Abstract
Upon exposure to UV radiation, Shigella flexneri SA100 displayed survival and mutation frequencies comparable to those of Escherichia coli AB1157, which contains a functional UmuDC error-prone DNA repair system. Survival of SA100 after UV irradiation was associated with the presence of the 220-kb virulence plasmid, pVP. This plasmid encodes homologues of ImpA and ImpB, which comprise an error-prone DNA repair system encoded on plasmid TP110 that was initially identified in Salmonella typhimurium, and ImpC, encoded upstream of ImpA and ImpB. Although the impB gene was present in representatives of all four species of Shigella, not all isolates tested contained the gene. Shigella isolates that lacked impB were more sensitive to UV radiation than isolates that contained impB. The nucleotide sequence of a 2.4-kb DNA fragment containing the imp operon from S. flexneri SA100 pVP was 96% identical to the imp operon from the plasmid TP110. An SA100 derivative with a mutation in the impB gene had reduced survival following UV irradiation and less UV-induced mutagenesis relative to the parental strain. We also found that S. flexneri contained a chromosomally encoded umuDC operon; however, the umuDC promoter was not induced by exposure to UV radiation. This suggests that the imp operon but not the umuDC operon contributes to survival and induced mutagenesis in S. flexneri following exposure to UV radiation.
Shigella flexneri, a facultative intracellular bacterium, causes bacterial dysentery in humans (13, 28, 39). This pathogen faces numerous potentially stressful environments during its life cycle. These environments are encountered as Shigella is exposed to the external environment, as it transits through the human gastrointestinal tract, and during its growth within colonic epithelial cells. However, little is known about the response of Shigella to these potentially stressful environments.
One stress response that has been studied in detail in bacteria is the SOS response to DNA damage. Upon exposure to UV radiation or chemicals that damage DNA, Escherichia coli and other bacteria express a specific set of genes that are normally repressed by the LexA repressor binding to an SOS box in each promoter (11, 54). The initial signal of DNA damage, which is thought to be single-stranded DNA, activates RecA to RecA* (11). RecA* mediates the self-cleavage of the LexA repressor, leading to derepression of more than 20 genes, including recA, lexA, excision DNA repair genes, and the error-prone DNA repair genes umuD and umuC (11). RecA* also mediates the self-cleavage of UmuD into UmuD′, which is the form that mediates error-prone DNA repair (5, 32, 46).
The E. coli umuDC promoter, which contains a highly conserved SOS box, has a strong affinity for LexA. Thus, UV induction of the umuDC operon occurs only after most of the LexA repressor has been cleaved. Induction of umuDC expression results in an increased frequency of mutagenesis (10, 47). Current genetic and biochemical evidence supports the model in which a mutasome, composed of UmuD′-UmuC, RecA*, and DNA polymerase III, allows the bypassing of potentially lethal lesions in the DNA during DNA replication (37). The interaction of the UmuD′-UmuC complex with DNA polymerase III is thought to result in relaxed fidelity, leading to the generation of mutations. Thus, the process of UV-induced mutagenesis is also known as error-prone DNA repair. Strains containing umuDC mutations are more sensitive to UV exposure than their isogenic parents, suggesting that error-prone DNA repair is important for survival following UV irradiation (3, 19).
The umuD and umuC genes constitute an operon located on the E. coli and Salmonella typhimurium chromosomes (20, 36, 49, 52). umuDC homologues also have been identified on several naturally occurring plasmids, and their gene products mediate error-prone DNA repair (56). These homologues include impAB, located on the large conjugative plasmid TP110, which was initially identified in Salmonella typhimurium (9, 26). Other plasmids from several different incompatibility groups also contain sequences with homology to the imp operon (27). The TP110 imp operon complements mutations in the E. coli umuDC operon, demonstrating that these operons are functionally similar (43).
Preliminary analysis of the response of Shigella growing in the intracellular environment led to the identification of an S. flexneri DNA fragment homologous to the 3′ end of the impB gene encoded on the plasmid TP110 (15). In this article, we describe the identification and characterization of the virulence plasmid-encoded impCAB genes and the chromosomally encoded umuDC genes in S. flexneri. Our data suggest that the imp operon but not the umuDC operon contributes to survival and induced mutagenesis in S. flexneri following UV irradiation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Bacterial strains and plasmids used in this work are listed in Table 1. All strains were maintained at −80°C in tryptic soy broth (TSB) plus 20% glycerol. E. coli strains were routinely grown in Luria broth (L broth) or on Luria agar (L agar) plus antibiotics at 37°C, and Shigella spp. strains were grown in L broth or on TSB agar plus 0.1% Congo red dye and antibiotics at 37°C. Strains containing gfp fusions were grown in low-salt L broth, which contains 5 g of NaCl/liter instead of 10 g/liter. Antibiotics were used at the following concentrations: 250 μg of carbenicillin/ml, 30 μg of chloramphenicol/ml, and 200 μg of streptomycin/ml.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Characteristics | Reference or sourcea |
|---|---|---|
| E. coli strains | ||
| DH5α | Standard lab cloning strain | 38 |
| SM10λpir | pirR6K | 51 |
| AB1157 | 16 | |
| 1107-81 | Enteroinvasive | J. H. Crosa, Oregon University of the Health Sciences |
| 550-3076 | Enteroinvasive | J. H. Crosa, Oregon University of the Health Sciences |
| Shigella strains | ||
| SA100 | S. flexneri wild-type serotype 2a | 35 |
| SA102 | SA100 derivative lacking the virulence plasmid pVP | T. Daskaleros |
| SM100 | Streptomycin-resistant SA100 derivative | S. Seliger |
| 2457 | S. flexneri serotype 2a | S. Formal |
| 8-2031 | S. flexneri serotype 2b | TDH |
| M90T | S. flexneri serotype 5 | P. Sansonetti |
| 229272 | S. flexneri serotype 5 | TDH |
| SD125 | S. dysenteriae serotype 1 | B. A. D. Stocker |
| O-4576 | S. dysenteriae serotype 1 | TDH |
| O-1392 | S. boydii serotype 5 | TDH |
| 224860 | S. boydii serotype 7 | TDH |
| PB66 | S. sonnei | D. Winsor |
| 1-1245 | S. sonnei | TDH |
| Plasmids | ||
| pVP | 220-kb virulence plasmid from SA100 | 35 |
| pWKS30 | Low-copy-number cloning vector | 55 |
| pIMP | pWKS30 containing the imp operon from SA100 on a 2.4-kb PCR product | This study |
| pGTXN3 | Transcriptional fusion vector containing the promoterless mutant 3 gfp gene from Cormack et al. | This study |
| pLR13 | pGTXN3 carrying a 479-bp DNA fragment containing the SA100 umuDC promoter | This study |
| pLR20 | pGTXN3 carrying a 479-bp DNA fragment containing the AB1157 umuDC promoter | This study |
TDH, Texas Department of Health.
Recombinant DNA methods.
