Summary
In mammals, nicotinamide (NAM) is the primary NAD precursor available in circulation, a signaling molecule, and a precursor for methyl-nicotinamide (M-NAM) synthesis. However, our knowledge about how the body regulates tissue NAM levels is still limited. Here we demonstrate that dietary vitamin B3 partially regulates plasma NAM and NAM-derived metabolites, but not their tissue levels. We found that NAD de novo synthesis from tryptophan contributes to plasma and tissue NAM, likely by providing substrates for NAD-degrading enzymes. We also demonstrate that tissue NAM is mainly generated by endogenous metabolism and that the NADase CD38 is the main enzyme that produces tissue NAM. Tissue-specific CD38-floxed mice revealed that CD38 activity on endothelial and immune cells is the major contributor to tissue steady-state levels of NAM in tissues like spleen and heart. Our findings uncover the presence of different pools of NAM in the body and a central role for CD38 in regulating tissue NAM levels.
Subject areas: Biological sciences, Biochemistry, Molecular biology, Immunology
Graphical abstract

Highlights
-
•
Different pools of nicotinamide (NAM) exist in the organism
-
•
NAD de novo synthesis and dietetic NAD precursors regulate the plasma NAM pool
-
•
CD38 in endothelial and immune cells regulates tissue NAM, but not plasma NAM pool
-
•
Tissue NAM used for M-NAM synthesis is a CD38-independent pool
Biological sciences; Biochemistry; Molecular biology; Immunology
Introduction
The discovery that NAD levels decline during aging and its association with many aging-related diseases (Camacho-Pereira et al., 2016; Chini et al., 2020; Covarrubias et al., 2020; dePicciotto et al., 2016; Gomes et al., 2013; Scheibye-Knudsen et al., 2014; Tarrago et al., 2018; Williams et al., 2017; Yoshino et al., 2011) raised interest to develop NAD-replacement therapies for human diseases (Chini et al., 2021; Radenkovic et al., 2020). Nonetheless, many aspects of the NAD precursors biochemistry are still obscure, limiting our capacity to design effective approaches for NAD precursors supplementation. Therefore, it is fundamental to understand the sources of NAD precursors, their transport, and metabolism within the body, including the involved cell types.
NAD-degrading enzymes like Poly (ADP-ribose) polymerases (PARPs), the protein deacetylases Sirtuins, and the ADP-ribose hydrolases such as CD38 degrade NAD molecules for signaling purposes (Katsyuba et al., 2020). Although PARPs are more active in specific contexts such as DNA damage (Brown et al., 2021; Cameron et al., 2019), sirtuins participate in stress responses (Kosciuk et al., 2019; Mao et al., 2011; Yang et al., 2016), and CD38 expression and activity increases in inflammatory conditions (Chini et al., 2020; Covarrubias et al., 2020; Piedra-Quintero et al., 2020). Although PARP1/2 and Sirtuins1/2 -account for most NAD-degrading activity in cultured cells under basal conditions (Liu et al., 2018), CD38 is the main NAD-degrading enzyme that regulates tissue NAD levels in physiological conditions in vivo (Aksoy et al., 2006). Therefore, organisms must constantly counterbalance NAD consumption with NAD synthesis to maintain NAD homeostasis.
A common product of these NADases is nicotinamide (NAM). In mammals, the NAD salvage pathway, in which the enzyme nicotinamide-phosphoribosyltransferase (NAMPT) is the rate-limiting step, recycles NAM generated by NAD breakdown or taken up from the circulation for NAD synthesis (Liu et al., 2018; Revollo et al., 2004; Rongvaux et al., 2002). NAD synthesis can also take place through the de novo pathway from tryptophan, the Preiss-Handler pathway from nicotinic acid, and the salvage pathway from nicotinamide mononucleotide (NMN) or nicotinamide riboside (NR) (Katsyuba et al., 2020). Although the body can use multiple precursors for NAD synthesis, NAM resulting from NAD breakdown in the liver – mainly coming from tryptophan – appears to be the primary circulating NAD precursor for most tissues (Liu et al., 2018). Thus, NAM is both a critical NAD precursor and a product of NAD degradation.
The roles of NAM appear to go beyond NAD synthesis. For instance, studies have proposed that NAM can inhibit sirtuins and PARPs in vitro (Bitterman et al., 2002; Dominguez-Gomez et al., 2015; Ungerstedt et al., 2003), in vivo (Green et al., 2008; Hathorn et al., 2011; Turunc Bayrakdar et al., 2014) and that it may possess anti-inflammatory activity (Hiromatsu et al., 1992, 1993; Kumakura et al., 2021; Ungerstedt et al., 2003). Recently, NAM has been recognized as a kinase inhibitor for p38 MAPK, Rho-associated protein kinase (ROCK), casein kinases (CK1), and others, regulating stem cell differentiation and survival (Meng et al., 2018, 2021). In addition, NAM is also a precursor for methyl-NAM (M-NAM) synthesis, a metabolite with immunomodulatory (Kilgour et al., 2021), vasoprotective (Bar et al., 2017), antithrombotic (Mogielnicki et al., 2007), and metabolic functions (Chen et al., 2021; Miwa, 2021; Takeuchi et al., 2018; Zhang et al., 2020). Therefore, NAM is not only a precursor for biologically active metabolites, but it also may directly act as a signaling molecule.
Here we explored currently underappreciated aspects of the NAM metabolism, identifying different pools of NAM in the body under physiological conditions. We demonstrate that although the diet regulates plasma levels of NAM, CD38 activity on endothelial and immune cells controls tissue levels of NAM in vivo. Our data indicate that NAM metabolism is complex and comprises the interplay between diet and tissue compartments.
Results
Dietary precursors regulate plasma levels, but not tissue levels of NAM
To address whether dietary NAD precursors regulate NAM levels or other NAD precursors in the body, we measured the levels of NAM, NAM-derived metabolites (M-NAM, pyridones N-methyl-2-pyridone-5-carboxamide (2-PY), and N-methyl-4-pyridone-3-carboxamide (4-PY)), NMN, and NR in plasma, urine, and tissues of mice fed a diet free of NAM (and other NAD precursors such as NMN and NR, or nicotinic acid) for 21 days. We observed that the levels of NAM and the NAM-derived metabolites M-NAM, 2-PY, and 4-PY were 55–67% lower in the plasma and 43–52% lower in the urine from mice fed a NAM-free diet compared to those fed a control diet from day 15 on (Figures 1A and 1B). However, tissue levels of NAM were similar in both diet groups after 21 days on diet (Figure 1C), except for the lung (with NAM and M-NAM levels that were on average 35 and 48% lower, respectively) and the spleen (with M-NAM levels on average 46% lower) in mice fed a NAM-free diet compared to those on control diet. In contrast, NAD levels were stable in all tissues (Figure 1C).
Figure 1.
Dietary precursors regulate plasma levels, but not tissue levels of NAM
(A–C) 3–5-month-old male mice were fed a control diet (CD) and a NAM-free diet for 21 days (n = 5 mice/group). (A) Plasma metabolites on days 8, 15, and 21. (B) Urine metabolites on day 15. (C) Tissues metabolites on day 21.
(D and E) 3-month-old hACMSD mice were fed a NAM-free diet in the presence of doxycycline (to induce hACMSD expression) for 3 and 13 weeks (n = 4–6 mice/group). (D) Levels of tissue metabolites relative to control diet (NMN was not detected in the spleen and NR was not detected in the liver and WAT). (E) NAM levels on the plasma.
(F) Expression of Parp1, Sirt1, and Cd38 by qPCR in tissues. Tissue abbreviations used in all figures are jejunum (JEJ), spleen (SPL), liver (LIV), white adipose tissue (WAT), lung (LUN), heart (HT), kidney (KID), skeletal muscle (MUS), and brain (BRA).
Error bars represent mean ± SD and asterisks indicate significant differences between control diet and NAM-free diet (t-test). In all figures: ∗, p < 0,05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. See also Figure S1.
An endogenous source of tissue NAM that could be compensating for a lack of dietary NAM is the NAD synthetic pathway from tryptophan. To investigate this possibility, we used a transgenic mouse model in which the human enzyme α-amino-β-carboxy-muconate-semialdehyde decarboxylase (hACMSD) is controlled by a tetracycline-inducible promoter (Palzer et al., 2018). Doxycycline (Dox)-induced overexpression of hACMSD in these mice converts the intermediate tryptophan metabolite ACMS into aminomuconic semialdehyde instead of quinolinic acid. It thereby removes this intermediate from the NAD de novo synthesis pathway (Palzer et al., 2018). In this model, hACMSD expression is only induced in tissues that naturally express this enzyme, such as the liver and kidney (Pucci et al., 2007) (Figure S1A), which are also the tissues that mostly perform NAD+de novo synthesis (Liu et al., 2018). Thus, hACMSD overexpression significantly impairs NAD synthesis from tryptophan, and we investigated if this was sufficient to cause a NAM-deficient state when combined with a NAM-free diet (Palzer et al., 2018).
We harvested tissues from hACMSD overexpressing mice that had been fed either a NAM-free or control diet for 3 weeks (21 days) and 13 weeks and measured NAD-related metabolite levels (Figure 1D). Of interest, in a tissue that perform NAD de novo synthesis – liver (Liu et al., 2018) – NAM levels decreased only after mice were fed a NAM-free diet for a long period (13 weeks), but not in the short-term (3 weeks). (Figure 1D). However, levels of these metabolites in white adipose tissue (WAT) – a tissue with low expression levels of de novo pathway genes (Figure 3A) – were similar in both groups even after 13 weeks of diet (Figure 1D). Analyzing plasma, we observed a decrease in NAM, M-NAM, NR, 2PY, and 4PY in NAM-free diet-fed mice (Figures 1E and S1B). In addition, hACMSD induction alone did not promote any changes in tissue NAM (Figure S1C), reinforcing that both a reduction in the NAD+ de novo synthesis and a NAM-free diet are required to decrease tissue NAM levels in mice. Thus, NAD synthesis from tryptophan seems to play a role in plasma and have limited effect in tissue NAM pools.
Figure 3.
