduction from NQF to HQF in vitro with the presence of NTR/NADH. a.u., arbitrary units. (C) UV-vis spectra and (D) fluorescence spectra of NQF before and after reduction. Excitation: 405 nm. (E) Frequency sweep of the dynamic storage modulus (G′) and loss modulus (G″) of an HQF hydrogel. (F) TEM image of nanofibers in an HQF hydrogel.
In addition, the conversion from NQF to HQF was quite specific upon NTR-catalyzed reduction rather than common biological reducing agents. As shown in fig. S5, NQF remained intact with Cys, glutathione (GSH), or dithiothreitol (DTT). The NTR-catalyzed reduction may also be inhibited by the NTR inhibitor dicuomarol. This reaction specificity allowed the selective formation of NTR-instructed self-assembly in biological environment.
In situ formation of NTR-instructed assemblies
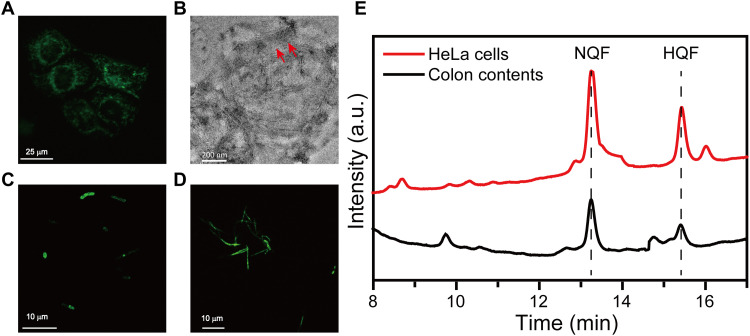
As mentioned above, the assemblies of HQF emitted green fluorescence. Thus, the fluorescence signal could conveniently determine the formation of assemblies in living system. Because the NTR activities were usually studied in hypoxic conditions (23–25), HeLa cells were first incubated under hypoxia condition for an optimized period to produce NTR (fig. S6) and then treated with NQF for a specific time (2, 4, and 8 hours). Confocal laser scanning microscopy (CLSM) images showed the green fluorescence emission in the hypoxic HeLa cells incubated with 500 μM NQF for 8 hours at 37°C, which indicated the formation of intracellular HQF assemblies (Fig. 4A). To determine the subcellular location of the assemblies, we used a MitoTracker to stain assemblies containing HeLa cells. A good overlap between green and red fluorescence implied the occurrence of assemblies around mitochondria (fig. S7). The addition of dicoumarol may prevent the formation of intracellular assemblies, as the fluorescence was absent in hypoxic HeLa cells (fig. S8). In addition, in HeLa cells under normoxic condition, which expressed a low level of NTR, negligible fluorescence emission was observed (fig. S9). This result indicated that NQF could undergo NTR catalysis to form peri-mitochondrial assemblies. Upon fractionation of a group of HeLa cells bearing HQF assemblies, plenty of nanofibers could be observed in the fraction containing mitochondria (Fig. 4B).
Fig. 4. NTR-instructed self-assembly in biological environment.
(A) CLSM of HeLa cells incubated with NQF under hypoxic condition. (B) TEM image of nanofibers (indicated by arrowheads) in the cellular fractions of treated HeLa cells. CLSM images of (C) E. coli and (D) F. nucleatum incubated with NQF. (E) Conversion of NQF to HQF in hypoxic HeLa cells and mouse colon contents.
Certain bacteria also overexpressed NTR, which may be used to instruct the in situ self-assembly process. For both Escherichia coli (a typical aerobic bacteria) and F. nucleatum (a typical anaerobic bacteria), coculturing with NQF made the bacterial cells to emit green fluorescence emission, which was similar to hypoxic HeLa cells (Fig. 4, C and D, and fig. S10). Apart from the living cells and bacteria, a suspension of fresh mouse colon contents was collected as the environment in real biological system. The colonic microbiota produces extensive amounts of enzymes, especially the oxidoreductases. Thus, the colonic enzymes were used for drug delivery design and diagnosis (26–28). After coculturing with colon contents under anaerobic incubator, the appearance of a peak at 15.44 min in HPLC indicated the reduction of NQF to HQF. Meanwhile, a similar enzymatic transformation was detected in hypoxic HeLa cell lysate (Fig. 4E). Overall, NTR-catalyzed reduction converted precursor NQF to hydrogelator HQF, which can self-assemble in various biological environments.
