Abstract
To evaluate the role of SigB in modulating the expression of virulence determinants in Staphylococcus aureus, we constructed a sigB mutant of RN6390, a prototypic S. aureus strain. The mutation in the sigB gene was confirmed by the absence of the SigB protein in the mutant on an immunoblot as well as the failure of the mutant to activate ςB-dependent promoters (e.g., the sarC promoter) of S. aureus. Phenotypic analysis indicated that both alpha-hemolysin level and fibrinogen-binding capacity were up-regulated in the mutant strain compared with the parental strain. The increase in fibrinogen-binding capacity correlated with enhanced expression of clumping factor and coagulase on immunoblots. The effect of the sigB mutation on the enhanced expression of the alpha-hemolysin gene (hla) was primarily transcriptional. Upon complementation with a plasmid containing the sigB gene, hla expression returned to near parental levels in the mutant. Detailed immunoblot analysis as well as a competitive enzyme-linked immunosorbent assay of the cell extract of the sigB mutant with anti-SarA monoclonal antibody 1D1 revealed that the expression of SarA was higher in the mutant than in the parental control. Despite an elevated SarA level, the transcription of RNAII and RNAIII of the agr locus remained unaltered in the sigB mutant. Because of a lack of perturbation in agr, we hypothesize that inactivation of sigB leads to increased expression of SarA which, in turn, modulates target genes via an agr-independent but SarA-dependent pathway.
Staphylococcus aureus is a major cause of human infections, such as superficial abscesses, pneumonia, endocarditis, and sepsis (6). The control of a multitude of extracellular and cell wall virulence determinants in S. aureus is growth phase dependent. In particular, cell wall proteins are normally synthesized in the logarithmic phase, while exoproteins are generally produced postexponentially. The growth phase dependence of these virulence factors is mediated in part by global regulatory loci, such as sar (12) and agr (22). These modulators may either interact with the target gene directly (e.g., RNAIII with hla [alpha-hemolysin gene] mRNA) or control another regulatory molecule (e.g., sar regulation of the agr gene product) which, in turn, alters the transcription of the target gene.
The sar locus is composed of three overlapping transcripts, designated sarA (0.56 kb), sarC (0.8 kb), and sarB (1.2 kb), initiated from the P1, P3, and P2 promoters, respectively. Because of this multiplicity of promoters, the activation of sar leading to the expression of SarA, the major sar regulatory molecule, is complex and may be growth phase dependent. Whereas the sarB transcript and the more abundant sarA transcripts are maximally expressed during the exponential phase, the transcription of sarC from the P3 promoter is most active during the postexponential phase (3). Additional transcriptional analysis indicated that the P3 promoter is ςB dependent (17, 20, 25).
In contrast to the primary sigma factor (ςA), which is required for the expression of housekeeping genes, SigB (ςB) is an alternate transcription factor that has been shown to respond to environmental stresses (e.g., stationary phase of growth) in gram-positive bacteria (20). The core RNA polymerase associated with a particular sigma factor recognizes a specific set of promoters with conserved sequence motifs to initiate the transcription of genes programmed to respond to certain environments (20, 22). For Bacillus subtilis, ςB activity is regulated posttranslationally by complex networks of protein-protein interactions governed by a variety of environmental stresses (1, 20). Because one of the promoters (P3) within the sar locus is ςB dependent, it is conceivable that the SigB protein influences sar expression. As the sar locus activates the synthesis of alpha-hemolysin at the transcriptional level, presumably in part through the interaction of SarA with the agr locus (15), we speculate that sigB may modulate sarA expression and the ensuing hla transcription.
In this study, we report the construction and characterization of a sigB mutant of RN6390, a prototypic S. aureus strain. The specificity of the mutation was confirmed by the absence of the SigB protein on an immunoblot, but the protein was restored in the mutant by a shuttle plasmid carrying the sigB gene. Phenotypic analysis revealed that the sigB mutant strain secreted more alpha-hemolysin than the parental strain, as determined by immunoblotting and Northern analysis. Complementation of the mutant with the sigB gene in trans reestablished alpha-hemolysin expression to near parental levels. Interestingly, the hyperproduction of alpha-hemolysin coincided with elevated SarA expression in the sigB mutant. Using the rabbit endocarditis model, we found that the sigB mutation was stable in vivo. We hypothesize that the hyperproduction of alpha-hemolysin in S. aureus as a result of the sigB mutation is mediated by an increase in the SarA level which, in turn, enhances the transcription of hla via a direct pathway (i.e., agr independent).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
The bacterial strains and plasmids used in this study are listed in Table 1. Phage φ11 was used as the transducing phage for S. aureus strains. CYGP, 0.3GL medium (26), and tryptic soy broth (TSB) were used for the growth of S. aureus strains, while Luria-Bertani medium was used for growing Escherichia coli. Antibiotics were used at the following concentrations: erythromycin at 5 μg/ml and ampicillin at 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Reference(s) or source | Description |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN6390 | 22, 26 | Laboratory strain that maintains its hemolytic pattern when propagated on sheep erythrocytes and has a genetic background similar to that of 8325-4 |
| RN4220 | 26 | Mutant of 8325-4 that accepts foreign DNA |
| COL | 30 | Methicillin-resistant S. aureus strain |
| RUSA168 | 30 | sigB mutant of COL (sigB::Tn551) |
| ALC1001 | This study | sigB mutant of RN6390 |
| ALC1497 | This study | ALC1001 complemented with shuttle plasmid pALC1496 (with the sigB gene) |
| E. coli | ||
| BL21 | Novagen | Host strain for pET14b |
| DU5384 | 27 | Host strain carrying pBR322 with a 3-kb EcoRI-HindIII fragment of the hla gene |
| Plasmids | ||
| pCR2.1 | Invitrogen | PCR cloning vector |
| pET14b | Novagen | E. coli expression vector |
| pALC103 | 3 | pSPT181 with a sar fragment from nucleotides 620 to 1349 |
| pALC1270 | This study | pET14b with the sigB coding region cloned into the NdeI-BamHI sites |
| pDG148 | 18 | B. subtilis-E. coli shuttle plasmid (8.2 kb) containing the pSpac promoter (IPTG inducible) followed by a polylinker site and lacI |
| pSK236 | 19 | S. aureus-E. coli shuttle vector with pUC19 cloned into the HindIII site of pC194 |
| pALC1456 | This study | pSK236 containing a 1.6-kb fragment derived from pDG148 and comprising, sequentially, the pSpac promoter, a polylinker site, and the lacI repressor |
| pALC1496 | This study | pALC1456 with the sigB gene (rbs + coding region) cloned into the polylinker site (SalI-PstI) derived from pDG148 |
Genetic manipulations in S. aureus.
