Summary
Background
Early detection and prognosis prediction of colorectal cancer (CRC) can significantly reduce CRC-related mortality. Recently, circulating tumour DNA (ctDNA) methylation has shown good application foreground in the early detection and prognosis prediction of multiple tumours.
Methods
This multicentre cohort study evaluated ctDNA methylation haplotype patterns based on archived plasma samples (collected between 2010 and 2018) from 1138 individuals at two medical centres: Fudan University Shanghai Cancer Center (Shanghai, China) and Southern Medical University Nanfang Hospital (Guangzhou, Guangdong, China), including 366 healthy individuals, 182 patients with advanced adenoma (AA), and 590 patients with CRC. Samples were processed using the ColonES assay, a targeted bisulfite sequencing method that detects ctDNA methylation haplotype patterns in 191 genomic regions. Among these 1138 samples, 748 were used to develop a classification model, and 390 served as a blinded cohort for independent validation. The study is registered at https://register.clinicaltrials.gov with the unique identifier NCT03737591.
Results
The model obtained from unblinded samples discriminated patients with CRC or AA from normal controls with high accuracy. In the blinded validation set, the ColonES assay achieved sensitivity values of 79.0% (95% confidence interval (CI), 66%–88%) in AA patients and 86.6% (95% CI, 81%–91%) in CRC patients with a specificity of 88.1% (95% CI, 81%–93%) in healthy individuals. The model area under the curve (AUC) for the blinded validation set was 0.903 for AA samples and 0.937 for CRC samples. Additionally, the prognosis of patients with high preoperative ctDNA methylation levels was worse than that of patients with low ctDNA methylation levels (p = 0.001 for relapse-free survival and p = 0.004 for overall survival).
Interpretation
We successfully developed and validated an accurate, noninvasive detection method based on ctDNA methylation haplotype patterns that may enable early detection and prognosis prediction for CRC.
Funding
The Grant of National Natural Science Foundation of China (No.81871958), National Natural Science Foundation of China (No. 82203215), Shanghai Science and Technology Committee (No. 19140902100), Scientific Research Fund of Fudan University (No.IDF159052), Shanghai Municipal Health Commission (SHWJRS 2021-99), and Shanghai Sailing Program (22YF1408800).
Keywords: ctDNA methylation, Early detection, Prognosis prediction, Precancerous adenomas, Colorectal cancer
Research in context.
Evidence before this study
Patients diagnosed with precancerous adenomas or at earlier stages of colorectal cancer (CRC) have a better prognosis than those diagnosed at advanced stages. Prognosis prediction is beneficial to the whole process of CRC patient management, and even helps clinicians formulate individualized treatment plans. Therefore, early detection and prognosis prediction can significantly improve patient survival but are limited by the development of effective noninvasive diagnostic methods.
Added value of this study
Here, we developed and validated a blood-based assay termed ColonES. This assay assesses multigene methylation by next-generation sequencing for noninvasive CRC detection and prognosis prediction. The ColonES assay achieved sensitivity values of 79.0% in advanced adenoma patients and 86.6% in CRC patients with a specificity of 88.1% in healthy individuals. In addition, significant differences in 5-year relapse-free survival (73.4% vs. 81.2%) and overall survival (64.0% vs. 79.0%) were noted between patients with preoperative high vs. low ctDNA methylation levels.
Implications of all the available evidence
The ColonES assay shows high sensitivity and specificity for the detection of advanced adenoma and early stages of CRC. If adopted as a precolonoscopy screening test, we expect that with the higher detection rate of advanced adenoma and early CRC as well as the benefit of prognostic prediction in strengthening the clinical management of CRC patients, CRC-related mortality may be greatly reduced. Further validation of this model is warranted in a general population study.
Introduction
Colorectal cancer (CRC) is the third most common solid malignant tumour worldwide.1, 2, 3, 4 It is a long process that precancerous lesions (adenomas) develop into malignant lesions,5,6 which allows CRC to be detected early by screening.7,8 Only four types of cancer, including CRC, can be effectively screened with screening guidelines according to the Centers for Disease Control and Prevention (CDC) and the United States Preventive Services Task Force (USPSTF).9, 10, 11 Through early detection and standard treatment, such as colonoscopy and surgery, the 5-year survival rate of CRC increases up to 90%.2,12 However, the survival rate decreases sharply, and the economic burden increases once the disease progresses into advanced stages of CRC.3,13
Early diagnosis and detection of recurrence or metastasis can significantly reduce CRC-related mortality and improve patient outcome. Colonoscopy is considered to be the gold standard for the diagnosis of CRC.14,15 During operation, neoplasms can be accurately examined, and precancerous lesions can be removed.6,16 However, its application is greatly limited with only 50% compliance due to the invasive procedure, bowel preparation and high cost.17,18 Faecal immunochemical testing (FIT), on the other hand, is noninvasive and economical, adopted as the main tool for large-scale population screening in most countries.6,19 However, FIT suffers from low accuracy in detecting advanced adenoma (AA).20,21 In a meta-analysis, the pooled sensitivity of FIT for CRC was 0.71 at a threshold of 20 μg/g, with a specificity of 0.95, and 0.91 at 10 μg/g with a specificity of 0.90. FIT sensitivity for AA was 0.40 at 10 μg/g and 0.25 at 20 μg/g.22 Besides, in another meta-analysis and modeling analysis, summary estimates (95% confidence intervals) of overall sensitivity for detecting CRC and AA were 65% (56–74%) and 27% (23–31%) for FIT.23 A single centre study24 showed that the FIT detected stage I cancers with 68% sensitivity (95% CI, 57%–78%), at the cutoff recommended by the manufacturer (17 μg/g feces). Although the FIT identifies CRC patients with overall high sensitivity, it can miss approximately one-third of stage I cases. Technique efforts are still needed to increase noninvasive detection rate of early-stage CRC.25 In an observational survey, patients greatly preferred noninvasive testing, but 83% preferred a blood-based test over a stool-based test.26 Serum carcinoembryonic antigen (CEA), a blood biomarker, has good specificity in the differential diagnosis of occult CRC, but its sensitivity is only 40%–60%.27,28 A high CEA level is most often detected in advanced CRC but less commonly found in early-stage CRC. All of these factors limit patient compliance with the recommended screenings, indicating an urgent need for effective noninvasive biomarkers for the early detection of CRC.
Circulating tumour DNA (ctDNA) is a DNA fragment derived from tumour with no cell component in peripheral blood.29,30 ctDNA is derived from tumour cells due to necrosis or apoptosis and carries tumour-specific genetic and epigenetic information.30 Therefore, ctDNA can be used as an alternative source of tumour DNA for tumour diagnosis and prognosis.30, 31, 32, 33 As one of the key targets of liquid biopsy, ctDNA has been studied in the aid for diagnosis, treatment evaluation, prognosis prediction, and relapse monitoring of a variety of cancers.34,35 DNA methylation is a major epigenetic modification that is closely related to the occurrence and development of malignant epithelial tumours.36 Aberrant methylation is one of the main features of the early stage of tumorigenesis, which is characterized by changes in DNA methylation status and disorders of gene expression.36,37 Some studies have identified specific DNA methylation sites, such as SEPT9, as biomarkers of CRC.38 EpiProColon was developed based on the methylation status of SEPT9 in plasma ctDNA and approved by the Food and Drug Administration (FDA).39,40 However, the USPSTF does not recommend it as a preliminary screening method due to its low sensitivity in AA (11.2%) and early-stage CRC (35%).41
Limitations in the early detection of tumours based on ctDNA methylation still exist. Only a limited amount of ctDNA is present in plasma, especially in patients with early-stage cancers, which makes ctDNA capture difficult for methylation detection.42 Second, current methods mainly focus on a few CRC-relevant loci,43 resulting in a risk of signal loss for early and precancerous stages. Therefore, the potential ctDNA methylation signatures carrying early tumour information remain to be further explored.