Plasmids smaller than 20 kb were isolated with the QIAprep Spin Miniprep kit (Qiagen, Santa Clarita, Calif.). The method of Kado and Liu (18) was used to isolate the 220-kb virulence plasmid from S. flexneri. Isolation of DNA fragments from agarose gels was performed with the QIAquick Gel Extraction kit (Qiagen) or the GeneClean kit (Bio 101, Vista, Calif.).
For matings, overnight cultures of the donor and recipient strains were washed once in phosphate-buffered saline (PBS) and resuspended at a concentration of approximately 1010 bacteria per ml. Twenty microliters of each culture was mixed, and the mixture was spotted onto Luria agar. After a 6-h incubation at 37°C, the cells from the plate were resuspended in 1 ml of PBS and plated on L agar or TSB agar plus 0.1% Congo red dye containing the appropriate antibiotics.
UV irradiation survival assays.
Bacteria were grown at 37°C with aeration in L broth containing the appropriate antibiotics to an optical density of 650 nm (OD650) of 0.5 to 0.9, pelleted by centrifugation, and resuspended in PBS at a concentration of 108 bacteria per ml. Aliquots of the concentrated bacteria (0.4 ml) in 16-mm petri dishes were irradiated with UV light at doses indicated in the figures (0 to 40 J/m2), and the number of cells that survived UV irradiation was quantitated by plate counts on L agar. Statistical analyses of the data were performed using the ANOVA statistics package in Microsoft Excel 97 (Microsoft Corporation, Redmond, Wash.).
Mutagenesis assays.
Bacteria grown to an OD650 of 0.5 to 0.9 were pelleted, resuspended in PBS to a concentration of 109 bacteria per ml, and irradiated with UV light at a fluence of 10 J/m2. The number of cells that survived UV irradiation was quantitated by plate counts on L agar, and the number of cells that acquired a UV-induced mutation conferring rifampin resistance was determined essentially as described by Sedgwick and Goodwin (41). Statistical analyses of the data were performed using the ANOVA statistics package in Microsoft Excel 97 (Microsoft Corporation).
PCR procedures.
All PCRs were carried out using Pfu polymerase (Stratagene Cloning Systems, La Jolla, Calif.) in the reaction buffer supplied by the manufacturer supplemented with 250 μM each dNTP and 1 μM primers. Bacterial cultures (1 to 2 μl) grown overnight, washed once in PBS, and diluted 10-fold were used as the template per 100-μl reaction. The reactions were initially incubated at 95°C for 5 min, and the PCR was then done according to the specific conditions for each reaction. Thirty cycles were carried out for all reactions. Each cycle consisted of a 1-min denaturation step at 95°C, a 1-min annealing step at 3 to 5°C below the melting temperature for the primers, and an extension step of 2 min per kb to be amplified at 72°C. The primers were impB1 (5′CACTCGATGAACTGAACC3′), impB2 (5′TTTCCCGTTTCATTTGCC3′), impB3 (5′CACAGCAGGCATACAGCC3′), upstream primer (5′AATTCTCCTCTCACATGCGG3′), downstream primer (5′GGTGCTTTGCAATCTGCTG3′), umuDCP1 (5′GATCTAATGCTCCATCTGCG3′), umuDCP2 (5′CGCGGAGATCCGCAGGC3′), and umuDC3 (5′GCCGCTATATTTATTTGACCC3′). For inverse PCR, plasmid DNA from SA100 was digested with HincII and EcoRV and ligated with T4 DNA ligase. The inverse PCR was carried out by using 2 μl of the ligation reaction and primers impB2 (5′TTTCCCGTTTCATTTGCC3′) and impB5 (5′TGCTCTCGCCTTCGTATA3′) at an annealing temperature of 50°C.
Sequence analysis of the imp operon.
For determining the nucleotide sequence of the imp operon from S. flexneri SA100 pVP, a 2.4-kb PCR product containing the operon and generated as described above was used as the template. For determining the nucleotide sequence of the 3′ end of the imp operon from S. flexneri 8-2031, a subclone of a cosmid that contained impB from 8-2031 (7) was used as the template for sequencing. Nucleotide sequences were determined by the Molecular Biology Sequencing Facility at the University of Texas at Austin. The DNA that was generated in the sequencing reactions was labeled with the dRhodamine Dideoxy-terminator Cycle Sequencing kit (Perkin-Elmer Co., Applied Biosystems Division, Foster City, Calif.) and analyzed with an ABI Prism 377 DNA sequencer (Perkin-Elmer Co., Applied Biosystems Division).
Construction of an impB mutation in S. flexneri by allelic exchange.
The allelic exchange vector pHM5 was constructed as follows. pGP704, which replicates in λpir lysogens but not in SM100 (29), was linearized with SmaI and ligated to a 1.9-kb EcoRV fragment containing the sacB gene, which encodes sucrose sensitivity, from pMTL-sac#4 (57). A 2-kb PCR product containing part of the imp operon and amplified with the primers impB3 and impB2 (see the description of the PCR procedures above) was cloned into the vector pWKS30 (55) linearized with EcoRV to generate pLR25. A 1.6-kb fragment containing a chloramphenicol resistance gene (cam) was isolated from pMA9, a derivative of pNK2884 (21), by digestion with HindIII followed by treatment with the Klenow fragment of DNA polymerase I. This cam cassette was inserted into the EcoRV site in the impB gene on pLR25 to generate pLR25::Cm. The imp operon with the cam resistance cassette was excised from pLR25::Cm as a XhoI-SmaI fragment and ligated into pHM5 digested with SalI and EcoRV to generate pLR27. pLR27 was mated from E. coli SM10λpir to S. flexneri SM100, and single crossovers in the impB gene were selected by plating on TSB agar plus 0.1% Congo red containing chloramphenicol and streptomycin. Double crossover recombinants were then selected on L agar containing 5% sucrose, chloramphenicol, and streptomycin and screened for carbenicillin sensitivity. PCR analysis, using primers that flank the cam insertion in impB, confirmed that the wild-type impB allele was replaced with the cam-disrupted impB allele in the chloramphenicol-resistant, sucrose-resistant, carbenicillin-sensitive recombinants. One recombinant, designated SM162, was used for further study.
Tissue culture cell invasion and plaque assays.
Henle cell monolayers were used in all experiments and were routinely maintained in Earle’s minimal essential medium plus 2 mM glutamine plus 10% fetal calf serum (Life Technologies, Grand Island, N.Y.) in a 5% CO2 atmosphere at 37°C. Plaque assays were done as described previously (34) with the following modifications. Confluent Henle cell monolayers in 35-mm plates were infected with 103 and 104 bacteria. After a 60-min incubation, the cells were washed four times with PBS and overlaid with fresh medium containing 0.45% (wt/vol) glucose, 0.5% agarose, and 20 μg of gentamicin/ml. Plaques were scored after 72 h.
Detection of GFP expression controlled by the umuDC promoter.
