Abstract
Visual transformers have recently gained popularity in the computer vision community as they began to outrank convolutional neural networks (CNNs) in one representative visual benchmark after another. However, the competition between visual transformers and CNNs in medical imaging is rarely studied, leaving many important questions unanswered. As the first step, we benchmark how well existing transformer variants that use various (supervised and self-supervised) pre-training methods perform against CNNs on a variety of medical classification tasks. Furthermore, given the data-hungry nature of transformers and the annotation-deficiency challenge of medical imaging, we present a practical approach for bridging the domain gap between photographic and medical images by utilizing unlabeled large-scale in-domain data. Our extensive empirical evaluations reveal the following insights in medical imaging: (1) good initialization is more crucial for transformer-based models than for CNNs, (2) self-supervised learning based on masked image modeling captures more generalizable representations than supervised models, and (3) assembling a larger-scale domain-specific dataset can better bridge the domain gap between photographic and medical images via self-supervised continuous pre-training. We hope this benchmark study can direct future research on applying transformers to medical imaging analysis. All codes and pre-trained models are available on our GitHub page https://github.com/JLiangLab/BenchmarkTransformers.
Keywords: Vision Transformer, Transfer learning, Domain-adaptive pre-training, Benchmarking
1. Introduction
Visual transformers have recently demonstrated the potential to be considered as an alternative to CNNs in visual recognition. Though visual transformers have attained state-of-the-art (SOTA) performance across a variety of computer vision tasks [11,20], their architectures lack convolutional inductive bias, making them more data-hungry than CNNs [7,31]. Given the data-hungry nature of transformers and the challenge of annotation scarcity in medical imaging, the efficacy of existing visual transformers in medical imaging is unknown. Our preliminary analysis revealed that on medical target tasks with limited annotated data, transformers lag behind CNNs in random initialization (scratch) settings. To overcome the challenge of annotation dearth in medical imaging, transfer learning from ImageNet pre-trained models has become a common practice [9,10,18,35]. As such, the first question this paper seeks to answer is: To what extent can ImageNet pre-training elevate transformers’ performance to rival CNNs in medical imaging?
Meanwhile, self-supervised learning (SSL) has drawn great attention in medical imaging due to its remarkable success in overcoming the challenge of annotation dearth in medical imaging [8,30]. The goal of the SSL paradigm is to learn general-purpose representations without using human-annotated labels [10,16]. Masked image modeling (MIM) methods, in addition to supervised pre-training, have recently emerged as promising SSL techniques for transformer models; the basic idea behind MIM-based methods is to learn representations by (randomly) masking portions of the input image and then recovering the input image at the masked areas. Recent advancements in MIM-based techniques have resulted in SSL techniques that outperform supervised pre-trained models in a variety of computer vision tasks [5,21]. As a result, the second question this paper seeks to answer is: How generalizable are MIM-based self-supervised methods to medical imaging in comparison to supervised ImageNet pre-trained models?
Furthermore, the marked differences between photographic and medical images [8,16,30] may result in a mismatch in learned features between the two domains, which is referred to as a “domain gap.” Hosseinzadeh Taher et al. [16] recently demonstrated that using a CNN as the backbone, a moderately-sized medical image dataset is sufficient to bridge the domain gap between photographic and medical images via supervised continual pre-training. Motivated but different from this work and given the data-hungry nature of transformers, we investigate domain-adaptive pre-training in an SSL setting. Naturally, the third question this paper seeks to answer is: How to scale up a domain-specific dataset for a transformer architecture to bridge the domain gap between photographic and medical images?
In addressing the three questions, we conduct a benchmarking study to assess the efficacy of transformer-based models on numerous medical classification tasks involving different diseases (thorax diseases, lung pulmonary embolism, and tuberculosis) and modalities (X-ray and CT). In particular, (1) we investigate the importance of pre-training for transformers versus CNNs in medical imaging; (2) we assess the transferability of SOTA MIM-based self-supervised method to a diverse set of medical image classification tasks; and (3) we investigate domain-adaptive pre-training on large-scale photographic and medical images to tailor self-supervised ImageNet models for target tasks on chest X-rays.
Our extensive empirical study yields the following findings: (1) In medical imaging, good initialization is more vital for transformer-based models than for CNNs (see Fig. 1). (2) MIM-based self-supervised methods capture finer-grained representations that can be useful for medical tasks better than supervised pre-trained models (see Table 1). (3) Continuous self-supervised pre-training of the self-supervised ImageNet model on large-scale medical images bridges the domain gap between photographic and medical images, providing more generalizable pre-trained models for medical image classification tasks (see Table 2). We will contrast our study with related works in each subsection of Sect. 3 to show our novelties.
Fig. 1.
