Abstract
The Vibrio cholerae genome contains a 5.4-kb pil gene cluster that resembles the Aeromonas hydrophila tap gene cluster and other type IV-A pilus assembly operons. The region consists of five complete open reading frames designated pilABCD and yacE, based on the nomenclature of related genes from Pseudomonas aeruginosa and Escherichia coli K-12. This cluster is present in both classical and El Tor biotypes, and the pilA and pilD genes are 100% conserved. The pilA gene encodes a putative type IV pilus subunit. However, deletion of pilA had no effect on either colonization of infant mice or adherence to HEp-2 cells, demonstrating that pilA does not encode the primary subunit of a pilus essential for these processes. The pilD gene product is similar to other type IV prepilin peptidases, proteins that process type IV signal sequences. Mutational analysis of the pilD gene showed that pilD is essential for secretion of cholera toxin and hemagglutinin-protease, mannose-sensitive hemagglutination (MSHA), production of toxin-coregulated pili, and colonization of infant mice. Defects in these functions are likely due to the lack of processing of N termini of four Eps secretion proteins, four proteins of the MSHA cluster, and TcpB, all of which contain type IV-A leader sequences. Some pilD mutants also showed reduced adherence to HEp-2 cells, but this defect could not be complemented in trans, indicating that the defect may not be directly due to a loss of pilD. Taken together, these data demonstrate the effectiveness of the V. cholerae genome project for rapid identification and characterization of potential virulence factors.
Vibrio cholerae is a gram-negative bacterial pathogen that causes the waterborne diarrheal disease cholera. Following ingestion by a host and entry into the upper intestine, V. cholerae colonizes the intestinal mucosa and begins to export enterotoxins, including the major virulence toxin, cholera toxin (CT). The activity of CT elicits severe diarrhea in the infected host, resulting in extreme dehydration, the hallmark of cholera.
Although extensive research has elucidated the key features of toxin production and regulation, the basic mechanism underlying the initial colonization of the intestine by V. cholerae remains elusive. Much work to date has focused on the identification of V. cholerae pili. V. cholerae produces at least three morphologically distinct types of pili (17). The first type, the toxin-coregulated pili (TCP), are bundle-forming pili that are coordinately regulated with CT (53). These pili are absolutely essential for colonization of the intestine by V. cholerae, both in the infant mouse colonization model and in human volunteers (2, 20, 40, 50, 53, 55). However, several observations suggest that TCP may not be directly required for adherence. Purified TCP do not bind to human intestinal epithelium, and the adherence of V. cholerae to intestinal epithelium and various epithelial cell lines is not blocked by growth under non-TCP-inducing conditions or by anti-TCP antibodies (3, 12, 48, 51). Further, when classical V. cholerae strains are grown under TCP-expressing conditions, the bacteria aggregate, suggesting that TCP plays a key role in bacterium-to-bacterium adhesion (9, 53). These observations indicate that other V. cholerae factors may mediate cellular adherence.
A second type of V. cholerae pili, mannose-sensitive hemagglutination (MSHA) pili, has also been investigated. These investigations have been done almost exclusively with El Tor strains because classical strains produce few or no pili (26). Within the El Tor strains, MSHA pili are essential for the hemagglutination of erythrocytes, although no other role in adherence or pathogenesis has been ascribed to these pili (24, 25). Disruption of the gene encoding the primary pilin subunit, mshA, has no effect on colonization of infant mice (2, 55) or on disease symptoms in human volunteers (50). Further, spontaneous mutants defective in hemagglutinating activity adhere well to intestinal tissue samples (42, 54). Recently, it was shown that MSHA pili mediate the adherence of V. cholerae El Tor strains to solid substrates, suggesting that they are important for survival in the environment rather than in the host (58).
Interestingly, the major pilin subunits of both TCP and MSHA pili, TcpA and MshA, respectively, are members of the type IV protein superfamily (25, 46). The type IV proteins all have recognizable N-terminal leader sequences that specify cleavage and N-methylation by specific prepilin leader peptidases (21). In addition to MshA and TcpA, V. cholerae contains at least eight other type IV proteins: EpsG, EpsH, EpsI, and EpsJ, which are part of a type II export machinery that secretes CT and other toxins; MshB, MshC, and MshD, which are part of the MSHA pilus assembly machinery; and TcpB, a protein essential for the production of TCP (25, 31, 40, 44). The tcpJ gene encodes a prepilin peptidase that processes TcpA into a form that can be assembled into TCP. Surprisingly, the disruption of tcpJ abolishes TCP assembly but does not affect either toxin secretion or hemagglutination of erythrocytes (28). This result suggests that a second prepilin peptidase is responsible for processing of the Eps and Msh proteins (28). The identification of a third type of pili by electron microscopy studies also suggests that another, uncharacterized pilus may be present in V. cholerae (17).
Recent progress by The Institute for Genomic Research (Gaithersburg, Md.) on the V. cholerae genome project has facilitated the rapid identification of genes of particular interest. In this paper, we describe the use of early data releases to identify a new gene cluster that is similar to type IV-A pilus assembly gene clusters. Proteins encoded by this gene cluster include a second prepilin peptidase that is important for toxin secretion, MSHA, and TCP production, as well as a new, putative type IV pilin protein. However, a role in pathogenesis for this pilin protein could not be established.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The V. cholerae strains used in this study are listed in Table 1. Escherichia coli DH5α and DH5αλpir were used for construction of plasmids. SM10λpir and β2115 were used to deliver plasmids to V. cholerae by conjugation. All strains were grown on Luria-Bertani (LB) medium at 37°C, except as noted otherwise. Antibiotics (micrograms per milliliter) used were as follows: ampicillin, 50; chloramphenicol, 4; kanamycin, 50; and streptomycin, 100.
TABLE 1.
V. cholerae strains used in this studya
| Strain | Description of genotype and construction | Reference(s) or source |
|---|---|---|
| O395 | Wild-type classical biotype; Smr | Laboratory strain |
| N16961Sm | Spontaneous Smr isolate of wild-type El Tor biotype N16961 | The Salk Institute and this study |
| O395N1 | O395 ΔctxA Smr | 33 |
| P4 (SM44) | P27459 (El Tor) ΔctxAB Smr Kmr | 14 |
| BGD4 | O395 ΔtcpA Smr | 9 |
| KFV5 | O395 pilD::kanπ; constructed with pKJF306; Smr Kmr | This study |
| KFV5R | KFV5 reverted to faster growth; Smr Kmr | This study |
| KFV5R(pKJF308) | KFV5R complemented with pilD in trans under the control of the arabinose-inducible promoter PBAD; Smr Kmr Apr | This study |
| KFV5R(pKJF308Cm) | KFV5R complemented with pilD in trans under the control of the arabinose-inducible promoter PBAD; Smr Kmr Cmr | This study |
| KFV6 | N16961Sm ΔpilA; constructed with pKJF313; Smr | This study |
| KFV8 | O395 ΔpilA; constructed with pKJF313; Smr | This study |
| KFV9 | O395N1 ΔpilA; constructed with pKJF313; Smr Kmr | This study |
| KFV10 | N16961Sm ΔlacZ; constructed with pDLT; Smr | This study |
| KFV11 | N16961Sm ΔmshA; constructed with pHT1; Smr | 58 |
| KFV12 | N16961Sm ΔpilA ΔmshA; constructed from KFV11 with pHT1; Smr | This study |
| KFV16 | O395N1 ΔlacZ; constructed with pDLT; Smr Kmr | This study |
| KFV18 | N16961Sm pilD::kanπ; constructed with pKJF306; Smr Kmr | This study |
| KFV18R | KFV18 reverted to faster growth; Smr Kmr | This study |
| KFV18R(pKJF308) | KFV18R complemented with pilD in trans under the control of the arabinose-inducible promoter PBAD; Smr Kmr Apr | This study |
| KFV26 | P4 ΔlacZ; constructed with pDLT; Smr Kmr | This study |
| KFV32 | O395 ΔpilD; constructed with pKJF324; Smr | This study |
| KFV33 | N16961Sm ΔtcpA; constructed with pHT3; Smr | This study |
| KFV36 | N16961Sm ΔpilA ΔpilD; constructed from KFV6 with pKJF324; Smr | This study |
| KFV38 | P4 ΔpilA; constructed with pKJF329; Smr Kmr | This study |
| KFV43 | N1691Sm ΔhapA; constructed with pCVDΔHapSal; Smr | This study |
| KFV44 | KFV18R ΔhapA; constructed from KFV18R with pCVDΔHapSal; Smr Kmr | This study |
| KP8.97 | O395 tcpB::TnphoA; Smr Kmr | 40 |
| KP9.79 | O395 tcpA::TnphoA; Smr Kmr | 40 |
| Lac1 | O395 ΔlacZ; reconstruction of CG842 (12); Smr | 56a |
| O395(CTXφ704A) | O395 carrying the replicative form of transducing phage CTXφ704A; Smr Apr | 30a |
| Peru-15(CTXφ-Km) | Peru-15 carrying the replicative form of transducing phage CTXφ-Km; Smr Kmr | 29, 57 |
For a more detailed description of strain construction, see Materials and Methods.
