Abstract
Novel therapeutic strategies are needed for paediatric patients affected by Acute Myeloid Leukaemia (AML), particularly for those at high-risk for relapse. MicroRNAs (miRs) have been extensively studied as biomarkers in cancer and haematological disorders, and their expression has been correlated to the presence of recurrent molecular abnormalities, expression of oncogenes, as well as to prognosis/clinical outcome. In the present study, expression signatures of different miRs related both to presence of myeloid/lymphoid or mixed-lineage leukaemia 1 and Fms like tyrosine kinase 3 internal tandem duplications rearrangements and to the clinical outcome of paediatric patients with AML were identified. Notably, miR-221-3p and miR-222-3p resulted as a possible relapse-risk related miR. Thus, miR-221-3p and miR-222-3p expression modulation was investigated by using a Bromodomain-containing protein 4 (BRD4) inhibitor (JQ1) and a natural compound that acts as histone acetyl transferase inhibitor (curcumin), alone or in association, in order to decrease acetylation of histone tails and potentiate the effect of BRD4 inhibition. JQ1 modulates miR-221-3p and miR-222-3p expression in AML with a synergic effect when associated with curcumin. Moreover, changes were observed in the expression of CDKN1B, a known target of miR-221-3p and miR-222-3p, increase in apoptosis and downregulation of miR-221-3p and miR-222-3p expression in CD34+ AML primary cells. Altogether, these findings suggested that several miRs expression signatures at diagnosis may be used for risk stratification and as relapse prediction biomarkers in paediatric AML outlining that epigenetic drugs, could represent a novel therapeutic strategy for high-risk paediatric patients with AML. For these epigenetic drugs, additional research for enhancing activity, bioavailability and safety is needed.
Keywords: acute myelogenous leukaemia, microRNAs, epigenetic drugs, biomarkers, children
Introduction
Acute Myeloid Leukaemia (AML) constitutes 20% of all paediatric leukaemia and is responsible for substantial mortality. Despite progress made over the past years in diagnosis, risk stratification and treatment of AML, survival remains suboptimal with a success rate of 60–70% (1–3), with relapse being the leading cause of death. Risk stratification in patients with AML is of paramount importance in order to deliver tailored therapy, enabling treatment intensification in high-risk patients (for example, allogenic stem cell transplantation in high-risk in first complete remission). Moreover, novel therapies are needed in order to further improve prognosis in these patients. Among prognostic factors, cytogenetic and molecular abnormalities, together with Minimal Residual Disease and treatment response, play a pivotal role in defining AML prognosis and treatment (4,5). Rearrangements involving Histone-lysine N-methyltransferase 2A (KMT2A) gene, formerly known as myeloid/lymphoid or mixed-lineage leukaemia 1 (MLL1) gene, as well as Fms like tyrosine kinase 3 (FLT3) internal tandem duplications (FLT3-ITD) represent useful prognostic factors and, possibly, therapeutic targets. Indeed, they still define an intermediate to high risk AML (1–3,6). In particular, FLT3-ITD mutations are associated with higher risk of relapse and dismal prognosis (1,3,7), whereas MLL-rearranged AML is an heterogeneous group of diseases with more than 100 rearrangements being described and with different outcome largely dependent on the fusion partner (8).
FLT3-ITD and MLL rearrangements have been functionally linked to dysregulation of expression of microRNAs (miRNAs or miRs) (9). miRs are small non-coding RNA molecules (~18-22 nucleotides long), involved in several cellular processes (10,11). In cancer, they are implicated both in promoting carcinogenesis (oncomiRs) and in suppressing tumour transformation (12). Their role in AML have been extensively investigated over the past years reviling promising data on diagnosis, prognostic stratification and, possibly, treatment in AML patients (13–15). Despite extensive research performed to understand the role of miRs in AML, the majority of studies are focused on adult patients, while a precise characterization of miRNAs expression in paediatric AML is less documented. Moreover, data on the role of different miRs are conflicting because of the variability of genetic abnormalities found in AML (16). In the present study, it was aimed to identify AML specific miR signatures in a cohort of patients harbouring molecular lesions (FLT3-ITD and MLL rearrangement), studying the expression of distinct miR sets in relation to relapse risk.
Epigenetic networks, including histone modification mechanisms, are involved in the regulation of both miRs expression and function. An increasing interest in the field of cancer therapeutic drugs is focused on small molecular compounds targeting epigenetic regulation (17).
Bromodomain and extra-terminal domain family of proteins (BET) and histone acetyl transferase (HAT) inhibitors proteins are the best characterized ones. BET are effective in preventing Bromodomain Containing protein 4 (BRD4) associated transcription of several oncogenes, reducing proliferation and increasing apoptosis in AML (18–22). BRD4 is a member of BET family proteins, characterized by the presence of functional structures called bromodomains which bind specific acetylated residues on histone tails to modulate transcription of target gene (23–25), enhancing transcription of several oncogenes (26).
Among BRD4 inhibitors, JQ1 was used as its activity on modulation of miRs was previously described (27). JQ1 [(S)-tert-butyl-2-(4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)acetate] is a small molecule belonging to the thienotriazolodiazepine group and it prevents the binding of BRD4 to acetylated residues on histone H3 tails, particularly H3AcK14 (20,28–30).
Furthermore, among HAT inhibitors, curcumin, a natural compound extracted from the root of Curcuma Longa has been shown to inhibit acetylation of histone tails, blocking the activity of the HAT p300 even causing its proteasomal degradation (31). This results in a global decrease of acetylation on histone tails and a consequent modulation of gene transcription (32,33).
BRD4 inhibitors have exhibited only moderate results in clinics and novel ways to increase their antitumour activity are needed (34). It was therefore hypothesized that the association with curcumin would increase JQ1 efficacy. The BET family are ‘readers’ of chromatin acetylation whereas HAT could be classified as a ‘writer’ of histone acetylation (34) thus offering a theoretical basis for JQ1 and curcumin synergic activity.
Moreover, it was previously showed that also JQ1, like curcumin, blocks p300-mediated acetylation (25,35). Thus, it was investigated in vitro whether a combination of BRD4 and HAT inhibitors have an effect in terms of modulation of miRs and antitumour effects on different AML cell-lines harbouring mutations resembling those present in our patients.
Materials and methods
Samples of patients
A total of 23 patients aged 1 to 18 years, who received a diagnosis of AML harbouring FLT3-ITD or MLL rearrangement (Table SI). Although not mutually exclusive, these rearrangements are not frequently found together. In the present study, none of the patients had both the rearrangements.
Bone marrow (BM) samples were collected from January 1st 2010 to December 31st 2016 at Bambino Gesù Children's Hospital in Rome and at Department of Paediatrics, University in Padua, at diagnosis and at disease recurrence from the 13 patients who underwent relapse (REL-D and REL-R groups, respectively) and at diagnosis from the 10 patients who did not display relapse (NREL group).
A total of 8 frozen age-matched BM samples from healthy children (HD) (unused aliquots from healthy BM donors) were retrieved from the tissue bank at Bambino Gesù Children's Hospital as a control population. Informed consent was obtained from either parents or legal guardians according to the Declaration of Helsinki (2008). The present study was approved by the Institutional Review Boards of Bambino Gesù Children's Hospital (Rome, Italy).
CD34+ cells isolation
Mononuclear cells were isolated by density gradient centrifugation at 400 g and 20°C for 30 min, diluted in 90% fetal bovine serum (FBS) plus 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen. CD34+ cells from BM samples of three patients randomly selected in our cohort, were magnetically separated using MACS CD34+ microbead kit (Miltenyi Biotech GmbH). In particular, the molecular analysis of these patients revealed a FLT3-ITD with normal karyotype and two MLL rearrangements [t(9;11) and t(10;11)]. The identity of CD34 cells was validated by flow cytometry using FACSCantoII equipped with FACSDiva 6.1 CellQuest software (Becton, Dickinson and Company) using 20 µl of CD34 PerCP antibody (cat. no 340666; BD Biosciences) with an incubation of 30 min at 4°C.
RNA extraction and miR profiling
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and purified using RNA Cleanup and Concentration kit according to the manufacturer's protocol (Norgen Biotek Corp.). RNA quantification was performed using Nanodrop 2000 at 260 nm wavelength (Thermo Fisher Scientific, Inc.) and RNA integrity and purity was assessed with RNA Bioanalyzer kit according to the manufacturer's protocol (Agilent Technologies, Inc.). miR expression profile was performed using the nCounter Human v2 miRNA Expression Assay and nCounter Nanostring platform according to manufacturer's protocol (NanoString Technologies).
