Summary
Large-scale, high-throughput specificity assays to characterize binding properties within a competitive and complex environment of potential binder-target pairs remain challenging and cost prohibitive. Barcode cycle sequencing (BCS) is a molecular binding assay for proteins, peptides, and other small molecules that is built on a next-generation sequencing (NGS) chip. BCS uses a binder library and targets labeled with unique DNA barcodes. Upon binding, binder barcodes are ligated to target barcodes and sequenced to identify encoded binding events.
For complete details on the use and execution of this protocol, please refer to Hong et al. (2022).
Subject areas: Genomics, Sequencing, High throughput screening, Molecular biology
Graphical abstract
Highlights
-
•
Binding events are encoded in DNA barcodes that identify both binders and targets
-
•
Binding events occur directly on a DNA sequencing chip for direct sequencing
-
•
Binding events can be multiplexed and scaled to record billions of events
-
•
Colocalization of a target and foundation allows multiple rounds of BCS on a chip
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Large-scale, high-throughput specificity assays to characterize binding properties within a competitive and complex environment of potential binder-target pairs remain challenging and cost prohibitive. Barcode cycle sequencing (BCS) is a molecular binding assay for proteins, peptides, and other small molecules that is built on a next-generation sequencing (NGS) chip. BCS uses a binder library and targets labeled with unique DNA barcodes. Upon binding, binder barcodes are ligated to target barcodes and sequenced to identify encoded binding events.
Before you begin
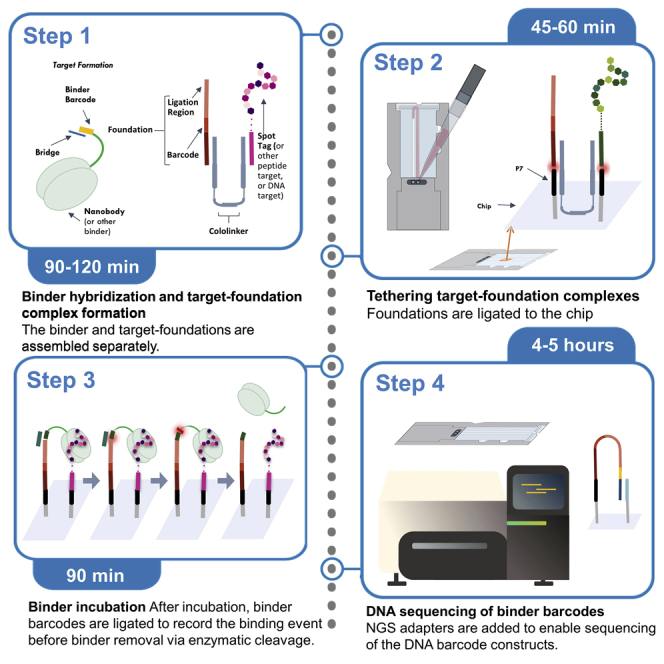
The concept of the BCS workflow is outlined in the Graphical Abstract. The key components required for BCS include: 1) preparation of binders and targets for use on a NGS chip, 2) attachment of targets to the NGS chip, 3) a mechanism for binders to deposit their unique DNA barcodes onto the chip at the binding site, and 4) DNA sequencing of the binder barcodes may be used to identify the binders. Multiple rounds of binding and ligation (step 3) may be performed before DNA sequencing. The mechanism for barcode deposition requires the presence of a short oligonucleotide sequence called a “foundation”, which is positioned beside each peptide target to act as a receiver for binder barcodes. Each foundation contains a DNA barcode unique to its corresponding target, which is used to identify the target.
This assay may be adapted for use with any molecular binder-target pair (ex. small molecule, quantum dot, protein and RNA, etc.) where both the binder and the target are amenable to conjugation with oligonucleotides, and where the presence of oligonucleotide tails does not significantly perturb binding affinity and specificity. The buffers described here were designed specifically to optimize binding dynamics between and structural stability of our binders and targets of interest, aptamers, and peptides. Because of the flexibility of the method to assay the binding properties of various types of molecules, it is important for users to adjust binding buffers and temperatures to facilitate binding and structural stability for their system. Users may choose to replace the recommended buffers and incubation parameters suitable to the user’s molecules of interest and the environment of their end application(s).
Here we demonstrate a single cycle of BCS using the Spot Tag system, a 12-amino acid fSpot-Tag and anti-Spot-Tag nanobody engineered to recognize the Spot-Tag specifically and with high affinity (dissociation constant Kd of approximately 6 nM) (Virant et al., 2018; Braun et al., 2016). The nanobody is approximately 10-fold smaller than an antibody (approximately 15 kDa vs 150 kDa), is a stable and robust reagent, is commercially available from Chromotek as a bivalent construct (two anti-Spot-Tag nanobodies genetically fused via a linker), and includes a C-terminal recognition motif LPETG for site-specific sortase-mediated conjugation of the protein to small molecules (Antos et al., 2017). Furthermore, conjugation of nanobodies to oligonucleotide tails has been reported and does not appear to affect nanobody functionality (Fabricius et al., 2018).
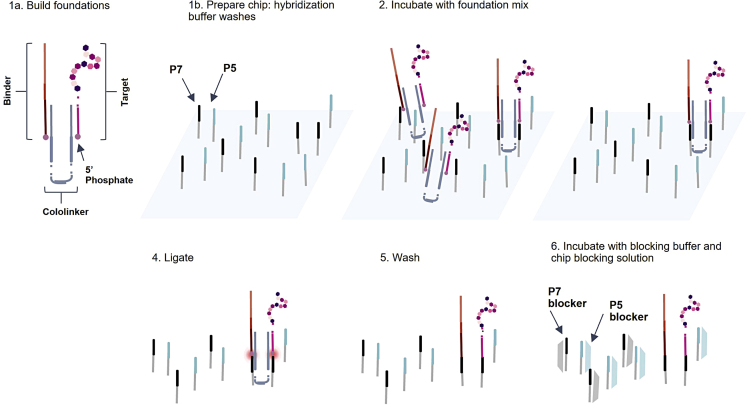
Peptide targets are displayed on the NGS chip through ligation to pre-existing P7 oligonucleotide adapters (Figure 1). Each peptide target exists as a peptide-oligonucleotide conjugate (POC), where a short, barcoded oligonucleotide is covalently attached to the C-terminus. Colocalization of the target and foundation on the chip is achieved by first forming a target-foundation complex in solution. Two short oligonucleotides called the “forward” and “reverse cololinkers” have regions of complementarity to the binder and target, as well as to pre-existing P7 oligonucleotide adapters on the Illumina DNA sequencing chip. The term cololinker is short for “colocalization linker,” since the cololinkers link together the POC and foundation (Figure 1, step 1a) so that they may colocalize on the chip. Experimental design can include negative control targets such as empty foundations (DNA foundation pieces without any targets colocalized) to measure signal-to-noise ratio and off-target binder-DNA binding, or scramble peptides to control for nonspecific, off-target binding.
Figure 1.
Ligating colocalized targets and foundations to pre-existing adapters on the NGS chip
The chip is rinsed with a buffer solution (Figure 1, step 1b) and the target-foundation complex is flowed onto the chip (Figure 1, step 2). The cololinkers facilitate binding of the target-foundation complex to P7 adapters on the chip, and unbound elements are washed away (Figure 1, step 3). The target and foundation are ligated to the 3′ ends of two adjacent P7 adapters (Figure 1, step 4). Unligated elements are washed away (Figure 1, step 5), and the single-stranded regions of P5 and P7 are blocked using complementary DNA (Figure 1, step 6).