Here, we describe the development and validation of ColonES, a high-throughput methylation-based blood test sensitive for the early detection and prognosis prediction of CRC. The ColonES test utilizes a highly efficient and multiplex targeted bisulfite sequencing method to simultaneously interrogate the methylation state of 191 genomic regions in plasma cell-free DNA. Using 1138 plasma samples from patients and healthy controls, we demonstrate that ColonES can be utilized to noninvasively detect early-stage CRC and AA with best-in-class sensitivity, paving the way for a preliminary noninvasive blood-based CRC screening assay.
Methods
Study design
This multicentre cohort study was based on well-characterized archived plasma samples from two medical centres: Fudan University Shanghai Cancer Center (Shanghai, China) and Southern Medical University Nanfang Hospital (Guangzhou, China). The study is registered at https://register.clinicaltrials.gov with the unique identifier NCT03737591.
Blood specimens were obtained from healthy individuals, advanced adenoma patients and CRC patients who were previously enrolled in Ethics Review Board or Institutional Review Board approved studies; informed written consent was obtained from all patients at the participating institutions to allow archival of their biospecimens and use in future studies. The study statistical plan incorporated both control individuals and case patients, which should be sufficient to detect a change in AUC of 0.1 (respective of a null hypothesis AUC of 0.7) with a power of 1-β = 80% and a significance level of α = 0.05. Additional case patients were incorporated into the study (as CRC and advanced adenoma patients were readily available and assay cost was low), increasing the power to 1-β = 97.5%. Eligible patients had blood drawn prior to any tumour-related treatment. Blood samples were collected in EDTA-containing blood tubes, and plasma samples were separated and archived (labelled, aliquoted and frozen, as stipulated by the standard protocols).
The target population of healthy controls was selected by each centre to roughly match cases by age and sex. The randomization process was stratified random sampling by age for training and validation dataset. The samples were grouped by age range for case, control and AA. Then, the samples in each subgroup (ie: case 30–40 or control 60–70) were randomly assigned to training or validation dataset to ensure that there is no difference in variables between the training and validation sets. Healthy controls were asymptomatic screening participants without colorectal neoplasms (polyps, adenomas or cancers) confirmed by colonoscopy. Participants who had a personal or family history of colorectal adenoma, colorectal neoplasia, digestive cancer, inflammatory bowel disease, or chronic complex organ disorders were excluded. Colonoscopy in conjunction with histopathology on all colorectal neoplasms served as the reference standard. In cases with multiple lesions, patients were classified based on their most advanced or largest neoplasm. All cases were classified as sporadic diseases, and Lynch syndrome patients were excluded from the study. The biopsy and surgical specimens underwent histopathological analysis and were reviewed by a gastrointestinal pathologist for confirmation at the laboratory at each site.
Advanced adenomas (precancerous lesions, high risk adenomatous polyps) were defined as high-grade dysplasia, villous or tubulovillous histologic features, or lesions measuring ≥1 cm in the greatest dimension according to the National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology Colorectal Cancer Screening.44
Based on clinical practice, the right colon was considered to include all segments proximal to the splenic flexure; the left colon was defined to include the splenic flexure and all other segments of the colon; and the rectum referred to the intestine within 12 cm from the anal verge. The clinical staging parameters were based on the American Joint Committee on Cancer's Cancer Staging Manual, 8th Edition.45,46
Postoperative follow-up was conducted according to hospital routines. Briefly, CEA and CA19-9 were tested every 3–6 months during the first 3 years and every 6 months thereafter until 5 years. Chest X ray or CT scans and abdominoperineal CT scans with contrast or MRI were performed every 6 months during the first 3 years and each year thereafter until 5 years. Additional blood or imaging tests were performed when clinically indicated. The treatment and outcome information were collected by phone and from outpatient records.
Two batches of blood samples, including samples from the unblinded group and the blinded group, were collected. All samples were sent to Singlera Genomics for processing after plasma separation; tumour pathology results were included for the unblinded group and omitted for the blinded group. Unblinded sample results were used for initial model training and testing; the model was then validated in the blinded group. The blinded group was single blinded to Singlera Genomics, and model results were sent back to Fudan University Shanghai Cancer Center and Southern Medical University Nanfang Hospital for comparison with pathology results.
The primary outcome of this retrospective study was the ability of ColonES to detect CRC and precancerous lesions (advanced adenomas). The secondary outcome was to compare ColonES to current stool or blood tests being run in clinics, including FIT and CEA.
FIT assay
A Faecal Occult Blood Gold Gel Stripe kit (W.H.P.M. Bioresearch & Technology Co, Ltd, Beijing) was used to examine stool samples provided by subjects according to the manufacturer's instructions, which was approved by the Chinese Food and Drug Administration Bureau. The cut-off value for a positive FIT result was 200 ng/mL haemoglobin in the stool.
CEA and CA199 biomarkers
The levels of serum tumour markers including CEA and CA19-9, were determined by an electrochemiluminescence immunoassay using a Roche Cobas 8000 immunoassay analyser (Roche Diagnostics, Mannheim, Germany). The normal upper limits of serum tumour markers were adopted as follows: CEA (5.2 ng/mL) and CA19-9 (27 U/mL).
Plasma collection and storage
At least 5 mL of blood was drawn into EDTA tubes. Immediately, the blood was centrifuged at 1600 g for 15 min. The plasma was then carefully separated from the red blood cells and buffy coat and placed in a new tube. The plasma was then centrifuged at 16000 g for 10 min, and once again transferred to a new tube. The plasma was then stored at −80 °C and shipped on dry ice to Singlera Genomics.
DNA extraction from plasma
Cell-free DNA (cfDNA) was extracted from 0.4 to 2.3 (median 1.2) mL plasma using a QIAamp Circulating Nucleic Acid kit (Qiagen, 55114) according to the manufacturer's instructions with the exception of a 1-h incubation period at 60 °C during the lysis step.
Singlera multigene methylation sequencing assay
All samples were sent to Singlera Genomics and processed using the Singlera multigene methylation next generation sequencing (NGS) assay according to the manufacturer's protocol. A total of 3.3–24.2 (median 16.4) ng cfDNA was input for the assay. Briefly, cfDNA was bisulfited and converted using the Methylcode Bisulfite Conversion Kit (ThermoFisher, MECOV50) according to the manufacturer's protocol. The bisulfite converted DNA was dephosphorylated and ligated to a universal adapter with a unique molecular identifier (UMI). Following a second strand synthesis and purification, the DNA underwent a semi-targeted amplification. Following a purification, a second PCR added sample specific barcodes and full-length sequencing adapters. The libraries were then quantified using the KAPA Library Quantification Kit for Illumina (KK4844) and sequenced on an Illumina NextSeq 500 in paired-end 300 bp mode requiring a minimum of 4 million reads per sample.
The targeted regions present in the Singlera multigene methylation NGS assay were previously identified and selected by Singlera from genomic regions differentially methylated between tumour tissues and adjacent normal tissues based on public The Cancer Genome Atlas (TCGA) Illumina 450k methylation array data and methylation markers listed in the literature,47 as well as in-house generated Reduced Representation Bisulfite Sequencing (RRBS) data to ensure low background signals in healthy plasma.
Sequencing data analysis
Data preprocessing was performed using the standard Singlera methylation sequencing preprocessing pipeline illustrated below: First, reads demultiplexing was done by the Illumina bcl2fastq software (https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html). To get the original insert DNA fragments, paired-end FASTQ data were merged into single reads with PEAR (https://sco.h-its.org/exelixis/web/software/pear/doc.html) for each sample. Adapters in the merged FASTQ data were trimmed using trim_galore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Unique molecular identifier sequence of each read was then moved to the read name using UMI_tools (https://github.com/CGATOxford/UMI-tools). Then rest part of reads were mapped to the CT and GA human reference genome (version hg19) with Bismark (https://www.bioinformatics.babraham.ac.uk/projects/bismark/) and Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml). From each aligned read BAM file, the MHF and uMHF measurements were then computed for each methylation haplotype in each target as follows:
where i is the current locus, h is the current haplotype, and Ni is the total number of reads covering the current locus. For MHF, Ni,h is the number of reads at the current locus containing a fully methylated haplotype. For uMHF, Ni,h is the number of reads at the current locus containing a fully unmethylated haplotype.
