Abstract
Since the initial emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron BA.1, several Omicron sublineages have emerged, leading to BA.5 as the current dominant sublineage. Here, we report the neutralization of different Omicron sublineages by human sera collected from individuals who had distinct mRNA vaccination and/or BA.1 infection. Four-dose-vaccine sera neutralize the original USA-WA1/2020, Omicron BA.1, BA.2, BA.2.12.1, BA.3, and BA.4/5 viruses with geometric mean titers (GMTs) of 1,554, 357, 236, 236, 165, and 95, respectively; two-dose-vaccine-plus-BA.1-infection sera exhibit GMTs of 2,114, 1,705, 730, 961, 813, and 274, respectively; and three-dose-vaccine-plus-BA.1-infection sera show GMTs of 2,962, 2,038, 983, 1,190, 1,019, and 297, respectively. Thus, the four-dose vaccine elicits the lowest neutralization against BA.5; the two-dose vaccine plus BA.1 infection elicits significantly higher GMTs against Omicron sublineages than the four-dose-vaccine; and the three-dose vaccine plus BA.1 infection elicits slightly higher GMTs (statistically insignificant) than the two-dose vaccine plus BA.1 infection. Finally, the BA.2.75 is more susceptible than BA.5 to four-dose-vaccine-elicited neutralization and three-dose-vaccine-plus-BA.1-infection-elicited neutralization.
Keywords: SARS-CoV-2, neutralization, mRNA vaccine, variants, breakthrough
Graphical abstract
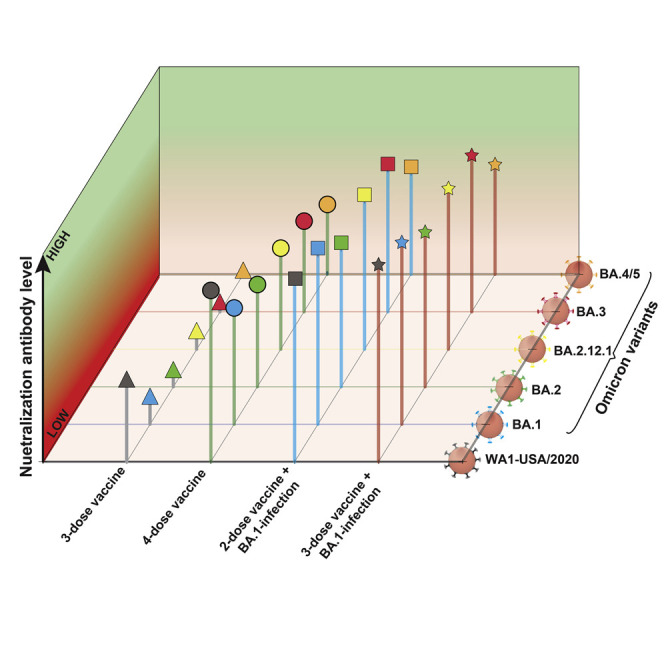
Xie at al. show that Omicron BA.5 evades vaccine-elicited neutralization most efficiently among all currently known SARS-CoV-2 variants. The neutralization against BA.5 remains low after four doses of mRNA vaccine, whereas two or three doses of vaccine plus BA.1 infection elicit high neutralization against BA.5. The results support a bivalent vaccine approach.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron variant was initially identified in South Africa in November 2021. Due to Omicron’s improved viral transmission and immune evasion,1 , 2 the World Health Organization designated it as the fifth variant of concern (VOC) after the previous Alpha, Beta, Gamma, and Delta VOCs. Since then, the Omicron variant has evolved to several sublineages, including BA.1, BA.2, BA.2.12.1, BA.3, BA.4, and BA.5. Among these sublineages, only Omicron BA.3 remained at a low frequency in circulation, most likely due to its low fitness, whereas other sublineages sequentially increased their prevalence over time. As of September 3, 2022, in the United States, sublineages BA.2.12.1, BA.4, and BA.5 accounted for 0.1%, 2.2%, and 87.5% of the total COVID-19 cases, respectively (https://covid.cdc.gov/covid-data-tracker/#variant-proportions). Besides the above Omicron sublineages, sublineage BA.2.75 emerged in late May 2022 and has increased its prevalence in many countries. It is thus important to determine the neutralization susceptibility of the ongoing Omicron sublineages, particularly the most prevalent, BA.5, to vaccination and previous infections.
Immediately after the emergence of the Omicron variant, BA.1 was found to evade vaccine-elicited neutralization more efficiently than any previous VOCs.2 , 3 , 4 , 5 , 6 , 7 Two doses of mRNA vaccine did not elicit robust neutralization against BA.1; three doses of vaccine were required to generate sufficient neutralization against BA.1.8 Non-Omicron infection did not elicit robust neutralization against BA.1 either.9 Among all the currently known Omicron sublineages, BA.5 exhibited the greatest evasion of vaccine-mediated neutralization; three doses of BNT162b2 mRNA vaccine elicited weak neutralization against BA.5.10 The latter result, together with the high prevalence of BA.5, underscores the urgency to examine the neutralization of BA.5 after four doses of mRNA vaccine. In addition, many people who had previously received two or three doses of vaccine contracted BA.1 breakthrough infection during the initial Omicron wave; it would be important to evaluate their antibody neutralization against BA.5. To address these key questions, we have characterized the neutralization profiles of sera, obtained from humans with distinct mRNA vaccination and/or BA.1 infection, against different Omicron sublineages.