Antibacterial activity of assemblies
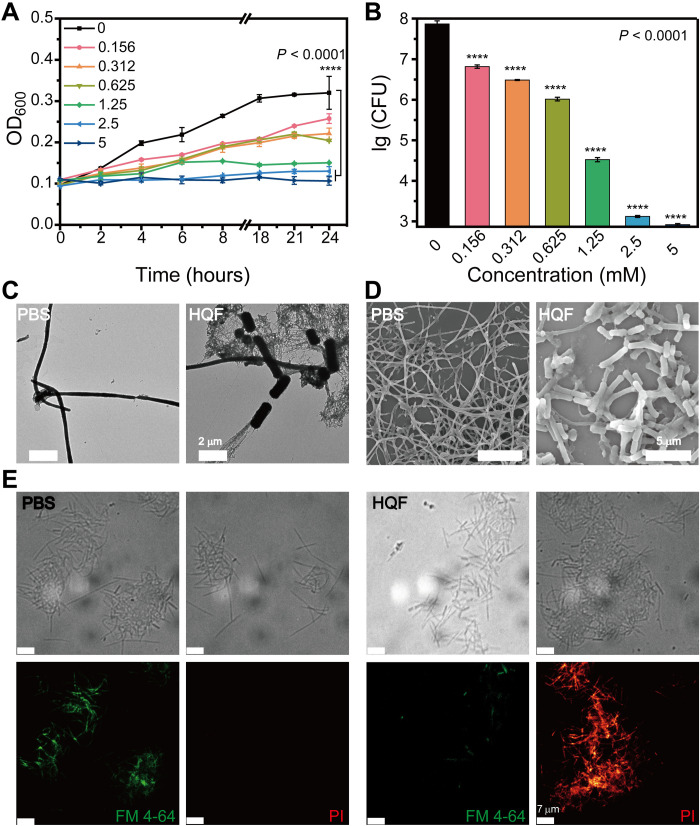
To verify the antibacterial activity of assemblies, we notably inhibited F. nucleatum by HQF at the concentration above 2 mM (fig. S11). A narrow concentration range showed the concentration- and time-dependent inhibition efficacy (Fig. 5A). At 24 hours, the population of F. nucleatum was quantified by plate counting (Fig. 5B and fig. S12). The quantity of F. nucleatum colonies remarkably decreased with a higher concentration of HQF, for instance, nearly five orders of magnitude decrease for treatments with 2.5 and 5 mM HQF. To elucidate the interaction of assemblies and F. nucleatum cells, we conducted a morphological study via transmission electron microscopy (TEM) and scanning electron microscopy (SEM). Wild-type F. nucleatum cells were fusiform rods and spindle-like shape while incubated in brain heart infusion (BHI) culture medium (fig. S13, A to C). After incubation with HQF assemblies for 24 hours, nanofibers formed around or stuck to F. nucleatum cells (fig. S13, D to I). The surrounding nanofibers made the bacterial cell membrane deformed, collapsed, and rough, which indicated the damage to F. nucleatum cells (Fig. 5C). Similar results could be observed in SEM images. The length of spindle shape was significantly shortened in the presence of assemblies (Fig. 5D and fig. S14).
Fig. 5. Antibacterial activity of HQF assemblies against F. nucleatum.
(A) Growth curves of F. nucleatum incubated with various concentrations of HQF and (B) plate counting for each sample at 24 hours. (C) TEM images and (D) SEM images of the morphological change of F. nucleatum incubated with/without HQF assemblies. (E) Live/dead staining of F. nucleatum with/without treatments of HQF assemblies. One-way analysis of variance (ANOVA) test was used in (A) and (B), while Tukey test was used to correct for multiple comparisons using statistical hypothesis. ****P < 0.0001.
To correlate the antibacterial activity with the formation of nanofibers, we challenged the growth of F. nucleatum with HQG, which will not form nanofibers (fig. S4D). F. nucleatum was incubated with 5 mM HQF and HQG for 24 hours. The inhibition efficacy for HQG was far lower than that of HQF against F. nucleatum (fig. S15). According to the live/dead staining by FM 4-64/propidium iodide (PI), F. nucleatum treated with 5 mM HQF displayed stronger red fluorescence (represented as PI) and weaker green fluorescence (represented as FM 4-64) compared to the phosphate-buffered saline (PBS) group, indicating the death of F. nucleatum (Fig. 5E). As for the F. nucleatum treated with 5 mM HQG, the CLSM images indicated nearly no cell death (fig. S16). These results confirmed that the HQF nanofibers were essential for the antibacterial activity.
Enhanced anticancer efficacy via F. nucleatum elimination
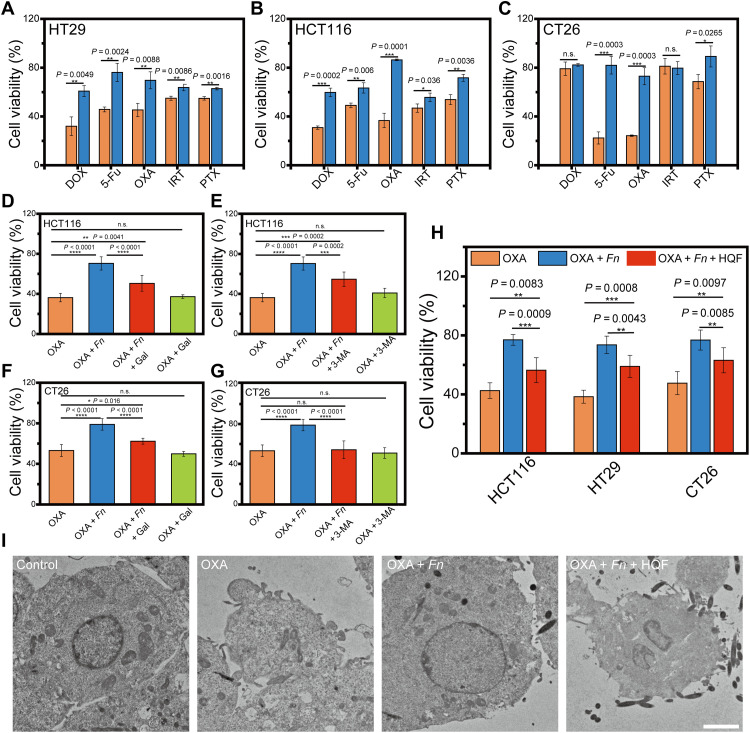
First, a series of multiplicity of infection (MOI) of F. nucleatum were used for the coculture with typical CRC cells (e.g., human colon cancer cells, HT29 and HCT116, and mouse colon cancer cells, CT26). Cell viability of all the tested cells was not influenced by the presence of F. nucleatum (fig. S17). Subsequently, each kind of CRC cells were cocultured with F. nucleatum (MOI = 100) and treated with certain anticancer drugs. For chemotherapy agents, OXA (128 μM), 5-fluorouracil (5-Fu; 800 μM), irinotecan (IRT; 30 μM), paclitaxel (PTX; 12 μM), and doxorubicin hydrochloride (DOX; 0.5 μM) were selected. For HT29 and HCT116 cells, F. nucleatum alleviated the cytotoxicity of all the five drugs to some extent (Fig. 6, A and B). For CT26 cells, F. nucleatum still alleviated the cytotoxicity of most drugs except IRT (Fig. 6C). The observation confirmed that the infection of F. nucleatum would elicit multidrug resistance for CRC cells.
