Abstract
O-GlcNAc transferase (OGT) is an essential enzyme that installs O-linked N-acetylglucosamine (O-GlcNAc) to thousands of protein substrates. OGT and its isoforms select from these substrates through the tetratricopeptide repeat (TPR) domain, yet the impact of truncations to the TPR domain on substrate and glycosite selection is unresolved. Here, we report the effects of iterative truncations to the TPR domain of OGT on substrate and glycosite selection with the model protein GFP-JunB and the surrounding O-GlcNAc proteome in U2OS cells. Iterative truncation of the TPR domain of OGT maintains glycosyltransferase activity but alters subcellular localization of OGT in cells. The glycoproteome and glycosites modified by four OGT TPR isoforms were examined on the whole proteome and a single target protein, GFP-JunB. We found the greatest changes in O-GlcNAc on proteins associated with mRNA splicing processes and that the first four TPRs of the canonical nucleocytoplasmic OGT had the broadest substrate scope. Subsequent glycosite analysis revealed that alteration to the last four TPRs corresponded to the greatest shift in the resulting O-GlcNAc consensus sequence. This dataset provides a foundation to analyze how perturbations to the TPR domain and expression of OGT isoforms affect the glycosylation of substrates, which will be critical for future efforts in protein engineering of OGT, the biology of OGT isoforms, and diseases associated with the TPR domain of OGT.
Keywords: O-GlcNAc, OGT, Tetratricopeptide repeats (TPRs), Glycoproteomics
Introduction
O-Linked N-acetyl glucosamine (O-GlcNAc) is a monosaccharide post-translational modification installed on serine or threonine residues of thousands of nucleocytoplasmic proteins across multiple species [1]. O-GlcNAc is a dynamic and reversible modification that is regulated by a pair of essential enzymes, the writer O-GlcNAc transferase (OGT) and the eraser O-GlcNAcase (OGA) [2, 3]. Regulatory maintenance of O-GlcNAc levels in the proteome is critical for normal cellular physiology as loss of either OGT or OGA disrupts cellular growth and limits organismal survival [4, 5], and disruptions to O-GlcNAc homeostasis have been linked to genetic diseases (e.g., X-linked intellectual disability) [6] and various chronic diseases, including neuro-degeneration [7], diabetes [8], and cancer [9].
Due to the essential requirement of OGT for substrate O-GlcNAcylation [10], deciphering the basis of OGT’s substrate selection mechanisms has attracted much interest [2, 11, 12]. The canonical nucleocytoplasmic form of OGT (ncOGT) is composed of two domains: an N-terminal tetratricopeptide repeat (TPR) domain and a C-terminal catalytic domain (Fig. 1A). The recognition and glycosylation of protein substrates are primarily mediated by the TPR domain, which consists of 13.5 (rounded to 13 hereafter) repetitive 34-amino-acid antiparallel alpha helices in ncOGT [13–15]. Two additional isoforms of OGT, the mitochondrial isoform (mOGT) and the short isoform (sOGT), have truncated TPR domains composed of 9.5 and 2.5 TPR units, respectively (rounded to 9 and 2 hereafter, Fig. 1A) [16–18]. Truncation of the TPR domain affects the OGT interactome [19–24] which may provide a molecular basis for observed cellular signaling and proliferation effects [10, 25, 26] or for human diseases associated with mutations in the TPR domain, such as X-linked intellectual disability (XLID) [6, 27–30]. The relationship of these OGT isoforms to substrate and glycosite selection has been explored in vitro, where variation of the TPR domain alters substrate selectivity and truncation of the TPR domain as in sOGT limits the resulting glycosyltransferase activity [11, 18, 23]. However, expression of sOGT in cells showed a robust increase in global O-GlcNAcylation [31, 32], indicative of differences between in vitro and cellular assays. Given the expression of OGT isoforms changes as a function of age [33] and their differential relationship to human disease [34], investigation of the substrate and glycosite selection of the OGT isoforms in cells is vital.
Fig. 1.
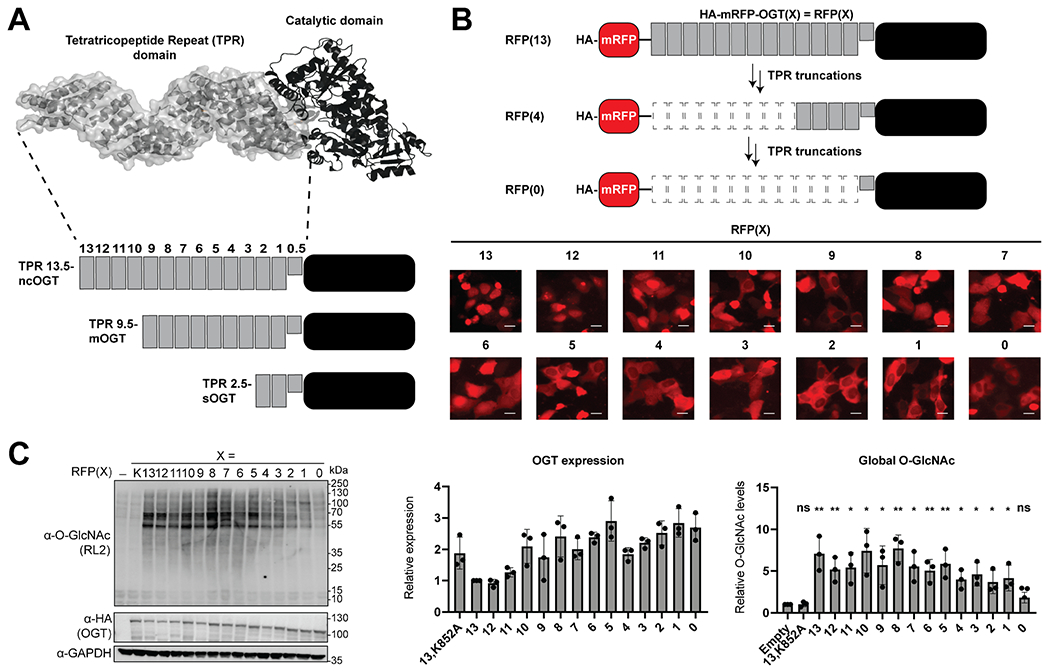
OGT TPR truncations localize differently and are active in cells. A Diagram of the OGT isoforms and TPR truncations generated. B Fluorescence localization of the RFP-OGT TPR truncation fusions [RFP(X)] with X representing the number of TPRs on OGT. The scale bar represents 10 μm. C Western blot of global O-GlcNAc and OGT expression levels from the different OGT truncations. – is no transfected OGT and X represents either the inactive K852A full-length OGT mutant (K) or the number of TPRs on OGT. Error bars represent the standard deviation. Under a two-tailed t test, ns represents p ≥ 0.05, one asterisk represents p ≤ 0.05, two asterisks represent p ≤ 0.01
In our efforts to better understand the regulatory role of O-GlcNAc on substrates in cells, we recently reported the development of a pair of complementary methods to write and erase O-GlcNAc on a target protein using nanobody fusions to OGT and split-OGA, respectively [35, 36]. The nanobody is a small single-domain protein binder that recruits the enzyme to the desired target protein in cells for selective manipulation of O-GlcNAc levels. To enhance selectivity for a desired substrate, these enzymes were engineered to reduce their innate substrate selection mechanisms, which were then replaced with a nanobody. Previously, we achieved enhanced protein selection through the nanobody with OGT by truncation of the TPR domain to four repeats [35]. However, whether further engineering of the TPR domain would afford even higher selectivity through the nanobody or enable enhanced selectivity for protein regions and glycosites was unknown.
Here, we report a systematic investigation of the TPR domain and its relationship to the protein substrate and glycosite selection of OGT in cells. We generated truncations of each of the TPRs of OGT to characterize subcellular localization and activity on a global scale, followed by a detailed survey of the substrates and glycosites produced by overexpression of RFP-tagged isoforms corresponding to ncOGT, mOGT, sOGT, and an OGT with 4 TPR repeats in U2OS cells. We found that most of these OGT isoforms were active in cells, which results in altered substrate and glycosite profiles and find that the first four TPRs afford the broadest substrate selection, while the last four TPRs mediate the glycosite selection. These data will enhance future engineering of the O-GlcNAc proteome and provide insight to the potential protein substrates mediated by different OGT isoforms.
Methods
Cell culture, transfection, immunofluorescence, and cell lysate collection
Methods outlined below were performed as previously reported, with modifications [37]. Unless otherwise noted, the experiments were performed with U2OS cells (a gift from the Choudhary Lab, Broad Institute). Cells were cultured in high glucose with pyruvate Dulbecco’s modified Eagle medium supplemented with 10% FBS and 1% penicillin-streptomycin at 37 °C in a humidified atmosphere with 5% CO2. Samples for western blot or immunofluorescence were prepared from cells seeded in a well of a sterile 6-well plate at a density ~ 0.4 × 106 cells/well and transfected at ~ 80% confluency the next day. For mass spectrometry–based glycoproteomics experiments, cells were seeded at the density ~ 7 × 106 cells/plate in sterile 150-mm tissue culture dishes (Corning, 25,383–103) and transfected at ~ 80% confluency the next day. Transient expression of the indicated proteins was performed by transfection using TransiT-PRO (Mirus Bio, MIR 5740) with the desired plasmids following the manufacturer’s protocol. Cells were incubated for 48 h after transfection for all experiments. For fluorescent imaging, cells were visualized with an ECHO microscope using the TX Red bandpass filter.
