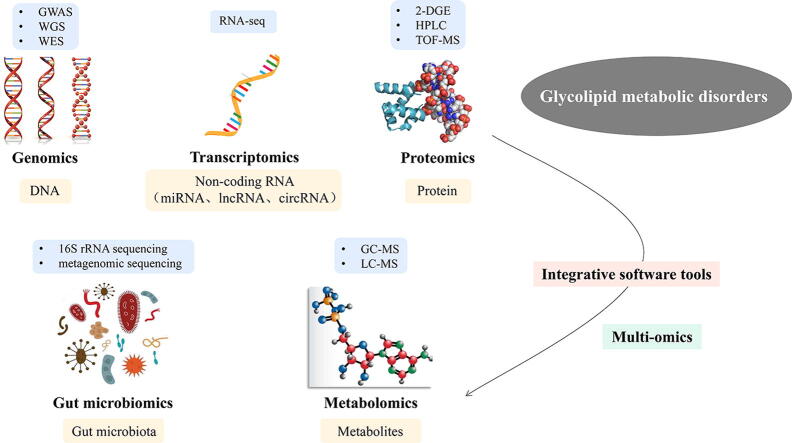
Graphical abstract
Keywords: Multi-omics, Glycolipid metabolism disorder, Biomarkers, Mechanism study
Abbreviations: 1,5-AG, 1,5-anhydroglucitol; 2-AAA, 2-aminoadipic acid; 2-DGE, two-dimensional gel electrophoresis; ABCC8, ATP binding cassette subfamily C member 8; ADA, American Diabetes Association; α-HB, α-hydroxybutyrate; AhR, aromatic hydrocarbon receptor; BA, bile acid; BCAA, branched-chain amino acid; BCKAs, branched chain keto acids; CE, cholesterol ester; CFL1, cofilin-1; circRNA, circular RNA; CRP, C reactive protein; CYP450, cytochrome P450; DAG, diacylglycerol; DPP-4, dipeptyl peptidase 4; EASD, European Association for the Study of Diabetes; FA, fatty acid; FFA, free fatty acid; FMT, fecal microbiota transplantation; FTO, fat mass and obesity-associated; GAS5, growth arrest-specific transcript 5; GC–MS/MS, gas chromatography-tandem mass spectrometry; GLP-1, glucagon-like peptide 1; GLP-1R, glucagon-like peptide 1 receptor; GSIS, glucose-stimulated insulin secretion; GWAS, genome-wide association study; hADCS, human adipose-derived stem cells; HbA1C, glycylated hemoglobin; HGF, hepatocyte growth factor; HMG-CoA, hydroxymethylglutaryl-coenzyme A; HMGCR, 3-hydroxy-methylglutaryl coenzyme A reductase; HPLC, high performance liquid chromatography; hsCRP, high-sensitivity C-reactive protein; IL-1ra, interleukin-1 receptor antagonist; IMP, imidazole propionate; IR, insulin resistance; JNK, c-Jun-N-terminal-kinase; KCNJ11, potassium inwardly rectifying channel subfamily J member 11; LC-MS/MS, liquid chromatography-tandem mass spectrometry; lnc-BATE1, brown adipose tissue enriched long non-coding RNA 1; lncRNA, long non-coding RNA; lncRNA SHGL, lncRNA suppressor of hepatic gluconeogenesis and lipogenesis; L-GPC, linoleoyl-glycerophosphocholine; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamines; LPS, lipopolysaccharide; MAPK, mitogen-activated protein kinase; MASP, mannose-binding lectin-associated serine protease 1; MATE1, multidrug and toxin extrusion protein 1; MDR1, multidrug resistance mutation 1; MGT, magnesium transporter; miRNA, micro RNA; mRNA, messenger RNAs; MS, Mass Spectrometry; mTORC1, mechanistic target of rapamycin complex 1; ncRNA, non-coding RNA; NK, natural killer; NMR, nuclear magnetic resonance; OCT, organic cationic transporter; OGTT, oral glucose tolerance test; PC-PL, phosphatidylcholine-plasmalogen; PE, phosphatidylethanolamines; PNPLA3, patatin-like phospholipase domain-containing protein 3; PPAR, peroxisome proliferator-activated receptor; PTBP1, polypyrimidine tract-binding protein 1; PTP, protein tyrosine phosphatase; RNS, reactive nitrogen species; ROS, reactive oxygen species; SCFA, short-chain fatty acid; SLC30A8, solute carrier family 30 member 8; SLC47A1, solute carrier family 47 member 1; SLC5A2, solute carrier family 5 member 2; SM, sphingomyelin; SNP, single-nucleotide polymorphism; SSPG, steady-state plasma glucose; SUR1, sulfonylurea receptor 1; T2DM, type 2 diabetes mellitus; TAG, triacylglycerol; TCA, tricarboxylic acid; TCF7L2, transcription factor 7-like 2; TF, transcription factor; TMA, trimethylamine; TMAO, trimethylamine oxide; TNF-α, tumor necrosis factor alpha; TOF-MS, time-of-flight mass spectrum; t-PA, tissue plasminogen activator; TUG1, taurine upregulated gene 1; USP20, ubiquitin-specificpeptidase 20; WBC, white blood cell
Abstract
Glycolipid metabolism disorder are major threats to human health and life. Genetic, environmental, psychological, cellular, and molecular factors contribute to their pathogenesis. Several studies demonstrated that neuroendocrine axis dysfunction, insulin resistance, oxidative stress, chronic inflammatory response, and gut microbiota dysbiosis are core pathological links associated with it. However, the underlying molecular mechanisms and therapeutic targets of glycolipid metabolism disorder remain to be elucidated. Progress in high-throughput technologies has helped clarify the pathophysiology of glycolipid metabolism disorder. In the present review, we explored the ways and means by which genomics, transcriptomics, proteomics, metabolomics, and gut microbiomics could help identify novel candidate biomarkers for the clinical management of glycolipid metabolism disorder. We also discuss the limitations and recommended future research directions of multi-omics studies on these diseases.
1. Introduction
Metabolic disease, such as type 2 diabetes mellitus (T2DM), hyperlipidemia, and obesity, are increasingly common owing to the change of lifestyle and the rapid development of industrialization. As an important feature of metabolic diseases, glycolipid metabolism disorder is silently threatening human health. It is estimated that in 2040, 642 million adults worldwide will have diabetes, and the vast majority of them will be T2DM [1]. Overweight is a risk factor for diabetes, dyslipidemia, and non-alcoholic fatty liver disease. According to an ecological study, about a third of the global population has been determined to be obese or overweight since 1980 [2]. The epidemiological findings of dyslipidemia are equally discouraging. Several researchers have found that total cholesterol decreased the most in high-income western regions and in central and eastern Europe, while increased the most in east and southeast Asia. Particularly, the population with high cholesterol has increased significantly in China, which now has one of the highest cholesterol levels in the world [3].
The harm of glycolipid metabolism disorder lies in the damage of general organ caused by long-term abnormal blood glucose and lipid levels, leading to the gradual decline of its function. Meanwhile, microvascular and macrovascular injury are regarded as the important cause of disability and mortality in patients. As reported, patients with concurrent T2DM, hypertension, and dyslipidemia are 6 times more likely to have cardiovascular disease compared with those with T2DM alone [4]. Currently, a single intervention of hyperglycemia or hyperlipidemia cannot effectively regulate multiple metabolic disorders, resulting in suboptimal lipid control and poor glycemic control. Glycolipid metabolism disorder are more complicated than single factor metabolic abnormalities, because of multi-factorial interaction [5]. Based on the complexity of the metabolic regulatory network, simultaneous regulation of different metabolic pathways can be more potent than regulation of a single pathway in the treatment of glycolipid metabolism disorder [6]. Combination therapy has been demonstrated to be superior to monotherapy in metabolic abnormalities [7], [8].
Owing to the complexity of the pathogenesis of glycolipid metabolism disorder, its underlying molecular mechanisms are so far unknown. A better understanding of the pathophysiology of glycolipid metabolism disorder using multi-omics will improve preventive, diagnostic, therapeutic and reparative strategies. In the pages that follow, we consider the progress made in genomics, transcriptomics, proteomics, metabolomics and gut microbiomics, and discuss how bringing data from these techniques together through integromics and systems biology. Multi-omics research sheds new light on revealing the potential pathogenic targets, pathophysiological mechanisms and biomarkers of therapeutic intervention for the occurrence and development of glycolipid metabolism disorder, and offers a fresh perspective on controlling the progression of it.
2. Pathophysiology of glycolipid metabolism disorder
Glycolipid metabolism disorder is a complex and systemic disease caused by multiple metabolic organ dysregulations. The pathogenesis of these conditions involves interactions among core pathological mechanisms such as neuroendocrine axis dysfunction, insulin resistance, oxidative stress, chronic inflammatory response, and gut microbiota dysbiosis. The foregoing processes are implicated in the occurrence and progression of these diseases.
The human body controls the release of neurotransmitters, hormones, and cytokines through the nervous and endocrine systems to maintain glycolipid metabolism homeostasis. The hypothalamus regulates energy metabolism by detecting signals from the peripheral tissues [9]. Tanycytes and 5-HT neurons are key cells and leptin are important signaling molecules regulating glycolipid metabolism along the neuroendocrine axis [10], [11], [12], [13]. Insulin resistance (IR) is characterized by declines in glucose uptake and insulin utilization efficiency and is a common symptom of glycolipid metabolism disorder. It occurs primarily in muscle, fat, and liver tissues [14]. The mechanism underlying the development of IR is associate with impaired insulin signal transduction, including accumulation of specific lipid mediators, abnormal features of mitochondrial function, and increases in stress-activated protein c-Jun-N-terminal-kinase (JNK) and inflammatory pathways [15], [16]. Oxidative stress is a central factor in the initiation and progression of glycolipid metabolism disorder and is characterized by the augmented generation or diminished elimination of reactive oxygen species (ROS) and reactive nitrogen species (RNS) because of an imbalance between pro-oxidant and antioxidant levels. Glucotoxicity and lipotoxicity promote IR through oxidative stress by damaging pancreatic islet cells, adipocytes, and their signaling pathways [17]. Chronic low-grade inflammation is a critical feature of glycolipid metabolism disorder and is characterized by massive infiltration of immunocytes including macrophages, natural killer (NK) cells, mast cells, and others [18], [19]. In chronic metabolic inflammation, inflammatory factors and cells regulate glycolipid metabolism in the liver, fat, muscle, pancreas, and other tissues and organs through an extensively interwoven immune network, thereby inducing IR and glycolipid metabolism disorder. Several recent studies showed that gut microbiota or the ‘second genome’ are implicated in the occurrence and development of multiple metabolic diseases. Gut microbiota can modulate nutrient metabolism upon dietary intake and produce many metabolites to interact with the host in a variety of ways, including regulating glucose and lipid metabolism pathways, influencing the differentiation and function of immune cells, affecting insulin sensitivity and so on [20]. In-depth research on the interactions between the gut microbiota and the host has revealed that the former execute vital functions in metabolic regulation via the gut-liver-brain axis [21].
3. Biomarkers of glycolipid metabolism disorder in multi-omics research
Biomarkers can be powerful tools in the management of diseases. For glycolipid metabolism disorder, plasma glucose (measured after fasting or a glucose tolerance test), glycylated hemoglobin, and plasma lipids are regarded as clinical biomarkers for diagnostic and screening. The future of medicine lies in individualized therapies, prospective tracking of individual health indicators, and critical attention on preventive measures [22]. Based on this, individualized and multidimensional biomarkers are urgently needed to reveal the prediction, diagnosis and prognostic features of glycolipid metabolism disorder, and provide more valuable reference information for drug development, clinical diagnosis and personalized treatment. Recently, with the development of omics technologies and bioinformatics, multi-omics research on glycolipid metabolism disorder has gradually increased, which is conducive to understanding the molecular mechanism of disease occurrence and evaluating biomarkers, promoting the process of precision medicine for glycolipid metabolism disorder (Supplementary Table 1).
3.1. Genomics
Genomics is a subdiscipline of genetics that characterizes and quantifies organism’s complete genome and studies the relationship between genes and their effects on organisms [23]. The etiology of glycolipid metabolism disorder is known to have a considerable genetic component. Over the past two decades, linkage analyses, candidate gene approaches, and large-scale genome-wide association study (GWAS) have successfully identified more than 100 genes that confer susceptibility to glycolipid metabolism disorder. Genomics explains the key genetic variants associated with the risk of glycolipid metabolism disorder, provides guidance for future studies, and helps formulate efficacious preventive and therapeutic measures for disease. Pharmacogenomics optimizes treatment schedules, improves the effectiveness of personalized therapy, and minimizes potential side effects during clinical treatment.
