Abstract
Filamentous fungi are ubiquitous and frequent components of biofilms. A means to visualize them and quantify their viability is essential for understanding their development and disruption. However, quantifying filamentous fungal biofilms poses challenges because, unlike yeasts and bacteria, they are not composed of discrete cells of similar size. This research focused on filamentous fungal biofilms that are representative of those in the built environment. The objective of this study was to develop a rapid method to examine biofilm structure and quantify live (metabolically active/ membrane undamaged) and dead (inactive/ membrane damaged) cells in Aspergillus niger biofilms utilizing a fluorescent probe staining method and confocal laser scanning microscopy (CLSM). For this, we compared two commercially available probe staining kits that have been developed for bacterial and yeast systems. One method utilized the classic cell stain FUN 1 that exhibits orange-red fluorescent intravacuolar structures in metabolically active cells, while dead cells are fluoresced green. The second method utilized a combination of SYTO9 and propidium iodide (PI), and stains cells based on their membrane morphology. SYTO9 is a green fluorescent stain with the capacity to penetrate the living cell walls, and PI is a red fluorescent stain that can only penetrate dead or dying cells with damaged cell membranes. Following staining, the biofilms were imaged using CLSM and biofilm volumes and thickness were quantified using COMSTAT, a computer program that measures biofilm accumulation from digital image stacks. The results were compared to independent measurements of live-dead cell density, as well as a classic cell viability assay-XTT. The data showed that the combination of SYTO9 and PI is optimal for staining filamentous fungal biofilms.
Keywords: Aspergillus niger, Cell viability, COMSTAT, Confocal laser scanning microscopy, Filamentous fungal biofilms, Fluorescent probes, XTT assay
1. Introduction
Biofilms are complex communities of microorganisms that are surface-associated or attached to one another and enclosed within a self-produced extracellular polymeric substance (EPS). The benefits of microbial biofilm formation include protection from environmental conditions, resistance development to physical and chemical stresses, metabolic cooperation between biofilm cells, and differential expression of genes and their regulation [[1], [2], [3]]. When in biofilms, microbial cells exhibit distinct physiological features that differ from their planktonic state. Hence, the establishment of biofilms is advantageous for processes such as fermentation and bioremediation [[4], [5], [6]], but at the same time disadvantages include the persistence of human disease, antimicrobial tolerance, and proliferation in medical devices [4,[7], [8], [9]]. In addition, biofilm cells can facilitate nutrient uptake, cell-to-cell signaling, and gene transfer [10]. Consequently, biofilm research on bacterial and yeast biofilms is well established [7,8,[11], [12], [13], [14], [15], [16], [17], [18], [19], [20]], but relatively fewer studies have focused on filamentous fungal biofilms.
Filamentous fungi are common contributors in robust biofilm formation because of their apical hyphal growth and surface-associated proliferation [2,21,22]. Biofilm formation by bacteria and yeast is similar, with stages that include surface adhesion, an initial proliferation of cells over the surface, microcolony formation, maturation, and dispersal of cells. The only exception for yeast is that a pseudo hyphal or hyphal growth is incorporated in the initial proliferation stage [2,22]. Biofilm formation by filamentous fungi is slightly different as they lack binary fission or budding processes, but instead, utilize hyphal tip growth for their biofilm establishment and have additional stages with mycelial development, hyphal layering, and hyphal bundling [2,23]. Here, the focus is on biofilms produced by the filamentous fungus Aspergillus niger, a model organism and an industrially and medically important species [4,6,[24], [25], [26], [27], [28]]. According to Harding et al. [2], the formation of filamentous fungal biofilms is divided into six distinct phases which includes adsorption, attachment to the surface, Microcolony formation I, Microcolony formation II or initial maturation, maturation or reproductive development, and dispersal or planktonic phase. These stages also apply to A. niger [[4], [5], [6],8,26]. Biotic and abiotic substrates with various environmental conditions also play a role in biofilm development [10,22]. In this study, A. niger biofilms were created to model those formed naturally under drip flow. A drip flow reactor (DFR) was used for biofilm production, as it creates a low shear environment that allows for liquid to flow along glass coupons and, imitates the conditions in industrial and household environments [19,29,30,55].
In previous studies, the assessment of cell viability in bacterial and fungal biofilms was determined by colorimetric assay methods such as 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl) 2H-tetrazolium-5-carboxanilide (XTT), and alamar blue. XTT and MTT assays involve the reduction of tetrazolium salts by mitochondrial dehydrogenase or ferric reductase. The cells are incubated with XTT or MTT assay which is converted by viable cells to a colored formazan product [31,32]. The metabolic activity of microbial cells is proportional to the formazan product’s color intensity, which is measured as the optical density of the assay [19,31,32]. For the XTT viability assessment, the amount of water-soluble formazan product is measured directly in the medium, while for the MTT assay, the treated cells are lysed with DMSO, before measurement of the optical density (OD) [12]. These assay methods are widely used to assess cell viability [12,[32], [33], [34], [35], [36], [37], [38], [39]]. The alamar blue cell viability indicator uses the natural reducing power of living cells to convert resazurin (blue compound) to resorufin which produces a bright pink-red fluorescence. The reduction of alamar blue is mediated by mitochondrial enzymes [[40], [41], [42]]. Although, this fluorogenic assay is mostly used as a viability assay for mammalian cells, it is also used for susceptibility testing of yeast and bacterial biofilms [40,42,43].
A reported drawback in some of these assays is that different yeast strains from the same species may show a marked variation in their ability to metabolize tetrazolium [12,18,19,32,44]. Moreover, during the different stages of biofilm formation, the metabolic state of microbes changes, which may lead to fluctuations in the ability of biofilm cells to metabolize chemical components during the assays. Vital stains (FUN 1, SYTO9, Propidium iodide) have been used as an alternative and have been found to have advantages over these assays. The main advantages include the possibility of multiple labeling with microbial cells and an extraordinary insight into the dynamics of labeled molecules within the microbial cells and their structures [3,12,19,32,35,37,40,42,45,46].
For visualizing and studying biofilm architectural features, fluorescence, scanning electron, and confocal microscopy techniques have been used. Fluorescence microscopy provides data on biofilm morphology and the emergence of the extracellular matrix during the biofilm formation, whereas scanning electron microscope (SEM) analysis allows the evaluation of elaborate topographical views of biofilm surface structure at high magnification. However, the fixation and dehydration steps performed during SEM sample preparation often degrade the biofilm matrix. This can be overcome by using confocal laser scanning microscopy (CLSM), which allows visualization of the three-dimensional structure at different biofilm depths. Moreover, it allows for measurements of the biofilm thickness without disrupting the biofilm architecture. Quantification of live and dead cells within the biofilm is ultimately important to better understand the dynamics of filamentous fungal biofilm formation, degradation, and prevention. To this end, fluorescent metabolic stains may be used with CLSM to evaluate these parameters while simultaneously examining the biofilm structure.