CD38 activity correlates with salvage pathway flux in different tissues
(A) Gene expression of the NAD degradation (Cd38, Parp1, Sirt1, Sarm1), salvage (Nampt, Nmnat1-3, Naprt, Nrk), excretion (Nnmt, Aox1, Aox3, Cyp2e1), de novo synthesis (Ido1, Ido2, Kynu, Haao, Qprt), and repair (Apoa1pb, Carkd) pathways in different tissues of 3-month-old WT male mice (average of n = 4). The data is expressed as log10 of the delta Cq relative to the Tbp gene.
(B–G) NADase activity measured in tissues of 3 months-old male WT mice was plotted against: (B) Cd38 mRNA expression, (C) NAD half-life (data from Liu et al., 2018), (D) NAD+ levels, (E) NAM levels, (F) NAM exchange flux (data from Liu et al., 2018), and (G) NAM recycling flux (data from Liu et al., 2018) (average of n = 4).
See also Figures S2, S3, and Table S2.
Of interest, ADP-ribose (ADPR) levels, a product of NADases, decreased in the liver and especially in the spleen of ACMSD-overexpressing mice fed a NAM-free diet compared to the control diet (Figure 1D). This result suggests that NAD-degrading enzymes may contribute to NAM levels in these tissues. To investigate this possibility, we analyzed tissue expression of Parp1, Sirt1, and Cd38 – the main NADases in the body. In spleen, we found that at 13 weeks Cd38, but not Parp1 and Sirt1 expression, was reduced by about 60% in hACMSD-overexpressing mice fed a NAM-free diet relative to control diet (Figure 1F), and this occurs while changes in ADPR and NAM were observed in this tissue. However, the expression of these genes was comparable in the liver and fat on either diet (Figure 1F). This may explain why NAM levels were more affected in the spleen compared to the liver and WAT.
Overall, these results indicate that dietary NAM regulates plasma levels of NAM, whereas other mechanisms involving the NAD de novo synthesis pathway from tryptophan in combination with a NAM-free diet and the NAD degradation pathway seem to maintain tissue levels of this metabolite. However, it appears that, at least in the case of the spleen, the effect could be mediated by changes in CD38 because both NAM and ADPR decreased. Therefore, next we explored the potential role of endogenous tissue metabolism as a source of tissue NAM.
The steady-state levels of NAM correlate with NAM exchange flux and NAD salvage pathway flux in multiple tissues
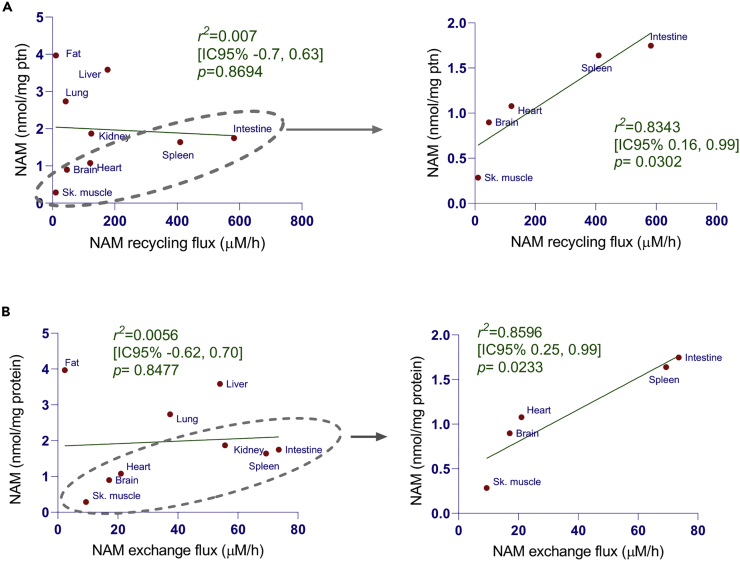
In tissues, a combination of three different mechanisms may regulate the steady-state levels of NAM: NAM production by NAD-degrading enzymes, its recycling flux through the salvage pathway, and its uptake from plasma. To address possible contributions of these different pathways to NAM steady-state levels, we made a set of observational analysis to find correlations between NAM levels and NAM fluxes through the NAD salvage pathway or NAM exchange (between tissues and plasma) in different tissues. For this, we plotted NAM concentrations measured in C57BL/6 mouse tissues against NAM fluxes through salvage and exchange pathways using data provided elsewhere (Liu et al., 2018) (Figure 2). Overall, we found no correlation between NAM levels and NAM fluxes through the salvage pathway (NAM recycling flux) (Figure 2A) and the NAM exchange flux (Figure 2B). However, NAM levels correlate with NAM fluxes through the salvage pathway and exchange pathway in a particular subset of tissues (muscle, brain, heart, spleen, and intestine), with r2 values of 0.83 (p = 0.0302) and 0.86 (p = 0.0233), respectively. Of interest, the tissues that do not fit the correlation (liver, fat, lung, and kidney) have the highest expression of NAM excretion pathway genes compared to other tissues (Figure 3A), suggesting that flux through M-NAM and N-oxide formation may influence the steady-state levels of NAM in these tissues.
Figure 2.
The steady-state levels of NAM correlate with NAM fluxes in some tissues
(A and B) NAM levels data measured in 3–5 months-old C57BL/6 male mice tissues (average of n = 4) against NAM fluxes through salvage and exchange pathways using data previously published (Liu et al., 2018). See also Table S2.
CD38 activity highly correlates with NAM flux through the salvage pathway in different tissues
To investigate how tissues regulate their steady-state levels of NAM, we did an observational analysis of gene expression related to the NAD metabolome in different tissues. Because NAD half-life seems to mainly reflect the activity of NAD consumers in different tissues (Liu et al., 2018; McReynolds et al., 2021), and because flux through different pathways may also influence the steady-state levels of NAM, we quantified mRNA expression of genes of the NAD degradation (Cd38, Parp1, Sirt1, Sarm1), salvage (Nampt, Nmnat1-3, Naprt, Nrk), excretion (Nnmt, Aox1, Aox3, Cyp2e1), de novo synthesis (Ido1, Ido2, Kynu, Haao, Qprt), and repair (Apoa1pb, Carkd) pathways. We used the expression levels of the housekeeping gene Tbp as a reference, because its expression had low variation among the different tissues tested (Figures S2A and S2B). To search for obvious correlations in this screening, we arranged the mRNA data from different tissues in order of NAD half-life, according to Liu et al. (2018). Note that we excluded skeletal muscle data specifically from the correlation analysis with NAD half-life (Figure 3C) because this tissue behaved as an outlier due to its very long NAD half-life. As shown in Figure 3A, Cd38 expression seem to inversely correlate with NAD half-life, indicating that this gene may be the primary driver for NAD turnover in tissues. Moreover, although Nampt expression appeared to correlate positively with NAD half-life (Figure 3A), further analysis revealed no correlation (Figure S3A). We confirmed a high correlation of Cd38 expression and NADase activity (r2 of 0.79, p = 0.0013, Figures 3B and S3B) and NAD half-life (r2 of 0.69, p = 0.0103 Figure 3C) in different tissues. However, NAD+ levels poorly correlate with NADase activity (Figure 3D), indicating that other factors may contribute to the steady-state levels of NAD+ in tissues.
To determine whether CD38 activity contributes to NAM steady-state levels, we plotted NAM concentration against NADase activity in different tissues (Figure 3E). Consistent with the data in Figure 2, NAM levels correlate well with NADase activity only if we exclude those tissues that highly express the NAM excretion pathway (r2 of 0.75, p = 0.0557, Figures 3E, S2C, and S2D). Also, NADase activity only moderately correlates with NAM exchange flux (r2 of 0.54, p = 0.0240 Figure 3F) but strongly correlates with NAM recycling flux through the NAD salvage pathway (r2 of 0.89, p = 0.0001 Figure 3G).
Overall, these data suggest that CD38 activity contributes to the NAD turnover in different tissues in young mice, promoting constitutive NAD breakdown, presumably counteracted by NAM flux through the salvage pathway. Also, CD38 expression and activity seem to correlate with the steady-state levels of NAM in most of the tissues under physiological conditions.
CD38 regulates tissue levels of NAM, but not plasma levels
As aforementioned, the expression and activity of the NADase CD38 correlate with tissue NAM levels. To confirm the relationship between CD38 and tissue NAM, we measured the concentrations of NAM and NAM-derived metabolites (M-NAM, 2-PY, 4-PY) in tissues, plasma, and urine of young wild type (WT) and CD38-KO mice. Our previous work on CD38-KO mice found low NADase activity and increased NAD+ levels in all tested tissues compared to WT mice (Aksoy et al., 2006; Chini et al., 2002). Surprisingly, CD38-KO mice contained 3-25 times (average 13 times) less NAM in different tissues than WT mice (Figure 4A), although plasma levels of NAM were similar (Figure 4B). In addition, urine from CD38-KO revealed around two times less NAM (Figure 4C). This difference may reflect the lower levels of NAM in the kidney (Figure 4A) or represent how the body regulates plasma NAM. Remarkably, levels of M-NAM, a NAM-derived metabolite, were comparable between WT and CD38-KO mice (Figure 4A), indicating that the NAM used for M-NAM synthesis comes from a CD38-independent source within the tissues. As a control for our M-NAM measurements, we measured M-NAM levels in WT and KO mice for the enzyme nicotinamide-N-methyl-transferase (NNMT) (Brachs et al., 2019). NNMT uses NAM and the universal methyl donor S-adenosyl-methionine (SAM) as substrates for M-NAM synthesis (Alston and Abeles, 1988; Scheller et al., 1996). In contrast to CD38-KO mice, we did not detect M-NAM in NNMT-KO mice (Figure S4A). Moreover, levels of M-NAM and derived metabolites (2-PY and 4-PY) were similar in tissues, plasma, and urine from WT and CD38-KO mice (Figures 4A–4C and S4B).
Figure 4.
Tissue levels of NAM are lower in the absence of CD38 activity
(A–C) Levels of NAM and NAM-derived metabolites (M-NAM, 2-PY, 4-PY) in: (A) tissues, (B) plasma, and (C) urine of 3–5-month-old male WT and CD38-KO mice (n = 4).
