Summary
Currently, there are no treatments that ameliorate cardiac cell death, the underlying basis of cardiovascular disease. An unexplored cell type in cardiac regeneration is cardiac Purkinje cells; specialized cells from the cardiac conduction system (CCS) responsible for propagating electrical signals. Purkinje cells have tremendous potential as a regenerative treatment because they may intrinsically integrate with the CCS of a recipient myocardium, resulting in more efficient electrical conduction in diseased hearts. This study is the first to demonstrate an effective protocol for the direct reprogramming of human cardiomyocytes into cardiac Purkinje-like cells using small molecules. The cells generated were genetically and functionally similar to native cardiac Purkinje cells, where expression of key cardiac Purkinje genes such as CNTN2, ETV1, PCP4, IRX3, SCN5a, HCN2 and the conduction of electrical signals with increased velocity was observed. This study may help to advance the quest to finding an optimized cell therapy for heart regeneration.
Subject areas: Bioengineering, Cell biology, Stem cells research
Graphical abstract
Highlights
-
•
Small molecule treatment of human cardiomyocytes leads to Purkinje differentiation
-
•
Small molecule differentiation results in key Purkinje gene expression
-
•
Differentiated Purkinje cells can conduct fast electrical signals
-
•
Differentiated Purkinje cells are comparable to native Purkinje cells
Bioengineering; Cell biology; Stem cells research
Introduction
Currently, cardiovascular diseases (CVD) are the primary cause of death of women, men, and people of most racial groups (Virani et al., 2020; Prevention, 1999). Devastatingly, every 36 s, CVD claims a person’s life in the US (Benjamin et al., 2017; Virani et al., 2020; Prevention, 1999). It is expected that by the year 2035, CVD will affect the lives of approximately 45% of the US population (Benjamin et al., 2017; Virani et al., 2020; Prevention, 1999). However, the catastrophic impacts of CVD do not stop there. CVD also poses an incredible burden to the economy (Benjamin et al., 2017; Virani et al., 2020; Prevention, 1999). It is expected that the financial cost of CVD, over the next two decades, will increase by $131.1 million, reaching an astounding $1.1 trillion (Benjamin et al., 2017).
There are several standard pharmacological treatments for heart failure (HF), including angiotensin-converting enzyme (ACE) inhibitors, β-adrenergic receptor (β-AR) blockers (carvedilol or metoprolol), and phosphodiesterase inhibitors. Unfortunately, many of these agents are either ineffective in some patients or have severe side effects (Pellicori et al., 2020; Jarjour et al., 2020). It is also important to note that the pharmacological treatments currently available for HF do not prevent or ameliorate myocardial cell death and fibrotic scar formation, which are the main contributors to provoking HF (Pellicori et al., 2020; Jarjour et al., 2020). To date, the only cure for end-stage HF is a heart transplant (Boorsma et al., 2020). Unfortunately, heart transplantation is risky, expensive, scarce, and not feasible for most patients (Moayedi et al., 2019). Thus, novel approaches to possibly cure CVD and HF are increasingly needed.
Regenerative medicine has gained attraction over the past few years as a prospective way to cure CVD and HF. This is because the major contributor to the pathophysiological profile of diseases such as CVD and HF stems from the death of millions of cells. Therefore, providing a means to regenerate cardiac tissue is of high importance in successfully finding a cure for HF and CVD (Terzic and Behfar, 2016; Xin et al., 2013). Multiple studies have proposed to regenerate failing hearts via direct implantation of stem cell-derived cardiomyocytes (Monteiro et al., 2017). Unfortunately, these cardiomyocytes are unable to be electrically activated synchronously by the recipient myocardium. As a result, the worsening of HF symptoms and the generation of arrhythmias are often observed (Xin et al., 2013; Monteiro et al., 2017). Alternatively, cells that can intrinsically synchronize with the myocardium and effectively propagate electrical signals may be a better approach as a treatment for HF and heart regeneration (Monteiro et al., 2017; Prabhu and Frangogiannis, 2016).
An example of such cells would be cardiac Purkinje cells, which are one of the building blocks of the cardiac conduction system (CCS), and natural function to propagate electrical signals throughout the heart (Cingolani et al., 2018). This is because an important feature of cardiac Purkinje cells is that they do not contract. They also have little automaticity potential, so they do not automatically fire electrical signals on their own, without prior stimulation (Monteiro et al., 2017). Thus, the risk for arrhythmias caused by the injection of exogenous cardiac Purkinje cells could possibly be lower compared to injections of exogenous working myocytes. Additionally, cardiac Purkinje cells have the native function of conducting electrical signals. Moreover, fibrotic scar tissue formation is commonly observed in failing and myocardial infarction (MI) damaged hearts, which impairs the cardiac conduction system’s ability to effectively propagate electrical signals to allow the heart to contract efficiently (Xin et al., 2013; Monteiro et al., 2017). Replacing the cardiac Purkinje cell mass would help overcome the barriers formed by the fibrotic scar tissue in propagating the electrical signals necessary for a controlled and efficient contraction-relaxation rhythm of the heart. This is important because the dysfunction of the cardiac electrical conduction system leads to arrhythmias, which exacerbate the effects of HF and MI, many times resulting in sudden cardiac death (SCD) in patients (Prabhu and Frangogiannis, 2016; Terzic and Behfar, 2016). Lastly, failing or post-MI hearts show myocardial conduction slowing which may be due to the remodeling of the remaining cardiac conduction system’s cells, including Purkinje cells (Wang and Hill, 2010). Remodeling of the cardiac conduction system’s cells due to HF and MI results in changes in the functional expression of ion channels, causing alterations in the action potential profiles of the cells (Wang and Hill, 2010). Remodeling also involves changes in the expression of gap junctions, such as Connexin 43, which are important for cell-to-cell coupling that mediates the flow of current required for heart rhythm to be synchronous (Wang and Hill, 2010). Therefore, restoring Purkinje cell mass in failing and infarcted hearts may be of high importance for the effective treatment of CVDs.
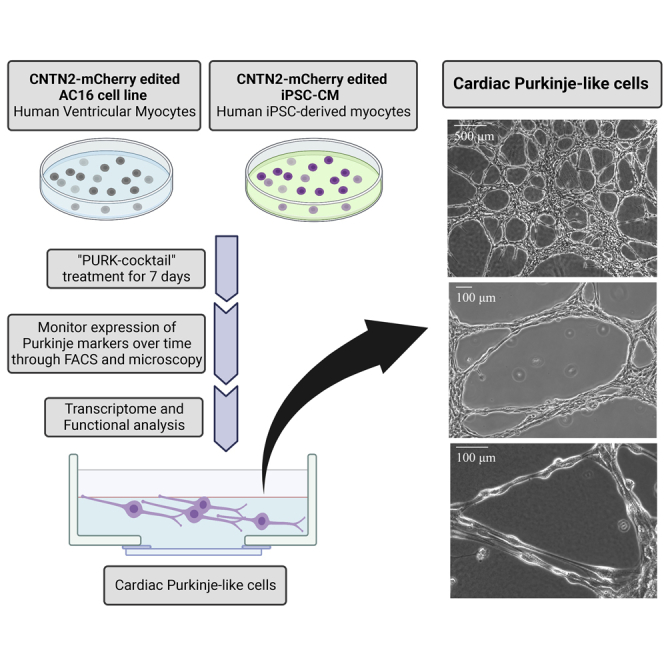
In this study, we generated cardiac Purkinje-like cells for potential use as a cell-based treatment for HF. Currently, the main HF therapy associated with CCS is the implantation of pacemakers (Sayers and Riley, 2021). In the last few years, however, the creation of biological pacemakers has been proposed and it has gained a fair amount of attraction (Komosa et al., 2021). Nonetheless, very little has been done in investigating the potential of cardiac Purkinje cells as a therapy, or the creation of such cells from human cell sources (Huang et al., 2018). Here we demonstrate the successful direct reprogramming of human cardiomyocyte cell lines (AC16-CMs and iPSC-CMs) into Purkinje-like cells using a unique cocktail of small molecules. Direct reprogramming is a technique that produces an epigenetically unstable “plastic” state on the cells to facilitate the conversion of a fully differentiated and matured cell into a different new cell type (Aydin and Mazzoni, 2019; Chambers and Studer, 2011; Zhou and Melton, 2008). Thus, direct reprogramming is a cell conversion method that bypasses the pluripotency state, offering the advantage of generating a diverse range of clinically relevant cell types via a much simpler and quicker protocol (Chambers and Studer, 2011). Upon the treatment of the CMs with our small molecule differentiation cocktail (“PURK-cocktail”), we observed tremendous morphological changes, such that the PURK-cocktail-treated cells differentiated and converted to closely morphologically resemble native cardiac Purkinje cells. Moreover, immunocytochemistry, FACS, and transcriptome analyses demonstrated that our generated Purkinje-like cells expressed a range of specific cardiac Purkinje genes, such as PCP4, ETV1, CNTN2, NKX2-5, and more. We were also able to demonstrate that the reprogrammed Purkinje-like cells were functionally similar to native cardiac Purkinje cells since they were able to rapidly conduct electrical impulses with a faster conduction velocity compared to the control CMs. Thus, our reprogrammed Purkinje-like cells displayed morphological, transcriptomic, and functional characteristics similar to that of native cardiac Purkinje cells.
Results
Creation of Contactin 2-mCherry reporter cell line
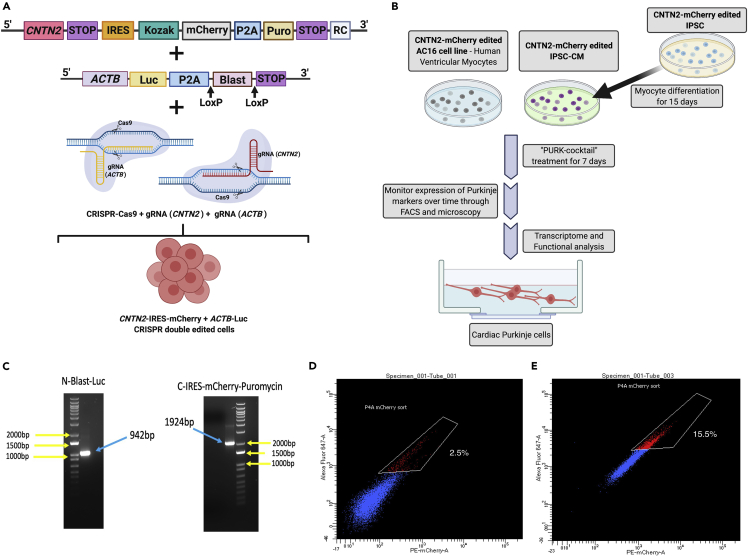
Our first step prior to differentiation was to genetically edit the cells so that differentiation could be tracked and monitored by the expression of a specific Purkinje reporter gene. An IRES-mCherry-Puromycin tag was introduced into the Contactin 2 (CNTN2) gene. Figure 1A depicts the overall approach to creating our co-gene-edited cells. CNTN2 is an adhesion molecule that can participate in migration, adhesion, neurite outgrowth and fasciculation, myelination, and axon pathfinding during brain development (Cohen et al., 1998). Pallante et al. were the first to identify CNTN2 as a specific marker of cardiac Purkinje cells (Pallante et al., 2010). Therefore, the expression of CNTN2 was used to indicate whether our treated cells were differentiating into Purkinje-like cells. To be able to select all of the CRISPR-edited cells, the cells were co-gene edited with a Blasticidin-Luciferase tag on the Beta-actin (ACTB) gene (Figure 1A). This allowed us to select the edited cells prior to differentiation since mCherry-Puromycin-CNTN2 would only be expressed on differentiated cells. Upon CRISPR-Cas9 knock-in co-edition of the CNTN2 and ACTB, the edited cells were selected with Blasticidin treatment and genotyped to confirm that the editing was successful in both genes. As shown in Figure 1C, genotyping confirmed that the cells were successfully edited to express both an IRES-mCherry-Puromycin tag on the CNTN2 gene and a BLAST-Luc tag on the ACTB. This was shown by the PCR products with the exact size of the corresponding tags.
Figure 1.
Strategy for creating Purkinje-like cells
(A) Co-gene edition approach using CRISPR-Cas9 knock-in system, where a mCherry-IRES-Puromycin tag is added at the c-terminal of CNTN2 gene at the same time that a Blasticidin-Luciferase (Blast-Luc) tag is added to the ACTB gene to allow the selection of the gene-edited cells prior to differentiation, and after Purkinje differentiation.
(B) Overall scheme for differentiating CMs into cardiac Purkinje-like cells using our small molecule cocktail (“PURK-cocktail”). In brief, the AC16-CM and iPSC were co-gene edited using our CRISPR-Cas9 Knock-in approach. The iPSCs were differentiated into myocytes (iPSC-CM). The AC16-CM and iPSC-CM were then treated with the PURK-cocktail and characterized by downstream transcriptome and functional analysis to determine if the differentiated cells were cardiac Purkinje-like.
(C) PCR genotyping confirms the cells were successfully gene-edited using CRISPR-Cas9 at the CTNT2 and ACTB genes.
(D and E) (D) FACS analysis of control (2.5 ± 0.9%) and (E) PURK-cocktail-treated cells (15.5 ± 1.2%) showed a significant difference in the amount of CNTN2-mCherry + cells (p < 0.005). The control (vehicle)-treated cell population showed minimal expression of CNTN2-IRES-mCherry + cells, whereas the PURK-cocktail-treated cells showed a significant amount of CNTN2-IRES-mCherry + cell population. Created using BioRender.com.