Singlera ColonES assay
Sixty CRC and 20 AA tissue samples were used as case samples, and 28 healthy colon tissue samples served as control samples.48 The number of CpGs is positively correlated with fragment size, whereas sequencing depth is negatively correlated with fragment size. To select the most informative methylation haplotypes, we only selected haplotypes with lengths ranging from 20 to 75 and greater than 3 CpGs, which balanced the information carried by the haplotype and sequence depth. Furthermore, we selected haplotypes, 90% of which were detected with a depth greater than 30 reads. Among these haplotypes, we selected those with a t test p value <0.003 as candidate tissue markers. We did Shapiro–Wilk test to the 239 markers and found that 202 markers in normal samples and 29 in cancer samples were from normal distribution. Although not all of the markers were from normal distribution, we used t test in the consideration of its higher power test to select candidate markers. We also performed Mann–Whitney U test to validate these markers and all the p values were less than 0.003 (the maximum p-value 5.61e-07). Given that some of these markers might be derived from the same target region, which are highly correlated with each other, will offer redundant information for further analysis. Candidate tissue markers were grouped to their corresponding target regions. Most significant (ranked by p-value) markers for each target region were selected as marker identified from tissue samples, which were incorporated into the panel designated as ColonES Assay.
Modeling and test
Due to the instability of DNA fragments in plasma and variation of amplification efficiency with plasma samples, some of the markers may not appear in all plasma samples. The 239 markers identified in tissue samples with missing values in any plasma sample were removed from further analysis. Unblinded samples were randomly split. Half of the healthy, CRC and AA samples were included in the training and validation datasets. We further filtered the selected markers using plasma samples in the training set, and only the markers with consistent methylation patterns as tissue samples were kept for model building (t test, p < 0.01). Using these markers logistic regression model was fitted with the L2 penalty and balanced class weight. The cutoff of the model was set at a specificity closest to 0.9 when greater than 0.9 in the training dataset. Then, the model and cutoff were applied to the validation dataset and blind test dataset.
Ethics statement
The Ethical Committee and Institutional Review Board of the Fudan University Shanghai Cancer Center and Southern Medical University Nanfang Hospital reviewed and approved this study protocol. All patients signed written informed consent. The study was conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects. The work has been reported in line with the STROBE criteria.49
Statistical analysis
Accuracy metrics were computed for each sample set and subset based on sample covariates. Sensitivity is defined as true positives/(true positives + false negatives). Specificity is defined as true negatives/(true negatives + false positives). Binomial 95% confidence intervals were computed using the Clopper-Pearson method. Survival curves were plotted using the Kaplan–Meier method with the log-rank test. The detection rate and sensitivity were compared using the chi-squared test with python command chi2_contingency in scipy package (v1.5.4). Continuous variables were presented as the mean ± SD, and the difference was analyzed using one-way ANOVA or Student's t-test. Cox regression model were used for univariate and multivariate analyses of prognostic factors. All statistical tests were 2-sided, and p values <0.05 were regarded as statistically significant.
Role of funding source
The funding resource of the study had no role in study design, data collection, data analysis, data interpretation, or the writing of this report. All authors read, discussed, and approved the final version of the manuscript. The co-corresponding authors (G.C., S.C., L.L., and Y.X.) had full access to the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis as well as the decision to submit the data for publication.
Results
Patient and sample characteristics
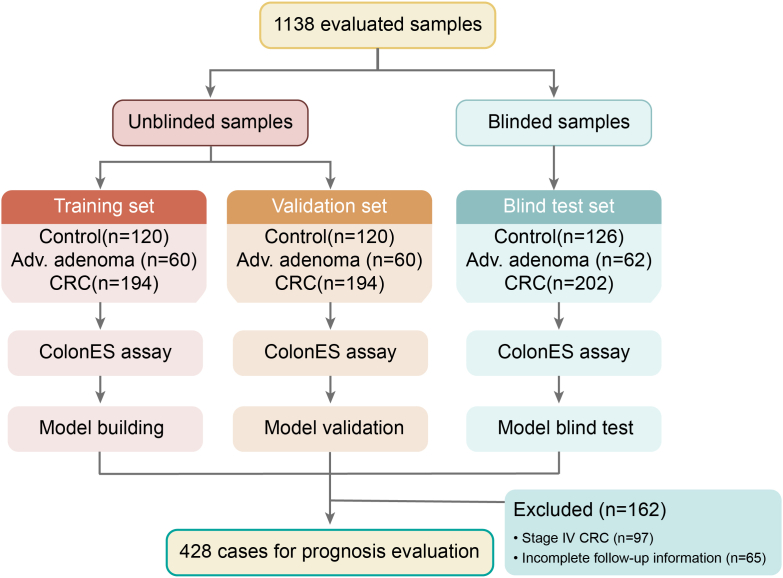
Patient enrolment and the study overview are presented in Fig. 1. In total, archived blood samples (collected between 2010 and 2018) from 1138 individuals were obtained for this study in two batches (Table 1 and Table S1, Supporting Information). The first batch consisted of 748 plasma samples, including 240 control healthy individuals, 120 patients with precancerous AAs, and 388 patients with colorectal adenocarcinoma diagnosed by colonoscopy and pathology. The median patient age was 59 years (ages 30–85) and was matched between cases and controls. Because this assay is intended to be used for early screening, our sample cohort was intentionally weighted to have a larger proportion of AA samples (120 cases) and early-stage CRC (187 cases of stage I, 48.2% of total CRC patients) and a smaller proportion of late-stage CRC samples (201 stage II–IV cases). These samples were randomly split into a training and validation set of 374 samples each; sets were balanced with respect to patient age, sex, and number of cases and controls (Table 1). In parallel, an independent set of 390 blood samples (consisting of 126 control asymptomatic healthy individuals, 62 case AA patients, and 202 case CRC patients) was utilized for blinded model validation.
Fig. 1.
An outline of the study design.
Table 1.
Summary of patient and tumor clinical information.