PCR and DNA sequencing.
All oligonucleotides used for PCR and DNA sequencing were obtained from Biosynthesis, Inc. (Lewisville, Tex.) or Operon Technologies (Alameda, Calif.) and are listed in Table 2. PCR was performed with TaKaRa Taq polymerase and reagents from Oncor (Gaithersburg, Md.), PCR Supermix from Gibco BRL (Gaithersburg, Md.), or Vent Taq polymerase and reagents from New England BioLabs (Beverly, Mass.). PCR templates were prepared by suspending a single bacterial colony in 200 μl of distilled H2O, of which 1 μl was used in either a 25- or a 50-μl PCR amplification. For sequencing of the pil operon and for preparation of all plasmids, colonies of V. cholerae N16961 or N16961Sm were used as the PCR template, except as noted otherwise. All PCRs were performed with standard amplification parameters and an MJ Research Thermocycler (model PTC-200). For direct sequencing, PCR products were purified by two passages over a QiaQuick PCR purification kit (Qiagen, Valencia, Calif.). Automated DNA sequencing was done at the Harvard Microbiology Core Sequencing Facility. The reported sequence has a minimum of fourfold coverage with at least two sequences for both the coding and the noncoding strands. The DNA sequence was assembled and open reading frames were assigned with the GeneWorks Analysis software package.
TABLE 2.
Oligonucleotides used in this study
| Oligo-nucleotide | Location (nt)a | Sequence (5′-3′)b |
|---|---|---|
| NADC1 | 9–31 | CTTGAGATGATGCGTGAAGCGGT |
| PRO1 | 435–418 | CATAATTTCCTCCGTAG |
| PILA1 | 869–891 | GTACTGCAGGTGCAACAATTAAC |
| PILA2 | 432–453 | AGTCATATGAAAGCGTATAAGAACAAA |
| PILA3 | 891–871 | GGTGAGCTCGTTAATTGTTGCACCTGCAGT |
| PILB1 | 894–914 | ACACCTACGGAGGAAATTATGCTCACCAAC CTTGTTGCT |
| PILB2 | 1520–1543 | CATAGCCTAGGCGGACGCGACGC |
| PILB3 | 1397–1421 | CTGCAATCCATTGAAGATCTCAGC |
| PILB4 | 1978–2000 | AATACCAAGTGCAAGTGCAGCCG |
| PILB5 | 1695–1672 | AATGGCGGTATCGCCCGTTAGGC |
| PILC1 | 3442–3420 | GAAGCCTTAGCGAGCACATTACC |
| PILC2 | 2905–2881 | CCAACTTGAGGGCTTGCACAATGG |
| PILC3c | 2385–2361 | AATAGCCGCCCGTACACTCGTTAC |
| PILC4 | 2664–2687 | TGGAAGGCATCAACAGCAACGG |
| PILC5 | 3192–3213 | ATGCTCACCATGGTCATCCCAG |
| PILC6 | 3911–3889 | CATTCTCTTGCTCTAAGCGAAT |
| PILD1 | 3891–3911 | GAATTCGCTTAGAGCAAGAGAATG |
| PILD2 | 4796–4773 | ACGCGTCGACAAAACTCATAACGGTTG |
| PILD3 | 4318–4327 | GCTATGTGTTGATTGCGGCA |
| PILD5 | 4045–4022 | CGTGATTCCGCGCGCCATTCACG |
| YACE1 | 4780–4802 | ATTCGCTTAGAGCAAGAGAATGAGTTTTGT CGTAGCATTGA |
| YACE2 | 5394–5371 | ATTCTCACAAATTTTGCCTGCTC |
According to the pil operon sequence determined for this paper.
Underlined nucleotides are not exact matches to the sequence and were altered either to add restriction enzyme sites or to facilitate crossover PCR.
Actually binds within the pilB open reading frame.
Construction of counterselectable plasmids.
pKJF306 is a streptomycin-based counterselectable plasmid which generates kanamycin insertions in pilD. Primers PILD1 and PILD2 were used in a Taq polymerase PCR to generate a 908-bp product encompassing the pilD gene; this fragment was ligated directly into the TA cloning vector pCR2.1 (Invitrogen, Carlsbad, Calif.) to create pKJF302. The fragment was then moved to pBluescript IIKS(+) (Stratagene, La Jolla, Calif.) as an EcoRI fragment to generate pKJF303. The kanamycin resistance cassette kanπ (Pharmacia, Piscataway, N.J.) as an HaeIII fragment and inserted into pilD on pKJF303 between two Eco47III sites found at nucleotides (nt) 294 and 334 of the pilD PCR product to generate pKJF305. The entire pilD::kanπ fragment was then moved into the streptomycin-based counterselectable plasmid pKAS32 (47) as an EcoRI fragment, generating plasmid pKJF306.
Counterselectable plasmids pKJF313 and pKJF329 were used to generate nonpolar deletions of pilA. The first step in the construction of these plasmids was crossover PCR. PCR with primers NADC1 and PRO1 amplifies the region 428 bp upstream of the putative translational start site of pilA, while PILB1 and PILB2 amplify the region 647 bp downstream of pilA. Primers PRO1 and PILB1 have 21 bp of overlapping nucleotide sequence, such that the crossover PCR brings the second codon of pilB to the AUG codon of pilA. PCR products resulting from Vent Taq polymerase PCR with primer pair NADC1-PRO1 or PILB1-PILB2 were purified by two passages over the QiaQuick PCR purification kit. Five microliters of each product was then added to 45 μl of PCR Supermix and treated in a 10-cycle reaction (92°C, 40 s; 50°C, 40 s; 72°C, 45 s) without primers to enhance crossover extension of the two products. One microliter of this reaction mixture was then used as a template with PCR Supermix and NADC1 and PILB2 as primers to amplify the 1,075-bp crossover product, which was then directly ligated into pCR2.1 to create pKJF310. The crossover fragment was then moved to the polylinkers of the streptomycin-based counterselectable plasmid pKAS46 (47) and the sucrose-based counterselectable plasmid pWM91 (34) as a SpeI-XhoI fragment to generate pKJF313 and pKJF329, respectively.
Counterselectable plasmid pKJF324 generates a nonpolar deletion of pilD. This plasmid was made following the crossover PCR protocol described above. In the first reaction, primers PILC5 and PILC6 amplify the region 850 bp upstream of pilD, while YACE1 and YACE2 amplify the region 750 bp downstream of pilD. YACE1 and PILC6 share 21 nt of homology such that the second codon of yacE is linked to the first AUG codon of pilD in the crossover PCR. Following crossover PCR, the 1,800-bp product was cloned into pCR2.1 and was then moved to the polylinker of sucrose-based counterselectable plasmid pWM91 (34) as a SpeI-XhoI fragment to create pKJF324.
Construction of pilD-complementing plasmids.
Primers PILD1 and PILD2 are designed to introduce EcoRI and SalI sites at the 5′ and 3′ ends of pilD, respectively. Following PCR amplification, the 908-bp product was treated for 20 min at 72°C with Taq polymerase to add an adenosine nucleotide to the termini for direct cloning into pCR2.1 to create pKJF304. The novel EcoRI and SalI sites were then used to move pilD into the pBAD18 (15) polylinker, placing pilD under the control of the arabinose-inducible promoter PBAD but with its own ribosome binding site. For alternative antibiotic selection, the pilD insert was moved from pBAD18 to pBAD18Cm (15) by use of a unique MluI site found on both plasmids and SalI from the polylinker. The resulting plasmids, pKJF308 and pKJF308Cm, were transferred to V. cholerae by electroporation.
Generation of mutations in V. cholerae strains by use of counterselectable markers.
Counterselectable plasmids pKJF306 (pKAS32 pilD::kanπ), pKJF313 (pKAS46 ΔpilA), pKJF324 (pWM91 ΔpilD), pKJF329 (pWM91 ΔpilA), pDLT (pCVD442 ΔlacZ [57a]), pHT1 (pCVD442 ΔmshA1 [55]), pHT3 (pCVD442 ΔtcpA10 [55]), and pCVDΔHapSal (pCVD442 ΔhapA [17a]) were introduced into V. cholerae by conjugation on an LB agar plate. Counterselections were done by the protocol of Metcalf et al. (34). Briefly, four cointegrants were purified by streaking one time under selection and then were passaged one time without selection to allow recombination to occur. Sixteen independent colonies were then streaked on the counterselection medium: LB medium containing streptomycin at 3 mg/ml for streptomycin-based selection and LB medium without NaCl but containing 6% sucrose for sucrose-based selection. Best results were achieved when counterselection plates were incubated at room temperature for 2 days. Isolated colonies were checked by PCR for gain of the mutation and loss of the wild-type gene.
Agg and CTXφ transduction.