3.1.1. Genes associated with susceptibility to glycolipid metabolism disorder
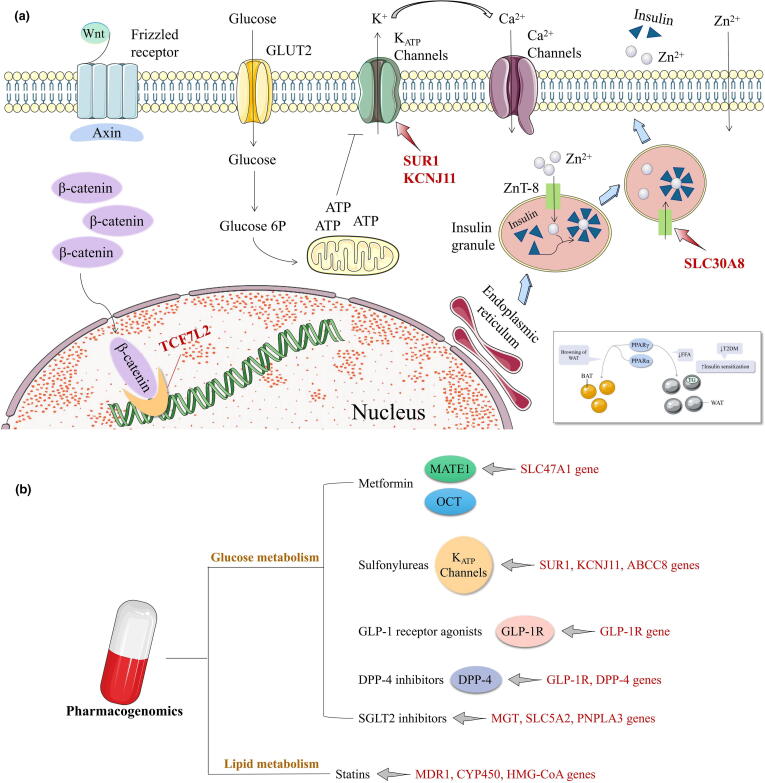
Genes determine individual susceptibility to diseases. Most genes determining susceptibility to glycolipid metabolism disorder regulate insulin secretion and sensitivity and pancreatic β cell function. These include TCF7L2 (transcription factor 7-like 2), PPARs (peroxisome proliferator-activated receptors), KCNJ11 (potassium inwardly rectifying channel subfamily J member 11), SLC30A8 (solute carrier family 30 member 8), FTO (fat mass and obesity-associated), and so on [24] (Fig. 1a). TCF7L2 is strongly associated with T2DM [25], is a major transcription factor (TF) in the canonical Wnt signaling pathway, regulates intrapancreatic glucose homeostasis, and is essential for glucose-stimulated insulin secretion (GSIS) maintenance and pancreatic β cell survival [26]. In 2006, Grant et al. reported that variation in TCF7L2 expression was closely associated with T2DM risk in a case-control study on Caucasians in Iceland, Denmark, and the United States [27]. The association of T2DM with single-nucleotide polymorphisms (SNPs) in TCF7L2 has raised global concern and was confirmed in ethnically diverse populations [28], [29], [30], [31]. PPARγ (peroxisome proliferative-activated receptor, gamma) is a member of the nuclear hormone receptor superfamily of TFs and the first screened candidate gene associated with glycolipid metabolism disorder [32]. PPAR activation regulates gene networks controlling various homeostatic processes involving inflammation, adipogenesis, lipid and glucose metabolism, and insulin resistance [33]. SLC30A8 is correlated with pancreatic function and is predominantly expressed in that organ. It encodes the endocrine pancreas-restricted zinc transporter ZnT8. Abnormal SLC30A8 and ZnT8 function affect insulin biosynthesis, storage, and secretion and hinder normal glucose metabolism. In 2007, SLC30A8 was identified as a novel T2DM susceptibility gene [34]. Subsequent studies verified the association between SLC30A8 SNPs and T2DM in different racial and ethnic groups [35], [36], [37]. FTO is the first candidate obesity gene to be recognized in the general population. It is highly expressed in hypothalamic nuclei and homeostatically controls the energy balance. Variations in FTO are associated with the risks of obesity and T2DM [38], [39], [40], [41]. Kir6.2 and SUR1 (sulfonylurea receptor 1) form the pancreatic β cell KATP channel. SUR1 is the site of sulfonylurea binding while Kir6.2 is an ion channel encoded by ABCC8 (ATP binding cassette subfamily C member 8) and KCNJ11 [42], [43]. Variations in KCNJ11 and SUR1 impede KATP channel function, impair insulin secretion, and increased susceptibility to T2DM [44], [45].
Fig. 1.
Genetic variants associated with glycolipid metabolic disorders. a. Overview of canonical signaling mechanisms involved in beta-cell glucose sensing and responses to secretory potentiators or inhibitors. TCF7L2 (Transcription factor 7-like 2) has a role in the canonical Wnt (Wingless-type MMTV integration site) pathway. SLC30A8 (Solute-linked carrier 30, member 8) encodes ZnT8 (Zinc transporter 8) which regulates the influx of zinc into intracellular vesicles of insulin is presumed to be critical for insulin storage and secretion. KCNJ11 (K+ inwardly rectifying channel, subfamily J, member 11) and SUR1 (Sulfonylurea receptor 1) encode KATP channel together, thus indirectly sensing blood glucose concentrations and controlling insulin release. PPARs (Peroxisome proliferator-activated receptors) in glycolipid metabolism disorder. BAT, brown adipose tissue; FFA, free fatty acids; ROS, reactive oxygen species; T2DM, type 2 diabetes mellitus; TG, triglycerides; WAT, white adipose tissue. b. Pharmacogenomics and its targets in glycolipid metabolic disorders. MATE1, multidrug and toxin extrusion 1; OCT, organic cation transporters; SLC47A1, solute carrier family 47 member 1; SUR1, Sulfonylurea receptor 1; KCNJ11, K+ inwardly rectifying channel, subfamily J, member 11; ABCC8, ATP-binding cassette, subfamily C, member 8; GLP-1, glucagon-like peptide-1; GLP-1R, glucagon-like peptide-1 receptor; DPP-4, dipeptidyl peptidase-4; SGLT2, sodium-glucose cotransporter 2; MGT, magnesium transporter; SLC5A2, solute carrier family 5 member 2; PNPLA3, patatin-like phospholipase domain-containing protein 3; MDR1, multidrug resistance gene 1; CYP450, cytochrome P450; HMG-CoA, 3-hydroxy-3-methylglutaryl coenzyme A.
As deep sequencing technologies continued to evolve, the focus of glycolipid metabolism disorder research gradually shifted from common genetic variations (GWAS (genome-wide association studies) era) to rare genetic variations (post-GWAS era). To date, the exact number of genes determining glycolipid metabolism disorder susceptibility and the precise mechanisms of their interactions have not been established. However, the results of recent GWAS have been encouraging. Xue et al conduct a meta-analysis of GWAS with 16 million genetic variants and identify 139 common and 4 rare variants associated with T2DM, 42 of which (39 common and 3 rare variants) are independent of the known variants [46]. Fuchsberger et al. reported that 126 variants in four genes (TCF7L2, ADCY5, CCND2, EML4) were significantly associated with the risk of T2DM [47]. In a large-scale genome-wide association study, Spracklen et al. identified new genetic links correlated with T2DM in 433,540 East Asians. They identified 301 distinct association signals at 183 loci, and 61 loci among that are newly implicated in predisposition to T2DM [48]. Locke et al conduct a genome-wide association study and Metabochip meta-analysis of body mass index (BMI) in up to 339,224 individuals. This analysis identifies 97 BMI-associated loci, 56 of which are novel [49].
It is well known that glycolipid metabolism disorder are polygenic disorders, but even combining with exons and whole-genome sequencing data, genetic variants might explain only about 10 % of the phenotypic variability in patients with glycolipid metabolism disorder [50]. Environmental factors such as lifestyle modification, nutritional imbalance and behaviour change might be more critical in the development of glycolipid metabolism disorder. Aging and genetic variation are both important contributors to the epigenetic variability seen in individuals affected by obesity or T2DM. Furthermore, the in utero environment and external factors such as physical activity and availability of nutrients affect the epigenome [51]. Consequently, further exploration of epigenetic factors and their mechanisms of glycolipid metabolism disorder will bring new ideas and opportunities for the prevention and treatment of glycolipid metabolism disorder.
3.1.2. Pharmacogenomics of glycolipid metabolism disorder
Metformin is prescribed as a first-line therapy for T2DM, as it is low-cost, has high efficacy, and is unlikely to induce hypoglycemia or other adverse reactions. When metformin monotherapy fails to provide satisfactory efficacy or provokes adverse effects, other hypoglycemic agents can be combined with metformin or substitute for it altogether. The latest American Diabetes Association/European Association for the Study of Diabetes (ADA/EASD) consensus reports indicate that sulfonylureas, thiazolidinediones, dipeptidyl peptidase 4 (DPP-4) and sodium-glucose cotransporter 2 (SGLT2) inhibitors, and glucagon-like peptide-1 (GLP-1) receptor agonists are reasonable as second-line treatment options [52]. Metformin is a hydrophilic organic cationic drug and it depends upon organic cation transporters (OCTs) to enter hepatocytes and renal epithelial cells where it is excreted through bile and urine, respectively, via multidrug and toxin extrusion protein 1 (MATE1). SLC47A1 (solute carrier family 47 member 1) encodes MATE1 and plays a key role in metformin transport and excretion [53]. Most OCT polymorphisms except MATE1 affect metformin pharmacokinetics and pharmacodynamics [54]. Sulfonylureas are insulin secretagogues and comprise an important class of oral hypoglycemic agents. They stimulate pancreatic β cells to release insulin by binding high-affinity plasma membrane receptors conjugated with KATP channels. The latter are regulated by SUR1, KCNJ11, and ABCC8 and their polymorphisms are associated with sulfonylurea efficacy [55], [56], [57]. GLP-1 receptor agonists exert their hypoglycemic effect by binding glucagon-like peptide 1 receptor (GLP-1R). GLP-1R polymorphisms are correlated with GLP-1 receptor agonists efficacy [58], [59]. DPP-4 inhibitors upregulate GLP-1 by retarding DPP-4 inactivation, activate intestinal GLP-1R, promote insulin release, and reduce glycemia [60]. Hence, DPP-4 inhibitor efficacy may be influenced by GLP-1R and DPP-4 polymorphisms [61], [62]. SGLT2 inhibitors constitute a novel class of antidiabetic drugs that lower plasma glucose by inhibiting renal glucose reabsorption and promoting urinary glucose excretion. Genes related to the clinical efficacy of SGLT2 inhibitors include MGT (magnesium transporter), SLC5A2 (solute carrier family 5 member 2), PNPLA3 (patatin-like phospholipase domain-containing protein 3), and others [63], [64], [65].
Polymorphisms associated with statins have become the focus of pharmacogenomics studies on lipid-lowering drugs. Candidate genes associated with the differential statin efficacy are divided into two main categories. Members of the first class such as CYP450 (cytochrome P450) and MDR1 (multidrug resistance mutation 1) regulate pharmacokinetics and encode drug-metabolizing enzymes and drug transport [66]. Members of the second class such as apolipoprotein and HMG-CoA (hydroxymethylglutaryl-coenzyme A) regulate pharmacodynamics and encode drug targets and lipid metabolism [67], [68], [69]. (Fig. 1b).
3.2. Transcriptomics
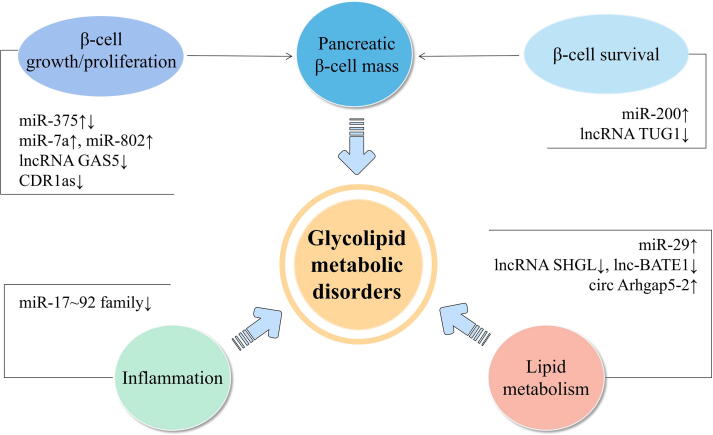
Transcriptomics is a discipline that studies the gene transcription and transcriptional regulation in cells at the overall level, which contributes to understand the gene expression profiles of diseases, and then reveals the metabolic network and regulatory mechanisms of life course from the transcriptional level. As is well known, there are some non-protein-coding genes within the organism, and the transcription products of these genes are known as non-coding RNAs (ncRNAs), mainly including long non-coding RNAs (lncRNAs), micro RNAs (miRNAs), circular RNAs (circRNAs). Non-coding RNAs plays a substantial regulatory role in the occurrence and development of glycolipid metabolism disorder, and could be useful as early molecule marker for the diagnosis of glycolipid metabolism disorder (Fig. 2).
Fig. 2.
Key RNAs affecting beta-cell mass, inflammation or lipid metabolism. lncRNA TUG1, taurine upregulated gene 1; lncRNA GAS5, growth arrest-specific transcript 5; lnc-BATE1, brown adipose tissue enriched long non-coding RNA 1; lncRNA SHGL, suppressor of hepatic gluconeogenesis and lipogenesis; CDR1as, hsa_circ_0001946, ciRS-7.
3.2.1. LncRNAs
LncRNAs account for more than 80 % of all non-coding RNAs, and its transcripts are widely involved with every aspect of cellular biological function. They regulate related protein-coding genes in numerous ways, and complement DNA bases to form stable triple-helix complexes, thus impairing the expression of target genes [70]. Altered expression of lncRNAs has been associated with poor glycemic control, insulin resistance, accelerated cellular senescence, and inflammation in diabetes patients [71]. Morán et al comprehensively reported the lncRNA expression profiles in human pancreatic β-cells, uncovered a high-confidence set of 1128 human islet-cell genes, and showed that they are an integral component of the β-cells differentiation and maturation program [72]. Several researchers have found that downregulation of lncRNA TUG1 (taurine upregulated gene 1) expression affected apoptosis and insulin secretion in pancreatic β-cells in vitro and in vivo, resulting in the occurrence of diabetes [73]. More recent research has revealed that the downregulation of the lncRNA GAS5 (growth arrest-specific transcript 5) is significantly associated with the occurrence and development of diabetes. Its downregulation can affect cell cycle and insulin secretion in pancreatic β-cells [74], [75]. Alvarez-Dominguez et al established the transcripome of mouse adipose tissues by RNA sequencing, identified 1500 lncRNAs, and located lnc-BATE1 (brown adipose tissue enriched long non-coding RNA 1) is the key lncRNA regulating brown fat, providing a new target for the treatment of obesity [76]. Recent study determined that a new lncRNA, lncRNA suppressor of hepatic gluconeogenesis and lipogenesis (lncRNA SHGL), is a novel insulin-independent suppressor of hepatic gluconeogenesis and lipogenesis [77].