To generate lysates, the transfected cells were collected and lysed by probe sonication in lysis buffer [150 μL for western blot samples or 800 μL for MS samples of 2% SDS + 1 × PBS + 50 μM Thiamet-G + 1 × protease inhibitors (cOmplete™, EDTAfree Protease Inhibitor Cocktail, Sigma Aldrich; 11,873,580,001)]. A BCA assay was performed to determine protein concentration, and the concentration was adjusted to 2.5 μg/μL with lysis buffer for western blot samples and 5 μg/μL for MS samples. One hundred micrograms of protein sample was prepared with 5 × BME buffer, and the sample was heated for 5 min at 98 °C and centrifuged prior to gel loading.
Western blotting
The protein sample (15 μL) was loaded on 6–12% Tris–glycine SDS-PAGE gels and run on a Mini-PROTEAN® BioRad gel system. Gels were transferred with the Invitrogen iBlot. Membranes were stained with Ponceau stain to verify transfer and equal protein loading and blocked with 3% BSA + 1 × TBST for 1 h at 24 °C. Primary antibodies and the following dilutions were incubated with the membranes for 12 h: anti-HA (1:1000; Cell Signaling; 3724S), anti-O-GlcNAc RL2 (1:1000; Abcam; ab2739), and anti-GAPDH (1:1000; Cell Signaling; D4C6R). The membranes were washed 3 × 5 min each wash with 1 × TBST and incubated with the following secondary antibodies and dilutions: anti-rabbit HRP (1:10,000; Rockland; 611–1302) and anti-mouse IR 800 (1:10,000; LI-COR; 925–32,210). The membranes were washed 3 × 5 min each wash with 1 × TBST, and results were obtained by chemiluminescence or IR imaging using Azure c600. The membranes were quantified using LI-COR Image Studio Lite, and graphs were made with GraphPad Prism 9.
Molecular cloning
The RFP-OGT TPR truncations were generated by restriction enzyme digestion of the previously developed plasmid HA-RFP-OGT(13) [35] using the BamHI and NotI enzymes to extract the OGT(13) sequence. OGT TPR truncation regions of interest were amplified from the HA-RFP-OGT(13) with forward primers containing a BamHI restriction site and reverse primers containing a NotI restriction site. Standard molecular cloning techniques were used to generate the final plasmids.
Chemical enrichment of glycoproteins and sample preparation for IsoTag
Chemical enrichment of glycoproteins for MS analysis was processed as previously described with minor modifications [35, 38]. After the collection of transfected cells and the generation of reduced and alkylated cell lysates, a BCA protein assay (Pierce) was performed on cell lysates and protein concentration was adjusted to 5 μg/μL with lysis buffer. The protein lysate (2 mg, 400 μL) was placed on ice and mixed with water (490 μL), GalT1 labeling buffer (800 μL, final concentrations: 50 mM NaCl, 20 mM HEPES, 2% NP-40, pH 7.9), 100 mM MnCl2 (110 μL), 500 μM UDP-GalNAz (100 μL), and 2 mg/mL GalT1 enzyme (100 μL). The sample reaction was then rotated for 16 h at 4 °C, and the GalT reaction was quenched by methanol–chloroform protein precipitation [methanol/chloroform=4:1 (v/v)]. The sample was allowed to air-dry for 5 min at 24 °C before resuspension in 2% SDS + 1 × PBS (400 μL). The sample was treated with a pre-mixed solution of the click chemistry reagents [100 μL; final concentration of 200 μM IsoTaG silane probe (3:1 heavy:light mixture), 500 μM CuSO4, 100 μM THPTA, 2.5 mM sodium ascorbate], and the reaction was incubated for 4 h at 37 °C. The click reaction was quenched by methanol–chloroform protein precipitation, and the protein pellet was allowed to air-dry for 5 min at 24 °C. The dried pellet was resuspended in 2% SDS + 1 × PBS (300 μL) by probe tip sonication and then diluted in 1 × PBS (3 mL) to a final concentration of 0.2% SDS. A 50% slurry of streptavidin–aga-rose resin [800 μL, washed with 1 × PBS (2 × 1 mL)] was added to the protein solution, and the resulting mixture was incubated for 12 h at 24 °C with rotation. The beads were washed using spin columns with 8 M urea (5 × 1 mL), 0.1% SDS + 1 × PBS (5 × 1 mL), 1 × PBS (5 × 1 mL), and water (5 × 1 mL). Spin columns were capped and the beads were resuspended in 470 μL 1 × PBS + 1 mM CaCl2, urea (8 M, 30 μL), and trypsin (2 μg), and protein digestion was performed for 16 h at 37 °C with rotation. Supernatant was collected and the beads were washed three times with 1 × PBS (2 × 200 μL) and MilliQ water (200 μL). The trypsin fraction for protein identification was formed by combining the washes and supernatant digest. To cleave glycopeptides from beads, the IsoTaG silane probe was cleaved with 5% formic acid/water (2 × 200 μL) for 1 h at 24 °C with rotation and the glycopeptide eluent was collected. The beads were washed with 50% acetonitrile–water + 1% formic acid (2 × 200 μL), and the washes were combined with the glycopeptide eluent to form the cleavage fraction for site-level identification. The trypsin and cleavage fractions were dried in a vacuum centrifuge and desalted using C18 tips following the manufacturer’s instructions. The trypsin fractions were resuspended in 50 mM TEAB (20 μL), and the samples were labeled with the corresponding amine-based TMT 16-plex (5 μL) and reacted for 1 h at 24 °C. The TMT reactions were quenched with 2 μL of a 5% hydroxylamine solution and combined before concentration and fractionation into six samples using a High pH Reversed-Phase Peptide Fractionation Kit (Thermo Fisher Scientific). All samples were stored at − 20 °C until analysis.
Mass spectrometry parameters used for glycoproteomics and data analysis
The following instruments and parameters were used as previously described [36]. A Thermo Scientific EASY-nLC 1000 system was connected to an Orbitrap Fusion Tribrid with a nano-electrospray ion source. The mobile phases A and B were water with 0.1% (vol/vol) formic acid and acetonitrile with 0.1% (vol/vol) formic acid, respectively. For the trypsin fractions, peptides were separated using a linear gradient from 4 to 32% B for 50 min, then increased to 50% B for 10 min and finally increased to 98% B for 10 min followed by re-equilibration. For the cleavage fractions, peptides were separated with a linear gradient from 5 to 30% B for 95 min, then increased to 50% B for 15 min and finally increased to 98% B for 10 min followed by re-equilibration. The instrument parameters were set as previously described with minor modifications [35, 38]. Briefly, the MS1 spectra were recorded from m/z 400–2000 Da and if the oxonium product ions (138.0545, 204.0867, 345.1400, 347.1530 m/z) were observed in the HCD spectra, then ETD (250 ms) with supplemental activation (35%) was performed in a subsequent scan on the same precursor ion selected for HCD. Other relevant parameters of EThcD include an isolation window (3 m/z), use calibrated charge-dependent ETD parameters (True), Orbitrap resolution (50 k), first mass (100 m/z), and inject ions for all available parallelizable time (True). The raw data was processed using Proteome Discoverer 2.4 (Thermo Fisher Scientific). For the trypsin fraction, the data were searched against the UniProt/SwissProt human (Homo sapiens) protein database (20,355 proteins, downloaded on Feb. 21, 2019) and contaminant proteins using the Sequest HT algorithm. The database was adjusted by removing the OGT and JunB entries and replacing with RFP-OGT and GFP-JunB. Searches were performed as previously described [36]. For the cleavage fraction, both HCD and EThcD spectra were searched against the proteome identified in the trypsin fraction using Byonic algorithms. The searches were performed with the following guidelines: trypsin as enzyme, 3 missed cleavages allowed; 10 ppm mass error tolerance on precursor ions; and 0.02 Da mass error tolerance on fragment ions. Intact glycopeptide searches allowed for tagged HexNAc modifications (HexNAc + 203.0794 on cysteine, serine, threonine). Methionine oxidation and cysteine carbaminomethylation were set as variable modifications. Glycopeptide spectral assignments passing a FDR of 1% at the peptide spectrum match level based on a target decoy database were kept. Singly modified glycopeptides assigned from EThcD spectra passing a 1% FDR and possessing a delta modification score of greater than or equal to ten from > 2 PSMs were considered unambiguous glycosites. The label-free quantification of unambiguous glycosite was performed as previously described [39].
Gene ontology analysis
Differentially abundant proteins (DAPs) across treatments were calculated in R using one-way ANOVA and an adjusted p < 0.01 after Benjamini–Hochberg FDR correction. The Database for Annotation, Visualization and Integrated Discovery (DAVID) [40] was used for gene ontology (GO) enrichment analysis of DAPs with the whole Homo sapiens genome as the background and with a significant enrichment threshold of a Benjamini correction of p < 0.05. The DAPs were also subjected to protein–protein interaction (PPI) analysis using the STRING database [41].