Fig. 6. Enhanced chemotherapy by inhibiting F. nucleatum in vitro.
Cell viability of (A) human HT29, (B) human HCT116, and (C) mouse CT26 cells upon anticancer drugs with/without F. nucleatum infection. The sensitization effect of certain inhibitors on (D and E) HCT116 cells and (F and G) CT26 cells with F. nucleatum infection. Gal, F. nucleatum cell adhesion inhibitor; 3-MA, autophagy inhibitor. (H) Sensitization effect of HQF assemblies on CRC cells with F. nucleatum infection. (I) TEM images of HCT116 cell sections with different treatments. Scale bars, 2 μm. All the data were presented as means ± SD. The significance was evaluated by unpaired two-tailed Student’s t test between two groups in (A) to (C). One-way ANOVA test was used in (D) to (H), while Tukey test was used to correct for multiple comparisons using statistical hypothesis. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. n.s., not significant.
The multidrug resistance in CRC was mainly attributed to autophagy in colorectal adenocarcinoma and cancer tissues (9). Those CRC cells usually overexpressed a glycan residue, Gal-GalNAc, which facilitated the Fap2-based adhesion and mediated F. nucleatum enrichment (29). We hypothesized that the inhibition of autophagy or the disruption of the interaction between F. nucleatum and CRC cells should enhance the chemotherapy efficacy. Accordingly, two inhibitors, galactosamine (Gal; to inhibit the enrichment of F. nucleatum) (30, 31) and 3-methyladenine (3-MA; a broad-spectrum autophagy inhibitor) (32), were used to test our hypothesis in HCT116 cells and CT26 cells. While Gal or 3-MA itself did not affect the viability of OXA-treated cells (Fig. 6, D to G), in the group of cells infected with F. nucleatum, both the treatment of Gal and 3-MA could enhance the efficacy of OXA in certain extent, confirming that the successful infection of F. nucleatum was critical for the induction of multidrug resistance.
Assemblies of HQF inhibited F. nucleatum; thus, they might disrupt the interaction between F. nucleatum and CRC cells and alleviate the multidrug resistance. To evaluate the effect of assemblies in F. nucleatum–induced chemoresistance, we cocultured CRC cells with infection solution containing F. nucleatum, OXA, and HQF assemblies. As shown in Fig. 6H and fig. S18, for all the three tested cell lines, F. nucleatum infection made the cells resistant to OXA, while the additional treatment of assemblies sensitized the cells to OXA. Furthermore, as the autophagy induced chemoresistance, we hypothesized that the inhibition of F. nucleatum would prevent the formation of autophagosomes. We used TEM images to characterize the morphology of cells and autophagy, which indicated similar results. The disturbed cell morphology revealed an abolished chemoresistance after introduction of HQF assemblies (Fig. 6I), while the quantity of autophagosomes was also significantly decreased (fig. S19).
In vivo anticancer studies
We established CRC xenograft mouse model to evaluate the anticancer efficacy in vivo, which was illustrated in Fig. 7A. Briefly, HCT116 cells were inoculated into nude mice and then followed by F. nucleatum infection and different treatments as stated. Initially, the effect of F. nucleatum in tumor growth was tested. The result indicated that the F. nucleatum–cocultured tumor shared almost the same volume as the F. nucleatum–negative tumor (fig. S20A). We further tested whether the chemoresistance induced by F. nucleatum was detected in vivo. F. nucleatum–positive or F. nucleatum–negative mice were treated with OXA for three courses. In agreement with the in vitro results discussed above, the efficacy of OXA was alleviated upon F. nucleatum infection in vivo (fig. S20B).
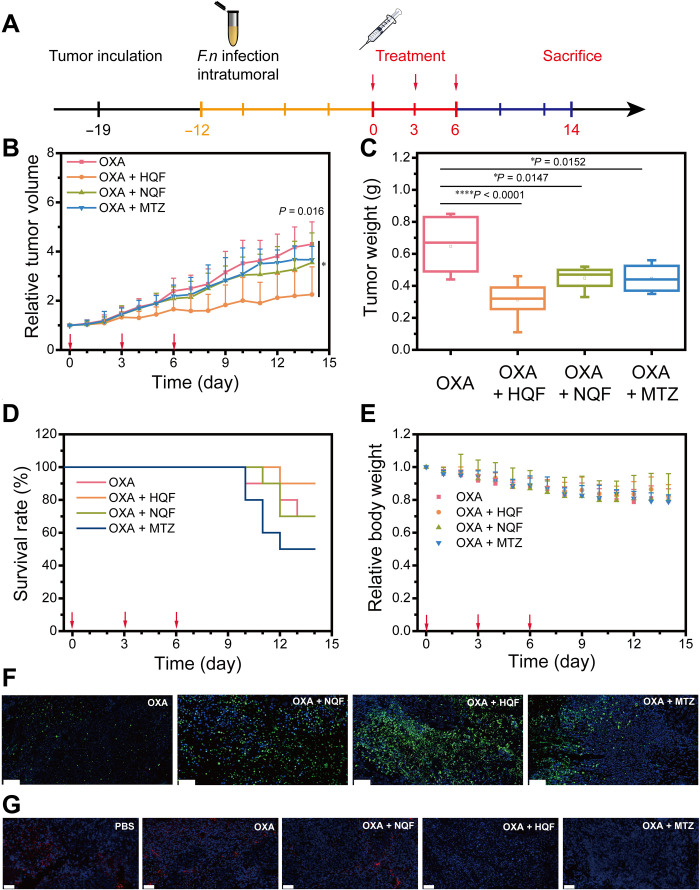
Fig. 7. Anticancer effects in subcutaneous tumor model.