3.2.2. MiRNAs
MiRNAs are small non-coding RNAs composed of 19–22 nucleotides that modulate gene expression by binding to the 3′ untranslated region of specific messenger RNAs (mRNAs) [78]. Impaired insulin secretion from the pancreatic β-cells is central in the pathogenesis of T2DM, and miRNAs are fundamental regulatory factors in this process [79]. The most abundant miRNA in the islet, miR-375, was also the first miRNA detected in pancreatic islet and may constitute a novel pharmacological target for the treatment of diabetes as a regulator of insulin secretion [80]. The miR-7 and the miR-200 family are other examples of islet abundant miRNAs. β-cell-specific overexpression of miR-7a in mice results in reduced insulin secretion [81]. Overexpression of miR-200 in mice is sufficient to induce beta cell apoptosis and lethal T2DM in mice [82]. High expression of miR-29 in liver, fat, and muscle tissue may trigger insulin resistance [83]. A growing body of evidence suggests that obesity-related and adipose tissue-derived circulating miRNAs are promising as novel therapeutic targets for obesity and related diseases [84]. The research group of Prof. Hu revealed that miRNAs of the miR-17 ∼ 92 family inhibit the inflammatory response of macrophages by maintaining the expression of IL-10, thus maintaining the homeostasis of adipose tissue macrophages and inhibiting obesity [85]. Several studies have shown that miR-802 is increased in the pancreatic islets of obese mouse models and inducible transgenic overexpression of miR-802 in mice causes impaired insulin transcription and secretion [86].
3.2.3. CircRNAs
Circular RNAs are covalently closed transcripts mostly generated from precursor-mRNA by a non-canonical event called back-splicing. They are highly stable, evolutionarily conserved, and widely distributed in eukaryotes [87]. Recently, mounting evidence suggests that the misregulation of circRNAs is among the first alterations in various metabolic disorders including obesity and diabetes mellitus (DM). So far, the best known endogenous circRNA related to diabetes is CDR1as (also termed as hsa_circ_0001946, ciRS-7) which can promotes islet β-cells proliferation and insulin secretion in diabetes as a powerful miR-7 inhibitor [88], [89]. Zhao et al found has_circ_0054633 differentially expressed in peripheral blood of patients with T2DM and healthy control [90]. Another study revealed that has_circ_0054633 can regulates high glucose-induced human vascular endothelial cell dysfunction [91]. Hence, hsa_circ_0054633 may be involved in the pathogenesis of diabetes and could be used as a biomarker for the diagnosis of T2DM. The past two years have witnessed a significant increase in the number of studies determining the function of circRNAs in human adipogenesis and obesity [92]. Researchers analyzed the transcriptome of human and mouse visceral and subcutaneous fat by RNA sequencing methods, found that the silencing of circArhgap5-2 in vivo resulted in inhibition of lipid droplet accumulation and downregulation of adipogenic markers [93]. However, the mechanism by which circArhgap5-2 modulates adipogenesis remains to be determined. Zhu’s experiments suggest that knockdown of hsa_circH19 promotes hADCSs (human adipose-derived stem cells) adipogenic differentiation via targeting of PTBP1 (polypyrimidine tract-binding protein 1), high levels of hsa_circH19 is an independent risk factor for the metabolic syndrome [94].
3.3. Proteomics
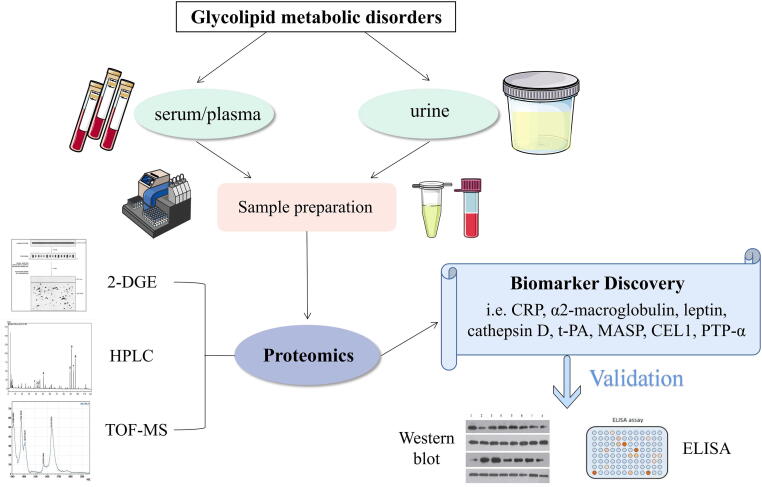
The essence of proteomics is to study proteins on a large scale, including protein expression, post-translational modifications, and protein–protein interactions [95]. The study of proteins, as the final product of genetic transcription and posttranscriptional modifications, has also played a pivotal role in the understanding of disease. At present, the proteomics research of glycolipid metabolism disorder mainly use two-dimensional gel electrophoresis (2-DGE), high performance liquid chromatography (HPLC), time-of-flight mass spectrum (TOF-MS) and other methods to explore the biomarkers for diagnosis and the pathways involved in disease pathogenesis (Fig. 3).
Fig. 3.
General scheme of current proteomic, for clinical metabolic research. Human body fluids (i.e. serum, urine and blood) have to be properly stored and prepared with optimised protocols. Subsequently, the proteins should be purifified and/or isolated to get digested peptides. The adequate 2-DE (two-dimensional gel electrophoresis), HPLC (high-performance liquid chromatography) or TOF-MS (time-of-flight mass spectrometer) based strategy is applied, and once we get the data (potential biomarkers), validation assays (i.e. ELISA and/or western blotting) can be carried out choosing specifific antibodies to identify real protein-biomarkers.
Shono et al. revealed the possible pathogenesis of T2DM by proteomics methods, which was specifically caused by alterations of protein secondary structure domains and post-translational modifications after the body received multiple pathogenic signals [96]. C reactive protein and α2-macroglobulin are clinically sensitive biomarkers of T2DM. Riaz et al. compared changes in serum differential protein levels in diabetic patients and healthy populations. Levels of C reactive protein (CRP) was found to increased by 872 % in the diabetic patients as compared to the controls, which supporting for the viewpoint that the occurrence of diabetes is related to inflammation [97]. Takada et al. found that serum monomeric α2-macroglobulin is highly expressed in many diabetic subjects by mass spectrometry analysis, and it might become an important biomarker for diagnosis of T2DM [98]. A Swedish study identified cathepsin D and confirmed six proteins (leptin, renin, interleukin-1 receptor antagonist [IL-1ra], hepatocyte growth factor, fatty acid-binding protein 4, and tissue plasminogen activator [t-PA]) as IR biomarkers [99]. Huth et al. identified proteins related to the prediction and early diagnosis of T2DM [100]. Mannose-binding lectin-associated serine protease 1 (MASP) levels were positively associated with both incident type 2 diabetes and prediabetes. Adiponectin was inversely associated with incident type 2 diabetes. MASP, adiponectin, apolipoprotein A-IV, apolipoprotein C-II, C reactive protein were associated with individual continuous outcomes. A recent example of MS (Mass Spectrometry) proteomics analysis, paired with 2-dimensional gel electrophoresis, showed higher levels of Alpha-1-antichymotrypsin, Alpha-1-antitrypsin, apolipoprotein A-I, haptoglobin, retinol-binding protein 4, transthyretin, and zinc-alpha-2-glycoprotein in those with abdominal adiposity or insulin resistance compared with normal individuals [101].
Benabdelkamel et al. compared the protein expression of mature adipocytes within subcutaneous adipose tissues and found that, compared to the healthy individuals, a total of 23 proteins specifically expressed were identified in obese subjects, which are mainly involved in glucose and lipid metabolism, energy regulation, cytoskeleton structure and redox reactions [102]. Bae et al. proposed that individuals who are obese harbor a large number of differential proteins in insulin-sensitive tissues such as liver, skeletal muscle and adipose tissue. Of these, leukocyte common antigen-related phosphatase, PTP-α (protein tyrosine phosphatase α), PTP-1B (protein tyrosine phosphatase 1B) are highly expressed. Follow-up studies demonstrated that these enzymes participate in insulin signaling [103]. It has been established that cofilin-1 (CFL1) has an inhibitory effect on brown adipocyte differentiation. The overexpression of CFL1 inhibited the brown fat deposition and repressed the brown marker genes UCP1, PRDM16, PGC-1α and PPARγ [104].
3.4. Metabolomics
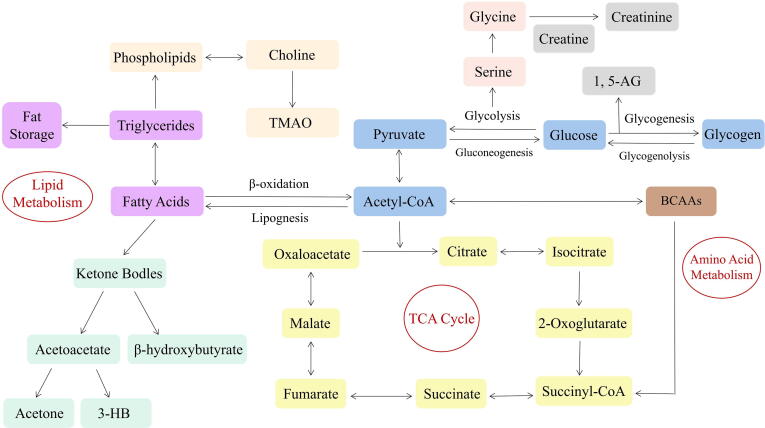
Metabolomics often utilizes approaches based on nuclear magnetic resonance (NMR) and/or various MS techniques to analyse the metabolites in biological samples, including low-molecular-weight compounds such as amino acids, organic acids, lipids, nucleotides, and sugars [105]. A review of recent research revealed that many studies found correlations between glycolipid metabolism disorder and metabolomics characteristics [106]. Thus, metabolomics research can be used to describe abnormal metabolism during the progression of glycolipid metabolism disorder, provide insight into disease mechanisms, and explore disease-associated biomarkers to assess the severity of disease and potential metabolic pathways (Fig. 4).
Fig. 4.
Schematic representation of the metabolic pathways in in the events of glycolipid metabolism disorder. Purple represents fatty acids metabolism; Flesh color represents choline metabolism; bisque represents phospholipids metabolism; green represents ketone metabolism; blue represents carbohydrate metabolism; yellow represents the tricarboxylic acid (TCA) cycle; pink, gray, and brown represent amino acid metabolism.
3.4.1. Amino acids metabolomics of glycolipid metabolism disorder
In recent years, numerous studies found that branched chain amino acids (BCAAs) are potential biomarkers of glycolipid metabolism disorder, including valine, leucine and isoleucine. Newgard et al confirmed that BCAAs in particular were higher in individuals that were obese in a cross-sectional metabolomics analysis of obese and lean individuals [107]. Guasch-Ferré et al meta-analyzed results from eight prospective studies that reported risk estimates for metabolites and T2DM, including 8,000 individuals of whom 1,940 had T2DM [108]. The results showed that BCAAs was positively associated with the risk of T2DM. Growing experimental evidence have posited potential mechanism of glycolipid metabolism disorder caused by up-regulation of BCAAs. BCAAs and their corresponding branched chain keto acids (BCKAs) can activate mTOR signaling, induce oxidative stress, cause mitochondrial dysfunction, and potentially contributing to the development of further insulin resistance [109], [110]. Analyses in a smaller cohort utilizing Mendelian randomization suggested that higher BCAAs levels do not have a causal effect on insulin resistance while increased insulin resistance drives higher circulating fasting BCAAs levels [111]. This findings point to elevated BCAAs as a downstream effect of adiposity and insulin resistance. Several studies have also found positive associations of aromatic amino acids, including tyrosine and phenylalanine, with future development of T2DM [112], while glycine and glutamine were negatively correlated with the development of T2DM [113], [114]. Higher levels of 2-aminoadipic acid (2-AAA), a lysine degradation product, were also found to be associated with increased risk for incident diabetes mellitus [115]. 2-AAA is associated with adipogenesis and insulin resistance, and can serve as a diabetes risk marker.
3.4.2. Lipids metabolomics of glycolipid metabolism disorder
Blood levels of free fatty acids (FFAs) rise slowly with elevated body mass, which is considered as an important feature of obesity-related metabolic diseases. Elevated clinical measures of lipids, specifically of bulk triglycerides, are considered a traditional risk factor for T2DM. The intracellular accumulation of fatty acid (FA) oxidation products such as diacylglycerols, triacylglycerols, and ceramides is linked with insulin resistance [116]. A large population-based study showed that fasting serum levels of glycerol, FFAs, monounsaturated FAs, saturated FAs, and n-7 and n-9 FAs are biomarkers for an increased risk of development of hyperglycemia and T2DM [117]. Lu et al. conducted metabolomics analysis of serum in the Chinese population and found that partial free fatty acid (palmitic acid, stearic acid, oleic acid and linoleic acid) and some ketone bodies (acetone and acetoacetic acid) in T2DM patients were significantly higher than those in healthy controls [118]. Ketone bodies are products of fat catabolism that are used as alternative substrates to glucose as sources of energy when carbohydrate intake is low and there is a surplus of circulating FFAs. It is considered to be a key metabolites in metabolism disruption. Several reports have shown that total ketone bodies were mildly elevated in patients with T2DM and were associated with fasting FFAs and inversely associated with triglycerides and insulin resistance [119]. Phospholipids are critical components of the cell lipid bilayer. Recent research has established that lysophosphatidylcholines (LPCs) and lysophosphatidylethanolamines (LPEs), phosphatidylcholine-plasmalogens (PC-PLs), sphingomyelins (SMs), and cholesterol esters (CEs) were inversely associated with risk of T2DM, while triacylglycerols (TAGs), diacylglycerols (DAGs), and phosphatidylethanolamines (PEs) were positively associated with T2DM risk [120].