A sigB mutant of RN6390 was constructed as described previously (12) by transducing the parental strain with a phage lysate of strain RUSA168 carrying the sigB mutation (30). Transductants were selected on agar containing erythromycin. Correct insertion of Tn551 into the sigB locus of RN6390 was confirmed by Southern blotting with Tn917 and sigB probes as described previously (12). One transductant, ALC1001, was chosen for further studies.
To complement the sigB mutation in ALC1001, we introduced into the shuttle plasmid pSK236 a 1.6-kb fragment (derived from pDG148) containing the pSpac promoter followed by the polylinker site and the lacI repressor of E. coli, yielding pALC1456. Plasmid pALC1496 was constructed by cloning the sigB open reading frame into the multiple cloning site of pALC1456. The shuttle plasmid was electroporated into RN4220 and then transduced into sigB mutant ALC1001 with phage φ11 as described previously (12). The presence of the recombinant plasmid was confirmed by restriction mapping.
Production of anti-SigB monoclonal antibodies.
Based on the published sequence (30), we amplified by PCR the 768-bp sigB gene with the following primers: 5′-GCCAT2687ATGGCGAAAGAGTCGAAATCAGCT2710-3′ (NdeI site underlined) and 5′-GCGGATCCCTA3454TTGATGTGCTGCTTCTTG3437-3′ (BamHI site underlined). The correct PCR product, verified by automated sequencing, was cloned into the NdeI-BamHI sites of expression vector pET14b (Novagen, Madison, Wis.) and transformed into E. coli BL21(DE3).pLys.S. The resulting plasmid, pALC1270, contained an N-terminal His tag and the entire sigB coding region.
Recombinant protein expression was induced by adding isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) to a growing culture (30°C) at an optical density at 600 nm (OD600) of 0.5. Three hours after induction, the cells were harvested, resuspended in binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]), and sonicated on ice. After removal of the cellular debris by centrifugation (15,000 × g for 15 min), the clarified supernatant was purified on a nickel affinity column in accordance with the manufacturer’s instructions. The protein (≈30,000 kDa) eluted from the column with elution buffer (1 M imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9]) appeared homogeneous on a sodium dodecyl sulfate gel (data not shown) and was authenticated by sequencing of the N-terminal 15 residues after the His tag had been removed by digestion with thrombin.
Purified SigB was used to immunize two (BALB/c × SJL/J) F1 mice (100 μg each) to obtain monoclonal antibodies as described by Jones et al. (21). The titers of the immune sera were determined by an enzyme-linked immunosorbent assay (ELISA) in which diluted sera were added to microtiter wells precoated with purified SigB (5 μg/ml). After hybridoma fusion, antibodies from limiting dilutions were screened by an ELISA with immobilized SigB protein. Monoclonal antibody 2D7 was purified from culture supernatants with a protein A affinity column (21) and tested for reactivity with purified SigB on immunoblots.
Phenotypic analysis of the sigB mutant and its isogenic parent.
Several virulence traits of the sigB mutant were evaluated. First, the production of alpha-, beta-, and delta-hemolysins in the sigB mutant and its parental counterpart was assessed by cross-streaking the tested strain with indicator strains as described previously (9). To verify the production of alpha-hemolysin on immunoblots, equivalent volumes of extracellular proteins that had been harvested at the stationary phase and concentrated 50-fold by use of a Centriprep concentrator (Amicon Inc., Beverly, Mass.) were blotted onto nitrocellulose and probed with rabbit anti-alpha-hemolysin antibody (a gift from B. Menzies, Nashville, Tenn.) diluted 1:2,000 and then with the F(ab)2 fragment of the goat anti-rabbit antibody–alkaline phosphatase conjugate (Jackson ImmunoResearch, West Grove, Pa.) as described previously (7). Reactive bands were visualized as described by Blake et al. (5).
Taking advantage of the cytolytic effects of alpha-hemolysin upon rabbit erythrocytes and platelets (2, 4), two functional assays for alpha-hemolysin production were performed. First, alpha-hemolysin levels were quantitated by assaying the hemolytic titers of serially diluted stationary-phase culture supernatants for 1% washed rabbit erythrocytes as described previously (2, 4). Purified alpha-hemolysin was used as the positive control. The data were expressed as mean units of hemolytic activity per milliliter of culture supernatant from two separate runs. The hemolytic units were defined as the reciprocal of the highest dilution of the culture supernatant causing 50% erythrocyte lysis. Second, the extent of alpha-hemolysin production was ascertained by measuring platelet lysis spectrophotometrically (at OD600) upon exposure to bacterial supernatants as described previously (2).