(A) Schematic illustration of tumor inoculation and treatment protocol for the HCT116 tumor-bearing mice. (B) Tumor growth curve in each group: (i) intraperitoneal injection of OXA (7.5 mg/kg), (ii) intraperitoneal injection of OXA (7.5 mg/kg) and then peritumoral injection of NQF solution, (iii) intraperitoneal injection of OXA (7.5 mg/kg) and then peritumoral injection of HQF assemblies, and (iv) intraperitoneal injection of OXA (7.5 mg/kg) and then oral administration of MTZ (200 mg/kg). (C) Tumor weight in each group. (D) Survival rate curve in each group. (E) Quantification of relative body weight in each group. (F) Representative TUNEL staining examination results of the dissected tumors. (G) Representative FISH staining to visualize F. nucleatum in tumor tissue sections. Scale bars, 50 μm. All the data were presented as means ± SD, while n = 10. Red arrows in (B), (D), and (E) indicated the day of treatment. The significance was evaluated by one-way ANOVA test in (B) and (C). *P < 0.05; ****P < 0.0001.
To determine whether antibacterial assemblies could enhance the anticancer chemotherapy, we used cotreatments of OXA with HQF assemblies or NQF solution. Meanwhile, MTZ was used as a broad-spectrum antibiotic against anaerobic species. After the volume of tumor reached approximately 200 mm3, all the mice were divided into four individual groups (n = 10). The tumor growth was remarkably inhibited in the OXA + HQF group. After three courses of treatments, the tumor volume reached only two times at day 14. However, the volume in the OXA group increased for four times (Fig. 7B). The cotreatment with NQF or MTZ also slightly decreased the tumor growth than that of OXA alone. Moreover, the tumor weight and typical tumor photographs at day 14 (Fig. 7C and fig S21) further confirmed that the HQF remarkably improved the therapy efficacy. Although the cotreatment with NQF or MTZ contributed to inhibit tumor growth in some extent, such treatment barely prolonged the overall survival rate. The cotreatment with HQF gave the highest survival rate (Fig. 7D). The body weight remained steady in all treatment groups (Fig. 7E). In addition, there was no inhibition of tumor growth in the groups with the treatment of NQF or HQF alone (fig. S22).
After day 14, all the mice were sacrificed. Typical major organs and tumors were dissected for further examinations. Terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick end labeling (TUNEL) staining showed the significant cell apoptosis in cotreatment groups than OXA alone, especially in the group treated with OXA + HQF assemblies (Fig. 7F). Furthermore, hematoxylin and eosin (H&E) staining of tumors indicated the lowest density of tumor cells and less tumor nests in the OXA + HQF group, where the cells held shortened cell nuclei. OXA + NQF or MTZ groups also presented enhanced therapy effect than the group with OXA alone, where the cells showed atypia, which was attributed to the chemoresistance induced by F. nucleatum (fig. S23). To verify the possible toxicity of NQF, HQF, or MTZ, the cotreatment groups exhibited insignificant difference compared to the OXA group in heart, liver, spleen, lung, and kidney based on H&E staining, which indicated the biocompatibility of the used cotreatments (fig. S24).
Next, we used fluorescence in situ hybridization (FISH) to visualize the F. nucleatum (red fluorescence) in the tumor sections. As shown in Fig. 7G and figs. S25 and S26, the group with HQF assemblies showed the least red fluorescence, indicating the efficient clearance of F. nucleatum, which was comparable to the group with oral administration of MTZ. The HQF assemblies successfully eliminated F. nucleatum in vivo, which eventually alleviated the chemoresistance and enhanced the chemotherapy efficacy.
DISCUSSION
In this work, we have demonstrated a process of NTR-instructed supramolecular self-assembly. The designed precursors underwent NTR/NADH-catalyzed transformation to yield hydrogelator, which self-assembled into nanofibers. These fluorescence-emitting nanofibers served as traps to disturb F. nucleatum. Because the F. nucleatum infection induced the chemoresistance in CRC therapy, the nanofibers with antibacterial activity could further eliminate the F. nucleatum and enhance the chemotherapy efficacy both in vitro and in vivo. Our strategy expanded the toolbox of EISA by the introduction of a new type of enzyme (oxidoreductase) besides well-established hydrolases. The formed assemblies served as physical traps to efficiently inhibit bacteria, providing an antibiotic-free strategy. Note that, for the in vivo anticancer efficacy, the utilization of NQF precursor was not as effective as HQF assemblies, which might be due to the slow NTR-catalyzed transformation from NQF to HQF. Efficient enzymatic transformation from precursors to hydrogelators, which allowed the local concentration of hydrogelators to exceed CAC, is critical for the successful EISA in nonequilibrium living systems (33, 34). At last, the promoted chemotherapeutic efficacy via the elimination of protumoral F. nucleatum may encourage the microbiome regulation for better CRC treatments.
