Abstract
Mitochondria generate heat due to H+ leak (IH) across their inner membrane1. IH results from the action of long-chain fatty acids on uncoupling protein 1 (UCP1) in brown fat2-6 and ADP/ATP carrier (AAC) in other tissues1,7-9, but the underlying mechanism is poorly understood. As evidence of pharmacological activators of IH through UCP1 and AAC is lacking, IH is induced by protonophores such as 2,4-dinitrophenol (DNP) and cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP)10,11. Although protonophores show potential in combating obesity, diabetes and fatty liver in animal models12-14, their clinical potential for treating human disease is limited due to indiscriminately increasing H+ conductance across all biological membranes10,11 and adverse side effects15. Here we report the direct measurement of IH induced by DNP, FCCP and other common protonophores and find that it is dependent on AAC and UCP1. Using molecular structures of AAC, we perform a computational analysis to determine the binding sites for protonophores and long-chain fatty acids, and find that they overlap with the putative ADP/ATP-binding site. We also develop a mathematical model that proposes a mechanism of uncoupler-dependent IH through AAC. Thus, common protonophoric uncouplers are synthetic activators of IH through AAC and UCP1, paving the way for the development of new and more specific activators of these two central mediators of mitochondrial bioenergetics.
Mitochondrial IH controls cellular bioenergetics by uncoupling H+ flow between the electron-transport chain and ATP synthase. IH causes thermogenesis, increases body energy expenditure and limits mitochondrial production of reactive oxygen species1,16. Pharmacological control of IH is central to the development of effective treatments for metabolic and age-related disorders. Unfortunately, the choice of small-molecule activators of IH through AAC and UCP1 is extremely limited. Physiologically, IH through UCP1 and AAC is activated by free long-chain fatty acids (FAs)3,16-18, but they cannot be used as drugs owing to their poor selectivity and bioavailability. Thus, protonophores such as DNP and FCCP have emerged as pharmacological tools to increase H+ conductance of the inner mitochondrial membrane (IMM). Protonophores are membrane-soluble weak acids that carry H+ across biological membranes without the need for membrane-transport proteins10,11 (Fig. 1e). This protein-independent mechanism of action is broadly accepted as universal for all biological and artificial lipid membranes, including the IMM.
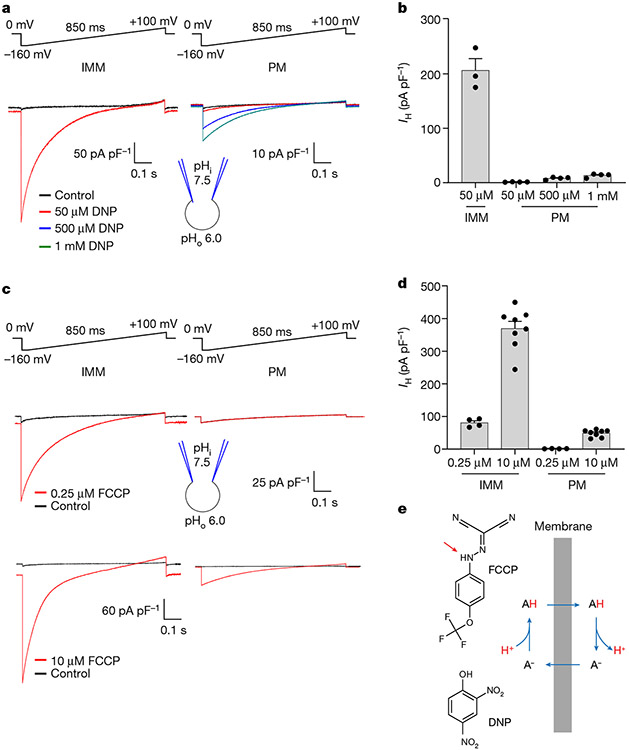
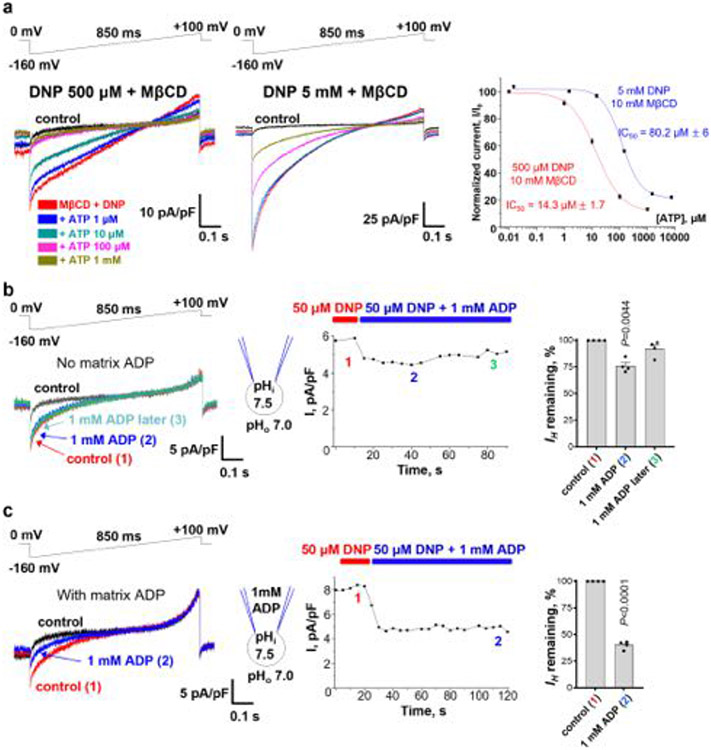
Fig. 1 ∣. H+ currents activated by DNP and FCCP in the IMM versus plasma membrane.
a, DNP-induced IH recorded across the whole IMM of skeletal muscle (IMM, left) or the whole plasma membrane of HEK293 cells (PM, right). b, Densities of DNP-induced IH in the IMM (n = 3) versus PM (n = 4 for each concentration), measured at −160 mV. Data are mean ± s.e.m. c, FCCP-induced IH recorded across the whole IMM of skeletal muscle (left) or the whole plasma membrane of HEK293 cells (right). d, Densities of FCCP-induced IH in the IMM (n = 4 and n = 8) versus PM (n = 4 and n = 8), measured at −160 mV. Data are mean ± s.e.m. e, Protonatable groups of DNP (pKa = 4.1) and FCCP (pKa = 6.2) indicated by red arrows (left). The mechanism of IH induced by protonophores (right). ‘A’ indicates protonatable weak acid.
Although DNP and FCCP have become indispensable tools for controlling energy metabolism at the mitochondrial, cellular and systemic levels, here we demonstrate that their mechanism of action was substantially misunderstood. Although these mitochondrial uncouplers do possess protonophoric activity, their major effect in the mitochondria is the activation of IH through AAC and UCP1.
Preference of DNP and FCCP towards the IMM
We used the patch-clamp technique to directly record IH induced by DNP and FCCP across the whole IMM of individual isolated mitochondria stripped of the outer membrane (mitoplasts)2,7,19,20. Application of DNP and FCCP to the cytosolic side of skeletal muscle mitoplasts resulted in a high IH. By contrast, H+ currents induced across the plasma membrane of HEK293 cells were considerably smaller even when much higher concentrations of DNP and FCCP were used (Fig. 1a-d). These first direct measurements of DNP- and FCCP-induced IH across the IMM suggest that the IMM has a unique ability to potentiate the action of protonophores, either due to its distinct biophysical properties or the presence of specific membrane proteins.
Protonophores induce IH through AAC
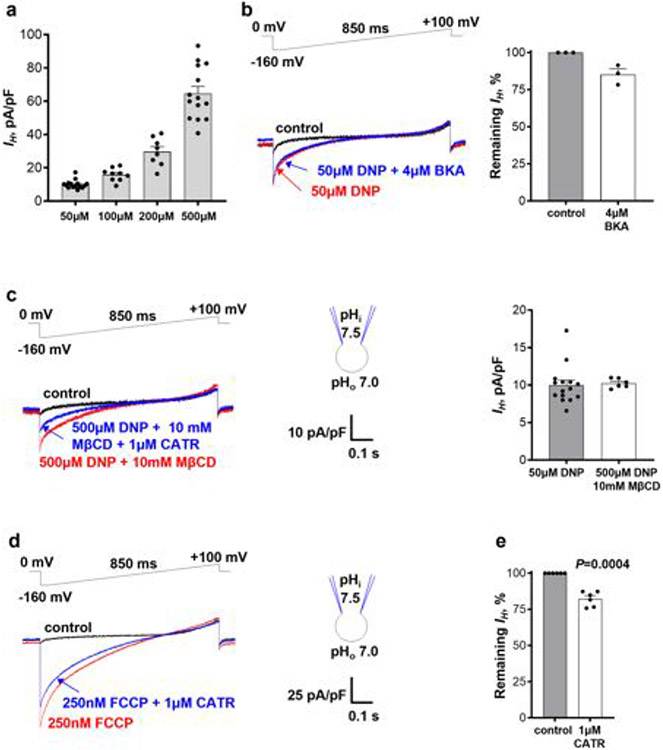
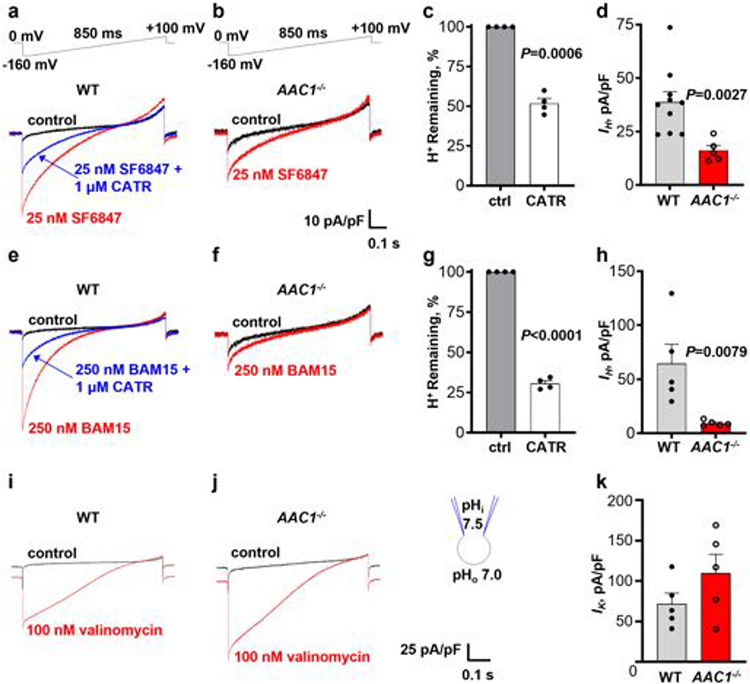
As AAC is responsible for FA-induced mitochondrial IH in tissues other than brown/beige fat7, we hypothesized that DNP and FCCP might activate IH through AAC. DNP induced IH in a dose-dependent manner (Fig. 2a and Extended Data Fig. 1a). The AAC inhibitor carboxyatractyloside (CATR) suppressed the current by around 60%(Fig. 2a,c),whereas another AAC inhibitor, bongkrekic acid (BKA; which inhibits the m-state of AAC versus the c-state for CATR), had only a small effect (around 15% inhibition; Extended Data Fig. 1b).