3.4.3. Carbohydrate metabolomics of glycolipid metabolism disorder
Elevated glucose level is an important metabolic feature of glycolipid metabolism disorder. Hexose sugars are the most frequently analyzed carbohydrate in metabolomics studies of incident diabetes mellitus. A prospective study revealed that hexose sugars was positively correlated with T2DM, whereas a species of mannitol and several deoxyhexose sugars were found to be inversely associated with diabetes mellitus risk [121]. Mack et al. conducted oral glucose tolerance test (OGTT) on healthy control populations, prediabetic populations and diabetic participants, and found that maltose, trehalose, fructose and mannose in plasma of prediabetic populations and diabetic participants were higher than those in healthy populations [122]. At the onset of glycolysis, glucose is converted to pyruvate inside the cell. Lu et al. found that serum pyruvate concentration was significantly higher in T2DM patients compared with normal controls, indicating increased glycolysis in T2DM patients [123]. A recent study based on 1H NMR found elevated levels of pyruvate, lactate, and citric acid in T2DM patients, as well as elevated serum levels of tricarboxylic acid (TCA) cycle intermediates, such as succinic acid, creatine, and creatinine, compared with healthy controls [124].
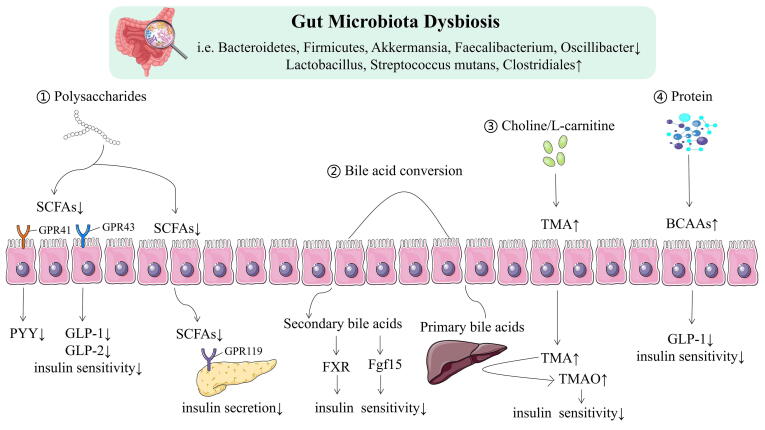
3.4.4. The regulatory role of metabolites of gut microbiota in glycolipid metabolism disorder
Gut microbiota is involved in the catabolism and anabolism of nutritional elements in daily foods. About 10 % of the circulating metabolites in the human body come from bacteria and participate in metabolic regulation in human [125]. The metabolic products of gut microbiota, such as short-chain fatty acids (SCFAs), BCAAs, trimethylamine oxide (TMAO), tryptophan and indole derivatives, are intimately correlated with the pathogenesis of glycolipid metabolism disorder. In the proximal colon, gut microbiota ferment carbohydrates to produce SCFAs (such as acetate, propionate, and butyrate). Numerous in vitro and in vivo studies revealed that SCFAs, as beneficial microbial metabolites for prevention and treatment of glycolipid metabolism disorder, participate in the maintenance of intestinal mucosa integrity, improve glycolipid metabolism, control energy expenditure and regulate the immune system and inflammatory responses [126], [127]. In contrast, in the distal colorectum, protein hydrolysis and fermentation can yield various harmful metabolites such as BCAAs (valine, isoleucine and leucine), phenols, and ammonia. BCAAs are involved in various bioprocess such as protein metabolism, gene expression, insulin resistance and proliferation of hepatocytes [128]. Gut microbiota also can directly modulate bile acid (BA) metabolism through the enterohepatic FXR-FGF15-FGFR4 axis. BA regulates cholesterol and triglyceride metabolism and maintains glucose and energy homeostasis [129], [130]. Additionally, the gut microbiota can metabolise choline and L-carnitine from dietary sources (eg, red meat, eggs and fish) to produce trimethylamine (TMA), and then convert into TMAO [131]. In humans, the level of TMAO increased in patients with diabetes or at risk of diabetes and in obesity [132], [133], [131]. Tryptophan is an essential aromatic amino acid acquired through common diet sources, including oats, poultry, fish, milk and cheese. In addition to kynurenine and serotonin, tryptophan can also be directly metabolized into indole and its derivatives by gut microbiota, some of which are available as aromatic hydrocarbon receptor (AhR) ligands [134]. It has previously been observed that metabolic disorders are characterised by a reduced capacity of the microbiota to metabolise tryptophan into AhR agonists [135]. It was recently shown that IMP (imidazole propionate), a metabolite produced by histidine utilisation of gut microbiota, was enhanced in T2DM and associated with insulin resistance [136].
3.4.5. Other metabolites
1,5-anhydroglucitol (1,5-AG) is the major polyol in vivo, which structure is similar to glucose. Most notably, 1,5-AG level is reflective of short-term glucose status, postprandial hyperglycemia, and glycemic variability which are not captured by HbA1c assay [137]. Studies found evidence that a single-nucleotide polymorphism in the CYP7A1 coding region associated with deoxycholic acid levels that was also associated with T2DM in published GWAS meta-analyses, and the metabolism of bile acids and phospholipids shares some common genetic origin with T2DM [138]. Ferrannini et al identified α-hydroxybutyrate (α-HB) and linoleoyl-glycerophosphocholine (L-GPC) as joint markers of IR and glucose intolerance [139]. α-HB is an organic acid positioned at an interesting crossroad of intermediary metabolism—amino acid catabolism and glutathione synthesis—and upstream to the TCA cycle. Prior studies that have noted that α-HB is derived from α-ketobutyrate and has the potential to identify IR and risk of impairment of glycemic control and conversion of prediabetes to an evident diabetic state [140], [141]. Metabolomics and lipidomics delineation by liquid/gas chromatography mass spectrometry was conducted on 115 middle-aged Dutch individuals (50 with MetS; 65 controls) in the Leiden Longevity Study [142]. They found that 9 metabolites were negatively correlated and 26 metabolites (mostly acyl carnitines, amino acids and keto acids) were positively correlated with the metabolic syndrome score. In addition, the metabolic syndrome (score) was associated with multiple individual metabolites (e.g., valeryl carnitine, pyruvic acid, lactic acid, alanine) and lipids in the univariate analyses [143]. Mainly, these molecules were intertwined with the metabolism of glucose, amino acid, and lipid.
3.5. Gut microbiomics
Intestinal flora is the microbiota colonized the gut. Its composition and function can be influenced by many factors, such as inheritance, living circumstances, lifestyle, dietary habits and drugs, and thus may impact the glucose and lipid metabolism in the host through inflammation and immune responses and metabolic pathways [144]. Genomics technologies, such as shotgun metagenomic sequencing and high-throughput sequencing of 16S rRNA gene, are by far commonly employed techniques of microbiome sequencing to determine the diversity, composition, structure distribution and function of gut microbiota. It has been reported that the gut microbiota of patients with glycolipid metabolism disorder is comprised primarily of opportunistic pathogens, accompanied by decreasing in beneficial microbes (Fig. 5).
Fig. 5.
Schematic overview of functions attributed to gut microbiota. SCFA, short chain fatty acids; GPR41, G-protein coupled receptor 41; GPR43, G-protein coupled receptor 43; GPR119, G-protein coupled receptor 119; GLP1, Glucagon Like Protein 1; GLP2, Glucagon Like Protein 2; PYY, peptide YY; TMA, trimethylamine; TMAO, Trimethylamine-N-oxide; FXR, farnesoid X receptor; Fgf15, fibroblast growth factor 15; BCCAs, branch Chain Amino Acids.
The gut microbiota plays a predominant role in host nutrient metabolism, xenobiotic and drug metabolism, maintenance of structural integrity of the gut mucosal barrier, immunomodulation, and protection against pathogens [145]. Perturbations in gut microbiota can have negative health consequences. Particularly, the gut microbiota has advanced as an important contributor to the development of glycolipid metabolism disorder. In diabetic humans, there is a lack of uniformity in gut microbiota profiles. A number of researchers have demonstrated that the relative abundances of the genus Lactobacillus is positively correlated with T2DM [146], [147]. Notably, the association of several species of Lactobacillus with T2DM is species-specific. For example, in T2DM patients, Lactobacillus acidophilus and Lactobacillus gasseri are decreased, while Lactobacillus xylosus is increased [148], [149]. These results suggested that this bacterial genus’ influence on host metabolism present highly diversity. Studies in different population have also shown that diabetic gut microbiota have lower concentrations of Roseburia intestinalis and Faecalibacterium prausnitzii (both butyrate-producing bacteria), and higher levels of Streptococcus mutans and members of Clostridiales [150].
In recent years, the influence of gut microbiota as a potential mechanism driving factors of obesity and its related comorbidity has become the focus of attention. Up to now, most studies displayed that obesity leads to low richness and diversity of gut microbiota [151], [152], [153]. Turnbaugh et al. demonstrated for the first time that transferring the gut microbiota from genetic obesity model (ob/ob mice) to germ-free mice by fecal microbiota transplantation (FMT) resulted in body fat accumulation and body weight gain in the latter [154]. In addition, comparisons of the distal gut microbiota of genetically obese mice and their lean littermates, as well as those of obese and lean human volunteers revealed that obesity is associated with changes in the relative abundance of the two dominant bacterial divisions, the Bacteroidetes and the Firmicutes. Liu et al found that the abundance of Bacteroides thetaiotaomicron, a glutamate-fermenting commensal, was markedly decreased in obese individuals [155]. Obesity was associated with notable changes in microbiome composition, such as Akkermansia, Faecalibacterium, Oscillibacter, and Alistipes, which show a significant decrease [156].
4. Recent advances in joint multi-omics analyses of glycolipid metabolism disorder
The initiation and progression of human diseases involve several pathological processes at the genome, transcriptome, proteome and metabolome levels. Single-omics data only reflect changes at one disease level and have limited effectiveness at screening disease targets. The comprehensive analysis of multilevel omics data is a more integrative and accurate approach towards individualized treatment, elucidation of the molecular mechanisms of disease, early clinical diagnostics, prognostics, and drug dosage and administration.
4.1. Transcriptomics and proteomics combination
Transcriptomics and proteomics combination simultaneously measure overall RNA and protein status, clarify their roles in various physiological processes, and reveal their mutual regulation and association. Transcriptomics does not fully reflect all biological characteristics while proteomics does not dynamically reflect gene expression. However, integrating both analytical methods mutually overcomes these deficiencies. Using a combination of transcriptomics and proteomics, Haythorne et al find significant dysregulation of major metabolic pathways in islets of diabetic βV59M mice. Multiple genes/proteins involved in glycolysis/gluconeogenesis are upregulated, whereas those involved in oxidative phosphorylation and branched chain amino acid metabolism are markedly downregulated. Indeed, aldolase B was the most upregulated of all proteins (65-fold) and there was also a dramatic increase in both mRNA (246-fold) and protein levels (40-fold) of the fructose/glucose transporter SLC5A10 [157]. Losko et al. revealed the role of MCPIP1 in adipogenesis and adipocyte metabolism by proteomics and transcriptomics [158]. RNA-Seq analysis followed by confirmatory Q-RT-PCR revealed that elevated MCPIP1 levels in 3T3-L1 adipocytes upregulated transcripts encoding proteins involved in signal transmission and cellular remodeling and downregulated transcripts of factors involved in metabolism. These data are consistent with proteomic analysis, which showed that MCPIP1 expressing adipocytes exhibit upregulation of proteins involved in cellular organization and movement and decreased levels of proteins involved in lipid and carbohydrate metabolism. Moreover, MCPIP1 adipocytes are characterized by decreased level of insulin receptor, reduced insulin-induced Akt phosphorylation, as well as depleted Glut4 level and impaired glucose uptake.
4.2. Metabolomics and proteomics combination
Metabolomics and proteomics identify disease biomarkers. A combination of proteomics and metabolomics may be used for simultaneous mechanistic and phenotypic studies, systematically describes the regulation of protein synthesis and metabolism, discloses the upstream and downstream regulatory pathways of key proteins and metabolites, and helps explain the signaling pathways and mechanisms associated with disease development. Researchers have used proteomics and metabolomics to elucidate the mechanism of food-induced cholesterol biosynthesis. They found that elevated postprandial blood glucose and insulin levels activate mTORC1 (mechanistic target of rapamycin complex 1), which stabilizes HMGCR (3-hydroxy-methylglutaryl coenzyme A reductase), phosphorylates USP20 (ubiquitin-specific peptidase 20), and upregulates cholesterol biosynthesis. Long-term high-sucrose, high-fat diets induce USP20 phosphorylation, stabilize HMGCR, increases serum cholesterol, and cause metabolic diseases. By contrast, USP20 inhibition promotes HMGCR degradation, reduces lipid biosynthesis, enhances succinate production, and promotes heat generation. Therefore, USP20 inhibition is potentially an effective therapeutic approach for metabolic disorders including hyperlipidemia, non-alcoholic fatty liver disease, obesity, and T2DM [159]. Wang et al. harvested small intestine tissue and collected serum samples from T2DM model Chinese hamsters, analyzed them by LC-MS/MS (liquid chromatography-tandem mass spectrometry) proteomics and GC–MS/MS (gas chromatography-tandem mass spectrometry) metabolomics, respectively, and performed joint analyses of the differentially expressed proteins and metabolites. Annotation by bioinformatics analysis revealed that these differentially abundant proteins in the small intestine were commonly associated with abnormal glucose and lipid metabolism, IR, impaired insulin secretion, amino acid metabolism disorders, and inflammatory dysregulation. Moreover, differentially abundant metabolites in the serum were amino acids and were related to diabetic IR. Combined analysis of metabolomics and proteomics revealed significant changes in glutathione metabolism, biosynthesis of phenylalanine, tyrosine and tryptophan, and arginine and proline metabolism in T2DM model Chinese hamsters [160].