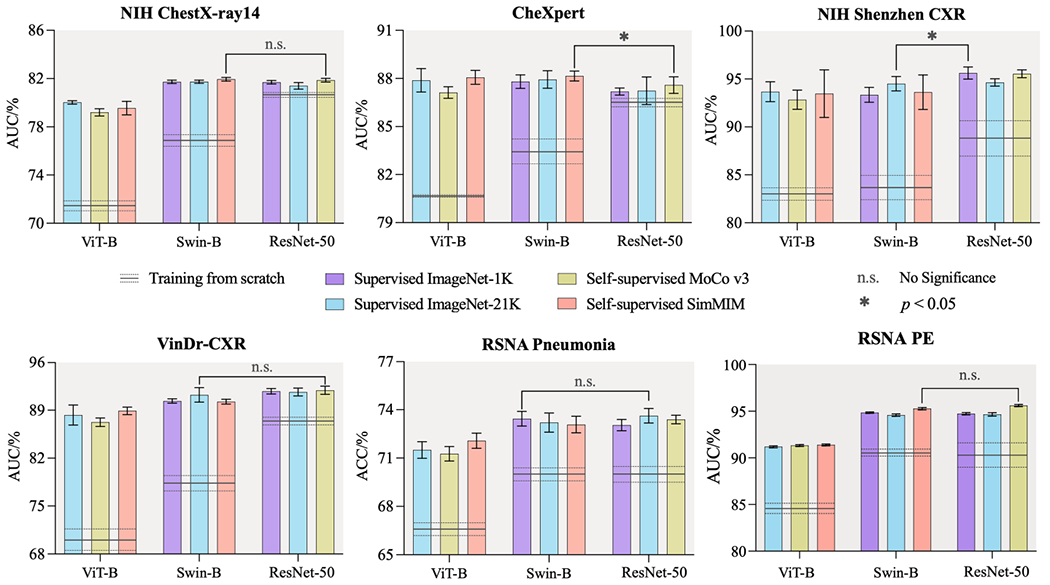
In medical imaging, good initialization is more vital for transformer-based models than for CNNs. When training from scratch, transformers perform significantly worse than CNNs on all target tasks. However, with supervised or self-supervised pre-training on ImageNet, transformers can offer the same results as CNNs, highlighting the importance of pre-training when using transformers for medical imaging tasks. We conduct statistical analysis between the best of six pre-trained transformer models and the best of three pre-trained CNN models.
Table 1.
Self-supervised SimMIM model with the Swin-B backbone outperforms fully-supervised baselines. The best methods are bolded while the second best are underlined. For every target task, we conduct statistical analysis between the best (bolded) vs. others. Green-highlighted boxes indicate no statistically significant difference at the p = 0.05 level.
| Initialization | Backbone | ChestX-ray14 | CheXpert | Shenzhen | VinDr-CXR | RSNA Pneumonia | RSNA PE |
|---|---|---|---|---|---|---|---|
| Scratch | ViT-B | 71.69±0.32 | 80.78±0.03 | 82.24±0.60 | 70.22±1.95 | 66.59±0.39 | 84.68±0.09 |
| Swin-B | 77.04±0.34 | 83.39±0.84 | 92.52±4.98 | 78.49±1.00 | 70.02±0.42 | 90.63±0.10 | |
|
| |||||||
| Supervised | ViT-B | 80.05±0.17 | 87.88±0.50 | 93.67±1.03 | 88.30±1.45 | 71.50±0.52 | 91.19±0.11 |
| Swin-B | 81.73±0.14 | 87.80±0.42 | 93.35±0.77 | 90.35±0.31 | 73.44±0.46 | 94.85±0.07 | |
|
| |||||||
| SimMIM | ViT-B | 79.55±0.56 | 88.07±0.43 | 93.47±2.48 | 88.91±0.55 | 72.08±0.47 | 91.39±0.10 |
| Swin-B | 81.95±0.15 | 88.16±0.31 | 94.12±0.96 | 90.24±0.35 | 73.66±0.34 | 95.27±0.12 | |
Table 2.
The domain-adapted pre-trained model which utilized a large number of in-domain data (X-rays(926K)) in an SSL manner achieves the best performance across all five target tasks. The best methods are bolded while the second best are underlined. For each target task, we conducted the independent two sample t-test between the best (bolded) vs. others. The absence of a statistically significant difference at the p = 0.05 level is indicated by green-highlighted boxes.
| Initialization | ChestX-ray14 | CheXpert | Shenzhen | VinDr-CXR | RSNA Pneumonia |
|---|---|---|---|---|---|
| Scratch | 77.04±0.34 | 83.39±0.84 | 83.92±1.19 | 78.49±1.00 | 70.02±0.42 |
|
| |||||
| ImageNet | 81.95±0.15 | 88.16±0.31 | 93.63±1.80 | 90.24±0.35 | 73.66±0.34 |
|
| |||||
| ChestX-ray14 | 78.87±0.69 | 86.75±0.96 | 93.03±0.48 | 79.86±1.82 | 71.99±0.55 |
| X-rays(926K) | 82.72±0.17 | 87.83±0.23 | 95.21±1.44 | 90.60±1.95 | 73.73±0.50 |
|
| |||||
| ImageNet→ChestX-ray14 | 82.45±0.15 | 87.74±0.31 | 94.83±0.90 | 90.33±0.88 | 73.87±0.48 |
| ImageNet→X-rays(926K) | 83.04±0.15 | 88.37±0.40 | 95.76±1.79 | 91.71±1.04 | 74.09±0.39 |
2. Benchmarking Setup
2.1. Transformer Backbones
In the target tasks in all experiments, we take two representative recent SOTA transformer backbones, including Vision Transformer (ViT) [7] and Swin Transformer (Swin) [22]. Visual transformer models, which have recently emerged as alternatives to convolutional neural networks (CNNs), have revolutionized computer vision fields. The groundbreaking work of ViT showcases how transformers can completely replace the CNNs backbone with a convolution-free model. Although ViT attains SOTA image classification performance, its architecture may not be suitable for use on dense vision tasks, such as object detection, segmentation, etc. Swin, a recent work, proposes a general-purpose transformer backbone to address this problem by building hierarchical feature maps, resulting in SOTA accuracy on object detection segmentation tasks. For transfer learning to the classification target tasks, we take the transformer pre-trained models and add a task-specific classification head. We assess the transfer learning performance of all pre-trained models by fine-tuning all layers in the downstream networks.