Cell culture methods
AML cell lines THP-1 (MLL-MLLT3; MLL-AF9), MOLM-13 (MLL-MLLT3; MLL-AF9; FLT3-ITD) and MV-4-11 (MLL-AFF1; MLL-AF4; FLT3-ITD) were obtained from DSMZ and cultured at 37°C using RPMI-1640 medium (Euroclone SpA) supplemented with 10% FBS (Thermo Fisher Scientific, Inc.) and 1% Penicillin-Streptomycin (Thermo Fisher Scientific, Inc.). Mycoplasma testing was performed for the cell lines used. Cell lines were treated with 250 nM JQ1 and 10 µM curcumin singularly alone or in association, for 48 h. JQ1 and Curcumin were obtained from Selleck Chemicals and resuspended in DMSO, following the manufacturer's protocol.
Western blot analysis
Whole-cell lysates were prepared with RIPA lysis buffer (Thermo Fisher Scientific, Inc.) supplemented with protease and phosphatase inhibitors (Thermo Fisher Scientific, Inc.). Cells were lysed by sonication, incubated for 30 min at 4°C and then obtained cells lysates were centrifuged at 13,000 × g for 20 min at 4°C. The protein concentration of the resulting supernatant was estimated by BCA assay. Then, 40 µg of sample was separated on Criterion TGX Precast Gels 4–20% (BioRad Laboratories, Inc.) and transferred to Hybond ECL nitrocellulose membranes (Amersham; Cytiva). Membranes were blocked at room temperature for 1 h in 5% non-fat milk in Tris buffered saline and 0,05% Tween-20 (TBS-T). Membranes were incubated at 4°C overnight with rabbit polyclonal anti-human CDKN1B (1:500; cat. no. sc-528; Santa Cruz Biotechnology, Inc.) and 1 h at room temperature with rabbit monoclonal anti-human GAPDH (1:1,000; cat. no. D16H11; Cell Signaling Technology, Inc.) primary antibodies. After incubation they were washed three times in TBS-T, then incubated with HRP-labelled goat anti-rabbit (1:5,000; cat. no. sc-2004) and goat anti-mouse (1:5,000; cat. no. sc-2005; both from Santa Cruz Biotechnology, Inc.) IgG secondary antibodies, respectively at room temperature for 1 h. Subsequently, they were washed an additional three times with TBS-T and then developed with ECL reagent (Western Lightning Plus; PerkinElmer, Inc.).
miR quantitative (q)PCR
Expression levels of hsa-miR-221-5p, hsa-miR-222-5p and U6 were measured using TaqMan microRNA assays (cat. nos. 000524, 002276 and 001973; Thermo Fisher Scientific, Inc.). Reverse transcription (RT) primer, preformulated forward/reverse primer and MGB probes for each assay were provided by the manufacturer. The TaqMan MiR Reverse Transcription kit was used for cDNA synthesis from 10 ng total RNA template according to the manufacturer's protocol. QuantStudio 12K Flex Real Time PCR System (Thermo Fisher Scientific, Inc.) was used for qPCR reactions with the following conditions: Enzyme activation 95°C for 20 sec and 40 cycles of denaturation (95°C for 1 sec) and annealing/extension (60°C for 20 sec) steps. miRNA expression data were normalized to U6 using the 2−ΔΔCq (36) method by the Relative Quantification module of Thermo Fisher Cloud Data Analysis Apps. At least two independent amplifications were performed for each probe on triplicate samples.
Apoptosis assay
Following treatment with 250 nM BRD4 and 10 µM curcumin, cells were washed twice with ice cold PBS and stained for 15 min at room temperature in calcium-binding buffer with PE-conjugated Annexin V (AnnV) and 7-Aminoactinomycin D (7-AAD) using the AnnV apoptosis detection kit (BD Pharmingen; BD Biosciences) according to the manufacturer's recommendations. Samples were analysed within 1 h by a fluorescence-activated cell sorting using a FACSCantoII equipped with FACSDiva 6.1 CellQuest software (Becton, Dickinson and Company).
Bioinformatics and statistical analyses
MicroRNA profiling normalization was performed using the nSolver Analysis Software (NanoString Technologies) as recommended by NanoString. P-values were calculated using the LIMMA (v.3.46.0) package (37) from the Bioconductor R (v.4.0.5) project. The P-values were adjusted for multiple testing using the Benjamini and Hochberg method to control the False Discovery Rate. An independent normalization phase for each comparison was performed, considering only the samples present in such comparison (for example, NREL vs. HD). Then, the miRNA expression of the HD group was specifically normalized in the comparisons in which the HD group was taken into consideration. Validated targets of miRs were reported in Table I according to miRWalk 2.0 online software analysis (http://mirwalk.umm.uni-heidelberg.de/). One-way ANOVA and post hoc comparison using Tukey's HSD Post Hoc or Dunnett's test were performed using SPSS software v19 (IBM Corp.) and GraphPad Prism v6 (GraphPad Software, Inc.). The heatmap was generated by using GenePattern tool (38), with Euclidean and Spearman correlation distances in columns and rows, respectively. Venn diagrams were created using web tool (39).
Table I.
Molecular based miR expression signature.
| FLT3-ITH vs. HD | Accession number | FC | Adjusted P-value |
|---|---|---|---|
| hsa-miR-10a-5p | MIMAT0000253 | 24.38 | 2×10−5 |
| hsa-miR-451a | MIMAT0001631 | 17.06 | 1×10−3 |
| hsa-miR-196b-5p | MIMAT0001080 | 4.31 | 5×10−4 |
| hsa-miR-34a-5p | MIMAT0000255 | 3.38 | 4×10−3 |
| hsa-let-7b-5p | MIMAT0000063 | 2.49 | 1×10−2 |
| hsa-miR-29c-3p | MIMAT0000681 | 2.12 | 1×10−2 |
| hsa-miR-520f-3p | MIMAT0002830 | −2.11 | 4×10−2 |
| hsa-miR-200c-3p | MIMAT0000617 | −2.16 | 4×10−2 |
| hsa-miR-421 | MIMAT0003339 | −2.34 | 2×10−2 |
| hsa-miR-6724-5p | MIMAT0025856 | −2.39 | 1×10−2 |
| hsa-miR-151a-3p | MIMAT0000757 | −2.45 | 1×10−2 |
| hsa-miR-4755-5p | MIMAT0019895 | −2.45 | 3×10−2 |
| hsa-miR-365a-3p + | MIMAT0000710 | −2.52 | 2×10−2 |
| hsa-miR-365b-3p | |||
| hsa-miR-520d-5p + | MIMAT0002855 | −2.73 | 4×10−3 |
| hsa-miR-527 + hsa-miR-518a-5p | |||
| hsa-miR-574-5p | MIMAT0004795 | −2.75 | 1×10−2 |
| hsa-miR-342-3p | MIMAT0000753 | −3.99 | 4×10−3 |
|
| |||
| FLT3-ITD vs. MLL | Accession number | FC | Adjusted P-value |
|
| |||
| hsa-miR-10a-5p | MIMAT0000253 | 34.87 | 1.44×10−7 |
| hsa-miR-99a-5p | MIMAT0000097 | 2.87 | 2.24×10−2 |
| hsa-miR-9-5p | MIMAT0000441 | −7.89 | 1.38×10−3 |
|
| |||
| MLL vs. HD | Accession number | FC | Adjusted P-value |
|
| |||
| hsa-miR-9-5p | MIMAT0000441 | 10.75 | 9.33×10−3 |
| hsa-miR-34a-5p | MIMAT0000255 | 5.50 | 8.78×10−5 |
| hsa-miR-196b-5p | MIMAT0001080 | 4.23 | 1.55×10−2 |
| hsa-miR-192-5p | MIMAT0000222 | −2.73 | 3.92×10−2 |
hsa, homo sapiens; miR, microRNA; FLT3-ITD; Fms like tyrosine kinase 3 (FLT3) internal tandem duplications; HD, healthy donors.
CD34+ cell culture
CD34+ cells were cultured using MethoCult H4434 methylcellulose medium (Stem Cell Technologies) supplemented with 250 nM JQ1 and 10 µM curcumin.