Treatment with the PURK cocktail provokes differentiation
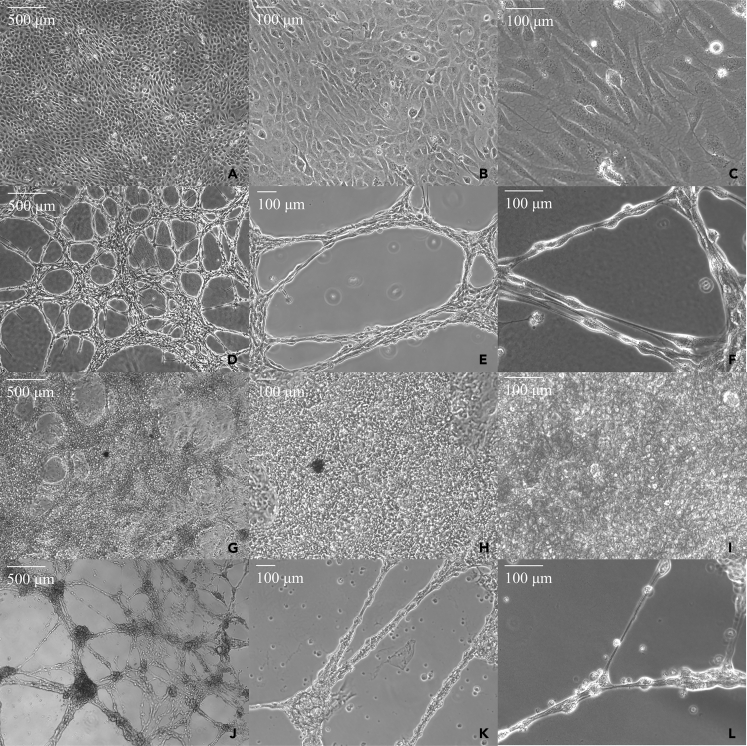
We tested various small molecule drug combinations and narrowed them down to 3 distinct small molecule drug cocktails. For the purposes of this article, we decided to focus on the data obtained from our top treatment (“PURK-cocktail”). Data in supplemental information – Figures S3 and S5 show the differences in RNA-seq gene expression between the 2 other cocktails and the PURK-cocktail. We discuss the cocktail further in the discussion. The overall scheme for differentiating the CMs into Purkinje-like cells using our small molecule differentiation cocktail (“PURK-cocktail”) is shown in Figure 1B. Upon the treatment of the AC16 and iPSC-CM cells with the PURK-cocktail tremendous morphological changes were observed (Figures 2D-2F and 2J-2L) compared to the control cells (Figures 2A-2C and 2G-2I). The cells treated with the PURK-cocktail displayed a Purkinje-like morphology, where they had round bodies and prolonged projections (Figures 2F and 2L). Moreover, it could be observed that the differentiating cells were highly adherent to one another and organized themselves in a “net-like” network (Figures 2D and 2J). Since the cells were grown and differentiated in a thick Fibrin matrix, it was observed that the cells freely moved within the Fibrin matrix as they differentiated, and their network was three-dimensional (3D). No obvious morphological changes were observed in the vehicle-treated control cells, despite them also growing on a Fibrin matrix. The iPSC-CMs were functionally active and were able to beat without extra stimulation upon transfer into a Fibrin matrix coated dish (supplemental information – Videos S1 (iPSC beating at 4X used for PURK differentiation) S2 (iPSC beating at 10X used for PURK differentiation) and S3 (iPSC beating at 20X used for PURK differentiation). The extreme changes in cell morphology were observed as early as day-2 of differentiation. As shown in the IncuCyte Movies – supplemental information (Videos S4 (IncuCyte video of Control cells in culture) and S5 (IncuCyte video of PURK cells in culture), the cells maintained their morphological profile even after the removal of treatment. The differentiating cells also noticeably stopped proliferating or proliferated very slowly compared to the control.
Figure 2.
Morphology of Purkinje-like cells differentiated with the “PURK-cocktail” compared to control CMs
(A-C) AC16-CMs treated with the vehicle at day-7 under 4X, 10X, and 20× magnification, respectively.
(D-F) AC16-Purkinje-like cells treated with the “PURK-cocktail” at day-7 under 4X, 10X, and 20× magnification, respectively.
(G-I) iPSC-CMs treated with the vehicle at day-7 under 4X, 10X, and 20× magnification, respectively.
(J-L) iPSC-Purkinje-like cells treated with the “PURK-cocktail” at day-7 under 4X, 10X, and 20× magnification, respectively.
This video shows the iPSC-CM cells used for PURK differentiation were functionally myocytes, that beat spontaneously, despite being cultured on a Fibrin matrix. The video magnification is at 4X.
This video shows the iPSC-CM cells used for PURK differentiation were functionally myocytes, that beat spontaneously, despite being cultured on a Fibrin matrix. The video magnification is at 10X.
This video shows the iPSC-CM cells used for PURK differentiation were functionally myocytes, that beat spontaneously, despite being cultured on a Fibrin matrix. The video magnification is at 20X.
This video shows the time-lapse progress of AC16-CM control cells being cultured on a Fibrin matrix for 7 days. No significant changes were observed in the cell’s morphology. The video magnification is at 10X.
This video shows the time-lapse progress of AC16-CM cells treated with PURK cocktail and cultured on a Fibrin matrix for 7 days. Significant changes were observed in the cell’s morphology and organization. The cells elongate and form “net-like” structures resembling Purkinje Fibers. The video magnification is at 10X.
Additionally, the differentiating cells samples were analyzed via Fluorescence-Activated Cell Sorting (FACS) (Figures 1D and 1E). A small group of CNTN2-mCherry + cells in the PURK-cocktail treatment was observed. It was observed that the optimal cell sorting day was day-7, where the highest number of CNTN2-mCherry + cells could be obtained via FACS. As shown in Figure 1E, the percentage of CNTN2-mCherry+ was 15.5 ± 1.2% for the “PURK-cocktail”-treated cells, compared to control-treated cells (Figure 1D), which showed a minimal amount of CNTN2-mCherry + cells (2.5 ± 0.9%, p < 0.005). All collected cells (n=3 – supplemental information – Figure S1) were then used for downstream experiments.
The differentiated cells express Purkinje-specific genes
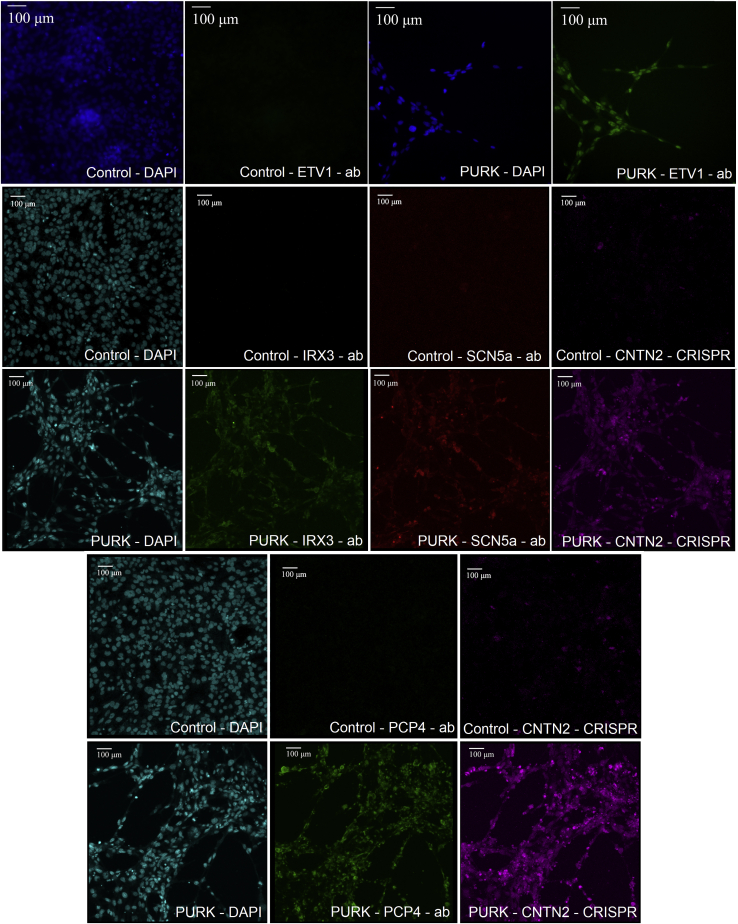
Next, to characterize the PURK-cocktail-treated cells and determine whether they expressed key cardiac Purkinje markers, we performed immunocytochemistry. Antibodies against ETV1, SCN5a, PCP4, and IRX3 were used. Additionally, the expression of the CRISPR-Cas9 knock-in CNTN2-IRES-mCherry in the samples was also evaluated through fluorescent microscopy (Figure 3). The control cells did not show any signal for the Purkinje-specific markers (ETV1, IRX3, SCN5a, or PCP4) and did not express the CNTN2-IRES-mCherry reporter gene (Figure 3). On the other hand, the PURK-cocktail-treated cells were stained for all Purkinje-specific markers (ETV1, IRX3, SCN5a, or PCP4) and highly expressed the CNTN2-IRES-mCherry reporter gene (Figure 3). SCN5a, IRX3, and PCP4 were shown to be expressed in the cell membrane of all differentiated cells. Whereas, ETV1, a nuclear Purkinje cell marker, was shown to be expressed in the cell’s nucleus. The CNTN2-IRES-mCherry reporter was seen in both the cell nucleus and cell surface.
Figure 3.
Evaluation of expression of key Purkinje markers in control vs PURK-cocktail-treated cells
Immunocytochemistry and fluorescent microscopy showed that the control cells did not express key Purkinje markers [ETV1, IRX3, SCN5a, or PCP4] or the CRISPR-Cas9 Knock-in CNTN2-IRES-mCherry. In contrast, the PURK-cocktail-treated cells strongly expressed ETV1, IRX3, SCN5a, or PCP4 and the CRISPR-Cas9 Knock-in CNTN2-IRES-mCherry reporter gene. DAPI was used to stain the cells' nuclei. The scale bar corresponds to 100 μm in all pictures.
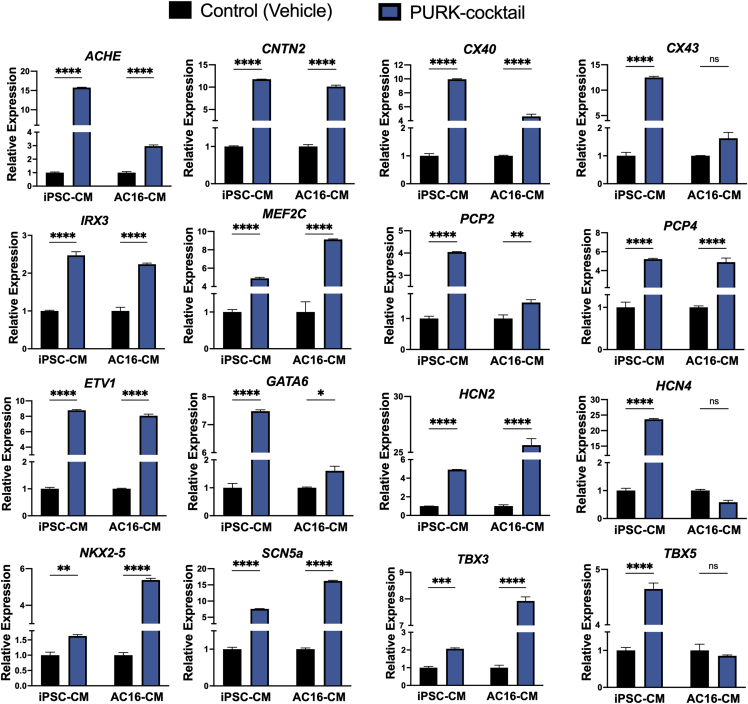
To further characterize the differentiated cells, the expression of 16 cardiac Purkinje-specific genes was analyzed through qRT-PCR. As shown in Figure 4, the PURK-cocktail induced a different transcriptomic profile in both AC16-CMs and iPSC-CMs compared to the control. In the PURK-cocktail-treated AC16 cells, 13 of the 16 genes evaluated were significantly upregulated (p < 0.05, n = 3). HCN4 and TBX5 appeared to be downregulated; however, the difference was not statistically significant. In the PURK-cocktail-treated iPSC-CMs, all 16 genes evaluated were significantly upregulated. The expression of key neuronal Purkinje markers was also evaluated. No expression of neuronal Purkinje markers, TUBB3, LHX5, SKOR2, and OLIG2 (Watson et al., 2018) was found in any of the samples (data not shown).
Figure 4.
qPCR data evaluating the expression of key Purkinje markers in control and PURK-cocktail-treated cells
The PURK-cocktail treatment induced a different genetic profile, which resembled closely to that of native cardiac Purkinje cells on both AC16-CM and iPSC-CM compared to the control. Data corresponds to mean ± SEM of n=3. ∗,∗∗,∗∗∗,∗∗∗∗ corresponds to p < 0.05, 0.01, 0.001, and 0.0001 in a two-Way ANOVA test. “ns” corresponds to “not significant.”
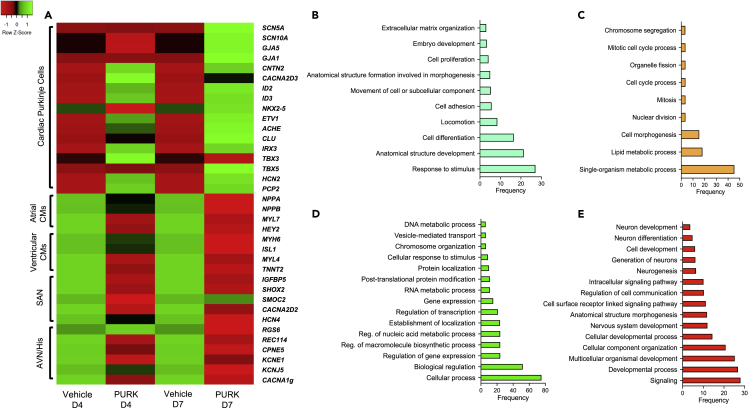
RNA-seq analysis was then performed to further evaluate the cells’ transcriptome. As shown in Figure 5A, signature genes of cardiac Purkinje cells were identified with significant expression changes (p < 0.05). Day-4 and day-7 differentiation samples showed slightly different expressions of these genes, with day-7 showing the most robust cardiac Purkinje gene expression profile. The data also shows a down-regulation of ventricular and atrial myocyte-specific genes. Furthermore, the Gene Ontology (GO) analysis shown in Figures 5B-5E suggests a major difference in the genes and pathways that are up- and down-regulated between day-4 and day-7 of differentiation.
Figure 5.
RNA-seq and GO-analysis of FACS Sorted PURK-cocktail-treated cells at distinct differentiation time points
(A) RNA-seq (HeatMap) showing the gene expression of PURK-cocktail-treated cells, compared to vehicle-treated control cells, on day-4 (D4) and day-7 (D7) of differentiation. PURK-cocktail treatment induced a genetic profile that closely resembled that of native cardiac Purkinje cells as compared to the control CMs. At day-4 (D4), the PURK-cocktail starts to induce a cardiac Purkinje-like genetic profile which becomes even more robust by day-7 (D7) of differentiation. Additionally, it can be observed that the expression of atrial or ventricular CM genes, as well as sinoatrial (SAN), atrioventricular (AVN) and His bundle-specific genes are down-regulated over time in the PURK-cocktail-treated cells, as the cells become more Purkinje-like.
(B and C) GO-analyses showing over-expressed and down-expressed genes, respectively, on day-4 (D4) of the differentiation of PURK-cocktail-treated cell.
(D and E) GO-analyses showing the over-expressed and down-expressed genes, respectively, on day-7 (D7) of the differentiation of PURK-cocktail-treated cells. The GO analyses collectively show the process of trans-differentiation from cardiomyocytes to cardiac Purkinje-like cells (PURK-cells).
