Abstract
Identification of Plasmodium-resistance genes in malaria vectors remains an elusive goal despite the recent availability of high-quality genomes of several mosquito vectors. Anopheles stephensi, with its three distinctly-identifiable forms at the egg stage, correlating with varying vector competence, offers an ideal species to discover functional mosquito genes implicated in Plasmodium resistance. Recently, the genomes of several strains of An. stephensi of the type-form, known to display high vectorial capacity, were reported. Here, we report a chromosomal-level assembly of an intermediate-form of An. stephensi strain (IndInt), shown to have reduced vectorial capacity relative to a strain of type-form (IndCh). The contig level assembly with a L50 of 4 was scaffolded into chromosomes by using the genome of IndCh as the reference. The final assembly shows a heterozygous paracentric inversion, 3Li, involving 8 Mbp, which is syntenic to the extensively-studied 2La inversion implicated in Plasmodium resistance in An. gambiae involving 21 Mbp. Deep annotation of genes within the 3Li region in the IndInt assembly using the state-of-the-art protein-fold prediction and other annotation tools reveals the presence of a tumor necrosis factor-alpha (TNF-alpha) like gene, which is the homolog of the Eiger gene in Drosophila. Subsequent chromosome-wide searches revealed homologs of Wengen (Wgn) and Grindelwald (Grnd) genes, which are known to be the receptors for Eiger in Drosophila. We have identified all the genes in IndInt required for Eiger-mediated signaling by analogy to the TNF-alpha system, suggesting the presence of a functionally-active Eiger signaling pathway in IndInt. Comparative genomics of the three type-forms with that of IndInt, reveals structurally disruptive mutations in Eiger gene in all three strains of the type-form, suggesting compromised innate immunity in the type-form as the likely cause of high vectorial capacity in these strains. This is the first report of the presence of a homolog of Eiger in malaria vectors, known to be involved in cell death in Drosophila, within an inversion region in IndInt syntenic to an inversion associated with Plasmodium resistance in An. gambiae.
Subject terms: Computational biology and bioinformatics, Evolution, Immunology
Introduction
Anopheles stephensi is an established urban malarial vector in India and South East Asia with expanding range into African cities. This vector quickly adapts to the local environment, survives extremely high temperatures, and establishes resistance to multiple insecticides (Vector alert: Anopheles stephensi invasion and spread, 2019)1. Based on the vectorial capacity of An. stephensi, there are three variants classified as type-form (high vectorial capacity), intermediate-form (moderate vectorial capacity), and mysorensis (low vectorial capacity)2,3. These forms are identifiable based on the number of ridges on eggs4. The genomes of three strains of the type-form have been reported recently5–7. A comparison of these three genomes revealed the presence of all three genotypes of the 2Rb inversion among the various strains of the type-form.
Sixteen paracentric inversions in the chromosomes of An. stephensi have been reported by visualizing loop formation in chromosomes from large heterozygous inversions. While chromosome X has no reported inversions, chromosomal arms 2R, 2L, 3R, and 3L have many paracentric inversions characterized by cytobands8,9. Moreover, each of these inversions is reported to impart varying resistance to insecticides10. However, during the maintenance of various strains of An. stephensi under laboratory conditions, several inversions are lost within the first few generations, such as 2Re, 3Rc, and 3Lc, whereas other inversions, such as 2Rb, 3Rb, and 3Li, persist through generations even in the absence of any environmental pressure, and in some cases, increasing in frequency with increasing generations8. Based on these reports and studies on other malaria vectors, it is now well established that chromosomal inversions are dynamically used by vectors to adapt to environmental changes while retaining fitness11.
An. gambiae, a malaria vector from Africa, is by far the most extensively-studied and interrogated using genomics. The inversions 2Rb and 2La in An. gambiae have been extensively characterized at the molecular and population levels. While the 2Rb inversion in An. gambiae, involving 8 Mbp, is associated with resistance to DDT and adaptation to climate11, the 2La inversion, encompassing 21 Mbp, is associated with resistance to Plasmodium and desiccation12,13. The breakpoints of 2La have been extensively studied by selecting BAC clones spanning breakpoints from strains with alternate arrangements of the 2La configuration14. A systems-level study of gene expression in An. gambiae between different configurations of a given inversion shows that a large number of genes within the inversion loci are differentially regulated15.
Comparative proteomics of D. melanogaster, Ae. aegypti, and An. gambiae revealed 91 gene trios with a total of 589 genes comprising the immune repertoire16. These are homologs of genes from pathways implicated in classical innate immunity or defense functions in insects. The pathways include apoptosis inhibitors (IAPs), oxidative defense enzymes [superoxide dismutases (SODs), glutathione peroxidases (GPXs), thioredoxin peroxidases (TPXs), and heme-containing peroxidases (HPXs)], and class A and B scavenger receptors (SCRs) common to all three species. Of these, more recently, homologs of 361 genes were reported in An. stephensi5, which is in the same range (285, 338, and 353, respectively) of immune-repertoire genes reported for D. melanogaster (Dm), An. gambiae (Ag), and Ae. aegypti (Aa), respectively.
Deep annotation of genes within inversion regions associated with resistance to pathogens offers an opportunity to discover additional immune repertoire genes that may have diverged beyond the point of recognition by standard sequence-based annotation tools. RoseTTaFold, a state-of-the-art protein-fold prediction tool, allows deeper annotation of uncharacterized or divergent proteins, thereby aiding the identification of hidden/missed immune-related genes in Anopheles17. The idea is that the diversity in proteins far exceeds the number of folds in nature, which is estimated to be as low as one thousand18. This is especially important in the identification of cytokine and lymphokine homologs, which are notorious for exhibiting low or undetectable homology at the sequence level19. For example, members of TNF and TNFR superfamilies, which has been identified and characterized in Drosophila, are missing in the collation of genes implicated in innate immunity in flies and in mosquito vectors5,16.
In order to understand the evolution of Plasmodium resistance in An. stephensi, we sequenced and assembled the genome of an isofemale line from a strain of intermediate-form from Bangalore (IndInt) displaying diminished susceptibility to Plasmodium20. We present the assembly of this strain and report an inversion not found in the genomes of the three strains of the type-form5–7,21. Comparative genomics with other Anopheles vectors and deep annotation of genes within this inversion, using fold-prediction algorithms, revealed missed or hidden genes likely to be relevant in Plasmodium resistance.
Results
Assembly of the genome of IndInt strain
Fourteen generations of isofemale lines from a parent T6 lab-colony of the intermediate-form of An. stephensi (IndInt) were grown, both to achieve homogeneity required for high-quality assembly from sequencing of pooled DNA from multiple insects and to enrich for Plasmodium resistant phenotype20. Multiple tools were used to obtain contig-level assembly of IndInt from 60X coverage of reads sequenced using PacBio Sequel II technologies. The assembly statistics obtained from the various tools are shown in Table 1. In the first round, the best assembly statistics were obtained using the WTDBG2 tool with a L50 value of 6 and N50 9.1 Mbp. The next best statistics was shown by the tool Flye assembler with a L50 of 15 and N50 value of 3.9 Mbp.
Table 1.