4.3. Gut microbiomics and metabolomics combination
Gut microbiota are vital to human metabolism. They provide enzymes for various biochemical and metabolic pathways in the host, participate in amino acid, bile acid, and carbohydrate metabolism, and form co-metabolic relationships with the host. Metabolomics is based on high-throughput analysis and bioinformatics technology and investigates variations and trends in overall endogenous metabolism. It can detect metabolites in gut microbiota, reflect the changes in gut microbiota function that occur under specific conditions, intuitively examine the relationships among gut microbiota and disease development and progression, and provide a research basis for disease prevention and treatment. The analysis of gut microbiota diversity based on shotgun metagenomic and 16S rRNA gene sequencing plus metabolomics comprehensively explores the relationships among gut microbiota disease occurrence, drug metabolism and pharmacodynamics, and gut microbiota structure and function. Pedersen et al found that the serum metabolome of insulin-resistant individuals is characterized by increased levels of BCAAs, which correlate with a gut microbiome that has an enriched biosynthetic potential for BCAAs and is deprived of genes encoding bacterial inward transporters for these amino acids. Prevotella copri and Bacteroides vulgatus are identified as the main species driving the association between biosynthesis of BCAAs and IR [161]. A metagenomic and targeted metabolomic analysis is conducted in 182 lean and abdominally obese individuals with and without newly diagnosed T2DM. The abundance of Akkermansia muciniphila (A. muciniphila) significantly decreases in lean individuals with T2DM than without T2DM. Its abundance correlates inversely with serum 3β-chenodeoxycholic acid (β CDCA) levels and positively with insulin secretion and fibroblast growth factor 15/19 (FGF15/19) concentrations [162].
4.4. Joint multi-omics analyses
To identify the early stages of T2DM, researchers obtained samples from 106 healthy and prediabetic individuals over approximately-four years and profiled their transcriptomes, metabolomes, cytokines, proteomes, and changes in their microbiomes [163]. Regression analyses of steady-state plasma glucose (SSPG) in insulin-resistant and insulin-sensitive subjects disclosed that IR was associated with elevated inflammation. It was also associated with altered lipid metabolism, and several long-chain polyunsaturated fatty acids were positively correlated with SSPG. Researchers also analyzed the relationships among the gut microbiota and host metabolites. In insulin-sensitive but not insulin-resistant subjects, Barnesiella spp. were positively correlated with IL-1β and Faecalibacterium spp. were negatively correlated with TNF-α. (tumor necrosis factor alpha). Butyricimonas spp. were negatively correlated with four lipids in insulin-resistant subjects. Multi-omics profile analyses revealed molecules that were unique to each individual and different from the cohort mean. One subject had abnormal levels of various metabolites and cytokines relative to the cohort average. Ten months after the final medical visit, the subject was diagnosed with T2DM and the multi-omics data indicated the dysregulation of T2DM related pathways. IL-1ra and high-sensitivity C-reactive protein (hsCRP) were highly elevated during the last three medical visits prior to the T2DM diagnosis. Researchers detected exogenous substances such as methyluric acid and methylxanthine among the molecules strongly associated with IL-1ra. The aforementioned substances are metabolites associated with glucose tolerance dysfunction and gut dysbiosis and were closely associated with the expression of host factors in the complement system, acute immune response signaling, and the lipopolysaccharide (LPS)-stimulated mitogen-activated protein kinase (MAPK) pathway. All of these are associated with the development of T2DM. Loss of gut microbial diversity and gain of body weight were observed even when subjects were diagnosed with T2DM. Several researchers used linear mixed models to examine the underlying relationships among glucose (FPG, HbA1C), inflammation (hsCRP) levels, and multi-omics measurements in healthy-baseline models and the relative changes at all time points in dynamic models [164]. The study indicated that both HbA1C and hsCRP were positively associated with total white blood cell (WBC), monocyte and neutrophil counts. Hepatocyte growth factor (HGF) was also associated with HbA1C and hsCRP which is consistent with its role in glucose metabolism and modulation of the inflammatory response. The authors also reported that FPG and HbA1C are associated with “leukotriene biosynthesis” which contributes to inflammation and leads to insulin resistance. HbA1C is also associated with other lipid metabolism-related pathways including “plasma lipoprotein assembly” and “chylomicron assembly”. The foregoing findings underscore the connections among inflammation and lipid and glucose metabolism as well as the regulation of these processes.
5. Discussion
The incidences of disorders of glycolipid metabolism such as T2DM, obesity, and hyperlipidemia have risen to epidemic proportions and pose serious threats to human health. Important objectives in medical research are the elucidation of the pathogenesis of glycolipid metabolism disorder and the development and implementation of efficacious prevention and treatment strategies. Neuroendocrine axis dysfunction, IR, oxidative stress, chronic inflammatory response, and gut microbiota dysbiosis are now considered the main pathological mechanisms of glycolipid metabolism disorder. They mutually interact in an interwoven network and initiate and cause the progression of disease. For most patients, existing glycolipid metabolism disorder prevention measures such as altering dietary habits or increasing exercise are mostly ineffective. Available therapeutic measures can improve patient health status to a certain extent. It is nonetheless difficult to restore metabolic levels to normal. Thus, further exploration into the pathogenesis of glycolipid metabolism disorder is necessary. Technological advances have led to the ‘omics era’, which is enabling the collection and integration of data and information at different molecular levels. The information obtained through omics techniques will contribute to a better understanding of glycolipid metabolism disorder pathophysiology, offer new opportunities for diagnosis and prognosis and lead to improved management of patients with glycolipid metabolism disorder. However, owing to the limitations of the development of omics technologies and the complexity of the research on glycolipid metabolism disorder, the multi-omics research on glycolipid metabolism disorder still faces numerous challenges.
5.1. Multi-omics studies reveal the pathophysiology of glycolipid metabolism disorder
Multi-omics studies usually examine genes (genomics), RNA (transcriptome), proteins (proteomics), and downstream metabolites (metabolomics) produced during DNA replication, transcription, translation, and post-translational modification, respectively. Multi-omics data provide evidence for pathogenesis, identify biomarkers, and reveal therapeutic targets for glycolipid metabolism disorder.
Research into the genes regulating susceptibility to glycolipid metabolism disorders is crucial. These include TCF7L2, PPARG, KCNJ11, SLC30A8, FTO. and others. Genetic polymorphisms affect glycolipid metabolism disorder mainly by decreasing pancreatic β cell function or increasing IR. GWAS have disclosed disease-related targets by contrasting genomic data for cases and controls at the population level. GWAS have returned encouraging results and helped direct and focus future research. The prediction of glycolipid metabolism disorder by screening susceptibility genes is in its infancy and few studies have been conducted in this area. They can nonetheless provide clues for exploring pathogenesis and searching for drug targets. Pharmacogenomics is the study of the interrelationships among genetic polymorphisms and drug effects and is based on genomics. Pharmacogenomics helps improve drug efficacy and safety, guides the research and development of new drugs, and provides a reference for the clinical administration of individualized medicine. Several studies demonstrated that individual differences in pharmacological glycolipid metabolism disorder therapy are closely associated with genetic polymorphisms in drug transporter and targets, drug catabolic enzymes, and genes related to the risk of developing glycolipid metabolism disorders.
Non-coding RNAs (ncRNAs) regulate gene expression at the transcriptional and post-transcriptional levels and affects the progression of glycolipid metabolism disorder. LncRNAs affect several molecular signaling pathways and participate in glycolipid metabolism disorder. Lnc-BATE1 establishes and maintains brown fat and its thermogenic capacity. Downregulation of lncRNA TUG1 and lncRNA GAS5 is connected to the occurrence of glycolipid metabolism disorder. MiRNAs regulate target gene expression at the post-transcriptional level and may serve as clinical diagnostic biomarkers. MiR-375 is specific to pancreatic β cells and its overexpression inhibits glucose-induced insulin secretion in them. Overexpressed miR-7, miR-200, miR-29 and miR-802 play vital roles in the pathogenesis of glycolipid metabolism disorder. CircRNAs regulate genes, compete with miRNAs for binding sites, and control glycolipid metabolism disorder. In islet cells, CDR1-as overexpression interferes with miR-7 function and improves the insulin level. Silencing circArhgap5-2 may inhibit lipid droplet accumulation and downregulate adipogenic markers.
Proteomics effectively identifies dysregulated proteins and pathways in cells under pathological conditions and helps discover disease-specific mutations and epigenetic alterations. C-reactive protein and α2-macroglobulin are sensitive markers of T2DM. MASP is positively correlated with T2DM and prediabetes mellitus, Adiponectin is negatively correlated with T2DM onset. Cathepsin D, leptins, renins, IL-1ra, and t-PA are IR biomarkers. Leukocyte common antigen-related phosphatase, PTP-α and PTP-1B are upregulated in the livers, skeletal muscle, and adipose tissue of obese persons. The proteomics of glycolipid metabolism disorder is in its infancy. Nevertheless, progress has been made in the proteomics study of β cells, skeletal muscle, and adipose tissue. Other novel biomarkers will eventually be discovered, and they might play pivotal roles in the analysis of clinical serum, plasma, and urine samples.
Metabolic markers are downstream genome outputs and upstream environmental inputs. Studies on metabolites and metabolomics can disclose gene-environment interactions [165]. Several studies demonstrated that BCAAs, tyrosine, phenylalanine, 2-AAA, FFAs, ceramides, TAGs, DAGs, PEs, hexose, maltose, trehalose, fructose, mannose, deoxycholic acid, 1,5-AG and LPS are associated with numerous disease pathways in glycolipid metabolism disorder. Gut microbiota produce various metabolites that function as signaling molecules and substrates for host metabolic responses. They affect both physiological and pathological processes in the host. Previous research showed that the gut metabolites SCFAs, BCAAs, bile acids, and TMAO are closely associated with the development of glycolipid metabolism disorder.
The gut microbiota are novel potential drivers of the pathophysiology of glycolipid metabolism disorder, interact with obesity, low-grade inflammation, IR, and T2DM, and might function as hubs. Human gut microbiota also affect the brain function and alter host behavior via the microbe-gut-brain axis [166]. They can promote host metabolic health by facilitating weight loss, improving blood glucose control and IR, and so on. Hence, gut microbiota are promising drug targets in the treatment of glycolipid metabolism disorder. Many patients with glycolipid metabolism disorder have moderate gut dysbiosis. The abundances of the Lactobacillus spp., Lactobacillus gasseri and Streptococcus mutans are elevated while those of Roseburia intestinalis, Faecalibacterium prausnitzii, Bacteroides thetaiotaomicron, Akkermansi spp., Faecalibacterium spp., Oscillibacter spp., and Alistipes spp. are reduced in patients with glycolipid metabolism disorder. As gut microbiota play vital roles in human health and disease, antibiotic, probiotic, and prebiotic administration might regulate the gut microbiota and, by extension, glycolipid metabolism. Several studies showed that reasonable probiotic or prebiotic supplementation can regulate the host gut microbiota, thereby ameliorating energy metabolism and controlling chronic low-level inflammation [167]. Probiotic therapy improved glucose intolerance, hyperlipidemia, and hyperinsulinemia ina glucose-induced diabetic mouse model [168].
Single-omics research lacks multilevel integration and has limited utility in determining the etiology of complex diseases. For these reasons, multi-omics is now widely applied in glycolipid metabolism disorder research. Multi-omics confirms pathogenesis through both macro- and micro-etiology, comprehensively and systematically investigates the roles of the environment and genetics in glycolipid metabolism disorder, and elucidates the pathogenic factors and molecular mechanisms of these diseases. Multi-omics also explores the factors mediating the association between the environment and lifestyle in glycolipid metabolism disorder and could, therefore, clarify pathogenic mechanisms. Multi-omics could also help develop risk prediction models for use in precision medicine, predict disease in high-risk individuals, and screen drug treatment subjects to monitor drug efficacy and adverse reactions.
5.2. Multi-omics studies exist limitations
Multi-omics studies of glycolipid metabolism disorder are gradually becoming more profound with the development of omics technology and bioinformatics. The precise diagnosis and treatment of glycolipid metabolism disorder based on joint multi-omics analyses could eventually dominate the field. However, several limitations remain. Early and timely diagnosis of glycolipid metabolism disorders could improve the control and prognosis of these diseases. GWAS expanded the identification of relevant gene loci associated with glycolipid metabolism disorder. In clinical practice, however, the applicability of known susceptibility polymorphisms is limited. Furthermore, existing gene loci only explain part of the genetic variation in glycolipid metabolism disorder and require validation in clinical samples and trials. Stable, detectable ncRNAs could serve as molecular markers for the clinical diagnosis and prognosis of glycolipid metabolism disorder. Nevertheless, there are few comprehensive studies on ncRNAs, their mechanisms are unclear, and big sample data are lacking for them. Proteomics has been widely applied in the study of diseases related to glycolipid metabolism disorder. As proteomics is relatively new, it is not yet optimally reproducible. Certain rare proteins are difficult to identify and studies on them are time-consuming and expensive. Current research still compares tissue and serum samples between healthy individuals and those diagnosed with glycolipid metabolic disorders. Metabolomics is an important technological approach in the study of glycolipid metabolism disorder. Its integration with multi-omics data has begun to elucidate the complex relationships among gut microbiota, host metabolism, and the pathogenesis of glycolipid metabolism disorder. This investigative strategy has also led to novel diagnostic and therapeutic approaches and has laid a solid foundation for precision medicine. However, metabolomics lacks a universal analytical platform and mature consistent operational methods. Moreover, its results are conflicting. Most studies have not been analytically or clinically validated. Hence, there are few examples of the highly efficient application of metabolomics in clinical research. Gut dysbiosis has been implicated in obesity, diabetes, other diseases, and their progression. Ameliorating gut dysbiosis might help treat glycolipid metabolism disorder. However, the precise components and metabolic activity of the gut microbiome associated with glycolipid metabolism disorder are unknown. Evidence from animal experimentation suggests that the gut microbiota is the key to the development of obesity, inflammation, insulin resistance, and intestinal barrier dysfunction. Nevertheless, there are few human clinical mechanistic studies, and randomized, large-sample, multicenter clinical trials are required. Though joint multi-omics analyses expand investigations into glycolipid metabolism disorder, they may lead to false discoveries because of the combined effects of multiple factors and high variability among individual datasets. Thus, it is difficult to interpret multi-omics data or use them to identify biologically relevant molecules. As international data become publicly available and analytical platforms and collaborative groups increase, available resources for multi-omics studies will become more abundant and the cost of research will dramatically decrease. On the other hand, long-term follow-up and laboratory tests are required for ongoing research and the ethical and data sharing issues related to the research are of great concern. GWAS has high throughput and low genome detection costs and has, therefore, been widely used in large-scale cohorts. By contrast, metabolomics and proteomics have relatively low throughput and high cost. For these reasons, it is still comparatively uncommon to apply omics in large-scale cohort assays. Integrative multi-omics data analysis is still in its infancy and universal data integration and analytical methods must be developed to make full and effective use of available multi-omics data.