The objective of this study was to develop a straightforward method to quantify live and dead cells in A. niger biofilms utilizing a fluorescent probe staining method and CLSM. For this, we used two different types of commercially available probe staining kits that were developed separately for bacterial and yeast systems. One method utilizes the classical cell stain, FUN 1, that exhibits orange-red fluorescent intravacuolar structures in metabolically active cells, while dead cells fluoresce green due to the lack of metabolically active components [47,48]. The second method uses a combination of SYTO9 and propidium iodide (PI). SYTO9 is a nucleic acid stain [40,47,49] with a capacity to penetrate cell walls with relative ease and stain the cells green, regardless of their viability. In comparison, PI is a red fluorescent stain that penetrates the damaged cell membranes of dead or dying cells, causing quenching of the SYTO9 green fluorescence when both stains are used together [[47], [48], [49]]. The two kits have been used in previous studies to investigate antimicrobial susceptibility or to differentiate between dead (inactive/membrane damaged) and live cells (metabolically active/membrane integrity) in both fungal and bacterial biofilms [19,47,[49], [50], [51], [52], [53]]. These two staining methods were tested as they target a specific metabolic or biological/morphological component (i.e. nucleic acid) and can be used to differentiate and illuminate the live and dead cells within the images of biofilm samples [47,53,54].
These two probe staining methods were compared to determine their staining efficiencies for the differentiation of live and dead cells in A. niger biofilms. Two regions of A. niger biofilms, the center and corner portions, were selected for comparison studies and to better understand biofilm dynamics. Cell viability assessments with the fluorescent probe stains were further compared to a standard cell viability assay, XTT.
2. Materials and methods
2.1. Fungus strain, culture conditions, and inoculum
Aspergillus niger van Tieghem (ATCC 6275) was maintained in petri plates (90 mm diam.) containing Sabouraud dextrose agar (SDA; Difco) sealed with Parafilm and maintained at 21–25 °C. The spore suspension was prepared from 7-day-old cultures. Spores were dislodged by pipetting sterile distilled water in 1 mL increments in five different places onto the surface of the culture. The petri plate was gently agitated to dislodge the spores, and the spores were transferred to a 50 mL centrifuge tube containing phosphate-buffered saline (PBS), pH 7.4 (Electron Microscopy Sciences). The spores were quantified with a Neubauer chamber and the concentration was adjusted with PBS to 105 spores/mL. This suspension was used as the inoculum.
2.2. Aspergillus niger biofilm formation in a drip flow reactor system
Biofilms were produced under low-shear conditions following the reproducible protocol developed by Kerrigan et al. [55]. Briefly, to initiate biofilm formation, 10 mL batch medium (sterile 3% wt/vol Sabouraud dextrose broth (SDB; Difco)) and 1 mL inoculum were added aseptically to each channel of the drip flow reactor (DFR; BioSurface Technologies) containing a glass coupon (22 × 22 mm). The reactor was sealed, set at a 10° angle backward, to prevent the fungal structures from clogging the effluent ports, and left at room temperature (∼23 °C) for 48 h with 12 h alternating intervals of artificial light (normal room light) and darkness. To prepare the low shear biofilm run, the reactor was positioned at a 10° angle forward. The system was then connected to a carboy which contained freshly prepared sterile SDB (0.03% wt/vol) and the media was pumped through the reactor using a peristaltic pump (Shenchen- Lab V series) at a rate of 3 mL/min. The effluent port was connected to a vessel to collect the used media. The run was continued for 24 h under the same light and temperature regime described above.
2.3. Fluorescent probe stains and confocal laser scanning microscopy
To quantify the live and dead cells in Aspergillus niger biofilms, two different viability staining kits were compared to determine the optimal method; One method utilized the LIVE/DEAD™ Yeast Viability Kit (Molecular Probes-Invitrogen, USA; Cat# L7009) containing FUN 1 cell stain. FUN 1 viability stain can be used alone or together with Calcofluor White M2R (which is part of the kit) to determine the metabolic activity of fungal cells by fluorescence microscopy and for detecting fungi in complex mixtures or pure cultures [48]. For this study, only the FUN 1 stain from the kit was used. The FUN 1 stain exhibits orange-red fluorescent intravacuolar structures in metabolically active cells, while dead cells fluoresce green-yellow [47,48]. For staining, the glass coupons containing the biofilm were transferred to a petri plate containing a sterile wet filter paper to prevent the desiccation of the biofilm sample during the staining incubation. For each biofilm area to be examined, 2 μL of FUN 1 stain was pipetted directly onto the center and corner portions of the A. niger biofilms. The biofilms were covered with aluminum foil and incubated at 30 °C for 60 min in the dark. According to the manufacturer’s instructions, the recommended incubation time for FUN 1 is 30 min; however, for A. niger biofilm staining, a 60 min incubation time produced better imaging results.
The second method utilized LIVE/ DEAD™ BacLight™ Bacterial Viability Kit (Molecular Probes - Invitrogen, USA; Cat# L7007) containing SYTO9, a green fluorescent stain with a capacity to penetrate both live and dead cells, and propidium iodide (PI), a red fluorescent stain that may only penetrate dead or dying cells with damaged cell membranes. The glass coupon covered with biofilm was transferred from the reactor to the petri plate as described above. For biofilm staining, a freshly prepared solution of 6 μL SYTO9 and 6 μL PI was mixed with 1 mL DI water, according to the manufacturer’s instructions, and pipetted so that the entire surface of the biofilm was covered. The petri plates were covered with aluminum foil and incubated for 30 min in the dark at room temperature per the manufacturer’s instructions.
Following incubation, the stained biofilms were imaged using a Leica TCS SPE CLSM (Buffalo Grove, IL, USA). The SYTO9-PI fluorescent probes were excited with 488 nm laser having an intensity of 17.2557%. An emittance from 490 nm to 550 nm was detected using a photomultiplier tube with a gain of 670. FUN 1 was excited with a 488 nm laser having an intensity of 30.4828%. An emittance from 577 nm to 633 nm was detected using a photomultiplier tube with a gain of 1085. For comparison, live and dead cells in the center and corner portions of the biofilm were selected for imaging. Due to its square shape, the center and corner portions of the biofilm could be readily located under the microscope. The biofilm images were captured using 10x, 0.3 numerical aperture (NA) dry objective.