OGT consensus sequence generation
Consensus sequences were generated by extraction of +/− 15 amino acid residues surrounding an unambiguous glycosite at S/T/C. Glycopeptides with only a single unambiguous glycosite were used for consensus sequence generation. Consensus sequences that were less than 31 amino acids were excluded. The final list of glycosites was entered into the Berkeley Weblogo program to generate the consensus sequences. We extracted amino acids around the glycosite in the − 5 and + 15 positions to afford the consensus sequence around these glycosites for clarity in figures.
Results
OGT TPR truncations exhibit altered localization, but most remain active in cells
We began our studies by generating sequential TPR truncations from ncOGT that are fused to the C-terminus of mRFP [RFP(X), X = number of TPRs], which we had previously characterized for high expression and activity (Fig. 1B) [35]. In addition, we included the catalytically inactive RFP(13,K852A) as a control. The resulting 14 RFP-fused OGT constructs with laddered TPR domains were transfected to U2OS cells, and the localization of the proteins was determined by fluorescence microscopy (Fig. 1B). We observed the localization of OGT transition from a primarily nuclear to a nucleocytoplasmic localization as the TPR domain was truncated, with a major inflection point occurring in constructs with five or fewer TPRs. The transfected cells were then collected to determine the levels of OGT expression and changes to the global O-GlcNAc proteome using the O-GlcNAc RL2 antibody (Fig. 1C). Excitingly, most of the TPR truncations exhibited elevated O-GlcNAc levels, with the exception of the catalytically inactive RFP(13,K852A) and RFP(0), which has the TPR domain completely removed. Interestingly, RFP(2) and RFP(1) displayed a reduced and altered O-GlcNAc band pattern relative to the other TPR truncations. The altered subcellular localization and global O-GlcNAc levels produced by the TPR truncations, particularly exhibited by RFP(2) and RFP(1), correspond with altered substrate selection previously shown by in vitro studies with single proteins and cell extracts [11, 18].
Overexpression of OGT isoforms enhances glycosylation of proteins associated with mRNA processing
To gain insight into the OGT substrates that are differentially modified by the TPR truncations, we next sought to gain deeper insight to the O-GlcNAc proteins and their modification sites by quantitative proteomics and glycoproteomics [35, 36]. We focused our efforts on the most studied OGT isoforms consisting of 13, 9, 4, and 2 TPRs. In addition, we co-expressed the RFP(X) construct with GFP-JunB to serve as an internal benchmark to determine the extent of modification on GFP-JunB by the different TPR truncations in comparison to endogenous OGT. U2OS cells co-transfected with GFP-JunB and the indicated OGT construct were collected, and the O-GlcNAcylated proteins were chemoenzymatically labeled for biotin-based enrichment. Following enrichment, the O-GlcNAcylated proteins were digested for protein identification and quantification by mass spectrometry (MS). The biotinylated glycopeptides were recovered by cleavage of the probe and analyzed separately. Over 1200 proteins were identified after O-GlcNAc enrichment (SI Table 1). To reduce complexity in the initial analysis and to identify the most significantly enriched proteins as a function of overexpression of at least one of the OGT isoforms, we performed a comparison of the samples transfected with an OGT isoform to the control by a one-way ANOVA, which revealed 32 significantly enriched proteins [fold enrichment ≥ 1.5, adjusted p value ≤ 0.01 (Fig. 2A, SI Table 1)]. As expected, JunB and OGT are the highest O-GlcNAcylated proteins in this dataset due to their overexpression. All 32 significantly enriched proteins are found in RFP(13) samples, and 24 are exclusively enriched with RFP(13). A subset of the other proteins are relatively insensitive to regulation through the TPR domain (i.e., SPART, ZC3HAV1, CEP170), while others are specifically modified by RFP(13) and RFP(9) (i.e., ANXA7, EIF2SI, MLF2, PHLDB1). A network analysis of the 32 proteins identified a highly enriched network cluster of 10 proteins involved in mRNA processes such as splicing, processing, and mRNA decay (Fig. 2B). The remaining proteins formed minor relationships related to lipid droplets, microtubules, or cellular signaling, or were not clustered to a network, and are not represented in the network analysis diagram for clarity.
Fig. 2.
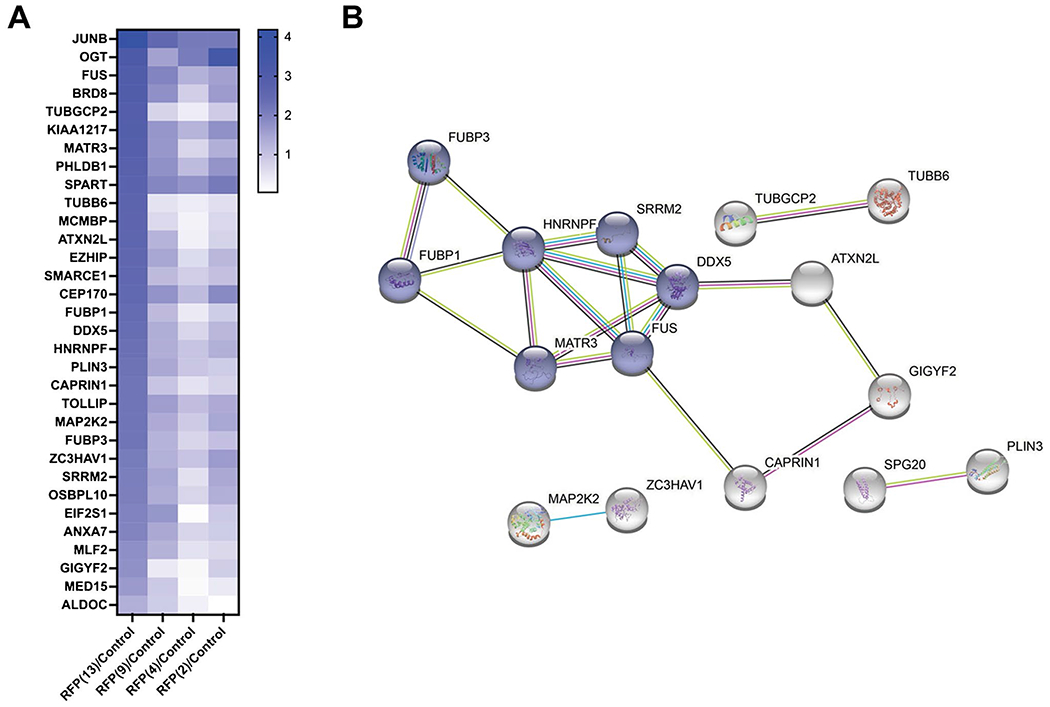
One-way ANOVA of the enriched glycoproteins and a network analysis of the highly enriched proteins. A A heat map representing the abundance of the 32 highly enriched glycoproteins p ≤ 0.01 after a one-way ANOVA comparison of the OGT transfected samples and the control. B The 32 proteins identified in A were analyzed for protein–protein interactions with a STRING database to identify highly enriched networks. Not all 32 proteins formed relationships with other proteins and are not represented in the network analysis for clarity. Nodes in blue represent a highly enriched network involved in mRNA splicing, and the remaining proteins formed minor relationships related to lipid droplets, microtubules, or cellular signaling
RFP(13) has the broadest substrate scope of all of the OGT isoforms
Having identified the most significantly enriched proteins and pathways shared between the OGT constructs, we then sought to analyze an expanded set of affected proteins and pathways enriched by OGT overexpression using a reduced cut-off criterion. We analyzed differences in the significantly enriched O-GlcNAcylated proteins between the co-transfected OGT constructs and the GFP-JunB only control [log2(RFP(X)/control) ≥ 1.5, p ≤ 0.05, Fig. 3A, SI Table 2]. Of the over 1200 proteins, 212 were significantly enriched in the RFP(13) sample, 124 with RFP(9), 114 with RFP(4), and 123 with RFP(2) in comparison to the control (Fig. 3B). Inherent glycosylation efficiency of the control protein GFP-JunB by each of the OGT isoforms was also variable, with the highest recognition by RFP(13) and the lowest glycosylation by RFP(4). In addition to GFP-JunB, a comparison of the significantly enriched proteins between the OGT constructs revealed a total of 59 highly enriched proteins across the four co-transfected samples, indicating that these proteins do not require the extended TPR domain for glycosylation. Conversely, the RFP(13) sample enhanced glycosylation of the most unique proteins at 71 proteins, which represents substrates that appear to require the full-length TPR domain (Fig. 3C). By contrast, selective O-GlcNAcylation of 3 proteins with RFP(9), 7 proteins with RFP(4), and 10 proteins with RFP(2) was observed (Fig. 3C).
Fig. 3.