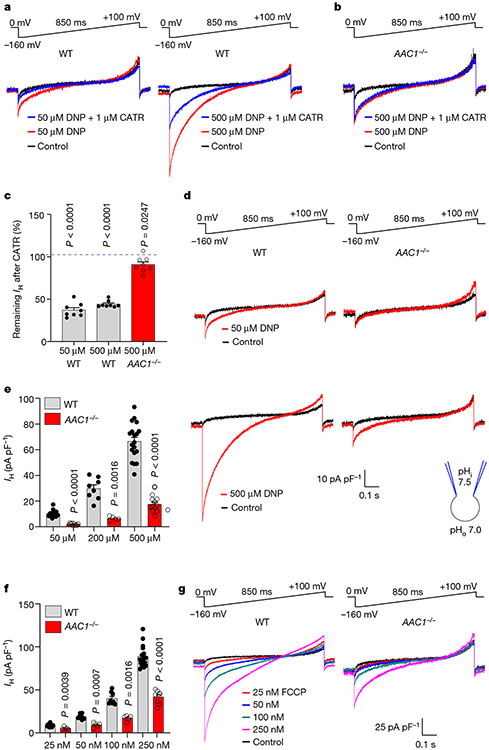
Fig. 2 ∣. DNP and FCCP activate IH through AAC.
a, IH induced by 50 μM (left) or 500 μM DNP (right) in heart IMM was inhibited by 1 μM CATR. b, IH induced by 500 μM DNP in AAC1−/− heart IMM before and after application of 1 μM CATR. c, The remaining fraction of DNP-induced IH at −160 mV after inhibition by 1 μM CATR in WT and AAC1−/−. Data are mean ± s.e.m. Statistical analysis was performed using two-tailed paired t-test, compared with IH before CATR treatment. n = 8 for all conditions. d, IH in WT and AAC1−/− heart IMM induced by 50 μM and 500 μM DNP. e, IH densities at −160 mV in WT and AAC1−/− heart IMM at different DNP concentrations. Data are mean ± s.e.m. Statistical analysis was performed using two-tailed Mann–Whitney U-tests, WT versus AAC1−/−. n = 21 (WT, 50 μM) and n = 10 (AAC1−/−, 50 μM); n = 8 (WT, 200 μM) and n = 5 (AAC1−/−, 200 μM); and n = 19 (WT, 500 μM) and n = 13 (AAC1−/−, 500 μM). f, IH densities at −160 mV in WT and AAC1−/− heart IMM at different FCCP concentrations. Data are mean ± s.e.m. Statistical analysis was performed using two-tailed Mann-Whitney U-tests, WT versus AAC1−/−. n = 12 (WT, 25 nM) and n = 5 (AAC1−/−, 25 nM); n = 10 (WT, 50 nM) and n = 5 (AAC1−/−, 50 nM); n = 8 (WT, 100 nM) and n = 5 (AAC1−/−, 100 nM); and n = 17 (WT, 250 nM) and n = 8 (AAC1−/−, 250 nM). g, IH induced by 25 nM, 50 nM, 100 nM and 250 nM FCCP in WT and AAC1−/− heart IMM.
There are two somatic AAC isoforms in mice, AAC1 and AAC221,22, with AAC1 being dominant in heart and skeletal muscle21,23. Accordingly, IH induced by DNP in cardiac IMMs from AAC1−/− mice 7,24 was reduced by around 80% (Fig. 2d,e) and was insensitive to CATR (Fig. 2b,c). Thus, DNP activates IH across the IMM primarily by targeting AAC. The AAC-independent component of DNP-induced IH (around 20%; Fig. 2d,e) may be associated with the protonophoric activity of DNP, its action on other IMM proteins or both.
We next investigated whether FCCP and other mitochondrial uncouplers also induce IH through AAC. In agreement with previous studies11,25, FCCP, SF684711,25 and BAM1525,26 were more potent than DNP and induced robust mitochondrial IH at nanomolar concentrations (Fig. 2g and Extended Data Fig. 2a,e). CATR inhibited IH induced by FCCP, SF6847 and BAM15 by approximately 20% (Extended Data Fig. 1d, e), 50% (Extended Data Fig. 2a, c) and 70% (Extended Data Fig. 2e,g), respectively. IH induced by FCCP, SF6847 and BAM15 in cardiac IMMs from AAC1−/− mice was reduced by around 50% (Fig. 2f,g), 60% (Extended Data Fig. 2a,b,d) and 85% (Extended Data Fig. 2e,f,h), respectively. In contrast to the uncoupler-induced IH, K+ current activated across the IMM by the K+-selective ionophore valinomycin27 was not reduced in cardiac IMMs from AAC1−/− mice (Extended Data Fig. 2i-k). Interestingly, for all uncouplers, AAC1 deficiency had a larger effect on IH than pharmacological inhibition with CATR (Fig. 2 and Extended Data Figs. 1 and 2). This could be due to competition between CATR and uncouplers for the AAC-binding site, or a CATR-insensitive AAC state induced by uncouplers (CATR binds only to the c-state of AAC28).
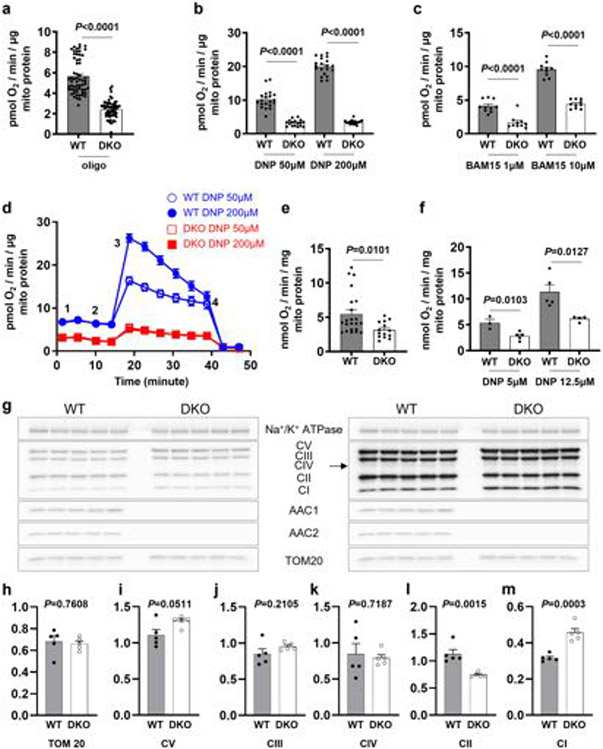
We next compared the rates of respiration induced by uncouplers in mitochondria isolated from wild-type (WT) and AAC1/AAC2 double-knockout (DKO) C2C12 cells7. DNP and BAM15 caused a dose-dependent increase in the rate of uncoupled respiration in mitochondria from WT C2C12 cells, which was substantially reduced in DKO mitochondria (Extended Data Fig. 3a-f).
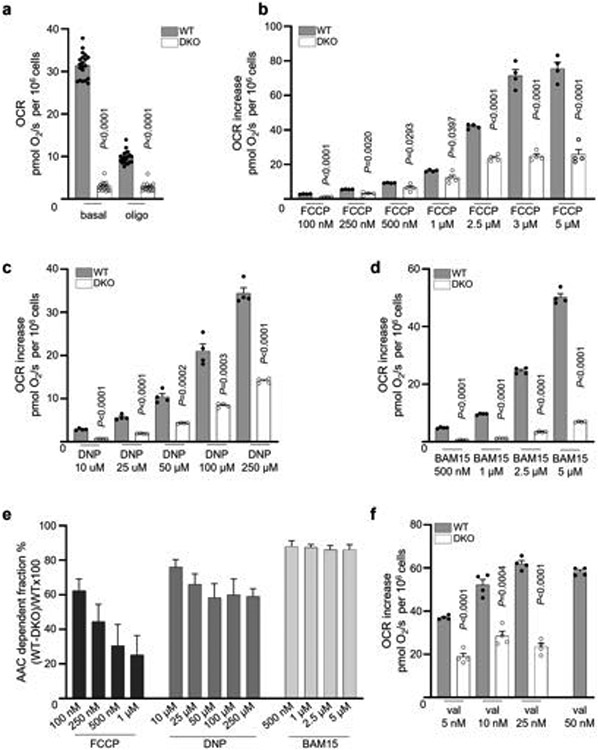
We also compared uncoupled respiration induced by FCCP, DNP and BAM15 in intact WT and DKO C2C12 cells. DKO cells have a significantly lower basal respiration rate than WT cells7 (Extended Data Fig. 4a). ATP synthase inhibitor oligomycin reduced respiration of WT cells but not DKO cells, as they lack mitochondrial ATP production7 (Extended Data Fig. 4a). Further application of increasing FCCP concentrations stimulated uncoupled respiration in both WT and DKO cells, but DKO respiration was lower and plateaued at around 20 pmol s−1 per 106 cells (Extended Data Fig. 4b), indicating that the DKO cells have a limited respiratory capacity. The limited respiration capacity of DKO cells was further confirmed using a combination of valinomycin and nigericin to induce AAC-independent mitochondrial uncoupled respiration (Extended Data Fig. 4f). As mitochondrial biomass and the expression of the electron-transport chain complexes was overall similar in DKO and WT cells (Extended Data Fig. 3g-m), reduced Krebs cycle activity or mitochondrial substrate availability in DKO cells (which fully rely on glycolysis for ATP production7) may limit maximal respiration.
Thus, to provide a valid comparison of uncoupler-induced respiration in WT and DKO cells, we analysed only lower concentrations of FCCP (100 nM to 1 μM), DNP (10 μM to 250 μM) and BAM15 (500 nM to 5 μM) that did not exceed the respiration ability of DKO cells (approximately 20 pmol s−1 per 106 cells). Respiration induced by all uncouplers was reduced in DKO cells, but FCCP showed an overall lower dependence on AAC compared with DNP and BAM15 (Extended Data Fig. 4b-e), correlating well with the electrophysiological data (Fig. 2 and Extended Data Fig. 2e-h). Moreover, as FCCP concentration increased, AAC dependence of the FCCP-induced respiration became even smaller (Extended Data Fig. 4e), probably due to the increased protonophoric component. By contrast, AAC dependence of BAM15-induced respiration remained constant (Extended Data Fig. 4e), perhaps due to its limited protonophoric activity26.
Thus, in addition to their protonophoric activity, widely used mitochondrial uncouplers such as FCCP, DNP and BAM15 activate IH through AAC. Notably, BAM15 is less toxic in cells and mice compared with FCCP and DNP29,30, which may correlate with its lower protonophoric activity26 and higher dependence on AAC (Extended Data Fig. 4e).
DNP and FCCP induce IH through UCP1
Being the dominant uncoupling protein in brown and beige fat2-5,19,31, UCP1 may be responsible for DNP- and FCCP-induced IH in these tissues. IMM-associated phospholipases result in substantial amounts of endogenous FAs in the IMM of brown fat. Thus, DNP and FCCP were applied together with 10 mM methyl-β-cyclodextrin (MβCD) to extract endogenous FA and therefore limit their effect on UCP12,19,20. Higher concentrations of DNP and FCCP were applied while using MβCD because it binds to hydrophobic compounds non-specifically, reducing their effective concentrations. For example, 500 μM DNP in the presence of 10 mM MβCD induces a similar IH amplitude as 50 μM DNP without MβCD in heart IMMs (Extended Data Fig. 1c).
Application of 500 μM DNP together with 10 mM MβCD induced robust IH in brown fat IMMs (Fig. 3a (left)). This current was strongly suppressed by 1 mM GDP (guanosine 5′-diphosphate), an established UCP1 inhibitor31 (Fig. 3a, b), and reduced in UCP1−/− IMMs (Fig. 3a,c). Thus, UCP1 is crucial for DNP-induced IH in brown fat mitochondria. Interestingly, DNP-induced IH carried by UCP1 had a linear current–voltage curve (Fig. 3a (left)) compared with the inwardly rectifying current–voltage relationship of DNP-induced IH carried by AAC in heart IMM, which showed a sharp increase in the amplitude of inward currents with membrane hyperpolarization (Fig. 2a and Extended Data Fig. 1c). In UCP1−/− brown fat IMMs, the residual IH was inwardly rectifying and GDP-insensitive (Fig. 3a,b), suggesting the involvement of AAC.