MATERIALS AND METHODS
Experimental materials and instruments
All common starting chemical reagents and solvents were used as purchased without further purification. NTRs, NADH, MTZ, and Gal were purchased from Sigma-Aldrich. Dicoumarol was purchased from Macklin. OXA, DOX, PTX, IRT, and 5-Fu were purchased from Aladdin Biotech Co. Ltd. 3-MA was purchased from Selleck Co. MitoTracker Red was purchased from Beyotime Biotechnology. MTT Cell Proliferation and Cytotoxicity Assay Kit was purchased from Beijing Solarbio Science & Technology. FM 4-64 and PI were purchased from Thermo Fisher Scientific for bacterial live/dead staining.
Nuclear magnetic resonance spectra were recorded on a Bruker 400-MHz Fourier transform spectrometer. TEM images were collected on a Tecnai G2 20 S-TWIN and HT7700 transmission electron microscope. SEM images were captured on a Merlin scanning electron microscope. MALDI-TOF mass spectra were recorded on a Bruker AutoFlex Max mass spectrometer. The fluorescence spectra were recorded on F98, and UV-vis spectra were obtained on Shimadzu UV-2600. The CLSM images were captured with a confocal microscope (Zeiss 710 and UltraVIEW VoX).
Cell lines
HeLa cells, HT29 cells, HCT116 cells, and CT26 cells were obtained from Procell Life Science & Technology Co. Ltd. (Wuhan, China). HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Procell) with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Gibco). HT29 and HCT116 cells were cultured in McCoy’s 5A medium (Procell) with 10% FBS (Gibco) and 1% penicillin-streptomycin (Gibco). CT26 cells were cultured in RPMI 1640 medium (Procell) with 10% FBS (Gibco) and 1% penicillin-streptomycin (Gibco). All the cells were incubated at 37°C with 5% CO2 for culture and passage unless otherwise stated. As for hypoxic condition, HeLa cells were cultured in a hypoxic incubator at 37°C with 1% O2 and 5% CO2.
Bacterial strains
F. nucleatum [American Type Culture Collection (ATCC) 25586] were cultured at 37°C under an anaerobic incubator in BHI broth (Qingdao Hope Bio-Technology, Qingdao, China). E. coli (ATCC 25922) were cultured in Luria-Bertani (LB) broth. All the strains were restored at −80°C before testing. The strains were melted to room temperature quickly and inoculated in culture medium at 1:100 volume ratio. As for F. nucleatum, the inoculated BHI culture medium was cultured at 37°C in an anaerobic incubator (YQX-II). The optical density at 600 nm (OD600) was used to monitor the growth of bacteria. To prepare the liquid culture medium, powder of the culture media (BHI and LB) was dissolved into a solution and sterilized at 121°C with a high pressure for about 1 hour and then cooled down to room temperature.
NTR-instructed supramolecular self-assembly
NQF (5 mM) was incubated with 10 eq of NADH and 10 μg of NTR and kept at 37°C overnight. The reduction of nitro group was detected by HPLC. For inhibition, 1 mM NQF solution was prepared with the same protocol containing 10 eq of NADH and 1 μg of NTR to obtain the working solution. Dicoumarol (500 μM) was added into the above working solution. As for the specific response to NTR, 1 mM different reducing agents (DTT, GSH, and Cys) were added into NQF solution and incubated at 37°C overnight for HPLC detecting.
For hydrogelation, 10 μg of NTR was added into a solution containing 5 mM NQF and NADH (10 eq) and kept at 37°C. A yellow and stable hydrogel of HQF formed. All the other concentrations of hydrogel were prepared with the same protocol. As for contrast, NQF was replaced by NQG to prepare the solution of NQG and HQG.
Oscillatory rheological test
The samples of 10 mM hydrogel were measured on Malvern Kinexus Pro+ at 25°C, with a 0.5-mm gap, 1-Hz frequency, and 0.5% strain. Each sample was repeatedly measured at least three times.
NTR activity assay
HeLa cells growing in log phase were incubated under normoxic conditions overnight at 37°C and then incubated under hypoxic conditions for 0, 2, 4, 8, and 12 hours. After washing with PBS for three times, the cells were disrupted by freezing-thawing cycle for three times. The NTR activity of HeLa cells was calculated by measuring the increased OD550 within 1 min using a commercially available kit.
Living cell and bacteria images
HeLa cells were first placed in glass chamber and cultured overnight under normoxia for attachment. Then, the cells were transformed to hypoxic incubator. After about 12 hours under hypoxia, the cells were treated with 500 μM NQF and placed under hypoxia for another 8 hours. Then, the cells were rinsed by PBS and treated with staining solution (MitoTracker Red) for about 15 min (according to the commercial protocol). For inhibition, hypoxic HeLa cells treated with NQF were incubated with 500 μM dicoumarol for another 8 hours. As for normal HeLa cells, the cells were treated with 500 μM NQF for 8 hours. The CLSM images were captured with a confocal microscope (Zeiss 710).
For bacterial images, the bacteria cultured in LB or BHI culture medium were maintained at 37°C for further test. Culture medium was used to dilute the solution with an OD600 at 0.1. Then, the species were treated with 500 μM NQF for 8 hours. After incubation, bacteria were collected by centrifugation and suspended in PBS. Ten microliters of the bacterial solution was dropped on glass slide and covered with coverslip. The CLSM images of bacteria were captured using a CLSM microscope with immersion oil (UltraVIEW VoX).