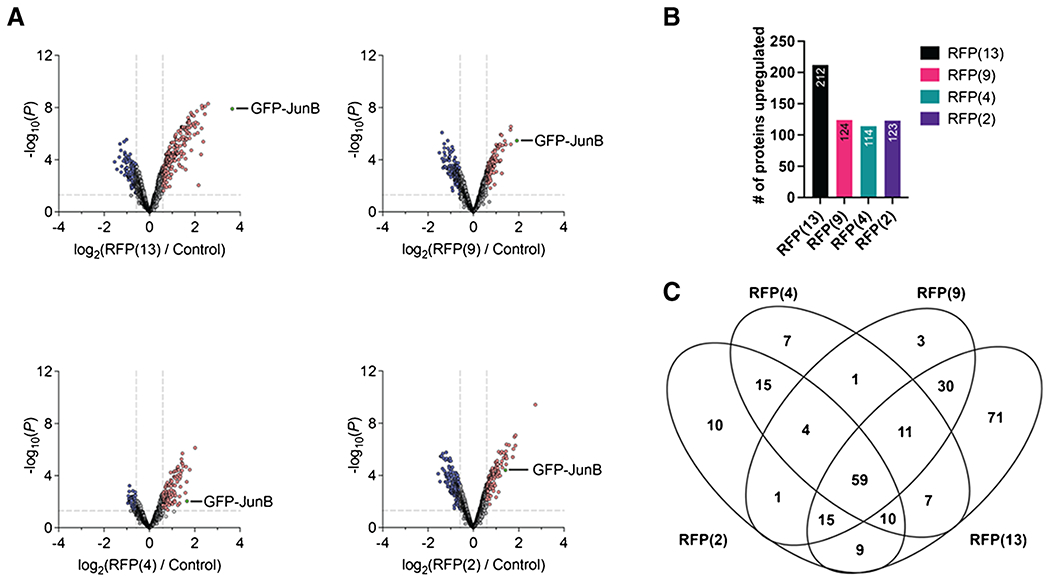
A comparison of each OGT condition to the control to identify differentially enriched glycoproteins. A Volcano plots of each of the OGT conditions to the control. B Bar graph of the number of upregulated proteins identified from the volcano plot. C A Venn diagram comparing all the upregulated proteins found in each of the OGT conditions to identify shared and unique proteins for each condition
To obtain a broad overview of the biological processes selectively enriched by expression of each of the OGT constructs, we performed a gene ontology (GO) analysis on all the enriched proteins from each condition. After a stringent filter for clusters of adjusted p value ≤ 0.05, two clusters involving the biological processes of cell–cell adhesion and regulation of translation initiation were shared between all the OGT constructs (Fig. 4, SI Table 3). As before, proteins that are O-GlcNAcylated by RFP(13) yielded the highest number of biological process clusters at 14, of which 9 of these processes involved disruptions to components of the nuclear pore complex. The mitotic nuclear envelope disassembly cluster was shared between proteins that are O-GlcNAcylated by RFP(13) and RFP(9), which represents the mitochondrial isoform of OGT.
Fig. 4.
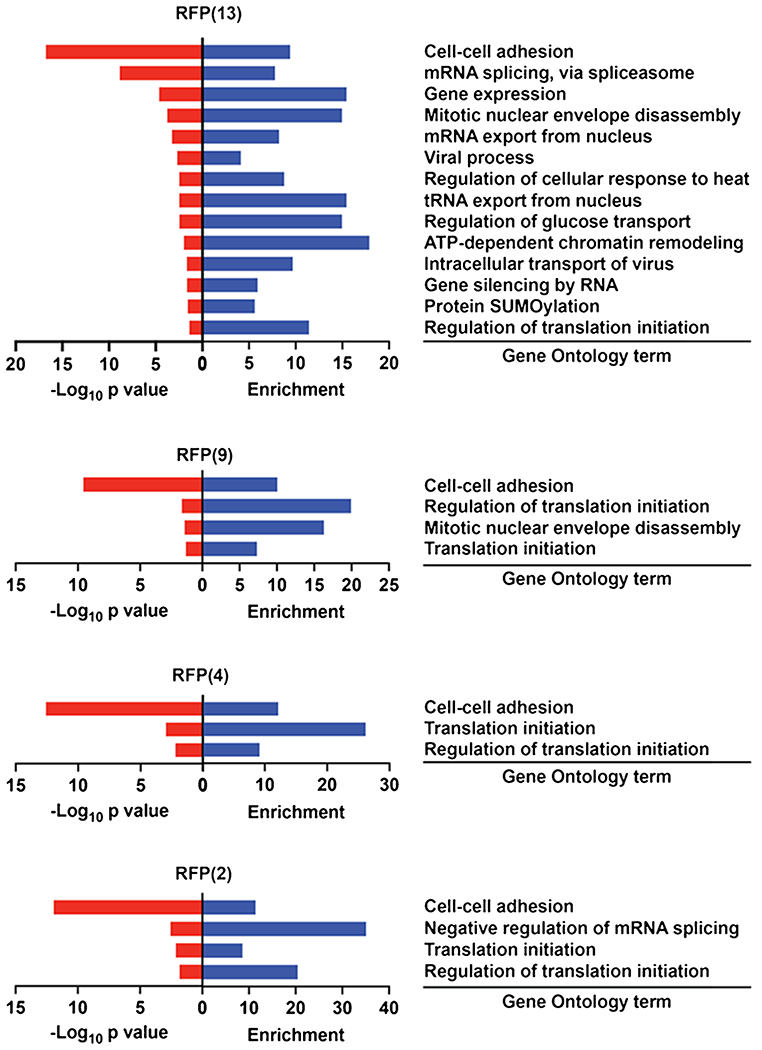
A gene ontology (GO) analysis of the upregulated proteins in each of the OGT conditions. Only biological pathways with a p ≤ 0.05 are represented
Interestingly, GO analysis of O-GlcNAcylated proteins found in the RFP(2) condition was the only set that enriched for the negative regulation of the mRNA splicing process. RFP(2) represents the sOGT isoform, which has been described as a negative regulator of ncOGT by disrupting the Ataxin-10–ncOGT interaction [42]. Of the 5 enriched proteins associated with the negative regulation of the mRNA splicing process in the RFP(2) condition, three (splicing factor U2AF, polypyrimidine tract-binding protein 1, and transformer-2 protein homolog beta) are shared between RFP(13) and RFP(2). The remaining two proteins, RNA-binding protein 10 and apoptotic chromatin condensation inducer in the nucleus (Acinus), are unique to the RFP(2) construct.
In sum, this quantitative proteomic analysis of the O-GlcNAcylated proteome produced by four OGT isoforms confirmed that the full-length OGT possesses the broadest swath of substrates across the greatest number of biological processes and revealed specific substrates and biological pathways that are shared and unique to each of the examined OGT isoforms. Furthermore, additional truncation of the TPR domain for higher protein selectivity through nanobody fusions may not be advantageous given the similar global O-GlcNAc levels and gain of specific substrates with RFP(2).
Overexpression of TPR truncations shows an altered consensus sequence at the glycosite
As the enrichment step of our proteomics workflow allows us to separate trypsin-digested glycopeptides from unmodified peptides, we next performed glycoproteomics on the recovered glycosites to determine the effects of overexpression of truncated OGT constructs on the site level. Mass spectra were searched against the over 1200 enriched proteins using Byonic, and the results were filtered based on ambiguous versus unambiguous glycosites (SI Table 4). We defined an unambiguous glycosite as a modified glycopeptide having a ≥ 10 delta modification score by EThcD fragmentation. All other glycopeptides were categorized as ambiguous. In total, we identified over 3900 peptide spectral matches (PSMs) that were assigned to 891 unique unambiguous glycopeptides from 210 proteins across 6 biological replicates for each condition and a total of 30 samples. To determine if any new unambiguous O-GlcNAc proteins had been identified, we searched for these 210 proteins on the recently published O-GlcNAc database [1] and found 6 new proteins (Table 1). Of these 6 proteins, three were modified by RFP(13), two by RFP(9), three by RFP(4), and four by RFP(2). Two proteins, PLCB3 and MAP2K1, were unique to RFP(2), and WBP2 was unique to RFP(4).
Table 1.
New glycoproteins identified in the OGT conditions. The > 200 glycoproteins with unambiguous glycosites were compared to the O-GlcNAc database, and 6 new glycoproteins were identified. The condition and glycosites in which the glycoprotein was identified are listed with the gene and protein name
| Entry | Entry name | Protein name | Gene name | Conditions | Glycosites |
|---|---|---|---|---|---|
| Q01970 | PLCB3_HUMAN | 1-Phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 (EC 3.1.4.11) | PLCB3 | RFP(2) | S931 |
| Q02750 | MP2K1_HUMAN | Dual specificity mitogen-activated protein kinase kinase 1 (MAP kinase kinase 1) | MAP2K1 | RFP(2) | T23 |
| O15479 | MAGB2_HUMAN | Melanoma-associated antigen B2 (cancer/testis antigen 3.2) | MAGEB2 | RFP(13), RFP(4), RFP(2) | T67 |
| Q7LG56 | RIR2B_HUMAN | Ribonucleoside-diphosphate reductase subunit M2 B (EC 1.17.4.1) | RRM2B | RFP(13), RFP(9), RFP(4), RFP(2) | S329 |
| Q9H6W3 | RIOX1_HUMAN | Ribosomal oxygenase 1 (60S ribosomal protein L8 histidine hydroxylase) | RIOX1 | RFP(13), RFP(9) | S126 |
| Q969T9 | WBP2_HUMAN | WW domain-binding protein 2 (WBP-2) | WBP2 | RFP(4) | T49, T53 |
To obtain a broad overview of how the glycosites on proteins changed with the different OGT constructs, we next generated consensus sequences of the unique glycosites found in each sample (Fig. 5). These consensus sequences were generated from unique unambiguous glycosites identified in each condition. We identified 113, 246, 179, 232, and 205 unique glycosites in the GFP-JunB control, RFP(13), RFP(9), RFP(4), and RFP(2) respectively. These unique glycosites were then used to obtain an O-GlcNAc consensus sequence for each of the OGT conditions that could be used to compare to the control, which contains glycosites introduced by the endogenous OGT (Fig. 5A). As the TPR domain positions substrates for glycosylation through interactions toward the C-terminus of the peptide sequence, we extracted amino acids around the glycosite in the − 5 and + 15 positions to afford the consensus sequence around these glycosites (SI Table 4). As expected, the consensus sequence local to the glycosite (position 0) is enriched for a PPV sequence at the − 3 to − 1 positions on the peptide, which assists in extending the peptide in the catalytic site of the OGT [43]. In general, glycosylated protein regions have a high abundance of polar uncharged and hydrophobic residues in the 1 to 15 position, with some enrichment for K that may form contacts with acidic residues in the TPR domain, as previously observed in vitro [44]. Unique sequences observed in the RFP(13) largely mimic the consensus sequence observed in the control conditions. However, overexpression of the additional OGT constructs, RFP(9), RFP(4), and RFP(2), increases the prevalence of additional polar uncharged and charged residues like Q, N, K, E, and D.