The promoterless gfp vector pGTXN3 was constructed as follows. The promoterless cat vector pKK232-8 (Pharmacia, Piscataway, N.J.) was digested with NcoI and HindIII to generate a 4.5-kb DNA fragment which is missing the 5′ 576 bp of the cat gene. The 4.5-kb fragment was treated with the Klenow fragment of DNA polymerase I, isolated by electrophoresis, and religated with T4 DNA ligase to generate pKK232-8Δ. This plasmid was digested with SalI, treated with the Klenow fragment of DNA polymerase I, and digested with BamHI. The promoterless gfp gene was isolated from pGFP3 (6) by digestion with HindIII and treated with the Klenow fragment of DNA polymerase I, followed by digestion with BamHI. The resulting 0.75-kb fragment was purified by electrophoresis and ligated to pKK232-8Δ isolated as described above. The resulting plasmid was designated pGTXN3.
A 479-bp PCR product containing the umuDC promoter was amplified from E. coli AB1157 by using PCR primers umuDCP1 and umuDCP2. This DNA fragment contained 420 bp 5′ of the transcriptional start site and 28 bp 3′ of the translational start codon and was cloned into the vector pGTXN3 cut with SmaI to generate pLR20. The same primers were used to amplify a 479-bp PCR product containing the umuDC promoter from SA100, which was cloned into the vector pKK232-8 cut with SmaI to generate pLR1R. The SA100 umuDC promoter was isolated from pLR1R by digestion with EcoRI and BamHI and cloned into pBSK− cut with EcoRI and BamHI to generate pLR7. The SA100 umuDC promoter was isolated from pLR7 by digestion with EcoRV and BamHI and cloned into pGTXN3 digested with XbaI, treated with the Klenow fragment of DNA polymerase, and digested with BamHI to generate pLR13.
umuDC promoter activity was assessed by quantitating the amount of the green fluorescent reporter protein with a Molecular Dynamics Fluorimager. Bacteria were grown in low-salt L broth containing the appropriate antibiotics to an OD650 of approximately 1.0, pelleted by centrifugation, and resuspended to a concentration of 109 bacteria per ml in PBS. One-half of each culture was exposed to 50 J/m2 of UV radiation. Each sample was then pelleted by centrifugation and resuspended to a concentration of 109 bacteria per ml in L broth. After a 2-h incubation at 37°C, the samples were concentrated to 1011 bacteria per ml by centrifugation. The levels of green fluorescent protein (GFP) were measured at 530 ± 15 nm. The background fluorescence of the cells containing the vector was subtracted from each sample. Specific activity of GFP for each sample was expressed in fluorescence units, which were calculated by dividing the relative fluorescence by the OD650 and volume of the sample.
Nucleotide sequence accession number.
The sequence data of a 2.4-kb DNA fragment containing the imp operon have been submitted to the GenBank database under the accession no. AF079316.
RESULTS
Sensitivity of S. flexneri isolates to UV radiation.
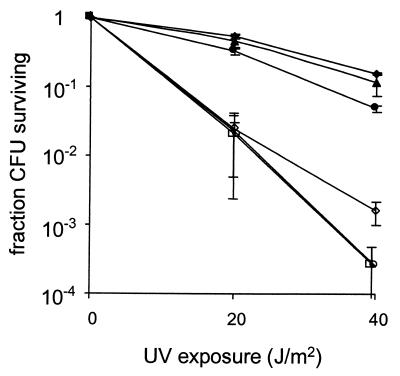
The ability of S. flexneri isolates to survive UV irradiation was assessed by measuring the fraction of cells that survived exposure to increasing doses of UV radiation. We used E. coli AB1157, a strain previously shown to be relatively UV resistant, as a reference (42). S. flexneri SA100 and 2457 were as UV resistant as AB1157 (Fig. 1). However, SA102, a derivative of SA100 lacking the 220-kb virulence plasmid pVP, was significantly more UV sensitive than SA100 (P < 0.05). At doses of UV radiation of 20 and 40 J/m2, the numbers of SA102 cells that survived exposure to UV radiation were 22- and 100-fold less than SA100, respectively (Fig. 1). S. flexneri M90T and 229272, both of which contain a virulence plasmid, were also significantly more UV sensitive than SA100 (P < 0.05). The numbers of M90T and 229272 cells that survived exposures to UV radiation of 20 and 40 J/m2 were 25- and 600-fold less, respectively, than the number of surviving SA100 cells (Fig. 1).
FIG. 1.
Survival of E. coli AB1157 and S. flexneri isolates after UV irradiation. Cells were subjected to varying doses of UV light, and the CFU were determined by plating onto L agar. The data are the means of three experiments, and the standard deviations of the means are indicated. Symbols: ●, AB1157; ▴, 2457; ⧫, SA100; ◊, SA102; ○, M90T; □, 229272.
impB is located on the virulence plasmid of S. flexneri SA100.
The different sensitivities to UV radiation of SA100 and SA102 suggested that survival following exposure to UV radiation was enhanced by the presence of pVP in SA100 (Fig. 1). Because a fragment of S. flexneri DNA homologous to the error-prone DNA repair gene impB had been identified in S. flexneri in our preliminary studies (15), it seemed likely that a pVP-encoded ImpB homologue contributes to resistance to UV radiation. To determine whether pVP encodes an ImpB homologue, we designed primers based on the plasmid TP110 impB sequence to amplify an 87-bp impB fragment from SA100 and SA102 by using PCR. A PCR product of this size was amplified from SA100 but not from SA102. Since the only known difference between SA100 and SA102 is that SA102 lacks pVP, it appeared that ImpB is encoded on pVP.
To provide further evidence that impB was located on the virulence plasmid in S. flexneri, the virulence plasmid was isolated from a derivative of S. flexneri SA100 (SM100/pVP::Cm) and transferred by electroporation into E. coli DH5α, which does not contain impB. An 87-bp PCR product corresponding to impB was amplified from these transformants but not from DH5α (data not shown); thus, we concluded that impB is located on pVP.
The presence of impB correlates with increased resistance to UV radiation.
We examined isolates of other Shigella spp. for the presence of impB and for their ability to survive UV irradiation. Although impB was present in representatives of all four species of Shigella, not all isolates tested contained the gene (Table 2). Within each species, isolates that contained impB were significantly more resistant to UV radiation than strains that lacked impB (P < 0.05) (Table 2). Eight isolates of enteroinvasive E. coli were also tested for the presence of the impB gene because enteroinvasive E. coli, which causes a disease that is similar to shigellosis, has a virulence plasmid that is related to the Shigella virulence plasmids (40). None of the eight isolates tested contained the impB gene (Table 2 and data not shown). We examined the ability of two enteroinvasive E. coli isolates to survive UV irradiation and found that they were significantly more sensitive to UV radiation than SA100 (P < 0.05) (Table 2).
TABLE 2.