Results
NanoString miR profiling reveals different expression signatures based on patient clinical and molecular features
A miR profiling analysis was first performed to verify whether the two distinct molecular subsets of the cohort of our patients (MLL rearranged and FLT3-ITD) showed different miR expression fingerprints. Comparing both MLL rearranged and FLT3-ITD sets with healthy donors (HDs), 4 and 16 significantly deregulated miRs were identified, respectively (Table I). miR-196b-5p and miR-34a-5p resulted upregulated in both AML sets. The comparison between the two molecular AML sets showed 3 differentially regulated miRs with miR-10a-5p and miR-99a-5p significantly higher in FLT3-ITD and miR-9a-5p with enhanced expression in MLL-rearranged sets (Table I). Other miRs such as miR-451a, miR-520d-5p/527/518a-5p, miR-574-5p and miR-192-5p were uniquely dysregulated in FLT3-ITD or MLL sets with respect to HDs (Table I). A miR expression profiling analysis was then performed based onto clinical outcomes of patients to identify those miRs associated with relapse.
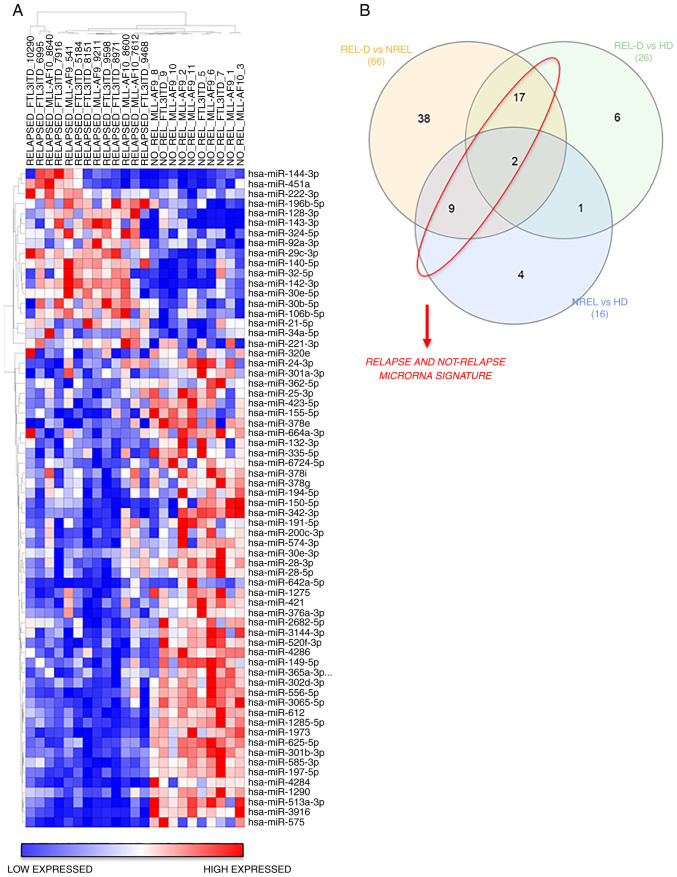
The hierarchical clustering results of miRNAs expression of REL-D vs. NREL groups is revealed in Fig. 1. A total of 18 miRs were broadly upregulated in the REL-D patients set with respect to the NREL set and 48 miRs displayed the opposite trend (Fig. 1A and Table II). To further identify and refine a signature that was associated with relapse, among these differentially expressed 66 miRs, only those shared in the REL-D vs. HD and NREL vs. HD comparison were subsequently considered (Fig. 1B and Table II). The resulted signature associated to relapse and not-relapse is listed in Table III. Validated targets of miRs were identified using miRWalk 2.0 online software (40) and are reported in Table III. CDKN1B, a key regulator of cell cycle which has been previously reported to be associated with prognosis in AML, resulted as primary target of miR-221-3p and miR-222-3p (41). To evaluate if the signature of miRs associated with relapse at diagnosis was maintained over time (if it is present also in the REL-R group), expression of miRs at diagnosis and at disease recurrence was compared between those patients who relapsed (REL-R vs. REL-D) and no significant differences were detected (Table II). Since our goal was to identify miRs associated to relapse with a significant prognostic value, the expression level of both miRs resulted overexpressed in REL-D vs. HD group and REL-D vs. NREL was first evaluated in the AML cells lines by qPCR (data not shown). High variability was obtained in the different cell lines. This result prompted the authors to focus only on hsa-miR-221-3p and hsa-miR-222-3p, presenting the same trend of expression in all the cell lines, as well as in vivo in the patients.
Figure 1.
NanoString Analysis reveals shared and non-shared miR expression signatures in pediatric patients with AML. (A) Supervised hierarchical clustering of REL-D vs. NREL analysis showed miR expression signature for relapse risk correlation at the time of diagnosis. Only miR with P≤0.05 were plotted. Benjamini-Hochberg multiple comparisons statistical method was used to control False Discovery Rate. (B) Venn diagrams reporting shared and non-shared dysregulated miRs between REL-D vs. NREL, REL-D vs. HD and NREL vs. HD analysis.
Table II.
Clinical outcome-based miR expression signature.
| REL-D vs. NREL | Accession number | FC | Adjusted P-value |
|---|---|---|---|
| hsa-miR-128-3p | MIMAT0000424 | 8,10 | 4×10−6 |
| hsa-miR-143-3p | MIMAT0000435 | 7,47 | 9×10−4 |
| hsa-miR-142-3p | MIMAT0000434 | 5,63 | 9×10−7 |
| hsa-miR-144-3p | MIMAT0000436 | 3,87 | 4×10−2 |
| hsa-miR-451a | MIMAT0001631 | 3,66 | 3×10−2 |
| hsa-miR-29c-3p | MIMAT0000681 | 3,24 | 4×10−6 |
| hsa-miR-140-5p | MIMAT0000431 | 2,53 | 2×10−5 |
| hsa-miR-196b-5p | MIMAT0001080 | 2,48 | 7×10−4 |
| hsa-miR-32-5p | MIMAT0000090 | 2,44 | 6×10−3 |
| hsa-miR-221-3p | MIMAT0000278 | 2,04 | 1×10−2 |
| hsa-miR-92a-3p | MIMAT0000092 | 1,93 | 4×10−3 |
| hsa-miR-222-3p | MIMAT0000279 | 1,92 | 5×10−3 |
| hsa-miR-21-5p | MIMAT0000076 | 1,88 | 1×10−2 |
| hsa-miR-324-5p | MIMAT0000761 | 1,84 | 5×10−3 |
| hsa-miR-34a-5p | MIMAT0000255 | 1,77 | 2×10−2 |
| hsa-miR-30b-5p | MIMAT0000420 | 1,67 | 6×10−3 |
| hsa-miR-30e-5p | MIMAT0000692 | 1,67 | 5×10−3 |
| hsa-miR-106b-5p | MIMAT0000680 | 1,55 | 2×10−2 |
| hsa-miR-30e-3p | MIMAT0000693 | −1,41 | 3×10−2 |
| hsa-miR-378i | MIMAT0019074 | −1,44 | 4×10−2 |
| hsa-miR-664a-3p | MIMAT0005949 | −1,46 | 2×10−2 |
| hsa-miR-24-3p | MIMAT0000080 | −1,49 | 4×10−2 |
| hsa-miR-25-3p | MIMAT0000081 | −1,54 | 3×10−2 |
| hsa-miR-2682-5p | MIMAT0013517 | −1,67 | 6×10−2 |
| hsa-miR-200c-3p | MIMAT0000617 | −1,67 | 4×10−2 |
| hsa-miR-320e | MIMAT0015072 | −1,67 | 5×10−2 |
| hsa-miR-191-5p | MIMAT0000440 | −1,68 | 3×10−2 |
| hsa-miR-423-5p | MIMAT0004748 | −1,69 | 3×10−3 |
| hsa-miR-362-5p | MIMAT0000705 | −1,71 | 2×10−2 |
| hsa-miR-149-5p | MIMAT0000450 | −1,72 | 2×10−3 |
| hsa-miR-194-5p | MIMAT0000460 | −1,75 | 1×10−2 |
| hsa-miR-6724-5p | MIMAT0025856 | −1,80 | 5×10−3 |
| hsa-miR-301a-3p | MIMAT0000688 | −1,84 | 3×10−2 |
| hsa-miR-378g | MIMAT0018937 | −1,88 | 5×10−3 |
| hsa-miR-132-3p | MIMAT0000426 | −1,89 | 9×10−3 |
| hsa-miR-28-3p | MIMAT0004502 | −1,97 | 6×10−4 |
| hsa-miR-28-5p | MIMAT0000085 | −2,01 | 9×10−4 |