Fig. 5.
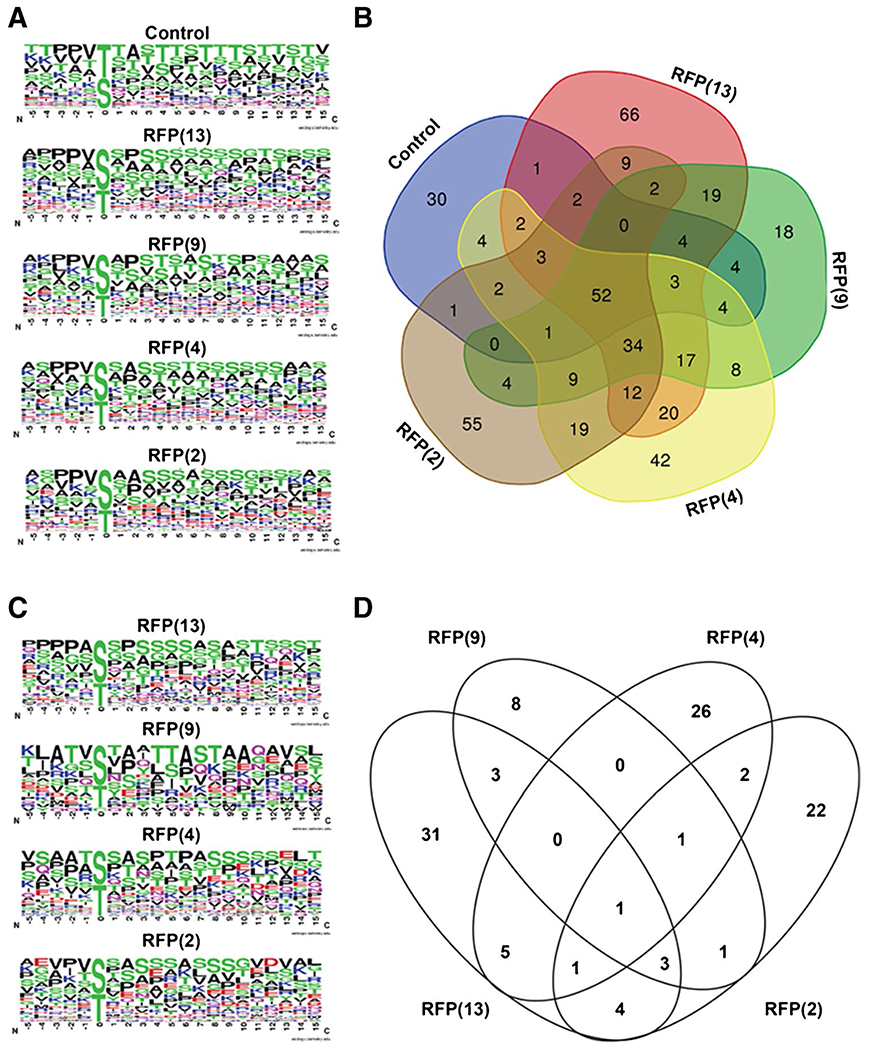
Glycosite analysis of TPR truncations. A Unique unambiguous glycosites from each of the conditions were used to generate OGT consensus sequences. B Venn diagram comparing unique unambiguous glycopeptides identified in each of the five conditions. C OGT consensus sequences of the unique glycosites identified from the Venn diagram in B of the overexpression of the indicated OGT construct. D Venn diagram of the proteins identified from the unique glycosites from the Venn diagram in B
We then generated a Venn diagram to visualize glycosites that are common between each OGT isoform and the GFP-JunB only control or unique to the overexpression of the indicated OGT construct. We found that the majority of glycosites are shared between two or more conditions, but that 66, 18, 42, and 55 glycosites from 48, 17, 36, and 35 proteins were identified only in the RFP(13), RFP(9), RFP(4), and RFP(2) conditions, respectively (Fig. 5B, SI Table 4). Most of these unique glycosites can be traced to a single protein though in some cases multiple unique glycosites are found on one protein (i.e., JunB, OGT, AHNAK, HCF-1, VIM, ZC3H14, PLEC). To identify glycosite features important for each OGT isoform, we generated an O-GlcNAc consensus sequence with these unique glycosites for each of the OGT conditions (Fig. 5C). Overexpression of the OGT constructs generally increases the prevalence of additional polar uncharged and charged residues like Q, N, K, E, and D. The consensus sequence from the RFP(2) condition was particularly enriched in negatively charged E residues. To determine if the changes in the O-GlcNAc consensus sequence were due to an altered glycosite selection or substrate preference, we generated a Venn diagram of the proteins identified from the unique glycosites in Fig. 5B. We found that the majority of these glycosites that were only observed in one condition also derived from proteins that were unique to that condition (Fig. 5D, SI Table 4). This analysis indicates that a major contributor to the altered O-GlcNAc consensus sequence in Fig. 5C derives from an altered substrate preference of the OGT isoforms. Collectively, the majority of glycosites detected during OGT overexpression are shared across two or more conditions and the unique glycosites identified can be traced to an altered substrate preference by the OGT isoforms.
Label-free quantification of GFP-JunB indicates increased glycosite abundance with RFP(13) expression
In addition to the glycosite level data obtained for the global O-GlcNAc proteome, co-transfection of GFP-JunB allowed us to compare all the OGT conditions and their effect on GFP-JunB glycosites. An analysis of the unambiguous peptide spectral matches (PSMs) obtained from GFP-JunB after a search of the MS data revealed that RFP(13) had the highest number of PSMs detected at 161 and the detection of PSMs decreased as the TPR was truncated with 96, 97, 69, and 20 for the RFP(9), RFP(4), RFP(2), and control conditions respectively (Table 2), indicating a reduced activity on GFP-JunB with the TPR truncations. To determine the relative abundance of the GFP-JunB glycosites across conditions, we employed label-free quantification as previously described [39], averaged the abundance across the three biological replicates for each condition, and generated a heatmap for the relative abundance of each glycosite (Fig. 6A). In line with the PSM data, all OGT conditions exhibited elevated levels of O-GlcNAc on GFP-JunB. Glycosites at positions 84, 85, 95, and 100 are modified by the endogenous OGT in the control condition, and the abundance and intensity of these glycosites increase with OGT overexpression indicating these are stable glycosites naturally found on GFP-JunB. Glycosites at positions 28, 84, 85, and 312 appear to increase in intensity with longer TPRs while glycosites 95 and 100 are relatively insensitive to the TPR length. Additional glycosites at positions 14, 28, 38, 76, 129, 251, and 312, and surrounding regions are more readily detected in OGT overexpression conditions. In general, all OGT isoforms can modify GFP-JunB, although a reduction to two TPRs appears to affect the efficiency and access to glycosites and at least four TPRs are necessary to modify similar glycosites as longer TPR isoforms. Thus, in terms of O-GlcNAc engineering, use of OGT isoforms with at least four TPRs appears to be optimal for generating a similar glycosite profile as ncOGT, but use of the sOGT isoform may access unique glycosites that cannot be targeted by the other constructs.
Table 2.
Total unambiguous peptide spectral matches (PSMs) obtained from GFP-JunB in each indicated condition
| Glycosite position | Control | RFP(13) | RFP(9) | RFP(4) | RFP(2) | Grand total |
|---|---|---|---|---|---|---|
| 14 | 6 | 3 | 2 | 11 | ||
| 16 | 2 | 1 | 1 | 4 | ||
| 18 | 1 | 1 | ||||
| 28 | 34 | 17 | 15 | 10 | 76 | |
| 38 | 10 | 2 | 3 | 3 | 18 | |
| 67 | 7 | 7 | ||||
| 70 | 4 | 1 | 1 | 1 | 7 | |
| 76 | 1 | 1 | ||||
| 84 | 2 | 28 | 20 | 6 | 3 | 59 |
| 85 | 2 | 10 | 7 | 4 | 3 | 26 |
| 95 | 15 | 40 | 40 | 48 | 40 | 183 |
| 100 | 1 | 3 | 1 | 5 | ||
| 117 | 4 | 4 | ||||
| 129 | 7 | 4 | 11 | |||
| 237 | 2 | 2 | ||||
| 251 | 4 | 4 | ||||
| 311 | 1 | 1 | ||||
| 312 | 5 | 5 | 5 | 8 | 23 | |
| Grand total | 20 | 161 | 96 | 97 | 69 | 443 |
Fig. 6.