6. Future perspectives
The innovation of high-throughput technologies and omics data including genomes, transcriptomes, proteomes and metabolomes, and so on has disclosed risk factors and helped develop novel biomarkers associated with glycolipid metabolism disorder. Disease biomarkers reveal specific pathological features and detect changes in the status of various medical conditions. Though they may have high predictive efficacy in clinical studies, they nonetheless have certain limitations. The reliability of a biomarker depends upon the genetic background of the study subject, the treatment regimen, the composition of the gut microbiome, and the diagnosis and intervention times. Moreover, biomarkers do not possess universal clinical value. Therefore, multicenter, large-scale, standardized clinical studies are required to improve the diagnostic efficacy and practical applicability of glycolipid metabolism disorder biomarkers.
The development of personalized treatment options for glycolipid metabolism disorder requires clinical feature information, the integration of complex multi-omics data, and a metabolite network map. In this manner, the expression patterns of key functional genes and signaling pathways regulating glycolipid metabolism disorders may be precisely plotted. For these reasons, multidimensional data sources must be integrated into big data studies to develop precision medicine for glycolipid metabolism disorder. A systematic and comprehensive understanding of the risk factors associated with glycolipid metabolism disorder is required to predict and prevent these medical conditions.
Prospective cohorts, multi-omics studies, high-quality baselines, follow-up information, biological samples, and multi-omics detection should be applied. In this way, novel risk factors associated with glycolipid metabolism disorders may be identified and their pathogenesis may be elucidated. Future multi-omics studies on glycolipid metabolism disorder, will require more clinical samples for verification and must develop and validate stratified risk models. The latter apply bioinformatics analysis and integrate high-throughput genomics, transcriptomics, proteomics, and metabolomics data to obtain comprehensive information regarding susceptibility genes, mechanistic pathways, and disease stage markers of glycolipid metabolism disorder. This information will facilitate early, precise intervention and enable accurate diagnosis and treatment in populations with high risk and incidence of glycolipid metabolism disorders.
7. Authors’ contributions
Jiaxing TIAN developed the review question. The initial literature review was performed by Xinyi FANG. The first draft of the manuscript was written by Xinyi FANG with all authors commenting on subsequent versions of the manuscript. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to thank all the authors for their contribution to the realization of this manuscript.
Availability of data and materials
The data and materials used are included in the review.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All the studies mentioned in this review were approved by the Ethics Committee, and written informed consent was obtained by all the participants.
Funding
This work was supported by the National Natural Science Foundation of China (81904187), Capital Health Development Research Project (CD2020-4-4155), CACMS Outstanding Young Scientific and Technological Talents Program (ZZ13-YQ-026), CACMS Innovation Fund (CI2021A01601), Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (ZYYCXTD-D-202001), Open Project of National Facility for Translational Medicine (TMSK-2021-407).
Human and animal rights and informed consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.10.030.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Zheng Y., Ley S.H., Hu F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat Rev Endocrinol. 2018;14:88–98. doi: 10.1038/nrendo.2017.151. [DOI] [PubMed] [Google Scholar]
- 2.Ataey A., Jafarvand E., Adham D., et al. The Relationship Between Obesity, Overweight, and the Human Development Index in World Health Organization Eastern Mediterranean Region Countries. J Prev Med Public Health. 2020;53(2):98–105. doi: 10.3961/jpmph.19.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NCD Risk Factor Collaboration (NCD-RisC) Repositioning of the global epicentre of non-optimal cholesterol. Nature. 2020;582(7810):73–77. doi: 10.1038/s41586-020-2338-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ji L, Hu D, Pan C, et al. Primacy of the 3B approach to control risk factors for cardiovascular disease in type 2 diabetes patients. Am J Med. 2013;126(10):925.e11-925.e9.25E22. 10.1016/j.amjmed.2013.02.035. [DOI] [PubMed]
- 5.Di S., Wang Y., Han L., et al. The Intervention Effect of Traditional Chinese Medicine on the Intestinal Flora and Its Metabolites in Glycolipid Metabolic Disorders. Evid Based Compl Alternat Med. 2019;2019:2958920. doi: 10.1155/2019/2958920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao J., Zhang M., Niu R., et al. The combination of cinnamaldehyde and kaempferol ameliorates glucose and lipid metabolism disorders by enhancing lipid metabolism via AMPK activation. J Funct Foods. 2021;83(7) doi: 10.1016/j.jff.2021.104556. [DOI] [Google Scholar]
- 7.Khadke S., Mandave P., Kuvalekar A., et al. Synergistic Effect of Omega-3 Fatty Acids and Oral-Hypoglycemic Drug on Lipid Normalization through Modulation of Hepatic Gene Expression in High Fat Diet with Low Streptozotocin-Induced Diabetic Rats. Nutrients. 2020;12(12):3652. doi: 10.3390/nu12123652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto T., Kiuchi S., Murase T. Synergistic activation of thermogenic adipocytes by a combination of PPARγ activation, SMAD3 inhibition and adrenergic receptor activation ameliorates metabolic abnormalities in rodents. Diabetologia. 2019;62(10):1915–1927. doi: 10.1007/s00125-019-4938-6. [DOI] [PubMed] [Google Scholar]
- 9.Matu J., Gonzalez J.T., Ispoglou T., et al. The effects of hypoxia on hunger perceptions, appetite-related hormone concentrations and energy intake: A systematic review and meta-analysis. Appetite. 2018;125:98–108. doi: 10.1016/j.appet.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 10.Gao Y., Vidal-Itriago A., Milanova I., et al. Corrigendum to “Deficiency of leptin receptor in myeloid cells disrupts hypothalamic metabolic circuits and causes body weight increase” [Molecular Metabolism 7 (2018) 155–160] Mol Metab. 2019;20:205. doi: 10.1016/j.molmet.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanfray D., Arthaud S., Ouellet J., et al. Gliotransmission and brain glucose sensing: critical role of endozepines. Diabetes. 2013;62(3):801–810. doi: 10.2337/db11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balland E., Dam J., Langlet F., et al. Hypothalamic tanycytes are an ERK-gated conduit for leptin into the brain. Cell Metab. 2014;19(2):293–301. doi: 10.1016/j.cmet.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGlashon J.M., Gorecki M.C., Kozlowski A.E., et al. Central serotonergic neurons activate and recruit thermogenic brown and beige fat and regulate glucose and lipid homeostasis. Cell Metab. 2015;21(5):692–705. doi: 10.1016/j.cmet.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mastrototaro L., Roden M. Insulin resistance and insulin sensitizing agents. Metabolism. 2021;125 doi: 10.1016/j.metabol.2021.154892. [DOI] [PubMed] [Google Scholar]
- 15.James D.E., Stöckli J., Birnbaum M.J. The aetiology and molecular landscape of insulin resistance. Nat Rev Mol Cell Biol. 2021;22(11):751–771. doi: 10.1038/s41580-021-00390-6. [DOI] [PubMed] [Google Scholar]
- 16.Roden M., Shulman G.I. The integrative biology of type 2 diabetes. Nature. 2019;576(7785):51–60. doi: 10.1038/s41586-019-1797-8. [DOI] [PubMed] [Google Scholar]
- 17.Korac B., Kalezic A., Pekovic-Vaughan V., et al. Redox changes in obesity, metabolic syndrome, and diabetes. Redox Biol. 2021;42 doi: 10.1016/j.redox.2021.101887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lackey D.E., Olefsky J.M. Regulation of metabolism by the innate immune system. Nat Rev Endocrinol. 2016;12(1):15–28. doi: 10.1038/nrendo.2015.189. [DOI] [PubMed] [Google Scholar]
- 19.Qin Q., Shou J., Li M., et al. Stk24 protects against obesity-associated metabolic disorders by disrupting the NLRP3 inflammasome. Cell Rep. 2021;35(8) doi: 10.1016/j.celrep.2021.109161. [DOI] [PubMed] [Google Scholar]
- 20.Wu J., Wang K., Wang X., et al. The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell. 2021;12(5):360–373. doi: 10.1007/s13238-020-00814-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S.Z., Yu Y.J., Adeli K. Role of Gut Microbiota in Neuroendocrine Regulation of Carbohydrate and Lipid Metabolism via the Microbiota-Gut-Brain-Liver Axis. Microorganisms. 2020;8(4):527. doi: 10.3390/microorganisms8040527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hampel H., Nisticò R., Seyfried N.T., et al. Omics sciences for systems biology in Alzheimer's disease: State-of-the-art of the evidence. Ageing Res Rev. 2021;69 doi: 10.1016/j.arr.2021.101346. [DOI] [PubMed] [Google Scholar]
- 23.Grozinger C.M., Zayed A. Improving bee health through genomics. Nat Rev Genet. 2020;21(5):277–291. doi: 10.1038/s41576-020-0216-1. [DOI] [PubMed] [Google Scholar]
- 24.Horikoshi M., Beaumont R.N., Day F.R., et al. Genome-wide associations for birth weight and correlations with adult disease. Nature. 2016;538(7624):248–252. doi: 10.1038/nature19806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis [published correction appears in Nat Genet. 2011 Apr;43(4):388]. Nat Genet. 2010;42(7):579-589. 10.1038/ng.609. [DOI] [PMC free article] [PubMed]
- 26.Nguyen-Tu M.S., da Silva X.G., Leclerc I., et al. Correction: Transcription factor-7-like 2 (TCF7L2) gene acts downstream of the Lkb1/Stk11 kinase to control mTOR signaling, β cell growth, and insulin secretion. J Biol Chem. 2018;293(47):18420. doi: 10.1074/jbc.AAC118.006541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant S.F., Thorleifsson G., Reynisdottir I., et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 28.Ding W., Xu L., Zhang L., et al. Meta-analysis of association between TCF7L2 polymorphism rs7903146 and type 2 diabetes mellitus. BMC Med Genet. 2018;19(1):38. doi: 10.1186/s12881-018-0553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noordam R., Zwetsloot C.P.A., de Mutsert R., et al. Interrelationship of the rs7903146 TCF7L2 gene variant with measures of glucose metabolism and adiposity: The NEO study. Nutr Metab Cardiovasc Dis. 2018;28(2):150–157. doi: 10.1016/j.numecd.2017.10.012. [DOI] [PubMed] [Google Scholar]
- 30.Hsiao T.J., Lin E. A common rs7903146 variant of the transcription factor 7-like 2 gene is associated with type 2 diabetes mellitus and fasting glucose in a Taiwanese population. Diabetes Metab. 2017;43(1):83–85. doi: 10.1016/j.diabet.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Plengvidhya N., Chanprasert C., Chongjaroen N., et al. Impact of KCNQ1, CDKN2A/2B, CDKAL1, HHEX, MTNR1B, SLC30A8, TCF7L2, and UBE2E2 on risk of developing type 2 diabetes in Thai population. BMC Med Genet. 2018;19(1):93. doi: 10.1186/s12881-018-0614-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vigouroux C., Fajas L., Khallouf E., et al. Human peroxisome proliferator-activated receptor-gamma2: genetic mapping, identification of a variant in the coding sequence, and exclusion as the gene responsible for lipoatrophic diabetes. Diabetes. 1998;47(3):490–492. doi: 10.2337/diabetes.47.3.490. [DOI] [PubMed] [Google Scholar]
- 33.Corrales P., Vidal-Puig A., Medina-Gómez G. PPARs and Metabolic Disorders Associated with Challenged Adipose Tissue Plasticity. Int J Mol Sci. 2018;19(7):2124. doi: 10.3390/ijms19072124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sladek R., Rocheleau G., Rung J., et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 35.Scott L.J., Mohlke K.L., Bonnycastle L.L., et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y., Li H., Loos R.J., et al. Common variants in CDKAL1, CDKN2A/B, IGF2BP2, SLC30A8, and HHEX/IDE genes are associated with type 2 diabetes and impaired fasting glucose in a Chinese Han population. Diabetes. 2008;57(10):2834–2842. doi: 10.2337/db08-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rong R., Hanson R.L., Ortiz D., et al. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes. 2009;58(2):478–488. doi: 10.2337/db08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerken T., Girard C.A., Tung Y.C., et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi: 10.1126/science.1151710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Magno F.C.C.M., Guaraná H.C., Fonseca A.C.P., et al. Influence of FTO rs9939609 polymorphism on appetite, ghrelin, leptin, IL6, TNFα levels, and food intake of women with morbid obesity. Diabetes Metab Syndr Obes. 2018;11:199–207. doi: 10.2147/DMSO.S154978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kamura Y., Iwata M., Maeda S., et al. FTO Gene Polymorphism Is Associated with Type 2 Diabetes through Its Effect on Increasing the Maximum BMI in Japanese Men. PLoS One. 2016;11(11):e0165523. doi: 10.1371/journal.pone.0165523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saravani R., Galavi H.R., Noorzehi N., et al. Common Variations in Perilipin rs1052700 and FTO rs3751812 Gene Variants, and Risk for Obesity and Type-2 Diabetes. Rep Biochem Mol Biol. 2017;6(1):80–87. [PMC free article] [PubMed] [Google Scholar]
- 42.Inagaki N., Gonoi T., Seino S. Subunit stoichiometry of the pancreatic beta-cell ATP-sensitive K+ channel. FEBS Lett. 1997;409(2):232–236. doi: 10.1016/s0014-5793(97)00488-2. [DOI] [PubMed] [Google Scholar]
- 43.Inagaki N., Gonoi T., Clement J.P., 4th, et al. Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270(5239):1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 44.Liu L., Nagashima K., Yasuda T., et al. Mutations in KCNJ11 are associated with the development of autosomal dominant, early-onset type 2 diabetes. Diabetologia. 2013;56(12):2609–2618. doi: 10.1007/s00125-013-3031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inoue H., Ferrer J., Welling C.M., et al. Sequence variants in the sulfonylurea receptor (SUR) gene are associated with NIDDM in Caucasians. Diabetes. 1996;45(6):825–831. doi: 10.2337/diab.45.6.825. [DOI] [PubMed] [Google Scholar]