The Purkinje-like cells are functionally similar to native Purkinje cells
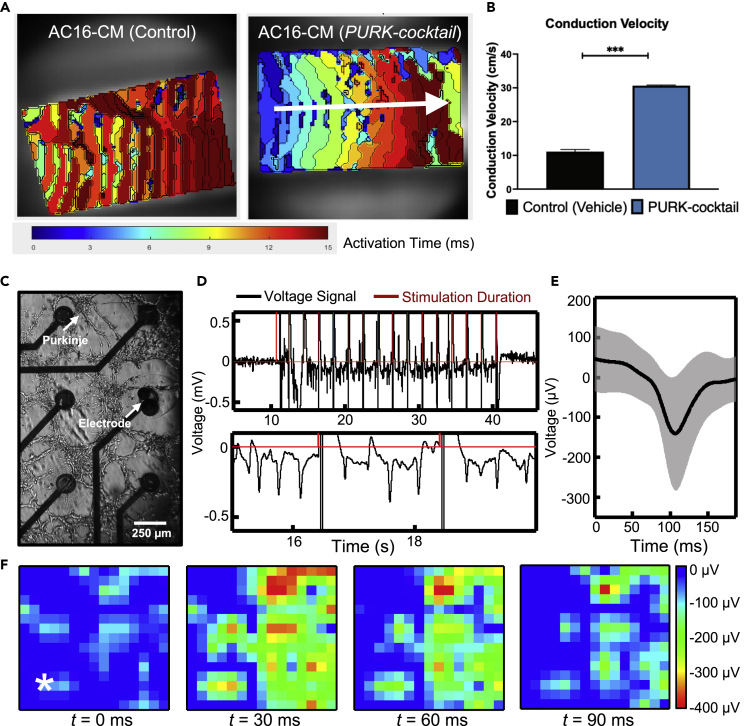
Our next step was to characterize the function of the differentiated cells via optical mapping and multielectrode array (MEA) electrophysiological studies. Based on optical mapping studies, the activation map of the control cells revealed a very slow pulsating-like activation from the cells, rather than the conduction of the electrical stimulation (Figure 6A and supplemental information – Video S6 (Optical mapping analysis of Control cells in culture)). In contrast, the activation map of the PURK-cocktail-treated cells showed that the cells were rapidly activated upon electrical stimulation and produced a very strong electrical signal propagation throughout the whole dish (Figure 6A and supplemental information – Video S7 (Optical mapping analysis of PURK cells in culture). The conduction velocity (CV) was almost 3-times faster in the PURK-cocktail-treated cells than in the control cells (Figure 6B).
Figure 6.
Electrophysiology of PURK-cocktail-treated cells
(A) The optical activation map shows an absence of electrical signal propagation in the control cells (left). The white arrow shows the direction of a uniform electrical signal in the PURK-cocktail-treated cells (right).
(B) The conduction velocity of the PURK-cocktail-treated cells was determined to be significantly faster than the control.
(C) PURK-cocktail cells cultured on the MEA on day-7.
(D) Voltage data from one channel of the MEA (top) with a zoomed-in view of the cellular response (bottom).
(E) Averaged spike waveform over all channels of the MEA in black with the SD of the signal shown in gray.
(F) Heatmaps of the voltage amplitude distribution from various time points across the entire MEA. The white star in the t = 0 ms heatmap indicates the stimulation electrode location. Warmer colors (red, orange) indicate higher spike amplitudes. A Welch’s T-Test was used to compare the conduction velocities. Data corresponds to mean ± SEM of n = 3. ∗∗∗ corresponds to p < 0.001.
The activation map of the control cells reveals a very slow pulsating-like activation from the cells, rather than the conduction of the electrical stimulation.
The activation map of the PURK-cocktail-treated cells show that the cells were rapidly activated upon electrical stimulation and produced a very strong electrical signal propagation throughout the whole dish.
Moreover, the PURK-cocktail-treated cells can be seen on the MEA after 7 days in culture (Figure 6C). The projections extend over a span of a few millimeters and are fully connected with the other Purkinje-like cells throughout the entirety of the MEA surface area (approximately 50 mm2). Electrophysiological recordings were performed to visualize the activity of the Purkinje-like cells. Electrical stimulation was applied through two of the electrodes on the MEA to stimulate cells. The single-channel recording (Figure 6D) shows that prior to the beginning of the stimulation (first spike in red), no electrical activity was evident. However, after the first few stimulation pulses, the Purkinje-like cells responded by emitting multiple pulse waves in a row (Figure 6D, bottom panel). After the stimulation concluded, no more electrical activities were observed. A single spike was isolated in the time domain and averaged across all 64 channels of the MEA. The average of the pulse waveform is shown in black in Figure 6E, and the gray area indicates the SD of the signal. The field potentials have an average magnitude of ∼155 μV with a SD of 144 μV. This deviation could be attributed to the varying proximity of the Purkinje-like cells to the electrodes of the MEA. The field potential duration is approximately ∼150 ms, which is on the order of that reported in other literature on human Purkinje cells (Washio et al., 2008; Dangman et al., 1982). It should be noted that the potentials recorded here are extracellular potentials and not membrane action potentials of cells. The heatmaps in Figure 6F demonstrate the spatial activations of the PURK-cocktail-treated cells across the MEA over time and indicate that the stimulation signal propagated from the bottom left corner of the MEA to the vast majority of the cells in the field. In addition, the activation delay time and conduction velocity were determined as shown in supplemental information – Figure S6. The heatmap shows the delay in propagation with the reference point indicated by the star (stimulation pulse) and the range of the delay is 5-20 ms, which is in agreement with the optical mapping results (Figure 6A). Furthermore, the conduction velocity vectors shown in the map (larger arrows indicate a higher magnitude of velocity) were averaged to be 41.8 ± 2.4 cm/s, which is similar to those reported from optical mapping (Figure 6B). Control (vehicle)-treated cells were not functionally active.
Discussion
CVD remains the leading cause of death globally (Gersh et al., 2010). Although increased advances in risk-factor management of CVDs have been made, which resulted in decreases in the rate of occurrence of MI and its resulting fatalities, finding a cure for the resulting myocardial damage has been challenging (Sadek and Olson, 2020). As a result, an increased prevalence of cardiomyopathies and HF has been observed. To date, there are no treatments that can ameliorate cardiomyocyte cell death, the underlying basis of CVDs (Sadek and Olson, 2020).
Over the past decades, the notion of cardiac regeneration upon injury has sparked significant interest in the scientific and medical community (Terzic and Behfar, 2016; Xin et al., 2013; Zhou and Melton, 2008). Most research has emphasized solely cardiomyocyte regeneration, which unfortunately has not yet been completely successful. The main challenge observed with the regeneration of the cardiomyocyte cell population is that the cardiomyocytes implanted do not seamlessly coupled with the recipient’s heart, resulting in arrhythmias. Thus, successful cardiac regeneration may require the implantation of numerous cell types (Terzic and Behfar, 2016; Xin et al., 2013; Zhou and Melton, 2008). Restoring the CCS in failing and infarcted hearts may be a better route to regenerating an injured myocardium, compared to the sole replacement of the lost myocyte cell mass (Xin et al., 2013). This is because the CCS is vital for generating coordinated contraction and relaxation rhythms of the heart. Therefore for the heart to properly operate, the CCS needs to also be fully functional (Xin et al., 2013).
The CCS is comprised of distinct cellular regions which are responsible for initiating and conducting electrical impulses across the cardiac tissue (Cingolani et al., 2018). Each region of the CCS consists of specialized cardiac cells with distinctive anatomical, functional, and molecular properties. The CCS regions are the sinoatrial node (SAN), the atrioventricular (AVN) conduction axis and node, the His bundle branches, and the Purkinje fiber (PF) network (Cingolani et al., 2018). Collectively, the particular element of each of these cell types allows them to build the electrical system of the heart (Cingolani et al., 2018).
Cardiac Purkinje cells are responsible for propagating electrical signals throughout the ventricles, which then allows the ventricles to contract and for blood to be pumped. Treating failing and infarcted hearts with cardiac Purkinje-like cells may restore the efficiency of electrical signal conduction, resulting in improvements in cardiac function, because cardiac Purkinje cells may intrinsically integrate with the CCS of a recipient myocardium (Xin et al., 2013). In this study, we presented evidence that we can generate cardiac Purkinje-like cells by applying a unique small molecule cocktail (“PURK-cocktail”) to human cardiomyocytes. The Purkinje-like cells created to exhibit a similar transcriptomic profile and electrophysiological functionality as native Purkinje cells.
Gourdie et al. showed through genetic lineage tracing techniques, that Purkinje cells have a myogenic origin (Gourdie et al., 1995). It has now been widely accepted that these cells are myogenic. In fact, Gourdie et al. also went on to show that Purkinje cells and working myocytes arise from a common myogenic precursor and that for Purkinje cells to form, the precursor myocyte must be induced by arterial vessel formation toward the Purkinje phenotype (Gourdie et al., 1995). The precursor myocyte that gets surrounded by vascular cells commits early on to a non-proliferation state and becomes mitotically quiescent, as opposed to precursor myocytes that do not get associated with arterial vessel formation (Cheng et al., 1999; Gourdie et al., 1995). These myocytes continue on to the working contractile myocyte lineage and continue to proliferate (Gourdie et al., 1995). Later studies have also determined that Purkinje cells do not share a parental cell lineage with the other CCS components, the SAN and AVN cells (van Weerd and Christoffels, 2016). Moreover, the ventricular conduction system (VCS), consisting of the cardiac Purkinje cells and His bundle cells, was shown to specifically develop from ventricular trabecular myocardium during the cardiac ventricular chamber formation (Miquerol et al., 2010). Some trabecular myocytes organize in a sheet-like layer associating themselves along the cardiac chamber endocardial surface and are induced to acquire fast conduction genes, subsequently differentiating from Purkinje cells (Dobrzynski et al., 2013; Sedmera and Gourdie, 2014). It is important to note, however, that not all trabecular myocytes are inclined to differentiate into the Purkinje phenotype. In reality, it seems that most of the precursor trabecular myocytes go on to form trabeculae carnae, which is mostly composed of working myocytes (Sedmera and Gourdie, 2014). Thus, there is still much to be elucidated on the mechanisms involved in the decision to commit to either the Purkinje cell fate or the working myocardium cell fate (Sedmera and Gourdie, 2014). Nonetheless, multiple studies have slowly been able to investigate key genetic aspects of Purkinje cell formation (van Weerd and Christoffels, 2016; Atkinson et al., 2011; Dobrzynski et al., 2013; Park and Fishman, 2017). Therefore, in line with the evidence of native cardiac Purkinje cell development from myogenic cells, we decided to use cardiomyocytes as the initial cell source for the induction of cardiac Purkinje differentiation in our protocol.
In order to determine what the “PURK-cocktail” should be comprised of, we did a thorough the investigation of the literature to pick the compounds we could start high-throughput testing. Modulation of the second messenger cAMP has been reported by many studies as essential in inducing cell reprogramming and differentiation (Qin et al., 2017; Ma et al., 2017). Increases in cAMP levels, via small molecule modulation, have been shown in many protocols to potentiate cell reprogramming and to facilitate the trans-differentiation of various cell types, such as neuronal and cardiac cells (Chen et al., 2006; Lepski et al., 2013). The mechanisms mainly implicated with cAMP’s effect on differentiation and reprogramming are the activation of exchange protein activated by cAMP (EPAC) and protein kinase A (PKA), which both may subsequently activate cAMP-response element binding protein (CREB) (Bos, 2006; Mei et al., 2002). In fact, EPAC activation has been shown to upregulate the transcription of MEF2c (Zobel et al., 2007), a key gene in cardiac Purkinje cell development. Additionally, Tsai et al. (2015) demonstrated that they were able to differentiate murine ESCs, via cAMP production from Sodium Nitroprusside treatment, and generate cardiac Purkinje-like cells. Sodium Nitroprusside is a potent relaxant and induces nitric oxide and iron production, which can activate cAMP production (Kim et al., 2006). Forskolin and Rolipram have been used in numerous protocols of cell reprogramming and differentiation, and have been reported as “reprogramming facilitators” since they have been shown to promote cell fate plasticity (Qin et al., 2017; Ma et al., 2017). Moreover, previous data from our lab (data not shown) suggested that the treatment of cardiac progenitor cells with Epinephrine resulted in the differentiation of the progenitor cells into cardiac conduction-like cells via the transcription activation of key CCS channel proteins including SCN5a (Islas et al., 2020).
Epigenetic regulation is also vital for cell differentiation and reprogramming (Boland et al., 2014; Wu and Sun, 2006). DNA methylation, histone modifications, and non-coding RNA-mediated regulatory events control the distinct gene expression patterns necessary for development, differentiation, and reprogramming (Boland et al., 2014; Wu and Sun, 2006). Demethylation of NKX2-5 and MEF2c has been associated with proper heart development (Moore-Morris et al., 2018; Chamberlain et al., 2014). Furthermore, studies by Stein et al. (2011) and Randall et al. (2009) showed that loss of H3K4 histone methylation leads to impairments in the formation of the cardiac conduction system by the destabilization of gene expression patterns. Whereas, Brunicka-Turek et al. (Burnicka-Turek et al., 2020) showed that the transcription of TBX5 is activated by H3K4 methylation. We tested multiple epigenetic regulators in our cocktail and saw the best results with RG108, Parnate, Valproic acid, and Resveratrol.
Retinoic acid-mediated signaling has been implicated in the activation of key Purkinje genes’ transcription, such as NKX2-5, CNTN1, CX43, and PCP2 (Balmer and Blomhoff, 2002; Lin et al., 2010; Matsui, 1997; Chen et al., 2013). Disruption of retinoic acid signaling has also been tied to cardiac conduction development abnormalities (Perl and Waxman, 2019). Moreover, Wobus et al. (1997) and Zhang et al. (2011) reported that the treatment of mouse and human embryonic stem cells, respectively, with Retinoic acid, resulted in the differentiation of ventricular conduction-like cells, as opposed to atrial or pacemaker-like cells.
Given the reported high influence of regulation on cardiac development, it is not surprising these drugs ended up comprising our “PURK-cocktail.”
The PURK-cocktail treatment induced a Purkinje-like morphology of elongated projections, a round cell body, and increased cell adhesion, which lead to the formation of “net-like” structures that resembled native PF networks. Anatomical studies conducted by Atkinson et al. (2011) showed PF network forms a complex 3D structure that is very similar to the structures observed formed by our cells (Atkinson et al., 2011). Additionally, the cells treated with the PURK-differentiation cocktail noticeably stopped proliferating and contracting upon differentiation, unlike the AC16-CM and iPSC-CM. This observation is in line with what has been reported in the literature (Gourdie et al., 1995). Precursor myocytes upon the commitment to the Purkinje cell lineage have been observed to enter a non-proliferation state and become mitotically quiescent (Gourdie et al., 1995).
Pallante et al. (2010) were the first to describe CNTN2 as a specific and novel marker of cardiac Purkinje cells due to the strictly high expression profile of these cells, compared to other cells in the heart (Pallante et al., 2010). Therefore, we expected that cardiac Purkinje differentiation would trigger the expression of our CNTN2-IRES-mCherry CRISPR/Cas9 reporter. Indeed, we saw through FACS analysis that a population of CNTN2-mCherry + cells could be identified in the PURK-cocktail treatment, suggesting some Purkinje differentiation occurred. Control-treated cells showed a minimal amount of CNTN2-mCherry + cells.