2.2. Target Tasks and Datasets
We consider a diverse suite of six common but challenging medical classification tasks including NIH ChestX-ray14 [32], CheXpert [17], VinDr-CXR [24], NIH Shenzhen CXR [19], RSNA PE Detection [6], and RSNA Pneumonia [1]. These tasks encompass various diseases (thorax diseases, lung pulmonary embolism, and tuberculosis) and modalities (X-ray and CT). We use official data split of these datasets if available; otherwise, we randomly divide the data into 80%/20% for training/testing. AUC (area under the ROC curve) is used to measure the performance of multi-label classification target tasks (NIH ChestX-ray14, CheX-pert, and VinDr-CXR) and binary classification target tasks (NIH Shenzhen CXR and RSNA PE). Accuracy is used to evaluate multi-class classification target task (RSNA Pneumonia) performance. The mean and standard deviation of performance metrics over ten runs are reported in all experiments, and statistical analyses based on an independent two sample t-test are presented.
3. Benchmarking and Boosting Transformers
3.1. Pre-training is More Vital for Transformer-Based Models than for CNNs in Medical Imaging
Transformers have recently attained SOTA results and surpassed CNNs in a variety of computer vision tasks [11,20]. However, the lack of convolutional inductive bias in transformer architectures makes them more data-hungry than CNNs [7,31]. Therefore, to rival CNNs in vision tasks, transformers require a millions or even billions of labeled data [7,28,34]. Given the data-hungry nature of transformers and the challenge of annotation scarcity in medical imaging [10,25,27,35], it is natural to wonder whether transformers can compute with CNNs if they are used directly on medical imaging applications. Our preliminary analysis showed that in random initialization (scratch) settings, transformers lag behind CNNs on medical target tasks with limited annotated data. Taken together, we hypothesize that in medical imaging, transformers require pre-trained models to rival with CNNs. To put this hypothesis to the test, we empirically validate how well transformer variants (ViT-B and Swin-B) that use various (supervised and self-supervised) pre-training methods compete with CNNs on a range of medical classification tasks. In contrast to previous work [23] which only compared one transformer model with a CNN counterpart, we benchmark six newly-developed transformer models and three CNN models.
Experimental Setup.
We evaluate the transferability of various popular transformer methods with officially released models on six diverse medical classification tasks. Our goal is to investigate the importance of pre-training for transformers versus CNNs in medical imaging. Given this goal, we use six popular transformer pre-trained models with ViT-B and Swin-B backbones and three standard CNNs pre-trained models with ResNet-50 backbones [15] that are already official and ready to use. Specifically, for supervised pre-training, we use official pre-trained ViT-B, Swin-B, and ResNet-50 on ImageNet-21K and pre-trained Swin-B and ResNet-50 on ImageNet-1K. For self-supervised pretraining, we use pre-trained ViT-B and Swin-B models with SimMIM [33] on ImageNet-1K, as well as pre-trained ViT-B and ResNet-50 models with MoCo v3 [4] on ImageNet-1K. The differences in pre-training data (ImageNet-1K or ImageNet-21K) are due to the availability of official pre-trained models.
Results and Analysis.
Our evaluations in Fig. 1 suggest three major results. Firstly, in random initialization (scratch) settings (horizontal lines), transformers (i.e., ViT-B and/or Swin-B) cannot compete with CNNs (i.e., ResNet50) in medical applications, as they offer performance equally or even worse than CNNs. We attribute this inferior performance to transformers’ lack of desirable inductive bias in comparison to CNNs, which has a negative impact on transformer performance on medical target tasks with limited annotated data. Secondly, Swin-B backbone consistently outperforms ViT-B across all target tasks. This reveals the importance of hierarchical inductive bias, which embedded in the Swin-B backbone, in elevating the performance of transformer-based models in medical image analysis. Thirdly, with supervised or self-supervised pre-training on ImageNet, transformers can offer competitive performance compare to CNNs, emphasizing the importance of pre-training when using transformers for medical imaging tasks. In particular, the best of six pre-trained transformer models outperform the best of three pre-trained CNN models in all target tasks, with the exception of NIH Shenzhen CXR, which can be attributed to a lack of sufficient training data (only 463 samples).