(D) NAM and M-NAM levels in tissues from 3–5-month-old male WT and CD38-CI mice (n = 5–8 mice/group).
(E and F) 3-month-old male WT mice were injected i.p. with vehicle (Ctrl, NMN groups), 5 mg/kg Ab68 (Ab68, Ab68 + NMN groups), with and without 500 mg/kg NMN. Tissue NAM and M-NAM levels expressed relative to control (Ctrl) (n = 5 mice per group).
(G) Total levels of NAM in all NAM-containing metabolites in tissues of 3–5-month-old WT and CD38-KO mice (n = 4).
(H) Fraction of NAM-containing metabolites compared to the sum of all NAM-containing metabolites.
Error bars represent mean ± SD and asterisks indicate significant differences (B–D, and G, t-test; E and F, one-way ANOVA, and Dunnett’s post-test). See also Figure S4.
We also measured the expression of several genes involved in the NAD metabolome in WT and CD38-KO mice. For most of these genes, we did not detect significant differences between WT and CD38-KO mice (Figure S4C). A few exceptions were some enzymes on the salvage pathway being decreased in a few tissues and some enzymes on the NAM excretion pathway showing an increase in some tissues of the CD38-KO mice. In conclusion, CD38 appears to regulate NAM in tissues without major alterations in gene expression of NAD metabolism.
We also observed that NAM levels, but not M-NAM, were lower in mice expressing the catalytically inactive form of CD38 (CD38-CI, Figure 4D). Our previous characterization of the CD38-CI mouse model found no NADase activity and increased tissue NAD+ levels (Tarrago et al., 2018). Hence, our present results in CD38-CI mice confirm that CD38 regulates tissue NAM through its enzymatic activity.
Because genetic models may be limited due to possible compensatory mechanisms adopted by the organism, we tested whether acute CD38 inhibition could decrease tissue NAM. For that, we injected mice (i.p.) with a neutralizing antibody against CD38 (Ab68) that inhibits its catalytic activity (Chini et al., 2020) and/or administered NMN, a CD38 substrate (Camacho-Pereira et al., 2016; Chini et al., 2020). In line with genetic models, treatment with Ab68 decreased NAM levels in different tissues (Figure 4E). NMN administration alone increased NAM levels in most tissues, and Ab68 treatment consistently reduced tissue NAM even in the presence of NMN (Figure 4E). These data reinforce that CD38 is the main NMNase in tissues and demonstrate that NMN is a CD38 substrate to generate NAM. Although tissue M-NAM levels increased with NMN treatment, CD38 inhibition did not influence M-NAM levels even in NMN-treated mice (Figure 4F). This evidence emphasizes that M-NAM synthesis occurs in a tissue compartment that generates or takes NAM up independent of CD38.
An unsolved question in the literature is the identity of the main liver NADase that produces NAM and allows its redistribution to other tissues within the body through the circulation (Liu et al., 2018). If CD38 is the main NADase involved in this process, we would expect that CD38-depleted mice fed a NAM-free diet would suffer from NAM deficiency in the plasma and show lower NAD levels systemically. To investigate this possibility, we fed CD38-CI mice a NAM-free diet for 21 days, similarly to Figure 1A, and measured metabolite levels in different tissues and plasma. Our results showed that the plasma and tissue NAD metabolome of CD38-CI mice is comparable during NAM-free or control diet (Figures S5A and S5B), further confirming that CD38 regulates tissue, but not plasma NAM.
Because NAM is a product of CD38-dependent NAD degradation, we investigated if CD38 inhibition induced a switch in the abundance of free versus bound forms of NAM in tissues. To address this, we calculated the total tissue NAM fractions as the sum of the free NAM amounts with the amounts of known NAM-containing intermediates: NAD, NMN, NR, and M-NAM. As shown in Figure 4G, the total NAM fractions were similar in WT and CD38-KO mice, except for WAT. Subsequently, we calculated how much of the total NAM fraction corresponded to the amounts of each metabolite in WT and CD38-KO tissues (except WAT). From all metabolites, free NAM and NAD-bound NAM represented the most abundant forms of tissue NAM, whereas NMN, M-NAM, and NR represented only a tiny fraction of the total tissue NAM forms (Figure 4H). We also detected a shift from free NAM to NAD-bound NAM abundance in CD38-KO tissues compared to WT (Figure 4H). Of interest, this shift in the NAD-bound NAM fraction was less prominent in tissues with lower CD38 expression (Figure 4H), reinforcing that CD38 is the main NADase regulating NAD and NAM tissue levels. In addition, it appears that CD38 regulates the ratio of the free versus incorporated NAM in tissues.
Other NAD-degrading enzymes do not regulate tissue NAM levels
We next examined whether other NADases contribute to the regulation of tissue NAM (Figure 5A). Measuring NAM levels in tissues of Sarm1-KO, Parp1-KO, Sirt1-KO and CD38-KO mice, we found reduced NAM levels only in CD38-KO mice, whereas those of WT, Parp1-KO, Sarm1-KO, and Sirt1-KO mice were similar (Figures 5A and S5C). Consistent with this finding, i.p. injections of olaparib, a Parp-1 and -2 inhibitor (Menear et al., 2008), did not significantly decrease tissue NAM in mice, whereas Ab68 treatment promoted a quick drop in tissue NAM, which was already evident 2 h after injection (Figure 5B). Thus, our data suggest that CD38, but not other NADases, regulates tissue NAM levels.
Figure 5.
CD38, but not of other NAD degrading enzymes, regulates tissue NAM levels
(A) NAM levels in 3–5-month-old male WT, PARP-KO, SARM1-KO, and CD38-KO mice (n = 3–5 mice/group). Values in KO mice were calculated relative to their respective control WT mice.
(B) NAD+ and NAM levels in 3-month-old WT male mice injected i.p. with 5 mg/kg Ab68 or Olaparib. Values were calculated relative to 0 h (n = 4 mice/time point).
Error bars represent mean ± SD, asterisks indicate significant differences using one-way ANOVA and Dunnett’s post-test (A) or two-way ANOVA and Šidák’s post-test (B).
See also Figure S5.
CD38 in endothelial and immune cells is the primary source of NAM in tissues
We next aimed to determine which cell types within the tissues generate NAM. CD38 expression is widely reported for immune and endothelial cells (Boslett et al., 2018; Chini et al., 2019, 2020; Covarrubias et al., 2020; Matalonga et al., 2017; Piedra-Quintero et al., 2020). Thus, we generated hematopoietic and endothelial cell-specific CD38-deficient mouse lines, namely CD38 Flox/Vav1-iCre and CD38 Flox/Tek-Cre. We bred CD38-floxed mice with two commercially available Cre recombinase lines: iCre mice, with improved Cre recombinase expression under control of the vav1 oncogene promoter to target deletion of loxP-flanked genes in hematopoietic cells (deBoer et al., 2003; Ogilvy et al., 1999; Shimshek et al., 2002), and TIE2Cre mice, with Cre-recombinase under the control of the receptor tyrosine kinase Tek promoter to target endothelial and hematopoietic cells (Koni et al., 2001) (Figure 6A). The CD38 Flox/Vav1-iCre line serves both to address the participation of immune cells in NAM generation and, by exclusion, to discriminate the participation of endothelial cells from immune cells in CD38 Flox/Tek-Cre mice.
Figure 6.
CD38 in endothelial cells is the main source of NAM in tissues
(A) Schematic representation of the generation of CD38 Flox/Vav1-iCre and CD38 Flox/Tek-Cre mouse models.
(B) Immunoblots of CD38 in 3–5-month-old CD38 Flox, CD38 Flox/Vav1-iCre, CD38 Flox/Tek-Cre, and CD38-KO male mice.
(C) Immunostaining of CD38, CD31, and CD45 in spleen and heart.
(D) NAM levels and NADase activity in CD38 Flox, CD38 Flox/Vav1-iCre, and CD38 Flox/Tek-Cre (n = 4 mice/group). Data are expressed relative to the control CD38 Flox mice for each tissue.
Error bars represent mean ± SD and asterisks indicate significant differences using one-way ANOVA and Dunnett’s post-test. See also Figure S6.
To characterize these new mouse models, we examined CD38 protein expression in tissues of CD38 Flox/Vav1-iCre and CD38 Flox/Tek-Cre mice by immunoblots (Figures 6B, S6A, and S6B). Within spleen, a tissue mainly constituted by immune cells, we virtually detected no CD38 in both lines, whereas in liver and heart CD38 expression was lower only in CD38 Flox/Tek-Cre samples (Figure 6B). This finding indicates that CD38 is mostly derived from immune cells in spleen, and endothelial cells in liver and heart. Of interest, in the intestine (jejunum), we only saw a minor decrease in CD38 levels in both mouse models (Figure S6A), indicating that most CD38 is present in other cell types. We also analyzed CD38 distribution in heart and spleen by imaging and flow cytometry (Figures 6C, S6C, and S6D). Both techniques confirmed the presence of CD38 in endothelial cells from the CD38 Flox/Vav1-iCre mice but not from CD38 Flox/Tek-Cre mice. We also corroborated CD38 deficiency in spleen, and its presence in the jejunum, in both mouse models by immunostaining (Figures 6C and S6D).
We measured NAM and NAD+ in different tissues from CD38 Flox/Vav1-iCre and CD38 Flox/Tek-Cre mice (Figures 6D and S6E). Except for the intestine and brain, CD38 deficiency of endothelial and immune cells (CD38 Flox/Tek-Cre) resulted in remarkably low tissue NAM levels, concomitant with a decrease in NADase activity (Figure 6D). On the other hand, mice depleted of CD38 in immune cells (CD38 Flox/Vav1-iCre) showed low NAM levels and NADase activity only in spleen, but not in the other tissues (Figure 6D), making it likely that immune cells are the main contributors to tissue NAM within the spleen. However, the involvement of splenic endothelial cells cannot be entirely excluded. Therefore, in young mice, under non-inflammatory conditions, endothelial cells appear to be the primary source of NAM in most tissues. These data are surprising because it has been assumed that the main source of CD38 enzymatic activity in vivo is the immune cells. However, our data provide clear evidence that endothelial cells are a significant source of CD38 NADase activity and key regulators of tissue NAM levels in young healthy mice. It does not exclude a possible role of CD38+ immune cells as important generators of NAM during aging or inflammation.