Statistics of contig-level assemblies by CANU, FALCON, FLYE, and WTDBG2 tools.
| Assembly size (Mbp) | Total contigs | Longest contig (Mbp) | Shortest contig (bp) | Median contig (Kbp) | N50 | L50 | |
|---|---|---|---|---|---|---|---|
| CANU | 405 | 3464 | 16.6 | 1055 | 44.2 | 48.7 Kbp | 115 |
| WTDBG | 205.4 | 789 | 33.08 | 1126 | 15.4 | 9.1 Mbp | 6 |
| FALCON | 293 | 851 | 11.4 | 5734 | 87.4 | 1.4 Mbp | 46 |
| Flye | 211.7 | 689 | 27.15 | 1007 | 14.6 | 3.9 Mbp | 15 |
| Flye + WTDBG | 213.3 | 521 | 45.56 | 1018 | 8.3 | 17.04 Mbp | 4 |
| (Flye + WTDBG) + SSPACE | 213.8 | 464 | 55.4 | 1018 | 5.9 | 36.9 Mbp | 3 |
To improve the contiguity of the assembly, the four different contig-level assemblies from CANU, FALCON, FLYE, and WTDBG222–25 were merged in different combinations using the tool Quickmerge26. Assembly statistics were computed for each of the merged combinations to determine the best merged assembly (Table 1). During the second round, the assembly with the best statistics was obtained by merging assemblies from FLYE and WTDBG2, with a L50 of 4 and a high N50 of 17 Mbp. The computed genome size of this merged assembly was comparable to the actual size of An. stephensi genome, and was used for scaffolding. The scaffolds of the error-corrected contigs were built using the SSPACE assembly tool with simulated mate-pairs of varying sizes from the assembly of IndCh strain as reference. The L50 value for the scaffold level assembly was 3 and the N50 value significantly increased from 17 to 37 Mbp (Table 1, last row).
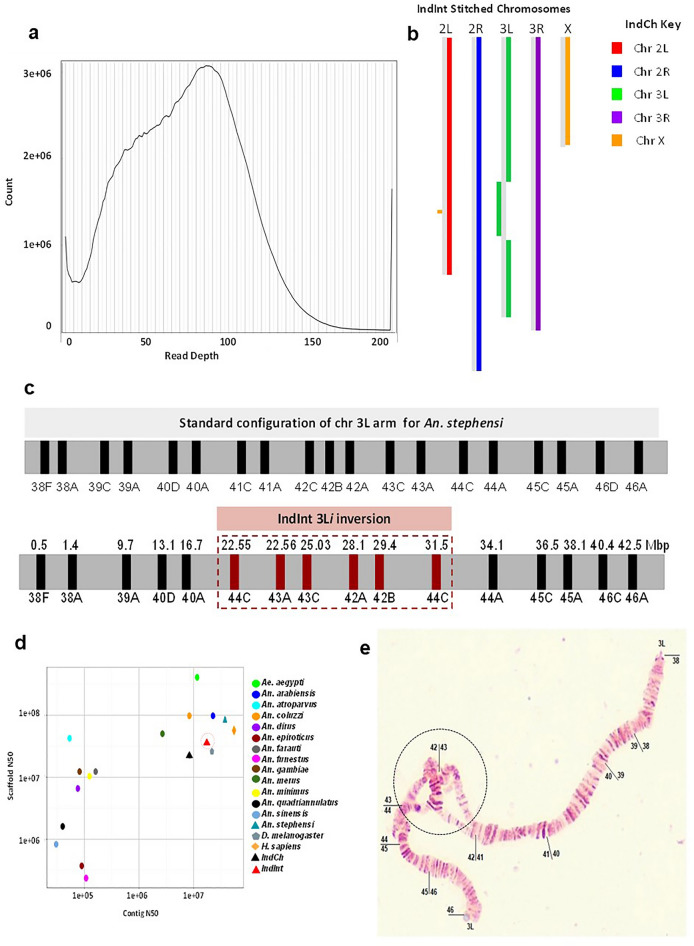
In order to check any heterozygosity from potential inversions, haplotype phasing of the assembly of the IndInt strain was done using a set of primary and associated contigs from FALCON and FALCON-unzip tools, respectively23. In the presence of heterozygosity, the mapped reads will be split between two contigs that represent the two forms of the heterozygous allele, resulting in reduced coverage. This creates two peaks in the histogram of read counts, one for the haploid level coverage from heterozygous regions and the other for the diploid level coverage from homozygous regions. A hump in the histogram indicated the presence of potential inversion heterozygosity in the genome of IndInt (Fig. 1a).
Figure 1.
(a) Histogram for the genome assembly of IndInt showing evidence of heterozygosity by Purge Haplotigs tool. (b) Block diagram of synteny between IndCh and IndInt assembly showing an inversion block in chromosome 3L of IndInt. (c) Top—Orientation of the physical markers as expected in the standard configuration for An. stephensi. Bottom—BLASTN analysis of the assembled IndInt strain revealing the orientation of the physical markers consistent with the 3Li inversion. (d) Quality of IndInt assembly as compared to that of other Anopheles species and completed gold-standard genomes of human and fly. (e) A sample photograph of polytene chromosomes from the IndInt strain showing the loop from the heterozygous inverted configuration of the 3Li.
Synteny between the assemblies of IndInt and IndCh strains is shown in Fig. 1b. The block view clearly suggests that IndInt is homozygous for the standard form of 2Rb (2R+b/2R+b) inversion similar to the IndCh strain. However, in the 3L arm of chromosome 3, IndInt shows the clear presence of an inversion not seen in the IndCh assembly. The locus of the physical markers revealed that the markers from 42A to 44C are reversed in the assembly, which matched with a reported breakpoint for the 3Li inversion in An. stephensi9. Based on the photomaps from several individuals of IndInt strain (see Fig. 1e), it is clear that 48% of the individuals are heterozygous for the 3Li inversion (3L+i/3Li), with the rest (52%) being homozygous for the standard form of 3Li (3L+i/3L+i) (Supplementary Fig. 1). It should be mentioned that the percentage heterozygosity reported here is from an outgrown generation maintained in the lab after the generation used for assembly. Markers 41A, 41B, 41C, and 46D shown in the standard form reported for 3Li (top Fig. 1c) are not detectable in the IndInt assembly using the BLASTN tool, perhaps because of limitations of the assembly tools in handling noise generated by reads representing the two arms near the inversion breakpoints from the heterozygous arms. Figure 1d suggests that the IndInt assembly is comparable to the high-quality genomes of other Anopheles species, Drosophila, and humans at both the contig and scaffold levels.
Validation of the 2Rb breakpoint region in IndInt
The IndInt strain is homozygous for the standard form of the 2Rb inversion, making it the second assembled genome for one of the three possible 2Rb genotypes, such as 2R+b/2R+b. This provides an opportunity to further refine and validate the breakpoint regions described previously by our group6. A BLASTN of the sequences from breakpoint regions from IndCh including 1000 bases to the left and right of both breakpoints, against the IndInt assembly, shows 100% identity for all the 2079 bases from the centromere-proximal breakpoint and 99.8% identity for all the 2009 bases from the centromere-distal breakpoint. We also provide the actual alignment with 50± nucleotides around the breakpoints in Supplementary Fig. 2 This confirms the 2Rb genotype to be 2R+b/2R+b for IndInt.