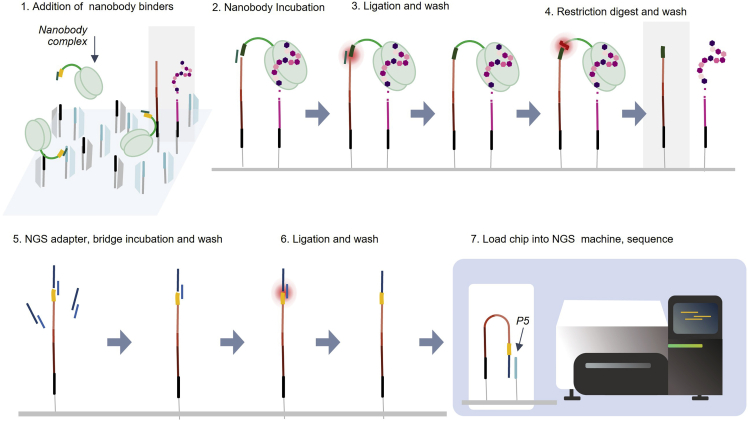
Each nanobody binder contains a DNA barcode whose purpose is to identify the binder. Binding events are recorded on the chip by transferring the binder’s DNA barcode onto the foundation (Figure 2). Before introducing binders to the NGS chip, a nanobody complex (Figure 2, step 1) is created by annealing the nanobody 5′ end to a short “universal bridge” oligonucleotide to form a ligation site. Upon binding the target (Figure 2, step 2), the 5′ binder barcode is ligated to the 3′ end of the foundation, and the universal bridge is washed away (Figure 2, step 3). The binder tail is cleaved downstream of the binder barcode and washed off the chip (Figure 2, step 4), leaving the binder barcode ligated to the foundation. Optionally, multiple cycles of binding, ligation, and cleavage may be performed, where each new binder barcode is ligated to the 3′ end of the previous binder barcode (not depicted). Assisted by a short universal NGS adapter bridge oligonucleotide, the universal NGS adapter for DNA sequencing is then ligated to the binder barcode (Figure 2, steps 5 and 6), and the DNA construct is sequenced (Figure 2, step 7).
Figure 2.
Capturing binding on the NGS chip via the ligation of binder barcodes to foundations
In steps 1–4, binder components include the universal bridge (dark green), binder barcode (yellow), and binder tail (light green). In steps 5 and 6, the NGS adapter and bridge are depicted in dark and light blue, respectively.
Designing DNA components of BCS
The following sections describe general principles which apply to designing DNA components of the BCS platform, including the foundation, POCs, cololinkers, binder barcodes, universal bridge, and blocking sequences.
All oligonucleotide components may be ordered at 100 nM scale with high-pressure liquid chromatography purification and resuspended at 100 μM concentration in nanopure, nuclease-free (NF) H2O. All DNA components may be stored at −20°C.
CRITICAL: Screen every designed sequence against other BCS DNA components to avoid undesired binding complexes. An online tool such as IDT Oligo Analyzer may be used for screening. We have observed as few as four consecutive complementary nucleotides between BCS components to induce off-target binding.
Designing the foundation
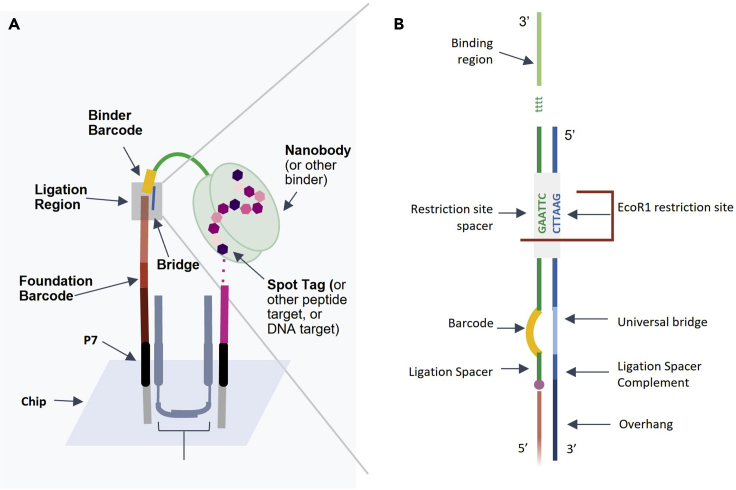
The purpose of this section is to design the foundation sequence. The foundation provides a site for a binder to deposit its barcode beside the peptide target. The 5′-phosphorylated end of the foundation is designed to be ligated to the existing P7 sequence on the chip. The foundation base is complementary to the 5′ end of the forward cololinker. The foundation barcode is a short oligonucleotide sequence unique to each peptide target. The bridge-binding sequence is complementary to the 3′ end of the universal bridge. The 3′ end of the foundation serves as the ligation site for a binder barcode (Figure 4A).
-
1.Design foundations.
-
a.Generate a foundation base sequence according to general principles of primer design (https://eu.idtdna.com/pages/education/decoded/article/designing-pcr-primers-and-probes). The 5′ end must be phosphorylated for successful ligation.
-
b.Generate a unique foundation barcode for each peptide target.
-
i.Barcodes should be selected such that they minimize the number of homopolymers, constrain the GC content to a desired range, and minimize similarity between sequences. In practice we used a max homopolymer length of 2, GC range of 0.4–0.6, and a minimum distance of 2 between all barcode pairs. Examples of validated barcodes are provided in Table 1.
-
i.
-
c.Generate a bridge-binding sequence according to general principles of primer design.
-
a.
Note: We observed that certain foundation barcodes affected the efficiency of peptide deposition onto the chip. To quantify these differences, we performed a single cycle of BCS on one target using every foundation barcode we designed. For each target-foundation pair, we measured (1) the seeding efficiency of the target-foundation pair onto the P7 sequences on the flow cell, and (2) the efficiency of binder barcode capture. We ultimately selected foundations that produced reliable seeding density and binder barcode capture, as well as a low incidence of nonspecific binding.
-
2.
Order the foundations and resuspend them at 100 μM in NF H2O.
Pause point: Store stably at −20°C.
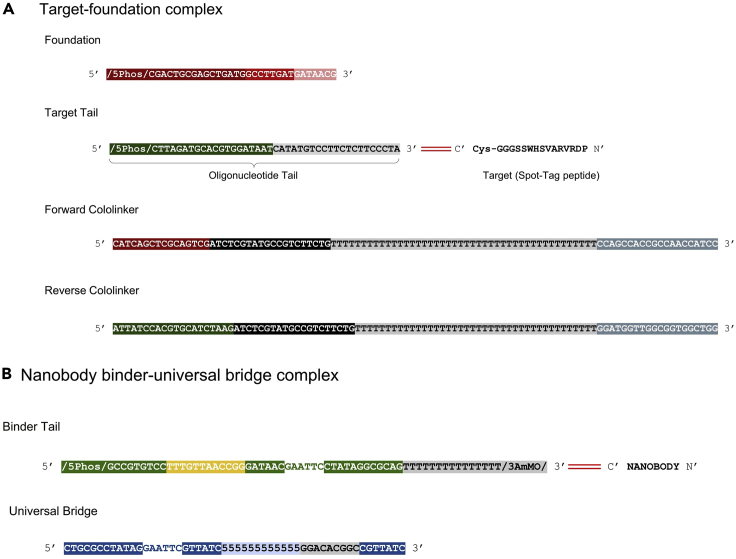
Figure 4.
DNA sequences of BCS components
(A) The foundation consists of a 16-nt foundation base, 8-nt foundation barcode, and 7-nt bridge-binding sequence. The target tail consists of a 5′ phosphate (represented as “/5Phos/”), 20-nt target base sequence, and a 21- or 25-nt spacer. Red bars symbolize covalent linkage to the target. The forward cololinker consists of 16-nt foundation base complement, 20-nt P7 complement, 44-nt T spacer, and 20-nt hybridization region. The reverse cololinker consists of a 20-nt target base complement, 20-nt P7 complement, 44-nt T spacer, and 20-nt hybridization region.