CD38 from immune cells also contributes to NAM generation
To confirm the contribution of immune cells to NAM generation, we transplanted bone marrow from WT and CD38-KO mice into irradiated CD38-KO mice (WT > KO and KO > KO, respectively). Afterward, we measured NAM levels in organs with high immune cell infiltration under physiological conditions, such as spleen and jejunum, and in organs with naturally low immune cell infiltration, such as heart, WAT, and liver. Bone marrow transplants from WT into CD38-KO mice resulted in an almost complete recovery of NAM levels in spleen, relative to WT, consistently with an increase of splenic NADase activity (Figure 7A). Jejunum showed a 55% recovery in NAM levels and a 21% recovery of NADase activity after transplant. The prominent recovery of NAM levels in these organs may be a result of a reminiscent inflammation and cellular senescence caused by the irradiation (necessary for the depletion of bone marrow cells in the recipient mice). Moreover, NAM levels and NADase activity recovered only slightly in other organs, agreeing with low immune cells infiltration in these tissues in young mice (Figure 7A). As expected, CD38-KO mice transplanted with CD38-KO bone marrow maintained low tissue NAM levels (Figure 7A). This reinforces that immune cell are the primary NAM generator in spleen and seem to contribute only marginally to NAM levels in other tissues.
Figure 7.
CD38 from immune cells contributes to NAM generation
(A) Sublethally irradiated 3–5-month-old male CD38 KO mice were subjected to bone marrow transplantation (BMT) with 1 × 106 bone marrow cells (BMCs) per animal from either WT (WT > KO) or CD38 KO donors (KO > KO). Twelve weeks after transplantation, tissues were harvested, and NAM levels and CD38 activity were measured. Graphs show data relative to WT mice (n = 4 mice per group in the BMT and n = 5–23 for WT control mice).
(B) 3–5-month-old male WT mice were treated with anti-CD4, CD8, and CD20 cytotoxic antibodies. Levels of NAM were measured in the spleen (n = 4).
(C–F) Single cell analysis of RNA-seq data for the spleen of 3 months-old C57BL/6 mice (data from Tabula Muris Consortium). (C) UMAP projections of RNA-seq data. (D) CD38 gene expression on UMAP projection. (E) Violin plot showing CD38 expression across different cell clusters. Expression value is shown as log normalized counts. (F) Dot plot showing CD38 expression across the cell clusters.
Error bars represent mean ± SD and asterisks indicate significant differences using one-way ANOVA and Dunnett’s post-test (A, compared to WT mice; B, compared to IgG control) or Šidák’s post-test (C). See also Figure S7.
To investigate which cell types within spleen, produce NAM, we treated mice with cytotoxic antibodies for 48 h to deplete cytotoxic T cells (CD8+), T-helper cells (CD4+), and B cells (CD20+), successfully depleting these lymphocyte populations from spleen (Figure S7). CD4+ cell depletion resulted in a slight decrease of NAM levels in the spleen (22%), whereas depletion of CD8+ and CD20+ cells resulted in a 68 and 74% reduction, respectively (Figure 7B). Thus, the main cell types generating NAM in the spleen appear to be B and CD8+T cells. Single cell RNA-sequencing analysis from the spleen of 3-month-old mice, using data provided in The Tabula Muris Consortium (Tabula Muris, 2020), shows a considerable proportion of B cells in the spleen expressing CD38 at high levels compared to other cell clusters in this organ (Figures 7C–7F). Nonetheless, only a small proportion of CD4+ and CD8+ cells express CD38, at low levels, compared to the other cell clusters identified (Figures 7C–7F). So, other NAD-degrading enzymes may account for NAM generation in splenic CD8+ cells. Therefore, our data supports the notion that the main source of CD38 mediated NAM production in the spleen are the B cells. The role of B cells in tissue nicotinamide metabolism deserves further investigation.
Discussion
Despite the importance of NAM as a fundamental NAD precursor (Liu et al., 2018), a substrate for M-NAM synthesis (Alston and Abeles, 1988; Scheller et al., 1996), and a signaling molecule (Hiromatsu et al., 1992, 1993; Meng et al., 2021; Papaccio et al., 1999; Ungerstedt et al., 2003), it is not well understood how the body controls the levels of NAM. Our study uncovers three different pools of NAM in mice: (1) The circulating pool, which mainly comes from NAD synthesis from tryptophan and from the diet; (2) the CD38-dependent tissue NAM pool; and (3) the CD38-independent tissue NAM used for M-NAM synthesis. Importantly, we reveal a significant role of endothelial CD38 in maintaining NAM levels in most tissues, whereas immune cells contribute to NAM generation in the spleen, at least partially via CD38, under physiological conditions.
Recent findings surprisingly demonstrate that NAD turnover is high, with NAD half-life ranging from 15 min to 15 h depending on the tissue (Liu et al., 2018). However, the molecular mechanism responsible for this fast NAD turnover remains unknown. Because NAD levels seem to decline with aging as a result of increased CD38-dependent NAD consumption (Camacho-Pereira et al., 2016; Chini et al., 2020; Covarrubias et al., 2020; Tarrago et al., 2018) without concomitant flux compensation through the salvage pathway (McReynolds et al., 2021), we investigated if CD38 contributes to the NAM fluxes under physiological conditions. Our results indicate that the NAD synthesis through the salvage pathway mainly counterbalances CD38 activity in young mice, explaining the high correlation between CD38 activity and NAM recycling flux. As previously proposed, these findings validate CD38 as the main NADase in tissues (Aksoy et al., 2006; Chini et al., 2020) and indicate that CD38 may be the primary contributor to the fast tissue NAD turnover under physiological conditions.
Although rodents are resistant to diet-induced pellagra (Fukuwatari et al., 2008; Palzer et al., 2018; Terakata et al., 2013), we demonstrate that dietary vitamin B3 deficiency affects plasma and urine levels of NAM and NAM-derived metabolites in mice. However, this decline of circulating NAM has no direct effect on the tissue NAD metabolome, including tissue NAM levels. This fact probably reflects the robustness by which the body performs NAD synthesis from tryptophan, as suggested by previous studies (Fukuwatari et al., 2008; Palzer et al., 2018; Terakata et al., 2013), and recycles NAM through the salvage pathway. Accordingly, only under conditions of a long-term NAM-free diet together with an impairment of the NAD de novo synthesis pathway (by overexpressing the hACMSD enzyme in mice), NAM levels decreased in some tissues. However, the molecular mechanism regulating the remaining plasma NAM (in mice fed a NAM-free diet) is still unclear, but it does not seem to require CD38.
Thus, although mice lacking CD38 activity have much less tissue NAM, only its free (unconjugated form) levels decrease. So, upon CD38 inhibition, NAM molecules remain in tissues primarily incorporated as NAD molecules moieties. This observation agrees with the role of CD38 as a regulator of NAD levels in the body (Aksoy et al., 2006; Camacho-Pereira et al., 2016; Chini et al., 2020). However, because the fraction of free (unconjugated NAM) is the precursor for M-NAM synthesis, it is unclear why M-NAM levels in tissues, plasma, and urine do not change upon CD38 inhibition. A potential explanation could be that different compartments exist within the tissues: a compartment where CD38 produces most tissue NAM and specific compartment(s) where M-NAM synthesis takes place using NAM that is not generated by CD38. Another possible explanation for the fact that the CD38 blockage does not affect M-NAM levels is that the amount of tissue NAM, although much lower in the absence of CD38, could still be sufficient to supply substrate for M-NAM synthesis. Indeed, tissue levels of M-NAM are on average 50 times lower than those of NAM, suggesting a low requirement for M-NAM synthesis. In addition, murine Nampt has a much higher affinity for NAM (KM = 0.92 μM) (Revollo et al., 2004) than Nnmt (KM = 370 μM) (Scheller et al., 1996), which could explain the low tissue levels of M-NAM. Therefore, it is unlikely that a drop in NAM levels would not impact M-NAM generation in tissues, considering the low affinity of NNMT for NAM. In conclusion, these observations support the idea of distinct compartments within the tissues where NAM pools are not interchangeable.
Further evidence supporting the existence of distinct tissue compartments for M-NAM synthesis is that the tissues with the highest NAM levels (liver, fat, and lung) express the highest levels of the excretion pathways genes, including Nnmt. Also, NAM levels in these tissues do not correlate with CD38 activity. Besides, NMN administration in mice promoted a 2- to 4-fold increase in M-NAM levels independently of CD38. Thus, it seems that tissues possess specific Nnmt-expressing compartments that do not take up CD38-generated NAM for M-NAM synthesis. Instead, they may generate their own NAM pool for M-NAM synthesis in a CD38-independent fashion and/or take up NAM from the circulation. However, the nature of this tissue compartment remains elusive. Whether endothelial cells accumulate CD38-generated NAM, or if this NAM is released to the extracellular space, is still an open question that we plan to address in future studies.
Our data demonstrate that immune cells produce NAM in the spleen. Considering the anti-inflammatory effects attributed to NAM, NAM released by immune cells could have immunomodulatory functions on the surrounding tissues during inflammation. For instance, NAM treatment was shown to (1) inhibit cytokine responses (IL-1β, IL-6, IL-8, TNFα) to endotoxin in whole human blood (Ungerstedt et al., 2003), (2) to diminish the IFN-γ-induced expression of adhesion proteins such as intercellular adhesion molecule-1 (ICAM-1) on thyroid and endothelial cells, which presumably reduces lymphocyte homing to inflammatory sites (Hiromatsu et al., 1992, 1993), and (iii) to inhibit cytokine-induced MHC class II antigens on pancreatic cells (Yamada et al., 1990). Administration of NAM in vivo attenuated cytokine induction in ischemia/reperfusion-induced lung injury in rats (Su et al., 2007); alleviated the progression of kidney disease in mouse models by reducing inflammation and fibrosis (Kumakura et al., 2021; Zhen et al., 2021); and reduced high-fat diet-induced steatosis and inflammation in mice (Mitchell et al., 2018). However, these studies did not investigate whether NAM has a direct effect as an anti-inflammatory molecule. Instead, a NAM-induced increase of M-NAM and/or NAD could be mediating this immunomodulatory function.