Interrogation of 3Li breakpoints in IndInt
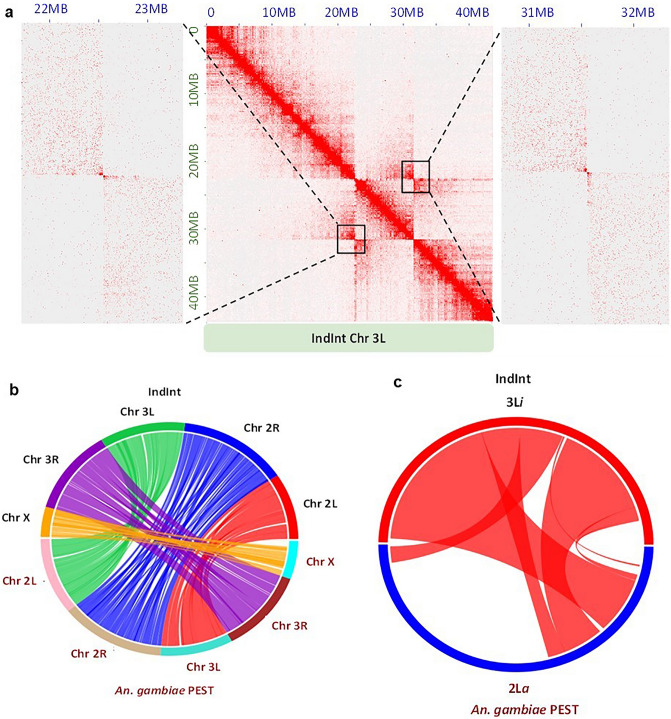
In order to define the breakpoints for the 3Li inversion, a contact map was generated by mapping the HiC reads from the UCI strain onto the final IndInt assembly. Since UCI is homozygous for the standard 3Li (3L+i/3L+i) configuration and IndInt is heterozygous (3L+i/3Li), two expected butterfly-like structures in the chromosome 3L were observed in the contact map (Fig. 2a) defining the location of both the proximal and distal breakpoints of the 3Li inversion in the IndInt assembly. By magnifying each of the butterfly structures to a resolution of 5 Kbp per dot, the proximal and distal breakpoints of the 3Li inversion were estimated to be between bp 22,545,000 to 22,550,000, and between bp 31,555,000 to 31,560,000 respectively.
Figure 2.
(a) Proximal and distal breakpoints obtained by mapping UCI HiC reads against the chromosome 3L of IndInt. The black boxes represent the centromere-proximal (left) and centromere-distal (right) breakpoints located between the ranges of bp 22,545,000 to 22,550,000 and bp 31,555,000 to 31,560,000, respectively, in the IndInt assembly. Adjacent to the contact map are the enlargements of the butterfly structures, for a resolution of 5 Kbp per dot. (b) Synteny between An. gambiae and IndInt assemblies. (c) Synteny between the An. gambiae 2La and the IndInt 3Li regions.
Synteny of 3Li inversion with 2La of An. gambiae
Synteny of the final IndInt assembly was also plotted against the An. gambiae PEST strain27. Figure 2b shows that chromosome X, 2R, and 3R of An. gambiae are homologous to that of IndInt, respectively. However, the synteny between chromosomal arms 2L and 3L of An. gambiae are switched to chromosomal arms 3L and 2L of IndInt, respectively, as reported elsewhere6. Figure 2c shows the synteny between the 2La inversion of An. gambiae with the 3Li region of IndInt. The 2La region of An. gambiae was taken from a previous study28. The circle view (Fig. 2c) of this synteny reveals that the entire 3Li inversion region of IndInt consisting of 8 Mbp is syntenic to a big part of the 2La inversion region of An. gambiae comprising 21 Mbp. Since the 2La inversion of An. gambiae is associated with Plasmodium sensitivity12,13, by analogy, we infer that the 3Li of IndInt may also be associated with the Plasmodium resistance observed for IndInt strain. Furthermore, it is reported that the strains of An. gambiae that are homozygous for the standard form of 2La (2L+a/2L+a) show higher vectorial capacity than the corresponding strains that are heterozygous for the inverted form. Similarly, the strain IndCh with higher vectorial capacity is homozygous for the standard form of 3Li (3L+i/3L+i) and IndInt is heterozygous for the 3Li inversion (3L+i/3Li), suggesting the potential role of the 3Li inversion in Plasmodium sensitivity.
Functional characterization of genes within the 3Li region
Within the 2La region of the An. gambiae PEST strain spanning 21 Mbp, there are 1243 genes of which there are 494 genes that are homologous/syntenic to the 3Li region of IndInt. Of these, 464 are functionally annotated in IndInt based on sequence level annotation tools, and the remaining 30 genes are uncharacterized due to a lack of sequence homology to known functional genes. Among functionally-characterized genes, we found 36 isoforms of cuticle-forming genes in IndInt. Interestingly, cuticle thickening is associated with pyrethroid & insecticide resistance in other Anopheles species29, while also resisting Plasmodium infection30. We found 99 missense mutations in the IndInt strain within 23 copies of the cuticle genes, as compared to the IndCh strain of the type form, which has a homozygous configuration for the standard form of 3Li, like the UCI strain.
In order to find genes implicated in immune response within the 3Li region of IndInt, we intersected the 494 genes within this region with a list of 589 genes implicated in immunity in insects16. Out of these 589 genes, we found only 17 genes within the 3Li region (Table 2) including NFkB, a transcription factor implicated in cytokine production in humans.
Table 2.
Genes within the 3Li region of IndInt that are reported to be implicated in immune response by Waterhouse et al. 16.
| IndInt Augustus ID |
IndInt Chr 3L start |
IndInt Chr 3L end |
Waterhouse et. al 16 Immune DB ID |
Alignment percentage | Function |
|---|---|---|---|---|---|
| g22033.t1 | 22194979 | 22196095 | AGAP005552 | 76.24% | Peptidoglycan recognition protein |
| g22086.t1 | 22559888 | 22560559 | AGAP007020 | 92.83% | Thioredoxin peroxidase 5 |
| g22107.t1 | 22657471 | 22661294 | AGAP006939 | 72.71% | Armitage protein |
| g22121.t1 | 22702737 | 22707661 | AAEL001675 | 60.41% | Clip domain, serine protease family A |
| g22273.t1 | 24373199 | 24379261 | AGAP006631 | 88.68% | Class A scavenger receptor (SRCR domain) with serine protease domain |
| g22562.t1 | 26855939 | 26862215 | AGAP006914 | 61.09% | Fibrinogen-related protein 1 |
| g22567.t1 | 26922106 | 26923713 | AGAP006911 | 91.98% | Serine protease inhibitor—Serpin 2 |
| g22568.t1 | 26925252 | 26932268 | AGAP006909 | 83.54% | Serine protease inhibitor—Serpin 1 |
| g22799.t1 | 28434986 | 28435854 | AGAP006790 | 72.73% | Fibrinogen C-terminal domain-containing protein |
| g22840.t1 | 28809510 | 28812635 | AGAP006761 | 83.96% | 3-Glucan binding protein |
| g22865.t1 | 28987304 | 28992526 | AGAP006747 | 79.51% | IMD pathway signaling NF-kappaB relish-like transcription factor |
| g22870.t1 | 29022137 | 29026993 | AGAP006743 | 64.44% |
Unspecified product BLASTP: similar to ficolin 1 |
| g22921.t1 | 29370252 | 29370571 | AGAP006722 | 81.25% |
Unspecified product BLASTP: similar to Cecropin B1 |
| g23080.t1 | 30464940 | 30470336 | AGAP005672 | 94.10% | Staphylococcal nuclease domain-containing protein 1 |
| g23109.t1 | 30680813 | 30689983 | AGAP005652 | 86.48% | ATP-dependent RNA helicase DDX5/DBP2 |
| g23151.t1 | 30998165 | 31003426 | AGAP005625 | 78.15% | Class A scavenger receptor (SRCR domain) with serine protease domain |
| g23220.t1 | 31575970 | 31576777 | CPIJ004568 | 68.64% | Similar to Cu-chaperone |
The genes implicated in Plasmodium resistance in An. gambiae31 LRIM1, APL1C, and TEP1 are not within the 3Li region in IndInt. The TEP1 protein in IndInt is located on chromosomal arm 2L. The LRIM1 gene is outside the 3Li region, while the gene APL1C is located immediately outside the distal breakpoint of the 3Li region. The LRIM1 gene has leucine-rich repeats and forms a disulfide-bonded complex with APL1C (PDB ID: 3OJA31), which stabilizes the mature TEP1 and promotes its binding to parasites to trigger pathogen destruction. Also, more recently, a knockout of LRIM1 in An. stephensi was shown to play a role in vector competence32. Furthermore, LRIM1 and APL1C are within the 2La region in An. gambiae, which is three times larger than the 3Li region in IndInt. In IndInt, the LRIM1 has four and six missense mutations compared to IndCh and UCI strains of type-form, respectively. However, since the gene APL1C shows no missense mutation in IndInt compared to IndCh, we expected other unannotated paralogs of LRIM1 within the 3Li region of IndInt. Deep annotation of the 30 functionally uncharacterized genes within the 3Li region shown in Supplementary Fig. 3 with fold-prediction algorithms, revealed two novel leucine-rich proteins, g22432 and g22212, similar in structure to LRIM1 (Supplementary Fig. 3a and 3b). These two LRIM1-like genes could potentially work with the intact APL1C gene in IndInt to launch a host defense system to antagonize the malaria parasite. The high conservation of these, otherwise uncharacterized genes across Anopheles species, shown in the multiple sequence alignments of g22432 and g22212, suggests functional conservation across species (Supplementary Fig. 4a and 4b).