3.2. Self-supervised Learning Based on Masked Image Modeling is a Preferable Option to Supervised Baselines for Medical Imaging
Visual transformer models, while powerful, are prone to over-fitting and rely heavily on supervised pre-training on large-scale image datasets [7,34], such as JFT-300M [29] and ImageNet-21K [26]. In addition to supervised pre-training, self-supervised learning (SSL) techniques account for a substantial part of pre-trained transformer models. Masked Image Modeling (MIM) - an approach in which portions of the input image signals are randomly masked and then the original input signals are recovered at the masked area - has recently received great attention in computer vision for pre-training transformers in a self-supervised manner [14,33]. MIM-based self-supervised methods are widely accepted to capture more task-agnostic features than supervised pre-trained models, making them better suited for fine-tuning on various vision tasks [5,21]. We hypothesize that existing self-supervised transformer models pre-trained on photographic images will outperform supervised transformer models in the medical image domain, where there is a significant domain shift between medical and photographic images. To test this hypothesis, we consider two recent SOTA transformer backbones, ViT-B and Swin-B, and compare their supervised and self-supervised pre-trained models for various medical image classification tasks.
Experimental Setup.
To investigate the efficacy of self-supervised and supervised pre-trained transformer models in medical imaging, we use existing supervised and SOTA self-supervised (i.e., SimMIM) pre-trained models with two representative transformer backbones, ViT-B and Swin-B; all pre-trained models are fine-tuned on six different medical classification tasks. To provide a comprehensive evaluation, we also include results for the training of these two architectures from scratch. We use SimMIM instead of the concurrent MAE [14] as the representative MIM-based method because SimMIM has been demonstrated superior performance to MAE in medical image analysis [5].
Results and Analysis.
As shown in Table 1, the self-supervised SimMIM model with the Swin-B backbone performs significantly better or on-par compared with both supervised baselines with either ViT-B or Swin-B backbones across all target tasks. The same observation of MIM-based models outperforming their supervised counterparts also exists in the finer-grained visual tasks, i.e., object detection [21] and medical image segmentation [5]; different from them, we focus on coarse-grained classification tasks. Furthermore, we observe that the SimMIM model with the Swin-B backbone consistently outperforms its counterpart with the ViT-B backbone in all cases, implying that the Swin-B backbone may be a superior option for medical imaging tasks to ViT-B. These findings suggest that the self-supervised SimMIM model with the Swin-B backbone could be a viable option for pre-training deep models in medical imaging applications.
3.3. Self-supervised Domain-Adaptive Pre-training on a Larger-Scale Domain-Specific Dataset Better Bridges the Domain Gap Between Photographic and Medical Imaging
Domain adaptation seeks to improve target model performance by reducing domain disparities between source and target domains. Recently, Hosseinzadeh Taher et al. [16] demonstrated that domain-adaptive pre-training can bridge the domain gap between natural and medical images. Particularly, Hosseinzadeh Taher et al. [16] first pre-trained a CNN model (i.e., ResNet-50) on ImageNet and then on domain-specific datasets (i.e., NIH ChestX-ray14 or CheXpert), demonstrating how domain-adaptive pre-training can tailor the ImageNet models to medical applications. Motivated by this work, we investigate domain-adaptive pre-training in the context of visual transformer architectures. Given the data-hungry nature of transformers and the annotation-dearth challenge of medical imaging, different from [16], we use the SSL pre-training approach to bridge the domain gap between photographic and medical images. Since no expert annotation is required in SSL pre-training, we are able to assemble multiple domain-specific datasets into a large-scale dataset, which is differentiated from Azizi et al. [2] who used only a single dataset.
Experimental Setup.
We evaluate the transferability of five different self-supervised SimMIM models with the Swin-B backbone by utilizing three different pre-training datasets, including ImageNet, ChestX-ray14, and X-rays(926K)— a large-scale dataset that we created by collecting 926,028 images from 13 different chest X-ray datasets. To do so, we use SimMIM released ImageNet model as well as two models pre-trained on ChestX-ray14 and X-rays(926K) using SimMIM; additionally, we created two new models that were initialized through the self-supervised ImageNet pre-trained model followed by self-supervised pre-training on ChestX-ray14 (ImageNet→ChestX-ray14) and X-rays(926 K) (ImageNet→X-rays(926 K)). Every pre-training experiment trains for 100 epochs using the default SimMIM settings.
Results and Analysis.
We draw the following observations from Table 2. (1) X-rays(926K) model consistently outperforms the ChestX-ray14 model in all cases. This observation suggests that scaling the pre-training data can significantly improve the self-supervised transformer models. (2) While the X-rays(926 K) model uses fewer images in the pre-training dataset than the ImageNet model, it shows superior or comparable performance over the ImageNet model across all target tasks. In line with Hosseinzadeh Taher et al. [16], this implies that, whenever possible, in-domain medical transfer learning should be preferred over ImageNet transfer learning. (3) The overall trend highlights the benefit of domain-adaptive pre-training, which leverages the ImageNet model’s learning experience and further refines it with domain-relevant data. Specifically, fine-tuning both domain-adapted models (ImageNet→ChestX-ray14 and ImageNet→X-rays(926 K)) outperforms ImageNet and corresponding in-domain models in all target tasks, with one exception; in the CheXpert, the ImageNet→ChestX-ray14 model performs worse in CheXpert than the corresponding ImageNet model. This exception, in line with Hosseinzadeh Taher et al. [16], suggests that the in-domain pre-training dataset should be larger than the target dataset. It is noteworthy that this gap was filled later by ImageNet→X-rays(926 K) model, which utilized more in-domain data. This highlights the significance of larger-scale medical data in improving the transformers’ ability to learn more discriminative representations.