Previous characterization of cardiac Purkinje cells in the literature showed that trabecular myocardium differentiates into Purkinje cells via gradient TBX3 downregulation (Bakker et al., 2008). As development progress, the expression of NKX2-5, TBX5, MEF2c, ETV1, IRX3, and other transcription factors enhance the expression of Gja5/Cx40, Gja1/Cx43, and SCN5a (van Eif et al., 2018; Miquerol et al., 2010, 2011; Park and Fishman, 2017). Moreover, the expression of these transcription factors dynamically represses the actions of TBX3, as the Purkinje cell continues to develop (Bakker et al., 2008; Hoogaars et al., 2004). Shekhar et al. (2016, 2018) also determined that ETV1 is highly expressed in fast-conduction cells, such as Purkinje cells. Loss of ETV1 provoked down-regulation of IRX3, SCN5a, NKX2-5, and Cx40. Acetylcholinesterase (ACHE) and PCP4 have also been shown to be highly expressed in cardiac Purkinje cells, but not other cells of the CCS (Johanson et al., 2000; Saimi and Kung, 2002; Mommaerts et al., 1953; Nakamura et al., 1994). Additionally, the expression of hyperpolarization-activated cyclic nucleotide-gated cation channels (HCN) within the CCS is highly important for its proper functioning (Scicchitano et al., 2012). “Funny” or “pacemaker” currents (If) can be observed in Purkinje cells, and are thought to be generated by both HCN2 and HCN4 (Boyden et al., 2010). Purkinje cells are also strongly coupled through gap junctions Cx40 and 43, which allows them to conduct fast electrical signals and rapidly activate the ventricles (Verheule and Kaese, 2013).
Through immunocytochemistry, fluorescent microscopy, qRT-PCR, and RNA-seq we were able to determine whether the cells expressed some of the aforementioned specific cardiac Purkinje markers. Indeed, the PURK-differentiation cocktail was positively stained for ETV1, SCN5a, PCP4, and IRX3. The control cells did not express or stain any of the cardiac Purkinje markers. It was also visually clear, through fluorescent microscopy, that the cells treated with the PURK-differentiation also expressed the CNTN2-IRES-mCherrry reporter gene. The qRT-PCR data showed that the cells treated with the cocktail significantly upregulated multiple key cardiac Purkinje markers such as NKX2-5, ACHE, MEF2C, PCP2, CX40, CX43, HCN2, HCN4, TBX3, and TBX5.
Moreover, given that cardiac Purkinje cells may share some genetic similarities with neuronal Purkinje cells, we also evaluated the expression of genes specific to neuronal Purkinje cells (Gorza et al., 1988; Watson et al., 2018). No expression of TUBB3, LHX5, SKOR2, and OLIG2 was found in any of the samples (data not shown). The lack of expression of neuronal Purkinje markers suggested that the Purkinje-like cells generated were indeed of cardiac origin and none of the drugs in our treatments upregulated the expression of the neuronal Purkinje cell markers tested.
RNA-sequencing data showed that PURK-cocktail treatment induced profound changes in gene expression, producing a robust Purkinje genetic profile on the cells compared to the control, which got stronger as differentiation progressed. We also determined that key known (Goodyer et al., 2019) Purkinje vs atrial and ventricular myocyte-specific genes were oppositely expressed in the control vs treated cells. Gene Ontology (GO) analysis data also showed that in the early days of differentiation (day-4), genes involved in embryo development, cell adhesion, differentiation, extracellular matrix organization, and response to stimulus were upregulated. Whereas, genes involved in cell morphogenesis and division were down-regulated. This is consistent with what has been reported in the literature about cardiac Purkinje cell development. At a later stage of cell differentiation (day-7), the upregulated genes were more involved with processes of gene expression and transcription, whereas, the down-regulated genes were mainly involved in neuron and nervous system development. These findings are consistent with what is expected of cell trans-differentiation processes. Taken together, the FACS, immunocytochemistry, RNA-seq, and qRT-PCR data strongly suggested that the cells generated via PURK-cocktail treatment were genetically similar to native cardiac Purkinje cells. The next step was to evaluate the electrophysiological functionality of the PURK-cocktail-treated cells.
Optical mapping and MEA electrophysiological studies revealed that the PURK-cocktail-treated cells were functionally similar to native Purkinje cells. Upon electrical stimulation, the control cells were not capable of propagating electrical signals. The activation map of the control cells revealed a very slow pulsating-like activation from the cells, rather than the conduction of the electrical stimulation. This was expected, as ventricular myocytes are not part of the CCS and do not conduct electrical signals (Cingolani et al., 2018). In contrast, the PURK-cocktail cells showed very fast and uniform conduction of electrical stimulation. The activation map of the PURK-cocktail cells showed that the cells were rapidly activated upon electrical stimulation and produced a very strong and fast electrical signal propagation throughout the whole dish. This behavior was in line with the functional behavior of native cardiac Purkinje cells (Cingolani et al., 2018).
Further evidence of our PURK-cocktail-treated cells' functionality was shown in the MEA experiments. MEA is an array of electrodes that are distributed over a small chip. Electroactive cells, such as cardiac and neuronal cells, can be cultured on top of the electrodes, and their electrophysiological activity can be measured and monitored to capture their functional capacity (Spira and Hai, 2013). MEA-based electrophysiological activity recordings have been particularly used to characterize neurons (McCready et al., 2022). We decided to investigate the functional phenotype of our PURK-cocktail-treated cells through MEA recordings because MEA systems can provide a great amount of spatiotemporal activity data from cell networks, not just individual cells so that we could understand and determine whether our cells collectively behaved similarly to a native Purkinje cell network. Moreover, MEA recordings are non-destructive to the cells or network, which allows for the culture to be monitored over time as they develop and mature (McCready et al., 2022).
Purkinje cells do not typically show much spontaneous firing, therefore, it was expected that during the periods without any stimulation that little to no spiking activity could be observed (Shi et al., 1999). Indeed, we observed that prior to the beginning of the stimulation, no spiking activity was evident. However, after the first few stimulation pulses, the PURK-cocktail-treated cells responded by emitting multiple spikes in a row. After the stimulation concluded, no more spikes were observed. These findings suggest that the PURK-cocktail-treated cells on the MEA behaved solely as conduction pathways when stimulated. The data gathered from the MEA and optical mapping studies clearly demonstrated the physiological functionality and electrophysiological activity of our Purkinje-like cells.
In conclusion, (1) this study demonstrates an innovative way to generate human Purkinje-like cells through direct reprogramming using a unique small molecule cocktail. (2) Our cocktail may be used, in the near future, to direct differentiate clinically relevant cells into Purkinje-like cells. (3) The functionality of the Purkinje-like cells is further verified by a collective characterization including electrical stimulation, optical mapping, and MEA recording. Thus, (4) this study may help to advance the quest to finding an optimized cell therapy that can aid in heart regeneration, potentially being further translated into the clinical setting in the upcoming years. Furthermore, (5) the cells generated by this study may be essential for tissue engineering artificial heart models in vitro. Moreover, (6) an in vitro heart model has the potential to be used for the development and investigation of new pharmacological therapies for heart diseases.
Limitations of the study
While conducting these experiments, it was observed that the percentage of mCherry + cells changed depending on the detachment method used (i.e., Papain, trypsin, EDTA, sodium citrate buffer, or Accutase). Detaching the cells with Papain yielded the highest percentage of mCherry + cells. This suggests that CNTN2 may be cleaved upon cell detachment, and different methods have varying degrees of CNTN2 destruction. This is not surprising, given that CNTN2 is a cell adhesion protein (Walsh and Doherty, 1997). Therefore, the efficiency of the treatment in generating Purkinje-like cells may be higher than the one reported here. Indeed, fluorescent microscopy pictures appear to indicate a much higher percentage of CNTN2-mCherry + than the one observed during FACS. To better determine the efficiency of generating Purkinje-like cells with the treatment, a different Purkinje cell marker may be chosen. Perhaps, ETV1, since it is a nuclear transcription factor and it would not be cleaved or degraded upon cell detachment.
Many studies have characterized the electrophysiological properties of native Purkinje cells using the whole-cell patch-clamp technique. While we do agree that collecting electrophysiological data through patch clamp is also important, it is beyond the scope of this study and is part of planned studies. This current study uses two independent approaches (i.e., optical mapping and MEA) to validate the electrophysiological function of the Purkinje-like cells. Nevertheless, the data we have gathered on the functionality of our cells through optical mapping and MEA measurements is highly relevant and valid, showing very similar conduction behavior to native Purkinje cells.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ETV1 | ThermoFisher | cat# PA589150; RRID:AB_2805388 |
| SCN5a | ThermoFisher | cat# MA1-27429; RRID:AB_1090515 |
| IRX3 | ThermoFisher | cat# PA5-111549; RRID:AB_2856959 |
| PCP4 | ThermoFisher | cat# 14705-1-AP; RRID:AB_2878075 |
| Alexa 488 | ThermoFisher | cat# A32731; RRID:AB_2633280 |
| Alexa 594 | ThermoFisher | cat# A-21044; RRID:AB_2535713 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM/F12 | SigmaAldrich | cat# D6434-500ML |
| Glutamax | Gibco | cat# 35050061 |
| FBS | Gibco | cat# 10082147 |
| 2-mercaptoethanol | Gibco | cat# 31350010 |
| Anti-anti | Gibco | cat#15240096 |
| mTeSRTM1 basal medium | StemCell Technologies | cat# 85850 |
| hESC-qualified Matrigel | Corning | cat# 354277 |
| Accutase | StemCell Technologies | cat# 07920 |
| Y-27632 | StemCell Technologies | cat# 72302 |
| STEMdiffTM Cardiomyocyte Dissociation Kit | StemCell Technologies | cat# 05025 |
| TrueCutTM Cas9 v2 | Invitrogen | cat# A36497 |
| Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent | Life Technologies | cat# CMAX00001 |
| Blasticidin | Gibco | cat# A1113902 |
| Rolipram | SigmaAldrich | cat# R6520-10MG |
| Forskolin | SigmaAldrich | cat# F3917-10MG |
| CHIR99021 | SigmaAldrich | cat# 361571-5MG |
| SB431542 | SigmaAldrich | cat# 616464-5MG |
| Valproic acid | SigmaAldrich | cat# P4543-10G |
| RG108 | SigmaAldrich | cat# R8279-10MG |
| Parnate | SigmaAldrich | cat# 616431-500MG |
| Resveratrol | SigmaAldrich | cat# R5010-100MG |
| Retinoic Acid | SigmaAldrich | cat# R2625-500MG |
| Neuregulin | StemCell Technologies | cat# 78071.1 |
| Epinephrine | SigmaAldrich | cat# E4250-5G |
| NeurobasalTM | Gibco | cat# 12348017 |
| KnockOutTM Serum Replacement | Gibco | cat# 10828010 |
| B-27TM supplement minus vitamin A | Gibco | cat# 12587010 |
| N-2 supplement | Gibco | cat# 17502048 |
| Trypsin-EDTA | Gibco | cat# 25200056 |
| Fibrinogen | SigmaAldrich | cat# F8630 |
| Thrombin | SigmaAldrich | T4648-10KU |
| D-PBS | Gibco | cat# 14190144 |
| 6-aminocaproic acid | Sigma Aldrich | cat# A2504 |
| SILAC DMEM Flex Media, no glucose, no phenol red | Gibco | cat# A2493901 |
| L-Glutamine | Gibco | cat# 25030081 |
| MEM Non Essential Amino Acids | Gibco | cat# 11140050 |
| Sodium DL Lactate | SigmaAldrich | cat# L4263-100ML |
| STEMdiffTM Cardiomyocyte Support Medium | StemCell Technologies | cat# 05027 |
| Papain from Carica papaya | SigmaAldrich | cat# 76216-50MG |
| PierceTM 16% Formaldehyde (w/v), methanol free | ThermoScientific | cat# 28908 |
| Triton X-100 | Sigma-Aldrich | cat# X100-100ML |
| 10% Normal Goat Serum | ThermoFisher | cat# 50062Z |
| DAPI solution | ThermoScientific | cat# 62248 |
| DEPC-treated water | Invitrogen | cat# AM9906 |
| PowerTrackTM SYBR Green Master Mix | Applied Biosystems | cat# A46109 |
| Di-4-ANEPPS | Invitrogen | cat#D1199 |
| Critical commercial assays | ||
| STEMdiffTM Cardiomyocyte Maintenance Kit | StemCell Technologies | cat# 05020 |
| TrueTagTM DNA Donor System | Life Technologies | cat# A42992 |
| STEMdiffTM Cardiomyocyte Differentiation Kit | StemCell Technologies | cat# 05010 |
| Quick-RNA Microprep kit | Zymo Research | cat# R1050 |
| RNA Clean & Concentrator-5 | Zymo Research | cat# R1013 |
| High-Capacity cDNA Reverse Transcriptase kit | Applied Biosystems | cat# 4368814 |
| Deposited data | ||
| Raw RNA-seq data Day 4 and Day 7 – PURK cocktail | This paper –Also shared in Figshare | https://doi.org/10.6084/m9.figshare.21290883 |
| Experimental models: Cell lines | ||
| AC16-Cardiomyocytes | SigmaAldrich | cat# SCC109 |
| Human iPSCs derived from cardiac fibroblasts | ATCC | cat# ATCC-CYS0105 |
| Oligonucleotides | ||
| Homology arms specific to CNTN2 | Life Technologies | This paper |
| Homology arms specific to ACTB | Life Technologies | This paper |
| Custom TrueGuideTM gRNA for CNTN2 | Life Technologies | This paper |
| Custom TrueGuideTM gRNA for ACTB | Life Technologies | This paper |
| GAPDH Human qPCR Primer Pair | OriGene | cat# HP205798 |
| CX40 Human qPCR Primer Pair | OriGene | cat# HP208435 |
| CX43 Human qPCR Primer Pair | OriGene | cat# HP200150 |
| ETV1 Human qPCR Primer Pair | OriGene | cat# HP208189 |
| CNTN2 Human qPCR Primer Pair | OriGene | cat# HP208284 |
| HCN2 Human qPCR Primer Pair | OriGene | cat# HP205124 |
| HCN4 Human qPCR Primer Pair | OriGene | cat# HP208604 |
| MEF2C Human qPCR Primer Pair | OriGene | cat# HP206089 |
| ACHE Human qPCR Primer Pair | OriGene | cat# HP231855 |
| TBX3 Human qPCR Primer Pair | OriGene | cat# HP231811 |
| TBX5 Human qPCR Primer Pair | OriGene | cat# HP200177 |
| SCN5a Human qPCR Primer Pair | OriGene | cat# HP234325 |
| IRX3 Human qPCR Primer Pair | OriGene | cat# HP214795 |
| GATA6 Human qPCR Primer Pair | OriGene | cat# HP208427 |
| NKX2-5 Human qPCR Primer Pair | OriGene | cat# HP207706 |
| PCP2 Human qPCR Primer Pair | OriGene | cat# HP218542 |
| PCP4 Human qPCR Primer Pair | OriGene | cat# HP209183 |
| TUBB3 Human qPCR Primer Pair | OriGene | cat# HP209096 |
| OLIG2 Human qPCR Primer Pair | Life Technologies | Watson et al. (2018) |
| SKOR2 Human qPCR Primer Pair | Life Technologies | Watson et al. (2018) |
| LHX5 Human qPCR Primer Pair | Life Technologies | Watson et al. (2018) |
| Recombinant DNA | ||
| Custom mCherry-puromycin vector DNA template | Life Technologies | This paper |
| Software and algorithms | ||
| MATLAB | Mathworks | https://www.mathworks.com/products/matlab.html |
| IncuCyte ZOOM | Sartorius | https://www.essenbioscience.com/en/resources/incucyte-zoom-resources-support/software-modules-incucyte-zoom/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| NIS Elements software v4.13 | Nikon Instruments | https://www.microscope.healthcare.nikon.com/products/software/nis-elements/nis-elements-advanced-research |
| Fluoview Software | Olympus | https://www.olympus-lifescience.com/en/laser-scanning/fv3000/super-resolution-software-module/ |
| HeatMapper | Babicki et al. (2016)https://doi.org/10.1093/nar/gkw419 | http://www.heatmapper.ca/ |
| ElectroMap | O’Shea et al. (2019a)https://doi.org/10.1038/s41598-018-38263-2 | |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Bradley K. McConnell, Ph.D. (bkmcconn@central.uh.edu).