Classical V. cholerae cultures were grown in 5 ml of LB broth at 30°C with rolling for optimal production of TCP (11). The autoagglutination (Agg) phenotype was scored visually. For CTXφ transductions, phage were prepared by filtering culture supernatant of Peru-15(CTXφ-Km) or O395(CTXφ704A) with a 0.2-μm-pore-size syringe filter. Fifty microliters of phage preparation was mixed with 50 μl of TCP-expressing culture, and the mixture was allowed to stand for 45 min at room temperature to allow for infection by the phage and expression of antibiotic resistance genes. The mixture was then diluted and plated. The transduction frequency is expressed as the CFU of transductants over the CFU of the recipient strain.
CT assay.
For optimal toxin production, classical strains and El Tor strains containing plasmid pBAD18Cm-ToxT (19) were grown at 30°C with rolling for 16 h. One milliliter of culture was spun down in a microcentrifuge, and the culture supernatant was set aside. The bacterial cells were resuspended in 1 ml of 10 mM Tris-HCl–1 mM EDTA–20% glucose (pH 7.5) and sonicated (Branson Sonifier 350) twice for 15 s each time on 20% power. CT present in 100 μl of culture supernatant or sonicate was measured by the GM1-ganglioside enzyme-linked immunosorbent assay with purified CT to generate a standard curve (11).
HAP assays.
The presence of hemagglutinin-protease (HAP) was detected on LB agar plates containing 1% nonfat milk (37). For some assays, 4 ml of agar was solidified in divided glass plates such that different quadrants contained different antibiotics and/or inducing compounds.
MSHA assays.
MSHA assays were performed as previously described, except that washed defibrinated sheep blood was substituted for human or chicken erythrocytes (12).
Infant mouse colonization.
Overnight cultures of V. cholerae strains were diluted 1:100 in fresh LB medium and grown with rolling to the mid-log to late log phase. Cultures were diluted in 0.15 M NaCl, and 8 μl of blue food coloring was added. Five- to 7-day-old infant mice were taken from their mothers at least 5 h prior to oral infection. Fifty microliters of inoculum was delivered orally to anesthetized mice. Mice were kept at 30°C for a full 24 h, at which time they were sacrificed and their intestines were dissected from above the cecum. The intestines were homogenized in 5 ml of LB broth, diluted, and plated on LB agar with streptomycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). For competition assays, CFU of blue (mutant) and white (wild-type) strains were counted, and the colonization index was calculated as the ratio of blue colonies to white colonies.
HEp-2 cell adherence assays.
Overnight cultures of V. cholerae strains were diluted 1:100 in fresh LB medium and grown at 37°C with rolling to the mid- to late-log phase. Bacterial cells were washed in phosphate-buffered saline (PBS) and adjusted to 109 bacteria per ml. Ten microliters was added to HEp-2 epithelial cells (ATCC F-13959) that had been grown in RPMI 1640 medium containing 10% fetal calf serum without antibiotics in 24-well dishes to 105 cells per well. For classical strains, plates were spun for 10 min at 300 × g and incubated at 37°C in 5% CO2 for 1 h. Cells were washed four times with PBS, and adherent bacteria were recovered in 1 ml of PBS–0.1% Triton X-100. For El Tor strains, plates were spun for 2 min and incubated for 15 min to reduce nonspecific adherence to the plastic. Cells were washed and recovered in 1 ml of PBS–1.0% Triton X-100. Percent adherence was calculated as CFU adherent divided by CFU added times 100.
Nucleotide sequence accession number.
The nucleotide sequence reported in this study was assigned GenBank accession no. AF109904.
RESULTS
Bioinformatics and sequencing of the pil gene cluster of V. cholerae.
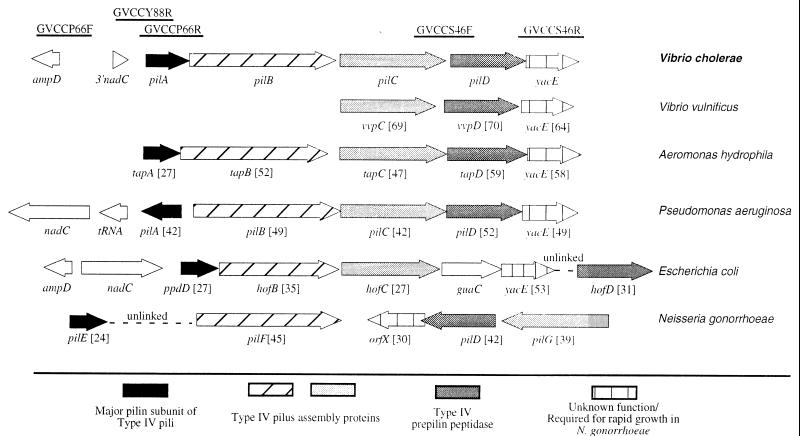
The publicly available sequences from the V. cholerae genome project (56a) were examined for the presence of open reading frames with known functions in pathogenesis. Interestingly, sequence runs GVCCS46F and GVCCS46R contain sequences with similarity to two genes found in type IV-A pilus assembly operons (Fig. 1). In addition, sequence runs GVCCP66R and GVCCY88R encode a novel type IV cleavable pilin protein that is closely related to the type IV-A pilin subunit of Pseudomonas aeruginosa (Fig. 1). In order to show that sequences GVCCP66R and GVCCS46F are linked on the V. cholerae genome, primers PILA1 and PILC1 were used in a PCR to amplify a 2.6-kb product representing the intervening region. Primers PILD1 and PILD2 were used to generate a 908-bp product to demonstrate the linkage of GVCCS46F and GVCCS46R (data not shown). The entire 5,401-bp gene cluster was sequenced directly from progressively smaller PCR products such that the operon was sequenced with a minimum of fourfold coverage with at least two sequences for each strand. All sequences from the original data release by The Institute for Genomic Research were also confirmed by the same method.
FIG. 1.
Organization of six representative type IV pilus assembly operons. The bars at the top indicate the approximate locations and designations of DNA sequences obtained from the publicly available genomic database. Numbers in brackets indicate the percentages of amino acid identity of deduced protein sequences to the translated open reading frame of the appropriate V. cholerae gene. Percentages of identity were determined by a ClustalW alignment done with a MacVector software package. Sequences were assembled from GenBank and other public databases as follows: V. vulnificus (accession no. AF070934) (38), A. hydrophila (accession no. U20255) (39), P. aeruginosa (accession no. M32066 and M14849) (23, 35) (genomic data from www.pseudomonas.com), and E. coli (accession no. L28105, AE000409, and D26562) (4, 59), and N. gonorrhoeae (accession no. U32588 and X66144) (10, 43, 56).
The V. cholerae pil gene cluster resembles other type IV-A pilus gene clusters.
The 5,401-bp region of V. cholerae contains five complete open reading frames, apparently organized into a single operon. The designation vcpD has been proposed for the fourth gene of this cluster (32), based on the nomenclature of the V. vulnificus operon (38). However, the name vvpA, appropriate for the first gene by this nomenclature, has already been used to describe a V. vulnificus protease gene (6). To avoid further confusion, the nomenclature for P. aeruginosa, the organism with the first identified type IV pilus operon, has been adopted for the first four open reading frames, pilABCD, and the fifth gene is referred to as yacE, consistent with the E. coli genome project designation. As a whole, the cluster is referred to as the pil gene cluster.
The organization of the V. cholerae pil gene cluster shows a striking similarity to that of the Aeromonas hydrophila tap gene cluster (Fig. 1). Although other organisms also tend to group these five genes, V. cholerae and A. hydrophila are apparently the only organisms which group these genes into a single operon. Presumably, the V. vulnificus operon is similarly arranged, although only vvpC, vvpD, and yacE have been identified to date. Interestingly, like the type IV-A pilus assembly operon from E. coli, the V. cholerae pil gene cluster is linked to ampD and nadC, demonstrating that the arrangement of these genes is well conserved between these two gram-negative species.
The first open reading frame, pilA, has two potential translation start sites. The second start site was selected because a consensus ribosome binding site is located just upstream of the AUG, whereas no such site is present upstream of the first potential start site. Further, consistent with the grouping of V. cholerae PilA into the type IV-A family, the use of the second translation start site would give a type IV signal sequence of 11 amino acids. Use of the first potential translation start site would give PilA a long signal sequence of 25 amino acids, a feature more common in type IV-B pilin subunits, such as V. cholerae TcpA (13).
Although the amino acid sequence similarity among the type IV-A pilins is generally localized to the first 30 amino acids, the sequence of the pilA gene from classical strain O395 is 100% identical at both the protein and the nucleotide levels to the El Tor strain N16961 gene sequence, indicating that the pilA gene is highly conserved in V. cholerae.
The next two open reading frames, pilB and pilC, are likely to encode proteins essential for the assembly of a new type IV pilus (1). Recent work with P. aeruginosa has shown that pilB and pilC are merely 2 of up to 30 genes essential for the assembly of type IV pili. These genes include pilT, pilE, and pilU (1), for which open reading frames are also present in the V. cholerae genome (56a).