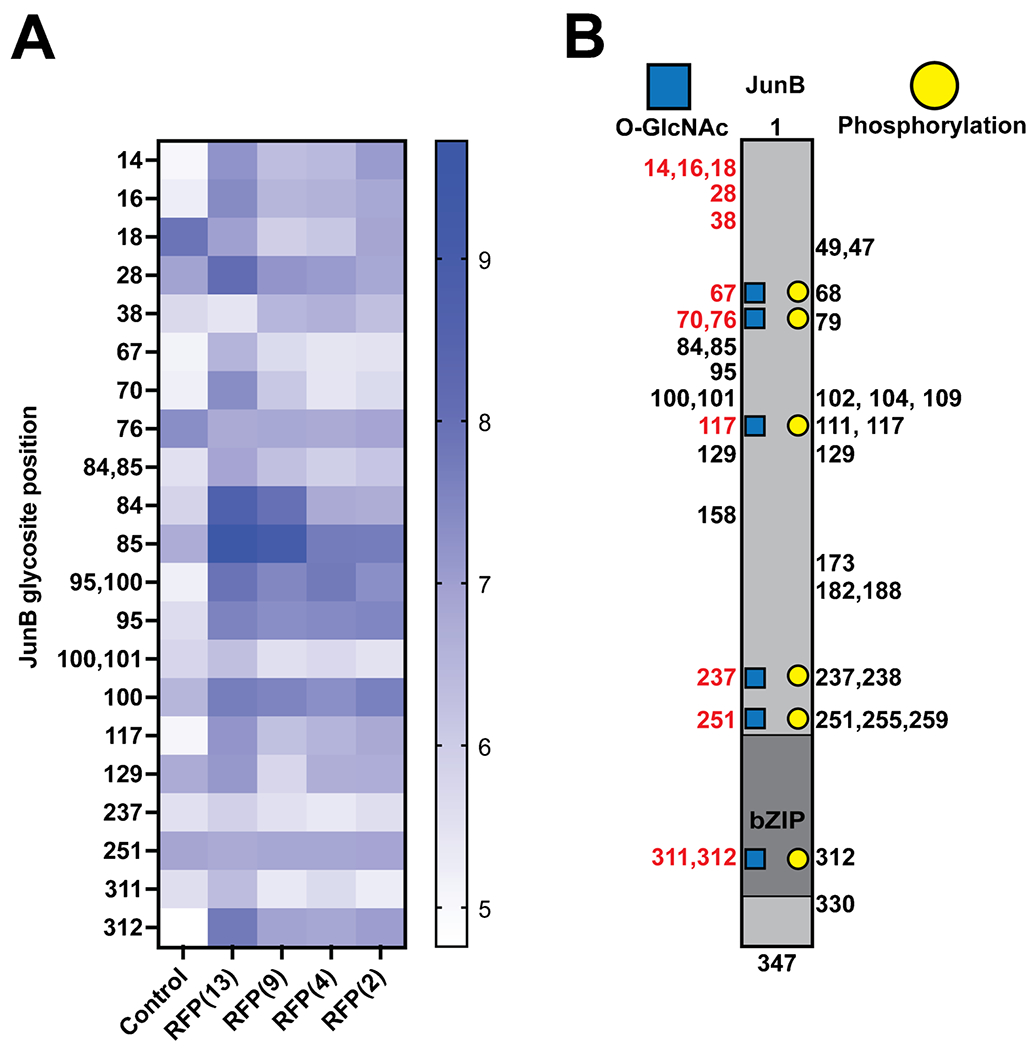
Glycosite analysis of GFP-JunB. A Label-free quantification heatmap of confident glycosites (right) identified in GFP-JunB labeled with the positions of JunB for each of the conditions (bottom). B Diagram of JunB glycosites identified in this study with new glycosites labeled in red. Phosphorylation sites from the PhosphoSite database were used to demonstrate regions of potential O-GlcNAc/phosphorylation crosstalk (blue squares and yellow circles)
Finally, we cross-referenced the identified glycosites on GFP-JunB with previously identified phosphorylation sites on JunB (Fig. 6B) [45]. Interestingly, we observed novel glycosites at positions 311 and 312 in the bZIP domain of JunB indicating a potential relationship between O-GlcNAc and DNA binding of JunB. Glycosites that also overlapped with phosphorylation at positions 68, 70, 102, 117, 237, 251, and 312 may also play a role in PTM crosstalk. Phosphorylation at positions 79 and 102 has been described as altering transcriptional activity [46], and phosphorylation in regions 251–259 has an effect on JunB degradation [47]. We have previously observed that JunB expression levels appear to increase as it is O-GlcNAcylated by a nanobody-OGT [35]. Further study of these intersections with JunB or other enriched glycoproteins obtained in our dataset may reveal additional differences between the glycosites installed by the OGT TPR truncations that may have implications on downstream regulation of the protein substrate.
Discussion
In this study, we report the first direct comparison of overexpressed OGT TPR truncations and their effect on the global O-GlcNAc proteome and a specific target protein GFP-JunB by quantitative proteomics and glycoproteomics. Consistent with prior reports, we find that OGT isoforms with a truncated TPR domain have intrinsic activity in cells [31, 32, 35], and now map this activity to substrates and their glycosites. We find that RFP(13), representing the full-length ncOGT, possesses the greatest substrate scope amongst the OGT isoforms examined, while the TPR truncations have more limited and sometimes unique access to substrates and glycosites. The differentiation is particularly accentuated by RFP(2), which represents sOGT, where the enriched glycoproteins and unique glycosites corresponded to a unique mRNA biological pathway and differentiated consensus sequence. These data could serve to increase glycosylation on desired substrates and influence glycosite selection while minimizing perturbations to the cell for O-GlcNAc engineering, in addition to the biological implications arising from differential substrate and site selection of the OGT TPR isoforms.
The relationship between the TPR isoforms of OGT and biological function is a growing area of investigation as interactions and substrate selection through the TPR domain may be of particular relevance to normal O-GlcNAc regulation [6, 10, 31, 48]. Comparison of glycoproteins in the presence of RFP(13) to a recently determined TPR interactome of ncOGT via a proximity-labeling approach [24] shows enrichment for similar biological processes, indicating that the TPR interactome and the enriched O-GlcNAcylated proteome correspond. Future studies may extend this comparison to the OGT isoforms mOGT and sOGT, in addition to characterizing whether these isoforms are important for cell viability and organismal function, although the isoforms of OGT are not required for cell proliferation in tissue culture [10]. Our data indicate that the isoforms of OGT possess both a complementary and unique substrate scope, accompanied by altered efficiency at modifying glycosites, which is potentially indicative of separable biological roles in cells if they are required for normal cellular function. These observations were made in the presence of endogenous OGT, which could mask more subtle differences between the OGT isoforms, and future studies in cell lines lacking endogenous OGT function may reveal additional differences. Further investigation of these OGT isoforms may yield essential substrates, binding partners, and glycosites required for cell proliferation and organismal development.
Analysis of the glycoproteome enriched by each of the OGT isoforms revealed differences in the biological processes that were conserved or unique to each isoform. One of the most enriched pathways across the OGT isoforms was the mRNA splicing pathway, which points to a possible reciprocal regulatory relationship between ncOGT and sOGT on mRNA splicing. Observation of the mRNA splicing pathway is particularly interesting given the highly regulated expression of OGT and OGA that maintain steady levels of O-GlcNAc by a detained intron-splicing mechanism [49, 50], although the exact mechanisms monitoring the levels of O-GlcNAc to trigger splicing have not been identified. If the OGT isoforms are differentially involved in the detained intron-splicing mechanism, the dataset reported here may assist in the evaluation of glycoproteins underlying this biological function. Indeed, one of the mRNA splicing proteins, Acinus, is regulated in Arabidopsis by O-GlcNAc or O-fucose and these distinct sugar modifications regulate which introns are spliced [51]. Separately, of the 14 enriched biological processes of RFP(13), 9 involved components of the nuclear pore complex (NPC). The biological role of O-GlcNAc on the NPC includes roles in the permeability barrier [52], transport rate [53], and stability of the nucleoporins to degradation [54–56]. Future study of these targets with nanobody-OGT [35] and nanobody-split-OGA [36] fusions for increasing or decreasing O-GlcNAc on specific target proteins may assist in the dissection of the role of O-GlcNAc in these proteins and their functions.
Of particular interest to our protein engineering efforts is that truncations of the TPR are still active in cells and modify similar glycosites as RFP(13) with a reduced substrate scope. The reduced substrate scope after truncation of the TPR domain to four repeats should indeed enhance selectivity for a desired target protein through the nanobody. Based on our data herein, further truncation of the TPR domain may not be advantageous given the similar global O-GlcNAc levels and gain of specific substrates and glycosites with RFP(2). However, the altered prevalence of polar and charged residues in the consensus sequence of truncated OGTs hints at the ability to limit access to certain glycosites and opens a pathway to creating glycosite-specific OGTs in the future. Undoubtedly, the full-length OGT is the construct that consistently generates the most detectable number of glycosites on overexpressed and endogenous protein substrates. Extension of efforts to determine how substrates engage the TPR domain of OGT would define the features of the extended TPR that promote this selection [11, 12]. Nevertheless, our dataset can shed light on the TPRs that are required to engage certain substrates in a global manner and the particular glycosites affected by TPR truncations.