2.4. COMSTAT analysis and biofilm quantification
Biofilms were analyzed using the computer program, COMSTAT, which was written as a script in MATLAB (MathWorks- R2018a) for quantification of biofilm structures [56]. Prior to quantification, each image stack was thresholded. In COMSAT, image stack thresholding is performed by applying a fixed threshold value which was determined manually. For thresholding, COMSTAT loads a black and white sequence of the images of interest, and a threshold value, ranging anywhere from 10 to 50 was set. Based on the entered threshold value, a binary image with a pixel intensity of the original image was created. This image was then compared to the original grey scale set of images that came up first. The threshold was adjusted until the two matched closely. Once the images were thresholded, the data volume was analyzed. The data volume to be analyzed depends on the variables that were selected and the number of image stacks that were acquired using CLSM [56,57]. For the quantification of cell viability, we used the biomass image analysis feature. Biomass was determined as the volume of all voxels that contain biomass pixels in all images of stacks divided by the substratum area of the image stack [56,57]. This is an expression of how much of the image stack is covered by microbial biofilm. The unit for biomass is μm3/μm2. We calculated the biomass of live and dead cells of each biofilm sample from their image stacks separately and calculated the viability using biomass of live cells/ (biomass of live cells + biomass of dead cells). Cell viability is a dimensionless quantity.
2.5. XTT assay
After biofilm production, without disrupting the structure of the biofilm, the center and the corner portions were cut into 1 cm × 1 cm (length x width) squares using a steel scalpel blade. These biofilm portions were carefully transferred to a small petri plate (60 × 15 mm) containing 4 mL of PBS (pH 7.2; Electron Microscopy Sciences). The XTT (2,3-bis(2-methoxy-4-nitro-5-sulfophenyl) 2H-tetrazolium-5-carboxanilide) assay was prepared according to the method previously described [12,32]. Briefly, 5 mg XTT (Santa Cruz Biotechnology, USA; Cat# SC-258336) was dissolved in 1 mL 37 °C PBS and from this a 1:5 (vol/vol) was made in 37 °C pre-warmed PBS. A 10 mM menadione stock solution contained, 0.017 g menadione (Vitamin K3, Santa Cruz Biotechnology, USA; Cat# SC-205990) dissolved in 10 mL 100% acetone (J.T. Baker-liquid chromatography grade). Menadione 1:10 (vol/vol) was diluted in PBS to obtain a 1 mM working solution before use. To each petri plate containing a 1 cm × 1 cm biofilm portion and PBS, 50 μL of 1 mg/mL XTT working solution, and 4 μL of 1 mM menadione (Vitamin K3) working solution were added. The solution was mixed gently, and the plate was covered with aluminum foil and incubated for 5, 6, 7, and 8hrs at 37 °C on a benchtop incubator shaker (New Brunswick™ Innova® 40) at a of speed 25 rpm. Blanks for the spectrophotometer were prepared with the same constituents and incubation times as the XTT assay, but no biofilms were included. The mixture containing biofilm and assay solution was transferred to a sterile 15 mL centrifuge tube and centrifuged at 3500 G for 5 min at 4 °C (Legend Micro-Thermo Scientific). The supernatant solution (1 mL) was transferred to a 1 mL cuvette and the absorbance was measured at a wavelength of 492 nm using a UV-spectrophotometer (Jasco® V-550). The experiment was repeated eight times with two locations (center and corner) of the biofilms. The results were compared statistically to the viability results obtained from the two fluorescent probes methods to determine their accuracy.
2.6. Statistical analysis
Statistical analysis (F-test) was performed using the SAS statistical program, JMP Pro 14 (SAS Institute Inc, NC, USA). To determine if there are any significant differences in the cell viability between fluorescent probes, locations, and their interactions (fluorescent probes-locations) in fluorescent probe staining methods, three F-tests were performed. Additionally, one F-test was performed for the XTT assay to determine if there are any significant differences between the locations. For investigating significant differences between the fluorescent probes/assay and locations, the least square mean student’s t-test was performed. Statistically significant results were depicted by P values of <0.05.
3. Results
3.1. Sensitivity of fluorescent probes and biofilm cell viability
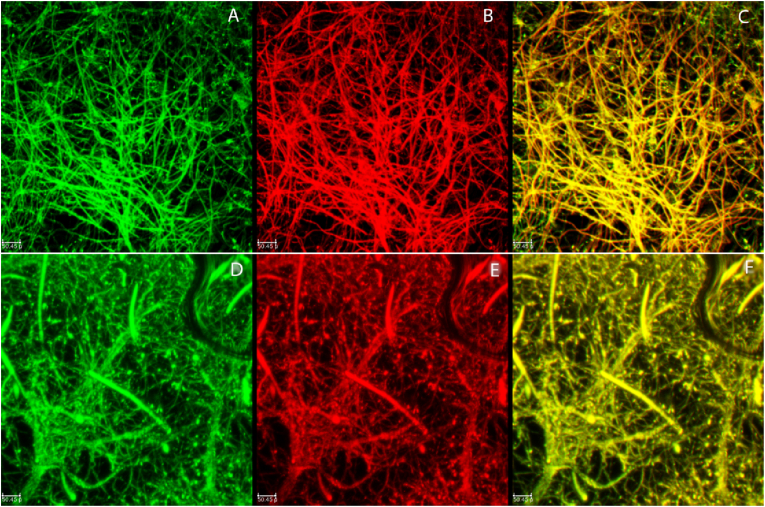
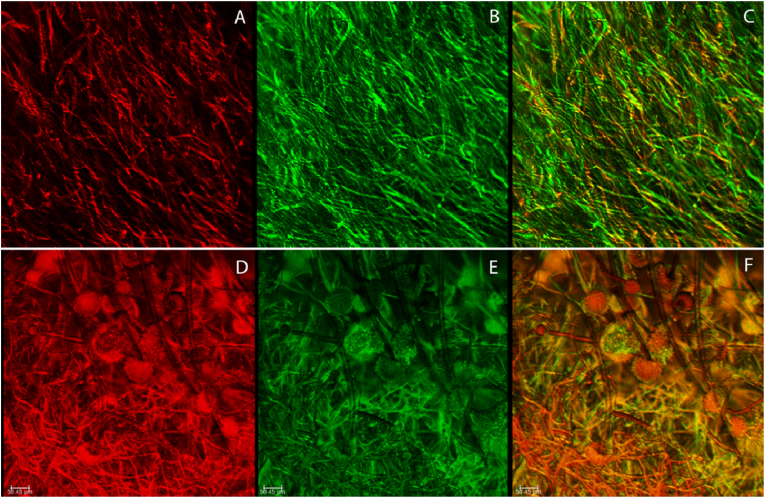
CLSM imaging was performed to compare the efficacy of two different live-dead fluorescent probe staining methods for use in determining cell viability in A. niger biofilms. The cell viability assessment was performed both at the center and the corner of the biofilm to understand the variation between the two locations (Fig. 1, Fig. 2, Fig. 3). The biofilm structures were dense from hyphae and the viability of the biofilm was not homogenous. From confocal images, the combination of the nucleic acid stains, SYTO9 and PI, showed a greater differentiation between the live and dead cells compared to the FUN 1 staining. FUN 1 stains metabolically active cells and emits an orange-red fluorescence (excitation/emission at 485/610 nm); however, in non-viable cells, the FUN 1 stain emission is blue-shifted and, green fluorescence is emitted (excitation/emission at 430/515 nm) (Fig. 1). In some cases, the orange-red fluorescence and the green fluorescence combine to form a yellow-orange color, which makes it difficult to distinguish between the live and dead cells (Fig. 1-center and Fig. 1-corner). As in Fig. 1, a masking effect is observed in the case of the FUN 1 stain. In fungal biofilms, the overestimation of live (metabolically active) cells was estimated based on multiple metabolic components present in the same cell, especially in high cell density. The z-stack images of FUN 1 support the overestimation of metabolically active cells. Due to this overlay in the image, it is difficult to distinguish distinct red and green location and this appears as a masking effect. However, in the SYTO9-PI combination, the live and dead cells can be easily differentiated as seen in Fig. 2. Also, visible in those figures is that the center portion of the biofilm contained more live cells compared to the corner portion of the biofilm. Notably the corner portion contained conidiogenous cells and conidiophores (Fig. 1A–F, Fig. 2A-F and Fig. 3A-F) that did not develop in the center portion.