| Training set (n = 374) | Validation set (n = 374) | Blind test set (n = 390) | Total (n = 1138) | |
|---|---|---|---|---|
| Age, years | ||||
| Median | 59 | 59 | 59 | 59 |
| Range | 30–87 | 30–85 | 31–92 | 30–92 |
| Sex, % | ||||
| Women | 49% | 56% | 52% | 52% |
| Men | 51% | 44% | 48% | 48% |
| Controls | ||||
| Healthy, n | 120 | 120 | 126 | 366 |
| Cases (colorectal tumor) | ||||
| Advanced adenoma | ||||
| n | 60 | 60 | 62 | 182 |
| Size, cm | ||||
| Median | 2.3 | 2.5 | 2.5 | 2.5 |
| Range | 1.0–15.0 | 1.0–10.0 | 0.6–11.5 | 0.6–15.0 |
| Site, n (%) | ||||
| Right colon | 12 (20%) | 8 (13%) | 11 (18%) | 31 (17%) |
| Left colon | 16 (27%) | 22 (37%) | 15 (24%) | 53 (29%) |
| Rectum | 32 (53%) | 30 (50%) | 36 (58%) | 98 (54%) |
| Histological, n (%) | ||||
| Tubular | 15 (25%) | 13 (22%) | 10 (16%) | 38 (21%) |
| Tubulovillous | 35 (58%) | 32 (53%) | 47 (76%) | 114 (63%) |
| Villous | 9 (15%) | 13 (22%) | 3 (5%) | 25 (14%) |
| Sessile serrated | 1 (2%) | 2 (3%) | 2 (3%) | 5 (3%) |
| Dysplasia, n (%) | ||||
| High grade | 27 (46%) | 21 (36%) | 20 (33%) | 68 (38%) |
| Low grade | 32 (54%) | 37 (64%) | 40 (67%) | 109 (62%) |
| Adenocarcinoma | ||||
| n | 194 | 194 | 202 | 590 |
| Size, cm | ||||
| Median | 3.5 | 3.5 | 3.5 | 3.5 |
| Range | 0.9–12.0 | 0.7–11.0 | 1–12.5 | 0.7–12.0 |
| Site, n (%) | ||||
| Right colon | 54 (28%) | 45 (23%) | 43 (21%) | 142 (24%) |
| Left colon | 46 (24%) | 39 (20%) | 46 (23%) | 131 (22%) |
| Rectum | 94 (48%) | 110 (57%) | 113 (56%) | 317 (54%) |
| Stage, n | ||||
| I | 94 (49%) | 79 (41%) | 88 (43%) | 261 (44%) |
| II | 36 (19%) | 40 (21%) | 40 (20%) | 116 (20%) |
| III | 32 (16%) | 44 (23%) | 40 (20%) | 116 (20%) |
| IV | 32 (16%) | 31 (16%) | 34 (17%) | 97 (16%) |
Development of a methylation haplotype-based classification model
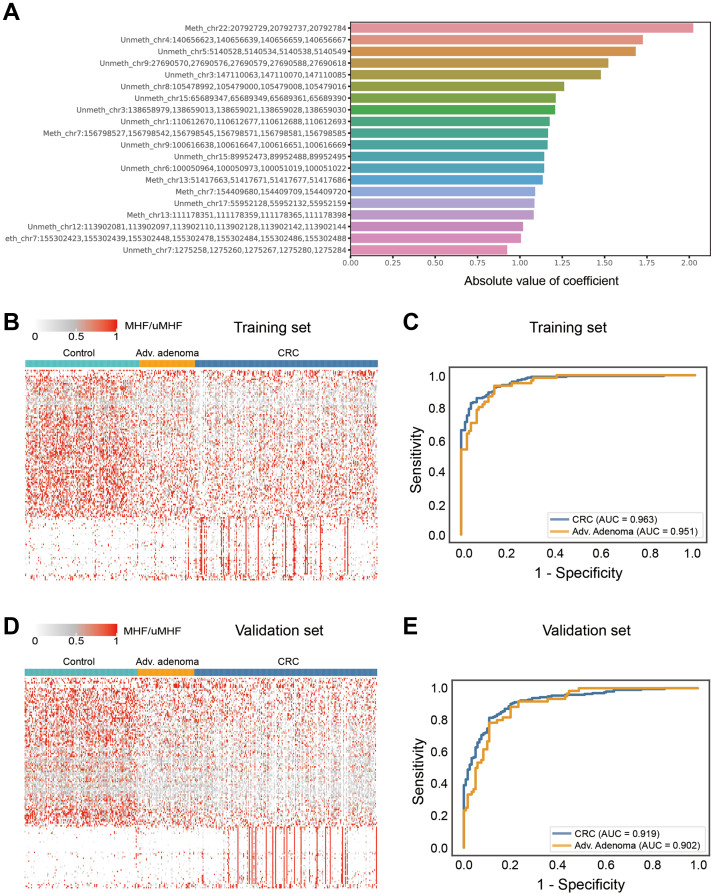
In total, 60 CRC, 20 AA, and 28 normal colon tissue samples were processed with the Panseer Assay, which utilized multiplex semi-targeted bisulfite PCR and high-throughput sequencing to interrogate the methylation state of 11,878 CpG sites across 595 genomic regions and was described in a previous study.50 The methylation sites have been identified based on differential methylation levels between tumour tissues and normal colon tissues; this ensures detectable methylation differences in circulating DNA when comparing plasma from tumour patients to that from healthy individuals.48,50 The 20 most effective MHFs are shown in Fig. 2A.
Fig. 2.
Development of a Methylation Haplotype-based Classification Model. (A) The top 20 absolute value of coefficient methylation regions identified from tissue samples. (B) Heatmap of methylation regions in model building of training data set including control, advanced adenoma and CRC plasmas. (C) ROC curve of training data set with LR score of 191 methylation haplotype markers. The AUCs were 0.951 for AA samples and 0.963 for CRC samples. (D) Heatmap of methylation regions in model building of validation data set including control, advanced adenoma and CRC plasmas. (E) ROC curve of validation data set with LR score of 191 methylation haplotype markers. The AUCs were 0.902 for AA samples and 0.919 for CRC samples.
We sought to develop a classification model capable of accurately separating CRC and AA from healthy plasma. After sequencing reads were aligned to the genome, unique molecular IDs (UMIs) were used to track the original molecules in the library. Reads with the same UMIs were merged to obtain original unique molecules. Methylation levels were expressed on a per-haplotype basis as the fraction of unique molecules containing each distinct methylation haplotype within each covered region; these values were referred to as methylated haplotype fractions (MHFs)/unmethylated haplotype fractions (uMHFs). We identified 239 haplotype markers with significantly different methylated level when comparing CRC/AA tissues with normal samples, 191 of which remained after marker filtering using training plasma samples (Table S2, Supporting Information). These 191 methylation haplotype markers were used to build a classification model with plasma samples and designated the ColonES panel. MHF values across the model regions in the 374 training set samples were used to train a logistic regression classifier (Fig. 2B), which computed a score corresponding to the probability of a sample being derived from a patient presenting with a tumour. The AUCs were 0.951 for AA samples and 0.963 for CRC samples (Fig. 2C). A score cut-off of 0.508822 was chosen to obtain a specificity greater than 90% in the training set.
ColonES detects early-stage neoplasms with high accuracy
We next examined the performance of the developed classifier in the 374 independent validation set samples. Validation set samples were processed in the same manner as the training set samples, and the methylation level of evaluated regions showed a similar pattern as the training set (Fig. 2D). The AUCs were 0.902 for AA samples and 0.919 for CRC samples (Fig. 2E).
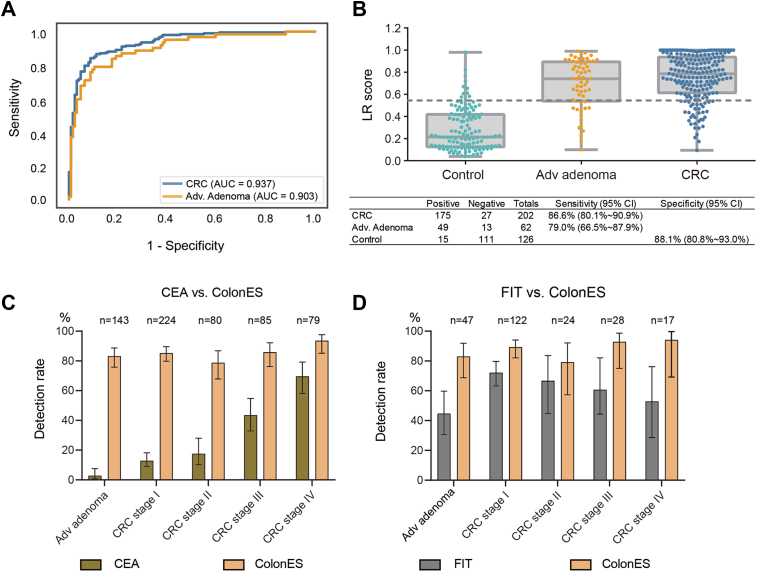
We further sought to validate the ColonES assay using a second independently collected batch of 390 plasma samples. These samples were processed in a blinded manner to accurately assess the performance of the ColonES classification model. After the samples were called using the haplotype classifier, the calls were compared to the true results for validation. In this blinded batch of samples, the model AUC for the blinded validation set was 0.903 for adenoma samples and 0.937 for CRC samples (Fig. 3A). The ColonES assay was able to detect 79.0% of AA patients and 86.6% of CRC patients with a specificity of 88.1% (Fig. 3B). All samples were combined for downstream analysis (comparison to other noninvasive CRC assays and influence of covariates on ColonES assay performance).
Fig. 3.
ColonES Detects Early-Stage Neoplasms with High Accuracy. (A) ROC curve of blind test data set. AUC for the blinded validation set was 0.903 for adenoma samples and 0.937 for CRC samples. (B) Logistic regression model score of control, advanced adenoma and CRC groups. The prediction table were also included. The sensitivity is 86.6% for CRC and 79.0% for AA with 88.1% specificity. (C) Bar plot with 95% confidence interval of the comparison between CEA and ColonES for different disease stages. (D) Bar plot with 95% confidence interval of the comparison between FIT and ColonES for different disease stages.