The fourth open reading frame encodes a putative member of the type IV prepilin peptidase family. These proteins recognize the type IV signal sequence and cleave the signal at a conserved glycine (36). The peptidase then modifies the next residue to the form that is assembled into either pili or other membrane structures (49). Like the pilA gene, the V. cholerae pilD gene is 100% conserved between the classical strain O395 and the El Tor strain N16961Sm.
The fifth open reading frame, yacE, is a member of a family of genes whose localization with type IV pilus assembly gene clusters is frequently conserved (Fig. 1), although its function is unknown.
Effect of various pilD mutations on growth patterns.
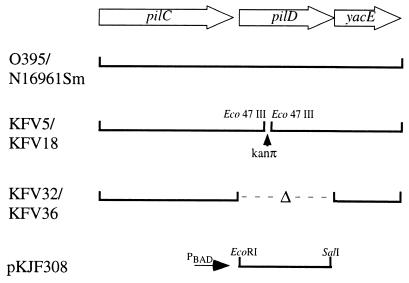
To test whether pilD encodes a prepilin peptidase essential for secretion and hemagglutination, we constructed a pilD mutant in which the kanamycin resistance cassette, kanπ, was inserted into the pilD gene by double homologous recombination with the streptomycin-based counterselectable plasmid pKJF306. Following mating into O395 and N16961Sm, cointegrants were counterselected on streptomycin and kanamycin to select for loss of the integrated plasmid and for maintenance of the kanπ cassette. However, following the selection process, only pinpoint colonies were recovered for both O395 and N16961Sm parental strains. These slowly growing mutants, designated KFV5 and KFV18 (Fig. 2), respectively, reverted to a faster growth pattern with a high frequency when grown in liquid cultures. Revertants for these strains were single colony purified and designated KFV5R and KFV18R, respectively.
FIG. 2.
Genetic organization of pilD mutants and the complementing plasmid. Construction of strains and plasmids is detailed in Materials and Methods. In addition to the pilD deletion, KFV36 also has a deletion in pilA.
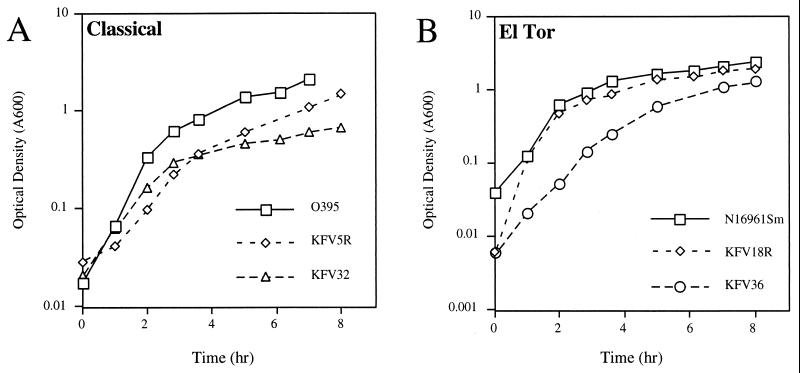
It was first surmised that the growth defect may be caused by the loss of the downstream gene yacE. Like the V. cholerae pilD insertion, transposon insertions in Neisseria gonorrhoeae pilD result in slow growth, but this slow growth is associated with the loss of the downstream gene orfX, a gene similar to V. cholerae yacE (10). A nonpolar pilD mutant of O395 was made by deleting the kanamycin cassette from KFV5 by use of the sucrose-based counterselectable plasmid pKJF324. As shown in Fig. 3A, the resulting strain, KFV32, grew slowly in cultures, demonstrating an apparent premature entry into the stationary phase. The revertant, the kanamycin insertion mutant KFV5R, although continuing to grow more slowly than the wild type, apparently escaped this premature stationary phase (Fig. 3A). In contrast, revertant KFV18R, an El Tor mutant, grew with a pattern similar to that of the wild type, suggesting that the mechanism of reversion in these two strains may differ.
FIG. 3.
Growth curves for pilD mutants. Overnight cultures were diluted 1:100 in 100 ml of LB broth with antibiotics and incubated with shaking at 37°C. At specific times, 1 ml of the cultures was removed and the optical density (A600) was measured. Time zero is the time at which the cultures exited the stationary phase.
Unfortunately, attempts to create a nonpolar deletion of pilD in El Tor strain N16961Sm by either deletion of pilD from the wild-type background or deletion of the kanπ insertion were not successful. However, it has been reported that slowly growing pilD mutants of N. gonorrhoeae can revert to faster growth by accumulating deletions in the pilin subunit gene, pilE (10). In accordance with this result, a nonpolar deletion of the pilD gene could be constructed in a pilA deletion background. However, as shown in Fig. 3B, this mutant, designated KFV36, grew more slowly than the wild type. Also, deletion of pilA from the classical deletion mutant KFV32 did not enhance its growth beyond that shown in Fig. 3A (data not shown). Thus, it seems that, in the absence of pilD, PilA precursor proteins are not processed and contribute in part, but not entirely, to slow growth.
V. cholerae pilD is essential for the secretion of CT and HAP.
The eps operon of V. cholerae encodes a type II secretion system that is essential for the export of CT, HAP, and other V. cholerae toxins (37, 44). Four of the proteins encoded by this operon, EpsG, EpsH, EpsI, and EpsJ, contain type IV signal sequences, although a gene for a type IV peptidase is absent from this operon (44). The signal sequence of EpsG is cleaved by N. gonorrhoeae PilD but not by V. cholerae TcpJ (44). These observations suggest that the newly recognized pilD gene of V. cholerae may encode a prepilin peptidase essential for toxin secretion.
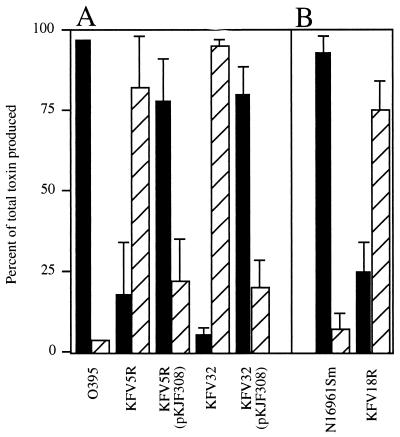
To determine if pilD of V. cholerae is essential for the processing of EpsG, EpsH, EpsI, and EpsJ, the various pilD mutants were analyzed for defects in the secretion of CT and HAP. As shown in Fig. 4A, wild-type classical strain O395 exported 97% of the CT that it produced. In contrast, most of the CT produced by mutants KFV5R and KFV32 accumulated within the bacteria and was not exported (Fig. 4A). Export could be restored by the introduction of pilD on a plasmid under the control of the arabinose-inducible promoter PBAD. The export of CT was not complemented either by the pBAD18 vector alone or in the absence of the inducer arabinose (data not shown).
FIG. 4.
Effect of pilD mutations on CT secretion. CT was measured as described in Materials and Methods. Strains carrying a pilD-complementing plasmid were grown in 0.05% arabinose to induce the expression of promoter PBAD, as were N16961Sm and KFV18R, which contain plasmid pBAD18Cm-ToxT for constitutive overexpression of CT. For both classical (A) and El Tor (B) strains, enzyme-linked immunosorbent assay plates were read with a microtiter reader, and amounts (nanograms) of intracellular (hatched bars) and extracellular (solid bars) toxins were determined by comparison against a standard curve. Results are presented as a percentage of the total toxin produced. Averages and standard deviations were determined from duplicates in a single experiment that is representative of at least two experiments for each sample.
El Tor pilD mutants also had defects in CT export. To increase the expression of CT, N16961Sm and KFV18R were transformed with plasmid pBAD18Cm-ToxT. The resulting strains overexpressed the regulatory protein ToxT and thereby had increased CT production in standard growth media. Even under these improved induction conditions, KFV18R produced about 10-fold less toxin than the wild type (data not shown). However, the majority of the toxin produced was not secreted, demonstrating that KFV18R had a generalized defect in CT secretion similar to that in the classical pilD mutants KFV5R and KFV32 (Fig. 4).
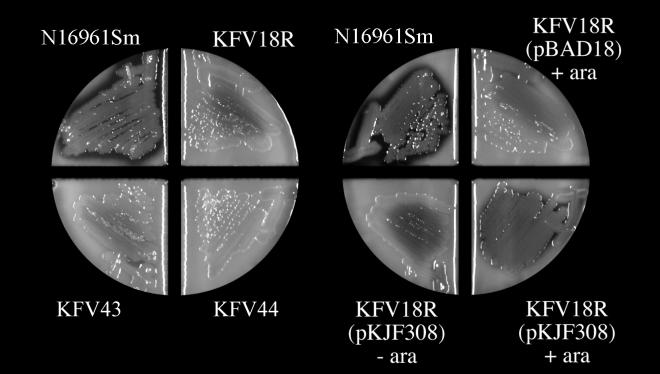
In addition to the loss of CT export, El Tor pilD mutants were also defective for the export of a second toxin, HAP. As shown in Fig. 5, HAP could be detected as a zone of clearing around colonies grown on agar plates containing 1% nonfat milk. The zone of clearing was decreased when the structural gene for HAP, hapA, was disrupted (Fig. 5, left). Similarly, pilD mutant KFV18R showed a decreased zone of clearing. Export could be restored by the introduction of pilD in trans only in the presence of the inducer arabinose and was not restored by the presence of the pBAD18 vector (Fig. 5, right). Similar results were found for the ΔpilA ΔpilD double mutant KFV36 (data not shown), although the small-colony phenotype of this strain alone could account for this defect, since HAP expression is proposed to be regulated by quorum sensing (22).