Materials availability
The plasmid and cell lines generated in this study are available from the lead contact on reasonable request.
Experimental model and subject details
Cell lines and cell culture
AC16-CM cells, a proliferating human cardiomyocyte cell line derived from the fusion of primary cells from adult human ventricular heart tissue with SV40 transformed uridine auxotroph human fibroblasts lacking mitochondrial DNA (SigmaAldrich; cat# SCC109) were cultured in DMEM/F12 (SigmaAldrich, cat# D6434-500ML) supplemented with 1% (v/v) 1X Glutamax (Gibco, cat# 35050061), 12% (v/v) FBS (Gibco, cat# 10082147), 0.05 mM 2-mercaptoethanol (Gibco, cat# 31350010), and 1X anti-anti (Gibco, cat#15240096).
Human Induced Pluripotent Stem cells derived from cardiac fibroblasts of a healthy 72 years old male donor (ATCC; cat# ATCC-CYS0105) were cultured in a feeder free manner using mTeSRTM1 basal medium (StemCell Technologies, cat# 85850) supplemented with 1X mTeSRTM1 supplement and 1X anti-anti (Gibco, cat#15240096). hESC-qualified Matrigel (Corning, cat# 354277) was used, according to the manufacturer’s protocol. Before downstream experiments (CRISPR-Cas9 editing and cell differentiation), the cells were single-cell passaged using the blend of proteolytic and collagenolytic enzymes Accutase (StemCell Technologies, cat# 07920), according to the manufacturer’s protocol. In the case of single-cell passage, 10 μM Y-27632 (StemCell Technologies, cat# 72302) was added to the media for the first 24 hours.
Human Induced Pluripotent Stem cells derived cardiomyocytes (iPSC-CM) were cultured in hESC-qualified Matrigel (Corning, cat# 354277) coated dishes. STEMdiffTM Cardiomyocyte Maintenance Kit (StemCell Technologies, cat# 05020) supplemented with 1X anti-anti (Gibco, cat# 15240096) was used to feed the cells for 1 month or longer. The iPSC-CMs were passaged and/or harvested for downstream experiments using STEMdiffTM Cardiomyocyte Dissociation Kit (StemCell Technologies, cat# 05025).
Method details
CRISPR-Cas9 gene editing and reporter cell line
Prior to differentiation, the cells were gene edited using a CRISPR-Cas9 knock-in approach to express a specific marker of cardiac Purkinje cells. The marker chosen was CNTN2, and a mCherry sequence was added at the c-terminal of this specific gene. The AC16-CM and iPSC cells were both genes edited in the same way. To co-gene edit the cells, we tagged the CNTN2 gene with the IRES and mCherry sequences, using the TrueTagTM DNA Donor System (Life Technologies, cat# A42992). Briefly, homology arms specific to the CNTN2 gene were added to the construct mCherry-puromycin vector DNA template (See sequence below) through custom target-specific oligos (See sequence below). Following column chromatographic purification of the PCR products, the DNA donors were then co-transfected using Lipofectamine™ CRISPRMAX™ Cas9 Transfection Reagent (Life technologies cat# CMAX00001) with TrueCutTM Cas9 v2 (Invitrogen, cat# A36497) protein and the two custom TrueGuideTM gRNAs for CNTN2 and ACTB genes into the AC16-CM and iPSC cells (See sequence below). A recombinant cell line was then generated and enriched for about 10 days using the antibiotic Blasticidin (Gibco, cat# A1113902). PCR reactions were performed using 200 ng of genomic DNA obtained from the recombinant cell lines to confirm the CRISPR-Cas9 co-gene editing was successful. mCherry-tagging was further confirmed by fluorescent microscopy.
Homology arm sequence specific to CNTN2 or ACTB
| Gene | Sequence |
|---|---|
| CNTN2 | TCCGTGGCGATGCTGATCCTCATAGGCTCCCTGGAGCTCGGAAG TGGCTCAGGTTCTGGA |
| CCAGCTGCGGCGCAGAGGGAGGGGTTCCAGGATCACTTGG CCGATCGCATACAGAG | |
| ACTB | AGCGCGCCCGGCTATTCTCGCAGCTCACCATGGGAGGT AAGCCCTTGCATTCG |
| TTGTCGACGACGAGCGCGGCGATATCATCATCACCGCTTCCA CTACCTGAACC |
Guide RNA sequences
| sgRNA | Sequence |
|---|---|
| CNTN2 | GTTCCAGGATCAGAGCTCCA |
| ACTB | GCTATTCTCGCAGCTCACCA |
Full sequence of the IRES-mCherry donor system
| CRISPR Donor True Tag System using IRES-mCherry system (FASTA Sequence) |
|---|
| GGAAGTGGCTCAGGTTCTGGATAAgagggcccggaaacctggccctgtcttcttgacgagcattcctaggggtctttcccctctcgccaaaggaat gcaaggtctgttgaatgtcgtgaaggaagcagttcctctggaagcttcttgaagacaaacaacgtctgtagcgaccctttgcaggcagcggaaccccccacctgg cgacaggtgcctctgcggccaaaagccacgtgtataagatacacctgcaaaggcggcacaaccccagtgccacgttgtgagttggatagttgtggaaagagtca aatggctctcctcaagcgtattcaacaaggggctgaaggatgcccagaaggtaccccattgtatgggatctgatctggggcctcggtgcacatgctttacatgtgtt tagtcgaggttaaaaaaacgtctaggccccccgaaccacggggacgtggttttcctttgaaaaacacgatgataaGatctgcgatctaagtaagcttggcattccg gtactgttggtaaaGCCGCCACCatggtgagcaagggcgaggaggataacatggccatcatcaaggagttcatgcgcttcaaggtgcacatggagggct ccgtgaacggccacgagttcgagatcgagggcgagggcgagggccgcccctacgagggcacccagaccgccaagctgaaggtgaccaagggtggccccc tgcccttcgcctgggacatcctgtcccctcagttcatgtacggctccaaggcctacgtgaagcaccccgccgacatccccgactacttgaagctgtccttccccgagg gcttcaagtgggagcgcgtgatgaacttcgaggacggcggcgtggtgaccgtgacccaggactcctccctgcaggacggcgagttcatctacaaggtgaagct gcgcggcaccaacttcccctccgacggccccgtaatgcagaagaagaccatgggctgggaggcctcctccgagcggatgtaccccgaggacggcgccctgaa gggcgagatcaagcagaggctgaagctgaaggacggcggccactacgacgctgaggtcaagaccacctacaaggccaagaagcccgtgcagctgcccggc gcctacaacgtcaacatcaagttggacatcacctcccacaacgaggactacaccatcgtggaacagtacgaacgcgccgagggccgccactccaccggcggcat ggacgagctgtacaagGccaccaacttctccctgctgaagcaggccggcgacgtggaggagaaccccggccccAtgaccgagtacaagcccacggtgcgcc tcgccacccgcgacgacgtcccccgggccgtacgcaccctcgccgccgcgttcgccgactaccccgccacgcgccacaccgtcgacccggaccgccacatcga gcgggtcaccgagctgcaagaactcttcctcacgcgcgtcgggctcgacatcggcaaggtgtgggtcgcggacgacggcgccgcggtggcggtctggacca cgccggagagcgtcgaagcgggggcggtgttcgccgagatcggcccgcgcatggccgagttgagcggttcccggctggccgcgcagcaacagatggaag gcctcctggcgccgcaccggcccaaggagcccgcgtggttcctggccaccgtcggcgtctcgcccgaccaccagggcaagggtctgggcagcgccgtcgtg ctccccggagtggaggcggccgagcgcgccggggtgcccgccttcctggagacctccgcgccccgcaacctccccttctacgagcggctcggcttcaccgtc accgccgacgtcgaggtgcccgaaggaccgcgcacctggtgcatgacccgcaagcccggtTAACTTGGCCGATCGCATACAGAG |
Differentiation
An overall scheme of the differentiation strategy and experimental procedure is shown in Figure 1B. Multiple studies have reported that small molecules can induce cell reprogramming and differentiation, generating distinct cell types including iPSC, cardiac, hepatic, and neuronal cells (Qin et al., 2017; Ma et al., 2017; Yuan et al., 2020; Xie et al., 2017). We tested various small molecule drug combinations. Our rationale for picking the small molecules that comprise our cocktail was through the identification of drugs that could modulate key pathways important for the development of cardiac Purkinje cells according to the information found in the literature. We narrowed it down to one small molecule drug cocktail, our “PURK-differentiation cocktail”.
Small molecule PURK-differentiation cocktail
Several combinations of small molecules were tested. The top combination of a small molecule for the PURK-differentiation cocktail was then identified. The PURK-cocktail is composed of 11 small molecules as shown below.
Detailed list of small molecules used in the PURK-cocktail
| Drug name | Final concentration | Stock vehicle | Manufacturer |
|---|---|---|---|
| Rolipram | 2 μM | DMSO | SigmaAldrich, cat# R6520-10MG |
| Forskolin | 10 μM | DMSO | SigmaAldrich, cat# F3917-10MG |
| CHIR99021 | 4 μM | DMSO | SigmaAldrich, cat# 361571-5MG |
| SB431542 | 2 μM | DMSO | SigmaAldrich, cat# 616464-5MG |
| Valproic acid | 2 μM | Water | SigmaAldrich, cat# P4543-10G |
| RG108 | 2 μM | DMSO | SigmaAldrich, cat# R8279-10MG |
| Parnate | 2 μM | Water | SigmaAldrich, cat# 616431-500MG |
| Resveratrol | 10 μM | DMSO | SigmaAldrich, cat# R5010-100MG |
| Retinoic Acid | 1 μM | DMSO | SigmaAldrich, cat# R2625-500MG |
| Neuregulin | 10 ng/mL | Water | StemCell Technologies, cat# 78071.1 |
| Epinephrine | 10 μM | 0.05 M HCl | SigmaAldrich, cat# E4250-5G |
Control cells were treated with the vehicle (DMSO, water, and 0.5 HCL) used to dissolve drugs. The small molecule cocktail or vehicle were further dissolved in 50% DMEM/F12 (Sigma Aldrich, cat# D6434-500ML) and 50% NeurobasalTM (Gibco, cat# 12348017) media supplemented with 1% (v/v) 1X Glutamax (Gibco, cat# 35050061), 5% (v/v) FBS (Gibco, cat# 10082147), 5% KnockOutTM Serum Replacement (Gibco, cat# 10828010), 0.05 mM 2-mercaptoethanol (Gibco, cat# 31350010), 1X B-27TM supplement minus vitamin A (Gibco, cat# 12587010), 1X N-2 supplement (Gibco, cat# 17502048) and 1X anti-anti (Gibco, cat# 15240096).
Differentiation of AC16 into Purkinje-like cells
The AC16-CM cells were plated one day before treatment with the PURK-differentiation cocktail (day -1). Trypsin-EDTA 0.25% (Gibco, cat# 25200056) was used to harvest the cells for plating. To neutralize the Trypsin-EDTA solution, 50% FBS (Gibco, cat# 10082147) and 50% DMEM/F12 (Sigma Aldrich, cat# D6434-500ML) was used. The cells were then centrifuged at 1,200 rpm for 2–3 minutes. The supernatant was removed and the cell pellet was resuspended in DMEM/F12 (Sigma Aldrich, cat# D6434-500ML) supplemented with 1% (v/v) 1X Glutamax (Gibco, cat# 35050061), 12% (v/v) FBS (Gibco, cat# 10082147), 0.05 mM 2-mercaptoethanol (Gibco, cat# 31350010), and 1X anti-anti (Gibco, cat# 15240096). The cells were plated on culture dishes coated with a fibrin gel matrix. The fibrin gel matrix consisted of a mixture of fibrinogen (SigmaAldrich, cat# F8630) and thrombin (SigmaAldrich, cat# T4648-10KU). Fibrinogen was diluted in D-PBS (Gibco, cat# 14190144) to a final concentration of 20 mg/mL. A stock solution of thrombin was first made in D-PBS + 0.1% BSA (w/v) to a final concentration of 200 U/mL. To prepare the fibrin gel matrix, thrombin was further diluted to 10 U/mL in ice-cold D-PBS and used to coat the cell culture dish. The fibrinogen solution was then mixed with the thrombin solution, at a ratio of 1:1.5, respectively. The coated dishes were incubated at room temperature for 15 minutes. After 15 minutes of incubation, the dishes were ready to be used. The cell suspension was then placed on top of the fibrin gel. The following day (day-0), the cells were checked for attachment to the fibrin gel matrix and treated with the small molecule PURK-differentiation cocktail. The PURK-differentiation cocktail was changed every 3–4 days. On day-3 of differentiation and onwards, the cell culture medium was supplemented with 0.1 mg/mL of 6-aminocaproic acid (Sigma Aldrich, cat# A2504). The morphology and expression of the fluorescent CNTN2-mCherry tag were checked daily on the Nikon Ti-E inverted microscope attached with a DS-Fi 1 5-megapixel color camera (Nikon Instruments).