Results and discussions
Experimental approach and rationale
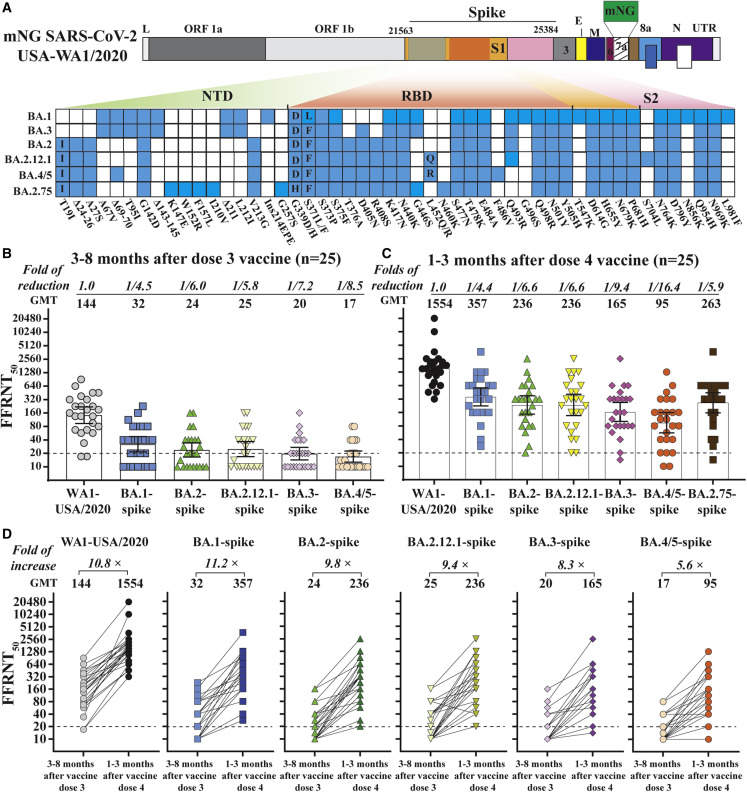
We used a set of previously established recombinant SARS-CoV-2s to determine the serum neutralization against different Omicron sublineages. Each recombinant SARS-CoV-2 contained a complete spike gene from BA.1, BA.2, BA.2.12.1, BA.3, or BA.4/5 in the backbone of USA-WA1/2020 (a virus strain isolated in January 2020) containing an mNeonGreen (mNG) reporter, resulting in BA.1-, BA.2-, BA.2.12.1-, BA.3-, or BA.4/5-spike mNG SARS-CoV-2.10 BA.4 and BA.5 have an identical spike sequence and are denoted as BA.4/5. Figure 1A summarizes the amino acid mutations of the spike protein from different Omicron sublineages. An mNG gene was engineered into open reading frame 7 (ORF7) of the viral genome to enable a fluorescent focus-reduction neutralization test (FFRNT) in a high-throughput format.9 The insertion of mNG reporter attenuated SARS-CoV-2 replication and pathogenesis.11 , 12The FFRNT has been reliably used to measure antibody neutralization for COVID-19 vaccine research and development.6 , 10
Figure 1.
Neutralization of Omicron sublineages before and after four doses of mRNA vaccine
(A) Construction of Omicron sublineage-spike mNG SARS-CoV-2. mNG USA-WA1/2020 SARS-CoV-2 was used to engineer Omicron-spike SARS-CoV-2s. The mNG reporter gene was engineered at open reading frame 7 (ORF7) of the USA-WA1/2020 genome. Amino acid mutations, deletions (Δ), and insertions (Ins) are indicated for variant spikes in reference to the USA-WA1/2020 spike. L, leader sequence; ORF, open reading frame; NTD, N-terminal domain of S1; RBD, receptor-binding domain of S1; S, spike glycoprotein; S1, N-terminal furin cleavage fragment of S; S2, C-terminal furin cleavage fragment of S; E, envelope protein; M, membrane protein; N, nucleoprotein; UTR, untranslated region. Twenty-five pairs of human sera were collected 3–8 months after dose three and 1–3 months after dose four of mRNA vaccine. The FFRNT50s for mNG BA.1-, BA.2-, BA.2.12.1, BA.3-, and BA.4/5-spike SARS-CoV-2s were determined in duplicate assays; the FFRNT50 for USA-WA1/2020 SARS-CoV-2 was determined in two independent experiments, each with duplicate assays.