Comparison to other noninvasive CRC assays
We sought to compare the ColonES assay to other noninvasive CRC detection methods. Among the 772 collected AA and CRC patient samples, many patients were simultaneously tested with other noninvasive cancer detection methods at the same time as plasma sample collection. In total, 238 samples were tested using a FIT stool screening assay,20 and 611 were tested for the presence of CEA glycoprotein in serum.51 The ColonES assay outperformed all other testing methods (Fig. 3C and D), especially in sensitivity for AA (83.2% for ColonES vs. 2.8% for plasma CEA and 83% for ColonES vs. 44.7% for FIT assays, respectively) and early-stage CRC samples (85.3% for ColonES vs. 12.9% for plasma CEA and 89.3% for ColonES vs. 72.1% for FIT assays, respectively).
Influence of covariates on ColonES assay performance
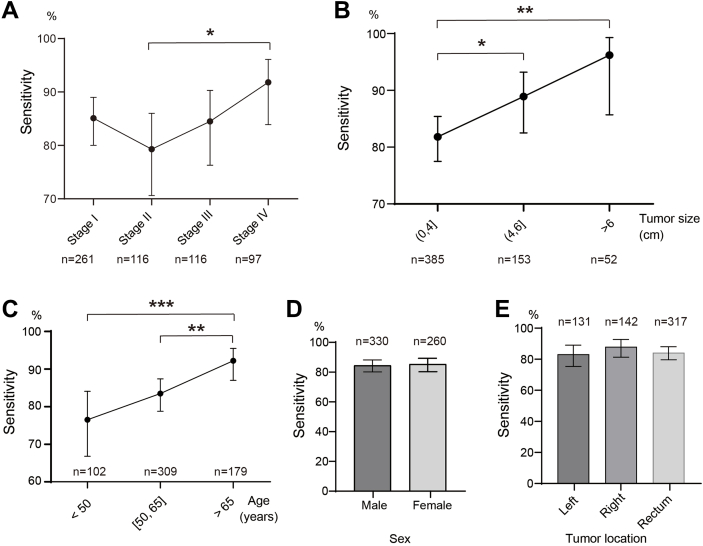
We next sought to determine whether sample covariates affected ColonES assay performance. Impressively, the detection rate of ColonES in stage I CRC reached 85.1%, and it increased from 79.3% in stage II to 91.8% in stage IV CRC patients (p ˂ 0.05, Fig. 4A). The CRC detection rate also increased with tumour size from 81.8% to 96.2% (p ˂ 0.05, Fig. 4B). The sensitivity of CRC detection increased from 76.5% in patients aged under 50 years to 92.2% in patients aged over 65 years (p ˂ 0.01, Fig. 4C). We did not observe any significant difference in the confidence intervals for assay sensitivity based on patient sex (Fig. 4D) or neoplasm site (Fig. 4E) by binomial comparison. Regarding the influence of covariates on ColonES assay performance in AA samples, the AA detection rate increased with tumour size from 75.7% to 87.7% (Fig. S1A, Supporting Information). No significant difference in assay sensitivity were noted based on patient age (Fig. S1B, Supporting Information), sex (Fig. S1C, Supporting Information) or neoplasm site (Fig. S1D, Supporting Information) in AA samples. The neoplasm detection rate of the ColonES assay remained high regardless of sample characteristics.
Fig. 4.
Influence of Covariates on ColonES Assay Performance. Covariate analysis of ColonES model for disease stages (A), tumor size of CRC patients (B), age of CRC patients (C), gender of CRC patients (D) and tumor location of CRC patients (E). The statical analysis of detection rate or sensitivity was done with z-test, Z-value was computed by the difference of two ratios and divided by the standard error of the overall ratio. The significant difference between any groups were labelled in figures. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Prognosis prediction for stage I–III CRC patients by ColonES assay
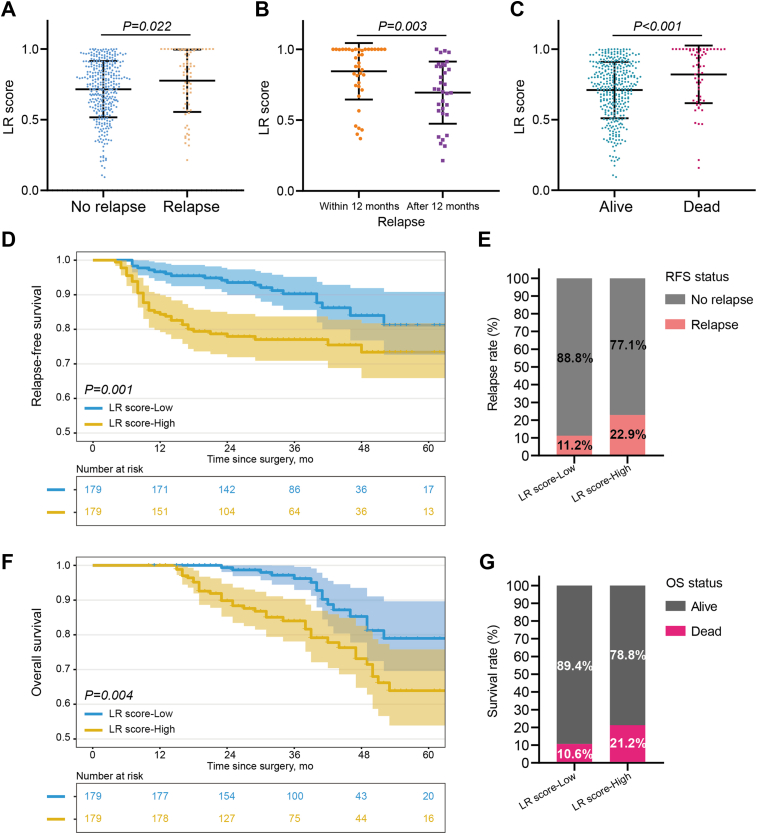
Prognosis prediction for stage I–III CRC patients was performed based on the preoperative ctDNA methylation level. Among all patients with stage I–III CRC (493 cases), 428 patients had complete follow-up information with a median (range) follow-up of 36 (10–120) months, and the 5-year relapse-free survival (RFS) and overall survival (OS) rates were 76.9% (Table S3, Supporting Information) and 72.6% (Table S4, Supporting Information), respectively. According to the prognostic status, all patients were divided into a postoperative relapse group (72 cases) and a non-relapse group (356 cases). We observed a significant difference (p = 0.022) in the preoperative ctDNA methylation level, as measured by logistic regression (LR) score, between the non-relapse and relapse groups (Fig. 5A). Then, we further divided the patients with postoperative relapse into an early relapse group (within 12 months) and a late relapse group (after 12 months). Interestingly, a significant difference (p = 0.003) in ctDNA methylation levels was noted between the two groups (Fig. 5B). A similar trend was also observed based on OS status after operation (p ˂ 0.001, Fig. 5C). Finally, we divided stage I–III CRC patients with preoperative ColonES assay-positive (358 cases) into LR score-High and -Low groups based on the median LR score (0.799754). The 5-year RFS and OS rates were 73.4% and 64.0% and 81.2% and 79.0% for the LR score-High and -Low groups, respectively. Patients in the LR score-Low group had significantly higher 5-year RFS and OS rate than that of patients in LR score-High group (p = 0.001 for RFS and p = 0.004 for OS) (Fig. 5D–G). Univariate and multivariate analyses showed that the preoperative ctDNA methylation level was an independent risk factor for RFS (HR, 2.136; 95% CI, 1.238–3.684; p = 0.006; Table 2) and OS (HR, 2.457; 95% CI, 1.398–4.317; p = 0.002; Table 3) in stage I–III CRC patients. Considering that stage III CRC patients are more likely to relapse after radical surgery, a blood test that can discriminate relapse in those patients would be valuable. Therefore, we conducted subgroup analysis and found that stage III CRC patients in the LR score-High group had significantly worse 5-year RFS and OS rate than that of patients in LR score-Low group (p = 0.005 for RFS and p ˂ 0.001 for OS, Figure S2, Supporting Information). Although this is a pilot study on CRC patients, our data suggest that the ColonES assay has the potential to monitor treatment response and cancer prognosis. Further validation of this model is warranted in prospective postoperative dynamic monitoring studies.