| hsa-miR-574-3p | MIMAT0003239 | −2,09 | 4×10−3 |
| hsa-miR-520f-3p | MIMAT0002830 | −2,10 | 5×10−5 |
| hsa-miR-3144-3p | MIMAT0015015 | −2,11 | 4×10−4 |
| hsa-miR-1290 | MIMAT0005880 | −2,22 | 5×10−5 |
| hsa-miR-421 | MIMAT0003339 | −2,24 | 6×10−4 |
| hsa-miR-1275 | MIMAT0005929 | −2,32 | 2×10−4 |
| hsa-miR-335-5p | MIMAT0000765 | −2,39 | 5×10−4 |
| hsa-miR-365a/b-3p | MIMAT0000710 | −2,42 | 2×10−4 |
| hsa-miR-625-5p | MIMAT0003294 | −2,67 | 9×10−7 |
| hsa-miR-612 | MIMAT0003280 | −2,70 | 2×10−6 |
| hsa-miR-302d-3p | MIMAT0000718 | −2,79 | 6×10−6 |
| hsa-miR-150-5p | MIMAT0000451 | −2,84 | 1×10−2 |
| hsa-miR-301b-3p | MIMAT0004958 | −2,90 | 8×10−7 |
| hsa-miR-155-5p | MIMAT0000646 | −2,95 | 3×10−3 |
| hsa-miR-556-5p | MIMAT0003220 | −3,04 | 8×10−7 |
| hsa-miR-3065-5p | MIMAT0015066 | −3,07 | 3×10−7 |
| hsa-miR-376a-3p | MIMAT0000729 | −3,15 | 6×10−4 |
| hsa-miR-342-3p | MIMAT0000753 | −3,16 | 5×10−4 |
| hsa-miR-197-5p | MIMAT0022691 | −3,30 | 1×10−7 |
| hsa-miR-4286 | MIMAT0016916 | −3,30 | 3×10−5 |
| hsa-miR-585-3p | MIMAT0003250 | −3,37 | 8×10−7 |
| hsa-miR-642a-5p | MIMAT0003312 | −3,41 | 5×10−3 |
| hsa-miR-513a-3p | MIMAT0004777 | −4,07 | 2×10−7 |
| hsa-miR-1285-5p | MIMAT0022719 | −4,10 | 1×10−7 |
| hsa-miR-4284 | MIMAT0016915 | −4,82 | 8×10−7 |
| hsa-miR-575 | MIMAT0003240 | −5,55 | 2×10−6 |
| hsa-miR-1973 | MIMAT0009448 | −6,57 | 4×10−8 |
| hsa-miR-3916 | MIMAT0018190 | −6,94 | 9×10−9 |
| hsa-miR-378e | MIMAT0018927 | −9,46 | 5×10−3 |
|
| |||
| REL-D vs. HD | Accession number | FC | Adjusted P-value |
|
| |||
| hsa-miR-451a | MIMAT0001631 | 10,46 | 1×10−2 |
| hsa-miR-196b-5p | MIMAT0001080 | 5,70 | 3×10−5 |
| hsa-miR-34a-5p | MIMAT0000255 | 5,29 | 3×10−4 |
| hsa-miR-221-3p | MIMAT0000278 | 2,14 | 2×10−2 |
| hsa-miR-222-3p | MIMAT0000279 | 2,09 | 2×10−2 |
| hsa-miR-29c-3p | MIMAT0000681 | 1,85 | 5×10−2 |
| hsa-miR-941 | MIMAT0004984 | −1,87 | 5×10−2 |
| hsa-miR-194-5p | MIMAT0000460 | −1,94 | 5×10−2 |
| hsa-miR-200c-3p | MIMAT0000617 | −1,96 | 5×10−2 |
| hsa-miR-192-5p | MIMAT0000222 | −2,01 | 5×10−2 |
| hsa-miR-1290 | MIMAT0005880 | −2,02 | 5×10−2 |
| hsa-miR-612 | MIMAT0003280 | −2,08 | 5×10−2 |
| hsa-miR-575 | MIMAT0003240 | −2,11 | 4×10−2 |
| hsa-miR-151a-3p | MIMAT0000757 | −2,11 | 4×10−2 |
| hsa-miR-520f-3p | MIMAT0002830 | −2,11 | 2×10−2 |
| hsa-miR-421 | MIMAT0003339 | −2,16 | 3×10−2 |
| hsa-miR-6724-5p | MIMAT0025856 | −2,26 | 1×10−2 |
| hsa-miR-4755-5p | MIMAT0019895 | −2,30 | 3×10−2 |
| hsa-miR-574-5p | MIMAT0004795 | −2,32 | 3×10−2 |
| hsa-miR-363-3p | MIMAT0000707 | −2,38 | 3×10−2 |
| hsa-miR-1285-5p | MIMAT0022719 | −2,39 | 3×10−2 |
| hsa-miR-4286 | MIMAT0016916 | −2,61 | 2×10−2 |
| hsa-miR-365a/b-3p | MIMAT0000710 | −3,00 | 3×10−3 |
| hsa-miR-520d-5p/527/518a-5p | MIMAT0002855 | −3,16 | 7×10−4 |
| hsa-miR-342-3p | MIMAT0000753 | −3,30 | 2×10−2 |
| hsa-miR-150-5p | MIMAT0000451 | −3,96 | 4×10−2 |
|
| |||
| NREL vs. HD | Accession number | FC | Adjusted P-value |
|
| |||
| hsa-miR-9-5p | MIMAT0000441 | 6,90 | 8×10−3 |
| hsa-miR-1973 | MIMAT0009448 | 3,10 | 5×10−3 |
| hsa-miR-4284 | MIMAT0016915 | 2,82 | 3×10−2 |
| hsa-miR-575 | MIMAT0003240 | 2,72 | 5×10−2 |
| hsa-miR-155-5p | MIMAT0000646 | 2,55 | 3×10−2 |
| hsa-miR-196b-5p | MIMAT0001080 | 2,37 | 5×10−2 |
| hsa-miR-191-5p | MIMAT0000440 | 2,27 | 2×10−2 |
| hsa-miR-324-5p | MIMAT0000761 | −2,50 | 8×10−3 |
| hsa-miR-140-5p | MIMAT0000431 | −2,53 | 1×10−2 |
| hsa-miR-450a-5p | MIMAT0001545 | −2,72 | 5×10−2 |
| hsa-miR-142-3p | MIMAT0000434 | −3,35 | 6×10−3 |
| hsa-miR-192-5p | MIMAT0000222 | −4,39 | 6×10−4 |
| hsa-miR-128-3p | MIMAT0000424 | −11,68 | 6×10−4 |
| hsa-miR-422a | MIMAT0001339 | −12,62 | 2×10−3 |
| hsa-miR-143-3p | MIMAT0000435 | −20,19 | 6×10−4 |
| hsa-miR-579-3p | MIMAT0003244 | −20,20 | 8×10−3 |
|
| |||
| REL-R vs. HD | Accession number | FC | Adjusted P-value |
|
| |||
| hsa-miR-451a | MIMAT0001631 | 18,17 | 1×10−3 |
| hsa-miR-144-3p | MIMAT0000436 | 7,97 | 5×10−3 |
| hsa-miR-34a-5p | MIMAT0000255 | 4,16 | 5×10−3 |
| hsa-miR-196b-5p | MIMAT0001080 | 4,22 | 1×10−2 |
| hsa-miR-520d-5p/527/518a-5p | MIMAT0002855 | −2,45 | 3×10−2 |
| hsa-miR-574-5p | MIMAT0004795 | −2,74 | 3×10−2 |
| hsa-miR-342-3p | MIMAT0000753 | −3,27 | 3×10−2 |
| hsa-miR-6724-5p | MIMAT0025856 | −2,50 | 4×10−2 |
|
| |||
| REL-R vs. REL-D | Accession number | FC | Adjusted P-value |
|
| |||
| hsa-miR-579-3p | MIMAT0003244 | 3,52 | 1 |
| hsa-miR-143-3p | MIMAT0000435 | 2,90 | 1 |
| hsa-miR-450a-5p | MIMAT0001545 | 2,30 | 1 |
| hsa-miR-378e | MIMAT0018927 | 2,27 | 1 |
| hsa-miR-145-5p | MIMAT0000437 | 2,19 | 1 |
| hsa-miR-4516 | MIMAT0019053 | 1,61 | 1 |
| hsa-miR-363-3p | MIMAT0000707 | 1,57 | 1 |
| hsa-miR-365a/b-3p | MIMAT0000710 | 1,46 | 1 |
| hsa-miR-664a-3p | MIMAT0005949 | −1,35 | 1 |
| hsa-let-7a-5p | MIMAT0000062 | −1,38 | 1 |
| hsa-miR-222-3p | MIMAT0000279 | −1,48 | 1 |
| hsa-let-7c-5p | MIMAT0000064 | −1,59 | 1 |
| hsa-miR-181a-5p | MIMAT0000256 | −1,60 | 1 |
| hsa-let-7b-5p | MIMAT0000063 | −1,84 | 1 |
| hsa-miR-100-5p | MIMAT0000098 | −1,86 | 1 |
| hsa-miR-1246 | MIMAT0005898 | −2,09 | 1 |
| hsa-miR-9-5p | MIMAT0000441 | −2,13 | 1 |
| hsa-miR-125b-5p | MIMAT0000423 | −2,20 | 1 |
| hsa-miR-10a-5p | MIMAT0000253 | −2,53 | 1 |
hsa, homo sapiens; miR, microRNA; REL-D, patient at the time of diagnosis; REL-R, relative samples at relapse; NREL, patients at diagnosis who never presented relapse; HD, healthy donors.