FIG. 5.
Effect of pilD mutations on HAP secretion. Strains were heavily streaked onto 1% nonfat milk agar in divided glass plates and incubated at 37°C for 24 h. The agar contained 0.05% arabinose (ara) as indicated. Plates were photographed with a Bio-Rad Fluor-S MultiImager.
Taken together, these data show that pilD mutants of both classical and El Tor strains have a generalized defect in toxin export that is due solely to pilD, since the mutant phenotype could be complemented in trans. This defect is probably directly due to the failure of a pilD-encoded peptidase to process EpsG, EpsH, EpsI, and EpsJ.
El Tor pilD mutants are defective in MSHA.
El Tor strains of V. cholerae produce a pilus that can hemagglutinate erythrocytes, but not in the presence of mannose (24). The major subunit of this pilus, MshA, and three accessory pilus assembly proteins, MshB, MshC, and MshD, have type IV cleavable sequences although, similar to the situation with the eps operon, a type IV prepilin peptidase is not encoded by the msh gene cluster (18, 25, 32). If pilD is essential for the construction of the MSHA pilus, then pilD mutants should be unable to hemagglutinate erythrocytes.
N16961Sm hemagglutinated sheep erythrocytes, and this hemagglutination was inhibited by both mannose and the nonhydrolyzable analog methyl-α-d-mannopyranoside but not by fucose or arabinose (Table 3). A deletion within the mshA gene in N16961Sm abolished this hemagglutination. Similarly, the El Tor pilD mutants KFV18R and KFV36 did not hemagglutinate erythrocytes. Hemagglutination was restored when these strains were complemented with pKJF308, which carries pilD in trans, but not when they carried the pBAD18 vector or were grown without arabinose to induce the expression of pilD on the plasmid (Table 3).
TABLE 3.
Effect of pilD mutations on MSHA activity
| Strain and growth condition | Hemagglutination activitya |
|---|---|
| N16961Sm | + |
| KFV11 (ΔmshA) | − |
| KFV18R (pilD::kanπ, reverted) | − |
| KFV18R (pBAD18) + 0.05% arabinose | − |
| KFV18R (pKJF308) + 0.05% arabinose | + |
| KFV18R (pKJF308) + no arabinose | − |
| KFV36 (ΔpilA ΔpilD) | − |
| KFV36 (pKJF308) + 0.05% arabinose | + |
| KFV36 (pKJF308) + no arabinose | − |
−, no hemagglutination; +, hemagglutination of sheep erythrocytes that was inhibited by mannose but not by fucose or arabinose.
These data show that pilD is essential for MSHA and that the defect is due solely to the loss of pilD, since the mutant phenotype could be rescued by the introduction of pilD in trans. This defect is presumably due to the loss of processing of the pilin subunit MshA as well as the three accessory proteins.
pilD mutants do not produce wild-type levels of TCP.
TCP are type IV pili of the B subfamily, members of which typically have longer type IV signal sequences than type IV-A pili and are carried on mobile elements, such as plasmids or pathogenicity islands (13). Since TcpJ processes the major TCP pilin subunit TcpA (28), it is not expected that pilD mutants should be defective for TCP production. However, it is possible that the essential accessory protein, TcpB (40), is processed by PilD. If this is the case, pilD mutants should be defective for TCP production.
When classical strain cultures are grown under conditions which favor TCP production (11), wild-type strain O395 aggregates, and this aggregation is dependent upon the presence of TCP, since mutants with mutations in tcpA, tcpB, and other tcp genes failed to aggregate (40) (Table 4). Similarly, pilD mutants KFV5R and KFV32 failed to aggregate in cultures, demonstrating a general defect in TCP assembly due to mutation of pilD.
TABLE 4.
Effect of pilD mutations on TCP production
| Recipient strain and growth condition | Agg | Transduction frequency witha:
|
|
|---|---|---|---|
| CTXφ704A | CTXφ-Km | ||
| O395 (TCP-inducing conditions) | + | 9.1 × 10−3 ± 2.0 × 10−3 | 9.1 × 10−3 ± 2.7 × 10−3 |
| O395 (non-TCP-inducing conditions) | − | <2.8 × 10−7 ± 1.1 × 10−7 | |
| BGD4 (ΔtcpA) | − | <6.0 × 10−8 ± 0.1 × 10−8 | <4.2 × 10−8 ± 0.1 × 10−8 |
| KP8.97 (tcpB::TnphoA) | − | <9.7 × 10−8 ± 0.1 × 10−8 | |
| KFV5R (pilD::kanπ, reverted) | − | 4.1 × 10−5 ± 0.3 × 10−5 | |
| KFV5R (pKJF308Cm) + 0.05% arabinose | + | 4.5 × 10−2 ± 1.7 × 10−2 | |
| KFV5R (pKJF308Cm) + no arabinose | − | 1.0 × 10−3 ± 0.4 × 10−3 | |
| KFV32 (ΔpilD) | − | 3.9 × 10−5 ± 0.4 × 10−5 | |
| KFV32 (pKJF308) + 0.05% arabinose | + | 1.4 × 10−2 ± 0.2 × 10−2 | |
| KFV32 (pKJF308) + no arabinose | − | 2.5 × 10−4 ± 0.8 × 10−4 | |
Transduction frequency is the CFU of transductants divided by the CFU of recovered recipients. Numbers given are averages ± standard deviations from duplicates in a single experiment.
TCP production can be quantitatively measured by transduction of CTXφ-Km (kanamycin resistance) and CTXφ704A (ampicillin resistance), filamentous transducing phages which adsorb to TCP during the infection process (57). Wild-type strain O395 was transduced by both phages at a frequency of 10−2, but tcpA and tcpB mutants could not be transduced (Table 4). pilD mutants KFV5R and KFV32 showed an intermediate phenotype. Although phage transduction was not completely abolished, pilD mutants KFV5R and KFV32 were transduced at a 150- to 240-fold-lower frequency (Table 4). Surprisingly, CTXφ transduction not only was restored but also was slightly enhanced by the presence of pilD in trans.
These data indicate that pilD is very important, although not absolutely essential, for TCP production. We thus propose that TcpA is processed primarily by TcpJ (28), while TcpB is processed primarily by PilD, although residual processing by TcpJ may occur. Further, the overproduction of TCP when the copy number of pilD is increased suggests that the processing of TcpB by PilD may be a limiting factor in TCP production.
pilD mutants do not colonize infant mice.
Since TCP production is absolutely essential for colonization, pilD mutants should not colonize infant mice. Indeed, strains KFV5R and KFV32 did not colonize infant mice when inoculated either by themselves or in competition with a lacZ derivative of O395 (Table 5).
TABLE 5.
Infant mouse colonization by pilD mutants of classical and El Tor V. cholerae strains
| Competing strainsa | Inoculum ratiob | In vitro ratioc | In vivo colonizationd
|
|||
|---|---|---|---|---|---|---|
| No. of mice | Avg CFU recovered
|
Ratio (mutant/ΔlacZ strain) | ||||
| Mutant | ΔlacZ strain | |||||
| KFV5R and Lac1 | 0.98 | 7.9 × 10−5 | 4 | 0 | 3.4 × 106 ± 4.2 × 106 | <2.9 × 10−7 |
| 3.8 | 1.3 × 10−3 | 5 | 0 | 2.3 × 106 ± 3.8 × 106 | <4.3 × 10−7 | |
| 41.3 | 0.056 | 4 | 0 | 5.4 × 105 ± 4.9 × 105 | <1.9 × 10−6 | |
| O395N1 alone | 5 | 1.6 × 107 ± 1.8 × 107 | ||||
| KFV5R alone | 5 | 0 | ||||
| KFV18R and KFV10 | 0.39 | 1.5 × 10−4 | 4 | 0 | 1.4 × 108 ± 0.9 × 108 | <7.3 × 10−9 |
| 2.5 | 4.0 × 10−4 | 4 | 0 | 1.2 × 108 ± 1.1 × 108 | <8.6 × 10−9 | |
| 20.9 | 0.028 | 5 | 1.6 × 104 ± 1.8 × 104 | 2.6 × 107 ± 2.6 × 107 | 6.2 × 10−4 | |
| P4 alone | 4 | 2.5 × 104 ± 1.2 × 104 | ||||
| KFV18R alone | 9 | ≤3.5 × 103 | ||||
For a description of strains, see Table 1.
For classical strains, inocula were adjusted to 1 × 106 to 2 × 106/ml (total dosage) at the ratios indicated, while El Tor strains were adjusted to 3 × 105 to 8 × 105/ml (total dosage) at the ratios indicated.