4. Conclusion and Future Work
We manifest an up-to-date benchmark study to shed light on the efficacy and limitations of existing visual transformer models in medical image classification when compared to CNN counterparts. Our extensive experiments yield important findings: (1) a good pre-train model can allow visual transformers to compete with CNNs on medical target tasks with limited annotated data; (2) MIM-based self-supervised methods, such as SimMIM, play an important role in pre-training visual transformer models, preventing them from over-fitting in medical target tasks; and (3) assembling multiple domain-specific datasets into a larger-scale one can better bridge the domain gap between photographic and medical imaging via continual SSL pre-training.
Future Work:
Recently, many transformer-based UNet architectures have been developed for 3D medical image segmentation [3,5,12,13]. To make a comprehensive benchmarking study of transformers for medical image analysis, we will extend the evaluation to more modalities and medical image segmentation tasks in future work.
Acknowledgments.
This research has been supported in part by ASU and Mayo Clinic through a Seed Grant and an Innovation Grant, and in part by the NIH under Award Number R01HL128785. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work has utilized the GPUs provided in part by the ASU Research Computing and in part by the Extreme Science and Engineering Discovery Environment (XSEDE) funded by the National Science Foundation (NSF) under grant numbers: ACI-1548562, ACI-1928147, and ACI-2005632. We thank Manas Chetan Valia and Haozhe Luo for evaluating the pre-trained ResNet50 models on the five chest X-ray tasks and the pre-trained transformer models on the VinDr-CXR target tasks, respectively. The content of this paper is covered by patents pending.
Appendix
A. Tabular results
In this section, tabulated results of different experiments are reported. All benchmarking results are presented in Table 1.
Table 1.
Benchmarking transfer learning with transformers (i.e., ViT-B and Swin-B) ImageNet pre-trained models on six medical image classification tasks. For comparison with CNNs, we evaluate four pre-trained models with ResNet-50 backbones. To provide a comprehensive evaluation, we also include results for the training of these three architectures from scratch.
| Backbone | Initialization | ChestX-ray14 | CheXpert | Shenzhen | VinDr-CXR | RSNA Pneumonia | RSNA PE |
|---|---|---|---|---|---|---|---|
| ResNet-50 | Scratch | 80.400.05 | 86.620.15 | 89.031.82 | 87.390.42 | 70.000.50 | 90.371.32 |
| Supervised[20] | 81.700.15 | 87.170.22 | 95.620.63 | 91.770.40 | 73.040.35 | 94.730.12 | |
| Sup.(IN21K)[21] | 81.400.27 | 87.230.86 | 94.640.39 | 91.660.56 | 73.630.45 | 94.660.18 | |
| DINO[6] | 81.410.35 | 87.370.45 | 96.380.48 | 90.960.68 | 73.580.35 | 95.600.10 | |
| MoCo-v3[8] | 81.870.15 | 87.590.51 | 95.550.40 | 91.910.59 | 73.390.27 | 95.610.12 | |
| ViT-B | Scratch | 71.690.32 | 80.780.03 | 82.240.60 | 70.221.95 | 66.590.39 | 84.680.09 |
| Sup. (in21k)[10] | 80.050.17 | 87.880.50 | 93.671.03 | 88.301.45 | 71.500.52 | 91.190.11 | |
| DeiT[23] | 79.460.24 | 87.490.43 | 95.350.80 | 89.642.97 | 72.930.62 | 91.950.07 | |
| DINO[6] | 78.370.47 | 87.010.62 | 90.394.29 | 82.891.10 | 71.270.45 | 88.990.08 | |
| MoCo-v3[8] | 79.200.30 | 87.120.36 | 92.851.00 | 87.250.63 | 72.790.52 | 91.330.10 | |
| BEiT(in21k)[3] | 79.910.24 | 87.770.38 | 92.871.08 | 85.931.98 | 72.780.37 | 91.310.10 | |
| MAE[12] | 79.010.58 | 87.120.54 | 92.524.98 | 87.001.74 | 72.850.50 | 91.960.12 | |
| SimMIM[27] | 79.550.56 | 88.070.43 | 93.472.48 | 88.910.55 | 72.080.47 | 91.390.10 | |
| Swin-B | Scratch | 77.040.34 | 83.390.84 | 92.524.98 | 78.491.00 | 70.020.42 | 90.630.10 |
| Supervised[18] | 81.730.14 | 87.800.42 | 93.350.77 | 90.350.31 | 73.440.46 | 94.850.07 | |
| Sup. (in21k)[18] | 81.740.13 | 87.940.54 | 94.211.25 | 91.231.06 | 73.200.59 | 94.580.13 | |
| SimMIM[27] | 81.950.15 | 88.160.31 | 94.120.96 | 90.240.35 | 73.660.34 | 95.270.12 | |
Abbreviations: Sup.: Supervised; IN21K: ImageNet-21K.
Unless mentioned, all models are pre-trained on ImageNet-1K.