- 46.Xue A., Wu Y., Zhu Z., et al. Genome-wide association analyses identify 143 risk variants and putative regulatory mechanisms for type 2 diabetes. Nat Commun. 2018;9(1):2941. doi: 10.1038/s41467-018-04951-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchsberger C., Flannick J., Teslovich T.M., et al. The genetic architecture of type 2 diabetes. Nature. 2016;536(7614):41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spracklen C.N., Horikoshi M., Kim Y.J., et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature. 2020;582(7811):240–245. doi: 10.1038/s41586-020-2263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Locke A.E., Kahali B., Berndt S.I., et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee D.H., Porta M., Jacobs D.R., Jr, et al. Chlorinated persistent organic pollutants, obesity, and type 2 diabetes. Endocr Rev. 2014;35(4):557–601. doi: 10.1210/er.2013-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ling C., Rönn T. Epigenetics in Human Obesity and Type 2 Diabetes. Cell Metab. 2019;29(5):1028–1044. doi: 10.1016/j.cmet.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buse JB, Wexler DJ, Tsapas A, et al. 2019 Update to: Management of Hyperglycemia in Type 2 Diabetes, 2018. A Consensus Report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD) [published correction appears in Diabetes Care. 2020 Jul;43(7):1670]. Diabetes Care. 2020;43(2):487-493. 10.2337/dci19-0066. [DOI] [PMC free article] [PubMed]
- 53.He R., Zhang D., Lu W., et al. SLC47A1 gene rs2289669 G>A variants enhance the glucose-lowering effect of metformin via delaying its excretion in Chinese type 2 diabetes patients. Diabetes Res Clin Pract. 2015;109(1):57–63. doi: 10.1016/j.diabres.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 54.Hou W., Zhang D., Lu W., et al. Polymorphism of organic cation transporter 2 improves glucose-lowering effect of metformin via influencing its pharmacokinetics in Chinese type 2 diabetic patients. Mol Diagn Ther. 2015;19(1):25–33. doi: 10.1007/s40291-014-0126-z. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H., Liu X., Kuang H., et al. Association of sulfonylurea receptor 1 genotype with therapeutic response to gliclazide in type 2 diabetes. Diabetes Res Clin Pract. 2007;77(1):58–61. doi: 10.1016/j.diabres.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 56.Feng Y., Mao G., Ren X., et al. Ser1369Ala variant in sulfonylurea receptor gene ABCC8 is associated with antidiabetic efficacy of gliclazide in Chinese type 2 diabetic patients. Diabetes Care. 2008;31(10):1939–1944. doi: 10.2337/dc07-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Javorsky M., Klimcakova L., Schroner Z., et al. KCNJ11 gene E23K variant and therapeutic response to sulfonylureas. Eur J Intern Med. 2012;23(3):245–249. doi: 10.1016/j.ejim.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 58.Lin C.H., Lee Y.S., Huang Y.Y., et al. Polymorphisms of GLP-1 receptor gene and response to GLP-1 analogue in patients with poorly controlled type 2 diabetes. J Diabetes Res. 2015;2015 doi: 10.1155/2015/176949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chedid V., Vijayvargiya P., Carlson P., et al. Allelic variant in the glucagon-like peptide 1 receptor gene associated with greater effect of liraglutide and exenatide on gastric emptying: A pilot pharmacogenetics study. Neurogastroenterol Motil. 2018;30(7):e13313. doi: 10.1111/nmo.13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waget A., Cabou C., Masseboeuf M., et al. Physiological and pharmacological mechanisms through which the DPP-4 inhibitor sitagliptin regulates glycemia in mice. Endocrinology. 2011;152(8):3018–3029. doi: 10.1210/en.2011-0286. [DOI] [PubMed] [Google Scholar]
- 61.Han E., Park H.S., Kwon O., et al. A genetic variant in GLP1R is associated with response to DPP-4 inhibitors in patients with type 2 diabetes. Medicine (Baltimore) 2016;95(44):e5155. doi: 10.1097/MD.0000000000005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Böhm A., Wagner R., Machicao F., et al. DPP4 gene variation affects GLP-1 secretion, insulin secretion, and glucose tolerance in humans with high body adiposity. PLoS ONE. 2017;12(7):e0181880. doi: 10.1371/journal.pone.0181880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hoeben E., De Winter W., Neyens M., et al. Population Pharmacokinetic Modeling of Canagliflozin in Healthy Volunteers and Patients with Type 2 Diabetes Mellitus. Clin Pharmacokinet. 2016;55(2):209–223. doi: 10.1007/s40262-015-0307-x. [DOI] [PubMed] [Google Scholar]
- 64.Zimdahl H., Haupt A., Brendel M., et al. Influence of common polymorphisms in the SLC5A2 gene on metabolic traits in subjects at increased risk of diabetes and on response to empagliflozin treatment in patients with diabetes. Pharmacogenet Genomics. 2017;27(4):135–142. doi: 10.1097/FPC.0000000000000268. [DOI] [PubMed] [Google Scholar]
- 65.Eriksson J.W., Lundkvist P., Jansson P.A., et al. Effects of dapagliflozin and n-3 carboxylic acids on non-alcoholic fatty liver disease in people with type 2 diabetes: a double-blind randomised placebo-controlled study. Diabetologia. 2018;61(9):1923–1934. doi: 10.1007/s00125-018-4675-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kajinami K., Brousseau M.E., Ordovas J.M., et al. CYP3A4 genotypes and plasma lipoprotein levels before and after treatment with atorvastatin in primary hypercholesterolemia. Am J Cardiol. 2004;93(1):104–107. doi: 10.1016/j.amjcard.2003.08.078. [DOI] [PubMed] [Google Scholar]
- 67.Chasman D.I., Posada D., Subrahmanyan L., et al. Pharmacogenetic study of statin therapy and cholesterol reduction. JAMA. 2004;291(23):2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 68.Lahoz C., Peña R., Mostaza J.M., et al. Apo A-I promoter polymorphism influences basal HDL-cholesterol and its response to pravastatin therapy. Atherosclerosis. 2003;168(2):289–295. doi: 10.1016/s0021-9150(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 69.Pedro-Botet J., Schaefer E.J., Bakker-Arkema R.G., et al. Apolipoprotein E genotype affects plasma lipid response to atorvastatin in a gender specific manner. Atherosclerosis. 2001;158(1):183–193. doi: 10.1016/s0021-9150(01)00410-5. [DOI] [PubMed] [Google Scholar]
- 70.Chu C., Spitale R.C., Chang H.Y. Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol. 2015;22(1):29–35. doi: 10.1038/nsmb.2921. [DOI] [PubMed] [Google Scholar]
- 71.Sathishkumar C., Prabu P., Mohan V., et al. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics. 2018;12(1):41. doi: 10.1186/s40246-018-0173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Morán I., Akerman I., van de Bunt M., et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435–448. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin D.D., Zhang E.B., You L.H., et al. Downregulation of lncRNA TUG1 affects apoptosis and insulin secretion in mouse pancreatic β cells. Cell Physiol Biochem. 2015;35(5):1892–1904. doi: 10.1159/000373999. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y., Zheng L., Wang Q., et al. Emerging roles and mechanisms of long noncoding RNAs in atherosclerosis. Int J Cardiol. 2017;228:570–582. doi: 10.1016/j.ijcard.2016.11.182. [DOI] [PubMed] [Google Scholar]
- 75.Jin F., Wang N., Zhu Y., et al. Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic β Cells. Cell Physiol Biochem. 2017;43(5):2062–2073. doi: 10.1159/000484191. [DOI] [PubMed] [Google Scholar]
- 76.Alvarez-Dominguez JR, Bai Z, Xu D, et al. De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Long Non-coding RNA Regulators of Brown Adipocyte Development [published correction appears in Cell Metab. 2015 Jun 2;21(6):918]. Cell Metab. 2015;21(5):764-776. 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed]
- 77.Wang J., Yang W., Chen Z., et al. Long Noncoding RNA lncSHGL Recruits hnRNPA1 to Suppress Hepatic Gluconeogenesis and Lipogenesis. Diabetes. 2018;67(4):581–593. doi: 10.2337/db17-0799. [DOI] [PubMed] [Google Scholar]
- 78.Mitchell P.S., Parkin R.K., Kroh E.M., et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eliasson L., Esguerra J.L.S. MicroRNA Networks in Pancreatic Islet Cells: Normal Function and Type 2 Diabetes. Diabetes. 2020;69(5):804–812. doi: 10.2337/dbi19-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poy M.N., Eliasson L., Krutzfeldt J., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432(7014):226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 81.Latreille M., Herrmanns K., Renwick N., et al. miR-375 gene dosage in pancreatic β-cells: implications for regulation of β-cell mass and biomarker development. J Mol Med (Berl) 2015;93(10):1159–1169. doi: 10.1007/s00109-015-1296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Belgardt B.F., Ahmed K., Spranger M., et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21(6):619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 83.Sun S.F., Tang P.M.K., Feng M., et al. Novel lncRNA Erbb4-IR Promotes Diabetic Kidney Injury in db/db Mice by Targeting miR-29b. Diabetes. 2018;67(4):731–744. doi: 10.2337/db17-0816. [DOI] [PubMed] [Google Scholar]
- 84.Ji C., Guo X. The clinical potential of circulating microRNAs in obesity. Nat Rev Endocrinol. 2019;15(12):731–743. doi: 10.1038/s41574-019-0260-0. [DOI] [PubMed] [Google Scholar]
- 85.Zhang X., Liu J., Wu L., et al. MicroRNAs of the miR-17∼9 family maintain adipose tissue macrophage homeostasis by sustaining IL-10 expression. Elife. 2020;9:e55676. doi: 10.7554/eLife.55676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang F., Ma D., Zhao W., et al. Obesity-induced overexpression of miR-802 impairs insulin transcription and secretion. Nat Commun. 2020;11(1):1822. doi: 10.1038/s41467-020-15529-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zaiou M. circRNAs Signature as Potential Diagnostic and Prognostic Biomarker for Diabetes Mellitus and Related Cardiovascular Complications. Cells. 2020;9(3):659. doi: 10.3390/cells9030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Memczak S., Jens M., Elefsinioti A., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 89.Xu H., Guo S., Li W., et al. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao Z., Li X., Jian D., et al. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54(3):237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pan L., Lian W., Zhang X., et al. Human circular RNA–0054633 regulates high glucose–induced vascular endothelial cell dysfunction through the microRNA–218/roundabout 1 and microRNA–218/heme oxygenase–1 axes. Int J Mol Med. 2018;42(1):597–606. doi: 10.3892/ijmm.2018.3625. [DOI] [PubMed] [Google Scholar]
- 92.Zaiou M. The Emerging Role and Promise of Circular RNAs in Obesity and Related Metabolic Disorders. Cells. 2020;9(6):1473. doi: 10.3390/cells9061473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Arcinas C., Tan W., Fang W., et al. Adipose circular RNAs exhibit dynamic regulation in obesity and functional role in adipogenesis. Nat Metab. 2019;1(7):688–703. doi: 10.1038/s42255-019-0078-z. [DOI] [PubMed] [Google Scholar]
- 94.Zhu Y., Gui W., Lin X., et al. Knock-down of circular RNA H19 induces human adipose-derived stem cells adipogenic differentiation via a mechanism involving the polypyrimidine tract-binding protein 1. Exp Cell Res. 2020;387(2) doi: 10.1016/j.yexcr.2019.111753. [DOI] [PubMed] [Google Scholar]
- 95.Mauvoisin D. Circadian rhythms and proteomics: It's all about posttranslational modifications! Wiley Interdiscip Rev Syst Biol Med. 2019;11(5):e1450. doi: 10.1002/wsbm.1450. [DOI] [PubMed] [Google Scholar]
- 96.Shono S., Kose H., Yamada T., et al. Proteomic analysis of a diabetic congenic rat identified age-dependent alteration of an acidic protein. J Med Invest. 2007;54(3–4):289–294. doi: 10.2152/jmi.54.289. [DOI] [PubMed] [Google Scholar]
- 97.Riaz S., Alam S.S., Akhtar M.W. Proteomic identification of human serum biomarkers in diabetes mellitus type 2. J Pharm Biomed Anal. 2010;51(5):1103–1107. doi: 10.1016/j.jpba.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 98.Takada T., Kodera Y., Matsubara M., et al. Serum monomeric α2-macroglobulin as a clinical biomarker in diabetes. Atherosclerosis. 2013;228(1):270–276. doi: 10.1016/j.atherosclerosis.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 99.Nowak C., Sundström J., Gustafsson S., et al. Protein Biomarkers for Insulin Resistance and Type 2 Diabetes Risk in Two Large Community Cohorts. Diabetes. 2016;65(1):276–284. doi: 10.2337/db15-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huth C., von Toerne C., Schederecker F., et al. Protein markers and risk of type 2 diabetes and prediabetes: a targeted proteomics approach in the KORA F4/FF4 study. Eur J Epidemiol. 2019;34(4):409–422. doi: 10.1007/s10654-018-0475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim S.W., Choi J.W., Yun J.W., et al. Proteomics approach to identify serum biomarkers associated with the progression of diabetes in Korean patients with abdominal obesity. PLoS ONE. 2019;14(9):e0222032. doi: 10.1371/journal.pone.0222032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Benabdelkamel H., Masood A., Almidani G.M., et al. Mature adipocyte proteome reveals differentially altered protein abundances between lean, overweight and morbidly obese human subjects. Mol Cell Endocrinol. 2015;401:142–154. doi: 10.1016/j.mce.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 103.Bae K.H., Kim W.K., Lee S.C. Involvement of protein tyrosine phosphatases in adipogenesis: new anti-obesity targets? BMB Rep. 2012;45(12):700–706. doi: 10.5483/bmbrep.2012.45.12.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kamal A.H., Kim W.K., Cho K., et al. Investigation of adipocyte proteome during the differentiation of brown preadipocytes. J Proteomics. 2013;94:327–336. doi: 10.1016/j.jprot.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 105.Zhang Y., Yang Y., Ding L., et al. Emerging Applications of Metabolomics to Assess the Efficacy of Traditional Chinese Medicines for Treating Type 2 Diabetes Mellitus. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.735410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jin Q., Ma R.C.W. Metabolomics in Diabetes and Diabetic Complications: Insights from Epidemiological Studies. Cells. 2021;10(11):2832. doi: 10.3390/cells10112832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance [published correction appears in Cell Metab. 2009 Jun;9(6):565-6]. Cell Metab. 2009;9(4):311-326. 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed]