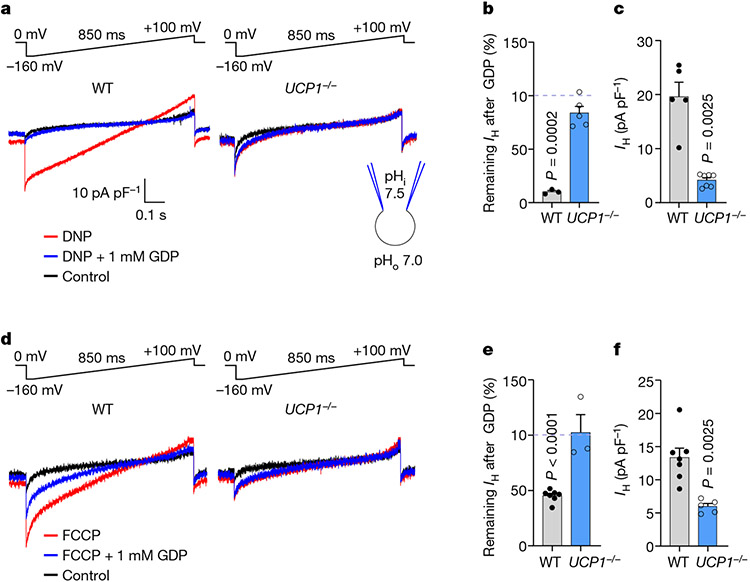
Fig. 3 ∣. DNP and FCCP induce IH through UCP1.
a, DNP-induced IH across the IMM of brown fat in WT and UCP1−/− before and after application of 1 mM GDP. DNP (500 μM) was applied in the presence of 10 mM MβCD to extract endogenous membrane FAs. b, The remaining fraction of DNP-induced IH at −160 mV after inhibition by 1 mM GDP in WT (n = 3) and UCP1−/− (n = 5). Statistical analysis was performed using two-tailed paired t-tests, IH without GDP versus with GDP in the WT. c, The densities of DNP-induced IH in WT (n = 5) and UCP1−/− (n = 7) as measured at −160 mV. Data are mean ± s.e.m. Statistical analysis was performed using two-tailed Mann–Whitney U-tests, WT versus AAC1−/−. d, FCCP-induced IH across the IMM of brown fat in WT and UCP1−/− before and after application of 1 mM GDP. FCCP (10 μM) was applied in the presence of 10 mM MβCD to extract endogenous membrane FAs. e, The remaining fraction of FCCP-induced IHat −160 mV after inhibition by 1 mM GDP in WT (n = 7) and UCP1−/− (n = 3). Statistical analysis was performed using paired two-tailed t-tests, IH without GDP versus with GDP in the WT. f, The densities of DNP-induced IH in WT (n = 7) and UCP1−/− (n = 5) mitoplasts. IH was measured at −160 mV. Data are mean ± s.e.m. Statistical analysis was performed using two-tailed Mann–Whitney U-tests, WT versus AAC1−/−.
FCCP-induced IH in brown fat IMMs was also inhibited by GDP and significantly reduced in UCP1−/− mice (Fig. 3d,f). IH remaining in UCP1−/− IMMs was insensitive to GDP (Fig. 3d,e). Interestingly, FCCP-induced IH was less dependent on UCP1 than DNP-induced IH (~50% versus ~80% reduction in UCP1−/− IMMs, respectively; Fig. 3c,f) similar to the relative AAC dependence of FCCP- and DNP-induced IH in heart IMMs (Fig. 2e,f). FCCP depends less on AAC and UCP1 than DNP due to either its higher protonophoric activity or the involvement of other IMM proteins.
Nucleotides inhibit DNP-induced IH
Mg2+-free ADP and ATP are negative physiological regulators of FA-induced IH through UCP1 and AAC2,7,32-34. Thus, we hypothesized that uncoupler-activated IH is also inhibited by adenine nucleotides.
ATP inhibited IH through UCP1 induced by 500 μM DNP with a half-maximum inhibitory concentration (IC50) of around 14 μM, but the IC50 increased to about 80 μM when 5 mM DNP was used (Extended Data Fig. 5a). Thus, ATP appears to compete with DNP for binding to UCP1, as previously observed for FA2,19. As the nucleotide-binding site of UCP1 is not selective for ATP31, other purine nucleotides, such as GDP (Fig. 3a), also inhibit DNP-induced IH through UCP1.
In contrast to UCP1, which binds only purine nucleotides on the cytosolic face of the IMM, AAC transports adenine nucleotides and binds them on both IMM faces, depending on the conformational state (c or m)28. As the vast majority of AAC proteins in isolated mitoplasts are initially in the c-state, with the nucleotide-binding site exposed to the cytosolic face of the IMM7, 1 mM ADP was first applied only to the cytosolic (bath) solution. We observed a small, transient inhibition of DNP-induced IH (Extended Data Fig. 5b), probably due to the binding of cytosolic ADP to the c-state of AAC, followed by a transition to the m-state and release of ADP to the matrix side. In this state, the translocation pathway is expected to become unobstructed as there is no ADP in the pipette solution, enabling DNP to rebind and allowing IH to recover (Extended Data Fig. 5b).
We next applied ADP on both IMM sides to study how continuous nucleotide exchange interferes with IH. The electroneutral ADP/ADP homoexchange was used to avoid IH contamination with ADP/ATP heteroexchange current7. The addition of 1 mM ADP to both sides of the IMM led to a greater and persistent inhibition of DNP-induced IH (compare Extended Data Fig. 5c with Extended Data Fig. 5b). This suggests that DNP cannot induce IH in a fraction of AAC transporters that are nucleotide bound during continuous nucleotide exchange, either because of nucleotide–DNP competition or simple obstruction of the translocation pathway. As 1 mM ADP is at the higher end of physiologically relevant Mg2+-free ADP concentrations, we propose that adenine nucleotides cannot completely suppress DNP-induced H+ transport through AAC under physiological conditions but can downregulate it. The inhibitory effects of one-sided and two-sided ADP application on DNP-induced IH are similar to those on FA-induced IH through AAC7 and suggest that DNP and FA induce IH after binding to the central translocation pathway of AAC and UCP12,7.
Uncouplers and FAs bind within AAC
We sought to identify putative binding sites for chemical uncouplers in AAC using in silico docking and molecular dynamics simulations. AAC1 is the only protein mediating IH for which a high-resolution structure has been determined. In the c-state, AAC1 forms a large water-accessible cavity facing the cytosolic side of the IMM, which is occupied by CATR in the crystal structures35-37. Previous studies have identified the base of the cavity as the binding site for transported nucleotides38,39. We targeted the entire cavity of bovine AAC1 (Protein Data Bank (PDB): 2C3E)36 for molecular docking with the CATR molecule removed.
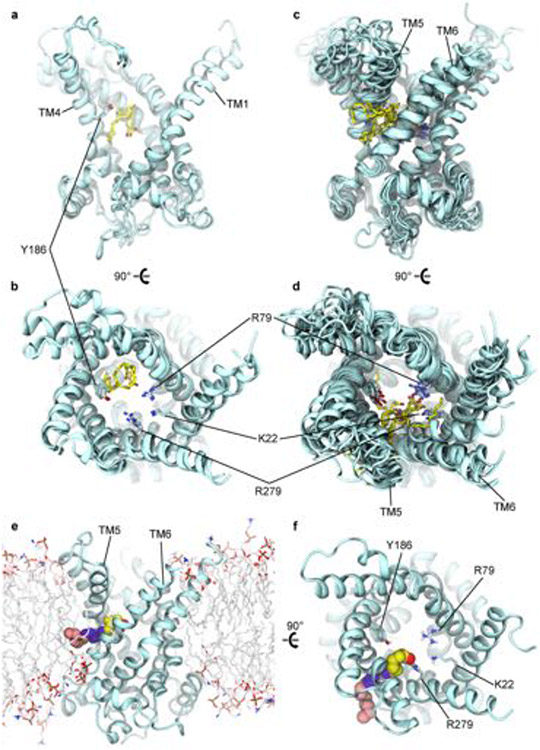
We identified highly reproducible binding modes for DNP deep in this cavity, about halfway across the IMM, which were unaffected by the protonation state of the compound (Fig. 4a-c). At this depth in the cavity, one side is highly charged and composed primarily of basic residues, but DNP packs against the more hydrophobic transmembrane (TM) helix 4 on the opposite side. The aromatic moiety of DNP contacts hydrophobic residue Ile183 and forms T-shaped aromatic stacking interactions with Tyr186. Moreover, one of the nitro groups forms electrostatic interactions with Arg234, whereas the hydroxyl group hydrogen bonds with Ser227. In this position, DNP occupies approximately the same space as the adenosine ring of ADP and the diterpene moiety of CATR (Extended Data Fig. 6a,b).
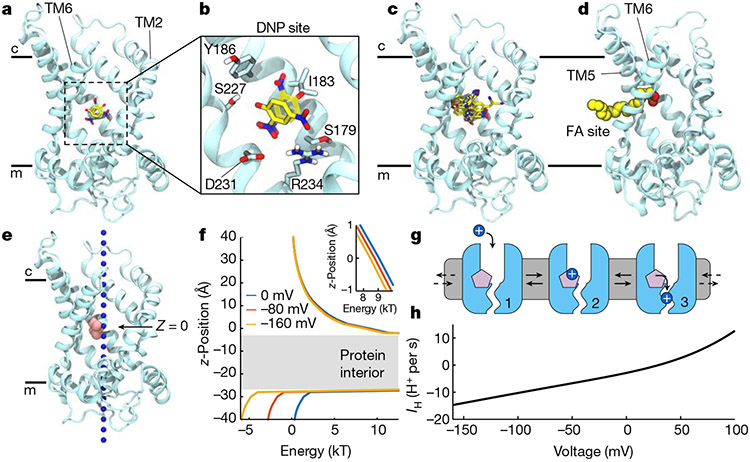
Fig. 4 ∣. Uncouplers and FAs bind within AAC.
a, Binding poses of deprotonated DNP in the cavity of the AAC1 c-state obtained from docking (solid) and a 500 ns molecular dynamics simulation (transparent) viewed from the membrane with TM1 hidden. AAC1 (cyan) is represented as ribbons, and DNP as sticks (carbon (yellow), oxygen (red) and nitrogen (blue)). b, Expanded view of DNP poses in a. c, Superposition of predicted binding poses for protonated and deprotonated forms of DNP, FCCP, BAM15 and SF6847. d, Arachidonic acid from a molecular dynamics simulation showing the carboxylate headgroup occupying the central cavity and aliphatic tail extending into the bilayer between TM5 and TM6. e, Hypothetical H3O+ (blue spheres) pathway from the intermembrane space (c) to the matrix (m). Pink spheres are titratable groups of docked uncouplers shown in c set to z = 0. f, Electrostatic energy profile computed along pathway in e using Poisson–Boltzmann continuum electrostatics. The grey region indicates high electrostatic barrier due to lack of solvation in the protein interior. The profiles correspond to applied voltages of 0 mV (blue), −80 mV (red) and −160 mV (yellow). Inset: magnified view of the profile near protonatable groups at z = 0. g, Cartoon model of the three-state H+ transport cycle: (1) the uncoupler (pink) bound to c-state of AAC1 (light blue); (2) cytoplasmic H+ (blue sphere) binds to the uncoupler; and (3) H+ transitions to the matrix pathway. H+ exits AAC1 to the matrix as the cycle resets back to state 1. Each step is reversible, but only H+ movement from the cytosol to matrix is shown because it is favoured under negative matrix voltages. h, The steady state single-transporter current–voltage relationship predicted from the model in g using the values in Supplementary Table 2 for a DNP-like uncoupler with cytosolic pH = 7 and matrix pH = 7.5. Model equations are provided in Supplementary Note 1.