Recently, members of LPS-induced TNF-alpha factors, LITAFs, have been shown to regulate anti-Plasmodium immunity in An. Gambiae33,34. A search in the assembled genome of An. stephensi identified four members of LITAF-like transmembrane genes (Supplementary Text 1). LITAFs are known to send signals to the nucleus to transcribe TNF-alpha molecules in humans35. In fact, LPS-induced animal models were used in developing drugs against TNF-alpha to treat auto-immune disorders. We were also able to find homologs of all the genes involved in this signaling pathway in IndInt (Supplementary Text 2).
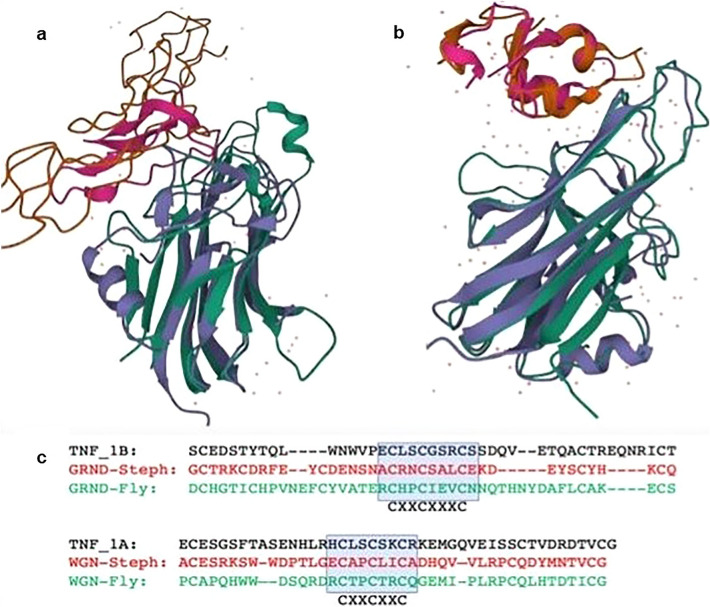
We found an Eiger homolog, g22826, which is within the 3Li region of IndInt and is missing in the reported list of 361 genes linked to the immune repertoire in An. stephensi5. A crystal structure of the complex of Eiger and its receptor from Drosophila has revealed that Eiger belongs to the TNF superfamily and its two receptors, Wengen and Grnd, are members of the TNFR superfamily (PDB: 6ZT036)37–39. A search for an active TNFR-like gene in An. stephensi, using the HMM model (PF00020) of the cysteine-rich domain (CRD) from PFAM database, identified two potential hits g1129 and g18030 in IndInt. It should be mentioned that while Wengen displays detectable homology with TNFRSF, predicted Grnd homologs from many sequenced species, including malaria vectors, remain uncharacterized by sequence-based annotation tools. The gene, g1129, from chromosome X of the IndInt strain encodes a Wengen homolog of Drosophila, with a well-defined extracellular CRD, and a transmembrane region. The Grnd homolog (g18030) in the IndInt strain consists of an intact CRD region sandwiched between a predicted signal sequence and transmembrane region (Supplementary Fig. 5c and 5d). We found that the Eiger gene has a frame-shift mutation at the tail-end of the very conserved folding domain in the IndCh strain. Also, in the UCI and STE2 strains, the predicted Eiger homologs are missing the critical N-terminal transmembrane region (Supplementary Fig. 6). In Fig. 3a, a 3D model of the Eiger gene from IndInt, built using RoseTTaFold tool17, is shown superimposed on the crystal structure of human TNF-TNFR complex (PDB: 1TNR40) on the left, along with a Wengen model built using homology to the second domain of TNFR-1A. Similarly, the Eiger model is shown superimposed on the crystal structure of the Eiger-Grnd complex (PDB: 6ZT036), along with a model of Grnd built using homology to Grnd of Drosophila in Fig. 3b. The choice of templates for comparative modeling of the CRDs of the two receptors is based on the conserved cysteine patterns of the ligand binding regions, which are different for the two receptors Wengen and Grnd. Interestingly, the Wengen binding domain has a cysteine pattern similar to TNFR-1A of humans and that of Grnd is similar to that from TNFR-1B, suggesting an evolutionary-conserved link between Eiger and TNF-alpha mediated signaling in arthropods and vertebrates, respectively.
Figure 3.
(a) De novo model of Eiger and comparative model of Wengen superimposed on the crystal structure of the complex of TNFb-TNFR-1A (1tnr). (b) De novo model Eiger and comparative model of Grnd superimposed on the crystal structure of the complex of Eiger and Grnd from Drosophila (6zt0). (c) Sequence alignment of Wengen and Grnd against TNFR-1A, -1B and Grnd from Drosophila used in comparative modeling.
A functional TNF-alpha/TNFR pathway in malaria vectors with lower vectorial capacity
It is now well established that the Drosophila genome harbors two functional TNFR receptors and a single ligand, Eiger, belonging to the TNF-TNFR system36. In order to predict whether the homologs of the Eiger and its cognate receptor genes in An. stephensi (IndInt) are potentially functional, we used signal peptide and transmembrane predictions on the respective protein sequences derived from IndInt (Supplementary Fig. 5). The Eiger homolog shows a transmembrane region at its N-terminus (Supplementary Fig. 5a) consistent with type II configuration common to all members of the TNF superfamily across organisms. The two receptor homologs show clear transmembrane regions (Supplementary Fig. 5b and 5c) following the cysteine-rich folding domain. However, only Grnd, but not Wengen, has a predicted signal peptide signature (Supplementary Fig. 5d) N-terminal to the CRD, consistent with the localization of these two genes in Drosophila, where Wengen is almost always localized in intracellular vesicles36. We also found the homolog of the TACE gene in IndInt, which is a TNF-alpha processing metalloprotease that converts the type-II membrane-anchored ligand into its soluble/active form in human41. In the Eiger gene from IndInt, we also found a cleavage site (AQ) N-terminal to the folding domain that is known to be specific to TACE42. The gene TACE itself is rendered active by genes called iRhom1 and iRhom2, which are actively being developed as potential targets to inhibit TNF-alpha processing. Interestingly, there are homologs of iRhoms in IndInt, albeit with a much longer N-terminal region than human iRhom1 and 2.