In addition to hemolysins, we also measured other putative virulence traits, such as fibronectin- and fibrinogen-binding capacities. The fibronectin-binding capacity of the isogenic pair was compared with an 125I fibronectin-binding assay as described previously (9). The fibrinogen-binding capacity was determined semiquantitatively by immunoblotting. Cell wall proteins from equivalent numbers of bacterial cells grown overnight were extracted as described previously (10). Equivalent volumes of cell wall extracts (10 μl) were resolved on sodium dodecyl sulfate gels and transferred to nitrocellulose. The nitrocellulose membranes were then blocked with blocking buffer (0.01 M Tris, 0.5 M NaCl, 0.5% Tween 20 [pH 8.2]) containing 1% bovine serum albumin for 1 h at 37°C, incubated at 37°C with fibrinogen (1 mg/ml) in the same buffer, and washed three times with blocking buffer and finally with goat antifibrinogen antibody conjugated to alkaline phosphatase (1:1,000) as described previously (13). As both coagulase and clumping factor bind fibrinogen (14, 29), we assayed for the presence of these proteins in cell wall extracts by immunoblotting. Nitrocellulose membranes containing cell wall extracts were incubated with blocking buffer containing 0.1% human serum (for the blocking protein A-Fc interaction) for 1 h at room temperature. Rabbit anticoagulase (1:1,000) and anti-ClfA (1:1,000) antibodies were then added to separate blots, followed by goat anti-rabbit antibody–alkaline phosphatase conjugate (1:10,000). All reactive bands were visualized as described by Blake et al. (5).
Isolation of RNA and Northern blot hybridization.
Overnight cultures of S. aureus were diluted 1:50 in CYGP and grown to the mid-log (OD650, 0.7), late log (OD650, 1.1), and postexponential (OD650, 1.7) phases. The cells were pelleted and processed with a FastRNA isolation kit (Bio 101, Inc., Vista, Calif.) in combination with 0.1-mm-diameter sirconia-silica beads in a FastPrep reciprocating shaker (Bio 101) as described previously (8). Ten micrograms of each sample was electrophoresed through a 1.5% agarose–0.66 M formaldehyde gel in morpholinepropanesulfonic acid (MOPS) running buffer (20 mM MOPS, 10 mM sodium acetate, 2 mM EDTA [pH 7.0]). Blotting of RNA onto Hybond N+ membranes (Amersham, Arlington Heights, Ill.) was performed with a Turboblotter alkaline transfer system (Schleicher & Schuell, Inc., Keene, N.H.). For detection of specific transcripts (agr, sar, and hla), gel-purified DNA probes were radiolabeled with [α-32P]dCTP by the random-primer method (Ready-To-Go labeling kit; Pharmacia) and hybridized under high-stringency conditions (7). The blots were subsequently washed and autoradiographed.
Preparation of cell extracts and immunoblot analysis of SigB and SarA in the sigB mutant.
Cell extracts were prepared for strain RN6390 and the corresponding sigB mutant. After being pelleted, the cells were resuspended in 1 ml of TEG buffer (25 mM Tris, 5 mM EGTA [pH 8]), and cell extracts were prepared from lysostaphin-treated cells as described previously (15).
Cell extracts were immunoblotted onto nitrocellulose membranes as described above. For the detection of SigB and SarA, monoclonal antibodies 2D7 (1:1,000 dilution) and 1D1 (1:2,500 dilution), respectively, were added to an immunoblot and allowed to incubate with the membrane for 3 h, followed by another h of incubation with a 1:10,000 dilution of goat anti-mouse antibody–alkaline phosphatase conjugate. The reactive bands were visualized as described previously (5).
Determination of SarA levels by a competitive ELISA.
Microtiter wells were coated with purified SarA protein (ca. 3 μg/ml) in 0.1 M Tris–0.3 mM MgCl2 (pH 10) for 3 h at 37°C. After being washed with phosphate-buffered saline (PBS)–Brij (pH 7.4), the wells were blocked with PBS-Brij containing 1% bovine serum albumin overnight at 4°C. The optimal dilution of antibody, defined as the dilution at which 50% of the antibody (i.e., 50% of the maximum amount of bound antibody) bound to the protein-coated wells, was determined to be 1:20,000. Using anti-SarA antibody 1D1 at the optimal dilution (1:20,000), we constructed a standardized curve by adding known quantities of purified SarA (0 to 25 μg/ml) to compete for binding to the anti-SarA antibody, resulting in inhibition of binding to the immobilized SarA protein. For the competitive assay, triplicate cell extracts prepared from individual sar mutant clones (see above for preparation methods) were substituted for purified SarA. The protein concentrations in the cell extracts were determined with a protein assay kit from Bio-Rad Laboratories, Hercules, Calif. By comparing the levels of inhibition to the standardized curve, the relative concentration of the SarA protein in each extract was derived.
Rabbit model of endocarditis.
To ascertain the stability of the sigB mutation in vivo, we chose the rabbit model of endocarditis. Briefly, a bacterial suspension harvested from an overnight culture was diluted in PBS, and bacterial numbers were confirmed by plate counting. Endocarditis on the aortic valves of New Zealand White rabbits (2 kg) was induced by catheterization as previously described (9). At 48 h postcatheterization, groups of animals (three each) were separately challenged intravenously with an inoculum of either 2 × 105 or 2 × 106 CFU. Catheters remained in place until animals were sacrificed by lethal intravenous injection of sodium pentobarbital (100 mg/kg of body weight). At the time of sacrifice (48 h postinfection), aortic valves and left ventricular vegetations from infected animals were removed, pooled, homogenized, and quantitatively cultured. Some colonies were then examined for the retention of the sigB mutation after in vivo passage by Southern (with a Tn551 probe) and Western blotting.