Differentiation of iPSC into cardiomyocytes
The iPSCs were differentiated into cardiomyocytes using STEMdiffTM Cardiomyocyte Differentiation Kit (StemCell Technologies, cat# 05010), according to the manufacturer’s instructions. In brief, 2 days before treatment, the iPSCs were dissociated into single cells using Accutase (StemCell Technologies, cat# 07920). The cells were then centrifuged at 1,200 rpm for 2–3 minutes. The supernatant was removed and the cell pellet was resuspended in mTeSRTM1 basal medium (StemCell Technologies, cat# 85850) supplemented with 1X mTeSRTM1 supplement, 1X anti-anti (Gibco, cat# 15240096), and 10 μM Y-27632 (SigmaAldrich, cat# 688002-1MG-M). The single IPS cells were then plated onto dishes coated with hESC-qualified Matrigel (Corning, cat# 354277). At day-1, the cell culture media was replaced with mTeSRTM1 basal medium (StemCell Technologies, cat# 85850) supplemented with 1X mTeSRTM1 supplement, 1X anti-anti (Gibco, cat# 15240096). By day-0, the cell confluence was >95%, and differentiation into myocytes started by treating the cells with STEMdiffTM Cardiomyocyte Differentiation Medium A supplemented with 1:100 dilution of hESC-qualified Matrigel (Corning, cat# 354277). On day-2, the media was changed to STEMdiffTM Cardiomyocyte Differentiation Medium B. On days 4 and 6, complete medium change into STEMdiffTM Cardiomyocyte Differentiation Medium C was used to treat the cells. From day-8 onwards, the cells were treated with STEMdiffTM Cardiomyocyte Maintenance Medium (StemCell Technologies, cat# 05020). By day-9, areas of beating IPS-CMs were visible.
On day-14, the beating myocytes were purified by metabolic selection according to the protocol described by Rupert et al. (2020). In brief, the STEMdiffTM Cardiomyocyte Maintenance Medium was replaced by a lactate purification medium (LPM) which consisted of SILAC DMEM Flex Media, no glucose, no phenol red (Gibco, cat# A2493901) supplemented with 4 mM L-Glutamine (Gibco, cat# 25030081), 1X MEM Non Essential Amino Acids (Gibco, cat# 11140050), 1X Glutamax (Gibco, cat# 35050061), 4 mM Sodium DL Lactate (SigmaAldrich, cat# L4263-100ML) and 1X anti-anti (Gibco, cat# 15240096). Cell death of undifferentiated IPS cells was observed after 2 days of treatment with the LPM. The IPS-CM cells were kept in LPM for 7 days to obtain purified myocytes. The purified IPS-CMs were then differentiated into Purkinje-like cells and used in downstream experiments.
Differentiation of iPSC into Purkinje-like cells
The iPSC-CM cells were plated one day before treatment with the PURK-differentiation cocktail (day-1). The iPSC-CM were washed twice with D-PBS and STEMdiffTM Cardiomyocyte Dissociation Kit (StemCell Technologies, cat# 05025) was used to harvest the cells for plating. STEMdiffTM Cardiomyocyte Support Medium (StemCell Technologies, cat# 05027) was added to dilute the dissociation medium. The cells were then centrifuged at 1,200 rpm for 5 minutes. The supernatant was removed and the cell pellet was resuspended in STEMdiffTM Cardiomyocyte Support Medium (StemCell Technologies, cat# 05027), 10 μM Y-27632 (StemCell Technologies, cat# 72302), and 1X anti-anti (Gibco, cat# 15240096). The cells were plated on culture dishes coated with a fibrin gel matrix. The fibrin gel matrix consisted of a mixture of fibrinogen (SigmaAldrich, cat# F8630) and thrombin (SigmaAldrich, cat# T4648-10KU). Fibrinogen was diluted in D-PBS (Gibco, cat# 14190144) to a final concentration of 20 mg/mL. A stock solution of thrombin was first made in D-PBS + 0.1% BSA (w/v) to a final concentration of 200 U/mL. To prepare the fibrin gel matrix, thrombin was further diluted to 10 U/mL in ice-cold D-PBS and used to coat the cell culture dish. The fibrinogen solution was then mixed with the thrombin solution, at a ratio of 1:1.5, respectively. The coated dishes were incubated at room temperature for 15 minutes. After 15 minutes of incubation, the dishes were ready to be used. The cell suspension was then placed on top of the fibrin gel. The following day (day-0), the cells were checked for attachment to the fibrin gel matrix and treated with the small molecule PURK-differentiation cocktail. Note: The PURK-cocktail recipe was the same as previously described, however, 5% FBS was NOT added to the media. The PURK-differentiation cocktail was changed every 3–4 days. On day-3 of differentiation and onwards, the cell culture medium was supplemented with 0.1 mg/mL of 6-aminocaproic acid (Sigma Aldrich, cat# A2504). The morphology and expression of the fluorescent CNTN2-mCherry tag were checked daily on the Nikon Ti-E inverted microscope attached with a DS-Fi 1 5-megapixel color camera (Nikon Instruments).
Fluorescence microscopy
To initially investigate the morphological changes, as well as expression of the CNTN2-mCherry tag introduced by CRISPR-Cas9 into the cells and determine if the cells have successfully differentiated into Purkinje-like cells, fluorescence microscopy was used. Brightfield and fluorescence images were taken using either a Nikon Ti-E inverted microscope attached with a DS-Fi 1 5-megapixel color camera (Nikon Instruments), or an FV3000 inverted confocal microscope (Olympus). NIS Elements software v4.13 (Nikon Instruments) or Fluoview Software (Olympus) were used to capture and analyze the images.
FACS sorting and analysis
Purkinje-like cells were FACS sorted for CNTN2-mCherry positive at different differentiation days to determine the optimal % of differentiation over different periods. The cells were first washed with D-PBS (Gibco, cat# 14190144), then dissociated using 2.5U/mL Papain from Carica papaya (SigmaAldrich, cat# 76216-50MG) for 2–3 minutes. This was followed by neutralization of Papain using 50% FBS (Gibco, cat# 10082147) and 50% DMEM/F12 medium (Sigma Aldrich, cat# D6434-500ML). The cells were then centrifuged at 1,200 rpm for 2–3 minutes. Subsequently, the supernatant was aspirated and the cell pellet was resuspended in DMEM/F12 supplemented with 1% FBS. The cells were counted with a cell counter and diluted to obtain a concentration between 1–1.5 × 106 cells/mL. The cells were dissociated into a single cell suspension by passing them through the cell strainer cap of a Falcon Round Bottom tube (StemCell Technologies, cat# 38030). FACS analysis was performed using a BD LSRII flow cytometer (BD Biosciences) which was followed by sorting on a FACS Aria Fusion flow cytometer (BD Biosciences). The sorted cells were collected in 50% DMEM/F12 supplemented with 50% FBS and used for downstream applications such as RNA extraction (qPCR and RNA-seq).
Immunostaining
Purkinje-like cells were immunostained for a series of Purkinje-specific markers at day-7 of differentiation. The cells were first differentiated using the PURK-differentiation cocktails. To stain the cells, the samples were first washed with D-PBS twice (Gibco, cat# 14190144). A 4% Formaldehyde fixing solution was made by diluting PierceTM 16% Formaldehyde (w/v), methanol free (ThermoScientificTM, cat# 28908) in D-PBS. The samples were incubated with the 4% Formaldehyde fixing solution for 10 minutes at room temperature. After 10 minutes of incubation, the samples were washed with ice-cold D-PBS three times. Depending on the antibody (shown below), the cells were permeabilized by incubation with D-PBS containing 0.2% Triton X-100 (Sigma-Aldrich, cat# X100-100ML) for 10 minutes at room temperature. The cells were then washed three times with D-PBS for 5 minutes each time. The permeabilization step was not performed when staining for membrane-associated proteins and was only used when investigating cytosolic, vesicular, or nuclear proteins. The next step was to block the unspecific binding of the antibodies by using 10% Normal Goat Serum (ThermoFisher Scientific, cat# 50062Z) for 30 minutes at room temperature. The samples were then incubated with the diluted antibody (dilution used shown below) for 1 hour at room temperature or overnight at 4°C. Next, the samples were washed three times with D-PBS for 5 minutes each time. After the last wash, the samples were subsequently incubated with the corresponding secondary antibody diluted in 10% Normal Goat Serum for 1 hour at room temperature in the dark. This was followed by washing the samples three times with D-PBS, for 5 minutes for each wash. The samples were then incubated with 1 μg/mL DAPI solution (ThermoScientific, cat# 62248) for 1 minute at room temperature. Finally, the samples were rinsed with D-PBS and were ready to be imaged using a fluorescent microscope.
List of antibodies used in immunocytochemistry experiments
| Antibody | Manufacturer | Dilution used | Condition |
|---|---|---|---|
| ETV1 | ThermoFisher (cat# PA589150) | 1:150 | Intracellular |
| SCN5a | ThermoFisher (cat# MA1-27429) | 1:100 | Cell Surface |
| IRX3 | ThermoFisher (cat# PA5-111549) | 1:250 | Cell Surface |
| PCP4 | ThermoFisher (cat# 14705-1-AP) | 1:250 | Cell Surface |
| Alexa 488 | ThermoFisher (cat# A32731) | 1:1,000 | Secondary AB |
| Alexa 594 | ThermoFisher (cat# A-21044) | 1:1,000 | Secondary AB |
RNA extraction and qRT-PCR
RNA was extracted from CNTN2-mCherry positive cells, that were FACS sorted, using a Quick-RNA Microprep kit (Zymo Research, cat# R1050) according to the manufacturer’s protocol. The extracted RNA was then cleaned and concentrated using RNA Clean & Concentrator-5 (Zymo Research, cat# R1013), according to the manufacturer’s protocol. The extracted RNA was resuspended in DEPC-treated water (Invitrogen, cat# AM9906) and stored at −80°C until its use in downstream applications. The quality and concentration of the RNA were then analyzed by Nanodrop. Subsequently, cDNA was synthesized from the sample RNA using a High-Capacity cDNA Reverse Transcriptase kit (Applied Biosystems, cat# 4368814), according to the manufacturer’s protocol. At least 1 μg of RNA was used in each cDNA synthesis reaction. The sample cDNAs were stored at −20°C until their use in downstream applications. Once the cDNA was ready, it was used in qRT-PCR experiments. The qRT-PCR mix consisted of cDNA, PowerTrackTM SYBR Green Master Mix (Applied Biosystems, cat# A46109), DEPC-treated water, and 10 μM Forward and Reverse Primers (sequences shown in Table S1). The qRT-PCR reaction was run on a CFX Opus Real-Time PCR System (Bio-Rad). Normalization was done based on GAPDH levels. The qPCR program is shown below.
qPCR program
| Step | Cycles | Temp | Time |
|---|---|---|---|
| Enzyme Activation | 1 | 95°C | 2 min |
| Denaturation | 40 | 95°C | 15 seconds |
| Annealing | 60°C | 1 min | |
| Final Hold | 1 | 4°C | HOLD |
All primers were custom synthesized by Life Technologies. The sequences are found in Table S1.
RNA-sequencing
RNA was extracted from the cells using a Quick-RNA Microprep kit (Zymo Research, cat# R1050) according to the manufacturer’s protocol. The extracted RNA was then cleaned, DNAse treated, and concentrated using RNA Clean & Concentrator-5 (Zymo Research, cat# R1013), according to the manufacturer’s protocol. The extracted RNA was resuspended in DEPC-treated water (Invitrogen, cat# AM9906) and stored at −80°C until its use in downstream applications. The quality and concentration of the RNA were then analyzed by Nanodrop, as well as an RNA nano 6000 chip through Bioanalyzer 2100 (Agilent). The RIN value of all RNA was >8, and concentrations were >100 ng/uL. The RNA library was prepared and sequenced at the University of Houston Seq-N-Edit Core (NuGen) using up to 1,000 ng of RNA. SPRIselect beads (Beckman Coulter) were used to size select the RNA libraries, which were then analyzed for their purity using a High Sensitivity DNA chip on a Bioanalyzer 2100 (Agilent). NextSeq 500 (Illumina) was then used to sequence the RNA libraries and approx. 20 million 2 × 76 bp paired-end reads per sample were generated. The raw RNA-seq fastq data were then processed using the RNA-seq Alignment feature of the Illumina BaseSpace. Differentially expressed genes, with an FDR adjusted p-value of at least 0.05, were identified between the PURK-cocktail treatment and the control (vehicle) treated cells. The online HeatMapper (Babicki et al., 2016) tool was used to generate the heatmaps containing genes of interest that were significantly different from the control. No clustering was used in the maps.
Optical mapping
Field stimulation was delivered to the recording chamber connected to a MyoPacer (IonOptix). The pacing protocol was used as: 1) Pacing frequency: 1 Hz, 2) Pacing voltage: 15 V and 3) Pulse width: 10 ms. The MyoPacer cable was connected to a chamber with built-in wire electrodes which were then placed inside the cell culture dish. The cells were cultured and differentiated for 7 days prior to the experiment. On the day of the experiment (day-7 of differentiation), the voltage-sensitive dye Di-4-ANEPPS (Scott et al., 2021) (10 μmol/L, Invitrogen, cat#D1199) was added to the cell culture media. Cells were incubated with the dye for 5 minutes in the incubator (37°C) and subsequently washed with D-PBS. The long-passed (>700 nm) Vm-signal was acquired by a MiCAM CMOS camera (SciMedia, USA) (1-kHz sampling rate, 100 μm/pixel). ElectroMap was used to generate activation maps and analyze conduction velocity (CV) (O’Shea et al., 2019a, 2019b).
IncuCyte
To temporally track changes in cell morphology and to monitor the differentiation process of the Purkinje-like cells, time-lapse live-cell images were collected using the IncuCyte® Live-Cell System (Sartorius). IncuCyte®, a microscopy system fully integrated within a cell culture incubator, allows for cell monitoring over long periods without disturbing the cells (Gordonov et al., 2016). Cells were cultured and placed inside the IncuCyte® incubator. Bright-field images were taken using the 10X objective, every 2 hours, over 12 days. The cell culture media was replaced every 2–3 days. A time-lapse movie was then created from the collection of pictures taken by the IncuCyte®.
MEA fabrication and electrophysiology data
The processes started with cleaning a glass substrate (2″ × 1″) with isopropyl alcohol and acetone, followed by oxygen plasma (CS-1701, Nordson) treatment. (3-Aminopropyl)triethoxysilane (1 wt%, Sigma) aqueous solution as the adhesion promoter was applied on the treated glass substrate and spin coated at 3,000 rpm. Afterward, the polyimide precursor solution (PI-2545, HD Microsystems) was coated on the glass substrate at 3,000 rpm and then cured at 250°C for 1 hour. To form the conductive traces, the metal layer (Cr/Au, 5/100 nm) was deposited by an e-beam evaporator and then patterned by lithography and wet etching. Thereafter, a second thin PI layer (encapsulation) was coated on top of the metallic traces, followed by wet etching to expose the sensing electrode array in the center part and contact pads at two ends. The PI encapsulation was first baked at 150°C for 10 minutes, then photoresist (AZ 5214, MicroChemicals) was coated on the PI and selectively exposed, developed (1.5 minutes in AZ 917 MIF, MicroChemicals), and removed by acetone. The device was then baked at 250°C for 1 hour to fully cure the PI.