Correlation between the presence of impB and resistance to UV radiation in Shigella spp. and enteroinvasive E. coli
| Strain | Sero-type | Presence of impBa | Resistance to UVb (% survival ± SD) |
|---|---|---|---|
| S. flexneri SA100 | 2a | + | + (15.4% ± 9.3%) |
| S. flexneri 2457 | 2a | + | + (11.8% ± 4.4%) |
| S. flexneri M90T | 5 | − | − (0.025% ± 0.023%) |
| S. flexneri 229272 | 5 | − | − (0.024% ± 0.024%) |
| S. dysenteriae SD125 | 1 | + | + (2.6% ± 2.0%) |
| S. dysenteriae O-4576 | 1 | + | + (2.4% ± 2.0%) |
| S. sonnei PB66 | + | + (20.4% ± 2.4%) | |
| S. sonnei 1-1245 | + | + (15.6% ± 1.4%) | |
| S. boydii 224860 | 7 | + | + (0.20% ± 0.10%) |
| S. boydii O-1392 | 5 | − | − (0.010% ± 0.003%) |
| Enteroinvasive E. coli 1107-81 | − | − (0.002% ± 0.002%) | |
| Enteroinvasive E. coli 550-3076 | − | − (<0.001%) |
The presence of impB was detected by PCR using primers impB1 and impB2, which are complementary to plasmid TP110 impB.
+ and − indicate that ≥0.1% and ≤0.1% of the cells, respectively, survived UV irradiation at a fluence of 40 J/m2.
Sequencing the imp operon from SA100 pVP.
We isolated the entire imp operon from S. flexneri SA100 pVP by PCR. The imp upstream primer was designed using the sequence upstream from the TP110-encoded imp operon. However, the DNA fragment containing the imp operon could not be amplified using the imp upstream primer and a primer with a nucleotide sequence based on the sequence downstream from the TP110 imp operon. Therefore, we used inverse PCR with primers corresponding to the coding sequence of impB to isolate a fragment downstream from the SA100 pVP imp operon. The nucleotide sequence of the inverse PCR product was determined, and a downstream primer was designed using this sequence. The imp upstream and downstream primers were used to amplify a 2.4-kb DNA fragment containing the imp operon, and the nucleotide sequence of this fragment was 96% identical to that of a DNA fragment containing the impCAB operon on the conjugative plasmid TP110.
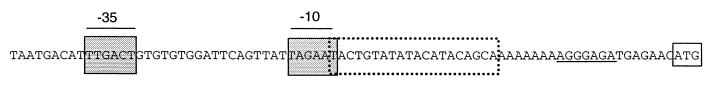
The nucleotide sequence of the 2.4-kb PCR product from SA100 pVP contained three overlapping open reading frames (ORFs) encoding putative proteins of 9.5, 16.2, and 47.6 kDa, which were designated ImpC, ImpA, and ImpB, respectively, based on their amino acid homologies (see below). The predicted pIs of these proteins were 5.13, 4.98, and 9.52, respectively. Because the overlapping arrangement of the three imp genes suggested that they constituted an operon, the region upstream of impC was examined for promoter-like features (Fig. 2). We identified a putative ς70 recognition sequence in which each of the −35 and −10 hexamers deviates in 1 of 6 nucleotides from the consensus binding sequence for ς70 (14). A 20-bp SOS box, which may function as a binding site for the LexA repressor, overlapped the putative −10 sequence by 1 nucleotide. Nucleotides 12 and 19 deviated from the consensus SOS box; however, these 2 nucleotides are less critical for LexA binding (11). We were unable to locate a Rho-independent terminator downstream of the SA100 pVP imp operon. This suggests that transcriptional termination occurs via a Rho-dependent manner or that this operon contains an additional downstream gene.
FIG. 2.
Nucleotide sequence of the impCAB promoter from S. flexneri. The dotted box indicates the putative SOS box for LexA binding, and the shaded box indicates the putative −10 and −35 hexamers for ς70. The proposed translational initiation codon is boxed, and the Shine-Delgarno sequence is underlined.
impC, the first gene in the SA100 pVP imp operon, was preceded by a potential ribosome binding site (AGGGAGA) located 8 nucleotides 5′ to the translational start codon. The deduced amino acid sequence of impC was used to conduct a BLASTX search of the National Center for Biotechnology Information nonredundant database (1). The predicted protein was 100% identical to the ImpC protein encoded on the plasmid TP110. ImpC has been postulated to be involved in the regulation of ImpA and ImpB expression by an undefined mechanism (26). Additionally, S. flexneri pVP ImpC was homologous to ORFf, encoded by the retronphage ΦR67 in E. coli (8, 17); Tum, encoded by prophage 186 in E. coli (4); and E. coli DinI (59) (Table 3). The function of ORFf has not been determined; however, Tum is an antirepressor that causes induction of prophage 186 by directly interfering with the ability of the phage repressor to bind DNA (45). DinI has been shown to inhibit RecA*-mediated self-cleavage of LexA and UmuD (58). Thus, ImpC may have a similar role in the regulation of ImpA self-cleavage to ImpA′.
TABLE 3.
Selected homologies of S. flexneri pVP-encoded ImpC, ImpA, and ImpB to other proteinsa
| Protein | Organism | Location | % Identity | % Similarityb | Reference |
|---|---|---|---|---|---|
| ImpC homologues | |||||
| ImpC | Salmonella typhimurium | Plasmid TP110 | 100 | 100 | 26 |
| ORFf | E. coli | Retronphage Φ67 | 34 | 64 | 17 |
| DinI | E. coli | Chromosome | 30 | 55 | 59 |
| Tum | E. coli | Coliphage 186 | 25 | 50 | 4 |
| ImpA homologues | |||||
| ImpA | Salmonella typhimurium | Plasmid TP110 | 100 | 100 | 26 |
| SamA | Salmonella typhimurium | 60-MDa virulence plasmid | 59 | 73 | 33 |
| MucA | Serratia marcescens | Plasmid R471a | 46 | 61 | 22 |
| MucA | Salmonella typhimurium | Plasmid R46 | 43 | 56 | GenBank accession no. X16596 |
| MucA | E. coli | Plasmid pKM101 | 44 | 57 | 36 |
| RumA | Proteus rettgeri | Plasmid R391 | 46 | 56 | 23 |
| RulA | Pseudomonas syringae | Plasmid pPSR1 | 28 | 43 | 50 |
| UmuD | E. coli | Chromosome | 43 | 58 | 36 |
| UmuD | Salmonella typhimurium | Chromosome | 42 | 57 | 49, 52 |
| ImpB homologues | |||||
| ImpB | Salmonella typhimurium | Plasmid TP110 | 97 | 98 | 26 |
| SamB | Salmonella typhimurium | 60-MDa virulence plasmid | 70 | 83 | 33 |
| MucB | Serratia marcescens | Plasmid R471a | 51 | 67 | 22 |
| MucB | Salmonella typhimurium | Plasmid R46 | 55 | 69 | GenBank accession no. X16596 |
| MucB | E. coli | Plasmid pKM101 | 55 | 69 | 36 |
| RumB | Proteus rettgeri | Plasmid R391 | 57 | 74 | 23 |
| RulB | Pseudomonas syringae | Plasmid pPSR1 | 42 | 61 | 50 |
| UmuC | E. coli | Chromosome | 57 | 72 | 36 |
| UmuC | Salmonella typhimurium | Chromosome | 58 | 71 | 49, 52 |
Homologies were calculated using the CLUSTAL W algorithm (53) in MacVector 6.0 (Oxford Molecular Group).
Includes conserved substitutions.