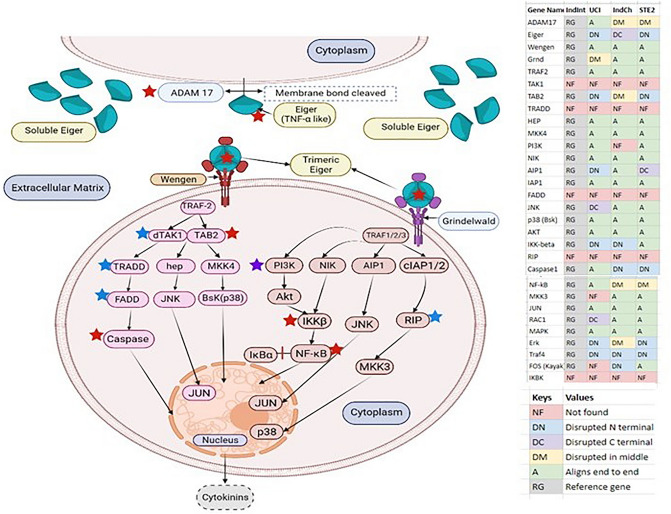
In order to show that all the genes in the cytoplasm necessary for Eiger-Wgn-Grnd mediated signaling are encoded by the genome of IndInt, we identified homologs of all the pathway genes from TNF-alpha and eiger signaling pathways from KEGG43–45 (Supplementary Text 2). Figure 4 shows genes implicated in Wengen-based (left) and Grnd-based (right, hypothesis) based on the homology of the receptors to TNFR-1A and -1B, respectively. Several genes are disrupted in the other strains of type-form viz IndCh, UCI and STE2, as shown in Fig. 4, Supplementary table 1 and Supplementary table 2. The homologs for genes marked by red and purple stars are disrupted and absent, respectively, in the IndCh genome, potentially disrupting the Eiger-mediated signaling pathway. These genes are Eiger, ADAM17, Tab2, IKKb, NFkB, and Caspase. Interestingly, genes with blue stars are missing in all 4 assembled genomes.
Figure 4.
Pathways leading to Eiger-Grnd/Wgn-mediated signaling collated from TNF-TNFR and Eiger-Wgn-mediated signaling pathways obtained from the KEGG database43–45 and validated using other sources46,47. Red stars represent the disrupted genes in the IndCh genome, purple star is the gene missing in IndCh but present in IndInt, blue stars are the missing pathway genes in all four assembled An. stephensi genomes. The inset table shows the status of the genes in all 4 strains (IndInt, IndCh, UCI and STE2) of An. stephensi for which a high-quality genome is available.
It should be mentioned that a majority of these genes including Eiger, Grnd, and Wengen are missing in the collated list of 589 immune related genes in insects16 and, more specifically, missing from a list of 361 immune related genes in An. stephensi reported recently in the UCI strain5.
Discussion
We report a high-quality genome assembly of an isofemale line of the intermediate-form of An. stephensi, IndInt, displaying low vectorial capacity relative to the strain IndCh of type-form20. A comparative analysis of this assembly with those of type-forms reveals a 3Li inversion (in a heterozygous configuration) that is uniquely present only in the IndInt strain. As shown in Fig. 2c, this inversion is syntenic to the 2La inversion region in An. gambiae, the heterozygous genotype of which is shown to be associated with Plasmodium resistance12; thus, providing an opportunity to zero-in on genes that may be offering resistance to Plasmodium.
Here, for the first time, we report the presence of an Eiger homolog, a member of the TNF family, within this inversion region (Fig. 3b). Considering that Eiger has been shown to cause cell death in Drosophila37, it is tempting to hypothesize apoptotic role for Eiger in defence of parasites. Furthermore, considering that the Eiger gene in the IndCh strain harbors a frameshift mutation within the structurally folding domain and considering the extensive role of TNF-alpha in human immunity, it is tempting to predict a role for the Eiger gene in the innate immune response of An. stephensi against Plasmodium in IndInt strain. Similarly, the predicted Eiger genes in the STE2 and UCI strains are both disrupted in the N-terminal segments lacking a transmembrane region and several genes in the Eiger pathway are disrupted in the IndCh strain (Fig. 4, Supplementary table 2). These observations suggest a testable correlation between high vectorial capacity in An. stephensi strains with a dysfunctional Eiger-mediated signaling pathway, which has a lower vectorial capacity for Plasmodium with a potentially functional Eiger-signaling pathway. Furthermore, the increased expression of Eiger-Wgn genes in response to infection by Nora virus in Drosophila48 demands an exploration of the role of Eiger mediated signalling in Plasmodium resistance in An. stephensi and in other malaria vectors, in general. More recently, in support of this, it has been shown that inhibitors of JNK49, a downstream gene of the Eiger/TNF-alpha pathway, abolish resistance to Plasmodium in both An. gambiae50 and An. stephensi51.
The gene TNF-alpha and its two cognate receptors in humans, TNFR-1A and -1B, are implicated both in adaptive and innate immune response. Indeed, drugs inhibiting this pathway are now available to treat various autoimmune diseases such as rheumatoid arthritis, Crohn’s disease, and psoriasis. The two cognate receptors of Eiger, Wengen and Grnd, show homology in their cysteine patterns to TNFR-1A and TNFR-1B respectively (Fig. 3c), suggesting evolutionary conservation of this signaling pathway. Consistent with the ability of the Eiger gene to participate in innate immune response analogous to signaling by TNF-alpha, we found genes in An. stephensi (IndInt) that are homologous to genes implicated in TNF-alpha mediated signaling (Fig. 4). Most interestingly, Eiger is not only a type-2 membrane protein like TNF-alpha, but the presence of homologs of ADAM17 and those of iRhoms in IndInt suggests that Eiger is rendered soluble/active similar to TNF-alpha, thus strengthening the evolutionary conservation of this entire pathway. Considering that many genes downstream of the TNF-alpha pathway are involved in multiple processes, it is surprising that a majority of these are missing from the list of genes with immune repertoire reported for insects16 (Supplementary Table 1).
It is known that human TNF-alpha reduces Plasmodium falciparum growth and activates calcium channels in human malarial parasites52. It is reasonable to hypothesize that mosquito vectors may also elicit similar responses via the Eiger-Wengen-Grnd signaling pathway in parasites to control their growth. Furthermore, the Eiger mediated cell-death in Drosophila could be reversed by JNK inhibitors36. Most importantly, JNK inhibitors reverse Plasmodium resistance in both An. gambiae50 and An. stephensi51. These observations support the hypothesis that a functional Eiger pathway in malaria vectors may reduce the growth of Plasmodium falciparum, similar to the action of TNF-alpha in the human host.
Drosophila and mosquitoes diverged some 350 million years ago, suggesting that the TNF-alpha based innate immunity may have evolved before this point of inflection between vertebrate and invertebrates. A search for TNF and TNFR-like molecules in the Alpha-fold database of C. elegans reveals ~ 14 TNF-like molecules without any TNFR-like homologs. The TNF-like molecules in C. elegans are actually cerebellin homologs. Interestingly, cerebellins are also both type 2-membrane proteins and form threefold functional oligomers just like members of the TNF superfamily. As proof of this, there are homologs of glutamate receptors and neurexin genes encoded by C. elegans, which are involved in forming synaptic structures in rats53. Since the TNF-alpha signaling system is conserved in flies, mosquitoes and vertebrates, missing/lost in nematodes, innate immunity by the TNF-alpha mediated signaling may predate the invertebrate-vertebrate split and TNF-like fold evolution may even be older.