B. The large-scale domain-specific dataset
Given the data-hungry nature of transformers and the annotation-deficiency challenge of medical imaging, we present a practical approach for bridging the domain gap between photographic and medical images by utilizing unlabeled large-scale in-domain data. We assembled 926,028 images from 13 chest X-ray datasets into a large-scale dataset. The datasets we used are listed in Table 2.
C. State-of-the-art performance
Our domain-adapted model pre-trained using the large-scale domain-specific dataset that we assembled, i.e., X-ray(926K), achieves SOTA performance when transfered to NIH ChestX-ray14 dataset. The result of our best model is present in Table 3 with the results of other SOTA methods.
Table 2.
We created a large-scale dataset, namely X-ray(926K), by collecting 926,028 images from 13 different chest X-ray datasets. For the datasets that have official training/testing data split, we only used the training set.
| Source Datasets | Number of Images |
|---|---|
| NIH ChestX-ray14 [26] | 86,524 |
| CheXpert [13] | 223,414 |
| VinDr-CXR [19] | 15,000 |
| NIH Shenzhen CXR [14] | 662 |
| RSNA Pneumonia Detection Challenge [1] | 26,684 |
| MIMIC-CXR database v2.0.0 [15] | 377,028 |
| PadChest [4] | 160,828 |
| COVID-19 Radiography Database [9] | 21,165 |
| Indiana ChestX-ray [2] | 7,883 |
| Mendeley-V2 [16] | 5,232 |
| COVIDx [25] | 1,223 |
| JSRT [22] | 243 |
| Montgomery County X-ray Set [5] | 138 |
| X-ray(926K) | 926,028 |
Table 3.
Continual pre-training on a large-scale domain-specific dataset (ImageNet→X-ray(926K)) via self-supervised SimMIM method achieves SOTA performance on the NIH ChestX-ray14 target task.
| Method | Backbone | Mean | Atel | Card | Effu | Infi | Mass | Nodu | Pne1 | Pne2 | Cons | Edem | Emph | Fibr | P.T. | Hern |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CXRDANet[7] | DN-121 | 81.9 | 75.9 | 89.8 | 82.7 | 71.0 | 83.2 | 75.3 | 73.6 | 87.6 | 75.1 | 86.4 | 89.0 | 83.6 | 78.9 | 94.0 |
| A3Net[24] | DN-121 | 82.6 | 77.9 | 89.5 | 83.6 | 71.0 | 83.4 | 77.7 | 73.7 | 87.8 | 75.9 | 85.5 | 93.3 | 83.8 | 79.1 | 93.8 |
| XProtoNet[17] | DN-121 | 82.2 | 78.0 | 88.7 | 83.5 | 71.0 | 83.1 | 80.4 | 73.4 | 87.1 | 74.7 | 84.0 | 94.1 | 81.5 | 79.9 | 90.9 |
| TransVW[11] | DN-121 | 82.5 | 78.7 | 88.1 | 84.1 | 69.9 | 83.1 | 79.2 | 73.1 | 87.8 | 75.4 | 85.6 | 93.5 | 84.4 | 80.2 | 92.2 |
| Ours | Swin-B | 83.2 | 79.3 | 90.7 | 84.6 | 71.5 | 85.3 | 76.6 | 74.5 | 88.5 | 76.8 | 86.3 | 92.9 | 84.2 | 80.4 | 93.5 |
Abbreviation of each pathology is as follow: Atel: Atelectasis; Card: Cardiomegaly; Effu: Effusion; Infi: Infiltration; Nodu: Nodule; Pne1: Pneumonia; Pne2: Pneumothorax; Cons: Consolidation; Edem: Edema; Emph: Emphysema; Fibr: Fibrosis; P.T.: Pleural Thickening; Hern: Hernia.
References
- 1.Rsna pneumonia detection challenge (2018), https://www.kaggle.com/c/rsna-pneumonia-detection-challenge
- 2.Chest x-rays (indiana university) (2020), https://www.kaggle.com/datasets/raddar/chest-xrays-indiana-university
- 3.Bao H, Dong L, Wei F: Beit: Bert pre-training of image transformers. arXiv preprint arXiv:2106.08254 (2021) [Google Scholar]
- 4.Bustos A, Pertusa A, Salinas JM, de la Iglesia-Vayá M: Padchest: A large chest x-ray image dataset with multi-label annotated reports. Medical image analysis 66, 101797 (2020) [DOI] [PubMed] [Google Scholar]
- 5.Candemir S, Jaeger S, Palaniappan K, Musco JP, Singh RK, Xue Z, Karargyris A, Antani S, Thoma G, McDonald CJ: Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE transactions on medical imaging 33(2), 577–590 (2013) [DOI] [PubMed] [Google Scholar]
- 6.Caron M, Touvron H, Misra I, Jégou H, Mairal J, Bojanowski P, Joulin A: Emerging properties in self-supervised vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 9650–9660 (2021) [Google Scholar]
- 7.Chen L, Mao T, Zhang Q: Identifying cardiomegaly in chest x-rays using dual attention network. Applied Intelligence pp. 1–10 (2022) [Google Scholar]
- 8.Chen X, Xie S, He K: An empirical study of training self-supervised vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 9640–9649 (2021) [Google Scholar]