The translational stop codon for ImpC overlapped the translational initiation codon for ImpA by 2 nucleotides. The deduced amino acid sequence of S. flexneri pVP impA was 100% identical to the TP110-encoded ImpA protein and was similar to other members of the UmuD family of error-prone DNA repair proteins (Table 3). Amino acid residues that are highly conserved among the UmuD family and are important in the RecA*-mediated self-cleavage reaction were conserved in S. flexneri pVP ImpA. These include the Ala-Gly or Cys-Gly residues (amino acids 24 and 25 in E. coli UmuD) where cleavage occurs; a serine residue (amino acid 60), which functions as a nucleophile; and a lysine residue (amino acid 97), which most likely functions as an activator (5, 32, 36, 46). Like other UmuD homologues, the N terminus of S. flexneri pVP ImpA was not as well conserved as the C terminus. Since the N terminus has been shown to be proteolytically removed in several UmuD homologues, it is probably not essential for ImpA function.
The translational stop site for ImpA overlapped the translational initiation site for ImpB by 1 nucleotide. The deduced amino acid sequence of S. flexneri pVP impB was 97% identical to the TP110-encoded ImpB protein and was similar to other members of the UmuC family of error-prone DNA repair proteins (Table 3). The nucleotide sequences of the 3′ ends and downstream regions of the TP110- and SA100 pVP-encoded impB genes were less conserved than the rest of the imp operon. The nucleotide sequences are highly conserved (98% identity) from 370 nucleotides 5′ to the impC translational start codon to 49 nucleotides 5′ to the impB translational stop codon. Beginning at this point and continuing downstream from impB, the nucleotide sequence is not as highly conserved between S. flexneri SA100 pVP and plasmid TP110 (45% identity). Additionally, the nucleotide sequences of the virulence plasmids from S. flexneri SA100 and 8-2031, another S. flexneri strain, were not as highly conserved in this region (47% identity). A sequence containing a direct repeat (GGCAAATGN4GGGAAATG [the underlining indicates the repeated sequence]) precedes the point of nucleotide divergence. The nucleotide sequence 3′ of impB in 8-2031 contained an 8-bp sequence (TTGTTATT) that was tandemly repeated six times, beginning 22 bp from the stop codon. The significance of these direct repeats is unknown.
Upstream from and in the opposite orientation of the imp operons on SA100 pVP and plasmid TP110, we identified a partial ORF with a deduced amino acid sequence that was 92% identical to that of an 8-kDa putative protein of unknown function encoded downstream of the sop locus. The sop locus controls partitioning of the F plasmid, but the 8-kDa putative protein is not required for plasmid partioning (30). On pVP, there was a stop codon in the ORF located 40 nucleotides downstream of the translational start codon. Additionally, on both pVP and TP110, there was a 1-bp insertion 76 nucleotides 5′ to the ORF translational stop codon that shifted the reading frame. Thus, it appears that this ORF does not encode a functional protein in strains carrying pVP or TP110.
Characterization of an ImpB mutant.
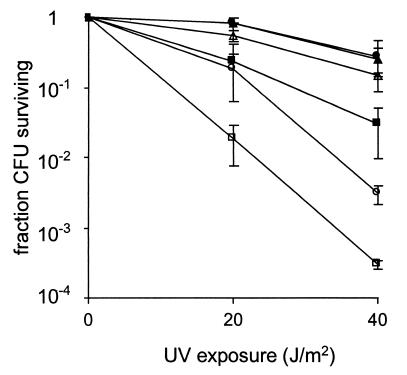
To determine whether any of the genes in the pVP imp operon was required for survival or induced mutagenesis in S. flexneri following UV irradiation, we constructed an impB mutation in which the wild-type allele was replaced with one containing a cam gene insertion. The mutation was constructed in SM100, a streptomycin-resistant derivative of SA100, and the resulting ImpB mutant was designated SM162. The ability of SM162 to survive UV irradiation was examined as described above. SM162 was significantly more sensitive to UV radiation than the parental strain SM100 (P < 0.05) (Fig. 3); however, SM162 was not as sensitive as SA102, which does not contain pVP (P < 0.05) (Fig. 3).
FIG. 3.
Effect of impB on survival after UV irradiation. Cells were subjected to varying doses of UV light, and the CFU were determined by plating on L agar. The data are the means of three experiments, and the standard deviations of the means are indicated. Symbols: ▵, SM100; ▴, SM100/pIMP; □, SA102; ■, SA102/pIMP; ○, SM162; and ●, SM162/pIMP.
A 2.4-kb PCR fragment containing the entire imp operon from S. flexneri SA100 pVP was cloned into pWKS30, a low-copy-number vector, to generate pIMP. SM100, SM162, and SA102 were transformed with pIMP, and the ability of the strains to survive UV irradiation was assessed (Fig. 3). Strains carrying the vector pWKS30 showed a response to UV irradiation that was identical to that of the same strain without the vector. The addition of pIMP to SM162 restored survival following UV irradiation to levels similar to that of the parental strain SM100 carrying pIMP. SA102 carrying pIMP showed an intermediate level of UV sensitivity that was between the sensitivity of SA102 and that of SM100 carrying pIMP.
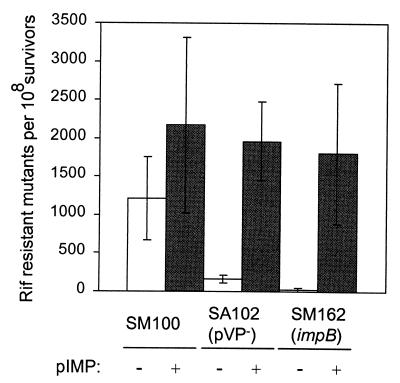
We examined the UV-induced mutagenesis phenotype of SM162 by measuring the frequency at which mutations that conferred rifampin resistance arose. SM100, SM162, and SA102, all of which are sensitive to rifampin, were exposed to 10 J/m2 of UV light and then tested for rifampin resistance. The UV-induced mutagenesis frequency was 40-fold lower in SM162 than in the parental strain SM100 (P < 0.05) (Fig. 4). Surprisingly, even though SA102 was more sensitive to UV exposure than SM162 (Fig. 3), the UV-induced mutagenesis frequency in SA102 was significantly higher than in SM162 (P < 0.05), although it was still sevenfold lower than that in the parental strain SM100 (P < 0.05) (Fig. 4). The addition of pIMP to SM162 and SA102 restored the UV-induced mutagenesis frequency to the levels of SM100/pIMP (Fig. 4).
FIG. 4.
Effect of impB on UV-induced mutagenesis. Rifampin-sensitive strains were grown to mid-logarithmic phase and irradiated with 10 J/m2 of UV light, and the number of rifampin-resistant colonies was determined. The data are graphed as rifampin-resistant mutants per 108 survivors, and the number of spontaneous mutations conferring rifampin resistance in the absence of UV irradiation has been subtracted. The data are the means of three experiments, and the standard deviations of the means are indicated.
Examination of the ability of SM162 to invade and spread in cultured epithelial cells.