In summary, we report here the first dataset directly comparing the effects of overexpression of OGT TPR truncations on the O-GlcNAc proteome. We find that truncation of the OGT isoforms alters the substrate and glycosite selection in cells and identify that the first four TPRs afford the broadest substrate selection, while the last four TPRs mediate the glycosite selection. These profiles of the substrate and glycosite selectivity of the OGT TPR truncations will assist with understanding the biological roles of the OGT isoforms. Future studies of the OGT isoforms will further enhance our understanding of the regulatory relationship between O-GlcNAc and human disease, and eventually mechanisms to introduce O-GlcNAc to desired target proteins and glycosites to influence specific biological pathways.
Supplementary Material
Funding
Support from the National Institutes of Health (U01CA242098-01, C.M. Woo), the Sloan Foundation (C.M. Woo), the Camille and Henry Dreyfus Foundation (C.M. Woo), and Harvard University is gratefully acknowledged.
Biographies

Daniel Ramirez PhD, is a recent PhD graduate of the Woo Lab at Harvard and a postdoctoral fellow in the Wang group at the Carnegie Institution for Science, Department of Plant Biology, located in Stanford University. Prior to his graduate work at Harvard, he worked at the intersection of the SUMOylation system and influenza with Dr. Germán Rosas-Acosta at the University of Texas at El Paso (UTEP) and in the structural biology of nucleoporins with Dr. Gunter Blobel at Rockefeller University. His current work at Carnegie focuses on the role sugar modifications play in integrating nutrient, hormone, and environmental signals in controlling plant growth and development.

Bo Yang PhD, received his PhD in bio-analytical chemistry from the University of Missouri-Columbia. He conducted research under the supervision of Professor C. Michael Greenlief and Professor Grace Y. Sun, focusing on the application of mass spectrometry–based metabolomics and proteomics, especially in the areas of phytochemicals and their impact on oxidative/inflammatory responses in the mammalian systems. Currently, he is a postdoctoral fellow studying glycoproteomics in the laboratory of Professor Christina M. Woo in the Department of Chemistry & Chemical Biology at Harvard University.

Alexandria D’Souza MSc, is currently working in Christina Woo’s Lab at Harvard University. She graduated from the University of Chicago with an MSc in Computer Science. Her interests include proteomics and data analysis—bioinformatics as well as statistics and pipeline development.

Dacheng Shen Dr. rer.nat., is a postdoctoral fellow in the Woo Lab at the Department of Chemistry and Chemical Biology at Harvard University. He has been in the field of sugar chemistry for years where he worked for his MSc in Biao Yu’s group at SIOC, Shanghai, China, and Dr. Rer. Nat. in Peter Seeberger’s group at MPIKG, Potsdam, Germany. He is currently focusing on the study of O-GlcNAcrelated chemical biology.

Christina Woo PhD, is Associate Professor in the Department of Chemistry and Chemical Biology at Harvard University and an affiliate member of the Broad Institute. The Woo Lab combines the rational design of small molecules and proteins with chemical proteomics to edit post-translational modifications and the signaling outcomes they produce in cells.
Footnotes
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s00216-021-03731-8.
Conflict of interest The authors declare no competing interests.
Data availability
The MS data were deposited at the ProteomeXchange Consortium [57] via the PRIDE partner repository, and are available with the identifier PXD028141.
References
- 1.Wulff-Fuentes E, Berendt RR, Massman L, Danner L, Malard F, Vora J, Kahsay R, Olivier-Van Stichelen S. The human O-GlcNA-come database and meta-analysis. Sci Data. 2021;8:25. 10.1038/s41597-021-00810-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joiner CM, Li H, Jiang J, Walker S. Structural characterization of the O-GlcNAc cycling enzymes: insights into substrate recognition and catalytic mechanisms. Curr Opin Struct Biol. 2019;56:97–106. 10.1016/j.sbi.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stephen HM, Adams TM, Wells L. Regulating the regulators: mechanisms of substrate selection of the O-GlcNAc cycling enzymes OGT and OGA. Glycobiology. 2021;1–5. 10.1093/glycob/cwab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–9. 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang YR, Song M, Lee H, Jeon Y, Choi EJ, Jang HJ, Moon HY, Byun HY, Kim EK, Kim DH, Lee MN, Koh A, Ghim J, Choi JH, Lee-Kwon W, Kim KT, Ryu SH, Suh PG. O-GlcNAcase is essential for embryonic development and maintenance of genomic stability. Aging Cell. 2012;11:439–48. 10.1111/10.1111/j.1474-9726.2012.00801.x. [DOI] [PubMed] [Google Scholar]
- 6.Pravata VM, Omelková M, Stavridis MP, Desbiens CM, Stephen HM, Lefeber DJ, Gecz J, Gundogdu M, Õunap K, Joss S, Schwartz CE, Wells L, van Aalten DMF. An intellectual disability syndrome with single-nucleotide variants in O-GlcNAc transferase. Eur J Hum Genet. 2020;28:706–14. 10.1038/s41431-020-0589-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuzwa SA, Vocadlo DJ. O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer’s disease and beyond. Chem Soc Rev. 2014;43:6839–58. 10.1039/c4cs00038b. [DOI] [PubMed] [Google Scholar]
- 8.Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev Proteomics. 2013;10:365–80. 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11:678–84. 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine ZG, Pottera SC, Joinerb CM, Feia GQ, Nabetc B, Sonnette M, Zacharaf NE, Grayc NS, Paulog JA, Walkera S (2021) Mammalian cell proliferation requires noncatalytic functions of O-GlcNAc transferase. Proc Natl Acad Sci U S A 118:2020.10.22.351288 . 10.1073/PNAS.2016778118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joiner CM, Hammel FA, Janetzko J, Walker S. Protein substrates engage the lumen of O-GlcNAc transferase’s tetratricopeptide repeat domain in different ways. Biochemistry. 2021. 10.1021/acs.biochem.0c00981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kositzke A, Fan D, Wang A, Li H, Worth M, Jiang J. Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe. Int J Biol Macromol. 2021;169:51–9. 10.1016/j.ijbiomac.2020.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kreppel LK, Hart GW. Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J Biol Chem. 1999;274:32015–22. 10.1074/jbc.274.45.32015. [DOI] [PubMed] [Google Scholar]
- 14.Lubas WA, Hanover JA. Functional expression of O-linked GlcNAc transferase. J Biol Chem. 2000;275:10983–8. [DOI] [PubMed] [Google Scholar]
- 15.Iyer SPN, Hart GW. Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. J Biol Chem. 2003;278:24608–16. 10.1074/jbc.M300036200. [DOI] [PubMed] [Google Scholar]
- 16.Hanover JA, Yu S, Lubas WB, Shin SH, Ragano-Caracciola M, Kochran J, Love DC. Mitochondrial and nucleocytoplasmic isoforms of O-linked GlcNAc transferase encoded by a single mammalian gene. Arch Biochem Biophys. 2003;409:287–97. 10.1016/S0003-9861(02)00578-7. [DOI] [PubMed] [Google Scholar]
- 17.Love DC. Mitochondrial and nucleocytoplasmic targeting of O-linked GlcNAc transferase. J Cell Sci. 2002;116:647–54. 10.1242/jcs.00246. [DOI] [PubMed] [Google Scholar]
- 18.Lazarus BD, Love DC, Hanover JA (2006) Recombinant O-GlcNAc transferase isoforms : identification of O-GlcNAcase , yes tyrosine kinase, and tau as isoform-specific substrates. 16:415–421 . 10.1093/glycob/cwj078 [DOI] [PubMed] [Google Scholar]
- 19.Zhang Q, Liu X, Gao W, Li P, Hou J, Li J, Wong J. Differential regulation of the ten-eleven translocation (TET) family of dioxygenases by O-linked β-N-acetylglucosamine transferase (OGT). J Biol Chem. 2014;289:5986–96. 10.1074/jbc.M113.524140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez M, Renuse S, Kreimer S, O’Meally R, Natov P, Madugundu AK, Nirujogi RS, Tahir R, Cole R, Pandey A, Zachara NE. Quantitative proteomics reveals that the OGT interactome is remodeled in response to oxidative stress. Mol Cell Proteomics. 2021;20: 100069. 10.1016/j.mcpro.2021.100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Yang Y, Qiu R, Zhang K, Teng X, Liu R, Wang Y. Proteomic analysis of the OGT interactome: novel links to epithelial-mesenchymal transition and metastasis of cervical cancer. Carcinogenesis. 2018;39:1222–34. 10.1093/carcin/bgy097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng RP, He X, Guo SJ, Liu WF, Tao Y, Tao SC. Global identification of O-GlcNAc transferase (OGT) interactors by a human proteome microarray and the construction of an OGT interactome. Proteomics. 2014;14:1020–30. 10.1002/pmic.201300144. [DOI] [PubMed] [Google Scholar]