Competitive growth in LB broth overnight at 37°C.
CFU recovered represent the total CFU in 5 ml of prepared intestinal extract ± standard deviations.
Like classical pilD mutants, El Tor pilD mutant KFV18R did not colonize infant mice reproducibly (Table 5). However, in some experiments, when the highest inocula were used or when the mutant was given at an excess in competition, some colonies of KFV18R could be recovered (Table 5). In similar assays, tcpA mutants also were frequently recovered (data not shown). Independent colonies of KFV18R recovered from five mice in two separate experiments were isolated and further analyzed. These isolates maintained the kanπ insertion in pilD and continued to be defective for HAP export and MSHA, indicating that they did not arise from in vivo selection for reversion or suppression of the original pilD mutations. Further, these mouse-recovered KFV18R isolates did not have enhanced infection capability when inoculated into new infant mice (data not shown). These observations demonstrate that while KFV18R fails to colonize well, the mutant is able to survive the in vivo environment. These observations support the notion that the loss of colonization is due either to the loss of TCP alone or to the loss of another pilD-processed protein as well.
pilA and mshA are not required for the colonization of infant mice.
If the loss of an additional pilD-processed protein contributes to the colonization defect, it is possible that the defect is due to pilA, which is linked to pilD. However, mutants with deletions of pilA, in both the classical and the El Tor backgrounds, colonized mice as well as the wild type (Table 6). Similarly, mutants with mutations in the MSHA pilus colonized infant mice as well as the wild type (2, 55) (Table 6). We considered that PilA and MshA might have redundant functions in colonization. However, double ΔpilA ΔmshA mutants also colonized mice as well as the wild type (Table 6). Since CT itself may play an important role in colonization (41), its presence might mask other adherence factors. However, pilA mutants of toxin-deficient strains O395N1 and P4 also showed no defect in colonization compared to the wild type (Table 6). Thus, we conclude that while pilA may encode a pilin protein for a new type IV pilus, this putative pilus is not required for colonization in the infant mouse model.
TABLE 6.
Infant mouse colonization by type IV pilin mutants of V. cholerae
| Competing strainsa | Inoculum ratiob | In vivo competitive indexb (no. of mice) |
|---|---|---|
| Classical O395 background | ||
| KFV8 (ΔpilA) × Lac1 (ΔlacZ) | 1.3 | 1.1 ± 0.1 (4) |
| KP9.79 (tcp) × Lac1 (ΔLacZ) | 1.14 | <2 × 10−6 (2) |
| El Tor N16961Sm background | ||
| KFV6 (ΔpilA) × KFV10 (ΔlacZ) | 1.17 | 0.49 ± 0.08 (7) |
| KFV11 (ΔmshA) × KFV10 (ΔlacZ) | 1.28 | 0.78 ± 0.24 (6) |
| KFV12 (ΔpilA ΔmshA) × KFV10 (ΔlacZ) | 1.57 | 0.76 ± 0.25 (5) |
| KFV33 (ΔtcpA) × KFV10 (ΔlacZ) | 1.15 | <3.3 × 10−6 (3) |
| Toxin-deficient background | ||
| KFV9 (O395N1 ΔpilA) × KFV16 (O395N1 ΔlacZ) | 0.37 | 1.4 ± 0.2 (4) |
| KFV38 (P4 ΔpilA) × KFV26 (P4 ΔlacZ) | 2.1 | 0.55 ± 0.08 (3) |
For toxin-producing backgrounds, a 50-μl dose at 1 × 104 to 5 × 104/ml for each strain was delivered by oral gavage and processed as described in Materials and Methods. O395N1-based strains were given at a 10-fold higher inoculum, while P4-based strains were given at a 1,000-fold higher inoculum, to counteract reduced adherence by toxin-deficient strains.
Expressed as CFU of lacZ+ colonies/CFU of lacZ colonies ± standard deviations.
Classical pilD mutants adhere poorly to HEp-2 cells due to mutation of an unknown gene.
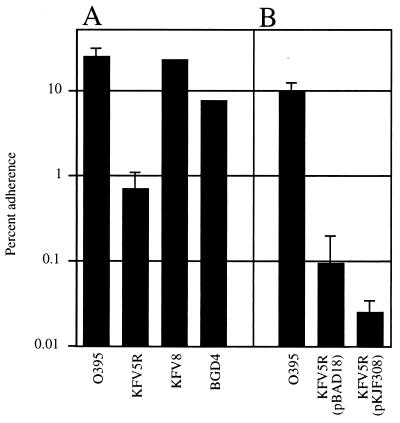
Given that PilD proteins in other organisms process proteins required for epithelial cell adherence (7, 16, 27, 38, 43), we examined the ability of V. cholerae mutants to adhere to HEp-2 cells to determine whether potential adherence factors other than TCP are absent in pilD mutants. When grown under conditions optimal for adherence, O395 adheres to HEp-2 cells. In contrast, the pilD mutant KFV5R adhered poorly compared to the wild type, showing up to a 10-fold defect in repeated assays (Fig. 6A). Surprisingly, unlike other pilD-dependent processes, this defect could not be restored by pilD in trans (Fig. 6B). This result shows that the loss of adherence is most likely due to a gene or protein that was terminally mutated during reversion of the pilD mutation. Unfortunately, attempts to demonstrate an adherence defect in deletion strain KFV32 could not be interpreted because wild-type strains grown to a low density also failed to adhere (data not shown).
FIG. 6.
Adherence of V. cholerae strains to HEp-2 cells. Assays were performed as described in Materials and Methods. KFV5R carrying either the pBAD18 vector or the pilD-complementing plasmid pKJF308 was grown in the presence of 0.05% arabinose to induce promoter PBAD. Arabinose does not inhibit the adherence of wild-type O395 (data not shown). Results are presented as the percentage of input bacteria that adhered to HEp-2 cells after 1 h of incubation. Numbers given are averages from triplicate samples in a single assay and are representative of at least two experiments. Error bars represent standard deviations. (A) Adherence by O395 mutants of V. cholerae. (B) Complementation of KFV5R by pilD in trans.
Several genes are clear candidates for the adherence gene lost in the reversion process. The terminal mutation may have resulted from the loss of one or more of the prepilin proteins, since evidence suggests that the slow growth of pilD strains is due in part to the accumulation of unprocessed prepilin proteins (10). However, deletion of pilA or tcpA had little effect on adherence to HEp-2 cells (Fig. 6A). These data show that these two type IV pilin proteins (PilA and TcpA) do not function in epithelial cell adherence and that the terminal loss of pilA or tcpA could not have caused the defect in adherence observed for pilD mutant strains.
Another candidate for the lost protein is OmpU, an outer membrane protein previously proposed to be required for the adherence of V. cholerae to HEp-2 cells (48). Sandkvist et. al (44) demonstrated that disruption of the type II secretion machinery encoded by the eps genes has several pleiotropic effects, including the rapid degradation of OmpU. Since pilD mutants are also secretion machinery mutants, they may have a similar defect. Indeed, KFV5R and KFV32 had reduced expression of OmpU, but the expression of OmpU was restored to wild-type levels when pilD was introduced in trans (data not shown). Thus, if OmpU is solely responsible for the adherence defect, complemented KFV5R should adhere to HEp-2 cells. Therefore, the failure of KFV5R to adhere to HEp-2 cells must be due to another protein terminally mutated during the reversion process. Thus, the nature of the adherence defect in classical strains cannot be fully determined at this time, although it is likely to involve either a previously unrecognized PilD-processed protein or another outer membrane protein which is terminally lost in the reversion process.
The adherence of El Tor strains to HEp-2 cells is not pilD dependent.
Determining the adherence of N16961Sm derivatives proved to be difficult, compared to that for the classical strains. We recently showed that V. cholerae El Tor and O139 strains produced a toxin that caused rounding and detachment of HEp-2 cells (30). The destruction of cellular integrity due to this toxin hampered interpretation of the results. Further, it was recently shown that MSHA pili mediated adherence to solid substrates, including laboratory plastic and glass items used for tissue culturing (58). To circumvent these technical difficulties, KFV18R with a disruption of the cytotoxin was compared to KFV11 (ΔmshA), which also had a disruption of the cytotoxin. Both strains showed 3% adherence to HEp-2 cells. This result shows that the loss of pilD and the subsequent reversion of growth defects to generate KFV18R did not permanently disrupt a key adhesin. However, the level of adherence was quite low compared to that observed for classical strains, suggesting that conditions favoring adherence by El Tor strains may be different from those for classical strains.