(B) The binder tail consists of a 9-nt ligation spacer, 12-nt binder barcode, 24-nt restriction site spacer (containing an EcoR1 site), and 15-nt T-spacer. Red bars indicate covalent linkage to the nanobody binder. The universal bridge consists of a 5′ region complementary to the restriction site spacer (containing the EcoRI restriction site), a region of 12 nucleotides of 5-nitroindole universal bases (represented as “5”) that can hybridize with any binder barcode, a ligation spacer complement, and a 3′ overhang complementary to the 5′ region of all foundations or previously ligated binder barcode sequences.
Table 1.
Examples of validated foundations
| Foundation name | Sequence |
|---|---|
| Fd7 | /5Phos/CGACTGCGAGCTGATGGCCTTGATGATAACG |
| Fd8 | /5Phos/CGACTGCGAGCTGATGCGTACTAGGATAACG |
| Fd11 | /5Phos/CGACTGCGAGCTGATGTGTACGCAGATAACG |
| Fd12 | /5Phos/CGACTGCGAGCTGATGCGTTTGCAGATAACG |
| Fd13 | /5Phos/CGACTGCGAGCTGATGTCTTTCCGGATAACG |
| Fd14 | /5Phos/CGACTGCGAGCTGATGTTGCTCACGATAACG |
| Fd15 | /5Phos/CGACTGCGAGCTGATGGAGTTACGGATAACG |
| Fd16 | /5Phos/CGACTGCGAGCTGATGTGATATAGGATAACG |
| Fd17 | /5Phos/CGACTGCGAGCTGATGACCTTAGAGATAACG |
| Fd18 | /5Phos/CGACTGCGAGCTGATGAGTTGCTTGATAACG |
| Fd19 | /5Phos/CGACTGCGAGCTGATGAGGTACCAGATAACG |
| Fd20 | /5Phos/CGACTGCGAGCTGATGCACTTACGGATAACG |
| Fd21 | /5Phos/CGACTGCGAGCTGATGTTGGGCAAGATAACG |
| Fd22 | /5Phos/CGACTGCGAGCTGATGTTGGGCAAGATAACG |
| Fd23 | /5Phos/CGACTGCGAGCTGATGTTCCACGTGATAACG |
| Fd24 | /5Phos/CGACTGCGAGCTGATGAGGAGCAAGATAACG |
| Fd25 | /5Phos/CGACTGCGAGCTGATGTTCCCTTCGATAACG |
| Fd26 | /5Phos/CGACTGCGAGCTGATGTCTGAGGTGATAACG |
| Fd27 | /5Phos/CGACTGCGAGCTGATGTCATGTGGGATAACG |
| Fd28 | /5Phos/CGACTGCGAGCTGATGCACCAAACGATAACG |
| Fd29 | /5Phos/CGACTGCGAGCTGATGATTGTCCCGATAACG |
| Fd31 | /5Phos/CGACTGCGAGCTGATGTGGCATCTGATAACG |
| Fd32 | /5Phos/CGACTGCGAGCTGATGCTTCTAGCGATAACG |
| Fd43 | /5Phos/CGACTGCGAGCTGATGCAGCACATGATAACG |
Designing and creating target-oligonucleotide conjugates
The purpose of this section is to design target POCs. Each target is displayed on the chip via covalent attachment to a short single-stranded oligonucleotide tail which is eventually ligated at the 5′-phosphorylated end to an existing P7 adapter on the chip.
-
3.Design the oligonucleotide tail (Figure 4A).
-
a.Generate the target base sequence according to general principles of primer design. The 5′ end must be phosphorylated for successful ligation.
-
b.Design a spacer region of approximately 21 nucleotides.
-
a.
-
4.Purchase or synthesize target-oligonucleotide conjugates.
-
a.Synthesis may be achieved through commercially available conjugation kits or chemical techniques such as click chemistry (Fantoni et al., 2021).
-
a.
Note: The Spot-tag peptide-oligonucleotide conjugates (POCs) used in this protocol were manufactured by GenScript. Users interested in creating their own POCs may contact lead contact to request further details about our in-house method developed for POC synthesis.
-
5.
Resuspend POCs in 100 μL and separate into single-use 10 μM aliquots to avoid freeze-thaw cycles.
Pause point: Store stably at −20°C.
Designing forward and reverse cololinkers
The forward and reverse cololinkers anneal to one another at their 3′ ends to form a U-shaped complex between the foundation, POC, and P7 adapters on the NGS chip. Both the forward and reverse cololinkers include a region of complementarity to P7 on the chip, allowing the forward cololinker in the assembled complex to bring the 3′ end of the foundation into proximity with a P7 adapter on the chip and the reverse cololinker to bring the 3′ end of the POC into proximity with another nearby P7 adapter (Figure 4A).
-
6.
Design the forward and reverse cololinkers with complementarity to the appropriate regions as depicted in Figure 4A.
Note: These oligonucleotides are not modified with 5′ phosphorylation (to avoid non-specific ligation to the cololinkers).
-
7.
Order the cololinkers and resuspend them at 100 μM in NF H2O.
-
8.
Create a cololinker stock solution containing a 3:1 ratio of forward to reverse cololinker in the Hybridization Buffer.
Pause point: Store stably at −20°C.
Designing the binder tail and binder barcodes
The purpose of this section is to design the binder tail that contains the DNA barcode used to identify the binder. On the 5′ phosphorylated end of the binder tail is a short “ligation spacer” complementary to the universal bridge. When the universal bridge anneals to the ligation spacer, a short double-stranded region is created to facilitate ligation between the binder tail and foundation. Following the ligation spacer is a 6- or 8-nt binder barcode unique to each binder and another short region of complementarity to the universal bridge. A downstream restriction enzyme site allows the binder to be cleaved and washed away, leaving the binder barcode attached to the foundation (Figure 4B). The 3′ end of the binder tail is conjugated to the C-terminus of the binder (anti-Spot-Tag nanobody).
-
9.
Design the binder barcodes (see designing the foundation section for barcode design guidelines).
Note: We noted similarities in performance between 6- and 8-nt barcodes. 8-nt barcodes may be useful in specific cases (ex. experiments with multiple binding cycles, NGS of the reverse read).
Note: We tested binder tail sequences with and without the ligation and restriction site spacers. The addition of spacers improved enzyme activity, presumably by extending the double-stranded region surrounding the single-stranded break (Doherty and Suh, 2000). A shorter T-spacer may be used.
-
10.Conjugate the anti-Spot-Tag nanobody to the binder tail.
-
a.Various conjugation methods may be used, including the SoluLINK Protein-Oligonucleotide Conjugation Kit (Vector Laboratories) and sortase-mediated conjugation (Antos et al., 2017). In Hong et al. (2022), we describe a conjugation method using the SoluLINK kit according to manufacturer instructions.
-
a.
Pause point: Store stably at −20°C.
Designing chip blocking components
Various DNA components of the BCS platform may unintentionally affect binder-target interactions. We found that blocking several ssDNA regions using complementary DNA reduced these effects. The blocked ssDDNA components included the POC oligonucleotide tail and the Illumina P7 and P5 adapters. These blocking oligonucleotides are used in the Chip Blocking Solution.
-
11.
Design and order blocking sequences complementary to the POC tail, Illumina P5 sequence, and Illumina P7 sequence.
-
12.
Resuspend each complementary sequence at 100 μM in NF H2O.
Pause point: Store stably at −20°C.