We subsequently docked FCCP, BAM15 and SF6487 into the c-state cavity, and found that they all bind in the pocket identified for DNP, with the best binding modes conserved between their neutral and negatively charged states (Fig. 4c and Extended Data Fig. 7a-d). Despite the chemical diversity of these compounds, their poses largely overlap such that key titratable atoms on all four molecules lie in the same spatial location (Extended Data Fig. 6e) with their aromatic rings against the hydrophobic side of the pocket.
We repeated the docking of DNP and other uncouplers using a homology model of the bovine transporter m-state based on a recent fungal AAC structure (PDB: 6GCI)40. This revealed an interaction landscape for the molecules that largely resembled that observed in the c-state (Extended Data Fig. 7), supporting our experimental observation that cytosolic DNP activates IH from both the c- and m-states of AAC (Extended Data Fig. 5b).
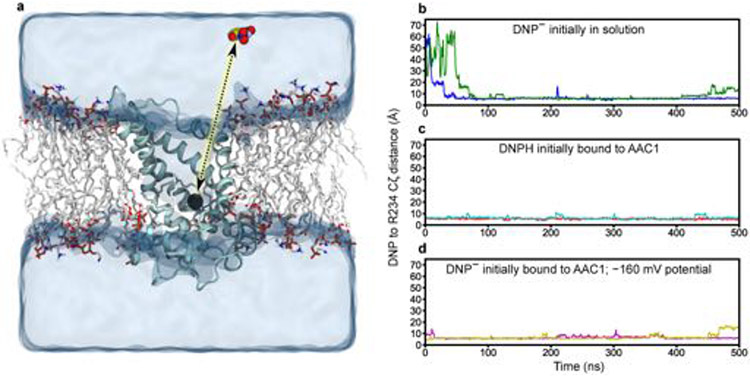
To validate this site, we performed molecular dynamics simulations of AAC1 in the c-state with charged DNP placed in solution away from the protein. Within 100 ns, DNP consistently entered the transporter driven by the strong positive potential in the cavity, and quickly moved to the location predicted from docking (Fig. 4b, Extended Data Fig. 8a,b and Supplementary Video 1). DNP in this site is dynamic, rapidly reorienting its hydroxyl and nitro substituents to interact with different protein moieties while largely remaining anchored to a single position on the protein through contact with its aromatic ring. We initiated additional simulations starting with DNP in the identified site either using neutral, protonated DNP or deprotonated DNP in a −160 mV membrane potential to bias negative charges toward the cytosol. In all simulations, DNP remained bound in the site for nearly the entire 500 ns duration (Extended Data Fig. 8b-d) and maintained interactions observed in the docking, as well as water-mediated contacts with Arg79, Arg187 and Asp231.
To determine whether FAs share any of the interactions predicted for chemical uncouplers, we initiated 14 molecular dynamics simulations of AAC1 in the c-state and 4 in the m-state with 16 FAs, either arachidonic or palmitic acid, placed randomly throughout the membrane (Supplementary Table 3). In 7 out of 14 c-state simulations, FAs from the outer leaflet entered the cavity through several distinct routes, while none bound to the m-state. In two trajectories, arachidonic acid initially occupies the chemical uncoupler site for over 1 μs (Extended Data Fig. 9a,b and Supplementary Video 2). However, in all seven binding events, both arachidonic and palmitic acid eventually occupy a fenestration between TM5 and TM6 from which they do not leave. In many instances, the acyl chain protrudes into the lipid bilayer while the carboxylate interacts with residues Lys22, Arg79 and Arg279–all predicted to engage nucleotide phosphate groups38,39 (Fig. 4d and Extended Data Figs. 6f and 9c,d). The TM5–6 fenestration is evident in X-ray structures of the c-state in which it is occupied by several water molecules35,36.
The configuration with the acyl tail anchored in the bilayer suggests a physical basis for our previous finding that a long hydrophobic tail is critical for FA activation of IH through AAC and UCP12,7. Our data also reveal that both mitochondrial uncouplers and FAs bind within the water-accessible translocation pathway, but to different sites (Extended Data Fig. 6g). The uncoupler site overlaps with a portion of the CATR-binding site and the adenosine ring of docked ADP, whereas the region occupied by the dynamic FA headgroup overlaps with the carboxylate groups of CATR and the ADP phosphate groups (Extended Data Fig. 6a-d). We cannot exclude the possibility that FA exerts its effect from the uncoupler site rather than the TM5/6 fenestration site, as we observed a FA in both sites in the simulations (Extended Data Fig. 9a-d). Moreover, a simulation with two FAs placed in the cavity, one in each site, remains stable for a full microsecond (Extended Data Fig. 6h and Supplementary Video 3), showing that the c-state can simultaneously accommodate more than one FA molecule.
Biophysical model of H+ permeation through AAC
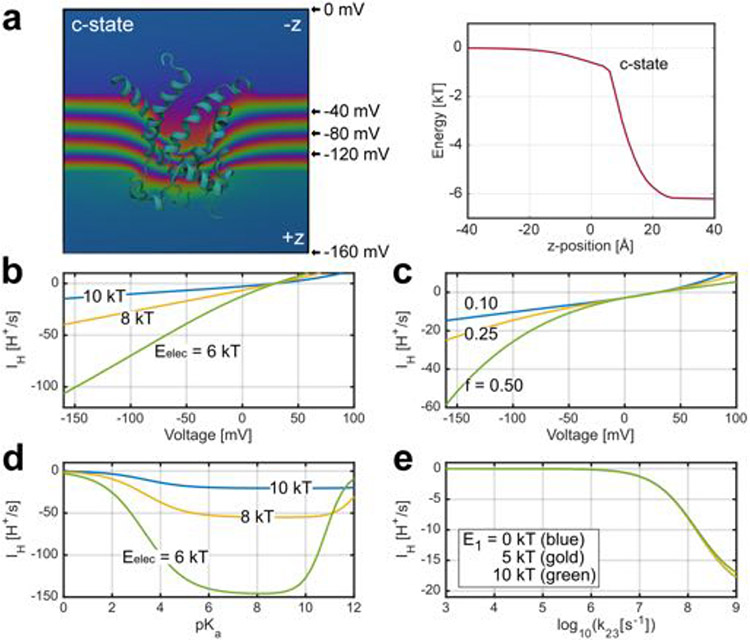
Continuum electrostatics calculations revealed a large, positive potential opposing H+ entry into the cytosolic cavity of the AAC1 c-state (Fig. 4e,f), consistent with its role as an anion transporter. Protons experience an electrostatic energy barrier (Eelec) of 10 kBT at the bottom of the cavity near the centre of the membrane (z = 0), where the protonatable groups of bound uncoupling agents are located (Fig. 4e and Extended Data Fig. 6e). Between z = −5 and −25 Å, there is no aqueous pathway through the closed gate on the matrix side and, therefore, another large energy barrier is present (Fig. 4f (grey region)). A negative membrane potential (normally present across the IMM) tilts the energy landscape in favour of H+ entry to the matrix, but does not significantly reduce the barrier (Fig. 4f and Extended Data Fig. 10a), consistent with a lack of IH through AAC or UCP1 in the absence of FA2,7,19 or uncoupling agents (Fig. 1).
FAs and uncouplers probably enable H+ permeation by reducing the positive charge in the cavity and providing a protonatable group that acts as a stepping stone, similar to the FA–cofactor model proposed for UCP16. However, overcoming the energetic barrier of the closed matrix gate requires a conformational change to open a H+ permeation pathway to the matrix. This pathway should be narrow to ensure selective H+ permeation, and would therefore require only a minor conformational change short of the full c-state to m-state transition required for ADP/ATP exchange. We assume that both FAs and chemical uncouplers induce such a change as suggested by transient pre-steady-state currents recorded for AAC (and UCP1) in the presence of low-pKa FA analogues after the application of a negative voltage2,7.
On the basis of these assumptions, we created a mathematical model to examine the possibility of H+ permeation through AAC once uncouplers bind and an H+-selective pathway to the matrix has opened. In this model, a proton first enters the transporter from the cytosol and binds to the uncoupler (pink molecule), followed by a transition to the narrow H+ permeation pathway and eventual delivery to the matrix (Fig. 4g and Supplementary Note 1). The true nature of the H+ pathway remains unclear as it may occur through a water wire, Brownian diffusion through a water filled passage, transfer to a protonatable site on the protein or another mechanism. Thus, we modelled this step as an Arrhenius rate, which can generally describe all of these processes at this coarse level.
We populated the rates in the model from the results of our structure-based computations on the c-state along with additional biophysical considerations (Supplementary Note 1). The model predicts a H+ flux rate of around 15 H+ per s at −160 mV in the presence of a DNP-like activator (pKa = 4.2 in solution) and cytosolic and matrix pH values of 7 and 7.5, respectively (Fig. 4h; using the values shown in Supplementary Table 2). The large positive electrostatic barrier (Eelec) is a key determinant of the flux explaining the small unitary current. Even reducing Eelec from 10 kBT to 6 kBT causes an eightfold increase in IH (Extended Data Fig. 10b). Thus, if a minor conformational change induced by the uncoupler/FA affects the shape of the cytosolic cavity and its electrostatic potential, it could significantly affect the H+ transport rate. Furthermore, the same conformational change could position the uncoupler/FA deeper into the membrane electric field, which would increase the fraction of the membrane voltage (f) that H+ move through as they bind from the cytosol. Increasing f from 0.1 (the computed value) to 0.5 causes a fourfold increase in the current (Extended Data Fig. 10c). Moreover, calculations with f = 0.5 better approximate the inward rectification observed in the experimental traces (Fig. 2 and Extended Data Fig. 2).
Tuning the pKa of the uncoupler reveals that IH is optimal above 5, and compounds with values under 2 do not induce current (Extended Data Fig. 10d), consistent with our earlier finding that low-pKa FA analogues cannot induce IH in either UCP1 or AAC2,7. Uncouplers with moderately high pKa values are still effective because the positive potential in the cavity essentially lowers their intrinsic pKa making them more likely to give up their proton.
The largest structural uncertainty involves the nature of the H+ pathway from the uncoupler to the matrix; however, the model places strong constraints on this rate (transition from state 2 to state 3 in Fig. 4g). The transition must occur in the upper nanosecond range or faster for efficient conduction or else H+ unbinding to the cytoplasm dominates due to the large positive potential in the cavity (Extended Data Fig. 10e). Moreover, IH is insensitive to the energy of the H+ in the pathway provided that the forward rate from the uncoupler is fast, consistent with a number of proton permeation mechanisms (further details are provided in Supplementary Note 1).
This model demonstrates the feasibility of the uncoupler-induced H+ transport through the translocation pathway of AAC, replicates some of the measured electrophysiological properties and provides a molecular-based hypothesis that can drive future studies.
Discussion
Our results challenge the dogma that mitochondrial uncouplers such as DNP, FCCP, SF6847 and BAM15 induce IH across the IMM through a protein-independent protonophoric mechanism41-45. We demonstrate that the dominant mechanism of action of these uncouplers in the mitochondria is to induce IH by activating AAC and UCP1, the same proteins that mediate the physiological, FA-induced mitochondrial IH. However, at the level of the whole cell and the whole organism, the protonophoric activity of the uncouplers can still have a considerable impact due to their broad, indiscriminate effect on various cell membranes.