Table III.
miR expression signatures related to relapse risk or not-relapse with validated targets.
| RELAPSE RISK | Validated target genes | NOT RELAPSE | Validated target genes |
|---|---|---|---|
| hsa-miR-128-3p | hsa-miR-200c-3p | ZEB1; ERRFI1 | |
| hsa-miR-143-3p | KRAS; FNDC3B; DNMT3A; MAPK7 | hsa-miR-191-5p | |
| hsa-miR-142-3p | hsa-miR-194-5p | ||
| hsa-miR-451a | MIF; ABCB1 | hsa-miR-6724-5p | |
| hsa-miR-29c-3p | hsa-miR-520f-3p | ||
| hsa-miR-140-5p | HDAC4; IGFBP5; VEGFA | hsa-miR-1290 | |
| hsa-miR-196b-5p | hsa-miR-421 | ||
| hsa-miR-221-3p | BMF; FOS; CDKN1B; KIT; ESR1; | hsa-miR-365a/b-3p | |
| CDKN1C; ICAM1; DDIT4 | |||
| hsa-miR-222-3p | CDKN1B; FOS; KIT; CDKN1C; | hsa-miR-612 | |
| ESR1; MMP1; SOD2; PPP2R2A | |||
| hsa-miR-324-5p | SMO; GLI1 | hsa-miR-150-5p | |
| hsa-miR-34a-5p | MAP2K1; E2F3; SIRT1; MYB; CDK6; | hsa-miR-155-5p | JARID2; IKBKE; ETS1; |
| DLL1; CCND1; NOTCH1; NOTCH2; | BACH1; TAB2; MEIS1; | ||
| JAG1; MET; BCL2; MYCN; WNT1; | MECP2; CEBPB; FOXO3; | ||
| AXIN2; VEGFA; MYC | FGF7; SOCS1; INPP5D; | ||
| AGTR1; SPI1; CYR61; | |||
| SMAD2; LDOC1; MATR3; | |||
| TM6SF1; RHOA | |||
| hsa-miR-342-3p | |||
| hsa-miR-4286 | |||
| hsa-miR-1285-5p | |||
| hsa-miR-4284 | |||
| hsa-miR-575 | |||
| hsa-miR-1973 |
hsa, homo sapiens; miR, microRNA.
Association of JQ1 and curcumin leads to miR-221-3p and miR-222-3p downregulation and CDKN1B upregulation in AML cell lines
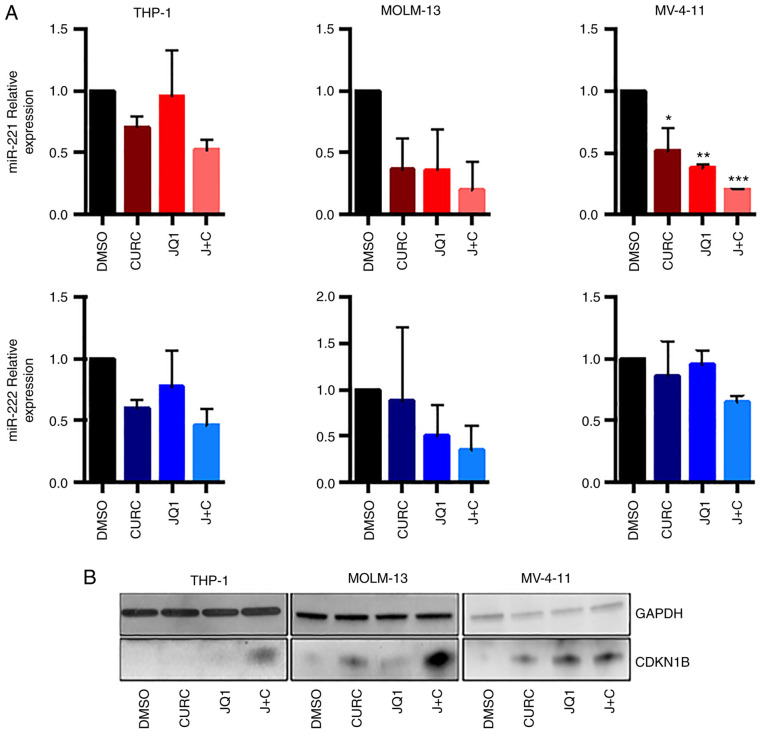
It was first analysed whether the combination of JQ1 and curcumin could modulate miR-221-3p and miR-222-3p expression in AML cell lines.
RT-qPCR experiments (Fig. 2A), revealed a trend towards downregulation of the two miRs in all the cell lines, featuring an enhanced effect with the drug combination. The expression of miR-221-3p expression showed a clear trend towards downregulation, reaching statistical significance in MV-4-11 cells when treated with JQ1, curcumin or the combination of both (Fig. 2A, upper panel). miR-222-3p demonstrated a trend towards downregulation with coupled treatment in all cell-lines, but it did not reach statistical significance (Fig. 2A, lower panel).
Figure 2.
Curcumin and JQ1 singular and coupled treatments impair protein and miR-221-3p and miR-222-3p expression in leukemia cell lines. (A) Reverse transcription-quantitative PCR showed a significant decrease of miR-221-3p in MV-4-11 cells after 48 h of coupled treatment (ANOVA followed by Tukey's post hoc test), while miR-222-3p was not significantly downregulated. (B) Western blot analysis revealing the expression of CDKN1B at protein level after single and coupled treatment with curcumin and JQ1 in protein lysates of AML cell lines. CDKN1B displayed a significant increase in its expression while treated with the combination drugs and mildly with singular drugs. All the western blots were performed after 48 h from treatment with 250 nM of JQ1 and 10 µM curcumin. *P≤0.05, **P≤0.01 and ***P≤0.005.
It was then analysed whether the combination of JQ1 and curcumin could modulate the CDKN1B protein level, that it is a miR-221-3p and miR-222-3p target, as before mentioned. As revealed in Fig. 2B, the treatment determined an increase in CDKN1B expression both in THP-1 and in MOLM-13 cells while in MV-411 cells the effect of drug combination on the upregulation of CDKN1B expression was comparable to that obtained by single treatment with JQ1.
Association of curcumin and JQ1 causes apoptosis
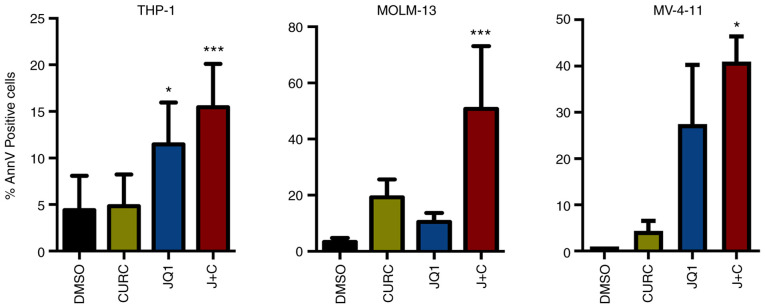
It was assessed whether these treatments could drive the cell lines toward an apoptotic response. A significant increase in AnnV positive cell percentage was identified in all the cell lines while treated with the drug combination compared with DMSO (Figs. S1 and 3). At a deeper glance, THP-1 and MV-4-11 cells, showing a minor increase in CDKN1B expression, displayed a milder apoptotic response compared with MOLM-13, characterized instead by higher changes in term of expression of CDKN1B (Figs. 2 and 3).