- 9.Chowdhury ME, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Al Emadi N, et al. Can ai help in screening viral and covid-19 pneumonia? IEEE Access 8, 132665–132676 (2020) [Google Scholar]
- 10.Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, et al. An image is worth 16×16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020) [Google Scholar]
- 11.Haghighi F, Taher MRH, Zhou Z, Gotway MB, Liang J: Transferable visual words: Exploiting the semantics of anatomical patterns for self-supervised learning. IEEE Transactions on Medical Imaging 40(10), 2857–2868 (2021). 10.1109/TMI.2021.3060634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He K, Chen X, Xie S, Li Y, Dollár P, Girshick R: Masked autoencoders are scalable vision learners. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. pp. 16000–16009 (2022) [Google Scholar]
- 13.Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of the AAAI conference on artificial intelligence. vol. 33, pp. 590–597 (2019) [Google Scholar]
- 14.Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G: Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quantitative imaging in medicine and surgery 4(6), 475 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson AE, Pollard TJ, Berkowitz SJ, Greenbaum NR, Lungren MP, Deng C.y., Mark RG, Horng S: Mimic-cxr, a de-identified publicly available database of chest radiographs with free-text reports. Scientific data 6(1), 1–8 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kermany D, Zhang K, Goldbaum M, et al. Labeled optical coherence tomography (oct) and chest x-ray images for classification. Mendeley data 2(2) (2018) [Google Scholar]
- 17.Kim E, Kim S, Seo M, Yoon S: Xprotonet: diagnosis in chest radiography with global and local explanations. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 15719–15728 (2021) [Google Scholar]
- 18.Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B: Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision. pp. 10012–10022 (2021) [Google Scholar]
- 19.Nguyen HQ, Lam K, Le LT, Pham HH, Tran DQ, Nguyen DB, Le DD, Pham CM, Tong HT, Dinh DH, et al. Vindr-cxr: An open dataset of chest x-rays with radiologist’s annotations. arXiv preprint arXiv:2012.15029 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L, Desmaison A, Kopf A, Yang E, DeVito Z, Raison M, Tejani A, Chilamkurthy S, Steiner B, Fang L, Bai J , Chintala S: Pytorch: An imperative style, high-performance deep learning library. In: Wallach H, Larochelle H, Beygelzimer A, dAlché-Buc F, Fox E, Garnett R (eds.) Advances in Neural Information Processing Systems 32, pp. 8024–8035. Curran Associates, Inc. (2019), http://papers.neurips.cc/paper/9015-pytorch-an-imperative-style-high-performance-deep-learning-library.pdf [Google Scholar]
- 21.Ridnik T, Ben-Baruch E, Noy A, Zelnik-Manor L: Imagenet-21k pretraining for the masses. arXiv preprint arXiv:2104.10972 (2021) [Google Scholar]
- 22.Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu K.i., Matsui M, Fujita H, Kodera Y, Doi K: Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. American Journal of Roentgenology 174(1), 71–74 (2000) [DOI] [PubMed] [Google Scholar]
- 23.Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H: Training data-efficient image transformers & distillation through attention. In: International Conference on Machine Learning. pp. 10347–10357. PMLR; (2021) [Google Scholar]
- 24.Wang H, Wang S, Qin Z, Zhang Y, Li R, Xia Y: Triple attention learning for classification of 14 thoracic diseases using chest radiography. Medical Image Analysis 67, 101846 (2021) [DOI] [PubMed] [Google Scholar]
- 25.Wang L, Lin ZQ, Wong A: Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Scientific Reports 10(1), 1–12 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM: Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition. pp. 2097–2106 (2017) [Google Scholar]
- 27.Xie Z, Zhang Z, Cao Y, Lin Y, Bao J, Yao Z, Dai Q, Hu H: Simmim: A simple framework for masked image modeling. arXiv preprint arXiv:2111.09886 (2021) [Google Scholar]
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/978-3-031-16852-9_2.