To confine the biological cells and medium on top of the sensing electrode array, a polydimethylsiloxane (PDMS) chamber that served as the culture well was sealed on top of the device. Briefly, PDMS precursor (10:1 (w/w) prepolymer/curing agent, Sylgard 184 silicone elastomer kit) was poured into a petri dish to achieve a thickness of 7 mm. After degassing and curing the PDMS in an oven at 90°C overnight, the PDMS was cut into square chambers. To ensure good bonding strength between the PDMS chamber and the PI surface, the PDMS chamber was treated with plasma oxidization and the top surface of the device was also treated with plasma oxidization and coated with (3-Aminopropyl)triethoxysilane. Afterward, the PDMS chamber was bonded to the device under pressure and heat for 10 minutes. The contact pads were interfaced with anisotropic conductive film (ACF, Elform Heat Seal Connectors) based ribbon cables under pressure at 170°C for 1 minute to two printed circuit boards (PCB), which were connected to an external amplifier (Recording controller, Intan Technologies) for signal acquisition.
The resulting 64-channel multielectrode array (MEA) was used to measure the functional capabilities of the cells. The cells were grown and differentiated into the MEA. On day-7 of differentiation, the recordings were conducted. For the stimulation experiments, a function generator (DG4062, RIGOL) was used. In each trial, spontaneous activity was first recorded for 10 s. This was followed by 30 s of stimulation and recording, and 10 s of spontaneous recording again. The recordings were done at 0.5, 1, 2, and 3 Hz at different pulse widths and amplitudes. As one example, the pulse parameters for the 0.5 Hz condition are shown below. The activation delay times were determined by identifying the locations of electrodes that showed obvious peaks and taking the time difference of those from that of the stimulus channel peak. The conduction velocities were calculated based on the spacing of the electrodes in the MEA and the activation delay times. The conduction velocities were calculated based on the spacing of the electrodes in the MEA and the activation delay times. The data were filtered and analyzed in MATLAB with custom code.
Different pulse conditions for MEA measurements
| Pulse amplitude | Pulse width |
|---|---|
| 5 Vpp | 6.25, 10, 20 ms |
| 10 Vpp | 6.25, 10, 20 ms |
| 15 Vpp | 6.25, 10, 20 ms |
Quantification and statistical analysis
The qPCR and optical mapping data were processed using GraphPad Prism 9 and Microsoft Excel. The data represent the mean ± standard deviation (S.D.). The qPCR data were analyzed with Two-Way ANOVA, where the treated samples were compared against the control (vehicle). ∗, ∗∗, ∗∗∗, ∗∗∗∗ shown in the graphs correspond to p < 0.05, 0.01, 0.001, and 0.0001, respectively. A Welch’s T-Test was used to compare the conduction velocities obtained from the optical mapping. ∗∗∗ corresponds to p < 0.001. All experiments presented in this manuscript were independently performed at least 3 times (at least 3 biological replicates). Each experiment had at least 3 technical replicates. All tests were considered significant where it was observed a p < 0.05 at least. Statistical details of experiments can be found in the figure legends and results section.
Acknowledgments
The authors would like to acknowledge Alon R. Azares (Texas Heart Institute, Houston TX) for his assistance with the FACS experiments and Dr. Jean J. Kim (Baylor College of Medicine, Houston TX) for her assistance in training N.P. on culturing iPSCs through the “Basics of Human Pluripotent Stem Cells” course. This article is in partial fulfillment of the requirements for a Ph.D. (Pharmacology) in the Department of Pharmacological and Pharmaceutical Sciences in the College of Pharmacy at the University of Houston (N.P.). Research reported in this article was supported in part by the National Heart, Lung, and Blood Institute (NHLBI) of the National Institutes of Health (NIH) under award numbers R15 HL141963 (B.K.M.), R01 HL136389, R01 HL163277 and R01 HL147108 (N.L.); the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the NIH under award numbers R21 EB026175 (C.Y.) and 1R21 EB030257-01 (C.Y.); the National Science Foundation under award number 1936151 (C.Y.); the American Heart Association (AHA) under award number 18AIREA 33960175 (B.K.M.) and EIA93611 (N.L.); and a grant from the Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation (B.K.M.). Also, the research reported in this article was supported in part by a University of Houston (UH)/Baylor College of Medicine (BCM) Collaborative Pilot Grant (B.K.M., N.L.). The funders had no role in the preparation of the article or the decision to publish this article.
Author contributions
N.P. has performed the conception and design of the study, planned, executed, and participated in all experiments, organized and analyzed the data, wrote the article. F.E, L.G., and Z.R. executed experiments and analyzed the MEA data. A.R.A planned and executed CRISPR-Cas9 editing of the cells. L.L. planned, executed, and analyzed the data for the optical mapping studies. B.M. planned, executed, and analyzed the RNA-sequencing studies. C.Y., P.H.G., and N.L. collaborated and provided technical expertise. R.J.S. and B.K.M. performed the conception and design of the study and B.K.M. oversaw the entire project and writing of the article. All authors critically revised and approved the article.
Declaration of interests
The authors declare that the following patent applications have been filed related to this work:
-
•
McConnell BK, Schwartz RJ, and Prodan N. Direct reprogramming of cells into cardiac Purkinje-like cells using a universal small molecule cocktail. US Provisional Patent Application Number: 63234399 - Filed Aug. 18, 2021.
-
•
McConnell BK, Schwartz RJ, and Prodan N. Direct reprogramming of cells into cardiac Purkinje-like cells using a universal small molecule cocktail. PCT Patent Application Number: PCT/US2022/039546 - Filed Aug. 5, 2022.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105402.
Supplemental information
Data and code availability
-
•
Data
Raw RNA-seq data of PURK-cocktail treated cells at day-4 and day-7 that supports the findings of this study has been deposited in the Figshare database, is publicly available, and can be accessed at (https://doi.org/10.6084/m9.figshare.21290883.v1).
-
•
Code
This paper does not report original code.
-
•
Additional Information
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Atkinson A., Inada S., Li J., Tellez J.O., Yanni J., Sleiman R., Allah E.A., Anderson R.H., Zhang H., Boyett M.R., Dobrzynski H. Anatomical and molecular mapping of the left and right ventricular His-Purkinje conduction networks. J. Mol. Cell. Cardiol. 2011;51:689–701. doi: 10.1016/j.yjmcc.2011.05.020. [DOI] [PubMed] [Google Scholar]
- Aydin B., Mazzoni E.O. Cell reprogramming: the many roads to success. Annu. Rev. Cell Dev. Biol. 2019;35:433–452. doi: 10.1146/annurev-cellbio-100818-125127. [DOI] [PubMed] [Google Scholar]
- Babicki S., Arndt D., Marcu A., Liang Y., Grant J.R., Maciejewski A., Wishart D.S. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–W153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker M.L., Boukens B.J., Mommersteeg M.T.M., Brons J.F., Wakker V., Moorman A.F.M., Christoffels V.M. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Balmer J.E., Blomhoff R. Gene expression regulation by retinoic acid. J. Lipid Res. 2002;43:1773–1808. doi: 10.1194/jlr.r100015-jlr200. [DOI] [PubMed] [Google Scholar]
- Benjamin E.J., Blaha M.J., Chiuve S.E., Cushman M., Das S.R., Deo R., De Ferranti S.D., Floyd J., Fornage M., Gillespie C., et al. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland M.J., Nazor K.L., Loring J.F. Epigenetic regulation of pluripotency and differentiation. Circ. Res. 2014;115:311–324. doi: 10.1161/CIRCRESAHA.115.301517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorsma E.M., Ter Maaten J.M., Damman K., Dinh W., Gustafsson F., Goldsmith S., Burkhoff D., Zannad F., Udelson J.E., Voors A.A. Congestion in heart failure: a contemporary look at physiology, diagnosis and treatment. Nat. Rev. Cardiol. 2020;17:641–655. doi: 10.1038/s41569-020-0379-7. [DOI] [PubMed] [Google Scholar]
- Bos J.L. Epac proteins: multi-purpose cAMP targets. Trends Biochem. Sci. 2006;31:680–686. doi: 10.1016/j.tibs.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Boyden P.A., Hirose M., Dun W. Cardiac Purkinje cells. Heart Rhythm. 2010;7:127–135. doi: 10.1016/j.hrthm.2009.09.017. [DOI] [PubMed] [Google Scholar]
- Burnicka-Turek O., Broman M.T., Steimle J.D., Boukens B.J., Petrenko N.B., Ikegami K., Nadadur R.D., Qiao Y., Arnolds D.E., Yang X.H., et al. Transcriptional patterning of the ventricular cardiac conduction system. Circ. Res. 2020;127:e94–e106. doi: 10.1161/CIRCRESAHA.118.314460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain A.A., Lin M., Lister R.L., Maslov A.A., Wang Y., Suzuki M., Wu B., Greally J.M., Zheng D., Zhou B. DNA methylation is developmentally regulated for genes essential for cardiogenesis. J. Am. Heart Assoc. 2014;3:e000976. doi: 10.1161/JAHA.114.000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Studer L. Cell fate plug and play: direct reprogramming and induced pluripotency. Cell. 2011;145:827–830. doi: 10.1016/j.cell.2011.05.036. [DOI] [PubMed] [Google Scholar]
- Chen X.R., Heck N., Lohof A.M., Rochefort C., Morel M.P., Wehrlé R., Doulazmi M., Marty S., Cannaya V., Avci H.X., et al. Mature Purkinje cells require the retinoic acid-related orphan receptor-alpha (RORalpha) to maintain climbing fiber mono-innervation and other adult characteristics. J. Neurosci. 2013;33:9546–9562. doi: 10.1523/JNEUROSCI.2977-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Shao J.Z., Xiang L.X., Guo J., Zhou Q.J., Yao X., Dai L.C., Lu Y.L. Cyclic adenosine 3', 5'-monophosphate induces differentiation of mouse embryonic stem cells into cardiomyocytes. Cell Biol. Int. 2006;30:301–307. doi: 10.1016/j.cellbi.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Cheng G., Litchenberg W.H., Cole G.J., Mikawa T., Thompson R.P., Gourdie R.G. Development of the cardiac conduction system involves recruitment within a multipotent cardiomyogenic lineage. Development. 1999;126:5041–5049. doi: 10.1242/dev.126.22.5041. [DOI] [PubMed] [Google Scholar]
- Cingolani E., Goldhaber J.I., Marbán E. Next-generation pacemakers: from small devices to biological pacemakers. Nat. Rev. Cardiol. 2018;15:139–150. doi: 10.1038/nrcardio.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen N.R., Taylor J.S., Scott L.B., Guillery R.W., Soriano P., Furley A.J. Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr. Biol. 1998;8:26–33. doi: 10.1016/s0960-9822(98)70017-x. [DOI] [PubMed] [Google Scholar]
- Dangman K.H., Danilo P., Jr., Hordof A.J., Mary-Rabine L., Reder R.F., Rosen M.R. Electrophysiologic characteristics of human ventricular and Purkinje fibers. Circulation. 1982;65:362–368. doi: 10.1161/01.cir.65.2.362. [DOI] [PubMed] [Google Scholar]
- Dobrzynski H., Anderson R.H., Atkinson A., Borbas Z., D'souza A., Fraser J.F., Inada S., Logantha S.J.R.J., Monfredi O., Morris G.M., et al. Structure, function and clinical relevance of the cardiac conduction system, including the atrioventricular ring and outflow tract tissues. Pharmacol. Ther. 2013;139:260–288. doi: 10.1016/j.pharmthera.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Gersh B.J., Sliwa K., Mayosi B.M., Yusuf S. Novel therapeutic concepts: the epidemic of cardiovascular disease in the developing world: global implications. Eur. Heart J. 2010;31:642–648. doi: 10.1093/eurheartj/ehq030. [DOI] [PubMed] [Google Scholar]
- Goodyer W.R., Beyersdorf B.M., Paik D.T., Tian L., Li G., Buikema J.W., Chirikian O., Choi S., Venkatraman S., Adams E.L., et al. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ. Res. 2019;125:379–397. doi: 10.1161/CIRCRESAHA.118.314578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordonov S., Hwang M.K., Wells A., Gertler F.B., Lauffenburger D.A., Bathe M. Time series modeling of live-cell shape dynamics for image-based phenotypic profiling. Integr. Biol. 2016;8:73–90. doi: 10.1039/c5ib00283d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorza L., Schiaffino S., Vitadello M. Heart conduction system: a neural crest derivative? Brain Res. 1988;457:360–366. doi: 10.1016/0006-8993(88)90707-x. [DOI] [PubMed] [Google Scholar]
- Gourdie R.G., Mima T., Thompson R.P., Mikawa T. Terminal diversification of the myocyte lineage generates Purkinje fibers of the cardiac conduction system. Development. 1995;121:1423–1431. doi: 10.1242/dev.121.5.1423. [DOI] [PubMed] [Google Scholar]
- Hoogaars W.M.H., Tessari A., Moorman A.F.M., de Boer P.A.J., Hagoort J., Soufan A.T., Campione M., Christoffels V.M. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Huang N.F., Serpooshan V., Morris V.B., Sayed N., Pardon G., Abilez O.J., Nakayama K.H., Pruitt B.L., Wu S.M., Yoon Y.S., et al. Big bottlenecks in cardiovascular tissue engineering. Commun. Biol. 2018;1:199. doi: 10.1038/s42003-018-0202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas J.F., Abbasgholizadeh R., Dacso C., Potaman V.N., Navran S., Bond R.A., Iyer D., Birla R., Schwartz R.J. beta-Adrenergic stimuli and rotating suspension culture enhance conversion of human adipogenic mesenchymal stem cells into highly conductive cardiac progenitors. J. Tissue Eng. Regen. Med. 2020;14:306–318. doi: 10.1002/term.2994. [DOI] [PubMed] [Google Scholar]
- Jarjour M., Henri C., De Denus S., Fortier A., Bouabdallaoui N., Nigam A., O'meara E., Ahnadi C., White M., Garceau P., et al. Care gaps in adherence to heart failure guidelines: clinical inertia or physiological limitations? JACC Heart Fail. 2020;8:725–738. doi: 10.1016/j.jchf.2020.04.019. [DOI] [PubMed] [Google Scholar]
- Johanson R.A., Sarau H.M., Foley J.J., Slemmon J.R. Calmodulin-binding peptide PEP-19 modulates activation of calmodulin kinase II in situ. J. Neurosci. 2000;20:2860–2866. doi: 10.1523/JNEUROSCI.20-08-02860.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Tsoy I., Park M.K., Lee Y.S., Lee J.H., Seo H.G., Chang K.C. Iron released by sodium nitroprusside contributes to heme oxygenase-1 induction via the cAMP-protein kinase A-mitogen-activated protein kinase pathway in RAW 264.7 cells. Mol. Pharmacol. 2006;69:1633–1640. doi: 10.1124/mol.105.020487. [DOI] [PubMed] [Google Scholar]
