Abstract
Sodium dodecyl sulfate-polyacrylamide gel analysis of lipooligosaccharide (LOS) from Neisseria meningitidis has demonstrated considerable microheterogeneity in the variable region of LOS due to the presence of novel glycoforms. As a step toward understanding the basis for the expression of these novel glycoforms, we have examined the LOS structures and UDP-glucose 4-epimerase (epimerase) activity levels in two strains (NMB and MA-1) and their respective galE mutants. Strain NMB was found to have low epimerase activity and to contain multiple glycoforms, some of which appear to contain only glucose sugars. The galE mutant had only the oligoglucose glycoforms. Strain MA-1 had higher epimerase activity at both log and stationary phases (2- and 12.5-fold, respectively) and one glycoform with a putative lactosyl structure. Strain MA-1 galE had two glycoforms that contained one or two glucose residues. To understand the molecular basis for the different epimerase activities, we examined the predicted amino acid sequences of the respective galE open reading frames and determined the relative amounts of GalE protein. We found no significant differences between the predicted amino acid sequence of the GalE protein in NMB and that in MA-1. We observed no significant differences in the level of GalE protein between MA-1 and NMB at exponential or stationary phase. We also observed an 8.2-fold drop in epimerase activity in NMB between the log and stationary phases that was not due to the GalE protein level or low glucose levels.
Pathogenic Neisseria species are gram-negative obligate human pathogens. Neisseria meningitidis, the causative agent of meningococcal meningitis, possesses a number of virulence factors. One of these virulence factors is the lipooligosaccharide (LOS). This molecule is composed of a variable oligosaccharide portion and a conserved core-lipid A structure (1).
The assembly of the LOS molecule is a complex anabolic process involving an array of biosynthetic enzymes including kinases, transferases, and isomerases. One of the isomerases is the UDP-glucose 4-epimerase (galactowaldenase, EC 5.1.3.2). This enzyme carries out the reversible epimerization of UDP-glucose to UDP-galactose, the cognate substrate for galactosyltransferases. The Escherichia coli UDP-glucose 4-epimerase has been purified and studied in detail at both the biochemical and the structural level. The holoenzyme is a homodimer held together by hydrophobic interactions and contains one NAD+ molecule per subunit (4).
The number of unique LOS species expressed by a meningococcal strain can vary widely (24, 29). We have previously reported on the LOS microheterogeneity of serogroup B N. meningitidis NMB and its galE mutant NMB-SS3 (14). The basis of this microheterogeneity was the presence of three novel glycoforms that contained two to four glucose residues. In the galE mutant NMB-SS3, only the oligoglucose glycoforms were detected, and the relative amounts of some of these novel glycoforms were significantly increased compared to those in NMB. Also, in N. meningitidis MC58 galE, a second glycoform with two glucose molecules was observed (33). It is not known if the diglucose glycoform was present in the parent strain. Pathogenic Neisseria species cannot utilize exogenous sources of galactose, and thus the only source of UDP-galactose is UDP-glucose. This suggests an important role for UDP-glucose 4-epimerase in determining the UDP-glucose and UDP-galactose concentrations and thereby perhaps influencing the activity of cognate glycosyltransferases. In this report, we present the LOS structures of MA-1 and its galE mutant MA-1 galE, and we compare the UDP-glucose 4-epimerase activity level of strain NMB to that of strain MA-1. We also present evidence demonstrating growth phase-dependent variation of epimerase activity levels in NMB.
MATERIALS AND METHODS
Materials.
Chemicals and antibiotics were obtained from Sigma Chemical Co. (St. Louis, Mo.). Restriction enzymes and DNA-modifying enzymes were purchased from New England Biolabs, Promega Co., and Boehringer Mannheim Biochemicals.
Bacterial strains and plasmids.
The bacteria and plasmids used in this study are described in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or selective marker | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1 BLUE | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15 Tn10(Tetr)](Con) | Stratagene |
| N. meningitidis NMB | Wild-type clinical isolate; serogroup B; L2, L3, L7, L9 | 14, 22, 25 |
| N. meningitidis NMB ΔgalEt::npt | This study | |
| N. meningitidis MA-1 | Wild-type clinical isolate; serogroup A; L8 | This study |
| N. meningitidis MA-1 ΔgalEt::npt | This study | |
| Plasmids | ||
| pDEADII | Kanamycin | pZERO (Invitrogen) modified by author |
| pBluescript II SK(−) | Ampicillin | Stratagene |
| pBSL14 | Ampicillin and kanamycin | 1 |
Growth of bacteria.
E. coli was grown at 37°C in Luria-Bertani medium with or without 1.5% agar and supplemented with antibiotics as needed. Wild-type N. meningitidis was grown either on gonococcal agar with 1% IsoVitaleX supplement (BBL Laboratories) or in brain heart infusion (BHI) broth supplemented with 2.5% heat-inactivated fetal calf serum and 1% IsoVitaleX. Kanamycin-resistant N. meningitidis was grown on supplemented BHI agar with 45 μg of kanamycin/ml or in supplemented BHI broth with 5 (for strain MA-1) or 25 (for strain NMB) μg of kanamycin/ml. N. meningitidis on agar plates was cultured in the presence of 5% CO2 at 85% relative humidity.
Recombinant DNA and transformation methods.
All recombinant DNA techniques were used as outlined elsewhere (23). Transformations of N. meningitidis were performed as previously described by Catlin (4) and modified by Stephens et al. (25). Electroporations were carried out by using the GIBCO-BRL Cell-Porator under the recommended conditions.
DNA sequencing.
The nucleotide sequences of cloned genomic DNA fragments were determined at the DNA Facility at the University of Iowa by using an ABI373A automated sequencer.
Hybridizations.
Analyses of Southern blots were carried out with radiolabelled random-primed probes at 65°C in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt’s solution, 20 mM Na3P2O7, 0.1% sodium dodecyl sulfate (SDS), and 100 μg of heterologous DNA/ml. The filters were washed twice at the hybridization temperature in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS for 15 min, followed by two washes in 0.1× SSC and 0.1% SDS for 15 min. The radioactive blots were exposed to Kodak XAR5 or BioMaxMR film at −70°C.
Hot-phenol LOS extraction.
LOS was purified from N. meningitidis by a modified hot-phenol method (34) as previously described (14). The purified LOS was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and visualized by silver staining as described elsewhere (30).
Structural characterization of LOS.
To determine the effects of UDP-glucose 4-epimerase activity on LOS biosynthesis, LOS from the wild-type strain MA1 and a galE mutant were partially characterized by mass spectrometry and composition analysis. Crude LOS preparations (∼1 mg each) from the two strains were first O deacylated by treatment with hydrazine under mild conditions (37°C for 30 min), followed by precipitation with chilled acetone (10). For mass spectrometric analysis, O-deacylated LOS was dissolved in a stock water solution (10 μg/μl) and analyzed by matrix-assisted laser desorption ionization (MALDI) (7) and electrospray ionization (ESI) (8) mass spectrometry as previously described. For the MALDI analyses, several dilutions were made of the original stock (1/10 to 1/100), and ∼0.1 μg or less of each O-deacylated LOS was added to 1 μl of a matrix solution of 2,5-dihydroxybenzoic acid (DHB) and spotted on a stainless-steel MALDI sample plate. Mass spectra were taken on a PerSeptive Biosystems Voyager MALDI-time-of-flight mass spectrometer (PerSeptive Biosystems, Framingham, Mass.) fitted with a N2 laser operating at 337 nm. Typically, 50 to 100 single laser shots were averaged to obtain a single spectrum by using a 100-ns delay time and a laser power setting of 1,800 to 2,000. Spectra were smoothed (Savitsky-Golay procedure; 2 by 19 point) and externally calibrated with a close proximity standard consisting of the commercial peptides bradykinin (Mavg = 1,060.2) and ACTH 1-24 (Mavg = 2,465.7). For ESI analyses, a stock solution of each O-deacylated LOS preparation was diluted fivefold in water-acetonitrile (1:1, vol/vol) containing 1% acetic acid to yield a final O-deacylated LOS concentration of approximately 2 μg/μl. Five microliters of each LOS sample was then injected via a Rheodyne valve and analyzed in the negative-ion mode on a Sciex API 300 triple quadrupole mass spectrometer (Perkin-Elmer SCIEX, Mississauga, Ontario, Canada) with a flow rate of 3 to 4 μl/min. Mass calibration was carried out with an external myoglobin reference by using the commercial software supplied.