Cell fractionation
The cellular fraction was used to detect the in situ assemblies according to previous report (35). Generally, 1 × 106 HeLa cells were seeded in culture flasks (75 cm2) under normoxic condition overnight first and then transformed into hypoxic incubator for about 12 hours. Ten milliliters of fresh DMEM containing 1 mM NQF was added and maintained under hypoxic condition for another 8 hours. After the treatment, the cells were washed with PBS for three times and harvested. All the cells were collected by centrifugation and suspended in 5 ml of deionized water for 30 min to lyse cells. Clumps of unbroken and ruptured cells were removed by centrifugation at 300g for 5 min. Aliquots of the supernatant were collected and then centrifuged at 600g for 10 min. The aliquots of supernatant were collected for further centrifugation. The remaining precipitate was collected and termed as Sample N representing the cell nuclei. Mitochondria, lysosomes, and peroxisomes were collected by centrifugation from the aliquot of Sample N at 15,000g for 5 min, termed as Sample M. Sample N and Sample M are used for TEM images.
Antibacterial effect
To investigate the antibacterial effect against F. nucleatum, F. nucleatum was cocultured with different concentrations of HQF. For higher concentrations, 10, 8, 6, 4, and 2 mM HQF were cocultured with F. nucleatum at 1:1 volume ratio. A narrow range of HQF was also used including 10, 5, 2.5, 1.25, 0.625, 0.312, and 0 mM. OD600 was measured over time to monitor the antibacterial effect. After 24 hours, the population of F. nucleatum was quantified by plate counting. A solution of 10 mM HQG was used to determine the essential of the formation of assemblies with the same protocol. The antibacterial effect of HQG was measured by OD600 at 24 hours. Each sample was set up with three parallels.
Trapping bacteria
F. nucleatum (0.5 ml; OD of 0.1 in BHI medium) were separately cultured with 10 mM HQF hydrogel (0.5 ml) in hypoxic incubator at 37°C. Equal volume of PBS was used as contrast. Later after 24 hours, the samples were collected by centrifugation and suspended in PBS. The collected samples were separated in two parts for TEM images and SEM images.
Live/dead staining
F. nucleatum in medium [0.3 ml; 107 colony-forming units (CFUs)/ml] were cocultured with PBS (0.3 ml), HQF (0.3 ml; 10 mM), and HQG (0.3 ml; 10 mM) in hypoxic incubator at 37°C. After 12 hours, the bacterial cells were collected by centrifugation and rinsed by PBS for three times. Samples were incubated with 100 μl of PI (20 μM) for 15 min in the dark or incubated with 100 μl of FM 4-64 (5 μg/ml) in ice bath. The fluorescence was observed with a confocal microscope (UltraVIEW VoX).
Cytotoxicity of F. nucleatum
The cytotoxicity of F. nucleatum was detected by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay according to the commercial protocol. HT29, HCT116, and CT26 cells were seeded on 96-well plates with a density of 5 × 103 cells per well overnight first and then cocultured with F. nucleatum (MOI = 0, 1, 10, 100, and 1000) for 48 hours. After removing the medium, 100 μl of MTT solution (MTT/medium = 1:9, v/v) was then added into each well and coincubated for another 4 hours. The cultured medium was then removed. A total of 110 μl of dimethyl sulfoxide was added into each well. The OD490 was measured using a plate reader (PerkinElmer).
Chemoresistance induced by F. nucleatum
F. nucleatum was prepared with an MOI of 100 (5 × 105 CFUs per well) suspended culture media with FBS. HT29, HCT116, and CT26 cells were seeded on 96-well plates with a density of 5 × 103 cells per well overnight first and then cocultured with F. nucleatum and chemotherapeutic agents [OXA (128 μM), 5-Fu (800 μM), IRT (30 μM), PTX (12 μM), and DOX (0.5 μM)] for 48 hours. Cell viability was detected by MTT assay to evaluate the chemoresistance.
Enhanced chemotherapy in vitro
The biological effect of two inhibitors (3-MA and Gal) was also investigated by cell viability. Generally, HCT116 and CT26 cells were seeded on a 96-well plate with the density of 5 × 103 cells per well overnight first. After that, the cells were cocultured with F. nucleatum (MOI = 100; 5 × 105 CFUs per well), OXA (128 μM), and a certain inhibitor [3-MA (60 μM) or Gal (33 mg/ml)] for 48 hours in culture medium containing 10% FBS. The OD490 value was detected to determine cytotoxicity.
To evaluate the effect of HQF assemblies, HT29, HCT116, and CT26 cells were seeded on 96-well plates with a density of 5 × 103 cells per well overnight. Then, the cells were cocultured with F. nucleatum (MOI = 100; 5 × 105 CFUs per well), OXA (128 μM), and 2 mM HQF assemblies for 48 hours in culture medium containing 10% FBS. Cell viability was detected by MTT assay.
TEM images of cell sections
TEM images were obtained to determine the autophagosomes after different treatments by a general protocol. HCT116 cells were seeded on six-well plates with a density of 1 × 105 per well overnight and then treated with PBS, OXA (128 μM), OXA (128 μM) + F. nucleatum (MOI = 100; 5 × 107 CFU per well), and OXA (128 μM) + F. nucleatum (MOI = 100; 5 × 107 CFU per well) + HQF assemblies (2 mM). After incubation for 6 hours, the culture medium was removed and the cells were washed by PBS for three times. After fixation with 2.5% glutaraldehyde, the cell samples were fixed with 1% osmium tetroxide followed by dehydration, embedding, sectioning, and staining for TEM images. TEM images were captured on an HT7700 transmission electron microscope.