(B) FFRNT50 of sera collected before dose four of the vaccine. The bar heights and the numbers above indicate neutralizing GMTs. The whiskers indicate 95% confidence interval (CI). The fold of GMT reduction against each Omicron sublineage, compared with the GMT against USA-WA1/2020, is shown in italic font. The dotted line indicates the limit of detection of FFRNT50. FFRNT50 <20 was treated as 10 for plot purposes and statistical analysis. The p values (Wilcoxon matched-pairs signed-rank test) for group comparison of GMTs are the following: USA-WA1/2020 versus all Omicron sublineage-spike: <0.0001; BA.1-spike versus BA.2-, BA.2.12.1-, BA.3-, and BA.4/5-spike: 0.004, 0.0336, <0.0001, and <0.0001, respectively; BA.2-spike versus BA.2.12.1-, BA.3-, and BA.4/5-spike: 0.5, 0.065, and 0.0083, respectively. BA.2.12.1-spike versus BA.3- and BA.4/5-spike: 0.0098 and 0.0002, respectively; BA.3-spike versus BA.4/5-spike: 0.156.
(C) FFRNT50 of sera collected after dose four of the vaccine. The p values (Wilcoxon matched-pairs signed-rank test) for group comparison of GMTs are the following: USA-WA1/2020 versus all Omicron sublineage-spike: <0.0001; BA.1-spike versus BA.2-, BA.2.12.1-, BA.3-, BA.4/5, and BA.2.75-spike: 0.008, 0.033, <0.0001, <0.0001, and 0.37, respectively; BA.2-spike versus BA.2.12.1-, BA.3-, BA.4/5, and BA.2.75-spike: 0.12, <0.0001, <0.0001, and 0.56, respectively; BA.2.12.1-spike versus BA.3-, BA.4/5, and BA.2.75-spike: 0.0002, <0.0001, and 0.94, respectively; BA.3-spike versus BA.4/5- and BA.2.75-spike: 0.0009 and 0.13, respectively; BA.4/5-spike versus BA.2.75-spike: 0.017.
(D) FFRNT50 values with connected lines for each serum pair before and after dose four of the vaccine. The GMT fold increase before and after dose four is shown in italic font. The p values of GMT (Wilcoxon matched-pairs signed-rank test) before and after dose four of the vaccines are all <0.0001. The p values (Friedman with Dunn’s multiple comparisons test) for group comparison of the increase in the neutralization of sera against different variants are the following: BA.4/5-spike (5.6-fold) versus WA1 (10.8-fold), BA.1-spike (11.2-fold), and BA.2-spike (9.8-fold): 0.042, 0.0007, and 0.048, respectively.
Using FFRNT, we measured the neutralization of three panels of human sera against the chimeric Omicron sublineage-spike mNG SARS-CoV-2s. The first panel consisted of 25 pairs of sera collected from individuals before and after dose four of Pfizer or Moderna’s original vaccine (Table S1). Those specimens tested negative against viral nucleocapsid protein, suggesting those individuals had not been infected by SARS-CoV-2. The second and third serum panels were collected from individuals who had received two (n = 29; Table S2) or three (n = 38; Table S3) doses of the original mRNA vaccine and subsequently contracted Omicron BA.1 breakthrough infection. The BA.1 breakthrough infection was confirmed for each patient by sequencing viral RNA collected from nasopharyngeal swab samples. Tables S1–S3 summarize (1) the serum information and (2) the 50% fluorescent focus-reduction neutralization titers (FFRNT50) against USA-WA1/2020, BA.1-, BA.2-, BA.2.12.1-, BA.3-, and BA.4/5-spike SARS-CoV-2s. The description and analysis of the FFRNT50 results against different Omicron sublineages are detailed in the following sections for each serum panel.
The booster effect by dose four of mRNA vaccine is less pronounced against BA.4/5 compared with USA-WA1/2020 and other Omicron sublineages
To measure four doses of vaccine-elicited neutralization, we collected 25 pairs of sera from individuals before and after dose four of Pfizer or Moderna mRNA vaccine. For each serum pair, one sample was collected 3–8 months after dose three of the vaccine; the other sample was obtained from the same individual 1–3 months after dose four of the vaccine (Table S1). Before the fourth dose of the vaccine, the three-dose-vaccine sera neutralized USA-WA1/2020, BA.1-, BA.2-, BA.2.12.1-, BA.3-, and BA.4/5-spike mNG viruses with low geometric mean titers (GMTs) of 144, 32, 24, 25, 20, and 17, respectively (Figure 1B), and after the fourth dose of the vaccine, the GMTs increased significantly to 1,554, 357, 236, 236, 165, and 95, respectively (Figure 1C), so the fourth dose of the vaccine significantly increased the neutralization against the corresponding viruses by 10.8-, 11.2-, 9.8-, 9.4-, 8.3-, and 5.6-fold, respectively (Figure 1D). Despite the significant increase in neutralization after the fourth dose of the vaccine, the GMTs against BA.1-, BA.2-, BA.2.12.1-, BA.3-, and BA.4/5-spike viruses were 4.4-, 6.6-, 6.6-, 9.4-, and 16.4-fold lower than the GMTs against the USA-WA1/2020, respectively (Figure 1C). These results support three conclusions. First, among the tested Omicron sublineages, BA.5 possesses the greatest evasion of vaccine-elicited neutralization. Our results are in agreement with other studies supporting that BA.5 and other Omicron sublineages efficiently evade vaccine-elicited neutralization.13 , 14 , 15 , 16 , 17 Second, the booster effect by the fourth dose is less pronounced against BA.4/5 compared with USA-WA1/2020 and other Omicron sublineages. It should be noted that dose four did increase the neutralizing GMTs against BA.4/5 from 17 (Figure 1B) to 95 (Figure 1C). A recent study reported a neutralizing titer of 70 as the threshold to prevent breakthrough infections of the Delta variant.18 Although the minimal neutralizing titer required to prevent BA.5 infection has not been determined, the low neutralization against BA.5 after dose three of the vaccine (GMT of 103 at 1 month post-dose three, reported by Kurhade et al.10) and dose four of the vaccine (GMT of 95 at 1 to 3 months post-dose four, reported here), together with the increased viral transmissibility, could account for the ongoing surge of BA.5 around the world. Third, an updated vaccine that matches the highly immune-evasive and prevalent BA.5 spike is needed to mitigate the current and future Omicron surges. Our results support the US Food and Drug Administration’s recommendation to include BA.5 spike for future COVID-19 vaccine booster doses.