Conclusion
The three forms of An. stephensi (type, intermediate and mysorensis) displaying varying vector competence, which are clearly identifiable by their number of egg ridges, offered an ideal system to discover genes implicated in Plasmodium resistance by interrogating genomes of strains with low vectorial capacity. Here, we have demonstrated that a bottom-up approach starting from phenotype to gene discovery, is more efficient in identifying hidden/divergent genes implicated in Plasmodium sensitivity. We have identified several genes in An. stephensi from gene families known to be involved in innate immunity including LITAFs, novel homologs of LRIM1 within the 3Li region, a homolog of TNF-alpha within the 3Li region, and majority of the genes from TNF-alpha signaling pathway. In particular. this is the first report of a homolog of Eiger in vectors with demonstrated role in innate immunity particularly apoptosis. This approach has opened up several novel avenues/molecular targets for advancing our understanding of the evolution of Plasmodium resistance in vectors and, perhaps, offer effective ways to manage malaria in the near future.
Materials and methods
Maintenance of An. stephensi in insectary
Larvae of An. stephensi were collected from Sriramanahalli area of Bangalore rural (13.4310° N, 77.3310° E) from the state of Karnataka, India, and successfully colonized in the insectary. Larvae were provided with larval food prepared by mixing Brewer’s yeast and dog biscuits at a ratio of 30:70. Pupae were bleached with 1% sodium hypochlorite for 1 min and kept in adult rearing cages (Bugdorm-4S3030) (W32.5 × D32.5 × H32.5 cm). The adults were fed on a mixture of 8% sucrose, 2% glucose solution mixed with 3% multivitamin syrup (Polybion LC®)54. Adults were maintained in the insectary at 28 ± 1 °C, RH 75 ± 5% and 12:12-h day and night photoperiod cycles. Adult females were provided with blood on day 7 or 8 post-emergence to obtain eggs for continuing next progeny. Eggs were collected on day 4 in an ovicup containing water and lined with filter paper at the inner margin. Larvae were hatched out after 36–48 h and transferred to the rearing trays (L39 × B30 cm; Polylab, Catalog no. 81702) with larval food.
Establishment of IndInt isofemale line of An. stephensi
About 10–15 gravid females were separated from the intermediate line55. On day 3, each gravid female was transferred carefully to a single ovicup. Each ovicup was covered with a nylon net. A cotton ball soaked with sugar solution was kept on the top of each ovicup. The line from single female (G0) was selected to generate the isofemale lines that laid the highest number of eggs and had highest percent hatchability with egg ridge numbers 15–17. The eggs thus collected from the single female were allowed to hatch, and the larvae were provided with larval food and reared to adults as per the protocol mentioned earlier. Emerged adult siblings (G1) were kept in a mosquito rearing cage and allowed to inbreed (sibling mating). Mated females were blood fed, and the same procedure was followed as mentioned earlier and continued till further generations. As of July 2022, the IndInt isofemale colony has been maintained in our insectary for 55 generations.
Karyotyping of chromosome 3 of IndInt strain
Polytene chromosomes were prepared from the ovarian nurse cells collected from semi-gravid females of the IndInt isofemale lines as per the method of Ghosh & Shetty, which was adapted from our recent publication6. The semi-gravid females were anesthetized and placed on a microslide in a drop of phosphate buffer. The ovaries were pulled out gently and fixed in modified Carnoy’s fixative (methanol: acetic acid, 3:1) for 2–3 min. The remaining mosquito body was preserved in an Eppendorf tube with a few drops of phosphate buffer solution and kept at − 20 °C for PCR analysis. After fixation, the ovaries were stained with lacto acetic orcein for 15–20 min. After staining, 60% acetic acid was added to it and a clean coverslip was placed on top of the stained sample. Gentle pressure was applied on the cover glass for squashing. The edges of the coverslip were sealed with nail polish to avoid evaporation. The slides were examined under a microscope using a 40X objective lens for chromosome inversions. The inversion nomenclature and their frequency were recorded56.
Assembly
PacBio reads from the IndInt strain were assembled independently using FLYE24 and WTDBG225 assemblers. The resulting assemblies were combined using Quickmerge26 where the FLYE assembly was taken as the query and the WTDBG2 assembly served as the reference. Two rounds of Arrow polishing (PacBio gcpp v2.0.2) using PacBio reads, followed by two rounds of Pilon polishing (v1.22) using 100 × Illumina reads, were carried out on the merged assembly. In order to proceed with reference based de novo assembly, simulated mate-pair reads were generated from the IndCh standard assembly (https://doi.org/10.1038/s41598-022-07462-3) using the tool WGSIM (https://github.com/lh3/wgsim) for the sizes 50 Kbp, 100 Kbp, 250 Kbp, 500 Kbp, 1 Mbp, 2.5 Mbp and 5 Mbp. These simulated mate-pair reads along with the contig level assembly from FLYE + WTDBG2 served as input to obtain a scaffold level assembly using the SSPACE57 tool. DNA physical marker sequences for each chromosome arm published by Jiang et al.21. were downloaded and aligned against the scaffolds using BLAST, in order to assign the chromosomal positions to the scaffolds. The scaffolds were stitched based on the order and orientation of the markers into pseudo-chromosomes. 100 N’s were added between two scaffolds during stitching. Physical marker sequences were realigned to the stitched chromosomes for validation.
Haplotype phasing
The assemblers FALCON and FALCON-Unzip23 were used to phase the IndInt genome into haplotypes. Raw IndInt PacBio reads were used by the FALCON assembler to produce a set of primary contig files and an associate contig file representing the divergent allelic variants. The output of primary and associate contig files were then used by FALCON-Unzip to produce partially phased primary contigs (all_p_ctg.fa) and fully phased haplotigs (all_h_ctg.fa), which represent divergent haplotypes. Polishing was performed on the phased contigs by FALCON-Unzip using Arrow. This procedure was adapted from our recent publication6.
Purge haplotigs
The tool ‘Purge Haplotigs’58 was used to determine the degree of heterozygosity in the IndInt strain. PacBio reads of IndInt strain were mapped to the consensus primary assembly (cns_p_ctg.fasta) obtained from FALCON-Unzip to obtain an aligned BAM file. Coverage analysis was performed on the BAM file to generate a read-depth histogram.
Identification of 3L breakpoints in IndInt
The tool HiC-Pro59 was used to generate a contact map by mapping the HiC reads from the UCI strain onto the genome of IndInt with different inversion genotype to produce a butterfly-like structure representing the breakpoints, which were visualized and extrapolated from the contact map of the tool Juicebox60. The breakpoints were validated by mapping the PacBio and Illumina reads onto the respective genomes. Alignment using ‘unimap’ was performed between the genomes of the IndInt and UCI strains, to determine the precise breakpoints for the 3Li inversion to consolidate with the findings from HiC-Pro.
Gene annotation
AUGUSTUS (version 3.2.3)61, an eukaryotic gene prediction tool, was used to find protein-coding genes for all the assembled genomes. The model organism closest to An. stephensi, Ae. aegypti, which was made available by AUGUSTUS, was used for gene prediction. This procedure was adapted from our recent publication6.