References
- 1.Rsna pneumonia detection challenge (2018). https://www.kaggle.com/c/rsna-pneumonia-detection-challenge
- 2.Azizi S, et al. Big self-supervised models advance medical image classification. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 3478–3488 (2021) [Google Scholar]
- 3.Cao H, et al. Swin-unet: Unet-like pure transformer for medical image segmentation. arXiv preprint arXiv:2105.05537 (2021) [Google Scholar]
- 4.Chen X, Xie S, He K: An empirical study of training self-supervised vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 9640–9649 (2021) [Google Scholar]
- 5.Chen Z, et al. Masked image modeling advances 3D medical image analysis. arXiv preprint arXiv:2204.11716 (2022) [Google Scholar]
- 6.Colak E, et al. The RSNA pulmonary embolism CT dataset. Radiol. Artif. Intell 3(2) (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dosovitskiy A, et al. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020) [Google Scholar]
- 8.Haghighi F, Hosseinzadeh Taher MR, Gotway MB, Liang J: DiRA: Discriminative, restorative, and adversarial learning for self-supervised medical image analysis. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 20824–20834 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haghighi F, Hosseinzadeh Taher MR, Zhou Z, Gotway MB, Liang J: Learning semantics-enriched representation via self-discovery, self-classification, and self-restoration. In: Martel AL (ed.) MICCAI 2020. LNCS, vol. 12261, pp. 137–147. Springer, Cham: (2020). 10.1007/978-3-030-59710-8_14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haghighi F, Taher MRH, Zhou Z, Gotway MB, Liang J: Transferable visual words: exploiting the semantics of anatomical patterns for self-supervised learning. IEEE Trans. Med. Imaging 40(10), 2857–2868 (2021). 10.1109/TMI.2021.3060634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han K, et al. A survey on vision transformer. IEEE Trans. Patt. Anal. Mach. Intell (2022) [DOI] [PubMed] [Google Scholar]
- 12.Hatamizadeh A, Nath V, Tang Y, Yang D, Roth H, Xu D: Swin UNETR: Swin transformers for semantic segmentation of brain tumors in MRI images. arXiv preprint arXiv:2201.01266 (2022) [Google Scholar]
- 13.Hatamizadeh A, et al. UNETR: Transformers for 3D medical image segmentation. In: Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, pp. 574–584 (2022) [Google Scholar]
- 14.He K, Chen X, Xie S, Li Y, Dollár P, Girshick R: Masked autoencoders are scalable vision learners. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 16000–16009 (2022) [Google Scholar]
- 15.He K, Zhang X, Ren S, Sun J: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016) [Google Scholar]
- 16.Hosseinzadeh Taher MR, Haghighi F, Feng R, Gotway MB, Liang J: A systematic benchmarking analysis of transfer learning for medical image analysis. In: Albarqouni S (ed.) DART/FAIR -2021. LNCS, vol. 12968, pp. 3–13. Springer, Cham: (2021). 10.1007/978-3-030-87722-4_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irvin J, et al. Chexpert: A large chest radiograph dataset with uncertainty labels and expert comparison. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 33, pp. 590–597 (2019) [Google Scholar]
- 18.Islam NU, Gehlot S, Zhou Z, Gotway MB, Liang J: Seeking an optimal approach for computer-aided pulmonary embolism detection. In: Lian C, Cao X, Rekik I, Xu X, Yan P (eds.) MLMI 2021. LNCS, vol. 12966, pp. 692–702. Springer, Cham: (2021). 10.1007/978-3-030-87589-3_71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaeger S, Candemir S, Antani S, Wáng YXJ, Lu PX, Thoma G: Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quant. Imaging Med. Surg 4(6), 475 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan S, Naseer M, Hayat M, Zamir SW, Khan FS, Shah M: Transformers in vision: A survey. ACM Computing Surveys (CSUR) (2021) [Google Scholar]
- 21.Li Y, Xie S, Chen X, Dollar P, He K, Girshick R: Benchmarking detection transfer learning with vision transformers. arXiv preprint arXiv:2111.11429 (2021) [Google Scholar]
- 22.Liu Z, et al. Swin transformer: Hierarchical vision transformer using shifted windows. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 10012–10022 (2021) [Google Scholar]
- 23.Matsoukas C, Haslum JF, Söderberg M, Smith K: Is it time to replace CNNs with transformers for medical images? arXiv preprint arXiv:2108.09038 (2021) [Google Scholar]
- 24.Nguyen HQ, et al. VinDR-CXR: An open dataset of chest x-rays with radiologist’s annotations. arXiv preprint arXiv:2012.15029 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parvaiz A, Khalid MA, Zafar R, Ameer H, Ali M, Fraz MM: Vision transformers in medical computer vision-a contemplative retrospection. arXiv preprint arXiv:2203.15269 (2022) [Google Scholar]
- 26.Russakovsky O, et al. ImageNet large scale visual recognition challenge. Int. J. Comput. Vision 115(3), 211–252 (2015) [Google Scholar]
- 27.Shamshad F, et al. Transformers in medical imaging: A survey. arXiv preprint arXiv:2201.09873 (2022) [Google Scholar]
- 28.Steiner A, Kolesnikov A, Zhai X, Wightman R, Uszkoreit J, Beyer L: How to train your viT? data, augmentation, and regularization in vision transformers. arXiv preprint arXiv:2106.10270 (2021) [Google Scholar]
- 29.Sun C, Shrivastava A, Singh S, Gupta A: Revisiting unreasonable effectiveness of data in deep learning era. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 843–852 (2017) [Google Scholar]
- 30.Taher MRH, Haghighi F, Gotway MB, Liang J: CAid: Context-aware instance discrimination for self-supervised learning in medical imaging. arXiv:2204.07344 (2022). 10.48550/ARXIV.2204.07344, https://arxiv.org/abs/2204.07344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touvron H, Cord M, Douze M, Massa F, Sablayrolles A, Jégou H: Training data-efficient image transformers & distillation through attention. In: International Conference on Machine Learning, pp. 10347–10357, PMLR; (2021) [Google Scholar]
- 32.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM: Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. pp. 2097–2106 (2017) [Google Scholar]
- 33.Xie Z, et al. SimMIM: A simple framework for masked image modeling. arXiv preprint arXiv:2111.09886 (2021) [Google Scholar]
- 34.Zhai X, Kolesnikov A, Houlsby N, Beyer L: Scaling Vision Transformers. arXiv preprint arXiv:2106.04560 (2021) [Google Scholar]
- 35.Zhou Z, Sodha V, Pang J, Gotway MB, Liang J: Models genesis. Med. Image Anal 67, 101840 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]