Many of the genes on the S. flexneri virulence plasmid are important for invasion of colonic epithelial cells, for intracellular survival and proliferation, and for spread to adjacent cells (28, 40). To determine whether ImpB was essential for any of these processes, we examined the ability of the ImpB mutant SM162 to form plaques on a Henle cell monolayer. SM162 formed plaques that were similar in size and number to the plaques formed by the parental strain SM100, suggesting that impB was not required for the invasion of Henle cells or for cell-to-cell spread in a plaque assay (data not shown).
Identification of the umuDC operon in S. flexneri SA100 and analysis of its induction by UV radiation.
The E. coli error-prone DNA repair system UmuDC is chromosomally encoded (20, 36). Although Shigella spp. are closely related to E. coli, S. flexneri appears to require the virulence plasmid-encoded impB gene for induced mutagenesis. This suggested that S. flexneri does not contain a functional, chromosomally encoded UmuDC system. To investigate this possibility, we designed the primers umuDCP1 and umuDC3 based on the nucleotide sequence of the E. coli umuDC operon to amplify a 2.1-kb fragment containing the umuDC operon from S. flexneri. A PCR product of this size was amplified from SA100 and SA102, which lacks the virulence plasmid. These data demonstrate that S. flexneri contains a umuDC operon and suggests that the operon is chromosomally encoded.
To begin characterization of the contribution of the SA100 umuDC operon to survival and induced mutagenesis following UV irradiation, we measured the level of induction of a umuDC promoter-gfp transcriptional fusion after exposure to UV radiation. pLR13, which contained the S. flexneri SA100 umuDC promoter-gfp transcriptional fusion, was transformed into E. coli AB1157 and S. flexneri SA100. Expression of gfp controlled by the S. flexneri umuDC promoter on pLR13 in either AB1157 or SA100 was not induced by UV irradiation. In contrast, expression of gfp controlled by the E. coli AB1157 umuDC promoter on pLR20 was induced 12-fold by UV irradiation. These data suggest that although S. flexneri SA100 contains the umuDC operon, it is not expressed in response to UV irradiation.
Sequence analysis of the S. flexneri umuDC promoter.
To verify that the lack of UV-induced expression from the S. flexneri SA100 umuDC promoter was not a result of a PCR-derived mutation in the promoter, we sequenced two independently isolated umuDC promoter clones and a umuDC promoter PCR product. Sequence analysis of these DNA fragments showed that all three sequences were identical, demonstrating that there was not a PCR-derived mutation in the umuDC promoter amplified from SA100.
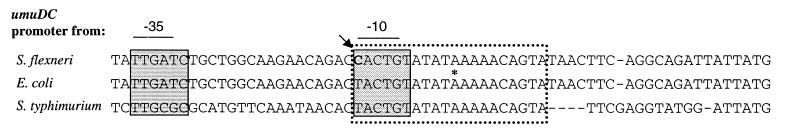
We compared the nucleotide sequence of the S. flexneri umuDC promoter with those of the E. coli and Salmonella typhimurium umuDC promoters. Over a 113-bp overlap, the S. flexneri umuDC promoter was 98% identical to the E. coli umuDC promoter and 56% identical to the Salmonella typhimurium umuDC promoter. In all three promoters, the SOS box and the −10 ς70 recognition sequence overlapped (Fig. 5). The first nucleotide of the SOS box corresponds to the first nucleotide of the −10 ς70 recognition sequence; however, this nucleotide was a T in E. coli and Salmonella typhimurium and a C in S. flexneri (Fig. 5). Although the T at position 1 in the SOS box is not essential for binding of the LexA repressor (11), it is a highly conserved nucleotide in E. coli −10 ς70 hexamers (14). Thus, the presence of a C at this position in the S. flexneri umuDC promoter may weaken the binding of RNA polymerase and contribute to the lack of UV-induced expression of the promoter.
FIG. 5.
Nucleotide sequence of the umuDC promoters from S. flexneri, E. coli, and Salmonella typhimurium. The dotted box indicates sequences corresponding to the SOS box for LexA binding, and the shaded boxes indicate sequences corresponding to the −10 and −35 hexamers for ς70 binding. An asterisk indicates the transcriptional initiation site in E. coli. The arrow points to the nucleotide that is altered in the S. flexneri umuDC promoter.
DISCUSSION
In the course of analyzing the response of S. flexneri to potentially stressful environments, including the intracellular environment, we discovered that S. flexneri contains a homologue of the impB gene, first identified on the plasmid TP110 in Salmonella typhimurium (9, 26). Our data suggest that, like the TP110-encoded impB gene, the S. flexneri impB gene is located on a large plasmid, the 220-kb virulence plasmid pVP. SA102, a derivative of S. flexneri SA100 that lacks pVP, does not contain the impB gene. Furthermore, the impB gene could be transferred to E. coli on pVP. We have not examined the location of the impB genes in other Shigella strains that contain impB; however, it is likely that impB is virulence plasmid encoded in these isolates since Shigella virulence plasmids have a high degree of similarity (40).
Based on amino acid sequence comparisons, it appears that two of the proteins encoded by the S. flexneri pVP imp operon (ImpA and ImpB) are members of the error-prone DNA repair family of proteins that contribute to survival after UV irradiation. Consistent with the sequence homology, S. flexneri SM162 and SA102, which contain an impB mutation and a deletion of the entire virulence plasmid, respectively, showed a reduced ability to survive UV irradiation. SA102 had a lower frequency of survival after UV irradiation than SM162, suggesting that pVP encodes another gene product that is important either specifically for resistance to UV irradiation or generally for survival under stressful conditions.
There is a precedent for other genes that contribute to resistance to UV irradiation. For instance, the uvr genes are important for surviving UV irradiation, but they are chromosomally encoded in E. coli (2, 3, 48). The plasmid pKM101, which encodes the MucAB error-prone DNA repair system, contains an additional uncharacterized gene which contributes to survival following UV irradiation (24). Salmonella typhimurium strains carrying a deletion derivative of pKM101, in which the mucAB genes were still present but the uncharacterized gene was absent, showed decreased survival following UV irradiation. Langer et al. (24) proposed that either this deletion in pKM101 resulted in overexpression of mucAB, leading to decreased survival following UV irradiation because of excessive levels of mutagenesis, or that the deletion removed a suppressor of a UV sensitization gene. Additionally, even among isolates of Shigella spp., there is a wide range of sensitivities to UV radiation (Table 2). For example, Shigella boydii 224860 is significantly less UV resistant than S. flexneri SA100 (P < 0.05), even though both contain impB, but is significantly more UV resistant than S. boydii O-1392 (P < 0.05), which does not contain impB. In general, among Shigella spp. isolates that contain impB, there is the following correlation between species type and UV resistance: S. flexneri = Shigella sonnei > Shigella dysenteriae > S. boydii. These observations emphasize the complexity of the UV sensitivity phenotype and suggest that although impB clearly contributes to UV resistance, Shigella spp. contain other genes that are also important for surviving UV irradiation.