Fig. 1.
CLSM images of the center portion (top) and corner portion (bottom) of the Aspergillus niger biofilms stained with FUN 1. (A & D) Green channel-Inactive/Dead cells, (B & E) Red channel metabolicaly active/Living cells (C & F) Overlay of both active/live and Inactive/dead cells. Scale bars = 50.45 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
CLSM images of the center portion (top) and corner portion* (bottom) of the Aspergillus niger biofilms stained with SYTO9 and PI. (A & D) Red channel - Inactive/Dead cells with PI (B & E) Green channel-Living/active cells with SYTO9. (C & F) Overlay of both active/live and Inactive/dead cells. Scale bars = 50.45μm.*- Presence of conidiogenous cells and conidiophore. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 3.
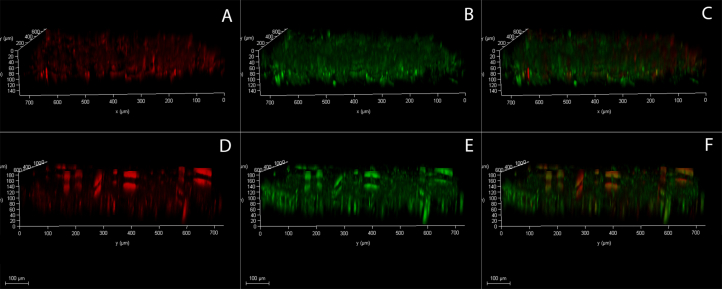
Vertical view of Aspergillus niger biofilm grown on a glass coupon center portion (top) and corner portion (bottom) stained with SYTO9 and PI. (A & D) Red channel - Inactive/Dead cells with PI (B & E) Green channel-Living/active cells with SYTO9. (C & F) Overlay of both active/live (SYTO9) and Inactive/dead (PI) cells. Scale bars = 200 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
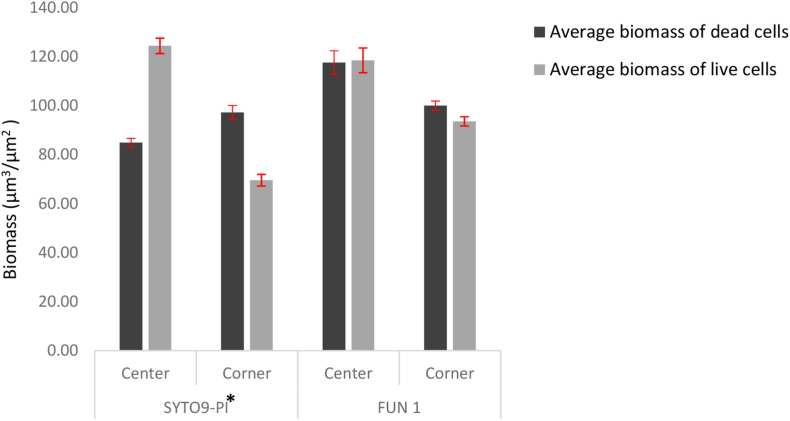
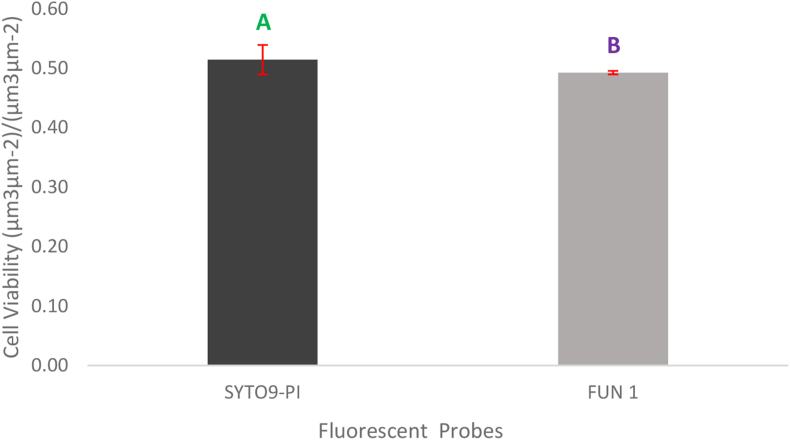
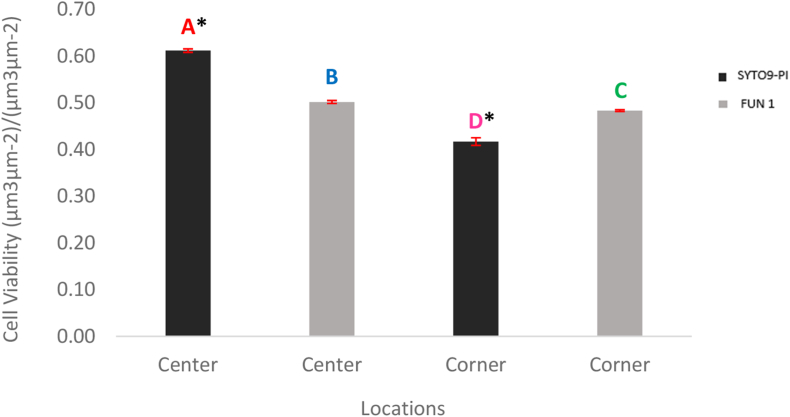
Live and dead cells biomass of biofilms were quantified using COMSTAT (Fig. 4). Three F-test were used for quantifying cell viability using fluorescent probe staining methods, two for the main effects (fluorescent probes and locations), and one for the interaction (fluorescent probes-locations). Based on the F-test, it was observed that there was a significant difference in the main two effects and the interaction. The F-test performed for the fluorescent probes - FUN 1 and the SYTO9-PI combination, was observed to have a value of p = 0.0005, meeting the criteria of p < 0.05 making the difference in the cell viability assessment statistically significant (Fig. 5). Also, a significant difference between the cell viability of the two locations of the biofilms was recorded (Fig. 6). Moreover, the F-test performed for the interaction was observed to have a value of P = 0.0001, meeting the criteria of P < 0.05 making it statistically significant. This indicates that the impact of cell viability assessment of the two fluorescent probes compared to each other was not consistent for the center and corner locations. Since the P-value from the F-test (fluorescent probes by locations interaction) came out as < 0.05, for further effect details, the least-square mean student’s t-test was performed. The least-square mean student’s t-test helps in understanding if there are any significant differences in the cell viability among the fluorescent probes with respect to their location interaction. From Fig. 6, a significant difference was observed between the cell viability in the center and the corner portion of A. niger biofilm stained with SYTO9-PI and FUN 1.