RESULTS
Construction of a sigB mutant of RN6390.
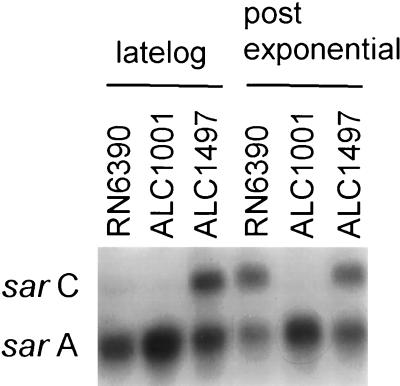
The mutation in a Tn551 insertion mutant, RUSA168, was previously mapped to the sigB gene of S. aureus COL (30). To assess the phenotypic effect of the sigB mutation in a genetic background with well-defined virulence determinants (9), we elected to transduce the mutation from RUSA168 to RN6390 to yield mutant ALC1001. Southern blot hybridization with a Tn917 probe which shows significant homology with Tn551 revealed that the transposon insertion in ALC1001 was analogous to that found in RUSA168 (data not shown). To further confirm the mutation, we made use of the observation that the P3 promoter of the sar locus of S. aureus is ςB dependent (17, 25). Northern analysis revealed that P3-initiated sarC transcription was absent in strain ALC1001, whereas the ςA-dependent P1 promoter of sar was unaffected (Fig. 1). Complementation studies with plasmid pALC1496, carrying the sigB gene, revealed that complemented mutant ALC1497 showed sarC transcription. Of note, the pSpac promoter was found to be active in S. aureus even in the absence of the inducing agent IPTG, suggesting that, contrary to observations made for B. subtilis (18), this promoter is leaky in the staphylococcal background.
FIG. 1.
Northern analysis of sar transcripts of S. aureus RN6390, its isogenic sigB mutant ALC1001, and complemented mutant ALC1497. Northern analysis revealed that the sigB gene was transcribed in ALC1497 (data not shown). Ten micrograms of total cellular RNA obtained at the stationary phase (OD650, 1.7, as determined with an 18-mm borosilicate glass tube) was applied to each lane. The probe was a 730-bp sarA fragment (nucleotides 620 to 1349, based on the published sequence) (3). Because the transcription of sarB, the largest transcript within the sar locus, was minimal during the late log and stationary phases, only data for sarA and sarC transcripts are shown.
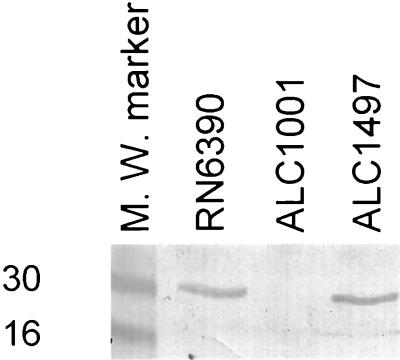
To ascertain if the mutation in ALC1001 was indeed due to sigB, we probed a cell lysate of the mutant with anti-SigB monoclonal antibody 2D7 in an immunoblot. As shown in Fig. 2, SigB was not detectable in the mutant, while the parental strain (RN6390) as well as the complemented mutant strain (ALC1497) were found to contain SigB, as confirmed by the reactivity of the cell extract with the anti-SigB antibody. Additionally, SigB was not found in the mutant strain carrying the vector alone (data not shown). Consistent with the observation that SigB, being a regulatory molecule, is present in a low quantity in cells, we were able to discern the presence of SigB only upon loading at least 50 μg of cellular proteins from the parental strain.
FIG. 2.
Immunoblot of cell extracts of the parental strain, sigB mutant strain ALC1001, and complemented mutant strain ALC1497 probed with anti-sigB monoclonal antibody 2D7 (1:1,000 dilution). About 50 μg of cellular proteins was applied to each lane. The experiment was repeated two times, with essentially the same results. M. W., molecular weight. Numbers at left are in thousands.
To determine the stability of the sigB mutation in vivo, we infected rabbits that had been previously catheterized to produce thrombotic endocarditis with ALC1001. After harvesting the bacteria obtained from the cardiac vegetations of this animal model, we confirmed by Southern and Western analyses that the sigB mutation remained stable after in vivo passage (data not shown).
Phenotypic characterization of sigB mutant ALC1001.