Structural prediction of genes of unknown functions
The genes obtained from AUGUSTUS were used as an input for RoseTTAFold (https://robetta.bakerlab.org/, version August 2021), a deep learning-based protein modeling method. High confidence models obtained from Robetta were used as an input for PyMOL(Version 2.0 Schrödinger, LLC). Regions conserved across the models were identified after structural alignment in PyMol as putative protein domains. Protein contact maps were generated using BIOVIA Discovery Studio Visualizer (v21.1.0.20298) to visually confirm the conserved regions and check for short and long-range interactions in the identified domains. The identified domains were given as input in both sequence and structure-based comparison softwares. Structural comparison was done using DALI (DaliLite.v5) and sequence comparison was done using online utilities such as SwissModel Expasy, Conserved Domain Database from NCBI, InterPro, HMMER Homology Search, and Pfam Sequence Search. Default parameters were used for all the tools. The DALI result showing optimal RMSD value, and seen as hits in the sequence-based tools as well, was chosen as the possible domain.
Supplementary Information
Acknowledgements
The authors thank Nucleome Informatics Pvt. Ltd. for generating Pacbio reads and Illumina reads for the IndInt strain. The authors thank Tata Institute for Genetics and Society (TIGS) India for funding researchers involved in this work and for sequencing and the Government of Karnataka for funding the computational infrastructure at IBAB. The authors would like to acknowledge one of the authors Ms. Apoorva Ganesh for drawing figure 4 using the Biorender tool https://app.biorender.com/biorender-templates.
Author contributions
S.Sri. for overseeing bioinformatics work, setting the tone and writing of the manuscript. S.D. for writing the first draft, taming the assembly and discovery of 3Li inversion. C.G. for creating the IndInt isofemale line and generating photomaps. SSingh for contig level assemblies using various methods. A.T. for characterizing candidate genes. A.G. characterizing breakpoints and TNF-TNFR pathway work. H.M., A.J., M.K., A.C., R.B., V.S. for deep annotation of 3Li region using RoseTTaFold prediction. S.M. Insectary work. N.K., S.K. insectary work and for vectorial capacity in both standard and heterozygous 3Li inverted individuals. S.Swa. for overseeing the entire insect work. S.S. for conceptualization and contributions to the manuscript.
Funding
This research is funded by internal grants from both the Institute of Bioinformatics and Applied Biotechnology and the Tata Institute for Genetics and Society in Bangalore, India.
Data availability
The raw PacBio/Illumina reads used in the assembly and the IndInt chromosome level assembly are uploaded onto the NCBI server under the BioProject ID: PRJNA757559.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Chaitali Ghosh, Shrestha Das and Aditi Thakare.
Contributor Information
Subhashini Srinivasan, Email: ssubha@ibab.ac.in.
Sunita Swain, Email: sunita.swain@tigs.res.in.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-23780-y.
References
- 1.Programme GM. Vector alert: Anopheles stephensi invasion and spread. World Health Organization; 26 Aug 2019 [cited 30 Sep 2022]. Available: https://www.who.int/publications/i/item/WHO-HTM-GMP-2019.09.
- 2.Shinzawa N, Ishino T, Tachibana M, Tsuboi T, Torii M. Phenotypic dissection of a plasmodium-refractory strain of malaria vector Anopheles stephensi: The reduced susceptibility to P. berghei and P. yoelii. PLoS One. 2013;8:e63753. doi: 10.1371/journal.pone.0063753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basseri HR, Mohamadzadeh Hajipirloo H, Mohammadi Bavani M, Whitten MMA. Comparative susceptibility of different biological forms of Anopheles stephensi to Plasmodium berghei ANKA strain. PLoS ONE. 2013;8:e75413. doi: 10.1371/journal.pone.0075413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chavshin AR, Oshaghi MA, Vatandoost H, Hanafi-Bojd AA, Raeisi A, Nikpoor F. Molecular characterization, biological forms and sporozoite rate of Anopheles stephensi in southern Iran. Asian Pac. J. Trop. Biomed. 2014;4:47–51. doi: 10.1016/S2221-1691(14)60207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chakraborty M, Ramaiah A, Adolfi A, Halas P, Kaduskar B, Ngo LT, et al. Author Correction: Hidden genomic features of an invasive malaria vector, Anopheles stephensi, revealed by a chromosome-level genome assembly. BMC Biol. 2022;20:96. doi: 10.1186/s12915-022-01314-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thakare A, Ghosh C, Alalamath T, Kumar N, Narang H, Whadgar S, et al. The genome trilogy of Anopheles stephensi, an urban malaria vector, reveals structure of a locus associated with adaptation to environmental heterogeneity. Sci. Rep. 2022;12:3610. doi: 10.1038/s41598-022-07462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chida AR, Ravi S, Jayaprasad S, Paul K, Saha J, Suresh C, et al. A near-chromosome level genome assembly of Anopheles stephensi. Front. Genet. 2020;11:565626. doi: 10.3389/fgene.2020.565626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahmood F, Sakai RK. Inversion polymorphisms in natural populations of Anopheles stephensi. Can. J. Genet. Cytol. 1984;26:538–546. doi: 10.1139/g84-086. [DOI] [PubMed] [Google Scholar]
- 9.Kamali M, Sharakhova MV, Baricheva E, Karagodin D, Tu Z, Sharakhov IV. An integrated chromosome map of microsatellite markers and inversion breakpoints for an Asian malaria mosquito, Anopheles stephensi. J. Hered. 2011;102:719–726. doi: 10.1093/jhered/esr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shetty NJ, Hariprasad TPN, Sanil D, Zin T. Chromosomal inversions among insecticide-resistant strains of Anopheles stephensi Liston, a malaria mosquito. Parasitol. Res. 2013;112:3851–3857. doi: 10.1007/s00436-013-3575-0. [DOI] [PubMed] [Google Scholar]
- 11.Ayala D, Ullastres A, González J. Adaptation through chromosomal inversions in Anopheles. Front. Genet. 2014;5:129. doi: 10.3389/fgene.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riehle MM, Bukhari T, Gneme A, Guelbeogo WM, Coulibaly B, Fofana A, et al. The 2La chromosome inversion is associated with susceptibility to in Africa. Elife. 2017 doi: 10.7554/eLife.25813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gray EM, Rocca KAC, Costantini C, Besansky NJ. Inversion 2La is associated with enhanced desiccation resistance in Anopheles gambiae. Malar. J. 2009;8:215. doi: 10.1186/1475-2875-8-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharakhov IV, White BJ, Sharakhova MV, Kayondo J, Lobo NF, Santolamazza F, et al. Breakpoint structure reveals the unique origin of an interspecific chromosomal inversion (2La) in the Anopheles gambiae complex. Proc. Natl. Acad. Sci. USA. 2006;103:6258–6262. doi: 10.1073/pnas.0509683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng C, Tan JC, Hahn MW, Besansky NJ. Systems genetic analysis of inversion polymorphisms in the malaria mosquito. Proc. Natl. Acad. Sci. USA. 2018;115:E7005–E7014. doi: 10.1073/pnas.1806760115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. 2021;373:871–876. doi: 10.1126/science.abj8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Proteins CC. One thousand families for the molecular biologist. Nature. 1992;357:543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- 19.Srinivasan, S. Homology Folding of Proteins: Application to Cytokine Engineering (Springer, 1998).