Fig. 5.
Prognosis Prediction for Stage I–III CRC Patients by ColonES Assay. (A) LR scores between relapse (n = 72) and non-relapse (n = 356) groups. (B) LR scores between early relapse group (within 12 months, n = 39) and late relapse group (after 12 months, n = 33). (C) LR scores between alive (n = 366) and dead (n = 62) groups. (D) Kaplan–Meier curves of RFS between LR score-High and -Low groups (p = 0.001). (E) Comparison of relapse rate for 358 stage I–III CRC patients stratified by pre-LR score status on average. (F) Kaplan–Meier curves of OS between LR score-High and -Low groups (p = 0.004). (G) Comparison of overall survival rate for 358 stage I–III CRC patients stratified by pre-LR score status on average.
Table 2.
Univariable and multivariable Cox regression analyses of relapse-free survival in stage I–III CRC patients.
| Variables | Univariable analyses |
multivariable analyses |
||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Gender | 0.023 | 0.018 | ||
| Female | Reference | Reference | ||
| Male | 1.9 (1.1–3.2) | 2.0 (1.1–3.6) | ||
| Age | 0.54 | |||
| <60 | Reference | |||
| ≥60 | 1.2 (0.7–2.0) | |||
| Tumor site | 0.011 | 0.75 | ||
| Rectum | Reference | Reference | ||
| Left | 2.3 (1.3–4.3) | 0.007 | 1.0 (0.5–2.0) | 0.93 |
| Right | 2.0 (1.1–3.7) | 0.023 | 1.3 (0.6–2.6) | 0.49 |
| Tumor size | 0.002 | 0.18 | ||
| <4 | Reference | Reference | ||
| ≥4 | 2.2 (1.3–3.8) | 1.5 (0.8–2.5) | ||
| Tumor stage | <0.001 | <0.001 | ||
| Stage I | Reference | Reference | ||
| Stage II | 2.2 (0.9–5.3) | 0.071 | 1.8 (0.7–4.5) | 0.20 |
| Stage III | 11.5 (6.0–21.9) | <0.001 | 10.4 (5.2–20.7) | <0.001 |
| LR score | 0.002 | 0.006 | ||
| Low | Reference | Reference | ||
| High | 2.3 (1.4–4.0) | 2.1 (1.2–3.7) | ||
CI, confidence interval; CRC, colorectal cancer; HR, hazard ratio; LR, logistic regression.
Table 3.
Univariable and multivariable Cox regression analyses of overall survival in stage I–III CRC patients.
| Variables | Univariable analyses |
multivariable analyses |
||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Gender | 0.057 | |||
| Female | Reference | |||
| Male | 1.7 (0.9–3.0) | |||
| Age | 0.41 | |||
| <60 | Reference | |||
| ≥60 | 0.8 (0.5–1.4) | |||
| Tumor site | 0.009 | 0.53 | ||
| Rectum | Reference | Reference | ||
| Left | 2.0 (1.1–4.0) | 0.039 | 0.8 (0.4–1.7) | 0.58 |
| Right | 2.5 (1.3–4.5) | 0.003 | 1.2 (0.6–2.4) | 0.58 |
| Tumor size | 0.002 | 0.079 | ||
| <4 | Reference | Reference | ||
| ≥4 | 2.3 (1.3–3.9) | 1.7 (0.9–2.9) | ||
| Tumor stage | <0.001 | <0.001 | ||
| Stage I | Reference | Reference | ||
| Stage II | 1.5 (0.7–3.6) | 0.32 | 1.3 (0.5–3.2) | 0.56 |
| Stage III | 6.6 (3.5–12.2) | <0.001 | 6.4 (3.1–13.0) | <0.001 |
| LR score | 0.005 | 0.002 | ||
| Low | Reference | Reference | ||
| High | 2.2 (1.3–3.9) | 2.5 (1.4–4.3) | ||
CI, confidence interval; CRC, colorectal cancer; HR, hazard ratio; LR, logistic regression.
Discussion
In CRC screening, high sensitivity to detect early-stage CRC and AA along with high specificity are of paramount importance to improve patient outcomes. In this study, we developed and validated a blood-based screening test with drastically improved accuracy compared to other noninvasive screening methods. In an independent single-blinded validation set of samples, we achieved sensitivity values of 86.6% for CRC and 79.0% for AA with a specificity of 88.1%. Furthermore, our study indicated that ctDNA methylation levels exhibit the potential to predict patient outcome. The prognosis of patients with high preoperative ctDNA methylation levels was poor, regardless of RFS or OS. Recent work has identified that an increase in the AA detection rate significantly decreases cancer risk and improves the survival rate of patients.52 However, current commercial tests have a low sensitivity for AA with reported sensitivities ranging from 11.2% to 42.4%.40,53 The AA detection rate obtained with the ColonES assay greatly outclasses other noninvasive tests, indicating a great potential to impact patient outcomes.
Cell-free DNA presents unique challenges in a screening assay, whereas epigenetic markers, such as methylated cytosines, have long been known as cancer biomarkers. Only a small number of circulating DNA molecules are derived from cancer tissue; the random drop-out of cancer signals is common, especially in early-stage patients. We therefore incorporated several key innovations into the ColonES assay to allow cell-free DNA methylation in plasma to be used to its fullest extent. First, the ColonES platform simultaneously interrogates the methylation status of 595 genomic regions; previous assays have interrogated only limited methylation markers.39,40,53, 54, 55 Second, the ColonES method analyzes the methylation haplotype that incorporates the linked methylation pattern of nearby CpG sites from the same DNA molecule56; this reduces background noise commonly noted in other methods such as quantitative PCR or microarrays. Finally, the ColonES classification model utilizes an ensembled score based on hundreds of targets simultaneously, which allows robust detection given the heterogeneity of cancer epigenomes from one person to another. Together, these innovations allow the ColonES assay to greatly outperform existing CRC screening tests.
The ColonES assay has several advantages compared with other noninvasive tests. The ColonES assay demonstrates a superior sensitivity for AA (79.0% in the blinded validation set) compared to the stool-based Cologuard (42.4%) or the blood-based EpiProColon (11.2%).39,40,53,54,57 In recent years, research centres have successively performed early screening studies of EpiProColon 2.0 with a specificity of 66.7%–94.5%,58,59 which is comparable to that of ColonES. However, its sensitivity to AA is only 17.1%–27.6%, and that of early CRC is 26.5%–52.6%, which is far lower than the detection rate of ColonES. Blood-based tests are preferred by patients over stool-based tests, although the stool DNA test is sensitive to early-stage CRC detection, as shown in previous studies.53,54 Although a blood-based test (EpiProColon) is FDA-approved, this SEPT9 promoter DNA methylation test is not currently recommended by the USPSTF due to its lower sensitivity.39 The ColonES assay overcomes all of these concerns with a simple blood-based single reaction workflow and a high sensitivity even for AA and CRC early stages. For the current CRC screening field, our preliminary idea on how to combine ColonES assay with FIT in the future comes from four aspects: (1) ColonES test can be further conducted as a supplementary means for people with negative FIT test but high risk clinical factors (family history, etc.); (2) ColonES test can be used as the basis for further screening for people with positive FIT test who refuse colonoscopy screening; (3) ColonES assay can be used as an alternative CRC screening means for people who refuse FIT test or cannot provide fecal samples for FIT test at the clinic; (4) ColonES assay can improve the detection rate of AAs and serve as a major screening method in the population with high risk of AAs, such as polyposis history.
ctDNA methylation has also been explored as a potential biomarker for predicting the prognosis and treatment response of CRC to guide treatment or postoperative follow-up monitoring.60 As for blood biomarkers, it has been reported that higher ctDNA levels (especially KRAS, APC and TP53 mutations) have a poor prognosis.61,62 Luo et al.63 found that the ctDNA methylation spectrum could be used for the early diagnosis, prognosis prediction and screening of CRC. However, CEA is the main serum biomarker used to assess relapse in current clinical practice, especially for stage II–III CRC.2 CEA is more sensitive to advanced CRC than early diseases, which limits its application in many surgical patients.64 Therefore, new prognostic and predictive biomarkers are still needed, such as methylated ctDNA, which is easy to detect. In this study, we found that the prognosis of patients with high ctDNA methylation levels was worse than that of patients with low ctDNA methylation levels, indicating that ctDNA methylation levels are of great value in monitoring CRC relapse. Prospective multicentre clinical trials are urgently needed to further consolidate the important role of ctDNA methylation in the dynamic monitoring of postoperative recurrence and metastasis.