Monosaccharide composition analyses were carried out on O-deacylated LOS preparations by two independent methods. For neutral sugar analysis, aliquots of each O-deacylated LOS pool were hydrolyzed in 2 M trifluoroacetic acid for 3 h at 100°C. Amino sugars were analyzed after hydrolysis in 6 N HCl for 3 h at 100°C. In both cases, aliquots of the final hydrolysates were evaporated to dryness, redissolved in 20 μl H2O, dried, and then reconstituted in water. A 5% aliquot (1 of 20 μl) of each sample was analyzed by high pH anion-exchange chromatography by using a Dionex high-pressure liquid chromatography system equipped with a PA1 column as previously described (20). To identify and quantify the monosaccharides from each hydrolysate, a standard mixture of monosaccharides containing equimolar amounts of fucose, galactosamine, glucosamine, galactose, glucose, and mannose was analyzed before and after each run.
Glucose supplement of stationary-phase culture.
A fresh overnight culture of N. meningitidis NMB grown in Morse’s defined medium with 20 mM glucose (18) was used to inoculate 50 ml of fresh medium and incubated at 37°C with shaking. Aliquots (15 ml) were removed at mid-log and early-stationary phases, and epimerase activities were determined. Glucose was added to the remaining culture to a final concentration of 20 mM, and it was incubated for one additional hour, after which the epimerase activity was determined.
Purification of recombinant meningococcal GalE.
The NMB galE open reading frame was cloned into plasmid pCYB2 of the IMPACT Protein Purification System (New England Biolabs), and the recombinant meningococcal GalE was purified according to the manufacturer’s recommendations. The purified protein was verified by amino-terminal sequencing at the Protein Structure Facility, University of Iowa.
Generation of polyclonal antibodies.
Mouse polyclonal antibodies to recombinant meningococcal GalE were raised by coinjection with the RIBI Adjuvant System (RIBI ImmunoChem Research, Inc.) according to the manufacturer’s recommendations. The ascites fluid was harvested and assayed for reactivity to the purified antigen by enzyme-linked immunosorbent assay.
SDS-PAGE and Western blot analysis.
Cultures of N. meningitidis MA-1 and NMB were harvested and lysed by sonication. Total protein concentrations were determined with the Bio-Rad Protein Assay Reagent system, and equivalent amounts of total protein were fractionated by 12.5% SDS–12.5% PAGE. The resolved proteins were transferred to nitrocellulose under standard conditions and reacted with mouse polyclonal antisera to recombinant meningococcal GalE that had been preabsorbed with paraformaldehyde-fixed MA-1 galE. The reacting bands were visualized with a horseradish peroxidase-conjugated anti-mouse immunoglobulin G secondary antibody (Bio-Rad, Inc.) and the SuperSignal Horseradish Peroxidase Detection System from Pierce Chemicals. The relative intensities of the bands were estimated with Eastman Kodak (Rochester, N.Y.) 1D Image Analysis Software.
Preparation of cell extract.
N. meningitidis strains were grown in supplemented BHI broth with kanamycin as required. For each strain, a 50-ml culture was inoculated with 0.01 volume of fresh overnight culture and incubated overnight at 37°C with shaking. The washed pellets were resuspended in 2 ml of buffer (0.125 M potassium bicinate [pH 8.5]–1 mM phenylmethylsulfonyl fluoride) and lysed by sonication. The bacterial debris was pelleted by centrifugation at 15,800 × g for 30 min at 4°C. The supernatants were transferred to prechilled microcentrifuge tubes and kept on ice. The total protein contents of the cleared cell extracts were determined by using the Bio-Rad Protein Assay Reagent system following the microassay protocol. Equal amounts of total protein were added to the two-step UDP-glucose 4-epimerase assay as described below.
Enzyme assays.
The standard UDP-glucose 4-epimerase assays have been published elsewhere (35). We modified the assay slightly to optimize it for meningococcal extracts. The first step of the two-step assay was carried out in a 500-μl reaction volume (0.125 M bicinate [pH 8.5]–0.45 mM UDP-galactose) at 37°C for 15 min. The reaction mixture was then placed in a boiling water bath for 90 s, chilled on ice for 5 min, and then centrifuged at 15,800 × g for 10 min at 4°C. A 400-μl aliquot of the supernatant was added to the second step of the assay in 600 μl (0.125 M bicinate [pH 8.5]–1.25 mM NAD+–0.02 U of UDP-glucose dehydrogenase). The reaction mixture was incubated at room temperature for 3 min in a methylacrylate cuvette, and the increase in absorbance was measured at 340 nm at 15-s intervals. We were able to achieve a twofold increase in sensitivity with the modifications compared to that of the standard two-step method using the glycine buffer. All extracts, including appropriate controls, were assayed in triplicate.
Determination of UDP-glucose 4-epimerase activity levels.
The net absorbance was determined after adjustment for endogenous UDP-galactose and UDP-glucose and for UDP-glucose contamination of exogenous UDP-galactose preparations. The initial velocities of the second reaction (UDP-glucose to UDP-glucuronic acid) were determined over the first 30 s. This initial reaction velocity (Vi) is a function of the initial UDP-glucose concentration, which in turn is a function of the UDP-glucose 4-epimerase activity level. These values were converted to nanomoles of NADH generated per minute by using the Beer-Lambert law and the equation ɛNADH = 6.2 × 103 · M−1 · cm−1. The assay results were analyzed by paired t tests using the Statview program (Abacus Concepts).
Nucleotide sequence accession number.
The nucleotide sequence the galE region of strain MA-1 is listed under accession no. AF083467.
RESULTS
Analysis of LOS from MA-1 and MA-1 galE by SDS-PAGE.
MA-1 LOS migrated faster than the major NMB LOS species on SDS-PAGE analysis and did not appear to be sialylated (Fig. 1). The MA-1 LOS also migrated as a single band, in contrast to NMB LOS. The MA-1 LOS also reacted with monoclonal antibody 4C4, which recognizes Gal-Glc-Hep or Glc-Hep structures (12) (data not shown).
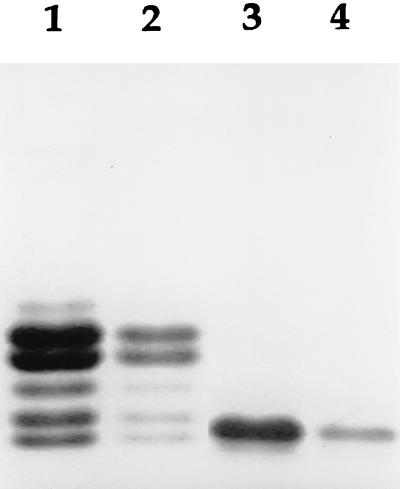
FIG. 1.