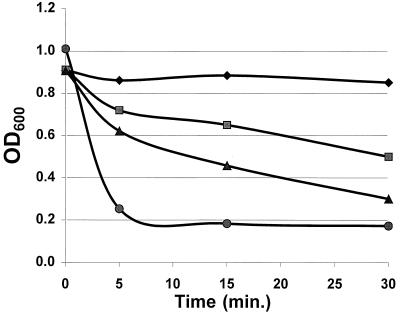
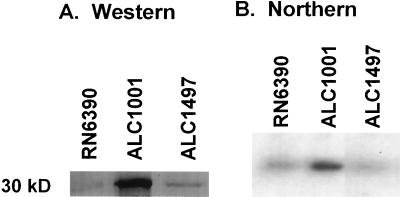
When ALC1001 and RUSA168 were streaked on blood agar plates, it was observed that the clear zone of hemolysis surrounding 24-h cultures was significantly enhanced in both sigB mutants compared with the respective parental strains. As the virulence determinants of RN6390 are well described (9), we chose to focus our analysis on sigB mutant ALC1001. In quantitative hemolysis assays for alpha-hemolysin, the culture supernatant of mutant ALC1001 yielded a mean hemolytic titer of 1,560 U/ml, while the corresponding parental strain titer was 490 U/ml. Purified alpha-hemolysin yielded a titer of 22,736 U/mg of protein. As an additional assay for the functional aspects of alpha-hemolysin, we also found that the culture supernatant of ALC1001 had a greater capacity to lyse platelets than the isogenic parental strain, as monitored by a decrease in the OD600 in a turbidimetric assay (Fig. 3). An immunoblot of equivalent amounts of extracellular proteins of ALC1001, RN6390, and complemented strain ALC1497 disclosed that alpha-hemolysin was synthesized at a higher level in the sigB mutant than in the parental strain but was present at near the parental level in ALC1497 (Fig. 4A). In measuring the transcriptional activity of hla by Northern blotting, we also found that the hla mRNA level of ALC1001 was higher than that of RN6390 (Fig. 4B). Complementation of sigB mutant ALC1001 with a sigB-carrying plasmid (pALC1496) expressing the sigB transcript (data not shown) restored hla transcription to the parental level (Fig. 4B). Similar results on the expression of alpha-hemolysin were observed with RUSA168 complemented with the sigB-carrying plasmid pALC1496 (data not shown). Collectively, these data imply that the sigB mutation is associated with enhanced hla expression initiated at the transcriptional level.
FIG. 3.
Lysis of platelets monitored on the basis of OD600. The supernatants of 18-h bacterial cultures in TSB were mixed with platelets (109 platelets/ml). Platelet lysis was monitored by measuring the decrease in the OD600 over time (2). The medium and purified alpha-toxin served as the negative and positive controls, respectively. Symbols: diamonds, TSB control plus washed platelets; squares, RN6390 (supernatant) plus washed platelets; triangles, SigB (supernatant) plus washed platelets; circles, purified alpha-toxin (1 mg/ml) plus washed platelets.
FIG. 4.
Western and Northern analyses of the alpha-hemolysin gene product. (A) An immunoblot of extracellular proteins of ALC1001, its isogenic parent RN6390, and complemented strain ALC1497 from the late log phase was probed with rabbit anti–alpha-hemolysin antibody (1:2,500 dilution). (B) Ten micrograms of total cellular RNA obtained from the late log to early stationary phases was applied to each lane. The probe was a 3-kb EcoRI-HindIII fragment of the alpha-hemolysin gene (7). Similar complementation results were obtained with another sigB mutant (RUSA168) and plasmid pALC1496. These experiments were repeated three times, with similar results. The results of a representative experiment are shown.
The effect of the sigB mutation on beta-hemolysin secretion, as assayed on immunoblots, was more equivocal, with only a marginal increase in ALC1001 and no change in RUSA168 compared with the results for the respective parental strains. In comparison to that in RN6390, the expression of delta-hemolysin (hld) in ALC1001 on cross-streaked blood agar plates was not altered (see below for the RNAIII transcript encoding hld).
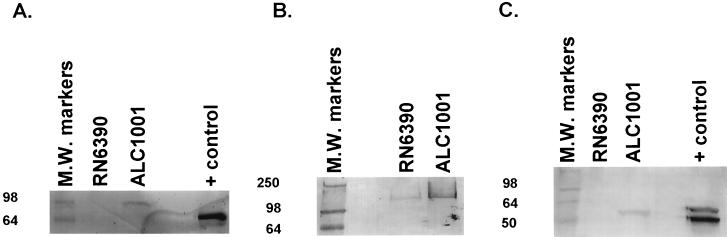
As cell wall-associated proteins such as fibronectin- and fibrinogen-binding proteins likely play a role in mediating the binding of S. aureus to catheters and host valvular tissues (9, 11, 24, 28), we also assessed the ability of the isogenic pair to bind fibronectin and fibrinogen in vitro. The binding of mutant strain ALC1001 to radiolabeled fibronectin was similar to that of parental strain RN6390 (11,480 ± 856 [mean ± standard error of the mean] cpm versus 11,455 ± 816 cpm). In contrast, the sigB mutant bound more fibrinogen than its parental counterpart (Fig. 5A). Recognizing that multiple fibrinogen-binding proteins may be affected by the sigB mutation, we probed immunoblots containing cell wall extracts of the isogenic pair with anti-clumping factor (ClfA) and anticoagulase (Coa) antibodies (Fig. 5B and 3C). In comparison to that in RN6390, the expression of ClfA and Coa in sigB mutant ALC1001 was augmented, confirming the observation that the sigB mutant strain has a higher fibrinogen-binding capacity than the isogenic parental strain.
FIG. 5.
Immunoblots of cell wall extracts of RN6390 and ALC1001 probed with fibrinogen (A), anti-ClfA antibody (B), and anti-Coa antibody (C). Equivalent amounts of cell extracts were applied to the lanes. The positive controls were fibrinogen (A) and purified coagulase (C). M.W., molecular weight. Numbers at left are in thousands.
Expression of agr and sar in sigB mutant ALC1001.
The expression of alpha-hemolysin is influenced by global regulatory loci such as agr and sar. To assess if the sigB mutation in ALC1001 affected the ability of these loci to modulate alpha-hemolysin expression, we performed Northern blotting and showed that the expression of RNAII and RNAIII of the agr locus was not altered in the mutant strain relative to the parental strain (data not shown).