Several limitations of this study should be acknowledged. First, this study was retrospective and only included patients already diagnosed with AA or CRC by colonoscopy. Although we were able to show that ColonES could identify early-stage cancer with a high accuracy, a prospective study is guaranteed in the future to determine the efficacy of ColonES to improve patient outcomes in the general population, including asymptomatic and symptomatic population cohorts. Second, the number of healthy controls was still limited in this study; additional healthy samples are needed in future studies to confirm the assay specificity. Third, as a complementary means of FIT detection, the sample size in the FIT negative population is relatively limited, resulting in a shortage of stratified analysis evidence. Relevant statistical analysis should be fully considered in the multicentre prospective clinicaltrial to ensure sufficient sample size for stratified analysis. Finally, we did not directly evaluate the ColonES assay in patients with other cancer types in this study, which may determine whether the ColonES classifier is specific for CRC.
In summary, we have successfully developed and validated an accurate, noninvasive detection method based on ctDNA methylation haplotype patterns, which accurately enables early detection and prognosis prediction for CRC. With a high patient compliance, we hope it can offer a more cost-effective tool for CRC screening prior to invasive colonoscopy for an average-risk population.
Contributors
G.C., S.C., L.L., and Y.X. were responsible for the overall study design. G.C., S.C., L.L., and Y.X. supervised the clinical sample collection and assessment. S.M., W.D., X.L., W.X., L.H., W.L., L.Z., R.W., Y.Z., W.Z., L.Y., R.L., L.G., Y.Z., and M.H. collected samples and organized clinical information at two clinical sites. H.W., C.M., and Z.S. performed experiments and data analysis. S.M., W.D., H.W., and G.C. wrote the manuscript with inputs from the other co-authors. All authors read, discussed, and approved the final version of the manuscript. The co-corresponding authors had full access to the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Data sharing statement
The data presented in this study are available on request from the corresponding author.
Declaration of interests
H.W., C.M. and Z.S. are employed by company Singlera Genomics (Shanghai). The other authors declare no competing interests.
Acknowledgments
The authors would like to thank all the medical workers who participated in this study. This study received funding from the Grant of National Natural Science Foundation of China (No. 81871958), National Natural Science Foundation of China (No. 82203215), Shanghai Science and Technology Committee (No. 19140902100), Scientific Research Fund of Fudan University (No. IDF159052), Shanghai Municipal Health Commission (SHWJRS 2021-99), and Shanghai Sailing Program (22YF1408800).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.eclinm.2022.101717.
Contributor Information
Ye Xu, Email: yexu@shmu.edu.cn.
Li Liang, Email: Lli@fimmu.com.
Sanjun Cai, Email: caisanjun_sh@163.com.
Guoxiang Cai, Email: gxcai@fudan.edu.cn.
Appendix A. Supplementary data
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Goding Sauer A., et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 3.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Chen W., Zheng R., Baade P.D., et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Nguyen L.H., Goel A., Chung D.C. Pathways of colorectal carcinogenesis. Gastroenterology. 2020;158(2):291–302. doi: 10.1053/j.gastro.2019.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–976. doi: 10.2147/CIA.S109285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Provenzale D., Ness R.M., Llor X., et al. NCCN guidelines insights: colorectal cancer screening, version 2.2020. J Natl Compr Canc Netw. 2020;18(10):1312–1320. doi: 10.6004/jnccn.2020.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dekker E., Tanis P.J., Vleugels J.L.A., Kasi P.M., Wallace M.B. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 9.Curry S.J., Krist A.H., Owens D.K., et al. Screening for cervical cancer: US Preventive Services Task Force recommendation statement. JAMA. 2018;320(7):674–686. doi: 10.1001/jama.2018.10897. [DOI] [PubMed] [Google Scholar]
- 10.Siu A.L. Screening for breast cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2016;164(4):279–296. doi: 10.7326/M15-2886. [DOI] [PubMed] [Google Scholar]
- 11.Moyer V.A. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med. 2014;160(5):330–338. doi: 10.7326/M13-2771. [DOI] [PubMed] [Google Scholar]
- 12.O'Connell J.B., Maggard M.A., Ko C.Y. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J Natl Cancer Inst. 2004;96(19):1420–1425. doi: 10.1093/jnci/djh275. [DOI] [PubMed] [Google Scholar]
- 13.Joranger P., Nesbakken A., Sorbye H., Hoff G., Oshaug A., Aas E. Survival and costs of colorectal cancer treatment and effects of changing treatment strategies: a model approach. Eur J Health Econ. 2020;21(3):321–334. doi: 10.1007/s10198-019-01130-6. [DOI] [PubMed] [Google Scholar]
- 14.Rex D.K., Boland C.R., Dominitz J.A., et al. Colorectal cancer screening: recommendations for physicians and patients from the U.S. Multi-Society Task Force on colorectal cancer. Gastroenterology. 2017;153(1):307–323. doi: 10.1053/j.gastro.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Regula J., Rupinski M., Kraszewska E., et al. Colonoscopy in colorectal-cancer screening for detection of advanced neoplasia. N Engl J Med. 2006;355(18):1863–1872. doi: 10.1056/NEJMoa054967. [DOI] [PubMed] [Google Scholar]
- 16.Levin B., Lieberman D.A., McFarland B., et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. CA Cancer J Clin. 2008;58(3):130–160. doi: 10.3322/CA.2007.0018. [DOI] [PubMed] [Google Scholar]
- 17.Gimeno García A.Z. Factors influencing colorectal cancer screening participation. Gastroenterol Res Pract. 2012;2012 doi: 10.1155/2012/483417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inadomi J.M., Vijan S., Janz N.K., et al. Adherence to colorectal cancer screening: a randomized clinical trial of competing strategies. Arch Intern Med. 2012;172(7):575–582. doi: 10.1001/archinternmed.2012.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helsingen L.M., Vandvik P.O., Jodal H.C., et al. Colorectal cancer screening with faecal immunochemical testing, sigmoidoscopy or colonoscopy: a clinical practice guideline. BMJ. 2019;367:l5515. doi: 10.1136/bmj.l5515. [DOI] [PubMed] [Google Scholar]
- 20.Quintero E., Castells A., Bujanda L., et al. Colonoscopy versus fecal immunochemical testing in colorectal-cancer screening. N Engl J Med. 2012;366(8):697–706. doi: 10.1056/NEJMoa1108895. [DOI] [PubMed] [Google Scholar]
- 21.Buskermolen M., Cenin D.R., Helsingen L.M., et al. Colorectal cancer screening with faecal immunochemical testing, sigmoidoscopy or colonoscopy: a microsimulation modelling study. BMJ. 2019;367:l5383. doi: 10.1136/bmj.l5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imperiale T.F., Gruber R.N., Stump T.E., Emmett T.W., Monahan P.O. Performance characteristics of fecal immunochemical tests for colorectal cancer and advanced adenomatous polyps: a systematic review and meta-analysis. Ann Intern Med. 2019;170(5):319–329. doi: 10.7326/M18-2390. [DOI] [PubMed] [Google Scholar]
- 23.Niedermaier T., Weigl K., Hoffmeister M., Brenner H. Diagnostic performance of flexible sigmoidoscopy combined with fecal immunochemical test in colorectal cancer screening: meta-analysis and modeling. Eur J Epidemiol. 2017;32(6):481–493. doi: 10.1007/s10654-017-0279-2. [DOI] [PubMed] [Google Scholar]
- 24.Niedermaier T., Tikk K., Gies A., Bieck S., Brenner H. Sensitivity of fecal immunochemical test for colorectal cancer detection differs according to stage and location. Clin Gastroenterol Hepatol. 2020;18(13):2920–2928.e6. doi: 10.1016/j.cgh.2020.01.025. [DOI] [PubMed] [Google Scholar]