SDS-PAGE analysis of LOS isolated from N. meningitidis MA-1 and NMB. This figure illustrates the multiple glycoforms observed in NMB LOS due to variations in the oligosaccharide or PEA composition and the apparent homogeneity of MA-1 LOS. Lanes 1 and 2, N. meningitidis NMB LOS (2.0 and 0.2 μg, respectively); lanes 3 and 4, N. meningitidis MA-1 LOS (2.0 and 0.2 μg, respectively).
A 4.2-kb DraI fragment that contained the galE gene and three other open reading frames was cloned from strain MA-1. The last three open reading frames were identified as galE, rfbB, and rfbA homologues. The first open reading frame (ORF1) was identical to the recently reported mynD (27). A FASTA search of the protein database indicated that ORF1 (mynD) had the highest similarity to a putative DNA-binding protein from yeast (P = 0.062 over 201 amino acids) (data not shown).
We placed a kanamycin cassette 647 bp into the galE open reading frame and introduced the mutation into the chromosome by allelic replacement. A kanamycin-resistant transformant designated MA-1 galE was screened by Southern hybridization (Fig. 2a), SDS-PAGE analysis (Fig. 2b), and epimerase enzyme assay (data not shown). The results confirmed the insertion of the kanamycin cassette into the meningococcal genome through allelic replacement, resulting in a significant decrease in UDP-glucose 4-epimerase activity. SDS-PAGE analysis of isolated MA-1 galE LOS indicated the presence of two bands. The majority of the LOS molecules appeared to be the expected truncated form, but there was a minor band which appeared to comigrate with wild-type LOS (Fig. 2b).
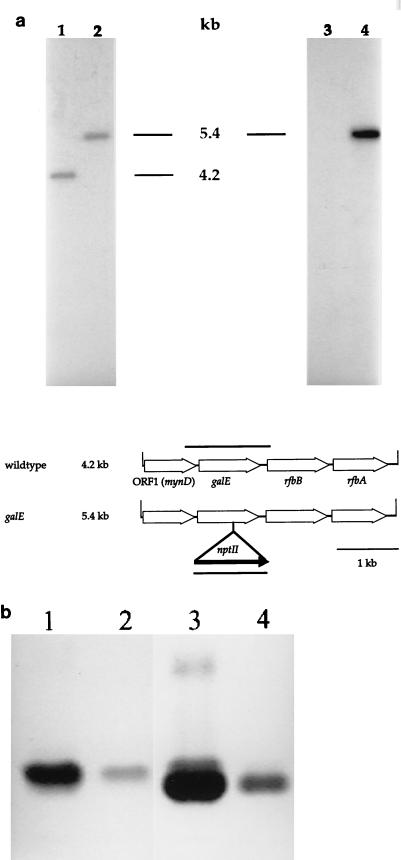
FIG. 2.
Analysis of N. meningitidis MA-1 galE. (a) Southern blot analysis of genomic DNA from N. meningitidis MA-1 galE. Genomic DNAs isolated from N. meningitidis MA-1 and MA-1 galE were digested with DraI and transferred to a nylon membrane after electrophoresis. Duplicate filters were probed with either a galE gene probe (lanes 1 and 2) or a kanamycin cassette probe (lanes 3 and 4). The hybridization patterns indicated the incorporation of the kanamycin cassette in the galE open reading frame. A genetic map of the N. meningitidis MA-1 galE mutant is shown below. The MA-1 galE gene was cloned on a 4.2-kb DraI fragment. This fragment had four open reading frames, as designated. To construct MA-1 galE, a kanamycin cassette from pBSL14 was inserted into the unique MunI site 647 bases into the galE coding region. This construct was introduced into the chromosome of MA-1 by allelic replacement. Open arrows, meningococcal open reading frames; solid arrow, kanamycin resistance cassette. (b) SDS-PAGE analysis of MA-1 and MA-1 galE LOS. The wild-type LOS migrated as a single band. The LOS from the galE mutant gave two distinct bands, of which the upper band comigrated with the wild-type LOS molecule. The lower band is the Hex-Hep2-GlcNAc-Kdo2-lipid A structure. Lanes 1 and 2, MA-1 LOS (2.0 and 0.2 μg, respectively); lanes 3 and 4, MA-1 galE LOS (2.0 and 0.5 μg, respectively).
Genetic analysis of the MA-1 galE gene.
The organization of the 4.2-kb fragment suggested that the MA-1 galE gene was within an operon, perhaps contiguous with the previously described myn operon (27). We introduced a kanamycin cassette in polar and nonpolar orientations upstream of the galE open reading frame at two different sites (Fig. 3). Analysis of these mutants by epimerase enzyme assay and SDS-PAGE of their LOS (Table 2) indicated that the presence of polar insertions at EcoRI and NdeI sites (RKR and NKR, respectively) abrogated the expression of the galE gene.
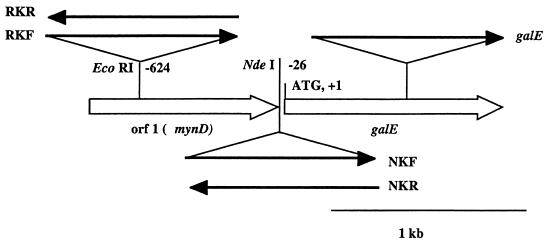
FIG. 3.
Positions and orientations of the kanamycin cassette (solid lines with arrows) in the region upstream of the galE open reading frame. The positions of the restriction sites are given relative to the galE gene start codon. Open arrows, open reading frames. RKR and NKR, polar insertions of the kanamycin cassette at the EcoRI and NdeI sites, respectively. RKF and NKF, respective nonpolar insertions.
TABLE 2.
Genetic analysis of MA-1 galE gene
| Strain, positiona | nmol of NADH generated/min (± SE) | LOS migration patternb (kDa) |
|---|---|---|
| MA-1 | 3.5 (0.2) | Wild type (3.4) |
| RKF, −624 | 2.7 (0.2)c | Wild type (3.4) |
| RKR, −624 | 0.4 (0.2)d | Truncated (3.2) |
| NKF, −26 | 21.3 (0.4)e | Wild type (3.4) |
| NKR, −26 | 0.0 (0.2)f | Truncated (3.2) |
| MA-1 galE | 0.2 (0.0)g | Truncated (3.2) |
Position of the kanamycin cassette relative to the galE gene start codon.
Relative to that of the wild type.
P = 0.0527 for this value versus that obtained with MA-1.
P = 0.0153 for this value versus that obtained with MA-1.
P = 0.0008 for this value versus that obtained with MA-1.
P = 0.0091 for this value versus that obtained with MA-1.
P = 0.0034 for this value versus that obtained with MA-1.
The promoter for the galE gene of a serogroup B strain, B1940, has been mapped previously (9). The nucleotide sequence upstream of the NMB galE gene is identical to the sequence from B1940 (data not shown).
ESI mass spectrometric analysis of LOS isolated from N. meningitidis MA-1 and MA-1 galE.