High neutralization against BA.5 and other Omicron sublineages after two or three doses of vaccine plus BA.1 infection
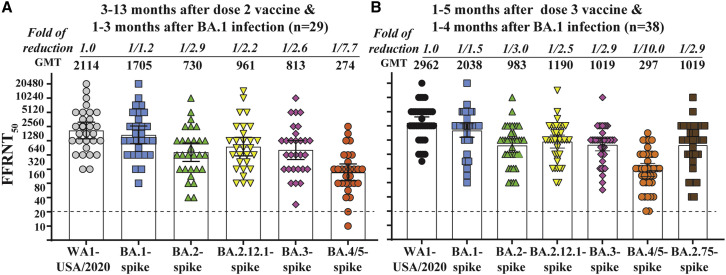
To compare with four-dose-vaccine sera, we measured the neutralization against Omicron sublineages using sera collected from individuals who had received two or three doses of the original mRNA vaccine and subsequently contracted BA.1 infection (Figure 2 ). Tables S2 and S3 summarize the FFRNT50 results for two-dose-vaccine-plus-BA.1-infection sera and three-dose-vaccine-plus-BA.1-infection sera, respectively. The two-dose-vaccine-plus-BA.1-infection sera neutralized BA.1, BA.2, BA.2.12.1, BA.3, and BA.4/5 with GMTs of 2,114, 1,705, 730, 961, 813, and 274, respectively (Figure 2A), and the three-dose-vaccine-plus-BA.1-infection sera showed slightly higher GMTs of 2,962, 2,038, 983, 1,190, 1,019, and 297, respectively (Figure 2B). So, the GMT ratios between the three-dose-vaccine-plus-BA.1-infection sera and the two-dose-vaccine-plus-BA.1-infection sera were 1.4, 1.2, 1.3, 1.2, 1.3, and 1.1 when neutralizing USA-WA1/2020, BA.1-, BA.2-, BA.2.12.1-, BA.3-, and BA.4/5-spike viruses, respectively; these GMT differences between the two serum groups were statistically insignificant, suggesting that the extra dose of vaccine does not significantly boost neutralization for the three-dose-vaccine-plus-BA.1-infection sera.
Figure 2.
Neutralization of Omicron sublineages after two or three doses of mRNA vaccine and BA.1 infection
(A) FFRNT50 of two-dose-vaccine-plus-BA.1-infection sera. Twenty-nine sera were collected from individuals who received three doses of vaccine and subsequently contracted BA.1 breakthrough infection. The GMT reduction fold against each Omicron sublineage and USA-WA1/2020 is shown in italic font. The dotted line indicates the limit of detection of FFRNT50. FFRNT50 <20 was treated as 10 for plot purposes and statistical analysis. USA-WA1/2020 versus BA.1-spike and other sublineage-spikes: 0.053 and <0.0001, respectively; BA.1-spike versus other sublineage-spike: <0.0001; BA.2-spike versus BA.2.12.1-, BA.3-, and BA.4/5-spike: 0.0006, 0.0215, and <0.0001, respectively; BA.2.12.1-spike versus BA.3- and BA.4/5-spike: 0.0309 and <0.0001, respectively; BA.3-spike versus BA.4/5-spike SARS-CoV-2: <0.0001.
(B) FFRNT50 of two-dose-vaccine-plus-BA.1-infection sera. Thirty-eight sera were collected from individuals who received three doses of vaccine and subsequently contracted BA.1 infection. The p values (Wilcoxon matched-pairs signed-rank test) for group comparison of GMTs are indicated below. USA-WA1/2020 versus all Omicron sublineage-spike: <0.0001; BA.1-spike versus other sublineage-spike: <0.0001; BA.2-spike versus Omicron BA.2.12.1-, BA.3-, BA.4/5, and BA.2.75-spike: 0.0082, 0.9095, <0.0001, and 0.093, respectively; BA.2.12.1-spike versus Omicron BA.3-, BA.4/5, and BA.2.75-spike: 0.0018, <0.0001, and 0.58, respectively; BA.3-spike versus BA.4/5- and BA.2.75-spike: <0.0001 and 0.062, respectively; BA.4/5-spike versus BA.2.75-spike: <0.0001.