In vivo antitumor effect in subcutaneous tumor model
Animal experiments were performed according to the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals. All the animal experimental protocols were approved by the Institutional Animal Care and Use Committee of the National Center for Nanoscience and Technology (NCNST21-2107-0612). The in vivo effect was assessed in subcutaneous xenograft HCT116 CRC tumor model. Five- to 6-week-old male BALB/c nude mice were fed in laminar flow cabinets under a specific pathogen–free condition with randomly provided food and water. A total of 100 μl of solution containing 5 × 106 HCT116 cells mixed with Matrigel was injected into the right axilla of each mouse to establish subcutaneous CRC model. About a week after inoculation, the mice were divided into seven groups randomly for different sets of experiments. F. nucleatum (100 μl; 1 × 108 CFUs) was given by multipoint intratumoral injection every 3 days for about 2 weeks. For treatments, HQF (2 mM, 100 μl), NQF (2 mM, 100 μl), and PBS (100 μl) were subcutaneously injected into the tumor; OXA (7.5 mg/kg) was intraperitoneally injected; and MTZ (200 mg/kg) was given by oral administration.
The longest length (termed as a) and the shortest length (termed as b) of the tumor were measured every day (represented in millimeters). The tumor volume was calculated using the formula V = 0.5 × a × b2. After 2 weeks, all the mice were sacrificed. The tumors and organs were collected.
Statistical analysis
Data performed as indicated were presented as means ± SD, while error bars in all the bar graphs, line graphs, and box graphs represented SD. Shapiro-Wilk test was used to determine whether the data were normally distributed. The comparisons between two groups were evaluated by independent sample two-tailed t test. Three or more groups were evaluated by one-way analysis of variance (ANOVA) test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (32122045 and 51873046 to Y.G. and 51903064 to Q.Y.).
Author contributions: J.C. and Y.G. conceived and designed experiments. J.C., R.Z., J.Z., and X.W. synthesized and characterized materials. J.C. and Y.Z. performed in vitro cell experiments. P.Z. and J.C. performed in vitro microbiological experiments and in vivo animal experiments. J.C., P.Z., and Y.Z. collected and analyzed the data. Q.Y. and R.C. took part in discussions. Y.G. supervised the project. J.C., Q.Y., and Y.G. cowrote the manuscript. All authors discussed the results and revised the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S33
Table S1
References
REFERENCES AND NOTES
- 1.Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., Bray F., Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Dekker E., Tanis P. J., Vleugels J. L. A., Kasi P. M., Wallace M. B., Colorectal cancer. Lancet 394, 1467–1480 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Dagogo-Jack I., Shaw A. T., Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Janney A., Powrie F., Mann E. H., Host-microbiota maladaptation in colorectal cancer. Nature 585, 509–517 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Castellarin M., Warren R. L., Freeman J. D., Dreolini L., Krzywinski M., Strauss J., Barnes R., Watson P., Allen-Vercoe E., Moore R. A., Holt R. A., Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22, 299–306 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostic A. D., Chun E., Robertson L., Glickman J. N., Gallini C. A., Michaud M., Clancy T. E., Chung D. C., Lochhead P., Hold G. L., El-Omar E. M., Brenner D., Fuchs C. S., Meyerson M., Garrett W. S., Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 14, 207–215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mima K., Nishihara R., Qian Z. R., Cao Y., Sukawa Y., Nowak J. A., Yang J. H., Dou R. X., Masugi Y., Song M., Kostic A. D., Giannakis M., Bullman S., Milner D. A., Baba H., Giovannucci E. L., Garraway L. A., Freeman G. J., Dranoff G., Garrett W. S., Huttenhower C., Meyerson M., Meyerhardt J. A., Chan A. T., Fuchs C. S., Ogino S., Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 65, 1973–1980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullman S., Pedamallu C. S., Sicinska E., Claney T. E., Zhang X. Y., Cai D. N., Neuberg D., Huang K., Guevara F., Nelson T., Chipashvili O., Hagan T., Walker M., Ramachandran A., Diosdado B., Serna G., Mulet N., Landolfi S., Ramon y Cajal S., Fasani R., Aguirre A. J., Ng K., Elez E., Ogino S., Tabernero J., Fuchs C. S., Hahn W. C., Nuciforo P., Meyerson M., Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 358, 1443–1448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N., Chen Y., Chen H., Hong J., Zou W., Fang J.-Y., Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song W. T., Tiruthani K., Wang Y., Shen L. M., Hu M. Y., Dorosheva O., Qiu K. Y., Kinghorn K. A., Liu R. H., Huang L., Trapping of lipopolysaccharide to promote immunotherapy against colorectal cancer and attenuate liver metastasis. Adv. Mater. 30, e1805007 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song W., Anselmo A. C., Huang L., Nanotechnology intervention of the microbiome for cancer therapy. Nat. Nanotechnol. 14, 1093–1103 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Clemente J. C., Ursell L. K., Parfrey L. W., Knight R., The impact of the gut microbiota on human health: An integrative view. Cell 148, 1258–1270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng D. W., Dong X., Pan P., Chen K. W., Fan J. X., Cheng S. X., Zhang X. Z., Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 3, 717–728 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Salick D. A., Kretsinger J. K., Pochan D. J., Schneider J. P., Inherent antibacterial activity of a peptide-based beta-hairpin hydrogel. J. Am. Chem. Soc. 129, 14793–14799 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu B., Owh C., Chee P., Leow W., Liu X., Wu Y.