Figure 3.
Curcumin and JQ1 treatments lead leukemia cell lines to apoptosis. Annexin V positive cells assessed by flow cytometry were significantly increased in all the cell lines after 48 h coupled treatment with 10 µM curcumin and 250 nM JQ1 (ANOVA followed by Tukey's post hoc test). *P≤0.05 and ***P≤0.005.
CD34+ primary cells from AML patients exhibit miR-221-3p and miR-222-3p downregulation after single and double treatments
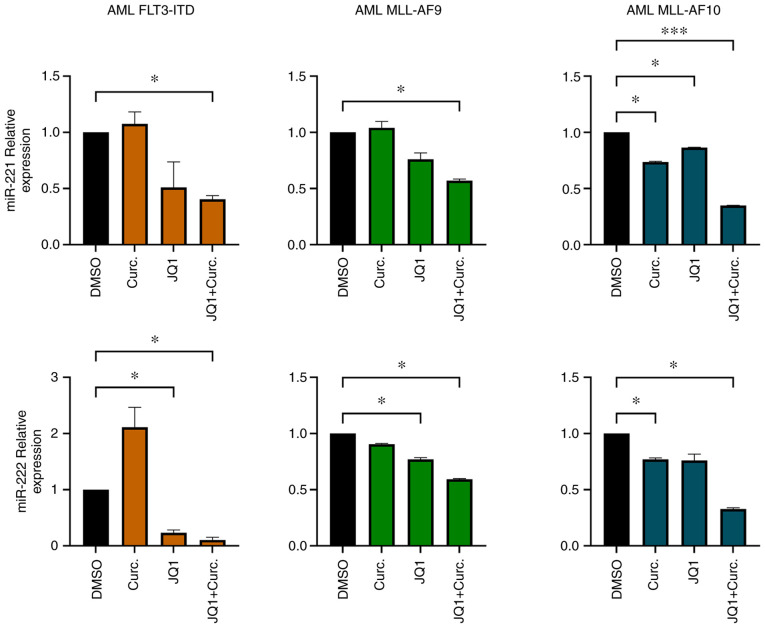
miR-221-3p and miR-222-3p expression was analysed in CD34+ cells isolated from BM samples from three randomly selected patients from our cohort. The purity of CD34+ cells after isolation was 89% (Fig. S2). After CD34+ cells were cultured with MethoCult methylcellulose medium supplemented with JQ1 and curcumin, RNA was extracted and miR-221-3p, miR-222-3p expression was evaluated. In all the samples analysed miR-221-3p and miR-222-3p were modulated when treated with JQ1 and curcumin. In particular, the double treatment led to a significant downregulation of their expression (Fig. 4).
Figure 4.
Primary CD34+ cultures cells exhibit a decrease of miR-221-3p and miR-222-3p expression after treatments. Expression of miR-221-3p and miR-222-3p was significantly downregulated in CD34+ primary cells when treated with both JQ1 and curcumin (ANOVA followed by Dunnett's test). *P≤0.05 and ***P≤0.005.
Discussion
Despite improvements in the treatment of paediatric patients affected by AML, novel therapeutic strategies are needed, particularly for paediatric high-risk AML, characterized by high relapse incidence. miRs have been extensively studied as potential biomarkers in adult AML and, recently, expression signatures of miRs in paediatric samples have been proposed (14,19,42).
Notably, few miRNA-based prognostic models have been proposed for paediatric cytogenetically normal AML (18,43), t(8;21) RUNX1-RUNX1T1 AML (44) and AML without considering the cytogenetics (18,45). However, conflicting results on the prognostic value of different miRs were reported, possibly due to the vast variability of miRs expression among different cytogenetic and molecular subtypes of AML (15). It was therefore decided, first, to characterize signature of miRs in patients harbouring FLT3-ITD and MLL rearrangement to verify whether distinct AML molecular subsets could affect different miR expression fingerprints. Moreover, to further narrow our miRs signature (identifying only AML associated miRs), HDs were used as a control group. Data on expression of miRs among different AML molecular subtypes are available (16). In accordance with literature data (46), miR-9-5p and miR-10a-5p were upregulated in MLL-rearranged and FLT3-ITD sets, respectively, even compared with HDs. In addition, miR-99a-5p was upregulated in FLT3-ITD patients as compared with MLL patients, and was already described in literature as a potential oncomiR in paediatric AML (42). miR-196b-5p and miR-34a-5p were both upregulated in MLL and FLT3-ITD with respect to HDs and they are possibly involved in the pathogenesis of both AML variants. miR-34a is a widely studied miR as a promising target and it has been considered as a preclinical and clinical model for the treatment of solid tumours, myeloma and B-cell lymphoma (47,48).
Despite the limited number of patients, to the best of our knowledge, this is the first study investigating the role of miRs as predictive biomarker of relapse in paediatric AML patients with FLT3-ITD or MLL rearrangement. A total of 28 miRs differentially expressed were identified between patients who relapsed and those who did not, being possibly associated with prognosis. Among them, the vast majority of miRs have already been reported as related to cancer. Comparing the signature of our miRs to those previously reported, a common dysregulation of mir-155 was identified, but displaying an opposite behaviour with a protective effect in the present study as opposed to a relapse association in others (19,49). miR-155 has led to conflicting results in previous studies, with certain groups reporting an anti-leukemic role (50) and others (13,51–53) reporting a role as oncomiR in AML. This is possibly explained by a different level of expression of miR-155, acting as a tumour suppressor when highly expressed and as an oncomiR when overexpressed to an intermediate level (54). miR-34a-5p expression was positively correlated with relapse. It was already described that patients with low miR-34a expression showed shorter overall and recurrence-free survival (55). Numerous of the identified miRs, have also been independently considered as promising tool to develop novel therapies; these include miR-200c (47,56), which was revealed to be associated to NREL patients.
miR-221-3p and miR-222-3p, both associated to relapse, gained the attention of the authors as they have been broadly reported in literature as oncomiRs both in hematologic malignancies such as chronic lymphocytic leukemia (57), myelodisplastic syndrome (58), acute lymphoblastic leukemia (59) and AML (60,61) as well as in solid tumours. These miRs have as their primary target CDKN1B, a master regulator of the cell cycle. CDKN1B is a well-known cyclin-dependent kinase inhibitor which regulates cell cycle progression at G1 stage, preventing the activation of cyclin E-CDK2 or cyclin D-CDK4 complexes, resulting in a blockade of cell division cycle (62).
BET and HAT inhibitors have already been associated with modulation of miRs in hematological malignances (63,64). The present findings revealed, for the first time, that JQ1 determines a clear trend towards downregulation of miR-221-3p expression in both MOLM-13 and MV-4-11AML cell lines with a synergic effect when associated with curcumin; reaching statistical significance in the second one. This is interesting also considering that for FLT3-ITD rearrangement MOLM-13 expresses both mutated and wild-type allele, while MV-4-11 expresses mutated allele only (65). It could be hypothesized that FLT3-ITD in both mutated alleles renders MV-4-11 cells more sensitive to the effect of drug on miR-221-3p modulation.
Moreover, following the combined treatment, an increase was demonstrated in CDKN1B expression and in apoptotic response in our AML cell lines. The present results were confirmed in cultures with primary leukemic cells, showing a marked reduction of miR-221-3p and miR-222-3p expression. These results supported the idea that BET inhibitors, along with curcumin, could regulate not only coding RNA transcription, but also non-coding RNA such as miRs.
Although the combination of JQ1 and curcumin synergistically reduced miR-221 and miR-222 expression and increased apoptosis in AML cells, a limitation to the present study was represented by insufficient patient samples. Direct regulation of CDKN1B by miR-221-3p and miR-222-3p should be confirmed by further experiments including western blot analysis to verify expression levels of CDKN1B in samples of patients and HDs and the use of inhibitors of miR-221 or miR-222 in a bigger cohort of patients.