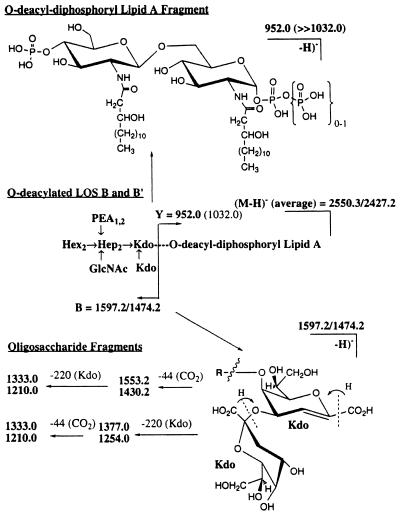
Mass spectrometric analyses of the O-deacylated LOS preparations from the wild-type strain, MA-1, and the galE mutant are shown in Fig. 4 for the corresponding MALDI spectra. For the wild-type strain, N. meningitidis MA-1, two prominent, singly deprotonated molecular-ion species were identified in the high-mass range (m/z >2,000) with m/z values of (M − H)− = 2,428.0 and 2,551.1, as well as peaks related to these two major species through either loss of water (−18 Da), β-elimination of phosphoric acid (−98 Da), or noncovalent addition of one or more sodium (+22 Da) or magnesium (+38 Da) ions or both. As shown in Table 3, these masses are consistent with a LOS composition of a Hep2-HexNAc-2 molecules of 3-deoxy-d-manno-octulosonic acid (Kdo2)-lipid A core structure substituted with two hexoses (galactose and glucose) and containing either one (LOS B; M calculated 2,428.2) or two (LOS B′; M calculated 2,551.3) phosphoethanolamine (PEA) groups (ΔM = 123 Da).
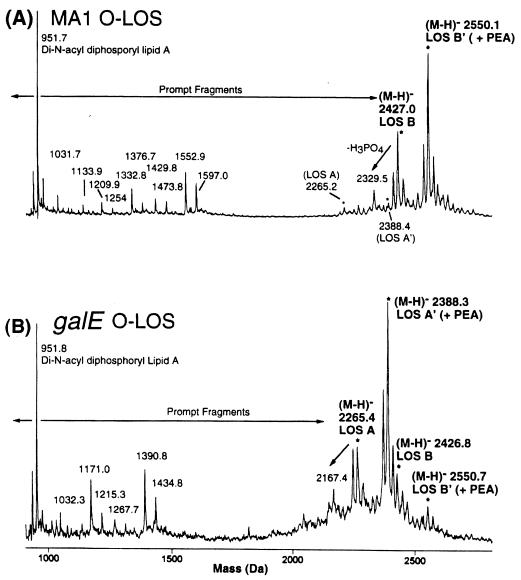
FIG. 4.
Negative-ion MALDI-time-of-flight mass spectra of O-deacylated LOS preparations of wild-type MA-1 (A) and MA-1 galE (B).
TABLE 3.
Molecular weight analysis of O-deacylated LOS by electrospray MS for MA-1 and MA-1 galE
| N. meningitidis strain | LOS | Observed (calculated) mol wta | Relative abundanceb by MALDI (ESI) | Proposed compositionc |
|---|---|---|---|---|
| MA-1 | LOS B | 2,428.0 (2,428.22) | 50 (36) | Hex2-Hep2-GlcNAc-PEA1-Kdo2-lipid A |
| LOS B′ | 2,551.1 (2,551.27) | 100 (100) | Hex2-Hep2-GlcNAc-PEA2-Kdo2-lipid A* | |
| MA-1 galE | LOS A | 2,266.4 (2,266.07) | 32 (43) | Hex-Hep2-GlcNAc-PEA1-Kdo2-lipid A* |
| LOS A′ | 2,389.3 (2,389.12) | 100 (100) | Hex-Hep2-GlcNAc-PEA2-Kdo2-lipid A* | |
| LOS B | 2,427.8 (2,428.22) | 20d (13) | Hex2-Hep2-GlcNAc-PEA1-Kdo2-lipid A* | |
| LOS B′ | 2,551.7 (2,551.27) | 11 (12) | Hex2-Hep2-GlcNAc-PEA2-Kdo2-lipid A* |
All molecular weights for O-deacylated LOS are reported from the MALDI data as their average mass values based on singly deprotonated charged molecular ions, (M − H)−.
Expressed as a percentage; determined from peak heights of the singly charged molecular ions for MALDI and of the doubly and triply charged ions for ESI.
After O deacylation, the lipid A moiety is converted into diphosphoryl diacyl lipid A, containing two N-linked β-hydroxymyristic acid chains with an average Mr of 953.0.
In the MALDI spectrum, this glycoform peak partially overlaps the potassium adduct of the molecular ion for the more abundant glycoform LOS A′ resulting in overestimation of this LOS B glycoform.
The MALDI spectra of the galE mutant LOS were markedly different from those of the wild-type MA-1 LOS. In the former case, the two most abundant molecular-ion species were seen at m/z∼2,265 and 2,388, approximately 1 hexose lower in mass (i.e., Hex = 162 Da) than the two base peaks in the wild-type MA-1 MALDI spectra. However, peaks corresponding to the two wild-type molecular ions at m/z ∼2,427 and 2,551 were also present in the mutant LOS preparations, but at considerably lower abundances. Electrospray analysis of the mutant and wild-type LOS preparations (see Table 3) also supported these assignments, containing both doubly and triply charged ions for the LOS A and B glycoforms (with and without the additional PEA) for the mutant, and primarily the two LOS B glycoforms in the wild type.
In addition to the prominent molecular-ion regions of these two spectra, the low-mass regions of the two MALDI spectra (m/z <1,600) contained ions whose relative abundance was dependent on the laser power. As reported previously (7), these ions are “prompt fragments” and are generated primarily from cleavage at the Kdo glycosidic bond to the lipid A moiety. Figure 5 shows the likely fragmentation pathways for these ions for MA-1, which give additional support for the assignment of the major glycoform species. For example, in the MA-1 spectra the peak at m/z 951.7 would correspond to the deprotonated lipid A species containing two phosphate groups. The small peak at m/z 1,031.7 suggests that a triphosphoryl lipid A species is also present, but at a much lower abundance (<10%) than the dominant diphosphoryl lipid A form. The peaks at m/z 1,597.0 and 1,473.8 can be assigned as originating from the same cleavage but with charge retention on the oligosaccharide fragments, which can undergo further fragmentation due to losses of 44 Da (−CO2; m/z 1,552.9 and 1,429.8) and 220 Da for the terminal Kdo moiety (m/z 1,376.7 and 1,254.0). An analogous set of assignments can be made for the galE strain; although in addition to the fragments arising from the less-abundant LOS B glycoforms as just described, ions are seen for the oligosaccharide fragments of the LOS A glycoforms that are now shifted down in mass by 1 hexose unit (−162 Da).
FIG. 5.
Fragmentation pathways of the two prominent singly charged ions corresponding to the LOS B and LOS B′ glycoforms. All masses are calculated masses. See the spectra (Fig. 4) for the actual experimental masses.