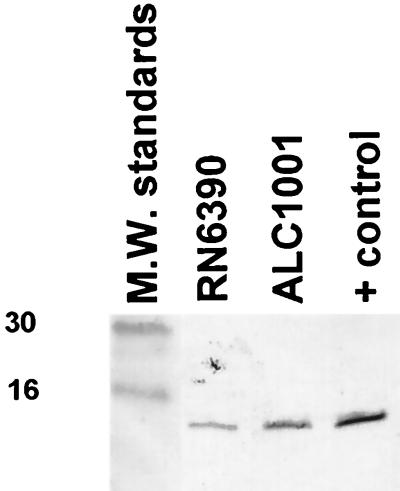
We have shown that the sarC transcript was absent in sigB mutant ALC1001 but was restored upon complementation (Fig. 1). Concomitant with the decrease in sarC transcription in sigB mutant ALC1001 was the repeated observation of an increase in sarA transcription in that strain (Fig. 1) as well as in RUSA168 (data not shown). Notably, sarA transcription was present at near the parental level in complemented mutant ALC1497. In contrast to RNA-mediated control of the agr locus, genetic analyses have indicated that the major regulatory molecule of sar is the SarA protein (7, 15). To assess SarA expression, we probed an immunoblot of cell extracts of the isogenic pair with anti-SarA monoclonal antibody 1D1. The binding epitope of this antibody was recently mapped to residues 16 to 43 of the N terminus of the SarA molecule (16). Our data indicated that the expression of SarA, as determined by immunoblotting, was higher in ALC1001 than in its isogenic parent RN6390 (Fig. 6). This pattern of SarA expression held true in three repetitions of the experiment. Using a recently described competitive ELISA (16) and the cell extracts of these two strains as competitors of 1D1 binding to immobilized purified SarA, we confirmed that the SarA level was elevated in mutant strain ALC1001 (280 ± 25 ng/mg of extract proteins) compared with the parental strain (170 ± 9 ng/mg). Collectively, these data support the notion that a sigB mutation results in enhanced SarA expression, thereby leading to an ensuing increase in alpha-hemolysin expression via an agr-independent mechanism.
FIG. 6.
Immunoblot of cell extracts of RN6390 and ALC1001 probed with anti-SarA monoclonal antibody 1D1 (1:2,500 dilution). About 30 μg of protein was applied to each lane. The positive control was purified SarA protein (14.5 kDa). M.W., molecular weight. Numbers at left are in thousands.
DISCUSSION
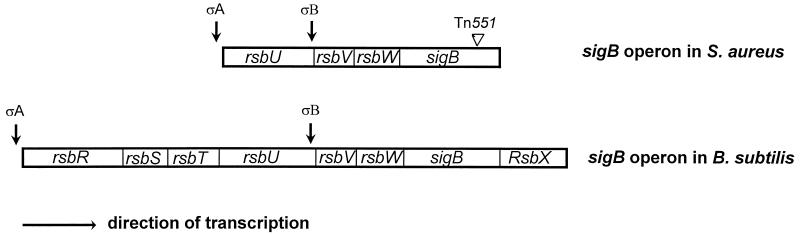
The sigB locus of S. aureus, contrary to the eight-gene sigB operon of B. subtilis (rsbR-rsbS-rsbT-rsbU-rsbV-rsbW-sigB-rsbX), comprises in sequential order four open reading frames which show sequence similarity to the rsbU-rsbV-rsbW-sigB gene cluster of B. subtilis (Fig. 7) (30). Transcriptional analysis indicated that the sigB operon in B. subtilis is transcribed from two distinct but convergent promoters, the ςA and ςB promoters upstream of rsbR and rsbV, respectively. Although sequence analysis of the sigB locus of S. aureus disclosed potential ςA- and ςB-like promoter sequences upstream of rsbU and rsbV, respectively, the transcription start sites attributed to these putative promoters have not been experimentally confirmed. Irrespective of whether these two putative promoters or possibly another or others are active, it is likely that sigB is the last gene encoded with the mRNA message.
FIG. 7.
Organization of the sigB operon in B. subtilis and S. aureus. In contrast to those in B. subtilis, the functions of the putative rsbU, rsbV, and rsbW genes in S. aureus have not been demonstrated. Similarly, the putative ςA and ςB promoters in S. aureus have not been experimentally confirmed.
Because the Tn551 insertion site in sigB mutant RUSA168 has been mapped to approximately nucleotide position 3254 (30) (or 189 residues from the initiation codon and 67 residues from the termination codon), we wanted to assess if the SigB protein was indeed absent in sigB mutant ALC1001. Immunoblot analysis with anti-SigB monoclonal antibody 2D7 revealed the absence of the SigB protein in the mutant strain compared with parental strain RN6390 (Fig. 2). This finding was also confirmed with an additional blot probed with mouse anti-SigB polyclonal antibody (data not shown), indicating that it is unlikely that SigB is synthesized as a truncated form. As the transposon insertion site in the sigB mutant is near the 3′ end of the sigB coding region and sigB may be the last gene transcribed in the polycistronic message, it is unlikely that the Tn551 insertion in the mutant significantly alters the transcription and translation of genes upstream (i.e., rsbU, rsbV, and rsbW). As an additional confirmation that sigB was altered in mutant ALC1001, we took advantage of the fact that the sarC promoter within the triple-promoter system of sar is ςB dependent (17, 25). Predictably, the sarC transcript was absent in mutant ALC1001 but was restored upon complementation with a plasmid carrying a functional copy of the sigB gene (Fig. 1). Collectively, these data clearly indicated that mutant ALC1001 is defective in the synthesis of SigB.