Fig. 4.
COMSTAT analysis of the live and dead cell biomass (μm3/μm2) of Aspergillus niger biofilms. Bars represent the average of the biomass measured with, two fluoresent probes (SYTO9-PI & FUN 1) at two locations (Center & Corner). Biomass average were calculated from eight replicates of each fluorescent probes at each location. *- In SYTO9 & PI stained biofilm, significant difference observed between the cellviability in the center and the corner portion.
Fig. 5.
A comparison of cell viability analysis of Aspergillus niger biofilms quantified with two fluorescent probes (SYTO9-PI & FUN 1). Bars represent the least square mean value and each error bar is constructed using one standard error from the mean. Bars represented by different letters are significantly different. Cell viability = Biomass of live cells/ (Biomass of live cells + Biomass of dead cells), calculated using COMSTAT.
Fig. 6.
The cell viability analysis of Aspergillus niger biofilm in fluorescent probes (SYTO9-PI & FUN 1)by location interaction model. Bars represent the least square mean value and each error bar is constructed using one standard error from the mean. Bars represented by different letters are significantly different. Cell viability = Biomass of live cells/ (Biomass of live cells + Biomass of dead cells), calculated using COMSTAT. *- SYTO9 & PI stained biofilm, the significant difference observed between the cell viability in the center and corner portion.
3.2. XTT viability for A. niger biofilms
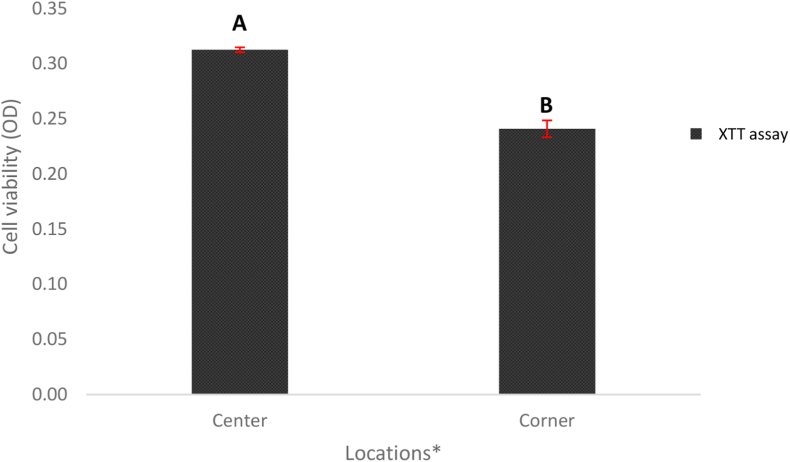
The metabolically active cells of the A. niger biofilms incubated with XTT assay yields a water-soluble formazan colored (gradient light orange) solution. Incubation times ranging from 5 to 8 h were tested for optimization, and optimum incubation time for A. niger biofilm was determined to be 7 h. The difference in OD values between the center and corner portion samples of XTT assay reflected what was recorded with the fluorescent probes (Fig. 7).
Fig. 7.
The cell viability of Aspergillus niger biofilms in the center and corner portions quantified with the XTT assay method. Bars represent the least square mean value and each error bar is constructed using one standard error from the mean. Bars represented by different letters are significantly different. Cell viability measured using UV-spectrophotometer. *- Significant difference observed between the cell viability in the center and the corner portion.
For the XTT assay, since only one assay is involved, the F-test models were performed on the cell viability assessment for the location effect. However, the F-test performed for the location (Fig. 7) was observed to have a value of P = 0.0001, meeting the criteria of P < 0.05 to make it statistically significant. This indicates that the cell viability quantified in A. niger biofilm using XTT assay has a significant difference between the center and the corner portion. This correlation supports the accuracy of fluorescent probes in cell viability assessment of A. niger biofilms.
4. Discussion and conclusion
The purpose of this study was to develop a straightforward, simple method for quantifying and visualizing filamentous fungal biofilms, specifically those of A.niger, in their physiological adherent state without any disturbance. This is possible with fluorescent probe stains followed CLSM. Two broad range fluorescence stains, differing in their target area and means of cell penetration, were used in biofilm cell viability differentiation. These methods involve staining the biofilm without physical disruption, which is desirable for live-cell imaging compared to assay methods.
The LIVE/DEAD BacLight™ Bacterial Viability Kit, which contains the fluorescent stains SYTO9 and PI, has been used in the past to evaluate the cell viability of bacteria and yeast [19,47,49,50,52,53], but to our knowledge has not been used to study A. niger biofilms. FUN 1 in LIVE/DEAD™ Yeast Viability kit is a fungal specific fluorescent stain, which is widely used for yeast viability staining. Only metabolically active cells are marked clearly with fluorescent intravacuolar structures, while the dead cells exhibit bright green fluorescence [47]. This means, the cells with an intact membrane with no or very little metabolic activity have green cytoplasmic fluorescence and lack fluorescent intravacuolar structures. FUN 1 detects the metabolically hyphal active phase of A. niger cells, whereas the combination of SYTO9 and PI stain the cells based on their membrane integrity instead of cell metabolism. Also, during the different stages of biofilm formation, the metabolic state of the organism can be modified, affecting the FUN 1 stain cell differentiation. During the metabolically active hyphal stage of A. niger, cells contain more than one intravascular structure [58,59], which leads to the overestimation of live cells (Fig. 4).