In contrast, the GMT ratios between the two-dose-vaccine-plus-BA.1-infection and the four-dose-vaccine sera were 1.4, 4.8, 3.1, 4.1, 4.9, and 3.9 when neutralizing USA-WA1/2020, BA.1-, BA.2-, BA.2.12.1-, BA.3-, and BA.4/5-spike viruses, respectively. The result suggests that, compared with the two extra doses of vaccine in the four-dose-vaccine sera, the BA.1 infection in the two-dose-vaccine-plus-BA.1-infection sera is more efficient in boosting both the magnitude and breadth of neutralization against all Omicron sublineages; however, the neutralization against BA.5 was still the lowest among all tested sublineages.
For the two-dose-vaccine-plus-BA.1-infection sera, the GMTs against BA.1-, BA.2-, BA.2.12.1-, BA.3-, and BA.4/5-spike viruses were 1.2-, 2.9-, 2.2-, 2.6-, and 7.7-fold lower than the GMT against the USA-WA1/2020, respectively (Figure 2A); similar results were observed for the three-dose-vaccine-plus-BA.1-infection sera, with GMTs against BA.1-, BA.2-, BA.2.12.1-, BA.3-, and BA.4/5-spike viruses that were 1.5-, 3.0-, 2.5-, 2.9-, and 10-fold lower than the GMT against the USA-WA1/2020, respectively (Figure 2B). The GMT decreases against Omicron sublineages for the two-dose-vaccine-plus-BA.1-infection sera and those for the three-dose-vaccine-plus-BA.1-infection sera are significantly less than those observed for the four-dose-vaccine sera (compare Figures 2A and 2B with Figure 1C). The results again indicate that BA.1 infection of vaccinated people efficiently boosts the breadth of neutralization against all tested Omicron sublineages. However, such BA.1-infection-mediated boost of neutralizing magnitude/breadth is dependent on previous vaccination. This is because BA.1 infection of unvaccinated people did not elicit greater neutralizing magnitude/breadth against Omicron sublineages than three doses of mRNA vaccine.10
Neutralization against Omicron sublineage BA.2.75
To assess the neutralization of sublineage BA.2.75, we engineered the complete spike gene of BA.2.75 (Figure 1A) into the backbone of mNG USA-WA1/2020, resulting in BA.2.75-spike mNG SARS-CoV-2. The BA.2.75-spike mNG SARS-CoV-2 was sequenced to ensure no undesired mutations. When tested with four-dose-vaccine sera, the neutralizing GMT against BA.2.75-spike virus was 2.8-fold higher than that against BA.5-spike virus (Figure 1C). Similarly, when tested with three-dose-vaccine-plus-BA.1-infection sera, the neutralizing GMT against BA.2.75-spike virus was 3.4-fold higher than that against BA.5-spike virus (Figure 2B). Collectively, the results indicate that BA.2.75 is less immune-evasive than BA.4/5.
Limitations of the study
Our study has several limitations. First, this study lacks analysis of sera from vaccinated people who were infected with Omicron sublineages other than BA.1. It would be particularly important to study vaccinated individuals who contracted BA.5 infection. Second, we have not analyzed T cells and non-neutralizing antibodies that have Fc-mediated effector functions. These two immune arms, together with neutralizing antibodies, protect patients from severe disease.19 Many T cell epitopes after vaccination or natural infection are preserved in Omicron spikes.20 Third, the relatively small sample size and heterogeneity in vaccination/infection time have weakened our ability to compare the results among the three distinct serum panels. Regardless of these limitations, our results consistently showed that (1) the booster effect by dose four of the mRNA vaccine is less pronounced against BA.4/5 compared with USA-WA1/2020 and other Omicron sublineages; (2) four doses of the current mRNA vaccine elicit low neutralization against BA.5 spike, rationalizing the need to update the vaccine sequence to match the highly immune-evasive and prevalent BA.5; and (3) vaccinated individuals with BA.1 breakthrough infection develop greater neutralizing magnitude/breadth against Omicron sublineages than those who have received four doses of mRNA vaccine. These laboratory investigations, together with real-world vaccine effectiveness data, will continue to guide vaccine strategy and public health policy.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli strain Top10 | ThermoFisher Scientific | Cat# C404006 |
| TransforMax™ EPI300™ Chemically Competent E. coli | Lucigen Corporation | Cat# C300C105 |
| mNG USA-WA1/2020 SARS-CoV-2 | Xie et al., 202021 | N/A |
| BA.1-, BA.2-, BA.2.12.1-, BA.3-, BA.4/5-spike mNG SARS-CoV-2s | Kurhade et al., 2022b10 | N/A |
| Biological samples | ||