Designing the universal bridge
The universal bridge anneals to the 3′ end of the foundation and the 5′ end of the binder to create a double-stranded region that facilitates ligation between the two. The universal bridge contains an 8- or 12-nt universal base region intended to anneal to the variable binder barcode, as well as the EcoRI restriction site to allow the binding region of the binder to be cleaved and eventually washed away for subsequent rounds of binding or NGS adaptor ligation for sequencing (Figure 4B).
-
13.Design the universal bridge.
-
a.Design complementarity to the foundation and binder tail as shown in Figure 3B.
-
a.
Note: The signal to noise ratio observed after DNA sequencing was comparable between runs using the universal 5-nitroindole bridge and runs using bridge sequences with complementarity to specific binder barcodes. The universal sequence was chosen for ease of use.
Note: We observed that ligation efficiency improved with universal bridges of longer length. Our bridge length was optimized for ligation efficiency balanced against purity and reagent cost.
-
14.
Order the universal bridge and resuspend at 100 μM in NF H2O.
Pause point: Store stably at −20°C.
Figure 3.
Composition of BCS target-foundation complex
(A) A target-foundation complex assembled on the NGS chip.
(B) Magnification of the ligation site between binder tail and foundation. The left-sided DNA strand depicts the 5′ end of the nanobody binder tail (green and yellow) with 5′ phosphorylation (pink circle) and 3′ end of the foundation (orange). The right-sided DNA strand (blue) depicts the universal bridge sequence.
Preparing the NGS sequencer and chip
Timing: varies
The purpose of this section is to describe considerations for NGS chip selection and sequencer reprogramming. When choosing an NGS chip, the user should consider the number of desired sites for target-foundation pairs and the number of successful reads balanced against the cost of each sequencing run. For experiments with read counts under one million, we recommend a V2 nano MiSeq chip. For experiments requiring greater read counts, we recommend V3 chips.
-
15.
Remove the initial washing and library loading steps from the cartridge to the sequencing chip on the MiSeq instrument.
CRITICAL: Modified run instructions on a commercial sequencer may no longer be appropriate for standard DNA sequencing experiments. Users should be familiar with storing a copy of the original code file and reverting the sequencer to run its original code if they wish to continue utilizing the sequencer for standard procedures.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Spot-Tag Experiment Spot-Tag∗ (peptide target) target called Spot-Tag.O1 (N-terminus)-PDRVRA VSHWSSGGG-Cys (C-terminus)-3′ATCCCTTCTCTTCC TGTATACTAATAGGTGCACGTAGATTC/5Phos/ |
Hong et al., 2022 | |
| Spot-Tag Experiment Bradykinin∗ (peptide target control for non-specific binding) target called Brady.O1 (N-terminus)-RPPGFSPFR-Cys (C-terminus)-3′ATCC CTTCTCTTCCTGTATACTAATAGGTGCACGTA GATTC/5Phos/ |
Hong et al., 2022 | |
| EcoRI | NEB | Cat#R0101S |
| Cutsmart buffer | NEB | Cat#B6004 |
| Hybridization buffer (0.025% TWEEN20 in no 1× PBS) | This paper | |
| Blocking buffer (0.025% TWEEN20 in 1× PBS + 10 mg/mL BSA) | This paper | |
| Chip blocking buffer (10 μM of P5 Complementary oligo (5′-TCTCGGTGGTCGCCGTATCATT-3′)/P7 Complementary oligo (5′-ATCTCGTATGCCGTCT TCTGCTTG-3′) sequences + 10 μM POC Tail blocking sequence (5′-TAGGGAAGAGAAGGACATATGATTATCC ACGTGCATCTAAG-3′) in 60 μL of Blocking Buffer) |
This paper | |
| Incubation buffer (0.025% TWEEN20 in 1× PBS + 0.1 mg/mL BSA) | This paper | |
| Bovine serum albumin (BSA) | Sigma-Aldrich | A2153-50G |
| Phosphate buffered saline (PBS) | Thermo Fisher Scientific | AM9624 |
| TWEEN20 | Sigma-Aldrich | P9416 |
| SimplyBlue SafeStain | Thermo Fisher Scientific | LC6060 |
| Glycerol | Sigma-Aldrich | G5516-500ML |
| Formamide | Sigma-Aldrich | 11814320001 |
| T4 DNA ligase | NEB | M0202S |
| HT1 buffer | Illumina | 20015892 |
| 70% ethanol | Fisher Scientific | BP8201-500 |
| Critical commercial assays | ||
| MiSeq Reagent Nano Kit v2 (300-cycles) | Illumina | MS-103-1001 |
| MiSeq Reagent Nano Kit v2 (500 cycles) | Illumina | MS-103-1003 |
| Miseq Reagents Kits v2 (50 Cycles) | Illumina | MS-102-2001 |
| MiSeq® Reagent Kit v3 (150 cycle) | Illumina | MS-102-3001 |
| PhiX Control v3 | Illumina | FC-110-3001 |
| Blunt/TA Ligase Master Mix | NEB | Cat#M0367L |
| SoluLINK Protein-Oligonucleotide Conjugation Kit | Vector Laboratories | S-9011-1 |
| Deposited data | ||
| Raw sequencing data for BCS | Mendeley | https://data.mendeley.com/datasets/f9hdn5xc3v/1 |
| Oligonucleotides | ||
| All oligonucleotide sequences are listed throughout the protocol where relevant | This paper | N/A |
| Software and algorithms | ||
| MiSeq Control Software | Illumina | https://www.illumina.com/systems/sequencing-platforms/miseq/products-services/miseq-control-software.html |
| Colab | https://colab.research.google.com/ | |
| Custom analysis code | GitHub | https://github.com/google-research/google-research/tree/master/protseq |
| Other | ||
| MiSeq 500 | Illumina | SY-410-1003 |
| 1.5 mL microfuge tubes, DNA LoBind | Eppendorf | cat#022431021 |
| 96-well plates, DNA Lo-Bind | Eppendorf | 0030129512 |
| Mastercycler® nexus gradient, 115 V/50–60 Hz (US) | Eppendorf | 6331000025 |
| Mastercycler® nexus eco, 115 V/50–60 Hz (US) | Eppendorf | 6332000029 |
| Mastercycler® nexus flat eco, 110 V/50–60 Hz (JP/South America/TW/US) | Eppendorf | 6330000021 |
| Adhesive PCR Plate Seals | Thermo Fisher Scientific | AB0558 |
CRITICAL: Formamide is toxic and all steps involving formamide should be performed in an appropriate fume hood.
Note: All oligonucleotides are suspended in nanopure, NF H2O and stored at −20°C prior to use.
Materials and equipment
Hybridization Buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| TWEEN20 | 0.025% | 1 μM |
| 1× PBS | 1× | 1,000 mL |
Store at 25°C for up to 1 month.
Blocking buffer
Dissolve the BSA in 1× PBS first before adding TWEEN20.
| Reagent | Final concentration | Amount |
|---|---|---|
| TWEEN20 | 0.025% | 25 μL |
| BSA | 10 mg/mL | 10 g |
| 1× PBS | 1× | 1,000 mL |
Store at 25°C for up to 1 month.
Chip Blocking Solution
| Reagent | Amount | Stock concentration | Final concentration |
|---|---|---|---|
| P5 Complementary oligo (5′-TCTCGGTGGTCGCCGTATCATT-3′)/P7 Complementary oligo (5′-ATCTCGTATGCCGTCTTCTGCTTG-3′) sequences | 10 μL | 100 μM | 10 μM |
| Peptide-Oligo Conjugate Tail blocking sequence (5′-TAGGGAAGAGAAGGACATATGATTATCCACG TGCATCTAAG-3′) |
10 μL | 100 μM | 10 μM |
| Blocking Buffer (see above) | 80 μL | 1× | N/A |
| Total | 100 μL |
Prepare a fresh stock for every experiment.