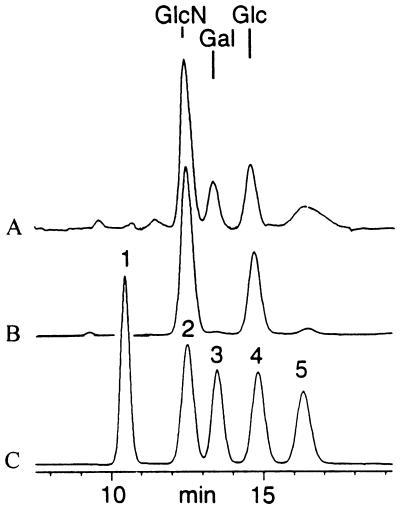
Composition analysis clearly supported the loss of galactose as underlying the shift in mass (MALDI spectra) and faster migration (SDS-PAGE) of galE LOS. As shown in Fig. 6, the MA-1 wild-type LOS preparation contained glucosamine (from the lipid A and oligosaccharide regions), galactose, and glucose (oligosaccharide branch). In contrast, the MA-1 galE mutant had significantly reduced levels of galactose, which was barely detectable. Both MA-1 and MA-1 galE LOS preparations had residual amounts of mannose. The mannose appears to be a contaminant in the LOS, since multiple washes of the LOS result in its removal.
FIG. 6.
Composition analysis of MA-1 (A) and MA-1 galE (B) under trifluoroacetic acid conditions, compared with standard mixture (profile C) containing galactosamine (peak 1), glucosamine (peak 2), galactose (peak 3), glucose (peak 4), and mannose (peak 5). The conserved core Kdo is destroyed under the hydrolysis conditions; the l-glycerol-d-manno-heptose peaks are not shown and elute much later (t > 25 min) under the gradient conditions (see Materials and Methods).
Comparison of GalE activity in N. meningitidis MA-1 and NMB.
The epimerase activity levels in MA-1 and NMB at the exponential and stationary phases were measured by the two-step assay. At the exponential phase, there was a twofold difference in the epimerase activity level between MA-1 and NMB (Table 4). In stationary-phase cultures, there was a 12.5-fold difference in the enzyme activity levels. This difference was not due to an increase in enzyme activity in MA-1, but rather to an 8.2-fold decrease in NMB epimerase activity from the exponential to the stationary phase. N. meningitidis MA-1 had similar activity levels at both phases of growth. The addition of 20 mM glucose to a stationary-phase culture of strain NMB did not increase the epimerase activity (1.7 ± 0.3 versus 2.7 ± 0.5 nmol of NADH generated per min [means ± standard errors] for NMB with and without glucose, respective; P = 0.0377).
TABLE 4.
UDP-glucose 4-epimerase activity levels in N. meningitidis MA-1 and NMB and their ΔgalEt::npt mutants
| Phase | nmol of NADH generated/min (± SE) in strain:
|
|||
|---|---|---|---|---|
| MA-1 | MA-1 ΔgalEt::npt | NMB | NMB ΔgalEt::npt | |
| Exponential | 22.2 (1.4) | 21.0 (1.6)a | 11.3 (0.6)b | 9.8 (1.0)c |
| Stationary | 17.2 (2.0)d | 12.0 (1.0)e | 1.4 (0.5)f,g | 3.4 (1.2)h |
P = 0.4178 for MA-1 versus MA-1 ΔgalEt::npt values in the exponential phase.
P = 0.0004 for MA-1 versus NMB values in the exponential phase.
P = 0.1352 for NMB versus NMB ΔgalEt::npt values in the exponential phase.
P = 0.0514 for MA-1 values in the exponential versus the stationary phase.
P = 0.1365 for MA-1 versus MA-1 ΔgalEt::npt values in the stationary phase.
P < 0.0001 for NMB values in the exponential versus the stationary phase.
P = 0.0002 for MA-1 versus NMB values in the stationary phase.
P = 0.009 for NMB versus NMB ΔgalEt::npt values in the stationary phase.
Comparison of predicted GalE amino acid sequences.
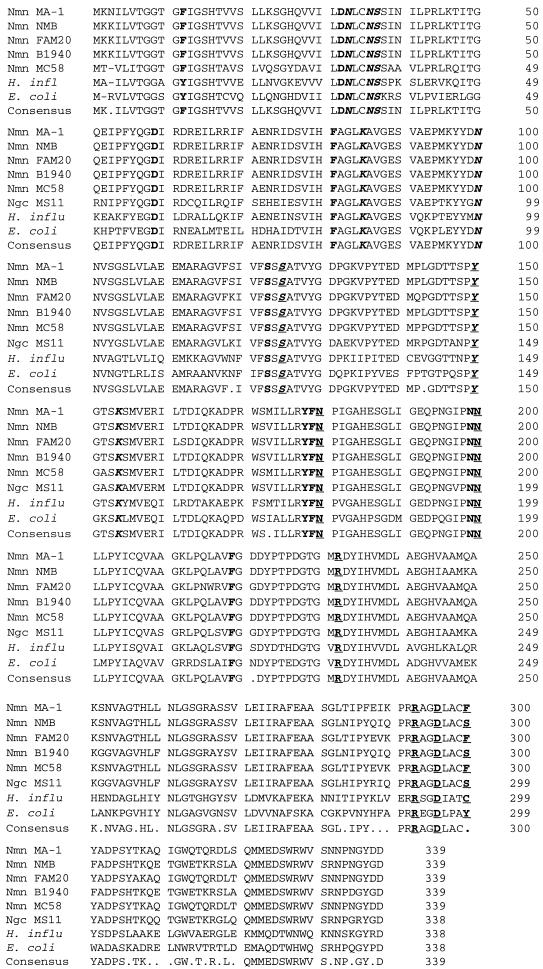
We cloned the galE genes from N. meningitidis MA-1 and NMB and compared the predicted amino acid sequences of the galE open reading frames alongside that of the previously cloned FAM20 galE gene (14). There was a high degree of identity among the putative NAD+ and UDP-sugar binding site residues (28) (mean = 95.65%; 22 of 23 amino acids) and 91% identity over the entire length of the polypeptide. There was one binding site residue change at amino acid 300 (F in MA-1, S in NMB, and F in FAM20) (Fig. 7). The corresponding residue is a tyrosine (position 299) in the E. coli GalE polypeptide, and this amino acid is not conserved among the other GalE proteins of gram-negative bacteria. In contrast, other binding site residues are highly conserved between meningococcal GalE and the E. coli GalE protein. Protein secondary-structure analysis indicated a high degree of conservation in the first 290 residues between MA-1 and NMB GalE polypeptides. This observation is consistent with the predicted amino acid sequence, where the first 290 residues are highly conserved (97.24% identity) and the last 49 residues are less conserved (75.51% identity). The last 49 residues contain 3 of the 21 active-site residues.
FIG. 7.
Predicted amino acid alignment of GalE proteins from various gram-negative bacteria. The consensus sequence is shown at the bottom. The meningococcal polypeptide is 1 amino acid longer than the protein from gonococci, Haemophilus influenzae, or E. coli. According to the structural data from the E. coli epimerase (3, 17, 26, 28), the amino acids that form the binding pockets of NAD+ and UDP-sugar are boldfaced. The amino acids that form hydrogen bonds with NAD+ are italicized, and those bonding with UDP-sugar are underlined. The residues involved in binding of both the cofactor and the substrate are italicized and underlined. S124 and Y149 of the E. coli protein are critical for catalysis, and these residues are conserved in both prokaryotes and eukaryotes. Nmn, N. meningitidis, Ngc, Neisseria gonorrhoeae; H. influ, H. influenzae.
Analysis of UDP-galactose epimerase activity in galEt deletion mutants of strains MA-1 and NMB.