- 23.Ortiz-Meoz RF, Merbl Y, Kirschner MW, Walker S. Microarray discovery of new OGT substrates: the medulloblastoma oncogene OTX2 is O-GlcNAcylated. J Am Chem Soc. 2014;136:4845–8. 10.1021/ja500451w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephen HM, Praissman JL, Wells L. Generation of an interactome for the tetratricopeptide repeat domain of O-GlcNAc transferase indicates a role for the enzyme in intellectual disability. J Proteome Res. 2021;20:1229–42. 10.1021/acs.jproteome.0c00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tseng TS, Swain SM, Olszewski NE. Ectopic expression of the tetratricopeptide repeat domain of SPINDLY causes defects in gibberellin response. Plant Physiol. 2001;126:1250–8. 10.1104/pp.126.3.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Izhaki A, Swain SM, Tseng TS, Borochov A, Olszewski NE, Weiss D. The role of SPY and its TPR domain in the regulation of gibberellin action throughout the life cycle of Petunia hybrida plants. Plant J. 2001;28:181–90. 10.1046/j.1365-313X.2001.01144.x. [DOI] [PubMed] [Google Scholar]
- 27.Vaidyanathan K, Niranjan T, Selvan N, Teo CF, May M, Patel S, Weatherly B, Skinner C, Opitz J, Carey J, Viskochil D, Gecz J, Shaw M, Peng Y, Alexov E, Wang T, Schwartz C, Wells L. Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J Biol Chem. 2017;292:8948–63. 10.1074/jbc.M116.771030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selvan N, George S, Serajee FJ, Shaw M, Hobson L, Kalscheuer V, Prasad N, Levy SE, Taylor J, Aftimos S, Schwartz CE, Huq AM, Gecz J, Wells L. O-GlcNAc transferase missense mutations linked to X-linked intellectual disability deregulate genes involved in cell fate determination and signaling. J Biol Chem. 2018;293:10810–24. 10.1074/jbc.RA118.002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willems AP, Gundogdu M, Kempers MJE, Giltay JC, Pfundt R, Elferink M, Loza BF, Fuijkschot J, Ferenbach AT, Van Gassen KLI, Van Aalten DMF, Lefeber DJ. Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J Biol Chem. 2017;292. 10.1074/jbc.M117.790097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gundogdu M, Llabrés S, Gorelik A, Ferenbach AT, Zachariae U, van Aalten DMF. The O-GlcNAc transferase intellectual disability mutation L254F distorts the TPR helix. Cell Chem Biol. 2018;1–6. 10.1016/jxhembiol.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu L, Li L, Ma C, Shi Y, Liu C, Xiao Z, Zhang Y, Tian F, Gao Y, Zhang J, Ying W, Wang PG, Zhang L. O-GlcNAcylation of Thr12/Ser56 in short-form O-GlcNAc transferase (sOGT) regulates its substrate selectivity. J Biol Chem. 2019;294:16620–33. 10.1074/jbc.RA119.009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng Z, Hui Y, Ling L, Xiaoyan L, Yuqiu W, Peng W, Lianwen Z. FBXW10 is negatively regulated in transcription and expression level by protein O-GlcNAcylation. Biochem Biophys Res Commun. 2013;438:427–32. 10.1016/j.bbrc.2013.07.091. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Li X, Yu Y, Shi J, Liang Z, Run X, Li Y, Dai C, ling, Grundke-Iqbal I, Iqbal K, Liu F, Gong CX, . Developmental regulation of protein O-GlcNAcylation, O-GlcNAc transferase, and O-GlcNAcase in mammalian brain. PLoS ONE. 2012;7:1–13. 10.1371/journal.pone.0043724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Konzman D, Abramowitz LK, Steenackers A, Mukherjee MM, Na H-J, Hanover JA. O-GlcNAc: Regulator of signaling and epigenetics linked to X-linked intellectual disability. Front Genet. 2020;11. 10.3389/fgene.2020.605263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramirez DH, Aonbangkhen C, Wu HY, Naftaly JA, Tang S, O’Meara TR, Woo CM. Engineering a proximity-directed O-GlcNAc transferase for selective protein O-GlcNAcylation in cells. ACS Chem Biol. 2020;15:1059–66. 10.1021/acschembio.0c00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge Y, Ramirez DH, Yang B, D’Souza AK, Aonbangkhen C, Wong S, Woo CM. Target protein deglycosylation in living cells by a nanobody-fused split O-GlcNAcase. Nat Chem Biol. 2021. 10.1038/s41589-021-00757-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez DH, Ge Y, Woo CM. O-GlcNAc Engineering on a target protein in cells with nanobody-OGT and nanobody-splitOGA. Curr Protoc. 2021;1:1–28. 10.1002/cpz1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darabedian N, Yang B, Ding R, Cutolo G, Zaro BW, Woo CM, Pratt MR. O-Acetylated chemical reporters of glycosylation can display metabolism-dependent background labeling of proteins but are generally reliable tools for the identification of glycoproteins. Front Chem. 2020;8:1–14. 10.3389/fchem.2020.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woo CM, Lund PJ, Huang AC, Davis MM, Bertozzi CR, Pitteri SJ. Mapping and quantification of over 2000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (Isotag). Mol Cell Proteomics. 2018;17:764–75. 10.1074/mcp.RA117.000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Von Mering C. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–13. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.März P, Stetefeld J, Bendfeldt K, Nitsch C, Reinstein J, Shoeman RL, Dimitriades-Schmutz B, Schwager M, Leiser D, Özcan S, Otten U, Özbek S. Ataxin-10 interacts with O-linked β-N-acetylglucosamine transferase in the brain. J Biol Chem. 2006;281:20263–70. 10.1074/jbc.M601563200. [DOI] [PubMed] [Google Scholar]
- 43.Pathak S, Alonso J, Schimpl M, Rafie K, Blair DE, Borodkin VS, Schüttelkopf AW, Albarbarawi O, Van Aalten DMF. The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat Struct Mol Biol. 2015;22:744–9. 10.1038/nsmb.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joiner CM, Levine ZG, Aonbangkhen C, Woo CM, Walker S. Aspartate residues far from the active site drive O-GlcNAc transferase substrate selection. J Am Chem Soc. 2019;141:12974–8. 10.1021/jacs.9b06061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hornbeck PV, Kornhauser JM, Latham V, Murray B, Nandhikonda V, Nord A, Skrzypek E, Wheeler T, Zhang B, Gnad F. 15 years of PhosphoSitePlus ® : integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res. 2019;47:D433–41. 10.1093/nar/gky1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narayanan K, Srinivas R, Peterson MC, Ramachandran A, Hao J, Thimmapaya B, Scherer PE, George A. Transcriptional regulation, of dentin matrix protein 1 by JunB and p300 during osteoblast differentiation. J Biol Chem. 2004;279:44294–302. 10.1074/jbc.M403511200. [DOI] [PubMed] [Google Scholar]
- 47.Pérez-Benavente B, García JL, Rodríguez MS, Pineda-Lucena A, Piechaczyk M, Font De Mora J, Farràs R. GSK3-SCF FBXW7 targets JunB for degradation in G2 to preserve chromatid cohesion before anaphase. Oncogene. 2013;32:2189–99. 10.1038/onc.2012.235. [DOI] [PubMed] [Google Scholar]
- 48.Sacoman JL, Dagda RY, Burnham-Marusich AR, Dagda RK, Berninsone PM. Mitochondrial O-GlcNAc transferase (mOGT) regulates mitochondrial structure, function, and survival in HeLa cells. J Biol Chem. 2017;292:4499–518. 10.1074/jbc.M116.726752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan ZW, Fei G, Paulo JA, Bellaousov S, Martin SES, Duveau DY, Thomas CJ, Gygi SP, Boutz PL, Walker S. O-GlcNAc regulates gene expression by controlling detained intron splicing. Nucleic Acids Res. 2020;48:5656–69. 10.1093/nar/gkaa263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park SK, Zhou X, Pendleton KE, Hunter OV, Kohler JJ, O’Donnell KA, Conrad NK. A conserved splicing silencer dynamically regulates O-GlcNAc transferase intron retention and O-GlcNAc homeostasis. Cell Rep. 2017;20:1088–99. 10.1016/j.celrep.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bi Y, Deng Z, Ni W, Shrestha R, Savage D, Hartwig T, Patil S, Hong SH, Zhang Z, Oses-Prieto JA, Li KH, Quail PH, Burlingame AL, Xu S, Wang Z. Arabidopsis ACINUS is O-glycosylated and regulates transcription and alternative splicing of regulators of reproductive transitions. Nat Commun. 2021;12:945. 10.1038/s41467-021-20929-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Labokha AA, Gradmann S, Frey S, Hülsmann BB, Urlaub H, Baldus M, Görlich D. Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 2013;32:204–18. 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yoo TY, Mitchison TJ. O -GlcNAc modification of nuclear pore complexes accelerates bidirectional transport. J Cell Biol. 2021;220. 10.1083/jcb.202010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Y, Liu TW, Madden Z, Yuzwa SA, Murray K, Cecioni S, Zachara N, Vocadlo DJ. Post-translational O-GlcNAcylation is essential for nuclear pore integrity and maintenance of the pore selectivity filter. J Mol Cell Biol. 2016;8:2–16. 10.1093/jmcb/mjv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruba A, Yang W. O-GlcNAc-ylation in the nuclear pore complex. Cell Mol Bioeng. 2016;9:227–33. 10.1007/s12195-016-0440-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Kohler JJ (2014) Glycosylation of the nuclear pore. Traffic [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolomé S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–6. 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS data were deposited at the ProteomeXchange Consortium [57] via the PRIDE partner repository, and are available with the identifier PXD028141.