| Human serum | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol™ LS Reagent | ThermoFisher Scientific | Cat# 10296028 |
| Critical commercial assays | ||
| T7 mMessage mMachine kit | ThermoFisher Scientific | Cat# AM1344 |
| Ingenio® Electroporation solution | Mirus Bio LLC | Cat# MIR 50117 |
| Direct-zol RNA Miniprep Plus kit | Zymo Research | Cat# R2072 |
| NEBuilder HiFi DNA Assembly kit | New England Biolabs | Cat#E5520S |
| SuperScript™ IV One-Step RT-PCR System | ThermoFisher Scientific | Cat#12594025 |
| Experimental models: Cell lines | ||
| Vero E6 cells | ATCC | Cat# CRL-1586, RRID:CVCL_0574 |
| Vero E6-TMPRSS2 cells | SEKISUI XenoTech, LLC | N/A |
| Experimental models: Organisms/strains | ||
| Omicron variant sublineage BA.1 | GISAID EPI_ISL_6640916 | N/A |
| Omicron variant sublineage BA.2 | GISAID EPI_ISL_6795834.2), | N/A |
| Omicron variant sublineage BA.2.12.1 | GISAID EPI_ISL_12115772) | N/A |
| Omicron variant sublineage BA.4 | GISAID EPI_ISL_11542270 | N/A |
| Omicron variant sublineage BA.2.75 | EPI_ISL_13521499 | N/A |
| Recombinant DNA | ||
| pUC57-CoV2-F1 | Xie et al., 202021 | N/A |
| pCC1-CoV2-F2 | Xie et al., 202021 | N/A |
| pCC1-CoV2-F3 | Xie et al., 202021 | N/A |
| pUC57-CoV2-F4 | Xie et al., 202021 | N/A |
| pCC1-F567-BA.2.75 spike | This paper | N/A |
| Software and algorithms | ||
| Prism 9.0 software | GraphPad | N/A |
| Illustrator CC | Adobe | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Pei-Yong Shi (peshi@utmb.edu).
Materials availability
The Omicron-spike SARS-CoV-2 has been deposited to the World Reference Center for Emerging Viruses and Arboviruses (https://ww.utmb.edu/wrceva) at UTMB for distribution. All reagents generated in this study are available from the lead contact with a completed Materials Transfer Agreement.
Data and code availablity
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Ethical statement
All virus work was performed in a biosafety level 3 (BSL-3) laboratory with redundant fans in the biosafety cabinets at The University of Texas Medical Branch at Galveston. All personnel wore powered air-purifying respirators (Breathe Easy, 3M) with Tyvek suits, aprons, booties, and double gloves.
The research protocol regarding the use of human serum specimens was reviewed and approved by the University of Texas Medical Branch (UTMB) Institutional Review Board (IRB number 20–0070). No informed consent was required since these deidentified sera were leftover specimens before being discarded. No diagnosis or treatment was involved either.
Cells
Vero E6 (ATCC® CRL-1586) was purchased from the American Type Culture Collection (ATCC, Bethesda, MD), and maintained in a high-glucose Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS; HyClone Laboratories, South Logan, UT) and 1% penicillin/streptomycin at 37°C with 5% CO2. Culture media and antibiotics were purchased from ThermoFisher Scientific (Waltham, MA). The cell line was tested negative for mycoplasma.
Recombinant SARS-CoV-2
Viruses were recovered from infectious cDNA clones of SARS-CoV-2 and propagated on the Vero-E6 TMPRSS2 cells in the DMEM medium (GIBCO) supplemented with 2% fetal bovine serum (FBS) and 1%penecillin/streptomycin (GIBCO).
Human serum
Three panels of human sera were used in the study. The first panel consisted of 25 pairs of sera collected from individuals 3–8 months after vaccine dose 3, and no more than 3 months after dose 4 of the Pfizer-BioNTech or Moderna vaccine. This panel had been tested negative for SARS-CoV-2 nucleocapsid protein expression using Bio-Plex Pro Human IgG SARS-CoV-2 N/RBD/S1/S2 4-Plex Panel (Bio-rad). The second serum panel (n = 29) was collected from individuals who had received 2 doses of mRNA vaccine and subsequently contracted Omicron BA.1. The third serum panel (n = 38) was collected from individuals who had received 3 doses of mRNA vaccine and subsequently contracted Omicron BA.1. The genotype of infecting virus was verified by the molecular tests with FDA’s Emergency Use Authorization and Sanger sequencing. The de-identified human sera were heat-inactivated at 56°C for 30 min before the neutralization test. The serum information is presented in Tables S1–S3. The age distribution of the human patients in each panel is the following: panel 1 aged 59–94 yrs with a medium of 80 yrs, panel 2 aged 18–96 yrs with a media of 46 yrs, panel 3 aged 26–84 yrs with a medium of 64 yrs. Both genders of human patients were included. The gender distribution of the human patients in each panel is the following: 52%/48% (female/male) in panel 1, 55%/45% in panel 2; 55%/45% in panel 3.