The meningococcal genome contains a second partially duplicated copy of the galE gene (9, 11, 14, 15) that is not present in the gonococcal genome. This second copy is a duplication of the last 621 bp of the full-length open reading frame and contains one of the two putative hydrophobic domains involved in homodimer formation (3). To determine the contribution of the duplicated galE gene to the UDP-glucose 4-epimerase activities in strains MA-1 and NMB, all but the first 20 bp of the second galE open reading frame was deleted and replaced with a kanamycin cassette from pBSL14 (1). The deletion was introduced into the chromosomes of N. meningitidis MA-1 and NMB by transformation. Kanamycin-resistant transformants were screened by Southern blot hybridization and SDS-PAGE analysis of LOS (data not shown).
Crude cell extracts from cultures of wild-type MA-1 and NMB and their ΔgalEt::npt mutants at exponential and stationary phases were prepared, and epimerase activity levels were measured. For both MA-1 and NMB, the UDP-glucose 4-epimerase activity levels were not significantly different in the wild type and the ΔgalEt::npt mutant at exponential phase (Table 4). The growth curves of the wild-type and ΔgalEt::npt strains were identical (data not shown). The deletion of the galEt gene in MA-1 had no significant effect on epimerase activity at stationary phase. In NMB there was a 2.5-fold increase in enzyme activity in the mutant compared to the wild type at stationary phase (Table 4). The removal of the galEt gene was not able to restore the 8.2-fold decrease in NMB epimerase activity between the exponential and stationary phases.
Western blot analysis of wild-type meningococcal strains.
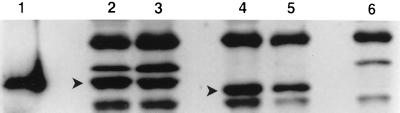
We analyzed whole-cell extracts from wild-type N. meningitidis MA-1 and NMB with polyclonal antisera in order to determine the amounts of GalE protein in the respective strains at the exponential and stationary phases. We detected a band that was specific to the GalE protein which was not present in the extract prepared from MA-1 galE. The Western blot analysis indicated that there were equivalent amounts of the GalE protein in N. meningitidis MA-1 and NMB at exponential phase. Similarly, at stationary phase, the amounts of GalE protein in MA-1 and NMB appeared to be equivalent. Within NMB, there was a slight increase in the amount of GalE protein at stationary phase compared to exponential phase. A similar observation was made in strain MA-1 (Fig. 8). Compared to the normalized band intensity of NMB GalE at log phase (taken as 1.0), the band intensity of NMB GalE at stationary phase was 1.3 (P = 0.0099), that of MA-1 GalE at log phase was 1.0 (P = 0.4639 for MA-1 GalE versus NMB GalE at log phase), and that of MA-1 GalE at stationary phase was 1.8 (P = 0.0025 for MA-1 GalE at log phase versus stationary phase; P = 0.1296 for MA-1 GalE versus NMB GalE at stationary phase). These values are means of three calculations.
FIG. 8.
Western blot analysis of meningococcal whole-cell lysates. Bacteria at either the exponential (lanes 2 and 4) or the stationary (lanes 3 and 5) phase were harvested and lysed by sonication. The protein concentrations were determined, and equal amounts of protein were loaded. The Western blots were incubated with preabsorbed mouse polyclonal antisera to recombinant meningococcal GalE protein. Lane 1, purified recombinant GalE; lanes 2 and 3, N. meningitidis NMB; lanes 4 and 5, N. meningitidis MA-1; lane 6, MA-1 galE. Solid arrowheads denote the band specific to GalE.
DISCUSSION
To further understand the role of UDP-glucose 4-epimerase in the biosynthesis of oligoglucose glycoforms found in meningococcal LOS, we examined the LOS from strain MA-1 and its galE mutant by mass spectrometry. We compared the results from these studies with the structure of LOS from NMB and NMB-SS3, which we had previously characterized (14). We also examined the epimerase activity levels in MA-1 and NMB at the log and stationary phases of growth. The LOS of MA-1 was composed of one glycoform with one or two PEA groups. The MA-1 galE LOS had two glycoforms with either one or two glucose residues and PEA substituted for HepII. The UDP-glucose 4-epimerase assay indicated a twofold difference in epimerase activity at log phase and a 12.5-fold difference at stationary phase between MA-1 and NMB (Table 3). Recently, Wakarchuk et al. (33) reported the presence of a glycoform with a diglucose structure in their galE strain. Pathogenic Neisseria species do not possess the accessory enzymes required for the utilization of exogenous galactose. This implies that UDP-glucose 4-epimerase is involved in controlling the ratio of UDP-galactose to UDP-glucose. The meningococcal epimerase, like the epimerases from yeast and E. coli, has an equilibrium constant favoring the formation of UDP-glucose (14). These observations suggest that the oligoglucose glycoforms in strain NMB and the absence of these glycoforms in strain MA-1 may be linked to the level of UDP-glucose 4-epimerase activity in the respective strains. The presence of the oligoglucose glycoforms in NMB-SS3 and the diglucose LOS structure in MA-1 galE and MC58 galE further support this interpretation.
The oligoglucose glycoforms in NMB-SS3 had up to two additional glucose molecules, while the oligoglucose glycoforms in MA-1 galE had one additional glucose. Similarly, MC58 galE had a novel glycoform containing an additional glucose (33). The transfer of the additional glucose residue(s) could occur by one of two pathways. The increased UDP-glucose concentration could either activate an unidentified glucosyltransferase or induce its expression. Alternatively, in the absence of UDP-galactose, galactosyltransferase could use UDP-glucose as a substrate. Based on our observations and that of Wakarchuk et al. (33), the first glucose residue after the Glc-HepI core structure could be added by the LgtE protein, a galactosyltransferase. The detection of the tetraglucose glycoform in NMB and NMB-SS3 suggests that the addition of the last two glucose residues to the core Glc-Hep structure in NMB may have been performed by a second glucosyltransferase, perhaps the recently identified glycosyl transferase, LgtG (2). This conclusion is supported by the observation that NMB LOS has a glucose residue attached to HepII through an α1-3 linkage in addition to the conserved N-acetylglucosamine (6, 22). These observations suggest that the appearance of the oligoglucose glycoforms in NMB but not in MA-1 may be a combinatorial effect of low UDP-glucose 4-epimerase activity and the presence of a second glucosyltransferase activity. This gene does not appear to be present in the genome of serogroup A meningococci. Recently, Kahler et al. (13) identified the lgtF locus in strain NMB. An LgtF− strain did not contain any detectable levels of glucose, suggesting that LgtF is responsible for adding the glucose to HepI. The second glucosyltransferase activity (GlcII to HepII; LgtG) requires the presence of the first glucose residue on HepI, analogous to the Kdo-dependent acyltransferases in E. coli lipid A biosynthesis (21).
Recently, further analysis of LOS from strain NMB was reported by Rahman et al. (22). They were unable to detect oligoglucose glycoforms in the LOS prepared from wild-type NMB. The detection of oligoglucose glycoforms in LOS preparations from galE strains of NMB, MA-1, and MC58 (14, 15, 33) suggests that they may be present in the wild type, albeit at low levels, perhaps below the detector sensitivity. It is also possible that different culture conditions led to differential expression of various LOS glycoforms. Such observations have been previously reported by other investigators (19, 29).