NAM was also shown to act as a signaling molecule by modulating protein kinases. NAM directly inhibits the activity of ROCK1/2, CK1 (α, β, and δ), and p38δ by interfering with their kinase-ligand interaction in vitro and promoting human embryonic stem cells differentiation (Meng et al., 2018, 2021). In addition, KINOMEscan assays revealed that NAM interacts with the active sites of multiple other kinases, including some involved in cellular inflammatory responses, such as IKKα, IKKβ, JAK2 JAK3, and JNK1 (Meng et al., 2018, 2021). Thus, the anti-inflammatory role of NAM may depend on its ability to inhibit protein kinases. However, irrespective of the mechanism, the possibility that immune cells may secrete NAM to prevent an exacerbated immune response and tissue damage during inflammation deserves further investigation.
In summary, we demonstrate the existence of distinct NAM pools in the body, which are differentially regulated. Although diet and NAD de novo synthesis regulate the plasma NAM pool, the NADase CD38 regulates the major tissue NAM pool. Furthermore, tissues possess a minor CD38-independent NAM pool dedicated to M-NAM synthesis. Finally, endothelial cells play a major role in regulating tissue NAM. These findings may pave the way for improved design of NAM-based therapies. For instance, many aspects of NAD precursor supplementation are currently attributed to NAD levels restoration. Instead, they may reflect increases in NAM or M-NAM levels. Therefore, it is essential to explore this subject further to identify the molecular pathways that regulate these different NAM pools, such as a putative NAM transporter.
Limitations of the study
In this study, we did not address if CD38-derived NAM accumulates intracellularly or if it accumulates in the extracellular space within the tissues. In other words, the nature of the tissue compartmentalization for NAM and M-NAM synthesis is still an open question. In addition, we have not done metabolite tracing analysis to address if CD38-generated NAM can produce M-NAM directly. Although we have identified the existence of different pools of NAM in a mouse model and that CD38 is an essential regulator of the tissue NAM pool, here, we did not explore the consequences of a disruption in CD38-generated tissue NAM levels in physiological or pathological conditions. We also have not investigated if CD38-derived NAM has some signaling or immunoregulatory role on endothelial, immune cells, or other cell types within the tissues. Finally, most of the experiments performed here were done in male mice only. So, we cannot exclude the possibility of sex differences.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse CD38 Affinity Purified Polyclonal Ab | R and D Systems | Cat# AF4947, RRID:AB_1241945 |
| CD45 antibody | Abcam | Cat# ab40763, RRID:AB_726545 |
| Phospholamban Monoclonal Antibody (2D12) | Thermo Fisher Scientific | Cat# MA3-922, RRID:AB_2252716 |
| CYP3A4 antibody | Proteintech | Cat# 18227-1-AP, RRID:AB_2090329 |
| β-Actin (D6A8) Rabbit mAb | Cell Signaling Technology | Cat# 8457, RRID:AB_10950489 |
| Mouse Anti-GAPDH Monoclonal Antibody, Unconjugated, Clone D4C6R | Cell Signaling Technology | Cat# 97166, RRID:AB_2756824 |
| Alexa Fluor 647 Rat Anti-Mouse CD38 | BD Biosciences | Cat# 562769, RRID:AB_2728651 |
| CD45 FITC | BD Biosciences | Cat# 553080, RRID:AB_394610 |
| Rat IgG2a, k | BD Biosciences | Cat# 557690, RRID:AB_396799 |
| Rat IgG2b, k | BD Biosciences | Cat# 553988, RRID:AB_479619 |
| Rabbit anti ACMSD | Novus Biologicals | Cat# NBP1-33499 |
| Anti-α-Tubulin antibody | Sigma Aldrich | Cat# T9026, RRID:AB_477593 |
| anti-mouse CD38 antibody Ab68 | TeneoBio | UniAb clone ID 337468 |
| anti-mouse CD8α | Bio X Cell | Cat# BE0061, RRID:AB_1125541 |
| anti-mouse CD4 | Bio X Cell | Cat# BE0003-1, RRID:AB_1107636 |
| αCD20Ab | Genentech, Roche Group | mlgG2a 5D2 |
| Chemicals, peptides, and recombinant proteins | ||
| Olaparib | LC Laboratories | Cat# O-9201 |
| Alcohol dehydrogenase (ADH) | Sigma-Aldrich | Cat# A3263 |
| Diaphorase | Sigma-Aldrich | Cat# A3263 |
| Resazurin sodium salt | Sigma-Aldrich | Cat# 199303 |
| ND1 – Niacin deficient diet | Teklad | Cat# TD140376 |
| CD1 – 10% casein diet | Teklad | Cat# TD140375 |
| Deposited data | ||
| Spleen AnnData (h5ad) | Tabula Muris Senis | https://cellxgene.cziscience.com |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | Dr.Eduardo N. Chini, Mayo Clinic | N/A |
| Mouse: C57BL/6 CD38KO | Dr.Eduardo N. Chini, Mayo Clinic (Partida-Sánchez et al., 2001) | N/A |
| Mouse: C57BL/6 CD38 catalytically-inactive (CI) | Dr. Eduardo Chini Mayo Clinic (Trans-Viragen) | N/A |
| Mouse: PARP1-KO: C57BL/6 | Dr. Eduardo Chini. Original from Jackson laboratory (129S-Parp1tm1Zqw/J) backcross to C57BL/6 for 10 generations | N/A |
| Mouse: SARM1-KO: C57BL/6 (B6.129X1-Sarm1tm1Aidi/J) | Jackson laboratory | RRID:IMSR_JAX:03439 |
| Mouse: SIRT1-KO tissues | Dr. Keir Menzies | N/A |
| Mouse: C57BL/6J-Gt(Rosa)26Sortm1(rTTa∗M2)JaeCol1a1tm6(tetO-hACMSD)MMF | Palzer et al. (2018) | N/A |
| Mouse: CD38 flox (Cd38fl/fl) | This study | N/A |
| Mouse: CD38 flox/Vav1-iCre | This study | N/A |
| Mouse: CD38 Flox/Tek-Cre | This study | N/A |
| Oligonucleotides | ||
| For primers, see Table S1 | Thermo Fisher Scientific | N/A |
Resource availability
Lead contact
Further information and requests for resources should be directed to the lead contact, Dr. Eduardo Chini (chini.eduardo@mayo.edu).
Materials availability
The original datasets and the tissue specific knockout mouse generated in this study can be made available to investigators based on reasonable requests and scientific collaborations.
Experimental model and subject details
Ethical compliance
All Protocols requiring the use of animals were approved by the Institutional Animal Care and Use Committee (IACUC) of Mayo Clinic and of Utah State University, and studies were conducted in adherence with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Mice were housed in standard cages at regulated temperature (ranging from 68°C to 72°C), and humidity (30–70%) with a 12 h light–dark cycles and were maintained on a standard chow diet (ND; PicoLab 5053 Rodent Diet 20; Lab Diets) ad libitum. Male mice were used for these experiments.
Mouse models
We performed the experiments using the following mouse strains: C57BL/6 (WT), CD38-KO, mice expressing the catalytically inactive form of CD38 (CD38-CI), 129S and C57BL/6, PARP1-KO, CD38Vav1−icre, and CD38Tek−Cre conditional-KO mice. Except for the PARP1-KO mice, which are on a mixed background (129S and C57BL/6), the other transgenic lines are on a C57BL/6 background. All animals used in this study were young adults (3–5 months). Mice received standard chow ad libitum, except for the NAM-free diet experiment, in which special diets were given (control diet – TD.140375 and NAM-free diet – TD.140376, both from Envigo). The CD38-KO mice (Aksoy et al., 2006; Partida-Sanchez et al., 2001), as well as CD38-CI mice (Tarrago et al., 2018), and hACMSD mice (Palzer et al., 2018) have been described previously. SARM1-KO mice and PARP1-KO mice were purchased from the Jackson Laboratories, and the tissue-specific CD38-KO is described below. All head-to-head comparisons were with the respective WT background mice.
Method details
Development of CD38 tissue-specific KO mice
C57-BL/6 mice with homozygous floxed (loxP-flanked) Cd38 alleles (Cd38fl/fl) were bred with commercially available Vav-iCre mice (JAX#008610) or Tie2-Cre mice (JAX#004128). Cd38fl/+ offspring that were positive for the Cre recombinase transgene were bred back with homozygous floxed CD38 mice. Resulting CD38 flox (Cd38fl/fl), CD38 flox/Vav1-iCre or CD38 Flox/Tek-Cre, which were our experimental mice. Control mice were Cd38fl/fl Cre-negative. Experimental mice were screened by PCR for the presence of recombination in blood and absence of recombination in ear punch samples.
Antibody and drug treatments in vivo
NAM-deficient diet
3-5-month-old C57BL/6 mice were fed a control diet (CD1 – 10% casein diet; TD140375, Teklad custom diet) and a NAM-free diet (ND1 – Niacin deficient diet, 10% casein; TD140376, Teklad custom diet) for 21 days (n = 5 mice/group). Plasma was collected on days 8, 15, and 21 and levels of NAM metabolites were determined. Urine was collected on day 15 and levels of metabolites were determined. At day 21, mice were euthanized, and tissues were collected for further analyses. Male mice were used for this experiment.
Induction of ACMSD with doxycycline
3-month-old hACMSD mice were fed a NAM-free diet in the presence of doxycycline for three months (n = 3–6 mice/group). Composition of defined diets and hACMSD gene induction was performed as described previously (Palzer et al., 2018). After 3 and 13 weeks on diets, mice were euthanized, tissues were harvested, and metabolites were measured for further analysis. Male mice were used for this experiment.