Method details
Recombinant Omicron sublineage spike mNG SARS-CoV-2
Recombinant Omicron sublineage BA.1-, BA.2-, BA.2.12.1-, BA.3-, BA.4/5-spike mNG SARS-CoV-2s that was constructed by engineering the complete spike gene from the indicated variants into an infectious cDNA clone of mNG USA-WA1/2020 were reported previously.10 , 21 BA.2.75-spike sequence was based on GISAID EPI_ISL_13521499. Figure 1A depicts the spike mutations from different Omicron sublineages. The full-length cDNA of the viral genome bearing the variant spike was assembled via in vitro ligation and used as a template for in vitro transcription. The full-length viral RNA was then electroporated into Vero E6-TMPRSS2 cells. On day 3–4 post electroporation, the original P0 virus was harvested from the electroporated cells and propagated for another round on Vero E6 cells to produce the P1 virus. The infectious titer of the P1 virus was quantified by fluorescent focus assay on Vero E6 cells and sequenced for the complete spike gene to ensure no undesired mutations. The P1 virus was used for the neutralization test. The protocols for the mutagenesis of mNG SARS-CoV-2 and virus production were reported previously.13
Fluorescent focus reduction neutralization test
A fluorescent focus reduction neutralization test (FFRNT) was performed to measure the neutralization titers of sera against USA-WA1/2020, BA.1-, BA.2-, BA.2.12.1-, BA.3-, and BA4/5-spike mNG SARS-CoV-2. The FFRNT protocol was reported previously.9 Vero E6 cells were seeded onto 96-well plates with 2.5×104 cells per well (Greiner Bio-one™) and incubated overnight. On the next day, each serum was 2-fold serially diluted in a culture medium and mixed with 100–150 focus-forming units of mNG SARS-CoV-2. The final serum dilution ranged from 1:20 to 1:20,480. After incubation at 37°C for 1 h, the serum-virus mixtures were loaded onto the pre-seeded Vero E6 cell monolayer in 96-well plates. After 1 h infection, the inoculum was removed and 100 μL of overlay medium containing 0.8% methylcellulose was added to each well. After incubating the plates at 37°C for 16 h, raw images of mNG foci were acquired using Cytation™ 7 (BioTek) armed with 2.5× FL Zeiss objective with a wide field of view and processed using the software settings (GFP [469,525] threshold 4000, object selection size 50–1000 μm). The fluorescent mNG foci were counted in each well and normalized to the non-serum-treated controls to calculate the relative infectivities. The FFRNT50 value was defined as the minimal serum dilution to suppress >50% of fluorescent foci. The neutralization titer of each serum was determined in duplicate assays, and the geometric mean was taken. Tables S1–S3 summarize the FFRNT50 results.
Quantification and statistical analysis
Statistical analysis
Data were plotted as scatted dots. The geometric mean with 95% confidence intervals was presented. Sample size (n represents the number of human serum samples) was indicated in the corresponding figures. The nonparametric Wilcoxon matched-pairs signed rank test was used to analyze the statistical significance in Figures 1 and 2. Friedman with Dunn’s multiple comparisons test was performed to assess the statistical significance of the increase in the neutralization of sera against variants in Figure 1D. Figures were initially plotted in GraphPad Prism and assembled using Adobe illustrator.
Acknowledgments
We thank colleagues at the University of Texas Medical Branch (UTMB) for helpful discussions. We thank Michael L O’Rourke from the Information System Department at UTMB for assisting with electronic medical record systems. P.-Y.S. was supported by NIH contract HHSN272201600013C and awards from the Sealy & Smith Foundation, the Kleberg Foundation, the John S. Dunn Foundation, The Amon G. Carter Foundation, the Summerfield Robert Foundation, and Edith and Robert Zinn.
Author contributions
Conceptualization, X.X., P.R., and P.-Y.S.; methodology, X.X., J.Z., C.K., M.L., P.R., and P.-Y.S.; investigation, X.X., J.Z., C.K., M.L., P.R., and P.-Y.S.; resources, X.X., P.R., and P.-Y.S.; data curation, X.X., J.Z., C.K., M.L., and P.R.; writing – original draft, X.X., J.Z., R.R., and P.-Y.S.; writing – review & editing, X.X., P.R., and P.-Y.S.; supervision, X.X., P.R., and P.-Y.S.; funding acquisition, P.-Y.S.
Declaration of interests
X.X. and P.-Y.S. have filed a patent on the reverse genetic system. X.X., J.Z., and P.-Y.S. received compensation from Pfizer for COVID-19 vaccine development.
Published: November 10, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111729.