In conclusion, the present study identified fingerprints of miRs related to relapse and non-relapse in paediatric FLT3-ITD- or MLL-rearranged AML. Numerous of these miRs are known to be involved in pathogenetic mechanisms of several haematological malignancies as well as solid tumours and represent both good candidates for targeted treatments and therapeutic tools in different neoplasms. The use of the well-known BRD4 inhibitor JQ1, as well as novel BRD inhibitors in the care of leukaemia could be potentiated by epigenetic drugs such as HATs inhibitors, antagomiR or miR mimic and could expand the therapeutic arsenal in HR-AMLs, particularly for paediatric patients.
Supplementary Material
Acknowledgements
The authors would like to thank Giuseppe Basso (1948–2021), MD, Department of Woman's and Children's Health, University of Padua (Padua, Italy) for his help in the study design and for providing samples.
Funding Statement
The present study was supported by Ministero della Salute (grant no. GR-2011-02350175) and by Fondazione Umberto Veronesi (Milano, Italy).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the GEO repository (accession no. GSE209871; http://www.ncbi.nlm.nih.gov/gds).
Authors' contributions
PPL, PV, AB, PM and DP designed the study. RR, AB, PPL and PV conceived and designed the experiments. PPL, PV and VT performed the experiments and data analyses. GN, DV, PV, PF, MS and SR performed Nanostring, data and statistical analyses. VP, FS, EM, AB, DP and PM enrolled patients and collected clinical samples. SR, PV, PPL, MM and PM contributed to manuscript preparation, editing and reviewing. GN and PPL confirm the authenticity of all the raw data. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Review Boards of Bambino Gesù Children's Hospital (Rome, Italy). Informed consent was obtained from either parents or legal guardians according to the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pui CH, Carroll WL, Meshinchi S, Arceci RJ. Biology, risk stratification, and therapy of pediatric acute leukemias: An update. J Clin Oncol. 2011;29:551–565. doi: 10.1200/JCO.2010.30.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creutzig U, van den Heuvel-Eibrink MM, Gibson B, Dworzak MN, Adachi S, de Bont E, Harbott J, Hasle H, Johnston D, Kinoshita A, et al. Diagnosis and management of acute myeloid leukemia in children and adolescents: Recommendations from an international expert panel. Blood. 2012;120:3187–3205. doi: 10.1182/blood-2012-03-362608. [DOI] [PubMed] [Google Scholar]
- 3.Rubnitz JE, Inaba H. Childhood acute myeloid leukaemia. Br J Haematol. 2012;159:259–276. doi: 10.1111/bjh.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elgarten CW, Aplenc R. Pediatric acute myeloid leukemia: Updates on biology, risk stratification, and therapy. Curr Opin Pediatr. 2020;32:57–66. doi: 10.1097/MOP.0000000000000855. [DOI] [PubMed] [Google Scholar]
- 5.Buldini B, Rizzati F, Masetti R, Fagioli F, Menna G, Micalizzi C, Putti MC, Rizzari C, Santoro N, Zecca M, et al. Prognostic significance of flow-cytometry evaluation of minimal residual disease in children with acute myeloid leukaemia treated according to the AIEOP-AML 2002/01 study protocol. Br J Haematol. 2017;177:116–126. doi: 10.1111/bjh.14523. [DOI] [PubMed] [Google Scholar]
- 6.Manara E, Basso G, Zampini M, Buldini B, Tregnago C, Rondelli R, Masetti R, Bisio V, Frison M, Polato K, et al. Characterization of children with FLT3-ITD acute myeloid leukemia: A report from the AIEOP AML-2002 study group. Leukemia. 2017;31:18–25. doi: 10.1038/leu.2016.177. [DOI] [PubMed] [Google Scholar]
- 7.Masetti R, Vendemini F, Zama D, Biagi C, Pession A, Locatelli F. Acute myeloid leukemia in infants: Biology and treatment. Front Pediatr. 2015;3:37. doi: 10.3389/fped.2015.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balgobind BV, Raimondi SC, Harbott J, Zimmermann M, Alonzo TA, Auvrignon A, Beverloo HB, Chang M, Creutzig U, Dworzak MN, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: Results of an international retrospective study. Blood. 2009;114:2489–2496. doi: 10.1182/blood-2009-04-215152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang X, Huang H, Li Z, Li Y, Wang X, Gurbuxani S, Chen P, He C, You D, Zhang S, et al. Blockade of miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for MLL-associated leukemia. Cancer Cell. 2012;22:524–535. doi: 10.1016/j.ccr.2012.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obulkasim A, Katsman-Kuipers JE, Verboon L, Sanders M, Touw I, Jongen-Lavrencic M, Pieters R, Klusmann JH, Michel Zwaan C, van den Heuvel-Eibrink MM, Fornerod M. Classification of pediatric acute myeloid leukemia based on miRNA expression profiles. Oncotarget. 2017;8:33078–33085. doi: 10.18632/oncotarget.16525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danen-van Oorschot AA, Kuipers JE, Arentsen-Peters S, Schotte D, de Haas V, Trka J, Baruchel A, Reinhardt D, Pieters R, Zwaan CM, van den Heuvel-Eibrink MM. Differentially expressed miRNAs in cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Pediatr Blood Cancer. 2012;58:715–721. doi: 10.1002/pbc.23279. [DOI] [PubMed] [Google Scholar]
- 16.Wallace JA, O'Connell RM. MicroRNAs and acute myeloid leukemia: Therapeutic implications and emerging concepts. Blood. 2017;130:1290–1301. doi: 10.1182/blood-2016-10-697698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D, Qiu Y, Jiao Y, Qiu Z, Liu D. Small molecules targeting HATs, HDACs, and BRDs in cancer therapy. Front Oncol. 2020;10:560487. doi: 10.3389/fonc.2020.560487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu R, Zhao W, Fan F, Tang L, Liu J, Luo T, Deng J, Hu Y. A 3-miRNA signature predicts prognosis of pediatric and adolescent cytogenetically normal acute myeloid leukemia. Oncotarget. 2017;8:38902–38913. doi: 10.18632/oncotarget.17151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lim EL, Trinh DL, Ries RE, Wang J, Gerbing RB, Ma Y, Topham J, Hughes M, Pleasance E, Mungall AJ, et al. MicroRNA expression-based model indicates event-free survival in pediatric acute myeloid leukemia. J Clin Oncol. 2017;35:3964–3977. doi: 10.1200/JCO.2017.74.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brondfield S, Umesh S, Corella A, Zuber J, Rappaport AR, Gaillard C, Lowe SW, Goga A, Kogan SC. Direct and indirect targeting of MYC to treat acute myeloid leukemia. Cancer Chemother Pharmacol. 2015;76:35–46. doi: 10.1007/s00280-015-2766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benetatos L, Vartholomatos G. MicroRNAs mark in the MLL-rearranged leukemia. Ann Hematol. 2013;92:1439–1450. doi: 10.1007/s00277-013-1803-4. [DOI] [PubMed] [Google Scholar]
- 22.Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849:506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Donati B, Lorenzini E, Ciarrocchi A. BRD4 and cancer: Going beyond transcriptional regulation. Mol Cancer. 2018;17:164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang G, Deng W, Liu Y, Wang C. General mechanism of JQ1 in inhibiting various types of cancer. Mol Med Rep. 2020;21:1021–1034. doi: 10.3892/mmr.2020.10927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu T, Kamikawa YF, Donohoe ME. Brd4′s bromodomains mediate histone H3 acetylation and chromatin remodeling in pluripotent cells through P300 and Brg1. Cell Rep. 2018;25:1756–1771. doi: 10.1016/j.celrep.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C, Portier BP, Iyer S, Bradner JE, Bhalla KN. BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD. Mol Cancer Ther. 2014;13:2315–2327. doi: 10.1158/1535-7163.MCT-14-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mio C, Conzatti K, Baldan F, Allegri L, Sponziello M, Rosignolo F, Russo D, Filetti S, Damante G. BET bromodomain inhibitor JQ1 modulates microRNA expression in thyroid cancer cells. Oncol Rep. 2018;39:582–588. doi: 10.3892/or.2017.6152. [DOI] [PubMed] [Google Scholar]