- 108.Guasch-Ferré M., Hruby A., Toledo E., et al. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care. 2016;39(5):833–846. doi: 10.2337/dc15-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bloomgarden Z. Diabetes and branched-chain amino acids: What is the link? J Diabetes. 2018;10(5):350–352. doi: 10.1111/1753-0407.12645. [DOI] [PubMed] [Google Scholar]
- 110.Siddik M.A.B., Shin A.C. Recent Progress on Branched-Chain Amino Acids in Obesity, Diabetes, and Beyond. Endocrinol Metab (Seoul) 2019;34(3):234–246. doi: 10.3803/EnM.2019.34.3.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mahendran Y., Jonsson A., Have C.T., et al. Genetic evidence of a causal effect of insulin resistance on branched-chain amino acid levels. Diabetologia. 2017;60(5):873–878. doi: 10.1007/s00125-017-4222-6. [DOI] [PubMed] [Google Scholar]
- 112.Tremblay F., Marette A. Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway. A negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem. 2001;276(41):38052–38060. doi: 10.1074/jbc.M106703200. [DOI] [PubMed] [Google Scholar]
- 113.Palmer N.D., Stevens R.D., Antinozzi P.A., et al. Metabolomic profile associated with insulin resistance and conversion to diabetes in the Insulin Resistance Atherosclerosis Study. J Clin Endocrinol Metab. 2015;100(3):E463–E468. doi: 10.1210/jc.2014-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang Q., Holmes M.V., Davey Smith G., et al. Genetic Support for a Causal Role of Insulin Resistance on Circulating Branched-Chain Amino Acids and Inflammation. Diabetes Care. 2017;40(12):1779–1786. doi: 10.2337/dc17-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang T.J., Ngo D., Psychogios N., et al. 2-Aminoadipic acid is a biomarker for diabetes risk. J Clin Invest. 2013;123(10):4309–4317. doi: 10.1172/JCI64801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shulman G.I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N Engl J Med. 2014;371(23):2237–2238. doi: 10.1056/NEJMc1412427. [DOI] [PubMed] [Google Scholar]
- 117.Mahendran Y., Cederberg H., Vangipurapu J., et al. Glycerol and fatty acids in serum predict the development of hyperglycemia and type 2 diabetes in Finnish men. Diabetes Care. 2013;36(11):3732–3738. doi: 10.2337/dc13-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lu Y., Wang Y., Ong C.N., et al. Metabolic signatures and risk of type 2 diabetes in a Chinese population: an untargeted metabolomics study using both LC-MS and GC-MS. Diabetologia. 2016;59(11):2349–2359. doi: 10.1007/s00125-016-4069-2. [DOI] [PubMed] [Google Scholar]
- 119.Garcia E, Shalaurova I, Matyus SP, et al. Ketone Bodies Are Mildly Elevated in Subjects with Type 2 Diabetes Mellitus and Are Inversely Associated with Insulin Resistance as Measured by the Lipoprotein Insulin Resistance Index. J Clin Med. 2020;9(2):321. Published 2020 Jan 23. 10.3390/jcm9020321. [DOI] [PMC free article] [PubMed]
- 120.Razquin C., Toledo E., Clish C.B., et al. Plasma Lipidomic Profiling and Risk of Type 2 Diabetes in the PREDIMED Trial. Diabetes Care. 2018;41(12):2617–2624. doi: 10.2337/dc18-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Drogan D., Dunn W.B., Lin W., et al. Untargeted metabolic profiling identifies altered serum metabolites of type 2 diabetes mellitus in a prospective, nested case control study. Clin Chem. 2015;61(3):487–497. doi: 10.1373/clinchem.2014.228965. [DOI] [PubMed] [Google Scholar]
- 122.Mack C.I., Ferrario P.G., Weinert C.H., et al. Exploring the Diversity of Sugar Compounds in Healthy, Prediabetic, and Diabetic Volunteers. Mol Nutr Food Res. 2020;64(9):e1901190. doi: 10.1002/mnfr.201901190. [DOI] [PubMed] [Google Scholar]
- 123.Lu J., Zhou J., Bao Y., et al. Serum metabolic signatures of fulminant type 1 diabetes. J Proteome Res. 2012;11(9):4705–4711. doi: 10.1021/pr300523x. [DOI] [PubMed] [Google Scholar]
- 124.Rawat A., Misra G., Saxena M., et al. 1H NMR based serum metabolic profiling reveals differentiating biomarkers in patients with diabetes and diabetes-related complication. Diabetes Metab Syndr. 2019;13(1):290–298. doi: 10.1016/j.dsx.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 125.Moran-Ramos S., López-Contreras B.E., Canizales-Quinteros S. Gut Microbiota in Obesity and Metabolic Abnormalities: A Matter of Composition or Functionality? Arch Med Res. 2017;48(8):735–753. doi: 10.1016/j.arcmed.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 126.Agus A., Clément K., Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 2021;70(6):1174–1182. doi: 10.1136/gutjnl-2020-323071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Canfora E.E., Meex R.C.R., Venema K., et al. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol. 2019;15(5):261–273. doi: 10.1038/s41574-019-0156-z. [DOI] [PubMed] [Google Scholar]
- 128.Tajiri K, Shimizu Y. Branched-chain amino acids in liver diseases. Transl Gastroenterol Hepatol. 2018;3:47. 10.21037/tgh.2018.07.06. [DOI] [PMC free article] [PubMed]
- 129.Houten S.M., Watanabe M., Auwerx J. Endocrine functions of bile acids. EMBO J. 2006;25(7):1419–1425. doi: 10.1038/sj.emboj.7601049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ðanić M., Stanimirov B., Pavlović N., et al. Pharmacological Applications of Bile Acids and Their Derivatives in the Treatment of Metabolic Syndrome. Front Pharmacol. 2018;9:1382. doi: 10.3389/fphar.2018.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dehghan P., Farhangi M.A., Nikniaz L., et al. Gut microbiota-derived metabolite trimethylamine N-oxide (TMAO) potentially increases the risk of obesity in adults: An exploratory systematic review and dose-response meta- analysis. Obes Rev. 2020;21(5):e12993. doi: 10.1111/obr.12993. [DOI] [PubMed] [Google Scholar]
- 132.Shan Z., Sun T., Huang H., et al. Association between microbiota-dependent metabolite trimethylamine-N-oxide and type 2 diabetes. Am J Clin Nutr. 2017;106(3):888–894. doi: 10.3945/ajcn.117.157107. [DOI] [PubMed] [Google Scholar]
- 133.Zhuang R., Ge X., Han L., et al. Gut microbe-generated metabolite trimethylamine N-oxide and the risk of diabetes: A systematic review and dose-response meta-analysis. Obes Rev. 2019;20(6):883–894. doi: 10.1111/obr.12843. [DOI] [PubMed] [Google Scholar]
- 134.Lavelle A., Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2020;17(4):223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 135.Natividad J.M., Agus A., Planchais J., et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018;28(5):737–749.e4. doi: 10.1016/j.cmet.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 136.Koh A., Molinaro A., Ståhlman M., et al. Microbially Produced Imidazole Propionate Impairs Insulin Signaling through mTORC1. Cell. 2018;175(4):947–961.e17. doi: 10.1016/j.cell.2018.09.055. [DOI] [PubMed] [Google Scholar]
- 137.Kim W.J., Park C.Y. 1,5-Anhydroglucitol in diabetes mellitus. Endocrine. 2013;43(1):33–40. doi: 10.1007/s12020-012-9760-6. [DOI] [PubMed] [Google Scholar]
- 138.Fall T., Salihovic S., Brandmaier S., et al. Non-targeted metabolomics combined with genetic analyses identifies bile acid synthesis and phospholipid metabolism as being associated with incident type 2 diabetes. Diabetologia. 2016;59(10):2114–2124. doi: 10.1007/s00125-016-4041-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ferrannini E., Natali A., Camastra S., et al. Early metabolic markers of the development of dysglycemia and type 2 diabetes and their physiological significance. Diabetes. 2013;62(5):1730–1737. doi: 10.2337/db12-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gall WE, Beebe K, Lawton KA, et al. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5(5):e10883. Published 2010 May 28. 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed]
- 141.Rosalki S.B., Wilkinson J.H. Reduction of alpha-ketobutyrate by human serum. Nature. 1960;188:1110–1111. doi: 10.1038/1881110a0. [DOI] [PubMed] [Google Scholar]
- 142.Kao T.W., Huang C.C. Recent Progress in Metabolic Syndrome Research and Therapeutics. Int J Mol Sci. 2021;22(13):6862. doi: 10.3390/ijms22136862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Surowiec I., Noordam R., Bennett K., et al. Metabolomic and lipidomic assessment of the metabolic syndrome in Dutch middle-aged individuals reveals novel biological signatures separating health and disease. Metabolomics. 2019;15(2):23. doi: 10.1007/s11306-019-1484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lazar V., Ditu L.M., Pircalabioru G.G., et al. Gut Microbiota, Host Organism, and Diet Trialogue in Diabetes and Obesity. Front Nutr. 2019;6:21. doi: 10.3389/fnut.2019.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jandhyala S.M., Talukdar R., Subramanyam C., et al. Role of the normal gut microbiota. World J Gastroenterol. 2015;21(29):8787–8803. doi: 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Candela M., Biagi E., Soverini M., et al. Modulation of gut microbiota dysbioses in type 2 diabetic patients by macrobiotic Ma-Pi 2 diet. Br J Nutr. 2016;116(1):80–93. doi: 10.1017/S0007114516001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Ni Y., Mu C., He X., et al. Characteristics of gut microbiota and its response to a Chinese Herbal Formula in elder patients with metabolic syndrome. Drug Discov Ther. 2018;12(3):161–169. doi: 10.5582/ddt.2018.01036. [DOI] [PubMed] [Google Scholar]
- 148.Graessler J., Qin Y., Zhong H., et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J. 2013;13(6):514–522. doi: 10.1038/tpj.2012.43. [DOI] [PubMed] [Google Scholar]
- 149.Karlsson F.H., Tremaroli V., Nookaew I., et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 150.Blandino G., Inturri R., Lazzara F., et al. Impact of gut microbiota on diabetes mellitus. Diabetes Metab. 2016;42(5):303–315. doi: 10.1016/j.diabet.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 151.Kasai C., Sugimoto K., Moritani I., et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15:100. doi: 10.1186/s12876-015-0330-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dao M.C., Everard A., Aron-Wisnewsky J., et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65(3):426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 153.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota [published correction appears in Nature. 2017 May 3;545(7652):116]. Nature. 2015;528(7581):262-266. 10.1038/nature15766. [DOI] [PMC free article] [PubMed]
- 154.Turnbaugh P.J., Ley R.E., Mahowald M.A., et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 155.Liu R., Hong J., Xu X., et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med. 2017;23(7):859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 156.Thingholm L.B., Rühlemann M.C., Koch M., et al. Obese Individuals with and without Type 2 Diabetes Show Different Gut Microbial Functional Capacity and Composition. Cell Host Microbe. 2019;26(2):252–264.e10. doi: 10.1016/j.chom.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Haythorne E., Rohm M., van de Bunt M., et al. Diabetes causes marked inhibition of mitochondrial metabolism in pancreatic β-cells. Nat Commun. 2019;10(1):2474. doi: 10.1038/s41467-019-10189-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Losko M., Dolicka D., Pydyn N., et al. Integrative genomics reveal a role for MCPIP1 in adipogenesis and adipocyte metabolism. Cell Mol Life Sci. 2020;77(23):4899–4919. doi: 10.1007/s00018-019-03434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lu X.Y., Shi X.J., Hu A., et al. Feeding induces cholesterol biosynthesis via the mTORC1-USP20-HMGCR axis. Nature. 2020;588(7838):479–484. doi: 10.1038/s41586-020-2928-y. [DOI] [PubMed] [Google Scholar]
- 160.Wang C., Yu J., Zhang R., et al. Small intestine proteomics coupled with serum metabolomics reveal disruption of amino acid metabolism in Chinese hamsters with type 2 diabetes mellitus. J Proteomics. 2020;223 doi: 10.1016/j.jprot.2020.103823. [DOI] [PubMed] [Google Scholar]
- 161.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 162.Zhang J., Ni Y., Qian L., et al. Decreased Abundance of Akkermansia muciniphila Leads to the Impairment of Insulin Secretion and Glucose Homeostasis in Lean Type 2 Diabetes. Adv Sci (Weinh) 2021;8(16):e2100536. doi: 10.1002/advs.202100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhou W., Sailani M.R., Contrepois K., et al. Longitudinal multi-omics of host-microbe dynamics in prediabetes. Nature. 2019;569(7758):663–671. doi: 10.1038/s41586-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Schüssler-Fiorenza Rose S.M., Contrepois K., Moneghetti K.J., et al. A longitudinal big data approach for precision health. Nat Med. 2019;25(5):792–804. doi: 10.1038/s41591-019-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat Rev Drug Discov. 2016;15(7):473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 166.Grenham S., Clarke G., Cryan J.F., et al. Brain-gut-microbe communication in health and disease. Front Physiol. 2011;2:94. doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Aw W., Fukuda S. Understanding the role of the gut ecosystem in diabetes mellitus. J Diabetes Investig. 2018;9(1):5–12. doi: 10.1111/jdi.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ejtahed H.S., Mohtadi-Nia J., Homayouni-Rad A., et al. Probiotic yogurt improves antioxidant status in type 2 diabetic patients. Nutrition. 2012;28(5):539–543. doi: 10.1016/j.nut.2011.08.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and materials used are included in the review.