Incubation buffer
Dissolve the BSA in 1× PBS first before adding TWEEN20.
| Reagent | Final concentration | Amount |
|---|---|---|
| TWEEN20 | 0.025% | 25 μL |
| BSA | 0.1 mg/mL | 100 mg |
| 1× PBS | 1× | 1,000 mL |
Store at 25°C for up to 1 month.
Step-by-step method details
Creating target-foundation complexes
Timing: 90–120 min
In this section, the cololinkers, foundation, and target are hybridized in solution to form a U-shaped complex (Figure 3A). The creation of this complex is required to ensure that targets and their respective foundations colocalize on the sequencing chip. In this protocol, the total concentration of foundations is 120 pM. Concentrations should be optimized according to the number of targets included (see troubleshooting section problem 1 for more details).
-
1.
Thaw cololinker stock solution, foundation, and Spot-tag peptide oligonucleotide conjugate on ice.
-
2.
Prepare buffers detailed in the materials and equipment section and the PhiX solution according to manufacturer instructions.
Note: Buffers may be prepared in bulk for subsequent runs, except for the chip blocking solution.
-
3.In a 96-well plate, combine the Hybridization Buffer, cololinker stock solution, and foundations in the order listed below. Add the target last, immediately prior to hybridization.
-
a.For positive controls, we used DNA targets (DNA Target 4.O1 and 6.O1), where binding occurs through complementary regions on the target and binder. For null control targets, we used a version of the peptide oligonucleotide tail without 5′ phosphorylation (CLR.Null.Block) and another without the peptide attached (5′Phos.01).
-
a.
| Reagent | Stock concentration | Amount | Final concentration |
|---|---|---|---|
| Hybridization Buffer | N/A | 91 μL | 91% |
| Cololinker mixture (1 μM stock has FC:RC 3:1) | 1 μM | 1 μL | 10 nM |
| Foundation | 1 μM | 5 μL | 50 nM |
| Target | 10 μM | 3 μL | 300 nM |
| Total | N/A | 100 μL | N/A |
| Target type | Target name | Sequence | Foundations used in replicates |
|---|---|---|---|
| Spot-Tag∗ (peptide target) | Spot-Tag.01 | (N-terminus)-PDRVRAVSHWSSGGG-Cys (C-terminus)-3′ATCCCTTCTCTTCCTGTAT ACTAATAGGTGCACGTAGATTC/5Phos/ |
Fd31, Fd19, Fd20, Fd27, Fd28, Fd29 |
| Bradykinin∗ (peptide target control for non-specific binding) |
Brady.01 | (N-terminus)-RPPGFSPFR-Cys (C-terminus)-3′ATCCCTTCTCTTCCTGTAT ACTAATAGGTGCACGTAGATTC/5Phos/ |
Fd12, Fd13, Fd14 |
| DNA∗∗ (null control) |
CLR.Null.Block | CTTAGATGCACGTGGATAAT | Fd24, Fd25, Fd26 |
| DNA∗∗ (null control) |
5′Phos.01 | /5Phos/CTTAGATGCACGTGGATA | Fd7, Fd8, Fd11 |
| DNA∗∗ (positive control) |
DNA Target 6.01 | /5Phos/CTTAGATGCACGTGGATAATCATA TGTCCTTCTCTTCCCTAATGAAGTAC TAA CCTGA |
Fd21, Fd22, Fd23 |
| DNA∗∗ (positive control) |
DNA Target 4.01 | /5Phos/CTTAGATGCACGTGGATAATCATA TGTCCTTCTCTTCCCTAATAGGATTCC | Fd15, Fd16, Fd17 |
∗The C-terminal of the peptide targets is directly conjugated to the 3′ end of one DNA tail via a cysteine.
∗∗Binding sequences and DNA tails of DNA targets are continuous oligos rather than conjugated through another chemical conjugation method.
Note: The final molecular ratios for individual components are 5:1 foundation to forward cololinker, 3:1 forward cololinker to reverse cololinker, and 10:1 Target to reverse cololinker.
-
4.
Thermocycle under the following conditions:
| Temperature | Time |
|---|---|
| 95°C | 5 min |
| 85°C | 1 min |
| 75°C | 2 min |
| 65°C | 3 min |
| 55°C | 5 min |
| 45°C | 5 min |
| 35°C | 5 min |
| 24°C | 40 min |
Note: While hybridizing target-foundation complexes on the thermocycler, begin preparing binders via hybridization to the universal bridge (see step 16).
Displaying target-foundation complexes on the NGS chip
Timing: 45–60 min
In this section, the target-foundation complex is ligated onto the chip, and the chip is blocked to decrease non-specific binding to the chip surface and ligated sequences (Figure 2).
-
5.Create the Foundation Mix.
-
a.Mix gently by pipetting up and down.
-
a.
| Reagent | Volume |
|---|---|
| Hybridization Buffer | 44 μL |
| 2× Blunt/TA Ligase Master Mix | 10 μL |
| Target-Foundation Complex | 1.14 μL (120 pM final concentration) |
CRITICAL: The overall concentration in solution should not exceed 120 pM for optimal clustering during sequencing runs.
-
6.
Dilute each target mixture to 0.5 nM.
-
7.
Carefully remove the chip from its original cylindrical container. Blot dry to remove salts left from the chip storage buffer.
Note: Retain the original storage buffer in the cylindrical container and store at 4°C until sequencing has been performed. This buffer can be used to temporarily store the chip if the user needs to pause the experiment. See Pause Point after step 43.
-
8.Wash the sequencing chip twice.
-
a.To wash, inject 100 μL of the Hybridization Buffer into either port on the sequencing chip (Figure 5).
-
a.
-
9.
Wash the sequencing chip with 30 μL of Foundation Mix twice.
Note: Blocking components should be added in greater than or equal to 2 times in excess of the component the user intends to block.
Note: The BSA in blocking solution is intended to block off-target binding to the glass surface of the sequencing chip.
-
10.
Incubate the sequencing chip at 28°C for 15 min on a hotplate.
-
11.
Wash the sequencing chip once with 100 μL of 100% formamide to remove unligated elements.
CRITICAL: All steps involving formamide should be performed in an appropriate certified chemical fume hood.
-
12.
Heat the sequencing chip at 40°C for 90 s on a hotplate.
-
13.
Wash the sequencing chip with 500 μL of Blocking Buffer.
-
14.
Wash the sequencing chip with 30 μL of Chip Blocking solution twice.
-
15.
Incubate the sequencing chip at 37°C for 15 min on a hot plate.
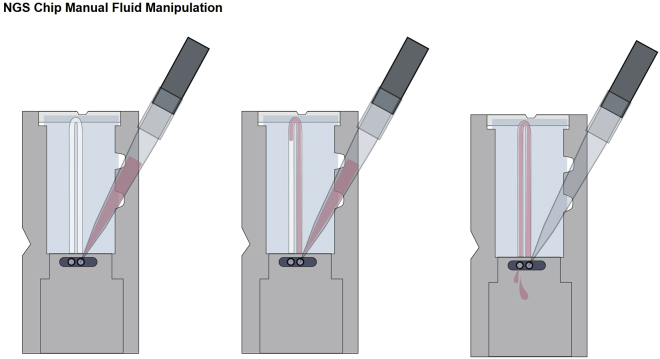
Figure 5.
NGS chip manual fluid manipulation
Fluid is pipetted into the chip through either of the ports, and exits through the other port. The last 5 μL of fluid remains inside the channel.