From this study, the combination of SYTO9 and PI was found to be a reliable staining method for the quantification of cell viability, and visualization of A. niger biofilm. Propidium iodide is generally used for staining dead cells in a population. As a counterstain in multicolor fluorescent techniques, it can only penetrate cells with disrupted membranes. SYTO9 can enter both live and dead cells [[47], [48], [49]]). The fluorescent signal of SYTO9 is strongly enhanced when bound to nucleic acids ([54]). When both fluorescent probes are present, in the case of damaged cell membranes, PI exhibits a stronger affinity towards nucleic acids than SYTO9 and is thus replaced by PI [50].
The confocal images showed that the cell viability is higher in the center portion of the biofilm when compared to the corner portion. This results from the media dripping to the surface of the biofilm coupons in a drip flow reactor creating a high nutrient concentration in the center portion when compared to the corner portion. Moreover, the corner portion of the biofilm is in contact with the glass coupon and the reactor surface. Previous studies [19,30] showed that biofilms in contact with different surfaces made of dissimilar materials (e.g. glass vs polysulfone) can affect the growth of biofilms. These conditions lead to faster maturation of the biofilm cells in the corner when compared to the center portion as indicated by the presence of conidiogenous cells and conidiophores (Fig. 2-bottom), representing the maturation or reproductive development stage of the biofilm.
From Fig. 4, it was observed that the SYTO9-PI stained A. niger biofilms showed a significant variation in the biomass between the center and the corner portion of the biofilm, whereas the FUN 1 stained A.niger biofilms have similar biomasses between the center and corner portions. This difference of biomass in the case of the SYTO9-PI stains and the similarity in the case of the FUN 1 stain is also evident from Figs.1 to 2, where one can observe that both center and corner portion of biofilm (Fig. 1), stained with FUN 1 looks similar with difficultly to differentiate between the live and dead cells, while in the case of Fig. 2 (both center and corner portions), stained with STYO9-PI, the live and dead cells can be easily differentiated. The statistical analysis shown in Fig. 6 also supports these findings. Even though both the probe methods can be used for labeling the live-dead cells, based on the confocal imaging, COMSTAT biomass estimation, and statistical analysis result, the combination of SYTO9-PI is found to be a more reliable method for A. niger biofilm.
Results generated with the fluorescent probes were, compared with XTT assay, a standard cell viability assessment method. From the statistical analysis of the XTT assay method, it was observed that there is a significant difference in the cell viability between the center and the corner of the biofilm. This was the same trend found with the fluorescent probe staining methods. The XTT assay method depends on the metabolic activity of the cell; therefore, any changes in the metabolic activity of the cell cause variation in the XTT assay results. Compared to the XTT assessment method, the fluorescent probe-COMSTAT analysis has the important advantage of analyzing biofilms without disrupting their structure [19,60,61]. Also, the COMSTAT analysis provided data on the A. niger biofilm architecture and allows for visualization of the three-dimensional structure of the biofilm community. In conclusion, the present study provides a fluorescent probe - COMSTAT cell viability assessment method for A. niger biofilms, which is an improvement over the XTT assay method.
Authors contributions
Aswathy Shailaja, Julia L. Kerrigan, Terri F. Bruce and Charles A. Pettigrew conceived and designed the study. Aswathy Shailaja, Julia L. Kerrigan, Terri F. Bruce, Rhonda R. Powell and Patrick Gerard performed the experiments and analyzed the data. Julia L. Kerrigan and Charles A. Pettigrew responsible for the funding acquisition. Aswathy Shailaja and Julia L. Kerrigan wrote and revised the manuscript.
Declaration of competing interest
The authors declaring no competing interests associated with this project.
Acknowledgements
This material is based upon the work supported by technical contribution No 7043 of the Clemson University Experiment Station and NIFA/USDA, under project number SC 1700560.
Data availability
No data was used for the research described in the article.
References
- 1.Córdova-Alcántara I.M., Venegas-Cortés D.L., Martínez-Rivera M.Á., Pérez N.O., Rodriguez-Tovar A.V. Biofilm characterization of Fusarium solani keratitis isolate: increased resistance to antifungals and UV light. J Microbiol. 2019;57:485–497. doi: 10.1007/s12275-019-8637-2. [DOI] [PubMed] [Google Scholar]
- 2.Harding M.W., Marques L.L.R., Howard R.J., Olson M.E. Can filamentous fungi form biofilms? Trends Microbiol. 2009;17(11):475–480. doi: 10.1016/j.tim.2009.08.007. https://doi-org.libproxy.clemson.edu/10.1016/j.tim.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 3.Ramage G., Rajendran R., Sherry L., Williams C. Fungal biofilm resistance. Internet J Microbiol. 2012;2012 doi: 10.1155/2012/528521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramage G., Rajendran R., Gutierrez-Correa M., Jones B., Williams C. Aspergillus biofilms: clinical and industrial significance. FEMS Microbiol Lett. 2011;324:89–97. doi: 10.1111/j.1574-6968.2011.02381.x. [DOI] [PubMed] [Google Scholar]
- 5.Villena G.K., Gutiérrez-Correa M. Morphological patterns of Aspergillus Niger biofilms and pellets related to lignocellulolytic enzyme productivities. Lett Appl Microbiol. 2007;45:231–237. doi: 10.1111/j.1472-765X.2007.02183.x. [DOI] [PubMed] [Google Scholar]
- 6.Villena G.K., Gutiérrez-Correa M. Production of cellulase by Aspergillus Niger biofilms developed on polyester cloth. Lett Appl Microbiol. 2006;43:262–268. doi: 10.1111/j.1472-765X.2006.01960.x. [DOI] [PubMed] [Google Scholar]
- 7.van Acker H., van Dijck P., Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014;22(6):326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Ramage G., Mowat E., Jones B., Williams C., Lopez-Ribot J. Our Current understanding of fungal biofilms. Crit Rev Microbiol. 2009;35:340–355. doi: 10.3109/10408410903241436. [DOI] [PubMed] [Google Scholar]