- 28.Schick M, Habringer S, Nilsson JA, Keller U. Pathogenesis and therapeutic targeting of aberrant MYC expression in haematological cancers. Br J Haematol. 2017;179:724–738. doi: 10.1111/bjh.14917. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. doi: 10.1038/s41392-018-0008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, Neckers L. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]
- 32.Kang SK, Cha SH, Jeon HG. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 2006;15:165–174. doi: 10.1089/scd.2006.15.165. [DOI] [PubMed] [Google Scholar]
- 33.Morimoto T, Sunagawa Y, Kawamura T, Takaya T, Wada H, Nagasawa A, Komeda M, Fujita M, Shimatsu A, Kita T, Hasegawa K. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest. 2008;118:868–878. doi: 10.1172/JCI33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spriano F, Stathis A, Bertoni F. Targeting BET bromodomain proteins in cancer: The example of lymphomas. Pharmacol Ther. 2020;215:107631. doi: 10.1016/j.pharmthera.2020.107631. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Yang Y, Ren D, Xia Y, He W, Wu Q, Zhang J, Liu M, Du Y, Ren C, et al. JQ1, a bromodomain inhibitor, suppresses Th17 effectors by blocking p300-mediated acetylation of RORγt. Br J Pharmacol. 2020;177:2959–2973. doi: 10.1111/bph.15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 39.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics. 2015;16:169. doi: 10.1186/s12859-015-0611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dweep H, Gretz N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 41.Haferlach C, Kern W, Schindela S, Kohlmann A, Alpermann T, Schnittger S, Haferlach T. Gene expression of BAALC, CDKN1B, ERG, and MN1 adds independent prognostic information to cytogenetics and molecular mutations in adult acute myeloid leukemia. Genes Chromosomes Cancer. 2012;51:257–265. doi: 10.1002/gcc.20950. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Luo XQ, Zhang P, Huang LB, Zheng YS, Wu J, Zhou H, Qu LH, Xu L, Chen YQ. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS One. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaur V, Chaudhary S, Tyagi A, Agarwal S, Sharawat SK, Sarkar S, Singh H, Bakhshi S, Sharma P, Kumar S. Dysregulation of miRNA expression and their prognostic significance in paediatric cytogenetically normal acute myeloid leukaemia. Br J Haematol. 2020;188:e90–e94. doi: 10.1111/bjh.16375. [DOI] [PubMed] [Google Scholar]
- 44.Zampini M, Bisio V, Leszl A, Putti MC, Menna G, Rizzari C, Pession A, Locatelli F, Basso G, Tregnago C, Pigazzi M. A three-miRNA-based expression signature at diagnosis can predict occurrence of relapse in children with t(8;21) RUNX1-RUNX1T1 acute myeloid leukaemia. Br J Haematol. 2018;183:298–301. doi: 10.1111/bjh.14950. [DOI] [PubMed] [Google Scholar]
- 45.Zhu R, Lin W, Zhao W, Fan F, Tang L, Hu Y. A 4-microRNA signature for survival prognosis in pediatric and adolescent acute myeloid leukemia. J Cell Biochem. 2019;120:3958–3968. doi: 10.1002/jcb.27679. [DOI] [PubMed] [Google Scholar]
- 46.Liu Y, Cheng Z, Pang Y, Cui L, Qian T, Quan L, Zhao H, Shi J, Ke X, Fu L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J Hematol Oncol. 2019;12:51. doi: 10.1186/s13045-019-0734-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 48.Beg MS, Brenner AJ, Sachdev J, Borad M, Kang YK, Stoudemire J, Smith S, Bader AG, Kim S, Hong DS. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest New Drugs. 2017;35:180–188. doi: 10.1007/s10637-016-0407-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan W, Xu L, Sun Z, Lin Y, Zhang W, Chen J, Hu S, Shen B. MicroRNA biomarker identification for pediatric acute myeloid leukemia based on a novel bioinformatics model. Oncotarget. 2015;6:26424–26436. doi: 10.18632/oncotarget.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palma CA, Al Sheikha D, Lim TK, Bryant A, Vu TT, Jayaswal V, Ma DD. MicroRNA-155 as an inducer of apoptosis and cell differentiation in acute myeloid leukaemia. Mol Cancer. 2014;13:79. doi: 10.1186/1476-4598-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrózek K, Nicolet D, Kohlschmidt J, Whitman SP, Mendler JH, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. 2013;31:2086–2093. doi: 10.1200/JCO.2012.45.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallace JA, Kagele DA, Eiring AM, Kim CN, Hu R, Runtsch MC, Alexander M, Huffaker TB, Lee SH, Patel AB, et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood. 2017;129:3074–3086. doi: 10.1182/blood-2016-09-740209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narayan N, Morenos L, Phipson B, Willis SN, Brumatti G, Eggers S, Lalaoui N, Brown LM, Kosasih HJ, Bartolo RC, et al. Functionally distinct roles for different miR-155 expression levels through contrasting effects on gene expression, in acute myeloid leukaemia. Leukemia. 2017;31:808–820. doi: 10.1038/leu.2016.279. [DOI] [PubMed] [Google Scholar]
- 55.Huang Y, Zou Y, Lin L, Ma X, Chen H. Identification of serum miR-34a as a potential biomarker in acute myeloid leukemia. Cancer Biomark. 2018;22:799–805. doi: 10.3233/CBM-181381. [DOI] [PubMed] [Google Scholar]
- 56.Chakraborty C, Sharma AR, Sharma G, Doss CGP, Lee SS. Therapeutic miRNA and siRNA: Moving from bench to clinic as next generation medicine. Mol Ther Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfò L, Rossi C, Ferrari G, Ghia P, Caligaris-Cappio F. MicroRNA and proliferation control in chronic lymphocytic leukemia: Functional relationship between miR-221/222 cluster and p27. Blood. 2010;115:3949–3959. doi: 10.1182/blood-2009-11-254656. [DOI] [PubMed] [Google Scholar]
- 58.Hussein K, Theophile K, Büsche G, Schlegelberger B, Göhring G, Kreipe H, Bock O. Significant inverse correlation of microRNA-150/MYB and microRNA-222/p27 in myelodysplastic syndrome. Leuk Res. 2010;34:328–334. doi: 10.1016/j.leukres.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 59.Moses BS, Evans R, Slone WL, Piktel D, Martinez I, Craig MD, Gibson LF. Bone marrow microenvironment niche regulates miR-221/222 in acute lymphoblastic leukemia. Mol Cancer Res. 2016;14:909–919. doi: 10.1158/1541-7786.MCR-15-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rommer A, Steinleitner K, Hackl H, Schneckenleithner C, Engelmann M, Scheideler M, Vlatkovic I, Kralovics R, Cerny-Reiterer S, Valent P, et al. Overexpression of primary microRNA 221/222 in acute myeloid leukemia. BMC Cancer. 2013;13:364. doi: 10.1186/1471-2407-13-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee YG, Kim I, Oh S, Shin DY, Koh Y, Lee KW. Small RNA sequencing profiles of mir-181 and mir-221, the most relevant microRNAs in acute myeloid leukemia. Korean J Intern Med. 2019;34:178–183. doi: 10.3904/kjim.2017.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.le Sage C, Nagel R, Agami R. Diverse ways to control p27Kip1 function: miRNAs come into play. Cell Cycle. 2007;6:2742–2749. doi: 10.4161/cc.6.22.4900. [DOI] [PubMed] [Google Scholar]
- 63.Kohnken R, McNeil B, Wen J, McConnell K, Grinshpun L, Keiter A, Chen L, William B, Porcu P, Mishra A. Preclinical targeting of MicroRNA-214 in cutaneous T-cell lymphoma. J Invest Dermatol. 2019;139:1966–1974.e3. doi: 10.1016/j.jid.2019.01.033. [DOI] [PubMed] [Google Scholar]
- 64.Mensah AA, Cascione L, Gaudio E, Tarantelli C, Bomben R, Bernasconi E, Zito D, Lampis A, Hahne JC, Rinaldi A, et al. Bromodomain and extra-terminal domain inhibition modulates the expression of pathologically relevant microRNAs in diffuse large B-cell lymphoma. Haematologica. 2018;103:2049–2058. doi: 10.3324/haematol.2018.191684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quentmeier H, Reinhardt J, Zaborski M, Drexler HG. FLT3 mutations in acute myeloid leukemia cell lines. Leukemia. 2003;17:120–124. doi: 10.1038/sj.leu.2402911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the GEO repository (accession no. GSE209871; http://www.ncbi.nlm.nih.gov/gds).