-L., Guo P., Loh X. J., Chen X., Supramolecular hydrogels for antimicrobial therapy. Chem. Soc. Rev. 47, 6917–6929 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Schnaider L., Brahmachari S., Schmidt N. W., Mensa B., Shaham-Niv S., Bychenko D., Adler-Abramovich L., Shimon L. J. W., Kolusheva S., DeGrado W. F., Gazit E., Self-assembling dipeptide antibacterial nanostructures with membrane disrupting activity. Nat. Commun. 8, 1365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang Z., Liu Y., Wang L., Ali A., Yao Q., Jiang X., Gao Y., Supramolecular assemblies mimicking neutrophil extracellular traps for mrse infection control. Biomaterials 253, 120124 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Kinouchi T., Manabe Y., Wakisaka K., Ohnishi Y., Biotransformation of 1-nitropyrene in intestinal anaerobic bacteria. Microbiol. Immunol. 26, 993–1005 (1982). [DOI] [PubMed] [Google Scholar]
- 19.Yao Q. X., Huang Z. T., Liu D. D., Chen J. L., Gao Y., Enzyme-instructed supramolecular self-assembly with anticancer activity. Adv. Mater. 31, 1804814 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Chagri S., Ng D. Y. W., Weil T., Designing bioresponsive nanomaterials for intracellular self-assembly. Nat. Rev. Chem. 6, 320–338 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Z., Yao Q., Chen J., Gao Y., Redox supramolecular self-assemblies nonlinearly enhance fluorescence to identify cancer cells. Chem. Commun. 54, 5385–5388 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Kuang Y., Long M. J. C., Zhou J., Shi J. F., Gao Y., Xu C., Hedstrom L., Xu B., Prion-like nanofibrils of small molecules (PriSM) selectively inhibit cancer cells by impeding cytoskeleton dynamics. J. Biol. Chem. 289, 29208–29218 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno S. N., Mason R. P., Docampo R., Reduction of nifurtimox and nitrofurantoin to free radical metabolites by rat liver mitochondria. Evidence of an outer membrane-located nitroreductase. J. Biol. Chem. 259, 6298–6305 (1984). [PubMed] [Google Scholar]
- 24.Chevalier A., Zhang Y., Khdour O. M., Kaye J. B., Hecht S. M., Mitochondrial nitroreductase activity enables selective imaging and therapeutic targeting. J. Am. Chem. Soc. 138, 12009–12012 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Zhang M. M., Guan Y., Dang Z., Zhang P. G., Zheng Z., Chen L., Kuang W., Wang C. C., Liang G. L., Directly observing intracellular nanoparticle formation with nanocomputed tomography. Sci. Adv. 6, eaba3190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sinha V. R., Kumria R., Colonic drug delivery: Prodrug approach. Pharm. Res. 18, 557–564 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Zhao X. B., Ha W., Gao K., Shi Y. P., Precisely traceable drug delivery of azoreductase-responsive prodrug for colon targeting via multimodal imaging. Anal. Chem. 92, 9039–9047 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Li M., Zhang Y., Ren X., Niu W., Yuan Q., Cao K., Zhang J., Gao X., Su D., Activatable fluorogenic probe for accurate imaging of ulcerative colitis hypoxia in vivo. Chem. Commun. 58, 819–822 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Abed J., Emgard J. E. M., Zamir G., Faroja M., Almogy G., Grenov A., Sol A., Naor R., Pikarsky E., Atlan K. A., Mellul A., Chaushu S., Manson A. L., Earl A. M., Ou N., Brennan C. A., Garrett W. S., Bachrach G., Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed gal-galnac. Cell Host Microbe 20, 215–225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolenbrander P. E., Andersen R. N., Inhibition of coaggregation between fusobacterium nucleatum and porphyromonas (bacteroides) gingivalis by lactose and related sugars. Infect. Immun. 57, 3204–3209 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coppenhagen-Glazer S., Sol A., Abed J., Naor R., Zhang X., Han Y. W., Bachrach G., Fap2 of fusobacterium nucleatum is a galactose-inhibitable adhesin involved in coaggregation, cell adhesion, and preterm birth. Infect. Immun. 83, 1104–1113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saini N. K., Baena A., Ng T. W., Venkataswamy M. M., Kennedy S. C., Kunnath-Velayudhan S., Carreno L. J., Xu J., Chan J., Larsen M. H., Jacobs W. R. Jr., Porcelli S. A., Suppression of autophagy and antigen presentation by mycobacterium tuberculosis pe_pgrs47. Nat. Microbiol. 1, 16133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He H., Tan W., Guo J., Yi M., Shy A., Xu B., Enzymatic noncovalent synthesis. Chem. Rev. 120, 9994–10078 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z. M., Xu K. M., Guo Z. F., Guo Z. H., Xu B., Intracellular enzymatic formation of nanofibers results in hydrogelation and regulated cell death. Adv. Mater. 19, 3152–3156 (2007). [Google Scholar]
- 35.Gao Y., Shi J., Yuan D., Xu B., Imaging enzyme-triggered self-assembly of small molecules inside live cells. Nat. Commun. 3, 1033 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Béreau V., Jubéra V., Arnaud P., Kaiba A., Guionneau P., Sutter J.-P., Modulation of the luminescence quantum efficiency for blue luminophor {Al(salophen)}+ by ester-substituents. Dalton Trans. 39, 2070–2077 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Moeity K., Tanizawa K., Kanaoka Y., Schiff base copper(II) chelate as a tool for immobilization of protein. Chem. Pharm. Bull. 37, 2849–2851 (1989). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S33
Table S1
References