- Komosa E.R., Wolfson D.W., Bressan M., Cho H.C., Ogle B.M. Implementing biological pacemakers: design criteria for successful. Circ. Arrhythm. Electrophysiol. 2021;14:e009957. doi: 10.1161/CIRCEP.121.009957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepski G., Jannes C.E., Nikkhah G., Bischofberger J. cAMP promotes the differentiation of neural progenitor cells in vitro via modulation of voltage-gated calcium channels. Front. Cell. Neurosci. 2013;7:155. doi: 10.3389/fncel.2013.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.C., Dollé P., Ryckebüsch L., Noseda M., Zaffran S., Schneider M.D., Niederreither K. Endogenous retinoic acid regulates cardiac progenitor differentiation. Proc. Natl. Acad. Sci. USA. 2010;107:9234–9239. doi: 10.1073/pnas.0910430107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Kong L., Zhu S. Reprogramming cell fates by small molecules. Protein Cell. 2017;8:328–348. doi: 10.1007/s13238-016-0362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T. Transcriptional regulation of a Purkinje cell-specific gene through a functional interaction between ROR alpha and RAR. Gene Cell. 1997;2:263–272. doi: 10.1111/j.1365-2443.1997.119gc0317.x. [DOI] [PubMed] [Google Scholar]
- McCready F.P., Gordillo-Sampedro S., Pradeepan K., Martinez-Trujillo J., Ellis J. Multielectrode arrays for functional phenotyping of neurons from induced pluripotent stem cell models of neurodevelopmental disorders. Biology. 2022;11:316. doi: 10.3390/biology11020316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei F.C., Qiao J., Tsygankova O.M., Meinkoth J.L., Quilliam L.A., Cheng X. Differential signaling of cyclic AMP: opposing effects of exchange protein directly activated by cyclic AMP and cAMP-dependent protein kinase on protein kinase B activation. J. Biol. Chem. 2002;277:11497–11504. doi: 10.1074/jbc.M110856200. [DOI] [PubMed] [Google Scholar]
- Miquerol L., Beyer S., Kelly R.G. Establishment of the mouse ventricular conduction system. Cardiovasc. Res. 2011;91:232–242. doi: 10.1093/cvr/cvr069. [DOI] [PubMed] [Google Scholar]
- Miquerol L., Moreno-Rascon N., Beyer S., Dupays L., Meilhac S.M., Buckingham M.E., Franco D., Kelly R.G. Biphasic development of the mammalian ventricular conduction system. Circ. Res. 2010;107:153–161. doi: 10.1161/CIRCRESAHA.110.218156. [DOI] [PubMed] [Google Scholar]
- Moayedi Y., Fan C.P.S., Cherikh W.S., Stehlik J., Teuteberg J.J., Ross H.J., Khush K.K. Survival outcomes after heart transplantation: does recipient sex matter? Circ. Heart Fail. 2019;12:e006218. doi: 10.1161/CIRCHEARTFAILURE.119.006218. [DOI] [PubMed] [Google Scholar]
- Mommaerts W.F., Khairallah P.A., Dickens M.F. Acetylcholinesterase in the conductive tissue of the heart. Circ. Res. 1953;1:460–465. doi: 10.1161/01.res.1.5.460. [DOI] [PubMed] [Google Scholar]
- Monteiro L.M., Vasques-Nóvoa F., Ferreira L., Pinto-do-Ó P., Nascimento D.S. Restoring heart function and electrical integrity: closing the circuit. NPJ Regen. Med. 2017;2:25. doi: 10.1038/s41536-017-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore-Morris T., Van Vliet P.P., Andelfinger G., Puceat M. Role of epigenetics in cardiac development and congenital diseases. Physiol. Rev. 2018;98:2453–2475. doi: 10.1152/physrev.00048.2017. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Ikeda T., Shimokawa I., Inoue Y., Suematsu T., Sakai H., Iwasaki K., Matsuo T. Distribution of acetylcholinesterase activity in the rat embryonic heart with reference to HNK-1 immunoreactivity in the conduction tissue. Anat. Embryol. 1994;190:367–373. doi: 10.1007/BF00187294. [DOI] [PubMed] [Google Scholar]
- O’Shea C., Holmes A.P., Yu T.Y., Winter J., Wells S.P., Correia J., Boukens B.J., De Groot J.R., Chu G.S., Li X., et al. ElectroMap: high-throughput open-source software for analysis and mapping of cardiac electrophysiology. Sci. Rep. 2019;9:1389. doi: 10.1038/s41598-018-38263-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea C., Holmes A.P., Yu T.Y., Winter J., Wells S.P., Parker B.A., Fobian D., Johnson D.M., Correia J., Kirchhof P., et al. High-throughput analysis of optical mapping data using ElectroMap. J. Vis. Exp. 2019:e59663. doi: 10.3791/59663. [DOI] [PubMed] [Google Scholar]
- Pallante B.A., Giovannone S., Fang-Yu L., Zhang J., Liu N., Kang G., Dun W., Boyden P.A., Fishman G.I. Contactin-2 expression in the cardiac Purkinje fiber network. Circ. Arrhythm. Electrophysiol. 2010;3:186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.S., Fishman G.I. Development and function of the cardiac conduction system in health and disease. J. Cardiovasc. Dev. Dis. 2017;4:7. doi: 10.3390/jcdd4020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellicori P., Khan M.J.I., Graham F.J., Cleland J.G.F. New perspectives and future directions in the treatment of heart failure. Heart Fail. Rev. 2020;25:147–159. doi: 10.1007/s10741-019-09829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perl E., Waxman J.S. Reiterative mechanisms of retinoic acid signaling during vertebrate heart development. J. Dev. Biol. 2019;7:E11. doi: 10.3390/jdb7020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S.D., Frangogiannis N.G. The biological basis for cardiac repair after myocardial infarction: from inflammation to fibrosis. Circ. Res. 2016;119:91–112. doi: 10.1161/CIRCRESAHA.116.303577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevention C.F.D.C.A. Underlying Cause of Death. Atlanta, GA: CDC WONDER Online Database. 1999 [Google Scholar]
- Qin H., Zhao A., Fu X. Small molecules for reprogramming and transdifferentiation. Cell. Mol. Life Sci. 2017;74:3553–3575. doi: 10.1007/s00018-017-2586-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall V., Mccue K., Roberts C., Kyriakopoulou V., Beddow S., Barrett A.N., Vitelli F., Prescott K., Shaw-Smith C., Devriendt K., et al. Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J. Clin. Invest. 2009;119:3301–3310. doi: 10.1172/JCI37561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert C.E., Irofuala C., Coulombe K.L.K. Practical adoption of state-of-the-art hiPSC-cardiomyocyte differentiation techniques. PLoS One. 2020;15:e0230001. doi: 10.1371/journal.pone.0230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadek H., Olson E.N. Toward the goal of human heart regeneration. Cell Stem Cell. 2020;26:7–16. doi: 10.1016/j.stem.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saimi Y., Kung C. Calmodulin as an ion channel subunit. Annu. Rev. Physiol. 2002;64:289–311. doi: 10.1146/annurev.physiol.64.100301.111649. [DOI] [PubMed] [Google Scholar]
- Sayers J.R., Riley P.R. Heart regeneration: beyond new muscle and vessels. Cardiovasc. Res. 2021;117:727–742. doi: 10.1093/cvr/cvaa320. [DOI] [PubMed] [Google Scholar]
- Scicchitano P., Carbonara S., Ricci G., Mandurino C., Locorotondo M., Bulzis G., Gesualdo M., Zito A., Carbonara R., Dentamaro I., et al. HCN channels and heart rate. Molecules. 2012;17:4225–4235. doi: 10.3390/molecules17044225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L., Jr., Fender A.C., Saljic A., Li L., Chen X., Wang X., Linz D., Lang J., Hohl M., Twomey D., et al. NLRP3 inflammasome is a key driver of obesity-induced atrial arrhythmias. Cardiovasc. Res. 2021;117:1746–1759. doi: 10.1093/cvr/cvab024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmera D., Gourdie R.G. Why do we have Purkinje fibers deep in our heart? Physiol. Res. 2014;63:S9–S18. doi: 10.33549/physiolres.932686. [DOI] [PubMed] [Google Scholar]
- Shekhar A., Lin X., Lin B., Liu F.Y., Zhang J., Khodadadi-Jamayran A., Tsirigos A., Bu L., Fishman G.I., Park D.S. ETV1 activates a rapid conduction transcriptional program in rodent and human cardiomyocytes. Sci. Rep. 2018;8:9944. doi: 10.1038/s41598-018-28239-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekhar A., Lin X., Liu F.Y., Zhang J., Mo H., Bastarache L., Denny J.C., Cox N.J., Delmar M., Roden D.M., et al. Transcription factor ETV1 is essential for rapid conduction in the heart. J. Clin. Invest. 2016;126:4444–4459. doi: 10.1172/JCI87968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Wymore R., Yu H., Wu J., Wymore R.T., Pan Z., Robinson R.B., Dixon J.E., Mckinnon D., Cohen I.S. Distribution and prevalence of hyperpolarization-activated cation channel (HCN) mRNA expression in cardiac tissues. Circ. Res. 1999;85:e1–e6. doi: 10.1161/01.res.85.1.e1. [DOI] [PubMed] [Google Scholar]
- Spira M.E., Hai A. Multi-electrode array technologies for neuroscience and cardiology. Nat. Nanotechnol. 2013;8:83–94. doi: 10.1038/nnano.2012.265. [DOI] [PubMed] [Google Scholar]
- Stein A.B., Jones T.A., Herron T.J., Patel S.R., Day S.M., Noujaim S.F., Milstein M.L., Klos M., Furspan P.B., Jalife J., Dressler G.R. Loss of H3K4 methylation destabilizes gene expression patterns and physiological functions in adult murine cardiomyocytes. J. Clin. Invest. 2011;121:2641–2650. doi: 10.1172/JCI44641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzic A., Behfar A. Stem cell therapy for heart failure: ensuring regenerative proficiency. Trends Cardiovasc. Med. 2016;26:395–404. doi: 10.1016/j.tcm.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai S.Y., Maass K., Lu J., Fishman G.I., Chen S., Evans T. Efficient generation of cardiac Purkinje cells from ESCs by activating cAMP signaling. Stem Cell Rep. 2015;4:1089–1102. doi: 10.1016/j.stemcr.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eif V.W.W., Devalla H.D., Boink G.J.J., Christoffels V.M. Transcriptional regulation of the cardiac conduction system. Nat. Rev. Cardiol. 2018;15:617–630. doi: 10.1038/s41569-018-0031-y. [DOI] [PubMed] [Google Scholar]
- van Weerd J.H., Christoffels V.M. The formation and function of the cardiac conduction system. Development. 2016;143:197–210. doi: 10.1242/dev.124883. [DOI] [PubMed] [Google Scholar]
- Verheule S., Kaese S. Connexin diversity in the heart: insights from transgenic mouse models. Front. Pharmacol. 2013;4:81. doi: 10.3389/fphar.2013.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virani S.S., Alonso A., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Delling F.N., et al. Heart disease and stroke statistics-2020 update: a report from the American Heart Association. Circulation. 2020;141:e139–e596. doi: 10.1161/CIR.0000000000000757. [DOI] [PubMed] [Google Scholar]
- Walsh F.S., Doherty P. Neural cell adhesion molecules of the immunoglobulin superfamily: role in axon growth and guidance. Annu. Rev. Cell Dev. Biol. 1997;13:425–456. doi: 10.1146/annurev.cellbio.13.1.425. [DOI] [PubMed] [Google Scholar]
- Wang Y., Hill J.A. Electrophysiological remodeling in heart failure. J. Mol. Cell. Cardiol. 2010;48:619–632. doi: 10.1016/j.yjmcc.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washio T., Okada J.i., Hisada T. A parallel multilevel technique for solving the bidomain equation on a human heart with Purkinje fibers and a Torso model. SIAM J. Sci. Comput. 2008;30:2855–2881. [Google Scholar]
- Watson L.M., Wong M.M.K., Vowles J., Cowley S.A., Becker E.B.E. A simplified method for generating Purkinje cells from human-induced pluripotent stem cells. Cerebellum. 2018;17:419–427. doi: 10.1007/s12311-017-0913-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus A.M., Kaomei G., Shan J., Wellner M.C., Rohwedel J., Ji G., Fleischmann B., Katus H.A., Hescheler J., Franz W.M. Retinoic acid accelerates embryonic stem cell-derived cardiac differentiation and enhances development of ventricular cardiomyocytes. J. Mol. Cell. Cardiol. 1997;29:1525–1539. doi: 10.1006/jmcc.1997.0433. [DOI] [PubMed] [Google Scholar]
- Wu H., Sun Y.E. Epigenetic regulation of stem cell differentiation. Pediatr. Res. 2006;59:21R–25R. doi: 10.1203/01.pdr.0000203565.76028.2a. [DOI] [PubMed] [Google Scholar]
- Xie X., Fu Y., Liu J. Chemical reprogramming and transdifferentiation. Curr. Opin. Genet. Dev. 2017;46:104–113. doi: 10.1016/j.gde.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Xin M., Olson E.N., Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z.D., Zhu W.N., Liu K.Z., Huang Z.P., Han Y.C. Small molecule epigenetic modulators in pure chemical cell fate conversion. Stem Cells Int. 2020;2020:8890917. doi: 10.1155/2020/8890917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Jiang J., Han P., Yuan Q., Zhang J., Zhang X., Xu Y., Cao H., Meng Q., Chen L., et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Melton D.A. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3:382–388. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Zobel C., Rana O.R., Saygili E., Bölck B., Saygili E., Diedrichs H., Reuter H., Frank K., Müller-Ehmsen J., Pfitzer G., Schwinger R.H.G. Mechanisms of Ca2+-dependent calcineurin activation in mechanical stretch-induced hypertrophy. Cardiology. 2007;107:281–290. doi: 10.1159/000099063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This video shows the iPSC-CM cells used for PURK differentiation were functionally myocytes, that beat spontaneously, despite being cultured on a Fibrin matrix. The video magnification is at 4X.
This video shows the iPSC-CM cells used for PURK differentiation were functionally myocytes, that beat spontaneously, despite being cultured on a Fibrin matrix. The video magnification is at 10X.
This video shows the iPSC-CM cells used for PURK differentiation were functionally myocytes, that beat spontaneously, despite being cultured on a Fibrin matrix. The video magnification is at 20X.
This video shows the time-lapse progress of AC16-CM control cells being cultured on a Fibrin matrix for 7 days. No significant changes were observed in the cell’s morphology. The video magnification is at 10X.
This video shows the time-lapse progress of AC16-CM cells treated with PURK cocktail and cultured on a Fibrin matrix for 7 days. Significant changes were observed in the cell’s morphology and organization. The cells elongate and form “net-like” structures resembling Purkinje Fibers. The video magnification is at 10X.
The activation map of the control cells reveals a very slow pulsating-like activation from the cells, rather than the conduction of the electrical stimulation.
The activation map of the PURK-cocktail-treated cells show that the cells were rapidly activated upon electrical stimulation and produced a very strong electrical signal propagation throughout the whole dish.
Data Availability Statement
-
•
Data
Raw RNA-seq data of PURK-cocktail treated cells at day-4 and day-7 that supports the findings of this study has been deposited in the Figshare database, is publicly available, and can be accessed at (https://doi.org/10.6084/m9.figshare.21290883.v1).
-
•
Code
This paper does not report original code.
-
•
Additional Information
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.