Supplemental information
References
- 1.Frederik Plesner Lyngse Transmission of SARS-CoV-2 omicron VOC subvariants BA.1 and BA.2: evidence from Danish households. Preprint at bioRxiv. 2022 doi: 10.1101/2022.1101.1128.22270044. [DOI] [Google Scholar]
- 2.Sandile Cele L.J., Khan D.S.K., Moyo-Gwete T., Tegally H., Scheepers C., Amoako D., Karim F., Bernstein M., Lustig G., et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. Preprint at medRxiv. 2021 doi: 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- 3.Liu Y., Liu J., Xia H., Zhang X., Fontes-Garfias C.R., Swanson K.A., Cai H., Sarkar R., Chen W., Cutler M., et al. Neutralizing activity of BNT162b2-elicited serum. N. Engl. J. Med. 2021;384:1466–1468. doi: 10.1056/NEJMc2102017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Liu J., Xia H., Zhang X., Zou J., Fontes-Garfias C.R., Weaver S.C., Swanson K.A., Cai H., Sarkar R., et al. BNT162b2-Elicited neutralization against new SARS-CoV-2 spike variants. N. Engl. J. Med. 2021;385:472–474. doi: 10.1056/NEJMc2106083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J., Liu Y., Xia H., Zou J., Weaver S.C., Swanson K.A., Cai H., Cutler M., Cooper D., Muik A., et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature. 2021;596:273–275. doi: 10.1038/s41586-021-03693-y. [DOI] [PubMed] [Google Scholar]
- 6.Kurhade C., Zou J., Xia H., Cai H., Yang Q., Cutler M., Cooper D., Muik A., Jansen K.U., Xie X., et al. Neutralization of Omicron BA.1, BA.2, and BA.3 SARS-CoV-2 by 3 doses of BNT162b2 vaccine. Nat. Commun. 2022;13:3602. doi: 10.1038/s41467-022-30681-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans J.P., Zeng C., Carlin C., Lozanski G., Saif L.J., Oltz E.M., Gumina R.J., Liu S.L. Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia H., Zou J., Kurhade C., Cai H., Yang Q., Cutler M., Cooper D., Muik A., Jansen K.U., Xie X., et al. Neutralization and durability of 2 or 3 doses of the BNT162b2 vaccine against Omicron SARS-CoV-2. Cell Host Microbe. 2022;30:485–488.e3. doi: 10.1016/j.chom.2022.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou J., Xia H., Xie X., Kurhade C., Machado R.R.G., Weaver S.C., Ren P., Shi P.Y. Neutralization against Omicron SARS-CoV-2 from previous non-Omicron infection. Nat. Commun. 2022;13:852. doi: 10.1038/s41467-022-28544-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurhade C., Zou J., Xia H., Liu M., Yang Q., Cutler M., Cooper D., Muik A., Sahin U., Jansen K.U., et al. Neutralization of Omicron sublineages and Deltacron SARS-CoV-2 by 3 doses of BNT162b2 vaccine or BA.1 infection. Emerg. Microbes Infect. 2022;11:1828–1832. doi: 10.1080/22221751.2022.2099305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson B.A., Zhou Y., Lokugamage K.G., Vu M.N., Bopp N., Crocquet-Valdes P.A., Kalveram B., Schindewolf C., Liu Y., Scharton D., et al. Nucleocapsid mutations in SARS-CoV-2 augment replication and pathogenesis. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y., Zhang X., Liu J., Xia H., Zou J., Muruato A.E., Periasamy S., Kurhade C., Plante J.A., Bopp N.E., et al. A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions. Nat. Commun. 2022;13:4337. doi: 10.1038/s41467-022-31930-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hachmann N.P., Miller J., Collier A.R.Y., Ventura J.D., Yu J., Rowe M., Bondzie E.A., Powers O., Surve N., Hall K., Barouch D.H. Neutralization escape by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4, and BA.5. N. Engl. J. Med. 2022;387:86–88. doi: 10.1056/NEJMc2206576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao Y., Yisimayi A., Jian F., Song W., Xiao T., Wang L., Du S., Wang J., Li Q., Chen X., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arora P., Kempf A., Nehlmeier I., Schulz S.R., Cossmann A., Stankov M.V., Jäck H.M., Behrens G.M.N., Pöhlmann S., Hoffmann M. Augmented neutralisation resistance of emerging omicron subvariants BA.2.12.1, BA.4, and BA.5. Lancet Infect. Dis. 2022;22:1117–1118. doi: 10.1016/S1473-3099(22)00422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan C.W., Lim B.L., Young B.E., Yeoh A.Y.Y., Yung C.F., Yap W.C., Althaus T., Chia W.N., Zhu F., Lye D.C., Wang L.F. Comparative neutralisation profile of SARS-CoV-2 omicron subvariants BA.2.75 and BA.5. Lancet Microbe. 2022 doi: 10.1016/S2666-5247(22)00220-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheward D.J., et al. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect. Dis. 2022;22:1421–1422. doi: 10.1016/S1473-3099(22)00524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou J., Xie X., Liu M., Shi P.Y., Ren P. Neutralization titers in vaccinated patients with SARS-CoV-2 Delta breakthrough infections. mBio. 2022;13 doi: 10.1128/mbio.01996-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barouch D.H. Covid-19 vaccines - immunity, variants, boosters. N. Engl. J. Med. 2022;387:1011–1020. doi: 10.1056/NEJMra2206573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Redd A.D., et al. Minimal crossover between mutations associated with omicron variant of SARS-CoV-2 and CD8(+) T-cell epitopes identified in COVID-19 convalescent individuals. mBio. 2022;13 doi: 10.1128/mbio.03617-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie X., Muruato A., Lokugamage K.G., Narayanan K., Zhang X., Zou J., Liu J., Schindewolf C., Bopp N.E., Aguilar P.V., et al. An infectious cDNA clone of SARS-CoV-2. Cell Host Microbe. 2020;27:841–848.e3. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.