Preparing binders via hybridization to universal bridge
Timing: 75–90 min
In this section, binder tails are hybridized to the universal bridge. The universal bridge is required to form a double-stranded nick between the binder tail and the foundation so the binder barcode can be ligated to the foundation. Positive control binders DNA Binder 4.2 and DNA Binder 6 bind to DNA Target DNA Target 4.O1 and 6.O1, respectively. The negative control DNA Binder 9 contains a binding region consisting of a scrambled DNA sequence that should bind to none of the targets present, serving as a control for noise.
-
16.Hybridize the positive and negative control binders to the universal bridge. This may be performed in a single reaction.
-
a.In one Eppendorf tube, combine:
-
a.
| Reagent | Amount | Stock concentration | Final concentration |
|---|---|---|---|
| Universal Bridge (5′-CTGCGCCTATAGGAATTCGT TATC/i5NitInd//i5NitInd//i5NitInd//i5NitInd// i5NitInd//i5NitInd//i5NitInd//i5NitInd//i5NitInd// i5NitInd//i5NitInd//i5NitInd/GGACACGGC CGTTATC-3′)∗ |
6 μL | 10 μM | 1,200 nM |
| DNA Binder 4.2 (ATACATGGAATCCTAT) | 1 μL | 10 μM | 200 nM |
| DNA Binder 6 (TCAGGTTAGTACTTCAT) | 1 μL | 10 μM | 200 nM |
| DNA Binder 9 (CTTGACTAGTACATGAC CACTTGA) |
1 μL | 10 μM | 200 nM |
| Hybridization Buffer | 41 μL | N/A | N/A |
| Total | 50 μL | N/A | N/A |
∗Each /i5NitInd/ is a 5-Nitroindole, a universal base analogue that exhibits high duplex stability and hybridizes indiscriminately with each of the four natural bases (Loakes and Brown, 1994).
-
17.
Heat the mixture of DNA binders to 95°C for 5 min.
-
18.
Cool to 25°C for 1 h.
-
19.While waiting for the controls to hybridize to the universal bridge, hybridize the anti-Spot-Tag nanobody to the universal bridge.
-
a.Combine the anti-Spot-Tag nanobody with 5 times excess universal bridge.
-
a.
Note: A 1:1 ratio of binder to universal bridge may also be used here. We used a 1:5 ratio of nanobody to universal bridge because the method used to conjugate the nanobody to the oligonucleotide tail made it possible for multiple oligonucleotide tails to attach to each POC and because excess oligonucleotide tail used during conjugation was not purified away.
| Reagent | Final concentration | Amount |
|---|---|---|
| Universal Bridge | 2 μM | 9.8 μL |
| Spot-Tag nanobody | 400 nM | 1.63 μL |
| Hybridization Buffer | N/A | 37.57 μL |
| Total | N/A | 49 μL |
-
20.
Heat the anti-Spot-Tag nanobody solution to 37°C for 30 min.
-
21.
Cool the anti-Spot-Tag nanobody solution to 25°C for 30 min.
-
22.
After cooling, combine the solutions to generate the Binder Incubation Solution as below.
| Reagents | Final concentration | Amount |
|---|---|---|
| Spot-Tag nanobody hybridized with Universal Bridge | 200 nM | 49 μL |
| DNA control binders hybridized with Universal Bridge | 300 nM | 50 μL |
| Blocking Buffer | N/A | 1 μL |
| Total | N/A | 100 μL |
Performing binding and barcode capture
Timing: 90 min
In this section, binders are incubated with targets on the NGS chip, binder barcodes are ligated to foundations to “record” the binding event, and restriction enzyme digestion is performed to remove binders from the platform.
-
23.
Wash the sequencing chip with 100 μL of Hybridization Buffer for 60 s twice.
-
24.
Wash the sequencing chip with 100 μL of Incubation Buffer for 60 s.
-
25.
Gently mix the prepared binder library by pipetting it up and down.
-
26.
Slowly load 30 μL of the Binder Incubation Solution onto the sequencing chip twice.
-
27.
Incubate the sequencing chip on a hotplate at 25°C for 30 min.
-
28.
Wash the sequencing chip with 100 μL of Incubation Buffer for 90 s three times.
-
29.
Dilute 7 μL of 2× Blunt/TA MM Ligase solution in 63 μL of Hybridization buffer and mix gently.
-
30.
Load 30 μL of the diluted ligase solution onto the sequencing chip twice.
-
31.
Incubate the chip for 5 min in a hotplate at 28°C.
-
32.
Terminate the ligation reaction by washing the sequencing chip with 100 μL of 1× CutSmart solution for 60 s three times.
-
33.Prepare the restriction enzyme mix by combining reagents in the order below.
Reagent Stock concentration Amount 77 μL of NF H2O N/A 77 μL 10× CutSmart solution N/A 10 μL Restriction bridge (5′-CTGCGCCTATACGAATTCGTTATC-3′) 10 μM 3 μL EcoRI 20 units/μL 10 μL Total N/A 100 μL -
a.Gently mix after combining.
-
a.
-
34.
Load 30 μL of the restriction enzyme mix onto the sequencing chip twice.
-
35.
Incubate the sequencing chip at 40°C on a hotplate for 30 min.
Note: Begin preparing reagents required for DNA sequencing of binder barcodes (see step 39).
-
36.
Terminate the restriction digestion reaction by loading the sequencing chip with 100 μL of 100% formamide.
CRITICAL: All steps involving formamide should be performed in a certified chemical fume hood.
-
37.
Incubate the sequencing chip at 40°C on a hotplate for 90 s.
-
38.
Wash the sequencing chip with 500 μL of Hybridization Buffer.
Optional: Additional cycles of binding and barcode capture (steps 16–38) may be repeated for a multi-cycle experiment.
DNA sequencing of binder barcodes
Timing: 4–5 h
In this section, a universal NGS adapter is ligated to the binder barcode with the assistance of a universal NGS adapter bridge (Figure 2, steps 5–7). The NGS adapter facilitates direct amplification and sequencing of the foundation with binder barcode.
-
39.
Thaw the sequencing cartridge at 25°C.
-
40.
Prepare the NGS ligation mix by combining the reagents below.
| Reagent | Stock concentration | Amount |
|---|---|---|
| 77 μL of NF H2O | N/A | 76 μL |
| 10× CutSmart solution | N/A | 10 μL |
| 2:1 1 μM Universal NGS Adapter + Universal NGS Adapter Bridge 9/5 | 10 μM | 12.5 μL |
| 2× Blunt/TA MM Ligase | N/A | 12.5 μL |
| Total | N/A | 100 μL |
-
41.
Load 30 μL of the NGS ligation mix onto the sequencing chip twice.
-
42.
Incubate the sequencing chip on a hotplate at 40°C for 2 min and 24 s.
-
43.Wash the sequencing chip twice with 500 μL of NF H2O.
-
a.Wait 90 s in between washes.
-
a.
Pause point: If users are unable to start the sequencing run after finishing NGS ligation, wash the chip with the buffer that the sequencing chip was originally stored in, return the chip to the cylindrical container, and keep at 4°C for no more than 24 h. When ready to sequence, wash the chip twice with 500 μL of NF H2O prior to continuing on to step 44.
-
44.
Dilute 20 μL of 20 pM PhiX in 580 μL of HT1 Buffer (supplied with MiSeq cartridges).
-
45.
Load the PhiX solution into the sample well of the sequencing cartridge.
Note: This protocol conducts a 45–600 cycle read using a V2 nano chip. If the pre-run check produces a flow error, exchange the plastic hinged piece that contains the gasket on the flow cell with the same piece from an old flow cell after thorough rinsing with 70% Ethanol and NF H2O.
-
46.
Load the sequencing chip, the cartridge, and the running buffer into the MiSeq according to the onscreen MiSeq sequencing run instructions.
-
47.
Start the sequencing run.