S. flexneri SM162 and SA102, which contain an impB mutation and a deletion of the entire virulence plasmid, respectively, also showed decreased levels of UV-induced mutagenesis. In contrast to the defect in survival following UV irradiation, which was greater in SA102, the defect in UV-induced mutagenesis was greater in SM162. One explanation for this result is that that expression of ImpA without ImpB in SM162 interferes with other systems that are mutagenic. This would not occur in SA102 because both ImpA and ImpB are absent. Although S. flexneri contains the UmuDC error-prone DNA repair system, the data presented in this article suggest that this system is not UV induced in S. flexneri. Furthermore, the S. flexneri umuDC operon does not appear to complement the defect in UV-induced mutagenesis in an E. coli umuDC mutant (12). However, S. flexneri may possess another mutagenesis system that is weakly induced by UV irradiation and has yet to be identified. Another possibility for the less severe defect in UV-induced mutagenesis in SA102 relative to that in SM162 is that pVP may encode a suppressor of another unidentified mutagenesis system which is activated upon deletion of pVP.
The analysis of the error-prone DNA repair imp operon presented here focused on the role of the operon in survival and induced mutagenesis after exposure to UV radiation. The UV inducibility of error-prone DNA repair operons is well conserved. To our knowledge the promoters of all error-prone DNA repair operons examined to date contain binding sites for the LexA repressor. Exposure to UV radiation induces LexA-repressed operons. Although we did not test directly whether expression of the pVP-encoded imp operon is UV inducible, the fact that a well-conserved LexA binding site overlaps the putative −10 sequence by 1 nucleotide in the imp promoter suggests that the imp operon is UV inducible. Additionally, the fact that UV exposure was required for ImpB-mediated induced mutagenesis supports this hypothesis. It is also possible that a signal other than or in addition to UV radiation may induce expression of the pVP-encoded imp operon in S. flexneri. Some of these signals may be encountered in the variety of stressful environments that Shigella encounters during its journey through the external environment and human host.
There are several possible molecular mechanisms by which Shigella spp. may have acquired the imp operon. The imp operon may have been acquired by fusion of the virulence plasmid or virulence plasmid progenitor with another plasmid, such as TP110, that contained the imp operon. Alternatively, the imp operon may have been acquired as part of a transposon or other mobile genetic element. There is evidence that error-prone DNA repair operons may have been located on transposable elements. Langer et al. (25) found that inverted repeats flank a 6-kb region that contains the mucAB genes on pKM101, suggesting that these genes were part of a transposon at one time. Additionally, direct repeats similar to the termini of the Tn3 group of transposases flank a 12- to 14-kb region that contains the umuDC genes in a variety of Escherichia spp. (44). Kulaeva et al. (22) found a retroelement encoding a putative reverse transcriptase located upstream of and an insertion sequence located downstream from the mucAB genes on plasmid R471a. Finally, the large conjugative transposon Tn5252 found in many clinical streptococci contains an error-prone DNA repair system (31). The fact that not all isolates of Shigella contain the imp operon suggests that the imp operon may have been acquired by some but not all phylogenetic lines of the virulence plasmid early in the evolution of the plasmid. Alternatively, the imp operon may have been part of the progenitor virulence plasmid and this operon was subsequently lost from the virulence plasmids in some strains.
Although it is clear that the impB gene is not an essential gene in S. flexneri, it is possible that the acquisition of the imp operon by some Shigella species gives those strains a selective advantage. The presence of the imp operon on pVP most likely compensates for the inefficient UV induction of the chromosomal umuDC genes in S. flexneri. In environments in which UV radiation or other DNA-damaging agents are encountered, such as the external environment, strains containing the imp operon may survive better than strains that do not have the imp operon. If the imp operon provides a selective advantage for maintaining the virulence plasmid in the external environment, strains containing the imp operon may be more likely to have the plasmid when Shigella reencounters a human host, where the organism needs other virulence plasmid-encoded genes for survival. Additionally, it is possible that the imp operon may be protective in other stressful environments, some of which may be encountered when Shigella is inside the human host.
ACKNOWLEDGMENTS
We gratefully thank the following individuals for their generous help: Elizabeth Wyckoff, Douglas Henderson, and Stephanie Reeves for their critical reading of the manuscript; Harsha Mistry and Adrienne Garcia for excellent technical help; Stefan Seliger for strain SM100; and James Walker for the use of equipment.
This work was supported by Public Health Service Grant AI09918 awarded to L.J.R.-J. and Public Health Service Grant AI16935 awarded to S.M.P.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Arthur H M, Eastlake P B. Transcriptional control of the uvrD gene of Escherichia coli. Gene. 1983;25:309–316. doi: 10.1016/0378-1119(83)90235-4. [DOI] [PubMed] [Google Scholar]
- 3.Bagg A, Kenyon C J, Walker G C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 1981;78:5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brumby A M, Lamont I, Dodd I B, Egan J B. Defining the SOS operon of coliphage 186. Virology. 1996;219:105–114. doi: 10.1006/viro.1996.0227. [DOI] [PubMed] [Google Scholar]
- 5.Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85:1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 7.Daskaleros, T. Unpublished data.
- 8.Dodd I B, Egan J B. The Escherichia coli retrons Ec67 and Ec86 replace DNA between the cos site and a transcriptional terminator of a 186-related prophage. Virology. 1996;219:115–124. doi: 10.1006/viro.1996.0228. [DOI] [PubMed] [Google Scholar]
- 9.Dowden S B, Glazebrook J A, Strike P. UV inducible UV protection and mutation functions on the I group plasmid TP110. Mol Gen Genet. 1984;193:316–321. doi: 10.1007/BF00330687. [DOI] [PubMed] [Google Scholar]
- 10.Elledge S J, Walker G C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983;164:175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 12.Garcia, A., and L. J. Runyen-Janecky. Unpublished data.