- 20.Ghosh C, Kumar N, Kushwah RBS, Soumya M, Joshi SG, Chethan KR, et al. Enrichment of phenotype among biological forms of Anopheles stephensi Liston through establishment of isofemale lines. bioRxiv. 2022 doi: 10.1101/2022.09.28.509862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang X, Peery A, Hall AB, Sharma A, Chen X-G, Waterhouse RM, et al. Genome analysis of a major urban malaria vector mosquito, Anopheles stephensi. Genome Biol. 2014;15:459. doi: 10.1186/s13059-014-0459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive -mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods. 2016;13:1050–1054. doi: 10.1038/nmeth.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 25.Ruan J, Li H. Fast and accurate long-read assembly with wtdbg2. Nat. Methods. 2020;17:155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakraborty M, Baldwin-Brown JG, Long AD, Emerson JJ. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016;44:e147. doi: 10.1093/nar/gkw654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharakhova MV, Hammond MP, Lobo NF, Krzywinski J, Unger MF, Hillenmeyer ME, et al. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 2007;8:R5. doi: 10.1186/gb-2007-8-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corbett-Detig RB, Said I, Calzetta M, Genetti M, McBroome J, Maurer NW, et al. Fine-mapping complex inversion breakpoints and investigating somatic pairing in the Anopheles gambiae species complex using proximity-ligation sequencing. Genetics. 2019 doi: 10.1101/662114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood O, Hanrahan S, Coetzee M, Koekemoer L, Brooke B. Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus. Parasit. Vectors. 2010;3:67. doi: 10.1186/1756-3305-3-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alout H, Djègbè I, Chandre F, Djogbénou LS, Dabiré RK, Corbel V, et al. Insecticide exposure impacts vector–parasite interactions in insecticide-resistant malaria vectors. Proc. R. Soc. B Biol. Sci. 2014 doi: 10.1098/rspb.2014.0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baxter RHG, Steinert S, Chelliah Y, Volohonsky G, Levashina EA, Deisenhofer J. A heterodimeric complex of the LRR proteins LRIM1 and APL1C regulates complement-like immunity in Anopheles gambiae. Proc. Natl. Acad. Sci. USA. 2010;107:16817–16822. doi: 10.1073/pnas.1010575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inbar E, Eappen AG, Alford RT, Reid W, Harrell RA, Hosseini M, et al. Knockout of Anopheles stephensi immune gene LRIM1 by CRISPR-Cas9 reveals its unexpected role in reproduction and vector competence. PLoS Pathog. 2021;17:e1009770. doi: 10.1371/journal.ppat.1009770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lombardo F, Christophides GK. Novel factors of Anopheles gambiae haemocyte immune response to Plasmodium berghei infection. Parasit. Vectors. 2016 doi: 10.1186/s13071-016-1359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith RC, Eappen AG, Radtke AJ, Jacobs-Lorena M. Regulation of anti-plasmodium immunity by a LITAF-like transcription factor in the malaria vector Anopheles gambiae. PLoS Pathogens. 2012 doi: 10.1371/journal.ppat.1002965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bushell KN, Leeman SE, Gillespie E, Gower AC, Reed KL, Stucchi AF, et al. LITAF mediation of increased TNF-α secretion from inflamed colonic lamina propria macrophages. PLoS ONE. 2011 doi: 10.1371/journal.pone.0025849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmerini V, Monzani S, Laurichesse Q, Loudhaief R, Mari S, Cecatiello V, et al. Drosophila TNFRs Grindelwald and Wengen bind Eiger with different affinities and promote distinct cellular functions. Nat. Commun. 2021 doi: 10.1038/s41467-021-22080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002;21:3009–3018. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanda H, Igaki T, Kanuka H, Yagi T, Miura M. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J. Biol. Chem. 2002;277:28372–28375. doi: 10.1074/jbc.C200324200. [DOI] [PubMed] [Google Scholar]
- 39.Andersen DS, Colombani J, Palmerini V, Chakrabandhu K, Boone E, Röthlisberger M, et al. The Drosophila TNF receptor Grindelwald couples loss of cell polarity and neoplastic growth. Nature. 2015;522:482–486. doi: 10.1038/nature14298. [DOI] [PubMed] [Google Scholar]
- 40.Banner DW, D’Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, et al. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 41.Black RA. Tumor necrosis factor-alpha converting enzyme. Int. J. Biochem. Cell Biol. 2002;34:1–5. doi: 10.1016/s1357-2725(01)00097-8. [DOI] [PubMed] [Google Scholar]
- 42.Mohan MJ, Seaton T, Mitchell J, Howe A, Blackburn K, Burkhart W, et al. The tumor necrosis factor-alpha converting enzyme (TACE): A unique metalloproteinase with highly defined substrate selectivity. Biochemistry. 2002;41:9462–9469. doi: 10.1021/bi0260132. [DOI] [PubMed] [Google Scholar]
- 43.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–D551. doi: 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Al-Lamki RS, Mayadas TN. TNF receptors: Signaling pathways and contribution to renal dysfunction. Kidney Int. 2015;87:281–296. doi: 10.1038/ki.2014.285. [DOI] [PubMed] [Google Scholar]
- 47.Hoareau L, Bencharif K, Rondeau P, Murumalla R, Ravanan P, Tallet F, et al. Signaling pathways involved in LPS induced TNFalpha production in human adipocytes. J. Inflamm. 2010;7:1. doi: 10.1186/1476-9255-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez W, Page AM, Carlson DJ, Ericson BL, Cserhati MF, Guda C, et al. Analysis of immune-related genes during Nora virus infection of using next generation sequencing. AIMS Microbiol. 2018;4:123–139. doi: 10.3934/microbiol.2018.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr. Biol. 2002;12:1263–1268. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- 50.Garver LS, de Almeida OG, Barillas-Mury C. The JNK pathway is a key mediator of Anopheles gambiae antiplasmodial immunity. PLoS Pathog. 2013;9:e1003622. doi: 10.1371/journal.ppat.1003622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Souvannaseng L, Hun LV, Baker H, Klyver JM, Wang B, Pakpour N, et al. Inhibition of JNK signaling in the Asian malaria vector Anopheles stephensi extends mosquito longevity and improves resistance to Plasmodium falciparum infection. PLoS Pathog. 2018;14:e1007418. doi: 10.1371/journal.ppat.1007418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cruz LN, Wu Y, Ulrich H, Craig AG, Garcia CRS. Tumor necrosis factor reduces Plasmodium falciparum growth and activates calcium signaling in human malaria parasites. Biochim. Biophys. Acta. 2016;1860:1489–1497. doi: 10.1016/j.bbagen.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng S, Seven AB, Wang J, Skiniotis G, Özkan E. Conformational plasticity in the transsynaptic neurexin-cerebellin-glutamate receptor adhesion complex. Structure. 2016;24:2163–2173. doi: 10.1016/j.str.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghosh, S. K., & Ghosh, C. New Ways to Tackle Malaria (IntechOpen, 2019). 10.5772/intechopen.89467. Available from: https://www.intechopen.com/online-first/new-ways-to-tackle-malaria.
- 55.Ghosh C, Shetty NJ. Mode of inheritance of fenitrothion resistance in Anopheles stephensi Liston. J. Cytol. Genet. 1999;342:141–146. [Google Scholar]
- 56.Sharakhova MV, Xia A, Tu Z, Shouche YS, Unger MF, Sharakhov IV. A physical map for an Asian malaria mosquito, Anopheles stephensi. Am. J. Trop. Med. Hyg. 2010;83:1023–1027. doi: 10.4269/ajtmh.2010.10-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 58.Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: Allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinform. 2018;19:460. doi: 10.1186/s12859-018-2485-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C-J, Vert J-P, et al. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanke M, Morgenstern B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw PacBio/Illumina reads used in the assembly and the IndInt chromosome level assembly are uploaded onto the NCBI server under the BioProject ID: PRJNA757559.