- 9.Shopova I., Bruns S., Thywissen A., Kniemeyer O., Brakhage A.A., Hillmann F. Extrinsic extracellular DNA leads to biofilm formation and colocalizes with matrix polysaccharides in the human pathogenic fungus Aspergillus fumigatus. Front Microbiol. 2013;4:141. doi: 10.3389/fmicb.2013.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donlan R.M. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogino P.C., de las Mercedes M., Sorroche F.G., Giordano W. The role of bacterial biofilms and surface components in plant-bacterial associations. Int J Mol Sci. 2013;14:15838–15859. doi: 10.3390/ijms140815838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra J., Mukherjee P.K., Ghannoum M.A. In vitro growth and analysis of Candida biofilms. Nat Protoc. 2008;3:1909–1924. doi: 10.1038/nprot.2008.192. [DOI] [PubMed] [Google Scholar]
- 13.Chandra J., Kuhn D.M., Mukherjee P.K., Hoyer L.L., McCormick T., Ghannoum M.A. biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183:5385. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costerton J.W., Cheng K.J., Geesey G.G., Ladd T.I., Nickel J.C., Dasgupta M., Marrie T.J. Bacterial biofilms in nature and disease. Annu Rev Microbiol. 1987;41:435–464. doi: 10.1146/annurev.mi.41.100187.002251. [DOI] [PubMed] [Google Scholar]
- 15.Dickschat J.S. Quorum sensing and bacterial biofilms. Nat Prod Rep. 2010;27:343–369. doi: 10.1039/B804469B. [DOI] [PubMed] [Google Scholar]
- 16.Hall-Stoodley L., Costerton J.W., Stoodley P. Bacterial biofilms: from the Natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 17.Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 18.Kuhn D.M., Chandra J., Mukherjee P.K., Ghannoum M.A. Comparison of biofilms formed by Candida albicans and Candida parapsilosis on bioprosthetic surfaces. Infect Immun. 2002;70:878. doi: 10.1128/IAI.70.2.878-888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seneviratne C.J., Silva W.J., Jin L.J., Samaranayake Y.H., Samaranayake L.P. Architectural analysis, viability assessment and growth kinetics of Candida albicans and Candida glabrata biofilms. Arch Oral Biol. 2009;54:1052–1060. doi: 10.1016/j.archoralbio.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Smirnova T.A., Didenko L. v, Azizbekyan R.R., Romanova YuM. Structural and functional characteristics of bacterial biofilms. Microbiology (N Y) 2010;79:413–423. doi: 10.1134/S0026261710040016. [DOI] [Google Scholar]
- 21.Blankenship J.R., Mitchell A.P. How to build a biofilm: a fungal perspective. Curr Opin Microbiol. 2006;9:588–594. doi: 10.1016/j.mib.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Chatterjee S., Das S. Developmental stages of biofilm and characterization of extracellular matrix of manglicolous fungus Aspergillus Niger BSC-1. J Basic Microbiol. 2020;60:231–242. doi: 10.1002/jobm.201900550. [DOI] [PubMed] [Google Scholar]
- 23.González-Ramírez A.I., Ramírez-Granillo A., Medina-Canales M.G., Rodríguez-Tovar A.V., Martínez-Rivera M.A. Analysis and description of the stages of Aspergillus fumigatus biofilm formation using scanning electron microscopy. BMC Microbiol. 2016;16 doi: 10.1186/s12866-016-0859-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhat M.K. Cellulases and related enzymes in biotechnology. Biotechnol Adv. 2000;18:355–383. doi: 10.1016/S0734-9750(00)00041-0. [DOI] [PubMed] [Google Scholar]
- 25.Gamarra N.N., Villena G.K., Gutiérrez-Correa M. Cellulase production by Aspergillus Niger in biofilm, solid-state, and submerged fermentations. Appl Microbiol Biotechnol. 2010;87:545–551. doi: 10.1007/s00253-010-2540-4. [DOI] [PubMed] [Google Scholar]
- 26.Gutiérrez-Correa M., Villena G.K. Surface adhesion fermentation: a new fermentation category. Rev Peru Biol. 2003 [Google Scholar]
- 27.Kimmerling E.A., Fedrick J.A., Tenholder M.F. Invasive Aspergillus Niger with fatal pulmonary oxalosis in chronic obstructive pulmonary disease. Chest. 1992;101(3):870–872. doi: 10.1378/chest.101.3.870. [DOI] [PubMed] [Google Scholar]
- 28.Priegnitz B.-E., Wargenau A., Brandt U., Rohde M., Dietrich S., Kwade A., Krull R., Fleißner A. The role of initial spore adhesion in pellet and biofilm formation in Aspergillus Niger. Fungal Genet Biol. 2012;49:30–38. doi: 10.1016/j.fgb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Goeres D.M., Hamilton M.A., Beck N.A., Buckingham-Meyer K., Hilyard J.D., Loetterle L.R., Lorenz L.A., Walker D.K., Stewart P.S. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat Protoc. 2009;4:783–788. doi: 10.1038/nprot.2009.59. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg M., Azevedo N.F., Ivask A. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci Rep. 2019;9:6483. doi: 10.1038/s41598-019-42906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandra J., Mukherjee P.K., Ghannoum M.A. In vitro growth and analysis of Candida biofilms. Nat Protoc. 2008;3:1909–1924. doi: 10.1038/nprot.2008.192. [DOI] [PubMed] [Google Scholar]
- 32.Pierce C.G., Uppuluri P., Tristan A.R., Wormley F.L., Mowat E., Ramage G., Lopez-Ribot J.L. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc. 2008;3:1494–1500. doi: 10.1038/nprot.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamid R., Rotshteyn Y., Rabadi L., Parikh R., Bullock P. Comparison of alamar blue and MTT assays for high through-put screening. Toxicol Vitro. 2004;18:703–710. doi: 10.1016/j.tiv.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Jin Y., Samaranayake L.P., Samaranayake Y., Yip H.K. Biofilm formation of Candida albicans is variably affected by saliva and dietary sugars. Arch Oral Biol. 2004;49:789–798. doi: 10.1016/j.archoralbio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Mowat E., Lang S., Williams C., McCulloch E., Jones B., Ramage G. Phase-dependent antifungal activity against Aspergillus fumigatus developing multicellular filamentous biofilms. J Antimicrob Chemother. 2008;62:1281–1284. doi: 10.1093/jac/dkn402. [DOI] [PubMed] [Google Scholar]
- 36.Mowat E., Butcher J., Lang S., Williams C., Ramage G. Development of a simple model for studying the effects of antifungal agents on multicellular communities of Aspergillus fumigatus. J Med Microbiol. 2007;56:1205–1212. doi: 10.1099/jmm.0.47247-0. [DOI] [PubMed] [Google Scholar]