Anti-CD38 mouse antibody and NMN
3-month-old C57BL/6 mice were treated with a single dose of PBS (vehicle) or Ab68 (i.p., 5mg kg−1). At day 3, mice were given an oral gavage with NMN (500mg kg−1) or vehicle PBS, euthanized 6 h later, and tissues and serum were collected for further analysis. Male and female mice were used for this experiment.
Time course with Ab68 and olaparib
3-month-old C57BL/6 mice were treated with Ab68 (i.p., 5mg kg−1 using PBS as vehicle) or olaparib (i.p., 10mg kg−1 using 5% DMSO, 15% PEG400, and 80% of 15% hydroxypropyl-γ-cyclodextrin, in citrate buffer pH 6.0 as vehicle) in a time course regimen of 2-, 4-, and 48 h. When treatment was completed, mice were euthanized, and tissues were harvested for further analysis. All mice used for this experiment were male.
Cytotoxic antibodies
3-5-month-old C57BL/6 mice were treated with a single injection of IgG control or cytotoxic antibodies – anti-CD20Ab (500ug; GeneTech), anti-CD4Ab (250ug; Bioxcell BE0003-1), or anti-CD8Ab (500ug; BioXcell BE0061) – by i.p. injection for 28 h. When treatment was completed, mice were euthanized, and tissues were harvested for further analysis. All mice used for this experiment were male.
Bone marrow transplant
3-4-month-old CD38 KO recipient mice were irradiated using a cesium irradiator with two dosages of 5 Gray (gY) radiation 24 h apart. Bone marrow cells were isolated from tibias and femurs of 6- to 8-week-old WT or CD38 KO donor mice. After the second irradiation dosage, recipient mice were injected with approximately 1×106 donor cells by retro-orbital injection. Transplant mice were given sulfamethoxazole (95mg kg−1 per 24 h) in drinking water starting at 3 days before the first irradiation through 2 weeks after transplant. Engraftment was confirmed by staining blood cells for CD38. Twelve weeks and five days after transplant, mice were euthanized, and tissues were harvested. Male and female mice were used for this experiment.
Immunoblotting
Immunoblotting assays for whole cell lysates of cells and tissues were performed as previously described (Tarrago et al., 2018). Tissues or cells were homogenized and lysed in NETN buffer (20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 1 mM EDTA and 0.5% Nonidet P-40) supplemented with 50 mM β-glycerophosphate, 5 mM NaF and a protease inhibitor cocktail (Roche). After 30 min of incubation at 4°C, the samples were centrifuged at 12,000 r.p.m. for 10 min at 4°C. Protein concentrations in the supernatants were determined by Bio-Rad protein assay. Lysates were separated by SDS–PAGE, and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore). Enhanced chemiluminescence detection was performed using Super-Signal West Pico or Femto Chemiluminescence Substrate (Thermo Scientific). Films were scanned and densitometry was performed using ImageJ. The following antibodies and their dilutions were used for immunoblotting: mouse CD38 (R&D Systems; AF4947, 1:1,000), CD45 (Abcam; AB40763; 1:1000), Phospholamban (PLN) (Invitrogen; MA3-922; 1:1000), CYP3a4 (Proteintech, 18227-1-AP; 1:1000), actin (Cell Signaling Technology; 8457, 1:5,000) and GAPDH (Cell Signaling Technology; 97166, 1:5,000).
Measurement of NAD levels by cycling assay
Detection of NAD+ was performed as described before using a cycling assay (Camacho-Pereira et al., 2016; Kanamori et al., 2018; Tarrago et al., 2018). In summary, to determine intracellular NAD+ levels approximately 20 mg of tissue was homogenized in 10% trichloroacetic acid (TCA). Samples were centrifuged at 12,000 r.p.m. for 2 min at 4°C. The supernatants were collected, and the pellets were resuspended in 0.2N NaOH for protein determination. TCA was removed with organic solvents (three volumes of 1,1,2-trichloro-1,2,2-trifluroethane: one volume of trioctylamine) in a ratio of two volumes of organic solvent to one volume of sample. After phase separation, the top aqueous layer containing NAD+ was recovered and the pH was corrected by addition of 1M Tris (pH 8.0). For the cycling assay, samples were diluted in 100 mM sodium phosphate buffer (pH 8) in a volume of 100 μL per well and added to white 96-well plates. Next, 100μL of reaction mix (0.76% ethanol, 4 μM flavin mononucleotide, 27.2U ml−1 alcohol dehydrogenase (ADH), 1.8U ml−1 diaphorase and 8 μM resazurin) was added to each well. Then, 96-well plates were read in a fluorescence plate reader (Molecular Devices, SpectraMax Gemini XPS) in an excitation wavelength of 544 nm and an emission wavelength of 590 nm. We have previously demonstrated that the NAD+ cycling assay is as sensitive and specific as the ultra-performance liquid chromatography (UPLC)-mass spectroscopy (MS) assay (Camacho-Pereira et al., 2016).
Metabolomics by HPLC-MS
NAD+ metabolome was measured as described in Tarrago et al. (2018). For nucleotide measurements, the HPLC was at a flow rate of 0.25 mLmin−1 with 99% buffer A from 0 to 3 min, a linear gradient to 99% buffer A/1% buffer B (100% methanol) from 3 to 20 min, 80% buffer A/20% buffer B from 20 to 21 min, a linear gradient to 30% buffer A/70% buffer B from 21 to 28 min at 0.35 mLmin−1, 99% buffer A/1% buffer B from 28 to 31 min and a linear gradient to 99% buffer A from 31 to 37 min at 0.25 mLmin−1. Concentrations were quantified based on the peak area compared to a standard curve and normalized to protein content in the tissue sample.
NADase activity
NADase activity was measured as described previously (deOliveira et al., 2018), where 50 μM nicotinamide 1,N6-ethenoadenine di-nucleotide (ε-NAD) was used as substrate in 0.25 mM sucrose and 40 mM Tris-Cl (pH 7.4).
Quantification of mRNA
RNA was isolated from cells and mouse tissues using TRIzol or Qiagen RNeasy kits. cDNA was synthesized using the Qiagen QuantiTect or ABI High-Capacity cDNA Reverse Transcription kit. Real-time qPCR was performed using commercially available TaqMan gene expression probes (Applied Biosystems), according to the manufacturer’s instructions on a Bio-Rad CFX384 thermal cycler. The relative mRNA abundance of target genes was calculated by the 2(−ΔΔCq) method. Expression changes were calculated relative to control. TaqMan probes used in this study are described in Table S1.
Immunostaining
OCT-embedded tissue was cryosectioned at 10-μm thickness and fixed in MeOH:acetone (1:1) for 5 min at −20°C. Sections were blocked with 10% normal donkey serum in PBS-T for 1 h at RT, and then incubated overnight at 4°C with fluorescently tagged primary antibodies in 1% donkey serum. The following antibodies from BD Biosciences were used: CD38 AF647 (562769), CD45 FITC (553080), IgG2aκ AF647 isotype control (557690) and IgG2bκ isotype control (553988), using a 1:100 dilution for all. Nuclei were stained with Hoechst 33342. Images were obtained with an LSM 780 confocal microscope, and image capture was performed with ZEN 2.1 black software (Zeiss) using standardized exposure settings.
Single-cell analysis
We downloaded the spleen (Smart-Seq2 RNA sequencing library) AnnData (h5ad) file generated by the Tabula Muris Senis (Tabula Muris, 2020) investigators from https://cellxgene.cziscience.com. We performed these analyses, filtering, and figure visualization using the Scanpy toolkit (Wolf et al., 2018) and considered for analysis only 3-month-old data. We filtered low-quality cells with fewer than 2,500 reads and 250 genes, as well as genes expressed in less than 3 cells. We normalize each cell to 10,000 read counts, and then log transformed the data. Finally, we scaled the dataset to unit variance and zero mean, and we truncated values with maximum value set to ten. After preprocessing, the number of retained cells were 1665.
Quantification and statistical analysis
Data are represented as mean ± SD, analyzed by two-sided Student’s t-test (for comparisons of two groups) or ANOVA(for more than two groups). The post-tests used and the value of n are indicated in the figure’s legends. Analyses were performed using GraphPad Prism 9 software.
Acknowledgments
The work in E.N.C.'s laboratory is supported in part by grants from the Ted Nash Long Life Foundation, and the Glenn Foundation for Medical Research via the Paul F. Glenn Laboratories for the Biology of Aging at the Mayo Clinic (to E.N.C.); sponsored research funding from Calico Life Sciences; the Mayo and Noaber Foundations; and NIH National Institute on Aging (NIA) grantsAG-26094, AG58812, and National Cancer Institute grant CA233790 (to E.N.C.); National Institute On Aging of the National Institutes of Health under Award Number R56AG069745 to E.N.C. and M.M.F, by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health underAward Number 1R15HD100970-01 to R.G.M. The content is solely the authors’ responsibility and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
Conceptualization, E.N.C., J.D.Z., and C.C.S.C.; Methodology, J.D.Z., C.C.S.C., K.S.K., G.M.W., K.M.T., and R.A.S.; Investigation, J.D.Z., C.C.S.C., K.S.K., J.M.E.N., K.T., G.W., F.S.C., T.R.P., L.S.G., S.A.L., M.K.W., R.A.S., and F.B.; Writing – original draft, J.D.Z., C.C.S.C., E.N.C., K.S.K., and G.W.; Writing – Review & Editing, C.C.S.C., E.N.C., S.B., M.M.F., R.G.M., and J.D.Z.; Funding acquisition, E.N.C.; Resources, E.N.C., M.L.M., S.B., J.M.R., R.G.M., M.M.F., K.R., and K.M.; Supervision, E.N.C. and C.C.S.C.
Declaration of interests
E.N.C. holds a patent on the use of CD38 inhibitors for metabolic diseases licensed by Elysium health. E.N.C. is a consultant for TeneoBio, Calico, Mitobridge, and Cytokinetics. E.N.C. is on the advisory board of Eolo Pharma. E.N.C. own stocks in TeneoBio. Research in the E.N.C. laboratory has been conducted in compliance with Mayo Clinic’s conflict of interest policies.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105431.