Electrophysiology and computational analysis suggest that chemical uncouplers and FAs induce IH in AAC and UCP1 through the same general mechanism: both are weak acids that bind within the central translocation pathway to facilitate H+ binding and permeation. Unexpectedly, FAs must be substantially more hydrophobic than chemical uncouplers to effectively activate IH. Only long-chain FAs containing more than 12 carbons (log[P] > 4.6) have the ability to activate AAC- and UCP1-dependent IH in the low micromolar range2,7. By contrast, DNP robustly activates IH despite its much lower hydrophobicity (6 carbon atoms, log[P] = 1.67). This indicates potential differences in the binding mechanisms of FAs and chemical uncouplers in the translocation pathways of AAC and UCP1, which are also confirmed by the molecular dynamics simulations and molecular docking (Supplementary Note 2).
All of the chemical uncouplers examined in this work have two mechanisms of action, a protonophoric activity affecting all cell membranes and an IMM-specific H+ leak through AAC and UCP1. Despite this, our data indicate that it might be possible to develop specific activators of mitochondrial IH through AAC (UCP1) that lack the protonophoric activity (Supplementary Note 1). Such next-generation anti-obesity and anti-diabetes drugs would activate native thermogenic pathways similar to their endogenous activators, long-chain FAs, and might have improved safety compared with general protonophores.
To date, UCP1 has been the primary pharmacological target to stimulate resting metabolic rate due to the thermogenesis that it induces in brown/beige fat. However, the impact of UCP1 in humans is limited because of a substantial reduction in brown adipose tissue with age and its near complete absence in certain individuals31,46. AAC-dependent IH can also increase energy expenditure by channelling the energy of mitochondrial substrate oxidation into heat. AAC is ubiquitously expressed and its contribution to overall body energy expenditure is therefore likely to exceed that of UCP1. By having two functions–ADP/ATP exchange and H+ leak–AAC functions as a master regulator that controls the partitioning of mitochondrial energy between ATP production and thermogenesis. Thus, this native, self-regulating pathway represents an attractive target for controlling energy expenditure.
Methods
Animals
Mice were maintained on a standard rodent chow diet under 12 h–12 h light–dark cycles. All of the animal experiments were performed with male mice according to procedures approved by the UCSF Institutional Animal Care and Use Committee. B6.129-Ucp1tm1Kz/J mice (abbreviated UCP1−/−) were obtained from the Jackson Laboratory. The AAC1-deficient mice on the C57BL/6J2z background were obtained from the laboratory of D. Wallace24. WT and UCPT−/− mice (B6.129-Ucp1tm1Kz/J) on the C56BL/6J background were obtained from the Jackson Laboratory. For the experiments with AAC1−/− mice, we used WT C57BL/6J2z and C56BL/6J control mice with the same results. For all of the other knockout mouse strains C56BL/6J mice were used as control. All mice were aged 2 months to 1 year.
C2C12 cell culture and generation of AAC1/AAC2 knockout cells
The original WT C2C12 cell line (immortalized mouse myoblast cell line) was purchased from ATCC (https://www.atcc.org/) and cultured in complete Dulbecco’s modified Eagle’s medium (DMEM; DMEM with 25 mM glucose, 10% fetal bovine serum (FBS), penicillin and streptomycin antibiotics). The C2C12 cell line was authenticated by ATCC. The AAC1/AAC2 DKO C2C12 cells were generated by Alstem (http://www.alstembio.com/.) using the CRISPR–Cas9 system7. Alstem provided initial authentication of AAC1/AAC2 DKO C2C12 cells using PCR. We provided further authentication of the DKO cells with western blotting. All cell lines were tested negative for mycoplasma contamination. Transfection of gRNA Cas9 plasmids was performed using the Invitrogen Neon transfection system. gRNA candidates were selected using PCR amplification and selected for single-cell clone isolation.
Isolation of mitochondria and mitoplasts from tissues
Mice were euthanized by CO2 asphyxiation followed by cervical dislocation. For the preparation of mitoplasts from heart, skeletal muscle, brown fat, kidneys and liver, the selected mouse tissues were isolated, rinsed and homogenized in ice-cold medium containing 250 mM sucrose, 10 mM HEPES, 1 mM EGTA and 0.1% bovine serum albumin (BSA) (pH adjusted to 7.2 with Trizma base) using a glass grinder with six slow strokes of a Teflon pestle rotating at 275 (soft tissues) or 600 (fibrous tissues) rotations per minute. The homogenate was centrifuged at 700g for 5–10 min to pellet nuclei and unbroken cells. For some tissues, the first nuclear pellet was resuspended in the same solution and homogenized again to increase the yield of mitochondria. Mitochondria were collected by centrifugation of the supernatant at 8,500g for 10 min.
Mitoplasts were produced from mitochondria using a French press. In brief, mitochondria were suspended in a solution containing 140 mM sucrose, 440 mM d-mannitol, 5 mM HEPES and 1 mM EGTA (pH adjusted to 7.2 with Trizma base) and then processed using a French press at 1,200–2,000 psi to rupture the outer membrane. Mitoplasts were pelleted at 10,500g for 15 min and resuspended for storage in 500 μl of solution containing 750 mM KCl, 100 mM HEPES and 1 mM EGTA (pH adjusted to 7.2 with Trizma base). Mitochondria and mitoplasts were prepared at 0–4 °C and stored on ice for up to 5 h. Immediately before the electrophysiological experiments, 15–50 μl of the mitoplast suspension was added to 500 μl solution containing 150 mM KCl, 10 mM HEPES and 1 mM EGTA (pH adjusted to 7.0 with Trizma base) and plated on 5 mm coverslips pretreated with 0.1% gelatin to reduce mitoplast adhesion.
Patch-clamp recordings
Whole-IMM patch-clamp recording was performed from isolated mitoplasts2,7,19. The mitoplasts used for patch-clamp experiments were 3–5 μm in diameter and typically had membrane capacitances of 0.3–1.2 pF. Gigaohm seals were formed in the bath solution containing 150 mM KCl, 10 mM HEPES and 1 mM EGTA (pH 7 adjusted using Trizma base). Voltage steps of 250–500 mV and 1–50 ms were applied to break-in into the mitoplast and obtain the whole-mitoplast configuration, as monitored by the appearance of capacitance transients. Mitoplasts were stimulated every 5 s. Currents were normalized to membrane capacitance to obtain current densities (pA pF−1).
All indicated voltages are on the matrix side of the IMM (pipette solution) as compared to the cytosolic side (bath solution, defined to be 0 mV). Normally, currents were induced by a voltage ramp from −160 mV to +100 mV to cover all physiological voltages across the IMM. In the figures, voltage protocols are shown above the current traces. Currents flowing into mitochondria are shown as negative, whereas those flowing out are positive. Membrane capacitance transients observed after the application of voltage steps were removed from current traces.
Both the bath and pipette solutions were formulated to record H+ currents and contained only salts that dissociate into large anions and cations normally impermeant through ion channels or transporters. The pipettes were filled with 130 mM tetramethylammonium hydroxide, 1.5 mM EGTA, 2 mM Tris chloride and 100 mM HEPES. pH was adjusted to 7.5 with d-gluconic acid, and tonicity was adjusted to around 360 mmol kg−1 with sucrose. Typically, pipettes had resistances of 25–35 MΩ. and the access resistance was 40–75 MΩ. Whole-mitoplast IH was recorded in the bath solution containing 100 mM HEPES and 1 mM EGTA (pH 7.0 with Trizma base, and tonicity ~300 mmol kg−1 with sucrose). Whole-mitoplast K+ current activated by valinomycin was recorded in the bath solution containing 50 mM KOH, 182 mM HEPES and 1 mM EGTA (pH 7.0 with HEPES, tonicity 300 mmol kg−1).
In the absence of significant cytosolic triglyceride storage in non-adipose tissues, membrane phospholipid hydrolysis may be an important source of free FA for IH activation in vivo. To record IH through UCP1, before the application of uncouplers, the endogenous membrane FA were removed by a 30–40 s pretreatment with 10 mM MβCD.
In the experiments that compared IH activated by protonophores in IMM versus plasma membrane, whole-IMM measurements were performed from skeletal muscle mitoplasts as described above. Whole-cell plasma membrane H+ current measurements were performed from HEK293 cells. Gigaohm seals with HEK293 cells were formed in a Ringer solution containing 150 mM NaCl, 4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, 5.6 mM glucose and 25 mM HEPES (pH 7.4 with NaOH). Whole-IMM and whole-cell IH measurements were performed under identical recording conditions. A large pH gradient was set up between the bath solution (pH 6.0) and pipette solution (pH 7.5) to facilitate the detection of small H+ currents. Currents were expressed as current densities (pA pF−1) to enable comparison between the small mitoplasts and much larger plasma membranes. The pipettes were filled with 130 mM tetramethylammonium hydroxide, 1.5 mM EGTA, 2 mM Tris chloride and 100 mM HEPES (pH 7.5 with d-gluconic acid, tonicity of around 360 mmol kg−1 with sucrose). The bath solution contained 100 mM MES and 1 mM EGTA (pH 6.0 with Trizma base, and tonicity of around 300 mmol kg−1 with sucrose).
All of the experiments were performed under continuous perfusion of the bath solution. All electrophysiological data presented were acquired at 10 kHz and filtered at 1 kHz.
C2C12 mitochondrial isolation
C2C12 cells (WT or DKO) from 10 15 cm dishes were trypsinized and collected with ice-cold PBS at 1,000g for 5 min. The cell pellet was incubated with hypotonic buffer (20 mM HEPES, pH 7.5, 5 mM KCl, 1.5 mM MgCl2 and 1 mg ml−1 essentially FA-free BSA) on ice for 10 min. Cells were then homogenized (25 strokes) using a tight-fitting glass dounce tissue grinder and rapidly made isotonic by adding the homogenate to 2/3 volume of 2.5× MSH (20 mM HEPES, pH 7.5, 525 mM mannitol, 175 mM sucrose, 5 mM EDTA, and 1 mg ml−1 essentially FA-free BSA). The homogenate was centrifuged at 600g for 10 min, and the supernatant was then centrifuged at 8,500g for 10 min to pellet mitochondria. Mitochondria were resuspended in MSH buffer (20 mM HEPES, pH 7.5, 210 mM mannitol, 70 mM sucrose and 2 mM EDTA). Mitochondrial protein was quantified using the bicinchoninic acid assay (Thermo Fisher Scientific).
Mitochondrial respiration using the Seahorse Extracellular Flux Analyzer
In a volume of 50 μl, 30 μg of mitochondrial protein was added to each well of a XF cell culture microplate (on ice). The mitochondria-loaded XF microplate was centrifuged at 2,000g for 20 min for adherence of organelles. Respiration buffer (20 mM Tris, pH 7.4, 210 mM mannitol, 70 mM sucrose, 0.1 mM EGTA, 3 mM MgCl2, 5 mM KH2PO4, 0.1% essentially FA-free BSA, 10 mM sodium pyruvate and 5 mM malate) was added to a final volume of 0.5 ml, and the XF microplate was warmed at 37 °C for 8 min before loading into a XFe24 Extracellular Flux Analyzer (Seahorse Bioscience). The OCRs were recorded using a mix (20 s) and measure (2 min) cycle.
Mitochondrial respiration using a Clark electrode
Mitochondrial respiration was assessed in a 0.4 ml continuously stirred chamber containing the standard incubation medium, containing 125 mM KCl, 0.5 mM MgCl2, 3 mM KH2PO4, 10 mM HEPES, pH 7.4, 10 μM EGTA and supplemented with 3 mM pyruvate plus 1 mM malate. The chamber was maintained at 37 °C and was equipped with a custom-made Clark-type oxygen electrode and a tightly sealed lid. The slope of the oxygen consumption trace corresponded to the respiratory rate. Linear regression was performed on sections of the respiration traces for the 60 s immediately preceding and immediately after the addition of DNP to the mitochondria. The linear equation yielded during each linear regression analysis was used to determine the change in volts per minute for the trace segment of interest. Using the total known scale in volts for each trace and an experimentally determined value, in nmol, for the amount of oxygen dissolved in an aqueous solution at 37 °C and 1 atm, electrical potential was converted to nmol O2. The known amount of dissolved O2 under normal air pressure in 0.15 M KCl solution at 37 °C has previously been determined to be 162.4 nmol O2 per ml (ref.47). Finally, the amount of nmol O2 consumed per minute was normalized to mitochondrial protein (275 μg mitochondria/400 μl volume of the chamber).