In analyzing some of the phenotypes related to virulence in sigB mutant ALC1001, we found that both extracellular and cell wall proteins were altered. In particular, the capacity to bind fibrinogen as well as the production of alpha-hemolysin were enhanced in the sigB mutant compared with the parent. The augmented fibrinogen-binding capacity was probably mediated by upregulation in the expression of both clumping factor and coagulase in sigB mutant ALC1001 (Fig. 5). Clearly, the sigB mutation was linked to elevated expression of functional alpha-hemolysin at the transcriptional level in two S. aureus strains (ALC1001 and RUSA168) that we examined (Fig. 4B). This linkage was confirmed by complementation studies showing that the hyperproduction of alpha-hemolysin in the mutant could be restored to near the parental level when sigB was supplied in trans on a shuttle plasmid. In addition to these phenotypic alterations, Kullik et al. (23) recently reported that a cytoplasmic protein identified as alkaline shock protein 23 was missing in a sigB deletion mutant, while staphylococcal thermonuclease, an extracellular protein, was secreted at a higher level than in the nonmutated controls. However, despite the pleiotropic nature of SigB in modulating a variety of genes, our results demonstrated that the effect of a sigB mutation on target gene expression can also be selective. For instance, a mutation in sigB in S. aureus can upregulate the expression of alpha-hemolysin but leave beta-hemolysin expression relatively unaltered. Similarly, such a mutation can enhance the expression of coagulase and clumping factor but not fibronectin-binding proteins.
It was recently shown that the SigB protein of S. aureus can activate ςB-dependent promoters in vitro (17). In addition, we have shown (25), as have others (17), that one of the promoters within the sar locus (i.e., the sarC, or P3, promoter) is ςB dependent. Because of the observation that a sigB mutant has phenotypes consistent with the upregulation of sar (increased alpha-hemolysin production and increased fibrinogen-binding capacity), it seemed reasonable to explore if SigB may directly or indirectly modulate global regulatory loci, such as sar and/or agr, to effect these phenotypic changes. However, scrutiny of the agr promoter region did not reveal any ςB-dependent promoter sequences. Additionally, Northern analysis indicated that the transcription of RNAII and RNAIII in the agr locus was not altered in ALC1001, implying that SigB does not modulate agr-related transcription.
The sar locus has a triple-promoter system for which the major regulatory element is the 14.5-kDa SarA protein (124 residues) (3, 7, 15). The sarA gene is encoded by each of three transcripts initiated from three distinct promoters, designated P1, P2, and P3 (3). In particular, the ςA-dependent proximal P1 and distal P2 promoters of sar are maximally expressed during the exponential phase, while ςB-dependent P3 promoter expression peaks postexponentially (7, 25). We speculate that differential promoter activation within sar may lead to disparate expression of the SarA protein (16, 25), the major regulatory molecule in the control of hemolysin production in S. aureus (7). To assess if a sigB mutation would lead to an increase in SarA expression and hence overproduction of alpha-hemolysin, we first established by immunoblotting and an ELISA that the SarA level was higher in the sigB mutant than in the parental control. Based on the phenotypes related to sar (9), a higher SarA protein level will likely enhance the expression of alpha-hemolysin (16). This notion was confirmed by Northern and Western analyses showing that the expression of hla was increased in a sigB mutant but was restored to normal levels upon complementation with a plasmid carrying sigB. Notably, the effect of a higher SarA level on target gene expression could not be mediated by agr, as evidenced by the fact that agr-related transcription was no different in mutant ALC1001 than in RN6390. In previous studies (9, 14), we inferred from phenotypic analysis that the sar locus may have its own direct effect on target gene expression (e.g., beta-hemolysin and fibrinogen-binding proteins). In addition, we demonstrated that the SarA protein may bind to target gene promoters (e.g., the fibronectin-binding protein gene) in gel shift assays (14). Collectively, these data suggested that an increase in alpha-hemolysin expression as well as an upregulation of fibrinogen-binding proteins may occur by an agr-independent but SarA-dependent mechanism. As agr is not substantially altered in the sigB mutant, it is likely that a factor(s) other than SarA also is involved in the expression of genes perturbed by SigB. Because SigB participates in the general stress response of gram-positive bacteria (20), it seems plausible that ςB-dependent genes act in concert with SarA to modulate target gene expression.
The mechanism by which inactivation of sigB leads to elevated SarA protein levels is not fully understood. Using sar promoter-XylE reporter fusions, we recently showed that a combined P3-P1 promoter is less active than a P1 promoter alone, suggesting that the P3 promoter may have a downregulatory effect on transcription from the P1 promoter of sar. A careful analysis of the sequence between the P3 and P1 promoters revealed a 16-bp inverted repeat. Gel shift studies revealed that a 12-kDa protein binds to this repeat (25). Additional genetic analysis indicated that the 12-kDa protein may be a repressor (unpublished data). Based on these data, we hypothesize that the P3 promoter (from which the sarC transcript is generated), upon activation by SigB, may undergo conformational changes to allow binding by the repressor protein. Presumably, a lack of SigB may interfere with the binding of this repressor protein, resulting in enhanced transcription from the P1 promoter and the ensuing elevated SarA expression. In concordance with this hypothesis is the observation that the level of sarA transcription was found to be consistently higher in sigB mutant strain ALC1001 than in the parental strain but returned to near the parental level upon complementation (Fig. 1). Additional experiments to address the interaction between SigB and this repressor protein are currently in progress.
ACKNOWLEDGMENTS
We thank Alexander Tomasz for providing the sigB mutant strain RUSA168; Bill Haldenwang for supplying pDG148; Tim Foster for providing anti-ClfA antibody; H. Igarishi for providing anti-coagulase antibody; Michael Yeaman for providing guidance in the platelet lysis assays; and Kelly Eberhardt, Cong Li, and Iri Kupferwasser for excellent technical assistance.