Supplemental information
Data and code availability
-
•
This article analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All data reported in this article will be shared by the lead contact on request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
References
- Aksoy P., White T.A., Thompson M., Chini E.N. Regulation of intracellular levels of NAD: a novel role for CD38. Biochem. Biophys. Res. Commun. 2006;345:1386–1392. doi: 10.1016/j.bbrc.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Alston T.A., Abeles R.H. Substrate specificity of nicotinamide methyltransferase isolated from porcine liver. Arch. Biochem. Biophys. 1988;260:601–608. doi: 10.1016/0003-9861(88)90487-0. [DOI] [PubMed] [Google Scholar]
- Bar A., Olkowicz M., Tyrankiewicz U., Kus E., Jasinski K., Smolenski R.T., Skorka T., Chlopicki S. Functional and biochemical endothelial profiling in vivo in a murine model of endothelial dysfunction; comparison of effects of 1-methylnicotinamide and angiotensin-converting enzyme inhibitor. Front. Pharmacol. 2017;8:183. doi: 10.3389/fphar.2017.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman K.J., Anderson R.M., Cohen H.Y., Latorre-Esteves M., Sinclair D.A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Boslett J., Hemann C., Christofi F.L., Zweier J.L. Characterization of CD38 in the major cell types of the heart: endothelial cells highly express CD38 with activation by hypoxia-reoxygenation triggering NAD(P)H depletion. Am. J. Physiol. Cell Physiol. 2018;314:C297–C309. doi: 10.1152/ajpcell.00139.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachs S., Polack J., Brachs M., Jahn-Hofmann K., Elvert R., Pfenninger A., Bärenz F., Margerie D., Mai K., Spranger J., Kannt A. Genetic nicotinamide N-methyltransferase (Nnmt) deficiency in male mice improves insulin sensitivity in diet-induced obesity but does not affect glucose tolerance. Diabetes. 2019;68:527–542. doi: 10.2337/db18-0780. [DOI] [PubMed] [Google Scholar]
- Brown E.E., Scandura M.J., Mehrotra S., Wang Y., Du J., Pierce E.A. Reduced nuclear NAD+ drives DNA damage and subsequent immune activation in the retina. Hum. Mol. Genet. 2022;31:1370–1388. doi: 10.1093/hmg/ddab324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Pereira J., Tarragó M.G., Chini C.C.S., Nin V., Escande C., Warner G.M., Puranik A.S., Schoon R.A., Reid J.M., Galina A., Chini E.N. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 2016;23:1127–1139. doi: 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A.M., Castoldi A., Sanin D.E., Flachsmann L.J., Field C.S., Puleston D.J., Kyle R.L., Patterson A.E., Hässler F., Buescher J.M., et al. Inflammatory macrophage dependence on NAD(+) salvage is a consequence of reactive oxygen species-mediated DNA damage. Nat. Immunol. 2019;20:420–432. doi: 10.1038/s41590-019-0336-y. [DOI] [PubMed] [Google Scholar]
- Chen Y., Zhang J., Li P., Liu C., Li L. N1methylnicotinamide ameliorates insulin resistance in skeletal muscle of type 2 diabetic mice by activating the SIRT1/PGC1alpha signaling pathway. Mol. Med. Rep. 2021;23:270. doi: 10.3892/mmr.2021.11909. [DOI] [PubMed] [Google Scholar]
- Chini C., Hogan K.A., Warner G.M., Tarragó M.G., Peclat T.R., Tchkonia T., Kirkland J.L., Chini E. The NADase CD38 is induced by factors secreted from senescent cells providing a potential link between senescence and age-related cellular NAD(+) decline. Biochem. Biophys. Res. Commun. 2019;513:486–493. doi: 10.1016/j.bbrc.2019.03.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini C.C.S., Peclat T.R., Warner G.M., Kashyap S., Espindola-Netto J.M., de Oliveira G.C., Gomez L.S., Hogan K.A., Tarragó M.G., Puranik A.S., et al. CD38 ecto-enzyme in immune cells is induced during aging and regulates NAD(+) and NMN levels. Nat. Metab. 2020;2:1284–1304. doi: 10.1038/s42255-020-00298-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini C.C.S., Zeidler J.D., Kashyap S., Warner G., Chini E.N. Evolving concepts in NAD(+) metabolism. Cell Metab. 2021;33:1076–1087. doi: 10.1016/j.cmet.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini E.N., Chini C.C.S., Kato I., Takasawa S., Okamoto H. CD38 is the major enzyme responsible for synthesis of nicotinic acid-adenine dinucleotide phosphate in mammalian tissues. Biochem. J. 2002;362:125–130. doi: 10.1042/0264-6021:3620125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias A.J., Kale A., Perrone R., Lopez-Dominguez J.A., Pisco A.O., Kasler H.G., Schmidt M.S., Heckenbach I., Kwok R., Wiley C.D., et al. Senescent cells promote tissue NAD(+) decline during ageing via the activation of CD38(+) macrophages. Nat. Metab. 2020;2:1265–1283. doi: 10.1038/s42255-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J., Williams A., Skavdis G., Harker N., Coles M., Tolaini M., Norton T., Williams K., Roderick K., Potocnik A.J., Kioussis D. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- de Oliveira G.C., Kanamori K.S., Auxiliadora-Martins M., Chini C.C.S., Chini E.N. Measuring CD38 hydrolase and cyclase activities: 1, N(6)-ethenonicotinamide adenine dinucleotide (epsilon-NAD) and nicotinamide guanine dinucleotide (NGD) fluorescence-based methods. Bio. Protoc. 2018;8:e2938. doi: 10.21769/BioProtoc.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Picciotto N.E., Gano L.B., Johnson L.C., Martens C.R., Sindler A.L., Mills K.F., Imai S.I., Seals D.R. Nicotinamide mononucleotide supplementation reverses vascular dysfunction and oxidative stress with aging in mice. Aging Cell. 2016;15:522–530. doi: 10.1111/acel.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domínguez-Gómez G., Díaz-Chávez J., Chávez-Blanco A., Gonzalez-Fierro A., Jiménez-Salazar J.E., Damián-Matsumura P., Gómez-Quiroz L.E., Dueñas-González A. Nicotinamide sensitizes human breast cancer cells to the cytotoxic effects of radiation and cisplatin. Oncol. Rep. 2015;33:721–728. doi: 10.3892/or.2014.3661. [DOI] [PubMed] [Google Scholar]
- Fukuwatari T., Wada H., Shibata K. Age-related alterations of B-group vitamin contents in urine, blood and liver from rats. J. Nutr. Sci. Vitaminol. 2008;54:357–362. doi: 10.3177/jnsv.54.357. [DOI] [PubMed] [Google Scholar]
- Gomes A.P., Price N.L., Ling A.J.Y., Moslehi J.J., Montgomery M.K., Rajman L., White J.P., Teodoro J.S., Wrann C.D., Hubbard B.P., et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green K.N., Steffan J.S., Martinez-Coria H., Sun X., Schreiber S.S., Thompson L.M., LaFerla F.M. Nicotinamide restores cognition in Alzheimer's disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J. Neurosci. 2008;28:11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hathorn T., Snyder-Keller A., Messer A. Nicotinamide improves motor deficits and upregulates PGC-1alpha and BDNF gene expression in a mouse model of Huntington's disease. Neurobiol. Dis. 2011;41:43–50. doi: 10.1016/j.nbd.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiromatsu Y., Sato M., Tanaka K., Ishisaka N., Kamachi J., Nonaka K. Inhibitory effects of nicotinamide on intercellular adhesion molecule-1 expression on cultured human thyroid cells. Immunology. 1993;80:330–332. [PMC free article] [PubMed] [Google Scholar]
- Hiromatsu Y., Sato M., Yamada K., Nonaka K. Inhibitory effects of nicotinamide on recombinant human interferon-gamma-induced intercellular adhesion molecule-1 (ICAM-1) and HLA-DR antigen expression on cultured human endothelial cells. Immunol. Lett. 1992;31:35–39. doi: 10.1016/0165-2478(92)90007-b. [DOI] [PubMed] [Google Scholar]
- Kanamori K.S., de Oliveira G.C., Auxiliadora-Martins M., Schoon R.A., Reid J.M., Chini E.N. Two different methods of quantification of oxidized nicotinamide adenine dinucleotide (NAD(+)) and reduced nicotinamide adenine dinucleotide (NADH) intracellular levels: enzymatic coupled cycling assay and ultra-performance liquid chromatography (UPLC)-Mass spectrometry. Bio. Protoc. 2018;8:e2937. doi: 10.21769/BioProtoc.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsyuba E., Romani M., Hofer D., Auwerx J. NAD(+) homeostasis in health and disease. Nat. Metab. 2020;2:9–31. doi: 10.1038/s42255-019-0161-5. [DOI] [PubMed] [Google Scholar]
- Kilgour M.K., MacPherson S., Zacharias L.G., Ellis A.E., Sheldon R.D., Liu E.Y., Keyes S., Pauly B., Carleton G., Allard B., et al. 1-Methylnicotinamide is an immune regulatory metabolite in human ovarian cancer. Sci. Adv. 2021;7:eabe1174. doi: 10.1126/sciadv.abe1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koni P.A., Joshi S.K., Temann U.A., Olson D., Burkly L., Flavell R.A. Conditional vascular cell adhesion molecule 1 deletion in mice: impaired lymphocyte migration to bone marrow. J. Exp. Med. 2001;193:741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosciuk T., Wang M., Hong J.Y., Lin H. Updates on the epigenetic roles of sirtuins. Curr. Opin. Chem. Biol. 2019;51:18–29. doi: 10.1016/j.cbpa.2019.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakura S., Sato E., Sekimoto A., Hashizume Y., Yamakage S., Miyazaki M., Ito S., Harigae H., Takahashi N. Nicotinamide attenuates the progression of renal failure in a mouse model of adenine-induced chronic kidney disease. Toxins. 2021;13:50. doi: 10.3390/toxins13010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Su X., Quinn W.J., 3rd, Hui S., Krukenberg K., Frederick D.W., Redpath P., Zhan L., Chellappa K., White E., et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 2018;27:1067–1080.e5. doi: 10.1016/j.cmet.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z., Hine C., Tian X., Van Meter M., Au M., Vaidya A., Seluanov A., Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matalonga J., Glaria E., Bresque M., Escande C., Carbó J.M., Kiefer K., Vicente R., León T.E., Beceiro S., Pascual-García M., et al. The nuclear receptor LXR limits bacterial infection of host macrophages through a mechanism that impacts cellular NAD metabolism. Cell Rep. 2017;18:1241–1255. doi: 10.1016/j.celrep.2017.01.007. [DOI] [PubMed] [Google Scholar]