Expected outcomes
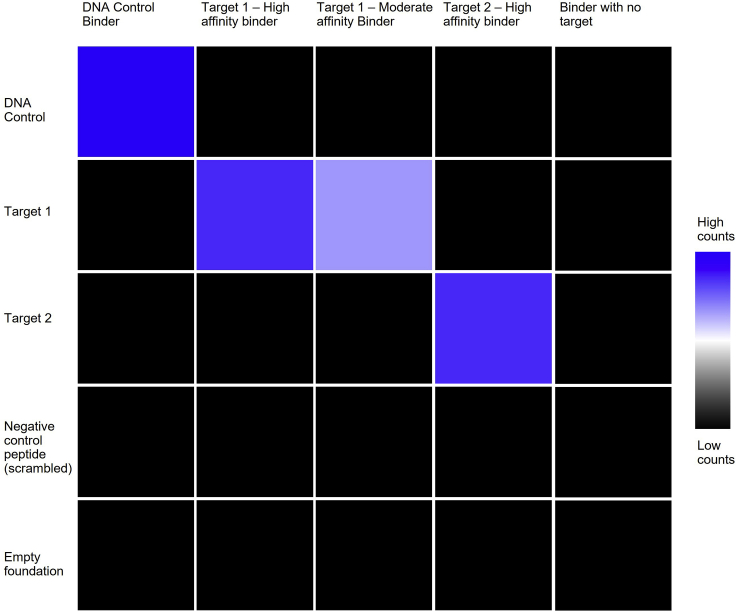
While we have seen consistent ranges of sequencing counts in repeated experiments, sequencing counts may vary depending on a multitude of factors, including the binding properties of binder-target pairs, sequencer type, chip capacity, and similarities between binders or targets in the same run. The expected counts of a positive binder-target pair may range from the low thousands to high millions depending on the sequencer used and total chip spot capacity, as well as the sequencing data analysis parameters. In Table 2, DNA Binder 6 and DNA Target 6 yielded reads in the 28-thousands, while the Spot-tag binding pairs yielded sequencing counts in the lower range (thousands). The ideal heat map in Figure 6 illustrates the expected distribution of counts for target-binder pairs of varying affinities. For multi-cycle experiments, the match rate is expected to undergo exponential decay with an increasing number of cycles.
Table 2.
Sequencing counts for Spot-Tag binder-target experiment
| Target | Target foundation | DNA binder 4.2 | DNA binder 6 | DNA binder 9 (Neg. Ctrl) | Binder Spot-Tag |
|---|---|---|---|---|---|
| 5Phos | Fd11 | 180 | 970 | 1 | 288 |
| 5Phos | Fd7 | 679 | 1269 | 4 | 611 |
| 5Phos | Fd8 | 91 | 523 | 0 | 175 |
| Brady | Fd12 | 24 | 116 | 0 | 42 |
| Brady | Fd13 | 516 | 1611 | 4 | 1663 |
| Brady | Fd14 | 222 | 1113 | 1 | 603 |
| CLR | Fd24 | 224 | 1061 | 2 | 405 |
| CLR | Fd25 | 233 | 930 | 3 | 558 |
| CLR | Fd26 | 81 | 361 | 0 | 92 |
| Empty | Empty | 0 | 0 | 0 | 0 |
| DNA Target 4 | Fd15 | 7201 | 397 | 0 | 207 |
| DNA Target 4 | Fd16 | 8399 | 447 | 2 | 107 |
| DNA Target 4 | Fd17 | 11355 | 441 | 1 | 188 |
| DNA Target 6 | Fd21 | 83 | 28641 | 3 | 129 |
| DNA Target 6 | Fd22 | 82 | 28871 | 1 | 148 |
| DNA Target 6 | Fd23 | 50 | 21007 | 0 | 96 |
| Spot-Tag | Fd19 | 132 | 485 | 2 | 7013 |
| Spot-Tag | Fd20 | 138 | 406 | 0 | 9825 |
| Spot-Tag | Fd27 | 114 | 453 | 38 | 3461 |
| Spot-Tag | Fd28 | 124 | 455 | 1 | 4976 |
| Spot-Tag | Fd29 | 161 | 458 | 1 | 10803 |
| Spot-Tag | Fd31 | 130 | 428 | 1 | 5256 |
Figure 6.
Example heat map of sequencing counts for binders (horizontal) and their respective targets (vertical), where red indicates high count number and black indicates low count number
Quantification and statistical analysis
The code used to analyze the sequencing data is on GitHub. The dataset is on Mendeley (link).
Limitations
Utility of this method toward certain applications may be limited by the binding kinetics, the physical constraints of the NGS chip, and interference of BCS DNA components with binding and DNA folding.
The presence of multiple DNA components and targets in close proximity necessitates use of binders with specificity high enough to overcome potential off-target effects and affinity high enough to overcome signal from noise. In practice, this means that certain binders (ex. low Kd aptamers) are challenging to use in this assay. The Kd of the Spot-tag-nanobody binding pair demonstrated here is approximately 6 nM (Virant et al., 2018). When considering potential peptide binders to proteins, antibodies and antibody-based binding domains such as scFvs and nanobodies (Götzke et al., 2019) have been developed to bind peptides and small molecules with high affinity (Tabares-da Rosa et al., 2011; Finlay and Almagro, 2012; Cobaugh et al., 2008). ClpS, which preferentially recognizes the N-terminal over internal residues within a peptide, represents another promising scaffold of a potential N-terminal amino acid binding reagent (Tullman et al., 2019).
While the chip-based platform allows for excellent spatial control of target-binder interaction, the number of target-binder pairs that can be screened at once is constrained by the need to maintain a certain distance between molecules on the chip. To maintain optimal clustering during sequencing, the concentration in solution cannot exceed 120 pM.
For applications where multiple cycles of BCS must be performed, one should consider that the binder barcode must be unique for each binder for each cycle. In other words, one binder may require multiple distinctive barcodes to differentiate between binding in different cycles. Thus, an application of BCS requiring a large binder library may be limited by the need to experimentally validate a large number of unique binder barcodes for interference with BCS components and between binders.
A final limitation is that targets used in BCS may not be recovered or removed from the chip after the assay is run.
Troubleshooting
Problem 1
Sequencing run failure.
There are multiple reasons for why a sequencing run may fail. We found the leading cause of failure to be a high loading concentration of targets (step 3 of ‘before you begin’) leading to overclustering during amplification on the sequencer. Other things to consider include the presence of large air bubbles during BCS incubation steps (steps 10, 15, 27, 31, 35, 37, and 42) or use of a degraded PhiX solution (step 2 of ‘before you begin’).
Potential solution
Both overclustering and under clustering can compromise data quality and output. Ideal cluster densities will vary by machine and should be confirmed in the Illumina user manual (Optimizing cluster density on Illumina sequencing systems). Our preferred method for assessing under- or overclustering is the Sequencing Analysis Viewer, which includes multiple other modalities for assessing cluster density.
Our constructs possess many shared sequences which leads to low nucleotide diversity that predispose the sequencing run to failure. Therefore, inclusion of a high-diversity library such as PhiX is especially important for sequencing run success. Regular preparations of fresh PhiX solution are necessary to avoid degradation from repeated freeze-thaw cycles.
The chip should be visualized before each incubation to ensure that no air bubbles are visible, as these will interfere with proper functioning of BCS components. Bubbles may be flushed out with an additional injection of a compatible buffer solution. Automation using a liquid handler may be used to help reduce air bubbles.
If successful colocalization of the foundation and target is questioned, one troubleshooting strategy is to couple BCS components with fluorescent probes and visualize their activity on the chip. We use single-molecule imaging to validate the physical colocalization of targets and their respective foundations in our previous paper (Hong et al., 2022). When interrogating more complex sources of failure or optimizing components at specific steps, couple the components with fluorescent probes and use an appropriate imaging modality to visualize each component.