DISCUSSION
In recent years, the new field of functional genomics has expanded for prokaryotic research. It has been predicted that most of the important human bacterial pathogens will be completely sequenced by the year 2000 (52). The new challenge presented to researchers concerns the use of this plethora of information in designing and performing informative experiments. While sequencing of the V. cholerae genome is not yet complete, preliminary data were successfully used to identify and characterize a new type IV prepilin peptidase of V. cholerae, PilD. This prepilin peptidase is distinctly different from a previously described peptidase, TcpJ, showing only 25% identity at the amino acid level. This difference reflects the specificity for protein substrates. TcpJ appears to be responsible solely for processing of the type IV-B pilin protein TcpA (28), while PilD has a multitude of type IV-A substrates, including Eps proteins, essential for CT and HAP secretion, Msh proteins, essential for hemagglutination, and possibly TcpB, a protein essential for TCP production. In this study, we demonstrated that the loss of pilD disrupts each of these processes and that these defects are due solely to the loss of pilD, since the defects could be complemented in trans.
Recently, Marsh and Taylor (32) directly demonstrated that V. cholerae pilD (vcpD) encodes a type IV prepilin peptidase. They showed that EpsI and MshA produced in E. coli are N-terminally processed only when coexpressed with PilD. This observation confirms our prediction that secretion and MSHA defects are due to the lack of processing of type IV secretion signals by PilD.
Like us, Marsh and Taylor (32) showed that pilD (vcpD) mutants are defective for toxin secretion and MSHA, as well as for infant mouse colonization. However, they were unable to distinguish the phenotypes observed from the growth defects exhibited by the pilD (vcpD) mutants (32). In this work, our isolation and characterization of mutants with enhanced growth rates enabled us to characterize the role of pilD with less consideration for growth defects. These mutants were compared to more slowly growing deletion mutants, with similar results, and we further demonstrated that the defects were complemented by the introduction of pilD on a plasmid. Thus, we can firmly conclude that pilD is essential for secretion, MSHA, and TCP production.
In addition to the characterization of pilD, this work sheds light on continuing questions regarding the mechanism of bacterial adherence. Since type IV pili are essential for adherence in P. aeruginosa and N. gonorrhoeae (7, 16, 27, 43), it seems plausible that PilA is assembled into a new pilus essential for adherence and colonization. Surprisingly, pilA mutations resulted in no defects in these processes either alone or in combination with a mutation in mshA. Further, pilA mutants are not required for adherence to solid substrates, disputing a role for adherence in the environment (58). At present there is no information about the role of the putative pilus. However, the location of its gene within an operon with pilD indicates that it is expressed both in vivo and ex vivo. pilD is required for CT secretion and TCP production and thus must be expressed in vivo, and it is essential for MSHA, a phenotype expressed by bacteria grown in cultures. Indeed, preliminary analysis of lacZ fusions to the pil operon showed that the expression of this operon is initiated in the mid-log phase, just as pilD mutants begin to show severe growth defects (Fig. 3 and data not shown). Thus, it is unlikely that pilA, pilB, and pilC are silent genes coincidentally present in an operon with pilD. The 100% conservation of the pilA sequence between the classical and El Tor biotypes also indicates that pilA is likely to be important for some aspect of V. cholerae biology.
What, then, is the nature of the V. cholerae adherence factors mediating adherence to HEp-2 cells? One possible hint comes from the analysis of the classical mutant KFV5R. This mutant is a spontaneously arising second-site suppressor of the slow-growth defect in pilD kanπ insertion mutant KFV5. It has a defect in HEp-2 cell adherence that cannot be complemented in trans by the wild-type pilD gene (Fig. 6B). Thus, an adhesion factor must have been disrupted during the reversion process, allowing faster growth. This lost protein may be a type IV protein. It is clear that the loss of pilin subunits can lead to enhanced growth of pilD mutants of N. gonorrhoeae (10), and our ability to produce a pilD mutation only in the ΔpilA background of N16961Sm suggests that a similar mechanism may allow enhanced growth of pilD mutants of V. cholerae. Analysis of more recent genomic data obtained from The Institute for Genomic Research shows that at least five additional type IV cleavable proteins are encoded by the V. cholerae genome (56a). Loss of any of these five proteins may have enhanced the growth of the pilD mutant KFV5 while decreasing HEp-2 cell adherence. Systematic deletion of each of the five corresponding genes will determine if any of these proteins are essential for HEp-2 cell adherence.
The adherence factor lost may also be an outer membrane protein. Secretion mutants of V. cholerae, including pilD mutants, lose outer membrane proteins, particularly the putative adhesin OmpU (32, 44). However, the loss of OmpU probably does not cause the HEp-2 cell adherence defect in KFV5R, since the reversion process and complementation restored OmpU expression to KFV5R. Thus, another outer membrane protein may be essential for HEp-2 cell adherence. Sengupta et al. (45) showed that rabbit antisera raised against major outer membrane proteins are partially protective against V. cholerae challenge in infant mice. This observation argues that outer membrane proteins may play a role in intestinal colonization and adherence. Attempts to isolate outer membrane protein mutants by signature-tagged mutagenesis and in vivo colonization repeatedly identified genes required for TCP production (8). Thus, the nature of outer membrane proteins that may be essential for adherence could probably be identified only by screening of transposon mutants for defects in adherence in vitro. Such an approach has identified a 40-kDa outer membrane protein from V. cholerae O139 that shows decreased adherence and infant mouse colonization, although the disrupted gene has not yet been identified (5).
This work has demonstrated that genomic data coupled with experimentation can rapidly expand the understanding of bacterial pathogenesis. However, the successful identification of a second prepilin peptidase is juxtaposed to the inaccurate supposition that pilA encodes a major adherence factor. These observations highlight the importance of genetic experimentation for complementing genomic data to fully enrich the fields of prokaryotic biology and eukaryotic biology.
ACKNOWLEDGMENTS
We thank Ronald Taylor for gifts of plasmids and strains. We thank Michael Starnbach and his research group for gracious sharing of tissue culture facilities, supplies, and knowledge. The members of the Mekalanos laboratory are thanked for their ideas and suggestions; in particular, we thank Barry Wanner for helpful hints on constructing the mutants for this paper. We thank the Microbiology Sequencing Core Facility for providing additional sequence data for the pil gene cluster.
This research was supported by grant AI-18045 from the National Institutes of Health, and K.J.F. was supported by National Research Service Award training grant AI-07410.
REFERENCES
- 1.Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 2.Attridge S R, Manning P A, Holmgren J, Jonson G. Relative significance of mannose-sensitive hemagglutinin and toxin-coregulated pili in colonization of infant mice by Vibrio cholerae El Tor. Infect Immun. 1996;64:3369–3373. doi: 10.1128/iai.64.8.3369-3373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benítes J A, Spelbrink R G, Silva A, Phillips T E, Stanley C M, Boesman-Finkelstein M, Finkelstein R A. Adherence of Vibrio cholerae to cultured differentiated human intestinal cells: an in vitro colonization model. Infect Immun. 1997;65:3474–3477. doi: 10.1128/iai.65.8.3474-3477.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett G I, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Bondre V P, Srivastava R, Sinha V B, Srivastava B S. Screening of TnphoA mutant of Vibrio cholerae O139 for identification of antigens involved in colonisation. J Med Microbiol. 1997;46:1007–1011. doi: 10.1099/00222615-46-12-1007. [DOI] [PubMed] [Google Scholar]
- 6.Cheng J-C, Shao C-P, Hor L-I. Cloning and nucleotide sequencing of the protease gene of Vibrio vulnificus. Gene. 1996;183:255–257. doi: 10.1016/s0378-1119(96)00488-x. [DOI] [PubMed] [Google Scholar]
- 7.Chi E, Mehl T, Nunn D, Lory S. Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infect Immun. 1991;59:822–828. doi: 10.1128/iai.59.3.822-828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang S L, Mekalanos J J. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 9.Chiang S L, Taylor R K, Koomey M, Mekalanos J J. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol Microbiol. 1995;17:1133–1142. doi: 10.1111/j.1365-2958.1995.mmi_17061133.x. [DOI] [PubMed] [Google Scholar]
- 10.Freitag N E, Seifert H S, Koomey M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol Microbiol. 1995;16:575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 11.Gardel C L, Mekalanos J J. Regulation of cholera toxin by temperature, pH, and osmolarity. Methods Enzymol. 1994;235:517–526. doi: 10.1016/0076-6879(94)35167-8. [DOI] [PubMed] [Google Scholar]
- 12.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girón J A, Gómez-Duarte O G, Jarvis K G, Kaper J B. Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili—a minireview. Gene. 1997;192:30–43. doi: 10.1016/s0378-1119(97)00039-5. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg I, Mekalanos J J. Cloning of the Vibrio cholerae recA gene and construction of a Vibrio cholerae recA mutant. J Bacteriol. 1986;165:715–722. doi: 10.1128/jb.165.3.715-722.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hahn H P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene. 1997;192:99–108. doi: 10.1016/s0378-1119(97)00116-9. [DOI] [PubMed] [Google Scholar]
- 17.Hall R H, Vial P A, Kaper J B, Mekalanos J J, Levine M M. Morphological studies on fimbriae expressed by Vibrio cholerae O1. Microb Pathog. 1988;4:257–265. doi: 10.1016/0882-4010(88)90086-1. [DOI] [PubMed] [Google Scholar]
- 17a.Häse, C., and J. J. Mekalanos. Unpublished data.