- McReynolds M.R., Chellappa K., Chiles E., Jankowski C., Shen Y., Chen L., Descamps H.C., Mukherjee S., Bhat Y.R., Lingala S.R., et al. NAD(+) flux is maintained in aged mice despite lower tissue concentrations. Cell Syst. 2021;12:1160–1172.e4. doi: 10.1016/j.cels.2021.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menear K.A., Adcock C., Boulter R., Cockcroft X.L., Copsey L., Cranston A., Dillon K.J., Drzewiecki J., Garman S., Gomez S., et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin- 1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J. Med. Chem. 2008;51:6581–6591. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- Meng Y., Ren Z., Xu F., Zhou X., Song C., Wang V.Y.F., Liu W., Lu L., Thomson J.A., Chen G. Nicotinamide promotes cell survival and differentiation as kinase inhibitor in human pluripotent stem cells. Stem Cell Rep. 2018;11:1347–1356. doi: 10.1016/j.stemcr.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Song C., Ren Z., Li X., Yang X., Ai N., Yang Y., Wang D., Zhan M., Wang J., et al. Nicotinamide promotes cardiomyocyte derivation and survival through kinase inhibition in human pluripotent stem cells. Cell Death Dis. 2021;12:1119. doi: 10.1038/s41419-021-04395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell S.J., Bernier M., Aon M.A., Cortassa S., Kim E.Y., Fang E.F., Palacios H.H., Ali A., Navas-Enamorado I., Di Francesco A., et al. Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Metab. 2018;27:667–676.e4. doi: 10.1016/j.cmet.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa T. Protective effects of N(1)-methylnicotinamide against high-fat diet- and age-induced hearing loss via moderate overexpression of sirtuin 1 protein. Front. Cell. Neurosci. 2021;15:634868. doi: 10.3389/fncel.2021.634868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogielnicki A., Kramkowski K., Pietrzak L., Buczko W. N-methylnicotinamide inhibits arterial thrombosis in hypertensive rats. J. Physiol. Pharmacol. 2007;58:515–527. [PubMed] [Google Scholar]
- Ogilvy S., Metcalf D., Gibson L., Bath M.L., Harris A.W., Adams J.M. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94:1855–1863. [PubMed] [Google Scholar]
- Palzer L., Bader J.J., Angel F., Witzel M., Blaser S., McNeil A., Wandersee M.K., Leu N.A., Lengner C.J., Cho C.E., et al. Alpha-amino-beta-carboxy-muconate-semialdehyde decarboxylase controls dietary niacin requirements for NAD(+) synthesis. Cell Rep. 2018;25:1359–1370.e4. doi: 10.1016/j.celrep.2018.09.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaccio G., Ammendola E., Pisanti F.A. Nicotinamide decreases MHC class II but not MHC class I expression and increases intercellular adhesion molecule-1 structures in non-obese diabetic mouse pancreas. J. Endocrinol. 1999;160:389–400. doi: 10.1677/joe.0.1600389. [DOI] [PubMed] [Google Scholar]
- Partida-Sánchez S., Cockayne D.A., Monard S., Jacobson E.L., Oppenheimer N., Garvy B., Kusser K., Goodrich S., Howard M., Harmsen A., et al. Cyclic ADP-ribose production by CD38 regulates intracellular calcium release, extracellular calcium influx and chemotaxis in neutrophils and is required for bacterial clearance in vivo. Nat. Med. 2001;7:1209–1216. doi: 10.1038/nm1101-1209. [DOI] [PubMed] [Google Scholar]
- Piedra-Quintero Z.L., Wilson Z., Nava P., Guerau-de-Arellano M. CD38: an immunomodulatory molecule in inflammation and autoimmunity. Front. Immunol. 2020;11:597959. doi: 10.3389/fimmu.2020.597959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucci L., Perozzi S., Cimadamore F., Orsomando G., Raffaelli N. Tissue expression and biochemical characterization of human 2-amino 3-carboxymuconate 6-semialdehyde decarboxylase, a key enzyme in tryptophan catabolism. FEBS J. 2007;274:827–840. doi: 10.1111/j.1742-4658.2007.05635.x. [DOI] [PubMed] [Google Scholar]
- Radenkovic D., Reason, Verdin E. Clinical evidence for targeting NAD therapeutically. Pharmaceuticals. 2020;13:E247. doi: 10.3390/ph13090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo J.R., Grimm A.A., Imai S.i. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rongvaux A., Shea R.J., Mulks M.H., Gigot D., Urbain J., Leo O., Andris F. Pre-B-cell colony-enhancing factor, whose expression is up-regulated in activated lymphocytes, is a nicotinamide phosphoribosyltransferase, a cytosolic enzyme involved in NAD biosynthesis. Eur. J. Immunol. 2002;32:3225–3234. doi: 10.1002/1521-4141(200211)32:11<3225::AID-IMMU3225>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M., Mitchell S.J., Fang E.F., Iyama T., Ward T., Wang J., Dunn C.A., Singh N., Veith S., Hasan-Olive M.M., et al. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab. 2014;20:840–855. doi: 10.1016/j.cmet.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller T., Orgacka H., Szumlanski C.L., Weinshilboum R.M. Mouse liver nicotinamide N-methyltransferase pharmacogenetics: biochemical properties and variation in activity among inbred strains. Pharmacogenetics. 1996;6:43–53. doi: 10.1097/00008571-199602000-00003. [DOI] [PubMed] [Google Scholar]
- Shimshek D.R., Kim J., Hübner M.R., Spergel D.J., Buchholz F., Casanova E., Stewart A.F., Seeburg P.H., Sprengel R. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. doi: 10.1002/gene.10023. [DOI] [PubMed] [Google Scholar]
- Su C.F., Liu D.D., Kao S.J., Chen H.I. Nicotinamide abrogates acute lung injury caused by ischaemia/reperfusion. Eur. Respir. J. 2007;30:199–204. doi: 10.1183/09031936.00025107. [DOI] [PubMed] [Google Scholar]
- Tabula Muris Consortium A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020;583:590–595. doi: 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi K., Yokouchi C., Goto H., Umehara K., Yamada H., Ishii Y. Alleviation of fatty liver in a rat model by enhancing N(1)-methylnicotinamide bioavailability through aldehyde oxidase inhibition. Biochem. Biophys. Res. Commun. 2018;507:203–210. doi: 10.1016/j.bbrc.2018.11.008. [DOI] [PubMed] [Google Scholar]
- Tarragó M.G., Chini C.C.S., Kanamori K.S., Warner G.M., Caride A., de Oliveira G.C., Rud M., Samani A., Hein K.Z., Huang R., et al. A potent and specific CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD(+) decline. Cell Metab. 2018;27:1081–1095.e10. doi: 10.1016/j.cmet.2018.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakata M., Fukuwatari T., Kadota E., Sano M., Kanai M., Nakamura T., Funakoshi H., Shibata K. The niacin required for optimum growth can be synthesized from L-tryptophan in growing mice lacking tryptophan-2, 3-dioxygenase. J. Nutr. 2013;143:1046–1051. doi: 10.3945/jn.113.176875. [DOI] [PubMed] [Google Scholar]
- Turunc Bayrakdar E., Uyanikgil Y., Kanit L., Koylu E., Yalcin A. Nicotinamide treatment reduces the levels of oxidative stress, apoptosis, and PARP-1 activity in Abeta(1-42)-induced rat model of Alzheimer's disease. Free Radic. Res. 2014;48:146–158. doi: 10.3109/10715762.2013.857018. [DOI] [PubMed] [Google Scholar]
- Ungerstedt J.S., Blömback M., Söderström T. Nicotinamide is a potent inhibitor of proinflammatory cytokines. Clin. Exp. Immunol. 2003;131:48–52. doi: 10.1046/j.1365-2249.2003.02031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P.A., Harder J.M., Foxworth N.E., Cochran K.E., Philip V.M., Porciatti V., Smithies O., John S.W.M. Vitamin B3 modulates mitochondrial vulnerability and prevents glaucoma in aged mice. Science. 2017;355:756–760. doi: 10.1126/science.aal0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Miyajima E., Nonaka K. Inhibition of cytokine-induced MHC class II but not class I molecule expression on mouse islet cells by niacinamide and 3-aminobenzamide. Diabetes. 1990;39:1125–1130. doi: 10.2337/diab.39.9.1125. [DOI] [PubMed] [Google Scholar]
- Yang W., Nagasawa K., Münch C., Xu Y., Satterstrom K., Jeong S., Hayes S.D., Jedrychowski M.P., Vyas F.S., Zaganjor E., et al. Mitochondrial sirtuin network reveals dynamic SIRT3-dependent deacetylation in response to membrane depolarization. Cell. 2016;167:985–1000.e21. doi: 10.1016/j.cell.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J., Mills K.F., Yoon M.J., Imai S.i. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Chen Y., Liu C., Li L., Li P. N(1)-Methylnicotinamide improves hepatic insulin sensitivity via activation of SIRT1 and inhibition of FOXO1 acetylation. J. Diabetes Res. 2020;2020:1080152. doi: 10.1155/2020/1080152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhen X., Zhang S., Xie F., Zhou M., Hu Z., Zhu F., Nie J. Nicotinamide supplementation attenuates renal interstitial fibrosis via boosting the activity of sirtuins. Kidney Dis. 2021;7:186–199. doi: 10.1159/000510943. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This article analyzes existing, publicly available data. These accession numbers for the datasets are listed in the key resources table. All data reported in this article will be shared by the lead contact on request.
-
•
This article does not report original code.
-
•
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.