- 37.Rajendran R., Mowat E., McCulloch E., Lappin D.F., Jones B., Lang S., Majithiya J.B., Warn P., Williams C., Ramage G. Azole resistance of Aspergillus fumigatus biofilms is partly associated with efflux pump activity. Antimicrob Agents Chemother. 2011;55:2092. doi: 10.1128/AAC.01189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramage G., vande Walle K., Wickes B.L., López-Ribot J.L. Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob Agents Chemother. 2001;45:2475. doi: 10.1128/AAC.45.9.2475-2479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Y., Liu X., Zhou P. In vitro antifungal effects of berberine against Candida spp. in planktonic and biofilm conditions. Drug Des Dev Ther. 2020;14:87. doi: 10.2147/DDDT.S230857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peeters E., Nelis H.J., Coenye T. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods. 2008;72:157–165. doi: 10.1016/j.mimet.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 41.Pettit R.K., Weber C.A., Pettit G.R. Application of a high throughput Alamar blue biofilm susceptibility assay to Staphylococcus aureus biofilms. Ann Clin Microbiol Antimicrob. 2009;8:28. doi: 10.1186/1476-0711-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Repp K.K., Menor S.A., Pettit R.K. Microplate Alamar blue assay for susceptibility testing of Candida albicans biofilms. Med Mycol. 2007;45:603–607. doi: 10.1080/13693780701581458. [DOI] [PubMed] [Google Scholar]
- 43.Mariscal A., Lopez-Gigosos R.M., Carnero-Varo M., Fernandez-Crehuet J. Fluorescent assay based on resazurin for detection of activity of disinfectants against bacterial biofilm. Appl Microbiol Biotechnol. 2009;82:773–783. doi: 10.1007/s00253-009-1879-x. [DOI] [PubMed] [Google Scholar]
- 44.Kuhn D.M., Balkis M., Chandra J., Mukherjee P.K., Ghannoum M.A. Uses and limitations of the XTT assay in studies of Candida growth and metabolism. J Clin Microbiol. 2003;41:506. doi: 10.1128/JCM.41.1.506-508.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henry-Stanley M.J., Garni R.M., Wells C.L. Adaptation of FUN-1 and Calcofluor white stains to assess the ability of viable and nonviable yeast to adhere to and be internalized by cultured mammalian cells. J Microbiol Methods. 2004;59:289–292. doi: 10.1016/j.mimet.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 46.Hickey P.C., Swift S.R., Roca M.G., Read N.D. Live-cell imaging of filamentous fungi using vital fluorescent dyes and confocal microscopy. Methods Microbiol. 2004;34:63–87. doi: 10.1016/S0580-9517(04)34003-1. [DOI] [Google Scholar]
- 47.Jin Y., Zhang T., Samaranayake Y.H., Fang H.H.P., Yip H.K., Samaranayake L.P. The use of new probes and stains for improved assessment of cell viability and extracellular polymeric substances in Candida albicans biofilms. Mycopathologia. 2005;159:353–360. doi: 10.1007/s11046-004-6987-7. [DOI] [PubMed] [Google Scholar]
- 48.Viability and Cytotoxicity Assay Kits for Diverse Cell Types - section 15.3 . Molecular ProbesTM handbook A guide to fluorescent probes and labeling Technologies. ThermoFischer Scientific; 2010. pp. 683–686. [Google Scholar]
- 49.Stiefel P., Schmidt-Emrich S., Maniura-Weber K., Ren Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015;15:36. doi: 10.1186/s12866-015-0376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao Y., Mai B., Wang A., Li M., Wang X., Zhang K., Liu Q., Wei S., Wang P. Antimicrobial properties of a new type of photosensitizer derived from phthalocyanine against planktonic and biofilm forms of Staphylococcus aureus. Photodiagnosis Photodyn Ther. 2018;21:316–326. doi: 10.1016/j.pdpdt.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 51.Qian W., Yang M., Li X., Sun Z., Li Y., Wang X., Wang T. Anti-microbial and anti-biofilm activities of combined chelerythrine-sanguinarine and mode of action against Candida albicans and Cryptococcus neoformans in vitro. Colloids Surf B Biointerfaces. 2020;191 doi: 10.1016/j.colsurfb.2020.111003. [DOI] [PubMed] [Google Scholar]
- 52.Takenaka S., Iwaku M., Hoshino E. Artificial Pseudomonas aeruginosa biofilms and confocal laser scanning microscopic analysis. J Infect Chemother. 2001;7:87–93. doi: 10.1007/s101560100014. [DOI] [PubMed] [Google Scholar]
- 53.Tawakoli P.N., Al-Ahmad A., Hoth-Hannig W., Hannig M., Hannig C. Comparison of different live/dead stainings for detection and quantification of adherent microorganisms in the initial oral biofilm. Clin Oral Invest. 2013;17:841–850. doi: 10.1007/s00784-012-0792-3. [DOI] [PubMed] [Google Scholar]
- 54.Assays for Cell Viability, Proliferation and Function - section 15.2 . Molecular probes handbook—a guide to fluorescent probes and labeling Technologies. ThermoFischer Scientific; 2010. pp. 664–665. [Google Scholar]
- 55.Kerrigan, J.L., Gruber, J., Pettigrew, C.A., Mulligan, D., Wright, K.I.T., n.d. Reproducible protocol for growing relevant filamentous fungal biofilms and phenotypic phases during biofilm maturation. Appl Environ Microbiol (in review).
- 56.Heydorn A., Nielsen A.T., Hentzer M., Sternberg C., Givskov M., Ersbøll B.K., Molin S. Quantification of biofilm structures by the novel computer program comstat. Microbiology (N Y) 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 57.Claus Sternberg . 2015. COMSTAT 2.1- Manual revision. [Google Scholar]
- 58.Arunmozhi B.S., A M.K. Conidial viability assay for rapid susceptibility testing of Aspergillus species. J Clin Microbiol. 2002;40:2741–2745. doi: 10.1128/JCM.40.8.2741-2745.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Essary B.D., Marshall P.A. Assessment of FUN-1 vital dye staining: yeast with a block in the vacuolar sorting pathway have impaired ability to form CIVS when stained with FUN-1 fluorescent dye. J Microbiol Methods. 2009;78:208–212. doi: 10.1016/j.mimet.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 60.Chandra J., Mukherjee P.K., Ghannoum M.A. In vitro growth and analysis of Candida biofilms. Nat Protoc. 2008;3:1909–1924. doi: 10.1038/nprot.2008.192. [DOI] [PubMed] [Google Scholar]
- 61.Villena G.K., Fujikawa T., Tsuyumu S., Gutiérrez-Correa M. Structural analysis of biofilms and pellets of Aspergillus Niger by confocal laser scanning microscopy and cryo scanning electron microscopy. Bioresour Technol. 2010;101:1920–1926. doi: 10.1016/j.biortech.2009.10.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.