Cell respiration analysis using the Oroboros O2k respirometer
C2C12 cell culture was maintained in DMEM supplemented with 25 mM glucose, 1 mM sodium pyruvate, 4 mM GlutaMAX and 10% FBS. The respiration medium was DMEM (containing 5.6 mM glucose, 1 mM sodium pyruvate, 4 mM l-glutamine, 20 mM HEPES, 10% FBS and no phenol red) with approximately 325 mOsm, at around pH 7.15. The cell concentration in each 2 ml respiration chamber was 5 × 106 cells per ml. The temperature was maintained at 37 °C and the cell suspension was stirred continuously at 750 rpm. After the oxygen consumption rate reached steady-state, 1 μM oligomycin was first applied to inhibit ATP synthase. Increasing concentrations of uncouplers (FCCP, 2,4-DNP and BAM15) or valinomycin were then added as indicated. Nigericin (K+/H+ antiporter, 1 μM) was added to reduce matrix K+ accumulation and mitochondrial swelling caused by valinomycin27. At the end of the experiments, 1 μM rotenone and 2.5 μM antimycin were applied to inhibit mitochondrial respiration and determine the non-mitochondrial respiration.
Immunoblot analysis
For western blot analysis, cells were lysed in RIPA buffer (1% igepal, 0.1% sodium dodecyl sulfate, 0.5% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 50 mM Tris-HCl (pH 7.4) and a cocktail of protease inhibitors). Lysates were resolved by SDS–PAGE, transferred to PVDF membrane (Millipore), and probed with anti-Na+/K+-ATPase (Abcam, ab76020, GR3237646–13, 1:10,000), anti-TOMM20 (Sigma Prestige, HPA011562, 000012083, 1:500), anti-AAC1 (Abcam, ab110322, R4759, 1:500) and anti-AAC2 (Cell Signaling Technology, CST 14671, 1:500) antibodies and OXPHOS antibody cocktail for mitochondrial respiratory content (Abcam, ab110413, MS604/Q5039, 1:500, NDUFB8 (complex I), SDHA (complex II), core 2 subunit (complex III), CIV-I subunit (complex IV) and ATP5A (complex V)).
Cell lines
The C2C12 WT cell line was purchased from ATCC (https://www.atcc.org/). These WT C2C12 cells were used to generate the double AAC1/AAC2 KO C2C12 (DKO) cell line. The DKO cell line was generated by Alstem (http://www.alstembio.com/) using the Crispr–Cas9 system7. The C2C12 cell line was authenticated by ATCC. The AAC1/AAC2 double KO C2C12 (DKO) cell line was generated and first authenticated by Alstem. We provided further authentication of the DKO cells with western blotting. All cell lines tested negative for mycoplasma contamination. The HEK293T cells (Homo sapiens) were provided and authenticated by the UCSF Cell Culture Facility (https://cgec.ucsf.edu/ cell- culture-and-banking-services).
Chemicals
The following chemicals were used in this study: carboxyatractyloside potassium salt (CATR, Sigma-Aldrich,4992), bongkrekic acid triammonium salt (BKA, Millipore 203671), guanosine 5′-diphosphate tris salt (GDP, Sigma-Aldrich, G7252), adenosine 5′-diphosphate sodium salt (ADP, Sigma-Aldrich, A2754), adenosine 5′-triphosphate disodium salt hydrate (ATP, Sigma-Aldrich, A6419), methyl-β-cyclodextrin (MβCD, Sigma-Aldrich, C4555), BAM15 (Sigma-Aldrich, SML1760) and SF6847 (CAS10537-47-0, Calbiochem, Sigma-Aldrich).
Statistical analysis
Data are presented as mean ± s.e.m. as specified in the figure legend. Statistical analysis was performed using GraphPad Prism 8. Statistical significance with exact P values was determined using the method indicated in the corresponding figure legend.
Docking methods
The Small Molecule Drug Discovery Suite 20191 (Schrödinger) was used for all docking calculations. The protein was prepared in a ready-to-dock-format with the Protein Preparation Wizard workflow. For docking to the c-state, PDB 2C3E (ref. 36) was used, whereas docking to the m-state used the homology model prepared for molecular dynamics simulations. Protein hydroxyl group orientations and protonation states were assigned at pH 7 using PROPKA48 in Epik v.4.7. Small molecules were drawn using Maestro v.11.9, and their protonation states and tautomers were assigned at pH 7.0 ± 3 in Epik v.4.7. Geometry was optimized and partial charges were assigned using Jaguar v.10.3 using the B3LYP-D3 density functional49 to account for non-covalent dispersion interactions with the 6-31G** and 6-31**+ basis sets for neutral states and anions, respectively, with ultra-fine level of accuracy of the calculation grid. Other parameters for proteins and small molecules used in the docking experiments came from the OPLS3e force-field50. A 30 × 32 × 30 Å docking grid was erected in the central cavity, and molecules docked in flexible mode allowing bond rotation while enforcing aromatic planarity restricting amide groups to trans, treating halogens as hydrogen-bond acceptors, and aromatic hydrogens as donors. For docking into the c-state, the protein was held rigid and the Glide XP51 scoring function was used, including strain energy for the ligands in the final score and returning a maximum of five optimal poses. For docking into the m-state, the induced fit workflow was applied, using the alanine trimming procedure along with the Glide SP scoring function for rescoring, and a maximum of 20 poses were returned. Final poses were picked on the basis of a combination of visual inspection, chemical intuition and score.
Molecular dynamics simulations
Simulations of the c-state of AAC1 were prepared using PDB 2C3E (ref. 36), while simulations of the m-state were prepared using a homology model based on PDB 6GCI (ref. 40), with the final simulation model in both cases consisting of residues 1–297 of bovine AAC1 and 3 bound cardiolipin lipids. Homology modelling of the bovine AAC1 m-state was performed using the automated SWISS-MODEL web server52, using the BKA-bound Thermothelomyces thermophilus AAC m-state structure (PDB: 6GCI) as a template (48% sequence identity to bovine AAC1). The inhibitors CATR and BKA found in the crystal structures were removed for all of the simulations. Four missing residues at the protein C terminus in PDB 2C3E were added manually in PyMOL (The PyMOL Molecular Graphics System, v.1.8 Schrödinger) as an extension of TM6. For both states, three tetra-linoleoyl cardiolipin lipids were built into lipid-binding sites manually in PyMOL using the partial lipid residues found in the crystal structures as templates. Structures were embedded in lipid bilayers using the membrane builder application of the CHARMM-GUI web server53. Some bilayers were built using POPC as the primary lipid, while others used a 1:1 mixture of PC:PE lipids with additional cardiolipin molecules to more closely mimic the composition of the mitochondrial inner membrane54 (Supplementary Table 3). The lipid composition of the bilayer had no apparent effect on the structure or ligand-binding properties of AAC1. For simulations using FA in the bilayer, FA molecules were also placed randomly using the CHARMM-GUI membrane builder. For simulations using DNP, the DNP molecule was placed either randomly in the aqueous portion of the simulation box, or in the AAC1-binding site on the basis of previous simulation runs. The force field parameters used for protein and lipid/FA were CHARMM36m55 and CHARMM3656, respectively. Water was represented with the TIP3P model57, and standard CHARMM ion parameters were used58. CHARMM-compatible parameters for neutral and deprotonated DNP were obtained for CgenFF (v.4.0)59 using the paramchem web server (https://cgenff.umaryland.edu/), and atomic partial charges were reoptimized using the Force Field Toolkit VMD plugin60 and gaussian09 (ref. 61). Additional simulation details are provided in Supplementary Table 3.
All molecular dynamics simulations were performed on local GPU computing resources using GROMACS (v.2018 or v.2020)62. Systems were initially minimized with 2,000 kJ mol−1 nm2 harmonic restraints on all protein heavy atoms and DNP or FA heavy atoms, when applicable, followed by a multistep equilibration in which protein restraints were gradually reduced over 10 ns. Production simulations were run using a 2 fs timestep, a Nose–Hoover (GROMACS v.2018) or stochastic-velocity rescaling (GROMACS v.2020) thermostat at 315 K (refs. 63-65), and a semi-isotropic Parrinello–Rahman barostat with a pressure of 1 atm (ref. 66). A 12 Å cut-off was used for Lennard–Jones interactions with force switching in the range of 10–12 Å, and long range electrostatics were calculated using the particle mesh Ewald method67. Bonds to hydrogen atoms were constrained using the LINCS algorithm68. Atomic coordinates were saved every 250 ps for all simulations. For systems simulated with an applied membrane potential, we used the constant electric field method where Eapplied = V/Lz where Lz is the length of the simulation box in the z dimension69, and the membrane potential was equilibrated at a rate of 10 mV per 5 ns. Simulation analysis was performed using the MDAnalysis Python package70. Molecular graphics were generated using the Visual Molecular Dynamics (VMD)71 and PyMOL software packages.
Electrostatic calculations
Continuum electrostatics calculations were carried out on a representative snapshot of bovine AAC1 extracted from a molecular dynamics simulation at 75 ns (ID 2 in Supplementary Table 3). The protein only was embedded in a membrane-like environment using APBSmem72 and calculations were performed using APBS73 with the PARSE parameter set for membrane voltage calculations and Swanson for ion solvation energy calculations (a list of additional parameters is provided in Supplementary Table 1; see ref. 72 for a more complete discussion of the electrostatic parameters, including the rationale for the standard dielectric values for the protein, membrane core and membrane headgroups used here). Ion solvation energies, accounting for all fixed and mobile charge interactions and the ion self-energy, were computed as described in ref.72 using nonlinear Poisson–Boltzmann theory (npbe), and the influence of the membrane potential was determined from separate calculations using the linearized form of the theory with appropriate boundary conditions (lpbe). Both calculations were added together to produce the composite curves shown in Fig. 4f.
Extended Data
Extended Data Fig. 1 ∣. DNP- and FCCP-induced mitochondrial IH.
a, IH densities induced by different DNP concentrations in heart IMM at −160 mV (n = 15 for 50 μM, n = 9 for 100 μM, n = 8 for 200 μM, n = 14 for 500 μM). Data are mean ± SEM. b, IH induced by 50 μM DNP in heart IMM before and after application 4 μM BKA (left). Remaining fraction of IH induced by 50 μM DNP at −160 mV after inhibition by 4 μM BKA, n = 3 (right). Data are mean ± SEM. c, IH induced by 500 μM DNP on the background of 10 mM MβCD in heart IMM before and after application of 1 μM CATR (left). IH densities induced by 50 μM DNP alone (n = 15)and 500 μM DNP on the background of 10 mM MβCD (n = 6) (right). Data are mean ± SEM. d, IH induced by 250 nM FCCP in heart IMM was inhibited by 1 μM CATR. e, Remaining fraction of FCCP-induced IHat −160 mV after inhibition by 1 μM CATR. Control (100% IH) is shown in gray. Data are mean ± SEM. Paired t-test, two-tailed, control vs. 1 μM CATR, n = 6.