- 13.Hale T L. Genetic basis of virulence in Shigella species. Microbiol Rev. 1991;55:206–224. doi: 10.1128/mr.55.2.206-224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong M, Altier C, Reed K, Maurer R, Payne S M. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. Differential expression of Shigella flexneri genes in vitro and in vivo, abstr. B-126; p. 187. [Google Scholar]
- 16.Howard-Flanders P, Simson E, Theriot L. A locus that controls filament formation and sensitivity to radiation in Escherichia coli K12. Genetics. 1964;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu M-Y, Inouye M, Inouye S. Retron for the 67-base multicopy single-stranded DNA from Escherichia coli: a potential transposable element encoding both reverse transcriptase and Dam methylase functions. Proc Natl Acad Sci USA. 1990;87:9454–9458. doi: 10.1073/pnas.87.23.9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kado C I, Liu S-T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato T, Shinoura Y. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol Gen Genet. 1977;156:121–131. doi: 10.1007/BF00283484. [DOI] [PubMed] [Google Scholar]
- 20.Kitagawa Y, Akaboshi E, Shinagawa H, Horii T, Ogawa H, Kato T. Structural analysis of the umu operon required for inducible mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 1985;82:4336–4340. doi: 10.1073/pnas.82.13.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 22.Kulaeva O I, Koonin E V, Wootton J C, Levine A S, Woodgate R. Unusual insertion element polymorphisms in the promoter and terminator regions of the mucAB-like genes of R471a and R446b. Mutat Res. 1998;397:247–262. doi: 10.1016/s0027-5107(97)00222-4. [DOI] [PubMed] [Google Scholar]
- 23.Kulaeva O I, Wootton J C, Levine A S, Woodgate R. Characterization of the umu-complementing operon from R391. J Bacteriol. 1995;177:2737–2743. doi: 10.1128/jb.177.10.2737-2743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langer P J, Perry K L, Walker G C. Complementation of a pKM101 derivative that decreases resistance to UV killing but increases susceptibility to mutagenesis. Mutat Res. 1985;150:147–158. doi: 10.1016/0027-5107(85)90112-5. [DOI] [PubMed] [Google Scholar]
- 25.Langer P J, Shanabruch W G, Walker G C. Functional organization of plasmid pKM101. J Bacteriol. 1981;145:1310–1316. doi: 10.1128/jb.145.3.1310-1316.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lodwick D, Owen D, Strike P. DNA sequence analysis of the imp UV protection and mutation operon of the plasmid TP110: identification of a third gene. Nucleic Acids Res. 1990;18:5045–5050. doi: 10.1093/nar/18.17.5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodwick D, Strike P. Distribution of sequences homologous to the impCAB operon of TP110 among bacterial plasmids of different incompatibility groups. Mol Gen Genet. 1991;229:27–30. doi: 10.1007/BF00264209. [DOI] [PubMed] [Google Scholar]
- 28.Menard R, Dehio C, Sansonetti P J. Bacterial entry into epithelial cells: the paradigm of Shigella. Trends Microbiol. 1996;4:220–226. doi: 10.1016/0966-842X(96)10039-1. [DOI] [PubMed] [Google Scholar]
- 29.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori H, Kondo A, Ohshima A, Ogura T, Hiraga S. Structure and function of the F plasmid genes essential for partitioning. J Mol Biol. 1986;192:1–15. doi: 10.1016/0022-2836(86)90459-6. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Najar U, Sampath J, Vijayakumar M N. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Characterization of a region conferring resistance to UV light in the conjugative transposon Tn5252, abstr. H-11; p. 278. [Google Scholar]
- 32.Nohmi T, Battista J R, Dodson L A, Walker G C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci USA. 1988;85:1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nohmi T, Hakura A, Nakai Y, Watanabe M, Murayama S Y, Sofuni T. Salmonella typhimurium has two homologous but different umuDC operons: cloning of a new umuDC-like operon (samAB) present in a 60-megadalton cryptic plasmid of S. typhimurium. J Bacteriol. 1991;173:1051–1063. doi: 10.1128/jb.173.3.1051-1063.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oaks E V, Wingfield M E, Formal S B. Plaque formation by virulent Shigella flexneri. Infect Immun. 1985;48:124–129. doi: 10.1128/iai.48.1.124-129.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Payne S M, Niesel D W, Peixotto S S, Lawlor K M. Expression of hydroxamate and phenolate siderophores by Shigella flexneri. J Bacteriol. 1983;155:949–955. doi: 10.1128/jb.155.3.949-955.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82:4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman M F, Echols H. Activity of the purified mutagenesis proteins UmuC, UmuD′, and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. Proc Natl Acad Sci USA. 1992;89:10777–10781. doi: 10.1073/pnas.89.22.10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sansonetti P J. Molecular and cellular biology of Shigella flexneri invasiveness: from cell assay systems to shigellosis. Curr Top Microbiol Immunol. 1992;180:1–19. doi: 10.1007/978-3-642-77238-2_1. [DOI] [PubMed] [Google Scholar]
- 40.Sasakawa C, Buysse J M, Watanabe H. The large virulence plasmid of Shigella. Curr Top Microbiol Immunol. 1992;180:21–44. doi: 10.1007/978-3-642-77238-2_2. [DOI] [PubMed] [Google Scholar]
- 41.Sedgwick S G, Goodwin P A. Differences in mutagenic and recombinational DNA repair in enterobacteria. Proc Natl Acad Sci USA. 1985;82:4172–4176. doi: 10.1073/pnas.82.12.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sedgwick S G, Ho C, Woodgate R. Mutagenic DNA repair in enterobacteria. J Bacteriol. 1991;173:5604–5611. doi: 10.1128/jb.173.18.5604-5611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sedgwick S G, Lodwick D, Doyle N, Crowne H, Strike P. Functional complementation between chromosomal and plasmid mutagenic DNA repair genes in bacteria. Mol Gen Genet. 1991;229:428–436. doi: 10.1007/BF00267466. [DOI] [PubMed] [Google Scholar]
- 44.Sedgwick S G, Robson M, Malik F. Polymorphisms in the umuDC region of Escherichia species. J Bacteriol. 1988;170:1610–1616. doi: 10.1128/jb.170.4.1610-1616.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shearwin K E, Brumby A M, Egan J B. The Tum protein of coliphage 186 is an antirepressor. J Biol Chem. 1998;273:5708–5715. doi: 10.1074/jbc.273.10.5708. [DOI] [PubMed] [Google Scholar]
- 46.Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85:1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shinagawa H, Kato T, Ise T, Makino K, Nakata A. Cloning and characterization of the umu operon responsible for inducible mutagenesis in Escherichia coli. Gene. 1983;23:167–174. doi: 10.1016/0378-1119(83)90048-3. [DOI] [PubMed] [Google Scholar]
- 48.Siegel E C. The Escherichia coli uvrD gene is inducible by DNA damage. Mol Gen Genet. 1983;191:397–400. doi: 10.1007/BF00425753. [DOI] [PubMed] [Google Scholar]
- 49.Smith C M, Koch W H, Franklin S B, Foster P L, Cebula T A, Eisenstadt E. Sequence analysis and mapping of the Salmonella typhimurium LT2 umuDC operon. J Bacteriol. 1990;172:4964–4978. doi: 10.1128/jb.172.9.4964-4978.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundin G W, Kidambi S P, Ullrich M, Bender C L. Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulAB genes. Gene. 1996;177:77–81. doi: 10.1016/0378-1119(96)00273-9. [DOI] [PubMed] [Google Scholar]
- 51.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thomas S M, Crowne H M, Pidsley S C, Sedgwick S G. Structural characterization of the Salmonella typhimurium LT2 umu operon. J Bacteriol. 1990;172:4979–4987. doi: 10.1128/jb.172.9.4979-4987.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walker G C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48:60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 56.Woodgate R, Sedgwick S G. Mutagenesis induced by bacterial UmuDC proteins and their plasmid homologues. Mol Microbiol. 1992;6:2213–2218. doi: 10.1111/j.1365-2958.1992.tb01397.x. [DOI] [PubMed] [Google Scholar]
- 57.Wyckoff, E. Unpublished data.
- 58.Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 1998;17:3207–3216. doi: 10.1093/emboj/17.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yasuda T, Nagata T, Ohmori H. Multicopy suppressors of the cold-sensitive phenotype of the pcsA68 (dinD68) mutation in Escherichia coli. J Bacteriol. 1996;178:3854–3859. doi: 10.1128/jb.178.13.3854-3859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]