- 25.Shaukat A., Levin T.R. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19(8):521–531. doi: 10.1038/s41575-022-00612-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler A., Geiger S., Keil A., et al. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol. 2014;14:183. doi: 10.1186/1471-230X-14-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmqvist R., Engarås B., Lindmark G., et al. Prediagnostic levels of carcinoembryonic antigen and CA 242 in colorectal cancer: a matched case-control study. Dis Colon Rectum. 2003;46(11):1538–1544. doi: 10.1007/s10350-004-6810-z. [DOI] [PubMed] [Google Scholar]
- 28.Macdonald J.S. Carcinoembryonic antigen screening: pros and cons. Semin Oncol. 1999;26(5):556–560. [PubMed] [Google Scholar]
- 29.Schwarzenbach H., Hoon D.S., Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 30.Alix-Panabières C., Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discov. 2016;6(5):479–491. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 31.Merker J.D., Oxnard G.R., Compton C., et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J Clin Oncol. 2018;36(16):1631–1641. doi: 10.1200/JCO.2017.76.8671. [DOI] [PubMed] [Google Scholar]
- 32.Ye Q., Ling S., Zheng S., Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer. 2019;18(1):114. doi: 10.1186/s12943-019-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hai L., Li L., Liu Z., Tong Z., Sun Y. Whole-genome circulating tumor DNA methylation landscape reveals sensitive biomarkers of breast cancer. MedComm (2020) 2022;3(3):e134. doi: 10.1002/mco2.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng M.L., Pectasides E., Hanna G.J., Parsons H.A., Choudhury A.D., Oxnard G.R. Circulating tumor DNA in advanced solid tumors: clinical relevance and future directions. CA Cancer J Clin. 2021;71(2):176–190. doi: 10.3322/caac.21650. [DOI] [PubMed] [Google Scholar]
- 35.Xie H., Kim R.D. The application of circulating tumor DNA in the screening, surveillance, and treatment monitoring of colorectal cancer. Ann Surg Oncol. 2021;28(3):1845–1858. doi: 10.1245/s10434-020-09002-7. [DOI] [PubMed] [Google Scholar]
- 36.Kulis M., Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 37.Klutstein M., Nejman D., Greenfield R., Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76(12):3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 38.Lam K., Pan K., Linnekamp J.F., Medema J.P., Kandimalla R. DNA methylation based biomarkers in colorectal cancer: a systematic review. Biochim Biophys Acta. 2016;1866(1):106–120. doi: 10.1016/j.bbcan.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 39.deVos T., Tetzner R., Model F., et al. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem. 2009;55(7):1337–1346. doi: 10.1373/clinchem.2008.115808. [DOI] [PubMed] [Google Scholar]
- 40.Church T.R., Wandell M., Lofton-Day C., et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut. 2014;63(2):317–325. doi: 10.1136/gutjnl-2012-304149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bibbins-Domingo K., Grossman D.C., Curry S.J., et al. Screening for colorectal cancer: US Preventive Services Task Force recommendation statement. JAMA. 2016;315(23):2564–2575. doi: 10.1001/jama.2016.5989. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Chang S., Li G., Sun Y. Application of liquid biopsy in precision medicine: opportunities and challenges. Front Med. 2017;11(4):522–527. doi: 10.1007/s11684-017-0526-7. [DOI] [PubMed] [Google Scholar]
- 43.Redshaw N., Huggett J.F., Taylor M.S., Foy C.A., Devonshire A.S. Quantification of epigenetic biomarkers: an evaluation of established and emerging methods for DNA methylation analysis. BMC Genomics. 2014;15(1):1174. doi: 10.1186/1471-2164-15-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Provenzale D., Gupta S., Ahnen D.J., et al. NCCN guidelines insights: colorectal cancer screening, version 1.2018. J Natl Compr Canc Netw. 2018;16(8):939–949. doi: 10.6004/jnccn.2018.0067. [DOI] [PubMed] [Google Scholar]
- 45.Weiser M.R. AJCC 8th edition: colorectal cancer. Ann Surg Oncol. 2018;25(6):1454–1455. doi: 10.1245/s10434-018-6462-1. [DOI] [PubMed] [Google Scholar]
- 46.Edge S.B., Compton C.C. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 47.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai G., Cai M., Feng Z., et al. A multilocus blood-based assay targeting circulating tumor DNA methylation enables early detection and early relapse prediction of colorectal cancer. Gastroenterology. 2021;161(6):2053–2056.e2. doi: 10.1053/j.gastro.2021.08.054. [DOI] [PubMed] [Google Scholar]
- 49.von Elm E., Altman D.G., Egger M., Pocock S.J., Gøtzsche P.C., Vandenbroucke J.P. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Int J Surg. 2014;12(12):1495–1499. doi: 10.1016/j.ijsu.2014.07.013. [DOI] [PubMed] [Google Scholar]
- 50.Chen X., Gole J., Gore A., et al. Non-invasive early detection of cancer four years before conventional diagnosis using a blood test. Nat Commun. 2020;11(1):3475. doi: 10.1038/s41467-020-17316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen C.C., Yang S.H., Lin J.K., et al. Is it reasonable to add preoperative serum level of CEA and CA19-9 to staging for colorectal cancer? J Surg Res. 2005;124(2):169–174. doi: 10.1016/j.jss.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 52.Corley D.A., Jensen C.D., Marks A.R., et al. Adenoma detection rate and risk of colorectal cancer and death. N Engl J Med. 2014;370(14):1298–1306. doi: 10.1056/NEJMoa1309086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Imperiale T.F., Ransohoff D.F., Itzkowitz S.H., et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 54.Ahlquist D.A., Zou H., Domanico M., et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142(2):248–256. doi: 10.1053/j.gastro.2011.10.031. quiz e25-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu R.H., Wei W., Krawczyk M., et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16(11):1155–1161. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 56.Guo S., Diep D., Plongthongkum N., Fung H.L., Zhang K., Zhang K. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat Genet. 2017;49(4):635–642. doi: 10.1038/ng.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warren J.D., Xiong W., Bunker A.M., et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9:133. doi: 10.1186/1741-7015-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma Z.Y., Law W.L., Ng E.K.O., et al. Methylated septin 9 and carcinoembryonic antigen for serological diagnosis and monitoring of patients with colorectal cancer after surgery. Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-46876-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun J., Fei F., Zhang M., et al. The role of (m)SEPT9 in screening, diagnosis, and recurrence monitoring of colorectal cancer. BMC Cancer. 2019;19(1):450. doi: 10.1186/s12885-019-5663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nassar F.J., Msheik Z.S., Nasr R.R., Temraz S.N. Methylated circulating tumor DNA as a biomarker for colorectal cancer diagnosis, prognosis, and prediction. Clin Epigenetics. 2021;13(1):111. doi: 10.1186/s13148-021-01095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu J.M., Liu X.J., Ge F.J., et al. KRAS mutations in tumor tissue and plasma by different assays predict survival of patients with metastatic colorectal cancer. J Exp Clin Cancer Res. 2014;33(1):104. doi: 10.1186/s13046-014-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J.Y., Hsieh J.S., Chang M.Y., et al. Molecular detection of APC, K- ras, and p53 mutations in the serum of colorectal cancer patients as circulating biomarkers. World J Surg. 2004;28(7):721–726. doi: 10.1007/s00268-004-7366-8. [DOI] [PubMed] [Google Scholar]
- 63.Luo H., Zhao Q., Wei W., et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. 2020;12(524) doi: 10.1126/scitranslmed.aax7533. [DOI] [PubMed] [Google Scholar]
- 64.Hauptman N., Glavač D. Colorectal cancer blood-based biomarkers. Gastroenterol Res Pract. 2017;2017:2195361. doi: 10.1155/2017/2195361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.