The putative structure of LOS from strain MA-1 was determined as Gal-Glc-Hep2-GlcNAc-Kdo2-lipid A based on ESI-mass spectrometric analysis. The absence of a sialylated derivative is consistent with the absence of sialic acid biosynthesis genes in serogroup A meningococci (16). Serogroup A strains are characterized by an (α1-6)-linked N-acetylmannosamine-1-phosphate capsule (16), rather than the sialic acid-containing capsules associated with other serogroups. It is interesting that the galE gene and the genes for production of the N-acetylmannosamine-1-phosphate capsule appear to be linked on the same operon. Indeed, polar insertion of a kanamycin marker into our ORF1 resulted in barely detectable levels of epimerase activity. This is not the organization found in serogroup B or C, where the capsule biosynthesis genes and the galE gene are separated by more than 1,000 nucleotides (5). Based on comparison of the nucleotide sequences upstream of galE genes from strains of serogroup B (B1940) and C (FAM20), it appears that the NMB galE gene is transcribed from its own promoter. The significance of this observation is not clear at the moment, but further investigation may yield explanations for the organization of these genes as a potential operon in serogroup A meningococci. The residual epimerase activity detected in MA-1 RKR and MA-1 galE was not statistically significant compared to that in MA-1 NKR. The high epimerase activity in MA-1 NKF is likely due to expression from the promoter for the kanamycin cassette.
The UDP-glucose 4-epimerase activity levels varied between MA-1 and NMB by as much as 12.5-fold in stationary cultures and as little as 2-fold in exponential cultures. This is the first report we are aware of documenting strain variability in the activity level of an enzyme important for the virulence of pathogenic Neisseria species. The UDP-galactose is not only used in LOS biosynthesis but also serves as the galactose donor in glycosylation of the pilin subunit of N. meningitidis (32). Although many LOS biosynthesis genes have been cloned and mutated, the regulation or strain variability of gene expression has not been observed, with the exception of the meningococcal rfaC gene (36). Our evidence strongly suggests that UDP-glucose 4-epimerase activity in N. meningitidis is different in different strains. In strain NMB, differences in epimerase activity were also observed between phases of growth.
The strain variability of epimerase activity could be due to a number of factors. The respective epimerases could be structurally different or could have different residues at the binding sites for the cofactor and substrate. This appears unlikely, since the putative active-site amino acids are nearly identical in the meningococcal GalE proteins (Fig. 7). It is also possible that the genes could be transcribed, or that galE mRNA could be translated, at different rates. In strain NMB, the galE gene appears to be the first gene of an operon containing rfb homologues, whereas in MA-1 (Fig. 2a), it is likely the fifth gene of a large operon containing the myn genes (27). We had noticed that the region immediately upstream of the NMB galE open reading frame start codon is very thymidine rich, whereas the corresponding region in front of the MA-1 galE gene has a few more adenines (Fig. 9). The 3′ end of the meningococcal 16S rRNA is purine rich, containing mostly guanines. The minor differences in the putative ribosome binding sites of the MA-1 and NMB galE genes do not appear to be biologically significant, since the difference in the relative amount of GalE protein between the strains at the exponential or stationary phase was not statistically significant.
FIG. 9.
Comparison of the nucleotide sequences of the region upstream of the galE open reading frame start codon in MA-1 and NMB. The sequence of the 3′ end of the meningococcal 16S rRNA is shown below for comparison.
In addition to the strain differences in epimerase activity, we observed an 8.2-fold decrease in enzyme activity in NMB between the two sampling time points. This difference was not due to the NMB GalE protein level at stationary phase, which was significantly higher than the protein level at exponential phase. This suggested the presence of a putative inactive enzyme at stationary phase. The putative inactive enzyme did not appear to involve a defective GalE protein from the second partial galE gene, since its deletion in NMB, although it resulted in a slight increase in the epimerase activity level, did not restore the stationary-phase epimerase activity to the exponential-phase level. Previously, the expression of the partial galE gene was not observed in Northern blot analysis (9). Similarly, our results suggest that the expression of the partial galE gene is minimal. These observations suggest a mechanism other than regulation of expression or defective dimer formation as a possible explanation for the low epimerase activity at stationary phase in strain NMB. Such a mechanism may be the formation of abortive enzyme complexes. Abortive UDP-glucose 4-epimerase complexes have been detected in E. coli (31). They are homodimers of full-length GalE protein that contain NADH and a uridine nucleotide (usually UTP) instead of NAD+ and UDP-sugar. It is possible that the physiological conditions in stationary-phase NMB favored abortive epimerase formation. The enzyme activity level was not dependent on the amount of glucose in the culture medium at stationary phase.
In this report we demonstrated that the absence of UDP-glucose 4-epimerase activity resulted in the expression of detectable levels of diglucose glycoforms in strain MA-1. Additionally, the level of epimerase activity may influence the expression of oligoglucose glycoforms in strain NMB. We also demonstrated the strain variability and growth phase-dependent variability of UDP-glucose 4-epimerase activity in meningococci. Based on our investigation, the difference in epimerase enzyme activity at the exponential and stationary phases appears not to be linked to the level of GalE protein. In strain NMB, the low epimerase activity may be due to allosteric inhibition through the formation of abortive epimerase complexes. Further biochemical analysis of UDP-glucose 4-epimerase from stationary-phase cultures of NMB is required to determine the nature of the putative abortive epimerase complexes. We do not know if the oligoglucose glycoforms are assembled throughout growth phase at a constant rate or assembled predominantly at late stages of growth.
ACKNOWLEDGMENTS
This research was supported in part by NIH grants AI18384 and AI38515 to M.A.A. and AI31254 to B.W.G. The mass spectrometry was performed at the UCSF Mass Spectrometry Facility, which is partially supported by a grant from the National Center for Research Resources (NCRR BRTP 01614).