- 18.Häse C C, Bauer M E, Finkelstein R A. Genetic characterization of mannose-sensitive (MSHA)-negative mutants of Vibrio cholerae derived by Tn5 mutagenesis. Gene. 1994;150:17–25. doi: 10.1016/0378-1119(94)90852-4. [DOI] [PubMed] [Google Scholar]
- 19.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 22.Jobling M G, Holmes R K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 23.Johnson K, Parker M L, Lory S. Nucleotide sequence and transcriptional initiation site of two Pseudomonas aeruginosa pilin genes. J Biol Chem. 1986;261:15703–15708. [PubMed] [Google Scholar]
- 24.Jonson G, Holmgren J, Svennerholm A-M. Identification of a mannose-binding pilus on Vibrio cholerae El Tor. Microb Pathog. 1991;11:433–441. doi: 10.1016/0882-4010(91)90039-d. [DOI] [PubMed] [Google Scholar]
- 25.Jonson G, Lebens M, Holmgren J. Cloning and sequencing of Vibrio cholerae mannose-sensitive haemagglutinin pilin gene: localization of mshA within a cluster of type 4 pilin genes. Mol Microbiol. 1994;13:109–118. doi: 10.1111/j.1365-2958.1994.tb00406.x. [DOI] [PubMed] [Google Scholar]
- 26.Jouravleva E A, McDonald G A, Marsh J W, Taylor R K, Boesman-Finkelstein M, Finkelstein R A. The Vibrio cholerae mannose-sensitive hemagglutinin is the receptor for a filamentous bacteriophage from V. cholerae O139. Infect Immun. 1998;66:2535–2539. doi: 10.1128/iai.66.6.2535-2539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Källström H, Liszewski M K, Atkinson J P, Jonsson A-B. Membrane cofactor protein (MCP or CD46) is a cellular pilus receptor for pathogenic Neisseria. Mol Microbiol. 1997;25:639–647. doi: 10.1046/j.1365-2958.1997.4841857.x. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman M R, Seyer J M, Taylor R K. Processing of TCP pilin by TcpJ typifies a common step intrinsic to a newly recognized pathway of extracellular protein secretion by gram-negative bacteria. Genes Dev. 1991;5:1834–1846. doi: 10.1101/gad.5.10.1834. [DOI] [PubMed] [Google Scholar]
- 29.Kenner J R, Coster T S, Taylor D N, Trofa A F, Barrera-Oro M, Hyman T, Adams J M, Beattie D T, Killeen K P, Spriggs D R, Mekalanos J J, Sadoff J C. Peru-15, an improved live attenuated oral vaccine candidate for Vibrio cholerae O1. J Infect Dis. 1995;172:1126–1129. doi: 10.1093/infdis/172.4.1126. [DOI] [PubMed] [Google Scholar]
- 30.Lin, W., K. J. Fullner, R. Clayton, J. Sexton, M. Rogers, K. Calia, S. Calderwood, C. Fraser, and J. J. Mekalanos. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 30a.Lin, W., and J. J. Mekalanos. Unpublished data.
- 31.Manning P A. The tcp cluster of Vibrio cholerae. Gene. 1997;192:63–70. doi: 10.1016/s0378-1119(97)00036-x. [DOI] [PubMed] [Google Scholar]
- 32.Marsh J W, Taylor R K. Identification of the Vibrio cholerae type 4 prepilin peptidase required for cholera toxin secretion and pilus formation. Mol Microbiol. 1998;29:1481–1492. doi: 10.1046/j.1365-2958.1998.01031.x. [DOI] [PubMed] [Google Scholar]
- 33.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:252–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 34.Metcalf W M, Jiang W, Daniels L L, Kim S-K, Haldimann A, Wanner B L. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 35.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosa pili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunn D N, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;88:3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overbye L J, Sandkvist M, Bagdasarian M. Genes required for extracellular secretion of enterotoxin are clustered in Vibrio cholerae. Gene. 1993;132:101–106. doi: 10.1016/0378-1119(93)90520-d. [DOI] [PubMed] [Google Scholar]
- 38.Paranjpye R, Lara J C, Pepe J C, Pepe C M, Strom M S. The type IV leader peptidase/N-methyltransferase of Vibrio vulnificus controls factors required for adherence to HEp-2 cells and virulence in iron-overloaded mice. Infect Immun. 1998;66:5659–5668. doi: 10.1128/iai.66.12.5659-5668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pepe C M, Eklund M W, Strom M S. Cloning of an Aeromonas hydrophila type IV pilus biogenesis gene cluster: complementation of pilus assembly functions and characterization of a type IV leader peptidase/N-methyltransferase required for extracellular protein secretion. Mol Microbiol. 1996;19:857–869. doi: 10.1046/j.1365-2958.1996.431958.x. [DOI] [PubMed] [Google Scholar]
- 40.Peterson K M, Mekalanos J J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988;56:2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce N F, Kaper J B, Mekalanos J J, Cray W C J. Role of cholera toxin in enteric colonization by Vibrio cholerae O1 in rabbits. Infect Immun. 1985;50:813–816. doi: 10.1128/iai.50.3.813-816.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Postnova T, Gómez-Duarte O G, Richardson K. Motility mutants of Vibrio cholerae O1 have reduced adherence in vitro to human small intestinal epithelial cells as demonstrated by ELISA. Microbiology. 1996;142:2767–2776. doi: 10.1099/13500872-142-10-2767. [DOI] [PubMed] [Google Scholar]
- 43.Rudel T, van Putten J P, Gibbs C P, Haas R, Meyer T F. Interaction of two variable proteins (PilE and PilC) required for pilus-mediated adherence of Neisseria gonorrhoeae to human epithelial cells. Mol Microbiol. 1992;6:3439–3450. doi: 10.1111/j.1365-2958.1992.tb02211.x. [DOI] [PubMed] [Google Scholar]
- 44.Sandkvist M, Michel L O, Hough L P, Morales V M, Bagdasarian M, Koomey M, DiRita V J, Bagdasarian M. General secretion pathway (eps) genes required for toxin secretion and outer membrane biogenesis in Vibrio cholerae. J Bacteriol. 1997;179:6994–7003. doi: 10.1128/jb.179.22.6994-7003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sengupta D K, Sengupta T K, Ghose A C. Major outer membrane proteins of Vibrio cholerae and their role in induction of protective immunity through inhibition of intestinal colonization. Infect Immun. 1992;60:4848–4855. doi: 10.1128/iai.60.11.4848-4855.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw C E, Taylor R K. Vibrio cholerae O395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilins. Infect Immun. 1994;58:3042–3049. doi: 10.1128/iai.58.9.3042-3049.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 48.Sperandio V, Girón J A, Silveira W D, Kaper J B. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect Immun. 1995;63:4433–4438. doi: 10.1128/iai.63.11.4433-4438.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strom M S, Nunn D N, Lory S. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc Natl Acad Sci USA. 1993;90:2404–2408. doi: 10.1073/pnas.90.6.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tacket C O, Taylor R K, Losonsky G, Lim Y, Nataro J P, Kaper J B, Levine M M. Investigation of the roles of toxin-coregulated pili and mannose-sensitive hemagglutinin pili in the pathogenesis of Vibrio cholerae O139 infection. Infect Immun. 1998;66:692–695. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamamoto T, Nakashima K, Nakasone N, Hanma Y, Higa N, Yamashiro T. Adhesive property of toxin-coregulated pilus of Vibrio cholerae O1. Microbiol Immunol. 1998;42:41–45. doi: 10.1111/j.1348-0421.1998.tb01967.x. [DOI] [PubMed] [Google Scholar]
- 52.Tang C M, Hood D W, Moxon E R. Haemophilus influence: the impact of whole genome sequencing on microbiology. Trends Genet. 1997;13:399–404. doi: 10.1016/s0168-9525(97)01245-6. [DOI] [PubMed] [Google Scholar]
- 53.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teppema J S, Guinée P A M, Ibrahim A A, Pâques M, Ruitenberg E J. In vivo adherence and colonization of Vibrio cholerae strains that differ in hemagglutinating activity and motility. Infect Immun. 1987;55:2093–2102. doi: 10.1128/iai.55.9.2093-2102.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thelin K H, Taylor R K. Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tonjum T, Freitag N E, Namork E, Koomey M. Identification and characterization of pilG, a highly conserved pilus assembly gene in pathogenic Neisseria. Mol Microbiol. 1995;16:451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- 56a.Vibrio cholerae Genome Project Website. [Online.] The Institute for Genomic Research. http://www.tigr.org. [June 1997 and October 1998, last dates accessed.]
- 57.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 57a.Waldor, M. Unpublished data.
- 58.Watnick, P., K. J. Fullner, and R. Kolter. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 59.Whitchurch C B, Mattick J S. Escherichia coli contains a set of genes homologous to those involved in protein secretion, DNA uptake and the assembly of type-4 fimbriae in other bacteria. Gene. 1994;150:9–15. doi: 10.1016/0378-1119(94)90851-6. [DOI] [PubMed] [Google Scholar]