This work was supported in part by grants-in-aid from the American Heart Association and the New York Heart Association and by NIH grants AI30061 and AI37142 to A.L.C. and by NIH grant AI39108 to A.S.B. Y.-T.C. was supported by a New York Heart Participatory Laboratory Award. A.L.C. is a recipient of the Irma T. Hirshl Career Scientist Award as well as the AHA-Genentech Established Investigator Award from the American Heart Association.
REFERENCES
- 1.Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. doi: 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 2.Bayer A S, Ramos M D, Menzies B E, Yeaman M R, Shen A, Cheung A L. Hyperproduction of alpha-toxin by Staphylococcus aureus results in paradoxically reduced virulence in experimental endocarditis—host defense role for platelet microbicidal protein. Infect Immun. 1997;65:4652–4660. doi: 10.1128/iai.65.11.4652-4660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernheimer A S W. Assay of hemolytic toxins. Methods Enzymol. 1988;165:213–217. doi: 10.1016/s0076-6879(88)65033-6. [DOI] [PubMed] [Google Scholar]
- 5.Blake M S, Johnston K H, Russell-Jones G J, Gotschlich E C. A rapid sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 6.Boyce J M. Epidemiology and prevention of nosocomial infections. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 309–329. [Google Scholar]
- 7.Cheung A L, Bayer M G, Heinrichs J H. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J Bacteriol. 1997;179:3963–3971. doi: 10.1128/jb.179.12.3963-3971.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A L, Eberhardt K, Fischetti V A. A method to isolate RNA from gram-positive bacteria and Mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 9.Cheung A L, Eberhardt K J, Chung E, Yeaman M R, Sullam P M, Ramos M, Bayer A S. Diminished virulence of a sar− agr− mutant of Staphylococcus aureus in the rabbit model of endocarditis. J Clin Investig. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung A L, Fischetti V A. Variation in the expression of cell wall proteins of Staphylococcus aureus grown on solid and liquid media. Infect Immun. 1988;56:1061–1065. doi: 10.1128/iai.56.5.1061-1065.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung A L, Fischetti V A. The role of fibrinogen in staphylococcal adherence to catheters in vitro. J Infect Dis. 1990;161:1177–1186. doi: 10.1093/infdis/161.6.1177. [DOI] [PubMed] [Google Scholar]
- 12.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung A L, Projan S J, Edelstein R, Fischetti V A. Cloning, expression, and nucleotide sequencing of a Staphylococcus aureus gene (fbpA) encoding a fibrinogen binding protein. Infect Immun. 1995;63:1914–1920. doi: 10.1128/iai.63.5.1914-1920.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung A L, Wolz C. Abstracts of the 37th Intersceince Conference on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1997. Regulation of fibronectin binding by the sar locus of Staphylococcus aureus, abstr. B-60; p. 37. [Google Scholar]
- 15.Chien Y, Cheung A L. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J Biol Chem. 1998;237:2645–2652. doi: 10.1074/jbc.273.5.2645. [DOI] [PubMed] [Google Scholar]
- 16.Chien Y T, Manna A C, Cheung A L. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol Microbiol. 1998;31:991–1001. doi: 10.1046/j.1365-2958.1998.01126.x. [DOI] [PubMed] [Google Scholar]
- 17.Deora R, Tseng T, Misra T K. Alternative transcription factor ςSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J Bacteriol. 1997;179:6355–6359. doi: 10.1128/jb.179.20.6355-6359.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dufour A, Voelker U, Voelker A, Haldenwang W G. Relative levels and fractionation properties of Bacillus subtilis sigma B and its regulators during balanced growth and stress. J Bacteriol. 1996;178:3701–3709. doi: 10.1128/jb.178.13.3701-9sigma.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaskill M E, Khan S A. Regulation of the enterotoxin B gene in Staphylococcus aureus. J Biol Chem. 1988;263:6276–6280. [PubMed] [Google Scholar]
- 20.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:506–531. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones K F, Manjula B N, Johnston K H, Hollingshead S K, Scott J R, Fischetti V A. Location of variable and conserved epitopes among the multiple serotypes of streptococcal M protein. J Exp Med. 1985;161:623–628. doi: 10.1084/jem.161.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornblum J, Kreiswirth B, Projan S J, Ross H, Novick R P. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 373–402. [Google Scholar]
- 23.Kullik I, Gianchino P, Fuchs T. Deletion of the alternative sigma factor ςB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuypers J M, Proctor R A. Reduced adherence to traumatized rat heart valves by a low-fibronectin-binding mutant of Staphylococcus aureus. Infect Immun. 1989;57:2306–2312. doi: 10.1128/iai.57.8.2306-2312.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manna A C, Bayer M G, Cheung A L. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J Bacteriol. 1998;180:3828–3836. doi: 10.1128/jb.180.15.3828-3836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novick R P. The staphylococcus as a molecular genetic system. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 1–40. [Google Scholar]
- 27.O’Reilly M, de Azavedo J C S, Kennedy S, Foster T J. Inactivation of the alpha-hemolysin of Staphylococcus aureus 8325-4 by site directed mutagenesis and studies on the expression of its haemolysins. Microb Pathog. 1986;1:125–138. doi: 10.1016/0882-4010(86)90015-x. [DOI] [PubMed] [Google Scholar]
- 28.Scheld W M, Valone J A, Sande M A. Bacterial adherence in the pathogenesis of endocarditis. Interaction of bacterial dextran, platelets and fibrin. J Clin Investig. 1978;61:1394–1404. doi: 10.1172/JCI109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolz C, McDevitt D, Foster T J, Cheung A L. The influence of agr on fibrinogen binding in Staphylococcus aureus Newman. Infect Immun. 1996;64:3142–3147. doi: 10.1128/iai.64.8.3142-3147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu S, de Lencastre H, Tomasz A. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]