REFERENCES
- 1.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–55. [PubMed] [Google Scholar]
- 2.Banerjee A, Wang R, Uljohn S N, Rice P A, Gotschlich E C, Stein D C. Identification of the gene (lgtG) encoding the lipooligosaccharide beta chain synthesizing glucosyl transferase from Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1998;95:10872–10877. doi: 10.1073/pnas.95.18.10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauer A J, Rayment I, Frey P A, Holden H M. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 A resolution. Proteins. 1992;12:372–381. doi: 10.1002/prot.340120409. [DOI] [PubMed] [Google Scholar]
- 4.Catlin B W. Transformation of Neisseria meningitidis by deoxyribonucleates from cells and from culture slime. J Bacteriol. 1960;79:579–590. doi: 10.1128/jb.79.4.579-590.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frosch M, Weisgerber C, Meyer T F. Molecular characterization and expression in Escherichia coli of the gene complex encoding the polysaccharide capsule of Neisseria meningitidis group B. Proc Natl Acad Sci USA. 1989;86:1669–1673. doi: 10.1073/pnas.86.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gamian A, Beurret M, Michon F, Brisson J R, Jennings H J. Structure of the L2 lipopolysaccharide core oligosaccharides of Neisseria meningitidis. J Biol Chem. 1992;267:922–925. [PubMed] [Google Scholar]
- 7.Gibson B W, Engstrom J J, John C M, Hines W, Falick A M. Characterization of bacterial lipooligosaccharides by delayed extraction matrix-assisted laser ionization time-of-flight mass spectrometry. J Am Soc Mass Spectrom. 1997;8:645–658. [Google Scholar]
- 8.Gibson B W, Melaugh W, Phillips N J, Apicella M A, Campagnari A A, Griffiss J M. Investigation of the structural heterogeneity of lipooligosaccharides from pathogenic Haemophilus and Neisseria species and of R-type lipopolysaccharides from Salmonella typhimurium by electrospray mass spectrometry. J Bacteriol. 1993;175:2702–2712. doi: 10.1128/jb.175.9.2702-2712.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammerschmidt S, Birkholz C, Zahringer U, Robertson B D, van Putten J, Ebeling O, Frosch M. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol Microbiol. 1994;11:885–896. doi: 10.1111/j.1365-2958.1994.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 10.Helander I M, Lindner B, Brade H, Altmann K, Lindberg A A, Rietschel E T, Zahringe U. Chemical structure of the lipopolysaccharide of Haemophilus influenzae strain I-69Rd/b+ Eur J Biochem. 1988;177:483–492. doi: 10.1111/j.1432-1033.1988.tb14398.x. [DOI] [PubMed] [Google Scholar]
- 11.Jennings M P, van der Ley P, Wilks K E, Maskell D J, Poolman J T, Moxon E R. Cloning and molecular analysis of the galE gene of Neisseria meningitidis and its role in lipopolysaccharide biosynthesis. Mol Microbiol. 1993;10:361–369. [PubMed] [Google Scholar]
- 12.John C M, Griffiss J M, Apicella M A, Mandrell R E, Gibson B W. The structural basis for pyocin resistance in Neisseria gonorrhoeae lipooligosaccharides. J Biol Chem. 1991;266:19303–19311. [PubMed] [Google Scholar]
- 13.Kahler C M, Carlson R W, Rahman M M, Martin L E, Stephens D S. Two glycosyltransferase genes, lgtF and rfaK, constitute the lipooligosaccharide ice (inner core extension) biosynthesis operon of Neisseria meningitidis. J Bacteriol. 1996;178:6677–6684. doi: 10.1128/jb.178.23.6677-6684.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee F K, Stephens D S, Gibson B W, Engstrom J J, Zhou D, Apicella M A. Microheterogeneity of Neisseria lipooligosaccharide: analysis of a UDP-glucose 4-epimerase mutant of Neisseria meningitidis NMB. Infect Immun. 1995;63:2508–2515. doi: 10.1128/iai.63.7.2508-2515.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee F K N. Ph.D. thesis. Iowa City: The University of Iowa; 1998. [Google Scholar]
- 16.Liu T-Y, Gotschlich E C, Jonssen E K, Wysocki J R. Studies on the meningococcal polysaccharides. 1. Composition and chemical properties of the group A polysaccharide. J Biol Chem. 1971;246:2849–2858. [PubMed] [Google Scholar]
- 17.Liu Y, Thoden J B, Kim J, Berger E, Gulick A M, Ruzicka F J, Holden H M, Frey P A. Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:10675–10684. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- 18.Morse S A, Bartenstein L. Purine metabolism in Neisseria gonorrhoeae: the requirement for hypoxanthine. Can J Microbiol. 1980;26:13–20. doi: 10.1139/m80-003. [DOI] [PubMed] [Google Scholar]
- 19.Morse S A, Mintz C S, Sarafian S K, Bartenstein L, Bertram M, Apicella M A. Effect of dilution rate on lipopolysaccharide and serum resistance of Neisseria gonorrhoeae grown in continuous culture. Infect Immun. 1983;41:74–82. doi: 10.1128/iai.41.1.74-82.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips N J, John C M, Reinders L G, Gibson B W, Apicella M A, Griffiss J M. Structural models for the cell surface lipooligosaccharides of Neisseria gonorrhoeae and Haemophilus influenzae. Biomed Environ Mass Spectrum. 1990;19:731–745. doi: 10.1002/bms.1200191112. [DOI] [PubMed] [Google Scholar]
- 21.Raetz C R H. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. [Google Scholar]
- 22.Rahman M M, Stephens D S, Kahler C M, Glushka J, Carlson R W. The lipooligosaccharide (LOS) of Neisseria meningitidis serogroup B strain NMB contains L2, L3, and novel oligosaccharides, and lacks the lipid-A 4′-phosphate substituent. Carbohydr Res. 1998;307:311–324. doi: 10.1016/s0008-6215(98)00012-3. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.Schneider H, Hale T L, Zollinger W D, Seid R C, Jr, Hammack C A, Griffiss J M. Heterogeneity of molecular size and antigenic expression within lipooligosaccharides of individual strains of Neisseria gonorrhoeae and Neisseria meningitidis. Infect Immun. 1984;45:544–549. doi: 10.1128/iai.45.3.544-549.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens D S, McAllister C F, Zhou D, Lee F K, Apicella M A. Tn916-generated, lipooligosaccharide mutants of Neisseria meningitidis and Neisseria gonorrhoeae. Infect Immun. 1994;62:2947–2952. doi: 10.1128/iai.62.7.2947-2952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson B A, Frey P A. Identification of lysine 153 as a functionally important residue in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1993;32:13231–13236. doi: 10.1021/bi00211a035. [DOI] [PubMed] [Google Scholar]
- 27.Swartley J S, Liu L-J, Miller Y K, Martin L E, Edupuganti S, Stephens D S. Characterization of the gene cassette required for biosynthesis of the (α1→6)-linked N-acetyl-d-mannosamine-1-phosphate capsule of serogroup A Neisseria meningitidis. J Bacteriol. 1998;180:1533–1539. doi: 10.1128/jb.180.6.1533-1539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thoden J B, Frey P A, Holden H M. Molecular structure of the NADH/UDP-glucose abortive complex of UDP-galactose 4-epimerase from Escherichia coli: implications for the catalytic mechanism. Biochemistry. 1996;35:5137–5144. doi: 10.1021/bi9601114. [DOI] [PubMed] [Google Scholar]
- 29.Tsai C M, Boykins R, Frasch C E. Heterogeneity and variation among Neisseria meningitidis lipopolysaccharides. J Bacteriol. 1983;155:498–504. doi: 10.1128/jb.155.2.498-504.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsai C M, Frasch C. A sensitive silver stain for detecting lipooligosaccharide in polyacrylamide gels. Anal Biochem. 1981;199:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 31.Vanhooke J L, Frey P A. Characterization and activation of naturally occurring abortive complexes of UDP-galactose 4-epimerase from Escherichia coli. J Biol Chem. 1994;269:31496–31504. [PubMed] [Google Scholar]
- 32.Virji M, Stimson E, Makepeace K, Dell A, Morris H R, Payne G, Saunders J R, Moxon E R. Posttranslational modifications of meningococcal pili. Identification of a common trisaccharide substitution on variant pilins of strain C311. Ann N Y Acad Sci. 1996;797:53–64. doi: 10.1111/j.1749-6632.1996.tb52949.x. [DOI] [PubMed] [Google Scholar]
- 33.Wakarchuk W, Martin A, Jennings M P, Moxon E R, Richards J C. Functional relationships of the genetic locus encoding the glycosyltransferase enzymes involved in expression of the lacto-N-neotetraose terminal lipopolysaccharide structure in Neisseria meningitidis. J Biol Chem. 1996;271:19166–19173. doi: 10.1074/jbc.271.32.19166. [DOI] [PubMed] [Google Scholar]
- 34.Westphal O, Jann K. Bacterial lipopolysaccharides. Extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 35.Wilson D B, Hogness D S. The enzymes of the galactose operon in Escherichia coli. II. The subunits of uridine diphosphogalactose 4-epimerase. J Biol Chem. 1969;244:2132–2136. [PubMed] [Google Scholar]
- 36.Zhou D, Zaleski A, Buscher B, Preston A, Apicella M A. Presented at the Tenth International Pathogenic Neisseria Conference. Baltimore, Md. 1996. [Google Scholar]