Problem 2
Off-target binding.
Sequencing data can reveal unexpected binding interactions such as a single binder candidate binding to a significant percentage of targets or to negative controls (relevant to step 27).
Potential solution
Components of the platform may have non-specific or off-target binding properties. Exposed adaptors or empty foundation bases are examples of off-target binding sites. The barcode designs of the targets and binders should be checked for unintended complementarity to self or other. As a first-line solution, sequences may be redesigned accordingly. Other areas to optimize through unit testing include incubation times and enzyme concentrations, especially for steps involving Blunt/TA MM Ligase.
The Chip Blocking Solution contains sequences that block the single-stranded P5 and P7 adapters on the Illumina chip (see the section "designing chip blocking components"). The concentration of blocking components may be optimized through unit testing, and further blocking components may be added (such as blocking the POC ssDNA region or blocking part of the foundation).
In cases involving DNA components, hybridization between individual components can be assessed using a Bioanalyzer RNA assay, which can differentiate between double and single stranded DNA (Unpublished data). In other cases, a Kd measurement assay between binder and target (Plach and Schubert, 2019) may be considered to quantify true off-target binding.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Annalisa Pawlosky apawlosky@google.com.
Materials availability
This study did not generate new unique reagents. Example buffers and sequences of oligos, peptides, and oligo-peptides used in this protocol are listed in the key resources table, though these should be interchanged with materials compatible with the user’s subject(s) of study.
Acknowledgments
The authors would like to thank John Coller for early-stage technical discussions and feedback. The authors acknowledge Philip Nelson, John Platt, Erica Brand, Jason Miller, and Patrick Riley for project support and management advice and Tiffany Ly and Amy Chung-Yu Chou for administrative support. In addition, the authors would like to thank Ali Bashir, Brian Williams, Joshua Cutts, Samuel Yang, and Marytheresa Ifediba for technical discussions and protocol feedback. The work presented here was funded through Google Research.
Author contributions
Conceptualization, A.P.; methodology, M.G., M.C., V.A.C., and A.P.; software validation, M.B.; formal analysis, M.G., M.F., M.C., M.B., P.J., and A.P.; investigation, M.G., M.F., M.C., S.S., and S.A.; protocol documentation, L.S.; writing - original draft, J.M.H., M.F., S.S., L.C., and A.P.; writing - reviews, J.M.H., M.F., L.C., and A.P.; visualization, J.M.H., S.S., and A.P.; supervision, A.P.; project administration, A.P.
Declaration of interests
M.B., P.J., and A.P. are employees and shareholders of Alphabet. Google has filed patent applications related to this work, including PCT/US2020/050574, PCT/US2020/053716, PCT/US2020/040130, PCT/US2020/053715, US20210079398A1, US20210079557A1, US20210102248A1, and WO2021051011A1. All work was completed while authors were affiliated with Alphabet. Current affiliations for authors who have moved on from Alphabet are the following: M.G., 10× Genomics, employee; J.H., University of California, Los Angeles (UCLA); M.F., Conception, employee; M.C., Encodia, Inc., employee; S.S., L.C., University of California, San Francisco (UCSF); V.A.C., Washington University School of Medicine, St. Louis; S.A., Alkahest, employee; L.S., Eikon Therapeutics, employee.
Data and code availability
All original code developed to analyze data acquired from the platform described in this protocol has been deposited on GitHub (https://github.com/google-research/google-research/tree/master/protseq).
The data are available on Mendeley at https://data.mendeley.com/datasets/f9hdn5xc3v.
They are publicly available as of the date of this article’s publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Antos J.M., Ingram J., Fang T., Pishesha N., Truttmann M.C., Ploegh H.L. Site-specific protein labeling via sortase-mediated transpeptidation. Curr. Protoc. Protein Sci. 2017;89:15.3.1–15.3.19. doi: 10.1002/cpps.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M.B., Traenkle B., Koch P.A., Emele F., Weiss F., Poetz O., Stehle T., Rothbauer U. Peptides in headlock – a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy. Sci. Rep. 2016;6:19211. doi: 10.1038/srep19211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaugh C.W., Almagro J.C., Pogson M., Iverson B., Georgiou G. Synthetic antibody libraries focused towards peptide ligands. J. Mol. Biol. 2008;378:622–633. doi: 10.1016/j.jmb.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty A.J., Suh S.W. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 2000;28:4051–4058. doi: 10.1093/nar/28.21.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabricius V., Lefèbre J., Geertsema H., Marino S.F., Ewers H. Rapid and efficient C-terminal labeling of nanobodies for DNA-paint. J. Phys. D Appl. Phys. 2018;51:474005. [Google Scholar]
- Fantoni N.Z., El-Sagheer A.H., Brown T. A hitchhiker’s guide to click-chemistry with Nucleic Acids. Chem. Rev. 2021;121:7122–7154. doi: 10.1021/acs.chemrev.0c00928. [DOI] [PubMed] [Google Scholar]
- Finlay W.J.J., Almagro J.C. Natural and man-made V-gene repertoires for antibody discovery. Front. Immunol. 2012;3:342. doi: 10.3389/fimmu.2012.00342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götzke H., Kilisch M., Martínez-Carranza M., Sograte-Idrissi S., Rajavel A., Schlichthaerle T., Engels N., Jungmann R., Stenmark P., Opazo F., et al. The Alfa-tag is a highly versatile tool for nanobody-based bioscience applications. Nat. Commun. 2019;10:4403. doi: 10.1038/s41467-019-12301-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.M., Gibbons M., Bashir A., Wu D., Shao S., Cutts Z., Chavarha M., Chen Y., Schiff L., Foster M., et al. ProtSeq: toward high-throughput, single-molecule protein sequencing via amino acid conversion into DNA barcodes. iScience. 2022;25:103586. doi: 10.1016/j.isci.2021.103586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loakes D., Brown D.M. 5-nitroindole as an universal base analogue. Nucleic Acids Res. 1994;22:4039–4043. doi: 10.1093/nar/22.20.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plach M., Schubert T. Biophysical characterization of aptamer-target interactions. Adv. Biochem. Eng. Biotechnol. 2019;174:1–15. doi: 10.1007/10_2019_103. [DOI] [PubMed] [Google Scholar]
- Tabares-da Rosa S., Rossotti M., Carleiza C., Carrión F., Pritsch O., Ahn K.C., Last J.A., Hammock B.D., González-Sapienza G. Competitive selection from single domain antibody libraries allows isolation of high-affinity Antihapten antibodies that are not favored in the llama immune response. Anal. Chem. 2011;83:7213–7220. doi: 10.1021/ac201824z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullman J., Callahan N., Ellington B., Kelman Z., Marino J.P. Engineering CLPS for selective and enhanced N-terminal amino acid binding. Appl. Microbiol. Biotechnol. 2019;103:2621–2633. doi: 10.1007/s00253-019-09624-2. [DOI] [PubMed] [Google Scholar]
- Virant D., Traenkle B., Maier J., Kaiser P.D., Bodenhöfer M., Schmees C., Vojnovic I., Pisak-Lukáts B., Endesfelder U., Rothbauer U. A peptide tag-specific nanobody enables high-quality labeling for dSTORM imaging. Nat. Commun. 2018;9:930. doi: 10.1038/s41467-018-03191-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All original code developed to analyze data acquired from the platform described in this protocol has been deposited on GitHub (https://github.com/google-research/google-research/tree/master/protseq).
The data are available on Mendeley at https://data.mendeley.com/datasets/f9hdn5xc3v.
They are publicly available as of the date of this article’s publication. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.