Extended Data Fig. 2 ∣. SF6847 and BAM15 induce IH via AAC.
a, IH induced by 25 nM SF6847 across WT heart IMM before and after application 1 μM CATR. b, IH induced by 25 nM SF6847 in AAC1−/− heart IMM. c, Remaining fraction of IH induced by 25 nM SF6847 at −160 mV after inhibition by 1 μM CATR. Data are mean ± SEM. Paired t-test, two-tailed, control vs CATR, n = 6. d, IH densities induced by 25 nM SF6847 at −160 mV in WT (n = 10) and AAC1−/− (n = 5) heart IMM. IH was measured at −160 mV. Data are mean ± SEM. Mann–Whitney U-test, two-tailed, WT vs AAC1−/−. e, IH induced by 250 nM BAM15 across WT heart IMM before and after application 1 μM CATR. f, IH induced by 250 nM BAM15 in AAC1−/− heart IMM. g, Remaining fraction of IH induced by 250 nM BAM15 at −160 mV after inhibition by 1 μM CATR. Data are mean ± SEM. Paired t-test, two-tailed, control vs CATR, n = 4. h, IH densities induced by 250 nM BAM15 at −160 mV in WT (n = 5) and AAC1−/− (n = 5) heart IMM. IH was measured at −160 mV. Data are mean ± SEM. Mann–Whitney U-test, two-tailed, WT vs AAC1−/−. i, j, Representative K+ currents induced by 100 nM valinomycin across heart IMM in WT (i) and AAC1−/− (j). k, K+ current densities induced by 100 nM valinomycin at −160 mV in WT andAAC1−/− heart IMM (n = 5). Data are mean ± SEM.
Extended Data Fig. 3 ∣. DNP- and BAM-induced mitochondrial uncoupled respiration require AAC.
a, Oxygen consumption rate (OCR) of mitochondria isolated from WT and DKO C2C12 cells in the presence of oligomycin as measured with the Seahorse Analyzer. Each point corresponds to an individual respiration well, n = −60 (WT), and n = 56 (DKO). Mann–Whitney U-test, two- tailed. Data represent mean ± SEM. b, c OCR increase observed in mitochondria isolated from WT and DKO C2C12 cells after addition of DNP, n = 20, WT and n = 17, DKO (b) or BAM15, n = 10, WT and n = 10, DKO (c) in the presence of oligomycin. Each point corresponds to OCR change in an individual well of the Seahorse Analyzer, n = 17 – 20 (DNP) and n = 10 (BAM15). Data are mean ± SEM. Mann–Whitney U-test, two-tailed. d, OCR time course of isolated mitochondria from WT (n = 20 wells, blue) and DKO C2C12 cells (n = 17 wells, red). Basal respiration (1), addition of oligomycin (2), addition of DNP (3) and addition of rotenone/antimycin (4). Data represent mean ± SEM. e, Basal OCR of mitochondria isolated from WT or DKO C2C12 cells measured with a Clark electrode (n = 22, WT and n = 14, DKO). Mann–Whitney U-test, two-tailed. Data represent mean ± SEM. f, OCR increase above basal, observed using a Clark electrode after addition of DNP to mitochondria isolated from WT and DKO C2C12 cells; n = 3 (WT) and n = 5 (DKO) for 5 μM; n = 5 (WT) and n = 4 (DKO) for 12.5 μM. Data are mean ± SEM. Mann–Whitney U-test, two-tailed. g, Left panel: Immunoblots of WT (n = 5) and DKO (n = 5) C2C12 cells showing expression of the electron-transport chain complexes complex I (CI) – complex V (CV), TOM20, and the loading control (plasma membrane Na+/K+ ATPase). Right panel: The same immunoblots at longer exposure for the bands corresponding to complexes I-V. h, TOM20 protein expression level relative to Na+/K+ ATPase for the immunoblot shown in (g). Data shown as mean ± SEM, n = 5. Two-tailed unpaired t-test, WT vs DKO. i–m, Protein expression levels for complexes I-V relative to TOM20 for the immunoblots shown in (g). Data shown as mean ± SEM, n = 5. Two-tailed unpaired t-test, WT vs DKO. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4 ∣. Respiration induced by mitochondrial uncouplers in intact cells depends on AAC.
a, Oxygen consumption rate (OCR) of WT (n = 20) and DKO C2C12 cells (n = 16) before and after addition of oligomycin (oligo). Two-tailed unpaired t-test, WT vs DKO. b–d, OCR increase observed in WT and DKO C2C12 cells upon application of various concentrations of FCCP (n = 4), DNP (n = 4), and BAM15 (n = 4). All OCRs were measured in the presence of oligomycin. Two-tailed unpaired t-test, WT vs DKO. e, AAC-dependent fraction of uncoupled respiration at various concentrations of FCCP, DNP, and BAM15. All data shown as mean ± SEM, n = 4. f, OCR increase observed in WT and DKO C2C12 cells upon application of various concentrations of valinomycin. All OCRs were measured in the presence of oligomycin. Nigericin (1 μM) was added to reduce matrix K+ accumulation and mitochondrial swelling caused by valinomycin27. Two-tailed unpaired t-test, WT vs DKO, n = 4.
Extended Data Fig. 5 ∣. Adenine nucleotides inhibit DNP-induced IH.
a, DNP-induced IH at various ATP concentrations on the cytosolic face of the IMM of brown fat. IH was activated with either 500 μM DNP (left panel) or 5 mM DNP (middle panel) applied in the presence of 10 mM MβCD. Control currents were recorded in 10 mM MβCD. Right panel is the dose-dependence of IH inhibition by ATP at 500 μM DNP (red, n = 5) and 5 mM DNP (blue, n = 6).Current amplitudes were measured at −160 mV. Data are mean ± SEM. b, Left panel, IH induced by 50 μM DNP in control (1), upon transient inhibition by bath ADP (2), and upon subsequent recovery in the presence ADP (3). Heart mitoplast. Middle panel, DNP-dependent IH time course of left panel. IH was measured at −160 mV. Right panel, ADP inhibition of IH at points (2) and (3) in left and middle panels. Data are mean ± SEM. Paired t-test, two-tailed, ADP (2) vs. control, n = 4. c, IH activated by 50 μM DNP was inhibited by bath ADP. Pipette solution contained 1 mM ADP. Heart mitoplast. Middle panel, IH time course of the left panel. Right panel, ADP inhibition of IH in point (2) as in left and middle panels. Data are mean ± SEM. Paired t-test, two-tailed, n = 4.
Extended Data Fig. 6 ∣. Binding mode comparisons of CATR, ADP, small molecule uncouplers, and fatty acid.
a–d, Comparison of binding poses in the AAC1 c-state (2C3E) cavity of (a) docked ADP and docked DNP; (b) crystallographic CATR and docked DNP; (c) FA (arachidonic acid) pose from MD simulation and docked ADP; and (d) FA pose from MD simulation and crystallographic CATR. TM helices 1 and 6 of AAC1 are hidden. ADP and CATR are shown as thinner sticks with mauve carbon atoms. DNP and FA are shown as thicker sticks with yellow carbon atoms. e, Full final docked DNP, FCCP, BAM15, and SF6847 poses in their deprotonated and protonated forms (left) as in Fig. 4c and titratable heavy atoms (pink spheres) from these poses (right). TM helix 1 of AAC1 is hidden. f, ADP binding mode to the c-state as predicted from docking. g, Simulation snapshot showing arachidonic acid (solid spheres) bound to the TM5/6 fenestration of AAC1 in the c-state superimposed with the docked pose of DNP (transparent spheres). h, Simulation snapshot showing two arachidonic acid molecules bound simultaneously to the AAC1 c-state cavity, one in the TM5/6 fenestration and the other in the small molecule uncoupler site.
Extended Data Fig. 7 ∣. Docking reveals a common binding site for small molecules and nucleotide in both c- and m-states of AAC1.
a–j, Final docking poses to the AAC1 c-state (a–e) and m-state (f–j) structures of deprotonated forms of DNP, FCCP, BAM15, and SF6847, as well as ADP, respectively. In e and j ADP phosphate groups are hidden for clarity but were included in the docking. The same protein sidechains are shown as sticks in all panels.
Extended Data Fig. 8 ∣. DNP binds to the c-state of AAC1 in MD simulations.
a, illustration of the simulation setup used to assess binding of negatively charged DNP to the c-state of AAC1. DNP is shown in sphere representation in the aqueous region of the simulation box. AAC1 is shown as a cyan ribbon, with TM1 and TM6 hidden, and lipid molecules are shown as sticks. The Cζ atom of AAC1 residue R234 at the base of the c-state cavity is shown as a black sphere. Binding was tracked in b–d by monitoring the distance from the DNP centre of mass to AAC1 R234 Cζ atom indicated by the dotted line. b, Trajectories of negatively charged DNP initially placed in solution far from the binding site. c, Trajectories of neutral DNP, initially placed in the binding site. d, Trajectories of negatively charged DNP initially in the binding site, with an applied −160 mV membrane potential. In each plot, the two different colored traces are measurements made from two independent simulation trajectories. Simulations in c were initiated from docking poses, while those under an applied −160 mV potential in d were initiated from the final snapshots of the DNP binding simulations in b.
Extended Data Fig. 9 ∣. Fatty acids bind to the c-state of AAC1 via a TM5/6 fenestration.
a, Arachidonic acid transiently bound to the AAC1 region identified as the DNP/small molecule binding site in contact with protein residue Y186; snapshots are from two independent simulation trajectories. View is from the membrane with TM5 & TM6 of AAC1 hidden to show the cavity. b, Top view of structures in panel a viewed from the cytoplasm. c, Final states of four arachidonic and three palmitic acids bound to the fenestration between AAC1 helices TM5 & TM6; snapshots are from 7 independent simulation trajectories. d, Top view of structures in panel c. in panels a–d, AAC1 is shown as a cyan ribbon and FAs are shown as sticks with yellow carbon and red oxygen atoms. e, Side view and f, top view of a single structure from panels c and d with FA atoms shown as spheres and bilayer lipids shown as sticks. Carbon atoms 1–6, 7–12, and 13–20 of the arachidonic acid are colored yellow, blue, and pink, to highlight the parts that are inside the AAC1 cavity, in the TM5/TM6 fenestration, and interacting with bilayer lipids, respectively.
Extended Data Fig. 10 ∣. Calculation of membrane potential profiles and mathematical model results for AAC1.
a, Membrane potential profile (V(z)) across c-state (left) and energy values along the central pore for a monovalent cation (∣e∣·V(z)) (right). The profiles were computed under a −160 mV applied voltage using Poisson-Boltzmann theory (see Materials and Methods) on a configuration extracted from the equilibrated MD simulations. Colour gradients represent isocontours of linearly spaced scalar values ranging from 0 mV (upper region of graphic) to −160 mV (lower region). The water filled cavity facing upward supports a minor fraction of the field with the majority of the field focused across the closed matrix gate of the c-state. These values are quantified in the right panel showing the energy of interaction for a monovalent cation moving through the membrane field. b–e, Steady state single-transporter currents as a function of voltage (b, c), pKa (d), and rate k23 of proton transfer from the uncoupler to matrix pathway (e) for the 3-state model shown in Fig. 4g using values in Supplementary Table 2 unless otherwise indicated in the panel. The electrostatic potential in the cavity (Eelec) was varied: 10 kBT (blue), 8 kBT (gold), 6 kBT (green) (b and d). The fraction of the membrane voltage traversed by a H+ entering from the cytosol was varied: f = 0.1 (blue), 0.25 (gold), and 0.5 (green) (c). The energy of the proton in the matrix pathway (E1) was varied: 0 kBT (blue), 5 kBT (gold), 10 kBT (green) (e).
Supplementary Material
Acknowledgements
We thank D. C. Wallace for providing AAC1-knockout mice; A. Angelin for performing pilot mitochondrial respiration studies; and B. M. Spiegelman for the help with Seahorse respiration studies. This work was supported by NIH grants R01GM107710, R01GM118939 and R35GM136415 to Y.K., R01GM089740 and R01GM137109 to M.G., R01NS098772 to N.B., as well as a grant from the UCSF Program for Breakthrough Biomedical Research (PBBR) to M.G. and Y.K. A.M.B. was supported by an American Heart Association Career Development Award 19CDA34630062 and NIH grant R35GM143097. P.B. was supported by an American Heart Association Post-Doctoral Fellowship 18POST33960587. Simulations were carried out, in part, at the UCSF Wynton Cluster on hardware made possible through NIH grants 1S10OD021596 and R01GM089740 to M.G.
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-022-04747-5.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Competing interests Y.K., M.G., P.B. and J.S. are shareholders of Equator Therapeutics. Y.K. is an advisor to Equator Therapeutics. Y.K. is shareholder of YourChoice Therapeutics. E.T.C. is a founder, board member and equity holder in Matchpoint Therapeutics. The other authors declare no competing interests.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-022-04747-5.
Data availability
All data and materials are available on request from the corresponding authors.
Code availability
The MATLAB code for the mathematical model, force field and docking parameters for DNP, FCCP, BAM15, SF6847, CATR and the homology model of m-state bovine AAC1 are available at Zenodo (https://doi.org/10.5281/zenodo.5058463).
References
- 1.Divakaruni AS & Brand MD The regulation and physiology of mitochondrial proton leak. Physiology 26, 192–205 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Fedorenko A, Lishko PV & Kirichok Y Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 151, 400–413 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholls DG A history of UCP1. Biochem. Soc. Trans 29, 751–755 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Aquila H, Link TA & Klingenberg M The uncoupling protein from brown fat mitochondria is related to the mitochondrial ADP/ATP carrier. Analysis of sequence homologies and of folding of the protein in the membrane. EMBO J. 4, 2369–2376 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouillaud F, Weissenbach J & Ricquier D Complete cDNA-derived amino acid sequence of rat brown fat uncoupling protein. J. Biol. Chem 261, 1487–1490 (1986). [PubMed] [Google Scholar]
- 6.Klingenberg M & Huang SG Structure and function of the uncoupling protein from brown adipose tissue. Biochim. Biophys. Acta 1415, 271–296 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Bertholet AM et al. H+ transport is an integral function of the mitochondrial ADP/ATP carrier. Nature 571, 515–520 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreyev A et al. The ATP/ADP-antiporter is involved in the uncoupling effect of fatty acids on mitochondria. Eur. J. Biochem 182, 585–592 (1989). [DOI] [PubMed] [Google Scholar]
- 9.Brustovetsky N & Klingenberg M The reconstituted ADP/ATP carrier can mediate H+ transport by free fatty acids, which is further stimulated by mersalyl. J. Biol. Chem 269, 27329–27336 (1994). [PubMed] [Google Scholar]
- 10.McLaughlin SG & Dilger JP Transport of protons across membranes by weak acids. Physiol. Rev 60, 825–863 (1980). [DOI] [PubMed] [Google Scholar]
- 11.Terada H Uncouplers of oxidative phosphorylation. Environ. Health Perspect 87, 213–218 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry RJ, Zhang D, Zhang XM, Boyer JL & Shulman GI Controlled-release mitochondrial protonophore reverses diabetes and steatohepatitis in rats. Science 347, 1253–1256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry RJ et al. Reversal of hypertriglyceridemia, fatty liver disease, and insulin resistance by a liver-targeted mitochondrial uncoupler. Cell Metab. 18, 740–748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tainter ML, Stockton AB & Cutting WC Use of dinitrophenol in obesity and related conditions. J. Am. Med. Assoc 101, 1472–1475 (1933). [Google Scholar]
- 15.Grundlingh J, Dargan PI, El-Zanfaly M & Wood DM 2,4-Dinitrophenol (DNP): a weight loss agent with significant acute toxicity and risk of death. J. Med. Toxicol 7, 205–212 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowell BB & Spiegelman BM Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Krauss S, Zhang CY & Lowell BB The mitochondrial uncoupling-protein homologues. Nat. Rev. Mol. Cell Biol 6, 248–261 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Skulachev VP Uncoupling: new approaches to an old problem of bioenergetics. Biochim. Biophys. Acta 1363, 100–124 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Bertholet AM et al. Mitochondrial patch clamp of beige adipocytes reveals UCP1-positive and UCP1-negative cells both exhibiting futile creatine cycling. Cell Metab. 25, 811–822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertholet AM & Kirichok Y Patch-clamp analysis of the mitochondrial H+ leak in brown and beige fat. Front. Physiol 11, 326 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy SE, Chen YS, Graham BH & Wallace DC Expression and sequence analysis of the mouse adenine nucleotide translocase 1 and 2 genes. Gene 254, 57–66 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Rodic N et al. DNA methylation is required for silencing of ant4, an adenine nucleotide translocase selectively expressed in mouse embryonic stem cells and germ cells. Stem Cells 23, 1314–1323 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Cho J et al. Mitochondrial ATP transporter Ant2 depletion impairs erythropoiesis and B lymphopoiesis. Cell Death Differ. 22, 1437–1450 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham BH et al. A mouse model for mitochondrial myopathy and cardiomyopathy resulting from a deficiency in the heart/muscle isoform of the adenine nucleotide translocator. Nat. Genet 16, 226–234 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Childress ES, Alexopoulos SJ, Hoehn KL & Santos WL Small molecule mitochondrial uncouplers and their therapeutic potential. J. Med. Chem 61, 4641–4655 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Kenwood BM et al. Identification of a novel mitochondrial uncoupler that does not depolarize the plasma membrane. Mol. Metab 3, 114–123 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernardi P Mitochondrial transport of cations: channels, exchangers, and permeability transition. Physiol. Rev 79, 1127–1155 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Klingenberg M The ADP and ATP transport in mitochondria and its carrier. Biochim. Biophys. Acta 1778, 1978–2021 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Alexopoulos SJ et al. Mitochondrial uncoupler BAM15 reverses diet-induced obesity and insulin resistance in mice. Nat. Commun 11, 2397 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Axelrod CL et al. BAM15-mediated mitochondrial uncoupling protects against obesity and improves glycemic control. EMBO Mol. Med 12, e12088 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cannon B & Nedergaard J Brown adipose tissue: function and physiological significance. Physiol. Rev 84, 277–359 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Bertholet AM & Kirichok Y UCP1: a transporter for H+ and fatty acid anions. Biochimie 134, 28–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholls DG & Lindberg O Brown-adipose-tissue mitochondria. The influence of albumin and nucleotides on passive ion permeabilities. Eur. J. Biochem 37, 523–530 (1973). [DOI] [PubMed] [Google Scholar]
- 34.Klingenberg M UCP1—a sophisticated energy valve. Biochimie 134, 19–27 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Pebay-Peyroula E et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Nury H et al. Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett. 579, 6031–6036 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Ruprecht JJ et al. Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc. Natl Acad. Sci. USA 111, E426–E434 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kunji ER & Robinson AJ The conserved substrate binding site of mitochondrial carriers. Biochim. Biophys. Acta 1757, 1237–1248 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Dehez F, Pebay-Peyroula E & Chipot C Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J. Am. Chem. Soc 130, 12725–12733 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Ruprecht JJ et al. The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 176, 435–447 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bielawski J, Thompson TE & Lehninger AL The effect of 2,4-dinitrophenol on the electrical resistance of phospholipid bilayer membranes. Biochem. Biophys. Res. Commun 24, 948–954 (1966). [DOI] [PubMed] [Google Scholar]
- 42.Skulachev VP, Sharaf AA & Liberman EA Proton conductors in the respiratory chain and artificial membranes. Nature 216, 718–719 (1967). [DOI] [PubMed] [Google Scholar]
- 43.Skulachev VP et al. The effect of uncouplers on mitochondria, respiratory enzyme complexes and artificial phospholipid membranes. Curr. Mod. Biol 2, 98–105 (1968). [DOI] [PubMed] [Google Scholar]
- 44.Mitchell P & Moyle J Stoichiometry of proton translocation through the respiratory chain and adenosine triphosphatase systems of rat liver mitochondria. Nature 208, 147–151 (1965). [DOI] [PubMed] [Google Scholar]
- 45.Mitchell P Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Biol. Rev. Camb. Philos. Soc 41, 445–502 (1966). [DOI] [PubMed] [Google Scholar]
- 46.Betz MJ & Enerback S Human brown adipose tissue: what we have learned so far. Diabetes 64, 2352–2360 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Reynafarje B, Costa LE & Lehninger AL O2 solubility in aqueous media determined by a kinetic method. Anal. Biochem 145, 406–418 (1985). [DOI] [PubMed] [Google Scholar]
- 48.Olsson MH, Sondergaard CR, Rostkowski M & Jensen JH PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput 7, 525–537 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Grimme S, Antony J, Ehrlich S & Krieg H A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys 132, 154104 (2010). [DOI] [PubMed] [Google Scholar]
- 50.Roos K et al. OPLS3e: extending force field coverage for drug-like small molecules. J . Chem. Theory Comput 15, 1863–1874 (2019). [DOI] [PubMed] [Google Scholar]
- 51.Friesner RA et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem 49, 6177–6196 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Waterhouse A et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jo S, Kim T & Im W Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2, e880 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Comte J, Maisterrena B & Gautheron DC Lipid composition and protein profiles of outer and inner membranes from pig heart mitochondria. Comparison with microsomes. Biochim. Biophys. Acta 419, 271–284 (1976). [DOI] [PubMed] [Google Scholar]
- 55.Huang J et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klauda JB et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW & Klein ML Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 926–935 (1983). [Google Scholar]
- 58.Beglov D & Roux B Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J. Chem. Phys 100, 9050–9063 (1994). [Google Scholar]
- 59.Vanommeslaeghe K et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mayne CG, Saam J, Schulten K, Tajkhorshid E & Gumbart JC Rapid parameterization of small molecules using the Force Field Toolkit. J. Comput. Chem 34, 2757–2770 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frisch MJ et al. Gaussian 16 rev. C.01 (Gaussian, 2016). [Google Scholar]
- 62.Abraham MJ et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 63.Hoover WG Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 64.Nosé S A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys 52, 255–268 (1984). [Google Scholar]
- 65.Bussi G, Donadio D & Parrinello M Canonical sampling through velocity rescaling. J. Chem. Phys 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Parrinello M & Rahman A Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys 52, 7182–7190 (1981). [Google Scholar]
- 67.Darden T, York D & Pedersen L Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys 98, 10089–10092 (1993). [Google Scholar]
- 68.Hess B, Bekker H, Berendsen HJC & Fraaije JGEM LINCS. J. Comput. Chem 18, 1463–1472 (1997). [Google Scholar]
- 69.Gumbart J, Khalili-Araghi F, Sotomayor M & Roux B Constant electric field simulations of the membrane potential illustrated with simple systems. Biochim. Biophys. Acta 1818, 294–302 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michaud-Agrawal N, Denning EJ, Woolf TB & Beckstein O MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem 32, 2319–2327 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Humphrey W, Dalke A & Schulten K VMD: visual molecular dynamics. J. Mol. Graph 14, 33–38 (1996). 27–38. [DOI] [PubMed] [Google Scholar]
- 72.Marcoline FV, Bethel N, Guerriero CJ, Brodsky JL & Grabe M Membrane protein properties revealed through data-rich electrostatics calculations. Structure 23, 1526–1537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials are available on request from the corresponding authors.
The MATLAB code for the mathematical model, force field and docking parameters for DNP, FCCP, BAM15, SF6847, CATR and the homology model of m-state bovine AAC1 are available at Zenodo (https://doi.org/10.5281/zenodo.5058463).